Abstract
Astrocyte elevated gene-1 (AEG-1), an oncogene whose overexpression promotes tumor cell proliferation, angiogenesis, invasion, and enhanced chemoresistance, is thought to function primarily as a scaffolding protein, regulating PI3K/Akt and Wnt/β-catenin signaling pathways. Here we report that AEG-1 is an endoplasmic reticulum (ER) resident integral membrane RNA-binding protein (RBP). Examination of the AEG-1 RNA interactome by HITS-CLIP and PAR-CLIP methodologies revealed a high enrichment for endomembrane organelle-encoding transcripts, most prominently those encoding ER resident proteins, and within this cohort, for integral membrane protein-encoding RNAs. Cluster mapping of the AEG-1/RNA interaction sites demonstrated a normalized rank order interaction of coding sequence >5′ untranslated region, with 3′ untranslated region interactions only weakly represented. Intriguingly, AEG-1/membrane protein mRNA interaction sites clustered downstream from encoded transmembrane domains, suggestive of a role in membrane protein biogenesis. Secretory and cytosolic protein-encoding mRNAs were also represented in the AEG-1 RNA interactome, with the latter category notably enriched in genes functioning in mRNA localization, translational regulation, and RNA quality control. Bioinformatic analyses of RNA-binding motifs and predicted secondary structure characteristics indicate that AEG-1 lacks established RNA-binding sites though shares the property of high intrinsic disorder commonly seen in RBPs. These data implicate AEG-1 in the localization and regulation of secretory and membrane protein-encoding mRNAs and provide a framework for understanding AEG-1 function in health and disease.
Keywords: RNA-binding protein, endoplasmic reticulum, RNA localization, signal sequence, transmembrane protein
INTRODUCTION
RNA-binding proteins (RBPs) serve diverse and often pleiotropic roles in the post-transcriptional regulation of gene expression. In addition to prominent nuclear functions in transcription, splicing and mRNA export, RBPs function in the cytosol to regulate RNA localization, translation, and stability (Keene 2007; Glisovic et al. 2008; Halbeisen et al. 2008). Although the primary biological principles of RBP function in RNA localization, translation and turnover are now established, little is known regarding the subcellular distributions and potential compartment-specific functions of RBPs.
Studies into the mechanism(s) of RNA localization to the endoplasmic reticulum (ER), combined with recent landmark surveys of protein–mRNA interactomes, have provided intriguing evidence for an RBP function in mRNA localization to the ER (Chen et al. 2011; Baltz et al. 2012; Castello et al. 2012, 2016a; Kwon et al. 2013; Jagannathan et al. 2014a; Reid and Nicchitta 2015). In regard to the former, biochemical studies of mRNA–ER interactions, conducted in the classical pancreas rough microsome model as well as tissue culture cells, have demonstrated that organelle resident protein-encoding mRNAs are bound to the ER via a direct, i.e., ribosome-independent, mechanism (Chen et al. 2011; Jagannathan et al. 2014a; Reid and Nicchitta 2015). Secretory protein-encoding mRNAs, in contrast, are largely ER-associated via their translation on ER-bound ribosomes and are efficiently released in high salt/EDTA buffers, a finding consistent with current models of signal recognition particle (SRP) pathway-mediated RNA localization to the ER (Blobel and Dobberstein 1975a,b; Shields 1979; Chen et al. 2011; Jagannathan et al. 2014a; Reid and Nicchitta 2015). These findings point to a mRNA selective direct membrane anchoring interaction, presumably mediated by integral membrane RNA-binding protein(s), and which might function to efficiently constrain translation of the associated mRNAs to the ER membrane.
The biochemical and RNA interactome studies noted above identified an unexpected diversity of candidate ER membrane RBPs. Although RBP functions for this cohort of RBPs are largely unvalidated, one candidate, the ribosome receptor protein p180 (RRBP1), was previously identified as a general enhancer of poly(A) mRNA association with the ER and specifically implicated in the ribosome-independent anchoring of placental alkaline phosphatase- and calreticulin-encoding mRNAs to the ER (Cui et al. 2012, 2013). A direct proteomic polyribosome interactor screen identified numerous candidate ER resident RNA-binding proteins, including p180, components of the protein translocation machinery, subunits of the N-linked oligosaccharyl transferase enzyme complex, reticulons, as well as other ER resident membrane proteins (Jagannathan et al. 2014a). Similarly, proteomic screens for the mRNA interactomes of HeLa, HEK293, and mouse embryonic stem cells (mESCs) identified a diversity of candidate ER RNA-binding membrane proteins (Baltz et al. 2012; Castello et al. 2012; Kwon et al. 2013), also including components of the ER protein translocation machinery, the N-linked oligosaccharyl transferase complex, and other previously identified ER membrane proteins, as candidate ER RNA-binding proteins. Given the previously established biochemical functions for many of these resident ER proteins in protein translocation and secretory/membrane protein biogenesis, candidate functions for these proteins in RNA binding to the ER membrane were not anticipated.
RNA-binding proteins (RBPs) typically possess canonical RNA-binding domains, such as the RNA recognition motif (RRM), the K homology domain (KH), the Pumilio homology domain (PUM-HD), or the double-stranded RNA-binding domain (dsRBD), and frequently such RNA-binding motifs are present as multidomain/tandem RNA interaction sites (Lunde et al. 2007; Glisovic et al. 2008; Gerstberger et al. 2014). The candidate ER RNA-binding membrane proteins noted above lack known RNA-binding domains and thus, as proposed for other RBPs lacking canonical RNA-binding motifs, alternative mechanisms of protein–RNA recognition likely contribute to potential functions in mRNA localization, anchoring, and/or translational regulation on the ER (Castello et al. 2012, 2015; Beckmann et al. 2015). Little is known regarding the mechanism(s) of RNA recognition by any of this newly discovered class of noncanonical RNA-binding proteins, though a recent proteomic analysis of RNA–protein interaction sites, captured via UV crosslinking, represents a substantial advance toward answering this fundamental question (Baltz et al. 2012; Castello et al. 2012, 2016a,b; Gerstberger et al. 2013; Kwon et al. 2013). Viewed from the perspective of emerging evidence that many previously identified RNA-binding proteins are multifunctional, a role for nonconventional modes of RNA localization and anchoring to the ER membrane can now be reasonably considered (Mangus et al. 2003; Sawicka et al. 2008; Markus and Morris 2009; Turner and Hodson 2012).
Here we report that AEG-1 (LYRIC, metadherin), an ER resident integral membrane protein previously identified as an oncoprotein (Emdad et al. 2009; Hu et al. 2009; Yoo et al. 2011a) and identified in recent RNA interactome screens, functions as a selective ER mRNA-binding protein (Chen et al. 2011; Baltz et al. 2012; Castello et al. 2012, 2016a; Kwon et al. 2013; Jagannathan et al. 2014a; Reid and Nicchitta 2015). Genome-scale analysis of the AEG-1 RNA interactome by HITS-CLIP (high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation) and PAR-CLIP (photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation) revealed that the AEG-1 RNA interactome is enriched in organelle protein-encoding transcripts. AEG-1-RNA interaction sites were enriched in coding regions, present to a lesser degree in the 5′ untranslated region (UTR), and largely absent from the 3′ UTR. In summary, these data identify AEG-1 as an ER resident RNA binding protein whose interactome is enriched for organelle resident protein-encoding mRNAs, most notably those encoding membrane proteins. In addition to membrane protein-encoding mRNAs, we also report that AEG-1 binds a subset of secretory protein- and cytosolic protein-encoding mRNAs, suggestive of a broad role for AEG-1 in the coordinate regulation of translation on the ER. These data provide a useful framework for examining how AEG-1 up-regulation in the 8q22 genomic gain characteristic of aggressive cancers contributes to enhanced proliferation, metastasis, angiogenesis, and chemoresistance phenotypes.
RESULTS
Identification of high confidence candidate endoplasmic reticulum (ER) RNA-binding proteins
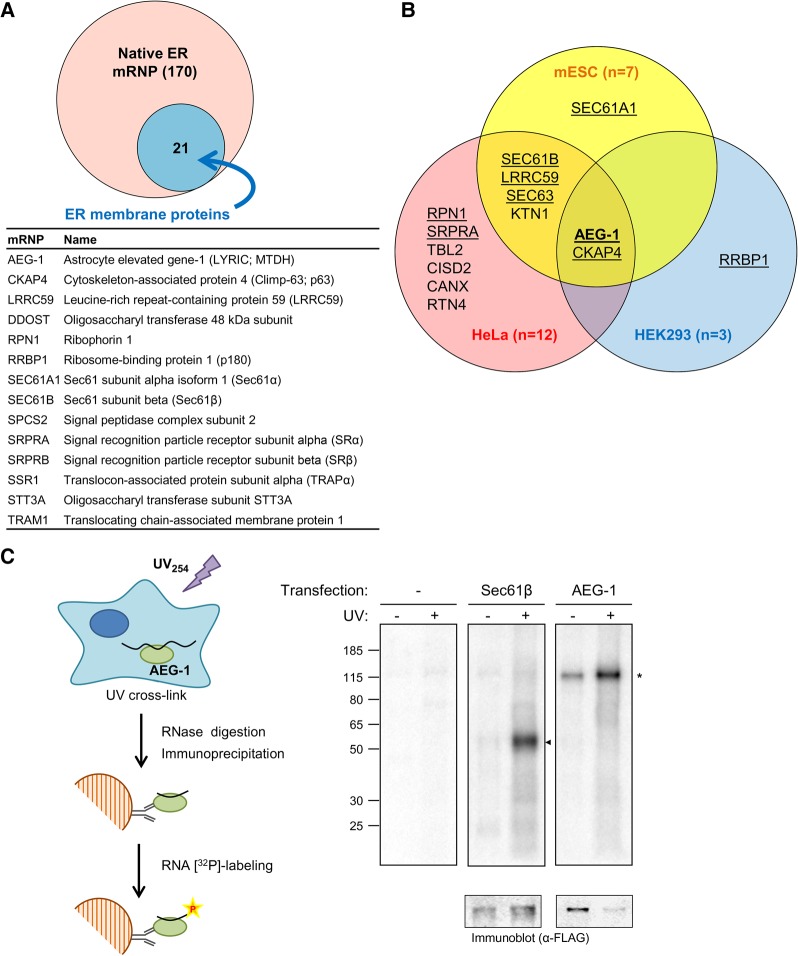
Recent poly(A) RNA interactome screens have identified numerous candidate ER integral membrane RNA-binding proteins (RBPs) including, unexpectedly, components of the protein translocation machinery (e.g., Sec61α, Sec61β, Sec63), the N-linked protein glycosylation machinery (e.g., ribophorin I) and ER-associated microtubule-binding proteins (e.g., CKAP4) (Baltz et al. 2012; Castello et al. 2012, 2016a; Kwon et al. 2013). At present, validation of RBP function for these candidate proteins is lacking, and the larger question of RBP function in mRNA localization, translation and/or stability on the ER has only recently come under investigation. Recent studies have implicated RBP(s) in the membrane association of mRNAs, both with respect to specific cohorts of mRNAs (Chen et al. 2011; Jagannathan et al. 2014a), and more broadly in the localization and association of poly(A) RNA to the ER (Cui et al. 2012, 2013). Complementing the global poly(A) RNA interactome screens, a recent proteomic interactor screen for ER poly(A) RNA-associated RBPs also revealed a diversity of candidate RBPs, including proteins with previously established functions in ER-associated processes, such as ribosome binding, protein translocation, and/or secretory/membrane protein processing (Fig. 1A; Jagannathan et al. 2014a).
