Abstract
The styrene monooxygenase (SMO) system from Pseudomonas sp. consists of two enzymes (StyA and StyB). StyB catalyses the reduction of FAD at the expense of NADH. After the transfer of FADH2 from StyB to StyA, reaction with O2 generates FAD‐OOH, which is the epoxidising agent. The wastage of redox equivalents due to partial diffusive transfer of FADH2, the insolubility of recombinant StyB and the impossibility of expressing StyA and StyB in a 1:1 molar ratio reduce the catalytic efficiency of the natural system. Herein we present a chimeric SMO (Fus‐SMO) that was obtained by genetic fusion of StyA and StyB through a flexible linker. Thanks to a combination of: 1) balanced and improved expression levels of reductase and epoxidase units, and 2) intrinsically higher specific epoxidation activity of Fus‐SMO in some cases, Escherichia coli cells expressing Fus‐SMO possess about 50 % higher activity for the epoxidation of styrene derivatives than E. coli cells coexpressing StyA and StyB as discrete enzymes. The epoxidation activity of purified Fus‐SMO was up to three times higher than that of the two‐component StyA/StyB (1:1, molar ratio) system and up to 110 times higher than that of the natural fused SMO. Determination of coupling efficiency and study of the influence of O2 pressure were also performed. Finally, Fus‐SMO and formate dehydrogenase were coexpressed in E. coli and applied as a self‐sufficient biocatalytic system for epoxidation on greater than 500 mg scale.
Keywords: asymmetric synthesis, biocatalysis, enzymatic fusion, epoxidation, styrene monooxygenases
Introduction
Chiral epoxides are important building blocks in organic synthesis because of their high versatility and reactivity towards a variety of reagents. Furthermore, they find wide application as intermediates for the synthesis of active pharmaceutical ingredients, natural products, flavours and fragrances, other fine chemicals and advanced polymeric materials.1
Chiral epoxides are classically synthesised by asymmetric epoxidation of alkenes in the presence of Ti(O‐iPr)4 (i.e., Katsuki–Sharpless)2 or salen‐MnIII (i.e., Jacobsen) complexes,3 which require stoichiometric amounts of a chemical oxidant. Hydrolytic kinetic resolution of racemic epoxides is also possible, either by chemical methods1b, 1h, 4 or by biocatalytic methods involving hydrolases.1c, 5 Other more recent methodologies involve iron‐based catalysts together with hydrogen peroxide6 or organocatalysts together with an oxidant such as hydrogen peroxide, oxone, hypochlorite salts, peroxides (e.g., TBHP, mCPBA), trichloroisocyanuric acid etc.7 Despite significant research efforts and progress in the fields of chemocatalytic and organocatalytic asymmetric epoxidation of terminal alkenes such as styrene and its derivatives, achieving elevated stereoselectivity (99 % ee or higher) still remains a challenge.7a, 7b, 7d, 8
Hence, the biocatalytic counterpart of this reaction has been investigated during the past 15 years by using either flavin‐ (FAD) or iron‐dependent monooxygenases.9 Enzymatic epoxidation is particularly attractive because epoxides are usually obtained with elevated enantiomeric excess (>99 %) by using molecular oxygen as oxidant. Among others, the bi‐enzymatic system of the FAD‐dependent styrene monooxygenase (SMO) from Pseudomonas sp. has been exploited for the production of enantiopure styrene oxide (and derivatives thereof) in the laboratory and in pilot‐scale production by using fermenting or resting recombinant Escherichia coli cells10 or crude enzyme preparations.11 A thorough comparison between the SMO enzymatic process and different chemical epoxidation processes showed that the former is the most advantageous when economic profitability and environmental impact are concomitantly considered.12 The potential of SMOs in chemical synthesis has also been demonstrated in the production of chiral vicinal diols, amino alcohols, α‐hydroxycarboxylic acids and α‐amino acids through one‐pot, concurrent multistep cascades.13
The current drawback relating to the use of the natural SMO enzymatic system, as in Pseudomonas sp., is the requirement for two separate enzymes (StyA and StyB) in order to promote efficient epoxidation activity.14 Hence, both enzymes are usually coexpressed in E. coli.10a StyB catalyses the reduction of FAD to FADH2 at the expense of NADH, whereas StyA utilises FADH2 and O2 to generate FAD‐OOH in its active site. The final epoxidation of the styrene substrate, performed by StyA, regenerates the oxidised FAD upon dehydration (Scheme 1).
Scheme 1.
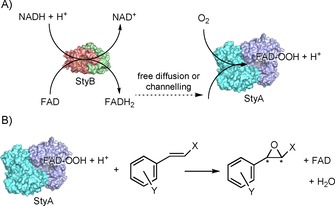
Simplified catalytic cycle of the flavin‐dependent SMO. A) Step 1: Reduction of FAD to FADH2 catalysed by StyB and further transfer to StyA, where oxidation to FAD‐OOH occurs. B) Step 2: Asymmetric epoxidation of styrene (or derivatives) by StyA with regeneration of oxidised FAD.
The actual mechanism of this bi‐enzymatic process is still a matter of debate. Early studies reported a purely diffusive transfer of FADH2 from StyB to StyA.15 Nonetheless, subsequent in‐depth studies strongly support the existence of a molecular interaction during the catalytic cycle between StyB and StyA from Pseudomonas sp., as well as in cases of other SMOs.16 Published kinetic data show: 1) the existence of two competitive mechanisms (diffusive and channelling) for the transfer of FADH2 from StyB to StyA,16 and 2) a variation in the epoxidation activity of StyA in the presence of different types of StyB, with the highest rate being observed in combination with the natural partner.16c A naturally occurring fused SMO (StyA2B) has been isolated, but its catalytic activity was from one to two orders of magnitude lower than that of the bienzymatic SMO system from Pseudomonas sp.17 Interestingly, the epoxidation activity of StyA2B increased when an additional epoxidase enzyme (StyA1) was included.16c
All of these findings reveal that the molecular interaction between the different enzymatic units has important synergistic effects on the overall catalytic cycle, besides mere improved transfer of FADH2 from one unit to the other one. However, StyA is also capable of catalysing epoxidation in the absence of StyB, as long as reduced FAD is supplied. This property has been exploited for the generation of hybrid chemo‐enzymatic and electro‐enzymatic systems.18 So far, the catalytic efficiencies of these “StyA hybrid” systems have been significantly lower than that of the natural bi‐enzymatic StyA/StyB system. This reduced efficiency may, in part, be attributed to the lack of catalytic activation on StyA effected by StyB. Additionally, it has been shown that: 1) the highest epoxidation activity is obtained when StyA and StyB are combined at about 1:1 ratio and at low FAD concentration (ca. 15 μm),15–, 16c and 2) the reduction of oxidised FAD by StyB is the rate‐limiting step.16b, 19 Obtaining a nearly 1:1 ratio mixture of recombinant StyA and StyB in active form in E. coli is still a challenging task. One issue is the difficulty inherent in balancing and regulating the expression of both genes. The second, more severe, issue is that recombinant StyB in E. coli is mainly obtained in the form of insoluble inclusion bodies (i.e., in a denatured form). Indeed, in vitro experiments have always required refolding of the inactive StyB, a lengthy and low‐yielding procedure.15, 16
Herein we present a chimeric SMO in which reductive (StyB) and epoxidation (StyA) enzymatic units are fused with a flexible linker of 30 amino acids20 in order: 1) to solve the insolubility issue of StyB, 2) to maximise the epoxidation activity (StyA/StyB 1:1 ratio), and 3) to improve FADH2 transfer and to find an optimum balance with coupling efficiency (NADH consumption vs. styrene epoxidation). A recent work reported a study on the catalytic mechanisms of other artificially fused SMOs.19 Nevertheless, these chimeric enzymes were either produced in insoluble (i.e., inactive) form or possessed lower catalytic rates for the epoxidation of styrene (ca. fivefold or less) than the natural system. The reason for this discrepancy in relation to our present work are also discussed.
