Abstract
Sea turtles are distributed in tropical and subtropical seas worldwide. They play several ecological roles and are considered important indicators of the health of marine ecosystems. Studying epibiotic diatoms living on turtle shells suggestively has great potential in the study of turtle behavior because diatoms are always there. However, diatom identification at the species level is time consuming, requires well-trained specialists, and there is a high probability of finding new taxa growing on turtle shells, which makes identification tricky. An alternative approach based on DNA barcoding and high throughput sequencing (HTS), metabarcoding, has been developed in recent years to identify species at the community level by using a DNA reference library. The suitabilities of morphological and molecular approaches were compared. Diatom assemblages were sampled from seven juvenile green turtles (Chelonia mydas) from Mayotte Island, France. The structures of the epibiotic diatom assemblages differed between both approaches. This resulted in different clustering of the turtles based on their diatom communities. Metabarcoding allowed better discrimination between turtles based on their epibiotic diatom assemblages and put into evidence the presence of a cryptic diatom diversity. Microscopy, for its part, provided more ecological information of sea turtles based on historical bibliographical data and the abundances of ecological guilds of the diatom species present in the samples. This study shows the complementary nature of these two methods for studying turtle behavior.
Introduction
Sea turtles play several ecological roles such as maintaining the health of seagrass beds and coral reefs; they also provide habitats for marine organisms, aid in maintaining balanced marine food webs, and promote nutrient cycling from marine to terrestrial ecosystems [1–4]. They are considered important indicators of the health of marine ecosystems [2, 5, 6]. In spite of this, turtle populations have declined significantly as consequence of human activities (e.g., habitat destruction, ocean pollution, poaching, and fishing) [7] and climate change [8]. Nowadays, six of the seven existing species of sea turtles are classified as threatened or endangered by the International Union for Conservation of Nature. Understanding sea turtle behavior (e.g., migration, feeding, and reproduction patterns) is important for their conservation and management [9, 10]. Several methods, such as aerial survey [10–14], snorkeling survey [14, 15] and telemetry [16] (e.g., very high frequency telemetry, sonic telemetry and satellite telemetry), have been used to study and monitor the population and distribution of sea turtles. Data acquisition systems such as GPS tracking, geolocating tags, time-temperature-depth recorders, and heart rate counters were also been used to monitor their behavior and physiology [16–20]. Another method suggestively having great potential for studying turtle behavior is to study the epibiota on shell turtles. Most of these studies have focused on macro-epibiota but, since the 70s (e.g., [21–25]), micro-epibiota has also received attention and is now the subject of growing interest given the diversity of organisms that live on(e.g., [26–28]). Among the micro-epibiotic organisms, diatoms (Bacillariophyta) receive the most attention because of their diversity and density on turtle shells [29–31]. Diatoms are a clade of microalgae ranging in size from a few to several hundreds of micrometers. Their most remarkable feature is their intricately ornamented siliceous exoskeleton, called frustule, which is used to characterize the various species, of which there is an estimated 100,000 species [32]. Recently, several new species have been described from marine [33–38] and freshwater [39–41] turtle shells. The capacity for diatom assemblages to change their species composition with changes in environmental conditions such as water turbulence [42], light intensity [43], and nutrient levels [44] is one of their characteristics.
These properties and their ubiquity make diatoms excellent ecological indicators. They are used worldwide to evaluate the ecological quality of rivers and lakes [44] and, more recently, of costal marine environments (e.g., [45]). They are ever-present on turtle shells [28]. Moreover, the species composition of epibiotic diatoms from freshwater turtles of the same species can differ based on turtle activities and their region of origin [46]. Similar differences have been observed with marine turtles [27]. Nevertheless, such differences were not observed between freshwater turtles (Emys orbicularis) coming from various ponds in South France (e.g., [47]) nor for certain marine turtles (Lepidochelys olivacea, [28]). Nevertheless, some authors acknowledged that their results should be confirmed [28]. For all these reasons, several researches have increasing diatom potential for studying turtle behavior [28].
Even if diatoms are excellent ecological indicators, the identification of individuals at the species level relies on the morphological criteria of their siliceous exoskeletons. This is time consuming, requires well-trained specialists spending hours under the microscope to establish floristic lists (usually a floristic list is based on 400 identified exoskeletons), and difficulties often appear when differentiating between morphologically near species (e.g., [48, 49]). Moreover, when working on diatom samples such as those of turtles shells, it is highly probable that new taxa to science will be observed, increasing the difficulty in identifying diatoms. In recent years, an alternative approach for identifying diatom species in environmental samples based on DNA has been developed. Metabarcoding [50] uses molecular techniques at the community level by combining DNA barcoding [51] with high throughput sequencing (HTS). DNA barcoding allows an accurate identification of an organism to species level from a short DNA fragment while HTS allows millions of DNA fragments from many samples to be sequenced simultaneously. Sequencing data is then used to obtain an accurate identification of diatom taxa at the species level by comparing to a DNA reference library. Several studies have already shown that metabarcoding has the potential to identify diatoms in freshwater [52–54]. The advantage of metabarcoding over microscopy in the identification of diatom samples is this high throughput.
The aim of this study was to determine whether metabarcoding carried out on epibiotic diatoms from sea turtle shells provide the same information about turtle behavior as does microscopy. In particular, we formed the following hypothesis:
Based on the composition of epibiotic diatom assemblages, turtles can be regrouped in a similar way using either microscopy or metabarcoding. If this hypothesis is correct, what information about the turtle behavior can epibiotic diatoms bring? If it is not correct, several additional questions are raised. Do both methods taxonomically identify the epibiotic diatoms with the same accuracy? Are diatom assemblages obtained with microscopy the same as those obtained with metabarcoding? Does the two methods discriminate epibiotic diatom assemblages equally well? Is there a cryptic diversity (which is difficult to observe in microscopy) that explains differences between microscopy and metabarcoding analyses?
Such questions were addressed for a population of seven juvenile green sea turtles, Chelonia mydas, feeding on a seagrass bed from the third largest coral atoll of the world, which is situated near the French Island Mayotte (Comoros archipelago) in the Northern Mozambique Channel.
Materials and methods
Sampling
Epibiotic samples used in this study were collected on 12 October 2015 from seven living green turtles Chelonia mydas at N'Gouja Bay in the Marine Nature Park of Mayotte, France (12°57’ 43.05” S, 45°5’ 6.49 E) (Fig 1). The entire shell of the turtle was scraped off using a clean toothbrush. This protocol was non-invasive and was limited to the external part of the turtle shell, and it does not harm or cause the animal suffering. All the procedures involved respect the ethical standards in the Helsinki Declaration of 1975, as revised in 2000 and 2008, as well as the applicable national law. All sampling procedures were carried out by A. Carpentier who was authorized by the Mayotte Prefecture to capture turtles and sample the epibiotic organisms.
Fig 1. Location of Mayotte Island (France) in the south west Indian Ocean.
The black square indicates where the Chelonia mydas were sampled.
Samples were fixed in ethanol (70% final concentration) according to European protocols [55] and kept cold (4–7°C) until molecular and microscopic treatments. Tag and physical details for each turtle are presented in Table 1.
Table 1. Details of the turtles sampled for this study.
| Turtle | Name | Age class | Tag left fin | Tag right fin | CCL [cm] |
SCL [cm] |
Weight [kg] |
|---|---|---|---|---|---|---|---|
| 1 | Zorro | Juvenile | yt5990A | yt4033B | 71.0 | 66.0 | 41.40 |
| 2 | Efe | Juvenile | yt4016B | yt5901A | 62.5 | 57.0 | 29.18 |
| 3 | Lost | Juvenile | yt4015B | yt5949A | 56.0 | 52.0 | 18.91 |
| 4 | Uhu | Juvenile | yt4019B | yt5943A | 58.0 | 54.0 | 21.96 |
| 5 | Mabawa | Juvenile | yt4020B | yt5945A | 74.0 | 68.0 | 45.40 |
| 6 | Digueline | Juvenile | yt4032B | yt5950A | 74.5 | 68.5 | 48.00 |
| 7 | Bertha | Juvenile | yt4021B | — | 73.5 | 69.0 | 46.26 |
CCL, curved carapace length; SCL, straight carapace length
Light and scanning electron microscopic analyses
Samples were cleaned with 40% H2O2 and HCl according to European standard EN 13946 [56]. After repeated rinsing and decantation with distilled water, air-dried aliquots were mounted on permanent glass slides using Naphrax®. At least 400 valves were identified and counted under the light microscope at a magnification of 1000X using a Zeiss Axio Imager A1® microscope according to European standard EN 14407 [57]. Taxonomic identifications were performed based on the specific marine diatom floras of [58–60], and also on the flora of [61]. Other papers dealing on marine species were also used (e.g., [62–64]). A list of the taxa and their relative abundances was produced for each of the 7 samples.
