SUMMARY
Clostridium (Clostridioides) difficile is a gastrointestinal pathogen that colonizes the intestinal tract of mammals and can cause severe diarrheal disease. Although C. difficile growth is confined to the intestinal tract, our understanding of the specific metabolites and host factors that are important for the growth of the bacterium is limited. In other enteric pathogens, the membrane-derived metabolite, ethanolamine (EA), is utilized as a nutrient source and can function as a signal to initiate the production of virulence factors. In this study, we investigated the effects of ethanolamine and the role of the predicted ethanolamine gene cluster (CD1907–CD1925) on C. difficile growth. Using targeted mutagenesis, we disrupted genes within the eut cluster and assessed their roles in ethanolamine utilization, and the impact of eut disruption on the outcome of infection in a hamster model of disease. Our results indicate that the eut gene cluster is required for the growth of C. difficile on ethanolamine as a primary nutrient source. Further, the inability to utilize ethanolamine resulted in greater virulence and a shorter time to morbidity in the animal model. Overall, these data suggest that ethanolamine is an important nutrient source within the host and that, in contrast to other intestinal pathogens, the metabolism of ethanolamine by C. difficile can delay the onset of disease.
Keywords: Clostridium difficile, Clostridioides, difficile, ethanolamine, metabolism, ethanolamine metabolism, ethanolamine utilization, eut, tcdA, tcdB, virulence, pathogenesis, branched-chain amino acids (BCAA)
INTRODUCTION
Clostridium (Clostridioides) difficile is a common nosocomial pathogen that can cause severe intestinal infections. C. difficile pathogenesis results primarily from the actions of two large exotoxins, TcdA and TcdB, that destroy host intestinal tissues (Lyerly et al., 1985; Lyras et al., 2009; Kuehne et al., 2010). This host cell destruction is thought to liberate cell contents that provide additional nutrients to support bacterial growth. However, little is known about how cell contents and other metabolites available in the intestine impact growth, or how the diverse metabolic pathways of C. difficile contribute to colonization and virulence.
A number of overlapping regulatory factors have been identified that work to govern C. difficile toxin expression (Martin-Verstraete et al., 2016). The uniting feature of many of these regulatory mechanisms is their connection with nutrient availability (Bouillaut et al., 2015). It is well established that the nutritional state of C. difficile has a major effect on the regulation of toxin expression, as well as the expression of nutrient acquisition mechanisms. Several studies have demonstrated that sugars and peptides suppress C. difficile toxin expression through the activity of nutrient-responsive regulators, such as CcpA and CodY (Dupuy and Sonenshein, 1998; Dineen et al., 2007; Antunes et al., 2011). When peptides and simple carbohydrates are depleted, the repression of toxin and nutrient acquisition mechanisms by CodY and CcpA is alleviated, and C. difficile utilizes alternative nutrients to sustain growth, though the details of this process in vivo are limited.
One particularly abundant potential nutrient in the intestine is phosphatidylethanolamine, a ubiquitous component of cell membranes that is readily hydrolyzed into ethanolamine and glycerol (Proulx and Fung, 1969; Larson et al., 1983). Ethanolamine is a simple amino alcohol that can serve as both a carbon and nitrogen source. Ethanolamine utilization genes have been identified and characterized in several pathogenic species that can inhabit the mammalian intestinal tract, including Salmonella enterica serovar Typhimurium, Escherichia coli, Enterococcus faecalis and Listeria monocytogenes (Faust et al., 1990; Stojiljkovic et al., 1995; Price-Carter et al., 2001; Del Papa and Perego, 2008; Toledo-Arana et al., 2009; Kendall et al., 2012). Though the predicted 19 gene cluster for ethanolamine utilization in C. difficile was identified when the first genome sequence was revealed, the functions and roles of these genes have not been investigated (Sebaihia et al., 2006).
As a strictly intestinal pathogen, the ability to utilize ethanolamine in the host could provide a growth advantage to C. difficile. In the current study, we examined the impact of ethanolamine and the putative ethanolamine utilization gene cluster, on the growth of C. difficile. In addition, we investigated the effect of ethanolamine metabolism on virulence in an animal model of infection. Using null mutants in the eutA and eutG genes, we demonstrate that C. difficile can utilize ethanolamine as a primary nutrient source and show that the proposed eut gene cluster is required for ethanolamine catabolism. Further, we found that the ability to utilize ethanolamine has a significant impact on the timing and intensity of disease in the hamster model of disease, indicating that ethanolamine utilization plays an important role in C. difficile pathogenesis.
