Abstract
During early development in placental mammals, proper trophoblast lineage development is essential for implantation and placentation. Defects in this lineage can cause early pregnancy failures and other pregnancy disorders. However, transcription factors controlling trophoblast development remain poorly understood. Here, we utilize Fosl1, previously implicated in trophoblast giant cell development as a member of the AP-1 complex, to trans-differentiate embryonic stem (ES) cells to trophoblast lineage-like cells. We first show that the ectopic expression of Fosl1 is sufficient to induce trophoblast-specific gene expression programs in ES cells. Surprisingly, we find that this transcriptional reprogramming occurs independently of changes in levels of ES cell core factors during the cell fate change. This suggests that Fosl1 acts in a novel way to orchestrate the ES to trophoblast cell fate conversion compared to previously known reprogramming factors. Mapping of Fosl1 targets reveals that Fosl1 directly activates TE lineage-specific genes as a pioneer factor. Our work suggests Fosl1 may be used to reprogram ES cells into differentiated cell types in trophoblast lineage, which not only enhances our knowledge of global trophoblast gene regulation but also may provide a future therapeutic tool for generating induced trophoblast cells from patient-derived pluripotent stem cells.
Keywords: Fosl1, Fra1, Trans-differentiation, Trophoblast, Trophectoderm, Pioneer factor
1. Introduction
During early embryo development, cells in inner cell mass (ICM) responsible for fetal development do not contribute to the trophectoderm (TE) or trophoblast lineages engendering placenta. Surprisingly, it has been shown that multiple TE lineage-specific transcription factors (TFs), such as Cdx2, Gata3, Hand1, and Tfap2c, are significantly up-regulated upon spontaneous differentiation of embryonic stem (ES) cells, an in vitro model for ICM (Hailesellasse Sene et al., 2007). Knockout (KO) or knockdown (KD) of a key pluripotency factor Oct4 (Pou5f1) in ES cells also induces multiple TE-specific marker genes (Niwa et al., 2000, 2005). Moreover, overexpression (OE) of individual TE-specific TFs, such as Cdx2 and Gata3 in ES cells, up-regulates TE lineage marker genes (Niwa et al., 2005; Ralston et al., 2010), revealing that trans-differentiation of ES cells towards trophoblast stem (TS)-like cells by modulating a single regulator or TF is feasible. More recent works have additionally showed that Arid3a, a previously known B-cell regulator, reprograms ES cells to TS-like cells upon OE (Rhee et al., 2017a, 2014). These Arid3a-OE cells can successfully be incorporated into the TE of developing embryos ex vivo. Subsequent study on the reprogramming mechanisms of ES cells to TS-like cell fate conversion further revealed that this process is achieved through a specific series of sequential epigenetic and transcriptional events. First, an initial suppression of the ES cell core pluripotency factors was observed, followed by a dramatic activation of TE lineage-specific genes (Rhee et al., 2014, 2017b). These findings demonstrate that ectopic expression of a single TE-specific transcription factor is sufficient to overcome the barrier between ES and TS cell identity. This implies that TE lineage-specific genes may exist in a poised configuration in terms of their proximal chromatin landscape, or that there exist additional factors sequestered in ES cells that may be liberated to activate the TE-specific transcriptional program. Therefore, ES cells can serve as a reliable model system to study important factors responsible for TE lineage development (Murry and Keller, 2008; Niwa, 2010).
Fosl1 (also known as Fra1) is a component of activator-protein 1 complex (AP-1), which comprises a heterodimer of Fos-Jun family proteins. The Fos family includes cFos, FosB, Fosl1, and Fosl2, whereas the JunB family comprises cJun, JunB, and JunD. The exact configuration of the heterodimer determines the cell-specific role of the AP-1 complex. For example, an AP-1 complex composed of cFos and JunB regulates cell proliferation and differentiation (Shaulian and Karin, 2002). Meanwhile, another AP-1 complex composed of Fosl1 and JunB is implicated in endocrine and invasive trophoblast differentiation (Kubota et al., 2015; Renaud et al., 2014). Fosl1 has numerous biological roles, highlighting its importance as a versatile transcription factor. Fosl1 can contribute significantly to tumorigenesis, cell invasion (Verde et al., 2007), bone development (Wagner, 2002), and somatic cell reprogramming processes (Chronis et al., 2017). Although Fosl1 null mice die due to placental defects at approximately E10.5 (Schreiber et al., 2000), the mechanisms through which Fosl1 regulates TE lineages have not been fully understood, and furthermore, whether the Fosl1 alone can induce TE lineage-specific gene expression programs in ES cells has not been tested.
In the current study, we tested the potential of Fosl1 in trans-differentiation of mouse ES cells to TS or TE lineage-like cells. We found that OE of Fosl1 in ES cells induces TE-specific gene expression programs, especially genes active in the later stage of TE lineage development or differentiated TS cells. Surprisingly, unlike Arid3a, Cdx2, and Gata3, OE of Fosl1 does not significantly repress core pluripotency factors. Rather, Fosl1 activates the genes involved in TE lineage development, in particular genes associated with terminal TE differentiation. This suggests that Fosl1-mediated reprogramming may be used in the future as a tool to directly establish patient-specific specialized cells in the TE lineage, such as trophoblast giant cells.