FIGURE 1.
RNA interactome screens identify candidate endoplasmic reticulum RNA-binding proteins (RBPs). (A) Proteomic interactor screens identify 170 poly(A) RNA-binding proteins (mRNPs) in canine pancreas rough microsomes and 21 candidate ER integral membrane mRNPs (Jagannathan et al. 2014a). A tabulated listing of 14 of the 21 candidates is provided. (B) Venn diagrams of ER-associated integral membrane mRNA-binding proteins identified in HeLa cells, HEK293 cells, and mouse embryonic stem cells (mESC) (Baltz et al. 2012; Castello et al. 2012; Kwon et al. 2013). The candidate mRBPs identified in the canine rough microsome screen in A are underlined. (C) Schematic of experimental protocol for identifying candidate RNA-binding activity in living cells by UV cross-linking and immunoprecipitation (CLIP). A representative phosphorimage depicting GFP-Sec61β and GFP-AEG-1 RNA complexes’ formation by UV cross-linking is depicted. The empty vector transfection condition is included to depict background labeling.
The Venn diagram depicted in Figure 1B depicts the common ER RBP candidates identified in the ER poly(A) RNA interactor screen of Jagannathan et al. (2014a), and three poly(A) RNA interactomes, conducted in HeLa, HEK293, or mouse embryonic stem cells (mESC), with AEG-1 and CKAP4 as the two candidate ER membrane RBPs common to the four studies (Baltz et al. 2012; Castello et al. 2012; Kwon et al. 2013; Jagannathan et al. 2014a). AEG-1 was previously reported to function as a scaffolding protein, interacting with PLZF, NF-κB, RXR, and the RNA-binding protein SND1 (Sarkar et al. 2008; Thirkettle et al. 2009; Yoo et al. 2011b; Meng et al. 2013; Srivastava et al. 2014) and demonstrated to associate with RBPs and ribosomal proteins in a nuclease-sensitive manner (Meng et al. 2013), a finding consistent with a direct or indirect mRNA binding function. CKAP4 is a microtubule binding protein functioning in the regulation of ER morphology and has also been implicated in the regulation of mRNA localization to the ER (Cui et al. 2012; Sandoz and van der Goot 2015). In this report, we focus on AEG-1.
To distinguish between a direct vs. indirect RNA-binding function for AEG-1, CLIP (crosslinking followed by immunoprecipitation) experiments were performed in tissue cell cultures transfected with constructs encoding FLAG epitope-tagged, cytosolic domains of AEG-1 or Sec61β, the latter of which was identified in three of the four RNA interactome screens noted here (Fig. 1A,B). In these experiments, tissue culture cells were UV-irradiated to introduce covalent protein-RNA cross-links, detergent-lysed, and cross-linked complexes isolated under stringent conditions. Following digestion with RNase I, the RNA moiety was radiolabeled with T4 polynucleotide kinase/[γ-32P]ATP (Fig. 1C). Mock transfection and (−) UV irradiation control conditions were included, to assess immunoprecipitation background and direct protein radiolabeling via protein kinase contaminants, respectively. As shown, prominent radiolabeled products were recovered in both the Sec61β and AEG-1 anti-FLAG immunoprecipitates, and in the transfected but not mock transfected samples (Fig. 1C). For both candidate RBPs, PNK labeling was largely UV irradiation-dependent, consistent with an RNA-binding function. Note that the extensive RNase digestion preceding PNK labeling eliminates the heterogeneity of RNA lengths in protein–RNA complexes to yield uniform protein–RNA complex mobility in SDS–PAGE (Konig et al. 2010). As previously reported, AEG-1 displays a lower than predicted mobility, a consequence of its highly basic pI (Lee et al. 2013).
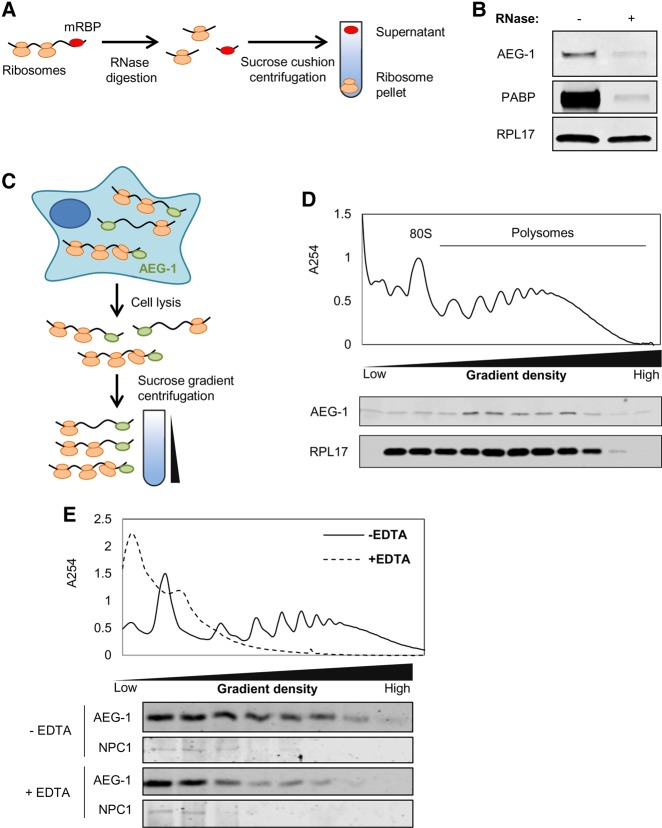
To determine if AEG-1 binds actively translating (i.e., ribosome-associated) mRNAs, cells were detergent-solubilized and the polyribosome fraction assayed for associated AEG-1 (Fig. 2). In one set of experiments, we sought to distinguish between mRNA-associated vs. ribosome-associated AEG-1. To this end, detergent extracts were treated with ribonuclease, subjected to ultracentrifugation, and AEG-1 levels in the ribosome pellet fractions determined by immunoblot (Fig. 2A,B). As shown in Figure 2B, in control samples lacking ribonuclease, AEG-1 was largely recovered in the ribosome pellet, which we confirmed by immunoblot analysis for RPL17 and poly(A)-binding protein (PABP). Following ribonuclease digestion, only background levels of AEG-1 and PABP were recovered in the ribosome pellet, indicating that AEG-1 stably interacts with mRNAs, but not ribosomes. To further examine this conclusion, sucrose density gradient sedimentation analyses were performed (Fig. 2C–E), and demonstrated that AEG-1 is present in the polysome fraction (Fig. 2D), with EDTA-elicited polysome resulting in a shift in AEG-1 migration to slower migrating fractions (Fig. 2E). As a further control, we also examined the distribution of NPC1, a lysosomal membrane protein. As a lysosomal membrane protein, NPC1 would not be expected to migrate with polysomes, which was confirmed by immunoblot analysis of the gradient fractions in the control and EDTA-treatment conditions (Fig. 2E). Combined, the data in Figures 1 and 2 identify AEG-1 as a poly(A) RNA-binding protein that can bind actively translating mRNAs.
FIGURE 2.
AEG-1 associates with actively translating mRNAs. (A) Schematic of experimental protocol for examining AEG-1 RNA-binding activity by ultracentrifugation analysis. Cell lysates were ultracentrifuged subsequent to RNase or control digestion. The polysome fraction (ribosome pellet) was analyzed by immunoblot. (B) Immunoblot analysis of AEG-1, poly(A)-binding protein (PABP), and ribosomal protein (RPL17) in the pellet fraction with/without RNase digestion. (C) Schematic of experimental protocol for the analysis of mRNA-binding activity of AEG-1 by polyribosome profiling. (D) Immunoblot analysis of AEG-1 and ribosomal protein (RPL17) in sucrose density gradient fractions. (E) Mobility of AEG-1 in sucrose density gradient velocity sedimentation in control (−EDTA) or after polyribosome disassembly (+EDTA). Immunoblot analysis of the lysosomal membrane protein NPC1 is illustrated as a membrane protein control.