Results and Discussion
Design of the fused SMO and initial tests for activity
The styA and styB genes belonging to the bi‐enzymatic system of the SMO from Pseudomonas sp. were genetically fused through a flexible linker,19 made up of 30 amino acid residues (for details, see Section S4.1 in the Supporting Information). The construct (Fus‐SMO) was designed in the following order: (N‐His6‐tag)‐StyA‐linker‐StyB. Positioning the His6‐tag DNA sequence downstream from the T7 promotor and upstream from the styA gene normally confers enhanced levels of enzyme expression. We positioned the gene for StyB at the end of the construct because this enzyme in its discrete form has always been expressed mainly as inclusion bodies (>95 % of inactive enzyme).16a E. coli BL21(D3) cells were transformed with the DNA encoding for the chimeric Fus‐SMO enzyme, and cells were grown on agar plates. Because of the different morphologies of the E. coli colonies obtained (i.e., various colours: pink, blue, white), four of them were selected for further testing of expression, solubility and activity (see Figure 1 for expression and solubility of colony 1; further information in Section S4). The generation of pigmented cells stems from the production of indigo (blue) or indirubin (red) during cultivation, which is enabled by the overexpressed Fus‐SMO.21
Figure 1.
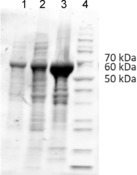
SDS‐PAGE for the expression and solubility of the chimeric fused SMO (Fus‐SMO, 69 kDa) col 1. Lane 1: free cell extract, lane 2: soluble protein fraction, lane 3: insoluble protein fraction, lane 4: PageRuler unstained protein ladder (ThermoFisher Scientific).
In these preliminary experiments, we determined the level of conversion for the epoxidation of styrene (1 a) after a specific reaction time by lyophilised E. coli whole cells overexpressing Fus‐SMO in a biphasic system (aqueous buffer/n‐decane, Table S2 in the Supporting Information). As reported in the literature,10b, 13b the organic phase acts as a styrene reservoir and reduces the molecular toxicity of the product styrene oxide. Interestingly, the quantity and solubility of the expressed Fus‐SMO into the cells did not seem to correlate with a particular pigmentation of the host organism (Figures S2 and S3), whereas a difference in the level of conversion was observed only in one case for colony 2 (Table S2). However, further optimisation of the expression conditions [i.e., isopropyl β‐d‐1‐thiogalactopyranoside (IPTG) concentration] revealed that all the E. coli colonies performed the epoxidation equally well, independently of their pigmentation. A glycerol stock solution prepared from the culture obtained from colony 1 was used for continuation of this study.
We also assayed the influence of the addition of exogenous FAD (50 μm) during the reaction. Biocatalytic reactions either in the presence or in the absence of FAD afforded statistically analogous results. Hence, E. coli is capable of producing sufficient amounts of cofactor in combination with Fus‐SMO to sustain the reaction (Table S2).
For the sake of reproducibility of the results, we decided to carry out this study by adding a minimal amount of FAD to each biocatalytic reaction mixture because long storage of E. coli/Fus‐SMO (frozen pellets or lyophilised cells) was possible, but data on the stability of FAD under such conditions were not available. Optimisation of the expression conditions (25 °C, 16 h, IPTG 0.1 mm) and further testing of activity led to the production of soluble and active Fus‐SMO in elevated amounts (Figures S2–S4 and Tables S3–S4). Because the biocatalytic epoxidation might also be influenced by the availability of dioxygen in the headspace, different reaction vessels had been considered previously (Table S3). Quantitative epoxidation of 1 a (10 mm) in 1 mL of biphasic reaction mixture (aqueous buffer/n‐decane 1:1, v/v) was achieved with use of 4 mL glass vials as reaction vessels. Employment of vials of smaller volume (2 mL) led to a maximum of 41 % conversion, likely due to insufficient availability of dioxygen. The use of vials of larger volume (e.g., 20 mL) is possible, although agitation must be carefully set in order to assure efficient mixing of the biphasic mixture.
The bi‐enzymatic StyA/StyB system has often been applied in biphasic systems in which the organic phase was a high‐boiling solvent such as hexadecane, bis‐(2‐ethylhexyl) phthalate, etc. In our preliminary experiments for the reaction on preparative scale, the difficult final evaporation of the high‐boiling organic solvent led to a troublesome and highly energy‐consuming workup procedure. Our experiments also showed that, if lyophilised cells are employed, it is possible to use low‐boiling organic solvents, such as n‐heptane or even neat styrene, without affecting the productivity (Table S5). Hence, a mixture of aqueous buffer (KPi, pH 8) and n‐heptane (1:1, v/v) was selected for further experiments.
Determination of the coupling efficiency of Fus‐SMO
The coupling efficiency of the StyA/StyB system (bi‐enzymatic or fused) is defined as the ratio between the quantity of substrate epoxidised and the reducing equivalents consumed. Detailed biochemical studies have revealed that the coupling efficiency is a function of: 1) the relative concentrations of StyA and StyB, 2) the substrate concentration, and 3) the FAD concentration.16a Coupling efficiency verging towards one was measured at low FAD concentrations (≤1 μm) and a StyA/StyB ratio of about 500. Under these conditions, StyB produces FADH2 in extremely low concentrations (nanomolar range) and it can be quantitatively transferred to StyA; hence, StyA produces FAD‐OOH that can be almost quantitatively consumed for the epoxidation of styrene. Therefore, virtually no FADH2 is wasted in the generation of H2O2 as by‐product.