RESULTS
C. difficile metabolizes ethanolamine as a primary nutrient source
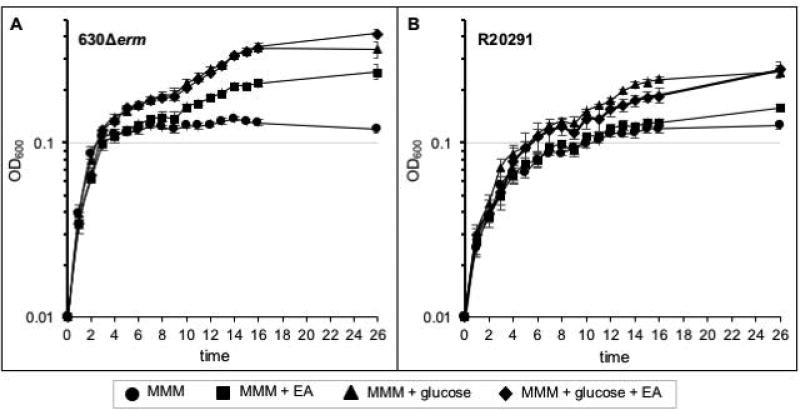
In order to determine if the predicted C. difficile eut genes can facilitate ethanolamine metabolism, we first examined the growth of epidemic and historical isolates of C. difficile (R20291 and 630Δerm strains, respectively) when ethanolamine is a major nutrient source present in the growth medium (Supplementary File S1). As shown in Figure 1, C. difficile strains 630Δerm and R20291 grown in a minimal defined medium with only amino acids present as a carbon source were able to replicate, but the bacteria reached stationary phase at a relatively low cell density. The addition of either ethanolamine or glucose to the medium produced a biphasic growth curve and increased the final density of the bacterial cultures, as well as the time needed to achieve maximum cell density. Notably, the 630Δerm strain grew faster and to higher cell density than the R20291 strain under all of the tested conditions (Figure 1 and S3). No apparent differences in cellular morphology were observed between strains grown in minimal medium or with the addition of ethanolamine (Figure S4). The similar slope of logarithmic growth in minimal medium with and without added carbon sources, followed by a protracted growth phase with the addition of either glucose or ethanolamine, suggests that C. difficile most efficiently utilizes amino acids for growth.
Figure 1. C. difficile metabolizes ethanolamine as a primary nutrient source.
Growth curve of strain A) 630Δerm or B) R20291 in modified minimal medium (MMM) or MMM supplemented with 15 mM ethanolamine (EA) and/or 5 mM D-glucose. The average of three independent biological replicates and standard deviations are shown.
eut gene expression is induced by ethanolamine, but is repressed by glucose and decreases during stationary phase growth in the absence of ethanolamine
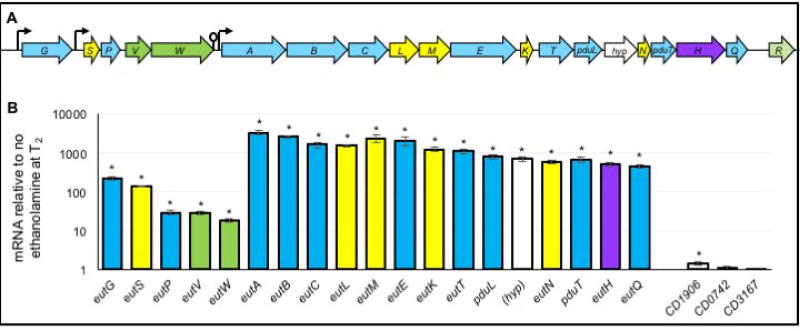
The C. difficile genes encoding the ethanolamine utilization pathway and microcompartment structural components were first identified in the genome of strain 630 (CD1907–CD1925; Figure 2) (Sebaihia et al., 2006; Pitts et al., 2012). Although the ethanolamine genes are located on a mobile genetic element, this region is highly conserved in C. difficile genomes (> 700 genomes at the time of publication), including strains of the epidemic 027 and 078 lineages. Similar to the Enterococcus faecalis ethanolamine cluster, the 19 genes of the C. difficile cluster include 9 enzymes that are involved in ethanolamine metabolism, 6 proteins that comprise the predicted ethanolamine microcompartment, a two-component regulatory system, a transporter, and 1 gene of unknown function (Del Papa and Perego, 2008). An additional MarR-family transcriptional regulator is encoded downstream of the eut gene cluster in most C. difficile genomes, but this gene is not present in strain 630 (Figure 2A). As the putative eut gene cluster was the most likely basis for ethanolamine metabolism, we examined the expression of the eut genes during growth with and without added ethanolamine. Expression of the predicted eut cluster genes (CD1907–CD1925) was induced in response to ethanolamine in the culture medium, and was most pronounced during stationary phase growth (Figure 2B). In addition, expression of the gene immediately upstream of eutG, CD1906, was marginally affected by ethanolamine (1.5-fold increase). Two other predicted ethanolamine-related genes located on different regions of the chromosome, CD0742 and CD3167, were also examined, and no change in expression was observed for either gene.
