Significance
Natural killer (NK) cells play an important role as a first line of immunological defense against tumor initiation. However, cancer cell intrinsic factors that regulate NK cell activity against cancer cells are not fully known. We performed a chemical genetics screen using small-molecule inhibitors of epigenetic factors and identified Enhancer of zeste homolog 2 (EZH2) as a key regulator of NK cell-mediated eradication of hepatocellular carcinoma cells. These results uncover a role of EZH2 in the regulation of NK cells as a mechanism to promote oncogenesis. EZH2 inhibitors are in clinical trials for treatment of EZH2 mutant lymphomas and solid tumors with INI1 deficiency. Our findings may guide uses of EZH2 inhibitors for cancer treatment and encourage their preclinical testing in immunocompetent hosts.
Keywords: EZH2, NK cells, HCC, NK cell ligands, DNMT3A
Abstract
Natural killer (NK) cell-mediated tumor cell eradication could inhibit tumor initiation and progression. However, the factors that regulate NK cell-mediated cancer cell eradication remain unclear. We determined that hepatocellular carcinoma (HCC) cells exhibit transcriptional down-regulation of NK group 2D (NKG2D) ligands and are largely resistant to NK cell-mediated eradication. Because the down-regulation of NKG2D ligands occurred at the transcriptional level, we tested 32 chemical inhibitors of epigenetic regulators for their ability to re-express NKG2D ligands and enhance HCC cell eradication by NK cells and found that Enhancer of zeste homolog 2 (EZH2) was a transcriptional repressor of NKG2D ligands. The inhibition of EZH2 by small-molecule inhibitors or genetic means enhanced HCC cell eradication by NK cells in a NKG2D ligand-dependent manner. Collectively, these results demonstrate that EZH2 inhibition enhances HCC eradication by NK cells and that EZH2 functions, in part, as an oncogene by inhibiting immune response.
Hepatocellular carcinoma (HCC) is the sixth most prevalent cancer worldwide and the third most common cause of cancer-related deaths (1, 2). Most HCC patients are diagnosed at an advanced stage, which contributes to the very low 5-y survival rate of ∼10% (3–5). Several viral (hepatitis B and C) and nonviral (fatty liver disease) etiological factors have been shown to drive HCC initiation and progression (1, 2). Current therapies include sorafenib and regorafenib, drugs approved by the Food and Drug Administration for the treatment of HCC (6, 7). However, sorafenib and regorafenib provide only marginal benefits to HCC patients and cause significant therapy-related side effects (6, 7). Similarly, although encouraging results have been obtained with immunotherapies in certain cancers, such as Hodgkin’s disease and melanoma (8, 9), success has been much more limited in other cancer types, such as HCC (10). Therefore, an urgent need exists to identify new therapies for the effective treatment of HCC.
Innate immunity plays an important role in inhibiting microbial infections and serves as a first line of defense against tumor development and progression (11–14). Among the cells of the innate immune system, natural killer (NK) cells are key mediators of the innate immune response (15). However, the potential utility of NK cell-based cancer therapies has not been tested in a clinical setting, and the factors that determine the sensitivity or resistance of cancer cells to NK cell-mediated eradication remain largely unknown. Nonetheless, a better understanding of the mechanisms underlying NK cell-mediated eradication of cancer cells will facilitate more efficient NK cell engineering and/or effective tumor cell targeting, which could enhance NK cell-mediated tumor cell clearance.
Here, we show that HCC cells display widespread down-regulation of NKG2D ligands that correlates with reduced NK cell-mediated cytotoxicity toward HCC cells. Additionally, using a chemical screen involving small-molecule inhibitors of epigenetic regulators, we determined that Enhancer of zeste homolog 2 (EZH2), a polycomb repressive complex 2 protein, is an important repressor of NK group 2D (NKG2D) ligands and regulator of NK cell-mediated eradication of HCC cells. Collectively, these studies reveal a function of EZH2 in regulating the ability of NK cells to eradicate HCC cells and suggest that EZH2 might function, in part, as an oncogene by regulating NK cell activity against HCC cells.
Results
Hepatocellular Carcinoma Cells Exhibit Down-Regulation of NKG2D Ligands.
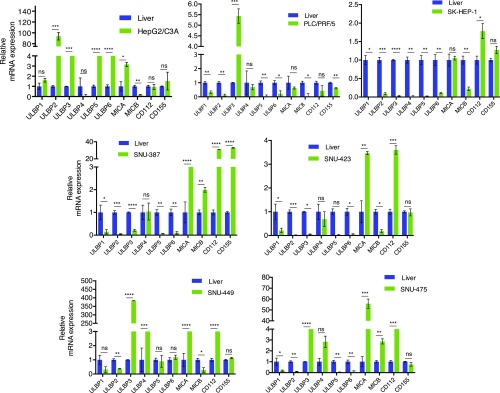
NK cells mediate innate immune responses against pathogens and cancer cells and encode NKG2D receptors, which are known to be important for the antitumor immunity exerted by NK cells (16–18). The ligands recognized by these receptors are called NKG2D ligands and include UL16-binding protein 1–6 (ULBP1-6), major histocompatibility complex class I chain-related gene A (MICA), major histocompatibility complex class I chain-related gene B (MICB), and two DNAX accessory molecule-1 (DNAM-1) ligands including CD112 and CD155 (19, 20). Therefore, as a first step toward understanding the mechanism by which HCC cells evade NK cell-mediated cytotoxicity, we measured the expression of these ligands in normal liver and in a panel of seven different human HCC cell lines (SI Appendix, Table S1). We found that a large majority of the HCC cell lines exhibited a significant down-regulation of several NKG2D ligands compared with normal liver samples (Fig. 1). For example, the HepG2/C3A primary HCC cell line expressed significantly higher levels of ULBP1, -2, -3, -5, and -6 and MICA, whereas the expression of ULBP4 and MICB were significantly lower in these cells compared with normal liver samples (Fig. 1). However, the metastatic SK-HEP-1 HCC cell line exhibited the down-regulation of all of the NKG2D ligands (Fig. 1). Similar to SK-HEP-1 cells, PLC/PRF/5 HCC cells exhibited the down-regulation of all NKG2D ligands except ULBP3. MICA was either up-regulated or normal in all HCC cell lines, except in PLC/PRF/5 cells, compared with normal liver samples (Fig. 1). Notably, ULBP1 expression was down-regulated in all HCC cell lines except HepG2/C3A cells (Fig. 1). Conversely, the DNAM1 ligands CD112 and CD155 were expressed in all of the HCC cell lines at levels comparable to the normal liver samples. SNU-387 showed higher fold-change in the expression of both DNAM1 ligands, whereas PLC/PRF/5 cells exhibited the down-regulation of the DNAM1 ligands (Fig. 1). Collectively, these results demonstrated that several NK cell ligands known to be necessary for NK cell-mediated cytotoxicity were down-regulated in HCC cells.
