SUMMARY
Sequence-specific pausing by RNA polymerase (RNAP) during transcription plays crucial and diverse roles in gene expression. In bacteria, RNA structures are thought to fold within the RNA exit channel of the RNAP and can increase pause lifetimes significantly. The biophysical mechanism of pausing is uncertain. We used single-particle cryo-EM to determine structures of paused complexes, including a 3.8 Å structure of an RNA hairpin-stabilized, paused RNAP that coordinates RNA folding in the his operon attenuation control region of E. coli. The structures revealed a half-translocated pause state (RNA post-translocated, DNA pre-translocated) that can explain transcriptional pausing, and a global conformational change of RNAP that allosterically inhibits trigger-loop folding and can explain pause hairpin action. Pause hairpin interactions with the RNAP RNA exit channel suggest how RNAP guides formation of nascent RNA structures.
eTOC Blurb
Cryo-EM structures of hairpin-stabilized and elemental paused transcription complexes reveal a biophysical basis of transcriptional pausing as formation of a half-translocated state (translocation of RNA, not DNA) that is unable to bind substrate NTP. The pause hairpin stabilizes rotation of a swivel module, which allosterically inhibits trigger-loop folding.
INTRODUCTION
Transcriptional pausing, in which RNA polymerase (RNAP) temporarily halts nucleotide addition by rearrangement into an inactive state, underpins regulatory mechanisms that control RNA chain initiation, elongation, folding, splicing, processing, and termination (Lerner et al., 2017; Mayer et al., 2017; Zhang and Landick, 2016). In bacteria, nascent RNA structures thought to form within the RNAP RNA exit channel can increase pause lifetimes ten-fold or more (Toulokhonov et al., 2001). In eukaryotes, specific interactions of RNA structure with RNAPs are undocumented, but pausing is increasingly understood to regulate RNA biogenesis (Mayer et al., 2017). The interplay between pause lifetimes and alternative folding patterns in the nascent RNA mediates both formation of biologically active RNA structures and diverse regulatory mechanisms (Hollands et al., 2014; Landick and Yanofsky, 1984; Pan et al., 1999; Steinert et al., 2017; Watters et al., 2016; Wickiser et al., 2005; Yakhnin et al., 2006; Zhang and Landick, 2016).
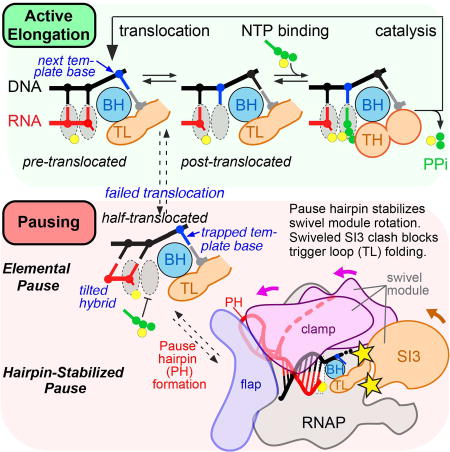
The mechanistic basis of pausing stimulated by nascent RNA structures (pause hairpins; PHs) has been studied extensively, establishing that pauses arise by rearrangement of the elongating transcription complex (EC) into a paused EC (PEC) at defined sequences with multipartite components (downstream DNA, downstream fork junction, RNA-DNA hybrid, upstream fork junction, and at some sites PH) that contact different RNAP surfaces (Chan et al., 1997; Larson et al., 2014; Wang et al., 1995). The PEC is an offline state, meaning that it forms in competition with nucleotide addition and that not every RNAP enters the paused state at a pause site. Initially, the PEC enters an elemental paused state (ePEC) with efficiency and duration that vary based on the sequences in contact with RNAP (Figure 1A). The ePEC can then undergo rearrangements that involve either (i) reverse threading of the RNA and DNA through RNAP to create a backtracked PEC, or (ii) formation of a PH whose interaction with RNAP creates a PH-stabilized PEC (hpPEC in Figure 1A).
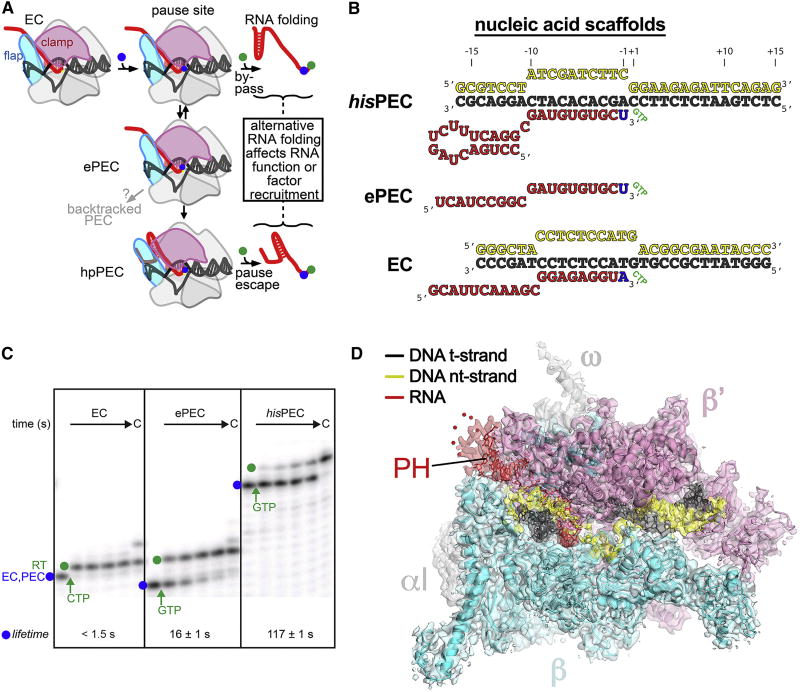
Figure 1. The mechanism of pausing and structure of the his RNA hairpin-stabilized PEC.
(A) Schematic illustration of the pause mechanism. Addition of a nucleotide that positions RNAP on a pause sequence (blue dot) permits formation of the offline elemental paused state (ePEC), which can either backtrack or be stabilized by PH formation (hpPEC). RNAP can bypass the pause by rapid addition of the next nucleotide (green dot), or add the next nucleotide more slowly if it enters the pause. The delay in RNA synthesis caused by the pause allows formation of biologically relevant RNA structures or recruitment of regulators.
(B) Nucleic acid scaffold sequences used for cryo-EM and pause assay experiments. The same t- and nt-strand DNA sequences are used in ePEC and hisPEC scaffolds.
(C) Lifetimes of elemental (ePEC) and hairpin-stabilized (hisPEC) pauses compared to the EC used for cryo-EM. ECs or PECs (blue dot) were pre-formed by addition of the relevant nucleic acid scaffold and then the rate of RNA extension was measured after addition of the next NTP (samples taken 5, 10, 20, and 40 s after NTP addition; see Methods and Figure S1 for quantitation; errors in lifetimes are s.d. of triplicates). RT, pause read-through RNA formed after NTP addition (green dot).
(D) The 3.8 Å resolution cryo-EM density map of the hisPEC is rendered as a transparent surface colored as labeled. Superimposed is the final refined model; the RNAP is shown as a backbone ribbon, the nucleic acids are shown in stick format.
See also Table S1 and Figures S1, S2.
Multiple PH-stabilized PECs have been characterized in diverse bacteria (Hollands et al., 2014; Landick and Yanofsky, 1984; Yakhnin et al., 2006). The best-studied example mediates RNA folding and synchronizes transcription of an attenuator with translation of a leader peptide coding region in the his biosynthetic operon of enteric bacteria (hisPEC; Chan and Landick, 1989; Zhang and Landick, 2016, and references therein). The PH-dependent increase in hisPEC lifetime (i) relies on the underlying elemental pause signal (Figure 1 B, C); (ii) is a function of hairpin stability and position (11 or 12 nt from pause RNA 3′ end) but not PH sequence (Chan and Landick, 1993; Chan et al., 1997; Toulokhonov et al., 2001); (iii) is stimulated by the NusA N-terminal domain (Ha et al., 2010); (iv) is suppressed by the RfaH N-terminal domain (Artsimovitch and Landick, 2002; Kolb et al., 2014); (v) requires PH contacts to and conformational changes in the flap tip and clamp of RNAP, which form the RNA exit channel (Hein et al., 2014; Toulokhonov and Landick, 2003); (vi) is modulated by spatially distant parts of RNAP that do not contact RNA or DNA, including a 188-residue insertion in the trigger loop (TL) of E. coli (Eco) RNAP called Sequence Insertion 3 (SI3, Hein et al., 2014; Windgassen et al., 2014; also called β′i6, Lane et al., 2010).
The extensive study of PH-stabilized pauses also has defined key questions that remain unanswered. How do RNA structures form within the RNA exit channel of RNAP? In what way is the active site altered to inhibit nucleotide addition in a paused RNAP? How do interactions of the PH in the exit channel prolong the inhibition of nucleotide addition?
Answering these questions and understanding how elongation factors modulate pausing by enhancing or inhibiting effects of the PH on RNAP requires a high-resolution structure of a PH-stabilized PEC. Here, we used single-particle cryo-electron microscopy (cryo-EM) to determine the Eco hisPEC structure at 3.8 Å resolution and used insights obtained by comparing this structure to a previously reported Eco EC structure (Kang et al., 2017) to guide biochemical tests of the mechanism of PH-stabilized pausing. The combination of the new hisPEC structure, the structure of a sub-population of particles lacking an ordered PH that may reflect the ePEC conformation, and the results of the biochemical experiments leads us to propose a model for pausing. In this model, pause sequences cause the RNA-DNA scaffold to enter an inactive configuration and the PH stabilizes a global conformational rearrangement of the EC that allosterically prolongs the pause by preventing the RNAP active site from assuming its optimal catalytic disposition.
RESULTS
A hisPEC cryo-EM structure reveals features implicated in transcriptional pausing
To validate a hisPEC suitable for cryo-EM structure determination, we tested pausing by Eco ΔαCTD-RNAP on an RNA-DNA scaffold based on his pause sequences within the context of a scaffold used previously for EC cryo-EM structures (Kang et al., 2017; Figure 1 B,C). The hisPEC obtained by direct reconstitution on the hisPEC scaffold (Figure 1B) in cryo-EM solution conditions (see Methods) exhibited a pause lifetime indistinguishable from hisPECs formed by nucleotide addition and on previously validated his pause scaffolds (Kyzer et al., 2007; Figures 1C, S1A–D).
We used single-particle cryo-EM to determine the Eco hisPEC structure to a nominal resolution of 3.8 Å (Figures 1D, S2, Table S1). Local resolution calculations indicate that much of the central core of the structure, including the active site and the RNA/DNA hybrid, was determined to 3.0–3.5 Å resolution (Figures S2G–I).
Initial examination of the cryo-EM map yielded several key observations. First, density for the 5-bp stem of the hisPH was identifiable within the RNAP RNA exit channel as an A-form double helix. The PH was connected by a single nucleotide, C−11(RNA), to the RNA in the RNA-DNA hybrid within the main RNAP cleft (Figure 2A). The 8-nucleotide loop of the PH (Figure 1B) and the flap tip were disordered and likely mobile in the hisPEC.
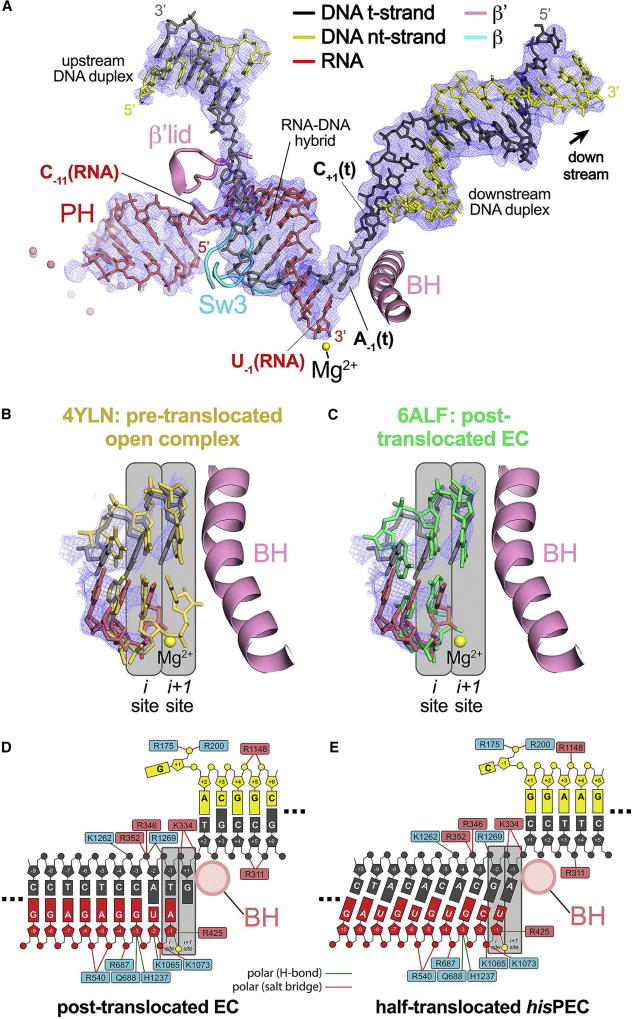
Figure 2. The hisPEC is trapped in a half-translocated state unable to load NTP substrate.
