Significance
During the biogenesis of light-harvesting complexes, the posttranslational chloroplast signal recognition particle (cpSRP) pathway is highly coordinated with chlorophyll (Chl) biosynthesis. To ensure the required supply of newly synthesized Chl for the assembly of light-harvesting Chl a/b-binding proteins (LHCPs) while avoiding the accumulation of photoreactive tetrapyrrole intermediates, Chl synthesis is tightly regulated at the level of glutamyl-tRNA reductase (GluTR), the initial enzyme of rate-limiting 5-aminolevolinic acid (ALA) synthesis. Here, we found that the chaperone for LHCPs, the chloroplast SRP43, directly binds to and prevents aggregation of GluTR, thereby enhancing the stability of active GluTR. This finding reveals a posttranslational control of ALA synthesis, which helps plants to adjust their Chl synthesis rate in response to fluctuating environmental conditions.
Keywords: chloroplast signal recognition particle, chaperone, tetrapyrrole biosynthesis, 5-aminolevulinic acid, chloroplast biogenesis
Abstract
Assembly of light-harvesting complexes requires synchronization of chlorophyll (Chl) biosynthesis with biogenesis of light-harvesting Chl a/b-binding proteins (LHCPs). The chloroplast signal recognition particle (cpSRP) pathway is responsible for transport of nucleus-encoded LHCPs in the stroma of the plastid and their integration into the thylakoid membranes. Correct folding and assembly of LHCPs require the incorporation of Chls, whose biosynthesis must therefore be precisely coordinated with membrane insertion of LHCPs. How the spatiotemporal coordination between the cpSRP machinery and Chl biosynthesis is achieved is poorly understood. In this work, we demonstrate a direct interaction between cpSRP43, the chaperone that mediates LHCP targeting and insertion, and glutamyl-tRNA reductase (GluTR), a rate-limiting enzyme in tetrapyrrole biosynthesis. Concurrent deficiency for cpSRP43 and the GluTR-binding protein (GBP) additively reduces GluTR levels, indicating that cpSRP43 and GBP act nonredundantly to stabilize GluTR. The substrate-binding domain of cpSRP43 binds to the N-terminal region of GluTR, which harbors aggregation-prone motifs, and the chaperone activity of cpSRP43 efficiently prevents aggregation of these regions. Our work thus reveals a function of cpSRP43 in Chl biosynthesis and suggests a striking mechanism for posttranslational coordination of LHCP insertion with Chl biosynthesis.
Photosynthesis is an indispensable process that provides chemical energy to photoautotrophs on Earth. At the initial step of photosynthesis in plant chloroplasts, specialized light-harvesting complexes (LHCs) contribute to the conversion of solar energy into electron excitation, which allows the reaction centers of photosystem (PS) I and PSII to initiate charge separation and fuel the photosynthetic electron transport chain in the thylakoid membranes (1, 2). In higher plants, the LHCs are composed of light-harvesting chlorophyll (Chl) a/b-binding proteins (LHCPs) bound to the Chl and carotenoid pigments of photosynthesis (3, 4). The availability of newly synthesized Chl is essential for the stabilization and integration of LHCPs into thylakoids (5, 6). However, free Chl and accumulating metabolic intermediate have a marked tendency to generate reactive oxygen species in the light (7). Thus, LHC biogenesis requires synchronized synthesis of LHCPs and Chls.
LHC biogenesis is a complex multistep process requiring not only gene expression and protein synthesis but plastid import, as well as the targeting and assembly of LHCPs into the photosynthetic membranes (1, 8). Within chloroplasts, the mature but unfolded LHCP is transported into the stroma and integrated into thylakoids via the chloroplast signal recognition particle (cpSRP) pathway (9, 10). The chloroplast-specific cpSRP43 interacts with LHCPs and cpSRP54 to form a soluble “transit complex” (11). Interaction of the GTPase domains of cpSRP54 with the SRP receptor cpFtsY targets the transit complex to the ALB3 translocase at the thylakoid membrane, which mediates the insertion of LHCPs (12). Remarkably, cpSRP43 coevolved with the emergence of LHCPs in green land plants (13, 14) and is suggested to be a dedicated chaperone that quantitatively prevents aggregation of the LHCP family in aqueous environments (15–17).
Chl is synthesized via the plastid-localized magnesium (Mg) branch of tetrapyrrole biosynthesis (TBS) (18). The first committed step in TBS is the formation of 5-aminolevulinic acid (ALA) (19, 20). In both prokaryotic and eukaryotic oxygenic photoautotrophs, ALA is synthesized from glutamyl-tRNAGlu by glutamyl-tRNA reductase (GluTR) and glutamate 1-semialdehyde aminotransferase (GSAAT). ALA synthesis is the rate-limiting step in TBS and relies specifically on tight control of GluTR stability and activity (21, 22). Six enzymatic steps convert eight units of ALA into protoporphyrin IX (Proto), which is then channeled into Chl and heme biosynthesis by chelation of Mg2+ and Fe2+, respectively.
Recent studies in the cyanobacterium Synechocystis PCC 6803 have demonstrated that Chl synthase (the last enzyme in Chl a biosynthesis) is located in close proximity to the cpSRP translocase ALB3 during the biogenesis of PS core subunits (23, 24), suggesting a potential link between Chl biosynthesis and LHCP biogenesis pathways. Nevertheless, how the assembly of Chl and LHCP into LHCs is regulated remains largely unknown. Here, we show that cpSRP43 efficiently chaperones and stabilizes GluTR. Hence, cpSRP43 not only acts as a chaperone for LHCP localization but also functions in the posttranslational control of GluTR to optimize ALA synthesis.
Results
Lack of cpSRP43 Reduces Levels of GluTR and Chl Biosynthesis.
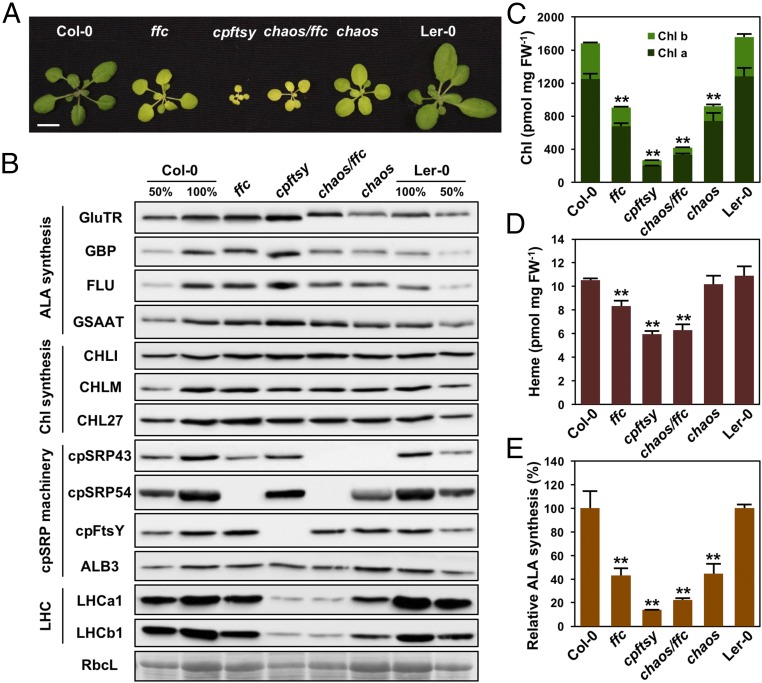
To explore potential links between LHCP translocation and Chl synthesis, we asked whether cpSRP components are involved in TBS. We measured the performance of the TBS pathway in various cpsrp mutants (Fig. 1A), including chaos [cpSRP43 null mutant (25)], ffc [cpSRP54 null mutant (26)], chaos/ffc (27), and cpftsy (28). All cpsrp mutants showed retarded growth, decreased leaf pigmentation, and reduced accumulation of Chl and LHCPs (Fig. 1 B and C), as reported previously (29). The pool of noncovalently bound heme was also significantly decreased in ffc, chaos/ffc, and cpftsy mutants, but not in the chaos single mutant (Fig. 1D). As the rate-limiting step of TBS, ALA synthesis was drastically decreased in all cpsrp mutants (Fig. 1E), and the flux of Chl precursors, including MgProto, MgProto monomethylester (MgProtoME), and protochlorophyllide (Pchlide), was correspondingly decreased (SI Appendix, Fig. S1 A–C). These results suggest that the dysfunction of the cpSRP machinery disrupts ALA synthesis, thus reducing levels of all other TBS intermediates.