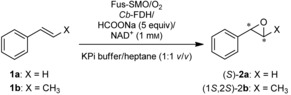
The logical drawback of this specific reaction condition is that the epoxidation activity is dramatically reduced, making it inapplicable for synthetic purposes. Therefore, the challenge is to maximise the coupling efficiency without affecting the overall epoxidation activity. We hypothesised that our Fus‐SMO might have improved coupling efficiency under reaction conditions that are suitable for a high epoxidation rate. In our synthetic set‐up, NAD+ was applied in a catalytic amount (1 mm) and recycled with the aid of a formate dehydrogenase from Candida boidinii (Cb‐FDH) and HCOONa. We performed a set of experiments in which the equivalents of HCOONa (i.e., the ultimate source of reducing equivalents) were gradually increased from 0 to 5. As depicted in Figure 2, only four equivalents of formate were required in order to reach full conversion of 1 a (20 mm). However, 20 % conversion of 1 a was observed even without any addition of HCOONa. Our hypothesis is that some endogenous enzymes from the lyophilised E. coli cells (10 mg mL−1) can somehow regenerate, in part, the NADH cofactor. If this background activity is taken into account, the remaining 80 % conversion of 1 a was driven by only 4 equivalents of HCOONa, corresponding to a remarkable estimated coupling efficiency of about 20 %. The product (S)‐2 a (Table 2, below) was obtained in enantiopure form (ee>99 %).
Figure 2.
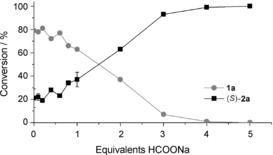
Conversion of 1 a (20 mm) into (S)‐2 a at various concentrations of HCOONa as final hydride donor. The bio‐transformations were performed in a biphasic system [KPi (pH 8, 50 mm)/n‐heptane (1:1, v/v, 1 mL total reaction volume)] containing E. coli/Fus‐SMO lyophilised cells (10 mg mL−1), NAD+ (1 mm), FAD (50 μm), HCOONa (0–100 mm) and Cb‐FDH (10 μm). The mixtures were incubated at 30 °C, 180 rpm for 24 h. Levels of conversion are the averages of two independent sets of experiments, both in duplicate (for a complete dataset including standard deviations see Section S6). The enantiomeric excess was determined by chiral HPLC to be >99 % S.
Table 2.
Upscaling for the biocatalytic synthesis of (S)‐2 a and (1S,2S)‐2 b by using lyophilised whole cells containing co‐expressed Fus‐SMO and Cb‐FDH (5 mg mL‐1); 200 mL total reaction volume (n‐heptane/KPi buffer 1:1, v/v).
| ||||||
|---|---|---|---|---|---|---|
| Substrate [mm] | Conversion [%] | Yield [%] | Purity [%] | ee [%] | de [%] | |
| 1 | 1 a (50) | >99 | 85 (+12)[a] | 99 | >99 | – |
| 2 | 1 b (50) | >99 | 90[b] | 95 | >99 | >98 |
[a] This isolated yield represents the amount of pure product that was isolated after simple evaporation of the n‐heptane as reaction phase. No further workup was required. The remaining amount of product (ca. 12 %) was recovered after extraction from the aqueous reaction phase. However, purity was lower (ca. 94 %), due to the generation of 2‐phenylethanol as a by‐product. [b] In this case, the purities of the isolated product obtained from the n‐heptane reaction phase and from the extraction of the aqueous phase were similar. Thus, the aliquots were combined and the total yield was reported.
Influence of the dioxygen pressure
Preliminary investigation has shown that the availability of dissolved molecular oxygen might become the limiting factor for the biocatalytic epoxidation by Fus‐SMO under particular reaction conditions (data not shown). It has been reported that the activity of some oxygenases can be strongly enhanced by using pure O2 in the headspace at atmospheric pressure or even under pressure.22 In order to assess the biocatalytic performance of our Fus‐SMO properly, we carried out a comparative study with the original bi‐enzymatic StyA/StyB construct (pSPZ10) developed by Panke and co‐workers.10a Expression of StyA/StyB with pSPZ10 plasmid was initially performed in E. coli JM101 according to the literature. We also tested expression in E. coli Arctic Express cells, because this strain is capable of improving expression of insoluble proteins such as StyB but was not commercially available at the time of Panke's study (Section S3.1). Although expression of soluble StyA was significantly superior in E. coli JM101 (Figure S1), the tests of conversion versus time with lyophilised cells of E. coli JM101 and E. coli Arctic Express provided similar results (Table S1, entry 3). This observation corroborates the assumption that the inefficient expression of soluble StyB in the bi‐enzymatic system is the limiting factor.
Finally, the rates of the epoxidation reaction in the presence of E. coli BL21(DE3)/Fus‐SMO (5 mg mL−1) and E. coli JM101/pSPZ10(StyA/StyB) (5 mg mL−1) were compared by measuring the levels of conversion after 20 min (linearity range for conversion vs. time) under the optimised reaction conditions, with styrene (50 mm) as substrate. Because the coupling efficiency of the two SMO systems is not perfect, formation of H2O2 during the reaction is to be expected. Hence, catalase (2 μm) was added. We conducted a set of experiments under air and O2 at atmospheric pressure (p rel=0 bar) as well as under pressurised O2 (p rel=1–4 bar). A pressurised closed system (Figure S5) might increase the concentration of O2 in the liquid phase and kinetically enhance the O2 transfer from the gas phase to the liquid phases.
Figure 3 shows that the addition of catalase had a significant influence on the rate of epoxidation. Independently of the composition and pressure of the gas phase, as well as of the type of SMO construct, reactions in the presence of catalase were accelerated. The chimeric Fus‐SMO system always performed better than the bi‐enzymatic StyA/StyB system in the presence of catalase at any composition and pressure of the gas phase. The maximum rate was obtained with Fus‐SMO with pure O2 at atmospheric pressure [(25±2) % conversion] with a productivity of 37.5 mm product h−1. Under the same conditions the bi‐enzymatic StyA/StyB gave a productivity of 25.5 mm product h−1. Supplying O2 under pressure was in general detrimental for the reaction. The influence of pressure in enzyme catalysis is still not a fully understood phenomenon.23 According to Le Châtelier's principle and the Eyring equation, an increase in pressure enhances the rates (k) of chemical reactions that have negative activation volumes (ΔV ≠).24 Typical ΔV ≠ values for enzymatic reactions are in the range of ±50 cm−3 mol−1, so the variation of the reaction rate constant (Δk) would be less than 1 % within the pressure range of our study (p abs=1–5 bar).23a, 25 On the other hand, variation of pressure can have a profound effect on enzyme structure and, therefore, activity. However, various studies have shown that significant structural changes in enzyme structure occur only at very high pressure, typically above 1 kbar.23a, 25a, 26 Hence, we can neglect both effects in our study because O2 was supplied at quite low pressure (p abs≤5 bar). Thus, we can speculate that the increased O2 pressure in our system might result in increased formation of FAD‐OOH, which cannot be entirely utilised for the epoxidation of styrene. Enhanced‐rate formation of FAD‐OOH would generate more H2O2, which would be deleterious for enzyme activity. The addition of catalase can only partially counteract this process as shown in Figure 3.
