In this study, Knuckles et al. identified Flacc/Zc3h13 as a novel interactor of m6A methyltransferase complex components in Drosophila and mice. They show that Flacc promotes the recruitment of the methyltransferase to mRNA by bridging Fl(2)d to the mRNA-binding factor Nito, providing novel insights into the conservation and regulation of the m6A machinery.
Keywords: Zc3h13, Flacc, m6A, methyltransferase complex, RNA modifications, sex determination
Abstract
N6-methyladenosine (m6A) is the most abundant mRNA modification in eukaryotes, playing crucial roles in multiple biological processes. m6A is catalyzed by the activity of methyltransferase-like 3 (Mettl3), which depends on additional proteins whose precise functions remain poorly understood. Here we identified Zc3h13 (zinc finger CCCH domain-containing protein 13)/Flacc [Fl(2)d-associated complex component] as a novel interactor of m6A methyltransferase complex components in Drosophila and mice. Like other components of this complex, Flacc controls m6A levels and is involved in sex determination in Drosophila. We demonstrate that Flacc promotes m6A deposition by bridging Fl(2)d to the mRNA-binding factor Nito. Altogether, our work advances the molecular understanding of conservation and regulation of the m6A machinery.
In the past years, N6-methyladenosine (m6A) RNA has emerged as an abundant and dynamically regulated modification throughout the transcriptome (Dominissini et al. 2012; Meyer et al. 2012). m6A affects almost every stage of mRNA metabolism, and its absence is associated with various defects in meiosis, embryonic stem cell (ESC) differentiation, DNA repair, circadian rhythm, neurogenesis, dosage compensation, and sex determination (for a recent review, see Roignant and Soller 2017; Zhang et al. 2017). Alteration of m6A levels also promotes glioblastoma progression and is linked to poor prognosis in myeloid leukemia (Barbieri et al. 2017; Cui et al. 2017; Jaffrey and Kharas 2017; Kwok et al. 2017; Li et al. 2017; Vu et al. 2017; Weng et al. 2018).
Formation of m6A is catalyzed by the activity of methyltransferase-like 3 (METTL3; also called MT-A70) (Bokar et al. 1997), which physically interacts with METTL14 (Liu et al. 2014; Ping et al. 2014; Schwartz et al. 2014; Wang et al. 2014), Wilms tumor 1-associated protein (WTAP) (Zhong et al. 2008; Agarwala et al. 2012), Vir-like m6A methyltransferase-associated (KIAA1429/VIRMA) (Schwartz et al. 2014), and RNA-binding motif 15 (RBM15) and its paralog, RBM15B (Patil et al. 2016). Drosophila has corresponding homologs Mettl3, Mettl14, Fl(2)d, Virilizer (Vir), and Spenito (Nito) (Lence et al. 2017). Recent crystal structural studies investigated the molecular activities of the two predicted methyltransferases METTL3 and METTL14 (Sledz and Jinek 2016; Wang et al. 2016a,b). Only METTL3 was shown to contain the catalytic activity and form a stable heterodimer with METTL14, which was required to enhance METTL3 enzymatic activity by binding substrate RNA and positioning the methyl group for transfer to adenosine. In addition, WTAP [Fl(2)d] ensures the stability and localization of the heterodimer to nuclear speckles (Ping et al. 2014; Lence et al. 2016). VIRMA (Vir) is essential for m6A deposition, but its molecular function is currently unknown. Last, RBM15 and RBM15B (Nito) have been suggested to recruit the methyltransferase complex to its target transcripts via direct binding to U-rich sequences on mRNA. In humans, this function is important to control m6A promoted X-chromosome inactivation via XIST-mediated transcriptional repression (Patil et al. 2016). In Drosophila, Nito promotes m6A function in the sex determination and dosage compensation pathways (Lence et al. 2016).
To date, it is unknown how Nito in Drosophila interacts with other members of the methyltransferase writer complex to ensure their recruitment to mRNA targets. Although, in human cells, RBM15/15B were reported to interact with METTL3 in a WTAP-dependent manner (Patil et al. 2016), it is unclear whether this interaction is conserved in other organisms. In order to address these questions, we performed interactome analyses from Mus musculus and Drosophila melanogaster cell extracts using Rbm15 and Nito as bait, respectively. We identified mouse zinc finger CCCH domain-containing protein 13 (Zc3h13) and its fly homolog, CG7358, which we named Fl(2)d-associated complex component (Flacc), as novel interactors of the m6A writer machinery. A lack of these proteins dramatically reduces m6A levels in both organisms. Consistent with the role of m6A in sex determination in Drosophila, Flacc depletion results in aberrant splicing of Sex lethal (Sxl) and leads to transformations of female into male-like structures. Moreover, we demonstrate that Flacc interacts with Nito and Fl(2)d and serves as an adaptor between these two proteins, thereby stabilizing the complex and promoting m6A deposition on mRNA.
Results
Zc3h13 interacts with the m6A machinery
In our recent work, we identified Nito as a novel interactor of the m6A methyltransferase complex (Lence et al. 2016). Because the role of the mouse Nito homolog protein Rbm15 appears to be evolutionarily conserved in regard to m6A deposition (Patil et al. 2016), we sought to identify interaction partners to obtain further insights into Rbm15's function. To this end, we tagged endogenous Rbm15 with the Flag-Avi tag using CRISPR–Cas9 genome editing in mouse ESCs (mESCs) that express the bacterial BirA ligase (Supplemental Fig. S1A, B; Flemr and Buhler 2015). Subsequently, we performed tandem affinity purification (TAP) coupled to liquid chromatography and mass spectrometry (TAP-LC-MS). We found that Rbm15 copurifies with Wtap, Virma, and Hakai (Fig. 1A) under stringent conditions (350 mM NaCl), demonstrating that these proteins stably interact with each other. Hakai was found recently to interact with other subunits of the m6A methyltransferase complex in plants (Ruzicka et al. 2017). Interestingly, we also observed Zc3h13 among the top hits. Although it was reported to interact with WTAP in human cells, it has not been linked previously to adenosine methylation (Horiuchi et al. 2013; Wan et al. 2015).
Figure 1.
Zc3h13/Flacc interacts with the m6A machinery. (A) TAP-LC-MS/MS of endogenously Flag-Avi-tagged Rbm15 mESCs. Parental cells were used as background control, and proteins were purified in the presence of 350 mM NaCl. Highlighted in the volcano plot are enriched proteins previously identified as Mettl3 interactors (red) as well as Zc3h13 (green). (B) Heat map comparing relative label-free quantification (LFQ) intensities of selected Mettl3-bound proteins across increasing NaCl concentrations. Statistical analysis was done with Perseus (see the Materials and Methods for details). MS raw data were deposited in ProteomeXchange. (C,D) Stable isotope labeling of amino acids in cell culture (SILAC) coupled to MS analysis using Nito-Myc as bait. Scatter plot of normalized forward versus inverted reverse experiments plotted on a log2 scale. The threshold was set to a 1.5-fold enrichment (red dashed line). Proteins in the top right quadrant of C are enriched in both replicates. Gene ontology (GO) term analysis (Tyanova et al. 2016) for enriched proteins is shown in D. (E) Coimmunoprecipitation experiments were carried out with lysates prepared from S2R+ cells transfected with FlagMyc-Flacc and HA-Nito. In control lanes, S2R+ cells were transfected with FlagMyc alone and an identical HA-containing protein. Extracts were immunoprecipitated with Myc antibody and immunoblotted using Flag and HA antibodies. Two percent of input was loaded. The same experiment was repeated in the presence of RNaseT1. Nito and Flacc interact with each other in an RNA-independent manner. (F) Table representing orthologous proteins of the m6A–METTL complex (MAC) and the m6A-METTL-associated complex (MACOM) in mice and flies.
Previous work suggested that the interaction of the heterodimer Mettl3/14 with Wtap, Virma, and Rbm15 is important to guide the methylation complex to its targets and correctly methylate mRNA (Ping et al. 2014; Schwartz et al. 2014; Patil et al. 2016). To test whether the Zc3h13-containing protein complex described above interacts with Mettl3/14 in mice, we also endogenously tagged Mettl3 with the Flag-Avi tag in mESCs (Supplemental Fig. S1A, B) and performed TAP-LC-MS. Consistent with previous reports, we found that Mettl3 copurifies with Mettl14, Wtap, Virma, Rbm15, and Hakai (Fig. 1B; Supplemental Fig. S2A). Importantly, we also recovered peptides from Zc3h13 (Fig. 1B; Supplemental Fig. S2A). Whereas these interactions resisted 350 mM NaCl, only the Mettl3/Mettl14 interaction remained at 500 mM NaCl (Fig. 1B). These results indicate the existence of two stable protein complexes (Mettl3/Mettl14 and Rbm15/Zc3h13/Wtap/Virma/Hakai), which we refer to as the m6A–METTL complex (MAC) and the m6A-METTL-associated complex (MACOM), respectively.
