Abstract
PsCYP15A is a cysteine protease from pea (Pisum sativum L.). It was first recognized as an up-regulated transcript in wilted shoots and subsequently in root nodules containing Rhizobium. Proteolytic activity of PsCYP15A in nodule extracts is now reported following immunopurification with polyclonal antiserum raised against recombinant antigen. Western-blot analysis indicated two forms of PsCYP15A, a pro-form (approximately 38 kD) and a mature form (approximately 30 kD). Both forms were present in most tissue samples, but only the mature form was isolated from cell-fractionated symbiosomes containing nitrogen-fixing bacteroids. Immunolabeling of nodule sections showed localization of PsCYP15A antigen in large vacuolar bodies, cytoplasmic vesicles, and the perisymbiont space. Immunolabeling of tissue sections from wilted shoots also indicated the presence of PsCYP15A in vacuoles and cytoplasmic vesicles. This protease may be involved in the adaptation to changes in cell turgor, both in wilted shoots and in nodule tissue. Additionally, the protease may be involved in protein turnover in the symbiosome compartment.
Proteolytic activity is ubiquitous in biological systems. In plants (Vierstra, 1996), proteolysis is a requirement for mobilization of seed storage proteins in germination, for the efficient recycling of amino acids in senescence and apoptosis, and also for “housekeeping” functions such as the activation of zymogens, removal of aberrant proteins, and degradation of proteins as a part of the homeostatic cycle of removal and renewal. The most common plant endopeptidases are Cys proteases, otherwise known as thiol proteases, which are characterized by an active site formed by conserved Cys and His residues (Barret et al., 1998).
PsCyp15a cDNA (accession no. X54358) encodes a Cys protease up-regulated in pea (Pisum sativum L.) stem tissue in response to water deficit (Guerrero et al., 1990; Jones and Mullet, 1995). However, PsCyp15a has also been identified as a transcript in the pea root nodule symbiosis with Rhizobium (Kardailsky and Brewin, 1996). The cellular and intracellular localization of PsCYP15A protease is therefore of interest from the point of view of nodule organogenesis and function. Previous investigations of PsCyp15a gene expression during nodule development (Kardailsky and Brewin, 1996) showed the highest levels of in situ hybridization in the apical region, a zone associated with meristematic activity and subsequent invasion of host cells by rhizobia. Expression was also observed throughout the central infected tissue, where rhizobia exist as endosymbionts. In the cytoplasm of host cells, these “bacteroids” are individually enclosed within membranous units called “symbiosomes,” where they differentiate into a nitrogen-fixing state (Brewin, 1991).
The deduced amino acid sequence for PsCYP15A indicates that it is synthesized as a pre-proprotein consisting of an N-terminal hydrophobic leader peptide, a 110-amino acid propeptide, and a 233-amino acid mature peptide containing conserved residues Cys-153 and His-299, which form the catalytic dyad. Sequence comparison with other Cys proteases places PsCYP15A in clan CA as a member of family C1, the papain family, as defined by Barret et al. (1998). Previous studies of this protease in pea using polyclonal antisera raised against recombinant antigens showed the presence of 30- and 38-kD bands in western blots (Jones and Mullet, 1995). These bands were shown to correspond to mature and pro-forms of the enzyme, respectively.
In the present study, we have used antibody probes to investigate the subcellular localization of PsCYP15A in nodule cells and have re-examined its distribution in wilt-induced stem tissue. In both cases, the protease antigen was found to accumulate in vacuoles and vesicles of the endomembrane system.
RESULTS
Western Analysis of Pea Protein Samples
Affinity purification of recombinant PsCYP15A was achieved from cell lysates of induced cultures of Escherichia coli expressing the PsCyp15a RT-PCR fusion construct (Fig. 1A). Following further purification, the identity of the major protein band was confirmed on western blots using a monoclonal antibody (T7 Tag, Invitrogen, Carlsbad, CA) specific to the fusion peptide from the pRSET vector (Fig. 1B). Following SDS-PAGE and electro-elution, this protein was used to immunize a rabbit to derive antiserum R79.
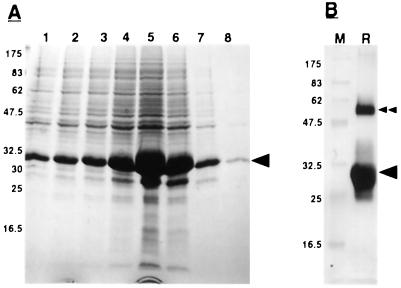
Figure 1.
Purification of recombinant PsCYP15A. A, SDS-PAGE of successive fractions from a nickel affinity column after the addition of elution buffer, pH 4.0; numbers to the side indicate molecular mass of marker proteins (kD); arrowhead indicates recombinant protein band that is the most intense in fraction 5. B, Western blot of electro-eluted recombinant PsCYP15A from fraction 5, using T7 Tag monoclonal antibody. M, Marker lane; R, recombinant protein lane; arrowhead, recombinant protein band; double arrowhead, higher Mr recombinant band, presumably resulting from dimerization of recombinant PsCYP15A monomer.
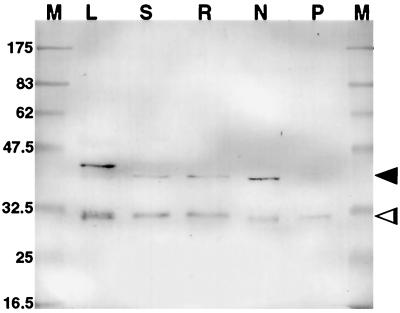
Figure 2 shows a western blot of pea protein samples following labeling with preabsorbed and immunopurified R79. Two major bands were observed at approximately 38 and 30 kD in all samples of tissue extracts, except in perisymbiont fluid, where only the approximately 30-kD species was identified. Preimmune serum used in identical western-blot labeling experiments did not produce any cross-reacting signal (data not shown). When compared with the predicted structure of PsCYP15A and the two main domains corresponding to pro- and mature peptides, it seems probable that the approximately 38-kD band relates to a pro-form of the antigen and the approximately 30-kD band relates to the processed, mature form. The reason for the slower mobility of one band in the leaf sample (40 kD compared with 38 kD in other lanes) has not been investigated. The presence of the antigen in the perisymbiont fluid sample suggests targeting to the symbiosome in nodule cells, and it is interesting that only the activated protease form (approximately 30 kD) was identified in this fraction.