2. Materials and methods
2.1. Cell culture
Mouse ES cell lines (J1 and E14) were maintained on 0.1% gelatin-coated plates in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 18% fetal bovine serum (FBS), 50 U/ml penicillin/streptomycin (Gibco), 2 mM L-glutamine (Gibco), 100 μM MEM nonessential amino acids (Gibco), nucleosides (Millipore), 100 μM β-mercaptoethanol (Sigma) and 1000 U/ml recombinant leukemia inhibitory factor (LIF, Millipore). Mouse TS cells were maintained at a ratio of 3:7 of TS medium to MEF-conditioned TS medium with 25 ng/ml Fgf4 and 1 μg/ml heparin. The TS medium consisted of RPMI 1640 (Gibco) supplemented with 20% FBS, 100 μM β-mercaptoethanol, 2 mM L-glutamine, 1 mM sodium pyruvate, 50 U/ml penicillin, and 50 mg/ml streptomycin. To make MEF-conditioned medium, mitomycin-treated MEF cells were cultured in TS medium for 3 days and the medium was collected every 3 days three times. To differentiate TS cells, we cultured the cells without Fgf4 and heparin.
2.2. Stable cell line generation
Full length Fosl1 cDNA was cloned into pEF1a-FLBIO (FB) vector (Kim et al., 2009, 2010). Primer sequences used for cloning are listed in Supplemental Table 1. Fosl1-containing vector (FB-Fosl1) was electroporated into BirA-expressing ES cells. Cells grew under puromycin and G418 selection for 9 days before picking colonies. OE of Fosl1 was confirmed by RT-qPCR and Western blotting. Fosl1 OE cells were maintained under ES cell culture conditions.
2.3. Generation of inducible cell lines
Lenti-X Tet-on 3G inducible expression system containing pLVX-Tet3G and pLVX-TRE3G-ZsGreen1 vectors was used following manufacturer’s instruction (Clontech). Fosl1 cDNA was prepared by PCR using primers listed in Supplemental Table 1 and cloned into the pLVX-TRE3G-ZsGreen1. The Tet3G or TRE3G-Fosl1 expression vectors were transfected with pCMV-Δ8.9 and VSV-G helper plasmids into 293T cells using Fugene (Promega), according to the manufacturer’s instruction. After 24 h, the 293T medium was replaced with ES medium. The supernatants containing virus particles were collected 48 h post transfection and filtered through 0.45 μm pore-size cellulose acetate filters (Pall). E14 ES cells were plated at ~2.5 × 105 cells per one well of 24-well plate with Tet3G virus-containing supernatant. After 24 h, the cells were selected with G418 for 2 days and re-infected with the TRE3G-Fosl1 virus. The co-infected E14 ES cells were placed under G418 and puromycin selection for 2 days. Fosl1 was induced by 500 ng/ml of doxycycline in ES cell culture media.
2.4. Western blotting
Cells were washed with PBS and lysed in 2× Laemmli sample buffer (Bio-Rad). Cell lysates were boiled at 100 °C for 15 min and centrifuged prior to loading. Proteins were separated on 4–20% gradient acrylamide gels (Bio-Rad) and transferred onto PVDF membranes using Trans-Blot® Turbo™ Transfer Starter System (Bio-Rad). After transfer, membranes were blocked with 5% BSA (Sigma) in TBST (20 mM Tris-HCl, pH 7.6, 13 mM NaCl, and 0.1% Tween-20) for an hour and incubated with primary antibody (or streptavidin-HRP) at 4 °C overnight. Membranes were then washed with TBST and incubated with secondary antibody for 1 h at room temperature. Antigens were detected using ECL reagents (GE Healthcare Amersham ECL prime) with Bio-Rad Molecular Imager® ChemiDoc™ XRS+ system. The antibodies used are streptavidin-HRP (1:2000, RPN1231V, GE Healthcare Life Sciences), anti-Fosl1 (1:1000, sc-183, Santa Cruz Biotechnology), and anti-β-actin (1:20,000, ab20272, Abcam).
2.5. Alkaline phosphatase (AP) staining
Alkaline phosphatase (AP) staining was performed according to manufacturer’s protocol using Alkaline Phosphatase Detection Kit (Millipore).
2.6. Real time-quantitative PCR (RT-qPCR)
Total RNA was isolated using RNeasy plus Mini Kit (Qiagen). 500 ng of total RNA was used for cDNA synthesis with ReadyScript® cDNA Synthesis Mix (Sigma). cDNA generated was diluted (20×) and RT-qPCRs were performed using 2 μl of diluted cDNA and PerfeCTa SYBR® Green FastMix, Low ROX™ (Quanta). RT-qPCR primers were designed to amplify exon junctions. Primer sequences are listed in Supplemental Table 1 and Gapdh was used as an internal control.