Fig. 1.
cpSRP43 modulates Chl biosynthesis in Arabidopsis. (A) Pale-green leaf phenotype of 21-d-old ffc, cpftsy, chaos/ffc, and chaos mutants and their WT plants (Col-0 ecotype for ffc and cpftsy and Ler-0 ecotype for chaos). (Scale bars: 1 cm.) (B) Steady-state levels of TBS proteins, cpSRP components, and LHC proteins in the WT and cpsrp mutants were detected by immunoblotting using the indicated antibodies. Ponceau S staining of the large subunit of RubisCo (RbcL) is shown as a loading control. Chl contents (C), heme contents (D), and relative ALA synthesis rates [E, 184.37 ± 21.20 nmol of ALA h−1⋅g−1 fresh weight (FW) for Col-0 and 121.39 ± 3.93 nmol of ALA h−1⋅g−1 FW for Ler-0] in WT and cpsrp mutants are shown. Data are mean ± SD (n = 4). Asterisks indicate significant differences compared with the WT plants (**P < 0.01, Student’s t test).
To gain more insight into TBS in cpsrp mutants, the steady-state levels of enzymes and regulators involved in the pathway were examined. Compared with the other TBS proteins, the GluTR level in chaos was specifically decreased to ∼76% of that in the corresponding WT (Ler-0; Fig. 1B). This change is modest, but reproducible and statistically significant (SI Appendix, Fig. S1D). In contrast, GluTR amounts were slightly increased in the ffc and cpftsy mutants; this led to the WT-like GluTR level in the chaos/ffc double mutant (Fig. 1B and SI Appendix, Fig. S1D). The altered level of GluTR in the chaos mutant was not due to perturbed transcriptional regulation, as amounts of HEMA1 mRNA (which encodes the dominant isoform of GluTR in the photosynthetic tissues) in chaos and WT plants were comparable (SI Appendix, Fig. S1E). In addition, the expression of GSAAT1 and CHL27 (which encode GSAAT and MgProtoME cyclase, respectively) were not significantly reduced in cpsrp mutants (SI Appendix, Fig. S1E). Together, these observations suggest that cpSRP43 has a specific function in the posttranslational control of GluTR.
Concurrent Lack of cpSRP43 and the GluTR-Binding Protein Drastically Reduces GluTR Levels.
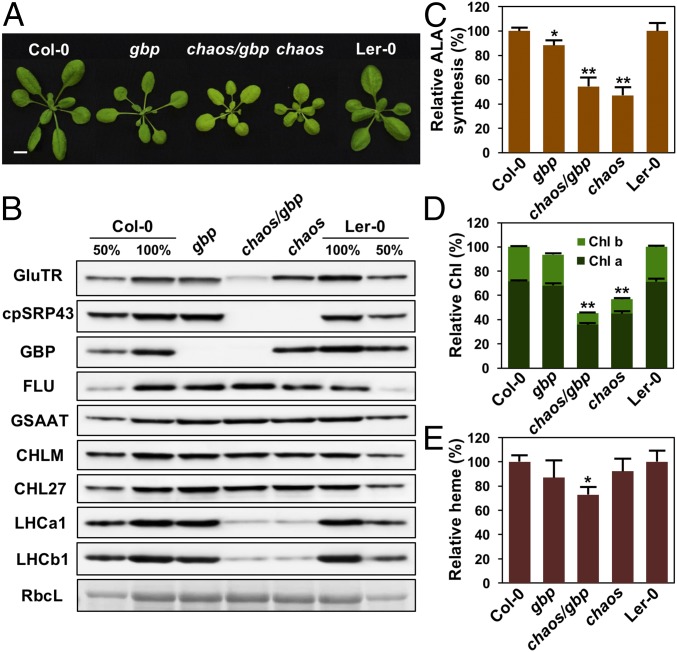
GluTR-binding protein (GBP) is suggested to function as a scaffold that binds GluTR, promotes synthesis of ALA, and preferentially directs it into heme synthesis (30). Previous studies have indicated that GBP interacts with and stabilizes GluTR, as reduced amounts of the enzyme are observed in gbp mutants (22, 30). We therefore quantified the contributions of cpSRP43 and GBP to the maintenance of GluTR using chaos, gbp, and homozygous chaos/gbp double mutants (Fig. 2 A and B). The chaos/gbp seedlings exhibited larger plant size than the chaos mutant (Fig. 2A), a chaos-like ALA synthesis rate (Fig. 2C), and an additive decrease in both Chl (Fig. 2D) and heme contents (Fig. 2E) in comparison to the single mutants. Remarkably, while deletion of either cpSRP43 or GBP shows only a modest effect, concurrent depletion of both proteins led to a pronounced decrease in GluTR level (from 76% to ∼37% of the WT amount; Fig. 2B). In contrast, simultaneous deletion of cpSRP54 and GBP in homozygous ffc/gbp mutants only showed a minor reduction of GluTR level compared with gbp (SI Appendix, Fig. S2), demonstrating the specificity of the cpSRP43 effect. It is worth noting that cpSRP54 allosterically enhances the chaperone activity of cpSRP43 (17), and therefore could indirectly impact GluTR stability. Taken together, these results reveal a synergistic effect of the chaos and gbp mutations on GluTR levels, suggesting that cpSRP43 and GBP play distinct roles in stabilizing GluTR.
Fig. 2.
cpSRP43 and GBP are cooperatively involved in the stabilization of GluTR in Arabidopsis. (A) Pale-green leaf phenotype of 24-d-old gbp, chaos, and chaos/gbp mutants and their WT plants (Col-0 ecotype for gbp and Ler-0 ecotype for chaos). (Scale bars: 1 cm.) (B) Steady-state levels of TBS enzymes, cpSRP components, and LHC proteins in seedlings of the genotypes mentioned above were quantified by immunoblotting using the indicated antibodies. Ponceau S-stained RubisCo (RbcL) is shown as a loading control. Relative ALA synthesis rates (C), Chl contents (D), and heme contents (E) are shown. Data are mean ± SD (n = 3). In C–E, asterisks indicate significant differences compared with WT plants (*P < 0.05, **P < 0.01; Student’s t test).
cpSRP43 Interacts Directly with GluTR.
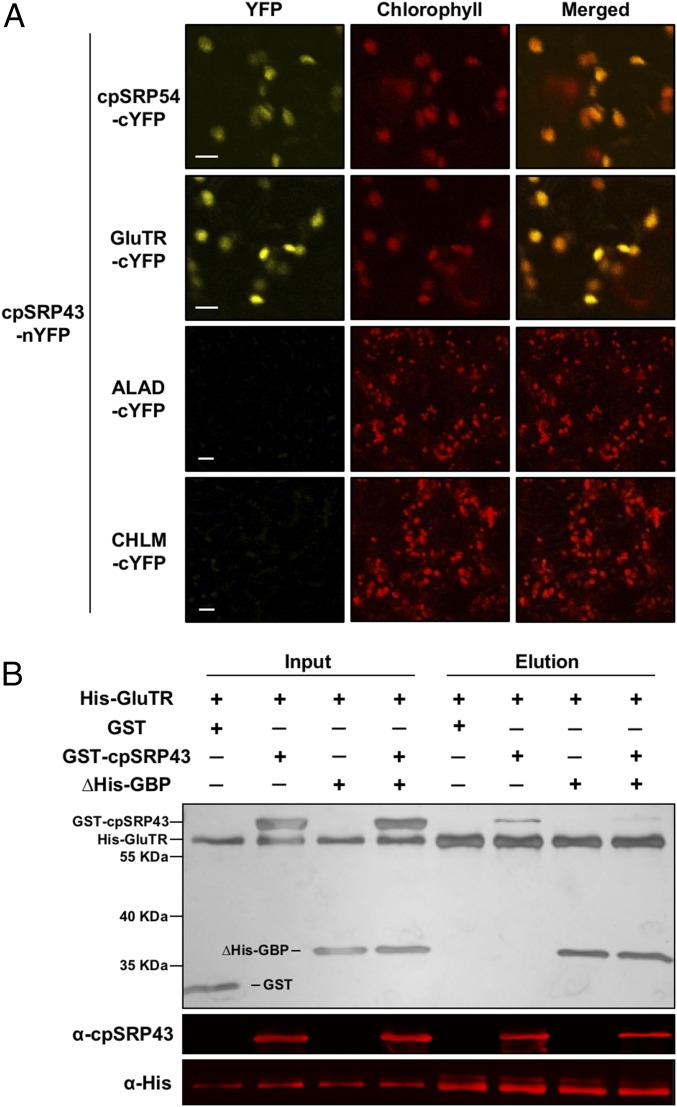
To test whether cpSRP43 physically interacts with TBS enzymes, we conducted the bimolecular fluorescence complementation (BiFC) assay. The N- and C-terminal halves of the split yellow fluorescent protein (nYFP and cYFP) were fused to cpSRP43 and target proteins of interest, respectively, and coexpressed in epidermal and parenchymal cells of Nicotiana benthamiana leaves. Complementation was observed between cpSRP43 and either GluTR or GBP, but not between cpSRP43 and the GluTR inhibitor FLUORESCENT [FLU (31)], ALA dehydratase (the TBS enzyme immediately downstream of GluTR), or MgProto methyltransferase (an enzyme in the Mg branch of TBS) (Fig. 3A and SI Appendix, Fig. S3 A–D). Furthermore, the interaction of cpSRP43 with GluTR and GBP was confirmed by in vivo pull-down and immunoprecipitation assays (SI Appendix, Fig. S3 E and F).