Figure 2. Expression of ethanolamine utilization genes in C. difficile.
A) Ethanolamine utilization (eut) gene cluster in C. difficile (eutG-eutQ). The eut gene cluster contains 19 predicted open reading frames (predicted functions listed in File S7). Genes in blue encode predicted metabolic enzymes, green encode regulatory factors, yellow are predicted microcompartment structural proteins, purple encode the putative transporter and white (hyp) is of unknown function. The putative factor downstream of eutQ (R, green stripes) is a MarR-family transcriptional regulator encoded in most sequenced C. difficile isolates downstream of eutQ, but is not found in strain 630. Suspected promoters and terminators are shown as arrows and lollipops, respectively. (B) qRT-PCR analysis of putative eut gene expression in strain 630Δerm grown in 70:30 liquid medium with and without the addition of 15 mM ethanolamine (EA). Samples were harvested for RNA isolation two hours after the entry into stationary phase (T2). The means and standard error of the means of three biological replicates are shown. *, p ≤ 0.05 by Student’s two-tailed t-test.
The expression analysis of the eut cluster revealed two distinct patterns of induction for these genes that suggests two polycistronic units are generated upon ethanolamine induction: a eutG-eutW transcript and a eutA-eutQ transcript (Figure 2B). Using cDNA generated from ethanolamine-induced cultures, we evaluated the entire eut gene cluster by performing PCR with primers designed to amplify across adjacent open reading frames to determine which genes were transcribed within the same mRNA (Figure S5). The predicted amplification products were generated across each adjacent eut gene, with the exception of the eutW-eutA segment, which contains a predicted transcriptional terminator that was previously identified (Fox et al., 2009; Ramesh et al., 2012). Altogether, these data demonstrate that the eut gene cluster is comprised of at least two polycistronic units, both of which are induced by ethanolamine.
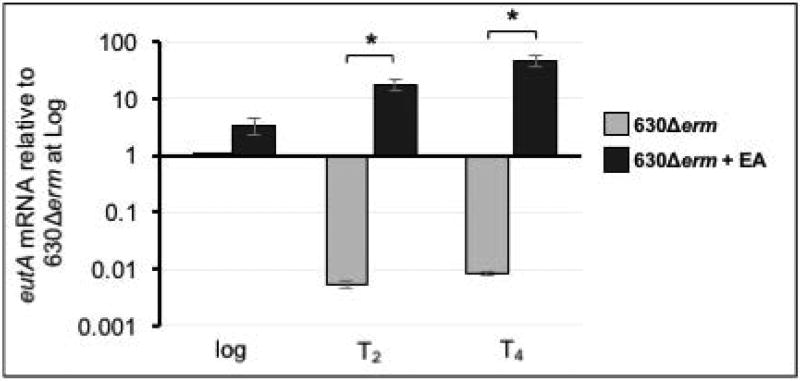
In E. faecalis, eut gene transcription is subject to positive regulatory control by the EutV-EutW two-component system, which disrupts a series of transcriptional terminators in the eut operon and allows transcription to progress (Del Papa and Perego, 2008; Fox et al., 2009; Garsin, 2010). Similar to E. faecalis, we observed basal expression of the eutG-eutQ genes during logarithmic growth in sporulation medium, in the absence of ethanolamine (Figure 3; eutA shown) (Fox et al., 2009). But, as cultures transitioned to stationary phase in sporulation medium, expression of all eut genes sharply decreased if ethanolamine was not present in the medium (Figure 3, eutA shown). The drastic decline in eut gene expression indicates that transcription is repressed during stationary phase, which is consistent with the previous observation that SigF-dependent repression of eut expression occurs in early sporulation (Fimlaid et al., 2013; Saujet et al., 2013).