2.7. bioChIP-seq
bioChIP assays were performed as previously described (Beck et al., 2014; Lee et al., 2015). Briefly, cells were cross-linked in 1% formaldehyde for 7 min at room temperature. The reaction was quenched for 5 min with 125 mM glycine followed by washing with PBS. Cells were centrifuged and the pellets were used immediately for experiments or stored at −80 °C. Cells were resuspended in ChIP buffer (1% TritonX-100, 2 mM EDTA, 20 mM TrisCl, pH 8.1, 150 mM NaCl, 0.1% SDS and protease inhibitor), sonicated for 30 min (30 s on/1 min off) and centrifuged at maximum speed for 10 min. The supernatant was pre-cleared with Protein A beads for 4 h, rotating in 4 °C. Samples were then centrifuged and the supernatant was incubated in 10 μg streptavidin beads (Roche) overnight. Beads were washed for 8 min, twice with 2% SDS, once with high salt buffer (0.1% Deoxycholate, 1% Triton X-100, 1 mM EDTA, 50 mM HEPES (pH 7.5), and 500 mM NaCl), once with LiCl wash buffer (250 mM LiCl, 0.5% NP40, 0.5% Deoxycholate, 1 mM EDTA, and10mM TrisCl pH 8.1), and twice with TE buffer. Samples were eluted by incubating the beads in SDS Elution buffer (1% SDS, 10 mM EDTA and 50 mM TrisCl pH 8.1) overnight at 65 °C. 200 μl of TE buffer with 1 μg RNase A was added and incubated for 30 min at 37 °C. 1 μg of Proteinase K was added and incubated for 2 h at 37 °C. ChIP-seq library prep kits (New England BioLabs) were used to generate ChIP-seq libraries and the libraries were sequenced using an Illumina HiSeq 2500 machine.
2.8. ChIP-seq data processes and analysis
75 bp reads from ChIP-seq were mapped onto the mouse genome assembly (mm9) using Bowtie2 (Langmead and Salzberg, 2012), allowing for 2 base pairs mismatch followed by peak calling with model-based analysis for ChIP-seq (MACS) (Zhang et al., 2008) with a default setting. To identify Fosl1 targets, Fosl1 binding sites were assigned to the region surrounding 8 Kb up- and 2 Kb down-stream of transcription start sites (TSSs) of all RefSeq genes. We used the following hierarchy to assign one binding site to one genomic feature: promoter > upstream > intron > exon > intergenic regions. A promoter and an upstream feature were defined as a region within ±2 kb from the TSS and as a region between 2 kb and 20 kb upstream from the TSS, respectively. Binding sites not belonging to promoter, upstream, in-tron, or exon were considered as intergenic target loci. Overlapping binding sites among ChIP-seq data were identified using a moving window. If the centers of peaks from different ChIP-seq data were found within a 500 bp window, we considered them common peaks. Peak calling followed by an overlap analysis identified the common binding sites of TFs to generate the binding site correlation map. Score 0 and 1 were assigned to unique and overlapped binding sites of two TFs, respectively. A paired-wise Pearson correlation coefficient between the binding sites of two TFs was calculated for each pair of TFs. Clustering analysis and visualization of the data were done by Cluster 3.0, and Java Treeview, respectively (de Hoon et al., 2004; Saldanha, 2004).
2.9. Library generation and data process of RNA-seq
Total RNA (1 μg) was used to prepare libraries for RNA-seq. mRNAs were isolated from total RNA using NEBNext Poly(A) mRNA Magnetic Isolation Module (E7490, New England BioLabs), and then RNA-seq libraries were constructed using NEBNext Ultra RNA Library Prep Kit for Illumina (E7530, New England BioLabs). RNA-seq libraries were sequenced using an Illumina HiSeq 2500 machine. Raw RNA-seq reads were mapped onto mm9 mouse genome using TopHat followed by analysis of differential gene expression using Cufflinks (Trapnell et al., 2012). We further narrowed down up- and down-regulated gene by applying cut-off criteria (absolute fold >0.7).
2.10. Data sets used for analysis
To perform GSEA analysis, we downloaded expression data from GSE12985 (time-course differentiation of TS cells) and GSE20177 (ES and TS cells). ATAC-seq data (GSE90752) was used to investigate chromatin accessibility in ES cells.
2.11. Data deposit
Raw and processed RNA-seq and ChIP-seq data have been deposited at the public server GEO database under accession number GSE100000.