Scanning electron microscopic examinations were carried out on an ultrahigh-resolution analytical field emission (FE) scanning electron microscope Hitachi® SU–70 (Hitachi High-Technologies Corporation, Japan) using an accelerated voltage of 5 kV. For these examinations, cleaned oxidized samples were concentrated on a polycarbonate membrane filter with a 3-μm mesh, attached to aluminum stubs, and sputtered with a 30-nm platinum layer.
In order to answer the question about the cryptic diversity of Nitzschia inconspicua Grunow, light and scanning electron microscopic photos were taken.
Molecular analysis
DNA extraction, PCR amplification, sample libraries preparation, and HTS followed the technical specifications given in [65]. Briefly, DNA extraction was based on Sigma-Aldrich GenElute™-LPA DNA precipitation, and PCR amplification was performed on the rbcL plastid gene, targeting a 312 bp barcode. To amplify the entire diatom diversity, equimolar mixes of 3 forward and 2 reverse primers were used as described previously [66]. For each DNA sample, PCR amplification was carried out in triplicate. To prepare sample libraries, the PCR products of each triplicate were pooled. After cleaning and checking for DNA purity and quantity, tags were added to each amplicon. Library preparation was performed as described in [65]. Libraries were sequenced on a PGM Ion Torrent machine by the “Plateforme Génome Transcriptome” (PGTB, Bordeaux, France) using the Ion 318™Chip Kit V2 (Life Technologies, Carlsbad, USA) on 7 December 2016.
Bioinformatic processing was performed according to the technical specifications given in [65]. In brief, after several steps of quality filtering, DNA reads were clustered into Operational Taxonomical Units (OTUs) using a distance similarity threshold of 95%. For each sample, a list of OTUs and their numbers of reads were obtained. Taxonomical identifications of the OTUs were obtained using the R-Syst::diatom library [67] (R-Syst::diatom v4, of 16-09-2015, http://www.rsyst.inra.fr/en).
Statistical analyses
Statistical analyses were carried out using XLSTAT version 2011.4.04 AddinsoftTM.
To test our hypothesis that based on the composition of epibiotic diatom assemblages, turtles can be regrouped similarly with microscopy and metabarcoding, a K-means clustering (using Bray-Curtis distances) was applied on the seven species lists (one for each sample), obtained using microscopy. The same was applied on the seven OTU lists, obtained using metabarcoding. For this statistical analysis (K-means), and those that followed, the OTUs and species were expressed in relative abundances (in percentages) for each sample. Given the number of samples, 3 groups were defined. Moreover, to visualize the epibiotic diatom assemblages, NMDS based on Bray-Curtis distances were drawn using XLSTAT software.
To determine whether the two methods discriminate epibiotic diatom assemblages equally well, a Student t test was performed to compare the Bray-Curtis distances calculated for the diatom assemblages obtained using microscopy with those obtained using metabarcoding.
To determine whether structure of diatom assemblages obtained using either method is the same, distance matrices (Bray-Curtis distances) were calculated between the species list (obtained using microscopy) and the OTU list (obtained using metabarcoding) for each sample. A Mantel test was then used to test the correlation between matrices.
If these three tests (K-means clustering, comparisons of Bray-Curtis distances, and the Mantel test) would show that both microscopical and molecular methods gave different results, then tests for cryptic diversity would be necessary. To this end, one of the most abundant species will be selected for carrying out further tests. A phylogeny will be calculated using reference sequences from R-Syst::diatom [66]. Then OTUs sequences corresponding to this species will be constrained by this phylogeny (see next section for detailed explanations), and co-occurrence in the samples of these OTUs will be tested. Correlations (Spearman coefficients) between the abundance of each OTUs in each samples would be calculated. If not significant, morphologic cryptic diversity can be argued, and metabarcoding can be used to detect cryptic diversity, whereas microscopy cannot. This would explain the differences between microscopy and metabarcoding in terms of community structure.
Phylogenetic analyses
To visualize the phylogenetic position of the 100 most abundant OTUs in the diatom phylogeny, a constraint phylogeny was carried out as explained in Rimet et al. [68]. Briefly, turtles OTU sequences were aligned with all the rbcL sequences in the R-Syst::diatom library using Muscle [69] in Seaview [70]. The best substitution model was then tested in MEGA7 [71]. A constraint phylogeny was then calculated in raxmlGUI [72] using the fast tree search option. The constraint sequences were the turtle OTUs sequences and the shortest sequences of R-Syst::diatom library. A tree was drawn using the online tool iTOL [73] (https://itol.embl.de).
A second phylogeny was carried out on Nitzschia inconspicua sequences from the R-Syst::diatom library and their neighbor OTU in the raxmlGUI software using the maximum likelihood and the thorough bootstrap option (see former section). A phylogeny was drawn using Mega7 [71].
Results
Microscopic analyses
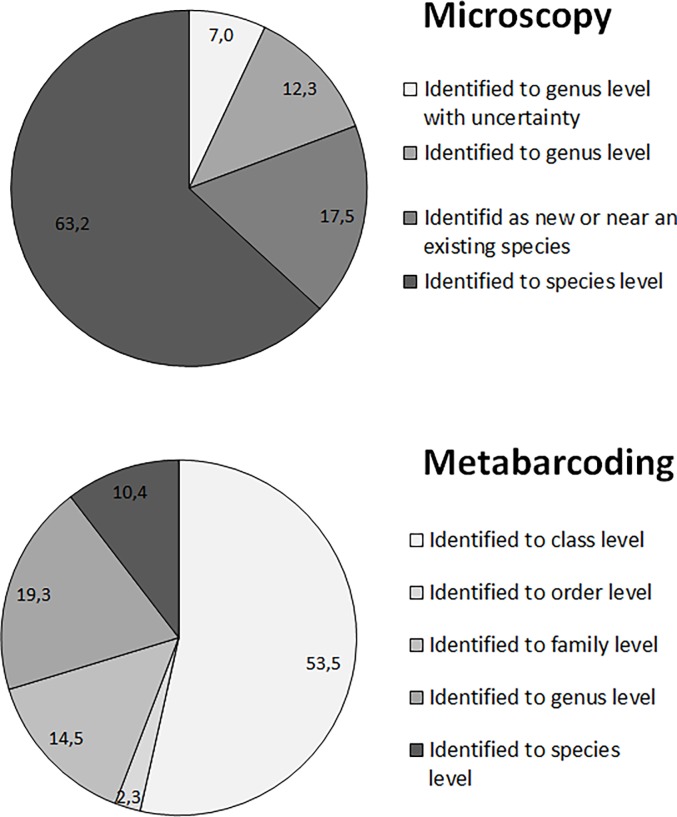
In total, 57 taxa were identified. The number of taxa per sample ranged from 15 to 24 with an average of 20 taxa per turtle. The most abundant taxon (average abundance across all the samples: 51%) was a small Labellicula (4 to 6 μm long and 1–1.5 μm wide) corresponding to the recently-described L. lecohuiana Majewska & Van de Vijver from sea turtles in Costa Rica. For this species, identification was possible only through electron microscopy because almost no visible features were observable using light microscopy. The second-most abundant species (average abundance across all samples: 14%) was Nitzschia inconspicua Grunow, a worldewide spread benthic euryhaline species. The third was Halamphora tenerrima (Aleem & Hustedt) Levkov (6%), also a widely distributed benthic marine species. For most (63%), species-level identification was reached (Fig 2), but an important part (37%) could not be identified precisely. Many of these taxa might be new to science.
Fig 2. Identification levels reached by microscopy and by metabarcoding.
Percentages of the identified taxa through microscopy and of the OTUs identified using R-Syst::diatom v4 (version of 16-09-2015) are given in the pie charts, respectively.
Fig 3 presents light and scanning electron microscopy photos of Nitzschia inconspicua from several turtles. The morphology of this species is homogeneous across all seven turtles.
Fig 3. Light and scanning electron microscopy images of Nitzschia inconspicua from each turtle.