Figure 2.
Western blot of tissue extracts (10 μg of protein per lane) after electrophoresis using immunopurified R79 as the primary antibody to identify PsCYP15A antigen. M, Marker; L, leaf; S, shoot; R, uninoculated root; N, nodule; P, perisymbiont fluid. Two bands are apparent in all lanes except P, indicating pro- (arrowhead) and mature (open arrowhead) forms of PsCYP15A. Note the marginally slower mobility of the pro-form in the leaf sample. Molecular masses of marker proteins are represented in kD.
Demonstration of Proteolytic Function
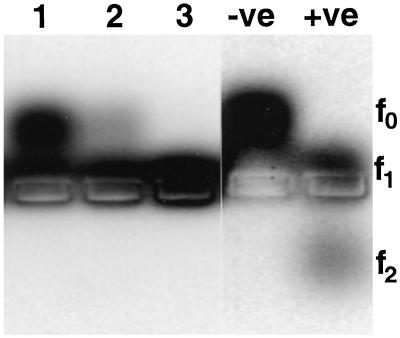
To demonstrate enzyme activity, a sample of native, nodule-derived PsCYP15A was immunopurified using paramagnetic beads charged with affinity-purified R79 antiserum. After washing with phosphate-buffered saline (PBS), varying amounts of the bead-bound material was used in the protease assay. Figure 3 shows cleavage of the synthetic peptide (f0) to the f1 state by PsCYP15A-mediated processing, whereas beads pretreated with preimmune serum and taken through the same procedure gave a negative result (data not shown). The assay monitors the products of a fluorescently labeled peptide substrate, which was resolved by electrophoresis into three states depending on whether there was no cleavage (f0), one cleavage event (f1), or an alternative cleavage event (f2).
Figure 3.
Agarose gel electrophoresis of protease assay reaction products. Lanes 1 to 3 show products following incubation with increasing amounts of plant-derived PsCYP15A bound to R79-linked paramagnetic beads. As controls, the negative lane (−ve) shows substrate with no protease added and the positive lane (+ve) shows the result of digestion with a bacterial alkaline protease. Different states of substrate cleavage are indicated to the right. Note that cleavage by PsCYP15A results in f1 state only, whereas cleavage by the control protease cleaves to both f1 and f2 states.
Confocal Microscopy
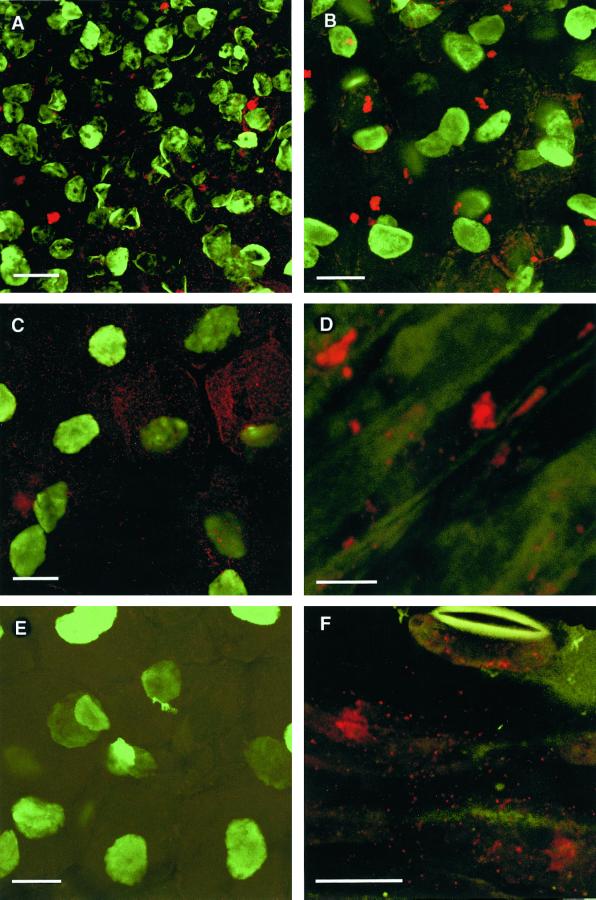
Tissue localization of PsCYP15A antigen was examined by using immunopurified R79 (or preimmune serum) with counter-immunolabeling by a mouse monoclonal antibody (1413) that reacts with nuclear material to immunolabel sections of pea tissue for laser scanning confocal microscopy (Fig. 4). Longitudinal sections of mature root nodules showed that PsCYP15A antigenicity was localized in large vacuolar bodies (approximately 3–5 μm in diameter) and cytoplasmic vesicles. An apparent increase in the number of R79-labeled vesicles and a decrease in the occurrence of R79-labeled vacuolar bodies was evident in the uninfected nodule apex relative to the central zone of the nodule. Sections treated with preimmune serum were not labeled. There was no evidence for the presence of antigen in the extracellular matrix or in the lumen of infection threads.
Figure 4.
