Abstract
We report here our combination of cryogenic, messenger-tagging, infrared (IR) spectroscopy with ion mobility spectrometry (IMS) and mass spectrometry (MS) as a way to identify and analyze a set of human milk oligosaccharides (HMOs) ranging from trisaccharides to hexasaccharides. The added dimension of IR spectroscopy provides a diagnostic fingerprint in the OH and NH stretching region, which is crucial to identify these oligosaccharides, which are difficult to distinguish by IMS alone. These results extend our previous work in demonstrating the generality of this combined approach for distinguishing subtly different structural and regioisomers of glycans of biologically relevant size.
INTRODUCTION
The multiple functional roles of glycans are directly linked to their structural diversity, but it is not well understood how these heterogeneous structures mediate different biological processes.1,2 This is mainly because structural analysis of glycans remains a major analytical challenge. The high degree of variation possible in the monosaccharide composition and sequence, the anomericity and location of the glycosidic linkages, as well as the possibility of branching, leads to the existence of many glycan isomers. Even for a simple hexasaccharide, the possible number of linear and branched isomers is calculated to be greater than 1.05 × 1012.3 This number emphasizes the obstacle that needs to be overcome by current analytical techniques before complete molecular characterization of glycans is achieved.3 The existence of different glycoforms when glycans are covalently linked to other biomolecules (proteins, lipids) further adds to this structural complexity.
Various analytical techniques,4–6 including nuclear magnetic resonance,7–9 capillary electrophoresis,10,11 chromatographic separation,12–14 and mass spectrometry (MS)15,16 are currently used for the analysis of glycans. Tandem MS-based methods, in particular, have evolved to be a major tool for glycan analysis due to their speed and sensitivity. However, these techniques still lack the ability to provide direct, unambiguous assignments of stereoisomers, linkage type, and monosaccharide composition. To overcome this limitation, different orthogonal approaches have been combined with MS. Ion mobility spectrometry (IMS), which separates ions based on their size-to-charge ratio on a millisecond timescale, is an example of one such approach that is easily coupled to various mass spectrometers. Several studies have shown that the rapid separation offered by IMS can sufficiently resolve many, but not all, glycan isomers that were previously indistinguishable by MS alone.17–19
Recently, we introduced a unique approach to identify isomeric glycans by combining cryogenic, messenger-tagging, infrared (IR) spectroscopy with IMS and MS.20,21 While IR spectroscopy provides important details about the structure of a molecule, its usefulness is often limited by the presence of broad bands that compromise the identification of closely related isomers.22–25 This drawback can be overcome by the use of cryogenic IR spectroscopy, which generates high-resolution vibrational spectra that are highly sensitive to minute structural differences.20,21,26 The enhanced resolution, which is a result of the decrease in the number of rovibrational states populated at low temperature, is crucial to distinguish various isomers with subtle structural variations. Our previous studies demonstrated the use of this approach in distinguishing disaccharide isomers that differed in anomericity, sequence, composition, glycosidic linkage, and functional sites, as well as a pair of pentasaccharide isomers with different branching patterns.20,21 The ability to distinguish the isomers spectroscopically allows us to identify them using a database approach.20,21 The fact that the linear vibrational spectrum is a fundamental property of the molecule should make it a robust measure for a database search.
While our previous work demonstrated the ability to distinguish all the various isomerisms in a series of disaccharides (and in the case of structural isomers, a pair of widely different pentasaccharides), one might question the generality and biological relevance of our approach. Here, we demonstrate the successful application of our method to larger glycans by distinguishing isomeric pairs of human milk oligosaccharides (HMOs) ranging from trisaccharides to hexasaccharides that are particularly difficult to distinguish using existing techniques. We have chosen pairs of isomers that are either subtly different structural isomers (in the case of tri-and hexasaccharides), or regioisomers (in the case of tetrasaccharides) in which the respective ion mobility drift-time distributions either partially or completely overlap. This emphasizes the sensitivity of cryogenic IR spectra to these subtle structural differences.