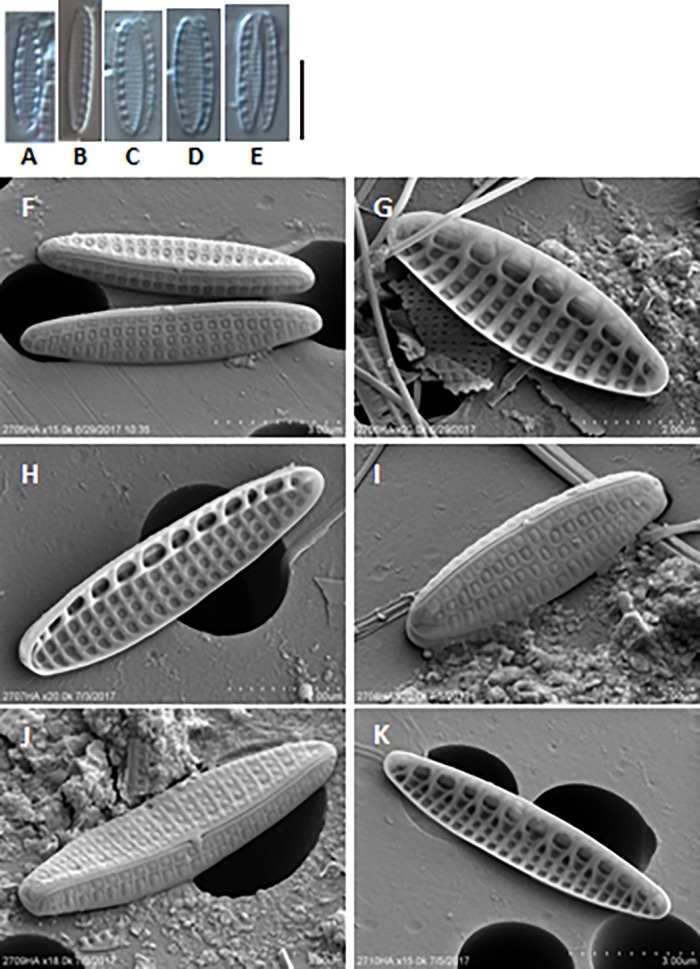
A―E. Light microscopy. A. Turtle 1. B. Turtle 2. C―D. Turtle 5. E. Turtle 6.F―K. Scanning electron microscopy. F. Turtle 1. G. Turtle 2. H. Turtle 3. I. Turtle 4. J. Turtle 5. K. Turtle 6. Scale bar: 10 μm.
Metabarcoding analyses
In total, 510,922 reads were obtained, averaging 72,989 reads per sample. After the various curation steps (i.e., length, quality, chimera, and alignment), 209,095 reads were conserved, averaging 29,871 reads per sample. Clustering reads at the 95% level resulted in 634 OTUs (range, 231–360 OTUs per sample; mean OTU in a sample, 280).
Taxonomic assignment of the OTUs using the R-Syst::diatom database v4 (16-09-2015) resulted in the identification of 19 species and 26 genera. Only 10% of the 634 OTUs could be assigned at the species level, 19% at the genus level, and 14,5% at the family level (Fig 2). More than 50% were identified at the class level only (diatoms are composed by 4 classes: Bacillariophyceae, Coscinodiscophyceae, Fragilariophyceae, and Mediophyceae).
Statistical analyses
K-means clustering and NMDS were carried out on the species lists established using microscopy and on the OTU lists established using metabarcoding. Cluster results (Fig 4) differed between both methods; therefore, we must reject our initial hypothesis and concluded that, based on the composition of epibiotic diatom assemblages, turtles cannot be regrouped similarly using microscopy versus metabarcoding.
Fig 4. NMDS ordination plots and cluster representations.
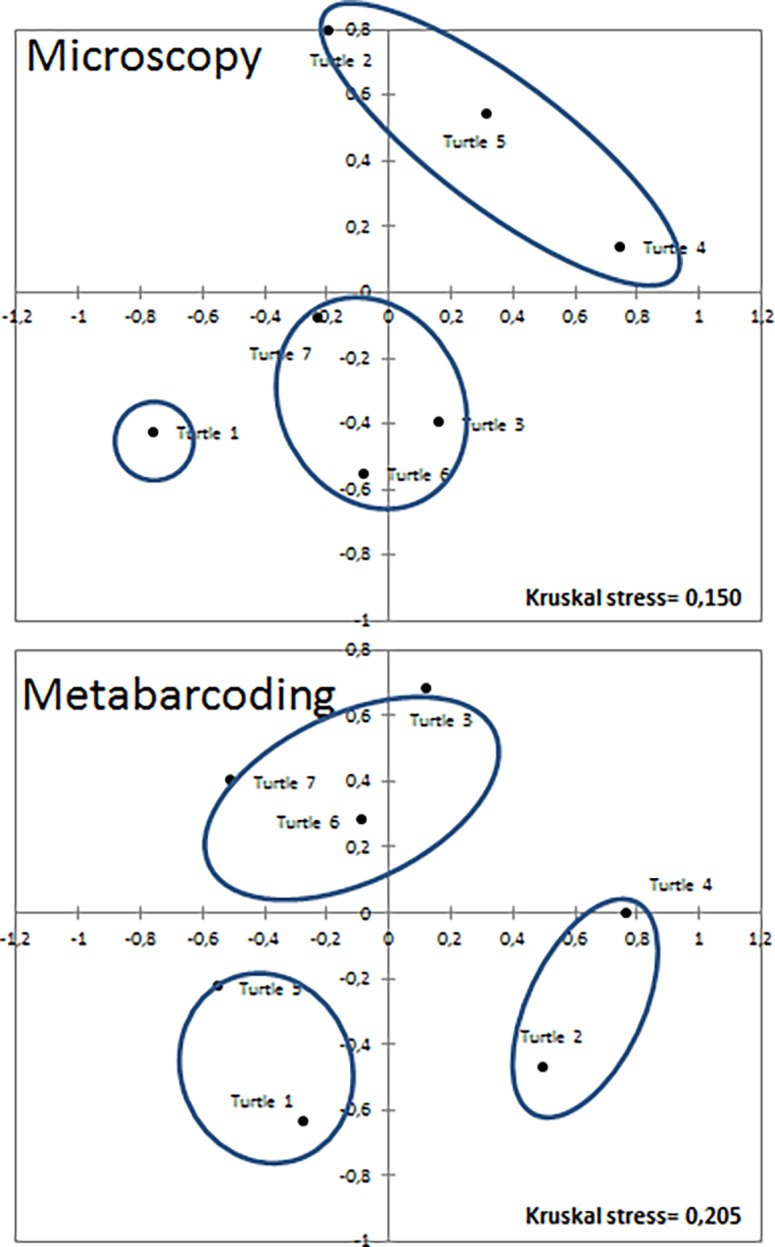
K-means and NMDSs were calculated based on the Bray-Curtis distances of the species lists and the OTU lists (each expressed in percentages) obtained using microscopy and metabarcoding, respectively.
The Mantel test carried out between the Bray-Curtis distance matrix of the microscopic species lists and the Bray-Curtis distance matrix of the OTUs lists was non-significant (p-value: 0.08), even though the distances matrices tented to be correlated (r2 = 15%). Based on these results, the structure of epibiotic diatom is not revealed in the same way by microscopy and by metabarcoding. Moreover, the Bray-Curtis distances revealed that we metabarcoding is significantly better than microscopy for discriminating diatom communities (0.55 vs. 0.27, p-value < 0.001, Student t test).
Phylogenetic analyses
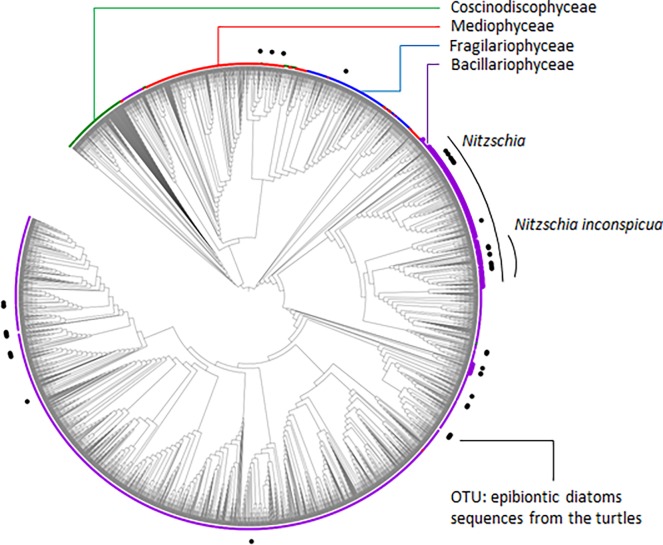
A constraint phylogeny was calculated using the fast tree search option in RaxML (Fig 5). In total, 2859 rbcL sequences were used in the alignment, and 813 were constrained among which the 100 most abundant OTU of the turtles samples. Phylogeny was carried out on 1532 nucleotides, using a GammaGTR substitution matrix (GAMMA rate heterogeneity model, ML estimate of the alpha-parameter). Most identified OTU were from the Bacillariophyceae family or of the Nitzschia genus. The species encompassing the highest number of OTUs was Nitzschia inconspicua (sensu lato).
