Abstract
Neurodegenerative disorders and type 2 diabetes are global epidemics compromising the quality of life of millions worldwide, with profound social and economic implications. Despite the significant differences in pathology – much of which are poorly understood – these diseases are commonly characterized by the presence of cross-β amyloid fibrils as well as the loss of neuronal or pancreatic β-cells. In this review, we document research progress on the molecular and mesoscopic self-assembly of amyloid-beta, alpha synuclein, human islet amyloid polypeptide and prions, the peptides and proteins associated with Alzheimer’s, Parkinson’s, type 2 diabetes and prions diseases. In addition, we discuss the toxicities of these amyloid proteins based on their self-assembly as well as their interactions with membranes, metal ions, small molecules and engineered nanoparticles. Through this presentation we show the remarkable similarities and differences in the structural transitions of the amyloid proteins through primary and secondary nucleation, the common evolution from disordered monomers to alpha-helices and then to β-sheets when the proteins encounter the cell membrane, and, the consensus (with a few exceptions) that off-pathway oligomers, rather than amyloid fibrils, are the toxic species regardless of the pathogenic protein sequence or physicochemical properties. In addition, we highlight the crucial role of molecular self-assembly in eliciting the biological and pathological consequences of the amyloid proteins within the context of their cellular environments and their spreading between cells and organs. Exploiting such structure-function-toxicity relationship may prove pivotal for the detection and mitigation of amyloid diseases.
Keywords: amyloid, oligomer, self-assembly, neurodegenerative disorders, Aβ, IAPP, α-synuclein, prion, theranostics
Graphical Abstract

1. Introduction
Molecular self-assembly is an ubiquitous phenomenon across all living systems: from the polymerization of tubulins and actins into microtubules and actin filaments, to the organization of lipids, transmembrane/peripheral proteins and ion channels into cell membranes, to the assembly of DNA and histones into chromatin fibers and solenoids, and to the aggregation of peptides and proteins intra- or extracellularly evolving from functional monomers to toxic oligomers, amyloid fibrils and plaques. Fundamental to these processes are interactions between the molecular constituents of the assemblies, as well as interactions between the molecular constituents and their associated chaperones, ligands, ions, molecular complexes and organizations, driven by kinetic and thermodynamic processes to elicit desirable biological functions or malfunctions and diseases.
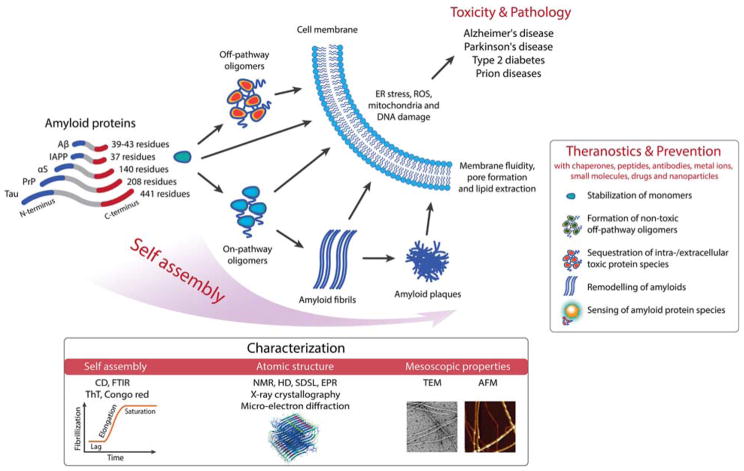
In this review, we attempt to draw parallels from the atomic and mesoscopic structures of five major classes of amyloid proteins in self-assembly, namely, amyloid-beta (Aβ), tau, alpha-synuclein (αS), prions, and human islet amyloid polypeptide (IAPP), as well as the biological and pathological endpoints these assemblies elicit in host systems (Fig. 1). The amyloid aggregation of these peptides has been implicated in Alzheimer’s, Parkinson’s, prion diseases and type 2 diabetes mellitus, or generically referred to as neurodegenerative disorders and T2D that debilitate hundreds of millions of people worldwide. In vitro, such peptides/proteins fibrillate on the timescales of tens of minutes for IAPP to days for Aβ and αS, characterized by a sigmoidal kinetic curve consisting of a lag phase, an elongation phase, and a saturation phase.1 The lag phase is where nucleation is initiated through protein misfolding and where intrinsic seeds and/or oligomers are formed, the elongation phase corresponds to the addition of monomers to growing protofilaments, while the saturation phase is where protofilaments associate through self-assembly to render amyloid fibrils. In addition to primary nucleation, secondary nucleation through the combination of both monomeric and aggregated species is also feasible.2, 3 It has been suggested that the amyloid state is perhaps available to any polypeptide chain4–9 and represents the energetically most favorable state even compared to native proteins.1 In vivo, however, the development of amyloids and plaques in the brain or pancreatic islets often takes decades, or ~10,000 times longer. Such drastic differences in fibrillization may originate from the crowded hierarchical cellular environments, where amyloid proteins are synthesized and then translocate and spread through inter- and intra-molecular assembly, chaperoned by proteins (e.g. insulin for IAPP) or modulated by pH and ionic strength. Accordingly, while the main purpose of this review is to highlight the structure-function-toxicity triangle of a selected few amyloid proteins, another goal of this presentation is to draw the readers’ attention from focusing exclusively on amyloid proteins to the environments of the culprits at large, which undoubtedly also contribute to the pathologies of the amyloid diseases. Such perspective may prove beneficial to the development of mitigation strategies and theranostics against amyloidogenesis that has become increasingly perilous to modern society.
Figure 1.
Scope of the present review, highlighting protein self-assembly, its biological and pathological implications, theranostics and prevention. Aβ: amyloid-beta; IAPP: islet amyloid polypeptide; αS: alpha-synuclein; PrP: prion protein; CD: circular dichroism spectroscopy; FTIR: Fourier transform infrared spectroscopy; ThT: thioflavin T assay; NMR: nuclear magnetic resonance; HD: hydrogen-deuterium exchange; SDSL: site directed spin labelling; EPR: electron paramagnetic resonance; TEM: transmission electron microscopy; AFM: atomic force microscopy. ER: endoplasmic reticulum; ROS: reactive oxygen species.
In terms of content, this review consists of 6 sections: section 1 offers an introduction to protein self-assembly and amyloid diseases; sections 2–5 review the structure, function and toxicity characteristics of Aβ, tau, IAPP, αS and prions, loosely following their increasing numbers of amino acids (residues); section 6 provides a summary. Aβ and tau, despite their great contrast in chain length, both contribute to the AD pathology and hence are presented together. Although Aβ is slightly longer than IAPP in chain length, Aβ is the most studied of all amyloid proteins10 and is therefore discussed in the early section of this review.
2. Aβ, tau and Alzheimer’s disease
The hallmark of Alzheimer’s disease (AD) is the accumulation of toxic aggregates that impair synaptic function and induce cognitive decline. The first reported occurrence of cognitive disorder linked to AD was in 1907 by Alzheimer, who observed two types of abnormality in a brain autopsy that he attributed to be the cause of an unusual type of dementia.11 The discovery of neuritic plaques (or miliary foci) and neurofibrillary tangles (NFTs) was immediately linked to the dystrophic neuronal process, and later Aβ fibrils12, 13 and hyperphosphorylated tau tangles14 were isolated and characterized (and proposed to cause dementia). Characterizations of the monomeric forms of the molecules found in these neurotoxic deposits have led to greater understanding of the pathways leading to AD, with a particular focus on the structure-function relationship between aggregates and neurotoxicity. However, almost all drugs tested thus far in clinical trials have failed or shown limited impact on AD.
Tau is a neuronal protein associated with microtubules and may regulate neuron morphology. There are six main tau isoforms in the brain and central nervous system (CNS). The longest human isoform has 441 residues with a high proportion of phosphorylable residues (serine and threonine) and a low proportion of hydrophobic amino acids. Tau protein in solution is considered an intrinsically disorder protein (IDP) and behaves as a random coil,15 although modifications by phosphorylation may lead to an increase in α-helix or β-sheet regions. Aggregation of totally or partially disordered proteins is associated with many neurodegenerative diseases, including AD.16 However, the molecular mechanism of aggregation and the structure of the aggregated form remain controversial. Tau is mainly an axonal protein but in AD and other tauopathies it is also present at dendritic spines and may play a toxic role. The tau hypothesis of AD considers that excessive phosphorylation of tau protein can result in the self-assembly of tangles of paired helical filaments (PHFs) and straight filaments which are involved in the pathogenesis of AD and other tauopathies. These neurofibrillary tangles are insoluble structures that impair axonal transport and lead to cell death. The molecular structures of PHFs and tau protein are not well defined.
NMR data17 have revealed that 343 of the 441 amino acids in tau are disordered with six segments of the sequence displaying propensity to form β-strands, three segments showing poly-Pro helices and two segments with a transient α-helix structure. In particular, aggregation of tau is believed to be strongly associated with two short residue sequences:18–21 the first in the third repeat fragment (R3, i.e. VQIVYKPVDLSKVTSKCGSLGNIHHK) of the microtubule binding domain of tau, VQIVYK, or the mutant VQIINK, in the second repeat fragment. The aggregation of the R3 fragment has been extensively studied in the presence of polyanions, such as heparin, pointing at the formation of fibrillar structures with similar features as those assembled from the pristine tau protein.22 In the absence of heparin, however, the same R3 fragment has been shown to self-assemble into giant amyloid ribbons of remarkable aspect ratios.23
The tau protein is a highly dynamic structure. An NMR study of a peptide derived from tau showed that phosphorylation stabilized the α-helix structure,24 suggesting a possible higher content of α-helices in hyperphosphorylated tau in PHFs. Tau protein can form dimers, oligomers and larger aggregates and fibrils. However, in this review we focus on the Aβ peptide rather than tau aggregates, as greater structural details are available for the former.
2.1 Role of APP and production of Aβ
Aβ peptides are produced by an intrinsic cleavage of the amyloid precursor protein (APP) that is an integral membrane protein encoded on chromosome 21 by the APP gene.25 It is accepted that patients with trisomy 21 (Down syndrome) overexpress APP and develop AD-like senile plaques in their brain.26, 27 Yet, the physiological function of APP remains uncertain, mostly because APP is part of a gene family with overlapping function (e.g. producing the amyloid precursor-like proteins APLP1 and APLP2) and is subject to various post-expression modifications.28 Still, only APP generates the Aβ fragment. APP modulates critical features in brain development since APP knock-out mice are viable but exhibit reduced body weight and brain mass29 with increased brain levels of copper,30 cholesterol and sphingolipid.31 Interestingly, reintroducing the APP ectodomain, which is produced by cleavage of the membrane-anchored APP, improved cognitive function and synaptic density32, 33 and acted as an apoptosis modulator through caspases activations.34 The intracellular C-terminal domain also has a functional role in sorting APP and, in particular, the highly conserved YENPTY cytoplasmic sequence is prone to interaction with other proteins, such as X11 and Fe65, which are postulated to regulate APP internalization.35
The location and sequence of the proteolytic cleavage of APP are critical to AD. APP is primarily translocated to the cell surface (short residence time) where α-secretase and then γ-secretase produce APPsα, p3 and AICD fragments, which are not amyloidogenic. However, when APP is relocated through endocytosis (rapid turnover due to the YENPTY sequence) into endosomes containing the β-secretase (also called BACE1) and the γ-secretase, then APPsβ, the toxic Aβ peptides and AICD fragments are produced (Fig. 2). BACE1 is an aspartyl protease that has optimum efficiency at pH 4.5.36 Interestingly, acid pH can promote greater aggregation rate of Aβ peptides37 due to the protonation state of the three histidines (His6, His13 and His14), and also attenuate lysosomal degradation of Aβ peptides.38 Furthermore, if APP is relocated to the trans Golgi network instead of the ER, BACE1 can produce N-truncated Aβ peptides which are prone to rapid pyroglutamylation.39 These species have been characterized as highly toxic40 and found in intracellular, extracellular and vascular Aβ deposits in AD brain tissue,41 while unmodified peptides are primarily located in endosomal compartments and are eventually exocytosed into the extracellular space.
Figure 2.
(a) Non-amyloidogenic pathway triggered by the location of APP at the plasma membrane interface; and (b) Amyloidogenic pathway induced through APP endocytosis into endosomal vesicles containing the protease BACE1. Aβ peptides are then prone to aggregation and can be either secreted extracellularly or remain in the intracellular space to target other organelles, such as mitochondria, or be degraded by proteases such as cathepsin B.
It is noteworthy that intracellular pools of Aβ peptides are pointed as the most toxic species causing the death of neurons.42, 43 The physiological function of these Aβ peptides, usually 39–43 residues in length, is unconfirmed. However, during excitatory neuronal activity, an increase in excretion of Aβ peptides is observed44 with the effect of downregulating excitatory synaptic transmission.45 Thus, Aβ peptides are an important modulator of memory, since inhibition of peptide production impairs learning.46 Aβ(1–40) is the most abundant Aβ isoform found in its soluble form in plasma, cerebrospinal fluid and brain interstitial fluid47 but is also a major component in amyloid plaques. Interestingly, the level of the fast-aggregating isoform Aβ(1–42) is a biomarker for detecting amyloid pathologic changes in the brain and cerebral vessels48 and, moreover, the relative Aβ(1–42)/Aβ(1–40) ratio is markedly increased in AD.49 Overall, both the concentration and location of Aβ peptides are critical for brain function, thereby complicating therapeutic strategies against AD.
2.2 Atomic structures of Aβ40 & Aβ42, post-modifications and amyloid fibrils
Aβ peptides vary in length due to the multiple cleavage sites recognized by the secretases, but the most abundant species are Aβ(1–40) and Aβ(1–42), whose sequences are shown in Fig. 3. Furthermore, Aβ peptides can be degraded by proteases such as insulin degrading enzymes,50 neprilysin51 and cathepsin B,52 which render the fragments non-amyloidogenic. Aβ peptides can be subject to post-translational modifications including pyroglutamate formation (Glu3, 11 and 22),53 phosphorylation (Ser8 and 26),54 dityrosine formation (Tyr10)55 and oxidation (Met35)56 (see Fig. 3).
Figure 3.
(a) Aβ peptide sequence (CINEMA color code), potential post-modification sites and physicochemical properties; and (b) Intramolecular interactions stabilizing the typical hairpin β-sheet structure. Red and orange dashes: molecular contacts. Blue dashes: side-chain packing. Green: hydrophobic residues. Black: salt bridge. Adapted from reference 57. Copyright Nature Publishing Group.
The Aβ peptide primary sequence exhibits two stretches of hydrophobic residues (17–21 and 32 until C-terminus), which are predicted to adopt a β-sheet conformation.57 Two turn regions are also predicted between residues His6 and Ser8, and between Asp23 and Asn27. Finally, the hydrophilic patches between Asp1 and Lys16 and between Glu22 and Lys28 have either β-sheet or α-helical propensity.58, 59
A missing piece in the AD puzzle is the secondary structures of Aβ peptides immediately after cleavage by the γ-secretase. Firstly, β-CTF is likely to remain structured after cleavage by BACE1, at least up to the transmembrane α-helical segment that contains part of Aβ sequence (Ala28 until C-terminus). Secondly, cleavage by the γ-secretase occurs at intra-membrane and Aβ peptides have a demonstrated affinity for lipid membranes. Thus immediate trafficking is unclear: do Aβ peptides remain in, on or away from the membrane interface? Since lipid membranes modulate the aggregation kinetics,60 this step could play a critical role in subsequent trafficking and AD pathology.
Several Aβ peptide structures have been compiled in the past two decades,61 with a consensus that an unstructured to β-sheet transition first occurs followed by a seeded aggregation process to form oligomeric structures that eventually proceeds to mature amyloid fibrils of 70–120 Å in diameter and an indeterminate length according to electron microscopy.62 Determination of the initial structure of Aβ peptides in native conditions is challenging since the rapid self-aggregation rate accompanied by poor solubility prevents the application of high-resolution techniques such as solution NMR. Nevertheless, several structures of the monomeric peptides have been determined in either organic solvents (dimethylsulfoxide, hexafluoroisopropanol, trifluoroethanol), aqueous solution or detergent micelles (sodium dodecyl sulfate or SDS). In general, Aβ peptides adopted helical conformations with unstructured termini and various turn regions in organic solvents,63–65 aqueous buffer66 and in membrane mimetic detergent micelles.67, 68 Interestingly, most structural studies show that physiological pH,69 low salt concentration70 and higher temperature71 could heavily modulate the peptide conformational transition to β-sheet structure and thereby promoting rapid self-aggregation. The α-helical conformation is proposed as a transient on-pathway intermediate during the complex amyloid fibril formation.72 Indeed, the multistep kinetics of amyloid assembly comprise a lag phase, during which little or no fibril material is formed, followed by an exponential growth of β-sheet-rich aggregates that propagate into amyloid fibrils.1 Increasing evidence suggests that the native partly helical intermediates form the early nucleation seeds during the lag phase.73 The intramolecular interactions stabilizing the β-sheet structure are shown in Fig. 3. In both Aβ isoforms, the turn conformation is stabilized by hydrophobic interactions and by a salt bridge between Asp23 and Lys28. Many side chain contacts are observed, in particular between Phe19 and Ile32, Leu34 and Val36, and between pairs Gln15 - Val36 and His13 -Val40.57,74
Phosphorylation of Aβ peptides, however, does not modify their primary unstructured conformation but leads to a 5-fold reduction in the lag phase due to a faster transition to β-sheet structures, more efficient nucleation and a greater number of oligomeric seeds.75 N-truncated and/or pyroglutamate-modified Aβ peptides form β-sheet structures76 with faster aggregation kinetics than the corresponding full-length peptides, which suggests they could be potential seeding species for aggregate formation. More dramatically, the pyroglutamate-modified Aβ peptides also inhibit the full-length peptide fibrillogenesis and lead to a greater content of small oligomeric species77 that have been demonstrated as the most toxic species.
2.3 Aβ aggregation kinetics and amyloid fibril formation
Knowledge of Aβ aggregation kinetics and mechanisms has been acquired mainly through in vitro studies using synthetic peptides. The kinetics of fibril formation depends on several intrinsic and extrinsic factors. The primary sequence of the peptide modulates the propensity to aggregate into mature fibrils. Post-modifications promote faster aggregation kinetics, as does the Aβ(1–42) sequence compared to the shorter Aβ(1–40) peptide. Extrinsic factors, such as interaction with lipid membranes, can have either a slowing or accelerating effect, rendering determination of a generic model nontrivial.
The lag phase is a period of slow self-aggregation and structural change, likely from helical to β-sheet structures, and characterized by a combination of multiple nucleation and elongation phases2,78, 79 leading to a large number of oligomeric species. Primary nucleation is a fast process (milliseconds) producing the first seeds that are elongated further into fibrils by the addition of monomers. The formation of new aggregates is thought to be dominated by a second nucleation phase where existing fibrils are fragmented to expose new seeds either co-aggregating or recruiting monomers. Interestingly, changes in the primary nucleation rate do not affect the elongation phase while secondary nucleation and fragmentation modify the lag and elongation phases.80 The difference in aggregation rate between the amyloid peptide species, however, may be related to their primary nucleation rate. In fact, Aβ(1–40) monomers, in comparison to Aβ(1–42), exhibit a slower nucleation rate inducing (or caused by) a shift towards nucleation on the fibril surface rather than accumulation of small oligomeric species.79 These fibril-catalyzed secondary nucleation and elongation processes could be a critical difference in relation to the trafficking and toxicity of the Aβ peptide variants.81 Notably, measuring the kinetics of aggregation is challenged by the difficulty in sample preparation,81 especially with regard to starting an experiment without any preformed seeds or a controlled amount of seeds.
The elongation phase is due to the addition of oligomers/monomers onto protofibrils (Fig. 4) or association of protofibrils, in competition with fragmentation of the protofibrils. It is often typified by the half time of the aggregation reaction where the monomers and protofibrils are near equimolar. However, intrinsic and extrinsic factors modulate the stability of the oligomeric species and can template seeds thereby shifting the kinetic rate towards primary nucleation with a faster aggregation rate. The stationary phase represents a steady state where the monomer concentration has reached an equilibrium value and the fibrils are the prevalent species. Notably, AFM studies show that fibrils of different amyloid-forming peptides with diverse macroscopic structures/polymorphism (i.e., ribbon-like versus nanotube-like packing) have a similar Young’s modulus; and thus all Aβ peptides are anticipated to exhibit similar mechanical strength.82
Figure 4.
Aβ aggregation pathways from monomer to fibril formation and toxic outcomes.
Intrinsic and extrinsic factors also play a critical role in the modulation of the lag and elongation phases by changing the concentration of free monomer in solution and/or acting as seeding interfaces. The molecular factors influencing the aggregation kinetic of Aβ peptides are various and difficult to assign to a particular microscopic event (primary versus secondary nucleation, fragmentation, etc.), although some properties are more straightforward to correlate, for instance, the effect of pH as electrostatic interactions mediate either attraction or repulsion of the monomers.78
2.4 Mesoscopic structures of Aβ amyloid fibrils
An original molecular model of Aβ(1–40) fibrils83 based on solid-state NMR data shows the first ~10 residues as structurally disordered while residues 12–24 and 30–40 adopt β-strand conformations and form parallel β-sheets through intermolecular hydrogen bonding. A bend at residues 25–29 brings the two β-sheets in contact through sidechain-sidechain interactions. The cross-β motif common to all amyloid fibrils is a double-layered structure, with in-register parallel β-sheets.83 However, several studies have shown that Aβ peptides form polymorphic fibrils that depend on growth conditions and various oligomeric aggregates. Thus it is unlikely that amyloid fibrils formed in vitro resemble those in the brain. Tycko and co-workers84 seeded fibril growth from brain extract and used solid-state NMR and electron microscopy to gain structural details of the Aβ fibrils. Using tissue from two AD patients they found a single Aβ40 fibril structure for each patient emphasizing the critical role of the seeding process. The molecular structure for Aβ40 fibrils from one patient (Fig. 5) revealed differences from in vitro fibrils. The authors proposed that fibrils may spread from a single nucleation site and that structural variations may correlate with variations in AD.
Figure 5.

Aβ40 structural polymorphism depending on experimental conditions. Rendered from PDB 1BA4, 1AML, 2MVX and 2MJ4.
In comparison with Aβ40, Aβ42 is more neurotoxic and their differences in behaviour may be due to intrinsic differences in structure. An atomic resolution structure of a single form of Aβ42 amyloid fibrils has been derived from high field magic angle spinning NMR spectra.85 The structure shows a dimer of Aβ42 molecules, each containing four β-strands in an S-shaped amyloid fold (Fig. 6). The dimer is arranged to form two hydrophobic cores, capped by a salt bridge at the end with a hydrophilic outer surface. The monomer interface within the dimer shows contacts between M35 of one molecule and L17 and Q15 of the second. Intermolecular constraints show that the amyloid fibrils are parallel in-register. Interestingly, Ishii and co-workers obtained a similar S-shape arrangement (Fig. 6) using ultra-fast spinning solid-state NMR techniques.86 Although knowing atomic details of the fibril may be useful for drug design, nevertheless, the oligomer species are generally accepted as the toxic species.87
Figure 6.
Two recent solid-state NMR Aβ42 fibril structures identifying different assemblies by (left) Griffin and co-workers (PDB: 5kk3)85 and (right) Ishii and co-workers (2MXU).86 High similarity is apparent with the β-sheet domain (purple ribbons) and the unstructured strand (gray ribbons) forming an S-shape. The hydrophobic surfaces are based on Kyte-Doolittle scale (red: hydrophobic, white: neutral, blue: hydrophilic).
2.5 Extrinsic factors modulating Aβ structure, aggregation kinetic and toxicity
2.5.1 Aβ-metal interactions
The role of transition metals in AD is highly debated and a recent literature search using meta-analysis and systemic review methodologies identified a widespread misconception that iron and, to a lesser degree, zinc and copper levels are increased in AD brain.88 Metals were primarily thought to be accumulated in AD brain tissue due to positive staining but quantitative analysis failed to confirm a significant increase,89 and more recent studies have confirmed the artefacts in quantitation due to tissue fixation prior to analysis.90 Qualitative ex vivo and in vitro studies have demonstrated that Aβ peptides recruit iron, zinc and copper with high affinity91 and, more dramatically, induce a redox complex with oxidative stress properties92 that may be related to the toxicity of Aβ peptides and which has been widely accepted as a potential toxic mechanism in AD.93 Two binding sites were identified: the Met35 mediating the Fenton reaction through the electron donor sulfide group;94 and the N-terminal region forming a chelating domain95 of Asp1, His6, His13 and His14, which undergoes a major structural rearrangement during the redox cycle of ROS production.96 Interestingly, in vitro experiments have also shown that metal binding noticeably extends the lag time by stabilizing oligomeric and amorphous aggregates,97 which may explain the poor in vivo detection of the peptide amyloids. Aβ-copper complexes have also been shown to promote lipid peroxidation, in particular within the polyunsaturated chains of membrane lipids, which is another potential toxic mechanism due to neuronal membrane disruption.98
2.5.2 Aβ-membrane interactions
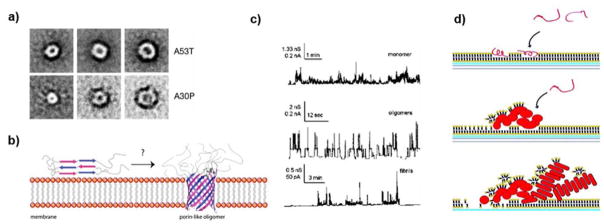
The role of lipids in AD was first suggested by Alzheimer when he discovered adipose inclusions and alterations of lipid composition in brain tissue.11 Several classes of lipids have been investigated for their specific interactions with Aβ peptides, such as cholesterol, gangliosides or anionic phospholipids.99 The lipid membrane interface itself is proposed to be a heterogeneous nucleation site, which modulates Aβ peptide folding kinetics and pathways by reducing the seeding mechanism to a two-dimensional system.100, 101 To date, there is a consensus that lipid bilayer plays a role in Aβ aggregation and may be involved in neurotoxicity. Different model membranes influence the structure and size of Aβ fibrils based on the charge and hydrophobicity of the membrane.60, 102 Membrane-attached oligomers of Aβ40 displayed a β-turn, flanked by two β-sheet regions or an anti-parallel beta-hairpin conformation by Raman spectroscopy and solid-state NMR.103 In contrast to the mature, less-toxic Aβ fibrils, the membrane-attached oligomer appeared to form a β-barrel or ‘porin’-like structure (also refer to Fig. 15b in section 4 for αS), which may account for a mechanism for Aβ toxicity.
Figure 15.
Proposed mechanisms of membrane damage induced by αS aggregation. (a) Projection averages of annular oligomers formed by αS mutants A53T and A30P.271 (b) αS oligomer spans the membrane in a porin-like fashion to induce toxicity.284 (c) Oligomers but not monomers or fibrils induced frequent channel formation in planar lipid bilayers formed from diphytanoylphosphatidylcholine dissolved in n-decane in 1 M KCl, at a bias of +100 mV.297 (d) (Top panel) Monomeric αS adsorbed to a lipid bilayer. (Middle panel) Aggregation of αS monomers causes membrane thinning and lipid extraction. (Lower panel) Further incubation results in assembly of mature αS fibrils and disassembly of the lipid membrane.298 Copyright Nature Publishing Group, Portland Press and The American Chemical Society.
