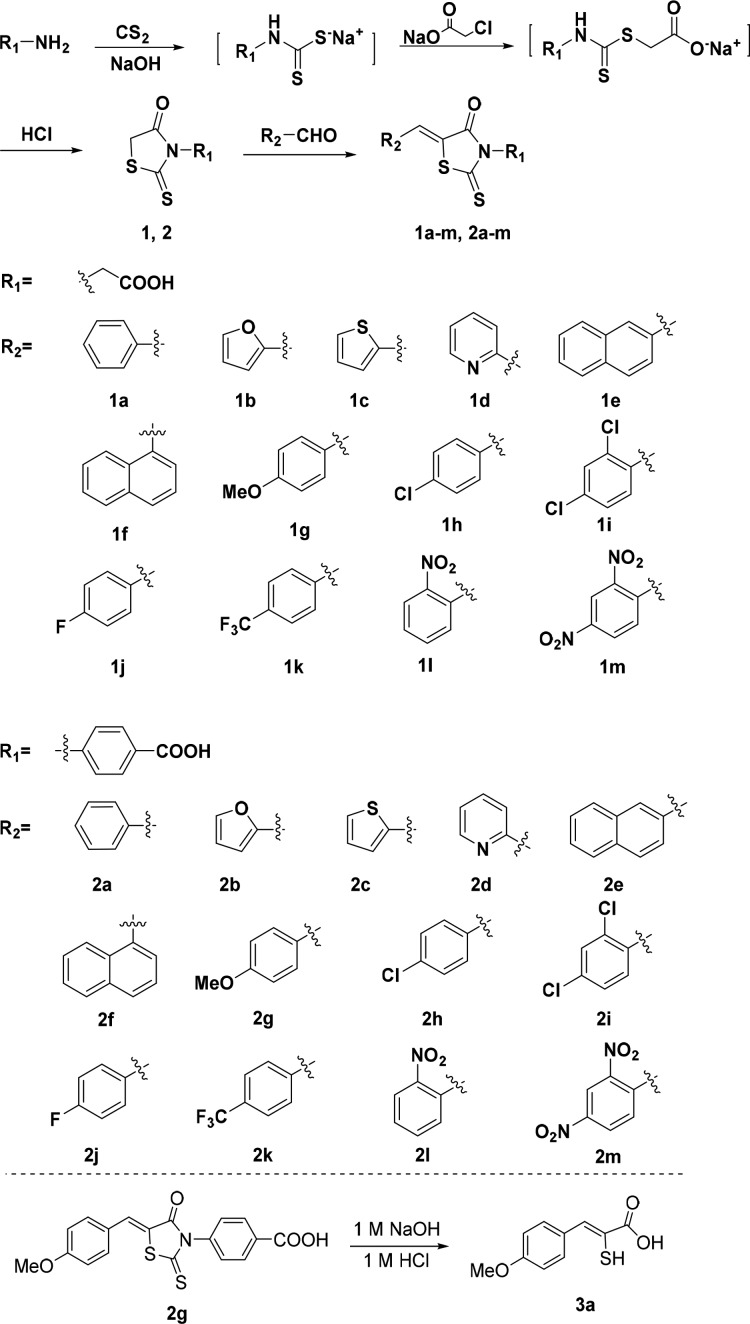
Abstract
A series of rhodanines was constructed, their Z-configuration was confirmed by small molecule X-ray crystal structures, and their activity against metallo-β-lactamases (MβLs) was measured. The obtained 26 molecules and a thioenolate specifically inhibited the MβL L1 with an IC50 range of 0.02–1.7 μM, and compounds 2h–m exhibited broad-spectrum inhibition of the MβLs NDM-1, VIM-2, ImiS, and L1 with IC50 values <16 μM. All inhibitors increased the antimicrobial effect of cefazolin against E. coli cells expressing L1, resulting in a 2–8-fold reduction in MIC. Docking studies suggested that the nitro (NDM-1, CphA, and L1) or carboxyl group (VIM-2) of 2l coordinates one or two Zn(II) ions, while the N-phenyl group of the inhibitor enhances its hydrophobic interaction with MβLs. These studies demonstrate that the diaryl-substituted rhodanines are good scaffolds for the design of future broad-spectrum inhibitors of MβLs.