To gain further insight into the relative amounts of MAC to MACOM, we performed intensity-based absolute quantification (iBAQ) analysis on TAP-LC-MS data from endogenously tagged Mettl3. We observed comparable stoichiometry between the bait (Mettl3) and Mettl14. In contrast, Wtap and other MACOM components were <1% abundant compared with Mettl3 and Mett14 (Supplemental Fig. S2B), an observation that we interpret as a sign of a weak and/or short-lived interaction. Alternatively, the abundance of MAC bound to MACOM could be very scarce relative to the level of each independent complex. Regardless of the precise mechanism, because components of both complexes are required to install m6A, we propose that MAC and MACOM interact with each other in order to deposit the methylation.
The Drosophila Zc3h13 homolog Flacc interacts with components of the m6A methyltransferase complex
To address whether Nito interacts with the same set of proteins that we identified in mice, we took an approach very similar to that described above using extracts from Drosophila S2R+ cells. We used stable isotope labeling of amino acids in cell culture (SILAC)-based quantitative proteomics. A Myc-tagged version of Nito was used to perform coimmunoprecipitation experiments from S2R+ cells. In total, we identified 40 factors that showed >1.5-fold enrichment in the Nito-Myc precipitate in comparison with control cells transfected with Myc alone (Fig. 1C; Supplemental Table 1). In agreement with mouse Rbm15 proteomic analysis, the homolog of Wtap in Drosophila Fl(2)d, was among the top candidates. We also found the previously reported m6A writers Vir and Hakai (refer to Fig. 1F for M. musculus and D. melanogaster nomenclature of m6A factors). We observed an overall enrichment for mRNA-binding proteins (Fig. 1D) and, importantly, Flacc, which is the closest homolog of Zc3h13. To confirm the interaction of this protein with Nito, we generated tagged proteins and performed coimmunoprecipitation assays. These experiments confirmed that Flacc interacts with Nito and that this occurs in an RNA-independent manner (Fig. 1E). To verify that Flacc interacts with other components of the m6A methyltransferase complex, we immunoprecipitated Flacc-Myc and tested for the presence of Vir and Fl(2)d. As shown in Supplemental Figure S3, A and B, Flacc interacts with both proteins independently of RNA, indicating that it might be a novel regulator of the m6A pathway. In contrast to mouse Zc3h13 (see below), however, Flacc does not contain a zinc finger motif (Supplemental Fig. S4).
Zc3h13/Flacc regulates the m6A pathway
To test whether Zc3h13 is necessary for adenosine methylation in mice, we measured global m6A levels by LC-MS/MS in Zc3h13 knockout mESCs (Supplemental Fig. S1C). We found an 80% reduction of m6A, similar to isogenic Mettl3 knockout mESCs (Fig. 2A). Consistent with a global reduction in m6A levels, Zc3h13 knockout cells displayed a drastic change in morphology, reminiscent of Mettl3 knockout, with loss of dome-shaped colony formation and an overall increase in cell size (data not shown). In addition, we performed m6A RNA immunoprecipitation coupled to deep sequencing (m6A-RIP-seq) on oligo-dT-selected mRNA from wild-type, Mettl3 knockout, and Zc3h13 knockout mESCs. As observed with Mettl3 knockout cells, ablation of Zc3h13 resulted in a drastic reduction of m6A enrichment, particularly at the 3′ end of target mRNAs (Fig. 2B,C). Hence, we conclude that Zc3h13 is essential for m6A installation in mESCs. To investigate evolutionary conservation of this activity in Drosophila, we quantified m6A levels using LC-MS/MS analysis upon Flacc depletion in Drosophila S2R+ cells. Similar to the reduction observed upon knockdown of other m6A components, depletion of Flacc also resulted in strongly reduced m6A levels (Fig. 2D; Supplemental Fig. S5A). This was not due to an indirect effect on expression of other components of the methyltransferase complex (Supplemental Fig. S5B–D). In agreement with decreased m6A levels, we found that binding of the reader protein Ythdc1 to its target transcripts was reduced in the absence of Flacc (Fig. 2E; Supplemental Fig. S3C,D). Together, these results demonstrate that Zc3h13 and its Drosophila ortholog, Flacc, are novel and essential components of the m6A pathways in mice and flies.
Figure 2.
Flacc/Zc3h13 regulates the m6A pathway. (A) LC-MS/MS quantification of m6A levels in mRNA extracts from wild-type mESCs, Mettl3 knockout and Mettl3 knockout plasmid rescue, and Zc3h13 knockout cells. The mean of two biological replicates and three independent measurements is shown. Errors bars indicate standard deviation (SD). (*) P < 0.01; (n.s.) not significant, Student's t-test. (B) University of California at Santa Cruz genome browser shots of m6A immunoprecipitation profiles of RNA isolated from Mettl3 knockout, Zc3h13 knockout, and wild-type cells and input samples for each genetic background at the Wtap-encoding locus. Scale is mapped reads in 100-base-pair (bp) bins normalized to mean library size. (C) Metaplot depicting reads from m6A immunoprecipitations at target genes (defined as genes overlapping or within 500 bp of MACS-identified peaks of m6A immunoprecipitation/input in wild-type cells) aligned to the coding sequence (“start” and “stop” refer to translation start and stop, respectively). (D) LC-MS/MS quantification of m6A levels in either control samples or mRNA extracts depleted for the indicated proteins in S2R+ cells. The bar chart shows the mean of three biological replicates and three independent measurements. Errors bars indicate SD. (*) P < 0.01, Student's t-test. Knockdown of the indicated proteins significantly reduces m6A levels. (E) Fold enrichment of m6A-regulated transcripts (Aldh-III and Dsp1) over input in Myc-Ythdc1 RIP after control or Flacc depletion. The bar chart shows the mean of three biological replicates. Errors bars indicate SD. (*) P < 0.01; (**) P < 0.001, Student's t-test. Loss of Flacc affects Ythdc1 binding. (F) Relative isoform quantification of m6A-regulated genes (Aldh-III, Hairless, and Dsp1) upon depletion of the indicated components. Flacc is required for m6A-dependent splicing events.
Flacc is required for pre-mRNA splicing
To further corroborate Flacc as a bona fide m6A writer, we tested whether it was required to control m6A splicing-related events. As reported previously, splicing of several transcripts, including AldhIII, Dsp1, and Hairless, is dependent on the m6A pathway (Lence et al. 2016). Remarkably, depletion of Flacc affected all of those transcripts (Fig. 2F; Supplemental Fig. S5E). We next expanded this analysis to a transcriptome-wide level, which revealed that depletion of Flacc in S2R+ cells leads to changes in gene expression and splicing that substantially overlap with changes observed upon knockdown of other m6A writers (Fig. 3A; Supplemental Fig. S6A; Supplemental Tables 3–8). In particular, the Flacc-depleted transcriptome clusters very closely with Fl(2)d- and Vir-depleted transcriptomes (Fig. 3B). Notably, Nito depletion induced greater changes and poorly clustered with the others, suggesting that Nito might be pleiotropic. Regardless of Nito's potential role in other pathways, common misregulated genes among components of the MACOM are larger than the average gene size (Fig. 3C) and are significantly methylated (61.5%; P = 6.94 × 10−31) (Fig. 3D). Importantly, differentially expressed genes generally change in the same direction upon the different knockdowns, confirming that MACOM components belong to the same complex and share similar functions (Fig. 3E). We noticed that common up-regulated genes tend to be larger (P = 2.9 × 10−40) and more methylated compared with down-regulated ones (78.2% [P = 6.12 × 10−31] vs. 44.5% [P = 0.086]) (Fig. 3C,D). Up-regulated genes were enriched for processes involved in embryonic development as well as epithelial cell differentiation and migration (Fig. 3F). Thus, it is possible that down-regulated genes, which are mostly enriched for metabolic processes, are affected indirectly (Fig. 3F).
Figure 3.