Figure 3. eut gene expression increases in response to ethanolamine, but expression is repressed during stationary phase if ethanolamine is absent.
qRT-PCR analysis of eutA expression of 630Δerm grown in 70:30 liquid medium with and without the addition of 15 mM ethanolamine (EA). Expression of eutA in 630Δerm grown in medium with and without EA was compared at each timepoint using the Student’s two-tailed t-test. The means and standard error of the means of three biological replicates are shown. * indicates p ≤ 0.05.
Similar to the results in sporulation medium, eut gene expression increased during log-phase growth in minimal medium containing EA (Figure S6A). Although eut genes were induced in strain 630Δerm and the epidemic isolate, R20291, the relative increase in eutA and eutG expression was lower in the epidemic strain. The difference in EA-dependent growth and eut gene induction between the strains suggests that the EA-dependent induction of the eut pathway varies within the species. However, both strains demonstrated no change in eut gene expression in the presence of glucose and intermediate induction of eut genes when both glucose and EA were available (Figure S6). The partial induction of eut genes with glucose and EA present is indicative of incomplete catabolite repression of eut gene expression. No change in tcdA or tcdB expression was observed for either strain when EA, glucose or the combination of EA and glucose were present in the medium (Figure S6B). The predicted regulator downstream of the eut operon in R20291, CDR20291_1847, also did not demonstrate differential expression in the presence of EA or glucose (Figure S6C).
Effects of eut gene disruption on ethanolamine metabolism
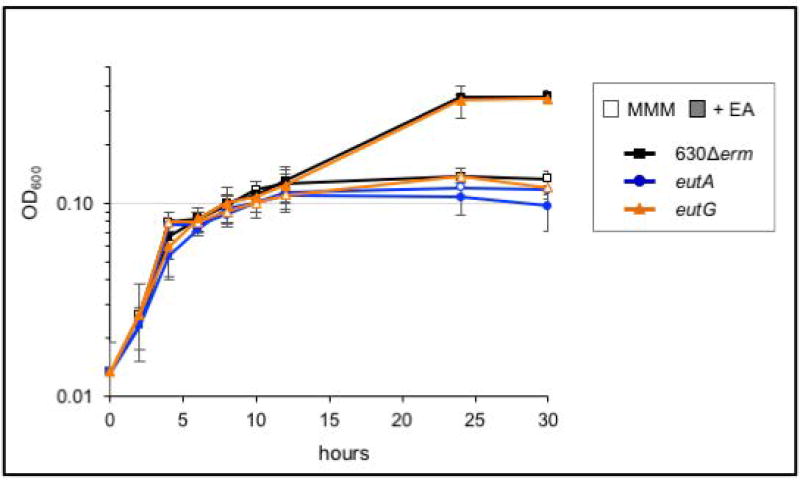
To determine the contribution of the predicted eut operons on ethanolamine metabolism, we generated null mutations via Targetron-based disruption of the upstream most genes of each operon: eutG, an alcohol dehydrogenase, and eutA, an ammonia lyase reactivating factor (Figures S7 and S8)(Karberg et al., 2001; Chen et al., 2005; Liu et al., 2007; Moore and Escalante-Semerena, 2016). The resulting strains were then tested for their ability to utilize ethanolamine as a primary nutrient source, as previously described (Figure 4). The eutG mutant retained the ability to utilize ethanolamine as a nutrient source, as evidenced by the wild-type-like, biphasic curve profile observed during growth in ethanolamine. However, the eutA strain displayed a single exponential growth phase in minimal medium, with or without the addition of ethanolamine, presumably because it was unable to metabolize ethanolamine (Figure 4). Further supporting the minimal medium studies, we observed that the eutA mutant did not produce a drop in the pH of TY medium supplemented with ethanolamine after 24 h growth, in contrast to the parent strain (Table 1). Hence, eutA is required for ethanolamine utilization, but eutG is not. We also observed that the genes downstream of eutA were not induced in the eutA mutant during growth in ethanolamine, implicating a promoter upstream of eutA in ethanolamine-dependent transcription of eutA-eutQ (not shown). Altogether, the data support that the CD1907–CD1925 eut gene cluster is the primary mechanism for ethanolamine utilization in C. difficile.