Fig. 3.
cpSRP43 interacts directly with GluTR. (A) BiFC analysis of interaction between cpSRP43 and GluTR. cpSRP43 fused to nYFP and cpSRP54/GluTR/ALA dehydratase (ALAD)/MgProto methyltransferase (CHLM) fused to cYFP were expressed in N. benthamiana after infiltration with appropriately transformed agrobacteria and analyzed by confocal microscopy 3 d after infiltration. The cpSRP54 was used as a positive control. The ALAD and CHLM were used as negative controls. (Scale bars: 10 μm.) (B) GluTR binding to cpSRP43 is detected by an in vitro His pull-down assay. Recombinant purified WT His-GluTR (5 μM) was used as bait and incubated with 10 μM GST, GST-cpSRP43, ∆His-GBP, or GST-cpSRP43 and ∆His-GBP. The proteins bound to His-GluTR were eluted with elution buffer containing 200 mM imidazole. Input and elution fractions were stained with silver (Upper), blotted, and probed with the indicated antibodies (Middle and Lower).
To examine the nature of the interactions of GluTR and GBP with cpSRP43, an in-vitro His pull-down assay was conducted with purified recombinant proteins. The results showed a specific interaction of cpSRP43 with GluTR, but not with GBP (Fig. 3B and SI Appendix, Fig. S4). Thus, the association of cpSRP43 with GBP observed in vivo likely results from the binding of cpSRP43 to GluTR that is itself associated with GBP. Intriguingly, the presence of GBP in the pull-down assay reduced binding of cpSRP43 to GluTR (Fig. 3B), indicating that GBP counteracts the GluTR–cpSRP43 interaction.
The Substrate-Binding Domain of cpSRP43 Binds GluTR and Is Important for GluTR Stability and Chl Biosynthesis in Planta.
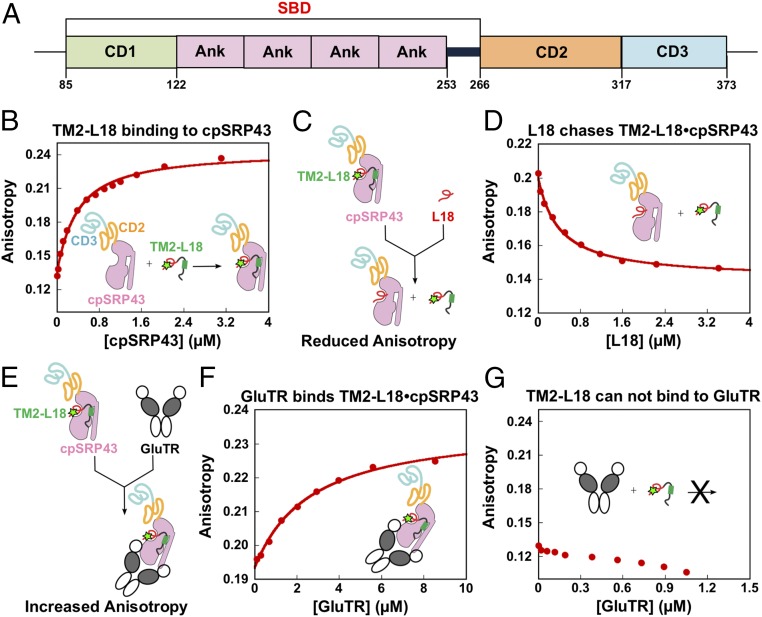
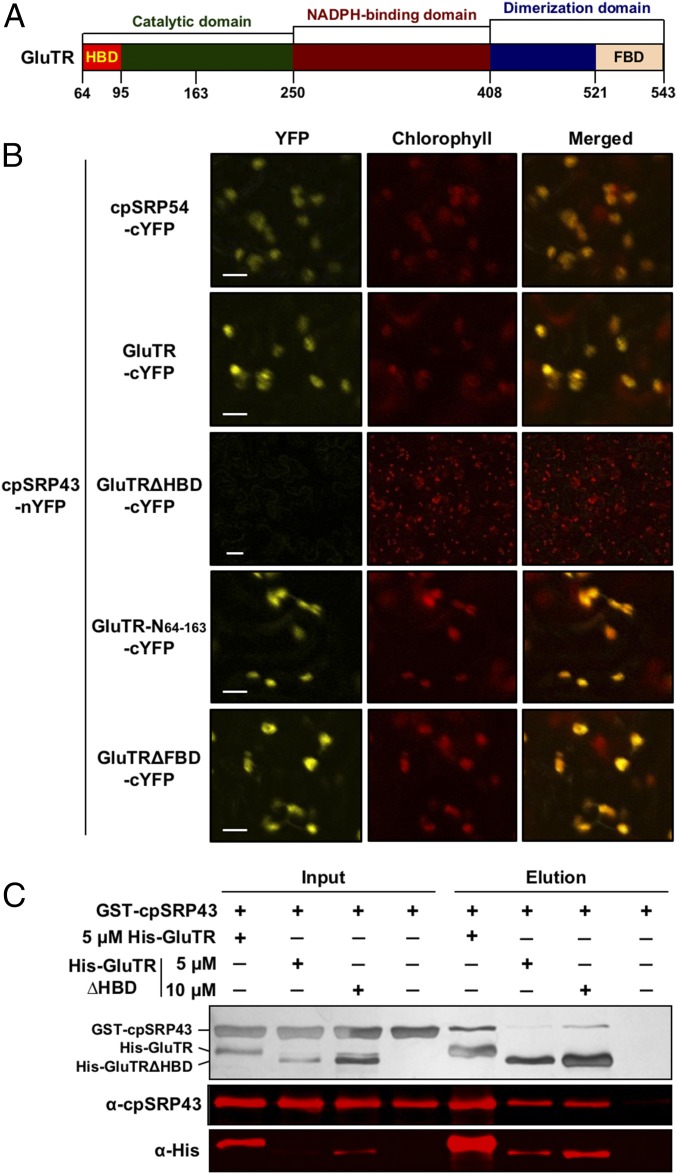
Crystallographic and biochemical analyses have revealed two types of protein interaction domains in cpSRP43: three chromodomains (CD1–CD3) and an ankyrin repeat domain (Ank1–Ank4) between CD1 and CD2 (9, 32) (Fig. 4A). These domains mediate different interactions with LHCPs and the other cpSRP components. The Ank3 recognizes an L18 motif, a conserved 18-aa sequence between the second and third transmembrane domains (TM2 and TM3) of LHCPs (16, 32, 33). CD1, Ank1–Ank4, and a 20-aa sequence located between Ank4 and CD2 form a substrate-binding domain (SBD), which is responsible for the chaperone activity of cpSRP43 that protects LHCPs from aggregating in an aqueous milieu (17). In parallel, cpSRP54 and ALB3 interact with CD2 and CD3, respectively (34–39).
Fig. 4.