Flacc regulates common transcripts with other components of the m6A complex. (A) Number of differentially expressed genes (5% false discovery rate [FDR]) upon knockdown of the indicated proteins (left) and common differentially expressed targets regulated by components of MACOM (right). (B) Scatter plot of the first two principal components of a principal component analysis (PCA) of the 500 most variable genes in all conditions. The biological replicates are indicated in the same color, with elliptical areas representing the SD of the two depicted components. (C) Gene length distribution for genes tested in the differential expression analysis and the differential expressed genes up-regulated or down-regulated in all conditions. The distributions were tested for difference using the Kolmogorov-Smirnov test. (D) Overlap between common up, down, or all differentially expressed genes and genes annotated to have m6A-modified transcripts (according to methylation individual nucleotide resolution cross-linking immunoprecipitation data from Kan et al. 2017). The significance of the overlap was tested using a hypergeometric test. (E) Fold change (log2) expression of commonly misregulated genes. The heat map is clustered according to rows and columns. The color gradient was adjusted to display the 1% lowest/highest values within the most extreme color (lowest values as the darkest blue and highest values as the darkest red). (F) The GO term analysis of common up-regulated and down-regulated genes performed using the package ClusterProfiler. The top 10 GO terms are displayed.
We next performed similar analysis with respect to splicing changes. We found that knockdown of each of the known m6A writer components, including Flacc, resulted in an increase of both alternative 5′ splice site usage and intron retention (Supplemental Fig. S6A,B). Moreover, most common misspliced transcripts upon knockdown of MACOM components are methylated (82.2%; P = 1.3 × 10−8), show similar splicing defects, and are enriched for neuronal processes, which is consistent with our previous findings (Supplemental Fig. S6C–E; Lence et al. 2016). Of note, knockdown of Mettl3/Mettl14 generally produces less effect compared with knockdown of MACOM components. This may be explained by residual m6A activity upon knockdown of the methyltransferases. Alternatively, MACOM components may have additional functions beyond m6A activity (see also the Discussion).
Flacc subcellular localization and expression through development
To further investigate the role of Flacc in Drosophila, we examined its subcellular localization as well as its developmental expression profile. We observed that Flacc is strictly localized in the nucleus in S2R+ cells (Supplemental Fig. S7A) and that its transcript is broadly expressed during embryogenesis but shows enrichment in the neuroectoderm (Supplemental Fig. S7B). Overall, flacc mRNA follows the same distribution as transcripts of other subunits of the methyltransferase complex (Lence et al. 2016) and as m6A levels in mRNA. An exception is the stage of maternal-to-zygotic transition (2 h after fertilization), where a boost of flacc expression is observed while m6A is rapidly decreasing (Supplemental Fig. S7C), suggesting that Flacc might have an additional function in early embryogenesis.
Flacc is required for sex determination and dosage compensation via Sxl alternative splicing
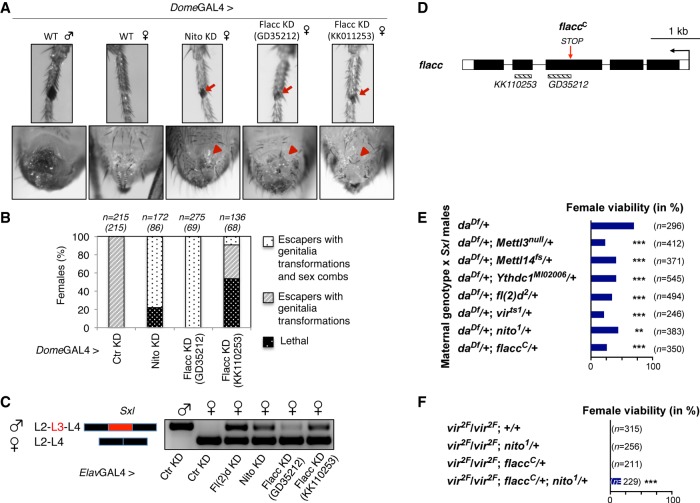
Components of the m6A machinery were shown previously to affect sex determination and dosage compensation in Drosophila via the control of Sxl alternative splicing (Haussmann et al. 2016; Lence et al. 2016; Kan et al. 2017). To address whether Flacc bears similar functions, we depleted its products by expressing corresponding dsRNA in both the legs and genital discs using the dome-GAL4 driver. Strikingly, these females displayed clear transformations into male structures, as shown previously for Nito (Fig. 4A; Yan and Perrimon 2015). This is illustrated by the appearance of sex combs in the forelegs of females that were depleted for Flacc. The phenotype was observed in ∼20% of females examined (Fig. 4B). Using a dsRNA that targets a distinct region of flacc (GD35212), the penetrance was even increased to all female escapers (Fig. 4B; Supplemental Fig. S7D). Furthermore, typical female external structures, such as vaginal bristles, were absent on the same individuals (Fig. 4A). Altogether, these data indicate that Flacc plays a major role in the control of sex determination in flies.
Figure 4.
Flacc is required for sex determination via control of Sxl alternative splicing. (A,B) dome-GAL4-driven expression of shRNA or dsRNA in genital discs and first pair of leg discs against Nito or Flacc, respectively. (Top) Forelegs of a wild-type male fly and female flies depleted for Nito or Flacc show the appearance of male-specific sex comb bristles (red arrow). (Bottom) Depletion of Nito or Flacc results in transformations of female genitalia and loss of vaginal bristles (red arrowhead). (B) Quantification of female survival and transformations in escapers upon depletion of Nito or Flacc using the dome-GAL4 driver. (n) The number of analyzed flies with the expected number of escapers in brackets. Depletion of Nito and Flacc results in a high level of transformation in female genitalia and the appearance of male-specific sex combs on forelegs. (C) Semiquantitative RT–PCR analysis of Sxl isoforms in male and female heads from flies depleted for Fl(2)d, Nito, or Flacc, respectively, using the ElavGAL4 driver. Inclusion of male-specific exon L3 is observed in flies lacking m6A components. (D) The flacc locus (flaccC) with a premature stop codon at amino acid Leu730. Sites of dsRNA fly lines KK110253 and GD35212 are shown below gene loci. (E) Viability of female flies from a cross of the indicated genotypes mated with Sxl7BO males. The loss of one copy of flacc significantly reduces female survival in a genetic background where one copy of Sxl and da are absent. The same compromised survival is observed for other m6A components [Mettl3, Mettl14, Ythdc1, fl(2)d, vir, and nito]. Viability was calculated from the numbers of females compared with males, and statistical significance was determined by a χ2 test (Graphpad Prism). (F) The viability of female flies with homozygous vir2F mutation can be rescued by the loss of a single copy of flacc and nito. Viability was calculated from the numbers of homozygous vir2F females compared with heterozygous balancer-carrying siblings, and statistical significance was determined by a χ2 test (Graphpad Prism).
To address how Flacc affects sex determination, we tested whether alternative splicing of Sxl, the master regulator of sex determination and dosage compensation, was affected. RNA extracts from fly heads depleted by RNAi for Fl(2)d, Nito, or Flacc were subjected to reverse transcription followed by PCR using primers spanning the common exons 2 and 4. While the male-specific exon 3 is absent in control female heads, it was clearly included upon the loss of components of the m6A machinery, including Flacc (Fig. 4C). This experiment indicates that Flacc regulates sex determination and dosage compensation via Sxl alternative splicing, as shown previously for other m6A writers.
To confirm the effect of Flacc on sex determination via Sxl alternative splicing observed when using RNAi, we analyzed a lethal flacc mutant allele harboring a stop codon at amino acid 730 (Fig. 4D). Reducing one copy of m6A components [Mettl3, Mettl14, fl(2)d, vir, nito, or Ythdc1] in a sensitized background (heterozygous for Sxl and daughterless) significantly alters female viability (Fig. 4E). We showed previously for the Mettl3 allele that this is due to activation of dosage compensation in females (Haussmann et al. 2016). Consistent with its role in N6-adenosine methylation, we found that removing one copy of the flacc allele results in female lethality (Fig. 4E). Likewise, the female-lethal single amino acid substitution allele vir2F interferes with Sxl recruitment, resulting in impaired Sxl autoregulation and inclusion of the male-specific exon (Hilfiker et al. 1995). We observed that female lethality of these alleles was rescued by flacc and nito double heterozygosity, further confirming the involvement of Flacc in Sxl alternative splicing (Fig. 4F).