Human milk oligosaccharides are known to shape the gut microbiota of infants and enhance their immune systems by inhibiting the virulence of pathogens.27,28 These oligosaccharides also serve as prebiotics by stimulating the growth of beneficial intestinal bacteria such as bifidobacteria and lactobacilli in neonates.27–29 Despite their importance, a clear link between the diverse structures of HMOs and their functional roles in shaping the immune response of infants has not been established, as structural variations between different HMO structures are difficult to characterize. Consequently, even though more than 200 HMOs have been identified, the structural details of only 115 of them have been fully characterized.28,30,31 Thus, it is crucial to develop techniques that are sensitive to structural details of different isomeric HMOs. In this work, we employ our combined approach to distinguish isomeric pairs of different HMOs. We examine isomeric pairs of trisaccharides 2′-fucosyllactose (2′-FL) and 3-fucosyllactose (3-FL), tetrasaccharides lacto-N-tetraose (LNT) and lacto-N-neotetraose (LNnT), and hexasaccharides lacto-N-difucohexaose I (LNDFH I) and lacto-N-difucohexaose II (LNDFH II). Our analysis shows that cryogenic IR spectra are particularly sensitive to the differences in various isomeric HMOs. The vibrational patterns of free and hydrogen-bonded hydroxyl groups that are unique to each isomer serve as identifying fingerprints. Our ultimate goal is to use this method to build a database of glycan standards and analyze biological samples through database screening based on collision cross section, mass, and IR spectrum.
EXPERIMENTAL APPROACH
We use a home-built instrument consisting of an ion-mobility drift tube, a cryogenic planar ion trap, and a time-of-flight (TOF) mass spectrometer to measure collisional cross sections, vibrational spectra, and mass for isomeric HMOs.32,33 Ions generated by nano-electrospray ionization (nESI) are pulsed into a 2-meter long drift tube maintained at ~10 V/cm and separated based on their mobility through helium buffer gas. A quadrupole mass filter with a channeltron detector at the end of the drift tube is used to detect the drift-time distributions of mass-selected species. Infrared spectroscopy on mass- and mobility-selected ions is performed in a planar, cryogenic ion trap filled with N2 buffer gas at ~42K in which the ions are collisionally cooled. This cooling also allows for the formation of weakly bound N2 adducts, which can be dissociated subsequent to resonant absorption of an IR photon. An infrared spectrum is recorded in the TOF mass spectrometer by measuring the depletion of selected N2 adducts as a function of the infrared frequency.34
Unlike our previous experiments that utilized H2 as the tagging gas,20,21 here we use N2 as a messenger tag, primarily because it sticks to the ions more efficiently due to its higher polarizability. This increased tagging efficiency also improves the signal-to-noise ratio in the IR spectrum, resulting in faster acquisition times. Our earlier studies demonstrated that IR spectra obtained with N2 at higher temperature retain highly resolved peaks and can equally well be used as an identifying fingerprint.20
The human milk oligosaccharides used for this study were purchased from Dextra Laboratories (United Kingdom) and used without further purification. Solutions of each HMO were prepared in 50:50 water:methanol with 2% acetic acid and 1–3 mM sodium chloride. The concentration of the HMOs ranged from 100 – 600 μM. Acetic acid (99%) was purchased from Sigma Aldrich. HPLC methanol and nuclease-free water were obtained from VWR International and Ambion, respectively. Metal-coated borosilicate emitters from Thermo Scientific were used for nano-electrospray ionization. We choose to probe the sodiated adduct of the glycans spectroscopically, which eliminates the need of additional steps for removing sodium that is naturally present in all samples. Cationization is easy to achieve, and it shifts some of the vibrational bands of hydroxyl groups outside the frequency region of our analysis, resulting in simpler spectra that can be used more effectively for diagnostic purposes. While complexation with a metal ion might lead to conformational compaction that minimizes differences in the measured drift times of isomeric species,18,19 it is the preferred option for spectroscopy, as it is significantly more efficient than protonation when no amino-sugars are present. While in principle one could measure vibrational spectra of negative ion species, which seem to exhibit greater diversity in measured cross sections,35 the presence of many weakly acidic hydroxyl groups is likely to lead to a wide distribution deprotonation sites. Such a heterogeneous mixture would likely result in a substantial increase in spectral congestion.
As our goal is to identify glycan isomers rather than probe their 3-dimensional structure, it does not pose a problem if there are multiple conformations present, as long as we measure the same conformer distribution each time. To help ensure this, we intentionally heat the molecules in the activation zone of our drift tube and then re-cool them to room temperature. This annealing process forms a quasi-equilibrium conformer distribution that should be insensitive to source conditions across different instrument platforms,20 which is important for our future goal of developing a broadly applicable database approach to identify glycans in a biological sample.