Immunofluorescence labeling of PsCYP15A antigen in plant sections visualized by laser scanning confocal microscopy. A, Apical region of a pea nodule treated with R79 and anti-rabbit Alexa Fluor 568 to visualize PsCYP15A (red); counter-labeled with antibody 1413 and anti-mouse Alexa Fluor 488 to visualize nuclei (green). Note the abundance of label in cytoplasmic vesicles and occasionally in small vacuoles. Bar = 20 μm. B, Central infected nodule cells showing R79 labeling (red) and counter-labeling with monoclonal anti-nuclear antibody 1413 (green). Vacuolar bodies are evident as well as punctate cytoplasmic labeling (vesicles or possibly symbiosomes). Bar = 20 μm. C, Same as for B except that cytoplasmic labeling is more evident. Note the clearly defined cell borders showing absence of PsCYP15A antigen in the cell walls/spaces. D, Immunolabeling of wilted stem tissue (nonelongating cortical area) treated with R79 to reveal PsCYP15A antigen (red) in vacuoles and cytoplasmic vesicles. (Green autofluorescence at 488 nm shows general tissue anatomy.) Bar = 10 μm. E, Control section taken from central infected nodule tissue challenged with preimmune serum and 1413 antibody. (Legend continues on facing page.)Only 1413-labeled nuclei are evident (green); no labeling by preimmune serum (red) is seen. Bar = 20 μm. F, Immuno- labeling of wilted stem tissue (epidermal region) treated with R79 to reveal PsCYP15A antigen (red) in cytoplasmic vesicles. Note the presence of vesicular labeling within the guard cell of an open stomate at the top of the frame. (Green autofluorescence at 488 nm shows general tissue anatomy.) Bar = 20 μm.
Expression of PsCYP15A in nodule tissues was compared with that in pea stems treated with 0.6 m mannitol to induce wilting alongside water-treated controls. Results confirmed the findings from nodule specimens that antigen was localized intracellularly in vacuoles and cytoplasmic vesicles, and that there was no evidence of antigen in the extracellular matrix. Patterns of localization were similar in unwilted specimens (data not shown), although a general increase in the abundance of antigen was observed in wilted tissue. Again, no labeling was achieved with the preimmune serum (data not shown).
Electron Microscopy
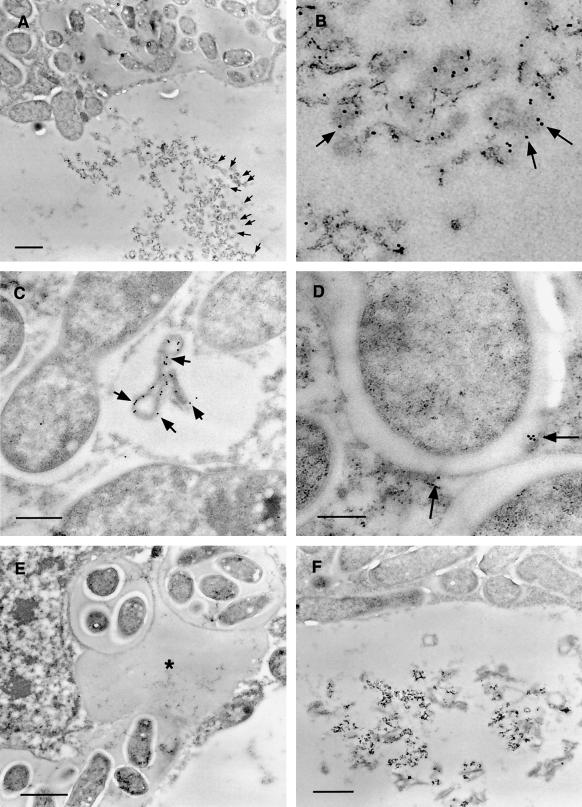
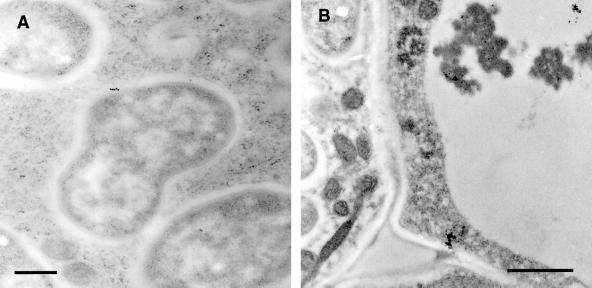
The results of immunogold localization (Figs. 5 and 6) confirmed and extended the results from confocal microscopy. Immunogold label from R79 was clearly associated with large vacuolar bodies of electron-dense material, apparently in aggregated form. Vesicular localization of antigen was also confirmed with R79 labeling of membrane-bound vesicles (approximately 500 nm in diameter) in the cytoplasm of infected cells. In addition, in infected cells, PsCYP15A antigen was found to be associated with the symbiosomal space but could not be identified in the extracellular matrix or cell wall, nor in the lumen of infection threads. Immunogold-labeled antigen was never abundant in the symbiosomal space and was not detectable in every symbiosome. Treatment of similar sections with preimmune serum gave no immunogold labeling above background levels.
Figure 5.
Immunogold-labeled nodule sections treated with R79 antiserum and visualized by transmission electron microscopy. A, Infected cell showing electron-dense aggregate within the vacuole heavily decorated with 15-nm immunogold particles (black dots indicated by arrows). Bar = 1 μm. B, Enlargement of vacuolar aggregate shown in A, with arrows indicating immunogold particles. C, R79-labeled infected cell showing cytoplasmic vesicle containing electron-dense material decorated with 15-nm immunogold. Bar = 500 nm. D, R79-labeled infected cell showing 15-nm immunogold particles associated with the perisymbiont area (arrows). Bar = 200 nm. E, R79-labeled infected cell showing cross-section through an infection thread and infection droplet. No immunogold label is seen associated with the infection thread matrix (asterisk). Bar = 1 μm. F, Infected cell challenged with preimmune serum, showing no immunogold labeling of perisymbiont spaces or aggregated vacuolar material. Bar = 1 μm.
Figure 6.
Immunogold-labeled nodule sections treated with R79 antiserum and visualized by transmission electron microscopy. A, Cytoplasm of an infected cell. The center of the frame shows a complete symbiosome that has been labeled with 15-nm colloidal gold conjugate (indicated by the black dots), showing antigen localized within the space between the perisymbiont membrane and the bacteroid. Bar = 500 nm. B, Central tissue of a nodule, showing an infected cell (left side) and an uninfected cell (right side) separated by their respective cell walls. To the bottom of the frame, a further junction of these two cells with a third cell produces a large intercellular space. Both the cell walls and the intercellular space are unlabeled, whereas gold particles are associated with the dense material visible in the vacuole of the uninfected cell. Bar = 1 μm.