GluTR and LHCb5 bind cpSRP43. (A) Schematic overview of cpSRP43 domains. (B) Binding of the loop1-TM2-L18 fragment of LHCb5 to cpSRP43. Binding was measured by monitoring changes in the fluorescence anisotropy of LHCb5 (G162C) labeled with fluorescein at position 162. The data were fit to SI Appendix, Eq. 1 and gave a Kd value of 338 nM. (C and E) Scheme illustrating the effects of a competitor (L18 in D) or cobinder (GluTR in F) on the fluorescence anisotropy of the LHCb5 fragment prebound to cpSRP43-SBD. The observed effect of the L18 peptide (D) or GluTR (F) on the fluorescence anisotropy of the LHCb5 fragment prebound to cpSRP43 is shown. The data in D were fit to the competition model described by SI Appendix, Eq. 2 and gave a value of 1 μM. The data in F were fit to SI Appendix, Eq. 1 and gave a Kd value of 2.6 μM. The SDs of the Kd and values were estimated to be ±10% based on at least two measurements (technical replicates). In G, GluTR does not bind to the fluorescein-labeled LHCb5 fragment.
We next asked whether the SBD of cpSRP43 is responsible for GluTR binding, and whether GluTR competes or cobinds with the known cpSRP43 substrate LHCP to GluTR. The interaction of GluTR with either WT cpSRP43 or its isolated SBD was assayed based on the change in anisotropy of a fluorescein-labeled loop1-TM2-L18 fragment derived from LHCb5. Incubation with cpSRP43 increased the fluorescence anisotropy of this LHCP fragment, indicating the formation of a complex between them (Fig. 4B). This complex was presented either with a nonfluorescent L18 peptide, which competes with the fluorescein-labeled LHCP fragment for binding cpSRP43 (17), or with GluTR. The cpSRP43-induced anisotropy enhancement of labeled loop1-TM2-L18 was effectively diminished by the L18 peptide, as predicted (Fig. 4 C and D). In contrast, the addition of GluTR further increased the fluorescence anisotropy of the preformed cpSRP43–loop1-TM2-L18 complex (Fig. 4 E and F). Notably, GluTR itself did not bind to the loop1-TM2-L18 fragment (Fig. 4G). These results indicate that GluTR binds to cpSRP43 in a manner that does not compete with the interaction of cpSRP43 with the LHCP fragment, suggesting the formation of a ternary complex between the three proteins. Importantly, the SBD of cpSRP43 binds GluTR with an affinity similar to that of the WT cpSRP43 (SI Appendix, Fig. S5), indicating that the SBD suffices for interaction with GluTR. Thus, GluTR directly binds to the SBD of cpSRP43, and GluTR and the L18 motif of LHCPs interact with a different binding site of the SBD of cpSRP43.
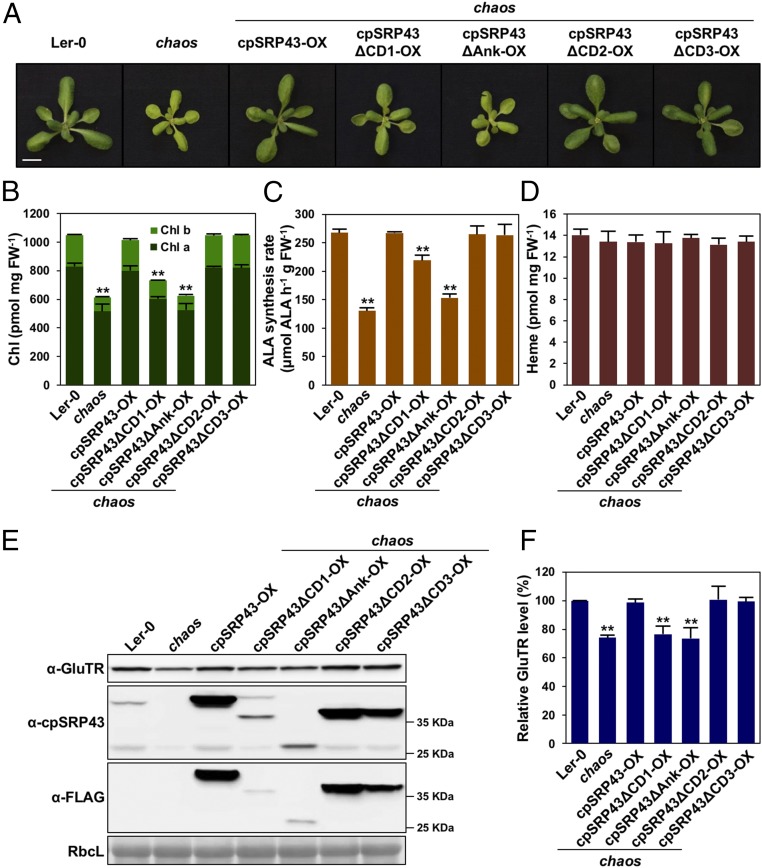
To genetically characterize the functional requirement of various cpSRP43 domains for LHCP biogenesis, Chl synthesis, and GluTR stability, a series of chaos complementation lines that constitutively express either WT cpSRP43 or truncated cpSRP43 variants lacking each of the four cpSRP43 domains was generated (SI Appendix, Fig. S6). Intriguingly, deletion of either CD1 or the ankyrin repeats in the SBD of cpSRP43 impaired the complementation of chaos by cpSRP43 (Fig. 5 A and B). In contrast, expression of either truncated cpSRP43∆CD2 or cpSRP43∆CD3 completely rescued the pale-green phenotype of chaos. The ALA synthesis rate of the chaos complementation lines correlated with their Chl contents (Fig. 5C). While the different chaos complementation lines displayed altered levels of Chl accumulation and ALA synthesis, their heme content remained the same as in WT plants (Fig. 5D), supporting a direct and specific impact of cpSRP43 on the control of Chl synthesis. Considering the fact that developing green seedlings require ∼100-fold more Chl than heme, it is understandable that heme synthesis would be much less affected than Chl synthesis when ALA synthesis is compromised. Consistently, the knockout mutant of protoporphyrinogen IX oxidase 1 (ppox1), which encodes the last enzyme in the common pathway to Chl and heme biosynthesis, exhibited a severe albino phenotype but had a WT-like heme level (40).
Fig. 5.
SBD of cpSRP43 is required for Chl biosynthesis and GluTR stability in Arabidopsis. (A) Phenotype of 24-d-old Ler-0, chaos, and various chaos complementation lines. (Scale bars: 1 cm.) The Chl level (B), ALA synthesis rate (C), and heme level (D) in Ler-0, chaos, and various chaos complementation lines are shown. (E) Steady-state levels of GluTR and cpSRP43 variants in Ler-0, chaos, and various chaos complementation lines were quantified by immunoblotting using the indicated antibodies. Ponceau S-stained RubisCo (RbcL) is shown as a loading control. (F) Semiquantitative analysis with Phoretix 1D software (Phoretix International) of the immunoblots in E from two biological replicates. The relative amounts of GluTR in chaos and various chaos complementation lines were normalized to the level in Ler-0. In B–D, data are mean ± SD (n = 3). Asterisks indicate significant differences compared with the WT (**P < 0.01, Student’s t test).
Furthermore, like chaos and chaos/gbp (Figs. 1B and 2B), the complementation lines cpSRP43∆CD1-OX(chaos) and cpSRP43∆Ank-OX(chaos) exhibited decreased GluTR contents (Fig. 5 E and F). The relative severity of the phenotypes of cpSRP43 truncation mutants correlated with the effects of these mutations on the chaperone activity of cpSRP43 (15), underlining the important role of cpSRP43’s chaperone function in ensuring normal leaf pigmentation and GluTR stabilization.
The canonical function of the SBD of cpSRP43 is to recognize and chaperone LHCPs in the stroma, as well as to facilitate their localization to the thylakoid membrane (9, 10). In accordance with this role and with our previous analyses (29), chaos showed a specific reduction in LHCP levels (SI Appendix, Fig. S7), while accumulation of core subunits of the four photosynthetic complexes (PSII, cytochrome b6f, PSI, and ATP synthase) was not affected. In addition, more LHCPs accumulated in cpSRP43∆CD1-OX(chaos) than in either the cpSRP43∆Ank-OX(chaos) or chaos mutant (SI Appendix, Fig. S7). Furthermore, 2D Blue Native (BN)-SDS/PAGE gel analysis confirmed the decrease in LHC contents in the chaos, cpSRP43∆CD1-OX(chaos), and cpSRP43∆Ank-OX(chaos) lines, as shown by the reduced accumulation of PSII–LHCII supercomplexes and PSI–LHCI complexes (SI Appendix, Fig. S8). In contrast, cpSRP43∆CD2-OX(chaos) and cpSRP43∆CD3-OX(chaos) accumulated WT-like amounts of LHCP, PSII–LHCII, and PSI–LHCI complexes in thylakoids. These observations are consistent with a previous report (28) and confirm the contribution of cpSRP43 to the insertion of LHCPs into thylakoids.
cpSRP43 Interacts with and Chaperones the N-Terminal Region of GluTR.