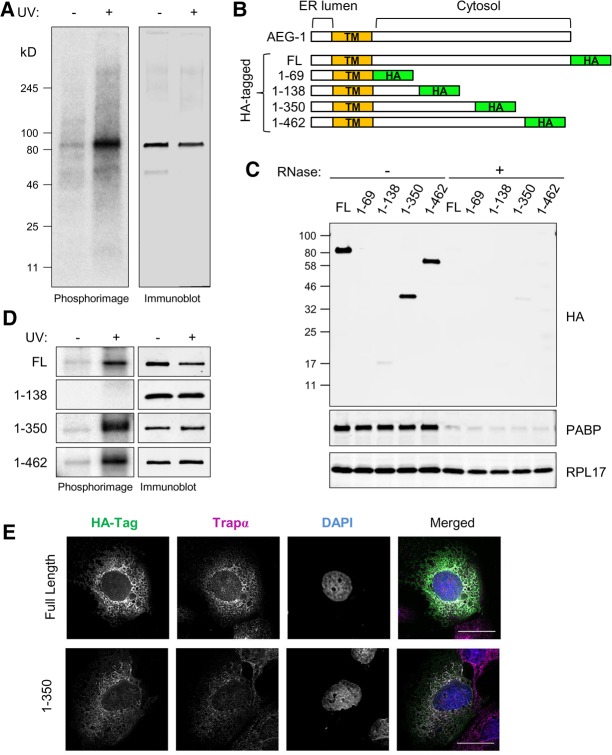
AEG-1 RNA-binding activity maps to the central disordered region
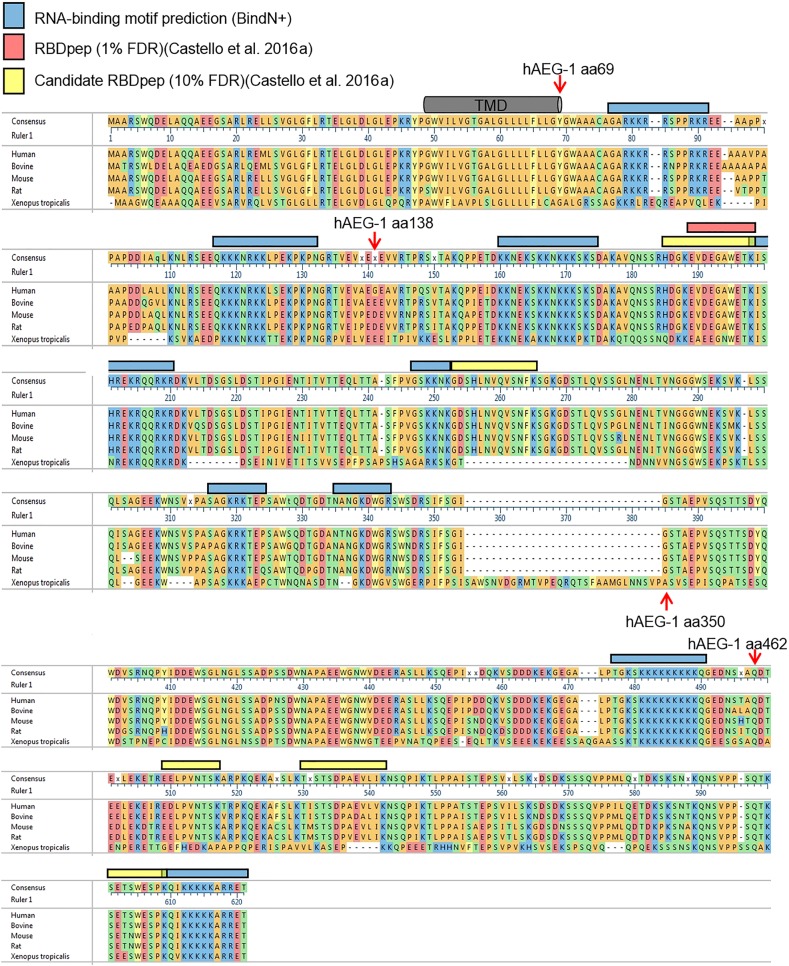
Poly(A) RNA interactome screens have identified over 1000 mRNA-binding proteins (RBPs), of which approximately 300 are common to the mammalian cell-derived data sets referenced above (Baltz et al. 2012; Castello et al. 2012; Kwon et al. 2013; Gerstberger et al. 2014). Approximately two-thirds of the candidate RBPs contain established RNA-binding motifs, e.g., RRM, DEAD, KH, zinc finger domain, frequently in self-tandem repeats or in combination (Gerstberger et al. 2014). AEG-1, however, lacks established RNA-binding domain motifs, and BLASTP searches against the cytosolic domain of the protein (aa70–582) did not reveal homologies with any proteins other than AEG-1 species variants (data not shown). To further validate a direct RNA-binding function for AEG-1, we first performed CLIP experiments on the native, endogenous AEG-1. As shown in Figure 3A, and similar to that depicted above for the FLAG-tagged AEG-1 cytosolic domain (Fig. 1C), UV-dependent RNA–protein crosslinks were readily identified with endogenous AEG-1. Having confirmed an apparent RNA-binding function for the native protein, we used HA-tagged AEG-1 constructs to map RNA-binding sites, using a nested deletion approach. AEG-1 is a Type I single transmembrane protein with a positively charged cytosolic domain (Britt et al. 2004; Sutherland et al. 2004; Kang et al. 2005). We prepared epitope-tagged C-terminal deletion mutants, guided by the RNA-binding prediction algorithm BindN+, AEG-1 RNA-peptide crosslink sites as reported in a recent global proteomic screen for RNA interaction sites (Castello et al. 2016b), and algorithmic analyses of AEG-1 structural disorder (Fig. 4; Wang et al. 2010). Individual members of the truncation series of epitope-tagged C-terminal deletion mutants (Fig. 3B) were expressed by transient transfection, ER localization confirmed by immunofluorescence imaging (Fig. 3E), and RNA-binding activity assessed by the polysome association/centrifugation assay described above (Figs. 2, 3D). We first confirmed that tagged, full-length AEG-1 bound to translating mRNAs and that its recovery in the polysome pellet was abolished by RNase-treatment (Fig. 3C, FL). Deletion mutants containing the first 350 or 462 amino acids of AEG-1 (AEG-1 [aa1–350] and AEG-1 [aa1–462]) retained RNA-binding activity and localized to the ER. Depicted in Figure 3E are representative micrographs depicting the ER localization of HA-tagged full length AEG-1 and AEG-1 (aa1–350), where the individual channels are depicted in gray scale and the merged channels in color, where white represents colocalization of AEG-1/AEG-1 (aa1–350) and the ER marker TRAPα. Two AEG-1 deletion mutants (AEG-1 [aa1–69] and AEG-1 [aa1–138]) were expressed but did not associate with translating mRNAs (Fig. 3C). To further substantiate the RNA-binding activity of the deletion mutants, we performed companion CLIP analyses (Fig. 3D). As shown, AEG-1 (aa1–350) and AEG-1 (aa1–462) formed protein–RNA complexes upon UV irradiation, whereas AEG-1 (aa1–138) lacked detectable RNA-binding activity. Though lacking primary sequence and RNA-binding domain homology with established RBPs, AEG-1 does share with other RBPs the structural property of high intrinsic disorder with these deletion mutant studies mapping the (a) AEG-1-RNA-binding region to the central disordered region (Fig. 4; Varadi et al. 2015; Järvelin et al. 2016). A more precise functional mapping of the AEG-1 RNA-binding domain(s) is currently in progress.
FIGURE 3.
Mutagenic domain mapping of AEG-1–RNA interaction. (A) Endogenous AEG-1 can be crosslinked to RNA in living cells. Cells were UV-irradiated and processed as described in the legend to Figure 1, using antibodies directed against native AEG-1. Depicted is a phosphorimage of the radiolabeled anti-AEG-1 immunoprecipitates and paired immunoblots in control (−UV) and experimental (+UV) conditions. (B) Schematic illustration of AEG-1 deletion mutants. HA-tagged full length (FL) and four C-terminal deletion mutants are shown. (TM) Transmembrane domain. (C) Immunoblot analysis of HA-tagged AEG-1 mutants, poly(A)-binding protein (PABP), and ribosomal protein (RPL17) in pellet fractions derived from ultracentrifugation analyses with/without RNase digestion. (D) Phosphorimager images depicting radiolabeled protein–RNA complex formation of AEG-1 mutants +/− UV crosslinking (left panels). Immunoblot analysis of HA-tagged AEG-1 truncation mutants +/− UV crosslinking, demonstrating recovery of HA-tagged AEG-1 forms in both fractions. (E) Immunofluorescence micrographs of full length (AEG-1) and truncation mutants illustrating ER localization of C-terminal truncated forms. Scale bar, 25 µm.
FIGURE 4.
AEG-1 primary amino acid sequence alignment, predicted RNA interaction sites, and candidate RNA interaction sites determined by proteomic mapping. Sequence alignment of AEG-1 homologs. Amino acid sequences of AEG-1 of human (NP_848927), bovine (NP_001039503), mouse (NP_080278), rat (NP_596889), and Xenopus tropicalis (NP_989164) were aligned by MegAlign Pro. A consensus sequence is shown as the uppermost sequence. The human AEG-1 C-terminal deletion mutants depicted in Figure 3B are indicated. Also indicated are candidate RNA-binding regions from the BindN(+) algorithm (blue bars), the location of a high-confidence RNA-binding region (peptide) identified in a proteomic screen for RNA-binding domains in HeLa cells (red bar) (Castello et al. 2016a), and two lower confidence RNA-binding regions (yellow bar). (TMD) Transmembrane domain.