3. Results and discussion
3.1. Overexpression (OE) of Fosl1 leads to differentiation of mouse ES cells
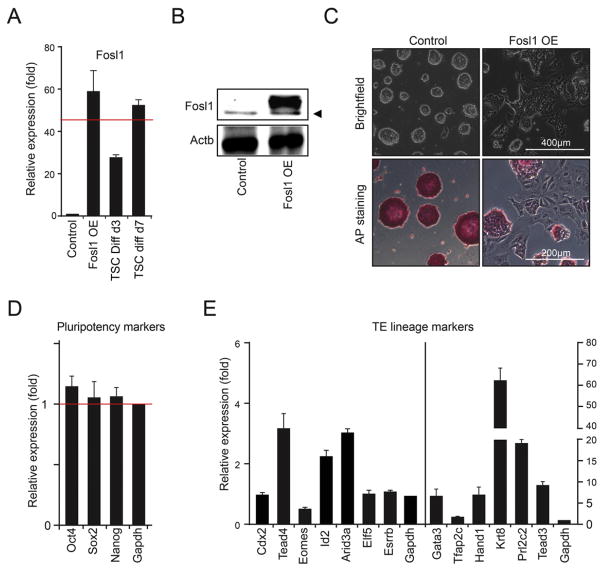
To examine whether Fosl1 can promote trans-differentiation of mouse ES cells to TS-like cells, we overexpressed Fosl1 using an FB vector system as previously described in mouse J1 ES cells (Kim et al., 2009). First, we confirmed the ectopic expression of Fosl1 using RT-qPCR (Fig. 1A) and Western blot (Fig. 1B). While the level of Fosl1 mRNA in Fosl1 OE cells even under the ES cell culture condition was comparable to the level in differentiated TS cells (for 7 days) (Fig. 1A), it was almost undetectable in control ES cells (Fig. 1A). Furthermore, compared to the typical round colony morphology of control ES cells, Fosl1 OE colonies were flattened with decreased AP activity, indicating exit from self-renewal (Fig. 1C). This aberrant morphology was consistently observed in another mouse ES cell line, E14, upon induction of Fosl1 by a doxycycline-inducible system under the ES cell culture condition (Supplemental Fig. 1).
Fig. 1.
Fosl1 overexpression induces trans-differentiation of ES cells to TS-like cells. A) Relative mRNA levels of Fosl1 in Fosl1 OE cells and differentiated TS cells (for 3 and 6 days) to the indigenous level of Fosl1 in ES cells. B) Western blots showing the protein levels of Fosl1 in Fosl1 OE and control cells. Arrowhead indicates non-specific. C) Bright field (upper panel) and AP activity (bottom panel) images in control and Fosl1 OE cells. D and E) Relative mRNA levels of the core pluripotency factors (D) and various TE lineage marker genes (E) in Fosl1 OE cells to control ES cells. Fosl1 OE cells were maintained under the ES cell culture condition and error bars in the bar graphs depict standard deviations of biological triplicates.
Interestingly, despite the differentiated morphology, expression of the core pluripotency genes remained almost undisturbed in Fosl1 OE cells (Fig. 1D and Supplemental Fig. 1D), suggesting that the morphology obtained upon OE of Fosl1 is not a direct outcome of downregulation of the core pluripotency factors. Since Fosl1 is implicated in trophoblast differentiation (Kubota et al., 2015), we monitored the levels of various TE lineage markers. Self-renewal markers of TS cells, such as Cdx2, Elf5, and Esrrb, showed marginal induction (Fig. 1E). Conversely, the genes associated with TS cell differentiation, including Krt8, Krt18, Prl2c2, and Hand1, were highly induced, regardless of whether OE of Fosl1 was constitutive (Fig. 1E) or inducible (Supplemental Fig. 1E). Thus, Fosl1 can induce differentiated TE lineage gene expression programs in ES cells.
3.2. Fosl1 OE globally induces the genes active in differentiated TS cells
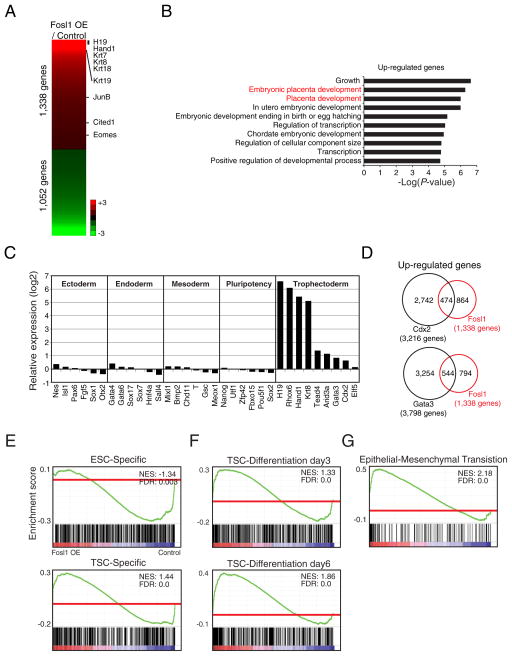
To investigate to what extent OE of Fosl1 affects the global gene expression programs in ES cells, we performed RNA-seq and found 1338 up-regulated as well as 1052 down-regulated genes compared to control cells (Supplemental Table 2). Up-regulated genes include many TE-related genes, such as H19, Eomes, Hand1, JunB, and Krt19 (Fig. 2A). Gene ontology (GO) analysis showed enrichment of genes involved in embryonic placenta development (Fig. 2B).