Figure 3.
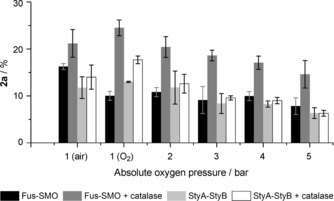
Conversion [%] of 1 a (50 mm) in the presence of lyophilised whole cells of E. coli BL21(DE3) expressing Fus‐SMO (5 mg mL−1) and E. coli JM101 expressing pSPZ10 (StyA/StyB, 5 mg mL−1). Reactions were carried out in glass vials introduced into a sealed pressurised chamber (Figure S5). The biotransformations were performed in a biphasic system [KPi (pH 8, 50 mm)/ n‐heptane (1:1, v/v, 1 mL total reaction volume)] containing NAD+ (1 mm), FAD (50 μm), HCOONa (250 mm, 5 equiv.) and Cb‐FDH (10 μm). Catalase (2 μm) was added in selected experiments. The mixtures were incubated at 30 °C, 200 rpm for 20 min. Two independent experiments were carried out, both in duplicate. Error bars represent the standard deviation.
Finally, the higher catalytic activity of the E. coli/Fus‐SMO cells relative to the E. coli cells expressing the bi‐enzymatic StyA/StyB was further confirmed in a study in which the conversion of styrene was monitored over time (10, 20, and 30 min). E. coli/Fus‐SMO cells showed about 50 % increased catalytic activity relative to E. coli cells expressing the bi‐enzymatic StyA/StyB (Figure S6 and Table S9).
Determination of the activity of purified Fus‐SMO and comparison with literature data for bi‐enzymatic StyA/StyB
Because the chimeric Fus‐SMO had been created with an N‐terminal His6‐tag, its purification was easily performed by Ni2+ affinity chromatography (Section S7.3.1). We carried out initial determinations of the enzymatic activity of Fus‐SMO for the epoxidation of 1 a at different pH values. Activity data showed negligible differences in the range between 6.5 and 9 (data not shown). Therefore, we selected Tris⋅HCl buffer (pH 8.5, 50 mm) for further determination. The determination of the epoxidation activity of Fus‐SMO was performed according to the general procedure reported by Otto et al.15 and Tischler et al.17 for the same experiment with bi‐enzymatic StyA/StyB. In this way, the new epoxidation activity data for Fus‐SMO can be compared with the data reported in the literature for the two‐component StyA/StyB system. Table 1 shows that the specific activities of Fus‐SMO and the StyA/StyB system are essentially identical for the epoxidation of styrene (1 a). However, Fus‐SMO showed a more than threefold increased activity relative to the StyA/StyB system for the epoxidation of para‐methylstyrene (1 c). We chose substrates 1 a and 1 c because data for the epoxidation activity for the two‐component StyA/StyB system were available in the literature. Besides the retained or improved epoxidation activity of Fus‐SMO, we point out another important aspect: the conditions for the maximum epoxidation activity of the two‐component StyA/StyB system are very complicated to reproduce in vitro and nearly impossible in a cell because a 1:1 mixture of StyA and StyB is required (also with consideration of the insolubility of StyB). In contrast, Fus‐SMO permits the highest rate to be set effortlessly, both in vitro and in a cell.
Table 1.
Specific activity for the epoxidation of 1 a and 1 c with Fus‐SMO (this study) in Tris⋅HCl buffer (pH 8.5, 50 mm), two‐component StyA/StyB (literature data) and natural fused StyA2B (literature data). For experimental details see Section S7.3.2.
| Sub. | Specific epoxidation activity [min−1] | |||
|---|---|---|---|---|
| Two‐component StyA/StyB | Natural fused StyA2B | Fus‐SMO[c] | ||
| 1 | 1 a | 97[a] | 1.3[b] | 95±5 |
| 2 | 1 c | 14[b] | 0.4[b] | 44±4 |
Finally, the activity of our artificial Fus‐SMO was about 70 and 110 times higher for 1 a and 1 c, respectively, than that of the naturally occurring fused SMO StyA2B.17
Self‐sufficient whole‐cell system for the epoxidation of styrene and derivatives
With the goal of enhancing the practical applicability, a whole‐cell system was created by simultaneously expressing Fus‐SMO and Cb‐FDH in E. coli BL21(DE3) as host organism. Under the optimised conditions for the co‐expression (i.e., 0.1 mm IPTG at 25 °C for 16 h), we observed expression of both enzymes in soluble form (Figure S9). Preliminary tests demonstrated that lyophilised whole cells containing co‐expressed Fus‐SMO and Cb‐FDH were highly active for the epoxidation of 1 a and 1 b (Table S10). It is known from the literature that the typical activity of Cb‐FDH for the recycling of NADH at the expense of formate is about 180 U μmolenzyme −1.27 Hence, cofactor regeneration is not the rate‐limiting step. Moreover, the perfect stereoselectivity of Fus‐SMO was retained [(S)‐2 a ee>99 %, (1S,2S)‐2 b ee>99 % and de>98 %]. We then monitored the progress of the conversion of 1 a and 1 b (50 mm) over time under the optimised reaction conditions (Figure 4 and Section S8.2). Lyophilised whole cells containing Fus‐SMO and Cb‐FDH (5 mg mL−1) had converted (27.1±1.2) % of 1 a at 50 mm scale within the first 30 min under air (Table S11). The epoxidation ran smoothly, reaching (93.8±1.3) % conversion after 6 h. Prolonging the reaction time permitted 99 % conversion to be achieved. The epoxidation of 1 b showed a similar trend, albeit it proceeded at a lower rate. This must be attributed to the lower intrinsic reactivity of SMO towards 1 b, as previously shown in Table 1. However, a maximum of (90.4±1.4) % conversion of (1S,2S)‐2 b was achieved (Table S11).
Figure 4.
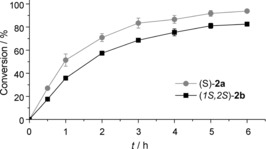
Progress of conversion over time for the epoxidation of 1 a and 1 b (50 mm) to give (S)‐2 a and (1S,2S)‐2 b, respectively, with the aid of lyophilised whole cells in which Fus‐SMO and Cb‐FDH were co‐expressed in E. coli BL21(DE3) (5 mg mL−1). Reactions were carried out in sealed 20 mL glass vials under air at atmospheric pressure. The biotransformations were performed in a biphasic KPi (pH 8, 50 mm)/n‐heptane (1:1, v/v, 1 mL total reaction volume) system containing NAD+ (1 mm), FAD (50 μm), HCOONa (250 mm, 5 equiv) and catalase (2 μm). Two independent experiments were carried out, both in duplicate. Error bars represent the standard deviation.