DISCUSSION
Isolation of the native, plant-derived PsCYP15A protein was achieved using the PsCYP15A-specific antibody R79 bound to paramagnetic beads. This material was shown to possess distinct proteolytic activity by cleavage of a synthetic substrate at acidic pH, thus demonstrating biochemically that the gene product of PsCyp15a is a functional protease.
Western blots using R79 on samples from pea tissue indicated antigenic bands migrating at approximately 38 and 30 kD, which is similar to those reported previously (Jones and Mullet, 1995). Using pro-peptide-specific anti-PsCYP15A antibodies, Jones and Mullet (1995) reported that only an approximately 38-kD band was recognized on western blots, whereas approximately 38- and 30-kD bands are defined by antibodies recognizing the mature peptide. Presence of the antigen was noted in all tissues tested, including the root nodule and perisymbiont fluid, where only the active approximately 30-kD form was apparent.
The use of transmission electron microscopy and immunogold localization in mature root nodules confirmed and extended the observations from laser scanning confocal microscopy. In this study, no antigen was observed in the extracellular matrix of nodule cells nor in the infection thread lumen, a matrix that is inherently apoplastic in nature (Rae et al., 1991). We identified low levels of antigen within the symbiosome compartment of infected cells, which is consistent with previous biochemical data from tissue fractionation studies suggesting that a protease activity was associated with this fraction (Kardailsky et al., 1996). Furthermore, it lends support to the proposal that proteins in the perisymbiont fluid may turn over very rapidly as a result of proteolytic activity (Dahiya et al., 1997; Balestrini et al., 1999).
Mellor (1989) suggested that the symbiosome unit could be considered a “temporary but independent organelle,” in which the presence of various lysosomal proteins such as α-mannosidase and acid protease indicates some similarity to the lysosome compartment. Thus, symbiosomes may represent prevacuolar structures accumulating without immediate fusion to the main lytic compartment (Marty, 1999). It is known that the perisymbiont membrane possesses tonoplast qualities, so it is not surprising to find a vacuolar protease also targeted to this compartment. In an earlier study of root nodule proteolysis (Vance et al., 1979), it was demonstrated that proteolytic activity was associated with the age-related senescence of alfalfa nodules. In subsequent studies, similar activities have been identified in the age-related senescence of soybean nodules (Pfeiffer et al., 1983) and French bean nodules (Pladys and Rigaud, 1985).
The first targets of these proteases are apparently nodule cytosolic proteins, especially leghemoglobin, and the degradation of bacteroids has also been demonstrated following rupture of the perisymbiont membrane. Increased Cys protease activity has been shown in early senescing nodules of alfalfa, indicating a specific role for such proteases in the senescent phase of nodule development (Pladys and Vance, 1993). However, in view of our immunolocalization studies and the in situ hybridization studies presented previously (Kardailsky and Brewin, 1996), it is likely that PsCYP15A delineates a new role for proteolysis within developing and functioning symbiosomes. It is interesting that Cys and Ser proteases are also expressed during invasion of non-legume actinorhizal nodules by Frankia (Goetting-Minesky and Mullin, 1994; Ribeiro et al., 1995), although the localizations have not been determined.
To date, a number of sequences homologous to PsCYP15A have been reported in the literature or sequence databases, but their proposed physiological functions show a somewhat varied repertoire. Seven of these sequences are involved in the germination process (accession nos. Z99953, Z30338, U59465, Z99172, Z99955, Z32795; Becker et al., 1994; Domoto et al., 1995; Nong et al., 1995). Four (including rd19 from Arabidopsis) are induced by water stress (accession nos. AF007215, D13043, X74359; Koizumi et al., 1993; Williams et al., 1994; Stroeher et al., 1997). Four are upregulated by other forms of stress such as wounding (accession nos. Z13959, Z13964; Linthorst et al., 1993a), Glc starvation (accession no. X82185; Chevalier et al., 1995) or pathogen attack (accession nos. AJ009878, Z14028; Linthorst et al., 1993b). One homolog (clone pLBPc13) is involved in leaf development (McKee et al., 1986) and another in leaf senescence (accession no. U68221; Buchanan-Wollaston and Ainsworth, 1997). Two more sequences are of unknown function in soybean leaves (accession nos. U71379, U71380).
PsCyp15a gene expression was shown to be drought inducible, yet absisic acid unresponsive (Jones and Mullet, 1995). Thus, it is possible that the function of PsCYP15A may be linked to stress adaptation in response to wilting, although the presence of lower quantities of PsCYP15A antigen in unstressed shoot cells could also point to a homeostatic role. Recreating the conditions of wilting of pea shoots reported previously (Jones and Mullet, 1995), we found by laser scanning confocal microscopy analysis that the PsCYP15A antigen was not associated with the cell walls or extracellular matrix, but was present in vacuolar bodies and vesicles (moreso in the water-stressed specimens). These observations support the immunolabeling data from sections of root nodules, but differ from the extracellular localization reported by Jones and Mullet (1995).
Thus, there is still no clear indication of the function of the PsCYP15A protease either in wilt-induced shoot cells or during nodule development. Vacuoles perform various functions, of which the most important are the storage and hydrolysis of macromolecules and the maintenance of cell turgor (Marty, 1999). It is possible that PsCYP15A-mediated proteolysis serves to increase the concentration of free amino acids and peptides, resulting in increased solute levels in vesicles and vacuoles, and leading to a lower internal water potential that might be adaptive to water stress. Alternatively, PsCYP15A may hydrolyze proteins no longer required by the cell in the new stress state or those proteins made structurally aberrant by the change in cell water chemistry. The hydrolysates from these proteins could be recycled by the cell catabolic machinery, providing raw material for new, stress-induced protein species. One such set of proteins may be the equivalent of TIP and PIP (tonoplast intrinsic protein and plasma membrane intrinsic protein) aquaporins (Kjellbom et al., 1999), examples of which have been identified in Rhizobium-infected host cells (Miao and Verma, 1993).