Keywords: Antibiotic resistance, metallo-β-lactamases, broad-spectrum inhibitor, rhodanine
The development of β-lactam antibiotics over the past 70 years has led to the availability of drugs to treat a wide range of bacterial infections. However, the widespread use of β-lactam containing antibiotics has resulted in a large number of bacteria that are resistant to almost all antibiotics. Most commonly, bacteria become resistant to β-lactam antibiotics by producing β-lactamases, which hydrolyze the C–N bond in the four-membered ring of β-lactam antibiotics. The β-lactamases have been categorized as serine β-lactamases (SβLs) and metallo-β-lactamases (MβLs), according to their mechanism of action.1 MβLs are further divided into subclasses B1, B2, and B3, based on amino acid sequence homology and Zn(II) content.1 The B1 and B3 subclasses hydrolyze almost all β-lactam antibiotics, including penicillins, cephalosporins, and carbapenems. In contrast, the B2 subclass enzymes preferentially hydrolyze carbapenems, which have been called “last resort” antibiotics.2
Facing the emergence of drug resistance mediated by MβLs, a large number of MβL inhibitors have been reported, such as β-lactam analogues,3 hydroxamic acid,4 azolylthioacetamides,5 and cyclic boronates.6 Ebselen7 and aspergillomarasmine A (AMA)8 have also been described to be inhibitors of MβLs.
The rhodanines, unique nontransitional state analogs that inhibit penicillin-binding proteins (PBPs)9 and SβLs,10 have recently been described to be inhibitors of MβLs. Spicer and co-workers reported that a rhodanine with a trichlorobenzylidene substituent (see Scheme S1) showed an inhibitory effect on Verona Integron-borne Metallo-β-lactamase 2 (VIM-2) and imipenemase 1 (IMP-1).11 Further mechanistic studies by Brem et al. indicated that the rhodanine hydrolysis product thioenolate (Scheme S1) also inhibited B1MβLs.12
Our goal is to develop specific or broad-spectrum inhibitors of MβLs and to use them in combination with β-lactams to combat bacterial infections, in which the bacteria produce MβLs. Based on the above information and to investigate whether the rhodanine alone is a potent inhibitor of MβLs, we constructed a series of novel rhodanines with benzyl, heterocyclic, naphthyl, aliphatic, and aromatic carboxyl substituents (Figure 1). These compounds were tested as inhibitors against the purified MβLs VIM-2, New Delhi Metallo-β-lactamase 1 (NDM-1), Imipenemase-1 from Aeromonas veronii bv. sobria (ImiS), and β-lactamase 1 from Stenotrophomonas maltophilia (L1), which are representative enzymes belonging to the B1, B2, and B3 subclasses of MβLs, respectively.13 Furthermore, the ability of these inhibitors to restore the antimicrobial activity of existing antibiotics against antibiotic-resistant strains was evaluated.
Figure 1.
Synthetic route and the structures of rhodanine derivatives and thioenolate 3a.
Twenty-six rhodanines 1a–m and 2a–m (Figure 1) were synthesized by a previously reported method. The synthetic route is shown in Figure 1. Briefly, amines reacted with carbon disulfide in NaOH aqueous solution at room temperature for 16 h, sodium chloroacetate was added and stirred for 3 h, and the resulting mixture was acidified with HCl and refluxed for 16 h to give N-substituted rhodanines 1 and 2.14 Knoevenagel condensation of aryl aldehyde and 1 or 2 in acetic acid offered the desired rhodanines.15,16
To confirm the molecular structures of the rhodanines, crystals suitable for X-ray analysis were obtained by slow evaporation of a solution of 1l and 2m in methanol-acetone. The crystal structures are given in Figure S1, and the resulting structures based on X-ray diffraction confirmed the expected structures. Coordinates of 1l and 2m in CIF format are available from the Cambridge Crystallographic Data Center (CCDC: 1480089 and 1479978, respectively). All synthesized rhodanines were characterized by 1H and 13C NMR and HRMS (see Supporting Information).
To test whether these rhodanines were MβL inhibitors, MβLs from subclasses B1 (VIM-2 and NDM-1), B2 (ImiS), and B3 (L1) were overexpressed and purified as previously described.17−20 The inhibition experiments with these compounds under steady-state conditions were conducted on an Agilent UV8453 spectrometer using cefazolin (50 μM, monitoring at 262 nm) as substrate for NDM-1, VIM-2, and L1 and imipenem (60 μM, monitoring at 300 nm) for ImiS. The concentrations of inhibitors were varied between 0 and 50 μM.
The concentrations of compounds 1a–m and 2a–m causing 50% decrease in enzyme activity (IC50) were determined in 50 mM Tris, pH 7.0 (at this pH, the rhodanine was not hydrolyzed, see the following discussion). The IC50 data (Table 1) indicate that all of these rhodanines exhibited excellent inhibition of L1 with IC50 values ranging from 0.02 to 1.7 μM, and 2m was found to be the most potent inhibitor (IC50 = 0.02 μM). For NDM-1, 1b, 1g, 1i–l, 2a–d, and 2g–m showed potency with IC50 values ranging from 0.69 to 47 μM, and 2b had the lowest IC50 (0.69 μM). Compounds 1f, 1h–l, 2d–e, and 2h–m inhibited VIM-2 with IC50 values ranging from 0.19 to 27.9 μM, and 2l was the most potent inhibitor (IC50 = 0.19 μM). ImiS was inhibited by 1d, 2a, and 2e–m with IC50 values ranging from 3.0 to 19.1 μM with 2l being the most potent inhibitor (3.0 μM).