(A) The 3.8 Å resolution hisPEC cryo-EM density map (blue mesh) with the superimposed model of only the hisPEC nucleic acids. Shown for reference are key RNAP structural elements (Sw3, lid, BH) and the RNAP active-site Mg2+-ion (yellow sphere).
(B) Comparison of the half-translocated hisPEC RNA-DNA hybrid (RNA, red; t-strand DNA, dark gray) proximal to the RNAP active site (active-site Mg2+-ion shown as a yellow sphere) with the pre-translocated hybrid of 4YLN (yellow; Zuo and Steitz, 2015). The hisPEC cryo-EM density map is superimposed (blue mesh). Also shown for reference is the hisPEC BH. The RNAP active site i and i+1 sites are denoted schematically (gray boxes).
(C) Same as (B) but the half-translocated hisPEC RNA-DNA hybrid is compared with the post-translocated hybrid of 6ALF (green; Kang et al., 2017).
(D) Schematic illustration showing the active-site proximal nucleic acids and key protein/nucleic acid interactions for the post-translocated EC (6ALF; Kang et al., 2017). RNAP β subunit residues are denoted in cyan boxes, β′ residues in pink boxes. The BH (pink circle), the active site Mg2+-ion (yellow circle), and the i and i+1 sites (gray boxes) are also denoted.
(E) Same as (D) but showing the active-site proximal nucleic acids and key protein/nucleic acid interactions for the half-translocated hisPEC.
See also Figure S3.
Second, a 10-bp RNA-DNA hybrid could be readily modeled into well-resolved density in the RNAP main cleft, but the hybrid was in neither the canonical pre-translocated nor post-translocated register (Figure 2). Instead, the RNA was post-translocated whereas the DNA was pre-translocated, with the incoming template strand (t-strand) cytosine, C+1(t), base-paired to the non-template strand (nt-strand) DNA in the downstream DNA duplex (Figure 2A).
Third, the overall conformation of RNAP exhibited significant rearrangements relative to the canonical EC cryo-EM structure (Kang et al., 2017; Figure 3). Most notably, a group of structural modules, including the clamp, dock, shelf, and SI3 domains plus a C-terminal segment of the β′ subunit (β′C-term; Figure 3D) exhibited a previously unseen rigid-body rotation or swivel roughly about an axis perpendicular to the plane defined by the helical axes of the downstream duplex DNA and the RNA-DNA hybrid (Figure 3C, D; we denote this RNAP structural collection the “swivel module”). We will describe each of these hisPEC structural features in the context of roles of RNAP in transcriptional pausing and nascent RNA folding, and present biochemical evidence for their roles in pausing.
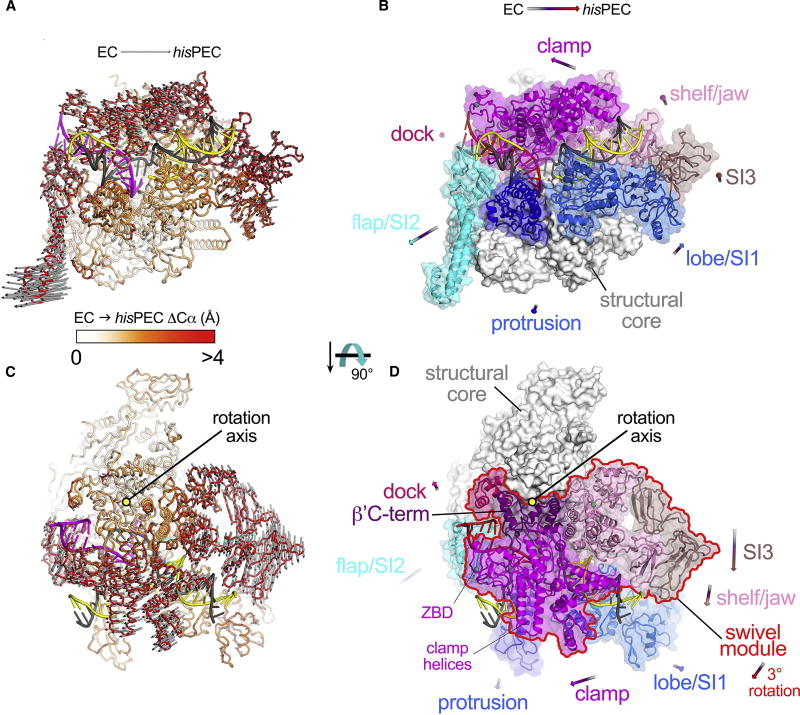
Figure 3. Global RNAP conformational changes: hisPEC vs. EC.
(A) The hisPEC structure is shown as an α-carbon backbone worm. The nucleic acids are shown in cartoon format (t-strand DNA, dark gray; nt-strand DNA, yellow; RNA, magenta). The protein is color-coded as a ramp (color-key shown below) according to the Cα(EC)-Cα(hisPEC) distance, where the two structures were superimposed via the structural core module (Table S2). The gray arrows denote the direction and distance (multiplied by a factor of 2) for the Cα(EC)-Cα(hisPEC) changes that are > 2 Å. The average ΔCα is 1.8 Å (± 1.7 Å standard deviation). The minimum and maximum ΔCα’s are 0.084 and 16 Å, with the largest ΔCα’s occurring in SI2.
(B) The hisPEC structure is shown [same view as (A)]. The structural core module (Table S2) is shown as a white molecular surface; mobile modules of the RNAP are shown as transparent surfaces with Cα cartoons and color-coded as labeled. The arrows denote the direction and length (multiplied by a factor of 5) of the changes in the center-of-mass positions for the mobile modules on going from the EC to the hisPEC conformation.
(C) Same as (A) but with a different view (rotated 90° about the horizontal axis) looking on top of the clamp module. The gray arrows show that a large portion of the structure rotates as a single unit clockwise about the indicated rotation axis.
(D) Same as (B) but with the rotated view on top of the clamp module. The rigid-body rotation [see (C)] includes the clamp/dock/shelf/jaw/SI3/β′C-term modules (outlined in red), which we term collectively the swivel module.
See also Table S2 and Figures S4, S5.
Substrate loading is blocked in the paused state by a half-translocated nucleic acid configuration
The half-translocated RNA-DNA hybrid (Figure 2), an unanticipated feature of the hisPEC, is most clearly defined by comparing it to the hybrids of pre-translocated (Figure 2B; PDB 4YLN; Zuo and Steitz, 2015) and post-translocated (Figure 2C; PDB 6ALF; Kang et al., 2017) Eco RNAP complexes. The hisPEC RNA is post-translocated because the 3′-nucleotide [U−1(RNA)] occupies the i site (product site) and not the i+1 site (NTP-binding site) as in the pre-translocated hybrid (Figure 2B). However, the hisPEC DNA is pre-translocated because the t-strand DNA base paired to the RNA 3′-nucleotide [A−1(t)] occupies the i+1 site, and the t-strand base for the incoming nucleotide substrate [C+1(t)] is base-paired in the downstream DNA duplex rather than occupying the i+1 site as in the post-translocated hybrid (Figure 2C). Overall, the hisPEC RNA contacts matched those of the post-translocated EC, especially for the 3′-proximal five nucleotides that make the most extensive RNAP contacts (Figures 2D, 2E, S3). However, the DNA contacts in the hisPEC were shifted away from the active site by one register. For example, β′R346 and R352 contact the −3 t-strand nucleotide in the post-translocated EC (Figure 2D) but contacted the −4 t-strand nucleotide in the hisPEC (Figure 2E). Translocation of the RNA but not the DNA causes the hybrid to be tilted, a geometry seen previously in yeast RNAPII initiating complexes with 4–6 nt RNAs (Cheung et al., 2011) and in a multi-nt backtracked yeast RNAPII EC (Cheung and Cramer, 2011).
The +1 t-strand DNA C that programs GTP binding in the hisPEC remained paired to the nt-strand in the downstream DNA duplex with RNAP contacts identical to the those of the +2 t-strand nucleotide in the post-translocated EC (Figure 2D, E). Thus, the hisPEC cannot template complementary substrate NTP binding, consistent with the inability of high NTP concentrations to affect disulfide-bond reporters of NTP-assisted TL folding in the hisPEC (Nayak et al., 2013). Other contacts to the downstream fork junction and downstream DNA duplex in the hisPEC were similar to those in an EC (but shifted), with two notable exceptions: (i) the +1 nt-strand C in the hisPEC did not make the characteristic RNAP contacts made by the core recognition-element as seen for +1G in the EC (Kang et al., 2017), and (ii) the downstream DNA duplex exhibited altered contacts to the clamp consistent with movement of the clamp in the hisPEC within the context of the swivel module (Figures 3, S3).
Taken together, the altered nucleic acid-RNAP contacts in the hisPEC suggest that the offline paused state of RNAP arises during translocation when nucleic acid sequences cause a modest remodeling of the RNAP-nucleic acid interface that stabilizes a half-translocated intermediate. Escape from the half-translocated state is inhibited because the pause sequence and its RNAP contacts resist completion of DNA translocation and NTP cannot bind the pre-translocated DNA.
The RNAP in the hisPEC is globally rearranged
To analyze conformational changes of the RNAP in the PH-stabilized state, we compared the Eco hisPEC structure with a canonical Eco EC structure (Kang et al., 2017; both determined by single-particle cryo-EM so unaltered by crystal packing forces). The two structures were superimposed by the structural core module (Table S2), comprising both α subunits, ω, and highly conserved β and β′ regions that form and surround the active center; these RNAP structural elements have not been observed to undergo significant conformational changes in dozens of existing RNAP structures. The structures superimposed this way with an rmsd of 0.675 Å over 1,322 aligned α-carbon (Cα) atoms of the structural core module, but an rmsd of 3.292 Å over the remaining 1,781 Cα atoms, indicating significant shifts of the mobile modules that surround the structural core (Figure 3, Table S2).
The movements of RNAP mobile modules can be divided into two main classes: (i) shifts away from the RNAP structural core, main cleft, and RNA exit channel by the flap, lobe, protrusion, SI1, and SI2, and (ii) a rotation of the swivel module around a central axis that roughly parallels the BH (Figures 3, S4; movie S1). The rotation of the swivel module has not previously been observed in RNAP structures. Instead, most previously observed clamp movements involve rotation of the clamp orthogonal to the main cleft rather than parallel to it, resulting in an “open” clamp (Darst et al., 2002; Feklistov et al., 2017; Tagami et al., 2010; Weixlbaumer et al., 2013; Zhang et al., 1999) that was postulated to occur in the hisPEC (Hein et al., 2014; Weixlbaumer et al., 2013). In the hisPEC, the swivel module rotates by ~3° about an axis centered in the shelf near a previously described hinge in TthRNAP (Tagami et al., 2010; Figures 3C, D). This rotation displaces the tip of the clamp helices ~3.5 Å and the zinc-binding domain (ZBD) ~3 Å toward the RNA exit channel (Figure 3D).
Although the width of the main cleft (clamp-to-protrusion/lobe distance) did not change appreciably as a consequence of the hisPEC conformational changes, in contrast to the large width increase of an “open” clamp conformation, the bend angle of the BH increased from 15° to 23° with a vertex near β′L788 (Figure S5). This change in BH bend angle does not occlude the i+1 template base position in the hisPEC, as was observed for a larger BH distortion in the Tth ePEC structure (Weixlbaumer et al., 2013). However, the hisPEC nucleic-acid scaffold also shifted in response to RNAP movements, with the downstream DNA duplex following the swivel module rotation (Figure 3) likely due to clamp-DNA contacts (Figure S3). The net result of these movements was that the angle between the downstream DNA and RNA-DNA hybrid decreased by about 2° in the hisPEC. Together, these changes in the BH and downstream DNA may help prevent the +1 t-strand DNA nucleotide to load into the RNAP active site.