AEG-1 CLIP-seq mapping reveals a high enrichment in coding sequence interactions
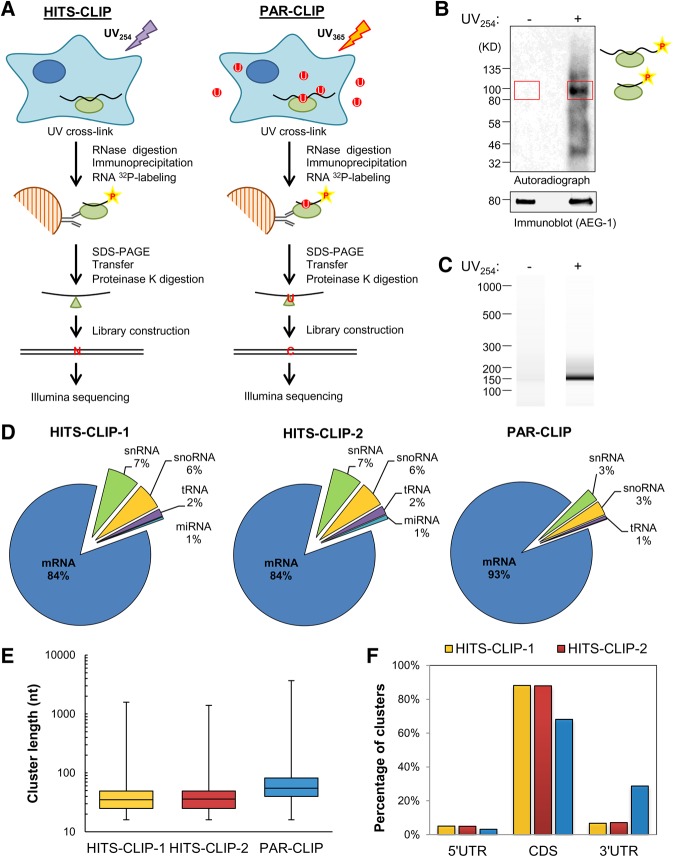
Genome-wide CLIP-seq experimental approaches have been recently applied to the study of RBP interactomes in living cells (Ule et al. 2003; Hafner et al. 2010; Konig et al. 2010). By these methods, bound RNA identities and single-nucleotide resolution mapping of target RNA interaction sequences can be obtained (Ule et al. 2003; Hafner et al. 2010; Konig et al. 2010). To identify the AEG-1 RNA interactome, we used two variations of this technique, high-throughput sequencing-CLIP (HITS-CLIP) and photoactivatable ribonucleoside-enhanced-CLIP (PAR-CLIP) (Darnell 2010; Hafner et al. 2010). The experimental approach is illustrated in Figure 5A. Cells expressing HA epitope-tagged AEG-1, were cultured in the absence (HITS-CLIP) or presence (PAR-CLIP) of 4SU and UV-irradiated to cross-link AEG-1-RNA complexes in vivo. The complexes were then partially RNase-digested, purified by immunoprecipitation, covalently associated RNA radiolabeled, resolved by gel electrophoresis, and transferred to nitrocellulose membranes. A representative autoradiograph of a transferred membrane is depicted in Figure 5B and shows prominent UV irradiation-dependent radiolabeled complexes at 90–100 kDa. These complexes have a molecular weight 10–20 kDa larger than that of AEG-1 protein (80 kDa), corresponding to AEG-1 protein cross-linked to small RNA fragments (Huppertz et al. 2014). The RNA fragments were released from the nitrocellulose membranes by proteinase K digestion, reverse transcribed for cDNA library construction (Fig. 5C), and both control (−UV irradiation) and experimental (+UV irradiation) cDNA libraries deep sequenced. As previously reported, proteinase K digestion leaves a small amino acid/peptide residue at the cross-linked site, resulting in a diagnostic point mutation during reverse transcription (Zhang and Darnell 2011; Sievers et al. 2012). One hundred to 120 million reads, with approximately 4 (HITS-CLIP) to 16 (PAR-CLIP) million reads uniquely mapped to the human transcriptome were obtained. Overall, sequence read distributions were highly enriched in mRNAs: 84%–93% mapped to mRNA, 3%–7% to snRNA, 3%–6% to snoRNA, 1%–2% to tRNA, and 1% to miRNA (Fig. 5D).
FIGURE 5.
CLIP-based AEG-1 RNA interactome analysis. (A) Schematic of experimental protocols for the identification of AEG-1-binding sites by HITS-CLIP and PAR-CLIP. (B) A representative autoradiogram of radiolabeled AEG-1-RNA complex formation obtained in the HITS-CLIP protocol. The red rectangles indicate the regions (∼85–110 kD) used for proteinase K digestion and RNA fragment purification. (C) A representative Bioanalyzer analysis for HITS-CLIP libraries. The libraries were further purified by PAGE electrophoresis and gel extraction. (D) Distributions of mapped RNA deep sequencing reads for the HITS-CLIP (HITS-CLIP-1 and HITS-CLIP-2) and one PAR-CLIP studies. (E) Cluster length distributions for the CLIP studies. (F) Non-normalized distributions of mapped clusters in the 5′ untranslated region (5′ UTR), coding sequence (CDS), and 3′ untranslated region (3′ UTR).
Groups of overlapping reads were considered AEG-1-binding clusters if they contained at least six unique reads and at least one mismatch for HITS-CLIP or at least one T-to-C conversion for PAR-CLIP. By these criteria, we obtained 15,998 and 17,484 clusters of 35 and 36 nt median length from the HITS-CLIP experiments and 99,064 clusters of 55 nt median length from the PAR-CLIP experiments (Fig. 5E). Of these clusters, 3%–5% mapped to the 5′ untranslated region (UTR), 68%–88% to the coding sequence (CDS), and 6%–29% to the 3′ UTR (Fig. 5F). Length-normalized cluster distributions were also calculated and revealed a similar distribution, with a region rank order of CDS >5′ UTR >>3′ UTR (HITS-CLIP = 26%, 5′ UTR, 67%, CDS, 6%, 3′ UTR; PAR-CLIP = 16%, 5′ UTR, 60%, CDS, 24%, 3′ UTR). The prominent cluster read enrichment in the CDS reveals an RNA-binding preference distinct from canonical UTR-targeting RNA-binding proteins (Gebauer et al. 2012; Szostak and Gebauer 2013).
The AEG-1 RNA interactome is enriched in transmembrane protein-encoding mRNAs
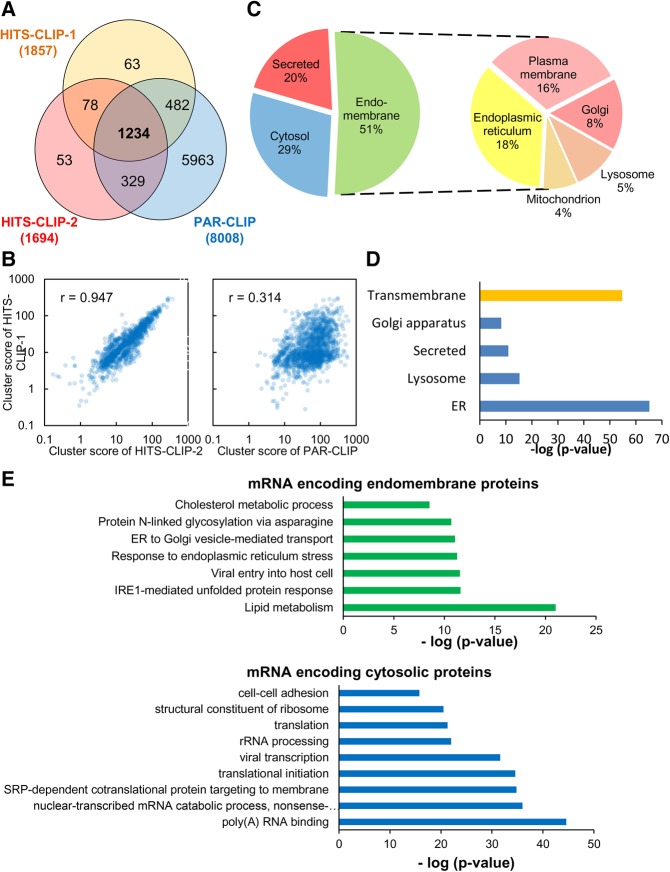
The AEG-1-bound clusters distributed over the 1234 mRNAs common to the two HITS-CLIP and single PAR-CLIP data sets were defined as AEG-1 bound mRNAs (Fig. 6A). In addition to a high shared mRNA composition, the HITS-CLIP replicates displayed a high cluster score reproducibility (Pearson correlation coefficient = 0.947) (Fig. 6B). The correlation between the HITS-CLIP and PAR-CLIP data sets was less robust. The approximately fourfold higher cluster depth in the PAR-CLIP data set however, compromises a sound statistical comparison (Fig. 6A,B). This data set was further analyzed for topology (integral membrane, secretory, or cytosolic), predicted subcellular localization, and enriched gene ontology (GO) terms (Huang et al. 2009a,b). Depicted in Figure 6C are the summary subcellular localization data, demonstrating that the majority (51%) of the clusters map to organelle resident protein-encoding mRNAs, with the remaining fraction to secretory (20%) and, intriguingly, cytosolic protein-encoding mRNAs (29%). The most abundant categories are proteins resident to the ER and the plasma membrane, with lysosome, Golgi apparatus, mitochondrial, and nuclear envelope mRNAs also represented, with ER enrichment being the most significant (P = 3.7 × 10−67) (Fig. 6C,D). In this mRNA cohort, GO analysis also demonstrated significant enrichments for mRNAs encoding lysosomal proteins (P = 4.8 × 10−17) (Fig. 6D). Intriguingly, the AEG-1 mRNA interactome displayed a very significant enrichment in transmembrane protein mRNAs (P = 3.5 × 10−57) (Fig. 6D). This cluster enrichment was evident in both the HITS-CLIP and PAR-CLIP data sets and was distinct from RNA-seq read distributions, indicating that the TMD downstream cluster enrichments were independent of library bias or related artifacts. This enrichment is further highlighted in comparisons of the RNA read distributions in the RNA-seq and the CLIP-seq data sets. Whereas genes encoding transmembrane proteins comprise approximately 26% of the human protein-coding genes (Almén et al. 2009; Fagerberg et al. 2010), only a small fraction of the RNA-seq reads map to transmembrane proteins mRNA (8.9%), indicating that expression levels of this gene superfamily are relatively low. Yet, more than 50% of the cluster values from the HITS-CLIP studies mapped to transmembrane protein mRNAs (data not shown).
FIGURE 6.
The AEG-1 RNA interactome is highly enriched in resident endomembrane protein- and transmembrane domain-encoding transcripts. (A) Venn diagram depicting common and unique mRNAs identified in the HITS-CLIP and PAR-CLIP studies. (B) Reproducibility of AEG-1 bound mRNAs, comparing HITS-CLIP duplicate libraries and HITS-CLIP vs. PAR-CLIP. (C) Subcellular locales of AEG-1 HITS-CLIP genes, depicting endomembrane organelle, cytosolic and secreted protein-encoding genes on the left. The composition of the category “endomembrane” is depicted on the right, including plasma membrane, endoplasmic reticulum, lysosome, Golgi apparatus, mitochondrion, and nuclear envelope. (D) Gene ontology analysis of AEG-1 associated mRNAs. The mRNAs were subjected to gene ontology analysis using DAVID (Huang et al. 2009a,b). Endomembrane organelle distributions of proteins encoded in the AEG-1 bound mRNAs are illustrated. (E) GO category enrichments of the AEG-1 RNA interactome in the categories “endomembrane proteins” and “cytosolic proteins.”
