Abstract
The cytokine interleukin-2 (IL-2) is critical for the functions of regulatory T cells (Tregs). The contribution of polymorphisms in the gene encoding the IL-2 receptor α subunit (IL2RA), which are associated with type 1 diabetes, is difficult to determine because autoimmunity depends on variations in multiple genes, where the contribution of any one gene product is small. We investigated the mechanisms whereby a modest reduction in IL-2R signaling selectively in T lymphocytes influenced the development of diabetes in the NOD mouse model. The sensitivity of IL-2R signaling was reduced approximately two- to three-fold in Tregs from mice that co-expressed wild-type IL-2Rβ and a mutant subunit (IL-2RβY3) with reduced signaling (designated NOD-Y3). Male and female NOD-Y3 mice exhibited accelerated diabetes onset due to intrinsic effects on multiple activities in Tregs. Bone marrow chimera and adoptive transfer experiments demonstrated that IL-2RβY3 Tregs resulted in impaired homeostasis of lymphoid-residing central Tregs and inefficient development of highly activated effector Tregs and that they were less suppressive. Pancreatic IL-2RβY3 Tregs showed impaired development into IL-10–secreting effector Tregs. The pancreatic lymph nodes and pancreases of NOD-Y3 mice had increased numbers of antigen-experienced CD4+ effector T cells, which was largely due to impaired Tregs, because adoptively transferred pancreatic autoantigen–specific CD4+ Foxp3− T cells from NOD-Y3 mice did not accelerate diabetes in NOD.SCID recipients. Our study indicates that the primary defect associated with chronic, mildly reduced IL-2R signaling is due to impaired Tregs that cannot effectively produce and maintain highly functional tissue-seeking effector Treg subsets.
Introduction
CD4+ Foxp3+ regulatory T cells (Tregs) maintain peripheral tolerance by suppressing autoreactive T cells that escape negative selection in the thymus. Interleukin-2 (IL-2), through its interaction with the high-affinity IL-2 receptor (IL-2R), which consists of IL-2Rα (CD25), IL-2Rβ (CD122), and γc (CD132), controls multiple critical activities in Tregs (1–4). IL-2R signaling is essential for thymic Treg development, promoting the maturation of CD4+ CD25lo Foxp3lo immature cells into functional Tregs (5–9). IL-2 also contributes to Treg peripheral homeostasis (10), in part by providing survival signals to central or resting CD62Lhi Tregs (11) and by supporting the development of terminally differentiated, short-lived Klrg1+ Tregs (12). Treg identity and suppressive function are enforced by IL-2 (13) through direct regulation of the expression of Foxp3. This property also influences the development of conventional T cells into peripherally induced Tregs (14–16). Because Foxp3 represses the expression of IL2, Tregs constitutively express the high-affinity trimeric IL-2R to respond to IL-2 produced by effector T cells and in some cases dendritic cells (17, 18).
Multiple single nucleotide polymorphisms (SNPs) in IL2, IL2RA, and IL2RB are associated with an increased risk for developing several human autoimmune diseases (19). On their own, these IL-2-related SNPs, some of which are common in the population, represent a small risk for autoimmunity, and most likely act in concert with SNPs in other genes as well as environmental factors to trigger autoimmunity. This complexity has made it difficult to determine how an individual SNP promotes autoimmunity. With respect to type one diabetes (T1D), individuals with susceptible SNPs in IL2RA (20) have a reduced abundance of CD25 on Treg and T memory cells, which leads to reduced IL-2R signaling (21). Some data have also associated reduced IL-2R signaling in Tregs with decreased fitness and suppressive function (22, 23). Nevertheless, we still poorly understand how a subtle reduction in IL-2R signaling represents a risk for autoimmunity, including T1D.
The non-obese diabetic (NOD) mouse has been widely used as a model for T1D, where Il2 is a genetic risk for development of diabetes. The importance of the insulin-dependent diabetes risk locus 3 (Idd3), which contains Il2, for diabetes is illustrated by the substantial reduction in diabetes occurrence in NOD congenic mice in which the Idd3 interval is derived from C57BL/6 mice (24). NOD Idd3 results in a two-fold reduction in IL-2 production by CD4+ T cells (25). Moreover, Il2 mRNA was selectively reduced in infiltrating cells in pancreatic islets of NOD mice when compared to that in peripheral immune tissues (26). Reduced IL-2 production is associated with a pancreas-specific decrease in the Treg to T effector (Teff) cell ratio, which might reflect impaired Treg homeostasis. Pancreatic Tregs in NOD mice have reduced amounts of CD25 and Bcl-2, but increased Ki67 abundance, the latter of which may reflect a compensatory proliferative response toward autoreactive T cells (26, 27). These studies concluded that the Il2-related genetic risk for diabetes in NOD mice is primarily due to a pancreas-specific impairment of Treg homeostasis that reduced the ability of these cells to suppress autoreactive T cells. However, these findings indirectly support these conclusions and the effect on IL-2R signaling as a risk for diabetes remains poorly understood. An additional complication in clearly defining the Il2-related risk for diabetes in NOD mice is that the Idd3 locus also contains Il21, which is closely linked to Il2. This polymorphism in Il21 results in increased secretion of IL-21, a pro-inflammatory cytokine, and this increase in IL-21 abundance is linked to diabetes susceptibility in NOD mice (28).
As discussed earlier, the direct contribution of the Il2-related risk for diabetes in NOD mice remains poorly understood. In addition, the reduced IL-2 production associated with the Idd3 locus does not directly test the consequences of altered IL-2R signaling, which is a risk for T1D and several other autoimmune diseases. To directly target IL-2R signaling, one must affect the activity of IL-2Rβ, because this subunit is responsible for the distinct signaling attributed to IL-2. Simply knocking out Il2rb in the germline or selectively in Tregs leads to the production of immature, nonfunctional Tregs, which results in rapid lethal systemic autoimmunity (29–31); this approach is not suitable to assess how subtle changes in IL-2R signaling might promote autoimmunity. For these reasons, we developed a model in which IL-2Rβ signaling was selectively reduced in all T cells of NOD mice. We reasoned that IL-2R-dependent processes in T cells relevant to diabetes development in NOD mice would be intensified and thus distinguished from other genetic risks in this model. Indeed, diabetes was accelerated in male and female NOD mice in which IL-2Rβ signaling was modestly and selectively reduced in T cells. Furthermore, this autoimmunity was directly related to substantial changes in Tregs that included, but were not limited to, altered homeostasis and function, whereas more modest alterations were noted in the Teff compartment.
Results
Expression of the IL-2RβY3 transgene in NOD mice reduces IL-2R signaling
We previously expressed a mutant IL-2Rβ, designated IL-2RβY3, in mice such that 3 critical cytoplasmic tyrosine residues, Tyr341, Tyr395, and Tyr498, were mutated to phenylalanines (Y341F, Y395F, and Y498F) (32). These mutations interfere with the association of the adaptor Shc and transcriptional regulator STAT5 to the cytoplasmic tail of IL-2Rβ and thus substantially reduce IL-2R signaling. IL-2RβY3 was targeted for exclusive expression by T lymphocytes using the Cd2 minigene. When crossed onto the IL-2Rβ−/− genetic background of C57BL/6 mice, the reduced IL-2Rβ signaling associated with IL-2RβY3 outwardly supported normal Treg development, homeostasis, and function, whereas Teff and T memory compartments were much more obviously impaired (32). However, in a competitive environment, IL-2RβY3 Tregs did not compete with wild-type Tregs, demonstrating that IL-2RβY3 Tregs are not fully functional when IL-2Rβ signaling is impaired (7).
The availability of mice that express IL-2RβY3 under the control of the Cd2 promoter, designated Y3 mice, provided an opportunity to directly model how impaired IL-2Rβ signaling in T cells represents a risk for autoimmunity. In the current study, we tested this concept for diabetes development by reducing IL-2Rβ signaling in conventional T cells and Tregs in NOD mice. C57BL/6-Y3 IL-2Rβ−/− mice were backcrossed to NOD IL-2Rβ+/+ mice for 12 generations to derive NOD-Y3 IL-2Rβ+/+ and NOD-Y3 IL-2Rβ+/− mice. SNP analysis of NOD-Y3 mice indicated that all chromosomal regions, including all Idd loci, were of NOD origin except for a region within chromosome 15 that surrounds Il2rb. We anticipated that when the IL-2RβY3 transgene was expressed in NOD mice on the IL-2Rβ+/+ or IL-2Rβ+/− genetic background, the competition for IL-2 between wild-type IL-2Rβ and IL-2RβY3 would lead to an overall reduction in IL-2Rβ signaling. Thus, all studies in this report (except when specified otherwise) used mice for which the IL-2RβY3 transgene was crossed onto IL-2Rβ+/+ or IL-2Rβ +/− NOD mice. As expected, IL-2RβY3 was detected on essentially all T lymphocytes (Fig. 1A), as indicated by the increased mean fluorescence intensity (MFI) on Treg and CD8+ T cells, which also had endogenous wild-type IL-2Rβ, and by essentially all conventional CD4+ T cells, which were mostly IL-2Rβneg in normal NOD mice. In contrast, the MFI of CD25 was reduced by 20 to 30% for Tregs in NOD-Y3 mice for all examined tissues (Fig. 1B). Because IL-2 increases the abundance of CD25 on the cell surface (33), this result suggests that IL-2R signaling is reduced in Tregs from NOD-Y3 mice.