RESULTS AND DISCUSSION
(i) Tetrasaccharide isomers that vary in a single glycosidic linkage
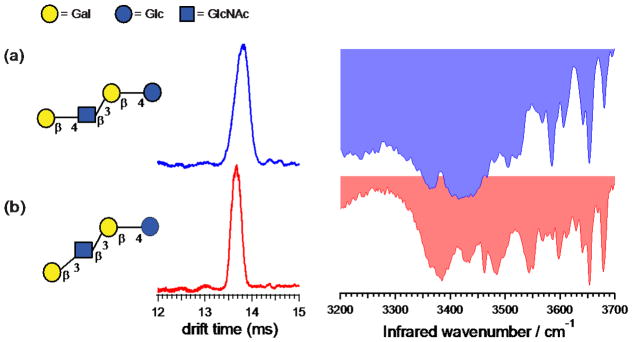
Tetrasaccharides LNT and LNnT are regioisomers that differ in the glycosidic linkage (β1-3 vs. β1-4) of the terminal galactose. Most HMOs share a common backbone that consist of a lactose core (Galβ1-4Glc) at the reducing end that can be further elongated by the addition of either lacto-N-biose (Galβ1-3GlcNAc-), as in the case of LNT, or by N-acetyllactosamine (Galβ1-4GlcNAc), as in the case of LNnT.27 Figure 1 shows the structure, ion mobility distributions, and the vibrational spectra for the singly sodiated adducts of LNT and LNnT (m/z 731). Overlapping drift times for LNnT (13.79 ms) and LNT (13.69 ms) suggest that a small difference in a single glycosidic linkage doesn’t influence the overall shape of the tetrasaccharide complexed with sodium.
Figure 1.
Ion mobility distributions and vibrational spectra of sodiated (a) LNnT and (b) LNT, each tagged with one molecule of N2. The symbols follow the Symbol Nomenclature for Glycans (SNFG) system.36 The drift times and peak widths are (a) 13.81 ms, FWHM=0.36 ms; and (b) 13.67 ms, FWHM=0.25 ms.
In contrast, the vibrational spectra measured for these two tetrasaccharides are distinctly different. The large number of OH oscillators results in rich and distinct IR spectra for these species. They both contain broad transitions in the hydrogen-bonding region (~ 3300–3550 cm−1) along with numerous sharp peaks in the free OH stretch region (~ 3550–3700 cm−1), all of which contribute to a fingerprint that can be used to distinguish these two molecules. In the lower-frequency region, three broad transitions (3385 cm−1, 3434 cm−1, and 3485 cm−1) as well as one sharp transition (3461 cm−1) are observed for LNT (Fig. 1b), while the spectrum of LNnT (Fig. 1a) contains only three broad transitions (3367 cm−1, 3420 cm−1, and 3505 cm−1) in the same region. In the region of free OH stretches, two sharp transitions (3653 cm−1 and 3678 cm−1) and one weak transition (3597 cm−1) are observed for LNT (Fig. 1b). Meanwhile, LNnT (Fig. 1a) shows three sharp transitions (3585 cm−1, 3652 cm−1, and 3680 cm−1) and one weak transition (3605 cm−1) in the same region. Considering the presence of many broad bands in the region of hydrogen-bonding interactions, it is possible that these tetrasaccharides exist as multiple structural isomers that are not distinguished by our current IMS setup. Nevertheless, we run our experiments under conditions in which the gas-phase conformer distribution is annealed to relax any kinetically trapped structures. In this way, we ensure that we observe the same conformational distribution each time, ensuring that the resulting spectroscopic fingerprint is robust.
(ii) Structural isomers related to fucose
Fucose is naturally present at the terminal end of many glycans and participates in various biological functions including the regulation of the immune system, neuron morphology, and cell-to-cell adhesion/recognition.37,38 Fucosylation is also a common type of modification prevalent in cancer cells. As a result, most cancer-related biomarkers are based on changes associated with the fucose residue.39,40 Furthermore, the stability and efficacy of monoclonal antibodies, which generate immune response against specific antigens, is associated with fucosylation.41–43 Considering the biological and pathological significance of fucose, it is crucial to identify isomeric structures associated with fucosylated glycans. However, the propensity of fucose to undergo rearrangement during MS/MS experiments (mostly in the case of protonated species)44–46 often leads to ambiguous structural assignments. Moreover, the mechanism of fucose migration is not well understood, which makes this rearrangement process highly unpredictable.45 Several methods, including permethylation and procainamide labeling, have been implemented to eliminate the migration of fucose during MS/MS; however, the additional sample preparation steps required for these chemical modifications are time consuming.47 It is thus of interest to see if fucose-based isomers can be distinguished by our method.