Movements of the BH and swivel module in the hisPEC also constrict a space between the BH and β that forms the binding site for the RNAP inhibitor streptolydigin (Stl; Tuske et al., 2005; Vassylyev et al., 2007b). This constriction would create steric clash with bound Stl, which explains why Stl fails to inhibit the hisPEC (Toulokhonov et al., 2007) and suggests that Stl binding cannot reverse the swivel module rotation in the hisPEC.
hisPEC conformational change inhibits trigger loop folding
The TL is a polymorphic structural element that is central to RNAP catalytic activity (Vassylyev et al., 2007b; Wang et al., 2006). In most structures, the TL is either disordered or is observed in an unfolded conformation. In response to proper Watson-Crick base pairing geometry between the template DNA base and the incoming NTP in the RNAP i+1 site, the TL folds into a helical hairpin that closes the active site and stimulates catalysis ~104-fold (Vassylyev et al., 2007b). In RNAPs from Eco and some other bacterial clades (Lane and Darst, 2010), an ~200-aa domain is inserted in the middle of the TL (β′i6; SI3 in EcoRNAP). When the TL is unfolded, SI3 has been observed in multiple EcoRNAP structures to reside outside the active-site cleft, interacting with the distal surface of the β′jaw and linked to the TL-helices by long, extended linkers (Figure 4A; Bae et al., 2013; Chen et al., 2017; Hudson et al., 2009; Kang et al., 2017; Opalka et al., 2010). This SI3 position is incompatible with the folded TL because folding shortens the linkers enough to require SI3 repositioning (Hudson et al., 2009; Opalka et al., 2010). Indeed, a structure of EcoRNAP holoenzyme with a folded TL revealed that SI3 moved to the top of the β′jaw and was accommodated in a complementary depression between the lobe, SI1, the β′jaw, the shelf, and the β′rim-helices (Figure 4B; PDB 4YLN; Zuo and Steitz, 2015).
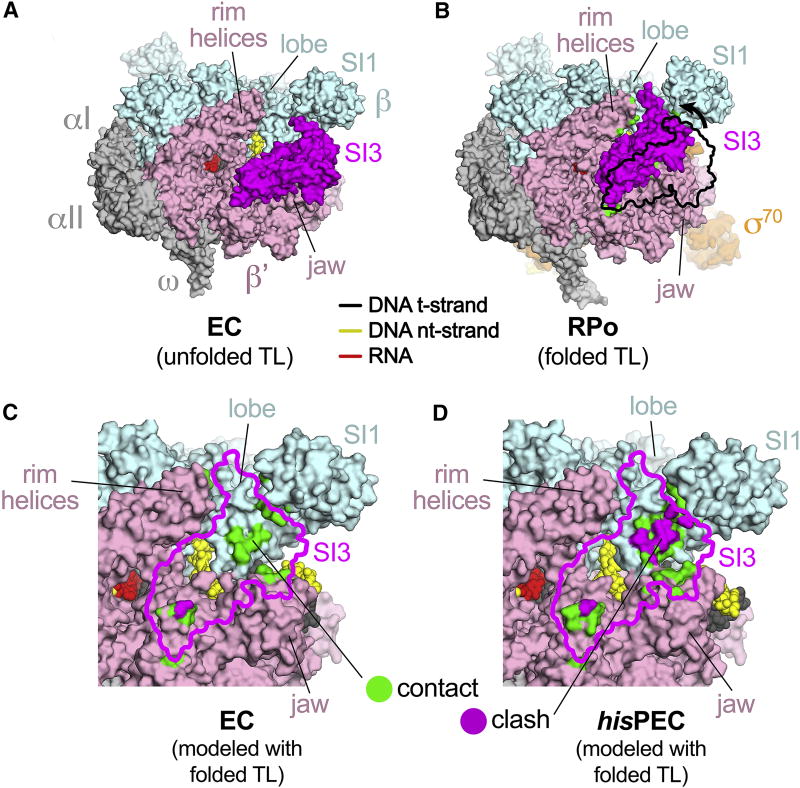
Figure 4. Inhibition of TL folding in the hisPEC.
(A) The Eco EC structure (6ALF; Kang et al., 2017), in which the TL is unfolded, is shown as a molecular surface color-coded as labeled. The nucleic acids are shown in CPK format and color-coded as shown in the key. The position of SI3 (magenta) is seen in numerous other Eco RNAP structures where the TL is unfolded (Bae et al., 2013; Chen et al., 2017; Hudson et al., 2009; Opalka et al., 2010).
(B) An Eco RNAP structure in which the TL is folded (4YLN; Zuo and Steitz, 2015). SI3 must move from its unfolded TL position (shown by the black outline) due to shortening of the TL-helices-SI3 linkers and is accommodated between the lobe, SI1, the shelf/jaw, and the rimhelices. New SI3-RNAP contacts upon TL folding are green (see panel C).
(C) The folded TL position of SI3 (magenta outline) was modeled onto the EC structure. RNAP surfaces that contact the modeled SI3 are colored green (4.5 Å > atomic distance > 3.0 Å). RNAP surfaces that clash with the modeled SI3 (atomic distance < 3.0 Å) are colored magenta.
(D) The folded TL position of SI3 (magenta outline) was modeled onto the hisPEC structure. RNAP surfaces contacting or clashing with the modeled SI3 are colored as in (C). Significant steric clash occurs between the modeled SI3 and the lobe/SI1.
Deletion of SI3 and small deletions within SI3 strongly reduce PH-dependent pause lifetime at the his pause (Artsimovitch et al., 2003; Conrad et al., 2010; Windgassen et al., 2014), but these observations have been difficult to explain since SI3 is nearly 100 Å away from the PH in the RNA exit channel. However, the rotation of the swivel module observed in the hisPEC structure distorts the SI3-complementary depression in the RNAP surface such that it can no longer accommodate the folded TL position of SI3 (Figure 4D; movie S2). This incompatibility explains how SI3-mediated inhibition of TL folding in the hisPEC is relieved by SI3 deletion (Windgassen et al., 2014). We conclude that the global conformational rearrangement stabilized by the PH in the hisPEC may increase pause duration by inhibiting SI3 movement required for TL folding.
The swivel module rotation can integrate inputs from a multipartite pause signal
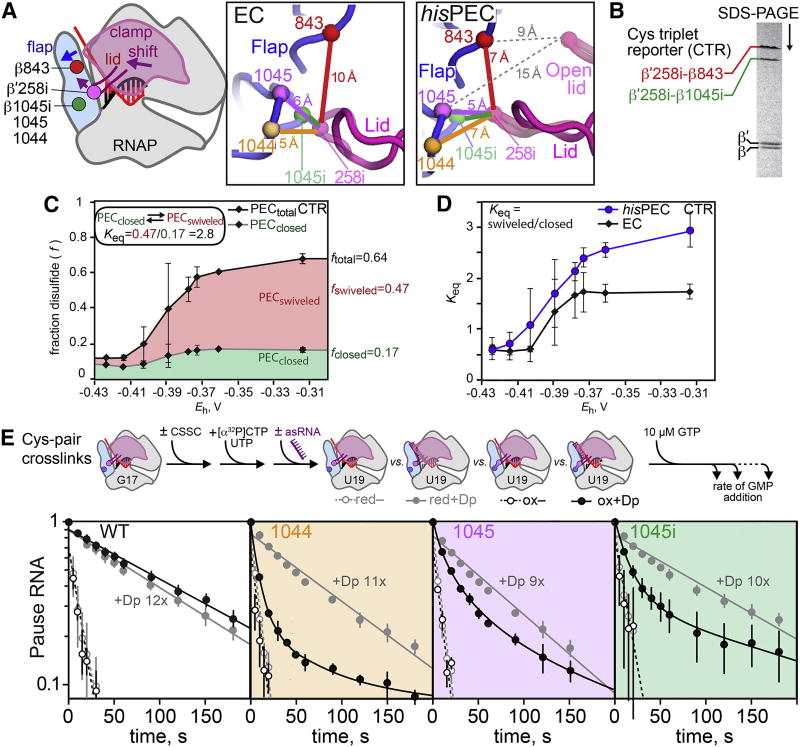
To investigate the role of the hisPEC conformational change in pausing, we extended experiments using engineered disulfide bonds in RNAP designed previously to test the role of clamp movement in pausing and PH formation. Hein et al. (2014) found that formation of a disulfide between a Cys inserted in the lid between G257 and G258 (β′C258i) and a second Cys substituted in the flap for βP1044 decreased the hisPEC lifetime before GMP addition by a factor of ~9, whereas a disulfide between β′C258i and a Cys substituted for flap βT843 increased pause lifetime ~18× in the absence of the PH. These data showed that biasing the clamp toward the closed position observed in an EC (258i-1044 disulfide) inhibited pausing, whereas biasing the clamp toward a swiveled or open position (258i-843 disulfide) promoted pausing.
To ask if the hisPEC caused the clamp to swivel as observed in the cryo-EM structure, we constructed a Cys-triplet reporter (CTR) containing β′C258i, βC843 (described above), and βC1045i in a single RNAP. Competition between the 258i-1045i vs. 258i-843 disulfides should depend on clamp conformation (Figure 5A). The Cα-Cα distances for the 258i-1045i and 258i-843 disulfides, which are distinguishable by SDS-PAGE (Figure 5B), are predicted to change from ~3 Å and ~10 Å, respectively, in an EC (Kang et al., 2017) to ~4 Å and ~7 Å in the hisPEC structure (Figure 5A). Thus the 258i-843 disulfide should be more favored in the swiveled conformation, although the ease of disulfide formation depends on multiple factors including Cys rotational freedom and chemical environment. Upon oxidation of the CTR-containing hisPEC with cystamine, we observed formation of both disulfides but an increase in the 258i-843 (swiveled) disulfide relative to the 258i-1045i (closed) disulfide (Figure 5C). The redox-potential-dependent shift toward the swiveled disulfide likely represents destabilization of the closed clamp due to the formation of mixed disulfides that sterically disfavor the closed clamp. Consistent with clamp swiveling in the hisPEC, we observed a significantly higher ratio of swiveled/closed disulfides in the hisPEC than in the EC (Figure 5D).
Figure 5. The PH induced modest clamp swiveling to increase pause lifetime.
(A) Disulfides used to probe changes in clamp position in the hisPEC shown on an EC cartoon, the EC exit channel (6ALF), and the hisPEC exit channel (6ASX). A single Cys insertion between β′257 and 258 in the lid (258i) was paired with different flap Cys substitutions (1044, 1045, and 1045i) and with two flap Cys residues (1045i and 843) in the Cys-triplet reporter (CTR). The distances between Cα atoms of the different Cys pairs are shown in colored lines, and the distance between 258i and flap Cys residues in the open clamp conformation of 4GZY (Weixlbaumer et al., 2013) are shown as grey dotted lines.
(B) The alternative disulfide bonds in the CTR can be distinguished by non-reducing SDS-PAGE.
C. Redox titration of the CTR on a hisPEC scaffold using DTT-cystamine mixtures (see Methods).
(D) The ratio of the swiveled (258i-843) disulfide to the closed (258i-1045i) disulfide in CTR RNAP on the hisPEC and EC scaffolds.
(E) The effect of disulfide bonds on RNA duplex-stimulation of pausing. An exit channel duplex formed with an 8mer antisense RNA is known to mimic the effect of the his PH (Dp, exit-channel duplex).
To determine the type of clamp movement (i.e., swiveling vs. opening) required for the PH to increase pause lifetime, we compared the effects of disulfides between 258i and 1044, 1045, or 1045i (Figure 5A); the 258i-1045 or 258i-1045i disulfides should accommodate the small movements associated with clamp swiveling, but not the larger clamp opening observed in the Tth ePEC crystal structure (Weixlbaumer et al., 2013; Cα-Cα ~15 Å for 1045; Figure 5A). We used an 8mer antisense RNA known to mimic the effect of the PH to enable direct comparisons of the same disulfide cross-linked PECs (Figure 5E; Hein et al., 2014). The 258i-1044 disulfide inhibited the PH effect, as seen previously (Hein et al., 2014). Strikingly, although multiphasic kinetics precluded precise quantitation, the 258i-1045 disulfide accommodated a nearly complete PH effect and the 258i-1045i disulfide accommodated most of the PH effect (Figure 5E). We conclude that only a small shift in clamp position, consistent with rotation of the swivel module by ~3° but not full clamp opening, is sufficient to enable PH function.