Table 1. Inhibitory Activity (IC50, μM) of Rhodanines against Four MβLs in 50 mM Tris-HCl, pH 7.0a.
| B1 |
B2 | B3 | B1 |
B2 | B3 | ||||
|---|---|---|---|---|---|---|---|---|---|
| compd | NDM-1b | VIM-2b | ImiSc | L1b | compd | NDM-1b | VIM-2b | ImiSc | L1b |
| 1a | 0.32 ± 0.02 | 2b | 0.69 ± 0.02 | 0.18 ± 0.02 | |||||
| 1b | 47 ± 1 | 1.7 ± 0.1 | 2c | 14.3 ± 0.4 | 0.59 ± 0.02 | ||||
| 1c | 0.77 ± 0.09 | 2d | 1.13 ± 0.06 | 0.92 ± 0.02 | 0.21 ± 0.02 | ||||
| 1d | 19.1 ± 0.6 | 0.16 ± 0.03 | 2e | 1.9 ± 0.3 | 16.5 ± 0.3 | 0.12 ± 0.01 | |||
| 1e | 0.37 ± 0.01 | 2f | 6.9 ± 0.2 | 0.10 ± 0.01 | |||||
| 1f | 27.9 ± 0.3 | 0.28 ± 0.01 | 2g | 12 ± 1 | 15.0 ± 0.3 | 0.31 ± 0.02 | |||
| 1g | 25.1 ± 0.8 | 0.6 ± 0.1 | 2h | 12.2 ± 0.9 | 3.6 ± 0.1 | 7.7 ± 0.9 | 0.070 ± 0.002 | ||
| 1h | 15.3 ± 0.8 | 0.22 ± 0.06 | 2i | 10.6 ± 0.8 | 3.5 ± 0.2 | 7.6 ± 0.2 | 0.030 ± 0.008 | ||
| 1i | 21.5 ± 0.8 | 25.1 ± 0.4 | 0.080 ± 0.002 | 2j | 8.4 ± 0.4 | 6.9 ± 0.2 | 3.6 ± 0.1 | 0.080 ± 0.003 | |
| 1j | 15.4 ± 0.9 | 14.4 ± 0.6 | 0.20 ± 0.02 | 2k | 10.6 ± 0.5 | 2.36 ± 0.09 | 7.8 ± 0.5 | 0.070 ± 0.008 | |
| 1k | 14.3 ± 0.9 | 12.7 ± 0.9 | 0.11 ± 0.02 | 2l | 1.31 ± 0.05 | 0.19 ± 0.03 | 3.0 ± 0.2 | 0.050 ± 0.003 | |
| 1l | 27.4 ± 0.4 | 16.8 ± 0.4 | 0.22 ± 0.01 | 2m | 6.7 ± 0.2 | 3.9 ± 0.3 | 15.9 ± 0.8 | 0.020 ± 0.004 | |
| 1m | 0.10 ± 0.01 | 3a | 4.7 ± 0.4 | 0.32 ± 0.01 | |||||
| 2a | 8.7 ± 0.9 | 18.3 ± 0.2 | 0.21 ± 0.01 | ||||||
Blank spaces represent percent inhibition under 50% at a concentration of 50 μM.
The antibiotic used was cefazolin.
The antibiotics used was imipenem.
It should be noted that 2h–m were potent broad-spectrum inhibitors of all four tested MβLs, exhibiting IC50 values <16 μM, and 2l was found to be the most potent broad-spectrum inhibitor with IC50 values ≤3 μM. Given the broad-spectrum potency of 2l, the inhibition curves of 2l against the four MβLs tested were analyzed in more detail (Figure 2). Compound 2l exhibited more than 95% inhibition (<5% residual activity) against NDM-1, VIM-2, ImiS, and L1 at concentrations of 20, 4, 40, and 0.8 μM, respectively.
Figure 2.
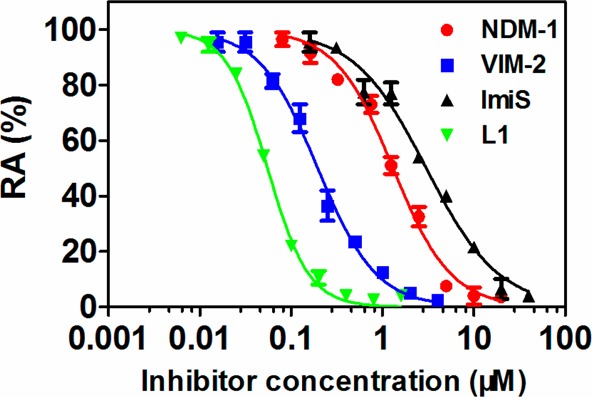
Inhibition curves of 2l against NDM-1, VIM-2, ImiS, and L1, where RA means residual activity.