Fig 5. Constraint phylogeny based on R-Syst::diatom sequences and on the OTU sequences of samples taken from the shells of sea turtles.
This tree was obtained using the fast tree search option in RaxML and was drawn using iTOL [73] http://itol.embl.de.
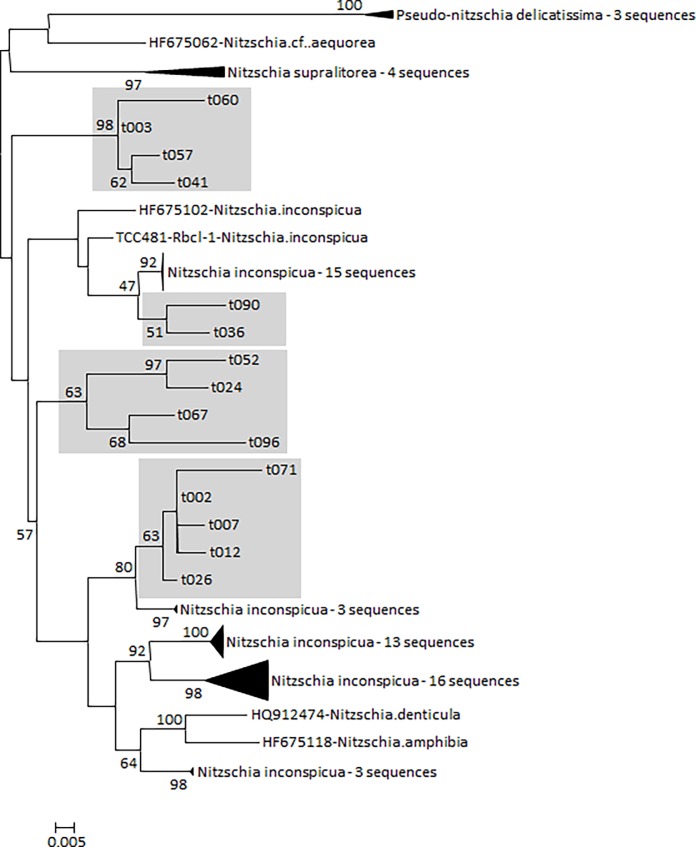
N. inconspicua was the species chosen for testing cryptic diversity because it is the second most abundant species observed in microscopy and because it encompasses the greatest number of OTUs. Even though Labellicula lecohuiana was the most abundant species observed in microscopy, it was not selected because its phylogenetic position is unknown. A detailed constraint phylogeny of Nitzschia inconspicua and its neighbor species (N. supralitorea Lange-Bertalot, N. amphibia Grunow, N. denticula Grunow) as well as the neighboring OTU was calculated using RAxML and the rapid bootstrapping and subsequent ML search option (Fig 6). Pseudo-nitzschia delicatissima (Cleve) Heiden was used to root the tree. The tree was drawn using MEGA7 [71]. The alignment used for this phylogeny is given in S1 Table. Four well-supported groups of OTUs are revealed by this phylogeny; three are embedded in Nitzschia inconspicua sensu stricto, and on its periphery.
Fig 6. Constraint phylogeny based on Nitzschia inconspicua sensu lato and its neighboring species from R-Syst::diatom sequences and on the neighboring OTU sequences of samples taken from the shells of sea turtles.
This tree was obtained using the rapid bootstrapping option in RaxML and was drawn using MEGA7. Only the OTUs were constrained. Grey squares delimit four groups of OTUs that co-occurred on the same turtles.
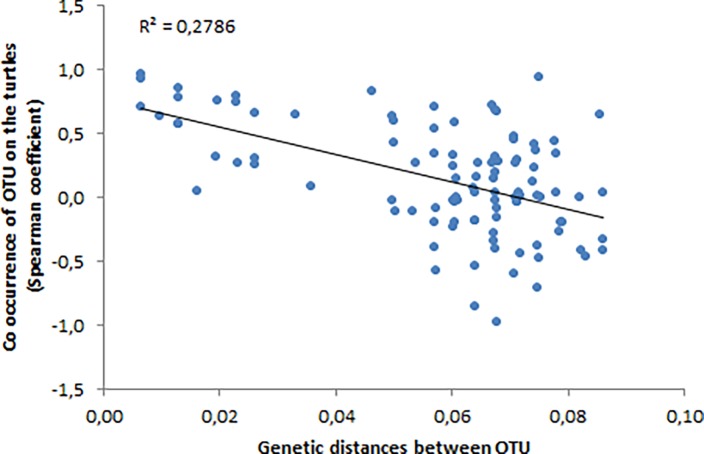
To determine whether these OTUs occurred on the same turtles, correlation coefficients (Spearman) between OTUs and their abundances in the samples were calculated. Results show that some OTUs always occur on the same turtles, but others do not (see S2 Table). We hypothesized that genetically similar OTUs occurred on the same turtles, signaling a relationship between genetic distance and the occurrence of these OTUs in the samples. To test this hypothesis, genetic distances were calculated using MEGA7, and the number of base substitutions per site between sequences, applying the maximum composite likelihood model. A correlation between the Spearman correlation coefficients (based on OTU abundances) and the genetic distance was tested and is represented in Fig 7. The tendency for OTUs to be genetically similar on the same turtle is significant. On the other hand, when OTUs were genetically different, they were not present on the same turtle. When genetic distances were less than 0.04 substitutions/site, OTUs co-occurred on the same turtle. At greater distances, they did not occur on the same turtle (Fig 7). When OTUs with distances less than 0.04 substitutions/site are clustered, they form four well-supported groups, as shown in Fig 6. This implies cryptic diversity inside Nitzschia inconspicua, comprising four groups that do not have similar ecologies.
Fig 7. Relationship between the genetic distances of OTUs and their occurrences on sea turtles.
Genetic distance is based on the number of base substitutions per site between sequences. Occurrence is based on the Spearman correlation coefficient calculated for the abundance of OTUs in the samples. The relationship is linear and significant (p-value < 0.001).
Discussion
Why the epibiotic diatom assemblages on sea turtles differ between metabarcoding and microscopy?
Results of metabarcoding and microscopic analyses are not comparable in several aspects: turtles are clustered differently based on their diatom assemblages, diatom assemblage structures differ, and metabarcoding yields better discrimination between turtles based on their diatom assemblages. This is unusual, because other studies [66, 74] comparing metabarcoding assemblages (expressed in OTUs abundances) with microscopic assemblages (expressed in species abundances) of benthic diatoms showed the methodologies to be similar in terms of diatom assemblages. As these authors, we chose to compare the microscopic assemblages expressed in species abundances to metabarcoding assemblages expressed in OTUs abundances. Metabarcoding is expressed in OTU abundances because expressing assemblages in species abundances, when an important part of the OTUs were not identified; would result in a comparison based on data truncated at least 64% for metabarcoding data (see Fig 2). Using OTU abundances without taxonomic assignation allows all data to be kept [75]. Several reasons can explain the differences between the methodologies.
First, a classical statistical problem exists. The number of samples is quite low (only seven turtles were sampled), and this can explain why the structure of epibiotic diatom assemblages obtained with both methods were not the same. The former studies of [74] and [66], obtained correlation factors between both methods of 18% and 20% across 90 samples and 66 samples, respectively, and the methods were found to be significantly correlated. Our results (15% correlation across 7 samples) are in the same range, but are slightly weaker.