Fig. 1.
HCC cell lines exhibit widespread down-regulation of the NKG2D ligands. The expression of NK cell ligands was analyzed in human HCC cell lines and normal liver samples using RT-qPCR. mRNA expression for indicated genes relative to normal liver mRNA in indicated HCC cell lines is shown. Data are presented as mean ± SEM; ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001.
HCC Cells Are Largely Resistant to NK Cell-Mediated Cytotoxicity.
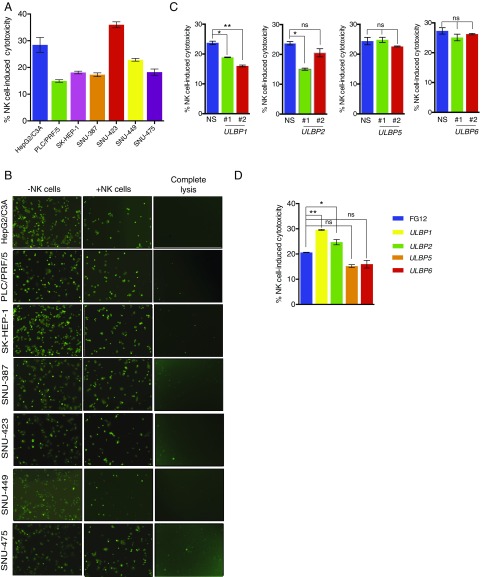
Several studies have shown that the expression of NKG2D ligands in cancer cells is associated with their eradication by NK cells (17, 18, 21). In addition, increased expression of ULBP1, MICA, and MICB correlates with better overall survival in HCC patients (22, 23). We found that a large majority of the HCC cell lines down-regulated NK cell ligands, including the NKG2D ligands (Fig. 1). To determine the correlation between NKG2D ligand expression and the sensitivity of HCC cells to NK cell-mediated cytotoxicity, we performed cell-based cytotoxicity assays using a panel of seven human HCC cell lines and two previously described methods for assessing NK cell-mediated cytotoxicity against cancer cells (24, 25). The first method was based on measuring lactate dehydrogenase (LDH) activity in the culture media after coculturing HCC cells with NK cells. The results presented in Fig. 2A show that the HCC cell lines exhibited varied sensitivity to NK cell-mediated cytotoxicity. HepG2/C3A and SNU-423 cells showed the highest sensitivity toward NK cell-mediated cytotoxicity (28.45 and 35.97%, respectively). However, the SK-HEP-1, PLC/PRF/5, SNU-387, SNU-475, and SNU-449 cells were almost twice as resistant to NK-cell mediated killing compared with HepG2/C3A and SNU-423 cells and showed only 18.1, 14.9, 17.28, 18.24, and 22.85% cytotoxicity, respectively, after incubation with NK cells (Fig. 2A).
Fig. 2.
NK cells exhibit differences in the ability to eradicate HCC. (A) HCC cell lines were incubated with NK cells in a 96-well plate at a 20:1 NK cell:cancer cell ratio. After incubating for 2 h, the supernatants were collected and LDH activity was measured. The percentage (%) of NK cell-induced cytotoxicity was calculated and plotted. (B) The cells were stained with Calcein AM and seeded with NK cells in 96-well plates at a 10:1 NK cell:cancer cell ratio. After incubation for 4 h, fluorescent images were captured using an inverted microscope. Images of indicated HCC cell lines showing loss of fluorescent cells (live cells) are presented. Calcein AM-stained cancer cells without NK cells served as the control. (C) HepG2 cells expressing shRNAs against ULBP1, -2, -5, or -6 and control nonspecific shRNAs were analyzed for NK cell cytotoxicity using an LDH activity cytotoxicity assay. The percentage (%) of NK cell-induced cytotoxicity in HepG2 cells was calculated and plotted for the indicated shRNAs. (D) ULBP1, -2, -5 or -6 ligands were ectopically expressed in SK-HEP-1 cells and analyzed for NK cell-mediated cytotoxicity using an LDH activity-based cytotoxicity assay. FG12 vector-transfected cells served as the negative control. The percentage (%) of NK cell-induced cytotoxicity in SK-HEP-1 cells was calculated and plotted for the indicated vector or ligand. Data are presented as mean ± SEM; ns, not significant; *P < 0.05; and **P < 0.01.
To validate these findings, we used a Calcein AM dye-based fluorescent imaging method to measure NK cell-mediated cytotoxicity. The HCC cell lines were first labeled with Calcein AM dye and then incubated with NK cells, and the resulting NK cell-mediated cytotoxicity was quantitated using fluorescent imaging. In accord with the LDH NK cell-mediated cytotoxicity assay, the results of the Calcein AM NK cell cytotoxicity assay revealed that SNU-423 and HepG2/C3A cells were more sensitive and the SK-HEP-1, PLC/PRF/5, SNU-387, and SNU-475 cells were less sensitive to NK cell-mediated cytotoxicity (Fig. 2B). Collectively, these results demonstrate that HCC cells are largely resistant to NK cell-mediated cytotoxicity.
ULBP1 Is both Necessary and Sufficient for NK Cell-Mediated Cytotoxicity Against HCC Cells.
After determining the expression of the NK cell ligands in HCC and NK cell-mediated cytotoxicity against HCC cells, we also assessed if NK cell-mediated cytotoxicity correlated with the expression of the NK cell ligands and found that HepG2/C3A cells exhibited the highest sensitivity to NK cell-mediated cytotoxicity and expressed ULBP1, -2, -3, -5, and -6 and MICA (Fig. 1). However, the HCC cell lines that were more resistant to NK cell-mediated cytotoxicity showed a general down-regulation of ULBP1, -2, -5, and -6 (Fig. 1). Based on these results, we hypothesized that NKG2D ligands, in particular ULBP1, -2, -5, and -6, might determine NK cell-mediated eradication of HCC cells. Our rationale was further supported by previous studies showing that reduced NKG2D ligand expression correlates with poor overall survival and higher rates of recurrence among HCC patients (22, 23).
However, only a few studies have directly tested the effect of individual NKG2D ligands on NK cell-mediated cancer cell eradication (21, 26, 27). Therefore, we performed experiments to determine the effect of ULBP1, -2, -5, and -6 on the ability of NK cells to eradicate HCC cells. To this end, we either knocked down the expression of these NKG2D ligands in HepG2/C3A cells that were relatively sensitive to NK cell-mediated cytotoxicity and expressed the ULBP1, -2, -5, and -6 ligands or overexpressed the ligands in SK-HEP-1 cells, which were relatively resistant to NK cell-mediated cytotoxicity and exhibited the down-regulation of these ligands. For the knockdown experiments, we analyzed two unrelated shRNAs directed against the same target gene. The knockdown of ULBP1, using either one of the two ULBP1 shRNAs, significantly decreased the ability of NK cells to eradicate HepG2/C3A cells (Fig. 2C and SI Appendix, Fig. S1 A and B). However, shRNAs directed against ULBP2, -5, or -6 did not lead to a reproducible significant decrease in NK cell-mediated killing of HepG2 cells (Fig. 2C and SI Appendix, Fig. S1 A and B).