Figure 4. The eutA operon, and not the eutG transcriptional unit, is essential for ethanolamine metabolism in C. difficile.
Growth curve with 630Δerm (black squares), eutA (MC394; blue circles) and the eutG strain (MC346; orange triangles) grown in modified minimal medium (MMM; open symbols) and with the addition of 15 mM ethanolamine (EA; filled symbols). Averages and standard deviations of three independent experiments shown.
Table 1.
pH of eut cultures after 24h growth
| Strain/condition | Final pHa |
|---|---|
| 630Δerm / TY | 7.41 ± 0.17 |
| 630Δerm / TY+EA 15 mM | 7.23 ± 0.16 |
| 630Δerm / TY+EA 30 mM | 7.05 ± 0.14 |
| 630Δerm / TY+EA 45 mM | 6.89 ± 0.13 |
|
| |
| eutA / TY | 7.37 ± 0.16 |
| eutA / TY+EA 15 mM | 7.32 ± 0.13 |
| eutA / TY+EA 30 mM | 7.33 ± 0.13 |
| eutA / TY+EA 45 mM | 7.30 ± 0.12 |
|
| |
| eutG / TY | 7.30 ± 0.13 |
| eutG / TY+EA 15 mM | 7.20 ± 0.14 |
| eutG / TY+EA 30 mM | 7.06 ± 0.12 |
| eutG / TY+EA 45 mM | 6.91 ± 0.10 |
mean pH of four biological replicates +/− standard deviation shown
bold indicates statistically significant difference between the mutant and parent strain for a given condition (p ≤ 0.05; ANOVA with Sidak’s multiple-comparison test)
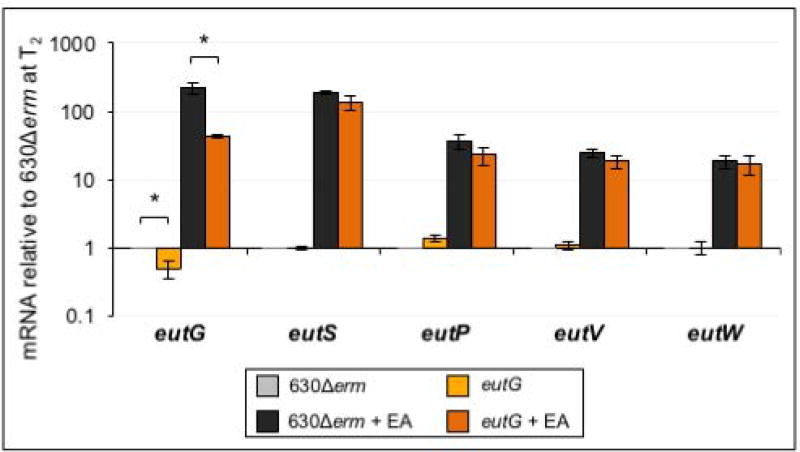
The products of the eutG operon are involved in multiple aspects of ethanolamine processing, including enzymatic functions (eutG and eutP), microcompartment structure (eutS) and transcriptional regulation (eutV and eutW). The absence of an ethanolamine phenotype in the eutG mutant raised the question of how disruption of eutG impacts transcription of the four downstream genes, and what the overall importance of these factors are in ethanolamine utilization. To answer these questions, we further examined the transcription of each gene in the eutG operon in the eutG null mutant. As shown in Figure 5, insertional disruption of eutG reduced eutG expression, relative to the parent strain. But, expression of eutS, eutP, eutV and eutW were still induced in the eutG mutant when ethanolamine was added to the medium, signifying that an additional, ethanolamine-inducible promoter element is present in the intergenic region between eutG and eutS. Thus, the eutG null mutant is deficient only for eutG expression, which has no apparent impact on the growth of C. difficile in ethanolamine (Figure 4).