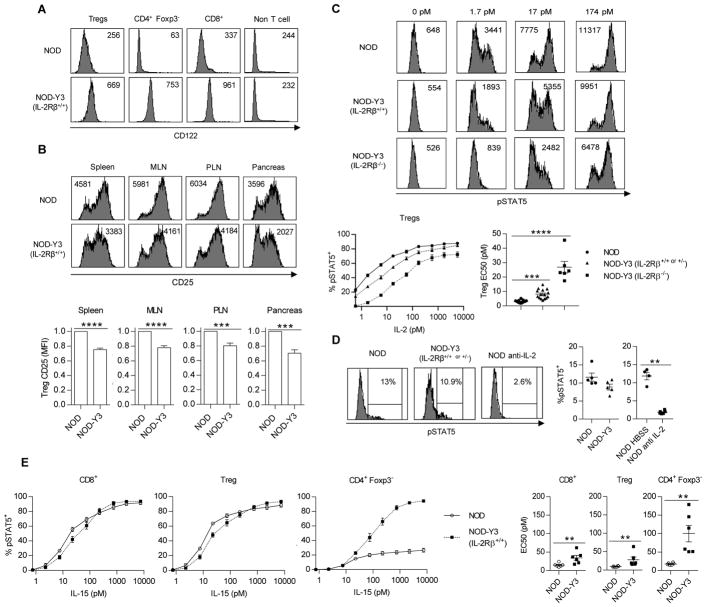
Fig. 1. Analysis of IL-2R abundance and STAT5 activation in lymphoid cells from NOD and NOD-Y3 mice.
(A and B) Flow cytometric analysis of the cell surface expression of CD122 (A) and CD25 (B) on the indicated cells from the spleens of the indicated mice (n = 15 to 18 mice). (B) Bottom: The MFI for CD25 expression by Tregs was assessed by the Wilcoxon signed-rank test. (C) Analysis of pSTAT5 abundance in CD4+ Foxp3+ gated Tregs after stimulation with the indicated concentrations of IL-2 for 15 min in vitro. Top: Representative flow cytometry plots with pSTAT5 MFI. Bottom: Dose-response curves (left), which were subjected to non-linear regression analysis to determine the EC50s (right). Data are from NOD IL-2Rβ+/+ or NOD IL-2Rβ +/− (n=15), NOD-Y3 IL-2Rβ+/+ or NOD−Y3 IL-2Rβ +/− (n=15), and NOD-Y3 IL-2Rβ−/− (n=6) mice and the resulting EC50 values were analyzed by the Kruskal-Wallis test. (D) Flow cytometric analysis of pSTAT5 abundance in Tregs directly after isolation from the indicated mice (n=5). In some cases, the NOD mice were treated with HBSS (n=4) or anti-IL-2- (n=6) for 24 hours before the cells were isolated and analyzed for pSTAT5. The resulting data were analyzed by Mann-Whitney test. (E) Flow cytometric analysis of pSTAT5 abundance in the indicated T cell populations stimulated in vitro with the indicated concentrations of IL-15 for 15 min. Dose-response curves (left) were subjected to non-linear regression analysis to determine the EC50 values (right), which were analyzed by the Mann-Whitney test (n = 6).
Dose-response studies that assessed the tyrosine-phosphorylation of STAT5 (pSTAT5) showed that Tregs expressing IL-2RβY3 had a reduced response to IL-2 (Fig. 1C). The EC50 value of IL-2 for wild-type Tregs was 3.1 pM, whereas the EC50 values for IL-2RβY3 Tregs on the IL-2Rβ+/+ and IL-2Rβ−/− background were 8.2 and 26.8 pM, respectively. Thus, Tregs that co-expressed the wild-type IL-2Rβ and IL-2RβY3 were 2.6-fold less responsive to IL-2 than were wild-type Tregs (Fig. 1C). In addition, the response by Tregs to IL-2 was impaired in NOD-Y3 mice, as demonstrated by the decreased percentage of NOD-Y3 cells that were pSTAT5+ (Fig. 1D). The reduced activation of STAT5 could be attributed to IL-2, because treatment of the mice with anti-IL-2 antibody for 24 hours substantially reduced the percentage of pSTAT5+ cells (Fig. 1D). Minimal IL-2-dependent STAT5 phosphorylation was stimulated in conventional CD4+ and CD8+ T cells from NOD and NOD-Y3 mice when assessed directly ex vivo or after in vitro challenge with IL-2, which was due to the lack of the high-affinity IL-2R on these cells (Fig. S1, A and B). To determine whether IL-2RβY3 altered IL-2Rβ signaling in Teff cells, IL-2-dependent pSTAT5 abundance was assessed for T cells pre-activated with anti-CD3 and anti-CD28 (Fig. S1, C and D). IL-2 signaling sensitivity was not altered by IL-2RβY3 under these conditions, because these cells had high amounts of all wild-type IL-2R subunits that likely outcompete the mutant IL-2RβY3 for IL-2. Nevertheless, because Teff cells in vivo transiently express the IL-2R at amounts typically lower than those of Tregs (34), we expected that IL-2RβY3 would also decrease IL-2R signaling in these cells by at least 2- to 3-fold as was detected for Tregs.
IL-2Rβ is also a subunit of the IL-15R. Thus, IL-2RβY3 is also expected to affect IL-15-dependent signaling. Indeed, IL-2RβY3 expression lowered IL-15-induced signaling in Tregs and CD8+ T cells by approximately 2.9 and 2.3-fold, respectively (Fig. 1E). This EC50 shift was similar to that noted for IL-2 on Tregs between NOD-Y3 and wild-type NOD mice (Fig. 1C). Because IL-2Rβ was not detected in CD4+ Foxp3− T cells, the response to IL-15 in NOD-Y3 mice was directly mediated by IL-2RβY3. The EC50 for CD4+ Foxp3− T cells from NOD-Y3 mice was 100 pM, which is approximately 10-fold higher than that measured for wild-type Tregs or CD8+ T cells. This high EC50 implies that IL-2RβY3 CD4+ Foxp3− T cells would only respond to IL-15 in environments that have a high concentration of this cytokine. Thus, IL-2RβY3 is more likely to potentially affect the responses of Tregs and CD8+ T cells to IL-15 than to affect those of conventional CD4+ T cells. This is an unavoidable potential off-target consequence of directly interfering with IL-2Rβ signaling.
IL-2RY3 accelerates the onset and increases the penetrance of diabetes in NOD-Y3 mice
IL-2RβY3 acted in a dominant-negative manner to reduce IL-2R signaling by 2- to 3-fold in NOD-Y3 mice. Therefore, we assessed the consequences of this modest reduction in IL-2R signaling by comparing diabetes onset in male and female NOD and NOD-Y3 mice (Fig. 2A) and NOD and NOD-Y3 BDC2.5 T cell receptor (TCR) transgenic mice (Fig. 2B), whose CD4+ T cells express a TCR specific for the chromogranin A autoantigen. In male mice, the average time to diabetes onset was 23 weeks for NOD-Y3 mice, but greater than 40 weeks for wild-type NOD mice. In female mice, the average diabetes onset was 18 weeks for NOD-Y3 mice, but was 27 weeks for wild-type NOD mice (Fig. 2A). Similarly, average diabetes onset was accelerated in both male and female NOD-Y3 BDC2.5 mice (Fig. 2B). Because of the unexpected break in tolerance in male mice, most of our further studies focused on male NOD and NOD-Y3 mice.
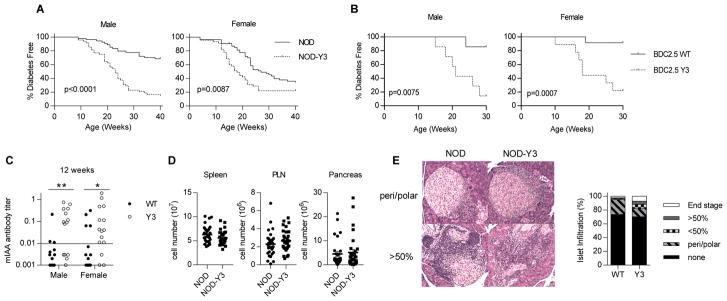
Fig. 2. The effect of IL-2RβY3 on diabetes in NOD mice.
(A and B) Diabetes onset was followed in male and female NOD and NOD-Y3 mice (n = 45 to 54 mice) for up to 40 weeks of age (A) and in male (n = 7) and female (n = 9 to 12) NOD BDC2.5 and NOD-Y3 BDC2.5 mice for up to 30 weeks of age (B). Data were analyzed by log-rank test and the resulting EC50 values were analyzed by Mann-Whitney test. (C and D) Insulin autoantibody (IAA) titers from male and female NOD and NOD-Y3 mice at 12 weeks of age (C) and analysis of cellularity of the indicated tissues from male mice (n=30) 8 to 16 weeks of age (D). Data were analyzed by Mann-Whitney test. (E) Pancreatic islet infiltration scoring by hematoxylin-eosin staining of pancreas sections from male mice at 7 to 12 weeks of age (>200 islets were evaluated).
In polyclonal mice, serum insulin autoantibody titers were also greater in male and female NOD-Y3 mice than in NOD mice when measured at 12 weeks of age (Fig. 2C). The overall cell numbers recovered from the pancreatic lymph node (PLN) and the pancreas were slightly, but not significantly, increased in male NOD-Y3 mice compared to those in male NOD mice (Fig. 2D). Male NOD and NOD-Y3 mice also had a similar percentage of islets showing lymphocytic infiltration; however, NOD-Y3 mice had more late stage infiltration compared to that of the wild-type controls (Fig. 2E). These data suggest that lymphoid cells present in the pancreas were somewhat more effective in attacking islet tissues in the NOD-Y3 pancreas and likely contributed to the acceleration of diabetes onset.