Second, each methodology measures something different. In microscopy, diatom extracellular skeletons (siliceous frustules) are counted even if they come from dead cells and stay glued to the biofilm without DNA in the sample. Metabarcoding won't detect these dead cells. This phenomenon has already been mentioned by others [52, 53, 66, 74]. Also, in microscopy, a single cell of a small species such as Labellicula lecohuiana can have a biovolume of 15 to 20 μm3, but has the same importance as a larger species such as Entomoneis punctulata (Grunow) Osada and Kobayasi, which has a biovolume of 1500–2000 μm3. In microscopy, skeletons are counted in the same way regardless of biovolume. On the other hand, the number of copies of rbcL genes in a cell depends on its biovolume [66, 76]. Therefore, for a similar number of large cells in a sample, a greater abundance of OTUs will be found, compared to small species. This has an obvious impact when comparing metabarcoding with microscopic diatom assemblage structures.
Third, samples were dominated by many very small species such as Labellicula spp. (average length, 5 μm), Halamphora spp. (length, 5–10 μm), and unidentified Navicula (length, 5–8 μm), all with cell widths around 1–2 μm. Such frustules were very difficult to identify and count precisely under light microscopy, and under scanning electron microscopy, a number of identification uncertainties could not be confirmed. Freshwater samples from Mayotte Island sampled and analyzed in microscopy for routine monitoring [77] and in metabarcoding (e.g., [66]) do not typically have so many small species. This is also the case for freshwater samples on mainland France (e.g., [78]). Identification using light microscopy reaches its limits when small species are too abundant; therefore, microscopic counts introduce significant uncertainty. A large set of epibiotic diatoms samples from turtles were analyzed using scanning electron microscopy, thus bypassing this problem and enabling robust identification [27]. Nevertheless, scanning electron microscopy is expensive and cannot be undertaken routinely. Use of the methodology is exceptional.
Fourth, many taxa examinated microscopically were probably new to science and this is a source of difficulty when establishing robust taxonomic lists. It reduces the ability of microscopy to differentiate turtles on the basis of their epibiotic diatom assemblages. More than 36% of the taxa from our samples were not identified with certainty at the species level. It is not unlikely that these unidentified diatoms represent probable new taxa; as many as 10 new taxa found on sea turtles have been described since 2015 [33–38]. In their paper, Majewska et al. [79], already observed that many new taxa remain undescribed in epibiotic samples and this is frequent in littoral marine habitats [32]. With metabarcoding, using diatom assemblages based on their OTUs abundances (with no taxonomic assignment) to compare sea turtles enables a much more objective and precise comparison than microscopy, even though metabarcoding includes bias (e.g., choice of the extraction method, and sequencing errors; see [74]). This bias should be the same among all samples.
Fifth, cryptic diversity in diatoms has been extensively studied, focusing on a few diatom species, and morphology has been shown to explain only part of the species diversity. Inside an apparently single morphological species, certain reproductive barriers exist, explaining why certain genetic groups exist while sometimes displaying slight morphological differences observables only after careful examination (e.g., Sellaphora pupula [80, 81]). This cryptic diversity explains differences between individuals in terms of ecological requirements for apparently cosmopolitan species (e.g., Pinnularia borealis and Hantzschia amphioxys, [82]). Moreover, these cryptic species can have different ecologies and can live in sympatry. This is the case for Navicula phyllepta, a marine species consisting of several cryptic taxa which have different tolerances in terms of salinity but which live in the same tidal flat in the Netherlands [83].
Nitzschia inconspicua was one the most abundant taxa observed in the turtles samples from Mayotte. The morphology of this species was homogeneous across the seven turtles. However it was made up of tens of OTUs which could be regrouped into four groups. Inside these four groups, OTUs occurred on a given turtle; conversely, these four groups did not occur together on a given turtle. This implies a cryptic diversity for N. inconspicua, which is usually described as cosmopolitan and euryhaline [84, 85]. We hypothesize that N. inconspicua comprises several cryptic taxa whose ecological requirements differ between each other. This could explain the apparent cosmopolitanism of this species as well as others (Pinnularia borealis, Hantzschia amphioxys, Sellaphora pupula, and Navicula phyllepta). Likewise, a study on diatom diversity in high altitude lakes [86] raised the hypothesis that HTS could detect cryptic diversity. Our study validate this hypothesis: cryptic diatom diversity is detectable in metabarcoding and surely makes a valuable contribution for the fine scale understanding of turtle behavior in Mayotte. This cryptic diversity also explains why metabarcoding enables better discrimination between sea turtles than microscopy.
The ecology and architecture of turtle biofilms can be characterized by microscopy, not by metabarcoding
More than 93% of the taxa could be identified at the species or genus level under microscopy. Even if species identifications based on microscopy were doubtful and yielded poor discrimination of turtles based on their epibiotic diatom assemblages, this was sufficient to give a rough but robust idea of the ecology of the diatoms living on these turtles. Sister species of diatoms were shown to typically have the same ecology [87], and that identification at the genus level enables a quick and robust ecological assessment [46, 88, 89]. Moreover, the ecological guilds used can be assigned to most of the taxa if their genera and sizes are known [77]. We could gain this information via microscopy.
From the abundance of ecological guilds, epibiotic diatoms are clearly from a benthic origin and are mostly loosely attached (see Table 2). In short, the turtles could be assumed to be slow mover. Nevertheless, some differences between turtles can be observed. For instance, Turtle 2 presented the lowest abundance of low-profile diatoms (adapted to resist water turbulences); therefore, it was likely a slow swimmer. On the other hand, Turtle 4 presented three times more low-profile diatoms and therefore probably swam faster.
Table 2. Abundance of diatom ecological guilds on turtle shells.
| Turtle | High profile [%] |
Low profile [%] |
Motile [%] |
Planktonic [%] |
|---|---|---|---|---|
| 1 | 5.9 | 12.9 | 80.7 | 0.5 |
| 2 | 12.5 | 5.2 | 81.2 | 1.2 |
| 3 | 4.0 | 13.2 | 82.8 | 0.0 |
| 4 | 11.5 | 17.6 | 70.0 | 0.9 |
| 5 | 6.5 | 9.2 | 84.3 | 0.0 |
| 6 | 3.9 | 14.2 | 80.0 | 1.9 |
| 7 | 3.9 | 12.7 | 83.5 | 0.0 |
High-profile encompasses cells attached to substrate but enable to resist turbulences. Low profile are cells firmly attached to substrate and resisting to turbulences. Motile are loosely attached cells moving in biofilms. Planktonic are free-floating cells.
All the taxa observed on these sea turtle shells were strictly marine. Nevertheless, one dominant species, Nitzschia inconspicua, is known to be a marine-brackish euryhaline taxon [90]. This probably reflects the travels of the turtles from water bodies of varying salinities or from various depths, showing salinities gradients in Mayotte [91].
The totality of the diatom taxa observed on these turtle shells are known to be from habitats other than turtles shells [40], except Tursicola sp. and Labellicula lecohuiana. Tursicola sp. is a genus known only from epibiotic habitats [40], but it was rare on Mayotte turtles (observed only on Turtle 1 at an abundance below 1%). Labellicula lecohuiana was dominant on all the turtle shells of Mayotte. This is a recently-described species also coming from shells of the same sea turtle species, Chelonia mydas, but from Costa-Rica [35]. Therefore, it is known to come only from epibiotic habitats. Nevertheless, the Labellicula genus is very closely related to Olifantiella (according to Majewska et al. [35]), which encompasses several species from the coral sands of various tropical islands [62]. Therefore, it would be necessary to sample the coral sands of Mayotte Island to be assure that Labellicula lecohuiana is not present on that substrate, making this species strictly epibiotic. We believe that the epibiotic diatoms living on turtle shells in Mayotte originate from benthic habitats: when turtles graze seagrass, they resuspend sand, which falls on their shells. Therefore, the shells are constantly seeded with benthic diatoms from their surrounding habitats. We believe that the hypothesis that these diatoms are strictly epizoic is weakly probable for the turtles we studied. We believe that diatom composition of the sea turtle shells is related to the diatom composition of its surrounding environment as it has been stated by Majewska et al. [27].
All this ecological information could not be gained only from metabarcoding, simply because some references in the R-Syst::diatom barcoding library are lacking. Only 29% of the OTUs could be identified at the species or genus level. Marine diatoms have been microscopically studied for than a century whereas sequencing diatom species is a much more recent development (since the late 90s; e.g. [92, 93]).