GluTR is composed of multiple functional domains, including the N-terminal heme-binding (HBD), catalytic, NADPH-binding, dimerization, and C-terminal FLU-binding (FBD) domains (22, 41) (Fig. 6A). Its N- and C-terminal domains interact with different regulators to effect various posttranslational control mechanisms (18). The HBD interacts with GBP and subunits of the caseinolytic peptidase (Clp) protease (22), and it has been suggested that GBP counteracts the proteolytic activity of Clp protease (22, 42, 43). FLU interacts with the FBD and efficiently represses ALA synthesis in the dark to avoid overaccumulation of Pchlide when the light-dependent Pchlide oxidoreductase is not active (31, 44).
Fig. 6.
cpSRP43 interacts with the N-terminal part of GluTR. (A) Schematic overview of GluTR domains. (B) BiFC assay reveals that cpSRP43 interacts with the N-terminal HBD of GluTR. The cpSRP43 fused to nYFP and cpSRP54/GluTR variants fused to cYFP were expressed in N. benthamiana following Agrobacterium infiltration and analyzed by confocal microscopy 3 d after infiltration. The cpSRP54 was used as a positive control. (Scale bars: 10 μm.) (C) Deletion of HBD in GluTR weakens the binding to cpSRP43, as shown by the in vitro His pull-down assay. Indicated concentrations of recombinant purified WT His-GluTR or His-GluTRΔHBD were used as bait and incubated with 10 μM GST-cpSRP43. Input and elution fractions were stained with silver (Upper), blotted, and probed with the indicated antibodies (Middle and Lower).
To identify which GluTR domain interacts with cpSRP43, we conducted BiFC assays. The cYFP was fused to the WT or truncated GluTR lacking either the HBD (GluTR∆HBD) or FBD (GluTR∆FBD), and transiently coexpressed with cpSPR43-nYFP in N. benthamiana (22) (Fig. 6B). Deletion of HBD abolished interaction of GluTR with cpSRP43 (Fig. 6B), while deletion of FBD had no effect on it. Conversely, cpSPR43 interacted specifically with GluTR-N64–163 containing the HBD and a part of the catalytic domain (Fig. 6B). In agreement with the in vivo observations, in vitro His pull-down assays confirmed that the truncated GluTR∆HBD has a lower affinity for cpSRP43 than WT GluTR (Fig. 6C).
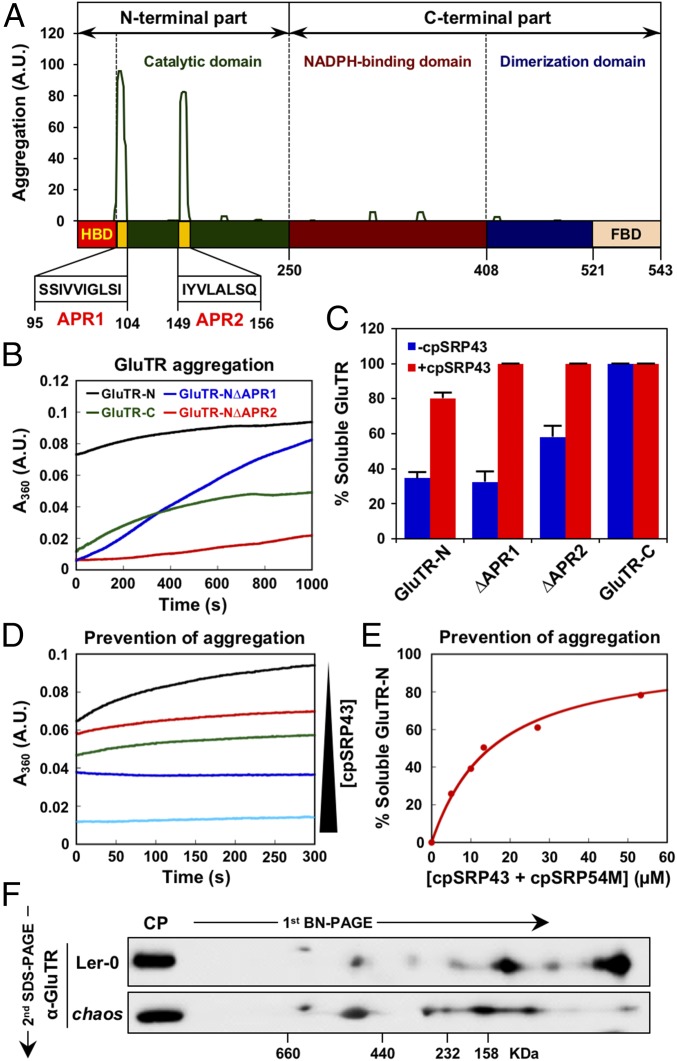
The SBD of cpSRP43 exhibits an ATP-independent chaperone function that prevents LHCP aggregation in aqueous milieu (15–17). Intriguingly, GluTR overexpression lines of Nicotiana tabacum or Arabidopsis thaliana did not display a corresponding increase in ALA synthesis rate; instead, GluTR formed aggregates (45). These aggregates remained insoluble even after leaf protein extracts had been boiled in Laemmli loading buffer. Excessive amounts of GluTR are apparently prone to form aggregates owing to an imbalance in protein homeostasis (46). Using the aggregation-predicting algorithm TANGO (47), two highly conserved aggregation-prone regions (APRs) predicted to form stable aggregates (48) were identified in the N-terminal catalytic domain of GluTR adjacent to the HBD (Fig. 7A and SI Appendix, Fig. S9).
Fig. 7.
cpSRP43 prevents the aggregation of GluTR. (A) GluTR sequence was analyzed with the TANGO algorithm, which identified regions that are predicted to be prone to aggregation. The sequences of two APRs (yellow) are indicated below. (B) Time courses for the aggregation of GluTR-N (black), GluTR-N∆APR1 (blue), GluTR-N ∆APR2 (red), and GluTR-C (green). (C) Semiquantitative analysis (legend for Fig. 5C) of the sedimentation assay (based on two biological replicates as shown in SI Appendix, Fig. S10) shows that cpSRP43 inhibits aggregation of GluTR. (D) Time courses for GluTR aggregation were measured at increasing cpSRP43 concentrations [5 (red), 10 (green), 27 (blue), and 53 (cyan) μM]. (E) Binding of GluTR-N to cpSRP43 was measured by the ability of cpSRP43 to prevent GluTR-N aggregation. The data were fit to SI Appendix, Eq. 3 and gave a Kapp value of 13 μM for GluTR-N. (F) Two-dimensional BN-SDS/PAGE analysis of GluTR allocation in chloroplasts isolated from WT (Ler-0) and chaos plants. GluTR monomers and oligomers were detected by immunoblotting analysis.
We therefore explored whether cpSRP43’s chaperone function acts to mask these APRs of GluTR. APR-dependent GluTR aggregation was directly monitored using a light-scattering assay in the presence or absence of cpSRP43. An N-terminal GluTR fragment (GluTR-N) containing the HBD and the catalytic domain rapidly formed large aggregates, whereas deletion of APR1 or APR2 reduced the propensity to aggregate (Fig. 7B). Moreover, the purified C-terminal half of GluTR (GluTR-C) is much more soluble in aqueous buffer than its N-terminal counterpart (Fig. 7B). The different propensities of the various GluTR fragments to aggregate were independently corroborated using a sedimentation assay (Fig. 7C and SI Appendix, Fig. S10). Importantly, the presence of cpSRP43 led to dose-dependent reductions in the aggregation of GluTR-N in the turbidity assay (Fig. 7 D and E) and shifted the majority of GluTR-N from the insoluble fraction into the soluble fraction in the sedimentation assay (Fig. 7C and SI Appendix, Fig. S10). Analysis of the light-scattering data yielded an apparent binding constant (Kapp) value of 13 μM for the interaction of cpSRP43 with GluTR-N (Fig. 7D), which is consistent with the Kd values determined above for the binding between cpSRP43 and GluTR using the fluorescence anisotropy assay (Fig. 4F).