Further examination of the endomembrane resident protein GO categories scoring as highly significant identified numerous categories linked to ER function, membrane biogenesis, and ER proteostatic stress response pathways (Fig. 6E). Also of interest, the AEG-1 cluster data sets included significant enrichments in cytosolic protein gene subsets, specifically the SRP pathway, nonsense-mediated decay (NMD), translational initiation, and poly(A) RNA binding (Fig. 6E). With recent studies having demonstrated that miRNA-mediated translational silencing occurs on the ER, that NMD is functionally linked to ER stress regulation and may occur on ER/nuclear envelope-associated ribosomes, and that ER-bound ribosomes participate in de novo translation initiation, these data suggest functional links between AEG-1 and ER-localized synthesis of cytosolic proteins, and in particular those linking to mRNA export, localization, quality control, and translation (Sakaki et al. 2012; Li et al. 2013; Popp and Maquat 2013; Stalder et al. 2013; Jagannathan et al. 2014a; Barman and Bhattacharyya 2015). These data corroborate recent reports that ER-bound ribosomes contribute significantly to cytosolic protein synthesis, and suggest a mechanism by which such noncanonical localization and translation of cytosolic protein-encoding RNAs could be mediated (Lerner et al. 2003; Pyhtila et al. 2008; Reid and Nicchitta 2012, 2015; Jagannathan et al. 2014b).
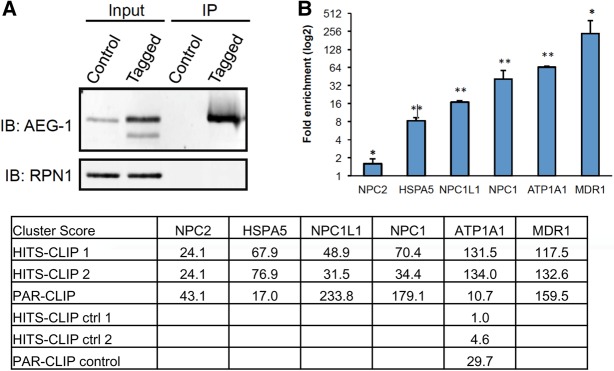
We used RNA-immunoprecipitation and qPCR (RIP-qPCR) as a complementary biochemical approach to validate AEG-1-mRNA interactions identified in the CLIP experiments. Here, HA-tagged AEG-1 and its bound mRNAs were immunoprecipitated from AEG-1-14 cells and the associated mRNAs interrogated by qPCR for mRNAs identified in the CLIP-seq study (Fig. 7A,B). Mock-transfected cells (PC-4) were used as negative controls. The RIP-qPCR data reveal significant enrichments of the AEG-1 bound mRNAs, MDR1, ATP1A1, HSPA5, NPC1, NPC2, and NPC1L1, confirming the specific association of AEG-1 with mRNAs encoding integral membrane and cytosolic proteins (Fig. 7B). Also illustrated are the cluster score enrichments from the two HITS-CLIP data sets, the single PAR-CLIP data set, and the control data sets, where it is evident that the overall rank order of enrichment seen in the RIP-qPCR data is evident in the cluster score enrichments. It was not possible to compare the fold-enrichment rank order for the two approaches because the majority of the genes assayed were absent from the controls. In the one case (ATP1A1) where CLIP clusters were identified in the controls, the fold-enrichment is similar to that seen by RIP-qPCR. These data further strengthen the validity of the primary finding in this study, that AEG-1 is an integral ER RNA binding protein whose interactome is enriched for transmembrane-encoding organelle resident protein mRNAs.
FIGURE 7.
Orthogonal validation and analysis of AEG-1 mRNA interactome. (A) Validation of CLIP results by RNA immunoprecipitation and RT-qPCR (RIP-qPCR) on RNA isolated from mock-transfected cells (PC-4) and HA-tagged AEG-1 transfected cells (AEG-1-14). Immunoblots of RIP-qPCR for AEG-1 and ribophorin I (RPN1). (B) RIP-qPCR fold enrichments for MDR1, ATP1A1, NPC1, NPC2, NPC1L1, HSPA5 mRNAs from AEG-1-14 over PC-4 cells. GAPDH mRNA was used for normalization. (*) P < 0.05, (**) P < 0.01. Included also are the cluster score enrichments for the indicated genes for the HITS-CLIP, PAR-CLIP, and control data sets, demonstrating generally similar rank order enrichments.
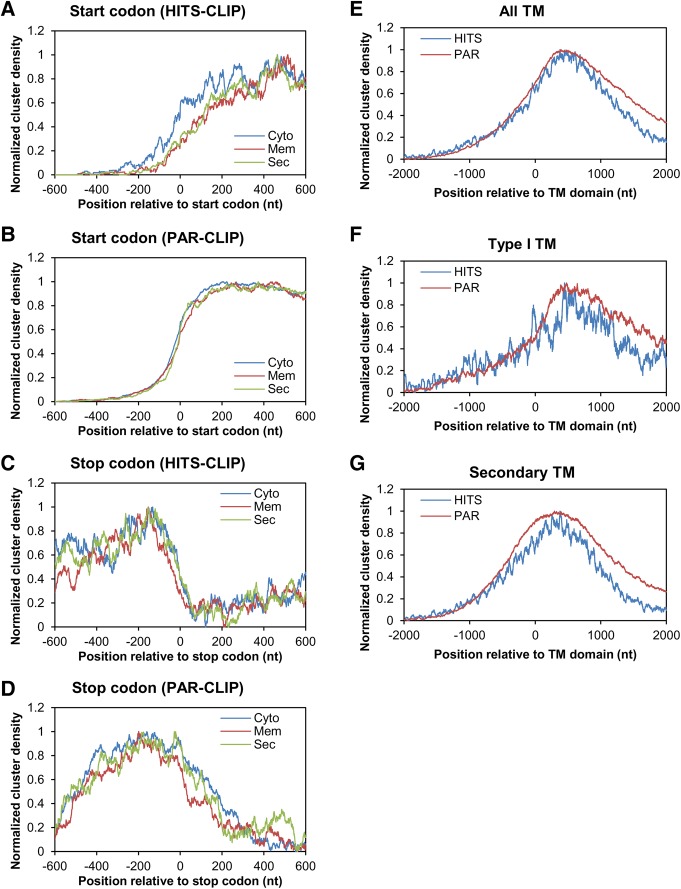
The location (UTR vs. CDS) of RBP-binding sites mirrors RBP function, be it mRNA processing, transport, degradation and/or translation. Thus, translation initiation factors bind to the 5′ UTR, regulating translation initiation and translation efficiency (Wilkie et al. 2003; Araujo et al. 2012); splicing factors recognize the intron-exon junctions (Glisovic et al. 2008; Gerstberger et al. 2014); and many mRBPs bind sites in the 3′ UTR functioning as regulatory signals for mRNA stability, transport, and degradation (Dreyfuss et al. 2002; Glisovic et al. 2008; Gerstberger et al. 2014). To gain insight into the possible molecular consequences of the AEG-1 mRNA interaction, we further analyzed intratranscript distributions of AEG-1 clusters in the integral membrane protein cohort, which is the most enriched in the data sets. Since AEG-1 cluster sites enrich in the CDS of actively translated mRNAs (Fig. 5F), we speculated that AEG-1 regulates translation initiation and/or termination by interacting with sequences flanking translation start/stop sites. We examined AEG-1 clusters within 200 nt flanking the start and stop codons; no significant over-representations of cluster density at either position were, however, observed (Fig. 8A–D). In contrast, we observed a significant over-representation of AEG-1 clusters downstream from encoded transmembrane domains, with total read densities increasing near encoded transmembrane domains and peaking from 100 to 250 codons downstream (Fig. 8E). This cluster enrichment was evident in both the HITS-CLIP and PAR-CLIP data sets and was distinct from RNA-seq read distributions, indicating that the TMD downstream cluster enrichments were independent of library bias or related artifacts. We also examined AEG-1 clusters proximal to the transmembrane domains of Type I membrane proteins as well as the transmembrane domains occurring subsequent to the first encoded transmembrane domain and found very similar read cluster distribution patterns, where AEG-1 binds proximal to transmembrane domains regardless of their processive order in the mRNA (Fig. 8E–G). Since AEG-1 interactions are enriched in the CDS of translating mRNAs, this transmembrane domain proximal association pattern suggests that AEG-1 may regulate the translation of transmembrane proteins, perhaps in a manner coupled to membrane insertion of transmembrane domains and/or topological assembly.
FIGURE 8.
Normalized distributions of AEG-1 clusters in transmembrane protein mRNAs. Plots depicting cluster distributions flanking the start codon (A,B) and stop codon (C,D) of mRNAs encoding cytosolic, transmembrane, or secretory proteins, in the HITS-CLIP (A,C) and PAR-CLIP (C,D) experiments. (E,F) Plots depicting cluster distributions flanking encoded transmembrane domains (E), the transmembrane domains of Type I membrane proteins (F), or relative to subsequent transmembrane domains (G), for the HITS-CLIP and PAR-CLIP data sets. Cluster enrichment data are plotted as cluster density where 0 indicates the first nucleotide of start codons, stop codons, or transmembrane domain-encoding regions.
In summary, the data included here demonstrate that AEG-1 functions in the association of select cohorts of mRNAs to the ER membrane. In the case of integral membrane protein mRNAs, the cluster enrichment downstream from encoded transmembrane domains suggests that AEG-1 may contribute to the dynamic regulation of transmembrane domain insertion, integration, and/or topological orientation. In the case of cytosolic protein-encoding mRNAs, interactions with AEG-1 may serve to localize translation to the ER and/or nuclear envelope membrane. More informed insights into AEG-1 function as an ER RNA anchoring protein await further study. In this regard, it should be noted that AEG-1 knockout mice are viable and thus AEG-1 is likely a member of the family of ER RNA-binding proteins that contribute redundant functionality to the localization and translational regulation of mRNAs on the ER (Fig. 1C; Lee et al. 2013; Robertson et al. 2015).
DISCUSSION
Here we report that AEG-1, an oncogene linked to the up-regulated proliferation, metastasis, angiogenesis, and chemoresistance phenotypes of aggressive cancers, is an ER resident RNA binding protein. In vivo genome-scale mapping of AEG-1/RNA interaction sites by HITS-CLIP and PAR-CLIP methods provided complementary experimental evidence that the RNA interactome of AEG-1 is highly enriched for organelle resident protein mRNAs, and in particular those encoding transmembrane proteins. Intriguingly, AEG-1/RNA-binding interactions are enriched in the coding sequence and although a clear consensus motif was not evident, a signature read cluster enrichment downstream from encoded transmembrane domains was identified. Also of significant interest, the AEG-1 interactome includes a cohort of cytosolic protein-encoding mRNAs, suggesting that AEG-1 contributes broadly to ER-localized mRNA translation.