We then tested the effects of the ectopic expression of ULBP1, -2, -5, or -6 in SK-HEP-1 cells. The results showed that the ectopic expression of ULBP1 or -2 increased NK cell-mediated cytotoxicity against SK-HEP-1 cells (Fig. 2D and SI Appendix, Fig. S1 C and D), whereas the ectopic expression of ULBP5 or ULBP6 had no significant effects (Fig. 2D and SI Appendix, Fig. S1 C and D). Collectively, our results demonstrate that, of the NKG2D ligands tested, ULBP1 is both necessary and sufficient to regulate NK cell-mediated cytotoxicity against HCC cells.
A Chemical Epigenetic Regulator Inhibitor Screen Identifies EZH2 as a Major Regulator of NK Cell Ligand Expression in HCC Cells.
The results of our experiments showed that NK cell ligands were transcriptionally repressed in HCC cells and that some of the ligands, such as ULBP1, were both necessary and sufficient for NK cell-mediated HCC cell eradication. Therefore, we rationalized that NK cell ligands could be re-expressed using pharmacological inhibitors targeting epigenetic regulators, which might subsequently increase NK cell-mediated eradication of HCC cells. Therefore, we tested 32 small-molecule inhibitors obtained from the Structural Genomics Consortium that specifically inhibit the activities of different epigenetic regulators (SI Appendix, Table S2). To perform the screen, we treated SK-HEP-1 cells with two different concentrations of the small-molecule inhibitors and measured NK cell ligand expression (Fig. 3 A and B and SI Appendix, Figs. S2 and S3 and Table S3). The results were then validated in PLC/PRF/5 cells as described below. The rationales for selecting SK-HEP-1 and PLC/PRF/5 cells for the screen and subsequent validation experiments were the following: (i) a large number of NKG2D ligands were down-regulated in these HCC cell lines compared with normal liver cells (Fig. 1) and (ii) these cell lines were among those that were more resistant to NK cell-mediated cytotoxicity than other HCC cell lines (Fig. 2). Our screen led to the identification of inhibitors that targeted eight different groups of epigenetic regulators (Baz2A/2B, BRPF1/2/3, EZH2/H1, an IDH1 mutant, LSD1, PAD4, PRMT5, and SMYD2) that upon treatment re-expressed seven or more NK cell ligands in HCC cells. The analysis of various patient samples in HCC gene expression datasets and The Cancer Genome Atlas (TCGA) HCC data revealed that of these nine groups of epigenetic regulators, four were also significantly overexpressed in the TCGA and other HCC datasets (SI Appendix, Fig. S4 A and B). These four chromatin regulators included EZH2, BRPF1/2/3, Baz2A/2B, and SMYD2. Based on clinical relevance, we prioritized five inhibitors that targeted EZH2, SMYD2, BAZ2A/2B, or BRPF1/2/3 for further studies.
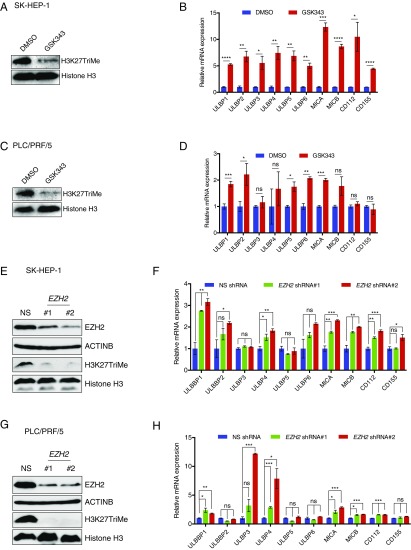
Fig. 3.
Pharmacological and genetic inhibition of EZH2 results in the up-regulation of NK cell ligands on HCC cells. (A) SK-HEP-1 cells were treated with DMSO or the EZH2 inhibitor GSK343 (3 μM) for 48 h. Immunoblotting for the EZH2-mediated H3K27TriMe mark was performed using DMSO or GSK343-treated cells. Histone H3 was used as a loading control. (B) SK-HEP-1 cells were treated with DMSO or the EZH2 inhibitor GSK343 (3 μM) for 48 h. NK cell ligand mRNA expression in GSK343-treated cells relative to DMSO-treated cells is shown. (C) PLC/PRF/5 cells were treated with DMSO or the EZH2 inhibitor GSK343 (3 μM) for 48 h. Immunoblotting for the EZH2-mediated H3K27TriMe mark was performed using DMSO- or GSK343-treated cells. Histone H3 was used as a loading control. (D) PLC/PRF/5 cells were treated with DMSO or the EZH2 inhibitor GSK343 (3 μM) for 48 h. NK cell ligand mRNA expression in GSK343-treated cells relative to DMSO-treated cells is shown. (E) SK-HEP-1 cells expressing either a nonsilencing (NS) shRNA or EZH2 shRNAs were analyzed for the indicated proteins by immunoblotting. (F) SK-HEP-1 cells expressing either a NS shRNA or EZH2 shRNAs were analyzed for the indicated ligands by RT-qPCR. NK cell ligand mRNA expression is plotted relative to NS shRNA-expressing cells. (G) PLC/PRF/5 cells expressing either a NS shRNA or EZH2 shRNAs were analyzed for the indicated proteins by immunoblotting. (H) PLC/PRF/5 cells expressing either a NS shRNA or EZH2 shRNAs were analyzed for the expression of the indicated ligands by RT-qPCR. NK cell ligand mRNA expression is plotted relative to NS shRNA-expressing cells. Data are presented as mean ± SEM; ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001.
To determine the ability of the inhibitors to re-express NK cell ligands in other HCC cell lines, we treated the PLC/PRF/5 HCC cell line with the six inhibitors and found that only inhibitors targeting EZH2 resulted in the re-expression of multiple NK cell ligands (Fig. 3 C and D and SI Appendix, Fig. S4C and Table S4).
After confirming that EZH2 was an important repressor of NK cell ligands, we tested the EZH2 inhibitor GSK126 and assessed the re-expression of the NK cell ligands. To this end, we treated both SK-HEP-1 and PLC/PRF/5 cells with GSK126 and found that GSK126 was also able to stimulate the expression of multiple NK cell ligands in both HCC cell lines (SI Appendix, Fig. S5 A and B).