We also investigated the possibility of increasing the substrate concentration from 50 mm up to 1 m for the epoxidation of 1 a and 1 b on analytical scale (total volume 1 mL) in the presence of the E. coli BL21(DE3)/Fus‐SMO/Cb‐FDH (5 mg mL−1) co‐expressed system in 20 mL reaction vessels (for details see Section S8.3). The results from this study confirmed that nearly quantitative conversion can be achieved at 50 mm substrate concentration by applying these reaction conditions and technical set‐up. In fact, repetition of the epoxidation of 1 a and 1 b (50 mm) afforded the corresponding products with (97.7±1.3) % and (96.1±2.8) % conversion, respectively. Increasing the substrate concentration resulted in a progressive decrease in conversion (Table S12). The productivity of the system (i.e., mmol of epoxide product obtained) was partially influenced by the substrate concentration and depended on the substrate tested. The highest productivity for the epoxidation of 1 a was observed at 150 mm substrate concentration, leading to the formation of (64.8±1.5) mm (S)‐2 a (Table S12, entry 4). For the epoxidation of 1 b, the highest productivity was observed at 250 mm substrate, leading to the formation of (98.3±1.3) mm (1S,2S)‐2 b (Table S12, entry 7). In addition, we demonstrated that the productivity of the system—with this reaction set‐up—was not limited by the volume of the gas headspace. In fact, increasing the volume of the reaction vessels up to 100 mL did not improve conversions and productivities (for comparison, see Tables S13 and S14).
Finally, we investigated the influence of different modes of dioxygen supply on the rate of the biocatalytic epoxidation. Hence, experiments were conducted by application either of a sealed system with a large volume of air in the headspace or of a system with a continuous flow of pure dioxygen (bubbling at about 1 mL min−1). In both cases the other reaction parameters were the same: E. coli cells co‐expressing Fus‐SMO and Cb‐FDH (5 mg mL−1), styrene (50 mm), catalase (2 μm), NAD+ (1 mm), FAD (50 μm) and HCOONa (5 equiv) in a stirred mixture of aqueous buffer (KPi, pH 8, 25 mL) and n‐heptane (25 mL) in round‐bottomed flasks (for details see Section S8.4). The difference in the rate of formation of styrene oxide for the two systems was minimal (ca. 10 %), but with the system consisting of air in the headspace performing better (Figure S10).
In general, we conclude that sufficient dioxygen supply (irrespective of the mode) as well as efficient mixing of the biphasic reaction mixture are crucial parameters for sustaining elevated epoxidation rate for longer times.
Epoxidation on a preparative scale
Biocatalytic epoxidation with the use of lyophilised whole cells containing co‐expressed Fus‐SMO and Cb‐FDH was performed on preparative scale for the two substrates 1 a (50 mm, 521 mg) and 1 b (50 mm, 591 mg). The reactions were run in a biphasic KPi (pH 8, 50 mm)/n‐heptane (1:1, v/v, 200 mL total reaction volume) system under the optimised reaction conditions. A large reaction vessel was used in order to assure a sufficient supply of molecular oxygen for the reaction as well as an efficient mixing of the biphasic reaction mixture.
The epoxidation of 1 a afforded quantitative conversion (Table 2, entry 1). The organic phase was separated from the aqueous buffer and the n‐heptane was evaporated, affording 509 mg of (S)‐2 a (equal to 85 % isolated yield) of elevated chemical purity (99 % measured by GC‐FID) as well as optical purity (ee>99 %). Hence, further purification of (S)‐2 a obtained from the evaporation of the organic phase was not required. The remaining aliquot of product (S)‐2 a (about 12 %) was recovered upon extraction from the aqueous reaction phase. In this case, the purity was determined to be 94 %. The by‐product was 2‐phenylethanol, as reported in the literature for the natural StyA/StyB enzymatic system.10a, 10c, 11, 12, 14 The epoxidation of 1 b afforded the product (1S,2S)‐2 b (Table 2, entry 2) with a total of >99 % conversion. In this case, the purities of the product isolated from the n‐heptane reaction phase and from the extraction of the aqueous phase were similar (ca. 95 %). 603 mg of (1S,2S)‐2 b (90 % isolated yield) were obtained with elevated diasteromeric (de>98 %) and enantiomeric excess (ee>99 %).
Conclusion
We have created a chimeric styrene monooxygenase (SMO) in which the two enzymatic units (StyA and StyB) are joined through a flexible linker. The fused SMO allowed a few long‐standing problems relating to its application in chemical synthesis to be solved. Firstly, the Fus‐SMO was expressed mainly in soluble form, whereas recombinant, discrete StyB is almost completely insoluble. Secondly, the activities of StyA and StyB are now properly balanced because they are produced at an exact ratio of 1:1. Thirdly, the flexible linker forces StyB and StyA to remain close to each other in solution, hence facilitating their contact. Therefore, channelling transfer of FAD from StyB to StyA might be favoured over diffusive transfer. These properties permit wastage of reducing equivalents during the overall process to be minimised. Moreover, the rates of the biocatalytic epoxidation catalysed by Fus‐SMO were comparable or superior to the value reported in the literature for the two‐component StyA/StyB system. Further comparison with a naturally occurring fused SMO system revealed our artificial Fus‐SMO to be about two orders of magnitude more active. Finally, a recent study showed that another artificially fused SMO showed an epoxidation rate more than five times lower than that of our Fus‐SMO.19 The different behaviour could be attributed to the different types of linkers used for the fusion. In our work we have used a longer (i.e., 30 amino acid residues) and flexible (i.e., containing 70 % glycine) linker, whereas in ref. 18 the linkers were shorter (i.e., three to six amino acid residues) and more rigid (i.e., no glycine residues).
When considering the application of the whole‐cell system, we can conclude that E. coli cells expressing Fus‐SMO show higher epoxidation activity than E. coli cells expressing separated StyA and StyB, thanks to a combination of: 1) balanced and improved expression levels of reductase and epoxidase units, and 2) intrinsically higher specific epoxidation activity of Fus‐SMO.
Another important aspect for future applications in chemical synthesis and biotechnology is that the His‐6‐tagged Fus‐SMO can now be easily purified. Individual expression of StyA and StyB—and, moreover, tedious and low‐yielding refolding of StyB—are no longer required. Hence, Fus‐SMO can now also be applied as an isolated enzyme construct in solution.
Finally, the genes coding for the Fus‐SMO and the formate dehydrogenase (Cb‐FDH) were co‐expressed in E. coli and applied as a self‐sufficient system for the epoxidation, at more than 500 mg scale, of two model substrates: styrene and β‐methylstyrene. The epoxide products were isolated in elevated yields and in perfect diasteromerically and enantiomerically pure form. Hence, Fus‐SMO retained the exquisite stereoselectivity of the parent bi‐enzymatic system.
In summary, this work should open new opportunities in organic synthesis for the exploitation of the asymmetric biocatalytic epoxidation of styrene derivatives. Furthermore, the same concept might be extended to other multi‐enzymatic flavindependent systems in order to extend the substrate scope of the reaction beyond styrene derivatives.