To obtain evidence of the chaperone function of cpSRP43 in the regulation of GluTR in planta, we examined the formation of GluTR oligomers and/or aggregates by 2D BN-SDS/PAGE combined with Western blot analysis. Intriguingly, these experiments demonstrated that GluTR forms detergent-soluble dimers and higher molecular-weight oligomers/aggregates in chloroplast extracts. Most importantly, we observed that GluTR-containing complexes tended to be larger in the chaos mutant than in WT plants (Fig. 7F). These data corroborate the idea that cpSRP43 interacts with the aggregation-prone N-terminal region of GluTR and inhibits its aggregation, thus enhancing the stability of the protein and promoting its localization to the thylakoid membrane.
Discussion
The assembly of LHCs requires the synchronization of Chl biosynthesis with cpSRP-mediated LHCP transport to ensure the supply of Chl and LHCPs in stoichiometric amounts, thus minimizing degradation of LHCPs and avoiding accumulation of free photoreactive tetrapyrrole intermediates (7). Previous studies in cyanobacteria revealed the close proximity of Chl synthase to the Alb3 translocase and raised the possibility of synchronized operation of the committed enzymes in Chl a synthesis and those auxiliary factors mediating cotranslational integration of PS core subunits (e.g., D1) into thylakoids (23, 24). In this study, the link between Chl biosynthesis and cpSRP-mediated LHCP transport within plastids was further investigated. Our data revealed a role of a cpSRP component in the posttranslational regulation of ALA synthesis in Arabidopsis.
cpSRP43 Is an Effector of GluTR.
To respond to dynamically changing environmental conditions, plants have evolved a multifaceted posttranslational regulatory mechanism that adjusts ALA synthesis to meet varying demands for Chl and heme (18, 49). GluTR catalyzes the initial rate-limiting step in ALA synthesis. The activity, stability, and allocation of GluTR are modulated by various posttranslational regulators, such as GBP, FLU, and the Clp protease, which determine the rates of ALA synthesis for Chl and heme biosynthesis (19, 20, 50). GBP and the Clp subunits interact with the N-terminal HBD of GluTR, and GBP counteracts the degradation of GluTR by Clp (22, 30, 42, 43). Furthermore, FLU interacts with the C-terminal end of GluTR and inhibits ALA synthesis in the dark (31, 44).
Multiple lines of evidence presented here indicate that cpSRP43 assists to regulate GluTR and ALA synthesis in Arabidopsis (Fig. 8): (i) Of the four cpSRP components, only cpSRP43 specifically and directly interacts with GluTR (Fig. 3 and SI Appendix, Fig. S3); (ii) concurrent deletion of cpSRP43 and GBP drastically reduces GluTR levels via a posttranslational mechanism (Fig. 2); (iii) cpSRP43 interacts with N-terminal sequences of GluTR containing the HBD, which also provides binding sites for GBP and components of the Clp system (Fig. 6); (iv) the SBD of cpSRP43, which is responsible for chaperoning LHCP (17), directly mediates its interaction with GluTR (Fig. 4 and SI Appendix, Fig. S5); and (v) the chaperone activity of cpSRP43 efficiently prevents aggregation of the APR-containing N-terminal part of GluTR (Fig. 7 and SI Appendix, Fig. S10). We therefore propose that cpSRP43 contributes to the control of GluTR content by modulating its folding and stability in plastids.
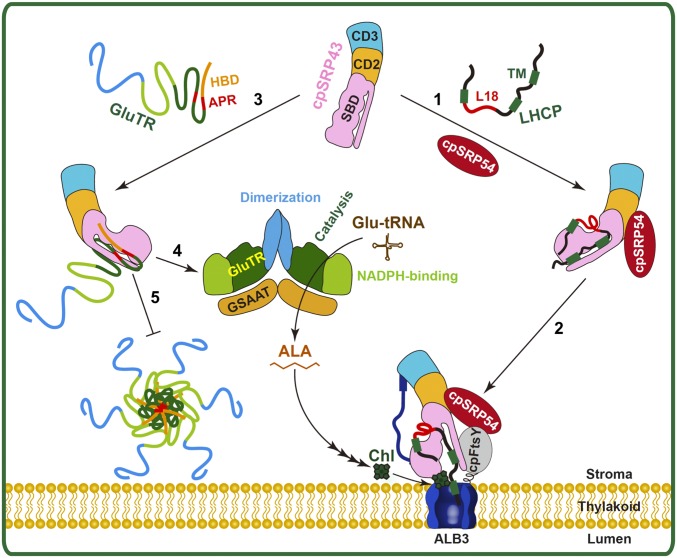
Fig. 8.
Model for the twofold function of cpSRP43 during LHCP membrane targeting and integration, as well as in the posttranslational control of GluTR. In step 1, cpSRP43 recognizes and chaperones newly imported LHCP. In step 2, LHCP is transported to the Alb3 translocase via the cpSRP pathway. In step 3, cpSRP43 binds to the N terminus of unfolded GluTR. In steps 4 and 5, the chaperone activity of cpSRP43 ensures proper folding of GluTR (step 4) and prevents GluTR aggregation (step 5).
A Function for cpSRP43 in Chaperoning GluTR.
To perform their cellular functions, most proteins must adopt a defined 3D structure. However, many proteins intrinsically tend to misfold and then aggregate (46). This applies particularly to nucleus-encoded plastid proteins that contain hydrophobic motifs or APRs, because they are translated on cytoplasmic ribosomes and imported into plastids in an unfolded state before they encounter the proper folding environment at the predestined subcompartment in plastids. This explains why a set of molecular chaperones is required to mediate the accurate targeting and proper folding of plastid-localized proteins (1, 51). To carry out its enzymatic function in ALA synthesis, GluTR forms a V-shaped homodimer (52). It was previously observed that in Arabidopsis and tobacco, overexpression of GluTR does not enhance ALA synthesis; instead, the excess GluTR forms aggregates (45). It can be deduced from this observation that folding of active GluTR requires chaperone activity, and that this functional capacity is limited in chloroplasts to maintain a native level of active GluTR.
The chaperone activity of cpSRP43 was long thought to be restricted to the LHCP family of proteins (Fig. 8, steps 1–2), based on its evolutionary coemergence with LHCs in terrestrial green plants (13, 14) and its recognition of the conserved L18 sequence motif in LHCPs (16, 32, 33). However, multiple observations presented here point to GluTR as another client protein for cpSRP43. First, the SBD of cpSRP43, which is necessary and sufficient for its chaperone activity (15–17), recognizes GluTR. Second, two APRs located in its N-terminal catalytic domain promote GluTR aggregation in vitro (Fig. 7 A–C), and cpSRP43 efficiently interacts with this same domain in vivo (Fig. 6B) and prevents its aggregation (Fig. 7 and SI Appendix, Fig. S10). Third, in the chaos mutant, which lacks cpSRP43, levels of GluTR monomers and dimers are reduced (relative to WT plants) in favor of larger oligomers (Fig. 7F). Finally, when cpSRP43 truncation mutants are introduced into chaos plants, mutants harboring deletions within the SBD fail to restore WT levels of GluTR (Fig. 5 E and F), ALA synthesis, or Chl accumulation (Fig. 5 B and C), whereas mutants lacking domains outside of the SBD still do so. Collectively, these results demonstrate that cpSRP43 can chaperone a client protein other than the LHCPs.
In properly folded GluTR, the two APRs are buried within the catalytic core of the HBD (41). However, GluTR molecules newly imported into chloroplast are much more susceptible to aggregation, as translocation across the chloroplast envelope requires unfolding of preproteins. The chaperone activity of cpSRP43 could protect the APRs in newly imported GluTR from exposure to the stroma, thus preventing its aggregation and facilitating adoption of its active conformation (Fig. 8, steps 3–4). Since the ternary complex composed of GluTR, cpSRP43, and the fluorescein-labeled loop1-TM2-L18 fragment derived from LHCb5 was detected in the anisotropy assay (Fig. 3F), we could not exclude the possibility that cpSRP43 could chaperone GluTR and LHCPs in a ternary complex. Furthermore, local conformational “breathing” or unfolding provides another plausible scenario in which the APRs in GluTR could be exposed, especially under stress conditions, and cpSRP43 could also protect GluTR from aggregation under these conditions (Fig. 8, step 5). The chaperone activity of cpSRP43 could contribute via either or both mechanisms to the control of the level of functionally folded GluTR.
cpSRP43 and GBP Cooperatively Preserve GluTR from Degradation in Plastids.