Paths of positive charge aid formation of an RNA duplex in the RNA exit channel
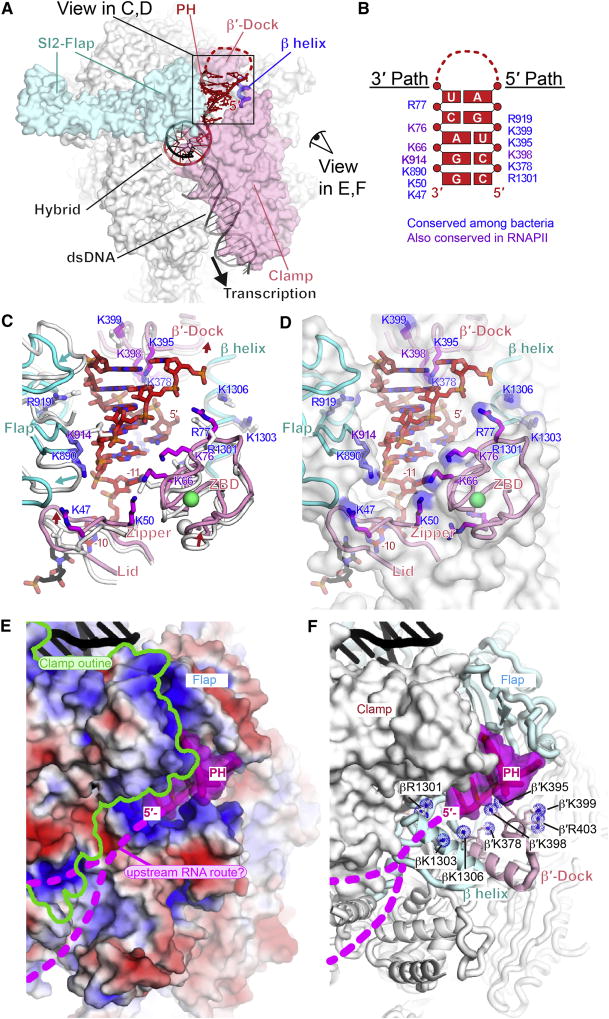
Studies of complementary RNA and DNA oligo pairing to nascent RNA in the sterically constrained exit channel suggest that bacterial RNAP may guide formation of nascent RNA structures (Hein et al., 2014; Kolb et al., 2014). Consistent with RNAP-guided structure formation, the hisPEC cryo-EM structure revealed a largely preformed, positively charged surface in the RNAP RNA exit channel that is complementary to the PH A-form helix and a route for RNA upstream from the PH that may facilitate its formation (Figure 6).
Figure 6. his PH-RNAP exit channel interaction.
(A) Interactions of PH in RNA exit channel. Flap, β′-dock, β helix, and clamp cradle the PH. Views in panels C-F are indicated.
(B) Map of side chains that form 3′ and 5′ paths complementary to PH. Purple, conserved among all RNAPs; blue, conserved only among bacterial RNAPs (Table S4).
(C) Movements in the RNA exit channel upon PH formation and key PH-interacting residues. The positions of exit channel modules (flap, lid, zipper, ZBD, β′-dock and β helix) in the hisPEC are shown as colored cartoons (cyan, β; pink, β′) and their corresponding positions in the EC are shown as white cartoons. The last bp of the RNA-DNA hybrid and the spacer −11 nt between the hybrid and the PH are shown. Positively charged residues that interact with the PH are shown as sticks (blue, β subunit; magenta, β′ subunit) with labels color-coded as in panel B.
(D) Same as panel C, except that the EC is not shown and the exit channel is shown as a semi-transparent, white surface with blue patches of positive charge.
(E) View from outside RNA exit channel and underside of the clamp with the surface of RNAP colored by electrostatic charge (−5 red to +5 blue). Upstream DNA is black. The PH is shown as a cartoon with a semi-transparent magenta surface and possible routes of ssRNA upstream from the PH are shown as dotted magenta lines (called “groove 1” in Cramer et al., 2001).
(F) Same view as panel E, but with a solid white RNAP surface except for the flap, β′-dock, and β-helix shown as cartoons and positive charges shown as blue dotted spheres.
Also see Table S4 and Figure S7.
Comparison of the positions of the flap, lid, clamp, and β′ dock that form the RNA exit channel in the hisPEC vs. EC revealed relatively minor movements upon formation of the PH (Figure 6A, C). The largest movement was a shift of the flap away from the exit channel accompanied by smaller shifts of the β′ N-terminal ZBD, zipper, and lid in the clamp parallel to the exit channel and away from the RNA-DNA hybrid (Figure 3B, 6C).
The positions of positive charge in the RNA exit channel roughly define two paths that appear ready-made to accommodate an A-form RNA duplex (Figures 6B, C, D). Phosphates on the 3′ arm of the PH duplex align with a path of positive charge on the clamp-side of the RNA exit channel generated by Arg and Lys residues in the ZBD, zipper, and flap. The 5′ arm of the PH duplex threads over a path of positive charge at the mouth of the exit channel generated by Lys and Arg residues in the flap, a β′-dock helix, and a β helix (Figure 6B, C, D). These positively charged paths create a template onto which the A-form helix of the PH stem can assemble in a complementary manner. Only the PH duplex lies within the exit channel; the PH loop may interact with the flap-tip helix (FTH), but both the loop and FTH are unresolved in the hisPEC cryo-EM structure.
The positively charged path in RNAP aligned with the PH 5′ arm guides the RNA chain out of the RNAP exit channel across the dock and β helices (Figures 6C, D). The location of the 5′-most PH nucleotide was close to that observed for the −14 nucleotide of single-stranded RNA exiting an EC crystal structure (Vassylyev et al., 2007a). From this point, single-stranded RNA upstream from the PH may traverse across the outside of the clamp domain, where additional positive charge could guide single-stranded RNA when it reverses direction during PH formation (upstream RNA route; Figure 6E, F). We conclude that charge-complementarity both inside and outside the RNA exit channel may explain how RNAP aids the formation of nascent RNA secondary structure despite the strong steric constraint of the narrow channel within which nascent RNA structures form.
A subpopulation of particles lacking an ordered PH may resemble the ePEC
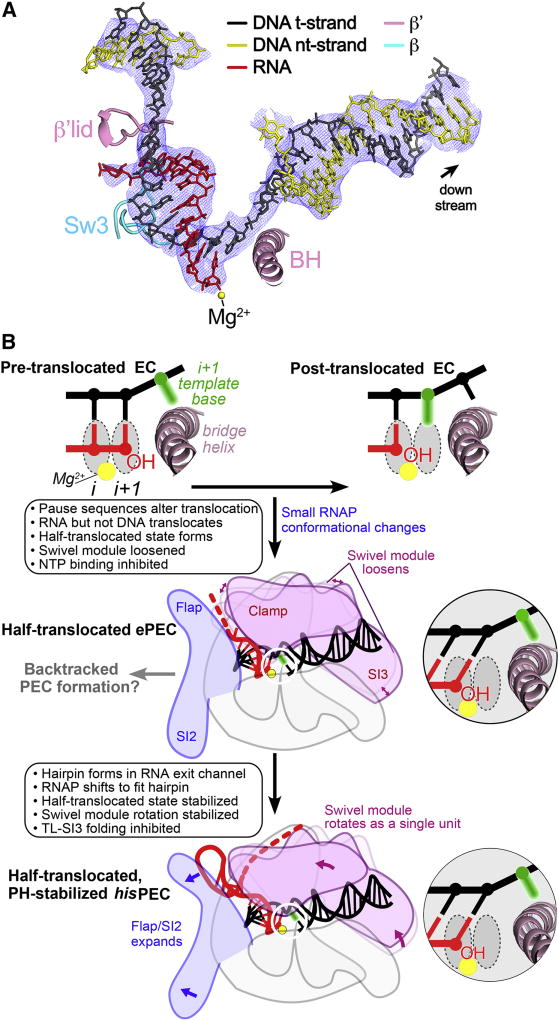
The poor resolution of the flap tip and PH loop in the hisPEC structure (Figure S2G–I) led us to perform a particle classification focused on this region. Although a class with an ordered flap-tip or PH loop did not emerge, a subpopulation of the particles (~12%) gave rise to a reconstruction that lacked density for a PH duplex (Figure S6). The nominal resolution was 5.5 Å (Figure S6B), but local resolution calculations (Cardone et al., 2013) indicated that the inner structural core around the active site and RNA-DNA hybrid was determined to about 4.3 Å resolution. In this population of particles (hisPEC-minus-PH), the PH was likely degraded, as observed previously in crystallization trials using hisPEC scaffolds (Weixlbaumer et al., 2013); however, we cannot exclude the possibility that the RNA was intact but the PH either was not formed or was disordered. Refinement into the 5.5 Å resolution cryo-EM density (Table S1) revealed an EC-like RNAP conformation (i.e., no swivel module rotation; Table S3, Figure S6E, F) but with a hisPEC-like half-translocated nucleic acid scaffold (Figure 7A). This half-translocated hisPEC-minus-PH structure formed on a scaffold with sequences known to form an ePEC (Kyzer et al., 2007; Weixlbaumer et al., 2013), suggesting that the hisPEC-minus-PH structure may correspond to an ePEC that can form on his pause sequences prior to PH formation. Thus, we propose that the sequence-dependent translocation block may be an initial step in pausing that occurs in an ePEC, which may become long-lived upon further RNAP conformational changes (i.e., swivel module rotation) stabilized by the PH (Figure 7B).
Figure 7. The half-translocated state arises in a PH-minus PEC.
(A) Shown is the 5.5 Å resolution (4.3 Å resolution around the active site and RNA-DNA hybrid) cryo-EM density map (blue mesh) with the superimposed model of the hisPEC-minus-PH nucleic acids. Shown for reference are key RNAP structural elements (Sw3, lid, BH) and the RNAP active-site Mg2+-ion (yellow sphere). Like the hisPEC (Figure 2A), the half-translocated RNA:DNA (shown), but not a pre-translocated or fully translocated RNA:DNA, fit the density map.
(B) A mechanistic model of pausing. The half-translocated state arises in the ePEC when RNAP conformation is loosened, making it susceptible to swiveling (Sekine et al., 2015), and is stabilized by global rearrangement of RNAP including swiveling in the PH-stabilized PEC. Insets depict template DNA and RNA near the active site.
See also Table S3 and Figure S6.
DISCUSSION
Understanding both the fundamental mechanism of transcriptional pausing and the interplay between saltatory 5′-to-3′ RNA synthesis and formation of biologically active RNA structures has grown in importance as the central roles of pausing and diverse RNAs in cellular regulation have come into focus (Mayer et al., 2017; Zhang and Landick, 2016). We report here key insights on both topics from a cryo-EM structure of the RNA hairpin-stabilized hisPEC. These insights resolve some questions but raise others, and include (i) identification of a halftranslocated RNA-DNA hybrid that can explain inhibition of substrate binding in PECs (Figure 2); (ii) discovery of a swiveled RNAP conformation that can explain how the PH inhibits TL folding (Figures 3, 4, 5); and (iii) the first view of how RNAP guides RNA structure formation via duplex accommodation in its RNA exit channel (Figure 6).
A translocation block followed by RNAP swiveling may explain elemental and PH-stabilized pausing
Our results suggest mechanistic hypotheses for inhibition of nucleotide addition in the sequentially formed ePEC and PH-stabilized PECs (Figure 7B). Formation of the ePEC is known to depend on multipartite sequences that, in aggregate, trigger entry into an altered, non-backtracked state that resists NTP binding (Larson et al., 2014; Vvedenskaya et al., 2014). Other than inhibition of translocation by GC bp at the upstream edges of the hybrid and downstream DNA, the causal effects of these sequences remain to be elucidated. Our observation of a half-translocated intermediate in the hisPEC-minus-PH structure on sequences known to generate an ePEC with only small changes in RNAP conformation (Table S3, Figure S6E) is consistent with a view that the ePEC may form when complete translocation fails (Imashimizu et al., 2015; Weixlbaumer et al., 2013). A better understanding will require a more complete structural study of ePECs by cryo-EM coupled with precise biochemical dissection of the multipartite pause signal.