RNA-binding proteins (RBPs) comprise a substantial fraction of the human proteome (approximately 7.5% of protein coding genes), with current estimates numbering close to 1000 RBPs binding the mRNA transcriptome (Baltz et al. 2012; Castello et al. 2012, 2016a; Kwon et al. 2013) Generally speaking, RBPs can be binned into two broad groups: canonical RBPs, which contain established RNA-binding motifs such as RRM, KH, dsRBD, and zinc finger, and whose binding sites are enriched in the untranslated regions (UTRs) of mRNAs, and noncanonical RBPs, generally multifunctional proteins which lack established RNA-binding motifs, and whose binding enrichments remain largely unknown. The latter category has recently gained considerable interest, in large part because of the diversity of candidate RNA-binding proteins revealed in recent large-scale RNA interactome studies (Baltz et al. 2012; Castello et al. 2012; Kwon et al. 2013). Particularly surprising has been the identification of housekeeping proteins, such as glycolytic enzymes, as RNA-binding proteins (Castello et al. 2012). The intriguing ramification of this broadening view of RNA-binding protein identity and functionality is the opportunity for higher order spatial organization of RNA localization and translation, and thus additional mechanisms for the coordinate regulation of related genes (Keene 2007; Kishore et al. 2010; Gebauer et al. 2012).
In contrast to many canonical RBPs, which display bioinformatically discernible and experimentally defined target recognition motifs, primarily UTR-enriched, AEG-1 binding sites are almost wholly located in the CDS of its mRNA interactome. This coding sequence enrichment likely precludes a distinct nucleotide level AEG-1 interaction motif, as that would also yield a primary amino acid sequence motif, which is not evident. Consistent with this view, analyses of the combined HITS-CLIP and PAR-CLIP cluster data sets did not reveal a consensus motif (data not shown). In the absence of discrete interaction sequences, we suggest that AEG-1 affects ER localization via low affinity (sequence diverse), high avidity interactions within the CDS. Such an avidity-based mechanism would be expected to yield stable membrane association. We speculate that an abundance of low affinity CDS-binding sites may help constrain the secondary structural space of the target mRNAs, thereby favoring a largely unstructured CDS that would serve as a more optimal translation substrate. Though there is extensive evidence for the broad utilization of low affinity, high avidity RNA–protein interactions (Williamson 2000; Leulliot and Varani 2001; Van Roey and Davey 2015; Varadi et al. 2015), the selectivity and enrichment for organelle protein-encoding mRNAs does imply the recognition of common features of this cohort of mRNAs, which we have yet to identify. Compounding this challenge, RBPs are noteworthy for their abundance of intrinsically disordered domains, as well as a diversity of potential induced fit interactions with RNA substrates (Williamson 2000; Leulliot and Varani 2001; Varadi et al. 2015; Järvelin et al. 2016). AEG-1 is marked by an extensive abundance of intrinsically disordered domains. Primary sequence analysis of intrinsically disordered domains in AEG-1, using the DISOPRED algorithm (Ward et al. 2004a,b), indicates that almost the entirety of the cytoplasmic domain of AEG-1 is of low complexity/highly disordered. The challenge ahead is to identify mechanistic and structural links between the intrinsically disordered AEG-1 cytoplasmic domain and its selective association with organelle resident membrane protein-encoding mRNAs. Deletion analyses (Fig. 4) highlight the region aa138–aa350, which includes a prominent region of low complexity/high disorder and which displays RNA binding activity in the cell, as a focus area for future study.
AEG-1 is highly conserved in vertebrates (Fig. 4; Lee et al. 2013), and so we hypothesized that the protein region(s) functioning in RNA binding would also be highly conserved. As illustrated in Figure 4, the region aa139 to aa350 is highly positively charged (27 Lys, 9 Arg residues) and the basic amino acid residues are both highly conserved and clustered in the region aa157 to aa209. Intriguingly, the primary sequence in this region is highly conserved from Xenopus to human (75.5% identity in 53 residues) although the overall sequence homology between Xenopus and human AEG-1 is significantly lower (46.3% identity in 587 residues). Also of high interest, a high-confidence RNA interaction site was identified in the region aa188–198, in a recent RNA interactome/binding site proteomic screen (Castello et al. 2016b). Such regionally high sequence conservation is consistent with an important cellular function for the central domain of the protein, which we propose to be RNA binding. These findings are also consistent with proposals regarding the role of disordered domains in RNA binding, in particular the view that such interactions may enable multiple binding interactions without the need for the constraints imposed by discrete, defined sequence motifs, both in coding and structural terms (Järvelin et al. 2016).
An important clue to a specificity in AEG-1/RNA interactions was revealed in the HITS-CLIP and PAR-CLIP data sets, where a signature high read cluster enrichment was identified in the region surrounding and downstream from encoded transmembrane domains (Fig. 8). Such enhanced interactions may be functionally linked to protein biogenesis, perhaps by enabling translational stalling, and thereby contributing to the coupling of protein synthesis and folding. We note that studies of transmembrane domain integration have demonstrated that both the primary and secondary structural characteristics of transmembrane domains within the ribosomal exit tunnel can serve as a signal regulating the assembly/insertion of upstream transmembrane domains in the translocon (Do et al. 1996; Daniel et al. 2008). The findings reported here may represent an additional molecular component of this ordered protein synthesis, signaling and translocation process (Cross and High 2009; Skach 2009; Shao and Hegde 2011; Mandon et al. 2013). Alternatively, such cluster read enrichments may be a consequence of the binding interactions between SRP and the nascent transmembrane domains, though how such long range interactions might influence AEG-1/mRNA interactions remains to be determined.
A particularly intriguing element of the data sets presented here is the identification of cytosolic protein-encoding mRNAs in the AEG-1 RNA interactome. GO analysis of this mRNA cohort revealed significant enrichments for genes functioning in translation, translational regulation, and RNA localization to the ER (Fig. 6C,E). These data corroborate prior studies reporting the localization and translation of cytoplasmic protein mRNAs on ER-bound ribosomes and provide further evidence for the emerging view that ER-localized translation contributes broadly to cellular proteome expression (Lerner et al. 2003; Pyhtila et al. 2008; Reid and Nicchitta 2012, 2015; Jagannathan et al. 2014b). These data, and specifically the GO analysis findings, link to recent studies demonstrating de novo translation initiation on the ER (Jagannathan et al. 2014b), as well as reports demonstrating that translational regulatory processes presumed to occur in the cytosol compartment are, at least in part, ER-localized (Sakaki et al. 2012; Li et al. 2013; Stalder et al. 2013; Barman and Bhattacharyya 2015). The functional significance of such noncanonical mRNA localization warrants further study, because such data reopen investigations into the mechanisms governing mRNA partitioning between the cytosol and the ER, and the contributions of ER-localized translation to post-transcriptional gene regulation.
In summary, we report that AEG-1, a resident ER membrane protein with a positively charged and largely disordered cytoplasmic domain, is an RNA-binding protein functioning in the selective localization and anchoring of mRNAs to the ER. This selective RNA binding/anchoring functionality expands the mechanisms used by eukaryotic cells to selectively localize translation to the ER and by virtue of the direct binding mechanism, serves to “imprint” the ER membrane with the information necessary for the synthesis of resident organelle proteins. From existing global RNA interactome studies, as well as prior reports, we speculate that AEG-1 is one of many ER membrane proteins that contribute to the ER-mRNA transcriptome and its composite regulation (Chen et al. 2011; Cui et al. 2012; Jagannathan et al. 2014a; Reid and Nicchitta 2015). Defining how these composite mRNA localization processes regulate proteome expression and secretory pathway functionality represents an intriguing new direction of research into the primary organizational principles governing eukaryotic cell structure and function.
MATERIALS AND METHODS
Cell lines and antibodies
AEG-1-14 and PC-4 cells were cultured as previously described (Yoo et al. 2009). AEG-1-14 cells are HepG3 cells stably transfected with a C-terminal HA-tag AEG-1 [pcDNA3.1/Hygro(+)] and express tagged AEG-1 at levels approximately 2.5-fold higher then parental lines (PC-4 cells). PC-4 cells are HepG3 cells stably transfected with empty vector pcDNA3.1/Hygro(+). Primary antibodies used were anti-AEG-1 (1:5000; chicken polyclonal) (Kang et al. 2005), anti-PABP (1:2000; rabbit polyclonal; kind gift of Dr. Jack Keene, DUMC), anti-α-tubulin (1:500; mouse monoclonal 12G10; obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA), anti-HA (1:10,000; mouse monoclonal; Thermo Fisher Scientific; 26183); FLAG (1:1000; rabbit polyclonal, AbboMax).
UV cross-linking and immunoprecipitation (CLIP)
AEG-1-14 cells were irradiated with 254 nm UV light at 400 mJ/cm2 (Stratagene Stratalinker) on ice, lysed with CLIP lysis buffer (50 mM Tris-HCl, pH 7.2, 100 mM NaCl, 1 mM EDTA, pH 8, 1% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS), nuclease-digested (Turbo DNase and RNase I; Thermo Fisher Scientific), and immunoprecipitated with anti-HA antibody and G protein Dynabeads at 4°C overnight. The beads were washed with high-salt buffer (50 mM Tris-HCl, pH 7.2, 1 M NaCl, 1 mM EDTA, pH 8, 1% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS), polynucleotide kinase (PNK) buffer (20 mM Tris-HCl, pH 7.2, 10 mM MgCl2, 0.2% Tween-20), and subsequently incubated with 0.5 U/µL of polynucleotide kinase (New England Biolabs) and 0.5 µCi/µL [γ-32P]ATP, in PNK buffer, to radiolabel RNA. The samples were eluted with HA peptide and analyzed by SDS–PAGE and phosphorimaging.