Cholesterol is proposed to be related to AD pathology although cholesterol stabilizes phospholipid bilayers against Aβ.104 Lipid ‘rafts’ or domains in the membrane enriched in cholesterol and sphingolipids could modulate Aβ production, aggregation and toxicity.105 Sanders and co-workers106 showed that the C99 segment of APP bound to cholesterol and proposed that APP might act as a cholesterol sensor critical for the trafficking of APP to cholesterol-rich membrane domains. Cholesterol increases the thickness of phospholipid bilayers and may influence the proteolytic processing of APP and proportion of Aβ40 to Aβ42 produced. Lipid membranes are also susceptible to oxidative stress, as mentioned above as a mechanism for neurodegeneration in AD.98
2.6 Aβ toxicity and Alzheimer’s disease
The physiological markers of AD are progressive cognitive decline, synaptic loss, presence of extracellular β-amyloid plaques and intracellular neurofibrillary tangles ultimately leading to neuronal cell death and a massive brain cell mass loss. To date, there is no drug that can prevent AD neurodegeneration probably because many pathways are activated during the uncontrolled production of Aβ peptides, although several candidates are in ongoing clinical trials. Indeed, it has been demonstrated that Aβ peptides accumulate at synapses, thereby disrupting the whole neuronal network.107 More specifically, complex interactions between Aβ peptides and both synaptic ion channels and mitochondria alter their physiological activities. Aβ peptides and, more particularly, the oligomers of Aβ have affinity for the glutamate108 and acetylcholine109 receptors, mediating the influx/efflux rate of critical mediators such as calcium ions. Aβ-mediated deregulation of these receptors – particularly NMDAR and AMPAR – has been linked to the impairment of plasticity and degeneration of synapses during AD.110
The observation that Aβ oligomers are able to co-localize within mitochondria has exposed another potential neurotoxic pathway.111 Aβ oligomers are able to alter the function of proteins involved in the mitochondrial fusion/fission process, which causes their fragmentation leading to the loss of neuron viability.112 Moreover, accumulation of Aβ peptides in synaptic mitochondria has been shown to decrease mitochondrial respiration and key respiratory enzyme activity, elevate oxidative stress, compromise calcium-handling capacity, and trigger apoptotic signals.113, 114 Finally, intracellular accumulation of Aβ peptides drastically reduces the lysosomal efficiency in removing damaged organelles and unfolded proteins, such as tau.115 Better understanding of the cell biology of the downstream effects of Aβ oligomers may uncover potential therapeutic targets for the prevention of AD.
2.7 Mitigation strategies and theranostics
With increased knowledge of the mechanism of fibril formation from the cleavage of APP to the kinetic modulation by extrinsic factors, several strategies to mitigate AD have emerged. Stabilizing the monomeric form of Aβ peptides is a direct strategy to limit the formation of oligomeric species. Peptides that specifically interact with the pro-aggregating domains have been developed, as recently shown with a cyclopeptide, to inhibit Aβ amyloidogenesis.116 Antibody-based immunotherapy is another strategy to mitigate AD. For instance, a promising candidate, aducanumab, has shown high selectively against aggregated Aβ, induced significant reduction of insoluble and insoluble Aβ population and slowed clinical decline; although the outcome of ongoing Phase 3 clinical trials are needed to confirm these promising observations 117. The affinity of Aβ peptides for transition metals was seen as another area for potential development of AD therapeutics, but so far chelators, such as D-penicillamine, have not produced any clinical improvement.118
After drugs (e.g. bapineuzumab and solanezumab) which seek to lower existing Aβ loads had failed, increasing attention was paid to BACE drugs that interfere with the process that creates Aβ. However, Merck recently closed its trial for the BACE inhibitor, verubecestat, in mild-to-moderate AD after concluding that the drug had little chance of success.119 A particular focus has been to decrease the production of apparently toxic Aβ peptides by inhibiting BACE1 activity.120 For instance, the cholesterol-rich endosomal environment, which promotes selective processing of APP by BACE1, has been pursued as a target using a membrane-anchored BACE1 transition-state inhibitor linked to a sterol moiety to generate highly effective BACE1 inhibitors.121 Treatment with BACE inhibitor IV, which does not change the APP concentration level, was shown to prevent mitochondrial abnormalities caused by Aβ.122 Reducing the activation of caspases, such as caspase 3, can improve neuronal growth and decrease abnormal tau species, which may be an interesting therapeutic pathway for the treatment of AD.123
Since the approval of memantine in 2003, no new AD drug candidate has passed the FDA approval, with an alarming failure rate of 99.6%, the highest in all serious disease research programs.124 A growing strategy in integrating therapeutics and personalized diagnostics has recently emerged as a promising route. Based on nanomedicine, small molecules – necessary to overcome prerequisite to cross the brain blood barrier – have been developed to label and simultaneously inhibit oligomerization of Aβ peptides.125 The term theranostic has thus been coined to characterize these new inhibitor-biomarkers, many based on scaffolds of fluorescent probes such as ThT, to detect fibril formation in vivo and alter their accumulation.126 These new strategies have been made possible by improved understanding of the assembly mechanism of Aβ at the molecular level, which will continue to guide rational drug design against AD.
3. IAPP and type 2 diabetes
3.1 Function of IAPP
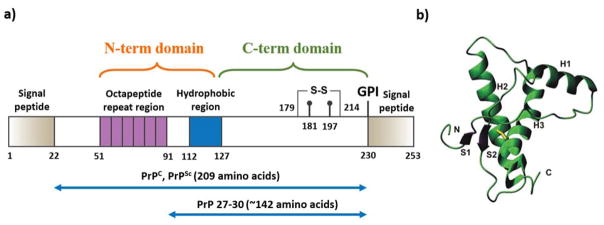
Human islet amyloid polypeptide (IAPP, a.k.a amylin) is a 37-residue peptide hormone co-secreted with insulin from pancreatic β-cell islets. The IAPP physiology has been recently reviewed by Westermark et al.127 Briefly, the peptide is synthesized from a 67-residue precursor peptide, proIAPP, by proteolysis and posttranslational modifications, such as the C-terminal amidation and a disulfide bond formation between residues 2 and 7 (Fig. 7a).128, 129 Both IAPP and insulin are regulated by similar factors with a common regulatory promoter motif.130 Before secreting to the circulation, IAPP is stored together with insulin inside the β-cell granules at high concentrations. IAPP functions as a synergistic partner of insulin to control the blood glucose level by slowing down gastric emptying, inhibiting digestive secretion, and promoting satiety.131, 132 IAPP is also known to play a role in bone metabolism along with calcitonin and calcitonin gene-related peptides.133
Figure 7.
Structural studies of IAPP. (a) The primary structure of IAPP peptide. Solution NMR structures of IAPP monomers stabilized by SDS micelle at (b) pH 4.2 (PDB: 2KB8) and (c) pH 7.3 (PDB: 2L86). (d) Solution NMR structure of IAPP whose aggregation is reduced at pH 5.3, 4 °C, and 100 μM in concentration (PDB: 5MGQ). Residues 1–19 are colored purple and His18 is in sticks. The overall U-shaped IAPP fibril models are derived from experimental constraints by (e) solid-state NMR134 and (f) X-ray crystallography of short peptides.135 In panels (e) and (f), two peptides in the fibril cross-section are shown in sticks viewed along the fibril axis. (g) EPR constraints were applied to reconstruct the fibril model of disulfide reduced IAPP. The sub-panels A and B correspond to views along and perpendicular to the fibril axis, and sub-panels C and D are the accordingly reconstructed fibril models with two different views perpendicular to the fibril axis.136 Copyright The American Chemical Society, John Wiley & Sons, and The American Society for Biochemistry and Molecular Biology.
A hallmark of type 2 diabetes (T2D) is the formation of IAPP-enriched amyloid plaques found in the pancreas of patients. Insulin resistance in T2D leads to increased production of insulin and also IAPP by β-cells because of their shared synthesis and secretion pathways. Since IAPP is one of the most amyloidogenic peptides known, over-production of IAPP in β-cells promotes the accumulation of toxic aggregates. Other studies also suggested that insufficient process of proIAPP and accumulation of intermediately processed peptides might promote the formation of amyloid fibrils, but the detailed molecular mechanisms remain unclear. The disease progression is marked by β-cell death and loss of β-cell functions, resulting in insulin deficiency and diabetic dependence on external insulin sources.
3.2 Atomic structures of IAPP and IAPP amyloid fibrils
Structural characterization of IAPP monomers is extremely challenging due to the high aggregation propensity of the peptide. By reducing IAPP aggregation with detergent micelles, solution NMR studies have been used to study the structure of IAPP monomers.137–139 It has been shown that SDS micelles stabilize IAPP in a highly helical form (Fig. 7b–d). At low pH, the peptide assumes an extended alpha-helix. At neutral pH, the peptide has been found to form a kinked helix around residue H18. Such structural difference is likely due to the electrostatic interaction of the protonated His18 at low pH with the anionic SDS. Combing low pH, low temperature, and low peptide concentrations to hinder IAPP aggregation in solution, an NMR study has recently revealed that the N-terminus of IAPP remains alpha-helical while the C-terminus is unstructured, which are consistent with molecular dynamics (MD) simulations of isolated IAPP monomers.140
The fibril aggregates of IAPPs share the same characteristic cross-beta structures of known fibrils.141 Although the atomic structure of full-length IAPP amyloid fibrils is not available, several model structures have been proposed based on various experimental methods. Using constraints derived from solid-state NMR, Tycko et al. proposed a U-shaped fibrils model where residues 8–17 and 26–37 form two beta-sheets (Fig. 7e).134 Based on X-ray microcrystallography structures of two short peptides, Eisenberg et al. reconstructed a similar fibril model with main differences in the side-chain packing (Fig. 7f).135 Recently, EPR studies of disulfide-reduced IAPP led to a different fibril model (Fig. 7g), where the peptide still adopts a U-shape with two strands separated by a longer distance.136 The two strands in a single peptide have to be staggered with respect to each other to have the appropriated inter β-sheet packing and distances.
3.3 Mesoscopic structure of IAPP amyloids
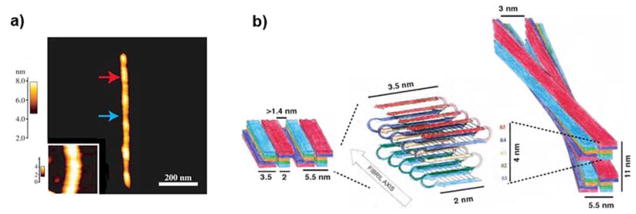
The morphology of IAPP amyloid fibril has been studied by both TEM and AFM.142, 143 IAPP fibrils at the mesoscopic scale displayed significant structural polymorphism, including ribbon-like, sheet-like and helical fibril morphologies (Fig. 8). The ribbons and sheets were formed by lateral association of 5-nm wide protofibrils (Fig. 8a). Most of the fibrils were found in left-handed coil morphology with cross-over periodicities of either ~25 nm or ~50 nm (Fig. 8b). Based on these observations, Goldsbury et al. proposed that the building block of IAPP fibrils is the 5 nm protofibril which can either self-assemble laterally into ribbon-like or sheet-like arrays or coiled fibrils.143 The atomic models of IAPP fibrils are consistent with these TEM and AFM observations.
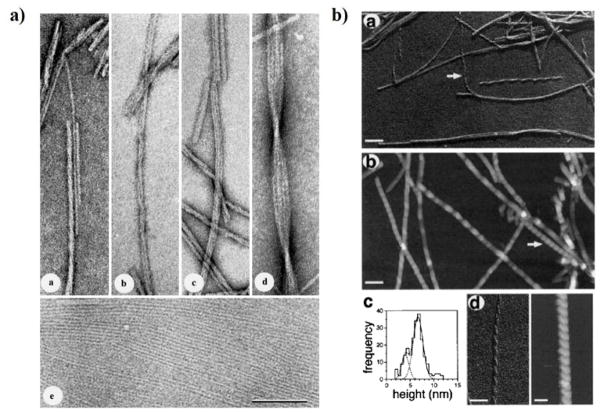
Figure 8.
Morphology of IAPP amyloid fibrils. (a) Lateral association of ribbon-like IAPP protofibrils revealed by TEM of freeze-dried tungsten-shadowed samples. Subpanels a–d depict ribbons assembled by lateral association of 1 to 4 protofibrils. Ribbons with multiple protofibrils often crossed over in a left-handed sense at moderately regular intervals. Subpanel e corresponds to lateral assembly of protofibrils into single-layered, sheet-like arrays. Scale bar: 100 nm. (b) IAPP fibrils with coiled morphologies.142, 143 Subpanels a and b denote coiled fibrils visualized by TEM and AFM, respectively. Arrows point to a left-handed fibril with a 25 nm cross-over periodicity. Longer periodicities of approximately 50 nm can also be seen in both subpanels. Subpanel c shows the AFM height distribution, and d compares the 25 nm periodicity fibril in TEM and AFM. Scale bars: 100 nm in subpanels a, b; and 50 nm in d. Copyright Elsevier.
3.4 IAPP toxicity and type 2 diabetes
Mounting studies suggest that IAPP aggregation and the related toxicity are associated with T2D. IAPP variants from diabetes-prone primates and cats form amyloid aggregates readily in vitro, while those from diabetes-free rodents and pigs feature significantly lower aggregation propensities.144 A naturally-occurring polymorphic S20G mutation renders IAPP more aggregation prone;145 and an Asian subpopulation carrying this mutation is subjected to early onset of T2D.146 IAPP aggregated rapidly upon transplanting human islets into nude mice, and the aggregation process occurred before the recurrence of hyperglycermia and was correlated with β-cell death.147, 148 Transgenic mice expressing human IAPP variant started to develop diabetes.149 Moreover, as with other amyloid proteins,150, 151 IAPP amyloid aggregates are toxic to pancreatic islet cells.152 Therefore, amyloid aggregation of IAPP is related to β-cell death in T2D.153
3.4.1 Oligomers vs amyloids
Amyloid aggregation is a nucleation process, featuring a characteristic all-or-none sigmoidal kinetics. The final mature amyloid fibrils have been found relatively inert and have no significant cell toxicity. In contrast, freshly dissolved IAPP have been found to be highly toxic to islet cells and also cause membrane instability in vitro,154 where the small and soluble aggregation intermediates of IAPP are expected to accumulate before the rapid fibril growth. IAPP oligomers have also been found to disrupt cell coupling, induce apoptosis, and impair insulin secretion in isolated human islets.155 Additional evidence include transgenic mice studies,149, 156 where amyloid deposits were not always observed under optical microscopy in animals starting to show diabetic symptoms, and there was a lack of autocorrelation between beta cell loss and amyloid deposits in these models.157 In addition, inhibition of the formation of insoluble IAPP aggregates but not oligomers by either small molecules158 or proteins159 did not reduce the cytotoxicity. Hence, these results among many others led to the toxic oligomer hypothesis in T2D.160, 161
As the aggregation intermediate species, IAPP oligomers are not well-defined and are extremely challenging to characterize due to their transient and heterogeneous nature. Many in vitro studies support the accumulation of helical intermediates populated along the aggregation pathway.162–164 It has been suggested that the N-terminal helixes of soluble IAPPs (Fig. 7d) are amphiphilic and hydrophobic interactions drive the helix association, which in turn increases the local concentration of the C-terminus containing the amyloidogenic sequence 20–29.165 Both discrete molecular simulations (DMD) of IAPP dimers140 and X-ray crystallography study of IAPP fused to a maltose-binding protein164 supported this scenario. On the other hand, ion mobility mass spectroscopy (IM-MS) combined with MD simulations pointed to a different model of early intermediate states with beta-hairpin dimers.166 The difference is possibly due to the enhanced sampling method - replica exchange167 - used in the MD study, which reduced the free energy barrier of the helix unfolding in the N-terminus. Further experimental and computational studies are necessary to fully understand the structure and dynamics of IAPP oligomers in order to identify the toxic species and the molecular mechanism of IAPP toxicity.
3.4.2 The endogenous inhibition of IAPP aggregation
IAPP is highly aggregation prone and readily forms amyloid fibrils in vitro at μM concentrations within hours.168 However, before its secretion to the bloodstream IAPP is stored inside β-cell granules at mM concentrations for hours without apparent formation of toxic aggregates in healthy individuals.169 Therefore, the physiological environment inside β-cell granules natively inhibits the formation of toxic IAPP aggregates while disruption to the native inhibition environment may lead to amyloid aggregation of IAPP, causing β-cell death.
Islet β-cell granules have a distinct cellular environment.170 First, the pH value inside the granules is 5.5, which is below the physiological pH of 7.4. Second, β-cell granules have one of the highest concentrations of Zn2+ ions in the entire human body. The high concentration of zinc in β-cell granules, maintained by a β-cell-specific zinc transporter — ZnT8,171 is believed to be important for the efficient storage of insulin in β-cell granules: zinc coordinates the formation of insulin hexamers, which form insulin crystals as the dense core of β-cell granules.172 Third, beside IAPP peptides β-cell granules also have other molecules in large quantities, including insulin and proinsulin C-peptide. Insulin is co-secreted with IAPP by β-cells at a ratio of ~100:1 in healthy individuals, and such a high insulin-to-IAPP ratio is reduced to ~20:1 in T2D patients.173 The C-peptide is a part of the proinsulin sequence connecting A- and B-chains of insulin. Protease-processing of proinsulin results into mature insulin and C-peptide with an equal molar concentration inside β-cell granules.
Low pH
Inhibition of IAPP aggregation at low pH has been observed in vitro. At low pH, an increase in the lag time and a decrease in the growth rate of IAPP fibrillization was observed.174, 175 The electrostatic repulsion between IAPPs with protonated histidine18 (His18) is responsible for inhibiting the self-association of IAPP at low pH,176 supported by DMD simulations of IAPP dimerization with and without protonation of His18.177 However, since the pH value inside β-cell granules is close to the isoelectric point of His18174, 178 and a significant portion of IAPP is still unprotonated, interactions of IAPP with other granule components are necessary for natively inhibiting the peptide amyloid aggregation at high concentrations.
Insulin
In vitro experiments have revealed that insulin is a potent IAPP aggregation inhibitor, which can significantly slow down aggregation at sub-stoichiometry concentrations.180 Several studies, including peptide mapping,181 IMS-MS combined with MD simulations,182 and DMD studies140 suggested that the B-chain of insulin can bind IAPP. Computational studies with atomistic DMD simulations showed that both insulin monomers and dimers (but not the zinc-bound hexamer as the IAPP-binding interface is buried) could bind IAPP monomer and inhibit IAPP self-association by competing with the amyloidogenic regions important for aggregation, subsequently preventing amyloid aggregation (Fig. 9a,b). The preferred binding of insulin with the amyloidogenic region in the beta-strand conformation (Fig. 9a) suggests that insulin can also cap the fibril growth, consistently with the observed sub-stoichiometric inhibition of IAPP aggregation by insulin. Comparing to high zinc concentrations where insulin is insoluble in the crystal form,183 zinc-deficiency due to loss-of-function mutations of ZnT8 shifts the insulin oligomer/crystallization equilibrium toward soluble monomers and dimers, which can efficiently inhibit IAPP aggregation and reduce T2D risk in the subpopulation carrying these mutations.184 However, since IAPP is found almost exclusively in the soluble halo fraction of β-cell granules while insulin is mostly insoluble in the core, the balance of other granule components such as Zn2+ and/or C-peptide co-localized with IAPP appears crucial for maintaining the native state of IAPP.
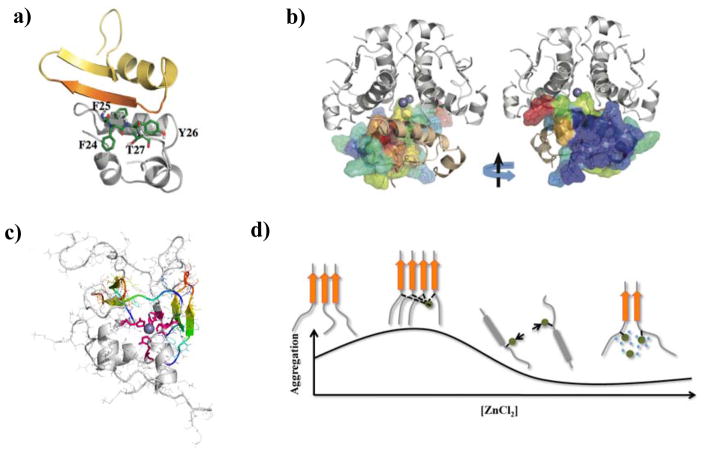
Figure 9.
Effects of β-cell granule components on IAPP aggregation. (a) A representative IAPP-insulin complex from DMD simulations,140 where the amyloidogenic residues of IAPP (residues 22–29) are shown in orange. The residues in the B-chain of insulin important for binding IAPP are highlighted in stick representation. (b) The residues of an insulin monomer are colored according to IAPP binding frequencies in the structure of an insulin hexamer. The view with an 180° rotation is also presented. The residues with strong IAPP-binding are located at the insulin monomer–monomer interface. (c) A representative IAPP tetramer with His18 (highlighted as sticks in pink) coordinated by a Zn2+ (blue sphere) from DMD simulations.179 The amyloidogenic sequences from each IAPP monomer are highlighted in rainbow colors. (d) A mechanistic scheme demonstrating the dependence of IAPP aggregation on relative zinc concentration. Copyright The American Chemical Society and Elsevier.
Zinc
In an early study by Steiner and co-workers where ZnCl2 was added to ~250 μM IAPP solution, aggregation promotion was observed.185 This promotion effect leveled off till ~1 mM zinc ion was added, but no data at higher salt concentrations was reported. In later experimental studies, IAPP aggregation inhibition was observed at low zinc concentrations (5 and 10 μM, but relatively high zinc/IAPP stoichiometry), followed by a partial recovery of aggregation at very high stoichiometry (~50–100).186, 187 A “two-site binding” model, where a high affinity binding with His18 stabilized non-aggregating oligomers but an unknown weaker secondary binding promoted amyloid fibril formation, was proposed.187 However, this model cannot account for aggregation-promotion at low ion/protein stoichiometry185 (e.g., in the case of 10 μM of IAPP there was a single data point with increased aggregation at ~25 μM of zinc.186). Combining DMD simulations with experimental characterizations179, Govindan et al. developed an alternative model that was consistent with the experimentally-observed concentration-dependent effect of zinc on IAPP aggregation. At low zinc/IAPP stoichiometry, the IAPP oligomers cross-linked by zinc were aggregation-prone due to high local peptide concentrations (Fig. 9c). As ion/protein stoichiometry increased, each IAPP tended to bind only one zinc ion at His18. The electrostatic repulsion between the bound zinc ions (+2e) inhibited IAPP aggregation, similarly to the low pH condition where IAPP aggregation was inhibited by protonated His18 (+1e).176 With zinc concentration kept increasing, the screening effect due to high salt concentrations reduced electrostatic repulsion, and allowed for the aggregation to recover (Fig. 9d).186, 187
C-peptide
Without zinc binding, C-peptide is disordered in water and weakly helical in trifluoroethanol (TFE) solution.188 The peptide contains five acidic amino acids. Alanine scan coupled with MS experiments suggest that all these acidic amino acids bind zinc ions and the binding is 1:1 in stoichiometry.189 It has been found that zinc-binding may induce structural changes.190 It was hypothesized that multiple negatively charged acidic amino acids in C-peptide allow the binding with multiple IAPP peptides, locally increase IAPP concentration and subsequently promote IAPP aggregation. Upon binding zinc, C-peptide adopts specific secondary and tertiary structures with reduced net charges, which might bind and stabilize IAPP peptides in the aggregation incompetent state. In addition, other granule molecules including proIAPP191 and proInsulin may also contribute to the native inhibition of IAPP aggregation and cytotoxicity in beta-cells and are subject to future investigations.
3.4.3 IAPP-membrane interactions
It has been proposed that IAPP exerts cytotoxicity by membrane disruption.154, 192, 193 The positively charged IAPP can bind anionic cell membranes and lipid micelles, and the peptide conformational and aggregation propensity change upon binding also depending on the membrane curvature.192 Binding of IAPP with small micelles was found to stabilize the peptide in helical conformation (Fig. 7), while absorption of IAPP on flat membrane accelerated the peptide aggregation.194 Using a lipophilic Laurdan dye for examining MIN6 cell membranes upon exposure to freshly dissolved IAPP as well as mature amyloid fibrils, Pilkington et al. found that all species, especially fresh IAPP, enhanced membrane fluidity and caused losses in cell viability.195 The cell generation of ROS, however, was the most pronounced with mature amyloid fibrils. This study suggested a correlation of cytotoxicity with changes in membrane fluidity rather than ROS production.
The exact mechanism by which IAPP oligomers disrupt the cell membrane is under active investigation. Pore formation by amyloid peptides has been suggested to be important for membrane disruption.196–198 The amyloid pore model is strongly supported by single channel recordings of IAPP on planar membranes.196, 199, 200 A detergent-like mechanism has also been advocated, where the mosaic-like opening and closing of transient defects within the membrane (also see Fig. 15d in section 4 for αS) was supported by AFM studies showing large-scale defects in the lipid bilayer upon prolonged exposure to IAPP.201 However, the strong correlation between fibril formation and membrane disruption by this mechanism202 is inconsistent with the toxic oligomer hypothesis. Recently, biophysical measurements in conjunction with cytotoxicity studies showed that nonamyloidogenic rat IAPP is as effective as IAPP at disrupting standard anionic model membranes under conditions where rat IAPP did not induce cellular toxicity, suggesting that there is no direct relationship between disruption of model membranes and induction of cellular toxicity.203 Therefore, the connection between IAPP cytotoxicity and membrane disruption remains inconclusive.
3.5 Mitigation strategies and theranostics
As with other amyloid diseases,204–206 inhibition of IAPP aggregation is an attractive therapeutic strategy to prevent β-cell death207 and halt the progression of diabetic conditions in T2D. Various approaches have been explored to reduce aggregation-induced IAPP cytotoxicity, through the use of peptides, peptide-mimetics,208–211 small molecules212–223, and nanoparticles (NPs)224–226. Non-amyloidogenic sequence variants of IAPP including rat IAPP227 have been found to inhibit the fibril formation of human IAPP,208, 209 and the inhibition efficacies can be improved by synthesizing peptide mimetics with conformational restraints.210, 211 Targeting the early helical intermediate states of IAPP aggregation162–164, small molecule peptidomimetics212, 213 have been designed to mimic helixes that complementarily bind to the N-terminal helix of IAPP. Another attractive set of amyloid aggregation inhibitors are small-molecule polyphenols221 such as epigallocatechin gallate (EGCG),228 curcumin,219, 220 and resveratrol,229 which inhibit aggregation and reduce the related cytotoxicity of IAPP230 as well as other proteins and peptides such as Aβ.231 These polyphenols have the advantage of being naturally occurring, and are non-toxic at moderate concentrations. Despite well-known therapeutic benefits of small molecules,232 however, pharmacological applications of these polyphenols are limited due to some common issues, such as their poor water solubility.233
Several studies have examined the anti-amyloid mechanisms of small molecules and NPs. For example, IMS-MS experiments showed that EGCG exerted an inhibitory effect on IAPP aggregation through direct binding of EGCG to the peptide215 and alternating the aggregation pathways.228 Using simulations of the amyloidogenic segment of IAPP, resveratrol was found to bind and prevent the lateral growth of the fibril-like β-sheets.235 In another work, resveratrol was found to bind weakly to IAPP and reduce inter-peptide contacts.236 A recent computational study showed that resveratrol altered the structure of an IAPP pentamer,237 which was modelled by the amyloid fibril structure derived from solid-state NMR.134 By modelling the effects of polyphenols like resveratrol and curcumin on the initial self-association and aggregation of IAPP in DMD simulation,234 Nedumpully-Govindan et al. showed that these polyphenols inhibited IAPP aggregation by promoting “off-pathway” oligomers with the hydrophobic polyphenols forming the core (Fig. 10a). The peptides were stabilized in the aggregation-incompetent helix-rich state by burying their hydrophobic residues inside the core and exposing the hydrophilic residues. Graphene oxide nanosheets displayed strong inhibition effects on IAPP aggregation and associated cytotoxicity because strong binding affinity rendered the peptides to bind with the nanosheets rather than between themselves.225 OH-terminated polyamyloidoamine (PAMAM-OH) dendrimers inhibited IAPP aggregation and cytotoxicity, where the polymeric NPs encapsulated and stabilized monomeric IAPP in their hydrophobic interior (Fig. 10b).224 In general, these inhibitors all reduced the population of the oligomeric species, thereby reducing IAPP toxicity.
Figure 10.
Inhibition of IAPP aggregation. (a) Left: High-throughput dynamic light scattering measurement of IAPP size distributions with and without resveratrol (2:1 ligand/IAPP ratio). Right: Distribution of IAPP aggregates of different molecular weights with and without resveratrol in silico. Stable IAPP/resveratrol oligomer has the resveratrol molecules forming a nano-sized core and IAPP peptides a corona, which prevents aggregation.234 (b) Left: a typical IAPP dimer in DMD simulations. Right: Binding to a PAMAM-OH dendrimer (spheres) inhibits self-association of the amyloidogenic sequences (yellow region) between two IAPP peptides.224 The peptides are shown in cartoon representation with rainbow color from blue (N-terminus) to red (C-terminus). Copyright Nature Publishing Group and John Wiley & Sons.