Control of protein stability provides another way to regulate ALA synthesis in chloroplasts. When 15N labeling was used to measure protein degradation rates in Arabidopsis rosette leaves, a correlation was observed between the elevated abundance of various TBS enzymes and their reduced degradation rates (53); this suggests that the regulation of degradation can modulate levels of GluTR and other TBS enzymes to tune the rates of Chl and heme biosynthesis. The Clp proteolytic system was found to mediate GluTR degradation (22, 42, 43). Its selector and chaperone subunits ClpS, ClpF, and ClpC interact with the N-terminal HBD of GluTR, but not with truncated GluTR∆HBD (22). Furthermore, GluTR∆HBD exhibited significantly enhanced stability in planta compared with WT GluTR (22). In addition, levels of GluTR are consistently higher in Clp protease mutants (42, 43). Taken together, these observations suggest that the HBD acts as an N degron (a Clp-specific degradation signal) that is recognized by the Clp protease to initiate GluTR degradation, possibly in response to the misfolding and aggregation of the HBD and nearby APRs (22).
The degradation of GluTR by Clp protease is counteracted by GBP, which also interacts with the HBD and stabilizes the active GluTR (22). Consequently, the greater accessibility of GluTR’s HBD to Clp protease can explain the lower GluTR levels seen in the gbp knockout mutant. Interestingly, cpSRP43 is shown here to also stabilize GluTR, and the synergistic effects of cpSRP43 and GBP deletions in reducing GluTR levels further indicate that the two proteins function in parallel to maintain GluTR stability (Fig. 2). As elevated levels of ClpC have been detected in mutants lacking a functional cpSRP pathway, including chaos and ffc (26, 54), the observed destabilization of GluTR in chaos may also be attributed to stress responses that up-regulate Clp levels to cope with the accumulation of misfolded and aggregated client proteins upon loss of cpSRP43.
Nevertheless, the observation that cpSRP43 also interacts with the N-terminal region of GluTR, including the HBD and APRs (Fig. 6), suggests a more direct mechanism in which cpSRP43 functions analogous to GBP and blocks the site recognized by the Clp protease system. This hypothesis is supported by in vitro studies showing an antagonistic effect of GBP on the cpSRP43–GluTR interaction (Fig. 3B), suggesting that GBP and cpSRP43 compete for overlapping binding sites in the N-terminal segment of GluTR. It is intriguing that the APRs, located near the HBD and buried in the core of folded GluTR, represent a recognition element that distinguishes the unfolded and misfolded conformations of GluTR from its active form, allowing its folding status, activity, and stability to be sensed and modulated by an intricate interplay between cpSRP43, GBP, and ClpC.
A scenario for the quantitative regulation of GluTR can be envisioned that involves the interaction of cpSRP43 with unfolded GluTR after its import into plastids, which prevents GluTR aggregation and ensures proper folding of the active enzyme. In contrast, GBP interacts with folded GluTR and forms a stable GBP–GluTR complex (41). In addition, a ternary GBP–GluTR–FLU complex has been proposed (55). Together, cpSRP43–GluTR and GBP–GluTR interactions provide a robust network to counteract the degradation of GluTR by the Clp protease.
Summary: cpSRP43 Coordinates LHCP Transport and Chl Biosynthesis
Multifaceted posttranslational regulation of GluTR is required to rapidly adjust the rate of ALA synthesis so as to provide adequate amounts of tetrapyrrole products in response to fluctuating environmental conditions such as changing light intensities. The dual role of cpSRP43 in transporting LHCP to the thylakoid membrane and controlling the amount of active GluTR represents a link between Chl biosynthesis and cpSRP-dependent processing of LHCPs (Fig. 8). We hypothesize that the ability of the cpSRP43 chaperone to enhance the proper folding and stability of GluTR ensures the supply of ALA for the synthesis of Chl during LHC biogenesis. Modulation of the level or activity of this chaperone could allow the cell to couple the rates of LHCP transport and Chl synthesis, both of which are enhanced by the presence of cpSRP43, and to ensure the supply of stoichiometric amounts of both components before their coordinated assembly into the thylakoid membrane. While we currently regard cpSRP43 as the sole source of this chaperone activity, the involvement of additional chloroplast auxiliary and regulatory factors in the quality control (activity and stability) of GluTR is not excluded and is worth examining in the future.
Materials and Methods
Plant materials and growth conditions used are described in SI Appendix, Materials and Methods and Table S1, together with details of all technical methods mentioned in the main text.
Supplementary Material
Acknowledgments
We thank Prof. Danja Schünemann (Ruhr University Bochum) for the help with Arabidopsis cpsrp mutants, GST-cpSRP43 expression vector, and antibodies against cpSRP components. We thank Dr. B. Hedtke and J. Apitz (B.G. laboratory) for help with research materials. This work was supported by a fellowship from the Alexander von Humboldt Foundation (to P.W.) and by grants from the Betty and Gordon Moore Foundation (Grant 94-3397785) and NIH (Grant R01 GM114390) (both to S.-o.S.) and a grant from the Deutsche Forschungsgemeinschaft (FOR2092, Grant GR 936/18-1 to B.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719645115/-/DCSupplemental.
References
- 1.Jarvis P, López-Juez E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol. 2013;14:787–802. doi: 10.1038/nrm3702. [DOI] [PubMed] [Google Scholar]
- 2.Allen JF, de Paula WB, Puthiyaveetil S, Nield J. A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci. 2011;16:645–655. doi: 10.1016/j.tplants.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Liu Z, et al. Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature. 2004;428:287–292. doi: 10.1038/nature02373. [DOI] [PubMed] [Google Scholar]
- 4.Wobbe L, Bassi R, Kruse O. Multi-level light capture control in plants and green algae. Trends Plant Sci. 2016;21:55–68. doi: 10.1016/j.tplants.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Plumley GF, Schmidt GW. Light-harvesting chlorophyll a/b complexes: Interdependent pigment synthesis and protein assembly. Plant Cell. 1995;7:689–704. doi: 10.1105/tpc.7.6.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Müller B, Eichacker LA. Assembly of the D1 precursor in monomeric photosystem II reaction center precomplexes precedes chlorophyll a-triggered accumulation of reaction center II in barley etioplasts. Plant Cell. 1999;11:2365–2377. [PMC free article] [PubMed] [Google Scholar]
- 7.Apel K, Hirt H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 8.Dall’Osto L, Bressan M, Bassi R. Biogenesis of light harvesting proteins. Biochim Biophys Acta. 2015;1847:861–871. doi: 10.1016/j.bbabio.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Richter CV, Bals T, Schünemann D. Component interactions, regulation and mechanisms of chloroplast signal recognition particle-dependent protein transport. Eur J Cell Biol. 2010;89:965–973. doi: 10.1016/j.ejcb.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 10.Akopian D, Shen K, Zhang X, Shan SO. Signal recognition particle: An essential protein-targeting machine. Annu Rev Biochem. 2013;82:693–721. doi: 10.1146/annurev-biochem-072711-164732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schüenemann D, et al. A novel signal recognition particle targets light-harvesting proteins to the thylakoid membranes. Proc Natl Acad Sci USA. 1998;95:10312–10316. doi: 10.1073/pnas.95.17.10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandrasekar S, Sweredoski MJ, Sohn CH, Hess S, Shan SO. Co-evolution of two GTPases enables efficient protein targeting in an RNA-less chloroplast signal recognition particle pathway. J Biol Chem. 2017;292:386–396. doi: 10.1074/jbc.M116.752931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dünschede B, et al. Chloroplast SRP54 was recruited for posttranslational protein transport via complex formation with chloroplast SRP43 during land plant evolution. J Biol Chem. 2015;290:13104–13114. doi: 10.1074/jbc.M114.597922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziehe D, Dünschede B, Schünemann D. From bacteria to chloroplasts: Evolution of the chloroplast SRP system. Biol Chem. 2017;398:653–661. doi: 10.1515/hsz-2016-0292. [DOI] [PubMed] [Google Scholar]