Figure 5. Insertional disruption of eutG does not affect transcription of downstream genes.
qRT-PCR analysis of eutG, eutS, eutP, eutV and eutW expression in 630Δerm and the eutG mutant (MC346) grown in 70:30 broth with and without the addition of 15 mM ethanolamine (EA). Samples for RNA isolation were collected two hours after the transition into stationary phase (T2). The means and standard error of the means for three biological replicates are shown. Gene expression of 630Δerm and the eutG mutant were compared for each condition using the Student’s two-tailed t-test. * indicates p ≤ 0.05.
An inability to utilize ethanolamine leads to increased virulence in the hamster model of infection
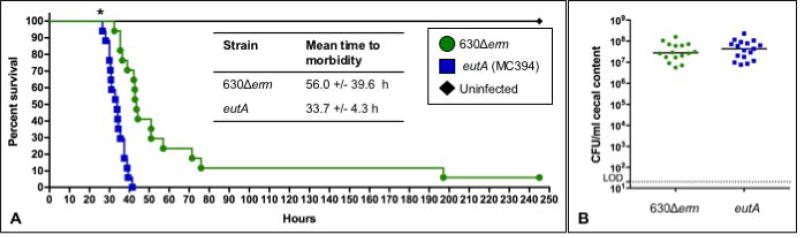
The regulation of C. difficile toxin expression is highly integrated with nutrient availability, alternative energy metabolism, and regulators of metabolic gene expression (Yamakawa et al., 1994; Dupuy and Sonenshein, 1998; Dineen et al., 2007; Karlsson et al., 2008; Dineen et al., 2010; Antunes et al., 2011; Antunes et al., 2012; Bouillaut et al., 2013; Edwards et al., 2014; Dubois et al., 2016; Nawrocki et al., 2016). Previous studies of C. difficile pathogenesis have demonstrated links between metabolism and virulence in animal models of disease, while ethanolamine metabolism is specifically associated with virulence in other enteric species (Joseph et al., 2006; Maadani et al., 2007; Thiennimitr et al., 2011; Kendall et al., 2012; Edwards et al., 2014; Girinathan et al., 2016; Kansau et al., 2016). To determine if ethanolamine utilization impacts virulence in C. difficile infections, we examined the effects of the eutA mutant in a hamster model of C. difficile infection (CDI). Syrian golden hamsters were infected with spores of either the 630Δerm parent strain or the eutA mutant, as described in the Methods. As shown in Figure 6A, hamsters infected with the eutA mutant succumbed to infection in less time than hamsters infected with the parent strain, and the variation in time to morbidity was significantly lower in eutA infections (630Δerm, 56.0 ± 39.6 h; eutA, 33.7 ± 4.3; P≤ 0.01, log rank test). Hamsters infected with the eutA mutant also lost weight more rapidly post-infection than 630Δerm-infected animals, consistent with the rapid time to morbidity observed (data not shown). Although eutA-infected animals demonstrated symptoms and became moribund more quickly, there was no significant difference in the number of C. difficile CFU recovered post-mortem (Figure 6B). These results suggest that the ability to utilize ethanolamine is not necessary for growth of the bacteria in vivo, but ethanolamine metabolism does delay pathogenesis during C. difficile infection.
Figure 6. A eutA mutant is more virulent in the hamster model of CDI.
A) Kaplan-Meier survival curve representing the results from three independent experiments of Syrian golden hamsters infected with C. difficile strain 630Δerm (n=17) or the eutA mutant (MC394; n=17). The mean times to morbidity were 56 +/− 39.6 h for 630Δerm and 33.7 +/− 4.3 h for the eutA mutant (P ≤ 0.01, log rank test). B) Total CFU of C. difficile were enumerated from hamster cecal samples collected post-mortem. Solid lines represent the median CFU for each strain and the dotted line denotes the limit of detection (2 × 101 CFU ml−1). CFU for the mutant and parent strains were compared using the Student’s two-tailed t test (no statistically significant differences were observed).