Finally, we used a genetic approach to further strengthen our EZH2 inhibitor findings (GSK343 and GSK126) and knocked down the expression of EZH2 using shRNAs in SK-HEP-1 and PLC/PRF/5 cells. SK-HEP-1 and PLC/PRF/5 cells expressing nonspecific shRNA were used as negative controls. We measured the expression of NK cell ligands in these cells and found that the knockdown of EZH2, similar to that of EZH2 inhibitors, resulted in the increased expression of NK cell ligands in both SK-HEP-1 and PLC/PRF/5 cells (Fig. 3 E–H). Collectively, these results demonstrate that EZH2 inhibition can be used as a strategy to re-express NK cell ligands, which might subsequently enhance NK cell-mediated eradication of HCC cells.
EZH2 Inhibition Enhances NK Cell-Mediated Eradication of HCC Cells.
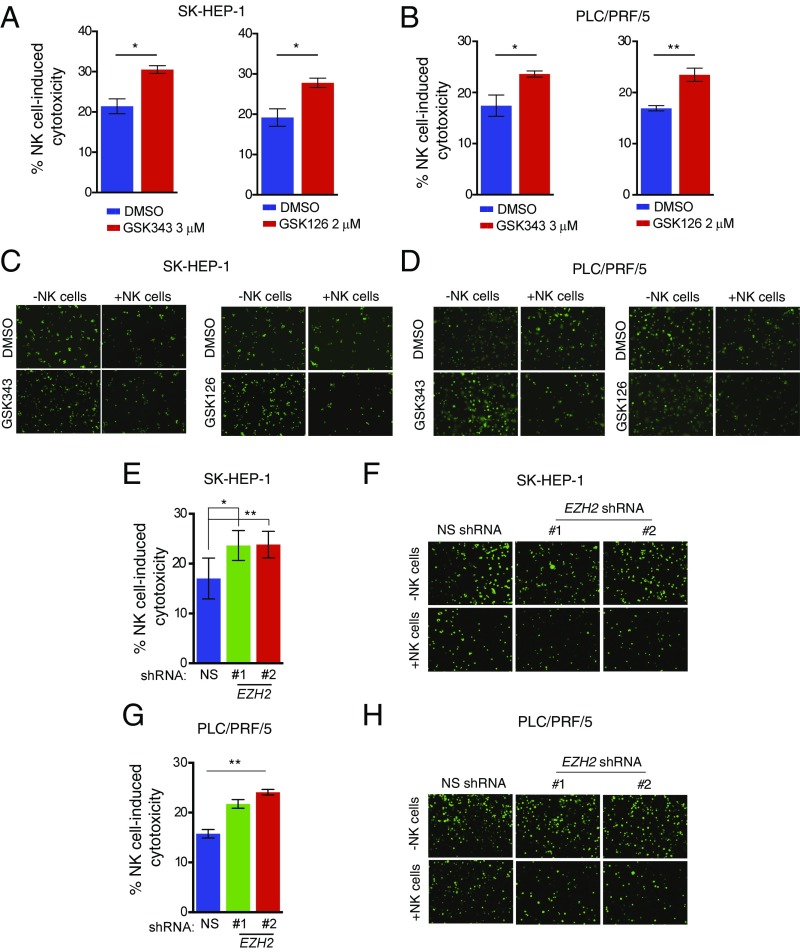
After confirming that both the pharmacological and the genetic inhibition of EZH2 resulted in the re-expression of NK cell ligands, we tested if the treatment of HCC cells with EZH2 inhibitors resulted in their enhanced eradication by NK cells. We treated the SK-HEP-1 or PLC/PRF/5 cells with the EZH2 inhibitors GSK343 and GSK126 and measured NK cell-mediated cytotoxicity using LDH- and Calcine AM-based methods, as described. Our results showed that the treatment of SK-HEP-1 and PLC/PRF/5 cells with EZH2 inhibitors resulted in a significant increase in NK cell-induced cytotoxicity against HCC cells (Fig. 4 A–D). To confirm the results obtained using EZH2 inhibitors, we also tested the effect of shRNA-mediated EZH2 knockdown on NK cell-mediated eradication of HCC cells, using SK-HEP-1 and PLC/PRF/5 cells expressing nonspecific shRNAs as controls. In agreement with the results observed with the EZH2 inhibitors, the shRNA-mediated knockdown of EZH2 also resulted in increased NK cell-induced cytotoxicity against HCC cells (Fig. 4 E–H).
Fig. 4.
Pharmacological and genetic inhibition of EZH2 enhances NK cell-mediated cytotoxicity against HCC cells. (A) SK-HEP-1 cells were treated with DMSO or the EZH2 inhibitors GSK343 (3 μM) or GSK126 (2 μM) for 48 h and incubated with NK cells at a ratio of 20:1. The percentage (%) of NK cell-induced cytotoxicity was calculated and plotted. (B) PLC/PRF/5 cells were treated with DMSO or the EZH2 inhibitors GSK343 (3 μM) or GSK126 (2 μM) for 48 h and incubated with NK cells at a ratio of 20:1. The percentage (%) of NK cell-induced cytotoxicity was calculated and plotted. (C) SK-HEP-1 cells were treated with DMSO or the EZH2 inhibitors GSK343 (3 μM) or GSK126 (2 μM) for 48 h, stained with Calcein AM, and incubated with NK cells at a ratio of 10:1. At 4 h post incubation, fluorescent images were captured using an inverted microscope. Images depicting the loss of fluorescent cells (live cells) under the indicated conditions are presented. Calcein AM-stained cancer cells without NK cells served as a negative control. (D) PLC/PRF/5 cells were treated with DMSO or the EZH2 inhibitors GSK343 (3 μM) or GSK126 (2 μM) for 48 h, stained with Calcein AM, and incubated with NK cells at a ratio of 10:1. At 4 h post incubation, fluorescent images were captured using an inverted microscope. Images depicting the loss of fluorescent cells (live cells) under the indicated conditions are presented. Calcein AM-stained cancer cells without NK cells served as the negative control. (E) SK-HEP-1 cells expressing NS or EZH2 shRNAs were analyzed for NK cell-induced cytotoxicity following incubation with NK cells at a ratio of 20:1. The percentage (%) of NK cell-induced cytotoxicity was calculated and plotted. (F) SK-HEP-1 cells expressing NS or EZH2 shRNAs were stained with Calcein AM and incubated with NK cells at a ratio of 10:1. Fluorescent images were captured 4 h post incubation using an inverted microscope. Images depicting the loss of fluorescent cells under the indicated conditions (live cells) are presented. Calcein AM-stained cancer cells without NK cells served as a negative control. (G) PLC/PRF/5 cells expressing NS or EZH2 shRNAs were analyzed for NK cell-induced cytotoxicity by incubation with NK cells at a ratio of 20:1. The percentage (%) of NK cell-induced cytotoxicity was calculated and plotted. (H) PLC/PRF/5 cells expressing NS or EZH2 shRNAs were stained with Calcein AM and incubated with NK cells at a ratio of 10:1. Fluorescent images were captured 4 h post incubation using an inverted microscope. Images depicting the loss of fluorescent cells (live cells) under the indicated conditions are presented. Calcein AM-stained cancer cells without NK cells served as a negative control. Data are presented as mean ± SEM; *P < 0.05 and **P < 0.01.