Since studies have shown that 35–50% of HMOs are fucosylated, they represent an ideal model system to investigate isomers related to fucose.27 The diversity associated with the fucosylation patterns in HMOs is dictated by the secretor and Lewis blood group of the mother.48 The α(1–2) glycosidic linkage between fucose and galactose of the lacto-N-biose is only present in the milk from secretor mothers as they have active fucosyltransferase FUT2 responsible for the linkage. On the other hand, the α(1–4) linkage between fucose and glucosamine of the lacto-N-biose is present only in Lewis positive mothers as they have active fucosyltransferase FUT3 responsible for the linkage. This FUT3 enzyme is also responsible for the addition of fucose to glucosamine in N-acetyllactosamine via an α(1–3) linkage. Thus, the milk from Le-positive Secretor (Se+Le+) mothers contains HMOs with α(1–2), α(1–3), and α(1–4) linkages associated with fucose, but these fucose linkages are absent in the milk from Le-negative non-Secretor (Se−Le−) mothers. To delineate the subtle differences in the position and the linkage of the fucose residue with our technique, we analyze the isomeric pair of trisaccharides, 2′-FL and 3-FL, along with the hexasaccharides LNDFH I and II.
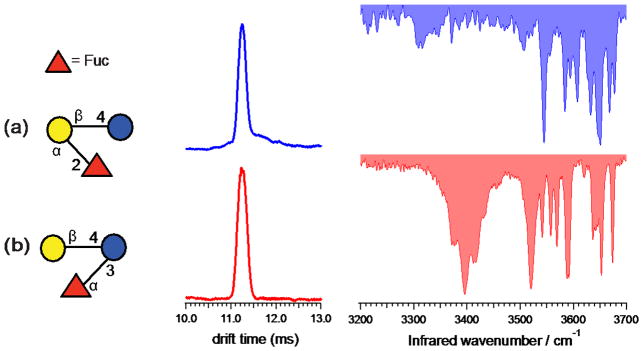
Figure 2 shows the structures, ion mobility distributions, and vibrational spectra for 2′-FL and 3-FL, which we study in their singly sodiated form (m/z 511). These trisaccharide isomers differ from each other in the position of the terminal fucose residue as well as the glycosidic linkage associated with it. In 2′-FL, fucose is attached to the galactose residue of the lactose core through an α(1–2) linkage, whereas 3-FL has fucose attached to glucose through an α(1–3) linkage. The measured drift times for 2′-FL and 3-FL are 11.24 ms and 11.23 ms, respectively, too close to allow them to be reliably distinguished by IMS.
Figure 2.
Ion mobility distributions and vibrational spectra of sodiated (a) 2′-FL and (b) 3-FL, each tagged with one molecule of N2. The drift times and peak widths are (a) 11.24 ms, FWHM=0.21 ms; and (b) 11.24 ms, FWHM=0.25 ms.
In contrast, the vibrational spectra of these two sodiated trisaccharides are clearly different, each serving as an identifying fingerprint. Both the broad feature in the hydrogen bonding region around 3395 cm−1 in the spectrum of 3-FL (Fig. 2b) as well as the different pattern of sharp lines in the higher frequency region serve as distinguishing features.