Fig. 2.
Global expression profiling reveals that Fosl1 OE induces genes implicated in the later stage of TE differentiation. A) Heatmap showing numbers of genes that are up- and down-regulated upon OE of Fosl1 in ES cells. Several TE lineage-specific genes are shown in the right side of the heatmap. B) Bar graphs presenting the enriched Gene Ontology (GO) terms of biological processes in up-regulated genes of Fosl1 OE cells. C) Relative transcript levels of various markers of lineages and pluripotency factors in Fosl1 OE cells to control cells. D) Venn diagrams showing overlaps of up-regulated genes between Fosl1 OE cells and Cdx2-OE cells (upper panel) or Gata3-OE cells (bottom panel). E–G) Gene Set Enrichment Analysis (GSEA) showing enrichment of gene sets such as ESC-specific genes as well as TSC-specific genes (E), active genes in differentiated TS cells (for 3 and 6 days) (F), and epithelial-mesenchymal transition in Fosl1- OE cells over control ES cells.
Notably, the levels of the core pluripotency, mesoendoderm, endoderm, ectoderm, and mesoderm genes were not significantly changed in Fosl1 OE cells (Fig. 2C). In drastic contrast, the genes more active in differentiated TS cells or later stage of TE lineage development, such as H19, Hand1, and Krt8 were dramatically induced (Fig. 2C). Since OE of a single factor, such as Cdx2 and Gata3, can induce ES cells to TS-like cell fate conversion (Ralston et al., 2010), we compared their expression patterns with that of Fosl1 OE cells. Approximately 35% and 40% of genes induced upon Cdx2 and Gata3 overlapped with the up-regulated genes in Fosl1 OE cells, respectively, suggesting that Fosl1 activates, to some degree, a different set of target genes from Gata3 or Cdx2 (Fig. 2D). Interestingly, prolactin (Prl) family genes, known to be highly expressed in trophoblast giant cells (Hamlin et al., 1994; Sahgal et al., 2005), were dramatically up-regulated only in Fosl1 OE cells (Supplemental Table 2), further indicating that Fosl1’s role is in TE lineage differentiation rather than self-renewal of TS cells.
We further performed Gene Set enrichment analysis (GSEA) to compare the gene expression profile of Fosl1 OE cells to those of ES cells and TS cells (Sakaue et al., 2010). As shown in Fig. 2E (top panel), ES cell-specific genes were not enriched in Fosl1 OE cells and there was only modest enrichment of TS cell-specific gene set in Fosl1 OE cells, showing that OE of Fosl1 in ES cells is sufficient to induce a moderate switch from an ES cell-specific to a TS cell-specific gene expression program (Fig. 2E, bottom). Then, we sought to investigate which stages of differentiated TS cells are similar to Fosl1 OE cells. For this, we used a published dataset obtained from time-course differentiation of TS cells for 6 days (Ralston et al., 2010) to perform GSEA (Fig. 2F). Remarkably, we found that the activity of the genes up-regulated upon Fosl1 OE gradually increases as differentiation of TS cells progresses, with the highest correlation seen on day 6. Thus, genes up-regulated upon differentiation of TS cells are also induced upon Fosl1 OE in ES cells. Since TS cells undergo epithelial-mesenchymal transition (EMT) as they differentiate (Sutherland, 2003), we additionally tested the EMT-associated gene set and found a strong correlation between EMT-associated genes, such as Serpine2, Col4a1, Itgb5, Ecm1, and Mmp14 and the genes up-regulated upon Fosl1 OE (Fig. 2G). The results clearly reveal that Fosl1 OE cells harbor expression programs of more differentiated TE lineages.
3.3. Global Fosl1 target occupancy patterns reflect its role in cell fate conversion and TE lineage development
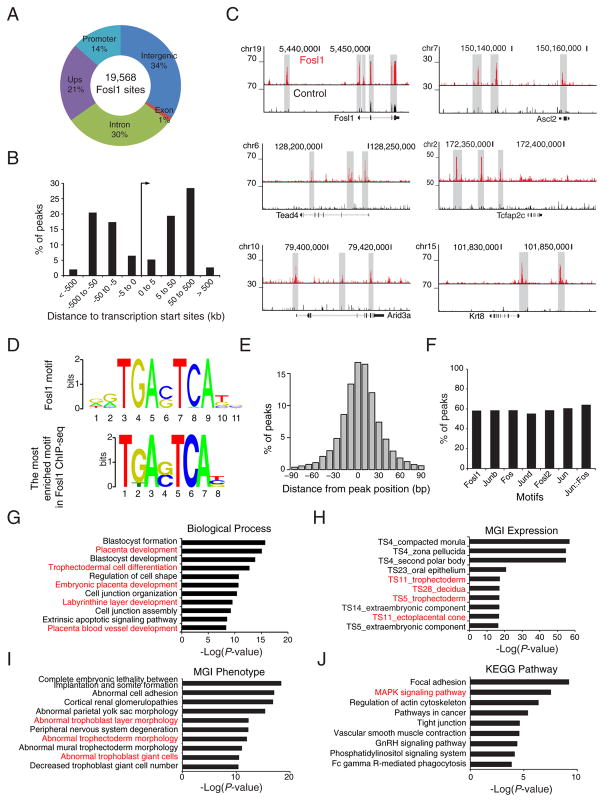
To elucidate the transcriptional regulatory mechanisms of Fosl1 during the cell fate conversion, we mapped global binding sites of Fosl1 using bioChIP followed by massive parallel sequencing (bioChIP-seq) (Beck et al., 2014), identifying 19,568 Fosl1 binding sites in Fosl1 OE cells (Supplemental Table 3). Most Fosl1 binding sites are located far away from the transcription start sties (TSSs) (Fig. 3A). In particular, Fosl1 tends to bind within 50 to 500 Kb from the TSS (Fig. 3B), suggesting that Fosl1 binds to distal enhancers. Notably, Fosl1 occupies its own promoter and distal enhancer regions (Fig. 3C), implying that Fosl1 forms an auto-regulatory loop to activate its own gene, indicative of master regulators.