4. Alpha-synuclein and Parkinson’s disease
4.1 Function of alpha-synuclein
Alpha synuclein (αS) is a 140-residue small protein highly concentrated in presynaptic terminals,238 making up as much as 1% of all proteins in the cytosol of brain cells. Small traces of αS are also found in the heart, gut,239 muscles and other tissues, reminiscent of the confounding bodily distributions of Aβ and IAPP beyond their purported origins. In the intra-neuronal space, αS assumes an equilibrium between an unfolded monomeric conformation and a membrane-bound state that is rich in alpha helices.240 The precise physiological role of αS is unclear, but is relevant to the modulation of neurotransmitter dopamine release, ER/Golgi trafficking, and synaptic vesicles.241 The membrane-bound αS influences lipid packing and induces vesicle clustering through physical and physicochemical interactions, while αS in the multimeric form has been shown to promote SNARE complex assembly during synaptic exocytosis.240
Aggregated αS mediates dopaminergic neurotoxicity in vivo.244 However, the precise mechanisms by which αS lends toxicity to host cells remain unclear. Pathologically, αS is a major component of Lewy bodies and neurites, the intracellular protein aggregates first identified by Spillantini et al. in 1997 (Fig. 11)242 and hallmarks of Parkinson’s disease (PD), Parkinson’s disease dementia (PDD) and dementia with Lewy bodies (DLB). Compared with the ambiguous pathology of αS, the neuritic pathology of β and γ synuclein homologs does not appear widespread, and both neuroprotective and neurotoxic potentials of β synuclein have been reported.245
Figure 11.
(a) (Large image) Pigmented nerve cells containing αS-positive Lewy body (thin arrows) and Lewy neurites (thick arrow).242 Small image: a pigmented nerve cell with two αS-positive Lewy bodies. Scale bar: 8 μm. (b) Hypothesized αS toxicity and spread of pathology in Parkinson’s disease (PD) and Parkinson’s disease dementia (PDD). UPS: Ubiquitin proteasome system.243 Copyright Nature Publishing Group.
4.2 Atomic structures of alpha-synuclein and alpha-synuclein amyloid fibrils
The sequence of αS is encoded by the SNCA gene and can be divided into three distinct domains: a) the amphipathic N-terminal domain (1–60), which contains consensus KTKEGV repeats and has alpha-helical propensity, b) the central domain (61–95) or the non-amyloid-beta component (NAC) that is highly hydrophobic and amyloidogenic, and c) the acidic C-terminal domain (96–140) which contains negatively charged and proline residues to afford protein flexibility but no apparent structural propensity.246 High resolution ion-mobility mass spectroscopy has revealed that HPLC-purified αS is autoproteolytic, giving rise to a number of small molecular weight fragments upon incubation. In particular, the fragment of residues 72–140 contains majority of the NAC region and aggregates faster than full-length αS.247 These autoproteolytic products may serve as intermediates or cofactors in the aggregation of αS in vivo.
The atomic structures of fragmental and full-length αS in the fibrillar form have been elucidated over the past decade (Fig. 12). Using quenched HD exchange Vilar et al. identified five β-strands within the fibril core comprising residues 35–96 and with solid-state NMR spectroscopy the presence of β-sheet secondary structure within the fibril core of residues 30–110.241 This study has further detailed the mesoscopic features of αS fibrils, as we will visit in the next sub-section.
Figure 12.
Landmark studies concerning the structures of αS fragments with respect to its full 140 residues consisting of N terminus, NAC and C terminus.248 The researcher teams are chronicled on the left while the employed techniques are abbreviated on the right. EPR: electron paramagnetic resonance; ssNMR: solid-state nuclear magnetic resonance; HD: hydrogen-deuterium exchange; SDSL: site directed spin labelling. Copyright Nature Publishing Group.
Based on micro-electron diffraction Rodriguez et al. revealed small crystal structures of the toxic NAC core (68–78, or NACore) and the preNAC segment (47–56) of αS, at spatial resolution of 1.4 Å (Fig. 13a).248 The NACore strands stacked in-register into β-sheets. The sheets were paired, forming steric-zipper protofilaments as observed for other types of amyloidogenic proteins. Notably, each pair of the sheets contained two water molecules, and each was associated with a threonine side chain within the interface. X-ray fiber diffraction patterns further revealed a similarity of the NACore to full-length αS fibrils.248
Figure 13.
(a) Top and side views of the structures of NACore (orange; residues 68–78, sequence also see Fig. 12) and PreNAC segments (blue; residues 47–56, sequence also see Fig. 12). The A53T mutation in PreNAC is shown in black.248 (b–e) Three-dimensional structure of a full αS fibril. (b) A central monomer from residues 44 to 96 looking down the fibril axis showing the Greek-key motif of the fibril core. (c) Stacked monomers showing the sidechain alignment between each monomer down the fibril axis. (d) Residues 25 to 105 of 8 monomers displaying the β-sheet alignment of each monomer in the fibril and the Greek-key topology of the core. (e) Overlaid ten lowest energy structures, showing sidechain positions within the core. Residues 51–57 are indicated in red with side chains removed.249 Copyright Nature Publishing Group.
In a more recent study, Tuttle et al. established the atomic structure of full-length αS fibrils based on 68 spectra, using 2D and 3D ssNMR.249 The fibrils were collected from cell culture and shown to adopt a β-serpentine arrangement (Fig. 13b–e). The fold exhibited hydrogen bonds in register along the fibril axis, orthogonal to the hydrogen bond geometry in a standard Greek-key motif unseen for other fibrils (Fig. 13d).249 The innermost β-sheet contained amyloidogenic residues 71–82, while the sidechains in the core were tightly packed (Fig. 13e). Compact residues facilitated a close backbone-backbone interaction: A69–G93 bridged the distal loops of the Greek key, and G47–A78 rendered a stable intermolecular salt bridge between E46 and K80. Hydrophobic sidechain packing among I88, A91 and F94 established the innermost portion of the Greek key. Residues 55–62 were disordered, consistent with that reported by Comellas et al.250 Collectively, the steric zippers, glutamine ladder and intermolecular salt bridge contributed to the structural complexity and stability of the fibril. However, it remains uncertain whether such atomic structure reflects that of αS fibrils extracted directly from PD patients.
4.3 Mesoscopic structure of alpha-synuclein amyloids
The morphology of αS fibrils has been examined with AFM251–253 and cryoelectron microscopy.241 A hierarchical assembly model (HAM) was proposed by Inonescu-Zanetti et al.254 to describe the architecture of immunoglobulin light-chain protein SMA fibrils assembled from smaller subspecies and has shown general applicability to the nanoscale assemblies of Aβ, αS and IAPP as well as SH3 domain, lysozyme, SMA, β2-microglobulin and beta-lactoglobulin.142, 251, 252, 255–260 Alternatively, a new packing model was proposed by Sweers et al.,261 in attempt to reconcile the morphological and mechanical data observed for two distinct fibril species of E46K, a mutant of αS. Nonetheless, according to the HAM, protofilaments are established by the nucleated polymerization kinetic model, in which the protofilaments elongate by the addition of monomeric, partially folded intermediates to their growing ends. The protofilaments then interact with each other to form protofibrils, each consisting of twisted 2–3 protofilaments, and two protofibrils entwine to form mature fibrils and, eventually, plaques. The driving force for such stepwise assembly is both electrostatic and hydrophobic.254
The HAM model predicts the occurrence of periodicity for protofilaments and fibrils, which assume twisted morphologies. Such periodicity is driven by a balance between mechanical forces dominated by the protofilament elasticity and electrostatic forces due to the distribution of hydrophobic regions and charge along the protofilament backbone,253 as well as by the inherent chirality of constituting amino acids and β-sheets/helices of the fibrils. The average heights of αS protofilaments and fibrils were 3.8 nm and 6.5 nm, respectively, while the periodicity of αS fibrils ranged from 100~150 nm as determined by AFM (Fig. 14a).253 These parameters are consistent with immunoelectron microscopy of filaments extracted from the brains of patients with DLB and multiple system atrophy,262 and agree with high-resolution cryoelectron microscopy where twisted protofilament of ~2×3.5 nm in boundaries and 120 ±10 nm in periodicity were observed leading to the proposal of a folded αS fibril model (Fig. 14b).241 The cross-section of individual αS monomers in the fold was trapezoid instead of circular, resulting in a two-fold increase in moment of inertia (Sweers 2012).263 Though not substantiated, such non-circular packing of monomers could also hold true for other β-sheet folded proteins.261, 263 In addition, curly αS fibrils prepared by filtration-like steps during aggregation possessed a persistence length of 170 nm, while straight αS fibrils from unperturbed aggregation displayed persistence lengths of up to 140 μm.264
Figure 14.
(A) AFM image showing a periodicity of 100–150 nm along an αS protofibril. The peak (red arrow) to trough (blue arrow) differs by ~1 nm in height. (Inset) A section of a protofilament with an average height of 3.8 nm.253 (B) Proposed fold of an αS fibril. A monomeric αS within a protofilament (center). Incorporation of protofilaments into a straight or twisted fibril is illustrated in the left and right panel, respectively.241 Copyright Cell Press and National Academy of Sciences.
4.4 Alpha-synuclein toxicity and Parkinson’s
4.4.1 Oligomers vs amyloids
Natively unfolded αS undergoes a transition to partially folded intermediates prior to fibril formation.265 Such partially folded conformations are favored by mutations266 or changes in pH, ionic strength and temperature and are thought to be critical intermediates in the transition to amyloid fibrils.253 Clearly, such dynamic transition has an important bearing on αS toxicity, as evidenced by a body of literature focused on the complex roles of αS oligomers and amyloids.
The aggregation of αS follows a nucleation polymerization pathway involving prefibrillar species of remarkable conformational plasticity,267 both transient and stable. Specifically, it is postulated that αS aggregation takes place in the cytoplasma or in association with the cellular membrane. In the cytosol, soluble monomers interact to form unstable dimers, which develop into oligomers and, subsequently, fibrils.268 The current understanding concerning αS toxicity follows the narrative of the “toxic oligomer hypothesis”,251 in that the oligomeric species are more toxic than the fibrillar form,251, 269–272 as similarly proposed for Aβ273–279 and IAPP.280 However, due to the different structural characteristics and aggregation rates, different cellular environments, as well as prion-like cell to cell spreading and crosstalk of proteins of different origins and pathologies, this generalization remains putative.281, 282
In an early in vitro study, Conway et al. compared the rates of disappearance of monomeric αS and appearance of fibrillar αS for wide-type and two mutant proteins A53T and A30P.251 The differences between the trends suggested the occurrence of nonfibrillar αS oligomers. Using sedimentation and gel filtration chromatography, the researchers identified spheres (range of heights: 2–6 nm), chains of spheres (protofibrils), and rings/annulars (heights: ~4 nm) from fibrils (~8 nm in diameter) by AFM. For a comprehensive account of αS oligomers and their in vitro preparation protocols, readers may refer to a recent review by van Diggelen et al.283
Using attenuated total reflection-Fourier-transform infrared (ATR-FTIR) spectroscopy Celej et al. revealed that isolated αS oligomers adopted an antiparallel β-sheet structure, whereas fibrils assumed a parallel arrangement.284 Notably, antiparallel β-sheet structures have also been reported for the oligomeric structures of Aβ, β2-microglobulin and human prion peptide PrP82–146.284 Such contrasting features in secondary structure between the oligomers and fibrils entail differences in conformational change, affinity and mode of interaction when binding with the cell membrane, further compounded by the differences in aspect ratio and surface hydrophobicity between the two species. The toxicities of αS oligomers and amyloid protein oligomers in general have been postulated as an inherent property.285 Unlike amyloid fibrils, the oligomers share similar structural properties273, 286 and possess higher portions of random coils and helical structures. Consequently, the exposed hydrophobic surfaces of the oligomers could mediate interactions with intracellular proteins to trigger aberrant cellular pathways.
Celej et al. found that purified αS oligomers spheroidal and polydisperse (10–60 nm), while αS fibrils were unbranched of 6–10 nm in diameter and micrometers long when examined under electron microscopy.287 These isolated oligomers were on-pathway intermediates sharing the same structural motif with other prefibrillar oligomers and possessing no canonical cross-β fibril structure.284 Curiously, the αS oligomers were recognized by A11 antibody, which also targeted the oligomeric but not monomeric or fibrillar forms of Aβ, prions, and IAPP.273, 288
While it remains debatable whether αS oligomers are intermediates in the process of amyloid formation, or precursors to fibrils, or byproducts of fibril elongation, or associated with a pathway of aggregation different from the standard amyloid fibrillization,269 there is little ambiguity that αS oligomers are toxic, as validated by in vitro and animal models.272, 273, 289, 290
4.4.2 Alpha-synuclein-membrane interactions
Towards understanding the origin of amyloid protein toxicity, much research over the past two decades has been focused on the interactions of the proteins as well as their aggregation products with cell membranes, model lipid vesicles, or lipid rafts. This focus is especially justified for αS considering its strong functional association with synaptic vesicles and its cell-to-cell spreading.291 From a biophysical standpoint such interactions may be understood as a manifestation of the structural attributes of αS (sequence, charge and hydrophobicity), as well as the changing properties of αS from soluble and disordered monomers to soluble and less random oligomers, and to waxy and highly ordered fibrils and plaques.
Research concerning protein-membrane interaction should take into consideration of two convoluting aspects: protein aggregation modulated by a model lipid bilayer or cell membrane, and membrane integrity perturbed by protein aggregation. Numerous studies have confirmed that lipid membranes can speed up the process of protein fibrillization due to the amphiphilicity of both interactants.292, 293 Specifically, the N-terminal region of αS, containing 7 amphiphilic imperfect repeats each of 11-residues, can initiate electrostatic interaction with anionic lipid head groups. The NAC region of the protein can establish hydrophobic interaction with lipid fatty acyl tails to promote membrane partitioning.294 Upon membrane exposure, the protein concentration at the membrane surface is abruptly increased due to the 3D to 2D transition. Consequently, protein conformational entropy is reduced to favor aggregation.294, 295 Specifically, the rate of αS primary nucleation was enhanced by three orders of magnitude when exposed to small unilamellar vesicles (SUVs, 20~100 nm in diameter) of 1,2-dimyristoyl-sn-glycerol-3-phospho-L-serine (DMPS).296
Upon adsorption onto lipid membranes, monomeric amyloid proteins adopt an α helical state, followed by a conversion to β-sheet rich oligomers and amyloid fibrils modulated by the curvature and charge of the membranes, presence of metal ions, peptide to lipid ratio, and ganglioside clusters, cholesterols and lipid rafts.299 αS assumes a fully extended α helical state coming into contact with larger vesicles, likely representative of the protein conformation in vivo.300, 301 In contrast, smaller vesicles with greater curvatures and smaller surface areas are associated with proteins in bent α helices or antiparallel helix-turn-helix conformation to maximize protein-membrane binding.293
A high peptide/lipid ratio favors protein crowding on the membrane surface to induce nucleation.302 Binding of αS (isoelectric point of 4.74)303 with membranes is elevated with increased acidic phospholipid content.304–306 αS oligomers also show propensity for the liquid disordered phase of anionic vesicles.307 The exact mechanism of αS association with lipid rafts is unclear, but is linked to the high lipid packing density of anionic head groups in the rafts. Such specific binding between αS and lipid rafts may be essential to both the normal cellular function of αS and its role in PD pathology.299
Elevated levels of metal ions have been found in the substantia nigra of PD patients.308 Addition of metal cations of Cu2+, Fe3+ or Co3+ induced secondary structure in αS and accelerated protein aggregation in vitro,265 through metal ion-mediated amyloid protein-membrane interaction. Although Ca2+ (of ~300 μM) in the ER serves to facilitate protein folding, addition of Ca2+ and other heavy meal ions to monomeric αS rapidly produced annular oligomers,309 while divalent metal ions also enabled the clustering of αS on the surfaces of anionic 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho(1′-rac-glycerol)/phosphatidylcholine bilayers.306 It is possible that metal cations enabled the interaction of the likely charged C-terminus of αS and membranes through charge neutralization. Such strong metal-hosting capacity of amyloid proteins has been utilized in entirely different contexts from amyloidogenesis, such as purification of wastewater and in vitro iron fortification using functional β-lactolglobulin amyloids.310–312
The adsorption of αS has been shown to compromise membrane permeability.282 One mechanism proposed for such perturbation is pore formation by the protein oligomers (Fig. 15a–c).271, 284, 293, 297 In combination with biochemical and ultrastructural analysis, Tsigelny et al. revealed through MD simulations and docking that αS monomers, upon adsorption onto lipid membranes through their N-termini, assembled into homodimers of both propagating (head to head) and non-propagating (head to tail) conformations. The propagating form docked on the membrane surface to recruit additional αS molecules, rendering pentamers and hexamers to form ring-like structures partitioning in the membrane.313 Consistently, addition of stable αS oligomers has been shown to induce ion-channel activity (Fig. 15c),297 while Ca2+ and dopamine exhibited much higher leakage rates than polymers of cytochrome c and fluorescein isothiocyanate-dextran from anionic vesicles in the presence of oligomeric A30P and A53T, two major αS mutations.314 Under conditions in which vesicular membranes were less stable due to the lack of counter-ion Ca2+, αS permeation was less size selective and monomeric αS permeated via a detergent-like mechanism.293
Another mechanism proposed for αS-membrane interaction is illustrated in Fig. 15d.298 Here the presence of a supported lipid bilayer facilitates the conversion of αS from randomly structured monomers to alpha helices (top panel), which further aggregate into oligomers and fibrils while stripping lipids off the bilayer (middle and lower panels). Membrane thinning and depolarization, changing fluidity, lipid flip-flop, calcium leakage, and disruption of ionic homeostasis are plausible consequences of αS membrane adsorption, αS self-assembly, and αS assembly with membrane lipids, through hydrophobic and electrostatic interactions as well as lipid micellar encasing of the protein species (i.e., the carpet model315). This mechanism is supported by experimental studies employing giant vesicles as well as reporters of ThT, calcein, Ca2+ and fluorescence recovery after photobleaching,316–319 to name a few.
In close connection with αS toxicity and αS-membrane interaction, a body of literature has revealed links between αS oligomers and mitochondrial dysfunction, cytoskeleton deformation, enhanced ROS production, neuroinflammation, ER stress, as well as impaired protein degradation systems.320–328 An analysis of wide-type αS and two mutational variants A30P and E46K interacting with synaptic-like SUVs suggested a mechanism by which a single αS binds to two different synaptic vesicles via the NAC to promote their assembly and vesicle clustering.329 In addition, promotion of SNARE-complex formation has been found to be associated with αS assembly into high-order multimers upon their binding with plasma membranes, suggesting that αS may act as a SNARE chaperone at the presynaptic terminal against neurodegeneration.240
4.4.3 Parkinson’s disease, mitigation strategies and theranostics
Synucleinopathies refer to a family of neurodegenerative diseases including PD, PDD and DLB, where inclusions of Lewy bodies and neurites are located within the neuronal cells (Fig. 11a). Multiple system atrophy (MSA) is a special type of synucleinopathy, since αS-positive inclusions are found in oligodendroglia instead of in neurons. In these diseases, αS pathology in the substantia nigra is closely correlated with motor symptoms and death of SN dopaminergic neurons stimulates the striatum.282 The PD pathology involves progressive neuronal accumulation of aggregated αS, and formation of Lewy bodies affects various functional structures throughout the human nervous system to compromise movement.330
Exogenous αS fibrils seeded Lewy body- and Lewy neurite-like inclusions in cell culture models, and direct neuron to neuron αS transmission throughout the brain propagated PD-like pathology.291, 331, 332 Failure of the protein quality control systems, especially lysosomes, promoted accumulation of transmitted αS and inclusion formation. Cells exposed to neuron-derived αS displayed signs of apoptosis, such as nuclear fragmentation and caspase 3 activation, both in vitro and in vivo.291 Inoculation of αS fibrils into wide-type non-transgenic mice seeded aggregation of endogenous mouse αS and reproduced key features of the neurodegenerative cascade.249 A molecular level understanding of the pathological spreading of αS in PD is lacking, but growing evidence suggests its origin lies in protein self-assembly through templated seeding, where the imported αS aggregates catalyze the conversion of local soluble protein molecules into their aggregated forms. A recent study has revealed regulation of motor deficits and neuroinflammation by intestinal microbiota in a PD model,239 suggesting a role for microbial signals in PD.333 Multiplication of the protein aggregates by recruiting additional αS en route has been proposed as an additional mechanism to templated seeding, to ensure sustainable concentration of the aggregates spreading from cell to cell.332 However, multiplication of αS at neutral pH has not been observed, pointing to the involvement of other cellular processes in enabling the prion-like αS spreading.
The ambiguities concerning the natural state, toxicity and aggregation pathway of αS have hindered the development of mitigation and theranostics against PD. The current approaches, still very much in the incubation stage, aim at exploiting the structural, functional and toxicological properties of αS, or the self-assembly of the protein and its structural and pathological characteristics for therapeutic efficacy.268
Stabilization of the native αS structure from misfolding is a logical strategy. This intervention may also help resolve the controversy concerning a tetrameric initial state of αS.334 One promising approach to slow down αS synucleinopathies is to limit the role of extracellular αS in disease progression, from interfering with αS secretion to neuronal uptake. Removal of αS from the extracellular space to minimize inflammations may be achieved with immunotherapy, as immunization with human αS suppressed protein aggregation and decreased neurodegeneration in transgenic mice overexpressing the protein.335–337 The use of small molecules and mutation is another feasible approach for stabilizing oligomeric species and ameliorating toxicity. The antioxidation and anti-inflammatory properties of the small molecules – often polyphenols or their structural derivatives with the capacity of interfering with protein aggregation through competing H-bonding, hydrophobic interaction and π-stacking with the protein – may counteract the toxicity elicited by the oligomeric species.338–340 The presence of small molecules and other aggregation antagonists may also reduce accessibility to the oligomers by environmental chaperones, ligands and molecular organizations, thereby driving the aggregation off pathway to halt αS pathology.
5. Prions and prion diseases
Transmissible spongiform encephalopathies (TSEs), also known as prion diseases, are a family of rare fatal neurodegenerative disorders associated with prion protein (PrP), and arise in several mammalian species by sporadic, inherited, or infectious means. Kuru, Creutzfeldt-Jakob disease (CJD), Gerstmann-Straussler-Scheinker (GSS), fatal familial insomnia (FFI) and fatal sporadic insomnia (FSI) are PrP-related human disorders,341 whereas scrapie, bovine spongiform encephalopathy (BSE) and chronic wasting disease (CWD) are known sheep,342 cattle343 and cevids344 prion diseases. The main characteristic symptoms of TSEs are brain vacuolation, astrogliosis and neuronal apoptosis,345, 346 which are associated with accumulation of extracellular PrP amyloid deposits in the CNS.347–350 Despite shared sequence between cellular non-pathological PrP (PrPC) and misfolded PrP (PrPSc),351 pathological PrPSc aggregates are proteinase K resistant352 and have a β-enriched secondary structure.353–355
The most distinct feature of TSEs, unique among diseases related to protein misfolding, is the infectivity of the pathogenic agent. Procedures that hydrolyze or modify proteins reduce scrapie infectivity, whereas procedures that alter nucleic acids have no effect.356–359 The “protein-only hypothesis” has now been widely accepted,360–363 contending that a protein structure can be replicated without the use of nucleic acids and the infectious pathogen is the misfolded PrPSc.356, 357, 360–365 In addition, prion diseases progress in host without any sign of immune responses to a “foreign infectious agent”.341 When the protein requirement for infectivity was established, prions were defined as proteinaceous infectious particles that resisted inactivation by procedures that modified nucleic acids.341
Since prion pathology and infectivity366 are closely related to a protein existing in two different conformations, much research in the last decade has been dedicated to understanding the structures of native PrPC 367 and pathological PrpSc.368 PrPSc is believed to act as a structural template, inducing conversion of other PrPC molecules into the pathological form.341 Understanding PrP conformational structures is therefore essential for describing protein misfolding and the specific role of PrP in prion pathology.
5.1 Function of PrP
PrP is encoded by gene (PRNP) found in chromosome 20 (in human)369 and expressed in many tissues, including the brain, circulating lymphocytes, heart, kidney, skin, digestive tract, endothelia, mammary gland and muscle. The physiological role of native PrPC remains unclear.370 It has been shown that the protein is involved in several cellular processes including neuroprotection against excitotoxicity and serum starvation,371 proliferation and cell-cell adhesion,370, 372 formation of synapses373 and ligand binding.374, 375 PrP can protect cells against heavy metal overloading and subsequent oxidative stress by binding divalent ions of copper, zinc, manganese and nickel.375 Due to the ability of PrP to modulate cell proliferation and apoptosis it is believed to play a role on cancer development.376 Indeed, increased PrPC level has been found in gastric cancer,377 colorectal cancer378 and skin cancer.379
A common approach to study the function of PrP is using PrP knockout transgenic mice (Prnp−/−). The major finding in Prnp−/− mice was the lack of developmental differences and resistance to prion diseases.380 However, Prnp−/− mice have shown cognitive abnormalities370 such as depressive-like behavior, anxiety-related disorders and alterations in circadian activity.381 In addition, decreased spatial learning of Prnp−/− mice has been noticed.382 Using three Prnp knockout mice lines Firestein and colleagues found that PrPC was important in the normal processing of sensory information by the olfactory system.383
5.2 Atomic structures of prions and prion amyloids
The atomic structures of full-length and truncated PrPC were mostly solved by NMR.384–388 Notably, X-ray crystallography studies were restricted to the C-terminal domain of PrP, suggesting an intrinsic tendency of the protein to avoid crystallization.389, 390
Proto-protein of human PrP (huPrP) is 253 residues long (Fig. 16a). After translation to mature form, the first 22 residues are removed and the last 23 residues are cleaved off prior to the addition of a glycosyl phosphatidylinositol (GPI) anchor to Ser230. PrP is attached to the outer surface of the cellular membrane by a GPI anchor within the raft domains. The sequence of PrP is highly conserved amongst mammals:391, 392 human PrP shares 99.2%, 94.9% and 92.8% of identical sequences with the proteins from chimpanzee, sheep and cow, respectively.
Figure 16.
(a) Overview of the PrP sequence and architecture.341, 389 The residue numbering refers to human PrP. (b) 3D representation of the secondary structure of mouse PrPC.384 The unordered N-terminus is omitted and the sulphur bridge between Cys179 and Cys214 is indicated in yellow. Copyright Nature Publishing Group.
PrPC has two regions with distinct structural and dynamical properties.367 In mammals, depending on the organism, the N-terminus contains a variable number of amino-terminal octapeptide repeats with sequence PHGGGWGQ. Each octarepeat is able to generate a divalent metals-binding domain via nitrogen atoms in the histidine imidazole side-chains.393 The N-terminus is up to residue 120 and this region has been shown to constitute a pH-dependent folding: at pH 4.5 it is flexibly disordered,394 however at pH 6.2 residues 61–84 of the octarepeats adopt a loop and a β-turn like conformation.386 In contrast, the C-terminus of PrPC is structured, containing three α-helices (H1, H2 and H3) and a short, two-stranded, antiparallel β-sheet (S1, S2)367 (Fig. 16b). A disulphide bridge is between Cys179 and Cys214, which anchors H2 and H3 helices. This disulphide bridge is one of the major determinants of the tertiary structure of PrP. FTIR study of PrP secondary structure revealed 42% of α-helices, 3% of β-sheets, 32% of turns and 23% of coils, respectively.395
The physicochemical properties of PrPSc and PrPC greatly differ. Spectroscopic measurements indicated that PrPSc contains about 34–43% of β-sheet structure,395, 396 significantly higher than that of PrPC.395 X-ray fiber diffraction of infectious prions revealed the presence of cross-β diffraction patterns. Meridional diffraction at 4.8 Å specified the presence of β-strands, characteristic of a stacked-sheet amyloid structure. Thus, β-enriched structure of PrPSc results from misfolding and self-assembly of protein PrPC into proteinase K-resistant amyloid-like aggregates. However, the high-resolution structures of infectious prions are not yet solved, as conventional structural methods have been hindered by the large and insoluble aggregates of PrP.