Sucrose cushion and density gradient polyribosome profiling
Cells grown to 80%–90% confluence were treated with 50 µg/mL cycloheximide (CHX) at 37°C for 10 min, washed with ice-cold DPBS, incubated with 50 µg/mL CHX/DPBS at 4°C for 20 min, and lysed by adding sucrose cushion lysis buffer (200 mM KCl, 25 mm KHEPES, pH 7.2, 50 mM EDTA, 1 mM DTT, 2% n-dodecyl-β-D-maltoside [DDM], and protease inhibitor cocktail) on ice for 15 min. The lysates were centrifuged at 14,000 rpm for 15 min to remove cell debris. The supernatants were incubated with either RNaseOUT or RNase Cocktail (4 U/mL of RNase A and 80 U/mL of RNase T1; Thermo Fisher Scientific) on ice for 30 min. The samples were loaded onto a sucrose cushion (500 mM sucrose, 200 mM KCl, 25 mm KHEPES, pH 7.2, 2 mM EDTA, 1 mM DTT). Ribosomes were collected by centrifugation at 90,000 rpm at 4°C for 15 min (TLA 100.2). Ribosome pellets were resuspended in SDS sample buffer and analyzed by SDS–PAGE and immunoblot analysis. For polysome profiling, cells were treated with CHX as above, washed in ice-cold DPBS, and lysed by addition of polysome lysis buffer (200 mM KCl, 25 mm KHEPES, pH 7.2, 10 mM MgCl2, 1 mM DTT, 2% DDM, RNaseOUT, and protease inhibitor cocktail) on ice for 15 min. Cell lysates were centrifuged at 14,000 rpm for 15 min to remove cell debris. Polyribosomes were resolved on 15%–50% sucrose gradients and fractionated as described previously (Stephens and Nicchitta 2007, 2008). The fractions were TCA precipitated, resuspended in SDS sample buffer, and analyzed by SDS–PAGE and immunoblotting.
Immunofluorescence microscopy
AEG-1-14 and PC-4 cell monolayers were fixed in 10% formalin for 10 minutes at room temperature. After permeabilization in 0.1% Triton X-100/PBS and blocking with 1% bovine serum albumin in PBS, the cells were incubated in a primary antibody solution for 1 h, extensively washed and subsequently incubated in secondary antibody solution for 45 min. Cell monolayers were again washed and the nuclei stained using DAPI. Coverslips were then mounted and imaged on a Deltavision Elite deconvolution microscope, using the 60×/1.42 oil PLAPON60XO WD 150 micron DIC ∞/0.17/FN26.5, UIS2, BFP1 objective. Light micrographs were uniformly size and contrast adjusted in Adobe Photoshop CS6.
CLIP-seq protocols
Cells were grown overnight with (PAR-CLIP), or without (HITS-CLIP), 100 µM 4-thiouridine. AEG-1-14 and PC-4 cells were UV cross-linked, nuclease-digested, and immunoprecipitated as described above. For RNA dephosphorylaton, samples were incubated with 0.5 U/µL of calf intestinal alkaline phosphatase (New England Biolabs) in dephosphorylation buffer (50 mM Tris-HCl, pH 8, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT) at 37°C for 25 min. For 5′-end radioisotope-labeling, the samples were incubated with 0.5 U/µL of polynucleotide kinase (New England Biolabs) and 0.5 µCi/µL [γ-32P]ATP in PNK buffer supplemented with 1 mM DTT. Protein-RNA complexes were eluted by 2× sample buffer at 95°C for 5 min, separated by SDS–PAGE, transferred to nitrocellulose membranes, and analyzed by phosphorimaging. RNA fragments were extracted from the nitrocellulose membranes by proteinase K digestion and purified by phenol/chloroform extraction. cDNA library construction was performed according to the manufacturer's instructions (New England Biolabs; E7330). cDNA libraries were analyzed by the Bioanalyzer DNA 1000 Kit (Agilent Technologies), Sanger sequencing, and Illumina HiSeq for 50-nucleotide single-end read runs.
Data analysis
Read mapping
A transcriptome was generated using RNA-seq data from HepG2 cells from the ENCODE project (GEO GSM958740). Tophat and Cufflinks were used to establish transcript abundance, and the most abundant isoform from each gene was selected for further analysis. PAR-CLIP and HITS-CLIP reads were trimmed using Cutadapt, then mapped to the reference transcriptome using Bowtie, allowing for one mismatch and one assigned location per read.
Cluster identification
Clusters were defined as an overlapping set of reads that contained at least one mismatch (T→C for PAR-CLIP, any mutation for HITS-CLIP) and contained at least six unique read starts or stops. A score for each cluster was also defined that favored clusters that had many unique reads and many mutations. The score was defined as log2[1000*(1–D)*M/R], where M is the number of mutations in the cluster and R is the abundance of the RNA from RNA-seq. D represents the breadth of read distribution, defined as (1/N)N, where N is the number of unique read starts or stops in the cluster. Clusters with scores of less than 0 were discarded.
Cluster analysis
For cluster positional analysis, total cluster score was summed relative to each landmark. For coding sequence starts and stops, cluster scores were summed relative to the Refseq annotations. For TM domain analysis, the cluster scores were summed relative to the first nucleotide of the first codon of a predicted point TM domain.
TM domains and signal sequences were identified algorithmically, using TMHMM and SignalP, respectively (Krogh et al. 2001). For analysis of the distribution of RNAs and clusters between different cohorts of mRNAs, cytosolic mRNAs were defined as lacking both TM domains and signal sequences, secretory mRNAs encoded a signal sequence but lacked a TM domain, and TM protein mRNAs encoded a TM domain with or without a signal sequence. Gene Ontology analyses were performed with the DAVID tools (Huang et al. 2009a,b).
RNA immunoprecipitation (RIP) and quantitative PCR
RIP was performed as described with minor modifications (Keene et al. 2006). PC-4 and AEG-1-14 cells were washed and harvested in ice-cold PBS and lysed in RIP lysis buffer (100 mM KCl, 10 mm KHEPES, pH 7.2, 5 mM MgCl2, 1 mM DTT, 400 µM VRC, 1 mM PMSF, 10 mM NaF, 2 mM sodium orthovanadate, 2 mM β-glycerophosphate, 0.5% IGEPAL CA-630, RNaseOUT, and protease inhibitor cocktail). The lysates were incubated on ice for 10 min and centrifuged at 14,000 rpm for 15 min to clear cell debris. The supernatants were supplemented with NT2 buffer (150 mM NaCl, 1 mM MgCl2, 50 mM Tris-HCl, pH 7.4, 0.05% IGEPAL CA-630), EDTA to 15 mM, DTT to 1 mM, RNaseOUT and VRC, and then immunoprecipitated with anti-HA antibody prebound Dynabeads Protein G beads (Thermo Fisher Scientific) at 4°C overnight. The samples were washed five times with NT2 buffer supplemented with 15 mM EDTA. RNA was eluted with TRIsure (Bioline) according to the manufacturer's instructions. The isolated RNA from RIP was analyzed by quantitative PCR. The results were shown as the relative fold enrichment of AEG-1-14 cell line over the PC-4 control signal.
DATA DEPOSITION
The sequencing data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Hsu et al. 2018) and are accessible through GEO Series accession number GSE110260 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110260).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jessica Child for critical reading and valuable criticism of the manuscript. The support of the Duke Sequencing and Genomic Technologies Core is gratefully acknowledged. The anti-tubulin monoclonal antibody 12G10, developed by J. Frankel and E.M. Nelsen, University of Iowa, was obtained from the Developmental Studies Hybridoma Bank, created by the National Institute of Child Health and Human Development of the National Institutes of Health (NIH) and maintained at The University of Iowa, Department of Biology, Iowa City, Iowa. This work was supported by grants from the National Institute of General Medical Sciences, NIH (GM101533-05A1 and GM118630-A1 to C.V.N.) and the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (1R01DK107451 to D.S.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.063313.117.