Ethanolamine and ethanolamine catabolism do not affect the expression of virulence factors, sporulation or germination in vitro
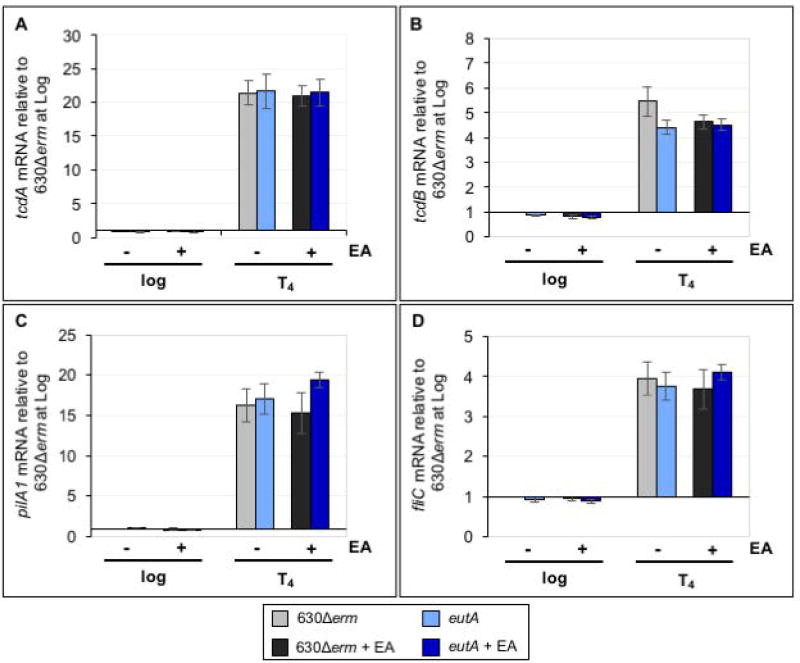
As mentioned, the virulence gene expression and ethanolamine utilization are correlated in other enteric pathogens, and there is evidence that ethanolamine can serve as a signal to induce pathogenesis in some species (Garsin, 2010; Garsin, 2012; Kendall et al., 2012). In view of the increased virulence observed with the eutA mutant, we investigated the effects of ethanolamine and the ability to utilize ethanolamine on genes associated with pathogenesis. Transcriptional analyses of the major toxins, tcdA and tcdB, revealed no change in toxin expression for C. difficile grown in ethanolamine in the eutA mutant or the parent strain in vitro (Figure 7A,B). Likewise, no difference in TcdA protein levels was observed for strains grown in TY medium (Figure S9). We also examined transcription of the major type IV pilin, pilA1, and the flagellar subunit, fliC, which are involved in swarming and swimming motility, respectively (Purcell et al., 2012; Purcell et al., 2016). C. difficile flagella and pilin are detected by the innate immune system and can elicit a response that may impact pathogen clearance or virulence (Pechine et al., 2005; Pechine et al., 2007; Yoshino et al., 2013; Maldarelli et al., 2014; Batah et al., 2016; Ghose et al., 2016; Pechine and Collignon, 2016). Additionally, a connection between ethanolamine and pilin expression has been reported in Escherichia coli O157:H7 (Gonyar and Kendall, 2014). Neither pilA1 nor fliC expression were impacted by the presence or ability to catabolize ethanolamine (Figure 7C,D). These data indicate that ethanolamine itself does not induce these common virulence factors in vitro and that the inability to utilize ethanolamine has no direct impact on the expression of the examined virulence genes.
Figure 7. Ethanolamine metabolism does not impact expression of known virulence factors in vitro.
qRT-PCR analysis of A) tcdA, B) tcdB, C) pilA1 and D) fliC expression in 630Δerm and the eutA mutant (MC394) grown in 70:30 broth with and without the addition of 15 mM EA. Samples for RNA isolation were collected during logarithmic growth (Log, OD600 of 0.5) and four hours after the transition into stationary phase (T4, late stationary). The means and standard error of the means of four biological replicates are shown. Gene expression of 630Δerm and the eutA mutant were compared at each timepoint and condition using the Student’s two-tailed t-test. * indicates p ≤ 0.05 (no significant differences were observed).
Nutrient deprivation and regulation of nutritional responses have also been connected to early entry into the sporulation pathway and increased sporulation frequency for C. difficile (Dineen et al., 2010; Saujet et al., 2011; Antunes et al., 2012; Edwards and McBride, 2014; Edwards et al., 2014; Nawrocki et al., 2016). To determine if ethanolamine or ethanolamine utilization influence sporulation, we assessed sporulation frequency and sporulation-specific gene expression in the eutA mutant and parent strain grown in sporulation medium, with and without ethanolamine. No significant differences were observed in spore formation, sporulation frequency, or transcription of early sporulation genes for the mutant or parent strain grown in 15 mM ethanolamine (Figure S10). Increasing the concentration of ethanolamine to 30 mM also had no effect on sporulation outcomes (data not shown). Moreover, the eutA and eutG mutant spores had no observable germination defect in vitro (Figure S11). Altogether, these results suggest that ethanolamine does not feed into the nutritional regulatory pathways that influence sporulation in C. difficile.