The similar half-translocated configuration of the hisPEC and hisPEC-minus-PH structures (putative ePEC structure; Figures 2A, 7A) is consistent with generation of the hisPEC from such an ePEC intermediate. Once formed, the PH appears to stabilize the RNAP in a conformation with a rotated swivel module and other changes relative to the conserved structural core of RNAP (Table S2, Figures 3, 7B). This hisPEC conformation explains inhibition of both substrate binding (incomplete translocation; Figure 2) and TL folding (steric clash of the swiveled SI3; Figure 4 and Nayak et al., 2013). The combined effects of a translocation block induced in the ePEC and stabilized further by these conformational changes in the hisPEC can explain the additive effects of the PH and hybrid, upstream fork junction, and downstream duplex DNA sequences on pause lifetime (Chan et al., 1997). In the unswiveled conformation, thermal fluctuation into the post-translocated register would allow NTP binding and immediate TL folding. In the swiveled conformation, such thermal fluctuation would allow NTP binding, but TL folding would remain inhibited due to swiveling. Thus, the swiveled conformation would give an SI3-dependent increase in pause dwell time, which matches experimental observations. Alternatively, formation of the PH in an ePEC could lock the half-translocated state by pulling the RNA and lid away from the main channel; however, this alternative is inconsistent with the finding that the his pause is lengthened by a factor of 2–3 when an extra nucleotide is added between the hybrid and PH (Toulokhonov et al., 2001), an observation we confirmed using the cryo-EM scaffold (Figure S1E).
In the hisPEC, the swivel module rotates in a plane orthogonal to that observed previously for clamp opening (Figure S4; Darst et al., 2002; Feklistov et al., 2017; Weixlbaumer et al., 2013). Swivel module rotation occurs roughly in the plane of the RNA-DNA hybrid and downstream duplex DNA helical axes (Figure S4) and maintains RNAP-downstream DNA contacts by shifting the downstream DNA position. In contrast, Weixlbaumer et al. (2013) observed loss of downstream DNA contacts accompanying clamp opening by orthogonal rotation in crystal structures of thermus RNAPs bound to incomplete hisPEC scaffolds lacking the PH. Thus, clamp opening may occur in paused complexes under some conditions, but our observation of clamp rotation without opening in a hisPEC cryo-EM structure in the absence of crystal packing forces, combined with our biochemical finding that only a small clamp motion is required for PH function (Figure 5E), indicates that clamp opening is not an obligate feature of transcriptional pausing. It is also possible that the swivel module rotation and clamp domain movement observed in the hisPEC could be relevant to other mechanisms that regulate RNAP, including steps that occur during initiation.
Weixlbaumer et al. (2013) also observed incomplete translocation in the TthRNAP ePEC with RNA and DNA positions similar to those in the hisPEC cryo-EM structure except that the +1 template dC was unpaired but blocked from entering the i+1 site by a bent BH rather than paired to the nt-strand DNA. A similar trapping of +1 dC was observed in amanitin-bound RNAPII ECs without significant clamp or BH movements (Brueckner and Cramer, 2008). Consistent with the idea that inhibition of DNA translocation may be an intrinsic consequence of ePEC sequence interactions with RNAP without requiring significant RNAP conformational changes, we observed formation of the half-translocated state on an ePEC sequence (hisPEC-minus-PH; Figures 7A, S6) without these conformational changes.
Although many aspects of the hisPEC structure are consistent with prior biochemical studies, the location of the RNA 3′ nucleotide is difficult to reconcile with crosslinking of 3′-(4-thio)U to hisPEC TL residues 933–936 (Toulokhonov et al., 2007). In the hisPEC structure, the TL appears trapped ≥19 Å from the 3′-U on the other side of the BH, in particular by interactions of R933 with F545 in fork loop 2, in a “wedged” conformation similar to that observed in the amanitin-bound RNAPII EC, which was also proposed to be a paused state (Brueckner and Cramer, 2008). The observed 3′-thioU crosslinking to the TL and β1086–1106 was rationalized by assuming 3′-U fraying from a pre-translocated position, but the hisPEC RNA 3′-U is clearly post-translocated (Figure 2). As noted by Toulokhonov et al. (2007), the crosslink could occur in a minor hisPEC population, and a contribution of 4-thioU to its formation cannot be excluded. Fluctuations among hisPEC conformations could also explain why both pre-translocated and post-translocated hisPEC states were detected in this study, but additional studies including time-resolved hisPEC formation may be needed to understand these discrepancies.
RNAP may guide nascent RNA structure formation
Elucidation of the hisPEC PH interaction with RNAP gives the first clear picture of how nascent RNA structures interact with RNAP and confirms that RNA duplexes can form within the RNAP RNA exit channel directly abutting the lid at the 3′-most duplex base pair (Figure 6). Biochemical evidence suggests that RNAP may not stabilize duplexes against melting (Hein et al., 2014), but may aid duplex formation within the sterically constrained exit channel. The hisPEC structure reveals two features that may explain the ease of duplex formation within the exit channel: (i) paths of positive charge in the exit channel that align with duplex phosphates, and (ii) a positively charged route outside the exit channel, corresponding to RNAPII “groove 1” (Cramer et al., 2001), that could guide upstream RNA into the PH during duplex formation (Figure 6). Thus, duplexes longer than the his PH may extend co-transcriptionally by corkscrewing out the exit channel along the paths of positive charge as upstream single-stranded RNA pairs with newly synthesized RNA emerging from the RNA-DNA hybrid. Any role of exit-channel positive charge in nucleating duplexes by loop formation remains to be tested, as experiments to date have employed unlinked antisense RNA to probe duplex formation and stability.
Implications of the hisPEC structure for transcriptional regulation
A key question is whether the mechanistic features of the hisPEC, both for formation of nascent RNA structures within the RNAP RNA exit channel and for transcriptional pausing, extend broadly in the domains of life. The specific effects on inhibition of TL folding mediated by Eco SI3 would not extend to bacterial lineages lacking β′i6/SI3 (e.g., gram-positives, thermus, and actinobacteria; Lane and Darst, 2010), archaea, or eukaryotes. Consistent with this conclusion, the his pause signal does not stimulate pausing by B. subtilis RNAP or mammalian RNAPII (Artsimovitch et al., 2000; Keene et al., 1999). However, this specificity may reflect a co-adaptation of the pause signal and a β′i6/SI3-containing RNAP; hairpin-stimulated pausing does occur in B. subtilis RNAP when the PH is spaced one nt further from the RNA 3′ end than the Eco his PH (Yakhnin et al., 2006). A more extensive exploration of RNA structure effects on pausing across diverse lineages is needed.
At least as importantly, do diverse RNAPs accommodate RNA duplexes in the RNA exit channel in a manner similar to EcoRNAP? The sequences that line the RNA exit channel and form the paths of positive charge seen in the hisPEC are conserved among bacterial RNAPs (Figure 6, Table S4). Thus, it seems highly likely that the bacterial enzymes all accommodate A-form RNA helices in their RNA exit channels and guide RNA into the structure from the outside of the clamp as proposed here. Alignment of a eukaryotic RNAPII EC with the hisPEC (Bernecky et al., 2016), as well as yeast RNAPI, yeast RNAPIII, and archaeal RNAP structures (Hoffmann et al., 2015; Neyer et al., 2016; Wojtas et al., 2012), reveal that the eukaryotic and archaeal enzymes also can accommodate an A-form exit channel duplex and contain a similar distribution of positive charge even though the secondary structures around the mouth of the exit channel are not conserved (Figure S7). Thus, the relationship between transcriptional pausing and the role of RNAP in formation of biologically active RNA structures requires more study.
Finally, the half-translocated nucleic-acid configuration defined here as a candidate for the offline elemental pause intermediate may have broader relevance. It is possible that normal translocation proceeds via a two-step mechanism of rapid RNA translocation followed by a DNA-template translocation, as detected for RNA-dependent RNAP (Shu and Gong, 2016), but that the half-translocated intermediate becomes long-lived only when trapped in the offline ePEC state. The ePEC could then enable backtracking or PH formation (Figure 7B), and could play similar roles in transcriptional pausing across biological kingdoms. The core structure of multisubunit RNAPs in contact with the RNA-DNA scaffold near the active site is conserved (Lane et al., 2010), and a partially translocated intermediate of yeast RNAPII is also proposed to be in a paused state (Brueckner and Cramer, 2008). Important questions now include how elongation factors like NusA, NusG, Spt4/5, Elf, and others may modulate formation of this offline intermediate.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
R. Landick, landick@bact.wisc.edu
METHODS DETAILS
RNAP expression and purification for Cryo-EM
Eco RNAP lacking the αCTDs was prepared as described previously (Twist et al., 2011). Glycerol was added to the purified RNAP to 15% (v/v), and the sample was aliquoted and flash-frozen in liquid nitrogen. The aliquots were stored at −80 °C until use.
hisPEC preparation for Cryo-EM
Synthetic DNA and RNA oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA). The hisPEC RNA was gel-purified before use. The nucleic acids for RNA hairpin-stabilized pause ECs (hisPECs) were dissolved in RNase-free water (Ambion/ThermoFisher Scientific, Waltham, MA) at 0.2–1 mM. Template DNA and RNA were annealed at a 1:1 ratio in a thermocycler (95 °C for 2 min, 75 °C for 2 min, 45 °C for 5 min, followed by a steady cooling to 25 °C at 1 °C/min). The annealed template RNA-DNA hybrid was stored at −80 °C until use. Purified Eco RNAP was buffer-exchanged over a Superose 6 INCREASE (GE Healthcare Life Sciences) column into 20 mM Tris-HCl, pH 8.0, 150 mM potassium glutamate, 5 mM MgCl2, 5 mM DTT. The eluted protein was mixed with template RNA-DNA hybrid at a molar ratio of 1:1.3 and incubated for 15 min at room temperature. Non-template DNA and additional 5 mM MgCl2 was added and incubated for 10 min. The complex was concentrated by centrifugal filtration (EMD Millipore, Billerica, MA) to 4.0–5.5 mg RNAP/ml concentration before grid preparation.
Cryo-EM grid preparation
Before freezing, CHAPSO was added to 8 mM final concentration to the samples. C-flat (Protochips, Morrisville, NC) CF-1.2/1.3 400 mesh gold grids were glow-charged for 15 s prior to the application of 3.5 µl of the complex sample and then plunge-frozen in liquid ethane using a Vitrobot mark IV (FEI, Hillsboro, OR) with 100% chamber humidity at 22 °C.
Cryo-EM data acquisition and processing
The grids were imaged using a 300 keV Titan Krios (FEI) equipped with a K2 Summit direct electron detector (Gatan, Pleasanton, CA). Images were recorded with Serial EM (Mastronarde, 2005) in super-resolution counting mode with a super resolution pixel size of 0.515 Å and a defocus range of 0.8 to 2.4 µm. Data were collected with a dose of 8 electrons/physical pixel/s (1.03 Å pixel size at the specimen). Images were recorded with a 10 s exposure and 0.2 s subframes (50 total frames) to give a total dose of 75.4 electrons/Å2. Dose fractionated subframes were 2 × 2 binned (giving a pixel size of 1.03 Å), aligned and summed using Unblur (Grant and Grigorieff, 2015). The contrast transfer function was estimated for each summed image using CTFFIND4 (Rohou and Grigorieff, 2015). From the summed images, particles were automatically picked in Gautomatch (Zhang, unpublished; see Key Resource Table), manually inspected, and then individually aligned using direct-detector-align_lmbfgs software (Rubinstein and Brubaker, 2015). The aligned particles were subjected to 2D classification in RELION specifying 100 classes (Scheres, 2012), and poorly populated classes were removed, resulting in 369,000 particles. These particles were 3D autorefined in RELION using a map of Eco elongation complex (EMD-8585; Kang et al., 2017), low-pass filtered to 60 Å resolution as an initial 3D template. With this initial model, 3D classifications were performed without alignment. Among the 3D classes, bad classes were excluded, and the rest were combined and subjected to second 3D classification without alignment. From this classification, the best-resolved class was 3D autorefined with solvent flattening, and post-processed in RELION. To resolve heterogeneity around RNA hairpin stem in RNA exit channel, focused 3D classification on the RNA hairpin and the surrounding helices was performed (Figure S2F). A soft map that excluded the PH and nearby protein regions was generated in Chimera and RELION. The mask was used to make a subtracted particle stack in RELION with the filtered map generated in the initial autorefinement. The subtracted particles were 3D classified into six classes without alignment, and one class lacked PH density. The original (unmasked) particles in this class were autorefined and post-processed in RELION, yielding the final reconstruction at 5.5 Å resolution (hisPEC-minus-PH). Local resolution calculation was performed using blocres (Cardone et al., 2013).