Zc3h13/Flacc stabilizes Wtap/Fl(2)d–Rbm15/Nito interaction
To obtain insights into the molecular function of Flacc, we investigated interactions between m6A writers in the absence of Flacc. We found previously that knockdown of Fl(2)d diminishes the interaction between Mettl3 and Mettl14 (Lence et al. 2016). Interestingly, we found that this interaction is not affected upon Flacc knockdown (Supplemental Fig. S8A,B). However, we observed that depleting Flacc almost completely abolished the association between Nito and Fl(2)d (Fig. 5A), whereas interactions between Vir and different isoforms of Fl(2)d as well as Vir and Nito were not affected (Supplemental Fig. S8C–F). Interactions between Nito and Mettl3/Mettl14 were also not compromised upon depletion of Flacc (Supplemental Fig. S8G,H). This indicates that Flacc stabilizes the complex and might serve as an adapter that connects the RNA-binding protein Nito to Fl(2)d. If this prediction was true, depletion of Flacc should prevent binding of Fl(2)d, but not Nito, to its mRNA targets. To test this hypothesis, we performed RNA immunoprecipitation (RIP) experiments to monitor the binding of these components to well-characterized m6A targets in the presence or absence of Flacc. As shown in Figure 5B, binding of Fl(2)d to AldhIII, Hairless, and Dsp1 mRNA was strongly decreased upon Flacc knockdown, whereas Nito binding was only slightly affected. Thus, we conclude that Flacc serves as an adapter between Fl(2)d and the mRNA-recruiting factor RBM15/Nito.
Figure 5.
Flacc bridges the methyltransferase complex to mRNA targets via binding to Nito. (A) Coimmunoprecipitation experiments were carried out with lysates prepared from S2R+ cells transfected with GFPMyc-Nito and Fl(2)d-HA. In control lanes, S2R+ cells were transfected with Myc alone and an identical HA-containing protein. Extracts were immunoprecipitated with Myc antibody and immunoblotted using Myc and HA antibodies. Two percent of input was loaded. The same experiment was repeated in Flacc knockdown conditions. Interaction between Nito and Fl(2)d is strongly reduced upon depletion of Flacc. (B) Fold enrichment of m6A-regulated transcripts (AldhIII, Hairless, and Dsp1) over input in Myc-Fl(2)d and Myc-Nito RIP upon depletion of Flacc or in control conditions. The bar chart shows the mean of three biological replicates. Errors bars indicate SD. (*) P < 0.01; (**) P < 0.001; (***) P < 0.0001; (n.s.) not significant, Student's t-test. Loss of Flacc strongly affects Fl(2)d binding and, to a milder extent, binding of Nito to m6A-regulated transcripts. (C,D) Coimmunoprecipitation experiments were carried out with lysates prepared from S2R+ cells transfected with either FlagMyc-Nito or Fl(2)d-HA. In control lanes, S2R+ cells were transfected with FlagMyc alone and an identical HA-containing protein. Extracts were immunoprecipitated with Flag antibody and immunoblotted using Myc and HA antibodies. Two percent of input was loaded. The same experiment was performed upon depletion of Flacc. Human ZC3H13 was transfected in an identical set of experiments. The interaction between Nito and Fl(2)d is strongly reduced upon loss of Flacc (lane 6) but can be rescued upon expression of human ZC3H13 protein (lane 8). Quantification of two replicates is shown in D.
To test functional conservation of Flacc, we cloned a human isoform of ZC3H13 and probed for the interaction between Nito and Fl(2)d upon depletion of endogenous Flacc protein in Drosophila S2R+ cells. Remarkably, expression of ZC3H13 was sufficient to re-establish the interaction between Nito and Fl(2)d (Fig. 5C,D; Supplemental Fig. S8I–K) even though the two orthologs bear low sequence similarity at the amino acid level (21% identity). These results hint at a conserved role of this newly characterized protein in stabilizing interactions within the MACOM. To address this more directly, we generated Zc3h13 knockout mESCs that express Flag-Avi-tagged Rbm15 and performed TAP-LC-MS/MS experiments. Starting with both whole-cell extracts and nuclear fractions, Rbm15 interaction with Wtap was markedly reduced (Fig. 6A,B; Supplemental Fig. S9A,B), which is consistent with observations in the fly knockdown experiments [Nito and Fl(2)d, respectively]. Furthermore, the reduced interaction was not attributable to a global decrease of Wtap or other components of MACOM (Supplemental Fig. S9C). As an alternative approach to test MACOM integrity, we used a protein fragment complementation assay (Dixon et al. 2016), generating fusion constructs of Rbm15 and Wtap to NanoBiT subunits. The optimal combination of fusions reconstituted the luciferase signal when transfected into wild-type cells (Wtap N-terminally tagged with the small nanoluciferase subunit and Rbm15 C-terminally tagged with the large nanoluciferase subunit) (Fig. 6C; Supplemental Fig. S9D–F). The relative luciferase signal intensity was strikingly reduced when fusion constructs were transfected in Zc3h13 knockout but not in Mettl3 knockout, discarding a secondary effect of global m6A loss (Fig. 6C). Taken together, these findings suggest that Zc3h13 acts as an adapter that connects the RNA-binding protein Rbm15 to Wtap also in mammals.
Figure 6.
Zc3h13 stabilizes the interaction between RBM15 and WTAP. (A,B) Comparison of TAP-LC-MS/MS of endogenously Flag-Avi-tagged Rbm15 mESCs in either a wild-type or a Zc3h13 knockout background. Rbm15 and associated proteins were purified in the presence of 350 mM NaCl. (A) Volcano plot showing enriched proteins in wild-type cells (right) versus Zc3h13 knockout cells (left). (B) Table of spectral counts, unique peptides, and percentage coverage of TAP-LC-MS/MS data in A. (C) Split luciferase NanoBiT assay examining the interaction of mouse Rbm15 and Wtap. (Left) Scheme representing luciferase reconstitution upon transfection of large (LgBit) and small (SmBit) NanoLuc subunit fusions and the interaction of Rbm15 and WTAP. (Right) Comparison of Rbm15–Wtap NanoBiT NanoLuc signal in wild-type and Zc3h13 and Mettl3 knockout cells. The mean of three independent experiments, three transfections each, is shown. Errors bars indicate SD. (*) P = 0.026, calculated using two-tailed Student's t-test.
Discussion
Our study identified a novel interactor of the m6A methyltransferase complex, which is conserved in Drosophila and mice. Its function in the m6A pathway is essential in both species, as its absence results in dramatic reduction of m6A levels. The facts that the human homolog was found recently in interactome studies with WTAP (Horiuchi et al. 2013; Wan et al. 2015) and that it can rescue the interaction between Fl(2)d and Nito in Drosophila suggest that it has a similar role in human cells. Despite this functional conservation, the protein sequence identity among different homologs is rather weak (Supplemental Fig. S4). Mouse Zc3h13 contains several additional domains as compared with Flacc. In particular, it differs by the presence of a zinc finger domain, which is present in a common ancestor but was lost in dipterian (Supplemental Fig. S4). Other species such as Ciona intestinalis also lack the zinc finger motif. In addition, the zinc finger motif can be found in two variants across evolution: one short and one long. As zinc finger motifs are commonly involved in nucleic acid binding or protein–protein interactions, it will be interesting to address the functional importance of this domain when present in the protein. Of note, Zc3h13 appears completely absent in nematodes, as is also the case for Mettl3 (Dezi et al. 2016), possibly indicating that these two proteins have coevolved for the regulation of adenosine methylation.
Our work strongly supports the existence of at least two distinct stable complexes that interact weakly to regulate m6A biogenesis. This result is consistent with earlier studies by Rottman and colleagues (Bokar et al. 1997), who isolated two protein components using an in vitro methylation assay and HeLa cell nuclear extracts, which are readily dissociable under nondenaturing conditions. Gel filtration and gradient glycerol sedimentation estimated molecular weights of 200 and 875 kDa (Bokar et al. 1997). While biochemical characterization will be required to address the exact identity of the different complex components, recent biochemical analysis suggests that the 200-kDa complex consists of Mettl3 and Mettl14 (Liu et al. 2014). Although the exact composition of the larger complex is currently unknown, we postulate that it is probably MACOM, consisting of Wtap, Virma, Hakai, Rbm15, and Zc3h13. The calculated total molecular weight of these proteins (600 kDa) is lower than that of the large complex (875 kDa), which suggests the presence of other factors or the inclusion of some subunits in multiple copies. For instance, recombinant WTAP can form aggregates, suggesting the possibility of higher complex organization (Liu et al. 2014). Finally, the existence of two complexes is also supported by our genetic analyses, which show that the knockout of Mettl3 and Mettl14 results in viable animals, while loss of function of fl(2)d, vir, nito, and flacc is lethal during development. This indicates that the MACOM acts beyond m6A methylation via Mettl3 (Fig. 7).
Figure 7.

Schematic representation of the role of the MACOM and the MAC. The MACOM can regulate gene expression in two ways: either on its own (MAC-independent functions) or by interacting with MAC components (m6A methylation). Flacc (Zc3h13) is a novel component of the MACOM that stabilizes the interaction between Fl(2)d and Nito (Wtap and Rbm15) proteins, thereby ensuring deposition of m6A to targeted transcripts.