Brem et al. reported that the rhodanine hydrolysis product thioenolate with a trichlorobenzylidene substituent (see Scheme S1) exhibited inhibition of MβLs.12 To investigate whether rhodanine alone inhibits MβLs in case it is not hydrolyzed, we assayed the stability of 2g (50 μM) in MES (pH 5.5, 6.0, and 6.5), Tris (pH 7.0, 7.5, 8.0, and 8.5), and Tris containing L1 (pH 7.0) through monitoring its absorbance change at 401 nm for 24 h. The results (Figure S3) show that 2g was hardly hydrolyzed at pH values ranging from 6.0 to 7.0 (Figure S3B–D), even in the presence of L1 enzyme (Figure S3H), perhaps because our rhodanines are not “activated” by multiple chlorines on the phenyl ring as in Brem et al.’s report. These results indicate that rhodanine 2g is not hydrolyzed by the MβL and that the rhodanine itself and not its hydrolysis product is responsible for the MβL inhibition observed.
To check whether the rhodanine hydrolysis product inhibits MβLs, 3a was prepared by hydrolysis of 2g with NaOH and acidification with HCl (Figure 1) and confirmed by 1H and 13C NMR and HRMS (see Supporting Information). Inhibition assays (in 50 mM Tris, pH 7.0) indicated that 3a inhibited L1 and VIM-2 with IC50 values of 0.32 and 4.7 μM, respectively, but not NDM-1 or ImiS at concentrations of up to 50 μM (Table 1), indicating that the rhodanine hydrolysis product thioenolate (3a) inhibits only specific MβLs, including VIM-212 and L1.
The IC50 data listed in Table 1 reveal a structure–activity relationship (SAR), which is that the diaryl-substituted rhodanines with electron-accepting atoms or groups (2h–m), such as chlorine, fluorine, nitro, and trifluoromethyl, exhibit broad-spectrum inhibition of MβLs, and the N-aromatic carboxyl (R1 in the 2 series) makes the inhibitors more potent than the aliphatic carboxyl (R1 in the 1 series) implying that the phenyl may interact with the hydrophobic pocket of MβLs.
The ability of the rhodanines to inhibit MβLs and to restore the antimicrobial activity of antibiotics was investigated by determining the minimum inhibitory concentrations (MICs) of existing antibiotics in the presence and absence of 1a–m, 2a–m, and 3a.21E. coli DH10B cells expressing VIM-2, NDM-1, ImiS, or L1 from pBCSK plasmids were used (Table 2A). The concentration of inhibitors used was kept constant at 32 μg/mL.
Table 2. Antibacterial Activities (MICs, μg/mL) of Cefazolin or Imipenem against E. coli DH10B Expressing MβLs in the Absence and Presence of Rhodanines at a Concentration of 32 μg/mL (A), and Cefazolin MICs against E. coli Expressing L1 at a 2l and 2m Concentration Range of 16–256 μg/mL (B)a.
| A | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| inhibitors | E. coli-NDM-1b | E. coli-VIM-2b | E. coli-ImiSc | E. coli-L1b | inhibitors | E. coli-NDM-1b | E. coli-VIM-2b | E. coli-ImiSc | E. coli-L1b |
| blank | 8 | 512 | 2 | 32 | 2a | 1 | 512 | 1 | 8 |
| 1a | 8 | 256 | 2 | 8 | 2b | 4 | 512 | 1 | 8 |
| 1b | 8 | 512 | 2 | 8 | 2c | 4 | 512 | 1 | 8 |
| 1c | 8 | 512 | 2 | 8 | 2d | 2 | 128 | 2 | 8 |
| 1d | 8 | 512 | 2 | 8 | 2e | >64 | 128 | 1 | 8 |
| 1e | 16 | 512 | 2 | 8 | 2f | 8 | 512 | 1 | 8 |
| 1f | 8 | 512 | 1 | 8 | 2g | 2 | >512 | 0.5 | 4 |
| 1g | 4 | 512 | 2 | 8 | 2h | 1 | 128 | 1 | 8 |
| 1h | 8 | 512 | 2 | 8 | 2i | 1 | 128 | 1 | 8 |
| 1i | 4 | 256 | 4 | 16 | 2j | 2 | 256 | 1 | 8 |
| 1j | 4 | 256 | 2 | 8 | 2k | 4 | 128 | 1 | 8 |
| 1k | 4 | 256 | 1 | 8 | 2l | 4 | 128 | 1 | 4 |
| 1l | 4 | 256 | 2 | 4 | 2m | 1 | 128 | 1 | 4 |
| 1m | 8 | 512 | 1 | 4 | 3a | 16 | 256 | 2 | 16 |
| B | ||||||
|---|---|---|---|---|---|---|
| compd\conc | 0 | 16 | 32 | 64 | 128 | 256 |
| 2l | 32 | 16 | 4 | 4 | 2 | 2 |
| 2m | 32 | 8 | 4 | 2 | 2 | 1 |
The MICs of cefazolin and imipenem alone against E. coli cells not expressing MβL were 1 and 0.125 μg/mL, respectively.