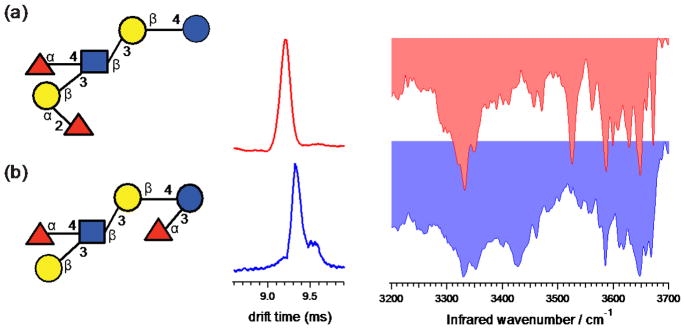
The structures, ion mobility distributions as well as vibrational spectra for LNDFH I and II are shown in Figure 3 for the doubly sodiated species, [M+2Na]2+ at m/z 523. Both LNDFH I and II have the same tetrasaccharide core as LNT (Galβ1-3GlcNAcβ1-3Galβ1-4Glc). The fucose residue that is attached to GlcNAc through an α(1–4) linkage is also present in both isomers. However, the position and linkage of the terminal fucose residue is different for these oligosaccharides: in LNDFH I it is attached to Gal through an α(1–2) linkage, while in LNDFH II it is attached to Glc through an α(1–3) linkage. The difference in the drift times measured for LNDFH I (9.20 ms) and LNDFH II (9.32 ms) just allows for these two species to be distinguished, although it would be difficult to resolve them in a mixture.
Figure 3.
Ion mobility distributions and vibrational spectra of sodiated (a) LNDFH I and (b) LNDFH II, each tagged with one molecule of N2. The drift times and peak widths are (a) 9.21 ms, FWHM=0.13 ms; and (b) 9.32 ms, FWHM=0.13 ms.
The IR spectra measured for these isomeric hexasaccharides are also very different from each other and can be used for identification. The spectrum of LNDFH I (Fig. 3a) displays a broad peak at 3332 cm−1 in the hydrogen-bonded region, while that of LNDFH II (Fig. 3b) consists of two broad transitions at 3330 cm−1 and 3426 cm−1. The number and pattern of free OH stretches observed for LNDFH I and II are also different: the latter contains sharp peaks at 3585 cm−1 and 3648 cm−1 as well as a number of weaker transitions, whereas LNDFH I displays four sharp peaks (~3525 cm−1, ~3587 cm−1, ~3648 cm−1, and ~3671 cm−1) and along with some weaker bands in this region.
Even though LNDFH II and I have the same tetrasaccharide core as that of LNT, it is interesting to note that the IR spectra of the hexasaccharides do not share any common peaks with the spectrum of LNT. It seems that the additional fucose residue in LNDFH II and I results in a totally different spectral pattern. This is not surprising, considering how isomers with a single change in the stereogenic center or the linkage type generated radically different spectra in our previous studies.20 Even LNT and LNnT, which differ by a single linkage, exhibit very different vibrational spectra. The distribution of shifts of the OH vibrational frequencies from those of a free alcohol emphasizes the fact that the OH oscillators in an oligosaccharide should be considered as a strongly coupled network. A slight change in structure, such as the inversion about a stereogenic center or the addition of an additional residue, can disrupt the network of hydrogen-bonding interactions and give rise to a radically different one, resulting in a completely different vibrational spectrum. While this feature makes it difficult to build up a spectrum from either measured or calculated spectra of root structures, it does provide unique spectroscopic fingerprints with which to identify all the isomers of a particular species if its spectrum has been previously measured and tabulated in a database.
The successful identification of various isomeric HMOs of different sizes demonstrates the robustness of this approach and addresses the question of its broader applicability for glycan analysis. We are able to show how the complexity of IR spectra changes when larger polysaccharides are probed. It is true that in comparison to the spectra obtained for various disaccharides,20,21 the spectra for larger polysaccharides are more complex, but the increased spectral complexity does not undermine its ability to serve as a spectroscopic fingerprint that is sensitive to all types of isomerization. It is important to realize that it is the entire spectral pattern that serves as to identify a particular glycan and not a single spectral feature.
CONCLUSIONS
In the present study, we measure drift time distributions and high-resolution vibrational spectra of isomeric HMOs, ranging from trisaccharides to hexasaccharides, using cryogenic IR spectroscopy together with IMS-MS. Even though most isomeric HMOs have similar drift times, their vibrational signatures in the hydrogen-stretching regions are distinctly different and serve as spectroscopic fingerprints. The highly structured nature of the vibrational spectra resulting from cryogenic cooling of the ions increases our ability to distinguish subtly different isomers with high specificity. This work demonstrates the immense potential of our approach as a practical tool for analysis of biologically relevant glycans. We ultimately aim to build a database containing the collision cross section, mass, and vibrational fingerprint spectrum of glycan standards, which can be used to identify glycans present in a biological sample.