Several structural models of PrPSc self-assembly have been proposed based on information derived from biophysical techniques. Parallel left-handed β-helical structure is the model proposed by Cohen and colleagues and based on electron microscopy analysis of 2D crystals368 (Fig. 17a). The authors constructed a trimeric model of PrP 27–30 from a study of 119 all-β folds globular proteins. PrP 27–30 is a protease-resistant 27–30 kDa core of PrPSc (Fig. 16), and it retains prion infectivity.353, 397 According to the β-helical model the N-terminal residues of PrP 27–30 form left-handed β-helices that are horizontally stacked, whereas the C-terminus maintains α-helical secondary structure as in native PrPC. Larger aggregates are formed by vertically stacking of PrP trimers along the β-helical axis (Fig. 17a).
Figure 17.
Structural models for the PrPSc aggregates: (a) In the β-helical model the N-terminal region (90–177 residues, light green) of PrP 27–30 refolds into a β-helix motif and the C-terminal region (residues 178–230, dark green) maintains α-helical secondary structure as in native PrPC.368 (b) The β-spiral model consists of a spiralling core of extended sheets consisting of short β-strands, comprising residues 116–119, 129–132 and 160–164. The three α-helices in C-terminus maintain this conformational motif.398 (c) The parallel in-register extended β-sheet model of PrPSc, where PrPC refolds into a structure consisting mainly of β-sheets.399 Copyright National Academy of Sciences.
Based on MD simulations of PrP 27–30 conformational fluctuations under amyloidogenic conditions, DeMarco and Daggett proposed the β-spiral model398 (Fig. 17b). Similarly to the β-helical model the C-terminal α-helical characteristics of PrPC remain unchanged and natively unfolded N-terminus adopt a β-structure. The core structure is comprised of three short β-strands spanning 116–119, 129–132 and 160–164 residues.
Surewicz and colleagues proposed the in-register β-sheet model of PrPSc using site-directed spin labelling and EPR spectroscopy399 (Fig. 17c). In contrast to the other models, they observed that the refolding of PrPC involved major refolding of the C-terminal α-helical region. According to this model PrPSc structure possesses no α-helices, consisting mainly of single molecules stacked on top of one another with parallel, in-register β-strands. Using MD simulations, Caughey and colleagues suggested that linear PrPSc fibrils possessed a parallel in-register β-sheet structure400 (Fig. 18e).
Figure 18.
Electron microscopy of prion fibrils. (a) Aggregates of wild-type 22L scrapie prion aggregates.414 (b) Prion rods of PrP 27–30.341 (c) Wild type RML scrapie prion structure obscured by non-fibrillar material, while (d) anchorless RML fibril morphology was much cleaner.414 (e) Celery stalk-like brain-derived GPI-anchorless 22L fibril414 and proposed parallel in-register β-sheet model of PrP (90–231) octametric segment.400 Scale bars: 100 nm. Copyright Elsevier, National Academy of Sciences and American Society for Biochemistry and Molecular Biology.
In addition, a number of structures have been proposed for mammalian401 and fungal402 prion protein segments. It is difficult to determine which of the proposed models is the closest to the PrPSc structure, as they were established based on low-resolution experimental data. The diversity of the models could originate from the specimens used. For example, Wille and colleagues compared natural brain-delivered PrPSc and synthetic bacteria-expressed recombinant PrP (with the same sequence) amyloid structure and found substantial differences in structure, heterogeneity and infectivity.403 In addition, the existence of PrP tertiary structural diversity and prion strains have been experimentally proven,404–408 including the formation of new strains during the passage of prions through animals with different PrP sequences.409, 410 For instance, multiple scrapie prion strains were isolated with different incubation times and neuropathology.411 However, these prion strains were encoded by the same PrP primary structure and were propagated in mice with the same PrP gene. Despite this, limited proteolysis generated different PrPSc fragments, suggesting that these prion strains possessed different conformations.352
5.3 Mesoscopic structures of prions
The morphology of prions has been examined with TEM412–414 and AFM.414 Usually PrPSc isolated from brain appears as large amorphous highly insoluble aggregates (Fig. 18a). Individual prion fibrils, termed prion rods (Fig. 18b), are not always visible probably because of heavy surface glycosylation415 (Fig. 18c). Each PrP monomer has up to two large sugar moieties linked to the N-terminus to obscure the fibril core. Deficiencies in glucans and GPI archorless PrP have been found suitable for analyzing the structural features of prion protofilaments (Fig. 18d), while neither glycosylation416, 417 nor the GPI anchor348, 418 is required for the infectivity of PrPSc.
Majority of PrP fibrils, either wild-type, anchorless, or of different strains, possess a twisted morphology. Prion fibrils can be either left-handed or right-handed414, consisting of two or more protofilaments.412 However, some fibrils also contain straight, parallel protofilaments.414 In addition, fibrils occasionally resemble celery stalks or half-pipes414 (Fig. 18c). In a recent study the gap previously thought to be the spacing between two protofilaments of celery stalk fibrils was assigned to represent the trough between the major hairpins. Accordingly, an in-register β-sheet PrP amyloid model was proposed.400 The periodicity of PrP fibrils, on the other hand, ranged between 40 nm to 133 nm412, 414, 419 while the width of each PrP protofilament varied from 3.1±0.7 nm414 to 6.9 nm.368
5.4 Transmission of prions
Epidemiological transmission of PrP diseases is via the exposure of PrPC to PrPSc. However, point mutations in the PRNP gene at K200E, D178N, L102P and V117A codons were observed in families initially diagnosed with vCJD and GSS.420–422 Linear transmission in human has an early history in the fore people of Papua New Guinea who suffered from Kuru: a human variant of PrP disease with clinical symptoms of ataxia, shivering and death within year of manifestation. Although the endemic is ceased by terminating the ritual cannibalism in 1950s, Kuru is waning gradually due to long sub-clinical incubation period i.e. > 50 years.423 In modern days, the inter-human transmission is via blood transfusions as person infected with vCJD carry PrPSc load in all blood components with transmission efficiency of WBCs > platelets > RBCs > plasma and shed PrPSc in saliva, urine and other bodily fluids.424, 425 Less common ways of transmission in humans are surgical instruments and human derived growth hormones.426
The clinical symptoms of PrP diseases originate from pathological changes in CNS such as vacuolization, astrogilosis and neuronal apoptosis. However, once prion replication in CNS reaches its peak, the PrPSc is disseminated centrifugally to the peripheral secretory organs and lymphoid tissues. PrPSc excretes are detected in blood, urine, saliva, milk and bone-meat meal (MBM) of infected animal even at sub-clinical stage. The titre from urine and saliva of CWD infected cervid was able to reproduce infectivity in naïve cervid and transgenic mice models.427–429 Salivary expression can contaminate drinking water and pose a risk of transmission to human and other animals.430 Scrapie infected sheep shed PrPSc in all components of colostrum and milk i.e. cells, cream and casein/whey proteins, which carried infectivity to lambs and dairy products.431, 432 The titre of infectivity per mL of milk was equivalent to 6 μg of brain homogenate from terminally scrapie-infected sheep.433 Bone and meat materials, either decaying in the soil or processed into MBM for cattle feeds, had PrPSc attached to its particles. PrPSc attached to MBM or soil particles had higher transmission efficiency.434 Infectivity was retained and transmissible to animals even after processing of MBM for biodiesel productions.435 Once attached to the soil particles, PrPSc not detachable via surfactants and soil could retain infectivity up to 19 years.436–438 However, hyperthermophilic bacteria were able to digest the PrPSc particles from soil by secreting keratins and β-sheets proteases.439
Rasmussen et al. first showed that hamster PrPSc were able to bind with wheat grass roots, from soil and brain homogenate, but neither absorbed in roots nor detected in areal parts of the plants.440 Pritzkow et al. used protein misfolded cyclic amplification (PMCA) as a more sensitive detection method and transmitted hamster 263K PrPSc to wheat grass roots via infected brain homogenate, excreta, contaminated soils and direct spray of PrP on areal parts.441 The 263K PrPSc were able to adsorb from the sources to the roots and travelled in the areal parts, which were further able to reproduce the infectivity in naïve hamster. Apart from the extraneous PrPSc in plants, a protein named luminidependens from Arabidopsis thaliana was transformed and propagated like PrP when injected in yeast cells.442
5.5 Conversion and replication of prions
The molecular interaction between PrPSc and PrPC is based on self-assembly driven by hydrogen bonding and π-stacking of the tyrosine residues. Two initial models described the mechanism of PrP replication.443 The “template directed” model described PrPSc as the more stable but thermodynamically inaccessible form of PrPC. In contrast, the “seeded nucleation” model described the contact of small oligomers of PrPSc with PrPC: the seeds of PrPSc recruit PrPC into conformationally changed form, and the growing fibril is broken down into various small seeds acting as nuclei for further recruitment. The PrPSc monomers are less stable but become stabilized when joined in the seeded oligomer form.444, 445 The seeded nucleation model was supported by later experiments where small amounts of preformed PrPSc oligomers converted large quantities of PrPC as in PMCA, where seeds shredding was induced by sonication and the conversion process was amplified.446 Makarava et al. refined the conversion phenomenon by studying the conformation switch (R and S) within single, mouse-hamster cross-seeded PrP amyloids and introduced the concepts of catalytic versus templated conversion and amyloid flexibility.447 Hamster PrP (S conformation), when incubated with mouse PrP monomers, catalyzed the conversion by accelerating the fibrillation rate and shortening the lag phase, but the newly formed daughter fibrils retained R conformation only. In contrast, when hamster PrP seeds were introduced to hamster PrP monomers, it accelerated fibrillation and templated the same S conformation in daughter fibrils. Molecular events occurring in template-directed PrP conversion started from π-π interaction of PrPC and PrPSc in 6 different binding and conversion domains (BCD) of PrP. In the absence of PrPSc, when human and hamster PrPC BCD were probed with monoclonal antibody (mAb), it resulted in structural denaturation of PrPC, regional loss of tertiary structure, dissociation of β-sheets, and exposure of bityrosine regions (YYR) at α1 and α2 helices. The exposure of YYR regions was confirmed with binding of anti-YYR mAb in these lose regions.448 In contrast to mAb, PrPSc binding induced melting and exposure of YYR regions from β2-α2, α2-α3 and α1 regions.448–450 The exposed YYR could be the site of further PrPC attachment and connected the oligomer and monomer.450 The loose structure induced by mAb was not able to acquire any conformation or secondary structure from mAb. However, when PrPSc oligomers induced this structural loosening, it acquired β-sheets from oligomer’s hydrogen bonded backbone and stabilized the whole fibril column.447
5.6 Co-factors in prion assembly
Co-factors, initially recognized as “Protein X” by Prusiner, stabilize the PrPC-PrPSc assembly and may further facilitate the spontaneous conversion of PrPC into protease resistant form.451 Biomacromolecules of polysaccharides, sphingolipids, phospholipids, cholesterol, detergents like SDS, lipopolysaccharides from bacterial membranes, and polyanions like RNA have been found to interact with PrPC and are co-localized with PrPSc from infected animals.452–454 The interaction with co-factors melted the secondary structure of PrPC and converted it into protease resistant but non-infectious β-sheet structure, differently from the β-sheets of PrPSc, even though PrPSc and co-factors exposed YYR from the same regions of PrPC.449, 450
5.7 Transmission barriers: sequence and conformation
A transmission barrier appears when there is no clinical neuropathology of spongiform encephalopathies upon inoculation of PrPSc from infected species A into the naïve species B. The molecular etiology for the transmission barrier is attributed to i) difference in the sequences of host and donor PrP, ii) conformational misfit during assembly, and iii) post conversion maturation in host. The exact residual regions responsible for the transmission barrier are i) 165–175 (β2-α2) with switches at 170 (S/N), 174 (N/T), 169 (Y/G), ii) 138–143 (β1-α1) with switches at 139 (M/I) and iii) 129 (β1) for M/V switch (Fig. 19).455–458 Sheep’s scrapie and cattle’s BSE are inter-transmissible with clinical symptoms as both are 170S homozygous. However, transmission between mice (170S) and hamster (170N) does not produce clinical disease.459, 460 Sigurdson et al. demonstrated the 170 S/N and 174 N/T switches at the molecular level.458 Inoculation of deer scrapie PrPSc (170N, 174N) into wild type tg20 mice (170S, 174N) didn’t express clinical symptoms at first passage but inoculation in rigid loop tg1020 mice (S170N, N174T) produced terminal symptoms in 74 days. Furthermore, hamster PrPSc (170N, 174T) accelerated clinical disease in tg1020 mice (S170N, N174T) but not in tg20 mice (170S, 174N). In contrast, cattle and sheep PrPSc (170S, 174N) produced disease in tg20 mice (170S, 174N) but not in tg1020 mice (S170N, N174T).458 Similarly, 139I in humans and mice PrPSc induced parallel β-sheet stacking and R conformation in fibril while 139M in hamster induced anti-parallel β-sheet stacking and S conformation.447, 456 The region 138–143 has been demonstrated as a steric zipper in stabilizing PrPSc fibril by hydrogen bonding between inter-monomeric β-sheets. Incompatibility at the steric zipper also erects a cross-species barrier.461 The tyrosine residue at 169 is responsible for initial π-π interaction between PrPSc oligomers and host PrPC monomers and is conserved in all mammalian PrP. Eliminating tyrosine or replacing it with glycine completely blocked PrPSc interaction with PrPC and halted the conversion.457 Another molecular switch between human TSE and mad cow BSE is at 129 residue (M/V). vCJD infected human possessed 129M homologues with bovine 129M. However, 129V PrP peptide was still converted to the PrPSc form in-vitro, but at a low efficiency. Hence the possibility of bovine 129M infection to 129V heterozygous human can not be excluded.455, 462
Figure 19.
Amino acid sequence and 3D structural comparison of β-sheet stacking from steric zones of PrPSc in different mammalian species. Superimposition of mouse (grey) and hamster (blue) PrPSc with 165–172 backbone fold (a). Amino acid sequence from 170–175 backbone region (b).463 Cyan highlights human while orange highlights elk specific residues. Stick representation of steric zipper interfacing β-sheet back bone region for human (c) and both alignments of elk (d, e). X-ray crystallographic atomic structures from barrier determining steric zippers from human, mouse and hamster, side view for single β-sheet stacking (f, g, h) and top view of steric zipper (i, j, k).456 Sequence differences at molecular switches, defining the conformational and transmission barrier between different species (l).456, 463 Grey and red indicate transmission and barrier while cyan at 139 presents molecular switch for parallel or anti-parallel sheet stacking in human, mouse and hamster. Copyright The American Chemical Society.
5.8 PrP evolution and conformational adaptation: sub-clinical stages
Cross-species transmission of PrPSc infection produced no clinical symptoms in first passage due to dissimilar residual sequence and thus conformation as discussed above. However, the clinically silent phase led to the understanding of i) long sub-clinical stage, with no disease symptoms but undergoing PrP replication in brain, spleen and lymphoid tissues and ii) concepts of PrPSc maturation, stability, selection and evolution.464, 465 First passage of cattle BSE to human PrPSc transgenic mice produced only a 0.6% attack rate after 739 days but a 75% attack rate after 639 days on second passage.466 The decrease in incubation time and increase in attack rate were also observed by passaging the inoculum in vitro with PCMA rounds, which led to the emergence of mutated and phenotypically distinct PrP strains.467–469 Kimberlin et al. inoculated 139A PrPSc from mouse to hamster and then back to mouse resulting in 139H/M strain.470 Colby et al. infected mice with rPrP of different conformational stability and resulted in phenotypically different strains with different physical morphologies, shorter incubation time, higher attack rate and varying clinical pathologies.471 It was hypothesized that the original inoculum consisted of different rPrP strains and upon infecting into the host, the strain having close conformational fit with host PrP replicated at a faster rate, induced the clinical disease and subsequently appeared in animal tissues.471 Similarly, Bruce et al. raised 22C natural PRNPa strain in PRNPb mice and ended up with 22H strain. However, upon passaging the recombinant and pure 22C strain, only 22C was resulted in the host indicating the presence of both 22C and 22H in natural inoculum but 22H with shorted incubation time and close fit with host PrP was able to replicate at a faster rate.472 Makarava et al. annealed hamster rPrPSc with normal hamster brain homogenate and inoculated in naïve hamster. On first passage, 50% attack was observed but all hamster succumbed to disease on second passage and clinical symptoms were different from original rPrPSc which was used to prepare the inoculum.464 The explanation is that i) original inoculum lacked the GPI anchor and failed to penetrate the cell to initiate neurotoxicity, ii) conformational adaptation and stability were observed upon serial PCMA and iii) rPrPSc-NBH failed to acquire co-factors, which it acquired on subsequent passages in the host.464 Serial passages of donor PrPSc with host PrPC by PCMA in vivo or in vitro also changed the biochemical parameters like electrophoretic mobility, protease digestion and degree of glycosylation.473 The efficiency and properties of evolutionary adaptation was also found to be tissue dependant, i.e., occurring at a faster rate in spleen than brain and cell and brain adapting different strains.465, 474
5.9 Toxicity of prions and mitigation
Two and a half decades on, the mechanism of prion toxicity has been narrowed down to the distortion of neuronal cell membranes due to the assembly of PrPSc oligomers with GPI anchored PrPC.475, 476 Deletion of GPI anchored PrPC from the membrane or expression of anchorless PrPC in transgenic mice resulted a minimum infectivity or reversal of clinical symptoms in infected mice, which implicated anchored PrPC for neurotoxicity.348, 477 However, the extracellular accumulation of PrPSc continued as plaques as in terminally-ill wild-type mice.78, 79 Apart from PrPSc, when anchorless PrPC was exposed to lipid membrane or expressed in transgenic mice they adhered to the membranes, underwent conformational changes into the protease resistant form, oligomerized locally, and caused membrane disruption and ion channel formation.478–480 Although PrPC has a neuroprotective role against cellular stress, it also intervenes toxic signals to neuronal cell and initiates an apoptotic cascade when probed by PrPSc, β-sheet conformers, yeast prions, Aβ or other amyloid oligomers and even anti-PrPC antibodies.481–483 The mechanism is postulated as either through blocking the physiological binding domains of PrPC or disruption of neuronal membranes by PrPC-PrPSc oligomer adducts.484, 485 The adduct formed on the membrane can be internalized and disrupt endosomal trafficking or distort the local fluidity, structure and function of lipid bilayers like channel formation in GSS.476, 486 In addition to adduct formation, PrPSc oligomers can independently interact with membranes via their own GPI anchors, which they tend to develop during sequel passages.487 The oligomer form of PrPSc has been shown to be the toxic species, other than the monomers or amyloids.480 PrPSc oligomers possess the necessary hydrogen bonding backbone running up and own in the column to induce conformational change in PrPC and recruit the latter at the growing end.447 PrPSc oligomers corrupt PrPC function and deliver a neurotoxic signal.476
Initial therapeutic strategies considered silencing of the PrPC gene. Silencing was well tolerated in animals apart from minor disturbance in sleep cycle and electrophysiology of hippocampus.500 However, as the PrPC is associated with neuroprotection excitotoxicity outcomes are being predicted for silencing PrPC in human.501 RNA is physiologically found co-localized with PrPSc which triggered research for RNA as an anti PrP drug.452 RNA based aptamers can bind PrPC to prevent PrPSc-induced conversion or with PrPSc oligomers to block their activity.502 The subsequent interaction of PrPC with the same aptamer increases the binding efficiency due to the adaptation flexibility of PrPC.503 Different ways of delivering aptamer RNA across the blood brain barrier (BBB) include conjugation with transferrin, cell penetrating peptides, NPs, liposomes and dendrimer.503 Polyethylene glycol-conjugated polycyanoacrylate NPs were able to penetrate the brain and spleen of scrapie infected animals, however their ability to delivery therapeutic cargo and mode of interaction with PrPSc fibrils are questioned.504 Branched polyamines degraded PrPSc amyloids to undetectable levels and reversed PrPSc toxicity in neuroblastoma cell culture (Fig. 20h).499 Pre-treatment of PrPSc amyloids with polyamines rendered them susceptible to proteolytic digestion. Tran et al. used polyallylamine (+) and polystyrenesulfonate (−) as two oppositely charged polyamines for layer-by-layer coating of gold NPs (AuNPs) (Fig. 20c). The AuNPs translocated across the BBB, disrupted the PrPSc amyloids, and mitigated the toxicity in scrapie-infected cells. Nanomolar concentrations of AuNPs, with poly(allylamine) as the outermost layer, prolonged incubation time and delayed the disease onset in infected mice.497 Polyamine-based dendrimers coated with maltose or maltotriose stimulated PrP fibrillization at lower concentrations by breaking long fibrils into small seeds, but at higher concentrations blocked the fibrillization by stabilizing individual seeds (Fig. 20h).491
Figure 20.
Prion diagnostics and therapeutics at the nano and medicinal chemistry fronts.488–499 Compilated from references 488–499. PrPSc can be sensed by turning on/off the fluorescence of fluorescein-AuNPs (a), free QDs (g) or QD-FeNP sandwiches (f), or by resonance light scattering (RLS) of lipoic acid-AuNP aggregates (b). Quantitative sensing can be performed by Raman spectroscopy of Au nanorods (j). PrPSc can be captured by AuNPs with polyamines and sulphonates surface layers of (c) or by FeNPs with mercaptopropionic acid, aspartic acid (d) or RNA aptamer surface layers (e). Cell bound PrPSc can be captured by aptamer-AgNP conjugates (i) while complete denaturation of PrPSc can be observed with 5G polyamine dendrimers (h). The TEM images in h are reproduced from ref. 488 with permission from the Royal Society of Chemistry. The TEM images in b and Raman spectra in j are copyrights of The American Society of Chemistry and National Academy of Science.
NP-PrP interactions have been explored for diagnostic and sensing applications. Monothiolation of RNA aptamers makes them a good ligand to cap AuNPs or AgNPs. These NPs then specifically interact with PrPSc and sequester the latter on their surfaces. The binding of PrPSc or cell bound PrPC is sensed in a concentration dependant manner via changes in the surface plasmon resonance or Raman signals of the AuNPs/nanorods (NRs) and aptamer ligated AgNPs (Fig. 20j).489, 495 PrP binding with aptamer-conjugated NPs induces controlled aggregation which can be sensed via resonance light scattering of metal NPs aggregation (Fig. 20b).488 Henry et al. employed fluorescence turn-on and turn-off sensing for PrP detection.498 The fluorescence of fluorescein-GABA-QYQRES-COOH peptide bound to antibody-conjugated AuNPs (turned-off) was turned-on by replacing the peptide with competitive binding of PrPSc with the antibody (Fig. 20a). The fluorescence property of biotin-avidin or monoclonal antibody bound quantum dots (QDs) has also been explored for the detection of PrP in vitro and in vivo (Fig. 20g).490 Xiao et al. used the dual-aptamer technique by ligating aptamer 1 on Fe magnetic NPs (MNPs) and aptamer 2 on QDs. MNPs and QDs sandwiched PrPSc or PrPC in between and the technique was used to detect and isolate PrP by fluorescent QDs and magnetic NPs even from 0.1 % infected brain homogenate (Fig. 20f).496 MNPs directly capped with aspartic acid or Au-mercaptopropionic acid were able to sequester PrP via carbodiimide coupling (Fig. 20d).505 Miller et al. engaged the aptamer-ligated MNPs to capture and clear PrPSc from solution. The sequestered PrPSc on MNPs were able to act as seeds in PCMA and enabled the detection and amplification of small quantities of PrPSc (Fig. 20e).506
On the medicinal chemistry front, amphotericin B,507, 508 quinacrine, dimeric analogues of statins, pyrazolones and pyridyl hydrozones are available drugs for prolonging the lifespan of PrP infected animals.493, 509 Tacrolimus and astemizole reduce the PrP expression on cell membranes and inhibit PrPSc replication.510 Drug discovery for small organic molecules led to 2-aminothiazoles which cap PrPSc seeds and inhibit their replication activity.511 Lipoic acid, an endogenous anti-oxidant compounds and when conjugated with acridine and quinolone, inhibited PrPSc fibril formation.494 A structure-activity relationship study of the pyrazole derivative of carbazole led to the understanding that a tricyclic aromatic ring with hydroxyl and amino groups inhibited PrPSc fibrillization in PrPSc-infected neuronal cells.492
5.10 Prions versus other neurodegenerative disorders
Cross-seeding and mutual stimulation of amyloid fibrils have revealed possible links between AD, PD and T2D.512, 513 Mougenot et al. injected the PrPSc from cattle BSE, human BSE and scrapie into mice over-expressing αS. The incubation time was reduced significantly and mice died of cerebral spongiform pathologies of PrPSc without accumulating insoluble fibrils of αS.514 AD and PD have different neuronal pathologies than PrP and their ability to transmit and infect like prions is inconclusive.515, 516 More in vitro and in vivo cross seeding studies are necessary to elucidate the mechanisms and relationships between these diseases of different origins.
6. Summary
A survey of the literature has revealed striking similarities in the cross-β motifs of amyloid fibrils held together by H-bonding, regardless of the sequence and origin of the proteins. However, a recent study reported a cross-α amyloid structure associated with PSMα3, a 22-residue functional amyloid peptide secreted by Staphylococcus aureus for inflammatory response stimulation, human cell lysis and biofilm formation, representing a surprising departure from the common amyloid structure.517 The suprastructure of amyloid fibrils – including that of all (S) Aβ1–40 and hen egg lysozyme – has been shown as predominantly left handed,518 originated from the inherent left-handed chirality of the (S) amino acids. However, right-handed amyloids have been reported for truncated serum amyloid A (SAA) peptides (< 12 residues), resulting from the occurrence of β helices in SAA protofilaments prior to their assembly into fibrils.519 A recent study on serum albumin amyloids, has revealed that handedness can be inverted from left to right handed, upon lateral addition of protofilaments of amyloid fibrils of a lower hierarchical level.520
There is compelling evidence that amyloid proteins can spread from cell to cell and cross talk in vivo to either speed up or slow down aggregation of the host protein.291, 519, 521–523 Furthermore, aggregates of non-amyloidogenic proteins, such as bovine PI3-SH3 domain and E. coli HypF domain, can serve as seeds to promote cytotoxicity in brain cells. This phenomenon suggests a generic origin of protein misfolding diseases resulting from the emergence of trace amounts of aggregates, either introduced intracellularly through misfolding or mutations or externally through cross seeding.285
The development of neurodegenerative disorders appears to be correlated with aging, where misfolding of proteins down the free energy landscape towards the amyloid state is likely prevented by metal ions (such as Ca2+ in the ER), molecular chaperones, ubiquitination enzymes and proteasomes, which kinetically trap the aggregating proteins off pathway.285, 524 Compared with Aβ or IAPP, the tremendous plasticity of αS and PrP may originate from their much longer chain lengths and, therefore, greater populations of misfolded intermediates.
Although controversies remain, the observations that oligomers are more toxic than their fibrillar counterparts appear pervasive to amyloid proteins. In addition, oligomer-specific antibody developed for Aβ also bound the oligomers of IAPP, lysozyme, prion106–126, human insulin, polyglutamine and αS, suggesting a common tertiary structure273 as well as a common mechanism of pathogenesis beyond the individuality of the proteins and compositions of their molecular chaperones and cellular environments. As amyloid proteins fibrillate along the kinetic pathway, both their solubility and reactivity appear to decline, consequently impacting protein self-assembly and their engagement with environmental ligands, proteins, cell membranes and organelles to elicit toxicity. While such conditions can be manipulated in vitro, such as through the regulation of temperature and pH or the introduction of metal ions, small molecules or engineered NPs, how to create in vivo conditions that prohibit trace amounts of aggregates from activating primary and/or secondary nucleation remains a tremendous challenge. Despite the complexity of protein structure, function and toxicity, as revealed by intensive research spanning the past two decades and highlighted in this review, protein aggregation through self-assembly and interaction with cellular environments constitutes hallmarks of neuronal and pancreatic β-cell degeneration. Consequently, understanding and exploiting molecular assembly under physiological conditions could make inroads on the development of therapeutics and diagnostics against amyloid diseases.