The physiological role of ZC3H13 in human cells has been poorly investigated. Recent reports suggest that mutant ZC3H13 facilitates glioblastoma progression and schizophrenia (Oldmeadow et al. 2014; Chow et al. 2017). It is possible that these diseases originate from misregulation of the m6A pathway upon ZC3H13 alteration. For instance, the association of m6A with cancer progression, in particular with glioblastoma and acute myeloid leukemia, has been demonstrated recently (Barbieri et al. 2017; Cui et al. 2017; Jaffrey and Kharas 2017; Kwok et al. 2017; Li et al. 2017; Vu et al. 2017; Zhang et al. 2017; Weng et al. 2018). Likewise, m6A plays an important role in cortical neurogenesis in the human forebrain (Yoon et al. 2017), a region of the brain that has been associated previously with schizophrenia (Heimer 2000). Hence, future studies should determine whether the role of ZC3H13 in these diseases is connected to its m6A-dependent function.
Materials and methods
mESC culture and genome editing
mESCs (129xC57Bl/6 genetic background; kindly provided by D. Schübeler of the Friedrich Miescher Institute for Biomedical Research) were cultured on gelatin-coated dishes in mES medium (DMEM; Gibco, 21969-035) supplemented with 15% FBS (Gibco), 1× nonessential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), 2 mM L-glutamine (Gibco), 0.1 mM 2-mercaptoethanol (Sigma), 50 mg/mL penicillin, 80 mg/mL streptomycin, MycoZap prophylactic, and LIF conditioned medium at 37°C in 5% CO2. Cultured cells were routinely tested for mycoplasma contamination using the VenorGeM mycoplasma detection kit according to manufacturer's recommendation (Sigma). For endogenous gene tagging using SpCas9-2A-mCherry (Knuckles et al. 2017), Rosa26:BirA-V5-expressing cells (RosaB) were transfected with 2 µg of SpCas9-sgRNA-2A-mCherry and 500 ng of ssODN as a donor when integration was desired. Small guide RNA constructs were generated as described in Knuckles et al. (2017). The ssODNs were synthesized as Ultramers by Integrated DNA Technologies, and their sequences are listed in Supplemental Table 2. All transfections were carried out using Lipofectamine 3000 reagent (Invitrogen). Twenty-four hours after transfection, mCherry-positive edited cells were sorted on a BD FACSAria III cell sorter (Becton Dickinson). mESCs were then sparsely seeded for clonal expansion, and then clones were individually picked, split, and screened by PCR for the desired mutation or integration. For tagging of Mettl3 and Rbm15, clones were subsequently screened by Western blotting using anti Flag (Sigma) or HRP-coupled streptavidin to confirm expression of endogenously tagged proteins. For Zc3h13 knockout lines, two independent single guide RNA (sgRNA) constructs were transfected to target sequences flanking exons 9–10, leading to a frameshift mutation and nonfunctional truncated protein. Deletion was confirmed via Western blotting using an anti-Zc3h13 antibody (Abcam, ab70802). Sequences of small guide RNAs are described in Supplemental Table 2.
Drosophila stocks and genetics
D. melanogaster w1118 was used as the wild-type control. Other fly stocks used were Fl(2)d shRNA (HMC03833; Bloomington Drosophila Stock Center [BDSC], 55674), Nito shRNA (HMS00166; obtained from Drosophila RNAi Screening Center [DRSC] [Harvard]), and Flacc dsRNA (GD35212 and KK110253; obtained from Vienna Drosophila Resource Center [VDRC]). For genetic interaction studies, we used Mettl3null (Haussmann et al. 2016), Mettl14fs (Lence et al. 2016), Ythdc1MI02006 (Bloomington), fl(2)d2 (Bloomington), virts1 (kind gift from Jamilla Horabin), nito1 (Yan and Perrimon 2015), and flacc mutant allele CG7358C (Bloomington). To remove daughterless, Df(2L)BSC209 (Bloomington) was used. Driver lines used in this study were dome-GAL4 (kind gift from Erika Bach, New York University Langone Medical Center) and elav-GAL4 (Bloomington). For the analysis of male-to-female transformations, flies of selected genotypes were chosen randomly.
Drosophila cell line
Drosophila S2R+ cells were embryonic-derived cells obtained from the Drosophila Genomics Resource Center (DGRC; at Indiana University; FlyBase accession FBtc0000150). Mycoplasma contamination was not present (verified by analyzing the RNA sequencing [RNA-seq] data from the cell line).
Cloning
The plasmids used for immunohistochemistry and coimmunoprecipitation assays in Drosophila S2R+ cells were constructed by cloning the corresponding cDNA in the pPAC vector (Lence et al. 2016) with an N-terminal Myc tag and the Gateway-based vectors with an N-terminal Flag-Myc tag (pPFMW) as well as a C-terminal HA tag (pPWH) (obtained from the DGRC at Indiana University).
TAP and MS
One confluent 15-cm dish of mESCs per sample was resuspended in 1 mL of ice-cold TAP lysis buffer (150–500 mM NaCl [depending on the experiment], 20 mM Tris-HCl at pH 7.5, 0.5% NP-40, 1 mM EDTA, 10% glycerol, 1 mM DTT supplemented with protease inhibitor cocktail [Roche]) after 0.25% trypsin/EDTA dissociation and PBS wash. Samples were shaken at 1000 rpm for 30 min at 4°C. Lysate was cleared by centrifugation at maximum speed at 4°C on a tabletop centrifuge. The protein concentration of each sample was determined using Bradford assay (Bio-Rad dye). Equal amounts of lysate (5 mg) from the control sample (parental untagged cells) and the bait protein sample (gene-tagged cells) were normalized by adding an appropriate amount of cold TAP lysis buffer to each sample to adjust the final sample concentration to ∼5 mg/mL. Equilibrated Flag M2 Dynabeads (10 µL of packed bead slurry per 5 mg of protein per sample; Sigma) previously washed twice with 1 mL of cold TAP lysis buffer were added to 5 mg/mL lysate. Tubes containing beads and lysates were incubated overnight at 4°C with end-over-end rotation.
The next day, Dynabeads were washed four times with TAP buffer for 10 min followed by three 15-min elutions of bound proteins with 3xFlag peptide (final concentration of 0.3 mg/mL in TAP buffer; Sigma). Next, elutions were pooled and added to the TAP-washed strepavidin Dynabeads (Thermo Fisher) and incubated overnight at 4°C with rotation. The next day, streptavidin Dynabeads were washed four times with TAP buffer for 10 min followed by a wash with TAP buffer without NP-40. The enriched proteins were digested directly on the Dynabeads with 0.1 mg/mL trypsin in digestion buffer (50 mM Tris at pH 8.0, 1 mM CaCl2, 1 mM TCEP).
The generated peptides were acidified with TFA to a final concentration of 0.8% and analyzed by capillary LC-MS/MS with an EASY-nLC 1000 using the two-column setup (Thermo Scientific). The peptides were loaded with 0.1% formic acid and 2% acetonitrile in H2O onto a peptide trap (Acclaim PepMap 100, 75 µm × 2 cm, C18, 3 µm, 100 Å) at a constant pressure of 800 bars. Peptides were separated at a flow rate of 150 nL/min with a linear gradient of 2%–6% buffer B (0.1% formic acid in acetonitrile) in buffer A (0.1% formic acid) for 3 min followed by a linear increase from 6% to 22% in 40 min, 22% to 28% in 9 min, 28% to 36% in 8 min, and 36% to 80% in 1 min, and the column was finally washed for 14 min at 80% B on a 50-µm × 15-cm ES801 C18, 2-µm, 100 Å column (Thermo Scientific) mounted on a DPV ion source (New Objective) connected to an Orbitrap Fusion mass spectrometer (Thermo Scientific). The data were acquired using 120,000 resolution for the peptide measurements in the Orbitrap and a top T (3-sec) method with HCD fragmentation for each precursor and fragment measurement in the ion trap according to the recommendations of the manufacturer (Thermo Scientific).
Protein identification and relative quantification of the proteins were done with MaxQuant version 1.5.3.8 using Andromeda as the search engine (Cox et al. 2011) and label-free quantification (LFQ) (Cox et al. 2014) as described in Hubner et al. (2010). The mouse subset of UniProt version 2015_01 combined with the contaminant database from MaxQuant was searched, and the protein and peptide false discovery rates (FDRs) were set to 0.01. All MaxQuant parameters are in the uploaded parameter file mqpar.xml.