The antibiotic used was cefazolin.
The antibiotics used was imipenem.
The MIC data indicate that all tested rhodanines 1a–m and 2a–m and the thioenolate 3a increased the antimicrobial effect of cefazolin against E. coli expressing L1, and the largest effect was observed to be from 1l, 1m, 2g, 2l, and 2m, resulting in an 8-fold reduction in MIC. Compounds 2a, 2d, 2i–g, and 2m increased the antimicrobial effect of cefazolin against E. coli producing NDM-1, resulting in a 4–8-fold reduction in MIC. Compounds 2d–e, 2h–i, and 2k–m also increased the antimicrobial effect of cefazolin against E. coli harboring VIM-2, resulting in a 4-fold reduction in MIC. Against E. coli expressing ImiS, only 2g had an effect larger than one dilution factor.
Given the excellent broad-spectrum inhibitory effect of 2l and 2m, dose-dependent MIC assays were performed for these compounds against cells expressing L1 (Table 2B). The MIC data indicate that the antimicrobial effect of cefazolin increased gradually with an increasing inhibitor dose. At the highest dose of 2l and 2m tested (256 μg/mL), the MICs of the antibiotic were decreased 16- and 32-fold, nearly and completely, respectively, restoring the antibacterial effect of cefazolin in the absence of MβLs. No antibacterial effect of the rhodanines alone against the E. coli with and without MβLs was observed at the same inhibitor doses, indicating that the rhodanines’ ability to restore antibiotic activity is due to their inhibitory effect on the MβLs. We further monitored the pH of the culture medium during MIC assays (Figure S4) and found that during the first 20 h it ranged from 6.6 to 7.1, a pH range at which the rhodamine was shown to be stable in the stability assays (Figure S3).
Also, the representative inhibitors 1l, 1m, 2l, 2m, and 3a were subjected to a cytotoxicity assay using mouse fibroblast cells (L929) with different working concentrations (12.5, 25, 50, 100, 200, and 400 μM). As shown in Figure S5, more than 80% of the cells tested maintained viability in the presence of the inhibitors at concentrations up to 200 μM (except the thioenolate 3a), indicating that these rhodanines have low cytotoxicity and are not (or only to a small degree) converted to thioenolates.
To explore potential binding modes, compound 2l was docked into the active sites of NDM-1, VIM-2, CphA (in lieu of ImiS, which has not been crystallized, yet, and with which it shares 96% sequence identity), and L1. The conformations shown in Figure 3 are the lowest-energy conformations of those clusters, with binding energies of −13.6, -13.1, −11.2, and −15.3 kcal/mol for the NDM-1/2l, VIM-2/2l, CphA/2l and L1/2l complexes, respectively. Views of 2l in complex with the four enzymes represented as a surface are shown in Figure S6. They indicate that 2l fits very tightly into the substrate binding sites of the four enzymes.
Figure 3.
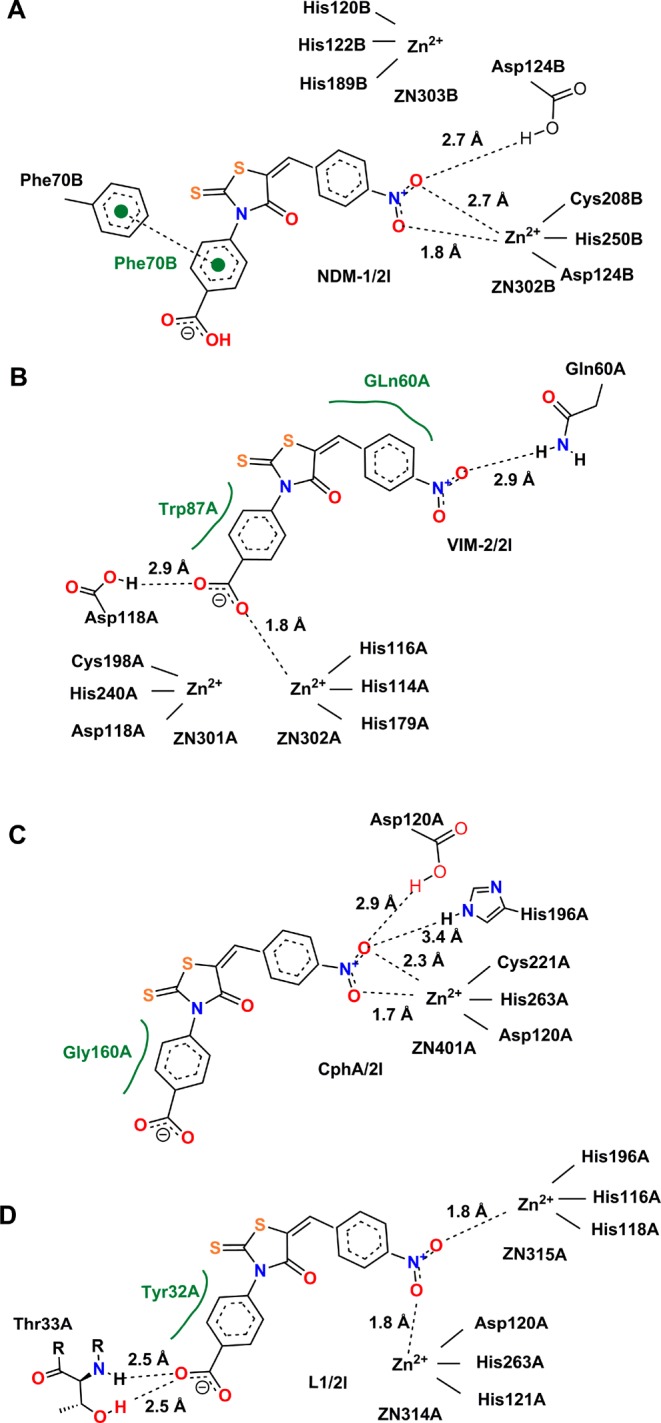