Experimental Section
For general information, materials, and expression and purification of enzymes, see Section S2.
General optimised procedure for the biocatalytic synthesis of (S)‐2 a and (1S,2S)‐2 b by use of lyophilised whole cells coexpressing Fus‐SMO and Cb ‐FDH (analytical scale): Lyophilised whole cells (5 mg) were rehydrated in KPi buffer (pH 8, 50 mm, 0.5 mL) in 4 mL or 20 mL glass vials, containing NAD+ (1 mm), HCOONa (5 equiv), FAD (50 μm) and catalase (2 μm). n‐Heptane (0.5 mL) and the substrate 1 a or 1 b (50 mm) were added. The concentration of the substrate is calculated based on the volume of the organic phase. The concentrations of NAD+ and HCOONa are calculated on the volume of the aqueous phase. The mixture was incubated at 30 °C and 180 rpm in an orbital shaker. At the end of the reaction, the organic phase was separated from the aqueous phase. The aqueous phase was extracted with tert‐butyl methyl ether (MTBE, 2×250 μL). The combined organic phases were dried with MgSO4. The levels of conversion were measured by GC‐FID, whereas the ee and de values were measured by HPLC. For analytical details see Sections S9 and S10.
General optimised procedure for the biocatalytic synthesis of (S)‐2 a by use of whole cells co‐expressing Fus‐SMO and Cb ‐FDH (scaled up): Lyophilised whole cells containing co‐expressed Fus‐SMO and Cb‐FDH (500 mg, 5 mg mL−1) were rehydrated in KPi buffer (pH 8, 50 mm, 100 mL) in a 1 L tri‐baffled flask. NAD+ (1 mm), FAD (50 μm), HCOONa (5 equiv) and catalase (2 μm) were added. n‐Heptane (100 mL) and the substrate 1 a (50 mm, 521 mg, 5.00 mmol) were added. The concentration of cells, NAD+ and HCOONa are calculated based on the volume of the aqueous phase. The concentration of the substrate is calculated based on the volume of the organic phase. The reaction mixture was incubated at 30 °C and 200 rpm in an orbital shaker for 16 h. After completion of the reaction was confirmed by GC‐FID, the n‐heptane phase was recovered, dried with MgSO4, and concentrated under reduced pressure, yielding 509 mg (85 %) of (S)‐2 a (99 % purity by GC‐FID; ee>99 % by chiral HPLC). Separately, the aqueous phase was extracted with MTBE (2×50 mL), the organic layer was dried with MgSO4, and the solvent was evaporated under reduced pressure, affording the remaining 12 % of product with 94 % chemical purity. The product was characterised by 1H NMR. For details see Sections S9 and S10.
General optimised procedure for the biocatalytic synthesis of (1S,2S)‐2 b by use of lyophilised whole cells co‐expressing Fus‐SMO and Cb‐FDH (scaled up): A procedure similar to that reported above was performed. Lyophilised whole cells containing co‐expressed Fus‐SMO and Cb‐FDH (500 mg, 5 mg mL−1) were rehydrated in KPi buffer (pH 8, 50 mm, 100 mL) in a 1 L tri‐baffled flask. NAD+ (1 mm), FAD (50 μm), HCOONa (5 equiv) and catalase (2 μm) were added. n‐Heptane (100 mL) and substrate 1 b (50 mm, 591 mg, 5.00 mmol) were added. The concentrations of cells, NAD+ and HCOONa are calculated based on the volume of the aqueous phase. The concentration of the substrate is calculated based on the volume of the organic phase. The reactions were incubated at 30 °C and 200 rpm in an orbital shaker for 16 h. After completion of the reaction was confirmed by GC‐FID, the n‐heptane phase was recovered and concentrated under reduced pressure. The aqueous phase was then extracted with MTBE (2×50 mL). In this case, the purity of the product (1S,2S)‐2 b was similar in the n‐heptane phase and in the extracted MTBE phase. Thus, the organic phases were combined and dried with MgSO4, and the solvent was evaporated. The final yield was 603 mg of (1S,2S)‐2 b (90 % isolated yield, 95 % purity by GC‐FID; ee>99 % and de>98 % by chiral HPLC). The product was characterised by 1H NMR. For details see Sections S9 and S10.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
This work was financed by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO), grant ECHO Chemistry in Relation to Technology and Sustainability 2013 CW, project number 717.014.007. F.G.M. and T.K. received funding from the European Research Council (ERC) Starting Grant (H2020, grant agreement 638271, BioSusAmin). Dutch funding from the NWO Sector Plan for Physics and Chemistry is also acknowledged. We thank Prof. Panke for the kind donation of the plasmid pSPZ10‐(StyA/StyB).
M. L. Corrado, T. Knaus, F. G. Mutti, ChemBioChem 2018, 19, 679.
References
- 1.For reviews see:
- 1a. Farina V., Reeves J. T., Senanayake C. H., Song J. J., Chem. Rev. 2006, 106, 2734–2793; [DOI] [PubMed] [Google Scholar]
- 1b. Kumar P., Gupta P., Synlett 2009, 1367–1382; [Google Scholar]
- 1c. Hwang S., Choi C. Y., Lee E. Y., J. Ind. Eng. Chem. 2010, 16, 1–6; [Google Scholar]
- 1d. Hughes D. L., Org. Process Res. Dev. 2016, 20, 2028–2042; [Google Scholar]
- 1e. Luo M., Zhang X. H., Darensbourg D. J., Acc. Chem. Res. 2016, 49, 2209–2219; [DOI] [PubMed] [Google Scholar]
- 1f. Dalpozzo R., Lattanzi A., Pellissier H., Curr. Org. Chem. 2017, 21, 1143–1191; [Google Scholar]
- 1g. Chung W. J., Vanderwal C. D., Angew. Chem. Int. Ed. 2016, 55, 4396–4434; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 4470–4510; For recent papers see: [Google Scholar]