Effect of IL-2RβY3 on Tregs and antigen-experienced T cells in NOD mice
Although the overall cellularity in male NOD-Y3 mice was relatively normal, the T lymphocyte composition was distinctive in lymphoid tissues and the pancreas. There were less Tregs in the spleen, PLN, and pancreas of NOD-Y3 mice compared to those of NOD mice (Fig. 3A). These Tregs exhibited a slight, but statistically significant reduction in Foxp3 abundance together with a reduced amount of CD25 (Fig. 1B). Conventional CD4+ T cells were slightly increased in number in the spleen and PLN, whereas antigen-experienced CD4+ Foxp3− CD44hi T cells were increased in number in the PLN and pancreas (Fig. 3B). In contrast, a reduced fraction of CD8+ T cells was noted for these tissues, whereas the proportion of antigen-experienced CD8+ CD44hi T cells remained largely unaltered (Fig. 3C). Most of these changes were also noted for female NOD-Y3 mice (Fig. S2). Two exceptions included a less striking effect on Treg and CD4+ Foxp3− CD44hi percentages in the pancreas of female NOD and NOD-Y3 mice. This result is most likely because inflammation leading to diabetes is normally more severe in female NOD mice than in male NOD mice, which might have obscured the effects of IL-2RβY3. For NOD-Y3 BDC2.5 CD4+ T cells, reducing IL-2Rβ signaling resulted in a consistent trend in decreased numbers of Tregs, but not conventional CD4+ T cells (Fig. S3), suggesting that reduced IL-2Rβ signaling was selective for Tregs. With respect to other lymphoid cell populations, the proportions of CD19+ B cells, Ly-6G+ granulocytes, CD11c+ dendritic cells, CD11b+ CD11c− macrophages, and CD49b+ NK cells were similar between male NOD and NOD-Y3 mice (Fig. S4), with the exception that the spleens of NOD-Y3 mice contained a reduced proportion of NK cells. These results, therefore, suggest that IL-2RβY3 primarily affects T cells and that these changes are sufficient to accelerate the onset of diabetes.
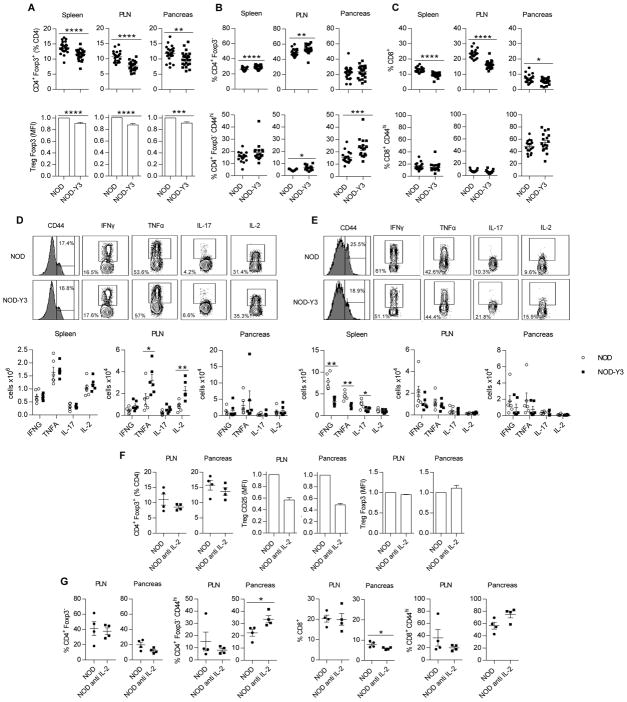
Fig. 3. The effect of IL-2RβY3 on Teff cells from male mice.
(A to C) Flow cytometric analysis of the proportions of (A) Tregs and their amount of Foxp3, (B) total CD4+ Foxp3− and CD4+ Foxp3− CD44hi antigen-experienced T cells, and (C) total CD8+ and CD8+ CD44hi antigen-experienced T cells in the indicated tissues. Data are from 22–25 male mice per group and were analyzed by Mann-Whitney or Wilcoxon signed-rank tests for the MFIs. (D and E) Flow cytometric analysis of cytokine production by CD4+ and CD8+ Teff cells. Representative flow cytometry profiles from the spleen (top) and cell numbers (bottom) of CD4+ CD44hi cells (D) and CD8+ CD44hi cells (E) from the indicated tissues that were positive for the indicated cytokines. Data were analyzed by Mann-Whitney test (n=6). (F and G) Analysis of the effect of anti-IL-2 on T cells in NOD mice. Mice received anti-IL-2 (S4B6) i.p., initially 1 mg on day 0, and 0.5 mg on days 2, 4, 7 and 9, and the indicated tissues were assessed one day after the last injection. (F) Effect on Tregs, including the amount of CD25 and Foxp3. (G) Effect on total CD4+ Foxp3− and CD8+ T conventional cells and CD44hi antigen-experienced CD4+ and CD8+ T cells. Data were analyzed by Mann-Whitney or Wilcoxon signed-rank tests for the MFIs (n=4).
IL-2RY3 increases the numbers of CD4+ Teff cells, but decreases the numbers of CD8+ Teff cells in NOD-Y3 mice
To further evaluate the effects of IL-2RβY3 on the conventional T cell compartment, we assessed cytokine production after stimulation of the T cells with PMA and ionomycin. Increased numbers of TNF-α– and IL-2–producing CD4+ CD44hi T cells were present in the PLNs of NOD-Y3 mice (Fig. 3D). These data associate the increased activity of CD4+ Teff cells with accelerated diabetes in NOD-Y3 mice. In contrast, a trend for reduced numbers of IFN-γ+, TNF-α+, and IL-17+ CD8+ CD44hi T cells was noted for NOD-Y3 mice, which was most obvious in the spleen (Fig. 3E). Thus, IL-2RβY3 did not substantially alter the frequency of Th1 and Th17 cells, which have been suggested to drive diabetes in NOD mice (35).
Reduced IL-2R signaling accounts for the altered numbers of CD4+, but not CD8+ T cells in NOD-Y3 mice
The expression of IL-2RβY3 resulted in a 2- to 3-fold reduction in IL-15R signaling CD8+ T cells. IL-15 is important for the persistence of CD8+ T memory cells (36, 37). Thus, the lower proportion (Fig. 3C) and functional activity (Fig. 3E) of CD8+ T cells in NOD-Y3 mice might reflect impaired IL-15 signaling. Therefore, we also examined the effect of acute treatment with anti-IL-2 on wild-type NOD mice for CD4+ Tregs and CD4+ and CD8+ T conventional cells. Similar to NOD-Y3 mice, anti-IL-2–treated NOD mice had reduced CD25 in Tregs and an increased proportion of antigen-experienced CD4+ Foxp3− CD44hi Foxp3− T cells in the pancreas (Fig. 3, F and G). In contrast, anti-IL-2 had minimal effect on the overall proportion of CD8+ T cells and distribution of CD8+ antigen-experienced cells in NOD mice (Fig. 3G). Thus, these data are consistent with the notion that IL-2RβY3 alters the CD4+ Treg and CD4+ Foxp3− antigen-experienced T cell compartments by reducing IL-2R signaling, whereas the reduction in the number of CD8+ T cells is likely a result of reduced IL-15R signaling.
Treg subsets from IL-2RβY3 mice have properties consistent with altered homeostasis
Tregs have been divided into subsets whereby increased cell surface expression of CD62L or CCR7 marks resting or central Tregs (cTregs), whereas reduced expression of these molecules marks activated or effector Tregs (eTregs) (11). Gene expression analysis indicates that eTregs are heterogeneous; eTregs that express CD69, CD103, or Klrg1 are more highly activated than their negative counterparts. In particular, these more-activated eTregs have increased amounts of Treg functional molecules and chemokine receptors that promote cell trafficking into non-lymphoid tissues, among other changes (38, 39).
Because Tregs were consistently decreased in number in NOD-Y3 mice, including those that expressed the BDC2.5 TCR, we further focused on how IL-2RβY3 might affect these cells. We assessed these Treg subsets based on their cell surface expression of CD62L and CD69 (Fig. 4A) and their expression of Ki67 (Fig. 4B) and Bcl-2 (Fig. 4C). This analysis revealed that NOD-Y3 mice had a reduced proportion of CD62Lhi CD69− cTregs compared to that of NOD mice (Fig. 4D). Accordingly, the spleen and pancreas contained an increased proportion of less activated CD69− eTregs (Fig. 4E), whereas the PLN contained more CD69+ eTregs (Fig. 4F). This altered distribution of Treg subsets in NOD-Y3 mice is accounted for in part by the altered homeostasis of these cells, as reflected by the increased abundance of Ki67 and the decreased abundance of Bcl-2. This phenotype was most apparent with cTregs (Fig. 4D), which depend on IL-2 for their survival (11). However, these changes were also noted for CD69− (Fig. 4E) and CD69+ (Fig. 4F) eTregs. The increased abundance of Ki67 likely reflects the increased proliferation of Tregs to compensate for their impaired survival.
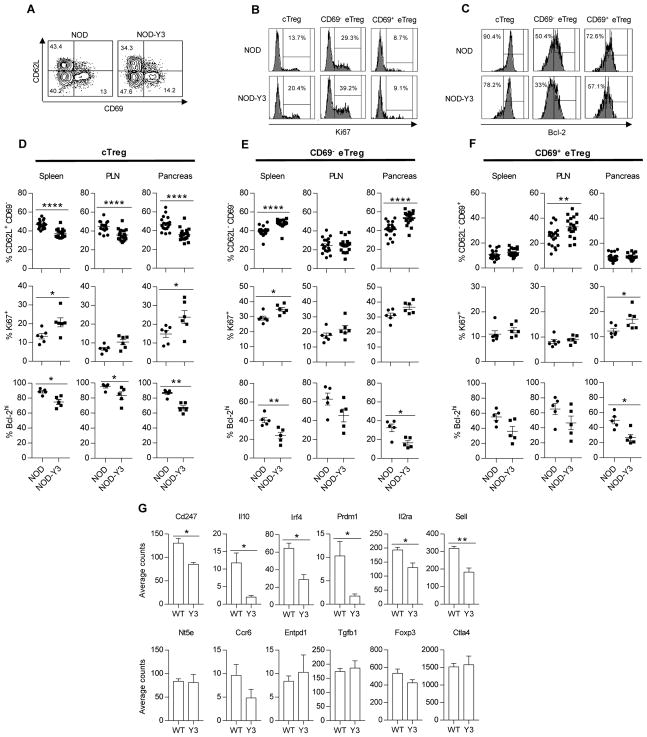
Fig. 4. Altered distribution of Treg subsets in male NOD-Y3 mice.
(A to C) Representative flow cytometry plots of Treg subsets from the spleens of male NOD and NOD-Y3 mice based on the expression of CD62L and CD69 (A), Treg subset Ki67 (B), or Treg subset Bcl-2 (C). (D to F) Analysis of the percentages of the designated subsets (n=19) and their expression of Ki67 (n=6) and Bcl-2 (n=5) for cTregs (D), CD69− eTregs (E), and CD69+ eTregs (F). Data were analyzed by Mann-Whitney test. (G) The abundances of the indicated mRNAs in pancreatic Tregs from male NOD and NOD-Y3 mice were assessed with the Nanostring platform. Data were analyzed by two-tail unpaired t-test with the nSolver analysis software package from Nanostring (n=3).