Lowest-energy conformations of 2l docked into the active sites of different enzymes. Graphs A–D show key electrostatic interactions of 2l with Zn(II) ions and residues of the MβLs NDM-1, VIM-2, CphA (as a proxy of ImiS), and L1, respectively, indicated by dashed lines, while hydrophobic interactions are shown in green. The capital letters A and B following the amino acid residue numbers show the protein chains A and B in the crystal structure. All 2D images were generated with PoseView (www.biosolveit.de/PoseView/) and redrawn with ChemBioDraw 14.0.
As shown in Figure 3A,C (as well as Figure S6A,C), 2l adopted similar binding modes to NDM-1 and CphA. The nitro group acted as a bidentate ligand of a Zn(II) ion, and one oxygen of the nitro group formed hydrogen bonds with Asp124 in NDM-1 and Asp120 and His196 in CphA. These residues are Zn(II) ligands in the two enzymes.22 The nitro group as a zinc ligand has been demonstrated previously in a crystal structure of Zn-dependent carboxypetidase A in complex with 2-benzyl-3-nitropropanoic acid.23 Also in the NDM-1/2l and CphA/2l complexes, the benzene ring at the R1 position formed a π–π stacking with Phe70 in NDM-1 and a hydrophobic interaction with Gly160 in CphA. In the complex VIM-2/2l (Figures 3B and S6B), the carboxyl group coordinated Zn2 (1.8 Å) and formed an H-bond with Asp118 (2.9 Å). Carboxylate as a Zn(II) ion ligand has been reported24 and is also seen in aspartate as a Zn(II) ligand in MβLs. In addition, the nitro oxygen interacted with Gln60 in VIM-2 (2.9 Å) via an H-bond, and the two benzene rings interacted with Trp87 and Gln60 via hydrophobic interactions. In complex L1/2l shown in Figures 3D and S6D, the nitro group bridged the two Zn(II) ions (1.8 Å), which is reminiscent of the binding mode of a micromolar inhibitor of the IMP-1 enzyme.25 Also, the carboxyl oxygen formed two hydrogen bonds with the backbone amide and side chain hydroxyl of Thr33 (2.5 Å), and the R1 benzene ring formed a hydrophobic interaction with Tyr32.
In summary, 26 rhodanines and one thioenolate were synthesized and characterized. Z-Configurations of rhodanine were confirmed by X-ray crystal structure resolution of 1l and 2m. Biochemical evaluation revealed that all rhodanines tested strongly inhibited L1, exhibiting IC50 values ranging from 0.02 to 1.7 μM. Specifically 2h–m showed broad-spectrum inhibition of all MβLs tested (NDM-1, VIM-2, ImiS, and L1), with IC50 values <16 μM. SAR studies revealed that the diaryl-substituted rhodanines with electron-accepting atoms or groups exhibited broad-spectrum MβL inhibition, and an N-aromatic carboxyl made the inhibitors more potent than an aliphatic carboxyl. MIC tests indicated that all rhodanines and the thioenolate tested enhanced the antimicrobial effect of cefazolin against E. coli expressing L1, and the largest effect was observed to be from 1l, 1m, 2g, 2l, and 2m, resulting in 8-fold reduction in MIC. Dose-dependency assays showed that the antimicrobial effect of cefazolin increased with increasing dose of inhibitors 2l or 2m. Docking studies suggest that the nitro group (NDM-1, CphA, and L1) or the carboxyl group (VIM-2) of 2l coordinates one or two Zn(II) ion(s), while the N-phenyl of the inhibitor enhances its hydrophobic interaction with the MβLs.