NK Cell-Mediated Cytotoxicity Stimulated by EZH2 Inhibition Is Dependent upon Re-Expression of NK Cell Ligands.
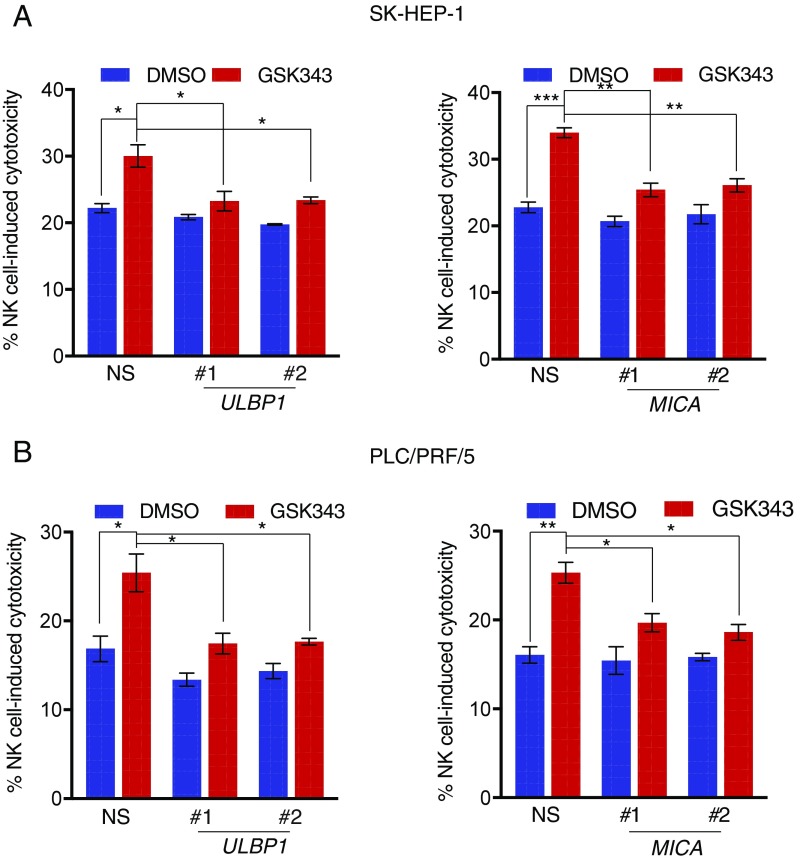
Next, we evaluated if the re-expression of NK cell ligands was required for the ability of EZH2 inhibitors or EZH2 shRNAs to enhance the ability of NK cells to eradicate HCC cells. We focused on ULBP1, MICA, and MICB ligands because these ligands were commonly up-regulated by both EZH2 inhibitors and EZH2 shRNAs in both SK-HEP-1 and PLC/PRF/5 cells (Fig. 3). We first knocked down the expression of ULBP1, MICA, and MICB in both SK-HEP-1 and PLC/PRF5 cells (SI Appendix, Fig. S5 C and D). Next, we tested if the knockdown of these ligands blocked the ability of EZH2 inhibitors to enhance NK cell-mediated eradication of HCC cells. Our results showed that the knockdown of either ULBP1 or MICA significantly inhibited NK cell-mediated cytotoxicity after GSK343 treatment in both SK-HEP-1 and PLC/PRF5 cells (Fig. 5), whereas MICB knockdown did not affect the EZH2 inhibitor-induced stimulation of NK cell-mediated HCC cell clearance (SI Appendix, Fig. S6A). Collectively, these results demonstrate that the up-regulation of ULBP1 and MICA following EZH2 inhibition is necessary for NK cell-mediated eradication of HCC cells.
Fig. 5.
ULBP1 and MICA are necessary for EZH2 inhibition-induced enhanced NK cell-mediated HCC cell eradication. (A) SK-HEP-1 cells expressing either NS shRNA, ULBP1 (Left), or MICA (Right) shRNAs were treated with GSK343 (3 μM) for 48 h and analyzed for NK cell-induced cytotoxicity. The percentage (%) of NK cell-induced cytotoxicity is shown. (B) PLC/PRF/5 cells expressing either NS shRNA, ULBP1 (Left), or MICA (Right) shRNAs were treated with GSK343 (3 μM) for 48 h and analyzed for NK cell-induced cytotoxicity. The percentage (%) of NK cell-induced cytotoxicity is shown. Data are presented as mean ± SEM; *P < 0.05; **P < 0.01; and ***P < 0.001.
EZH2 Enhances ULBP1 Promoter DNA Methylation by Promoting DNA Methyltransferase 3A.
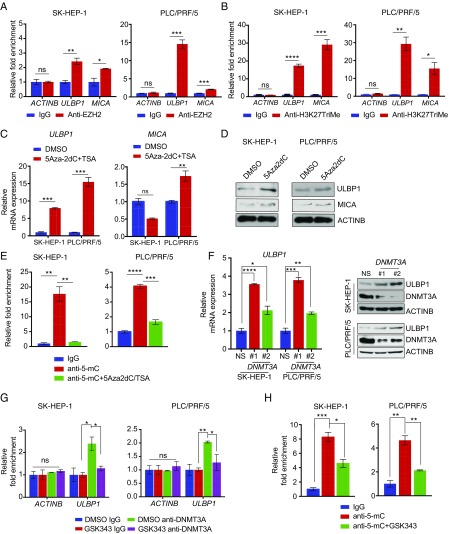
Finally, we aimed to determine the mechanism underlying ULBP1 and MICA ligand re-expression after treatment with the EZH2 inhibitor. We performed chromatin immunoprecipitation (ChIP) assays to measure the recruitment of EZH2 on ULBP1 and MICA promoters in HCC cell lines. Our results show that EZH2 was significantly enriched on ULBP1 and MICA promoters compared with the control ACTINB gene promoter. Furthermore, to determine if the increased recruitment of EZH2 also correlated with its histone mark, we performed a ChIP assay to measure the H3K27TriMe mark. The results indicated that EZH2 directly associated with the promoters of NK cell ligands, such as ULBP1 and MICA (Fig. 6A), and that its association with these promoters correlated with increases in the H3K27TriMe histone mark on these promoters (Fig. 6B).