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Bacterial and Virus Strains | ||
|
| ||
| E. coli BL21(λDE3)T-H/pEcrpo(HX-)ABCZ | (Twist et al., 2011) | N/A |
|
| ||
| E. coli BL21(λDE3)T-H/pRM1182 | (Kohler et al., 2017) | N/A |
|
| ||
| E. coli BLR λDE3 | Novagen | |
|
| ||
| Chemicals, Peptides, and Recombinant Proteins | ||
|
| ||
| 3-([3-Cholamidopropyl]dimethylammonio)-2-hydroxy-1-propanesulfonate (CHAPSO) | Sigma-Aldrich | Cat# C4695 |
|
| ||
| Cystamine dihydrochloride | Mpbio.com | Cat# ICN10049205 |
|
| ||
| Dithiothreitol (DTT) | Gold Biotechnology | Cat# DTT100 |
|
| ||
| [α-32P]CTP | Perkin Elmer | Cat# BLU008H |
|
| ||
| [γ-32P]ATP | Perkin Elmer | Cat# BLU002Z |
|
| ||
| Potassium glutamate | Sigma-Aldrich | Cat# G1501 |
|
| ||
| Iodoacetamide, SigmaUltra | Sigma-Aldrich | Cat# I1149 |
|
| ||
| Polyethyleneimine, ~M.W.60,000, 50% wt.% aqueous solution, branched, Acros Organics 178572500 | Fisher Scientific | Cat# AC178572500 |
|
| ||
| Bio-Rex 70 cation exchange resin, analytical grade, 100–200 mesh | Bio-Rad Cat# | 1425842 |
|
| ||
| HiLoad 26/600 Superdex 200 pg | GE Healthcare Life Sciences | Cat# 28989336 |
|
| ||
| HiTrap IMAC HP | GE Healthcare Life Sciences | Cat# 17092003 |
|
| ||
| Superose 6 INCREASE 10/300 GL | GE Healthcare Life Sciences | Cat# 29091596 |
|
| ||
| HiTrap Heparin HP column | GE Healthcare | Cat# 17-0406-01 |
|
| ||
| HisTrap HP column | GE Healthcare | Cat# 17-5247-01 |
|
| ||
| Streptactin 5 ml High Capacity Column | IBA | Cat# 2-1238-001 |
|
| ||
| ΔαCTD E. coli RNAP polymerase (cryo-EM samples) | (Twist et al., 2011) | N/A |
|
| ||
| ΔαCTD E. coli RNAP polymerase (transcription assays) | (Kohler et al., 2017) | N/A |
|
| ||
| Critical Commercial Assays | ||
|
| ||
| RNaseAlert QC system, Ambion | ThermoFisher Scientific | Cat# AM1966 |
|
| ||
| Deposited Data | ||
|
| ||
| Coordinates of E. coli RNAP | (Bae et al., 2013) | PDB: 4LJZ |
|
| ||
| Coordinates of E. coli ternary elongation complex | (Kang et al., 2017) | PDB: 6ALF |
|
| ||
| Coordinates of E. coli ternary elongation complex with HK022 Nun | (Kang et al., 2017) | PDB: 6ALG |
|
| ||
| Coordinates of E. coli his paused elongation complex (PEC) | This paper | PDB: 6ASX |
|
| ||
| Coordinates of E. coli hisPEC-minus-PH | This paper | PDB: 6BJS |
|
| ||
| Coordinates of Thermus Thermophilus hisPEC-minus-PH (downstream fork-junction scaffold) | (Weixlbaumer et al., 2013) | PDB: 4GZY |
|
| ||
| Coordinates of bovine RNA polymerase II | (Bernecky et al., 2016) | PDB: 5FLM |
|
| ||
| Coordinates of S. cerevisiae RNA polymerase I | (Neyer et al., 2016) | PDB: 5M3F |
|
| ||
| Coordinates of S. cerevisiae RNA polymerase III | (Hoffmann et al., 2015) | PDB: 5FJ8 |
|
| ||
| Coordinates of S. shibatae RNA polymerase | (Wojtas et al., 2012) | PDB: 4V8S |
|
| ||
| Oligonucleotides | ||
|
| ||
| hisPEC non-template DNA (6ASX) GCGTCCTATCGATCTTCGGAAGAGATTCAGAG | IDT | Lab stock #10924 |
|
| ||
| hisPEC template DNA (6ASX) CTCTGAATCTCTTCCAGCACACATCAGGACGC | IDT | Lab stock #10919 |
|
| ||
| hisPEC RNA (6ASX) CCUGACUAGUCUUUCAGGCGAUGUGUGCU | IDT | Lab stock #4865 |
|
| ||
| hisPEC C28 RNA CCUGACUAGUCUUUCAGGCGAUGUGUGC | IDT | Lab stock #4866 |
|
| ||
| hisPEC G27 RNA CCUGACUAGUCUUUCAGGCGAUGUGUG | IDT | Lab stock #4867 |
|
| ||
| hisPEC 2 nt spacer RNA CCUGACUAGUCUUUCAGGUCGAUGUGUGCU | IDT | Lab stock #11754 |
|
| ||
| his ePEC RNA UCAUCCGGCGAUGUGUGCU | IDT | Lab stock #7418 |
|
| ||
| his ePEC C18 RNA UCAUCCGGCGAUGUGUGC | IDT | Lab stock #12192 |
|
| ||
| his ePEC G17 RNA UCAUCCGGCGAUGUGUG | IDT | Lab stock #6593 |
|
| ||
| his 8-nt antisense RNA CCGGAUGA | IDT | Lab stock #6598 |
|
| ||
| EC non-template DNA (6ALF) GGGCTACCTCTCCATGACGGCGAATACCC | IDT | Lab stock #12198 |
|
| ||
| EC template DNA (6ALF) GGGTATTCGCCGTGTACCTCTCCTAGCCC | IDT | Lab stock #12199 |
|
| ||
| EC RNA (6ALF) GCAUUCAAAGCGGAGAGGUA | IDT | Lab stock #12197 |
|
| ||
| EC U19 RNA GCAUUCAAAGCGGAGAGGU | IDT | Lab stock #12291 |
|
| ||
| Recombinant DNA | ||
|
| ||
| pACYCDuet-1_Ec_rpoZ | (Bae et al., 2013) | N/A |
|
| ||
| pEcrpo(HX-)ABCZ | Twist et al., 2011 | N/A |
|
| ||
| pRM843 | (Hein et al., 2014) | Lab stock #5143 |
| Wildtype RNAP overexpression plasmid. HMK-Strep tag at rpoC-C-terminus, His10-ppx tag at rpoB N-terminus | ||
|
| ||
| pRM918 | (Hein et al., 2014) | Lab stock #5218 |
| RNAP overexpression plasmid. β′258iC. β1044C. HMK-Strep tag at rpoC-C-terminus, His10-ppx tag at rpoB N-terminus | ||
|
| ||
| pRM902 | This work | Lab stock #5202 |
| RNAP overexpression plasmid. β′258iC. β1045C HMK-Strep tag at rpoC-C-terminus, His10-ppx tag at rpoB N-terminus | ||
|
| ||
| pRM903 | This work | Lab stock #5203 |
| RNAP overexpression plasmid. β′258iC. β1045iC. HMK-Strep tag at rpoC-C-terminus, His10-ppx tag at rpoB N-terminus | ||
|
| ||
| pRM950 | This work | Lab stock #5250 |
| RNAP overexpression plasmid. β′258iC. β1045iC 843C. HMK-Strep tag at rpoC-C-terminus, His10-ppx tag at rpoB N-terminus | ||
|
| ||
| Software and Algorithms | ||
|
| ||
| Blocres | (Cardone et al., 2013) | https://lsbr.niams.nih.gov/bsoft/programs/blocres.html |
|
| ||
| Chimera | (Pettersen et al., 2004) | https://www.cgl.ucsf.edu/chimera |
|
| ||
| COOT | (Emsley and Cowtan, 2004) | https://www2.mrclmb.cam.ac.uk/personal/pemsley/coot |
|
| ||
| CTFFIND4 | (Rohou and Grigorieff, 2015) | http://grigoriefflab.janelia.org/ctffind4 |
|
| ||
| Direct-detector-align_lmbfgs | (Rubinstein and Brubaker, 2015) | https://sites.google.com/site/rubinsteingroup/direct-detectoralign_lmbfgs |
|
| ||
| Gautomatch | Zhang, unpublished | http://www.mrclmb.cam.ac.uk/kzhang/Gautomatch/ |
|
| ||
| MolProbity | (Chen et al., 2010) | http://molprobity.biochem.duke.edu |
|
| ||
| PHENIX | (Adams et al., 2010) | https://www.phenixonline.org/documentation/index.html |
|
| ||
| Pymol | Schrödinger, LLC | http://www.pymol.org |
|
| ||
| RELION | (Scheres, 2012) | http://www2.mrclmb.cam.ac.uk/relion |
|
| ||
| Serial EM | (Mastronarde, 2005) | http://bio3d.colorado.edu/SerialEM |
|
| ||
| Unblur & Summovie | (Grant and Grigorieff, 2015) | http://grigoriefflab.janelia.org/unblur |
|
| ||
| Other | ||
|
| ||
| C-flat CF-1.2/1.3 400 mesh gold grids | Electron Microscopy Sciences | CF413-100-Au |
Model building, refinement and validation
To build initial models, Eco core enzyme (PDB ID 4LJZ with σ70 removed; Bae et al., 2013) and nucleic acids (PDB ID 6ALF; Kang et al., 2017) were fitted into the electron density maps using Chimera (Pettersen et al., 2004). These initial models were real-space refined against the working half map using Phenix (Adams et al., 2010). In the refinement, domains in the core and nucleic acids were rigid-body refined, then subsequently refined with secondary structure restraints. At the end of refinement, Fourier shell correlations (FSC) were calculated between the refined model and the half map used for refinement (work), the other half map (free), and the full map to assess over-fitting (Figure S2E).
RNAP expression and purification for in vitro biochemical experiments
Wild-type and variant RNAPs were overexpressed from plasmids (Key Resources Table). Eco RNAP lacking the αCTDs was prepared as described previously (Kohler et al., 2017). pRM843 and mutant derivatives are T7 RNAP-based overexpression plasmids containing Eco rpoA, rpoZ, rpoB and rpoC and yields core RNAP with a His10 tag on the N terminus of β and protein kinase A (PKA) and a strep tag on the C terminus of β′. RNAPs were purified from Eco BLR λDE3 cells transformed with the appropriate plasmids as previously described (Hein et al., 2014). Briefly, after lysis and PEI precipitation and extraction, protein was precipitated with ammonium sulfate. After re-suspension, the protein solution was applied to a HisTrap column, washed, and eluted with a gradient of increasing imidazole concentration. Protein-containing fractions were pooled and dialyzed into 100 mM Tris-HCl, pH 7.9, 150 mM NaCl, 0.1 mM EDTA, and 5 mM β-mercaptoethanol for 4 h at 4 °C, and then loaded onto a StrepTactin column. After washing, the protein was step-eluted with the same buffer containing 2.5 mM D-desthiobiotin. Fractions with RNAP were identified, pooled, and loaded onto a HiTrap Heparin HP column. After elution, the protein was dialyzed into RNAP storage buffer (20 mM Tris-HCl, pH 8.0, 250 mM NaCl, 20 µM ZnCl2, 1 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, and 20% glycerol) and stored as aliquots at −80 °C until use.