Conclusions and perspectives
The two methods are complementary. The strength of microscopy is its ability to identify a large majority of the taxa, bringing valuable ecological information based on the historical bibliographical data and ecological guild abundances. But, its weakness is imprecise identifications, particularly for the small taxa that were numerous in our samples, making comparison of turtles based on their epibiotic diatoms assemblages uncertain. An additional weakness is that microscopy requires well trained people to identify diatoms, and each sample needs to be examined for several hours under the microscope. If large sets of turtles were studied routinely, microscopy would be too demanding in terms of manpower.
On the other hand, metabarcoding enables the analysis of many samples at low cost (e.g. [94],). Sample treatments (e.g., extraction, PCR, sequencing, and bioinformatics) do not require rare experts in diatom taxonomy. Moreover also allows cryptic diatom diversity to be revealed, a difficult undertaking using microscopy. This allows better discrimination between turtles based on their epibiotic diatom assemblages. Therefore, we recommend using metabarcoding for massive comparisons of sea turtles based on their diatom assemblages. But these comparisons must be performed based on OTU abundances without taxonomic assignation because barcoding libraries are not yet complete enough. We discourage use of this methodology if ecological information is needed and if reference barcoding libraries do not cover sufficient marine diatom diversity.
Supporting information
(TXT)
(XLSX)
Acknowledgments
We thank the French Agency for Biodiversity (AFB, Agence Française pour la Biodiversité) for its support.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the French Agency for Biodiversity (AFB, Agence Française de la Biodiversité). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bjorndal KA. Foraging ecology and nutrition of sea turtles In: Lutz P, Musick J, editors. The Biology of Sea Turtles. Boca Raton: Florida, CRC Press; 1997. pp. 199–231. [Google Scholar]
- 2.Bjorndal KA, Jackson JB. Roles of sea turtles in marine ecosystems: reconstructing the past In: Lutz P, Musick J, Wyneken J, editors. The Biology of Sea Turtles. Boca Raton:Florida, CRC Press; 2003. pp. 259–273. [Google Scholar]
- 3.Bouchard SS, Bjorndal KA. Sea turtles as biological transporters of nutrients and energy from marine to terrestrial ecosystems. Ecology. 2000;81: 2305–2313. [Google Scholar]
- 4.Hannan LB, Roth JD, Ehrhart LM, Weishampel JF. Dune vegetation fertilization by nesting sea turtles. Ecology. 2007;88: 1053–1058. [DOI] [PubMed] [Google Scholar]
- 5.Frazier J. Community based conservation In: Eckert KA, Bjorndal KA, Abreu-Grobois FA, Donnelly M, editors. Research and management techniques for the conservation of sea turtles. Washington, USA: IUCN/SSC Marine Turtle Specialist Group, Publication No.4; 1999. [Google Scholar]
- 6.Bugoni L, Krause L, Petry MV. Marine debris and human impacts on sea turtles in southern Brazil. Mar Pollut Bull. 2001;42: 1330–1334. [DOI] [PubMed] [Google Scholar]
- 7.Bourjea J, Ciccione S, Ratsimbazafy R. Marine turtles surveys in Nosy Iranja Kely, north-western Madagascar. West Indian Ocean J Mar Sci. 2006;5: 209–212. [Google Scholar]
- 8.Pike DA. Climate influences the global distribution of sea turtle nesting. Glob Ecol Biogeogr. 2013;22: 555–566. [Google Scholar]
- 9.Gerrodette T, Taylor B. Estimating Population Size. Research and Management Techniques for the Conservation of Sea Turtles. Washington, USA: IUCN/SSC Marine Turtle Specialist Group, Publication No.4; 1999. [Google Scholar]
- 10.Jean C, Ciccione S, Ballorain K, Georges J-Y, Bourjea J. Ultralight aircraft surveys reveal marine turtle population increases along the west coast of Reunion Island. Oryx. 2010;44: 223–229. [Google Scholar]
- 11.Epperly SP, Braun J, Chester AJ. Aerial surveys for sea turtles in North Carolina inshore waters. Oceanogr Lit Rev. 1996;1: 61. [Google Scholar]
- 12.Henwood TA, Epperly SP. Aerial surveys in foraging habitats In: Eckert KL, Bjorndal KA, Abreu-Grobois FA, Donnelly M, editors. Research and Management Techniques for the Conservation of Sea Turtles. Washington, USA: IUCN/SSC Marine Turtle Specialist Group, Publication No.4; 1999. [Google Scholar]
- 13.Schroeder B, Murphy S. Population surveys (ground and aerial) on nesting beaches In: Eckert KL, Bjorndal KA, Abreu-Grobois FA, Donnelly M, editors. Research and management techniques for the conservation of sea turtles. Washington, USA: IUCN/SSC Marine Turtle Specialist Group, Publication No.4; 1999. [Google Scholar]
- 14.Roos D, Pelletier D, Ciccione S, Taquet M, Hughes G. Aerial and snorkelling census techniques for estimating green turtle abundance on foraging areas: a pilot study in Mayotte Island (Indian Ocean). Aquat Living Resour. 2005;18: 193–198. [Google Scholar]
- 15.Ballorain K, Ciccione S, Bourjea J, Grizel H, Enstipp M, Georges J-Y. Habitat use of a multispecific seagrass meadow by green turtles Chelonia mydas at Mayotte Island. Mar Biol. 2010;157: 2581–2590. [Google Scholar]
- 16.Eckert SA. Data acquisition systems for monitoring sea turtle behavior and physiology In: Eckert KL, Bjorndal KA, Abreu-Grobois FA, Donnelly M, editors. Research and Management Techniques for the Conservation of Sea Turtles. Washington, USA: IUCN/SSC Marine Turtle Specialist Group, Publication No.4; 1999. pp. 88–93. [Google Scholar]
- 17.Enstipp MR, Ciccione S, Gineste B, Milbergue M, Ballorain K, Ropert-Coudert Y, et al. Energy expenditure of freely swimming adult green turtles (Chelonia mydas) and its link with body acceleration. J Exp Biol. 2011;214: 4010–4020. doi: 10.1242/jeb.062943 [DOI] [PubMed] [Google Scholar]
- 18.Ballorain K, Bourjea J, Ciccione S, Kato A, Hanuise N, Enstipp M, et al. Seasonal diving behaviour and feeding rhythms of green turtles at Mayotte Island. Mar Ecol Prog Ser. 2013;483: 289–302. [Google Scholar]
- 19.Fukuoka T, Narazaki T, Sato K. Summer-restricted migration of green turtles Chelonia mydas to a temperate habitat of the northwest Pacific Ocean. Endanger Species Res. 2015;28: 1–10. [Google Scholar]
- 20.Robinson DP, Jabado RW, Rohner CA, Pierce SJ, Hyland KP, Baverstock WR. Satellite tagging of rehabilitated green sea turtles Chelonia mydas from the United Arab Emirates, including the longest tracked journey for the species. PloS One. 2017;12: e0184286 doi: 10.1371/journal.pone.0184286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz FJ. Behavioral and tolerance responses to cold water temperatures by three species of sea turtles (Reptilia, Cheloniidae) in North Carolina. Fla Mar Res Publ. 1978;33: 16–18. [Google Scholar]
- 22.Hayashi R. A checklist of turtle and whale barnacles (Cirripedia: Thoracica: Coronuloidea). J Mar Biol Assoc U K. 2013;93: 143–182. [Google Scholar]
- 23.Frazier JG, Goodbody I, Ruckdeschel CA. Epizoan communities on marine turtles: II. Tunicates. Bull Mar Sci. 1991;48: 763–765. [Google Scholar]
- 24.Frazier JG, Winston JE, Ruckdeschel CA. Epizoan communities on marine turtles. III. Bryozoa. Bull Mar Sci. 1992;51: 1–8. [Google Scholar]