Fig. 3.
Mapping of global binding sites of Fosl1 unveils that Fosl1 directly activates TE lineage-specific genes by occupying their distal enhancers. A) A pie chart presenting the distribution of Fosl1 binding sites across the mouse genome. Promoters: regions within ±2 K from the TSSs; Upstream: regions between 2 K and 20 K upstream of the TSSs; Intergenic: regions except promoters, upstream, exons, and introns. B) Percentage of Fosl1 binding sites discovered in a given distance from transcription start sites of genes. C) ChIP-seq track images showing Fosl1 occupancy near TE lineage-specific genes in Fosl1 OE cells. D) Consensus sequence of Fosl1 motif (upper panel) and sequence that are the most enriched in Fosl1 binding sites. E) A histogram presenting the distribution of percentage peaks harboring the Fosl1 motif in a given distance from the center of Fosl1 binding sites. F) Percentage of Fosl1 binding sites containing consensus motifs of Jun or Fos family transcription factors. G–J) GO terms of biological processes (G), MGI expression (H), MGI phenotype (I), and KEGG pathway (J) enriched in Fosl1 binding sites.
Consistent with the expression data, we found that Fosl1 directly regulates TE lineage-specific genes at distal regulatory regions (Fig. 3C). Moreover, the most strongly enriched motif in Fosl1 binding sites is almost identical to the known consensus Fosl1 motif (Fig. 3D), which tends to be found near the center of the Fosl1 peaks (Fig. 3E), and the motifs of Fos and Jun family member genes were also enriched within the Fosl1 peaks (Fig. 3F). Fosl1 also occupies regulatory regions of JunB (Supplemental Table 3), collectively suggesting that Fosl1 collaborates with Jun family, particularly with JunB, to form an AP-1 complex during cell fate conversion.
We also evaluated the enrichment of Fosl1 target genes in biological processes, mouse development, and disease phenotypes using Genomic Regions Enrichment of Annotations Tool (GREAT). We found that Fosl1 target genes are strongly enriched in placenta development-related GO terms, such as placenta development, trophectodermal cell differentiation, and labyrinthine layer development (Fig. 3G). Fosl1 target genes are highly expressed in TE, decidua, and ectoplacental cone that constitute placenta (Fig. 3H). Aberrant expression of Fosl1 target genes is also associated with trophoblast abnormality and trophoblast giant cell deficiency (Fig. 3I). Additional analyses disclosed that the Fosl1 targets are strongly enriched in the MAPK signaling pathway (Fig. 3J) which is implicated in TE formation from ES cells and mouse embryo (Lu et al., 2008). Altogether, our analyses reveal not only the mode of target gene regulation of Fosl1 during the cell fate conversion, but also the putative involvement of the Fosl1 in TE lineage development and placentation.