Transcriptional changes imposed by IL-2RβY3 for Tregs from the pancreas were determined using the Nanostring platform, which measures the expression of 547 immune-related genes. We found that 236 mRNAs were expressed by these Tregs and 32 mRNAs were significantly differentially expressed (p≤0.05) (table S1, P < 0.05). Consistent with our flow cytometric analysis, Tregs from NOD-Y3 mice expressed less Il2ra (CD25) and Sell (CD62L) mRNAs than did Tregs from NOD mice (Fig. 4G). A group of differentially expressed genes related to eTregs exhibited decreased expression in IL-2RβY3 Tregs, including Irf4, Prdm1, Il10, and Cd247 (CD3ζ). IRF4 and TCR signaling are essential for eTreg development (40, 41), whereas Blimp-1 promotes IL-10 production by eTregs (40). Although Il10 expression was decreased, gene expression of other Treg functional molecules, such as Ctla4, Ccr6, Entpd1, Nt5e and Tgfb1, were not differentially expressed between NOD and NOD-Y3 mice. Thus, these data suggest that in addition to leading to altered Treg homeostasis, IL-2RβY3 also leads to a pancreatic Treg signature of reduced functional activity due in part to the impaired development of the more highly activated, IL-10–producing eTregs.
IL-2RβY3 intrinsically alters the activity of CD4+ Tregs
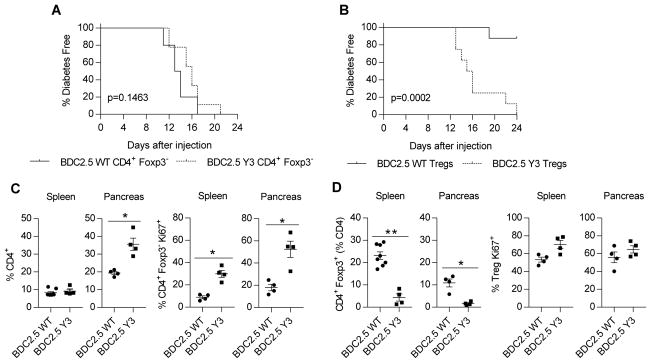
The increase in the number of CD4+ Teff cells associated in NOD-Y3 mice might reflect a direct effect of impaired IL-2R signaling, or alternatively it may be caused by an indirect effect due to an impaired Treg compartment. To compare the diabetogenic capacity of autoantigen-specific CD4+ T cells, we transferred wild-type or IL-2RβY3 CD4+ Foxp3− BDC2.5 TCR transgenic T cells into NOD.SCID recipient mice to induce diabetes (Fig. 5A). We found that 50% of the recipient mice became diabetic between 13 and 16 days after the transfer, with a slight, but statistically non-significant delay, apparent in NOD.SCID recipient mice that received CD4+ T cells that expressed IL-2RβY3. These data suggest that IL-2RβY3 does not accelerate diabetes by enhancing the function of autoreactive T cells.
Fig. 5. Functional activity of Teff and Treg cells that express IL-2RβY3 on the NOD BDC2.5 background.
(A) Analysis of the onset of diabetes induced by CD4+ Teff cells. CD4+ Foxp3− T cells (5 × 104) from NOD BDC2.5 or NOD-Y3 BDC2.5 mice were transferred into NOD.SCID recipients, which were monitored for 24 days. (B) Analysis of the suppression of diabetes by Tregs. Purified wild-type or IL-2RβY3 BDC2.5 Tregs (2.5 × 104) were co-transferred with wild-type CD4+ Foxp3− BDC2.5 T cells (5 × 104) into NOD.SCID mice, which were monitored for 24 days. Data in (A) and (B) were analyzed by log-rank test (n=9–10). (C and D) Analysis of the properties of Teff cells (C) and Tregs (D) from the mice shown in (B), where BDC2.5-WT or BDC2.5-Y3 on the x-axis refer to the type of Tregs transferred into the NOD.SCID recipients. Mice were analyzed when the mice were determined to be diabetic or 24 days after transfer. Data were analyzed by Mann-Whitney test (n=4–8).
To directly test the effect of IL-2RβY3 on Treg function, we co-transferred wild-type or IL-2RβY3 BDC.2.5 Tregs with wild-type BDC2.5 CD4+ Foxp3− T cells into NOD.SCID recipients. In this case, only the recipients that received the wild-type BDC2.5 Tregs were protected from diabetes (Fig. 5B). For recipients that received BDC2.5 IL-2RβY3 Tregs, donor wild-type BDC2.5 CD4+ Foxp3− T cells minimally increased in number in the spleen but substantially increased in the pancreas because of increased proliferation as determined by measuring Ki67 abundance (Fig. 5C). Furthermore, NOD-Y3 BDC2.5 mouse Tregs were found at a much lower proportion than those of NOD BDC2.5 mice, even though the NOD-Y3 BDC2.5 Tregs exhibited increased proliferation, consistent with an inability to suppress the autoreactive BDC2.5 T cells (Fig. 5D). These data suggest that IL-2RβY3 directly dysregulates Treg function.
IL-2RβY3 impairs the homeostasis of Treg subsets
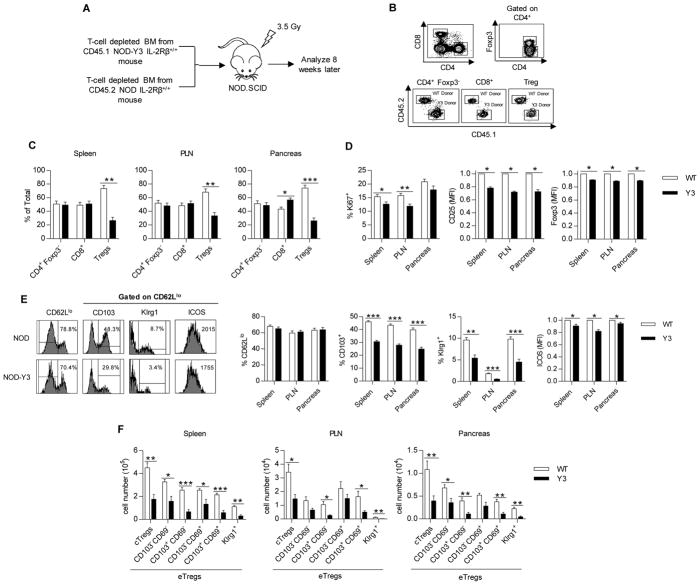
To assess the possibility that altered Treg homeostasis also contributed to the acceleration of diabetes in NOD-Y3 mice, mixed bone marrow chimeric mice were prepared such that irradiated NOD.SCID recipients received a 1:1 mixture of T cell-depleted bone marrow from NOD and NOD-Y3 mice (Fig. 6A). Eight weeks after transplantation, the distribution of donor T cells was assessed for the spleen, PLN, and pancreas (Fig. 6B). Tregs of IL-2RβY3 origin were substantially underrepresented in these recipients, whereas equivalent proportions of CD4+ Foxp3− and CD8+ T cells of wild-type and IL-2RβY3 origin were found in these tissues (Fig. 6C). In addition, IL-2RβY3 Tregs had decreased amounts of CD25 and Foxp3, consistent with reduced IL-2R signaling, and had decreased Ki67 abundance, consistent with impaired proliferation (Fig. 6D).
Fig. 6. Selective inability of IL-2RβY3 Tregs to compete with wild-type Tregs in vivo.
(A) Experimental scheme for establishing the mixed bone marrow chimeras. (B) Representative flow cytometry plots of the donor cell populations that were isolated from NOD.SCID recipient mice 8 weeks after transfer. (C) Percentages of the donor cells in the indicated tissues. Data were analyzed by Mann-Whitney test (n=7). (D) Flow cytometric analysis of the expression of Ki67, CD25, and Foxp3 in Tregs from the indicated tissues. Data analyzed by Mann-Whitney test and Wilcoxon signed-rank test for MFIs (n=7). (E) Representative flow plots from the spleen (left) and analysis of the percentage of CD62Llo Tregs and their expression of CD103, Klrg1 and ICOS (right) from the indicated tissues. Data for ICOS (MFI) were analyzed by Wilcoxon signed-rank test; the remaining data were analyzed by Mann-Whitney test (n=7). (F) Analysis of the numbers of the indicated Treg subsets from the indicated tissues based on the expression of CD62L, CD69, CD103, and Klrg1. Data were analyzed by Mann-Whitney test (n=7).
Although cTregs were reduced in number in NOD-Y3 mice compared to those in NOD mice (Fig. 4D), the proportions of cTregs and eTregs did not substantially vary between Tregs of NOD and NOD-Y3 origin in the bone marrow chimeras (Fig. 6E). However, the number of NOD-Y3 eTregs that expressed CD103, ICOS, and Klrg1 was statistically significantly reduced, consistent with the impaired development of highly activated and terminally differentiated Tregs. If cell numbers rather than percentages are considered, the numbers of IL-2RβY3 cTregs and most eTreg subsets were markedly reduced (Fig. 6F). These data indicate that IL-2RβY3 cTregs did not effectively compete with wild-type NOD cTregs, consistent with the impaired homeostasis of this Treg subset, and suggest that IL-2RβY3 Tregs cannot properly develop into highly activated subsets.