In vitro transcription assays
Nucleic acid scaffolds used to reconstitute RNA hairpin-stabilized pause ECs (hisPECs) were assembled in 20 mM Tris-HCl, pH 8.0, 150 potassium glutamate, 5 mM MgCl2, and 5 mM DTT in a thermocycler, as described for the cryo-EM scaffold preparation. Oligonucleotides for these experiments (Key Resources Table) were obtained from Integrated DNA Technologies (Coralville, IA) and gel-purified before use. The scaffolds contained 5 µM RNA and 10 µM template DNA. RNAs containing the hairpin were 5′-labeled prior to scaffold assembly, in a T4 PNK-catalyzed reaction with [γ-32P]ATP. The hisPECs were formed by incubating 1.5 µM RNAP (wild-type or αCTD RNAP as appropriate) with 0.5 µM (based on RNA concentration) scaffold for 15 min at room temperature, followed by an additional 10 min at room temperature in the presence of 2.5 µM non-template DNA. These reconstitution conditions were designed to mirror those used during cryo-EM sample preparation. CHAPSO (final concentration 8 mM, as in cryo-EM buffer) and heparin (final 0.1 mg/ml) were added to the reconstituted complexes, followed by incubation for 3 min at 37 °C. The ECs formed 1-nt upstream of the pause (C28) were then walked to the pause (U29) in a reaction with 100 µM UTP for 3 min at 37 °C. Kinetics of hisPEC escape from the pause was measured by reacting hisPECs with 10 µM GTP and 100 µM ATP at 37 °C. Reaction samples were removed at various time points and quenched with an equal volume of 2× urea stop buffer (8 M urea, 50 mM EDTA, 90 mM Tris-borate buffer, pH 8.3, 0.02% each bromophenol blue and xylene cyanol). All active PECs were chased out of the pause with 1 mM GTP for 5 min at 37 °C. RNAs in each quenched reaction sample were separated on a 15% urea-PAGE gel. The gel was exposed to a PhosphorImager screen, and the screen was scanned using Typhoon PhosphorImager software and quantified in ImageQuant (Figure 1B). The fraction of RNA at the position of the pause over time was fitted to a single-exponential in KaleidaGraph, to extract pause efficiencies (amplitude) and rate constants of pause escape for each RNAP (Figure S1A–F). We observed that the hisPEC escaped the pause site 10 times slower than the ePEC (Figure S1A); that CHAPSO modestly reduced pause lifetime (by a factor of ~2; Figure S1B); that neither the cryoEM scaffold nor the deletion of αCTD altered pause lifetimes (Figure S1C); that hisPECs formed by nucleotide addition (C28→U29; Figure S1D) were kinetically indistinguishable from those formed by direct reconstitution (U29, Figure S1C; see also Kyzer et al., 2007); and that a 1-nt extension of the spacer between the PH and the RNA-DNA hybrid increased pause lifetime (~2-fold, Figure S1E).
Cys Triplet Reporter (CTR) assays (Figures 5C and 5D)
Nucleic-acid scaffolds used to reconstitute hisPEC or ECs for Cys triplet reporter cross-linking assays were assembled on purified DNA and RNA oligonucleotides as described previously (Hein et al., 2014; Kyzer et al., 2007). Briefly, 10 µM RNA, 12 µM template DNA, and 15 µM non-template DNA (Resource Table) were annealed in reconstitution buffer (RB; 20 mM Tris-HCl, pH 7.9, 20 mM NaCl, and 0.1 mM EDTA). To assemble complexes, scaffold (2 µM) was mixed with limiting CTR RNAP (1 µM; CTR RNAP: β′1045iC 258iC, β843C) in transcription buffer (50 mM Tris-HCl, pH 7.9, 20 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, 5% glycerol, and 2.5 µg of acetylated bovine serum albumin/ml) and added to mixtures of cystamine and DTT to generate redox potentials that ranged from −0.314 to −0.424. Complexes were incubated for 60 min at room temperature and then were quenched with the addition of iodoacetamide to 15 mM. The formation of cysteine-pair cross-links was then evaluated by non-reducing SDS-PAGE (4–15% gradient Phastgel; GE) as described previously (Nayak et al., 2013). Gels were stained with Coomassie Blue and imaged with a CCD camera. The fraction cross-linked was quantified with ImageJ software. The experimental error was determined as the standard deviation of measurements from three or more independent replicates.
Cys-pair RNAP pause assays (Figure 5E)
Nucleic-acid scaffold used to reconstitute elemental paused ECs (ePECs; minus-PH) for Cys-pair crosslinking experiments was assembled as previously described (Hein et al., 2014). Briefly, PAGE-purified G17 RNA (2 nt upstream of the pause site, 10 µM), template DNA (15 µM), and non-template DNA (20 µM) were annealed in reconstitution buffer (RB; 10 mM Tris-HCl, pH 7.9, 40 mM KCl, and 5 mM MgCl2). The sequences of the nucleic acids and their corresponding stock numbers are listed in the Key Resources Table. The G17 ePECs containing limiting Cys-pair reporter RNAP (1 µM) were reconstituted on this scaffold (4 µM, based on RNA concentration) in Elongation Buffer (EB; 25 mM HEPES-KOH, pH 8.0, 130 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, DTT, 0.15 mM EDTA, 5% glycerol, and 25 µg of acetylated bovine serum albumin/ml), for 15 min at 37 °C. Wild-type RNAP was tested as a control side-by-side with Cys-pair reporter RNAPs. Crosslinking of 1 µM ePECs was performed in the presence of 1 mM cystamine as the oxidant and 0.8 mM DTT, for 15 min at 37 °C. An aliquot of the crosslinking reaction was quenched with 15 mM iodoacetamide (final concentration) and analyzed by non-reducing SDS-PAGE for the formation of the crosslink. PAGE separation of the crosslinked from non-crosslinked β-β′ RNAP subunits was performed on an 8% Bolt Bis-Tris gel (ThermoFisher Scientific), at 200 V for 1 hour. The gel was Coomassie-stained, destained, and visualized with a CCD camera to quantify the fraction of the RNAP with a disulfide bond crosslink.
The remaining crosslinked ePECs were diluted to 0.2 µM with EB (without DTT, for crosslinked samples) and incubated with heparin (0.1 mg/ml final) for 3 min at 37 °C. The ePECs were then radiolabeled by incorporation of trace [α-32P]CMP and 2 µM unlabeled CMP for 1 min at 37 °C, and walked to the position of the pause, U19, in the reaction with 100 µM UTP for an additional 3 min at 37 °C. The resulting U19 complexes were incubated without and with 1 µM 8mer antisense RNA (asRNA), to form an RNA-duplex mimic of the hisPEC hairpin. PECs were assayed for pause-escape kinetics by addition of 10 µM GTP in EB (without DTT, for crosslinked samples) at 37 °C. Reaction samples were removed at various time points and quenched with an equal volume of 2× urea stop buffer. All active PECs were chased out of the pause with 500 µM GTP for 5 min at 37 °C. RNAs in each quenched reaction. RNAs in each quenched reaction sample were separated on a 15% urea-PAGE gel. The gel was visualized and quantified as described for in vitro transcription assays.
QUANTIFICATION AND STATISTICAL ANALYSIS
To measure pausing kinetics (Figures 1C, 5E, and S1), RNAs were quantified after electrophoretic separation in polyacrylamide gels using a Typhoon PhosphorImager and ImageQuant software (GE Healthcare Life Sciences). Mean values and standard deviations from at least three independent experiments were calculated to create plots of RNA levels versus time, unless otherwise noted in the figure legends. The resulting data were used to calculate pause lifetimes, where reported, by non-linear regression of data from each experiment, and then reported as the mean and standard deviation of the calculated lifetimes. Measurements of disulfide crosslinking (Figure 5C and 5D) were obtained from Coomassie Blue R-stained polyacrylamide gels using a Protein Simple CCD camera and software, and then quantified using Imagequant. Mean values and standard deviations from at least three independent experiments were calculated to create plots of crosslink levels versus redox potential. The local resolution of the cryo-EM maps was estimated using blocres (Cardone et al., 2013) with the following parameters: (hisPEC, Figures 1D and S2G–I: box size 15, verbose 7, cutoff 0.5; hisPEC-minus-PH, Figure 7A: box size 22, verbose 7, cutoff 0.5). The quantification and statistical analyses for model refinement and validation were integral parts of the software algorithms used [MolProbity: (Chen et al., 2010); PHENIX: (Adams et al., 2010)].
DATA AND SOFTWARE AVAILABILITY
The cryoEM density maps have been deposited in the EM Data Bank with accession codes EMD-7002 (hisPEC) and EMD-7103 (hisPEC-minus-PH). Atomic coordinates have been deposited in the Protein Data Bank with accession codes 6ASX (hisPEC) and 6BJS (hisPEC-minus-PH).
Supplementary Material
In the first part of the movie, the RNA exit channel is widened to accommodate the pause RNA hairpin (PDB 6ASX) in comparison to the exit channel in an EC structure (PDB 6ALF). The RNA hairpin loop structure in the hisPEC was modeled using MCFold (http://www.major.iric.ca/MC-Pipeline/) because it was disordered in the cryo-EM density map. The RNA hairpin loop is connected to the pause hairpin stem that is positioned as observed in the cryo-EM map. The single-stranded RNA within and outside the EC exit channel is modeled arbitrarily (also disordered in the cryo-EM map). In the second part of the movie, the components of the swivel module (clamp, dock, β′ C-term, shelf, jaw, and SI3) are depicted and the rotation of the swivel module between the conformations observed in the hisPEC and EC structures is shown.
Movement of SI3 (magenta) between its position in EC (PDB 6ALF; unfolded trigger loop) and its position in an initiating transcription complex with bound NTP (PDB 4YLN; folded trigger loop) is shown. To aid comparison, sigma factor was removed from the initiating complex and a complete RNA:DNA hybrid was added. The upstream DNA duplex was removed from both models. When the trigger loop (TL) is unfolded in the EC, SI3 is in the OUT position (SI3 is a 188-aa insertion into the TL). When the TL folds into the trigger helices (TH) in the NTP-bound complex, SI3 moves toward the bridge helix into the IN position because the unfolded polypeptide linker segments between the TL and SI3 are shortened due to TH formation. In the hisPEC, however, rotation of the swivel module, which includes SI3, repositions SI3 and the shelf in ways that would generate steric clash if SI3 were to shift to the IN position (yellow stars; upstream DNA was also removed from this hisPEC model). The apparent rotation of downstream DNA during swivel rotation is an artifact of the morphing software used to generate the movie; the downstream DNA does not rotate, rather its position shifts slightly during swivel module rotation.
HIGHLIGHTS.