- 1h. Tak R., Kumar M., Menapara T., Gupta N., Kureshy R. I., Khan N.-u. H., Suresh E., Adv. Synth. Catal. 2017, 359, 3990–4001; [Google Scholar]
- 1i. El Assal M., Peixoto P. A., Coffinier R., Garnier T., Deffieux D., Miqueu K., Sotiropoulos J. M., Pouysegu L., Quideau S., J. Org. Chem. 2017, 82, 11816–11828; [DOI] [PubMed] [Google Scholar]
- 1j. Sharma A., Agarwal J., Peddinti R. K., Org. Biomol. Chem. 2017, 15, 1913–1920; [DOI] [PubMed] [Google Scholar]
- 1k. Radha Krishna P., Manikanta G., Nagaraju T., Synthesis 2016, 48, 4213–4220; [Google Scholar]
- 1l. Zhou Y., Yang P., Li S., Wang L., Yin J., Zhong J., Dong Y., Liu S., Wang M., Bian Q., Tetrahedron: Asymmetry 2017, 28, 338–343; [Google Scholar]
- 1m. Lienard P., Gradoz P., Greciet H., Jegham S., Legroux D., Org. Process Res. Dev. 2017, 21, 18–22; [Google Scholar]
- 1n. Gaikwad R. D., Kabiraj S. S., Bhat S. V., Flavour Fragrance J. 2016, 31, 350–355; [Google Scholar]
- 1o. Lee W., Kang S., Jung B., Lee H. S., Kang S. H., Chem. Commun. 2016, 52, 3536–3539; [DOI] [PubMed] [Google Scholar]
- 1p. Veerasamy N., Ghosh A., Li J., Watanabe K., Serrill J. D., Ishmael J. E., McPhail K. L., Carter R. G., J. Am. Chem. Soc. 2016, 138, 770–773; [DOI] [PubMed] [Google Scholar]
- 1q. García-Ruiz C., Cheng-Sanchez I., Sarabia F., Org. Lett. 2015, 17, 5558–5561; [DOI] [PubMed] [Google Scholar]
- 1r. McCarthy J. R., Tetrahedron Lett. 2015, 56, 6846–6847; [Google Scholar]
- 1s. Lalwani K. G., Sudalai A., Eur. J. Org. Chem. 2015, 7344–7351; [Google Scholar]
- 1t. Rodriguez A. R., Spur B. W., Tetrahedron Lett. 2015, 56, 5811–5815; [Google Scholar]
- 1u. Das S., Ramana C. V., Tetrahedron 2015, 71, 8577–8584; [Google Scholar]
- 1v. Ogata T., Tanaka M., Ishigaki M., Shimizu M., Nishiuchi A., Inamoto K., Kimachi T., Tetrahedron 2015, 71, 6672–6680. [Google Scholar]
- 2. Katsuki T., Sharpless K. B., J. Am. Chem. Soc. 1980, 102, 5974–5976. [Google Scholar]
- 3. Zhang W., Loebach J. L., Wilson S. R., Jacobsen E. N., J. Am. Chem. Soc. 1990, 112, 2801–2803. [Google Scholar]
- 4. Tokunaga M., Larrow J. F., Kakiuchi F., Jacobsen E. N., Science 1997, 277, 936–938. [DOI] [PubMed] [Google Scholar]
- 5.
- 5a. Spelberg J. H. Lutje, de Vries E. J. in Enzyme Catalysis in Organic Synthesis, 3rd ed. (Eds.: K. Drauz, H. Groeger, O. May), Wiley-VCH, Weinheim, 2012, pp. 363–416; [Google Scholar]
- 5b. Lin H., Liu J.-Y., Wang H.-B., Ahmed A. A. Q., Wu Z.-L., J. Mol. Catal. B 2011, 72, 77–89; [Google Scholar]
- 5c. Yildirim D., Tukel S. S., Alagoz D., Alptekin O., Enzyme Microb. Technol. 2011, 49, 555–559; [DOI] [PubMed] [Google Scholar]
- 5d. Choi W. J., Appl. Microbiol. Biotechnol. 2009, 84, 239–247; [DOI] [PubMed] [Google Scholar]
- 5e.M. P. Kamble, G. D. Yadav, Catal. Today 2017, DOI: https://dx.doi.org/10.1016/j.cattod.2017.06.013.
- 6.
- 6a. Fingerhut A., Serdyuk O. V., Tsogoeva S. B., Green Chem. 2015, 17, 2042–2058; [Google Scholar]
- 6b. Gelalcha F. G., Bitterlich B., Anilkumar G., Tse M. K., Beller M., Angew. Chem. Int. Ed. 2007, 46, 7293–7296; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2007, 119, 7431–7435; [Google Scholar]
- 6c. Collman J. P., Wang Z., Straumanis A., Quelquejeu M., Rose E., J. Am. Chem. Soc. 1999, 121, 460–461. [Google Scholar]
- 7.
- 7a. Davis R. L., Stiller J., Naicker T., Jiang H., Jorgensen K. A., Angew. Chem. Int. Ed. 2014, 53, 7406–7426; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 7534–7556; [Google Scholar]
- 7b. Zhu Y., Wang Q., Cornwall R. G., Shi Y., Chem. Rev. 2014, 114, 8199–8256; [DOI] [PubMed] [Google Scholar]
- 7c. Wong O. A., Ramirez T. A., Shi Y. in Comprehensive Chirality, Vol. 6 (Eds.: E. M. Carreira, H. Yamamoto), Elsevier, 2012, pp. 528–553; [Google Scholar]
- 7d. Xia Q. H., Ge H. Q., Ye C. P., Liu Z. M., Su K. X., Chem. Rev. 2005, 105, 1603–1662; [DOI] [PubMed] [Google Scholar]
- 7e. Shi Y., Acc. Chem. Res. 2004, 37, 488–496; [DOI] [PubMed] [Google Scholar]
- 7f. Wang Z.-X., Tu Y., Frohn M., Zhang J.-R., Shi Y., J. Am. Chem. Soc. 1997, 119, 11224–11235. [Google Scholar]
- 8.
- 8a. Ottenbacher R. V., Talsi E. P., Bryliakov K. P., Catal. Today 2016, 278, 30–39; [Google Scholar]
- 8b. Day D. P., Sellars P. B., Eur. J. Org. Chem. 2017, 1034–1044; [Google Scholar]
- 8c. Irie R., Uchida T., Matsumoto K., Chem. Lett. 2015, 44, 1268–1283; [Google Scholar]
- 8d. Burke A. J., Carreiro E. P. in Comprehensive Inorganic Chemistry II (Eds.: J. Reedijk, K. Poeppelmeier), Elsevier, 2013, pp. 309–382; [Google Scholar]
- 8e. Gelalcha F. G., Adv. Synth. Catal. 2014, 356, 261–299; [Google Scholar]
- 8f. Shi Q.-P., Shi Z.-H., Li N.-G., Tang Y.-P., Li W., Tang H., Wei Z., Shen M.-Z., Duan J.-A., Curr. Org. Chem. 2013, 17, 2936–2970; [Google Scholar]
- 8g. Matsumoto K., Sawada Y., Katsuki T., Pure Appl. Chem. 2008, 80, 1071–1077; [Google Scholar]
- 8h. Chatterjee D., Coord. Chem. Rev. 2008, 252, 176–198; [Google Scholar]
- 8i. Li Z., Yamamoto H., Acc. Chem. Res. 2013, 46, 506–518; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8j. Brandes B. D., Jacobsen E. N., Tetrahedron: Asymmetry 1997, 8, 3927–3933; [Google Scholar]
- 8k. Palucki M., McCormick G. J., Jacobsen E. N., Tetrahedron Lett. 1995, 36, 5457–5460. [Google Scholar]
- 9. Li A. T., Li Z. in Science of Synthesis Biocatalysis in Organic Synthesis 2 (Eds.: K. Faber, W.-D. Fessner, N. J. Turner), Thieme, Stuttgart, 2015, pp. 479–505. [Google Scholar]
- 10.