Statistical analysis was done in Perseus (version 1.5.2.6) (Cox et al. 2011, 2014; Tyanova et al. 2016). Results were filtered to remove reverse hits, contaminants, and peptides found in only one sample. Missing values were imputed, and potential interactors were determined using t-test and visualized by a volcano plot. Significance lines corresponding to a given FDR were determined by a permutation-based method (Tusher et al. 2001). Threshold values (FDR) were selected between 0.005 and 0.05 and SO (curve bend) between 0.2 and 2 and are shown in the corresponding figures. Results were exported from Perseus and visualized using statistical computing language R.
All MS raw data were deposited in ProteomeXchange (data set PXD00811).
iBAQ analysis
iBAQ was performed as described in Schwanhausser et al. (2011) to evaluate protein abundances in the MAC and the MACOM in Mettl3 TAP-LC-MS experiments.
m6A-RIP
Total RNA from mESCs was isolated using Absolutely RNA Microprep kit (Stratagene) followed by mRNA selection using double Oligo d(T)23 (New England Biolabs) purification. Five micrograms of mRNA was incubated with 4 µg of anti-m6A antibody (polyclonal rabbit; Synaptic Systems, catalog no. 202 003) in m6A immunoprecipitation buffer (150 mM NaCl, 10 mM Tris-HCl at pH 7.4, 0.1% NP-40) supplemented with 5 U/mL murine RNase inhibitor (New England Biolabs) for 2 h at 4°C. Ten microliters of protein G magnetic beads (Invitrogen) was added to all m6A immunoprecipitation samples for 2 h at 4°C. On-bead digestion with RNase T1 (Thermo Fisher) at a final concentration 0.1 U/mL was performed for 15 min at room temperature. Beads with captured RNA fragments were then immediately washed twice with 500 µL of ice-cold m6A immunoprecipitation buffer and twice with room temperature m6A immunoprecipitation buffer and further eluted with 100 µL of elution buffer (20 mM DTT, 150 mM NaCl, 50 mM Tris-HCl at pH 7.4, 1 mM EDTA, 0.1% SDS, 5 U/mL proteinase K) for 5 min at 42°C. The elution step was repeated four times, and 600 µL of acidic phenol/chloroform (pH 4.5) (Ambion) was added to 400 µL of the combined eluate per sample in order to extract captured RNA fragments. Samples were mixed and transferred to Phase Lock Gel Heavy tubes (5Prime) and centrifuged at 12,000g for 5 min. Aqueous phase was precipitated overnight at −80°C. On the following day, samples were centrifuged, washed twice with 80% EtOH, and resuspended in 15 µL of RNase-free H2O (Ambion). Recovered RNA was analyzed on RNA Pico Chip (Agilent), and concentrations were determined with RNA HS Qubit reagents. Since no RNA was recovered in the m6A immunoprecipitation no-antibody control samples, libraries were prepared with 30 ng of two independent m6A immunoprecipitations performed on RNA from wild-type and Mettl3 and Zc3h13 knockout cells. For every condition, input material (200 ng of mRNA) was also sequenced. Both m6A immunoprecipitations and inputs were sequenced using the NEBNext RNA directional library preparation kit.
m6A RIP sequencing analysis
MACS2 was used to call peaks of m6A enrichment for wild-type immunoprecipitation versus input samples using the default parameters. Peaks were assigned to overlapping gene bodies within 500 base pairs (bp). The intersection of the resulting gene lists (3285 genes) was taken as the set of m6A target genes.
BigWig files for each sample were created using the qExportWig function from the QuasR package in R (Cox et al. 2011). Read counts were binned in 50-bp windows, and counts for each sample were scaled to the mean aligned library size of all samples. The deepTools suite was used for metagene analysis (Ramirez et al. 2016). The bigwigCompare function was used to calculate the log2 ratio between each wild-type or Zc3h13 knockout sample and the Mettl3 knockout samples. m6A target CDS regions were scaled to 5 kb, and m6A enrichment versus Mettl3 knockout was calculated in 50-bp bins across scaled target regions as well as 2 kb upstream and downstream using the computeMatrix command.
NanoBiT protein complementation assay
Fusion constructs of mouse Rbm15 and Wtap to NanoBiT subunits were generated as follows: Full-length Rbm15- and Wtap-coding sequences were amplified with the oligonucleotides indicated in Supplemental Table 2 from poly-A-selected mRNA using NEBNext High-Fidelity 2X PCR master mix (New England BioLabs). Overhangs with homology to destination vectors (pBiT1.1-C, pBiT2.1-C, pBiT1.1-N, and pBiTN.1-C; Promega) were included in oligonucleotide sequences. Gel-purified PCR products were cloned into EcoRI sites using NEBuilder HiFi DNA assembly kit (New England BioLabs) following the manufacturer's recommendations. The optimal combinations of N-terminal- or C-terminal-tagged fusions to small or large subunits were determined through transfection of 20,000 wild-type mESCs per well with Lipofectamine 3000 reagent (Invitrogen) seeded in 96-well tissue culture plates (Corning, catalog no. 3917). Measurements were performed using the Nano-Glo live-cell assay system (Promega) and measured in a microplate luminometer (Berthold, LB960). The Rbm15/Wtap fusion combination yielding the highest luciferase activity was then transfected into distinct mESC genetic backgrounds, and the expression level of the fusion construct was quantified via RT-qPCR using oligonucleotides described in Supplemental Table 2.
Drosophila staging
The staging experiment was performed as described previously (Lence et al. 2016) using D. melanogaster w1118 flies. A total of three independent samples was collected for each Drosophila stage as well as for heads and ovaries. Samples from the staging experiment were used for RNA extraction to analyze m6A abundance in mRNA and expression levels of different transcripts during Drosophila development.
RNA isolation and mRNA purification
Total RNA from S2R+ cells was isolated using Trizol reagent (Invitrogen), and DNA was removed with DNase I treatment (New England Biolabs). Fly heads from 3- to 5-d-old flies were separated and homogenized in Trizol prior to RNA isolation. mRNA was isolated by two rounds of purification with Dynabeads Oligo d(T)25 (New England Biolabs).
RT–PCR
cDNA was prepared using M-MLV reverse transcriptase (Promega). Transcript levels were quantified using Power SYBR Green PCR master mix (Invitrogen) and the oligonucleotides indicated in Supplemental Table 2. RT–PCR was performed using the oligonucleotides described in Supplemental Table 2 to analyze Sxl splicing.
RNA in situ hybridization
In situ hybridization was performed as described previously (Lence et al. 2016). In situ probes were prepared with DIG RNA-labeling kit (Roche) following the manufacturer's protocol. Oligos used for the probes are listed in Supplemental Table 2.
RIP
S2R+ cells were transfected with Myc-tagged constructs using Effectene reagent. Seventy-two hours after transfection, cells were washed with ice-cold PBS and collected by centrifugation at 1000g for 5 min. The cell pellet was lysed in 1 mL of lysis buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 0.05% NP-40) supplemented with protease inhibitors, rotated head over tail for 30 min at 4°C, and centrifuged at 18,000g for 10 min at 4°C to remove the remaining cell debris. Protein concentrations were determined using Bradford reagent (Bio-Rad). For RIP, 2 mg of protein was incubated with 2 µg of anti-Myc antibody coupled to protein G magnetic beads (Invitrogen) in lysis buffer and rotated head over tail for 4 h at 4°C. The beads were washed three times with lysis buffer for 5 min. One-fourth of the immunoprecipitated protein–RNA complexes were eluted by incubation in 1× NuPAGE LDS buffer (Thermo Fisher) for 10 min at 70°C for protein analysis. RNA from the remaining protein–RNA complexes was further isolated using Trizol reagent. qPCR was performed with the oligos listed in Supplemental Table 2.
Immunostaining
For staining of Drosophila S2R+ cells, cells were transferred to the polylysine-pretreated eight-well chambers (Ibidi) at a density of 2 × 105 cells per well. After 30 min, cells were washed with 1× DPBS (Gibco), fixed with 4% formaldehyde for 10 min, and permeabilized with PBST (0.2% Triton X-100 in PBS) for 15 min. Cells were incubated with mouse anti-Myc (1:2000; Enzo, 9E10) in PBST supplemented with 10% donkey serum overnight at 4°C. Cells were washed three times for 15 min in PBST and then incubated with secondary antibody and 1× DAPI solution in PBST supplemented with 10% donkey serum for 2 h at 4°C. After three 15-min washes in PBST, cells were imaged with a Leica SP5 confocal microscope using a 63× oil immersion objective.