Acknowledgments
The authors thank the support of ARC Project CE140100036 (Davis), DP160100959 (Separovic), NSF CAREER CBET-1553945 (Ding), NIH MIRA 1R35GM119691 (Ding) and Monash Institute of Pharmaceutical Sciences (Ke). Davis is thankful for an Australian Laureate Fellowship from the ARC. We apologize to those whose works are not cited in this review due to the tremendous amount of the relevant literature.
List of abbreviations
- Aβ
amyloid-beta
- AD
Alzheimer’s disease
- AFM
atomic force microscopy
- APP
amyloid precursor protein
- αS
alpha-synuclein
- ATR-FTIR
attenuated total reflection-Fourier-transform infrared
- BBB
blood brain barrier
- BCD
binding and conversion domains
- BSE
bovine spongiform encephalopathy
- CD
circular dichroism spectroscopy
- CJD
Creutzfeldt-Jakob disease
- CNS
central nervous system
- CWD
chronic wasting disease
- DLB
dementia with Lewy bodies
- DMD
discrete molecular simulations
- EGCG
epigallocatechin gallate
- EPR
electron paramagnetic resonance
- ER
endoplasmic reticulum
- FFI
fatal familial insomnia
- FSI
fatal sporadic insomnia
- FTIR
Fourier transform infrared spectroscopy
- GPI anchor
glycosyl phosphatidylinositol
- GSS
Gerstmann-Straussler-Scheinker
- HAM
hierarchical assembly model
- HD
hydrogen-deuterium exchange
- huPrP
human PrP
- IAPP
islet amyloid polypeptide
- IDP
intrinsically disorder protein
- IM-MS
ion mobility mass spectroscopy
- mAb
monoclonal antibody
- MBM
bone-meat meal
- MD
molecular dynamics
- MNPs
Fe magnetic NPs
- NAC
non-amyloid-beta component
- NFTs
neurofibrillary tangles
- NMR
nuclear magnetic resonance
- NSF
N-ethylmaleimide-sensitive factor
- NPs
nanoparticles
- PD
Parkinson’s disease
- PDD
Parkinson’s disease dementia
- PHFs
paired helical filaments
- PMCA
protein misfolded cyclic amplification
- PrP
prion protein
- PrPC
non-pathological PrP
- PrPSc
misfolded PrP
- ROS
reactive oxygen species
- SAA
serum amyloid A
- SDS
sodium dodecyl sulfate
- SDSL
site directed spin labelling
- SNARE
soluble NSF attachment protein receptor
- ssNMR
solid-state nuclear magnetic resonance
- SUVs
small unilamellar vesicles
- T2D
type 2 diabetes
- TEM
transmission electron microscopy
- ThT
thioflavin T assay
- TSEs
transmissible spongiform encephalopathies
- UPS
Ubiquitin proteasome system
- YYR
bityrosine regions
References
- 1.Knowles TPJ, Vendruscolo M, Dobson CM. Nat Rev Mol Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 2.Cohen SIA, Linse S, Luheshi LM, Hellstrand E, White DA, Rajah L, Otzen DE, Vendruscolo M, Dobson CM, Knowles TPJ. Proc Natl Acad Sci USA. 2013;110:9758–9763. doi: 10.1073/pnas.1218402110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knowles TPJ, Waudby CA, Devlin GL, Cohen SIA, Aguzzi A, Vendruscolo M, Terentjev EM, Welland ME, Dobson CM. Science. 2009;326:1533–1537. doi: 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- 4.Guijarro JI, Sunde M, Jones JA, Campbell ID, Dobson CM. Proc Natl Acad Sci USA. 1998;95:4224–4228. doi: 10.1073/pnas.95.8.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, Dobson CM. Proc Natl Acad Sci USA. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urbanc B, Cruz L, Yun S, Buldyrev SV, Bitan G, Teplow DB, Stanley HE. Proc Natl Acad Sci USA. 2004;101:17345–17350. doi: 10.1073/pnas.0408153101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auer S, Meersman F, Dobson CM, Vendruscolo M. PLoS Comp Biol. 2008;4:e1000222. doi: 10.1371/journal.pcbi.1000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ratha BN, Ghosh A, Brender JR, Gayen N, Ilyas H, Neeraja C, Das KP, Mandal AK, Bhunia A. J Biol Chem. 2016;291:23545–23556. doi: 10.1074/jbc.M116.742460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamley IW. Angew Chem Int Ed. 2007;46:8128–8147. doi: 10.1002/anie.200700861. [DOI] [PubMed] [Google Scholar]
- 10.Hamley IW. Chem Rev. 2012;112:5147–5192. doi: 10.1021/cr3000994. [DOI] [PubMed] [Google Scholar]
- 11.Alzheimer A, Stelzmann RA, Schnitzlein HN, Murtagh FR. Clinical anatomy. 1995;8:429–431. doi: 10.1002/ca.980080612. [DOI] [PubMed] [Google Scholar]
- 12.Glenner GG, Wong CW. Biochemical and biophysical research communications. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 13.Blessed G, Tomlinson BE, Roth M. The British journal of psychiatry : the journal of mental science. 1968;114:797–811. doi: 10.1192/bjp.114.512.797. [DOI] [PubMed] [Google Scholar]
- 14.Grundke-Iqbal I, Iqbal K, Tung YC, Quinlan M, Wisniewski HM, Binder LI. Proc Natl Acad Sci USA. 1986;83:4913–4917. doi: 10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skrabana R, Sevcik J, Novak M. Cellular and molecular neurobiology. 2006;26:1085–1097. doi: 10.1007/s10571-006-9083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avila J, Pallas N, Bolos M, Sayas CL, Hernandez F. Expert opinion on therapeutic targets. 2016;20:653–661. doi: 10.1517/14728222.2016.1131269. [DOI] [PubMed] [Google Scholar]
- 17.Mukrasch MD, Bibow S, Korukottu J, Jeganathan S, Biernat J, Griesinger C, Mandelkow E, Zweckstetter M. PLoS biology. 2009;7:e34. doi: 10.1371/journal.pbio.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganguly P, Do TD, Larini L, LaPointe NE, Sercel AJ, Shade MF, Feinstein SC, Bowers MT, Shea JE. The journal of physical chemistry B. 2015;119:4582–4593. doi: 10.1021/acs.jpcb.5b00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W, Lee VM. Biochemistry. 2006;45:15692–15701. doi: 10.1021/bi061422+. [DOI] [PubMed] [Google Scholar]
- 20.Mandelkow E, von Bergen M, Biernat J, Mandelkow EM. Brain pathology. 2007;17:83–90. doi: 10.1111/j.1750-3639.2007.00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Bergen M, Friedhoff P, Biernat J, Heberle J, Mandelkow EM, Mandelkow E. Proc Natl Acad Sci USA. 2000;97:5129–5134. doi: 10.1073/pnas.97.10.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minoura K, Yao TM, Tomoo K, Sumida M, Sasaki M, Taniguchi T, Ishida T. European journal of biochemistry. 2004;271:545–552. doi: 10.1046/j.1432-1033.2003.03956.x. [DOI] [PubMed] [Google Scholar]
- 23.Adamcik J, Sanchez-Ferrer A, Ait-Bouziad N, Reynolds NP, Lashuel HA, Mezzenga R. Angewandte Chemie. 2016;55:618–622. doi: 10.1002/anie.201508968. [DOI] [PubMed] [Google Scholar]
- 24.Sibille N, Huvent I, Fauquant C, Verdegem D, Amniai L, Leroy A, Wieruszeski JM, Lippens G, Landrieu I. Proteins. 2012;80:454–462. doi: 10.1002/prot.23210. [DOI] [PubMed] [Google Scholar]
- 25.Sandbrink R, Masters CL, Beyreuther K. Neurobiology of disease. 1994;1:13–24. doi: 10.1006/nbdi.1994.0003. [DOI] [PubMed] [Google Scholar]
- 26.Tanzi RE, McClatchey AI, Lamperti ED, Villa-Komaroff L, Gusella JF, Neve RL. Nature. 1988;331:528–530. doi: 10.1038/331528a0. [DOI] [PubMed] [Google Scholar]
- 27.Burger PC, Vogel FS. The American journal of pathology. 1973;73:457–476. [PMC free article] [PubMed] [Google Scholar]
- 28.Selkoe DJ. Annual review of cell biology. 1994;10:373–403. doi: 10.1146/annurev.cb.10.110194.002105. [DOI] [PubMed] [Google Scholar]
- 29.Zheng H, Jiang M, Trumbauer ME, Sirinathsinghji DJ, Hopkins R, Smith DW, Heavens RP, Dawson GR, Boyce S, Conner MW, Stevens KA, Slunt HH, Sisoda SS, Chen HY, Van der Ploeg LH. Cell. 1995;81:525–531. doi: 10.1016/0092-8674(95)90073-x. [DOI] [PubMed] [Google Scholar]
- 30.White AR, Reyes R, Mercer JF, Camakaris J, Zheng H, Bush AI, Multhaup G, Beyreuther K, Masters CL, Cappai R. Brain research. 1999;842:439–444. doi: 10.1016/s0006-8993(99)01861-2. [DOI] [PubMed] [Google Scholar]
- 31.Grimm MO, Grimm HS, Patzold AJ, Zinser EG, Halonen R, Duering M, Tschape JA, De Strooper B, Muller U, Shen J, Hartmann T. Nature cell biology. 2005;7:1118–1123. doi: 10.1038/ncb1313. [DOI] [PubMed] [Google Scholar]
- 32.Meziane H, Dodart JC, Mathis C, Little S, Clemens J, Paul SM, Ungerer A. Proc Natl Acad Sci USA. 1998;95:12683–12688. doi: 10.1073/pnas.95.21.12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roch JM, Masliah E, Roch-Levecq AC, Sundsmo MP, Otero DA, Veinbergs I, Saitoh T. Proc Natl Acad Sci USA. 1994;91:7450–7454. doi: 10.1073/pnas.91.16.7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikolaev A, McLaughlin T, O’Leary DD, Tessier-Lavigne M. Nature. 2009;457:981–989. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Ring S, Weyer SW, Kilian SB, Waldron E, Pietrzik CU, Filippov MA, Herms J, Buchholz C, Eckman CB, Korte M, Wolfer DP, Muller UC. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:7817–7826. doi: 10.1523/JNEUROSCI.1026-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 37.Olubiyi OO, Strodel B. J Phys Chem B. 2012;116:3280–3291. doi: 10.1021/jp2076337. [DOI] [PubMed] [Google Scholar]
- 38.Caglayan S, Takagi-Niidome S, Liao F, Carlo AS, Schmidt V, Burgert T, Kitago Y, Fuchtbauer EM, Fuchtbauer A, Holtzman DM, Takagi J, Willnow TE. Science translational medicine. 2014;6:223ra220. doi: 10.1126/scitranslmed.3007747. [DOI] [PubMed] [Google Scholar]
- 39.Huse JT, Liu K, Pijak DS, Carlin D, Lee VM, Doms RW. J Biol Chem. 2002;277:16278–16284. doi: 10.1074/jbc.M111141200. [DOI] [PubMed] [Google Scholar]
- 40.Liu K, Solano I, Mann D, Lemere C, Mercken M, Trojanowski JQ, Lee VM. Acta neuropathologica. 2006;112:163–174. doi: 10.1007/s00401-006-0077-5. [DOI] [PubMed] [Google Scholar]
- 41.Hosoda R, Saido TC, Otvos L, Jr, Arai T, Mann DM, Lee VM, Trojanowski JQ, Iwatsubo T. Journal of neuropathology and experimental neurology. 1998;57:1089–1095. doi: 10.1097/00005072-199811000-00012. [DOI] [PubMed] [Google Scholar]
- 42.Gomez-Ramos P, Asuncion Moran M. Journal of Alzheimer’s disease : JAD. 2007;11:53–59. doi: 10.3233/jad-2007-11109. [DOI] [PubMed] [Google Scholar]
- 43.Wirths O, Multhaup G, Czech C, Blanchard V, Moussaoui S, Tremp G, Pradier L, Beyreuther K, Bayer TA. Neuroscience letters. 2001;306:116–120. doi: 10.1016/s0304-3940(01)01876-6. [DOI] [PubMed] [Google Scholar]
- 44.Cirrito JR, Yamada KA, Finn MB, Sloviter RS, Bales KR, May PC, Schoepp DD, Paul SM, Mennerick S, Holtzman DM. Neuron. 2005;48:913–922. doi: 10.1016/j.neuron.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 45.Kamenetz F, Tomita T, Hsieh H, Seabrook G, Borchelt D, Iwatsubo T, Sisodia S, Malinow R. Neuron. 2003;37:925–937. doi: 10.1016/s0896-6273(03)00124-7. [DOI] [PubMed] [Google Scholar]
- 46.Morley JE, Farr SA, Banks WA, Johnson SN, Yamada KA, Xu L. Journal of Alzheimer’s disease : JAD. 2010;19:441–449. doi: 10.3233/JAD-2009-1230. [DOI] [PubMed] [Google Scholar]
- 47.Ghiso J, Frangione B. Advanced drug delivery reviews. 2002;54:1539–1551. doi: 10.1016/s0169-409x(02)00149-7. [DOI] [PubMed] [Google Scholar]
- 48.Strozyk D, Blennow K, White LR, Launer LJ. Neurology. 2003;60:652–656. doi: 10.1212/01.wnl.0000046581.81650.d0. [DOI] [PubMed] [Google Scholar]
- 49.Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, Bird TD, Hardy J, Hutton M, Kukull W, Larson E, Levy-Lahad E, Viitanen M, Peskind E, Poorkaj P, Schellenberg G, Tanzi R, Wasco W, Lannfelt L, Selkoe D, Younkin S. Nature medicine. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 50.Kurochkin IV, Goto S. FEBS letters. 1994;345:33–37. doi: 10.1016/0014-5793(94)00387-4. [DOI] [PubMed] [Google Scholar]
- 51.Iwata N, Tsubuki S, Takaki Y, Watanabe K, Sekiguchi M, Hosoki E, Kawashima-Morishima M, Lee HJ, Hama E, Sekine-Aizawa Y, Saido TC. Nature medicine. 2000;6:143–150. doi: 10.1038/72237. [DOI] [PubMed] [Google Scholar]
- 52.Mueller-Steiner S, Zhou Y, Arai H, Roberson ED, Sun B, Chen J, Wang X, Yu G, Esposito L, Mucke L, Gan L. Neuron. 2006;51:703–714. doi: 10.1016/j.neuron.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 53.Tekirian TL, Saido TC, Markesbery WR, Russell MJ, Wekstein DR, Patel E, Geddes JW. Journal of neuropathology and experimental neurology. 1998;57:76–94. doi: 10.1097/00005072-199801000-00009. [DOI] [PubMed] [Google Scholar]
- 54.Rezaei-Ghaleh N, Amininasab M, Kumar S, Walter J, Zweckstetter M. Nature communications. 2016;7:11359. doi: 10.1038/ncomms11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Al-Hilaly YK, Williams TL, Stewart-Parker M, Ford L, Skaria E, Cole M, Bucher WG, Morris KL, Sada AA, Thorpe JR, Serpell LC. Acta neuropathologica communications. 2013;1:83. doi: 10.1186/2051-5960-1-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naslund J, Schierhorn A, Hellman U, Lannfelt L, Roses AD, Tjernberg LO, Silberring J, Gandy SE, Winblad B, Greengard P, et al. Proc Natl Acad Sci USA. 1994;91:8378–8382. doi: 10.1073/pnas.91.18.8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahmed M, Davis J, Aucoin D, Sato T, Ahuja S, Aimoto S, Elliott JI, Van Nostrand WE, Smith SO. Nature structural & molecular biology. 2010;17:561–567. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kirschner DA, Inouye H, Duffy LK, Sinclair A, Lind M, Selkoe DJ. Proc Natl Acad Sci USA. 1987;84:6953–6957. doi: 10.1073/pnas.84.19.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soto C, Branes MC, Alvarez J, Inestrosa NC. Journal of neurochemistry. 1994;63:1191–1198. doi: 10.1046/j.1471-4159.1994.63041191.x. [DOI] [PubMed] [Google Scholar]
- 60.Sani MA, Gehman JD, Separovic F. FEBS letters. 2011;585:749–754. doi: 10.1016/j.febslet.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 61.Serpell LC. Biochimica et biophysica acta. 2000;1502:16–30. doi: 10.1016/s0925-4439(00)00029-6. [DOI] [PubMed] [Google Scholar]
- 62.Shirahama T, Cohen AS. The Journal of cell biology. 1967;33:679–708. doi: 10.1083/jcb.33.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tomaselli S, Esposito V, Vangone P, van Nuland NA, Bonvin AM, Guerrini R, Tancredi T, Temussi PA, Picone D. Chembiochem : a European journal of chemical biology. 2006;7:257–267. doi: 10.1002/cbic.200500223. [DOI] [PubMed] [Google Scholar]
- 64.Barrow CJ, Zagorski MG. Science. 1991;253:179–182. doi: 10.1126/science.1853202. [DOI] [PubMed] [Google Scholar]
- 65.Crescenzi O, Tomaselli S, Guerrini R, Salvadori S, D’Ursi AM, Temussi PA, Picone D. Eur J Biochem. 2002;269:5642–5648. doi: 10.1046/j.1432-1033.2002.03271.x. [DOI] [PubMed] [Google Scholar]
- 66.Vivekanandan S, Brender JR, Lee SY, Ramamoorthy A. Biochemical and biophysical research communications. 2011;411:312–316. doi: 10.1016/j.bbrc.2011.06.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coles M, Bicknell W, Watson AA, Fairlie DP, Craik DJ. Biochemistry. 1998;37:11064–11077. doi: 10.1021/bi972979f. [DOI] [PubMed] [Google Scholar]
- 68.Shao H, Jao S, Ma K, Zagorski MG. Journal of molecular biology. 1999;285:755–773. doi: 10.1006/jmbi.1998.2348. [DOI] [PubMed] [Google Scholar]
- 69.Wood SJ, Maleeff B, Hart T, Wetzel R. Journal of molecular biology. 1996;256:870–877. doi: 10.1006/jmbi.1996.0133. [DOI] [PubMed] [Google Scholar]
- 70.Narayanan S, Reif B. Biochemistry. 2005;44:1444–1452. doi: 10.1021/bi048264b. [DOI] [PubMed] [Google Scholar]
- 71.Danielsson J, Jarvet J, Damberg P, Graslund A. The FEBS journal. 2005;272:3938–3949. doi: 10.1111/j.1742-4658.2005.04812.x. [DOI] [PubMed] [Google Scholar]
- 72.Fezoui Y, Teplow DB. J Biol Chem. 2002;277:36948–36954. doi: 10.1074/jbc.M204168200. [DOI] [PubMed] [Google Scholar]
- 73.Abedini A, Raleigh DP. Physical biology. 2009;6:015005. doi: 10.1088/1478-3975/6/1/015005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tycko R. Quarterly reviews of biophysics. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 75.Kumar S, Rezaei-Ghaleh N, Terwel D, Thal DR, Richard M, Hoch M, Mc Donald JM, Wullner U, Glebov K, Heneka MT, Walsh DM, Zweckstetter M, Walter J. The EMBO journal. 2011;30:2255–2265. doi: 10.1038/emboj.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.He W, Barrow CJ. Biochemistry. 1999;38:10871–10877. doi: 10.1021/bi990563r. [DOI] [PubMed] [Google Scholar]
- 77.D’Arrigo C, Tabaton M, Perico A. Biopolymers. 2009;91:861–873. doi: 10.1002/bip.21271. [DOI] [PubMed] [Google Scholar]
- 78.Hortschansky P, Schroeckh V, Christopeit T, Zandomeneghi G, Fandrich M. Protein science : a publication of the Protein Society. 2005;14:1753–1759. doi: 10.1110/ps.041266605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meisl G, Yang X, Hellstrand E, Frohm B, Kirkegaard JB, Cohen SI, Dobson CM, Linse S, Knowles TP. Proc Natl Acad Sci USA. 2014;111:9384–9389. doi: 10.1073/pnas.1401564111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arosio P, Knowles TP, Linse S. Physical chemistry chemical physics : PCCP. 2015;17:7606–7618. doi: 10.1039/c4cp05563b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meisl G, Yang X, Frohm B, Knowles TP, Linse S. Scientific reports. 2016;6:18728. doi: 10.1038/srep18728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Adamcik J, Lara C, Usov I, Jeong JS, Ruggeri FS, Dietler G, Lashuel HA, Hamley IW, Mezzenga R. Nanoscale. 2012;4:4426–4429. doi: 10.1039/c2nr30768e. [DOI] [PubMed] [Google Scholar]
- 83.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. Proc Natl Acad Sci USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Colvin MT, Silvers R, Ni QZ, Can TV, Sergeyev I, Rosay M, Donovan KJ, Michael B, Wall J, Linse S, Griffin RG. Journal of the American Chemical Society. 2016;138:9663–9674. doi: 10.1021/jacs.6b05129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiao Y, Ma B, McElheny D, Parthasarathy S, Long F, Hoshi M, Nussinov R, Ishii Y. Nature structural & molecular biology. 2015;22:499–505. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Walsh DM, Klyubin I, Fadeeva JV, Rowan MJ, Selkoe DJ. Biochem Soc Trans. 2002;30:552–557. doi: 10.1042/bst0300552. [DOI] [PubMed] [Google Scholar]
- 88.Schrag M, Mueller C, Oyoyo U, Smith MA, Kirsch WM. Progress in neurobiology. 2011;94:296–306. doi: 10.1016/j.pneurobio.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hallgren B, Sourander P. Journal of neurochemistry. 1960;5:307–310. doi: 10.1111/j.1471-4159.1960.tb13369.x. [DOI] [PubMed] [Google Scholar]
- 90.Schrag M, Dickson A, Jiffry A, Kirsch D, Vinters HV, Kirsch W. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine. 2010;23:1123–1127. doi: 10.1007/s10534-010-9359-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bush AI, Pettingell WH, Multhaup G, Paradis Md, Vonsattel JP, Gusella JF, Beyreuther K, Masters CL, Tanzi RE. Science. 1994;265:1464–1467. doi: 10.1126/science.8073293. [DOI] [PubMed] [Google Scholar]
- 92.Markesbery WR. Archives of neurology. 1999;56:1449–1452. doi: 10.1001/archneur.56.12.1449. [DOI] [PubMed] [Google Scholar]
- 93.Bush AI. Current opinion in chemical biology. 2000;4:184–191. doi: 10.1016/s1367-5931(99)00073-3. [DOI] [PubMed] [Google Scholar]
- 94.Butterfield DA, Sultana R. Journal of amino acids. 2011;2011:198430. doi: 10.4061/2011/198430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cassagnes LE, Herve V, Nepveu F, Hureau C, Faller P, Collin F. Angew Chem Int Ed. 2013;52:11110–11113. doi: 10.1002/anie.201305372. [DOI] [PubMed] [Google Scholar]
- 96.Parthasarathy S, Yoo B, McElheny D, Tay W, Ishii Y. J Biol Chem. 2014;289:9998–10010. doi: 10.1074/jbc.M113.511345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Keskitalo S, Farkas M, Hanenberg M, Szodorai A, Kulic L, Semmler A, Weller M, Nitsch RM, Linnebank M. Frontiers in aging neuroscience. 2014;6:237. doi: 10.3389/fnagi.2014.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Smith DG, Cappai R, Barnham KJ. Biochimica et biophysica acta. 2007;1768:1976–1990. doi: 10.1016/j.bbamem.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 99.Eckert GP, Wood WG, Muller WE. Current protein & peptide science. 2010;11:319–325. doi: 10.2174/138920310791330668. [DOI] [PubMed] [Google Scholar]
- 100.Aisenbrey C, Borowik T, Bystrom R, Bokvist M, Lindstrom F, Misiak H, Sani MA, Grobner G. European biophysics journal : EBJ. 2008;37:247–255. doi: 10.1007/s00249-007-0237-0. [DOI] [PubMed] [Google Scholar]
- 101.Bystrom R, Aisenbrey C, Borowik T, Bokvist M, Lindstrom F, Sani MA, Olofsson A, Grobner G. Cell biochemistry and biophysics. 2008;52:175–189. doi: 10.1007/s12013-008-9033-4. [DOI] [PubMed] [Google Scholar]
- 102.Weber DK, Gehman JD, Separovic F, Sani MA. Aus J Chem. 2012;65:472–479. [Google Scholar]
- 103.Bhowmik D, Mote KR, MacLaughlin CM, Biswas N, Chandra B, Basu JK, Walker GC, Madhu PK, Maiti S. ACS nano. 2015;9:9070–9077. doi: 10.1021/acsnano.5b03175. [DOI] [PubMed] [Google Scholar]
- 104.Lau TL, Gehman JD, Wade JD, Masters CL, Barnham KJ, Separovic F. Biochimica et biophysica acta. 2007;1768:3135–3144. doi: 10.1016/j.bbamem.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 105.Hicks DA, Nalivaeva NN, Turner AJ. Frontiers in physiology. 2012;3:189. doi: 10.3389/fphys.2012.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Beel AJ, Mobley CK, Kim HJ, Tian F, Hadziselimovic A, Jap B, Prestegard JH, Sanders CR. Biochemistry. 2008;47:9428–9446. doi: 10.1021/bi800993c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lacor PN, Buniel MC, Chang L, Fernandez SJ, Gong Y, Viola KL, Lambert MP, Velasco PT, Bigio EH, Finch CE, Krafft GA, Klein WL. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24:10191–10200. doi: 10.1523/JNEUROSCI.3432-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Garaschuk O, Schneggenburger R, Schirra C, Tempia F, Konnerth A. The Journal of physiology. 1996;491( Pt 3):757–772. doi: 10.1113/jphysiol.1996.sp021255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang HY, Lee DH, D’Andrea MR, Peterson PA, Shank RP, Reitz AB. J Biol Chem. 2000;275:5626–5632. doi: 10.1074/jbc.275.8.5626. [DOI] [PubMed] [Google Scholar]
- 110.Snyder EM, Nong Y, Almeida CG, Paul S, Moran T, Choi EY, Nairn AC, Salter MW, Lombroso PJ, Gouras GK, Greengard P. Nature neuroscience. 2005;8:1051–1058. doi: 10.1038/nn1503. [DOI] [PubMed] [Google Scholar]
- 111.Hansson Petersen CA, Alikhani N, Behbahani H, Wiehager B, Pavlov PF, Alafuzoff I, Leinonen V, Ito A, Winblad B, Glaser E, Ankarcrona M. Proc Natl Acad Sci USA. 2008;105:13145–13150. doi: 10.1073/pnas.0806192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Manczak M, Calkins MJ, Reddy PH. Human molecular genetics. 2011;20:2495–2509. doi: 10.1093/hmg/ddr139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Du H, Guo L, Yan S, Sosunov AA, McKhann GM, Yan SS. Proc Natl Acad Sci USA. 2010;107:18670–18675. doi: 10.1073/pnas.1006586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.D’Amelio M, Cavallucci V, Middei S, Marchetti C, Pacioni S, Ferri A, Diamantini A, De Zio D, Carrara P, Battistini L, Moreno S, Bacci A, Ammassari-Teule M, Marie H, Cecconi F. Nature neuroscience. 2011;14:69–76. doi: 10.1038/nn.2709. [DOI] [PubMed] [Google Scholar]
- 115.Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, Schmitt I, Wullner U, Evert BO, O’Kane CJ, Rubinsztein DC. Human molecular genetics. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 116.Wen G, Qin W, Chen D, Wang Y, Yue X, Liu Z, Cao Y, Du J, Zhou B, Bu X. Chemical communications. 2017;53:3886–3889. doi: 10.1039/c7cc00506g. [DOI] [PubMed] [Google Scholar]
- 117.Sevigny J, Chiao P, Bussiere T, Weinreb PH, Williams L, Maier M, Dunstan R, Salloway S, Chen T, Ling Y, O’Gorman J, Qian F, Arastu M, Li M, Chollate S, Brennan MS, Quintero-Monzon O, Scannevin RH, Arnold HM, Engber T, Rhodes K, Ferrero J, Hang Y, Mikulskis A, Grimm J, Hock C, Nitsch RM, Sandrock A. Nature. 2016;537:50–56. doi: 10.1038/nature19323. [DOI] [PubMed] [Google Scholar]
- 118.Squitti R, Lupoi D, Pasqualetti P, Dal Forno G, Vernieri F, Chiovenda P, Rossi L, Cortesi M, Cassetta E, Rossini PM. Neurology. 2002;59:1153–1161. doi: 10.1212/wnl.59.8.1153. [DOI] [PubMed] [Google Scholar]