In contrast to a previous report,12 hydrolysis of the rhodanines reported herein and MβL inhibition by the hydrolysis product thioenolate do not seem to play a major role. These studies support Spicer et al.’s original work11 and demonstrate that the diaryl-substituted rhodanines are a good scaffold for the future design of broad-spectrum inhibitors of the MβLs.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.7b00548.
Synthesis and characterization of compounds, X-ray crystallography, methods for enzyme expression and purification, rhodanine stability assays, inhibition kinetic studies, MIC assays including pH monitoring, cytotoxicity assay, docking studies, and graphical views of MβL/2l complexes (PDF)
Spectra (PDF)
Author Contributions
§ These authors contributed equally to this work. The manuscript was written through contributions of all authors.
This work was supported by grants 81361138018 and 21572179 (to K.W.Y.) and 31400663 (to Y.H.) from the National Natural Science Foundation of China and by grant 2014JQ3090 from Natural Science Foundation of Shaanxi Science and Technology Department.
The authors declare no competing financial interest.
Supplementary Material
References
- Bush K.; Jacoby G. A. Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–76. 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp-Wallace K. M.; Endimiani A.; Taracila M. A.; Bonomo R. A. Carbapenems: Past, present, and future. Antimicrob. Agents Chemother. 2011, 55, 4943–60. 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlaes D. M. New β-lactam-β-lactamase inhibitor combinations in clinical development. Ann. N. Y. Acad. Sci. 2013, 1277, 105–14. 10.1111/nyas.12010. [DOI] [PubMed] [Google Scholar]
- Liénard B. M. R.; Garau G.; Horsfall L.; Karsisiotis A. I.; Damblon C.; Lassaux P.; Papamicael C.; Roberts G. C. K.; Galleni M.; Dideberg O.; Frère J. M.; Schofield C. J. Structural basis for the broad-spectrum inhibition of metallo-β-lactamases by thiols. Org. Biomol. Chem. 2008, 6, 2282–94. 10.1039/b802311e. [DOI] [PubMed] [Google Scholar]
- Xiang Y.; Chang Y. N.; Ge Y.; Kang J. S.; Zhang Y. L.; Liu X. L.; Oelschlaeger P.; Yang K. W. Azolylthioacetamides as a potent scaffold for the development of metallo-β-lactamase inhibitors. Bioorg. Med. Chem. Lett. 2017, 27, 5225–9. 10.1016/j.bmcl.2017.10.038. [DOI] [PubMed] [Google Scholar]
- Brem J.; Cain R.; Cahill S.; Mcdonough M. A.; Clifton I. J.; Jiménezcastellanos J. C.; Avison M. B.; Spencer J.; Fishwick C. W. G.; Schofield C. J. Structural basis of metallo-β-lactamase, serine-β-lactamase and penicillin-binding protein inhibition by cyclic boronates. Nat. Commun. 2016, 7, 12406–13. 10.1038/ncomms12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou J.; Wan S.; Chan K. F.; So P. K.; He D.; Chan E. W.; Chan T. H.; Wong K. Y.; Tao J.; Chen S. Ebselen as a potent covalent inhibitor of new delhi metallo-β-lactamase (NDM-1). Chem. Commun. 2015, 51, 9543–6. 10.1039/C5CC02594J. [DOI] [PubMed] [Google Scholar]
- King A. M.; Reid-Yu S. A.; Wang W.; King D. T.; De Pascale G.; Strynadka N. C.; Walsh T. R.; Coombes B. K.; Wright G. D. Aspergillomarasmine a overcomes metallo-β-lactamase antibiotic resistance. Nature 2014, 510, 503–6. 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervosen A.; Lu W. P.; Chen Z.; White R. E.; Demuth T. P.; Frère J. M. Interactions between penicillin-binding proteins (PBPs) and two novel classes of PBP inhibitors, arylalkylidene rhodanines and arylalkylidene iminothiazolidin-4-ones. Antimicrob. Agents Chemother. 2004, 48, 961–9. 10.1128/AAC.48.3.961-969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant E. B.; Guiadeen D.; Baum E. Z.; Foleno B. D.; Jin H.; Montenegro D. A.; Nelson E. A.; Bush K.; Hlasta D. J. The synthesis and SAR of rhodanines as novel class C β-lactamase inhibitors. Bioorg. Med. Chem. Lett. 2000, 10, 2179–82. 10.1016/S0960-894X(00)00444-3. [DOI] [PubMed] [Google Scholar]