IL-2RβY3 impairs the development of eTreg subsets
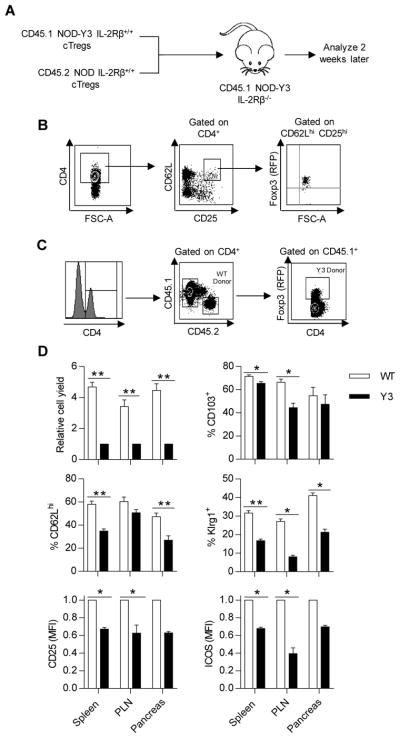
To directly assess the development of eTregs from cTregs, CD62Lhi cTregs were isolated from NOD and NOD-Y3 mice and transferred at a 1:1 ratio into NOD-Y3 IL-2Rβ−/− recipient mice (Fig. 7A). We have previously used these type of recipients, but on the C57BL/6 genetic background, to study Treg subset development (38). Tregs from C57BL/6-Y3 IL-2Rβ−/− mice have a more substantial block in IL-2Rβ signaling than Tregs from C57BL/6-Y3 IL-2Rβ+/+ mice (32), but they are not lymphopenic. These properties were also found for NOD-Y3 IL-2Rβ+/+ and NOD-Y3 IL-2Rβ−/− mice (Fig. 1C). When C57BL/6-Y3 IL-2Rβ−/− mice were used as recipients, reduced IL-2Rβ signaling associated with recipient-derived Tregs was favorable for engraftment of relatively small numbers of donor cTregs. This environment makes it feasible to study the development of Treg subpopulations in vivo without any conditioning regimen to make space for donor Tregs.
Fig. 7. IL-2RβY3 cTregs show impaired development into activated eTreg subsets.
(A) Experimental scheme showing how purified Tregs (8–15 × 104) from NOD and NOD-Y3 mice were premixed at a 1:1 ratio and verified by flow cytometry analysis before being transferred to NOD-Y3 IL-2Rβ−/− recipients. (B) Representative sorting strategy to purify donor-derived cTregs. (C) Representative flow plots to identify donor populations in male NOD-Y3 IL-2Rβ−/− hosts 2 weeks after transfer. (D) Relative donor Treg yields from the indicated tissues (n=5–6) and their expression of CD62L (n=5–6), CD25 (n=5–6), CD103 (n=3–4), Klrg1 (n=4–6), and ICOS (n=5–6) from the indicated tissues. Data were analyzed by Wilcoxon signed-rank test for MFIs, whereas the Mann-Whitney test was used for the remaining samples.
We purified cTregs based on CD4+ T cells that co-expressed CD25 and CD62L (Fig. 7B). Donor wild-type cells were >93% Foxp3+. Engrafted wild-type donor Tregs were identified solely based on their expression of CD45.2, whereas donor IL-2RβY3 Tregs were identified based on their expression of CD45.1 and RFP (Fig. 7C). These cells were distinguished from recipient Tregs, which were CD45.1+ but RFP−. This analysis revealed that the persistent engrafted wild-type and IL-2RβY3 donor cells were 95% Foxp3+. However, we cannot assess the fraction, if any, of donor IL-2RβY3 Tregs that may have converted to Foxp3− Teff cells.
Two weeks after transfer, the donor Tregs were approximately 80% of wild-type (CD45.2+) origin in the spleen, PLN, and pancreas (Fig. 7D). Tregs of NOD-Y3 origin expressed less CD25 and CD62L, the latter indicative of a reduced proportion of cTregs (Fig. 7D). These data further demonstrate the impaired homeostasis of cTregs with reduced IL-2Rβ signaling associated with IL-2RβY3. Furthermore, the development of eTregs was also abnormal. Compared to wild-type Tregs, IL-2RβY3 Tregs had reduced amounts of ICOS, CD103, and Klrg1 (Fig. 7D). These data indicate that IL-2RβY3 Tregs are impaired in their ability to generate highly activated eTreg subsets. Together, these data suggest that reducing IL-2R signaling in Tregs leads to compromised homeostasis of cTregs and the altered development of eTregs.
Discussion
Past studies provided some initial information consistent with the notion that variation in the genes encoding IL-2 or the IL-2R in mice and humans reduces the activity of Tregs (21, 24, 26), but little is understood regarding how this genetic variation represents a risk for autoimmunity. In the current study, we directly reduced IL-2Rβ signaling in T cells and showed that modestly, but chronically, reducing IL-2 responsiveness by ~2.6-fold accelerated diabetes in male and female NOD mice. Our findings demonstrate that accelerated diabetes occurs due to a multiplicity of effects on Tregs that affect their homeostasis and function. Altered Treg homeostasis by IL-2RβY3 was evident because Tregs from NOD-Y3 mice were broadly characterized by having increased amounts of Ki67 but decreased Bcl-2 abundance. This phenotype is consistent with Tregs that undergo increased proliferation to counterbalance enhanced cell death. Reduced IL-2R signaling affected Treg homeostasis in two distinct ways. First, cTregs (as defined by high expression of CD62L) were found at a reduced proportion under several different experimental conditions when they expressed IL-2RβY3 and this effect may reflect impaired survival, because IL-2 promotes the survival of this Treg subset (11). Second, the development of more highly activated eTregs [identified based on the expression of CD103 and Klrg1 by CD62Llo Tregs (38)], was less efficient for Tregs with reduced IL-2R signaling. This was most obviously seen in experiments with mixed bone marrow chimeras and the direct development of eTregs from purified adoptively transferred cTregs. The expression of Klrg1 by eTregs represents a terminally differentiated state of activation and occurs only after >8 or 9 cell divisions as cTregs develop into eTregs (12). The observation that reduced IL-2R signaling led to fewer Klrg1+ eTregs is likely explained by eTregs that die before reaching this state of activation.
In other systems, blockade of IL-2R signaling destabilizes Tregs so that they become IFNγ+ and IL-17+ Teff cells; this process is most evident for peripherally derived Tregs (42, 43). Our experimental design did not enable us to directly assess this process, and some of the decrease in Treg number that we noted might be due to their development into Teff cells. However, several observations suggest that this mechanism does not fully account for the accelerated onset of diabetes in NOD-Y3 mice. First, we did not detect increases in the numbers of IFN-γ– and IL-17–producing CD4+ T cells in NOD-Y3 mice. If Treg conversion to Th1 and Th17 Teff cells was a main mechanism, these cells should have increased in number, particularly in the pancreas. Second, TCR repertoire studies indicate that most Tregs in the peripheral immune tissues are thymic-derived cells (44) and have a stable phenotype (45). Thus, the poor persistence of adoptively transferred IL-2RβY3 Tregs is unlikely to be due to the few peripherally derived Tregs that may be part of the donor inoculum.
This study also begins to provide a molecular underpinning for the dysregulation associated with IL-2RβY3 Tregs. IRF4 is required for the cTreg-to-eTreg developmental step (40). Blimp-1 is selectively expressed by eTregs and contributes to the production of IL-10 (40). Broad immune mRNA profiling by Nanostring analysis showed that all of these mRNAs were less abundant in pancreatic Tregs of NOD-Y3 mice than in those of WT mice. These data are consistent with reduced IL-2R signaling interfering with the development of eTregs through an IRF4–Blimp-1–IL-10 axis. Activated STAT5 directly regulates the expression of Il2ra, Foxp3, Irf4, and Prdm1 (46–48). In addition, Il2ra, Irf4 and Prdm1 are direct targets of Foxp3 (49). Thus, subtly and chronically reducing both IL-2R signaling and the abundance of Foxp3 by IL-2RβY3 might cooperatively act to impair eTreg development. TCR signaling also importantly contributes to the development of eTregs (41). Thus, the reduced abundance of CD3ζ in pancreatic IL-2RβY3 Tregs raises the possibility that IL-2RβY3 might also influence eTreg development by reducing TCR signaling, but this requires further investigation.
We showed that IL-2RβY3 directly reduced functional activity by demonstrating the inability of Tregs from NOD-Y3 BDC2.5 mice to suppress autoreactive BDC2.5 T cells and diabetes development in a NOD.SCID transfer model. The effect of IL-2RβY3 on Tregs is predicted to reduce Treg function in several important ways. The impaired homeostasis of cTregs and eTregs likely diminishes the capacity of Tregs that reside in lymph nodes and tissue sites, respectively, to properly balance their numbers with autoreactive T cells. Furthermore, Klrg1+ and CD103+ eTregs are characterized by having increased amounts of several Treg functional molecules and tissue-targeting chemokine receptors (38, 39). Thus, IL-2–dependent alterations in eTreg development affect Treg function indirectly by reducing the proportions of Tregs with enhanced suppressive function, as well as directly by decreasing the amount of IL-10.
Previous studies designed to ascertain the contribution of IL-2R signaling to diabetes in NOD mice have been indirect in that they assessed the potential effects of reduced IL-2 production, especially by pancreatic T cells, on Tregs due to the Idd3 polymorphism (26). These studies implied that NOD Treg homeostasis was impaired as reflected by their altered amounts of Ki67 and Bcl-2, which we confirmed. A complication of the past results is that Idd3 not only reduces IL-2, but also increases the production of the proinflammatory cytokine IL-21 (28), which might also contribute the development of diabetes. Thus, an advantage of IL-2RβY3 is that IL-2R signaling is directly targeted. The caveat with IL-2RβY3 is that IL-15R signaling is also reduced. This was indeed detected in Tregs and CD8+ T cells. However, the Treg compartment is largely normal in IL-15-deficient mice (6) and we showed that in situ pSTAT5 activation by Tregs was inhibited by anti-IL-2, indicating that impaired IL-2R signaling largely accounted for the intrinsic effects of IL-2RβY3 on Tregs. In contrast, the lower proportion of CD8+ T cells associated with NOD-Y3 mice was unaffected by anti-IL-2. These results, in conjunction with the known role of IL-15 in CD8+ T cell homeostasis (50, 51), suggest that the decrease in the number of CD8+ T cells is accounted for by reduced IL-15R signaling. CD8+ T cells and increased IL-15 signaling have been implicated in promoting diabetes in NOD mice (52). Thus, a reduction in these two parameters by IL-2RβY3 is not expected to be a contributing factor to the accelerated disease in NOD-Y3 mice.