- 119.Carroll J. Science. 2017 doi: 10.1126/science.aal0759. [DOI] [Google Scholar]
- 120.Vassar R, Kuhn PH, Haass C, Kennedy ME, Rajendran L, Wong PC, Lichtenthaler SF. Journal of neurochemistry. 2014;130:4–28. doi: 10.1111/jnc.12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rajendran L, Schneider A, Schlechtingen G, Weidlich S, Ries J, Braxmeier T, Schwille P, Schulz JB, Schroeder C, Simons M, Jennings G, Knolker HJ, Simons K. Science. 2008;320:520–523. doi: 10.1126/science.1156609. [DOI] [PubMed] [Google Scholar]
- 122.Wang X, Su B, Siedlak SL, Moreira PI, Fujioka H, Wang Y, Casadesus G, Zhu X. Proc Natl Acad Sci USA. 2008;105:19318–19323. doi: 10.1073/pnas.0804871105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Noble W, Garwood C, Stephenson J, Kinsey AM, Hanger DP, Anderton BH. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2009;23:739–750. doi: 10.1096/fj.08-113795. [DOI] [PubMed] [Google Scholar]
- 124.Cummings JL, Morstorf T, Zhong K. Alzheimer’s research & therapy. 2014;6:37. doi: 10.1186/alzrt269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fu H, Cui M, Zhao L, Tu P, Zhou K, Dai J, Liu B. Journal of medicinal chemistry. 2015;58:6972–6983. doi: 10.1021/acs.jmedchem.5b00861. [DOI] [PubMed] [Google Scholar]
- 126.Bolognesi ML, Gandini A, Prati F, Uliassi E. J Med Chem. 2016;59:7759–7770. doi: 10.1021/acs.jmedchem.6b00151. [DOI] [PubMed] [Google Scholar]
- 127.Westermark P, Andersson A, Westermark GT. Physiol Rev. 2011;91:795–826. doi: 10.1152/physrev.00042.2009. [DOI] [PubMed] [Google Scholar]
- 128.Marzban L, Trigo-Gonzalez G, Zhu X, Rhodes CJ, Halban PA, Steiner DF, Verchere CB. Diabetes. 2004;53:141–148. doi: 10.2337/diabetes.53.1.141. [DOI] [PubMed] [Google Scholar]
- 129.Wang J, Xu J, Finnerty J, Furuta M, Steiner DF, Verchere CB. Diabetes. 2001;50:534–539. doi: 10.2337/diabetes.50.3.534. [DOI] [PubMed] [Google Scholar]
- 130.German MS, Moss LG, Wang J, Rutter WJ. Mol Cell Biol. 1992;12:1777–1788. doi: 10.1128/mcb.12.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lutz TA. Physiology & Behavior. 2006;89:465–471. doi: 10.1016/j.physbeh.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 132.Young A. Adv Pharmacol. 2005;52:99–121. doi: 10.1016/S1054-3589(05)52006-4. [DOI] [PubMed] [Google Scholar]
- 133.Naot D, Cornish J. Bone. 2008;43:813–818. doi: 10.1016/j.bone.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 134.Luca S, Yau W-M, Leapman R, Tycko R. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wiltzius JJW, Sievers SA, Sawaya MR, Cascio D, Popov D, Riekel C, Eisenberg D. Protein science : a publication of the Protein Society. 2008;17:1467–1474. doi: 10.1110/ps.036509.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bedrood S, Li Y, Isas JM, Hegde BG, Baxa U, Haworth IS, Langen R. J Biol Chem. 2012;287:5235–5241. doi: 10.1074/jbc.M111.327817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Patil SM, Xu S, Sheftic SR, Alexandrescu AT. J Biol Chem. 2009;284:11982–11991. doi: 10.1074/jbc.M809085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nanga RPR, Brender JR, Vivekanandan S, Ramamoorthy A. Biochimica et biophysica acta. 2011;1808:2337–2342. doi: 10.1016/j.bbamem.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Camargo DCR, Tripsianes K, Buday K, Franko A, Göbl C, Hartlmüller C, Sarkar R, Aichler M, Mettenleiter G, Schulz M, Böddrich A, Erck C, Martens H, Walch AK, Madl T, Wanker EE, Conrad M, Angelis MHd, Reif B. Scientific reports. 2017;7:44041. doi: 10.1038/srep44041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nedumpully-Govindan P, Ding F. Scientific reports. 2015;5:8240. doi: 10.1038/srep08240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sumner Makin O, Serpell LC. Journal of molecular biology. 2004;335:1279–1288. doi: 10.1016/j.jmb.2003.11.048. [DOI] [PubMed] [Google Scholar]
- 142.Goldsbury C, Kistler J, Aebi U, Arvinte T, Cooper GJS. Journal of molecular biology. 1999;285:33–39. doi: 10.1006/jmbi.1998.2299. [DOI] [PubMed] [Google Scholar]
- 143.Goldsbury CS, Cooper GJS, Goldie KN, Müller SA, Saafi EL, Gruijters WTM, Misur MP, Engel A, Aebi U, Kistler J. Journal of Structural Biology. 1997;119:17–27. doi: 10.1006/jsbi.1997.3858. [DOI] [PubMed] [Google Scholar]
- 144.Chakraborty S, Chatterjee B, Basu S. Biophys Chem. 2012;168–169:1–9. doi: 10.1016/j.bpc.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 145.Sakagashira S, Hiddinga HJ, Tateishi K, Sanke T, Hanabusa T, Nanjo K, Eberhardt NL. The American journal of pathology. 2000;157:2101–2109. doi: 10.1016/S0002-9440(10)64848-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Seino S Study Group of Comprehensive Analysis of Genetic Factors in Diabetes M. Diabetologia. 2001;44:906–909. doi: 10.1007/s001250100531. [DOI] [PubMed] [Google Scholar]
- 147.Westermark P, Eizirik DL, Pipeleers DG, Hellerström C, Andersson A. Diabetologia. 1995;38:543–549. doi: 10.1007/BF00400722. [DOI] [PubMed] [Google Scholar]
- 148.Udayasankar J, Kodama K, Hull RL, Zraika S, Aston-Mourney K, Subramanian SL, Tong J, Faulenbach MV, Vidal J, Kahn SE. Diabetologia. 2009;52:145–153. doi: 10.1007/s00125-008-1185-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Janson J, Soeller WC, Roche PC, Nelson RT, Torchia AJ, Kreutter DK, Butler PC. Proc Natl Acad Sci USA. 1996;93:7283–7288. doi: 10.1073/pnas.93.14.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yankner BA, Dawes LR, Fisher S, Villa-Komaroff L, Oster-Granite ML, Neve RL. Science. 1989;245:417–420. doi: 10.1126/science.2474201. [DOI] [PubMed] [Google Scholar]
- 151.Yankner BA, Duffy LK, Kirschner DA. Science. 1990;250:279–282. doi: 10.1126/science.2218531. [DOI] [PubMed] [Google Scholar]
- 152.Lorenzo A, Razzaboni B, Weir GC, Yankner BA. Nature. 1994;368:756–760. doi: 10.1038/368756a0. [DOI] [PubMed] [Google Scholar]
- 153.Lorenzo A, Yankner BA. Ann N Y Acad Sci. 1996;777:89–95. doi: 10.1111/j.1749-6632.1996.tb34406.x. [DOI] [PubMed] [Google Scholar]
- 154.Janson J, Ashley RH, Harrison D, McIntyre S, Butler PC. Diabetes. 1999;48:491–498. doi: 10.2337/diabetes.48.3.491. [DOI] [PubMed] [Google Scholar]
- 155.Ritzel RA, Meier JJ, Lin C-Y, Veldhuis JD, Butler PC. Diabetes. 2007;56:65–71. doi: 10.2337/db06-0734. [DOI] [PubMed] [Google Scholar]
- 156.Wong WPS, Scott DW, Chuang C-L, Zhang S, Liu H, Ferreira A, Saafi EL, Choong YS, Cooper GJS. Diabetes. 2008;57:2737–2744. doi: 10.2337/db06-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Butler AE, Janson J, Soeller WC, Butler PC. Diabetes. 2003;52:2304–2314. doi: 10.2337/diabetes.52.9.2304. [DOI] [PubMed] [Google Scholar]
- 158.Meier JJ, Kayed R, Lin C-Y, Gurlo T, Haataja L, Jayasinghe S, Langen R, Glabe CG, Butler PC. Am J Physiol Endocrinol Metab. 2006;291:E1317–1324. doi: 10.1152/ajpendo.00082.2006. [DOI] [PubMed] [Google Scholar]
- 159.Pilkington EH, Xing Y, Wang B, Kakinen A, Wang M, Davis TD, Ding F, Ke PC. Scientific reports. 2017 doi: 10.1038/s41598-017-02597-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Haataja L, Gurlo T, Huang CJ, Butler PC. Endocrine Reviews. 2008;29:303–316. doi: 10.1210/er.2007-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zraika S, Hull RL, Verchere CB, Clark A, Potter KJ, Fraser PE, Raleigh DP, Kahn SE. Diabetologia. 2010;53:1046–1056. doi: 10.1007/s00125-010-1671-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Williamson JA, Miranker AD. Protein science : a publication of the Protein Society. 2007;16:110–117. doi: 10.1110/ps.062486907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Williamson JA, Loria JP, Miranker AD. Journal of molecular biology. 2009;393:383–396. doi: 10.1016/j.jmb.2009.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wiltzius JJW, Sievers SA, Sawaya MR, Eisenberg D. Protein science : a publication of the Protein Society. 2009;18:1521–1530. doi: 10.1002/pro.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cao P, Abedini A, Raleigh DP. Curr Opin Struct Biol. 2013;23:82–89. doi: 10.1016/j.sbi.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dupuis NF, Wu C, Shea J-E, Bowers MT. Journal of the American Chemical Society. 2011;133:7240–7243. doi: 10.1021/ja1081537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Okamoto Y. Journal of Molecular Graphics and Modelling. 2004;22:425–439. doi: 10.1016/j.jmgm.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 168.Padrick SB, Miranker AD. Biochemistry. 2002;41:4694–4703. doi: 10.1021/bi0160462. [DOI] [PubMed] [Google Scholar]
- 169.Hutton JC. Diabetologia. 32:271–281. doi: 10.1007/BF00265542. [DOI] [PubMed] [Google Scholar]
- 170.DeToma AS, Salamekh S, Ramamoorthy A, Lim MH. Chem Soc Rev. 2012;41:608–621. doi: 10.1039/c1cs15112f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chimienti F, Favier A, Seve M. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine. 2005;18:313–317. doi: 10.1007/s10534-005-3687-9. [DOI] [PubMed] [Google Scholar]
- 172.Lemaire K, Ravier MA, Schraenen A, Creemers JWM, Van de Plas R, Granvik M, Van Lommel L, Waelkens E, Chimienti F, Rutter GA, Gilon P, in’t Veld PA, Schuit FC. Proc Natl Acad Sci USA. 2009;106:14872–14877. doi: 10.1073/pnas.0906587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hull RL, Westermark GT, Westermark P, Kahn SE. J Clin Endocrinol Metab. 2004;89:3629–3643. doi: 10.1210/jc.2004-0405. [DOI] [PubMed] [Google Scholar]
- 174.Jha S, Snell JM, Sheftic SR, Patil SM, Daniels SB, Kolling FW, Alexandrescu AT. Biochemistry. 2014;53:300–310. doi: 10.1021/bi401164k. [DOI] [PubMed] [Google Scholar]
- 175.Khemtémourian L, Doménech E, Doux JPF, Koorengevel MC, Killian JA. Journal of the American Chemical Society. 2011;133:15598–15604. doi: 10.1021/ja205007j. [DOI] [PubMed] [Google Scholar]
- 176.Abedini A, Raleigh DP. Biochemistry. 2005;44:16284–16291. doi: 10.1021/bi051432v. [DOI] [PubMed] [Google Scholar]
- 177.Nedumpully-Govindan P, Jemec DB, Ding F. J Chem Inf Model. 2015 doi: 10.1021/acs.jcim.5b00303. [DOI] [PubMed] [Google Scholar]
- 178.Hutton JC. Biochem J. 1982;204:171–178. doi: 10.1042/bj2040171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nedumpully-Govindan P, Yang Y, Andorfer R, Cao W, Ding F. Biochemistry. 2015;54:7335–7344. doi: 10.1021/acs.biochem.5b00891. [DOI] [PubMed] [Google Scholar]
- 180.Larson JL, Miranker AD. Journal of molecular biology. 2004;335:221–231. doi: 10.1016/j.jmb.2003.10.045. [DOI] [PubMed] [Google Scholar]
- 181.Gilead S, Wolfenson H, Gazit E. Angew Chemie Int Ed. 2006;45:6476–6480. doi: 10.1002/anie.200602034. [DOI] [PubMed] [Google Scholar]
- 182.Susa AC, Wu C, Bernstein SL, Dupuis NF, Wang H, Raleigh DP, Shea J-E, Bowers MT. Journal of the American Chemical Society. 2014;136:12912–12919. doi: 10.1021/ja504031d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Dodson G, Steiner D. Curr Opin Struct Biol. 1998;8:189–194. doi: 10.1016/s0959-440x(98)80037-7. [DOI] [PubMed] [Google Scholar]
- 184.Flannick J, Thorleifsson G, Beer NL, Jacobs SBR, Grarup N, Burtt NP, Mahajan A, Fuchsberger C, Atzmon G, Benediktsson R, Blangero J, Bowden DW, Brandslund I, Brosnan J, Burslem F, Chambers J, Cho YS, Christensen C, Douglas DA, Duggirala R, Dymek Z, Farjoun Y, Fennell T, Fontanillas P, Forsén T, Gabriel S, Glaser B, Gudbjartsson DF, Hanis C, Hansen T, Hreidarsson AB, Hveem K, Ingelsson E, Isomaa B, Johansson S, Jørgensen T, Jørgensen ME, Kathiresan S, Kong A, Kooner J, Kravic J, Laakso M, Lee J-Y, Lind L, Lindgren CM, Linneberg A, Masson G, Meitinger T, Mohlke KL, Molven A, Morris AP, Potluri S, Rauramaa R, Ribel-Madsen R, Richard A-M, Rolph T, Salomaa V, Segrè AV, Skärstrand H, Steinthorsdottir V, Stringham HM, Sulem P, Tai ES, Teo YY, Teslovich T, Thorsteinsdottir U, Trimmer JK, Tuomi T, Tuomilehto J, Vaziri-Sani F, Voight BF, Wilson JG, Boehnke M, McCarthy MI, Njølstad PR, Pedersen O, Go TDC, Groop L, Cox DR, Stefansson K, Altshuler D. Nat Genet. 2014;46:357–363. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Westermark P, Li Z-C, Westermark GT, Leckström A, Steiner DF. FEBS letters. 1996;379:203–206. doi: 10.1016/0014-5793(95)01512-4. [DOI] [PubMed] [Google Scholar]
- 186.Brender JR, Hartman K, Nanga RPR, Popovych N, de la Salud Bea R, Vivekanandan S, Marsh ENG, Ramamoorthy A. Journal of the American Chemical Society. 2010;132:8973–8983. doi: 10.1021/ja1007867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Salamekh S, Brender JR, Hyung S-J, Nanga RPR, Vivekanandan S, Ruotolo BT, Ramamoorthy A. Journal of molecular biology. 2011;410:294–306. doi: 10.1016/j.jmb.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Munte CE, Vilela L, Kalbitzer HR, Garratt RC. The FEBS journal. 2005;272:4284–4293. doi: 10.1111/j.1742-4658.2005.04843.x. [DOI] [PubMed] [Google Scholar]
- 189.Keltner Z, Meyer JA, Johnson EM, Palumbo AM, Spence DM, Reid GE. Analyst. 2010;135:278–288. doi: 10.1039/b917600d. [DOI] [PubMed] [Google Scholar]
- 190.Giebink AW, Vogel PA, Medawala W, Spence DM. Diabetes. 2013;29:44–52. doi: 10.1002/dmrr.2359. [DOI] [PubMed] [Google Scholar]
- 191.Meng F, Abedini A, Song B, Raleigh DP. Biochemistry. 2007;46:12091–12099. doi: 10.1021/bi7004834. [DOI] [PubMed] [Google Scholar]
- 192.Brender JR, Salamekh S, Ramamoorthy A. Acc Chem Res. 2012;45:454–462. doi: 10.1021/ar200189b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Khemtémourian L, Killian JA, Höppener JWM, Engel MFM. Exp Diabetes Res. 2008;2008:421287. doi: 10.1155/2008/421287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hebda JA, Miranker AD. Annu Rev Biophys. 2009;38:125–152. doi: 10.1146/annurev.biophys.050708.133622. [DOI] [PubMed] [Google Scholar]
- 195.Pilkington EH, Gurzov EN, Kakinen A, Litwak SA, Stanley WJ, Davis TP, Ke PC. Scientific reports. 2016;6:21274. doi: 10.1038/srep21274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mirzabekov TA, Lin MC, Kagan BL. J Biol Chem. 1996;271:1988–1992. doi: 10.1074/jbc.271.4.1988. [DOI] [PubMed] [Google Scholar]
- 197.Last NB, Rhoades E, Miranker AD. Proc Natl Acad Sci USA. 2011;108:9460–9465. doi: 10.1073/pnas.1102356108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Anguiano M, Nowak RJ, Lansbury PT. Biochemistry. 2002;41:11338–11343. doi: 10.1021/bi020314u. [DOI] [PubMed] [Google Scholar]
- 199.Hirakura Y, Yiu WW, Yamamoto A, Kagan BL. Amyloid. 2000;7:194–199. doi: 10.3109/13506120009146834. [DOI] [PubMed] [Google Scholar]
- 200.Quist A, Doudevski I, Lin H, Azimova R, Ng D, Frangione B, Kagan B, Ghiso J, Lal R. Proc Natl Acad Sci USA. 2005;102:10427–10432. doi: 10.1073/pnas.0502066102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Green JD, Kreplak L, Goldsbury C, Li Blatter X, Stolz M, Cooper GS, Seelig A, Kistler J, Aebi U. Journal of molecular biology. 2004;342:877–887. doi: 10.1016/j.jmb.2004.07.052. [DOI] [PubMed] [Google Scholar]
- 202.Engel MFM, Khemtémourian L, Kleijer CC, Meeldijk HJD, Jacobs J, Verkleij AJ, de Kruijff B, Killian JA, Höppener JWM. Proc Natl Acad Sci USA. 2008;105:6033–6038. doi: 10.1073/pnas.0708354105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Cao P, Abedini A, Wang H, Tu L-H, Zhang X, Schmidt AM, Raleigh DP. Proc Natl Acad Sci USA. 2013;110:19279–19284. doi: 10.1073/pnas.1305517110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Cohen FE, Kelly JW. Nature. 2003;426:905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 205.Klabunde T, Petrassi HM, Oza VB, Raman P, Kelly JW, Sacchettini JC. Nat Struct Biol. 2000;7:312–321. doi: 10.1038/74082. [DOI] [PubMed] [Google Scholar]
- 206.Wang H, Duennwald ML, Roberts BE, Rozeboom LM, Zhang YL, Steele AD, Krishnan R, Su LJ, Griffin D, Mukhopadhyay S, Hennessy EJ, Weigele P, Blanchard BJ, King J, Deniz AA, Buchwald SL, Ingram VM, Lindquist S, Shorter J. Proc Natl Acad Sci USA. 2008;105:7159–7164. doi: 10.1073/pnas.0801934105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Meng F, Abedini A, Plesner A, Middleton CT, Potter KJ, Zanni MT, Verchere CB, Raleigh DP. Journal of molecular biology. 2010;400:555–566. doi: 10.1016/j.jmb.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Abedini A, Meng F, Raleigh DP. Journal of the American Chemical Society. 2007;129:11300–11301. doi: 10.1021/ja072157y. [DOI] [PubMed] [Google Scholar]
- 209.Sellin D, Yan L-M, Kapurniotu A, Winter R. Biophys Chem. 2010;150:73–79. doi: 10.1016/j.bpc.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 210.Yan L-M, Velkova A, Tatarek-Nossol M, Andreetto E, Kapurniotu A. Angew Chemie Int Ed. 2007;46:1246–1252. doi: 10.1002/anie.200604056. [DOI] [PubMed] [Google Scholar]
- 211.Yan L-M, Tatarek-Nossol M, Velkova A, Kazantzis A, Kapurniotu A. Proc Natl Acad Sci USA. 2006;103:2046–2051. doi: 10.1073/pnas.0507471103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Saraogi I, Hebda JA, Becerril J, Estroff LA, Miranker AD, Hamilton AD. Angew Chemie Int Ed. 2010;49:736–739. doi: 10.1002/anie.200901694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Hebda JA, Saraogi I, Magzoub M, Hamilton AD, Miranker AD. Chemistry & Biology. 2009;16:943–950. doi: 10.1016/j.chembiol.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Meng F, Abedini A, Plesner A, Verchere CB, Raleigh DP. Biochemistry. 2010;49:8127–8133. doi: 10.1021/bi100939a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Young LM, Saunders JC, Mahood RA, Revill CH, Foster RJ, Tu LH, Raleigh DP, Radford SE, Ashcroft AE. Nat Chem. 2015;7:73–81. doi: 10.1038/nchem.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Noor H, Cao P, Raleigh DP. Protein science : a publication of the Protein Society. 2012;21:373–382. doi: 10.1002/pro.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zelus C, Fox A, Calciano A, Faridian BS, Nogaj LA, Moffet DA. Open Biochem J. 2012;6:66–70. doi: 10.2174/1874091X01206010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Tu L-H, Noor H, Cao P, Raleigh DP. ACS Chem Biol. 2014;9:1632–1637. doi: 10.1021/cb500162w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Daval M, Bedrood S, Gurlo T, Huang C-J, Costes S, Butler PC, Langen R. Amyloid. 2010;17:118–128. doi: 10.3109/13506129.2010.530008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sparks S, Liu G, Robbins KJ, Lazo ND. Biochemical and biophysical research communications. 2012;422:551–555. doi: 10.1016/j.bbrc.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 221.Cheng B, Gong H, Li X, Sun Y, Zhang X, Chen H, Liu X, Zheng L, Huang K. Biochemical and biophysical research communications. 2012;419:495–499. doi: 10.1016/j.bbrc.2012.02.042. [DOI] [PubMed] [Google Scholar]
- 222.Rigacci S, Guidotti V, Bucciantini M, Parri M, Nediani C, Cerbai E, Stefani M, Berti A. The Journal of Nutritional Biochemistry. 2010;21:726–735. doi: 10.1016/j.jnutbio.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 223.Mishra R, Sellin D, Radovan D, Gohlke A, Winter R. ChemBioChem. 2009;10:445–449. doi: 10.1002/cbic.200800762. [DOI] [PubMed] [Google Scholar]
- 224.Gurzov EN, Wang B, Pilkington EH, Chen P, Kakinen A, Stanley WJ, Litwak SA, Hanssen EG, Davis TP, Ding F, Ke PC. Small. 2016;12:1615–1626. doi: 10.1002/smll.201502317. [DOI] [PubMed] [Google Scholar]
- 225.Nedumpully-Govindan P, Gurzov EN, Chen P, Pilkington EH, Stanley WJ, Litwak SA, Davis TP, Ke PC, Ding F. Physical chemistry chemical physics : PCCP. 2016;18:94–100. doi: 10.1039/c5cp05924k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Cabaleiro-Lago C, Quinlan-Pluck F, Lynch I, Dawson KA, Linse S. ACS Chem Neurosci. 2010;1:279–287. doi: 10.1021/cn900027u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Middleton CT, Marek P, Cao P, Chiu C-c, Singh S, Woys AM, de Pablo JJ, Raleigh DP, Zanni MT. Nat Chem. 2012;4:355–360. doi: 10.1038/nchem.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ehrnhoefer DE, Bieschke J, Boeddrich A, Herbst M, Masino L, Lurz R, Engemann S, Pastore A, Wanker EE. Nature structural & molecular biology. 2008;15:558–566. doi: 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- 229.Ladiwala ARA, Lin JC, Bale SS, Marcelino-Cruz AM, Bhattacharya M, Dordick JS, Tessier PM. J Biol Chem. 2010;285:24228–24237. doi: 10.1074/jbc.M110.133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Mishra R, Bulic B, Sellin D, Jha S, Waldmann H, Winter R. Angew Chemie Int Ed. 2008;47:4679–4682. doi: 10.1002/anie.200705372. [DOI] [PubMed] [Google Scholar]
- 231.Yang F, Lim GP, Begum AN, Ubeda OJ, Simmons MR, Ambegaokar SS, Chen PP, Kayed R, Glabe CG, Frautschy SA, Cole GM. J Biol Chem. 2005;280:5892–5901. doi: 10.1074/jbc.M404751200. [DOI] [PubMed] [Google Scholar]
- 232.Adessi C, Soto C. Curr Med Chem. 2002;9:963–978. doi: 10.2174/0929867024606731. [DOI] [PubMed] [Google Scholar]
- 233.Santos AC, Veiga F, Ribeiro AJ. Expert Opin Drug Deliv. 2011;8:973–990. doi: 10.1517/17425247.2011.581655. [DOI] [PubMed] [Google Scholar]
- 234.Nedumpully-Govindan P, Kakinen A, Pilkington EH, Davis TP, Chun Ke P, Ding F. Scientific reports. 2016;6:19463. doi: 10.1038/srep19463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Jiang P, Li W, Shea JE, Mu Y. Biophys J. 2011;100:1550–1558. doi: 10.1016/j.bpj.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Lolicato F, Raudino A, Milardi D, La Rosa C. Eur J Med Chem. 2015;92:876–881. doi: 10.1016/j.ejmech.2015.01.047. [DOI] [PubMed] [Google Scholar]
- 237.Wang QQ, Ning LL, Niu YZ, Liu HX, Yao XJ. J Phys Chem B. 2015;119:15–24. doi: 10.1021/jp507529f. [DOI] [PubMed] [Google Scholar]
- 238.Maroteaux L, Campanelli J, Scheller R. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1988;8:2804–2815. doi: 10.1523/JNEUROSCI.08-08-02804.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, Chesselet M-F, Keshavarzian A, Shannon KM, Krajmalnik-Brown R, Wittung-Stafshede P, Knight R, Mazmanian SK. Cell. 2016;167:1469–1480.e1412. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Burré J, Sharma M, Südhof TC. Proc Natl Acad Sci USA. 2014;111:E4274–E4283. doi: 10.1073/pnas.1416598111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Vilar M, Chou H-T, Lührs T, Maji SK, Riek-Loher D, Verel R, Manning G, Stahlberg H, Riek R. Proc Natl Acad Sci USA. 2008;105:8637–8642. doi: 10.1073/pnas.0712179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Spillantini MG, Schmidt ML, Lee VMY, Trojanowski JQ, Jakes R, Goedert M. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 243.Irwin DJ, Lee VMY, Trojanowski JQ. Nat Rev Neurosci. 2013;14:626–636. doi: 10.1038/nrn3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Periquet M, Fulga T, Myllykangas L, Schlossmacher MG, Feany MB. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:3338–3346. doi: 10.1523/JNEUROSCI.0285-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Stefanis L. Cold Spring Harb Perspect Med. 2012;2:a009399. doi: 10.1101/cshperspect.a009399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Villar-Piqué A, Lopes da Fonseca T, Outeiro TF. Journal of neurochemistry. 2016;139:240–255. doi: 10.1111/jnc.13249. [DOI] [PubMed] [Google Scholar]