- 25.Frazier J, Margarttoulis D, Muldoon K, Potter C., Rosewater J, Ruckdeschel C, et al. Epizoan communities on marine turtles. Mar Ecol. 1985;6: 127–140. [Google Scholar]
- 26.Majewska R, De Stefano M, Bolaños F, Van de Vijver B. Hitchhinking on sea turtles: epizoic diatoms on olive ridleys (Lepidochelys olivacea) from the Pacific coast of Costa Rica. Diatomededelingen. 2016;40: 35–42. [Google Scholar]
- 27.Majewska R, Van de Vijver B, Nasrolahi A, Ehsanpour M, Afkhami M, Bolaños F, et al. Shared epizoic taxa and differences in diatom community structure between green turtles (Chelonia mydas) from distant habitats. Microb Ecol. 2017;74: 969–978. doi: 10.1007/s00248-017-0987-x [DOI] [PubMed] [Google Scholar]
- 28.Robinson NJ, Majewska R, Lazo-Wasem EA, Nel R, Paladino FV, Rojas L, et al. Epibiotic diatoms are universally present on all sea turtle species. PloS One. 2016;11 (6): e0157011 doi: 10.1371/journal.pone.0157011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayashi R, Tsuji K. Spatial distribution of turtle barnacles on the green sea turtle, Chelonia mydas. Ecol Res. 2008;23: 121. [Google Scholar]
- 30.Fuller WJ, Broderick AC, Enever R, Thorne P, Godley BJ. Motile homes: a comparison of the spatial distribution of epibiont communities on Mediterranean sea turtles. J Nat Hist. 2010;44: 1743–1753. [Google Scholar]
- 31.Casale P, D’addario M, Freggi D, Argano R. Barnacles (Cirripedia, Thoracica) and associated epibionts from sea turtles in the central Mediterranean. Crustaceana. 2012;85: 533–549. [Google Scholar]
- 32.Mann DG, Vanormelingen P. An inordinate fondness? The number, distributions, and origins of diatom species. J Eukaryot Microbiol. 2013;60: 414–420. doi: 10.1111/jeu.12047 [DOI] [PubMed] [Google Scholar]
- 33.Majewska R, Kociolek JP, Thomas EW, De Stefano M, Santoro M, Bolanos F, et al. Chelonicola and Poulinea, two new gomphonemoid diatom genera (Bacillariophyta) living on marine turtles from Costa Rica. Phytotaxa-Auckl NZ 2009 Currens. 2015;233: 236–250. [Google Scholar]
- 34.Majewska R, De Stefano M, Ector L, Bolaños F, Frankovich TA, Sullivan MJ, et al. Two new epizoic Achnanthes species (Bacillariophyta) living on marine turtles from Costa Rica. Bot March 2017;60: 303–318. [Google Scholar]
- 35.Majewska R, Stefano D, Vijver M. Labellicula lecohuiana, a new epizoic diatom species living on green turtles in Costa Rica. NOVA Hedwig. 2017;146: 221–231. [Google Scholar]
- 36.Frankovich TA, Ashworth MP, Sullivan MJ, Vesela J, Stacy NI. Medlinella amphoroidea gen. et sp. nov.(Bacillariophyta) from the neck skin of Loggerhead sea turtles (Caretta caretta). Phytotaxa. 2016;272: 101–114. [Google Scholar]
- 37.Frankovich TA, Sullivan MJ, Stacy N. Three new species of Tursiocola (Bacillariophyta) from the skin of the West Indian manatee (Trichechus manatus). Phytotaxa. 2015;204: 33–48. [Google Scholar]
- 38.Frankovich TA, Sullivan MJ, Stacy NI. Tursiocola denysii sp. nov.(Bacillariophyta) from the neck skin of Loggerhead sea turtles (Caretta caretta). Phytotaxa. 2015;234: 227–236. [Google Scholar]
- 39.Wetzel CE, Van de Vijver B, Ector L. Luticola deniseae sp. nov. a new epizoic diatom from the Rio Negro (Amazon hydrographic basin). Vie MiLieu-Life Environ. 2010;60: 177. [Google Scholar]
- 40.Wetzel CE, Van de Vijver B, Cox EJ, Bicudo D de C, Ector L. Tursiocola podocnemicola sp. nov., a new epizoic freshwater diatom species from the Rio Negro in the Brazilian Amazon Basin. Diatom Res. 2012;27: 1–8. [Google Scholar]
- 41.Pavlov A, Jovanovska E, Wetzel CE, Ector L, Levkov Z. Freshwater Mastogloia (Bacillariophyceae) taxa from Macedonia, with a description of the epizoic M. sterijovskii sp. nov. Diatom Res. 2016;31: 85–112. [Google Scholar]
- 42.Biggs BJ, Gerbeaux P. Periphyton development in relation to macro-scale (geology) and micro-scale (velocity) limiters in two gravel-bed rivers, New Zealand. N Z J Mar Freshw Res. 1993;27: 39–53. [Google Scholar]
- 43.Cox EJ. Observations on some benthic diatoms from North German lakes: The effect of substratum and light regime: With 3 figures in the text. Int Ver Für Theor Angew Limnol Verhandlungen. 1984;22: 924–928. [Google Scholar]
- 44.Rimet F. Recent views on river pollution and diatoms. Hydrobiologia. 2012;683: 1–24. [Google Scholar]
- 45.Semary E. Diatoms as bioindicators of littoral zone: a case study. Bangladesh J Bot. 2016;45: 1113–1121. [Google Scholar]
- 46.Wu SC, Bergey EA. Diatoms on the carapace of common snapping turtles: Luticola spp. dominate despite spatial variation in assemblages. PloS One. 2017;12: e0171910 doi: 10.1371/journal.pone.0171910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fayolle S, Moriconi C, Oursel B, Koenig C, Suet M, Ficheux S, et al. Epizoic algae distribution on the carapace and plastron of the European pond turtle (Emys orbicularis, Linnaeus, 1758): A study from the Camargue, France. Cryptogam Algol. 2016;37 (4): 221–232. [Google Scholar]
- 48.Kahlert M, Albert R-L, Anttila E-L, Bengtsson R, Bigler C, Eskola T, et al. Harmonization is more important than experience—results of the first Nordic–Baltic diatom intercalibration exercise 2007 (stream monitoring). J Appl Phycol. 2009;21: 471–482. [Google Scholar]
- 49.Kahlert M, Albert R-L, Anttila E-L, Bengtsson R, Bigler C, Eskola T, et al. Harmonization is more important than experience—results of the first Nordic–Baltic diatom intercalibration exercise 2007 (stream monitoring). J Appl Phycol. 2009;21: 471–482. [Google Scholar]
- 50.Taberlet P, Coissac E, Pompanon F, Brochmann C, Willerslev E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol Ecol. 2012;21: 2045–2050. doi: 10.1111/j.1365-294X.2012.05470.x [DOI] [PubMed] [Google Scholar]
- 51.Hebert PD, Cywinska A, Ball SL. Biological identifications through DNA barcodes. Proc R Soc Lond B Biol Sci. 2003;270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kermarrec L, Franc A, Rimet F, Chaumeil P, Frigerio J-M, Humbert J-F, et al. A next-generation sequencing approach to river biomonitoring using benthic diatoms. Freshw Sci. 2014;33: 349–363. [Google Scholar]
- 53.Visco JA, Apothéloz-Perret-Gentil L, Cordonier A, Esling P, Pillet L, Pawlowski J. Environmental monitoring: inferring the diatom index from next-generation sequencing data. Environ Sci Technol. 2015;49: 7597–7605. doi: 10.1021/es506158m [DOI] [PubMed] [Google Scholar]
- 54.Zimmermann J, Glöckner G, Jahn R, Enke N, Gemeinholzer B. Metabarcoding vs. morphological identification to assess diatom diversity in environmental studies. Mol Ecol Resour. 2015;15: 526–542. doi: 10.1111/1755-0998.12336 [DOI] [PubMed] [Google Scholar]
- 55.CEN. CEN/TC 230 N 1008, Water quality. Technical report for the routine sampling of benthic diatoms from rivers and lakes adapted for metabarcoding analyses. 2016: 7.
- 56.CEN. EN 13946. Water quality-Guidance standard for the routine sampling and pretreatment of benthic diatoms from rivers. European Commitee for Standardisation. 2002: 718.
- 57.CEN. EN 14407. Water quality-Guidance standard for the identification, enumeration and interpretation of benthic diatom samples from running waters. 2003: 13.