Upon reducing IL-2R signaling in T cells, Tregs were intrinsically affected in a manner that would reduce peripheral tolerance, as discussed earlier. Reduced IL-2R signaling is also expected to reduce Th1 and Th2 responses (53–56), which should not accelerate diabetes. However, decreased IL-2R signaling could intrinsically increase the concentration of IL-2, Th17 cells, and T follicular helper (Tfh) cells (57, 58), whereas increased IL-17 has been linked to diabetes in NOD mice (59). We indeed observed an increase in antigen-experienced CD4+ T cells in NOD-Y3 mice, which included increased numbers of IL-2–producing, but not IL-17–producing, T cells in the PLN. Increased anti-insulin autoantibodies were also observed, which may reflect an increase in the number of Tfh cells due to reduced STAT5 and Blimp-1-mediated repression (57). However, transfer studies of CD4+ Foxp3− T cells into NOD.SCID mice did not provide clear evidence for there being enhanced autoreactive T cells associated with NOD-Y3 mice. A more difficult question to resolve is whether accelerated diabetes in NOD-Y3 mice is due to the reduced intrinsic IL-2Rβ signaling by autoreactive T cells that enhances their activity or is simply the result of reduced Treg activity.
Although male NOD mice are relatively resistant to diabetes, a particularly striking result was that NOD-Y3 males developed diabetes at a similar rate to that of NOD females. Sex-biased development of T1D in NOD mice has been attributed to differences in microbiota and sex hormones (60–62). Indeed, differences in sex hormones have been suggested to influence the type of microbiota in male and female NOD mice (62), raising the possibility that these two factors are interrelated. Although our study did not address the factors responsible for the lack of sex-bias in NOD-Y3 mice, we speculate that altering the number of eTregs (which dominate the gut mucosa) through reduced IL-2R signaling distinctively shapes immune homeostasis in the gut mucosa to influence the microbiota in a way that promotes diabetes in both male and female mice. Alternatively, decreased Treg function as a result of reduced IL-2R signaling in NOD-Y3 mice may be sufficient to counteract the protective effects in male NOD mice that are attributed to microbiota and sex hormones. Experimentation is required to assess these points.
The IL-2–IL-2R axis has been implicated as a risk factor not only for T1D, but also for other autoimmune diseases, including multiple sclerosis, celiac disease, and rheumatoid arthritis. IL-2RβY3 reduced IL-2R signaling in all T cells, and this situation is predicted to mimic genetic variation due to SNPs within the noncoding region of IL2RA that may alter gene expression. Thus, the effects we noted in NOD-Y3 mice might also be detected in humans that carry some IL2RA risk alleles. In addition, low-dose IL-2 represents a promising strategy to boost Tregs in patients with autoimmunity (63). Our study raises the possibility that along with evaluating the capacity of low-dose IL-2 to increase the total number of Tregs, one may also wish to assess how this treatment affects particular subsets of Tregs in order to maximize therapeutic benefits.
Materials and Methods
Mice
NOD CD45.2 congenic (NOD.B6-Ptprcb/6908MrkJacJ) and NOD.SCID (NOD.CB17-PrkdcSCID/J) mice were purchased from Jackson Laboratories. NOD-Y3 IL-2Rβ+/+, NOD-Y3 IL-2Rβ +/−, NOD-Y3 IL-2Rβ−/− mice, where Y3 refers to the mutant IL-2Rβ transgene, were developed by backcrossing C57BL/6-Y3 IL-2Rβ−/− mice (32) to NOD/ShiLtJ mice for 12 generations. All Idd loci were confirmed to be of NOD origin. NOD/Foxp3-RFP reporter (FIR) mice were previously described (64) and crossed to NOD BDC2.5 and NOD-Y3 IL-2Rβ +/+ mice to yield NOD-Y3-FIR, NOD BDC2.5-FIR, and NOD-Y3 BDC2.5-FIR mice. All mice were housed in a specific pathogen-free animal facility at the University of Miami. The Institutional Animal Care and Use Committee at the University of Miami reviewed and approved all animal studies conducted in this study.
Assessment of diabetes, insulin autoantibodies, and islet infiltration scoring
Diabetes was determined by following urine glucose 2–3 times per week using Diastix strips (Bayer). After 2 positive urine tests (>250 mg/dL), blood glucose was assessed using OneTouch Ultra strips. Mice were considered diabetic when blood glucose readings were >250 mg/dL. Serum was analyzed for insulin autoantibodies by radioimmunoassay as previously described (65). To assess islet inflammation, the pancreas was fixed in 10% neutral buffered formalin, embedded in paraffin, cut into 5 micron sections, and stained with hematoxylin-eosin. Lymphocytic infiltration was scored for each islet and assessed blindly by light microscopy. Insulitis scoring was conducted per the following criteria: 0, no insulitis (free of infiltration); 1, peri/polar-insulitis (infiltration confined to the periphery of islets); 2, mild insulitis (<50% of the islet area infiltrated); 3, severe insulitis (≥50% of the islet area infiltrated); and 4, massive insulitis (≥90% of the islet area infiltrated) (66).
FACS analysis
Cell staining for FACS analysis was performed as previously described (67). Monoclonal antibodies (mAbs) specific for the following targets were used for flow cytometry: CD25 (PC61), pSTAT5 (pY641), Ki67 (B56), and CD11c (HL3) were purchased from BD bioscience; Foxp3 (FJK-16s), CD62L (MEL-14), IL-2 (JES6-5H4), CD103 (2E7), Klrg1 (2F1), ICOS (15F9), and streptavidin were purchased from eBioscience. Antibodies against CD122 (Tm-b1), CD4 (RM4-5), CD8 (53-6.7), IFN-γ (XMG1.2), TNF-α (MPG-XT22), IL-17 (TC11-18H10.1), CD69 (H1.2F3), Bcl-2 (BCL/10C4), CD45.1 (A20), CD45.2 (104), CD19 (GD5), CD49b (DX5), Ly6G (1A8), and CD11b (M1/70) and streptavidin were purchased from Biolegend. Antibodies against CD4 (GK1.5) and CD44 (PGP-1) were purified and conjugated in house. Intracellular staining for Foxp3 (eBioscience) was performed as recommended by the manufacturer. For intracellular staining of cytokines, the BD Cytofix/Cytoperm kit was used according to the manufacturer’s protocol. Samples were analyzed on a BD LSR-Fortessa-HTS or a BD LSRII flow cytometer. Typically, 200,000 total events were collected per sample. Flow cytometry data were analyzed with BD FACSDiva software (version 8.0.1).
Cell isolation and sorting
To isolate lymphoid cells from the mouse pancreas, pancreases were minced into ~2 mm2 pieces in Hank’s Balanced Salt Solution (HBSS) containing 50% fetal calf serum (FCS) and incubated at 37°C for 3 hours in a 7% CO2 incubator. The total pancreas pieces were then passed through a 70-μm nylon strainer (Falcon). The resulting single-cell suspension was centrifuged at 480 g for 5 min, pelleted, and then washed 1x with HBSS. To prepare T cell-depleted bone marrow cells, mAbs against CD4 (0.2 mL culture supernatant; RL1724), CD8 (0.2 mL culture supernatant; HO22), Thy-1.2 (10 μg purified antibody; 30-H12 Sigma Aldrich), DNAse (10 μg; Sigma Aldrich), and rabbit complement (0.12 mL; Cedarlane) were added to 1 mL of bone marrow cells (25 × 106/mL in RPMI 1640 containing 5% FCS). This mixture was incubated on ice for 15 min and then incubated 37°C for 30 min. After this time, the cells were washed 1x in RPMI 1640. In most experiments, CD4+ Tregs and T conventional cells from the spleen or pancreas were surface stained and sorted based on the presence or absence, respectively, of the Foxp3-RFP reporter dye with FACS-Aria-II or Beckman Coulter Moflo Astrios EQ cell sorters. Cell purity was usually greater than 95%. For the adoptive transfer of Tregs into NOD-Y3 IL-2Rβ−/− mice, where some donor mice were not on the Foxp3-RFP reporter background, Tregs were sorted based on CD4+ CD62Lhi CD25hi expression. This population was greater than 93% Foxp3+ cells.
Functional and molecular analyses
To measure intracellular cytokines by flow cytometry, cells (1–2 × 106) were cultured in 24 well flat bottom plates with RPMI 1640 complete medium (68) containing PMA (50 ng/mL), ionomycin (1 μM), and brefeldin A (5 μg) (Biolegend) for 4 hours. To assess mRNA abundance, cell lysates from FACS-purified cells were prepared with RLT lysis buffer (Qiagen). These lysates (10,000 cell equivalents) were subjected to Nanostring analysis using the nCounter Mouse Immunology Gene Expression Code set (Nanostring). Expression data were analyzed by normalizing counts to housekeeping genes and positive and negative controls using nSolver version 2.5 (38). Normalized counts were log2-transformed and the data were analyzed by Student’s two-tailed t test. IL-2- (eBioscience) and IL-15- (Peprotech) dependent pSTAT5 activation was assessed by flow cytometry as previously described (7), either directly ex vivo or after stimulation in vitro with the appropriate cytokine for 15 min. The resulting dose-response curve data were subjected to non-linear regression analysis using GraphPad prism software to determine the EC50.
In vivo studies
For adoptive transfer studies, the appropriate cell populations were transferred i.v. through the tail vein into NOD.SCID or NOD-Y3 IL-2Rβ−/− recipient mice. Glucose concentrations were monitored in the NOD.SCID recipients for up to 24 days after transfer, as required. For bone marrow chimeras, one day before cell transfer, NOD.SCID recipients were subjected to 3.5 Gy of gamma irradiation. Donor T cell–depleted bone marrow was mixed, as indicated in the Results, and injected i.v. into the tail vein. Blood was collected to monitor donor cell reconstitution before the recipient mice were sacrificed 8 weeks after transfer.