Fig. 6.
ULBP1 is repressed by EZH2 in a DNMT3A-mediated DNA methylation-dependent manner. (A) SK-HEP-1 or PLC/PRF/5 cells were analyzed for EZH2 recruitment on the ULBP1 or MICA promoter by ChIP analysis. The ACTINB promoter was used as a negative control. The ChIP results are shown as fold-change relative to the IgG control. (B) SK-HEP-1 or PLC/PRF/5 cells were analyzed for the H3K27TriMe mark on the ULBP1 or MICA promoter by ChIP analysis. The ACTINB promoter was used as a negative control. The ChIP results are shown as fold-change relative to the IgG control. (C) SK-HEP-1 or PLC/PRF/5 cells were treated with either DMSO or 5Aza2dC (5 μM) and TSA (1 μM) for 72 h. The expression of ULBP1 or MICA mRNA was analyzed by RT-qPCR. ULBP1 or MICA mRNA expression relative to DMSO-treated cells is shown. (D) SK-HEP-1 or PLC/PRF/5 cells were treated with either DMSO or 5Aza2dC (5 μM) and TSA (1 μM) for 72 h. The expression of the indicated proteins was analyzed by immunoblotting. (E) SK-HEP-1 or PLC/PRF/5 cells were treated with either DMSO or 3Aza2DC (5 μM) and TSA (1 μM) for 72 h. The ULBP1 promoter was analyzed for DNA methylation using the MeDIP method. The MeDIP results are shown as the fold-change in DNA methylation relative to IgG in either DMSO or 5Aza2dC+TSA-treated HCC cells. (F) SK-HEP-1 or PLCPRF/5 cells expressing either a NS shRNA or DNMT3A shRNAs were analyzed for ULBP1 mRNA expression (Left) or for the expression of the indicated proteins by immunoblotting (Right). (G) SK-HEP-1 or PLCPRF/5 cells were analyzed for DNMT3A recruitment on the ULBP1 promoter using a ChIP assay. The ACTINB promoter was used as a negative control. Relative fold-change compared with IgG is shown. (H) SK-HEP-1 or PLC/PRF/5 cells were treated with either DMSO or GSK343 (3 μM) for 48 h. The ULBP1 promoter was analyzed for DNA methylation using the MeDIP method. The MeDIP results are shown as fold-change in DNA methylation relative to IgG in either DMSO- or GSK343-treated HCC cells. Data are presented as mean ± SEM; ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001.
Next, we deduced that the EZH2-mediated repression of NK cell ligands was dependent upon DNA methylation in NK cell ligand promoters. This rationale was based on previous studies showing that EZH2 interacts with DNA methyltransferases (DNMTs) and is necessary for the recruitment of DNMTs on gene promoters and consequential promoter DNA methylation (28–30). To test our theory, we first analyzed the ULBP1 and MICA promoter regions. Furthermore, the promoter sequence analysis predicted distinct CpG islands on both the ULBP1 and the MICA promoters (SI Appendix, Fig. S6B). Based on the collective evidence, we tested the effects of 5-Aza-2-deoxyCytidine (5Aza2dC) and Trichostatin A (TSA) on the induction of ULPB1 and MICA expression in HCC cell lines. We found that the treatment of SK-HEP-1 and PLC/PRF/5 cells with 5Aza2dC resulted in the re-expression of ULPB1 in both cell lines, whereas MICA was only re-expressed in PLC/PRF/5 cells (Fig. 6 C and D). These results suggested that ULBP1 expression is largely regulated by promoter DNA methylation in HCC cell lines. Therefore, to confirm that the ULBP1 promoter is indeed subject to direct promoter DNA methylation, we performed a methylated-DNA immunoprecipitation (MeDIP) analysis using the 5-methylcytosine antibody to evaluate ULBP1 promoter DNA methylation. The MeDIP results revealed significantly higher ULPB1 promoter DNA methylation in HCC cell lines, which was reduced following 5Aza2dC and TSA treatment (Fig. 6E). Collectively, these results demonstrated that the ULBP1 promoter is methylated in SK-HEP-1 and PLC/PRF/5 cells.
To determine whether ULBP1 promoter methylation is dependent on EZH2-mediated DNMT recruitment, we evaluated the expression of DNMT1, DNMT3A, and DNMT3B. We found that DNMT1 and DNMT3A were expressed in HCC cells, but DNMT3B was undetectable (SI Appendix, Fig. S6C). Therefore, we focused on DNMT1 and DNMT3A for further studies. First, we knocked down the expression of DNMT1 and DNMT3A in SK-HEP-1 and PLC/PRF/5 cells using shRNAs (SI Appendix, Fig. S7 A and B). Our results showed that DNMT3A knockdown significantly up-regulated the expression of ULBP1 in SK-HEP-1 and PLC/PRF/5 cells, whereas DNMT1 knockdown had a significant effect only in PLC/PRF/5 cells but not in SK-HEP-1 cells (Fig. 6F and SI Appendix, Fig. S7 C and D). The results also indicated that DNMT3A is an important regulator of ULBP1 expression downstream of EZH2 in multiple HCC cells, whereas DNMT1 might play a more limited role in some HCC cells. Furthermore, to determine whether DNMT3A recruitment to the ULBP1 promoter was dependent on EZH2 activity, we performed a ChIP analysis of SK-HEP-1 and PLC/PRF/5 cells treated with the EZH2 inhibitor GSK343 or DMSO. We found that treatment with the EZH2 inhibitor resulted in reduced DNMT3A recruitment to the ULBP1 ligand promoter (Fig. 6G), reduced promoter DNA methylation, and increased ULBP1 expression (Fig. 6H). Collectively, these results indicated that EZH2-induced recruitment of DNMT3A to the ULBP1 promoter caused a subsequent increase in ULBP1 promoter DNA methylation.