- Spicer T.; Minond D.; Enogieru I.; Saldanha S. A.; Mercer B. A.; Allais C.; Liu Q.; Roush W. R.. ML302: A novel β-lactamase (bla) inhibitor. In Probe Reports from the NIH Molecular Libraries Program; NCBI, 2012. [PubMed] [Google Scholar]
- Brem J.; van Berkel S. S.; Aik W.; Rydzik A. M.; Avison M. B.; Pettinati I.; Umland K. D.; Kawamura A.; Spencer J.; Claridge T. D.; McDonough M. A.; Schofield C. J. Rhodanine hydrolysis leads to potent thioenolate mediated metallo-β-lactamase inhibition. Nat. Chem. 2014, 6, 1084–90. 10.1038/nchem.2110. [DOI] [PubMed] [Google Scholar]
- Bush K. The ABCD’s of β-lactamase nomenclature. J. Infect. Chemother. 2013, 19, 549–59. 10.1007/s10156-013-0640-7. [DOI] [PubMed] [Google Scholar]
- Bernardo P. H.; Sivaraman T. K.; Wan K.; Xu F. J.; Krishnamoorthy J.; Song C. M.; Tian L.; Chin J. S.F.; Chai C. L. L. Synthesis of a rhodanine-based compound library targeting Bcl-XL and Mcl-1. Pure Appl. Chem. 2011, 83, 723–31. 10.1351/PAC-CON-10-10-29. [DOI] [Google Scholar]
- Harada K.; Kubo H.; Abe J.; Haneta M.; Conception A.; Inoue S.; Okada S.; Nishioka K. Discovery of potent and orally bioavailable 17b-hydroxysteroid dehydrogenase type 3 inhibitors. Bioorg. Med. Chem. 2012, 20, 3242. 10.1016/j.bmc.2012.03.052. [DOI] [PubMed] [Google Scholar]
- Sing W. T.; Lee C. L.; Yeo S. L.; Lim S. P.; Sim M. M. Arylalkylidene rhodanine with bulky and hydrophobic functional group as selective HCV NS3 protease inhibitor. Bioorg. Med. Chem. Lett. 2001, 11, 91–4. 10.1016/S0960-894X(00)00610-7. [DOI] [PubMed] [Google Scholar]
- Yang H.; Aitha M.; Hetrick A. M.; Richmond T. K.; Tierney D. L.; Crowder M. W. Mechanistic and spectroscopic studies of metallo-β-lactamase NDM-1. Biochemistry 2012, 51, 3839–47. 10.1021/bi300056y. [DOI] [PubMed] [Google Scholar]
- Aitha M.; Marts A. R.; Bergstrom A.; Møller A. J.; Moritz L.; Turner L.; Nix J. C.; Bonomo R. A.; Page R. C.; Tierney D. L.; Crowder M. W. Biochemical, mechanistic, and spectroscopic characterization of metallo-β-lactamase VIM-2. Biochemistry 2014, 53, 7321–31. 10.1021/bi500916y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford P. A.; Sharma N.; Chandrasekar S.; Sigdel T.; Walsh T. R.; Spencer J.; Crowder M. W. Over-expression, purification, and characterization of metallo-β-lactamase imis from aeromonas veronii bv. Sobria. Protein Expression Purif. 2004, 36, 272–9. 10.1016/j.pep.2004.04.017. [DOI] [PubMed] [Google Scholar]
- Crowder M. W.; Walsh T. R.; Banovic L.; Pettit M.; Spencer J. Overexpression, purification, and characterization of the cloned metallo-β-lactamase L1 from stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 1998, 42, 921–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill F. R.Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard; Clinical and Laboratory Standards Institute, 2000. [Google Scholar]
- Chen J.; Chen H.; Zhu T.; Zhou D.; Zhang F.; Lao X.; Zheng H. Asp120Asn mutation impairs the catalytic activity of NDM-1 metallo-β-lactamase: Experimental and computational study. Phys. Chem. Chem. Phys. 2014, 16, 6709–16. 10.1039/c3cp55069a. [DOI] [PubMed] [Google Scholar]
- Wang S. H.; Wang S. F.; Xuan W.; Zeng Z. H.; Jin J. Y.; Ma J.; Tian G. R. Nitro as a novel Zinc-binding group in the inhibition of carboxypeptidase A. Bioorg. Med. Chem. 2008, 16, 3596–601. 10.1016/j.bmc.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Christopeit T.; Yang K. W.; Yang S. K.; Leiros H. K. The structure of the metallo-β-lactamase VIM-2 in complex with a triazolylthioacetamide inhibitor. Acta Crystallogr., Sect. F: Struct. Biol. Commun. 2016, 72, 813–9. 10.1107/S2053230X16016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCuran A. E.; Pegg K. M.; Liu E. M.; Bethel C. R.; Ai N.; Welsh W. J.; Bonomo R. A.; Oelschlaeger P. Elucidating the role of residue 67 in imp-type metallo-β-lactamase evolution. Antimicrob. Agents Chemother. 2015, 59, 7299–307. 10.1128/AAC.01651-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.