Supplementary Material
Acknowledgments
TRR would like to thank the EPFL and the Swiss National Science Foundation (grant number 200020_165908) for their financial support of this work. DEC would like to thank the National Institutes of Health for partial support of this work through grant (R01 GM117207-01). NK was supported by a Robert & Marjorie Mann Fellowship from Indiana University.
Footnotes
CONFLICTS OF INTEREST
There are no conflicts to declare.
References
- 1.Varki A, et al. Essentials of Glycobiology. 2. Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- 2.Raman R, Raguram S, Venkataraman G, Paulson JC, Sasisekharan R. Glycomics: an integrated systems approach to structure-function relationships of glycans. Nature Methods. 2005;2:817–824. doi: 10.1038/nmeth807. [DOI] [PubMed] [Google Scholar]
- 3.Laine RA. A calculation of all possible oligosaccharide isomers both branched and linear yields 1.05 × 10(12) structures for a reducing hexasaccharide: the Isomer Barrier to development of single-method saccharide sequencing or synthesis systems. Glycobiology. 1994;4:759–767. doi: 10.1093/glycob/4.6.759. [DOI] [PubMed] [Google Scholar]
- 4.Rudd PM, Dwek RA. Rapid, sensitive sequencing of oligosaccharides from glycoproteins. Current opinion in biotechnology. 1997;8:488–497. doi: 10.1016/s0958-1669(97)80073-0. [DOI] [PubMed] [Google Scholar]
- 5.Marino K, Bones J, Kattla JJ, Rudd PM. A systematic approach to protein glycosylation analysis: a path through the maze. Nat Chem Biol. 2010;6:713–723. doi: 10.1038/nchembio.437. [DOI] [PubMed] [Google Scholar]
- 6.Pabst M, Altmann F. Glycan analysis by modern instrumental methods. Proteomics. 2011;11:631–643. doi: 10.1002/pmic.201000517. [DOI] [PubMed] [Google Scholar]
- 7.Vliegenthart JF, Dorland L, van Halbeek H. High-resolution, 1 H-nuclear magnetic resonance spectroscopy as a tool in the structural analysis of carbohydrates related to glycoproteins. Advances in carbohydrate chemistry and biochemistry. 1983;41:209–374. [Google Scholar]
- 8.Battistel MD, Azurmendi HF, Yu B, Freedberg DI. NMR of glycans: shedding new light on old problems. Progress in nuclear magnetic resonance spectroscopy. 2014;79:48–68. doi: 10.1016/j.pnmrs.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Jiménez-Barbero J, Asensio JL, Cañada FJ, Poveda A. Free and protein-bound carbohydrate structures. Current opinion in structural biology. 1999;9:549–555. doi: 10.1016/s0959-440x(99)00004-4. [DOI] [PubMed] [Google Scholar]
- 10.Mechref Y, Novotny MV. Glycomic analysis by capillary electrophoresis–mass spectrometry. Mass spectrometry reviews. 2009;28:207–222. doi: 10.1002/mas.20196. [DOI] [PubMed] [Google Scholar]
- 11.Campa C, Coslovi A, Flamigni A, Rossi M. Overview on advances in capillary electrophoresis-mass spectrometry of carbohydrates: a tabulated review. Electrophoresis. 2006;27:2027–2050. doi: 10.1002/elps.200500960. [DOI] [PubMed] [Google Scholar]
- 12.Wuhrer M, de Boer AR, Deelder AM. Structural glycomics using hydrophilic interaction chromatography (HILIC) with mass spectrometry. Mass spectrometry reviews. 2009;28:192–206. doi: 10.1002/mas.20195. [DOI] [PubMed] [Google Scholar]
- 13.Guile GR, Rudd PM, Wing DR, Prime SB, Dwek RA. A rapid high-resolution high-performance liquid chromatographic method for separating glycan mixtures and analyzing oligosaccharide profiles. Analytical biochemistry. 1996;240:210–226. doi: 10.1006/abio.1996.0351. [DOI] [PubMed] [Google Scholar]
- 14.Saito Y, et al. Miniaturized sample preparation and separation methods for environmental and drug analyses. Analytical sciences: the international journal of the Japan Society for Analytical Chemistry. 2002;18:7–17. doi: 10.2116/analsci.18.7. [DOI] [PubMed] [Google Scholar]