- 247.Vlad C, Lindner K, Karreman C, Schildknecht S, Leist M, Tomczyk N, Rontree J, Langridge J, Danzer K, Ciossek T, Petre A, Gross ML, Hengerer B, Przybylski M. ChemBioChem. 2011;12:2740–2744. doi: 10.1002/cbic.201100569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Rodriguez JA, Ivanova MI, Sawaya MR, Cascio D, Reyes FE, Shi D, Sangwan S, Guenther EL, Johnson LM, Zhang M, Jiang L, Arbing MA, Nannenga BL, Hattne J, Whitelegge J, Brewster AS, Messerschmidt M, Boutet S, Sauter NK, Gonen T, Eisenberg DS. Nature. 2015;525:486–490. doi: 10.1038/nature15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Tuttle MD, Comellas G, Nieuwkoop AJ, Covell DJ, Berthold DA, Kloepper KD, Courtney JM, Kim JK, Barclay AM, Kendall A, Wan W, Stubbs G, Schwieters CD, Lee VMY, George JM, Rienstra CM. Nature structural & molecular biology. 2016;23:409–415. doi: 10.1038/nsmb.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Comellas G, Lemkau LR, Nieuwkoop AJ, Kloepper KD, Ladror DT, Ebisu R, Woods WS, Lipton AS, George JM, Rienstra CM. Journal of molecular biology. 2011;411:881–895. doi: 10.1016/j.jmb.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Conway KA, Lee S-J, Rochet J-C, Ding TT, Williamson RE, Lansbury PT. Proc Natl Acad Sci USA. 2000;97:571–576. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Rochet J-C, Conway KA, Lansbury PT. Biochemistry. 2000;39:10619–10626. doi: 10.1021/bi001315u. [DOI] [PubMed] [Google Scholar]
- 253.Khurana R, Ionescu-Zanetti C, Pope M, Li J, Nielson L, Ramírez-Alvarado M, Regan L, Fink AL, Carter SA. Biophys J. 2003;85:1135–1144. doi: 10.1016/S0006-3495(03)74550-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Ionescu-Zanetti C, Khurana R, Gillespie JR, Petrick JS, Trabachino LC, Minert LJ, Carter SA, Fink AL. Proc Natl Acad Sci USA. 1999;96:13175–13179. doi: 10.1073/pnas.96.23.13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Harper JD, Lieber CM, Lansbury PT. Chemistry & Biology. 1997;4:951–959. doi: 10.1016/s1074-5521(97)90303-3. [DOI] [PubMed] [Google Scholar]
- 256.Stine WB, Snyder SW, Ladror US, Wade WS, Miller MF, Perun TJ, Holzman TF, Krafft GA. J Prot Chem. 1996;15:193–203. doi: 10.1007/BF01887400. [DOI] [PubMed] [Google Scholar]
- 257.Blackley HKL, Sanders GHW, Davies MC, Roberts CJ, Tendler SJB, Wilkinson MJ. Journal of molecular biology. 2000;298:833–840. doi: 10.1006/jmbi.2000.3711. [DOI] [PubMed] [Google Scholar]
- 258.Chamberlain AK, MacPhee CE, Zurdo J, Morozova-Roche LA, Hill HA, Dobson CM, Davis JJ. Biophys J. 2000;79:3282–3293. doi: 10.1016/S0006-3495(00)76560-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Kad NM, Thomson NH, Smith DP, Smith DA, Radford SE. Journal of molecular biology. 2001;313:559–571. doi: 10.1006/jmbi.2001.5071. [DOI] [PubMed] [Google Scholar]
- 260.Adamcik J, Jung J-M, Flakowski J, De Los Rios P, Dietler G, Mezzenga R. Nat Nanotech. 2010;5:423–428. doi: 10.1038/nnano.2010.59. [DOI] [PubMed] [Google Scholar]
- 261.Sweers KKM, Segers-Nolten IMJ, Bennink ML, Subramaniam V. Soft Matter. 2012;8:7215–7222. [Google Scholar]
- 262.Crowther RA, Daniel SE, Goedert M. Neuroscience letters. 2000;292:128–130. doi: 10.1016/s0304-3940(00)01440-3. [DOI] [PubMed] [Google Scholar]
- 263.Sweers KKM, Bennink ML, Subramaniam V. J Phys Condens Matter. 2012;24:243101. doi: 10.1088/0953-8984/24/24/243101. [DOI] [PubMed] [Google Scholar]
- 264.Bhak G, Lee S, Park JW, Cho S, Paik SR. Biomat. 2010;31:5986–5995. doi: 10.1016/j.biomaterials.2010.03.080. [DOI] [PubMed] [Google Scholar]
- 265.Uversky VN, Li J, Fink AL. J Biol Chem. 2001;276:10737–10744. doi: 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]
- 266.Xu L, Ma B, Nussinov R, Thompson D. ACS Chem Neurosci. 2017;8:837–849. doi: 10.1021/acschemneuro.6b00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Deleersnijder A, Gerard M, Debyser Z, Baekelandt V. Trends Mol Med. 2013;19:368–377. doi: 10.1016/j.molmed.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 268.Lashuel HA, Overk CR, Oueslati A, Masliah E. Nat Rev Neurosci. 2013;14:38–48. doi: 10.1038/nrn3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Lorenzen N, Nielsen SB, Buell AK, Kaspersen JD, Arosio P, Vad BS, Paslawski W, Christiansen G, Valnickova-Hansen Z, Andreasen M, Enghild JJ, Pedersen JS, Dobson CM, Knowles TPJ, Otzen DE. Journal of the American Chemical Society. 2014;136:3859–3868. doi: 10.1021/ja411577t. [DOI] [PubMed] [Google Scholar]
- 270.Giehm L, Svergun DI, Otzen DE, Vestergaard B. Proc Natl Acad Sci USA. 2011;108:3246–3251. doi: 10.1073/pnas.1013225108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT. Nature. 2002;418:291–291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 272.Winner B, Jappelli R, Maji SK, Desplats PA, Boyer L, Aigner S, Hetzer C, Loher T, Vilar M, Campioni S, Tzitzilonis C, Soragni A, Jessberger S, Mira H, Consiglio A, Pham E, Masliah E, Gage FH, Riek R. Proc Natl Acad Sci USA. 2011;108:4194–4199. doi: 10.1073/pnas.1100976108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 274.Walsh DM, Selkoe DJ. Journal of neurochemistry. 2007;101:1172–1184. doi: 10.1111/j.1471-4159.2006.04426.x. [DOI] [PubMed] [Google Scholar]
- 275.Luheshi LM, Tartaglia GG, Brorsson A-C, Pawar AP, Watson IE, Chiti F, Vendruscolo M, Lomas DA, Dobson CM, Crowther DC. PLoS biology. 2007;5:e290. doi: 10.1371/journal.pbio.0050290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Dobson CM. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 277.Lesné S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 278.Nilsberth C, Westlind-Danielsson A, Eckman CB, Condron MM, Axelman K, Forsell C, Stenh C, Luthman J, Teplow DB, Younkin SG, Naslund J, Lannfelt L. Nature neuroscience. 2001;4:887–893. doi: 10.1038/nn0901-887. [DOI] [PubMed] [Google Scholar]
- 279.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea J-E, Ruotolo BT, Robinson CV, Bowers MT. Nat Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Haataja L, Gurlo T, Huang CJ, Butler PC. Endocrine Rev. 2008;29:303–316. doi: 10.1210/er.2007-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Krotee P, Rodriguez JA, Sawaya MR, Cascio D, Reyes FE, Shi D, Hattne J, Nannenga BL, Oskarsson ME, Philipp S, Griner S, Jiang L, Glabe CG, Westermark GT, Gonen T, Eisenberg DS. eLife. 2017;6:e19273. doi: 10.7554/eLife.19273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Roberts H, Brown D. Biomolecules. 2015;5:282–305. doi: 10.3390/biom5020282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.van Diggelen F, Tepper AWJW, Apetri MM, Otzen DE. Isr J Chem. 2017 doi: 10.1002/ijch.201600116. [DOI] [Google Scholar]
- 284.Celej María S, Sarroukh R, Goormaghtigh E, Fidelio Gerardo D, Ruysschaert J-M, Raussens V. Biochem J. 2012;443:719–726. doi: 10.1042/BJ20111924. [DOI] [PubMed] [Google Scholar]
- 285.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 286.Bolognesi B, Kumita JR, Barros TP, Esbjorner EK, Luheshi LM, Crowther DC, Wilson MR, Dobson CM, Favrin G, Yerbury JJ. ACS Chem Biol. 2010;5:735–740. doi: 10.1021/cb1001203. [DOI] [PubMed] [Google Scholar]
- 287.Serpell LC, Berriman J, Jakes R, Goedert M, Crowther RA. Proc Natl Acad Sci USA. 2000;97:4897–4902. doi: 10.1073/pnas.97.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Glabe CG. J Biol Chem. 2008;283:29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Danzer KM, Haasen D, Karow AR, Moussaud S, Habeck M, Giese A, Kretzschmar H, Hengerer B, Kostka M. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:9220–9232. doi: 10.1523/JNEUROSCI.2617-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Karpinar DP, Balija MBG, Kügler S, Opazo F, Rezaei-Ghaleh N, Wender N, Kim H-Y, Taschenberger G, Falkenburger BH, Heise H, Kumar A, Riedel D, Fichtner L, Voigt A, Braus GH, Giller K, Becker S, Herzig A, Baldus M, Jäckle H, Eimer S, Schulz JB, Griesinger C, Zweckstetter M. The EMBO journal. 2009;28:3256–3268. doi: 10.1038/emboj.2009.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Desplats P, Lee H-J, Bae E-J, Patrick C, Rockenstein E, Crews L, Spencer B, Masliah E, Lee S-J. Proc Natl Acad Sci USA. 2009;106:13010–13015. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Relini A, Cavalleri O, Rolandi R, Gliozzi A. Chem Phys Lipids. 2009;158:1–9. doi: 10.1016/j.chemphyslip.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 293.Butterfield SM, Lashuel HA. Angew Chem. 2010;49:5628–5654. doi: 10.1002/anie.200906670. [DOI] [PubMed] [Google Scholar]
- 294.Aisenbrey C, Borowik T, Byström R, Bokvist M, Lindström F, Misiak H, Sani M-A, Gröbner G. European biophysics journal : EBJ. 2008;37:247–255. doi: 10.1007/s00249-007-0237-0. [DOI] [PubMed] [Google Scholar]
- 295.Hebda JA, Miranker AD. Annu Rev Biophys. 2009;39:125–152. doi: 10.1146/annurev.biophys.050708.133622. [DOI] [PubMed] [Google Scholar]
- 296.Galvagnion C, Buell AK, Meisl G, Michaels TCT, Vendruscolo M, Knowles TPJ, Dobson CM. Nat Chem Biol. 2015;11:229–234. doi: 10.1038/nchembio.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Kim H-Y, Cho M-K, Kumar A, Maier E, Siebenhaar C, Becker S, Fernandez CO, Lashuel HA, Benz R, Lange A, Zweckstetter M. Journal of the American Chemical Society. 2009;131:17482–17489. doi: 10.1021/ja9077599. [DOI] [PubMed] [Google Scholar]
- 298.Reynolds NP, Soragni A, Rabe M, Verdes D, Liverani E, Handschin S, Riek R, Seeger S. Journal of the American Chemical Society. 2011;133:19366–19375. doi: 10.1021/ja2029848. [DOI] [PubMed] [Google Scholar]
- 299.Fortin DL, Troyer MD, Nakamura K, Kubo S-i, Anthony MD, Edwards RH. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24:6715–6723. doi: 10.1523/JNEUROSCI.1594-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Georgieva ER, Ramlall TF, Borbat PP, Freed JH, Eliezer D. Journal of the American Chemical Society. 2008;130:12856–12857. doi: 10.1021/ja804517m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Trexler AJ, Rhoades E. Biochemistry. 2009;48:2304–2306. doi: 10.1021/bi900114z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Wong PT, Schauerte JA, Wisser KC, Ding H, Lee EL, Steel DG, Gafni A. Journal of molecular biology. 2009;386:81–96. doi: 10.1016/j.jmb.2008.11.060. [DOI] [PubMed] [Google Scholar]
- 303.Morris AM, Finke RG. Biophys Chem. 2009;140:9–15. doi: 10.1016/j.bpc.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 304.Kjaer L, Giehm L, Heimburg T, Otzen D. Biophys J. 2009;96:2857–2870. doi: 10.1016/j.bpj.2008.12.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Smith DP, Tew DJ, Hill AF, Bottomley SP, Masters CL, Barnham KJ, Cappai R. Biochemistry. 2008;47:1425–1434. doi: 10.1021/bi701522m. [DOI] [PubMed] [Google Scholar]
- 306.Pandey AP, Haque F, Rochet J-C, Hovis JS. Biophys J. 2009;96:540–551. doi: 10.1016/j.bpj.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.van Rooijen BD, Claessens MMAE, Subramaniam V. FEBS letters. 2008;582:3788–3792. doi: 10.1016/j.febslet.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 308.Hirsch EC, Brandel JP, Galle P, Javoy-Agid F, Agid Y. Journal of neurochemistry. 1991;56:446–451. doi: 10.1111/j.1471-4159.1991.tb08170.x. [DOI] [PubMed] [Google Scholar]
- 309.Pountney DL, Voelcker NH, Gai WP. Neurotox Res. 2005;7:59–67. doi: 10.1007/BF03033776. [DOI] [PubMed] [Google Scholar]
- 310.Bolisetty S, Mezzenga R. Nat Nanotech. 2016;11:365–371. doi: 10.1038/nnano.2015.310. [DOI] [PubMed] [Google Scholar]
- 311.Shen Y, Posavec L, Bolisetty S, Hilty FM, Nyström G, Kohlbrecher J, Hilbe M, Rossi A, Baumgartner J, Zimmermann MB, Mezzenga R. Nat Nanotech. 2017 doi: 10.1038/nnano.2017.58. advance online publication. [DOI] [PubMed] [Google Scholar]
- 312.Wei G, Su Z, Reynolds NP, Arosio P, Hamley IW, Gazit E, Mezzenga R. Chem Soc Rev. 2017 doi: 10.1039/C6CS00542J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Tsigelny IF, Bar-On P, Sharikov Y, Crews L, Hashimoto M, Miller MA, Keller SH, Platoshyn O, Yuan JXJ, Masliah E. The FEBS journal. 2007;274:1862–1877. doi: 10.1111/j.1742-4658.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- 314.Volles MJ, Lansbury PT. Biochemistry. 2002;41:4595–4602. doi: 10.1021/bi0121353. [DOI] [PubMed] [Google Scholar]
- 315.Bechinger B, Lohner K. Biochem Biophys Acta Biomembr. 2006;1758:1529–1539. doi: 10.1016/j.bbamem.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 316.Kiskis J, Horvath I, Wittung-Stafshede P, Rocha S. Quarterly Rev Biophys. 2017:50. doi: 10.1017/S0033583517000026. [DOI] [PubMed] [Google Scholar]
- 317.Chaudhary H, Stefanovic AND, Subramaniam V, Claessens MMAE. FEBS letters. 2014;588:4457–4463. doi: 10.1016/j.febslet.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 318.Monsellier E, Bousset L, Melki R. Scientific reports. 2016;6:19180. doi: 10.1038/srep19180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Iyer A, Schilderink N, Claessens MMAE, Subramaniam V. Biophys J. 2016;111:2440–2449. doi: 10.1016/j.bpj.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Devi L, Raghavendran V, Prabhu BM, Avadhani NG, Anandatheerthavarada HK. J Biol Chem. 2008;283:9089–9100. doi: 10.1074/jbc.M710012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Luth ES, Stavrovskaya IG, Bartels T, Kristal BS, Selkoe DJ. J Biol Chem. 2014;289:21490–21507. doi: 10.1074/jbc.M113.545749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Chen L, Jin J, Davis J, Zhou Y, Wang Y, Liu J, Lockhart PJ, Zhang J. Biochemical and biophysical research communications. 2007;356:548–553. doi: 10.1016/j.bbrc.2007.02.163. [DOI] [PubMed] [Google Scholar]
- 323.Cremades N, Cohen Samuel I, Deas E, Abramov Andrey Y, Chen Allen Y, Orte A, Sandal M, Clarke Richard W, Dunne P, Aprile Francesco A, Bertoncini Carlos W, Wood Nicholas W, Knowles Tuomas P, Dobson Christopher M, Klenerman D. Cell. 2012;149:1048–1059. doi: 10.1016/j.cell.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Junn E, Mouradian MM. Neuroscience letters. 2002;320:146–150. doi: 10.1016/s0304-3940(02)00016-2. [DOI] [PubMed] [Google Scholar]
- 325.Choi D-H, Cristóvão AC, Guhathakurta S, Lee J, Joh TH, Beal MF, Kim Y-S. Antioxid Redox Signal. 2012;16:1033–1045. doi: 10.1089/ars.2011.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Lashuel HA, Petre BM, Wall J, Simon M, Nowak RJ, Walz T, Lansbury PT., Jr Journal of molecular biology. 2002;322:1089–1102. doi: 10.1016/s0022-2836(02)00735-0. [DOI] [PubMed] [Google Scholar]
- 327.Hoozemans JJM, van Haastert ES, Nijholt DAT, Rozemuller AJM, Scheper W. Neurodegener Dis. 2012;10:212–215. doi: 10.1159/000334536. [DOI] [PubMed] [Google Scholar]
- 328.Snyder H, Mensah K, Theisler C, Lee J, Matouschek A, Wolozin B. J Biol Chem. 2003;278:11753–11759. doi: 10.1074/jbc.M208641200. [DOI] [PubMed] [Google Scholar]
- 329.Fusco G, Pape T, Stephens AD, Mahou P, Costa AR, Kaminski CF, Kaminski Schierle GS, Vendruscolo M, Veglia G, Dobson CM, De Simone A. Nat Comm. 2016;7:12563. doi: 10.1038/ncomms12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Forno LS. Journal of neuropathology and experimental neurology. 1996;55:259–272. doi: 10.1097/00005072-199603000-00001. [DOI] [PubMed] [Google Scholar]
- 331.Recasens A, Dehay B, Bové J, Carballo-Carbajal I, Dovero S, Pérez-Villalba A, Fernagut P-O, Blesa J, Parent A, Perier C, Fariñas I, Obeso JA, Bezard E, Vila M. Ann Neurol. 2014;75:351–362. doi: 10.1002/ana.24066. [DOI] [PubMed] [Google Scholar]
- 332.Iljina M, Garcia GA, Horrocks MH, Tosatto L, Choi ML, Ganzinger KA, Abramov AY, Gandhi S, Wood NW, Cremades N, Dobson CM, Knowles TPJ, Klenerman D. Proc Natl Acad Sci USA. 2016;113:E1206–E1215. doi: 10.1073/pnas.1524128113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Malkki H. Nat Rev Neurol. 2017;13:66–67. doi: 10.1038/nrneurol.2016.195. [DOI] [PubMed] [Google Scholar]
- 334.Bartels T, Choi JG, Selkoe DJ. Nature. 2011;477:107–110. doi: 10.1038/nature10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Marques O, Outeiro TF. Cell Death Dis. 2012;3:e350. doi: 10.1038/cddis.2012.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Masliah E, Rockenstein E, Adame A, Alford M, Crews L, Hashimoto M, Seubert P, Lee M, Goldstein J, Chilcote T, Games D, Schenk D. Neuron. 46:857–868. doi: 10.1016/j.neuron.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 337.Valera E, Masliah E. Pharmacol Ther. 2013;138:311–322. doi: 10.1016/j.pharmthera.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Di Giovanni S, Eleuteri S, Paleologou KE, Yin G, Zweckstetter M, Carrupt P-A, Lashuel HA. J Biol Chem. 2010;285:14941–14954. doi: 10.1074/jbc.M109.080390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Masuda M, Suzuki N, Taniguchi S, Oikawa T, Nonaka T, Iwatsubo T, Hisanaga S-i, Goedert M, Hasegawa M. Biochemistry. 2006;45:6085–6094. doi: 10.1021/bi0600749. [DOI] [PubMed] [Google Scholar]
- 340.Porat Y, Abramowitz A, Gazit E. Chem Biol Drug Des. 2006;67:27–37. doi: 10.1111/j.1747-0285.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- 341.Prusiner SB. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Huang Z, Prusiner SB, Cohen FE. Fold Des. 1996;1:13–19. doi: 10.1016/S1359-0278(96)00007-7. [DOI] [PubMed] [Google Scholar]
- 343.Campbell PN. IUBMB Life. 2005;57:273–276. doi: 10.1080/15216540500097152. [DOI] [PubMed] [Google Scholar]
- 344.Sigurdson CJ, Aguzzi A. Biochimica et biophysica acta. 2007;1772:610–618. doi: 10.1016/j.bbadis.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Westaway D, Cooper C, Turner S, Da Costa M, Carlson GA, Prusiner SB. Proc Natl Acad Sci USA. 1994;91:6418–6422. doi: 10.1073/pnas.91.14.6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Mastrianni J, Nixon F, Layzer R, DeArmond SJ, Prusiner SB. Neurology. 1997;48:A296. [Google Scholar]
- 347.Westermark P, Benson MD, Buxbaum JN, Cohen AS, Frangione B, Ikeda S-I, Masters CL, Merlini G, Saraiva MJ, Sipe JD. Amyloid. 2005;12:1–4. doi: 10.1080/13506120500032196. [DOI] [PubMed] [Google Scholar]
- 348.Chesebro B, Trifilo M, Race R, Meade-White K, Teng C, LaCasse R, Raymond L, Favara C, Baron G, Priola S, Caughey B, Masliah E, Oldstone M. Science. 2005;308:1435. doi: 10.1126/science.1110837. [DOI] [PubMed] [Google Scholar]
- 349.Aguzzi A. Science. 2005;308:1420. doi: 10.1126/science.1114168. [DOI] [PubMed] [Google Scholar]
- 350.Aguzzi A, Heikenwalder M. Nat Rev Microbiol. 2006;4:765–775. doi: 10.1038/nrmicro1492. [DOI] [PubMed] [Google Scholar]
- 351.Baskakov IV, Legname G, Baldwin MA, Prusiner SB, Cohen FE. J Biol Chem. 2002;277:21140–21148. doi: 10.1074/jbc.M111402200. [DOI] [PubMed] [Google Scholar]
- 352.Bessen RA, Marsh RF. J Virol. 1994;68:7859–7868. doi: 10.1128/jvi.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.McKinley MP, Meyer RK, Kenaga L, Rahbar F, Cotter R, Serban A, Prusiner SB. J Virol. 1991;65:1340–1351. doi: 10.1128/jvi.65.3.1340-1351.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Sunde M, Blake CC. Quarterly reviews of biophysics. 1998;31:1–39. doi: 10.1017/s0033583598003400. [DOI] [PubMed] [Google Scholar]
- 355.Leffers K-W, Wille H, Stöhr J, Junger E, Prusiner Stanley B, Riesner D. Biol Chem. 2005;386:569. doi: 10.1515/BC.2005.067. [DOI] [PubMed] [Google Scholar]
- 356.Safar JG, Kellings K, Serban A, Groth D, Cleaver JE, Prusiner SB, Riesner D. J Virol. 2005;79:10796–10806. doi: 10.1128/JVI.79.16.10796-10806.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Caughey B, Baron GS. Nature. 2006;443:803–810. doi: 10.1038/nature05294. [DOI] [PubMed] [Google Scholar]
- 358.Manuelidis L. Virulence. 2013;4:373–383. doi: 10.4161/viru.24838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Bastian FO. Arch Pathol Lab Med. 1979;103:665–669. [PubMed] [Google Scholar]
- 360.Griffith JS. Nature. 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 361.Prusiner SB. Annu Rev Microbiol. 1989;43:345–374. doi: 10.1146/annurev.mi.43.100189.002021. [DOI] [PubMed] [Google Scholar]
- 362.Soto C, Castilla J. Nature medicine. 2004;10:63–67. doi: 10.1038/nm1069. [DOI] [PubMed] [Google Scholar]
- 363.Abid K, Soto C. Cell Mol Life Sci. 2006;63:2342–2351. doi: 10.1007/s00018-006-6140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Stefani M, Dobson CM. J Mol Med. 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 365.Novitskaya V, Bocharova OV, Bronstein I, Baskakov IV. J Biol Chem. 2006;281:13828–13836. doi: 10.1074/jbc.M511174200. [DOI] [PubMed] [Google Scholar]
- 366.Chiesa R, Piccardo P, Quaglio E, Drisaldi B, Si-Hoe SL, Takao M, Ghetti B, Harris DA. J Virol. 2003;77:7611–7622. doi: 10.1128/JVI.77.13.7611-7622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Wüthrich K, Riek R. Adv Protein Chem. 2001;57:55–82. doi: 10.1016/s0065-3233(01)57018-7. [DOI] [PubMed] [Google Scholar]
- 368.Govaerts C, Wille H, Prusiner SB, Cohen FE. Proc Natl Acad Sci USA. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Sparkes RS, Simon M, Cohn VH, Fournier RE, Lem J, Klisak I, Heinzmann C, Blatt C, Lucero M, Mohandas T. Proc Natl Acad Sci USA. 1986;83:7358–7362. doi: 10.1073/pnas.83.19.7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Linden R, Martins VR, Prado MAM, Cammarota M, Izquierdo I, Brentani RR. Physiol Rev. 2008;88:673–728. doi: 10.1152/physrev.00007.2007. [DOI] [PubMed] [Google Scholar]
- 371.Llorens F, del Río JA. Prion. 2012;6:245–251. doi: 10.4161/pri.19639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Aguzzi A, Heikenwalder M, Polymenidou M. Nat Rev Mol Cell Biol. 2007:8. doi: 10.1038/nrm2204. [DOI] [PubMed] [Google Scholar]
- 373.Kanaani J, Prusiner SB, Diacovo J, Baekkeskov S, Legname G. Journal of neurochemistry. 2005;95:1373–1386. doi: 10.1111/j.1471-4159.2005.03469.x. [DOI] [PubMed] [Google Scholar]
- 374.Brown DR, Qin K, Herms JW, Madlung A, Manson J, Strome R, Fraser PE, Kruck T, von Bohlen A, Schulz-Schaeffer W, Giese A, Westaway D, Kretzschmar H. Nature. 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 375.Prčina M, Kontseková E, Novák M. Acta Virol. 2015;59:179–184. doi: 10.4149/av_2015_02_179. [DOI] [PubMed] [Google Scholar]
- 376.Mehrpour M, Codogno P. Cancer Letters. 2010;290:1–23. doi: 10.1016/j.canlet.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 377.Du J, Pan Y, Shi Y, Guo C, Jin X, Sun L, Liu N, Qiao T, Fan D. Int J Cancer. 2005;113:213–220. doi: 10.1002/ijc.20570. [DOI] [PubMed] [Google Scholar]
- 378.Li Q-Q, Sun Y-P, Ruan C-P, Xu X-Y, Ge J-H, He J, Xu Z-D, Wang Q, Gao W-C. Cancer Sci. 2011;102:400–406. doi: 10.1111/j.1349-7006.2010.01811.x. [DOI] [PubMed] [Google Scholar]
- 379.de Wit M, Jimenez CR, Carvalho B, Belien JAM, Delis-van Diemen PM, Mongera S, Piersma SR, Vikas M, Navani S, Pontén F, Meijer GA, Fijneman RJA. Gut. 2012;61:855–864. doi: 10.1136/gutjnl-2011-300511. [DOI] [PubMed] [Google Scholar]
- 380.Büeler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 381.Steele AD, Lindquist S, Aguzzi A. Prion. 2007;1:83–93. doi: 10.4161/pri.1.2.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Criado JR, Sánchez-Alavez M, Conti B, Giacchino JL, Wills DN, Henriksen SJ, Race R, Manson JC, Chesebro B, Oldstone MBA. Neurobiology of disease. 2005;19:255–265. doi: 10.1016/j.nbd.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 383.Le Pichon CE, Valley MT, Polymenidou M, Chesler AT, Sagdullaev BT, Aguzzi A, Firestein S. Nature neuroscience. 2009;12:60–69. doi: 10.1038/nn.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wuthrich K. Nature. 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 385.Zahn R, Liu A, Lührs T, Riek R, von Schroetter C, López García F, Billeter M, Calzolai L, Wider G, Wüthrich K. Proc Natl Acad Sci USA. 2000;97:145–150. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Zahn R, Güntert P, von Schroetter C, Wüthrich K. Journal of molecular biology. 2003;326:225–234. doi: 10.1016/s0022-2836(02)01332-3. [DOI] [PubMed] [Google Scholar]
- 387.Calzolai L, Zahn R. J Biol Chem. 2003;278:35592–35596. doi: 10.1074/jbc.M303005200. [DOI] [PubMed] [Google Scholar]