In the root nodule, two distinct arenas for PsCYP15A involvement have been recognized (Kardailsky and Brewin, 1996): the apex/meristem region (where the most intense levels of gene expression are observed) and the infected cell/symbiosome region. It is likely that PsCyp15a expression in the infected host cells mirrors that of the shoot cortex, where the same large vacuolar bodies are observed and antigenic vesicles are seen in the cytoplasm. However, in the apical region of the nodule, where the cells are small and there are very few vacuoles, the antigen accumulates almost exclusively in cytoplasmic vesicles. A prerequisite for cell expansion is the need to develop a positive turgor pressure (Kutschera and Kende, 1988) and, in this respect, the meristematic and post-meristematic cells of the nodule apex may resemble the wilt-induced cells in mature tissues. Incidentally, by using the PsCyp15a promoter fused to a uidA reporter gene, we have recently demonstrated expression of PsCyp15a in other meristems of the plant, namely those of lateral roots (J.L. Vincent and N.J. Brewin, unpublished results).
In conclusion, we have demonstrated that PsCYP15A is a Cys protease associated with the vacuole, cytoplasmic vesicles, and non-senescent symbiosomes in pea. The number of homologs so far studied provides a broad forum for speculation about possible functional roles for this class of proteases. Further study of PsCyp15a within the root nodule provides an opportunity to correlate its role in symbiosis to functionality in the whole plant. In keeping with this aim, a homolog of PsCyp15a has recently been isolated from Medicago sativa (accession no. AJ245868; Vincent et al., 2000), enabling further molecular genetic investigations to be conducted in the model legume Medicago truncatula.
MATERIALS AND METHODS
Plant Materials
Pea (Pisum sativum var Wisconsin Perfection) plants were cultivated as described previously (Kardailsky et al., 1996). Root nodules were induced by inoculation with Rhizobium leguminosarum bv viciae strain 3841 (Wang et al., 1982), and mature root nodules were collected 4 weeks postinoculation. Perisymbiont fluid was obtained from nodule homogenates by differential centrifugation, as described previously (Dahiya et al., 1997). Tissue intended for biochemical purification was frozen in liquid nitrogen and stored at −70°C until required. Experiments on wilted pea shoots were carried out as described by Guerrero et al. (1990), taking hydroponically grown pea plants and excising the shoot at the base of the stem before immersion of the lower part of the stem in hypertonic solution or water. The stem was cut again while immersed to prevent air-locking, and the wilt-induction period was maintained in light conditions for 4 h. This study used 0.6 m mannitol solution as the osmoticum.
Protein Extraction and Estimation
Frozen plant tissue was ground to powder with a mortar and pestle and reconstituted in PBS at 4°C. Insoluble material was removed by centrifugation at 20,000g in a J2-21 centrifuge (Beckman Instruments, Fullerton, CA) at 4°C, and the concentration of soluble protein was estimated using a protein quantitation assay (Bio-Rad Laboratories, Hercules, CA) with bovine serum albumin (BSA) as a standard.
DNA and RNA Analysis
Total RNA from mature root nodules was prepared using the RNeasy Kit (Qiagen, Crawley, UK) as outlined by the manufacturer. Poly(A+) mRNA was purified from total RNA using oligo(dT)n-coated paramagnetic beads (Dynal, Bromborough, UK), following the manufacturer's instructions. Purification of plasmid DNA, subcloning, and restriction endonuclease analyses were performed as described by Sambrook et al. (1989).
Reverse Transcriptase (RT)-PCR
RT-PCR was used to amplify an approximately 700-bp fragment of PsCyp15a (corresponding to bases 481–1,146 of the cDNA) from 1 μg of root nodule mRNA. The translated product represents approximately 220 residues of the mature PsCYP15A peptide. The gene-specific primers were JLV2 (5′- GAC TGG TAC CAG CAT CCC TG –3′, in which the underlined bases represent a KpnI site) and JLV4 (5′-GTG AGC TCC GTC AAG GAC CAG GG-3′, in which the underlined bases represent a SacI site). AmpliTaq DNA polymerase (Perkin-Elmer-Applied Biosystems, Foster City, CA) used to amplify the 700-bp product from a cDNA population representing PsCyp15a transcripts primed in a Superscript (Life Technologies/Gibco-BRL, Basingstoke, UK) reverse transcription reaction using the gene-specific primer JLV2. A PTC thermocycler (MJ Research, Dunmow, UK) was used for all stages of the RT-PCR. Conditions for reverse transcription closely followed recommendations by Byrne et al. (1988), with the extension temperature set at 47°C for 1 h. PCR conditions closely followed recommendations by Saiki et al. (1988), cycle parameters being 94°C hot-start for 5 min followed by 55°C annealing step for 1 min and a 72°C extension step of 3.5 min. To complete the cycle, a 94°C denaturation step of 30 s was added. These steps were repeated 40 times to a 4°C hold. Confirmation of PsCyp15a identity was made by automated sequencing of both strands using a cycle sequencing kit (Perkin-Elmer-Applied Biosystems) and a sequencer (ABI 373, Perkin-Elmer-Applied Biosystems).
Purification of Recombinant Protein
Recombinant PsCYP15A protein was produced in E. coli using the pRSET expression system (Invitrogen, San Diego). pRSETB was linearized at the SacI and KpnI sites of the polylinker, and the 700-bp RT-PCR product was digested with the same enzymes to produce compatible termini. Ligation of these fragments resulted in an in-frame fusion of the PsCyp15a RT-PCR product with the pRSET B fusion tag under control of the T7 promoter. The resulting expression plasmid was named pRSETCyp7 and was transformed into the E. coli expressor strain K38 harboring the plasmid pGP1-2, necessary for heat-inducible T7 RNA polymerase expression (Russel and Model, 1984; Tabor and Richardson, 1985).