- 15.Kailemia MJ, Ruhaak LR, Lebrilla CB, Amster IJ. Oligosaccharide analysis by mass spectrometry: a review of recent developments. Anal Chem. 2013;86:196–212. doi: 10.1021/ac403969n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaia J. Mass spectrometry of oligosaccharides. Mass Spectrom Rev. 2004;23:161–227. doi: 10.1002/mas.10073. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Clemmer DE. Characterizing oligosaccharides using injected-ion mobility/mass spectrometry. Anal Chem. 1997;69:2504–2509. doi: 10.1021/ac9701344. [DOI] [PubMed] [Google Scholar]
- 18.Lee S, Wyttenbach T, Bowers MT. Gas phase structures of sodiated oligosaccharides by ion mobility/ion chromatography methods. International journal of mass spectrometry and ion processes. 1997;167:605–614. [Google Scholar]
- 19.Gray CJ, et al. Applications of ion mobility mass spectrometry for high throughput, high resolution glycan analysis. Biochimica et biophysica acta. 2016;1860:1688–1709. doi: 10.1016/j.bbagen.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Masellis C, Khanal N, Kamrath MZ, Clemmer DE, Rizzo TR. Cryogenic Vibrational Spectroscopy Provides Unique Fingerprints for Glycan Identification. Journal of the American Society for Mass Spectrometry. 2017;28:2217–2222. doi: 10.1007/s13361-017-1728-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khanal N, Masellis C, Kamrath MZ, Clemmer DE, Rizzo TR. Glycosaminoglycan Analysis by Cryogenic Messenger-Tagging IR Spectroscopy Combined with IMS-MS. Anal Chem. 2017;89:7601–7606. doi: 10.1021/acs.analchem.7b01467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan Y, et al. Vibrational Signatures of Isomeric Lithiated N-acetyl-D-hexosamines by Gas-Phase Infrared Multiple-Photon Dissociation (IRMPD) Spectroscopy. J Am Soc Mass Spectrom. 2017;28:539–550. doi: 10.1007/s13361-016-1575-x. [DOI] [PubMed] [Google Scholar]
- 23.Schindler B, et al. Distinguishing isobaric phosphated and sulfated carbohydrates by coupling of mass spectrometry with gas phase vibrational spectroscopy. Physical chemistry chemical physics: PCCP. 2014;16:22131–22138. doi: 10.1039/c4cp02898h. [DOI] [PubMed] [Google Scholar]
- 24.Schindler B, et al. IRMPD Spectroscopy Sheds New (Infrared) Light on the Sulfate Pattern of Carbohydrates. The journal of physical chemistry A. 2017;121:2114–2120. doi: 10.1021/acs.jpca.6b11642. [DOI] [PubMed] [Google Scholar]
- 25.Polfer NC, et al. Differentiation of isomers by wavelength-tunable infrared multiple-photon dissociation-mass spectrometry: application to glucose-containing disaccharides. Anal Chem. 2006;78:670–679. doi: 10.1021/ac0519458. [DOI] [PubMed] [Google Scholar]
- 26.Boyarkin OV, Mercier SR, Kamariotis A, Rizzo TR. Electronic spectroscopy of cold, protonated tryptophan and tyrosine. Journal of the American Chemical Society. 2006;128:2816–2817. doi: 10.1021/ja058383u. [DOI] [PubMed] [Google Scholar]
- 27.Smilowitz JT, Lebrilla CB, Mills DA, German JB, Freeman SL. Breast milk oligosaccharides: structure-function relationships in the neonate. Annual review of nutrition. 2014;34:143–169. doi: 10.1146/annurev-nutr-071813-105721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niñonuevo MR, Lebrilla CB. Mass spectrometric methods for analysis of oligosaccharides in human milk. Nutrition reviews. 2009;67:S216–S226. doi: 10.1111/j.1753-4887.2009.00243.x. [DOI] [PubMed] [Google Scholar]
- 29.Bode L. Human milk oligosaccharides: prebiotics and beyond. Nutrition reviews. 2009;67:S183–S191. doi: 10.1111/j.1753-4887.2009.00239.x. [DOI] [PubMed] [Google Scholar]