Statistical analysis
All data are represented as the mean ± SEM. Statistical tests employed are listed in the figure legends. Data were analyzed using Wilcoxon signed-rank tests for FACS expression based on the mean fluorescent intensity (MFI), where the MFI of cells from wild-type NOD mice were normalized to 1. The log-rank test was used to assess diabetes instances, the Kruskal-Wallis test was used to assess multi-group comparisons, and the Mann-Whitney test was used to assess autoantibody titers and two group comparisons. Two-tail unpaired t-tests were used to analyze pancreatic Treg Nanostring data. Statistical comparisons were performed using GraphPad Prism (version 7.01). Statistical significance is indicated in the figure legends as follows: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Supplementary Material
Fig. S1. Analysis of STAT5 activation in conventional T cells from male NOD-Y3 mice.
Fig. S2. Flow cytometric analysis of T lymphocytes from female NOD-Y3 mice.
Fig. S3. Analysis of IL-2RβY3 expression on T lymphocytes from mice on the NOD BDC2.5 background.
Fig. S4. Analysis of cell distribution in the non–T cell compartment in male NOD-Y3 mice.
Table S1. Nanostring analysis of mRNA expression in pancreatic Tregs from male NOD and NOD-Y3 mice.
Editor’s summary.
IL-2 signaling and autoimmunity
Regulatory T cells (Tregs) suppress autoreactive effector T cells to prevent the occurrence of autoimmune diseases, such as type I diabetes (T1D). The cytokine interleukin-2 (IL-2) is critical for the development and homeostasis of Treg subsets. Polymorphisms in the genes encoding IL-2 and its receptor subunits are associated with an increased risk of developing autoimmunity. To examine the effect of decreased IL-2 signaling, Dwyer et al. expressed a signaling-defective mutant IL-2 receptor (IL-2RβY3) in T cells in NOD mice, a model of T1D. Compared to NOD mice expressing wild-type IL-2Rβ, those expressing IL-2RβY3 in their T cells had accelerated onset of T1D. This was associated with a decrease in the numbers and suppressive activity of different Treg subsets and in the infiltration of autoreactive effector T cells into the pancreas. Together, these data suggest that the use of low-dose IL-2 to therapeutically modulate different Treg subsets in the context of autoimmune disease should be evaluated.
Acknowledgments
We thank the flow cytometry cores of the Diabetes Research Institute and the Sylvester Comprehensive Cancer Center at the University of Miami for help with cell sorting and flow cytometry analysis; K. Johnson for his preparation of pancreatic sections; and the Oncogenomics Core Facility at the University of Miami for help with Nanostring sample preparation.
Funding: This research was supported by the National Institutes of Health grant R01DK093866.
Footnotes
Author contributions: TRM conceived and supervised the study; TRM, ZC and CJD, designed experiments; ALB and CC-K developed the NOD-Y3 mice; CJD performed most experiments with help from CF in assessing islet inflammation, LY in determining insulin autoantibody titers, NCW with the anti-IL-2 in vivo experiments, and KHT in assessing the development of eTregs from cTregs; CJD and TRM analyzed the data; and CJD and TRM wrote the paper with edits from ALG, CF, KHT, and ZC.
Competing interests: The authors declare that they have no competing interests.
References and Notes
- 1.Fehervari Z, Sakaguchi S. CD4+ Tregs and immune control. J Clin Invest. 2004;114:1209–1217. doi: 10.1172/JCI23395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gavin M, Rudensky A. Control of immune homeostasis by naturally arising regulatory CD4+ T cells. Curr Opin Immunol. 2003;15:690–696. doi: 10.1016/j.coi.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Malek TR. The biology of interleukin-2. Annu Rev Immunol. 2008;26:453–479. doi: 10.1146/annurev.immunol.26.021607.090357. [DOI] [PubMed] [Google Scholar]
- 4.Malek TR, Bayer AL. Tolerance, not immunity, crucially depends on IL-2. Nat Rev Immunol. 2004;4:665–674. doi: 10.1038/nri1435. [DOI] [PubMed] [Google Scholar]
- 5.Burchill MA, Yang J, Vang KB, Moon JJ, Chu HH, Lio CW, Vegoe AL, Hsieh CS, Jenkins MK, Farrar MA. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 2008;28:112–121. doi: 10.1016/j.immuni.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 7.Cheng G, Yu A, Dee MJ, Malek TR. IL-2R signaling is essential for functional maturation of regulatory T cells during thymic development. J Immunol. 2013;190:1567–1575. doi: 10.4049/jimmunol.1201218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28:100–111. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bayer AL, Yu A, Adeegbe D, Malek TR. Essential role for interleukin-2 for CD4+CD25+ T regulatory cell development during the neonatal period. J Exp Med. 2005;201:769–777. doi: 10.1084/jem.20041179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malek TR, Castro I. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity. 2010;33:153–165. doi: 10.1016/j.immuni.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smigiel KS, Richards E, Srivastava S, Thomas KR, Dudda JC, Klonowski KD, Campbell DJ. CCR7 provides localized access to IL-2 and defines homeostatically distinct regulatory T cell subsets. J Exp Med. 2014;211:121–136. doi: 10.1084/jem.20131142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng G, Yuan X, Tsai MS, Podack ER, Yu A, Malek TR. IL-2 receptor signaling is essential for the development of Klrg1+ terminally differentiated T regulatory cells. J Immunol. 2012;189:1780–1791. doi: 10.4049/jimmunol.1103768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat Immunol. 2005;6:1142–1151. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- 14.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou L, Lopes JE, Chong MM, Ivanov, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, Ziegler SF, Littman DR. TGF-β-induced Foxp3 inhibits TH17 cell differentiation by antagonizing RORγt function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pandiyan P, Zheng L, Ishihara S, Reed J, Lenardo MJ. CD4+CD25+Foxp3+ regulatory T cells induce cytokine deprivation-mediated apoptosis of effector CD4+ T cells. Nat Immunol. 2007;8:1353–1362. doi: 10.1038/ni1536. [DOI] [PubMed] [Google Scholar]
- 18.Weist BM, Kurd N, Boussier J, Chan SW, Robey EA. Thymic regulatory T cell niche size is dictated by limiting IL-2 from antigen-bearing dendritic cells and feedback competition. Nat Immunol. 2015;16:635–641. doi: 10.1038/ni.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium WTCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vella A, Cooper JD, Lowe CE, Walker N, Nutland S, Widmer B, Jones R, Ring SM, McArdle W, Pembrey ME, Strachan DP, Dunger DB, Twells RC, Clayton DG, Todd JA. Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76:773–779. doi: 10.1086/429843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garg G, Tyler JR, Yang JH, Cutler AJ, Downes K, Pekalski M, Bell GL, Nutland S, Peakman M, Todd JA, Wicker LS, Tree TI. Type 1 diabetes-associated IL2RA variation lowers IL-2 signaling and contributes to diminished CD4+CD25+ regulatory T cell function. J Immunol. 2012;188:4644–4653. doi: 10.4049/jimmunol.1100272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindley S, Dayan CM, Bishop A, Roep BO, Peakman M, Tree TI. Defective suppressor function in CD4+CD25+ T-cells from patients with type 1 diabetes. Diabetes. 2005;54:92–99. doi: 10.2337/diabetes.54.1.92. [DOI] [PubMed] [Google Scholar]
- 23.Long SA, Cerosaletti K, Bollyky PL, Tatum M, Shilling H, Zhang S, Zhang ZY, Pihoker C, Sanda S, Greenbaum C, Buckner JH. Defects in IL-2R signaling contribute to diminished maintenance of FOXP3 expression in CD4+CD25+ regulatory T-cells of type 1 diabetic subjects. Diabetes. 2010;59:407–415. doi: 10.2337/db09-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wicker LS, Todd JA, Prins JB, Podolin PL, Renjilian RJ, Peterson LB. Resistance alleles at two non-major histocompatibility complex-linked insulin-dependent diabetes loci on chromosome 3, Idd3 and Idd10, protect nonobese diabetic mice from diabetes. J Exp Med. 1994;180:1705–1713. doi: 10.1084/jem.180.5.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamanouchi J, Rainbow D, Serra P, Howlett S, Hunter K, Garner VE, Gonzalez-Munoz A, Clark J, Veijola R, Cubbon R, Chen SL, Rosa R, Cumiskey AM, Serreze DV, Gregory S, Rogers J, Lyons PA, Healy B, Smink LJ, Todd JA, Peterson LB, Wicker LS, Santamaria P. Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nat Genet. 2007;39:329–337. doi: 10.1038/ng1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Q, Adams JY, Penaranda C, Melli K, Piaggio E, Sgouroudis E, Piccirillo CA, Salomon BL, Bluestone JA. Central role of defective interleukin-2 production in the triggering of islet autoimmune destruction. Immunity. 2008;28:687–697. doi: 10.1016/j.immuni.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sgouroudis E, Albanese A, Piccirillo CA. Impact of protective IL-2 allelic variants on CD4+ Foxp3+ regulatory T cell function in situ and resistance to autoimmune diabetes in NOD mice. J Immunol. 2008;181:6283–6292. doi: 10.4049/jimmunol.181.9.6283. [DOI] [PubMed] [Google Scholar]
- 28.McGuire HM, Vogelzang A, Hill N, Flodstrom-Tullberg M, Sprent J, King C. Loss of parity between IL-2 and IL-21 in the NOD Idd3 locus. Proc Natl Acad Sci U S A. 2009;106:19438–19443. doi: 10.1073/pnas.0903561106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malek TR, Yu A, Vincek V, Scibelli P, Kong L. CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rβ-deficient mice. Implications for the nonredundant function of IL-2. Immunity. 2002;17:167–178. doi: 10.1016/s1074-7613(02)00367-9. [DOI] [PubMed] [Google Scholar]