- 58.Witkowski A, Lange-Bertalot H, Metzeltin D. Diatom Flora Ofmarine Coasts I Iconographia Diatomologica 7. Rugell. ARG Gantner Verlag KG; 2000. [Google Scholar]
- 59.Hein MK, Winsborough BM, Sullivan MJ. Bacillariophyta (diatoms) of the Bahamas. Gantner Publishing; 2008. [Google Scholar]
- 60.Richard M. Atlas du phytoplankton marine: Diatomophyceae. Natl Recherce Paris. 1987;285. [Google Scholar]
- 61.Levkov Z. Amphora sensu lato Diatoms of Europe 5. Koeltz; 2009. p. 916. [Google Scholar]
- 62.Riaux-Gobin C, Al-Handal AY. New species in the marine diatom genus Olifantiella (Bacillariophyta, Biraphidineae) from Rodrigues Island (Western Indian Ocean). Fottea. 2012;12: 199–217. [Google Scholar]
- 63.Riaux-Gobin C, Witkowski A, Compère P, Romero OE. Cocconeis Ehrenberg taxa (Bacillariophyta) with a marginal row of simple processes: relationship with the valvocopula system and distinctive features of related taxa. Fottea Praha. 2015;15: 139–154. [Google Scholar]
- 64.Riaux–Gobin C, Witkowski A, Chevallier D, Daniszewska–Kowalczyk G. Two new Tursiocola species (Bacillariophyta) epizoic on green turtles (Chelo-nia mydas) in French Guiana and Eastern Caribbean. 2017;
- 65.Vasselon V, Domaizon I, Rimet F, Kahlert M, Bouchez A. Application of high-throughput sequencing (HTS) metabarcoding to diatom biomonitoring: Do DNA extraction methods matter? Freshw Sci. 2017;36: 162–177. [Google Scholar]
- 66.Vasselon V, Rimet F, Tapolczai K, Bouchez A. Assessing ecological status with diatoms DNA metabarcoding: scaling-up on a WFD monitoring network (Mayotte island, France). Ecol Indic. 2017;82: 1–12. [Google Scholar]
- 67.Rimet F, Chaumeil P, Keck F, Kermarrec L, Vasselon V, Kahlert M, et al. R-Syst:: diatom: an open-access and curated barcode database for diatoms and freshwater monitoring. Database. 2016;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rimet F, Abarca N, Bouchez A, Kusber WH, Jahn R, Kahlert M, et al. The potential of high throughput sequencing (HTS) of natural samples as a source of primary taxonomic information for reference libraries of diatom barcodes. Fottea. 2018;18(1): 37–54. [Google Scholar]
- 69.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32: 1792–1797. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2009;27: 221–224. doi: 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- 71.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33: 1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Silvestro D, Michalak I. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 2012;12: 335–337. [Google Scholar]
- 73.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44: W242–W245. doi: 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rivera SF, Vasselon V, Jacquet S, Bouchez A, Ariztegui D, Rimet F. Metabarcoding of lake benthic diatoms: from structure assemblages to ecological assessment. Hydrobiologia. 2018;807: 37–51. [Google Scholar]
- 75.Apothéloz-Perret-Gentil L, Cordonier A, Straub F, Iseli J, Esling P, Pawlowski J. Taxonomy-free molecular diatom index for high-throughput eDNA biomonitoring. Mol Ecol Resour. 2017;17: 1231–1242. doi: 10.1111/1755-0998.12668 [DOI] [PubMed] [Google Scholar]
- 76.Vasselon V, Bouchez A, Rimet F, Jacquet S, Trobajo R, Corniquel M, et al. Avoiding quantification bias in metabarcoding: application of a cell biovolume correction factor in diatom molecular biomonitoring. Methods Ecol Evol. 2018; doi: 10.1111/2041-210X.12960 [Google Scholar]
- 77.Tapolczai K, Bouchez A, Stenger-Kovács C, Padisák J, Rimet F. Taxonomy-or trait-based ecological assessment for tropical rivers? Case study on benthic diatoms in Mayotte island (France, Indian Ocean). Sci Total Environ. 2017;607: 1293–1303. doi: 10.1016/j.scitotenv.2017.07.093 [DOI] [PubMed] [Google Scholar]
- 78.Tison J, Park Y-S, Coste M, Wasson JG, Ector L, Rimet F, et al. Typology of diatom communities and the influence of hydro-ecoregions: a study on the French hydrosystem scale. Water Res. 2005;39: 3177–3188. doi: 10.1016/j.watres.2005.05.029 [DOI] [PubMed] [Google Scholar]
- 79.Majewska R, Santoro M, Bolaños F, Chaves G, De Stefano M. Diatoms and other epibionts associated with olive ridley (Lepidochelys olivacea) sea turtles from the Pacific Coast of Costa Rica. PloS One. 2015;10: e0130351 doi: 10.1371/journal.pone.0130351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mann DG, McDonald SM, Bayer MM, Droop SJ, Chepurnov VA, Loke RE, et al. The Sellaphora pupula species complex (Bacillariophyceae): morphometric analysis, ultrastructure and mating data provide evidence for five new species. Phycologia. 2004;43: 459–482. [Google Scholar]
- 81.Evans KM, Wortley AH, Simpson GE, Chepurnov VA, Mann DG. A molecular systematic approach to explore diversity within the Sellaphora pupula species complex (Bacillariophyta). J Phycol. 2008;44: 215–231. doi: 10.1111/j.1529-8817.2007.00454.x [DOI] [PubMed] [Google Scholar]
- 82.Souffreau C, Vanormelingen P, Van de Vijver B, Isheva T, Verleyen E, Sabbe K, et al. Molecular evidence for distinct Antarctic lineages in the cosmopolitan terrestrial diatoms Pinnularia borealis and Hantzschia amphioxys. Protist. 2013;164: 101–115. doi: 10.1016/j.protis.2012.04.001 [DOI] [PubMed] [Google Scholar]
- 83.Vanelslander B, Creach V, Vanormelingen P, Ernst A, Chepurnov VA, Sahan E, et al. Ecological differentiation between sympatric pseudocryptic species in the estuarine benthic diatom Navicula phyllepta (Bacillariophyceae). J Phycol. 2009;45: 1278–1289. doi: 10.1111/j.1529-8817.2009.00762.x [DOI] [PubMed] [Google Scholar]
- 84.Rovira L, Trobajo R, Sato S, Ibáñez C, Mann DG. Genetic and physiological diversity in the diatom Nitzschia inconspicua. J Eukaryot Microbiol. 2015;62: 815–832. doi: 10.1111/jeu.12240 [DOI] [PubMed] [Google Scholar]
- 85.Kelly MG, Trobajo R, Rovira L, Mann DG. Characterizing the niches of two very similar Nitzschia species and implications for ecological assessment. Diatom Res. 2015;30: 27–33. [Google Scholar]
- 86.Rimet F, Vasselon V, Barbara A, Bouchez A. Do we similarly assess diversity with microscopy and high-throughput sequencing? Case of microalgae in lakes. Org Divers Evol.: 1–12. [Google Scholar]
- 87.Keck F, Rimet F, Franc A, Bouchez A. Phylogenetic signal in diatom ecology: perspectives for aquatic ecosystems biomonitoring. Ecol Appl. 2016;26: 861–872. [DOI] [PubMed] [Google Scholar]
- 88.Rimet F, Bouchez A. Biomonitoring river diatoms: implications of taxonomic resolution. Ecol Indic. 2012;15: 92–99. [Google Scholar]
- 89.Chessman B, Growns I, Currey J, Plunkett-Cole N. Predicting diatom communities at the genus level for the rapid biological assessment of rivers. Freshw Biol. 1999;41: 317–331. [Google Scholar]
- 90.Trobajo R, Rovira L, Ector L, Wetzel CE, Kelly M, Mann DG. Morphology and identity of some ecologically important small Nitzschia species. Diatom Res. 2013;28: 37–59. [Google Scholar]
- 91.Guilcher A, Berthois L, Le Calvez Y, Battistini R, Crosnier A. Les récifs coralliens et le lagon de l’île Mayotte (archipel des Comores, océan Indien): géomorphologie, sédimentologie, hydrologie, foraminifères ORSTOM; Paris: 1965; [Google Scholar]
- 92.Medlin LK, Elwood HJ, Stickel S, Sogin ML. Morphological and genetic variation within the diatom Skeletonema costatum (Bacillariophyta): evidence for a new species, Skeletonema pseudocostatum. J Phycol. 1991;27: 514–524. [Google Scholar]
- 93.Medlin LK, Kooistra WH, Potter D, Saunders GW, Andersen RA. Phylogenetic relationships of the ‘golden algae’(haptophytes, heterokont chromophytes) and their plastids. Origins of algae and their plastids Springer; 1997. pp. 187–219. [Google Scholar]
- 94.Keck F, Vasselon V, Tapolczai K, Rimet F, Bouchez A. Freshwater biomonitoring in the information age. Front Ecol Environ. 2017;15: 266–274. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TXT)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.