Western blotting
Proteins were extracted for 30 min on ice, the lysates were centrifuged at 16,000g for 5 min at 4°C, and protein concentration in the supernatant was determined using the Bio-Rad protein assay. Protein samples were separated on NuPAGE-Novex Bis-Tris 4%–12% gradient gels (Invitrogen) in MOPS buffer for 40 min at 200 V. Semidry transfer to nitrocellulose membrane (Whatman) was performed for 40 min at 15 V. Membranes were blocked for 30 min in 2% nonfat dry milk and TBS–0.05% Tween 20 (TBST) and incubated with primary antibodies overnight at 4°C (Mettl3 [Protein Tech, 15073], Rbm15 [Abcam, ab70549], Zc3h13 [Abcam, ab70802], Hakai [Aviva Systems Biology, Cbll1 ARP39622], Wtap [Protein Tech, 60188], and Tubulin [Abcam, clone YL1/2]). Signal was detected with corresponding HRP-conjugated secondary antibodies and Immobilon Western Chemiluminiscent HRP substrate (Millipore).
Cell culture, RNAi, and transfection
Drosophila S2R+ cells were grown in Schneider's medium (Gibco) supplemented with 10% FBS (Sigma) and 1% penicillin–streptomycin (Sigma). For RNAi experiments, PCR templates were prepared using the oligonucleotides indicated in Supplemental Table 2. dsRNA were prepared using T7 megascript kit (New England Biolabs). dsRNA against the bacterial β-galactosidase gene (lacZ) was used as a control for all RNAi experiments. S2R+ cells were seeded at a density of 106 cells per milliliter in serum-free medium, and 7.5 µg of dsRNA was added to 106 cells. After 6 h of cell starvation, serum-supplemented medium was added to the cells. dsRNA treatment was repeated after 48 and 96 h, and cells were collected 24 h after the last treatment. Effectene (Qiagen) was used to transfect vector constructs in all overexpression experiments following the manufacturer's protocol.
Coimmunoprecipitation assay and Western blot analysis
For the coimmunoprecipitation assay, different combinations of vectors with the indicated tags were cotransfected in S2R+ cells. Forty-eight hours after transfection, cells were collected, washed with DPBS, and pelleted by centrifugation at 400g for 10 min. The cell pellet was lysed in 1 mL of lysis buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 0.05% NP-40) supplemented with protease inhibitors and rotated head over tail for 30 min at 4°C. Nuclei were collected by centrifugation at 1000g for 10 min at 4°C, resuspended in 300 µL of lysis buffer, and sonicated with five cycles of 30 sec on and 30 sec off at the low power setting. Cytoplasmic and nuclear fractions were joined and centrifuged at 18,000g for 10 min at 4°C to remove the remaining cell debris. Protein concentrations were determined using Bradford reagent (Bio-Rad). For immunoprecipitation, 2 mg of proteins was incubated with 2 µg of anti-Myc antibody coupled to protein G magnetic beads (Invitrogen) in lysis buffer and rotated head over tail overnight at 4°C. The beads were washed three times for 15 min with lysis buffer, and immunoprecipitated proteins were eluted by incubation in 1× NuPAGE LDS buffer (Thermo Fisher) for 10 min at 70°C. Eluted immunoprecipitated proteins were removed from the beads, and DTT was added to 10% final volume. Immunoprecipitated proteins and input samples were analyzed by Western blot after incubation for an additional 10 min at 70°C.
For Western blot analysis, proteins were separated on a 7% SDS-PAGE gel and transferred to a nitrocellulose membrane (Bio-Rad). After blocking with 5% milk in 0.05% Tween in PBS for 1 h at room temperature, the membrane was incubated with primary antibody in blocking solution overnight at 4°C. Primary antibodies used were mouse anti-Flag (1:2000; Sigma, M2-F1804), mouse anti-Myc (1:2000; Enzo, 9E10), mouse anti-HA (1:1000; Covance, 16B12), mouse anti-Tubulin (1:2000; Biolegend, 903401), mouse anti-Fl(2)d (1:500; Developmental Studies Hybridoma Bank, 9G2), and rabbit anti-Mettl14 and guinea pig anti-Mettl3 (1:500) (Lence et al. 2016). The membrane was washed three times in PBST for 15 min and incubated for 1 h at room temperature with secondary antibody in blocking solution. Protein bands were detected using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific).
SILAC experiment and LC-MS/MS analysis
For SILAC experiments, S2R+ cells were grown in Schneider medium (Dundee Cell) supplemented with either heavy (Arg8 and Lys8) (Cambridge Isotope Laboratories) or light (Arg0 and Lys0) (Sigma) amino acids. For the forward experiment, Myc-Nito was transfected in heavy-labeled cells, and Myc alone was transfected in light-labeled cells. The reverse experiment was performed vice versa. The coimmunoprecipitation experiment was done as described earlier. Before elution, beads of the heavy and light lysates were combined in a 1:1 ratio and eluted with 1× NuPAGE LDS buffer that was subjected to MS analysis as described previously (Bluhm et al. 2016). Raw files were processed with MaxQuant (version 1.5.2.8) and searched against the UniProt database of annotated Drosophila proteins (D. melanogaster: 41,850 entries, downloaded January 8, 2015).
LC-MS/MS analysis of m6A levels
Three-hundred nanograms of purified mRNA was digested using 0.3 U of nuclease P1 from Penicillum citrinum (Sigma-Aldrich) and 0.1 U of snake venom phosphodiesterase from Crotalus adamanteus (Worthington). RNA and enzymes were incubated in 5 mM ammonium acetate (pH 5.3) for 2 h at 37°C. The remaining phosphates were removed by 1 U of FastAP (Thermo Scientific) in a 1-h incubation at 37°C in 10 mM ammonium acetate (pH 8). The resulting nucleoside mix was then spiked with 13C stable isotope-labeled nucleoside mix from Escherichia coli RNA as an internal standard (SIL-IS) to a final concentration of 6 ng/µL for the sample RNA and 2 ng/µL for the SIL-IS. For analysis, 10 µL of the previously mentioned mixture was injected into the LC-MS/MS machine. Generation of technical triplicates was obligatory. Mouse mRNA samples were analyzed in biological duplicates, and fly samples were analyzed in triplicates. LC separation was performed on an Agilent 1200 series instrument using 5 mM ammonium acetate buffer as solvent A and acetonitrile as buffer B. Each run started with 100% buffer A, which was decreased to 92% within 10 min. Solvent A was further reduced to 60% within another 10 min. Until minute 23 of the run, solvent A was increased to 100% again and kept at 100% for 7 min to re-equilibrate the column (Synergi Fusion; 4 µM particle size, 80 Å pore size, 250 × 2.0 mm; Phenomenex). The ultraviolet signal at 254 nm was recorded via a DAD detector to monitor the main nucleosides.
MS/MS was then conducted on the coupled Agilent 6460 triple-quadrupole (QQQ) mass spectrometer equipped with an Agilent JetStream ESI source that was set to the following parameters: gas temperature, 350°C; gas flow, 8 L/min; nebulizer pressure, 50 psi; sheath gas temperature, 350°C; sheath gas flow, 12 L/min; and capillary voltage, 3000 V. To analyze the mass transitions of the unlabeled m6A and all 13C m6A simultaneously, we used the dynamic multiple reaction monitoring mode. The quantification was conducted as described previously (Kellner et al. 2014).
RNA-seq and computational analysis
Illumina TruSeq sequencing kit (Illumina) was used for RNA-seq and computational analysis. The RNA libraries were sequenced on a NextSeq500 with a read length of 85 bp. The data were mapped against Ensembl release 90 of D. melanogaster using STAR (version 2.5.1b). Counts per gene were derived using featureCounts (version 1.5.1). Differential expression analysis was performed using DESeq2 (version 1.16.1) and filtered for an FDR <5%. Differential splicing analysis was performed using rMATS (version 3.2.5) and filtered for an FDR <10%. Sequencing depth-normalized coverage tracks were generated using Bedtools (version 2.25.0), Samtools (version 1.3.1), and Kentutils (version 302). The heat map of the fold change (log2) of commonly misregulated genes was clustered according to rows and columns. The color gradient was adjusted to display the 1% lowest/highest values within the most extreme color (lowest values as the darkest blue and highest values as the darkest red). Splice events for different knockdown conditions are represented by pie charts. “Control” depicts the detected splice events, on average, in all of the comparisons of control versus knockdown. The pie charts for the individual knockdowns depict the amount of significantly different splicing events with a FDR value <10%. The gene ontology (GO) term analysis was performed using the package ClusterProfiler. The GO terms were semantic similarity-reduced using the “simplify” function of the package. The genes tested in all of the conditions were used as a background gene set. Default parameters were used for the analysis. We defined the set of genes that was analyzed for differential expression (in any condition) and whose transcripts contained m6A (according to Kan et al. 2017). The significance of the overlap of these genes with the genes commonly differentially regulated in the knockdowns (either commonly up-regulated, commonly down-regulated, or misregulated in all conditions) or commonly misspliced in all conditions was tested using a hypergeometric test.