- 30.Chinen T, Kannan AK, Levine AG, Fan X, Klein U, Zheng Y, Gasteiger G, Feng Y, Fontenot JD, Rudensky AY. An essential role for the IL-2 receptor in Treg cell function. Nat Immunol. 2016;17:1322–1333. doi: 10.1038/ni.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki H, Kundig TM, Furlonger C, Wakeham A, Timms E, Matsuyama T, Schmits R, Simard JJ, Ohashi PS, Griesser H, et al. Deregulated T cell activation and autoimmunity in mice lacking interleukin-2 receptor beta. Science. 1995;268:1472–1476. doi: 10.1126/science.7770771. [DOI] [PubMed] [Google Scholar]
- 32.Yu A, Zhu L, Altman NH, Malek TR. A low interleukin-2 receptor signaling threshold supports the development and homeostasis of T regulatory cells. Immunity. 2009;30:204–217. doi: 10.1016/j.immuni.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malek TR, Ashwell JD. Interleukin 2 upregulates expression of its receptor on a T cell clone. J Exp Med. 1985;161:1575–1580. doi: 10.1084/jem.161.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castro I, Yu A, Dee MJ, Malek TR. The basis of distinctive IL-2- and IL-15-dependent signaling: weak CD122-dependent signaling favors CD8+ T central-memory cell survival but not T effector-memory cell development. J Immunol. 2011;187:5170–5182. doi: 10.4049/jimmunol.1003961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haskins K, Cooke A. CD4 T cells and their antigens in the pathogenesis of autoimmune diabetes. Curr Opin Immunol. 2011;23:739–745. doi: 10.1016/j.coi.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carrio R, Bathe OF, Malek TR. Initial antigen encounter programs CD8+ T cells competent to develop into memory cells that are activated in an antigen-free, IL-7- and IL-15-rich environment. J Immunol. 2004;172:7315–7323. doi: 10.4049/jimmunol.172.12.7315. [DOI] [PubMed] [Google Scholar]
- 37.Manjunath N, Shankar P, Wan J, Weninger W, Crowley MA, Hieshima K, Springer TA, Fan X, Shen H, Lieberman J, von Andrian UH. Effector differentiation is not prerequisite for generation of memory cytotoxic T lymphocytes. J Clin Invest. 2001;108:871–878. doi: 10.1172/JCI13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toomer KH, Yuan X, Yang J, Dee MJ, Yu A, Malek TR. Developmental Progression and Interrelationship of Central and Effector Regulatory T Cell Subsets. J Immunol. 2016;196:3665–3676. doi: 10.4049/jimmunol.1500595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feuerer M, Hill JA, Kretschmer K, von Boehmer H, Mathis D, Benoist C. Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proc Natl Acad Sci U S A. 2010;107:5919–5924. doi: 10.1073/pnas.1002006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cretney E, Xin A, Shi W, Minnich M, Masson F, Miasari M, Belz GT, Smyth GK, Busslinger M, Nutt SL, Kallies A. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat Immunol. 2011;12:304–311. doi: 10.1038/ni.2006. [DOI] [PubMed] [Google Scholar]
- 41.Levine AG, Arvey A, Jin W, Rudensky AY. Continuous requirement for the TCR in regulatory T cell function. Nat Immunol. 2014;15:1070–1078. doi: 10.1038/ni.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey-Bucktrout SL, Martinez-Llordella M, Zhou X, Anthony B, Rosenthal W, Luche H, Fehling HJ, Bluestone JA. Self-antigen-driven activation induces instability of regulatory T cells during an inflammatory autoimmune response. Immunity. 2013;39:949–962. doi: 10.1016/j.immuni.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol. 2007;8:277–284. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- 44.Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat Immunol. 2006;7:401–410. doi: 10.1038/ni1318. [DOI] [PubMed] [Google Scholar]
- 45.Rubtsov YP, Niec RE, Josefowicz S, Li L, Darce J, Mathis D, Benoist C, Rudensky AY. Stability of the regulatory T cell lineage in vivo. Science. 2010;329:1667–1671. doi: 10.1126/science.1191996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez-Rodriguez J, Meylan F, Handon R, Hayes ET, Anderson SM, Kirby MR, Siegel RM, Schwartzberg PL. Itk is required for Th9 differentiation via TCR-mediated induction of IL-2 and IRF4. Nature communications. 2016;7:10857. doi: 10.1038/ncomms10857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villarino A, Laurence A, Robinson GW, Bonelli M, Dema B, Afzali B, Shih HY, Sun HW, Brooks SR, Hennighausen L, Kanno Y, O’Shea JJ. Signal transducer and activator of transcription 5 (STAT5) paralog dose governs T cell effector and regulatory functions. eLife. 2016;5 doi: 10.7554/eLife.08384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xin A, Masson F, Liao Y, Preston S, Guan T, Gloury R, Olshansky M, Lin JX, Li P, Speed TP, Smyth GK, Ernst M, Leonard WJ, Pellegrini M, Kaech SM, Nutt SL, Shi W, Belz GT, Kallies A. A molecular threshold for effector CD8+ T cell differentiation controlled by transcription factors Blimp-1 and T-bet. Nat Immunol. 2016;17:422–432. doi: 10.1038/ni.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- 50.Berard M, Brandt K, Bulfone-Paus S, Tough DF. IL-15 promotes the survival of naive and memory phenotype CD8+ T cells. J Immunol. 2003;170:5018–5026. doi: 10.4049/jimmunol.170.10.5018. [DOI] [PubMed] [Google Scholar]
- 51.Sandau MM, Kohlmeier JE, Woodland DL, Jameson SC. IL-15 regulates both quantitative and qualitative features of the memory CD8 T cell pool. J Immunol. 2010;184:35–44. doi: 10.4049/jimmunol.0803355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bobbala D, Chen XL, Leblanc C, Mayhue M, Stankova J, Tanaka T, Chen YG, Ilangumaran S, Ramanathan S. Interleukin-15 plays an essential role in the pathogenesis of autoimmune diabetes in the NOD mouse. Diabetologia. 2012;55:3010–3020. doi: 10.1007/s00125-012-2675-1. [DOI] [PubMed] [Google Scholar]
- 53.Cote-Sierra J, Foucras G, Guo L, Chiodetti L, Young HA, Hu-Li J, Zhu J, Paul WE. Interleukin 2 plays a central role in Th2 differentiation. Proc Natl Acad Sci U S A. 2004;101:3880–3885. doi: 10.1073/pnas.0400339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao W, Lin JX, Wang L, Li P, Leonard WJ. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat Immunol. 2011;12:551–559. doi: 10.1038/ni.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao W, Schones DE, Oh J, Cui Y, Cui K, Roh TY, Zhao K, Leonard WJ. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor α-chain expression. Nat Immunol. 2008;9:1288–1296. doi: 10.1038/ni.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Villarino AV, Stumhofer JS, Saris CJ, Kastelein RA, de Sauvage FJ, Hunter CA. IL-27 limits IL-2 production during Th1 differentiation. J Immunol. 2006;176:237–247. doi: 10.4049/jimmunol.176.1.237. [DOI] [PubMed] [Google Scholar]
- 57.Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, Shevach EM, O’Shea JJ. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 59.Vukkadapu SS, Belli JM, Ishii K, Jegga AG, Hutton JJ, Aronow BJ, Katz JD. Dynamic interaction between T cell-mediated β-cell damage and β-cell repair in the run up to autoimmune diabetes of the NOD mouse. Physiol Genomics. 2005;21:201–211. doi: 10.1152/physiolgenomics.00173.2004. [DOI] [PubMed] [Google Scholar]
- 60.Makino S, Kunimoto K, Muraoka Y, Katagiri K. Effect of castration on the appearance of diabetes in NOD mouse. Jikken Dobutsu. 1981;30:137–140. doi: 10.1538/expanim1978.30.2_137. [DOI] [PubMed] [Google Scholar]
- 61.Fox HS. Androgen treatment prevents diabetes in nonobese diabetic mice. J Exp Med. 1992;175:1409–1412. doi: 10.1084/jem.175.5.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, Antonopoulos D, Umesaki Y, Chervonsky AV. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klatzmann D, Abbas AK. The promise of low-dose interleukin-2 therapy for autoimmune and inflammatory diseases. Nat Rev Immunol. 2015;15:283–294. doi: 10.1038/nri3823. [DOI] [PubMed] [Google Scholar]
- 64.Miska J, Abdulreda MH, Devarajan P, Lui JB, Suzuki J, Pileggi A, Berggren PO, Chen Z. Real-time immune cell interactions in target tissue during autoimmune-induced damage and graft tolerance. J Exp Med. 2014;211:441–456. doi: 10.1084/jem.20130785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu L, Robles DT, Abiru N, Kaur P, Rewers M, Kelemen K, Eisenbarth GS. Early expression of antiinsulin autoantibodies of humans and the NOD mouse: evidence for early determination of subsequent diabetes. Proc Natl Acad Sci U S A. 2000;97:1701–1706. doi: 10.1073/pnas.040556697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Faleo G, Fotino C, Bocca N, Molano RD, Zahr-Akrawi E, Molina J, Villate S, Umland O, Skyler JS, Bayer AL, Ricordi C, Pileggi A. Prevention of autoimmune diabetes and induction of β-cell proliferation in NOD mice by hyperbaric oxygen therapy. Diabetes. 2012;61:1769–1778. doi: 10.2337/db11-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bayer AL, Yu A, Malek TR. Function of the IL-2R for thymic and peripheral CD4+CD25+ Foxp3+ T regulatory cells. J Immunol. 2007;178:4062–4071. doi: 10.4049/jimmunol.178.7.4062. [DOI] [PubMed] [Google Scholar]
- 68.Dalyot-Herman N, Bathe OF, Malek TR. Reversal of CD8+ T cell ignorance and induction of anti-tumor immunity by peptide-pulsed APC. J Immunol. 2000;165:6731–6737. doi: 10.4049/jimmunol.165.12.6731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Analysis of STAT5 activation in conventional T cells from male NOD-Y3 mice.
Fig. S2. Flow cytometric analysis of T lymphocytes from female NOD-Y3 mice.
Fig. S3. Analysis of IL-2RβY3 expression on T lymphocytes from mice on the NOD BDC2.5 background.
Fig. S4. Analysis of cell distribution in the non–T cell compartment in male NOD-Y3 mice.
Table S1. Nanostring analysis of mRNA expression in pancreatic Tregs from male NOD and NOD-Y3 mice.