Discussion
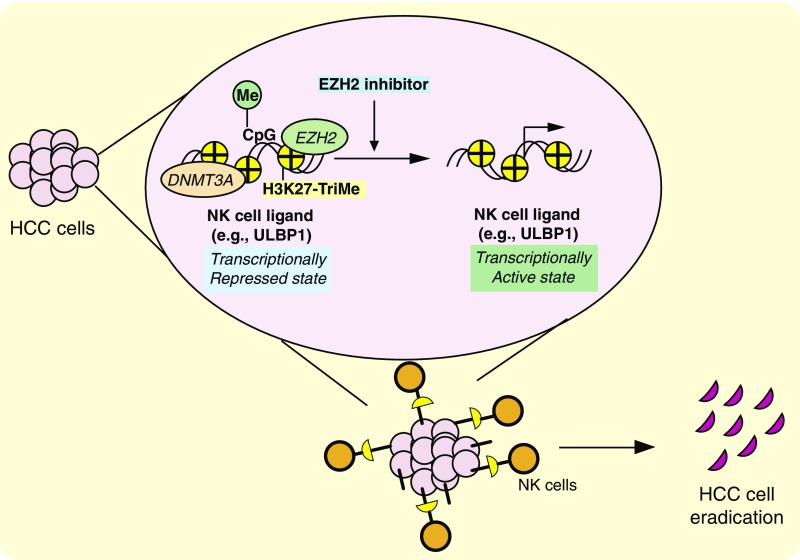
HCC is the most common and deadliest form of liver cancer. However, current therapies provide negligible clinical benefits and cause a large number of therapy-related life-threatening side effects. Here, we investigated the epigenetic factors that regulate NK cell-mediated cytotoxicity against HCC cells and determined that EZH2 is an important regulator of NK cell-mediated clearance of HCC cells. Our results allowed us to draw several important conclusions. First, we found that HCC cells exhibit transcriptional down-regulation of NK cell ligands, which could serve as a possible mechanism for evading NK cell-mediated cytotoxicity and subsequent immune invasion in HCC cells. Second, we demonstrated that HCC cells are largely resistant to NK cell-mediated eradication and that enhancing the effectiveness of NK cell-mediated eradication of HCC cells could be of significant therapeutic benefit to HCC patients. Finally, using an epigenetic regulator targeting-based chemical screen, we determined and validated that EZH2 is an important regulator of NK cell-mediated eradication of HCC cells that functions, in part, by regulating NK cell ligands, including ULBP1 promoter DNA methylation via DNMT3A in an EZH2-dependent manner. Our results are summarized in Fig. 7 and discussed below.
Fig. 7.
Model of EZH2-mediated regulation of HCC cell eradication. The model indicates that EZH2 represses the expression of NK cell ligands and that the pharmacological inhibition of EZH2 results in the re-expression of NK cell ligands and increases NK cell-mediated cytotoxicity against HCC cells.
NK Cell Ligands and Their Role in HCC.
NK cells recognize and bind to NK cell ligands expressed on target cells and induce their lysis (17, 18, 31). Several prior studies involving mouse models have shown an important role for NK cell ligands in tumor suppression (16, 17, 19). The down-regulation of NK cell ligands has also been shown to correlate with reduced overall survival and poor prognosis in cancer patients (22, 23). However, no study to date has comprehensively analyzed NK cell-mediated cytotoxicity against HCC cells and the epigenetic factors that regulate NK cell-mediated HCC cell eradication. In this study, we analyzed the expression of NK cell ligands in a panel of human HCC cell lines and found that several of the NKG2D ligands were down-regulated in a majority of the HCC cell lines.
Previous studies have also demonstrated that NKG2D ligand levels define the degree of cytotoxicity against target cells and that high NKG2D ligand levels correlate with NK cell-dependent tumor elimination (17, 20). Our NK cell-mediated cytotoxicity experiments involving a panel of HCC cell lines showed that the expression of NKG2D ligands generally correlated with the ability of NK cells to induce cytotoxicity against the HCC cells. For example, the PLC/PRF/5 and SK-HEP-1 HCC cells, which exhibited widespread down-regulation of NKG2D ligands, were relatively more resistant to NK cell-mediated cytotoxicity compared with cells such as HepG2/C3A, which expressed several of the ligands. Additionally, our studies measuring the contribution of individual NKG2D ligands revealed that ULBP1 significantly increased NK cell-mediated eradication of HCC cells. Collectively, these results demonstrated that specific approaches that allow increased NKG2D ligand expression could be used as a therapeutic approach to treat HCC.
EZH2 as a Regulator of NK Cell Cytotoxicity Against HCC.
Epigenetic alterations, such as changes in DNA methylation and/or modification of histone proteins, can consequently result in changes in gene expression and influence several aspects of cellular physiology and function. The role of epigenetic changes in host immune system regulation has been established in several prior studies (32–34). However, most of these studies have focused on events that cause these epigenetic alterations in immune cells and how they influence the functionality of immune cells. Furthermore, studies that have comprehensively measured the impact of epigenetic changes in target cells (such as in pathogens and cancer cells) on the functionality of immune cells are limited, and, in most cases, the molecular underpinnings for the phenomena reported are not established.
Using a chemical screen that targeted epigenetic regulators, we determined that EZH2 is a major modulator of NK cell-mediated eradication of HCC cells. Likewise, we showed that the genetic and pharmacological inhibition of EZH2 results in the re-expression of several NKG2D ligands and that their re-expression correlates with increased cytotoxicity of NK cells toward HCC cells. Our results also suggest that the oncogenic function of EZH2 might also be mediated, in part, by its ability to regulate immune cell function, such as that of NK cells. Notably, EZH2 overexpression is reportedly associated with tumor progression and aggressiveness in HCC (35, 36). Therefore, our results are in accord with the proposed role of NK cells in preventing tumor progression in an EZH2-dependent manner.
Interestingly, another study that investigated the impact of EZH2 on NK cells found that genetic or pharmacological inhibition of EZH2 increases the generation of the IL-15 receptor CD122+ NK precursors as well as mature NK cell progeny from both mouse and human hematopoietic stem and progenitor cells (37). This study also found that enhanced NK cell expansion and cytotoxicity against tumor cells were associated with the up-regulation of CD122 and the C-type lectin receptor NKG2D (37).
Therefore, based on this previous study and our current results, we predict that EZH2 inhibitors will offer a twofold benefit: they will function by increasing NK cell expansion and cytotoxicity and render HCC cells more susceptible to NK-mediated cytotoxicity. Additionally, these studies also reinforce the importance of testing EZH2 inhibitors in an immune-competent background to fully assess the effects of EZH2 inhibition.
Materials and Methods
Cell Lines and Cell Culture.
The human HCC panel containing the SNU-387, SNU-423, SNU-449, SNU-475, HepG2/C3A, SK-HEP-1, and PLC/PRF/5 cell lines was purchased from the American Type Culture Collection (TCP-1011; ATCC). The 293T cells were purchased from the ATCC. The HepG2/C3A, SK-HEP-1, and PLC/PRF/5 cells were grown in Dulbecco’s Modified Eagle Medium supplemented with 10% FBS (Sigma-Aldrich) and 1× penicillin–streptomycin (Invitrogen). The SNU-387, SNU-423, SNU-449, and SNU-475 cells were grown in RPMI-1640 supplemented with 10% FBS and penicillin–streptomycin. The NK92MI cells were obtained from ATCC and grown in Alpha Minimum Essential Medium with 2 mM L-glutamine, 1.5 g/L sodium bicarbonate, 0.2 mM inositol, 0.1 mM 2-mercaptoethanol, 0.02 mM folic acid, 12.5% horse serum, and 12.5% FBS, but without ribonucleosides and deoxyribonucleosides.