Phylogenetic analysis
The phylogenetic tree was constructed with ClustalX from multiple sequence alignments generated with MUSCLE of the Drosophila sequence with orthologs from human and other representative species.
Statistics
For m6A level measurements, data sets were compared using two-tailed Student's t-test for unequal variances. Normality was verified, and homogeneity of variances was analyzed with Levene's test. RIP-qPCR, ZC3H13 rescue quantification, split luciferase NanoBiT assay, and in vivo Flacc knockdown validation data sets were compared using two-tailed Student's t-test for equal variances. Statistical significance of fly viability was determined by a χ2 test (GraphPad Prism). Statistical tests used for RNA-seq and m6A-RIP data analysis, TAP-MS analysis, iBAQ analysis, and SILAC experiments are described in detail in the relevant sections of the Materials and Methods.
Data availability
The data that support the findings of this study have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE106614. All other relevant data are available from the corresponding author.
Supplementary Material
Acknowledgments
We thank the Bloomington Drosophila Stock Center and the Vienna Drosophila Resource Center for fly reagents, the Drosophila Genomics Resource Center at Indiana University for plasmids, members of the Bühler and Roignant laboratories for helpful discussion, Vytautas Iesmantavicius from the Protein Analysis Facility at Friedrich Miescher Institute for help with MS data analysis, and the Genomics and Bioinformatics Institute of Molecular Biology (IMB) Core Facilities for great support. Support by the IMB Proteomics Core Facility is gratefully acknowledged (the instrument is funded by Deutsche Forschungsgemeinschaft INST 247/766-1 FUGG). In particular, we thank Anja Freiwald from the IMB Proteomics Core Facility for sample preparation, and Dr. Mario Dejung from the Proteomics Core Facility for data processing and generating the scatter plot. Research in the laboratory of J.-Y.R. is supported by the Deutsche Forschungsgemeinschaft (DFG) RO 4681/5-1 and the Epitran COST action (CA16120). Work performed in M. Bühler's laboratory was supported by the Novartis Research Foundation and the Swiss Science Foundation National Centre of Competence in Research RNA and Disease (grant no. 141735). Work performed in M.S.’s laboratory was supported by the Biotechnology and Biological Sciences Research Council. The project was also supported by the DFG SPP1784 grant to M.H.
Author contributions: P.K., T.L., M. Bühler, and J.-Y.R. conceived the study. P.K., T.L., I.U.H., D.J., I.M., T.H., R.V., D.H., M. Biggiogera, M.H., M.S., M. Bühler, and J.-Y.R. performed the methodology. N.K. performed the computational analysis of the RNA-seq data. S.H.C. performed the computational analysis of the methylated RIP sequencing data. M.A.A.-N. performed the phylogenetic analysis. P.K. and T.L. wrote the draft of the manuscript. All authors reviewed and edited the manuscript. M. Bühler, and J.-Y.R. supervised the study.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.309146.117.
Freely available online through the Genes & Development Open Access option.
References
- Agarwala SD, Blitzblau HG, Hochwagen A, Fink GR. 2012. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet 8: e1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC, Aspris D, Migliori V, Bannister AJ, Han N, et al. 2017. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 552: 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluhm A, Casas-Vila N, Scheibe M, Butter F. 2016. Reader interactome of epigenetic histone marks in birds. Proteomics 16: 427–436. [DOI] [PubMed] [Google Scholar]
- Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. 1997. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3: 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Chow RD, Guzman CD, Wang G, Schmidt F, Youngblood MW, Ye L, Errami Y, Dong MB, Martinez MA, Zhang S, et al. 2017. AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat Neurosci 20: 1329–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10: 1794–1805. [DOI] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13: 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang CG, et al. 2017. m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep 18: 2622–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezi V, Ivanov C, Haussmann IU, Soller M. 2016. Nucleotide modifications in messenger RNA and their role in development and disease. Biochem Soc Trans 44: 1385–1393. [DOI] [PubMed] [Google Scholar]
- Dixon AS, Schwinn MK, Hall MP, Zimmerman K, Otto P, Lubben TH, Butler BL, Binkowski BF, Machleidt T, Kirkland TA, et al. 2016. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem Biol 11: 400–408. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206. [DOI] [PubMed] [Google Scholar]
- Flemr M, Buhler M. 2015. Single-step generation of conditional knockout mouse embryonic stem cells. Cell Rep 12: 709–716. [DOI] [PubMed] [Google Scholar]
- Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, Soller M. 2016. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540: 301–304. [DOI] [PubMed] [Google Scholar]
- Heimer L. 2000. Basal forebrain in the context of schizophrenia. Brain Res Brain Res Rev 31: 205–235. [DOI] [PubMed] [Google Scholar]
- Hilfiker A, Amrein H, Dubendorfer A, Schneiter R, Nothiger R. 1995. The gene virilizer is required for female-specific splicing controlled by Sxl, the master gene for sexual development in Drosophila. Development 121: 4017–4026. [DOI] [PubMed] [Google Scholar]
- Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T, Hamakubo T. 2013. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem 288: 33292–33302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner NC, Bird AW, Cox J, Splettstoesser B, Bandilla P, Poser I, Hyman A, Mann M. 2010. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol 189: 739–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey SR, Kharas MG. 2017. Emerging links between m6A and misregulated mRNA methylation in cancer. Genome Med 9: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan L, Grozhik AV, Vedanayagam J, Patil DP, Pang N, Lim KS, Huang YC, Joseph B, Lin CJ, Despic V, et al. 2017. The m6A pathway facilitates sex determination in Drosophila. Nat Commun 8: 15737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner S, Ochel A, Thuring K, Spenkuch F, Neumann J, Sharma S, Entian KD, Schneider D, Helm M. 2014. Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. Nucleic Acids Res 42: e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckles P, Carl SH, Musheev M, Niehrs C, Wenger A, Buhler M. 2017. RNA fate determination through cotranscriptional adenosine methylation and microprocessor binding. Nat Struct Mol Biol 24: 561–569. [DOI] [PubMed] [Google Scholar]
- Kwok CT, Marshall AD, Rasko JE, Wong JJ. 2017. Genetic alterations of m6A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol 10: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho CH, Kreim N, Andrade-Navarro MA, Poeck B, Helm M, et al. 2016. m6A modulates neuronal functions and sex determination in Drosophila. Nature 540: 242–247. [DOI] [PubMed] [Google Scholar]
- Lence T, Soller M, Roignant JY. 2017. A fly view on the roles and mechanisms of the m6A mRNA modification and its players. RNA Biol 14: 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, et al. 2017. FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell 31: 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. 2014. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10: 93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. 2012. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149: 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldmeadow C, Mossman D, Evans TJ, Holliday EG, Tooney PA, Cairns MJ, Wu J, Carr V, Attia JR, Scott RJ. 2014. Combined analysis of exon splicing and genome wide polymorphism data predict schizophrenia risk loci. J Psychiatr Res 52: 44–49. [DOI] [PubMed] [Google Scholar]
- Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. 2016. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537: 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. 2014. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 24: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant JY, Soller M. 2017. m6A in mRNA: an ancient mechanism for fine-tuning gene expression. Trends Genet 33: 380–390. [DOI] [PubMed] [Google Scholar]
- Ruzicka K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, Eeckhout D, El-Showk S, Li H, Zhong S, et al. 2017. Identification of factors required for m6 A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol 215: 157–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. 2014. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep 8: 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledz P, Jinek M. 2016. Structural insights into the molecular mechanism of the m6A writer complex. eLife 5: e18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13: 731–740. [DOI] [PubMed] [Google Scholar]
- Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al. 2017. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med 23: 1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C, Borgeson B, Phanse S, Tu F, Drew K, Clark G, Xiong X, Kagan O, Kwan J, Bezginov A, et al. 2015. Panorama of ancient metazoan macromolecular complexes. Nature 525: 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. 2014. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol 16: 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Doxtader KA, Nam Y. 2016a. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell 63: 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al. 2016b. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 534: 575–578. [DOI] [PubMed] [Google Scholar]
- Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, Shi H, Skibbe J, Shen C, Hu C, et al. 2018. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell 22: 191–205 e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Perrimon N. 2015. spenito is required for sex determination in Drosophila melanogaster. Proc Natl Acad Sci 112: 11606–11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, Su Y, Kim NS, Zhu Y, Zheng L, et al. 2017. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell 171: 877–889.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bogler O, et al. 2017. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31: 591–606.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, Fray RG. 2008. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20: 1278–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE106614. All other relevant data are available from the corresponding author.