REFERENCES
- Almén MS, Nordström KJ, Fredriksson R, Schiöth HB. 2009. Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol 7: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo PR, Yoon K, Ko D, Smith AD, Qiao M, Suresh U, Burns SC, Penalva LOF. 2012. Before it gets started: regulating translation at the 5′ UTR. Comp Funct Genomics 2012: 475731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. 2012. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell 46: 674–690. [DOI] [PubMed] [Google Scholar]
- Barman B, Bhattacharyya SN. 2015. mRNA targeting to endoplasmic reticulum precedes Ago protein interaction and microRNA (miRNA)-mediated translation repression in mammalian cells. J Biol Chem 290: 24650–24656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann BM, Horos R, Fischer B, Castello A, Eichelbaum K, Alleaume AM, Schwarzl T, Curk T, Foehr S, Huber W, et al. 2015. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat Commun 6: 10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Dobberstein B. 1975a. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol 67: 835–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Dobberstein B. 1975b. Transfer of proteins across membranes II. Reconstitution of functional rough microsomes from heterologous components. J Cell Biol 67: 852–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt DE, Yang DF, Yang DQ, Flanagan D, Callanan H, Lim YP, Lin SH, Hixson DC. 2004. Identification of a novel protein, LYRIC, localized to tight junctions of polarized epithelial cells. Exp Cell Res 300: 134–148. [DOI] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149: 1393–1406. [DOI] [PubMed] [Google Scholar]
- Castello A, Hentze MW, Preiss T. 2015. Metabolic enzymes enjoying new partnerships as RNA-binding proteins. Trends Endocrinol Metab 26: 746–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Frese CK, Horos R, Alleaume AM, Foehr S, Curk T, Krijgsveld J, Hentze MW. 2016a. Comprehensive identification of RNA-binding domains in human cells. Mol Cell 63: 696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Horos R, Strein C, Fischer B, Eichelbaum K, Steinmetz LM, Krijgsveld J, Hentze MW. 2016b. Comprehensive identification of RNA-binding proteins by RNA interactome capture. Methods Mol Biol 1358: 131–139. [DOI] [PubMed] [Google Scholar]
- Chen Q, Jagannathan S, Reid DW, Zheng T, Nicchitta CV. 2011. Hierarchical regulation of mRNA partitioning between the cytoplasm and the endoplasmic reticulum of mammalian cells. Mol Biol Cell 22: 2646–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross BC, High S. 2009. Dissecting the physiological role of selective transmembrane-segment retention at the ER translocon. J Cell Sci 122(Pt 11): 1768–1777. [DOI] [PubMed] [Google Scholar]
- Cui XA, Zhang H, Palazzo AF. 2012. p180 promotes the ribosome-independent localization of a subset of mRNA to the endoplasmic reticulum. PLoS Biol 10: e1001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui XA, Zhang Y, Hong SJ, Palazzo AF. 2013. Identification of a region within the placental alkaline phosphatase mRNA that mediates p180-dependent targeting to the endoplasmic reticulum. J Biol Chem 268: 29633–29641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel CJ, Conti B, Johnson AE, Skach WR. 2008. Control of translocation through the Sec61 translocon by nascent polypeptide structure within the ribosome. J Biol Chem 283: 20864–20873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. 2010. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA 1: 266–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do H, Falcone D, Lin JL, Andrews DW, Johnson AE. 1996. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell 85: 369–378. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Bio 3: 195–205. [DOI] [PubMed] [Google Scholar]
- Emdad L, Lee SG, Su ZZ, Jeon HY, Boukerche H, Sarkar D, Fisher PB. 2009. Astrocyte elevated gene-1 (AEG-1) functions as an oncogene and regulates angiogenesis. Proc Natl Acad Sci 106: 21300–21305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L, Jonasson K, von Heijne G, Uhlén M, Berglund L. 2010. Prediction of the human membrane proteome. Proteomics 10: 1141–1149. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Preiss T, Hentze MW. 2012. From cis-regulatory elements to complex RNPs and back. Cold Spring Harb Perspect Biol 4: a012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. 2013. Learning the language of post-transcriptional gene regulation. Genome Biol 14: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. 2014. A census of human RNA-binding proteins. Nat Rev Genet 15: 829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. 2008. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 582: 1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbeisen RE, Galgano A, Scherrer T, Gerber AP. 2008. Post-transcriptional gene regulation: from genome-wide studies to principles. Cell Mol Life Sci 65: 798–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Wei Y, Kang Y. 2009. The multifaceted role of MTDH/AEG-1 in cancer progression. Clin Cancer Res 15: 5615–5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Huppertz I, Attig J, D'Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, Tajnik M, König J, Ule J. 2014. iCLIP: protein-RNA interactions at nucleotide resolution. Methods 65: 274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan S, Hsu JC, Reid DW, Chen Q, Thompson WJ, Moseley AM, Nicchitta CV. 2014a. Multifunctional roles for the protein translocation machinery in RNA anchoring to the endoplasmic reticulum. J Biol Chem 289: 25907–25924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan S, Reid DW, Cox AH, Nicchitta CV. 2014b. De novo translation initiation on membrane-bound ribosomes as a mechanism for localization of cytosolic protein mRNAs to the endoplasmic reticulum. RNA 20: 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järvelin AI, Noerenberg M, Davis I, Castello A. 2016. The new (dis)order in RNA regulation. Cell Commun Signal 14: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DC, Su ZZ, Sarkar D, Emdad L, Volsky DJ, Fisher PB. 2005. Cloning and characterization of HIV-1-inducible astrocyte elevated gene-1, AEG-1. Gene 353: 8–15. [DOI] [PubMed] [Google Scholar]
- Keene JD. 2007. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8: 533–543. [DOI] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB. 2006. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc 1: 302–307. [DOI] [PubMed] [Google Scholar]
- Kishore S, Luber S, Zavolan M. 2010. Deciphering the role of RNA-binding proteins in the post-transcriptional control of gene expression. Brief Funct Genomics 9: 391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. 2010. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17: 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580. [DOI] [PubMed] [Google Scholar]
- Kwon SC, Yi H, Eichelbaum K, Föhr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, Kim VN. 2013. The RNA-binding protein repertoire of embryonic stem cells. Nat Struct Mol Biol 20: 1122–1130. [DOI] [PubMed] [Google Scholar]
- Lee SG, Kang DC, DeSalle R, Sarkar D, Fisher PB. 2013. AEG-1/MTDH/LYRIC, the beginning: initial cloning, structure, expression profile, and regulation of expression. Adv Cancer Res 120: 1–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner RS, Seiser RM, Zheng T, Lager PJ, Reedy MC, Keene JD, Nicchitta CV. 2003. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA 9: 1123–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulliot N, Varani G. 2001. Current topics in RNA–protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry 40: 7947–7956. [DOI] [PubMed] [Google Scholar]
- Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, Mo B, et al. 2013. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153: 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G. 2007. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 8: 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandon EC, Trueman SF, Gilmore R. 2013. Protein translocation across the rough endoplasmic reticulum. Cold Spring Harb Perspect Biol 5: a013342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus MA, Morris BJ. 2009. RBM4: a multifunctional RNA-binding protein. Int J Biochem Cell Biol 41: 740–743. [DOI] [PubMed] [Google Scholar]
- Meng X, Thiel KW, Leslie KK. 2013. Drug resistance mediated by AEG-1/MTDH/LYRIC. Adv Cancer Res 120: 135–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW, Maquat LE. 2013. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet 47: 139–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyhtila B, Zheng T, Lager PJ, Keene JD, Reedy MC, Nicchitta CV. 2008. Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA 14: 445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DW, Nicchitta CV. 2012. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem 287: 5518–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DW, Nicchitta CV. 2015. Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat Rev Mol Cell Biol 16: 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson CL, Srivastava J, Siddiq A, Gredler R, Emdad L, Rajasekaran D, Akiel M, Shen XN, Corwin F, Sundaresan G, et al. 2015. Astrocyte elevated gene-1 (AEG-1) regulates lipid homeostasis. J Biol Chem 290: 18227–18236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki K, Yoshina S, Shen X, Han J, DeSantis MR, Xiong M, Mitani S, Kaufman RJ. 2012. RNA surveillance is required for endoplasmic reticulum homeostasis. Proc Natl Acad Sci 109: 8079–8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoz PA, van der Goot FG. 2015. How many lives does CLIMP-63 have? Biochem Soc Trans 43: 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D, Park ES, Emdad L, Lee SG, Su ZZ, Fisher PB. 2008. Molecular basis of nuclear factor-κB activation by astrocyte elevated gene-1. Cancer Res 68: 1478–1484. [DOI] [PubMed] [Google Scholar]
- Sawicka K, Bushell M, Spriggs KA, Willis AE. 2008. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem Soc Trans 36: 641–647. [DOI] [PubMed] [Google Scholar]
- Shao S, Hegde RS. 2011. Membrane protein insertion at the endoplasmic reticulum. Annu Rev Cell Dev Biol 27: 25–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields D. 1979. Association of messenger ribonucleic acid with mammalian microsomal membranes: characterization by analysis of cell-free translation products. Biochemistry 18: 2622–2627. [DOI] [PubMed] [Google Scholar]
- Sievers C, Schlumpf T, Sawarkar R, Comoglio F, Paro R. 2012. Mixture models and wavelet transforms reveal high confidence RNA-protein interaction sites in MOV10 PAR-CLIP data. Nucleic Acids Res 40: e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skach WR. 2009. Cellular mechanisms of membrane protein folding. Nat Struct Mol Biol 16: 606–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava J, Robertson CL, Rajasekaran D, Gredler R, Siddiq A, Emdad L, Mukhopadhyay ND, Ghosh S, Hylemon PB, Gil G, et al. 2014. AEG-1 regulates retinoid X receptor and inhibits retinoid signaling. Cancer Res 74: 4364–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder L, Heusermann W, Sokol L, Trojer D, Wirz J, Hean J, Fritzsche A, Aeschimann F, Pfanzagl V, Basselet P, et al. 2013. The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. EMBO J 32: 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens SB, Nicchitta CV. 2007. In vitro and tissue culture methods for analysis of translation initiation on the endoplasmic reticulum. Method Enzymol 431: 47–60. [DOI] [PubMed] [Google Scholar]
- Stephens SB, Nicchitta CV. 2008. Divergent regulation of protein synthesis in the cytosol and endoplasmic reticulum compartments of mammalian cells. Mol Biol Cell 19: 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland HG, Lam YW, Briers S, Lamond AI, Bickmore WA. 2004. 3D3/lyric: a novel transmembrane protein of the endoplasmic reticulum and nuclear envelope, which is also present in the nucleolus. Exp Cell Res 294: 94–105. [DOI] [PubMed] [Google Scholar]
- Szostak E, Gebauer F. 2013. Translational control by 3′-UTR-binding proteins. Brief Funct Genomics 12: 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirkettle HJ, Mills IG, Whitaker HC, Neal DE. 2009. Nuclear LYRIC/AEG-1 interacts with PLZF and relieves PLZF-mediated repression. Oncogene 28: 3663–3670. [DOI] [PubMed] [Google Scholar]
- Turner M, Hodson DJ. 2012. An emerging role of RNA-binding proteins as multifunctional regulators of lymphocyte development and function. Adv Immunol 115: 161–185. [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. 2003. CLIP identifies Nova-regulated RNA networks in the brain. Science 302: 1212–1215. [DOI] [PubMed] [Google Scholar]
- Van Roey K, Davey NE. 2015. Motif co-regulation and co-operativity are common mechanisms in transcriptional, post-transcriptional and post-translational regulation. Cell Commun Signal 13: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi M, Zsolyomi F, Guharoy M, Tompa P. 2015. Functional advantages of conserved intrinsic disorder in RNA-binding proteins. PloS One 10: e0139731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Huang C, Yang MQ, Yang JY. 2010. BindN+ for accurate prediction of DNA and RNA-binding residues from protein sequence features. BMC Syst Biol 4(Suppl 1): S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT. 2004a. The DISOPRED server for the prediction of protein disorder. Bioinformatics 20: 2138–2139. [DOI] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. 2004b. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 337: 635–645. [DOI] [PubMed] [Google Scholar]
- Wilkie GS, Dickson KS, Gray NK. 2003. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci 28: 182–188. [DOI] [PubMed] [Google Scholar]
- Williamson JR. 2000. Induced fit in RNA-protein recognition. Nat Struct Biol 7: 834–837. [DOI] [PubMed] [Google Scholar]
- Yoo BK, Emdad L, Su Z, Villanueva A, Chiang DY, Mukhopadhyay ND, Mills AS, Waxman S, Fisher RA, Llovet JM, et al. 2009. Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. J Clin Invest 119: 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BK, Emdad L, Lee SG, Su ZZ, Santhekadur P, Chen D, Gredler R, Fisher PB, Sarkar D. 2011a. Astrocyte elevated gene-1 (AEG-1): a multifunctional regulator of normal and abnormal physiology. Pharmacol Ther 130: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BK, Santhekadur PK, Gredler R, Chen D, Emdad L, Bhutia S, Pannell L, Fisher PB, Sarkar D. 2011b. Increased RNA-induced silencing complex (RISC) activity contributes to hepatocellular carcinoma. Hepatology 53: 1538–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Darnell RB. 2011. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol 29: 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.