3.4. Fosl1 functions as a pioneer factor and activates TE lineage genes during cell fate conversion
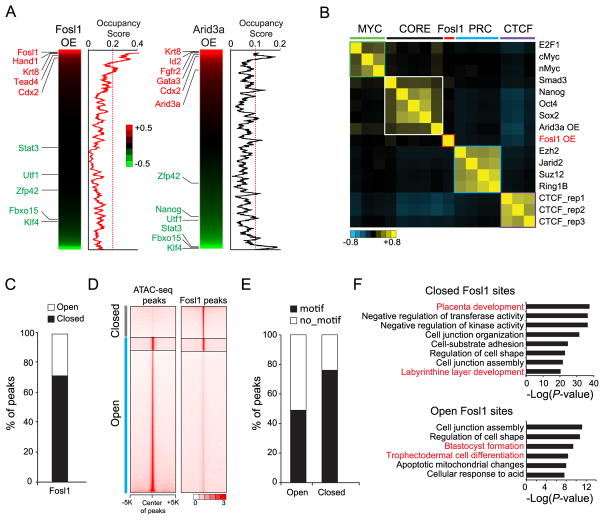
Recently, we reported that TE lineage-specific TFs, such as Cdx2, Gata3, and Arid3a, function as both activators and repressors during reprogramming (Rhee et al., 2017b). To interrogate how OE of Fosl1 induces cell fate changes, we integrated global gene expression analysis with target occupancy of Fosl1. We found that Fosl1 strongly occupies most of the genes that are up-regulated upon its own OE but not the down-regulated genes as Arid3a does (Fig. 4A). Thus, Fosl1 acts mainly as an activator, not as a repressor, unlike other TFs that can mediate ES to TS-like cell reprogramming processes. Additionally, target correlation analysis revealed that Fosl1’s targets are distinct from those of both ES cell factors and Arid3a (Fig. 4B). These results additionally support that the trans-differentiation mechanism of Fosl1 might be distinct from the other known TFs-mediated ES to TS-like cell fate conversion and Fosl1 may act independently of the Oct4 depletion-mediated differentiation towards TE lineages.
Fig. 4.
Fosl1 functions as an activator as well as a pioneer factor. A) Heatmaps presenting ranked relative gene expression of Fosl1 OE cells (left panel) and Arid3a-OE cells (right panel) to control cells. Beside the heatmaps, line graphs showing occupancy scores of Fosl1 and Arid3a that are averaged by moving window average (a window size of 100). B) Heatmap showing the similarity of their binding sites among diverse factors that belong to three modules (MYC, CORE, and PRC) and insulator binding protein CTCF tested in ES cells (Beck et al., 2014) as well as Arid3a OE (Rhee et al., 2014) and Fosl1 OE in ES cells. Arid3a and Fosl1 binding sites were obtained when they were overexpressed in ES cells. C) A bar graph showing percentage of peaks that are open or closed. D) Heatmaps depicting occupancy signals of ATAC-seq in ES cells (left panel) and Fosl1 in Fosl1 OE cells (right panel) within ±5 K from the center of peaks. E) A bar graph presenting percentage of peaks harboring motifs in open Fosl1 peaks or closed Fosl1 peaks. F) GO terms of biological processes enriched in the closed (upper panel) and the open (bottom panel) Fosl1 binding sites.
Since reprogramming factors, such as Oct4 (for the generation of induced pluripotent stem cells) and Gata3 (for TS-like cells) function as pioneer factors that can bind to closed chromatin and promote gene activation (Soufi et al., 2015; Takaku et al., 2016), we investigated whether Fosl1 can do the same by comparing the Fosl1 binding sites with the chromatin accessibility measured by ATAC-seq in ES cells (Rhee et al., 2017b). We found that approximately 29% and 71% of Fosl1 binding sites belong to open and closed chromatin, respectively (Fig. 4C and D). Fosl1 motif is preferentially observed in the closed Fosl1 binding sites compared to the open Fosl1 sites (Fig. 4E). Closed Fosl1 sites are enriched with the genes implicated in placenta development. Open Fosl1 sites are associated with blastocyst formation as well as TS cell differentiation, suggesting that some portion of genes implicated in placenta development is already open in ES cells (Fig. 4F). Collectively, these results suggest that Fosl1 can bind onto its own motif in closed chromatin as a pioneer factor to activate placenta-related genes during the cell fate conversion.
4. Conclusion
In summary, we show that the ectopic expression of Fosl1 in ES cells is sufficient to induce TE lineage-specific gene expression programs and trans-differentiate mouse ES cells to differentiated TS-like cells. Our data show that Fosl1 directly induces genes implicated in placental development and trophectodermal differentiation, acting primarily as a transcriptional activator unlike other reprogramming factors, such as Arid3a, Cdx2, and Gata3. We further reveal that Fosl1 functions as a pioneer factor, binding to closed chromatin of TE lineage-specific genes to directly transcriptionally activate them.
Supplementary Material
Acknowledgments
We thank the Genome Sequencing and Analysis Facilities (GSAF) at The University of Texas at Austin for ChIP-seq and RNA-seq sample processing. This work was partially supported by the NIH (R01GM112722) and JK holds a 2017 Preterm Birth Research Grant from the Burroughs Wellcome Fund.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2017.12.004.
Conflict of interest
We report no conflict of interest in conducting the work within this manuscript.