Cryo-EM structures of hairpin-stabilized and elemental paused RNAP
Transcriptional pausing can be explained by uncoupling of RNA and DNA translocation
Positive charges in the RNA exit channel guide formation of nascent RNA hairpins
Hairpin-stabilized allosteric changes inhibit trigger-loop folding and pause RNAP
Acknowledgments
We thank K. Uryu and D. Acehan at The Rockefeller University Electron Microscopy Resource Center for help with EM sample preparation, M. Ebrahim and J. Sotiris at The Rockefeller University Cryo-EM Resource Center for help with data collection, X. Guo and A. Weixlbaumer for helpful discussion and sharing results prior to publication, and members of our research groups for helpful comments on the manuscript. This work was supported by Burroughs Wellcome Grant 1016946 to T.V.M. and by NIH grants R01 GM38660 to R.L. and R35 GM118130 to S.A.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization, S.A.D. and R.L.; Investigation, J.Y.K., T.V.M., M.J.B., R.A.M; Analysis, J.Y.K., T.V.M., R.A.M., R.L., S.A.D.; Writing, J.Y.K., R.L., S.A.D.; Supervision, R.L., S.A.D.; Funding Acquisition, R.L., S.A.D.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Artsimovitch I, Svetlov V, Anthony L, Burgess RR, Landick R. RNA polymerases from Bacillus subtilis and Escherichia coli differ in recognition of regulatory signals in vitro. J Bacteriol. 2000;182:6027–6035. doi: 10.1128/jb.182.21.6027-6035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Svetlov V, Murakami K, Landick R. Co-overexpression of E. coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J Biol Chem. 2003;278:12344–12355. doi: 10.1074/jbc.M211214200. [DOI] [PubMed] [Google Scholar]
- Bae B, Davis E, Brown D, Campbell EA, Wigneshweraraj S, Darst SA. Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma70 domain 1.1. Proc Natl Acad Sci U S A. 2013;110:19772–19777. doi: 10.1073/pnas.1314576110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernecky C, Herzog F, Baumeister W, Plitzko JM, Cramer P. Structure of transcribing mammalian RNA polymerase II. Nature. 2016;529:551–554. doi: 10.1038/nature16482. [DOI] [PubMed] [Google Scholar]
- Brueckner F, Cramer P. Structural basis of transcription inhibition by alphaamanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol. 2008;15:811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- Cardone G, Heymann JB, Steven AC. One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J Struct Biol. 2013;184:226–236. doi: 10.1016/j.jsb.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CL, Landick R. The Salmonella typhimurium his operon leader region contains an RNA hairpin-dependent transcription pause site. Mechanistic implications of the effect on pausing of altered RNA hairpins. J Biol Chem. 1989;264:20796–20804. [PubMed] [Google Scholar]
- Chan CL, Landick R. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J Mol Biol. 1993;233:25–42. doi: 10.1006/jmbi.1993.1482. [DOI] [PubMed] [Google Scholar]
- Chan CL, Wang D, Landick R. Multiple interactions stabilize a single paused transcription intermediate in which hairpin to 3' end spacing distinguishes pause and termination pathways. J Mol Biol. 1997;268:54–68. doi: 10.1006/jmbi.1997.0935. [DOI] [PubMed] [Google Scholar]
- Chen J, Wassarman K, Feng S, Leon K, Feklistov A, Winkelman J, Li Z, Walz T, Campbell E, Darst S. RNA mimics B-form DNA to regulate Escherichia coli RNA polymerase. Mol Cell. 2017 doi: 10.1016/j.molcel.2017.09.006. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- Cheung AC, Sainsbury S, Cramer P. Structural basis of initial RNA polymerase II transcription. EMBO J. 2011;30:4755–4763. doi: 10.1038/emboj.2011.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T, Frazier M, Joyce A, Cho B, Knight E, Lewis N, Landick R, Palsson B. RNA polymerase mutants found through adaptive evolution re-program Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci U S A. 2010;107:20500–20505. doi: 10.1073/pnas.0911253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Bushnell D, Kornberg R. Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- Darst SA, Opalka N, Chacon P, Polyakov A, Richter C, Zhang G, Wriggers W. Conformational flexibility of bacterial RNA polymerase. Proc Natl Acad Sci U S A. 2002;99:4296–4301. doi: 10.1073/pnas.052054099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Feklistov A, Bae B, Hauver J, Lass-Napiorkowska A, Kalesse M, Glaus F, Altmann KH, Heyduk T, Landick R, Darst SA. RNA polymerase motions during promoter melting. Science. 2017;356:863–866. doi: 10.1126/science.aam7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. Elife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha KS, Toulokhonov I, Vassylyev DG, Landick R. The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. J Mol Biol. 2010;401:708–725. doi: 10.1016/j.jmb.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein PP, Kolb KE, Windgassen T, Bellecourt MJ, Darst SA, Mooney RA, Landick R. RNA polymerase pausing and nascent-RNA structure formation are linked through clamp-domain movement. Nat Struct Mol Biol. 2014;21:794–802. doi: 10.1038/nsmb.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann NA, Jakobi AJ, Moreno-Morcillo M, Glatt S, Kosinski J, Hagen WJ, Sachse C, Muller CW. Molecular structures of unbound and transcribing RNA polymerase III. Nature. 2015;528:231–236. doi: 10.1038/nature16143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollands K, Sevostiyanova A, Groisman EA. Unusually long-lived pause required for regulation of a Rho-dependent transcription terminator. Proc Natl Acad Sci U S A. 2014;111:E1999–2007. doi: 10.1073/pnas.1319193111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson BP, Quispe J, Lara-Gonzalez S, Kim Y, Berman HM, Arnold E, Ebright RH, Lawson CL. Three-dimensional EM structure of an intact activator-dependent transcription initiation complex. Proc Natl Acad Sci U S A. 2009;106:19830–19835. doi: 10.1073/pnas.0908782106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imashimizu M, Takahashi H, Oshima T, McIntosh C, Bubunenko M, Court DL, Kashlev M. Visualizing translocation dynamics and nascent transcript errors in paused RNA polymerases in vivo. Genome Biol. 2015;16:98. doi: 10.1186/s13059-015-0666-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Olinares PD, Chen J, Campbell EA, Mustaev A, Chait BT, Gottesman ME, Darst SA. Structural basis of transcription arrest by coliphage HK022 nun in an Escherichia coli RNA polymerase elongation complex. Elife. 2017;6:e25478. doi: 10.7554/eLife.25478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene RG, Mueller A, Landick R, London L. Transcriptional pause, arrest and termination sites for RNA polymerase II in mammalian N- and c-myc genes. Nucleic Acids Res. 1999;27:3173–3182. doi: 10.1093/nar/27.15.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler R, Mooney RA, Mills DJ, Landick R, Cramer P. Architecture of a transcribing-translating expressome. Science. 2017;356:194–197. doi: 10.1126/science.aal3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb KE, Hein PP, Landick R. Antisense oligonucleotide-stimulated transcriptional pausing reveals RNA exit channel specificity of RNA polymerase and mechanistic contributions of NusA and RfaH. J Biol Chem. 2014;289:1151–1163. doi: 10.1074/jbc.M113.521393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyzer S, Ha KS, Landick R, Palangat M. Direct versus limited-step reconstitution reveals key features of an RNA hairpin-stabilized paused transcription complex. J Biol Chem. 2007;282:19020–19028. doi: 10.1074/jbc.M701483200. [DOI] [PubMed] [Google Scholar]
- Landick R, Yanofsky C. Stability of an RNA secondary structure affectsin vitrotranscription pausing in the trp operon leader region. J Biol Chem. 1984;259:11550–11555. [PubMed] [Google Scholar]
- Lane WJ, Darst SA. Molecular evolution of multisubunit RNA polymerases: sequence analysis. J Mol Biol. 2010;395:671–685. doi: 10.1016/j.jmb.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, Block SM, Greenleaf WJ, Landick R, Weissman JS. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner E, Ingargiola A, Lee JJ, Borukhov S, Michalet X, Weiss S. Different types of pausing modes during transcription initiation. Transcription. 2017;8:242–253. doi: 10.1080/21541264.2017.1308853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Mayer A, Landry HM, Churchman LS. Pause & go: from the discovery of RNA polymerase pausing to its functional implications. Curr Opin Cell Biol. 2017;46:72–80. doi: 10.1016/j.ceb.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak D, Voss M, Windgassen T, Mooney RA, Landick R. Cys-pair reporters detect a constrained trigger loop in a paused RNA polymerase. Mol Cell. 2013;50:882–893. doi: 10.1016/j.molcel.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyer S, Kunz M, Geiss C, Hantsche M, Hodirnau VV, Seybert A, Engel C, Scheffer MP, Cramer P, Frangakis AS. Structure of RNA polymerase I transcribing ribosomal DNA genes. Nature. 2016;540:607–610. doi: 10.1038/nature20561. [DOI] [PubMed] [Google Scholar]
- Opalka N, Brown J, Lane WJ, Twist KA, Landick R, Asturias FJ, Darst SA. Complete structural model of Escherichia coli RNA polymerase from a hybrid approach. PLoS Biol. 2010;8:e1000483. doi: 10.1371/journal.pbio.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T, Artsimovitch I, Fang XW, Landick R, Sosnick TR. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc Natl Acad Sci U S A. 1999;96:9545–9550. doi: 10.1073/pnas.96.17.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Richardson JS, Schneider B, Murray LW, Kapral GJ, Immormino RM, Headd JJ, Richardson DC, Ham D, Hershkovits E, Williams LD, et al. RNA backbone: consensus all-angle conformers and modular string nomenclature (an RNA Ontology Consortium contribution) RNA. 2008;14:465–481. doi: 10.1261/rna.657708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Rubinstein JL, Brubaker MA. Alignment of cryo-EM movies of individual particles by optimization of image translations. J Struct Biol. 2015;192:188–195. doi: 10.1016/j.jsb.2015.08.007. [DOI] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine S, Murayama Y, Svetlov V, Nudler E, Yokoyama S. The ratcheted and ratchetable structural states of RNA polymerase underlie multiple transcriptional functions. Mol Cell. 2015;57:408–421. doi: 10.1016/j.molcel.2014.12.014. [DOI] [PubMed] [Google Scholar]
- Shu B, Gong P. Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proc Natl Acad Sci U S A. 2016;113:E4005–4014. doi: 10.1073/pnas.1602591113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert H, Sochor F, Wacker A, Buck J, Helmling C, Hiller F, Keyhani S, Noeske J, Grimm S, Rudolph MM, et al. Pausing guides RNA folding to populate transiently stable RNA structures for riboswitch-based transcription regulation. Elife. 2017;6 doi: 10.7554/eLife.21297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami S, Sekine SI, Kumarevel T, Hino N, Murayama Y, Kamegamori S, Yamamoto M, Sakamoto K, Yokoyama S. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature. 2010;468:978–982. doi: 10.1038/nature09573. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Landick R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol Cell. 2003;12:1125–1136. doi: 10.1016/s1097-2765(03)00439-8. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Tuske S, Sarafianos SG, Wang X, Hudson B, Sineva E, Mukhopadhyay J, Birktoft JJ, Leroy O, Ismail S, Clark AD, Jr, et al. Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation. Cell. 2005;122:541–552. doi: 10.1016/j.cell.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twist KA, Husnain SI, Franke JD, Jain D, Campbell EA, Nickels BE, Thomas MS, Darst SA, Westblade LF. A novel method for the production of in vivoassembled, recombinant Escherichia coli RNA polymerase lacking the alpha C-terminal domain. Protein Sci. 2011;20:986–995. doi: 10.1002/pro.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev D, Vassylyeva M, Perederina A, Tahirov T, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007a;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- Vassylyev D, Vassylyeva M, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007b;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- Vvedenskaya IO, Vahedian-Movahed H, Bird JG, Knoblauch JG, Goldman SR, Zhang Y, Ebright RH, Nickels BE. Transcription. Interactions between RNA polymerase and the "core recognition element" counteract pausing. Science. 2014;344:1285–1289. doi: 10.1126/science.1253458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Bushnell D, Westover K, Kaplan C, Kornberg R. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Meier TI, Chan CL, Feng G, Lee DN, Landick R. Discontinuous movements of DNA and RNA in RNA polymerase accompany formation of a paused transcription complex. Cell. 1995;81:341–350. doi: 10.1016/0092-8674(95)90387-9. [DOI] [PubMed] [Google Scholar]
- Watters KE, Strobel EJ, Yu AM, Lis JT, Lucks JB. Cotranscriptional folding of a riboswitch at nucleotide resolution. Nat Struct Mol Biol. 2016;23:1124–1131. doi: 10.1038/nsmb.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A, Leon K, Landick R, Darst SA. Structural basis of transcriptional pausing in bacteria. Cell. 2013;152:431–441. doi: 10.1016/j.cell.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Windgassen T, Mooney RA, Nayak D, Palangat M, Zhang J, Landick R. Trigger-helix folding pathway and SI3 mediate catalysis and hairpin-stabilized pausing by Escherichia coli RNA polymerase. Nucleic Acids Res. 2014;42:12707–12721. doi: 10.1093/nar/gku997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtas MN, Mogni M, Millet O, Bell SD, Abrescia NG. Structural and functional analyses of the interaction of archaeal RNA polymerase with DNA. Nucleic Acids Res. 2012;40:9941–9952. doi: 10.1093/nar/gks692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin AV, Yakhnin H, Babitzke P. RNA polymerase pausing regulates translation initiation by providing additional time for TRAP-RNA interaction. Mol Cell. 2006;24:547–557. doi: 10.1016/j.molcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- Zhang J, Landick R. A two-way street: Regulatory interplay between RNA polymerase and nascent RNA structure. Trends Biochem Sci. 2016;41:293–310. doi: 10.1016/j.tibs.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Steitz TA. Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol Cell. 2015;58:534–540. doi: 10.1016/j.molcel.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In the first part of the movie, the RNA exit channel is widened to accommodate the pause RNA hairpin (PDB 6ASX) in comparison to the exit channel in an EC structure (PDB 6ALF). The RNA hairpin loop structure in the hisPEC was modeled using MCFold (http://www.major.iric.ca/MC-Pipeline/) because it was disordered in the cryo-EM density map. The RNA hairpin loop is connected to the pause hairpin stem that is positioned as observed in the cryo-EM map. The single-stranded RNA within and outside the EC exit channel is modeled arbitrarily (also disordered in the cryo-EM map). In the second part of the movie, the components of the swivel module (clamp, dock, β′ C-term, shelf, jaw, and SI3) are depicted and the rotation of the swivel module between the conformations observed in the hisPEC and EC structures is shown.
Movement of SI3 (magenta) between its position in EC (PDB 6ALF; unfolded trigger loop) and its position in an initiating transcription complex with bound NTP (PDB 4YLN; folded trigger loop) is shown. To aid comparison, sigma factor was removed from the initiating complex and a complete RNA:DNA hybrid was added. The upstream DNA duplex was removed from both models. When the trigger loop (TL) is unfolded in the EC, SI3 is in the OUT position (SI3 is a 188-aa insertion into the TL). When the TL folds into the trigger helices (TH) in the NTP-bound complex, SI3 moves toward the bridge helix into the IN position because the unfolded polypeptide linker segments between the TL and SI3 are shortened due to TH formation. In the hisPEC, however, rotation of the swivel module, which includes SI3, repositions SI3 and the shelf in ways that would generate steric clash if SI3 were to shift to the IN position (yellow stars; upstream DNA was also removed from this hisPEC model). The apparent rotation of downstream DNA during swivel rotation is an artifact of the morphing software used to generate the movie; the downstream DNA does not rotate, rather its position shifts slightly during swivel module rotation.