- 30.Asakuma S, et al. Physiology of Consumption of Human Milk Oligosaccharides by Infant Gut-associated Bifidobacteria. Journal of Biological Chemistry. 2011;286:34583–34592. doi: 10.1074/jbc.M111.248138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barile D, Rastall RA. Human milk and related oligosaccharides as prebiotics. Curr Opin Biotechnol. 2013;24:214–219. doi: 10.1016/j.copbio.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 32.Masson A, et al. Infrared Spectroscopy of Mobility-Selected H+-Gly-Pro-Gly-Gly (GPGG) J Am Soc Mass Spectrom. 2015;26:1444–1454. doi: 10.1007/s13361-015-1172-4. [DOI] [PubMed] [Google Scholar]
- 33.Koeniger SL, et al. An IMS-IMS analogue of MS-MS. Anal Chem. 2006;78:4161–4174. doi: 10.1021/ac051060w. [DOI] [PubMed] [Google Scholar]
- 34.Kamrath MZ, et al. Vibrational characterization of simple peptides using cryogenic infrared photodissociation of H2-tagged, mass-selected ions. Journal of the American Chemical Society. 2011;133:6440–6448. doi: 10.1021/ja200849g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofmann J, Pagel K. Glycan Analysis by Ion Mobility-Mass Spectrometry. Angewandte Chemie-International Edition. 2017;56:8342–8349. doi: 10.1002/anie.201701309. [DOI] [PubMed] [Google Scholar]
- 36.Varki A, et al. Symbol Nomenclature for Graphical Representations of Glycans. Glycobiology. 2015;25:1323–1324. doi: 10.1093/glycob/cwv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider M, Al-Shareffi E, Haltiwanger RS. Biological functions of fucose in mammals. Glycobiology. 2017 doi: 10.1093/glycob/cwx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sturla L, et al. Differential terminal fucosylation of N-linked glycans versus protein O-fucosylation in leukocyte adhesion deficiency type II (CDG IIc) Journal of Biological Chemistry. 2003;278:26727–26733. doi: 10.1074/jbc.M304068200. [DOI] [PubMed] [Google Scholar]
- 39.Miyoshi E, et al. Fucosylation is a promising target for cancer diagnosis and therapy. Biomolecules. 2012;2:34–45. doi: 10.3390/biom2010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nwosu C, Yau HK, Becht S. Assignment of core versus antenna fucosylation types in protein N-glycosylation via procainamide labeling and tandem mass spectrometry. Anal Chem. 2015;87:5905–5913. doi: 10.1021/ac5040743. [DOI] [PubMed] [Google Scholar]
- 41.Shields RL, et al. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human FcγRIII and antibody-dependent cellular toxicity. Journal of Biological Chemistry. 2002;277:26733–26740. doi: 10.1074/jbc.M202069200. [DOI] [PubMed] [Google Scholar]
- 42.Yamane-Ohnuki N, Satoh M. MAbs. Taylor & Francis; pp. 230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeitlin L, et al. Enhanced potency of a fucose-free monoclonal antibody being developed as an Ebola virus immunoprotectant. Proceedings of the National Academy of Sciences. 2011;108:20690–20694. doi: 10.1073/pnas.1108360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wuhrer M, Koeleman CA, Hokke CH, Deelder AM. Mass spectrometry of proton adducts of fucosylated N-glycans: fucose transfer between antennae gives rise to misleading fragments. Rapid communications in mass spectrometry. 2006;20:1747–1754. doi: 10.1002/rcm.2509. [DOI] [PubMed] [Google Scholar]
- 45.Harvey DJ, et al. “Internal residue loss”: rearrangements occurring during the fragmentation of carbohydrates derivatized at the reducing terminus. Anal Chem. 2002;74:734–740. doi: 10.1021/ac0109321. [DOI] [PubMed] [Google Scholar]
- 46.Wuhrer M, Deelder AM, van der Burgt YE. Mass spectrometric glycan rearrangements. Mass Spectrom Rev. 2011;30:664–680. doi: 10.1002/mas.20337. [DOI] [PubMed] [Google Scholar]
- 47.Desai N, Thomas DA, Lee J, Gao J, Beauchamp J. Eradicating mass spectrometric glycan rearrangement by utilizing free radicals. Chemical Science. 2016;7:5390–5397. doi: 10.1039/c6sc01371f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. 2012;22:1147–1162. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.