For production of the recombinant protein, 500-mL cultures in Luria-Bertani broth, containing kanamycin (20 μg mL−1) and ampicillin (100 μg mL−1), were incubated at 30°C with vigorous shaking until the OD600 was 0.8 to 1.0. Expression was induced by the addition of 1 mm isopropylthio-β-galactoside and by increasing the temperature to 42°C. Incubation of the cultures under these conditions was maintained for 1.5 h, at which point the culture was centrifuged at 4°C to pellet the bacteria, and the supernatant was removed. The pellet was washed with PBS and lysed with 6 m guanidium chloride. Recombinant protein was purified from the lysate by affinity chromatography through a nickel resin column (ProBond, Invitrogen), due to the presence of a hexa-His nickel-binding motif in the fusion tag. Elution of recombinant protein was effected by a decreasing gradient of pH in a urea buffer, with the optimal elution point being pH 4.0.
Recombinant protein was further purified by large-scale SDS-PAGE. After staining with Coomassie Blue, the recombinant protein band was excised from the gel and electroeluted under denaturing conditions using an electroelution module (model 422 Electro-Eluter, Bio-Rad Laboratories). Recombinant protein was dialyzed against PBS at 4°C to remove SDS, and then concentrated by centrifugation in a protein concentrator (Centricon, Amicon, Beverly, MA).
Development of Polyclonal Antiserum
Purified recombinant PsCYP15A was used to generate antiserum in a New Zealand White rabbit. Recombinant antigen (100 μg) in PBS was emulsified with an equal volume of Freund's complete adjuvant (Sigma-Aldrich, St. Louis) and injected subcutaneously. Subsequent boosts of equivalent dosage were made at 4-week intervals in Freund's incomplete adjuvant (Sigma-Aldrich). A preimmune bleed was taken before immunization commenced, and test bleeds were taken for analysis 10 d after injections. To improve the immune response, the final two injections were made with a mixture of antigens that had been cross-linked with 2,4-dinitrobenzene sulfonic acid (Sigma-Aldrich) and heat treated (Amkraut et al., 1966; Harlow and Lane, 1988). The final bleed was taken 2 weeks after the last injection.
Antibodies that cross-reacted with bacterial antigens were removed from crude preparations of immune and preimmune antisera by pre-absorption against E. coli and R. leguminosarum bv viciae strain 3841 antigens (Gruber and Zingales, 1995). Further purification of PsCYP15A-specific antibodies from the pre-absorbed immune serum was performed by immunoadsorption (Smith and Fisher, 1984) using immobilized recombinant PsCYP15A on reinforced nitrocellulose membranes (Optitran, Schleicher & Schuell, Keene, NH). Pre-absorbed preimmune serum was also processed in this fashion. PsCYP15A-specific serum was named R79.
Immunoblotting
Following SDS-PAGE (MiniProtean II system, Bio-Rad Laboratories), western blotting of proteins was performed as described previously (Dahiya et al., 1997) using nitrocellulose membranes (Schleicher & Schuell), and 3-[cyclohexylamino]-1-propanesulfonic acid (CAPS) transfer buffer. Immunolabeling of western blots was performed as described previously (Perotto et al., 1991) using alkaline phosphatase-conjugated secondary antibodies and nitroblue tetrazolium as a substrate.
Native Protease Purification and PepTag Assay
Immunopurified R79 (or preimmune serum) was bound to anti-rabbit IgG-conjugated paramagnetic beads (Dynal). To isolate plant-derived protein antigen, the antibody-bound beads were incubated with total nodule soluble protein at 4°C, separated using a magnet, washed in PBS at 4°C, and stored at −70°C until use.
Proteolytic activity of immunopurified, native plant protein was measured using a protease assay (PepTag, Promega, Madison, WI) performed in sodium phosphate buffer at pH 5.3 (Yu and Greenwood, 1994) at room temperature for 16 h. Fluorochrome products with different electrophoretic mobilities were identified by electrophoresis in Tris-buffered 1% (w/v) agarose gels, with applied voltage of 100 V for 40 min. Separated products were imaged and recorded using UV illumination (GelDoc1000, Bio-Rad Laboratories).
Immunolocalization
Immunolocalization by laser-scanning confocal microscopy was performed on 100-μm sections of plant material that had been fixed in 4% (w/v) formaldehyde and stored in pH 6.9 PEM buffer (50 mm PIPES/KOH, 5 mm EGTA, and 5 mm MgSO4)/0.02% (w/v) NaN3, at 4°C. Sections were prepared on a vibratome (series 1000, TAAB, Allermarston, UK) and mounted on microscope slides coated with γ-aminopropyltriethoxysilane (APTES, Sigma-Aldrich). Sections were treated with 0.05% (w/v) cellulase (Onozuka, Yakult Honsha, Tokyo) for 10 min before blocking with 3% (w/v) BSA in PEM for 1.5 h. Primary antiserum (previously purified by immunoadsorption to gel-purified PsCYP15A) was applied in a 1% (w/v) BSA solution in PEM and incubated overnight at 4°C. Sections were then washed in excess PEM for 1 h, followed by another blocking step in 1% (w/v) BSA/PEM for 30 min. Secondary antibodies were applied as 1:50 to 1:100 dilutions in 1% (w/v) BSA solutions in PEM and incubated for 2 h at 37°C. Fluorescent conjugates of secondary antibodies were anti-rabbit Alexa Fluor 568 and anti-mouse Alexa Fluor 488 supplied by Molecular Probes Europe (Cambridge, UK). Washing of specimens was then completed over 2 d at room temperature in excess PEM before mounting in fluorescent mounting medium (VectaShield, Vector Laboratories, Burlingame, CA) and viewing on a laser-scanning confocal microscope (TCS-SP, Leica Microsystems, Wetzlar, Germany). Composite images were generated using processing software (TCS-NT, Leica Microsystems) and arranged for publication using Photoshop 5.0 (Adobe Systems, Mountain View, CA).