References
- Beck S, Lee BK, Rhee C, Song J, Woo AJ, Kim J. CpG island-mediated global gene regulatory modes in mouse embryonic stem cells. Nat Commun. 2014;5:5490. doi: 10.1038/ncomms6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronis C, Fiziev P, Papp B, Butz S, Bonora G, Sabri S, Ernst J, Plath K. Cooperative binding of transcription factors orchestrates reprogramming. Cell. 2017;168:442–459. e420. doi: 10.1016/j.cell.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailesellasse Sene K, Porter CJ, Palidwor G, Perez-Iratxeta C, Muro EM, Campbell PA, Rudnicki MA, Andrade-Navarro MA. Gene function in early mouse embryonic stem cell differentiation. BMC Genomics. 2007;8:85. doi: 10.1186/1471-2164-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin GP, Lu XJ, Roby KF, Soares MJ. Recapitulation of the pathway for trophoblast giant cell differentiation in vitro: stage-specific expression of members of the prolactin gene family. Endocrinology. 1994;134:2390–2396. doi: 10.1210/endo.134.6.8194465. [DOI] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bio-informatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Kim J, Cantor AB, Orkin SH, Wang J. Use of in vivo biotinylation to study protein-protein and protein-DNA interactions in mouse embryonic stem cells. Nat Protoc. 2009;4:506–517. doi: 10.1038/nprot.2009.23. [DOI] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota K, Kent LN, Rumi MA, Roby KF, Soares MJ. Dynamic regulation of AP-1 transcriptional complexes directs trophoblast differentiation. Mol Cell Biol. 2015;35:3163–3177. doi: 10.1128/MCB.00118-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BK, Shen W, Lee J, Rhee C, Chung H, Kim KY, Park IH, Kim J. Tgif1 counterbalances the activity of core pluripotency factors in mouse embryonic stem cells. Cell Rep. 2015;13:52–60. doi: 10.1016/j.celrep.2015.08.067. [DOI] [PubMed] [Google Scholar]
- Lu CW, Yabuuchi A, Chen L, Viswanathan S, Kim K, Daley GQ. Ras-MAPK signaling promotes trophectoderm formation from embryonic stem cells and mouse embryos. Nat Genet. 2008;40:921–926. doi: 10.1038/ng.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Niwa H. Mouse ES cell culture system as a model of development. Develop Growth Differ. 2010;52:275–283. doi: 10.1111/j.1440-169X.2009.01166.x. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137:395–403. doi: 10.1242/dev.038828. [DOI] [PubMed] [Google Scholar]
- Renaud SJ, Kubota K, Rumi MA, Soares MJ. The FOS transcription factor family differentially controls trophoblast migration and invasion. J Biol Chem. 2014;289:5025–5039. doi: 10.1074/jbc.M113.523746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee C, Lee BK, Beck S, Anjum A, Cook KR, Popowski M, Tucker HO, Kim J. Arid3a is essential to execution of the first cell fate decision via direct embryonic and extraembryonic transcriptional regulation. Genes Dev. 2014;28:2219–2232. doi: 10.1101/gad.247163.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee C, Edwards M, Dang C, Harris J, Brown M, Kim J, Tucker HO. ARID3A is required for mammalian placenta development. Dev Biol. 2017a;422:83–91. doi: 10.1016/j.ydbio.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee C, Lee BK, Beck S, LeBlanc L, Tucker OH, Kim J. Mechanisms of transcription factor-mediated direct reprogramming of mouse embryonic stem cells to trophoblast stem-like cells. Nucleic Acids Res. 2017b;45:10103–10114. doi: 10.1093/nar/gkx692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahgal N, Canham LN, Konno T, Wolfe MW, Soares MJ. Modulation of trophoblast stem cell and giant cell phenotypes: analyses using the Rcho-1 cell model. Differentiation. 2005;73:452–462. doi: 10.1111/j.1432-0436.2005.00044.x. [DOI] [PubMed] [Google Scholar]
- Sakaue M, Ohta H, Kumaki Y, Oda M, Sakaide Y, Matsuoka C, Yamagiwa A, Niwa H, Wakayama T, Okano M. DNA methylation is dispensable for the growth and survival of the extraembryonic lineages. Curr Biol. 2010;20:1452–1457. doi: 10.1016/j.cub.2010.06.050. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Schreiber M, Wang ZQ, Jochum W, Fetka I, Elliott C, Wagner EF. Placental vascularisation requires the AP-1 component fra1. Development. 2000;127:4937–4948. doi: 10.1242/dev.127.22.4937. [DOI] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland A. Mechanisms of implantation in the mouse: differentiation and functional importance of trophoblast giant cell behavior. Dev Biol. 2003;258:241–251. doi: 10.1016/s0012-1606(03)00130-1. [DOI] [PubMed] [Google Scholar]
- Takaku M, Grimm SA, Shimbo T, Perera L, Menafra R, Stunnenberg HG, Archer TK, Machida S, Kurumizaka H, Wade PA. GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol. 2016;17:36. doi: 10.1186/s13059-016-0897-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde P, Casalino L, Talotta F, Yaniv M, Weitzman JB. Deciphering AP-1 function in tumorigenesis: fraternizing on target promoters. Cell Cycle. 2007;6:2633–2639. doi: 10.4161/cc.6.21.4850. [DOI] [PubMed] [Google Scholar]
- Wagner EF. Functions of AP1 (Fos/Jun) in bone development. Ann Rheum Dis. 2002;61(Suppl 2):ii40–42. doi: 10.1136/ard.61.suppl_2.ii40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.