- 388.Calzolai L, Lysek DA, Güntert P, von Schroetter C, Riek R, Zahn R, Wüthrich K. Proc Natl Acad Sci USA. 2000;97:8340–8345. doi: 10.1073/pnas.97.15.8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Pastore A, Zagari A. Prion. 2007;1:185–197. doi: 10.4161/pri.1.3.5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Knaus KJ, Morillas M, Swietnicki W, Malone M, Surewicz WK, Yee VC. Nature structural & molecular biology. 2001;8:770–774. doi: 10.1038/nsb0901-770. [DOI] [PubMed] [Google Scholar]
- 391.Wopfner F, Weidenhöfer G, Schneider R, von Brunn A, Gilch S, Schwarz TF, Werner T, Schätzl HM. Journal of molecular biology. 1999;289:1163–1178. doi: 10.1006/jmbi.1999.2831. [DOI] [PubMed] [Google Scholar]
- 392.Rivera-Milla E, Oidtmann B, Panagiotidis CH, Baier M, Sklaviadis T, Hoffmann R, Zhou Y, Solis GP, Stuermer CAO, Málaga-Trillo E. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2005 doi: 10.1096/fj.05-4279fje. [DOI] [PubMed] [Google Scholar]
- 393.Millhauser GL. Annu Rev Phys Chem. 2007;58:299–320. doi: 10.1146/annurev.physchem.58.032806.104657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Donne DG, Viles JH, Groth D, Mehlhorn I, James TL, Cohen FE, Prusiner SB, Wright PE, Dyson HJ. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Safar J, Roller PP, Gajdusek DC, Gibbs CJ. J Biol Chem. 1993;268:20276–20284. [PubMed] [Google Scholar]
- 397.McKinley MP, Prusiner SB. Int Rev Neurobiol. 1986;28:1–57. doi: 10.1016/s0074-7742(08)60105-1. [DOI] [PubMed] [Google Scholar]
- 398.DeMarco ML, Daggett V. Proc Natl Acad Sci USA. 2004;101:2293–2298. doi: 10.1073/pnas.0307178101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Cobb NJ, Sönnichsen FD, McHaourab H, Surewicz WK. Proc Natl Acad Sci USA. 2007;104:18946–18951. doi: 10.1073/pnas.0706522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Groveman BR, Dolan MA, Taubner LM, Kraus A, Wickner RB, Caughey B. J Biol Chem. 2014;289:24129–24142. doi: 10.1074/jbc.M114.578344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, Madsen AO, Riekel C, Eisenberg D. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 402.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 403.Wille H, Bian W, McDonald M, Kendall A, Colby DW, Bloch L, Ollesch J, Borovinskiy AL, Cohen FE, Prusiner SB, Stubbs G. Proc Natl Acad Sci USA. 2009;106:16990–16995. doi: 10.1073/pnas.0909006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Dickinson AG, Meikle VMH, Fraser H. J Comp Pathol. 1968;78:293–299. doi: 10.1016/0021-9975(68)90005-4. [DOI] [PubMed] [Google Scholar]
- 405.Bruce ME, Dickinson AG. J Gen Virol. 1987;68:79–89. doi: 10.1099/0022-1317-68-1-79. [DOI] [PubMed] [Google Scholar]
- 406.Bessen RA, Marsh RF. J Gen Virol. 1992;73:329–334. doi: 10.1099/0022-1317-73-2-329. [DOI] [PubMed] [Google Scholar]
- 407.Monari L, Chen SG, Brown P, Parchi P, Petersen RB, Mikol J, Gray F, Cortelli P, Montagna P, Ghetti B. Proc Natl Acad Sci USA. 1994;91:2839–2842. doi: 10.1073/pnas.91.7.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Collinge J, Clarke AR. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 409.Scott MR, Groth D, Tatzelt J, Torchia M, Tremblay P, DeArmond SJ, Prusiner SB. J Virol. 1997;71:9032–9044. doi: 10.1128/jvi.71.12.9032-9044.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Kimberlin RH, Walker CA, Fraser H. J Gen Virol. 1989;70:2017–2025. doi: 10.1099/0022-1317-70-8-2017. [DOI] [PubMed] [Google Scholar]
- 411.Fraser H, Dickinson AG. J Comp Pathol. 1973;83:29–40. doi: 10.1016/0021-9975(73)90024-8. [DOI] [PubMed] [Google Scholar]
- 412.Merz PA, Somerville RA, Wisniewski HM, Iqbal K. Acta neuropathologica. 1981;54:63–74. doi: 10.1007/BF00691333. [DOI] [PubMed] [Google Scholar]
- 413.Prusiner SB, McKinley MP, Bowman KA, Bolton DC, Bendheim PE, Groth DF, Glenner GG. Cell. 1983;35:349–358. doi: 10.1016/0092-8674(83)90168-x. [DOI] [PubMed] [Google Scholar]
- 414.Sim VL, Caughey B. Neurobiol Aging. 2009;30:2031–2042. doi: 10.1016/j.neurobiolaging.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 415.Kascsak RJ, Rubenstein R, Merz PA, Carp RI, Robakis NK, Wisniewski HM, Diringer H. J Virol. 1986;59:676–683. doi: 10.1128/jvi.59.3.676-683.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Neuendorf E, Weber A, Saalmueller A, Schatzl H, Reifenberg K, Pfaff E, Groschup MH. J Biol Chem. 2004;279:53306–53316. doi: 10.1074/jbc.M410796200. [DOI] [PubMed] [Google Scholar]
- 417.Zanusso G, Polo A, Farinazzo A, et al. Consortium Td-G. Archives of neurology. 2007;64:595–599. doi: 10.1001/archneur.64.4.595. [DOI] [PubMed] [Google Scholar]
- 418.Lewis Patrick A, Properzi F, Prodromidou K, Clarke Anthony R, Collinge J, Jackson Graham S. Biochem J. 2006;395:443–448. doi: 10.1042/BJ20051677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Stack MJ, Aldrich AM, Kitching AD, Scott AC. Res Vet Sci. 1995;59:247–254. doi: 10.1016/0034-5288(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 420.Hsiao K, Prusiner SB. Neurology. 1990;40:1820–1820. doi: 10.1212/wnl.40.12.1820. [DOI] [PubMed] [Google Scholar]
- 421.Prusiner SB. Archives of neurology. 1993;50:1129–1153. doi: 10.1001/archneur.1993.00540110011002. [DOI] [PubMed] [Google Scholar]
- 422.Gajdusek D. Eur J Epidemiol. 1991;7:567–577. doi: 10.1007/BF00143141. [DOI] [PubMed] [Google Scholar]
- 423.Collinge J, Whitfield J, McKintosh E, Beck J, Mead S, Thomas DJ, Alpers MP. The Lancet. 2006;367:2068–2074. doi: 10.1016/S0140-6736(06)68930-7. [DOI] [PubMed] [Google Scholar]
- 424.McCutcheon S, Blanco ARA, Houston EF, de Wolf C, Tan BC, Smith A, Groschup MH, Hunter N, Hornsey VS, MacGregor IR. PloS One. 2011;6:e23169. doi: 10.1371/journal.pone.0023169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Andréoletti O, Litaise C, Simmons H, Corbière F, Lugan S, Costes P, Schelcher F, Vilette D, Grassi J, Lacroux C. PLoS Pathog. 2012;8:e1002782. doi: 10.1371/journal.ppat.1002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Bonda DJ, Manjila S, Mehndiratta P, Khan F, Miller BR, Onwuzulike K, Puoti G, Cohen ML, Schonberger LB, Cali I. Neurosurg Focus. 2016;41:E10. doi: 10.3171/2016.5.FOCUS15126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Mathiason CK, Powers JG, Dahmes SJ, Osborn DA, Miller KV, Warren RJ, Mason GL, Hays SA, Hayes-Klug J, Seelig DM. Science. 2006;314:133–136. doi: 10.1126/science.1132661. [DOI] [PubMed] [Google Scholar]
- 428.Mathiason CK, Hays SA, Powers J, Hayes-Klug J, Langenberg J, Dahmes SJ, Osborn DA, Miller KV, Warren RJ, Mason GL. PLoS One. 2009;4:e5916. doi: 10.1371/journal.pone.0005916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Haley NJ, Seelig DM, Zabel MD, Telling GC, Hoover EA. PLoS One. 2009;4:e4848. doi: 10.1371/journal.pone.0004848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.DeJoia C, Moreaux B, O’Connell K, Bessen RA. J Virol. 2006;80:4546–4556. doi: 10.1128/JVI.80.9.4546-4556.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Konold T, Moore SJ, Bellworthy SJ, Simmons HA. BMC Vet Res. 2008;4:14. doi: 10.1186/1746-6148-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Ligios C, Sigurdson CJ, Santucciu C, Carcassola G, Manco G, Basagni M, Maestrale C, Cancedda MG, Madau L, Aguzzi A. Nature medicine. 2005;11:1137–1139. doi: 10.1038/nm1105-1137. [DOI] [PubMed] [Google Scholar]
- 433.Lacroux C, Simon S, Benestad SL, Maillet S, Mathey J, Lugan S, Corbière F, Cassard H, Costes P, Bergonier D. PLoS Pathog. 2008;4:e1000238. doi: 10.1371/journal.ppat.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Johnson CJ, McKenzie D, Pedersen JA, Aiken JM. J Toxicol Environ Health Part A. 2011;74:161–166. doi: 10.1080/15287394.2011.529066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Bruederle CE, Hnasko RM, Kraemer T, Garcia RA, Haas MJ, Marmer WN, Carter JM. PloS One. 2008;3:e2969. doi: 10.1371/journal.pone.0002969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Miller MW. Emerg Infect Diseases. 2004;10:1003–1006. doi: 10.3201/eid1006.040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Georgsson G, Sigurdarson S, Brown P. J Gen Virol. 2006;87:3737–3740. doi: 10.1099/vir.0.82011-0. [DOI] [PubMed] [Google Scholar]
- 438.Saunders SE, Bartz JC, Bartelt-Hunt SL. Environ Sci Technol. 2009;43:5242. doi: 10.1021/es900502f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Tsiroulnikov K, Rezai H, Bonch-Osmolovskaya E, Nedkov P, Gousterova A, Cueff V, Godfroy A, Barbier G, Métro F, Chobert J-M. J Agric Food Chem. 2004;52:6353–6360. doi: 10.1021/jf0493324. [DOI] [PubMed] [Google Scholar]
- 440.Rasmussen J, Gilroyed BH, Reuter T, Dudas S, Neumann NF, Balachandran A, Kav NN, Graham C, Czub S, McAllister TA. Prion. 2014;8:136–142. doi: 10.4161/pri.27963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Pritzkow S, Morales R, Moda F, Khan U, Telling GC, Hoover E, Soto C. Cell Rep. 2015;11:1168–1175. doi: 10.1016/j.celrep.2015.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Chernoff YO. Proc Natl Acad Sci USA. 2016;113:6097–6099. doi: 10.1073/pnas.1605671113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Brandner S, Klein MA, Frigg R, Pekarik V, Parizek P, Raeber A, Glatzel M, Schwarz P, Rülicke T, Weissmann C. Exp Physiol. 2000;85:705–712. doi: 10.1111/j.1469-445x.2000.02091.x. [DOI] [PubMed] [Google Scholar]
- 444.Aguzzi A, Polymenidou M. Cell. 2004;116:313–327. doi: 10.1016/s0092-8674(03)01031-6. [DOI] [PubMed] [Google Scholar]
- 445.Aguzzi A, Calella AM. Physiol Rev. 2009;89:1105–1152. doi: 10.1152/physrev.00006.2009. [DOI] [PubMed] [Google Scholar]
- 446.Soto C, Estrada L, Castilla J. Trends Biochem Sci. 2006;31:150–155. doi: 10.1016/j.tibs.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 447.Makarava N, Ostapchenko VG, Savtchenko R, Baskakov IV. J Biol Chem. 2009;284:14386–14395. doi: 10.1074/jbc.M900533200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Li L, Guest W, Huang A, Plotkin SS, Cashman NR. Protein Eng Des Sel. 2009;22:523–529. doi: 10.1093/protein/gzp038. [DOI] [PubMed] [Google Scholar]
- 449.Legname G. Prion. 2012;6:37–39. doi: 10.4161/pri.6.1.18425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Paramithiotis E, Pinard M, Lawton T, LaBoissiere S, Leathers VL, Zou W-Q, Estey LA, Lamontagne J, Lehto MT, Kondejewski LH. Nature medicine. 2003;9:893–899. doi: 10.1038/nm883. [DOI] [PubMed] [Google Scholar]
- 451.Kaneko K, Zulianello L, Scott M, Cooper CM, Wallace AC, James TL, Cohen FE, Prusiner SB. Proc Natl Acad Sci USA. 1997;94:10069–10074. doi: 10.1073/pnas.94.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Geoghegan JC, Valdes PA, Orem NR, Deleault NR, Williamson RA, Harris BT, Supattapone S. J Biol Chem. 2007;282:36341–36353. doi: 10.1074/jbc.M704447200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Baskakov IV, Breydo L. BBA Mol Basis Dis. 2007;1772:692–703. doi: 10.1016/j.bbadis.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 454.Caughey B. Br Med Bull. 2003;66:109–120. doi: 10.1093/bmb/66.1.109. [DOI] [PubMed] [Google Scholar]
- 455.Bishop M, Hart P, Aitchison L, Baybutt H, Plinston C, Thomson V, Tuzi N, Head M, Ironside J, Will R. Lancet Neurol. 2006;5:393–398. doi: 10.1016/S1474-4422(06)70413-6. [DOI] [PubMed] [Google Scholar]
- 456.Apostol MI, Wiltzius JJ, Sawaya MR, Cascio D, Eisenberg D. Biochemistry. 2011;50:2456. doi: 10.1021/bi101803k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Kurt TD, Bett C, Fernández-Borges N, Joshi-Barr S, Hornemann S, Rülicke T, Castilla J, Wüthrich K, Aguzzi A, Sigurdson CJ. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:1022–1027. doi: 10.1523/JNEUROSCI.4636-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Sigurdson CJ, Nilsson KPR, Hornemann S, Manco G, Fernández-Borges N, Schwarz P, Castilla J, Wüthrich K, Aguzzi A. J Clin Invest. 2010;120:2590–2599. doi: 10.1172/JCI42051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Hill AF, Collinge J. Trends Microbiol. 2003;11:578–584. doi: 10.1016/j.tim.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 460.Thuring C, Erkens J, Jacobs J, Bossers A, Van Keulen L, Garssen G, Van Zijderveld F, Ryder S, Groschup M, Sweeney T. J Clin Microbiol. 2004;42:972–980. doi: 10.1128/JCM.42.3.972-980.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Jones EM, Surewicz WK. Cell. 2005;121:63–72. doi: 10.1016/j.cell.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 462.Jones M. Emerg Infect Diseases. 2009 [Google Scholar]
- 463.Liu H, Farr-Jones S, Ulyanov NB, Llinas M, Marqusee S, Groth D, Cohen FE, Prusiner SB, James TL. Biochemistry. 1999;38:5362–5377. doi: 10.1021/bi982878x. [DOI] [PubMed] [Google Scholar]
- 464.Makarava N, Kovacs GG, Bocharova O, Savtchenko R, Alexeeva I, Budka H, Rohwer RG, Baskakov IV. Acta Neurol. 2010;119:177–187. doi: 10.1007/s00401-009-0633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Béringue V, Herzog L, Jaumain E, Reine F, Sibille P, Le Dur A, Vilotte J-L, Laude H. Science. 2012;335:472–475. doi: 10.1126/science.1215659. [DOI] [PubMed] [Google Scholar]
- 466.Padilla D, Béringue V, Espinosa JC, Andreoletti O, Jaumain E, Reine F, Herzog L, Gutierrez-Adan A, Pintado B, Laude H. PLoS Pathog. 2011;7:e1001319. doi: 10.1371/journal.ppat.1001319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Castilla J, Gonzalez-Romero D, Saá P, Morales R, De Castro J, Soto C. Cell. 2008;134:757–768. doi: 10.1016/j.cell.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Asante EA, Linehan JM, Desbruslais M, Joiner S, Gowland I, Wood AL, Welch J, Hill AF, Lloyd SE, Wadsworth JD. The EMBO journal. 2002;21:6358–6366. doi: 10.1093/emboj/cdf653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Wadsworth JD, Asante EA, Desbruslais M, Linehan JM, Joiner S, Gowland I, Welch J, Stone L, Lloyd SE, Hill AF. Science. 2004;306:1793–1796. doi: 10.1126/science.1103932. [DOI] [PubMed] [Google Scholar]
- 470.Kimberlin RH, Cole S, Walker CA. J Gen Virol. 1987;68:1875–1881. doi: 10.1099/0022-1317-68-7-1875. [DOI] [PubMed] [Google Scholar]
- 471.Colby DW, Giles K, Legname G, Wille H, Baskakov IV, DeArmond SJ, Prusiner SB. Proc Natl Acad Sci USA. 2009;106:20417–20422. doi: 10.1073/pnas.0910350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Bruce ME. Br Med Bull. 2003;66:99–108. doi: 10.1093/bmb/66.1.99. [DOI] [PubMed] [Google Scholar]
- 473.Barria MA, Telling GC, Gambetti P, Mastrianni JA, Soto C. J Biol Chem. 2011;286:7490–7495. doi: 10.1074/jbc.M110.198465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C. Science. 2010;327:869–872. doi: 10.1126/science.1183218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Caughey B, Baron GS, Chesebro B, Jeffrey M. Annu Rev Biochem. 2009;78:177–204. doi: 10.1146/annurev.biochem.78.082907.145410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Aguzzi A, Falsig J. Nature neuroscience. 2012;15:936–939. doi: 10.1038/nn.3120. [DOI] [PubMed] [Google Scholar]
- 477.Mallucci G, Dickinson A, Linehan J, Klöhn P-C, Brandner S, Collinge J. Science. 2003;302:871–874. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- 478.Bahadi R, Farrelly PV, Kenna BL, Kourie JI, Tagliavini F, Forloni G, Salmona M. Am J Physiol-Cell Ph. 2003;285:C862–C872. doi: 10.1152/ajpcell.00077.2003. [DOI] [PubMed] [Google Scholar]
- 479.Sanghera N, Pinheiro TJ. Journal of molecular biology. 2002;315:1241–1256. doi: 10.1006/jmbi.2001.5322. [DOI] [PubMed] [Google Scholar]
- 480.Simoneau S, Rezaei H, Salès N, Kaiser-Schulz G, Lefebvre-Roque M, Vidal C, Fournier J-G, Comte J, Wopfner F, Grosclaude J. PLoS Pathog. 2007;3:e125. doi: 10.1371/journal.ppat.0030125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Resenberger UK, Harmeier A, Woerner AC, Goodman JL, Müller V, Krishnan R, Vabulas RM, Kretzschmar HA, Lindquist S, Hartl FU. The EMBO journal. 2011;30:2057–2070. doi: 10.1038/emboj.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Um JW, Nygaard HB, Heiss JK, Kostylev MA, Stagi M, Vortmeyer A, Wisniewski T, Gunther EC, Strittmatter SM. Nature neuroscience. 2012;15:1227–1235. doi: 10.1038/nn.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Linden R, Martins VR, Prado MA, Cammarota M, Izquierdo I, Brentani RR. Physiol Rev. 2008;88:673–728. doi: 10.1152/physrev.00007.2007. [DOI] [PubMed] [Google Scholar]
- 484.Kristiansen M, Deriziotis P, Dimcheff DE, Jackson GS, Ovaa H, Naumann H, Clarke AR, van Leeuwen FW, Menéndez-Benito V, Dantuma NP. Mol Cell. 2007;26:175–188. doi: 10.1016/j.molcel.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 485.Westergard L, Christensen HM, Harris DA. BBA Mol Basis Dis. 2007;1772:629–644. doi: 10.1016/j.bbadis.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Jeffrey M, McGovern G, Goodsir CM, González L. Brain Pathol. 2009;19:1–11. doi: 10.1111/j.1750-3639.2008.00150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Caughey B. Trends Biochem Sci. 2001;26:235–242. doi: 10.1016/s0968-0004(01)01792-3. [DOI] [PubMed] [Google Scholar]
- 488.Zhang H-J, Lu Y-H, Long Y-J, Wang Q-L, Huang X-X, Zhu R, Wang X-L, Liang L-P, Teng P, Zheng H-Z. Anal Method. 2014;6:2982–2987. [Google Scholar]
- 489.Zhan L, Peng L, Huang C-Z. Chin Sci Bull. 2014;59:964–970. [Google Scholar]
- 490.Sobrova P, Blazkova I, Chomoucka J, Drbohlavova J, Vaculovicova M, Kopel P, Hubalek J, Kizek R, Adam V. Prion. 2013;7:349–358. doi: 10.4161/pri.26524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Ottaviani MF, Cangiotti M, Fiorani L, Fattori A, Wasiak T, Appelhans D, Klajnert B. Curr Med Chem. 2012;19:5907–5921. doi: 10.2174/092986712804143259. [DOI] [PubMed] [Google Scholar]
- 492.Kimura T, Hosokawa-Muto J, Asami K, Murai T, Kuwata K. Eur J Med Chem. 2011;46:5675–5679. doi: 10.1016/j.ejmech.2011.08.039. [DOI] [PubMed] [Google Scholar]
- 493.Gallardo-Godoy A, Gever J, Fife KL, Silber BM, Prusiner SB, Renslo AR. J Med Chem. 2011;54:1010. doi: 10.1021/jm101250y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Bongarzone S, Tran HNA, Cavalli A, Roberti M, Rosini M, Carloni P, Legname G, Bolognesi ML. ChemMedChem. 2011;6:601–605. doi: 10.1002/cmdc.201100072. [DOI] [PubMed] [Google Scholar]
- 495.Alvarez-Puebla RA, Agarwal A, Manna P, Khanal BP, Aldeanueva-Potel P, Carbó-Argibay E, Pazos-Pérez N, Vigderman L, Zubarev ER, Kotov NA. Proc Natl Acad Sci USA. 2011;108:8157–8161. doi: 10.1073/pnas.1016530108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Xiao SJ, Hu PP, Wu XD, Zou YL, Chen LQ, Peng L, Ling J, Zhen SJ, Zhan L, Li YF. Anal Chem. 2010;82:9736–9742. doi: 10.1021/ac101865s. [DOI] [PubMed] [Google Scholar]
- 497.Tran HNA, Sousa F, Moda F, Mandal S, Chanana M, Vimercati C, Morbin M, Krol S, Tagliavini F, Legname G. Nanoscale. 2010;2:2724–2732. doi: 10.1039/c0nr00551g. [DOI] [PubMed] [Google Scholar]
- 498.Henry J, Anand A, Chowdhury M, Coté G, Moreira R, Good T. Anal Biochem. 2004;334:1–8. doi: 10.1016/j.ab.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 499.Supattapone S, Wille H, Uyechi L, Safar J, Tremblay P, Szoka FC, Cohen FE, Prusiner SB, Scott MR. J Virol. 2001;75:3453–3461. doi: 10.1128/JVI.75.7.3453-3461.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Rangel A, Madroñal N, Gavín R, Llorens F, Sumoy L, Torres JM, Delgado-García JM, Del Río JA. PloS One. 2009;4:e7592. doi: 10.1371/journal.pone.0007592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Khosravani H, Zhang Y, Tsutsui S, Hameed S, Altier C, Hamid J, Chen L, Villemaire M, Ali Z, Jirik FR. The Journal of cell biology. 2008;181:551–565. doi: 10.1083/jcb.200711002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Weiss S, Proske D, Neumann M, Groschup MH, Kretzschmar HA, Famulok M, Winnacker E-L. J Virol. 1997;71:8790–8797. doi: 10.1128/jvi.71.11.8790-8797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Boese AS, Majer A, Saba R, Booth SA. Exp Op Drug Disc. 2013;8:1265–1284. doi: 10.1517/17460441.2013.818976. [DOI] [PubMed] [Google Scholar]
- 504.Calvo P, Gouritin B, Brigger I, Lasmezas C, Deslys J-P, Williams A, Andreux JP, Dormont D, Couvreur P. J Neurosc Method. 2001;111:151–155. doi: 10.1016/s0165-0270(01)00450-2. [DOI] [PubMed] [Google Scholar]
- 505.Kouassi GK, Wang P, Sreevatan S, Irudayaraj J. Biotechnol Progr. 2007;23:1239–1244. doi: 10.1021/bp0602101. [DOI] [PubMed] [Google Scholar]
- 506.Miller MB, Supattapone S. J Virol. 2011;85:2813–2817. doi: 10.1128/JVI.02451-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Mangé A, Milhavet O, McMahon HEM, Casanova D, Lehmann S. Journal of neurochemistry. 2000;74:754–762. doi: 10.1046/j.1471-4159.2000.740754.x. [DOI] [PubMed] [Google Scholar]
- 508.Mangé A, Nishida N, Milhavet O, McMahon HEM, Casanova D, Lehmann S. J Virol. 2000;74:3135–3140. doi: 10.1128/jvi.74.7.3135-3140.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Mangé A, Nishida N, Milhavet O, McMahon HE, Casanova D, Lehmann S. J Virol. 2000;74:3135–3140. doi: 10.1128/jvi.74.7.3135-3140.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Karapetyan YE, Sferrazza GF, Zhou M, Ottenberg G, Spicer T, Chase P, Fallahi M, Hodder P, Weissmann C, Lasmézas CI. Proc Natl Acad Sci USA. 2013;110:7044–7049. doi: 10.1073/pnas.1303510110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Ghaemmaghami S, May BC, Renslo AR, Prusiner SB. J Virol. 2010;84:3408–3412. doi: 10.1128/JVI.02145-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Oskarsson ME, Paulsson JF, Schultz SW, Ingelsson M, Westermark P, Westermark GT. The American journal of pathology. 2015;185:834–846. doi: 10.1016/j.ajpath.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 513.Ono K, Takahashi R, Ikeda T, Yamada M. Journal of neurochemistry. 2012;122:883–890. doi: 10.1111/j.1471-4159.2012.07847.x. [DOI] [PubMed] [Google Scholar]
- 514.Mougenot A-LJ, Bencsik A, Nicot S, Vulin J, Morignat E, Verchère J, Bétemps D, Lakhdar L, Legastelois S, Baron TG. J Neuropath Exp Neur. 2011;70:377–385. doi: 10.1097/NEN.0b013e318217d95f. [DOI] [PubMed] [Google Scholar]
- 515.Olanow CW, Prusiner SB. Proc Natl Acad Sci USA. 2009;106:12571–12572. doi: 10.1073/pnas.0906759106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Beekes M, Thomzig A, Schulz-Schaeffer WJ, Burger R. Acta Neurol. 2014;128:463–476. doi: 10.1007/s00401-014-1324-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Tayeb-Fligelman E, Tabachnikov O, Moshe A, Goldshmidt-Tran O, Sawaya MR, Coquelle N, Colletier J-P, Landau M. Science. 2017;355:831–833. doi: 10.1126/science.aaf4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Rubin N, Perugia E, Goldschmidt M, Fridkin M, Addadi L. Journal of the American Chemical Society. 2008;130:4602–4603. doi: 10.1021/ja800328y. [DOI] [PubMed] [Google Scholar]
- 519.Rubin N, Perugia E, Wolf SG, Klein E, Fridkin M, Addadi L. Journal of the American Chemical Society. 2010;132:4242–4248. doi: 10.1021/ja909345p. [DOI] [PubMed] [Google Scholar]
- 520.Usov I, Adamcik J, Mezzenga R. ACS nano. 2013;7:10465–10474. doi: 10.1021/nn404886k. [DOI] [PubMed] [Google Scholar]
- 521.Lee S-J, Desplats P, Sigurdson C, Tsigelny I, Masliah E. Nat Rev Neurol. 2010;6:702–706. doi: 10.1038/nrneurol.2010.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Jackson K, Barisone GA, Diaz E, Jin L-w, DeCarli C, Despa F. Ann Neurol. 2013;74:517–526. doi: 10.1002/ana.23956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Horvath I, Wittung-Stafshede P. Proc Natl Acad Sci USA. 2016;113:12473–12477. doi: 10.1073/pnas.1610371113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Horwich A. J Clin Invest. 2002;110:1221–1232. doi: 10.1172/JCI16781. [DOI] [PMC free article] [PubMed] [Google Scholar]