Immunolocalization for transmission electron microscopy was carried out as previously described (Dahiya et al., 1997) using antiserum purified by immunoadsorption to gel-purifed PsCYP15A.
NOTE ADDED IN PROOF
Using a proteomics approach, another possible ortholog of PsCyp15a (SwissProt accession no. P25804) has recently been identified as a component of isolated symbiosomal membranes from soybean nodules (Panter et al., 2000); this provides further evidence that this class of protease is widespread in legume nodules and that it is associated with the symbiosome compartment.
ACKNOWLEDGMENTS
We thank Preeti Dahiya and Brian Wells for guidance in immunolabeling for electron microscopy and Ali Beven and Peter Shaw for advice on fluorescent probes and for the kind gift of monoclonal antibody 1413.
Footnotes
This work was supported by the UK Biotechnology and Biological Research Council and by a Biotechnology and Biological Science Research Council-funded post-graduate studentship (to J.L.V.).
LITERATURE CITED
- Amkraut AA, Garvey JS, Campbell DH. Competition of haptens. J Exp Med. 1966;124:293–306. doi: 10.1084/jem.124.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini B, Perotto N, Gasverde E, Dahiya P, Guldmann L-LG, Brewin NJ, Bonfante P. Transcription of a gene encoding a lectin-like glycoprotein is induced in root cells harbouring arbuscular mycorrhizal fungi in Pisum. Mol Plant Microbe Interact. 1999;12:785–791. [Google Scholar]
- Barret A, Rawlings ND, Woessner JF. Handbook of Proteolytic Enzymes. San Diego: Academic Press; 1998. [Google Scholar]
- Becker C, Fischer J, Nong VH, Munitz K. PCR cloning and expression analysis of cDNAs encoding cysteine proteinases from germinating seeds of Vicia sativa. Plant Mol Biol. 1994;26:1207–1212. doi: 10.1007/BF00040701. [DOI] [PubMed] [Google Scholar]
- Brewin NJ. Development of the legume root nodule. Annu Rev Cell Biol. 1991;7:191–226. doi: 10.1146/annurev.cb.07.110191.001203. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Ainsworth C. Leaf senescence in Brassica napus: cloning of senescence related genes by subtractive hybridisation. Plant Mol Biol. 1997;33:821–834. doi: 10.1023/a:1005774212410. [DOI] [PubMed] [Google Scholar]
- Byrne BC, Li JJ, Sninsky J, Poiesz BJ. Detection of HIV-1 RNA sequences by in vitro DNA amplification. Nucleic Acids Res. 1988;16:4165. doi: 10.1093/nar/16.9.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C, Bourgeois E, Pradet A, Raymond P. Molecular cloning and characterization of 6 cDNAs expressed during glucose starvation in excised maize (Zea mays) root tips. Plant Mol Biol. 1995;28:473–485. doi: 10.1007/BF00020395. [DOI] [PubMed] [Google Scholar]
- Dahiya P, Kardailsky IV, Brewin NJ. Immunolocalization of PsNLEC-1, a lectin-like glycoprotein expressed in developing pea nodules. Plant Physiol. 1997;115:1431–1442. doi: 10.1104/pp.115.4.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domoto C, Watanabe H, Abe M, Abe K, Arai S. Isolation and characterization of 2 distinct cDNA clones encoding corn seed cysteine proteinases. Biochim Biophys Acta Gene Struct Exp. 1995;1263:241–244. doi: 10.1016/0167-4781(95)00138-7. [DOI] [PubMed] [Google Scholar]
- Goetting-Minesky MP, Mullin BC. Differential gene-expression in an actinorhizal symbiosis: evidence for a nodule-specific cysteine proteinase. Proc Natl Acad Sci USA. 1994;91:9891–9895. doi: 10.1073/pnas.91.21.9891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A, Zingales B. Alternative method to remove antibacterial antibodies from antisera used for screening of expression libraries. Biotechniques. 1995;19:28–30. [PubMed] [Google Scholar]
- Guerrero FD, Jones JT, Mullet JE. Turgor-responsive gene-transcription and RNA levels increase rapidly when pea shoots are wilted: sequence and expression of 3-inducible genes. Plant Mol Biol. 1990;15:11–26. doi: 10.1007/BF00017720. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Jones JT, Mullet JE. A salt-inducible and dehydration-inducible pea gene, Cyp15a, encodes a cell-wall protein with sequence similarity to cysteine proteases. Plant Mol Biol. 1995;28:1055–1065. doi: 10.1007/BF00032666. [DOI] [PubMed] [Google Scholar]
- Kardailsky IV, Brewin NJ. Expression of cysteine protease genes in pea nodule development and senescence. Mol Plant-Microbe Interact. 1996;9:689–695. doi: 10.1094/mpmi-9-0689. [DOI] [PubMed] [Google Scholar]
- Kardailsky IV, Sherrier DJ, Brewin NJ. Identification of a new pea gene, PsNlec1, encoding a lectin-like glycoprotein isolated from the symbiosomes of root nodules. Plant Physiol. 1996;111:49–60. doi: 10.1104/pp.111.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjellbom P, Larsson C, Johansson I, Karlsson M, Johanson U. Aquaporins and water homeostasis in plants. Trends Plant Sci. 1999;4:308–314. doi: 10.1016/s1360-1385(99)01438-7. [DOI] [PubMed] [Google Scholar]
- Koizumi M, Yamaguchi-Shinozaki K, Tsuji H, Shinozaki K. Structure and expression of 2 genes that encode distinct drought-inducible cysteine proteinases in Arabidopsis thaliana. Gene. 1993;129:175–182. doi: 10.1016/0378-1119(93)90266-6. [DOI] [PubMed] [Google Scholar]
- Kutschera U, Kende H. The biophysical basis of elongation growth in internodes of deep-water rice. Plant Physiol. 1988;88:361–366. doi: 10.1104/pp.88.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linthorst HJM, Vanderdoes C, Brederode FT, Bol JF. Circadian expression and induction by wounding of tobacco genes for cysteine proteinase. Plant Mol Biol. 1993a;21:685–694. doi: 10.1007/BF00014551. [DOI] [PubMed] [Google Scholar]