- 10a. Panke S., Wubbolts M. G., Schmid A., Witholt B., Biotechnol. Bioeng. 2000, 69, 91–100; [DOI] [PubMed] [Google Scholar]
- 10b. Panke S., Held M., Wubbolts M. G., Witholt B., Schmid A., Biotechnol. Bioeng. 2002, 80, 33–41; [DOI] [PubMed] [Google Scholar]
- 10c. Park J. B., Buhler B., Habicher T., Hauer B., Panke S., Witholt B., Schmid A., Biotechnol. Bioeng. 2006, 95, 501–512; [DOI] [PubMed] [Google Scholar]
- 10d. Schmid A., Hofstetter K., Feiten H.-J., Hollmann F., Witholt B., Adv. Synth. Catal. 2001, 343, 732–737. [Google Scholar]
- 11. Hofstetter K., Lutz J., Lang I., Witholt B., Schmid A., Angew. Chem. Int. Ed. 2004, 43, 2163–2166; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2004, 116, 2215–2218. [Google Scholar]
- 12. Kuhn D., Kholiq M. A., Heinzle E., Bühler B., Schmid A., Green Chem. 2010, 12, 815. [Google Scholar]
- 13.
- 13a. Xu Y., Jia X., Panke S., Li Z., Chem. Commun. 2009, 1481–1483; [DOI] [PubMed] [Google Scholar]
- 13b. Wu S., Chen Y., Xu Y., Li A., Xu Q., Glieder A., Li Z., ACS Catal. 2014, 4, 409–420; [Google Scholar]
- 13c. Wu S., Zhou Y., Wang T., Too H. P., Wang D. I., Li Z., Nat. Commun. 2016, 7, 11917; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13d. Zhou Y., Wu S., Li Z., Angew. Chem. Int. Ed. 2016, 55, 11647–11650; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 11819–11822. [Google Scholar]
- 14. Panke S., Witholt B., Schmid A., Wubbolts M. G., Appl. Environ. Microbiol. 1998, 64, 2032–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Otto K., Hofstetter K., Rothlisberger M., Witholt B., Schmid A., J. Bacteriol. 2004, 186, 5292–5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.
- 16a. Kantz A., Chin F., Nallamothu N., Nguyen T., Gassner G. T., Arch. Biochem. Biophys. 2005, 442, 102–116; [DOI] [PubMed] [Google Scholar]
- 16b. Morrison E., Kantz A., Gassner G. T., Sazinsky M. H., Biochemistry 2013, 52, 6063–6075; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16c. Tischler D., Kermer R., Groning J. A., Kaschabek S. R., van Berkel W. J., Schlomann M., J. Bacteriol. 2010, 192, 5220–5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tischler D., Eulberg D., Lakner S., Kaschabek S. R., van Berkel W. J., Schlomann M., J. Bacteriol. 2009, 191, 4996–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.
- 18a. Hollmann F., Lin P. C., Witholt B., Schmid A., J. Am. Chem. Soc. 2003, 125, 8209–8217; [DOI] [PubMed] [Google Scholar]
- 18b. Hollmann F., Hofstetter K., Habicher T., Hauer B., Schmid A., J. Am. Chem. Soc. 2005, 127, 6540–6541; [DOI] [PubMed] [Google Scholar]
- 18c. Ruinatscha R., Dusny C., Buehler K., Schmid A., Adv. Synth. Catal. 2009, 351, 2505–2515. [Google Scholar]
- 19. Heine T., Tucker K., Okonkwo N., Assefa B., Conrad C., Scholtissek A., Schlomann M., Gassner G., Tischler D., Appl. Biochem. Biotechnol. 2017, 181, 1590. [DOI] [PubMed] [Google Scholar]
- 20. Chen X., Zaro J. L., Shen W. C., Adv. Drug Delivery Rev. 2013, 65, 1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.
- 21a. Bae J. W., Park M., Jeong Y. J., Park S., Lee S.-G., J. Ind. Eng. Chem. 2009, 15, 520–523; [Google Scholar]
- 21b. O'Connor K. E., Dobson A. D. W., Hartmans S., Appl. Environ. Microbiol. 1997, 4287–4291; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21c. Berry A., Dodge T. C., Pepsin M., Weyler W., J. Ind. Microbiol. Biotechnol. 2002, 28, 127–133. [DOI] [PubMed] [Google Scholar]
- 22.
- 22a. Lara M., Mutti F. G., Glueck S. M., Kroutil W., Eur. J. Org. Chem. 2008, 3668–3672; [Google Scholar]
- 22b. Rajagopalan A., Schober M., Emmerstorfer A., Hammerer L., Migglautsch A., Seisser B., Glueck S. M., Niehaus F., Eck J., Pichler H., Gruber K., Kroutil W., ChemBioChem 2013, 14, 2427–2430. [DOI] [PubMed] [Google Scholar]
- 23.
- 23a. Eisenmenger M. J., Reyes-De-Corcuera J. I., Enzyme Microb. Technol. 2009, 45, 331–347; [Google Scholar]
- 23b. Chakraborty S., Kaushik N., Rao P. S., Mishra H. N., Compr. Rev. Food Sci. Food Saf. 2014, 13, 578–596; [DOI] [PubMed] [Google Scholar]
- 23c. Hay S., Sutcliffe M. J., Scrutton N. S., Proc. Natl. Acad. Sci. USA 2007, 104, 507–512; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23d. Hay S., Scrutton N. S., Nat. Chem. 2012, 4, 161–168. [DOI] [PubMed] [Google Scholar]
- 24.Le Châtelier's principle: “At equilibrium a system tends to minimize the effect of any external factor by which it is perturbed. Consequently, elevated pressures favor changes that reduce a system's overall volume”. The Eyring equation: (∂ln k/∂p)T=−ΔV ≠/RT.
- 25.
- 25a. Mozhaev V. V., Heremans K., Frank J., Masson P., Balny C., Proteins Struct. Funct. Bioinf. 1996, 24, 81–91; [DOI] [PubMed] [Google Scholar]
- 25b. Northrop D. B., Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 2002, 1595, 71–79; [DOI] [PubMed] [Google Scholar]
- 25c. Groß M., Auerbach G., Jaenicke R., FEBS Lett. 1993, 321, 256–260. [DOI] [PubMed] [Google Scholar]
- 26.
- 26a. Dirix C., Duvetter T., Loey A. V., Hendrickx M., Heremans K., Biochem. J. 2005, 392, 565–571; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26b. Knez Z., Laudani C. G., Habulin M., Reverchon E., Biotechnol. Bioeng. 2007, 97, 1366–1375. [DOI] [PubMed] [Google Scholar]
- 27. Schutte H., Flossdorf J., Sahm H., Kula M.-R., Eur. J. Biochem. 1976, 62, 151–160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