Statistical Analysis.
All of the experiments were conducted in triplicate. The results of individual experiments are expressed as mean ± SEM. The P values were calculated by t tests using GraphPad Prism version 6.0h for Macintosh GraphPad Software (www.graphpad.com).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R01CA195077-01A1 (to N.W.) and R01CA200919-01 (to N.W.); Research Scholar Grant 128347-RSG-15-212-01-TBG from the American Cancer Society (to N.W.); and grant support from the Elsa U. Pardee Foundation and Beatrice Kleinberg Neuwirth Fund (to N.W.) M.R.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802691115/-/DCSupplemental.
References
- 1.Llovet JM, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341 e23. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jemal A, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Cannon MS, Hostetler JR. The anatomy of the parotoid gland in Bufonidae with some histochemical findings. II. Bufo alvarius. J Morphol. 1976;148:137–160. doi: 10.1002/jmor.1051480202. [DOI] [PubMed] [Google Scholar]
- 5.Sutherland LM, et al. Radiofrequency ablation of liver tumors: a systematic review. Arch Surg. 2006;141:181–190. doi: 10.1001/archsurg.141.2.181. [DOI] [PubMed] [Google Scholar]
- 6.Llovet JM, et al. SHARP Investigators Study Group Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 7.Bruix J, et al. RESORCE Investigators Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56–66. doi: 10.1016/S0140-6736(16)32453-9. [DOI] [PubMed] [Google Scholar]
- 8.Wolchok JD, et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2017;377:1345–1356. doi: 10.1056/NEJMoa1709684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ansell SM, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372:311–319. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Khoueiry AB, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389:2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medzhitov R, Janeway C., Jr Innate immune recognition: mechanisms and pathways. Immunol Rev. 2000;173:89–97. doi: 10.1034/j.1600-065x.2000.917309.x. [DOI] [PubMed] [Google Scholar]
- 12.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 13.Marcus A, et al. Recognition of tumors by the innate immune system and natural killer cells. Adv Immunol. 2014;122:91–128. doi: 10.1016/B978-0-12-800267-4.00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woo SR, Corrales L, Gajewski TF. Innate immune recognition of cancer. Annu Rev Immunol. 2015;33:445–474. doi: 10.1146/annurev-immunol-032414-112043. [DOI] [PubMed] [Google Scholar]
- 15.Hamerman JA, Ogasawara K, Lanier LL. NK cells in innate immunity. Curr Opin Immunol. 2005;17:29–35. doi: 10.1016/j.coi.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Guerra N, et al. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 2008;28:571–580. doi: 10.1016/j.immuni.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diefenbach A, Jensen ER, Jamieson AM, Raulet DH. Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature. 2001;413:165–171. doi: 10.1038/35093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerwenka A, Baron JL, Lanier LL. Ectopic expression of retinoic acid early inducible-1 gene (RAE-1) permits natural killer cell-mediated rejection of a MHC class I-bearing tumor in vivo. Proc Natl Acad Sci USA. 2001;98:11521–11526. doi: 10.1073/pnas.201238598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tahara-Hanaoka S, et al. Tumor rejection by the poliovirus receptor family ligands of the DNAM-1 (CD226) receptor. Blood. 2006;107:1491–1496. doi: 10.1182/blood-2005-04-1684. [DOI] [PubMed] [Google Scholar]
- 20.Raulet DH, Gasser S, Gowen BG, Deng W, Jung H. Regulation of ligands for the NKG2D activating receptor. Annu Rev Immunol. 2013;31:413–441. doi: 10.1146/annurev-immunol-032712-095951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauer S, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 22.Kamimura H, et al. Reduced NKG2D ligand expression in hepatocellular carcinoma correlates with early recurrence. J Hepatol. 2012;56:381–388. doi: 10.1016/j.jhep.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 23.Fang L, et al. MICA/B expression is inhibited by unfolded protein response and associated with poor prognosis in human hepatocellular carcinoma. J Exp Clin Cancer Res. 2014;33:76. doi: 10.1186/s13046-014-0076-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurisić V, Spuzić I, Konjević G. A comparison of the NK cell cytotoxicity with effects of TNF-alpha against K-562 cells, determined by LDH release assay. Cancer Lett. 1999;138:67–72. doi: 10.1016/s0304-3835(99)00011-7. [DOI] [PubMed] [Google Scholar]
- 25.Somanchi SS, McCulley KJ, Somanchi A, Chan LL, Lee DA. A novel method for assessment of natural killer cell cytotoxicity using image cytometry. PLoS One. 2015;10:e0141074. doi: 10.1371/journal.pone.0141074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerwenka A, et al. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity. 2000;12:721–727. doi: 10.1016/s1074-7613(00)80222-8. [DOI] [PubMed] [Google Scholar]
- 27.Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat Immunol. 2000;1:119–126. doi: 10.1038/77793. [DOI] [PubMed] [Google Scholar]
- 28.Viré E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 29.Wajapeyee N, Malonia SK, Palakurthy RK, Green MR. Oncogenic RAS directs silencing of tumor suppressor genes through ordered recruitment of transcriptional repressors. Genes Dev. 2013;27:2221–2226. doi: 10.1101/gad.227413.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lakshmikanth T, et al. NCRs and DNAM-1 mediate NK cell recognition and lysis of human and mouse melanoma cell lines in vitro and in vivo. J Clin Invest. 2009;119:1251–1263. doi: 10.1172/JCI36022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sellars M, et al. Regulation of DNA methylation dictates Cd4 expression during the development of helper and cytotoxic T cell lineages. Nat Immunol. 2015;16:746–754. doi: 10.1038/ni.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiappinelli KB, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemos B, Branco AT, Hartl DL. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc Natl Acad Sci USA. 2010;107:15826–15831. doi: 10.1073/pnas.1010383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sasaki M, et al. The overexpression of polycomb group proteins Bmi1 and EZH2 is associated with the progression and aggressive biological behavior of hepatocellular carcinoma. Lab Invest. 2008;88:873–882. doi: 10.1038/labinvest.2008.52. [DOI] [PubMed] [Google Scholar]
- 36.Sudo T, et al. Clinicopathological significance of EZH2 mRNA expression in patients with hepatocellular carcinoma. Br J Cancer. 2005;92:1754–1758. doi: 10.1038/sj.bjc.6602531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin J, et al. Ezh2 regulates differentiation and function of natural killer cells through histone methyltransferase activity. Proc Natl Acad Sci USA. 2015;112:15988–15993. doi: 10.1073/pnas.1521740112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.