- Linthorst HJM, Vanderdoes C, Vankan JAL, Bol JF. Nucleotide sequence of a cDNA clone encoding tomato (Lycopersicon esculentum) cysteine proteinase. Plant Physiol. 1993b;101:705–706. doi: 10.1104/pp.101.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty F. Plant vacuoles. Plant Cell. 1999;11:587–599. doi: 10.1105/tpc.11.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee RA, Adams S, Matthews JA, Smith CJ, Smith H. Molecular cloning of 2 cysteine proteinases from paw-paw (Carica papaya) Biochem J. 1986;237:105–110. doi: 10.1042/bj2370105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor RB. Bacteroids in the Rhizobium-legume symbiosis inhabit a plant internal lytic compartment: implications for other microbial endosymbioses. J Exp Bot. 1989;40:831–839. [Google Scholar]
- Miao GH, Verma DPS. Soybean nodulin-26 gene encoding a channel protein is expressed only in the infected cells of nodules and is regulated differently in roots of homologous and heterologous plants. Plant Cell. 1993;5:781–794. doi: 10.1105/tpc.5.7.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nong VH, Becker C, Muntz K. cDNA cloning for a putative cysteine proteinase from developing seeds of soybean. Biochim Biophys Acta Gene Struct Exp. 1995;1261:435–438. doi: 10.1016/0167-4781(95)00038-i. [DOI] [PubMed] [Google Scholar]
- Panter S, Thompson R, de Bruxelles G, Laver D, Trevaskis B, Udvardi M. Identification with proteomics of novel proteins associated with the peribacteroid membrane of soybean root nodules. Mol Plant-Microbe Interact. 2000;13:325–333. doi: 10.1094/MPMI.2000.13.3.325. [DOI] [PubMed] [Google Scholar]
- Perotto S, Vandenbosch KA, Butcher GW, Brewin NJ. Molecular composition and development of the plant glycocalyx associated with the peribacteroid membrane of pea root nodules. Development. 1991;112:763–773. [Google Scholar]
- Pfeiffer NE, Torres CM, Wagner FW. Proteolytic activity in soybean root nodules: activity in host-cell cytosol and bacteroids throughout physiological development and senescence. Plant Physiol. 1983;71:797–802. doi: 10.1104/pp.71.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pladys D, Rigaud J. Senescence in french bean nodules: occurrence of different proteolytic activities. Physiol Plant. 1985;63:43–48. [Google Scholar]
- Pladys D, Vance CP. Proteolysis during development and senescence of effective and plant gene-controlled ineffective alfalfa nodules. Plant Physiol. 1993;103:379–384. doi: 10.1104/pp.103.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae AL, Perotto S, Knox JP, Kannenberg EL, Brewin NJ. Expression of extracellular glycoproteins in the uninfected cells of developing pea nodule tissue. Mol Plant-Microbe Interact. 1991;4:563–570. [Google Scholar]
- Ribeiro A, Akkermans ADL, Vankammen A, Bisseling T, Pawlowski K. A nodule-specific gene encoding a subtilisin-like protease is expressed in early stages of actinorhizal nodule development. Plant Cell. 1995;7:785–794. doi: 10.1105/tpc.7.6.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel M, Model P. Replacement of the fip gene of Escherichia coli by an inactive gene cloned on a plasmid. J Bacteriol. 1984;159:1034–1039. doi: 10.1128/jb.159.3.1034-1039.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Smith DE, Fisher PA. Identification, developmental regulation, and response to heat-shock of 2 antigenically related forms of a major nuclear-envelope protein in Drosophila embryos: application of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J Cell Biol. 1984;99:20–28. doi: 10.1083/jcb.99.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroeher VL, Maclagan JL, Good AG. Molecular cloning of a Brassica napus cysteine protease gene inducible by drought and low temperature stress. Physiol Plant. 1997;101:389–397. [Google Scholar]
- Tabor S, Richardson CC. A bacteriophage T7 RNA polymerase promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance CP, Heichel GH, Barnes DK, Bryan JW, Johnson LE. Nitrogen fixation, nodule development, and vegetative regrowth of alfalfa (Medicago sativa L.) following harvest. Plant Physiol. 1979;64:1–8. doi: 10.1104/pp.64.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. Proteolysis in plants: mechanisms and functions. Plant Mol Biol. 1996;32:275–302. doi: 10.1007/BF00039386. [DOI] [PubMed] [Google Scholar]
- Vincent JL, Knox MR, Ellis THN, Kaló P, Kiss GB, Brewin NJ (2000) Nodule-expressed Cyp15a cysteine protease genes map to synthetic genome regions in Pisum and Medicago spp. Mol Plant-Microbe Interact (in press) [DOI] [PubMed]
- Wang TL, Wood EA, Brewin NJ. Growth regulators, Rhizobium and nodulation in peas: indole-3-acetic acid from the culture medium of nodulating and non-nodulating strains of R. leguminosarum. Planta. 1982;155:345–349. doi: 10.1007/BF00429463. [DOI] [PubMed] [Google Scholar]
- Williams J, Bulman M, Huttly A, Phillips A, Neill S. Characterization of a cDNA from Arabidopsis thaliana encoding a potential thiol protease whose expression is induced independently by wilting and abscisic acid. Plant Mol Biol. 1994;25:259–270. doi: 10.1007/BF00023242. [DOI] [PubMed] [Google Scholar]
- Yu WJ, Greenwood JS. Purification and characterization of a cysteine proteinase involved in globulin hydrolyzation in germinated Vicia faba. J Exp Bot. 1994;45:261–268. [Google Scholar]