- 15.Jaru-Ampornpan P, et al. ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP subunit. Nat Struct Mol Biol. 2010;17:696–702. doi: 10.1038/nsmb.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaru-Ampornpan P, et al. Mechanism of an ATP-independent protein disaggregase: II. Distinct molecular interactions drive multiple steps during aggregate disassembly. J Biol Chem. 2013;288:13431–13445. doi: 10.1074/jbc.M113.462861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang FC, et al. Conformational dynamics of a membrane protein chaperone enables spatially regulated substrate capture and release. Proc Natl Acad Sci USA. 2016;113:E1615–E1624. doi: 10.1073/pnas.1524777113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brzezowski P, Richter AS, Grimm B. Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochim Biophys Acta. 2015;1847:968–985. doi: 10.1016/j.bbabio.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka R, Tanaka A. Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol. 2007;58:321–346. doi: 10.1146/annurev.arplant.57.032905.105448. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka R, Kobayashi K, Masuda T. Tetrapyrrole metabolism in Arabidopsis thaliana. Arabidopsis Book. 2011;9:e0145. doi: 10.1199/tab.0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apitz J, Schmied J, Lehmann MJ, Hedtke B, Grimm B. GluTR2 complements a hema1 mutant lacking glutamyl-tRNA reductase 1, but is differently regulated at the post-translational level. Plant Cell Physiol. 2014;55:645–657. doi: 10.1093/pcp/pcu016. [DOI] [PubMed] [Google Scholar]
- 22.Apitz J, et al. Posttranslational control of ALA synthesis includes GluTR degradation by Clp protease and stabilization by GluTR-binding protein. Plant Physiol. 2016;170:2040–2051. doi: 10.1104/pp.15.01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chidgey JW, et al. A cyanobacterial chlorophyll synthase-HliD complex associates with the Ycf39 protein and the YidC/Alb3 insertase. Plant Cell. 2014;26:1267–1279. doi: 10.1105/tpc.114.124495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knoppová J, et al. Discovery of a chlorophyll binding protein complex involved in the early steps of photosystem II assembly in Synechocystis. Plant Cell. 2014;26:1200–1212. doi: 10.1105/tpc.114.123919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klimyuk VI, et al. A chromodomain protein encoded by the arabidopsis CAO gene is a plant-specific component of the chloroplast signal recognition particle pathway that is involved in LHCP targeting. Plant Cell. 1999;11:87–99. doi: 10.1105/tpc.11.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pilgrim ML, van Wijk KJ, Parry DH, Sy DA, Hoffman NE. Expression of a dominant negative form of cpSRP54 inhibits chloroplast biogenesis in Arabidopsis. Plant J. 1998;13:177–186. doi: 10.1046/j.1365-313x.1998.00021.x. [DOI] [PubMed] [Google Scholar]
- 27.Hutin C, et al. Double mutation cpSRP43–/cpSRP54– is necessary to abolish the cpSRP pathway required for thylakoid targeting of the light-harvesting chlorophyll proteins. Plant J. 2002;29:531–543. doi: 10.1046/j.0960-7412.2001.01211.x. [DOI] [PubMed] [Google Scholar]
- 28.Tzvetkova-Chevolleau T, et al. Canonical signal recognition particle components can be bypassed for posttranslational protein targeting in chloroplasts. Plant Cell. 2007;19:1635–1648. doi: 10.1105/tpc.106.048959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang P, Grimm B. Comparative analysis of light-harvesting antennae and state transition in chlorina and cpSRP mutants. Plant Physiol. 2016;172:1519–1531. doi: 10.1104/pp.16.01009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Czarnecki O, et al. An Arabidopsis GluTR binding protein mediates spatial separation of 5-aminolevulinic acid synthesis in chloroplasts. Plant Cell. 2011;23:4476–4491. doi: 10.1105/tpc.111.086421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meskauskiene R, et al. FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2001;98:12826–12831. doi: 10.1073/pnas.221252798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stengel KF, et al. Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43. Science. 2008;321:253–256. doi: 10.1126/science.1158640. [DOI] [PubMed] [Google Scholar]
- 33.DeLille J, et al. A novel precursor recognition element facilitates posttranslational binding to the signal recognition particle in chloroplasts. Proc Natl Acad Sci USA. 2000;97:1926–1931. doi: 10.1073/pnas.030395197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis NE, et al. A dynamic cpSRP43-Albino3 interaction mediates translocase regulation of chloroplast signal recognition particle (cpSRP)-targeting components. J Biol Chem. 2010;285:34220–34230. doi: 10.1074/jbc.M110.160093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falk S, Ravaud S, Koch J, Sinning I. The C terminus of the Alb3 membrane insertase recruits cpSRP43 to the thylakoid membrane. J Biol Chem. 2010;285:5954–5962. doi: 10.1074/jbc.M109.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dünschede B, Bals T, Funke S, Schünemann D. Interaction studies between the chloroplast signal recognition particle subunit cpSRP43 and the full-length translocase Alb3 reveal a membrane-embedded binding region in Alb3 protein. J Biol Chem. 2011;286:35187–35195. doi: 10.1074/jbc.M111.250746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holdermann I, et al. Chromodomains read the arginine code of post-translational targeting. Nat Struct Mol Biol. 2012;19:260–263. doi: 10.1038/nsmb.2196. [DOI] [PubMed] [Google Scholar]
- 38.Horn A, et al. Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction. Nat Commun. 2015;6:8875. doi: 10.1038/ncomms9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao F, et al. Regulation of structural dynamics within a signal recognition particle promotes binding of protein targeting substrates. J Biol Chem. 2015;290:15462–15474. doi: 10.1074/jbc.M114.624346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang F, et al. Tetrapyrrole biosynthetic enzyme protoporphyrinogen IX oxidase 1 is required for plastid RNA editing. Proc Natl Acad Sci USA. 2014;111:2023–2028. doi: 10.1073/pnas.1316183111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao A, et al. Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its stimulator protein. Proc Natl Acad Sci USA. 2014;111:6630–6635. doi: 10.1073/pnas.1400166111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishimura K, et al. Discovery of a unique Clp component, ClpF, in chloroplasts: A proposed binary ClpF-ClpS1 adaptor complex functions in substrate recognition and delivery. Plant Cell. 2015;27:2677–2691. doi: 10.1105/tpc.15.00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishimura K, et al. ClpS1 is a conserved substrate selector for the chloroplast Clp protease system in Arabidopsis. Plant Cell. 2013;25:2276–2301. doi: 10.1105/tpc.113.112557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goslings D, et al. Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J. 2004;40:957–967. doi: 10.1111/j.1365-313X.2004.02262.x. [DOI] [PubMed] [Google Scholar]
- 45.Schmied J, Hedtke B, Grimm B. Overexpression of HEMA1 encoding glutamyl-tRNA reductase. J Plant Physiol. 2011;168:1372–1379. doi: 10.1016/j.jplph.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 46.Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- 47.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 48.Meric G, Robinson AS, Roberts CJ. Driving forces for nonnative protein aggregation and approaches to predict aggregation-prone regions. Annu Rev Chem Biomol Eng. 2017;8:139–159. doi: 10.1146/annurev-chembioeng-060816-101404. [DOI] [PubMed] [Google Scholar]
- 49.Czarnecki O, Grimm B. Post-translational control of tetrapyrrole biosynthesis in plants, algae, and cyanobacteria. J Exp Bot. 2012;63:1675–1687. doi: 10.1093/jxb/err437. [DOI] [PubMed] [Google Scholar]
- 50.Mochizuki N, et al. The cell biology of tetrapyrroles: A life and death struggle. Trends Plant Sci. 2010;15:488–498. doi: 10.1016/j.tplants.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 51.Flores-Pérez Ú, Jarvis P. Molecular chaperone involvement in chloroplast protein import. Biochim Biophys Acta. 2013;1833:332–340. doi: 10.1016/j.bbamcr.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 52.Moser J, et al. V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis. EMBO J. 2001;20:6583–6590. doi: 10.1093/emboj/20.23.6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li L, et al. Protein degradation rate in Arabidopsis thaliana leaf growth and development. Plant Cell. 2017;29:207–228. doi: 10.1105/tpc.16.00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amin P, et al. Arabidopsis mutants lacking the 43- and 54-kilodalton subunits of the chloroplast signal recognition particle have distinct phenotypes. Plant Physiol. 1999;121:61–70. doi: 10.1104/pp.121.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fang Y, et al. The Arabidopsis glutamyl-tRNA reductase (GluTR) forms a ternary complex with FLU and GluTR-binding protein. Sci Rep. 2016;6:19756. doi: 10.1038/srep19756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.