DISCUSSION
Ethanolamine is an abundant membrane-derived compound present in the mammalian intestinal tract (Dowhan, 1997; Vance, 2008). Ethanolamine catabolism provides niche-specific growth advantages to select enteric bacteria, providing these species with an additional source of carbon and nitrogen, as well as extra energy generating potential in the intestinal environment. The ability to utilize ethanolamine has been shown to provide a nutritional growth advantage to other enteric pathogens, and in some cases, ethanolamine amplifies pathogenic phenotypes by inducing the expression of virulence genes (Joseph et al., 2006; Del Papa and Perego, 2008; Srikumar and Fuchs, 2011; Kendall et al., 2012; Luzader et al., 2013; Anderson et al., 2015). Prior analyses of C. difficile gene expression during animal infections indicated that the ethanolamine utilization gene cluster is induced in vivo, but the effects of ethanolamine on growth and virulence were not known (Janoir et al., 2013). In the current work, we demonstrate that the predicted ethanolamine utilization gene cluster is necessary for ethanolamine utilization and this ability has a significant impact on the virulence of C. difficile in an animal model of infection.
As anticipated from the identification of the eut gene homologs on the chromosome and previous assessments of in vivo transcription and metabolic phenotyping, we demonstrated that C. difficile can utilize ethanolamine as a primary nutrient source (Sebaihia et al., 2006; Janoir et al., 2013; Scaria et al., 2014; Theriot and Young, 2014). The addition of ethanolamine to minimal medium supported additional growth and increased cell density, indicating that ethanolamine is catabolized (Figure 1). The expression of eut genes when glucose is present also suggests that eut gene expression can overcome catabolite repression. We also observed that C. difficile grew surprisingly well in base minimal medium with only amino acids as the primary nutrient source. This rapid, peptide-fueled growth was followed by a more protracted growth phase when glucose or ethanolamine were supplemented in the medium, suggesting that C. difficile preferentially catabolizes amino acids over sugars and ethanolamine. If C. difficile also prefers to metabolize amino acids over other carbon sources in vivo, the abundant peptide sources in the intestine could promote the rapid expansion of the population in the intestine (Smith and Macfarlane, 1998). Once the bulk of intestinal peptides are limiting, this dramatic population growth would presumably give way to a slower growth phase with a large population of bacteria to support, and less desired nutrient sources available. Unfortunately, it is difficult to definitively verify whether amino acids are the preferred nutrient source because C. difficile is auxotrophic for many amino acids and therefore, cannot grow without amino acid supplementation (Karasawa et al., 1995). But, it is clear that C. difficile encodes a remarkable repertoire of metabolic capabilities and can readily utilize other intestinal metabolites to generate energy, including a variety of carbohydrates (glucose, lactose, fructose, succinate, sucrose, lactose, trehalose, mannose, cellobiose, galactitol, tagatose, raffinose, stachyose, sorbitol, N-acetylglucosamine and sialic acid), peptides and amino acids via Stickland fermentation, and may even be capable of generating energy by fixation of CO2 (Wilson and Perini, 1988; Karlsson et al., 2000; Jackson et al., 2006; Antunes et al., 2012; Bouillaut et al., 2013; Kopke et al., 2013; Ferreyra et al., 2014; Larocque et al., 2014; Theriot et al., 2014; Theriot and Young, 2015; Kansau et al., 2016).
Despite the large variety of energy sources that C. difficile can utilize, the importance of individual nutrient sources during infection is not well understood. Based on data from other enteric pathogens, we hypothesized that ethanolamine metabolism would be important for the growth of C. difficile in vivo and therefore, the ethanolamine mutant would be less fit for growth in the host than the parent strain (Stojiljkovic et al., 1995; Joseph et al., 2006; Maadani et al., 2007; Luzader et al., 2013; Anderson et al., 2015). Instead, the eutA mutant readily colonized hamsters, achieved equivalent cell densities, and caused more rapid morbidity than animals infected with the parent strain, 630Δerm. These findings suggested that ethanolamine utilization by C. difficile might delay the onset of virulence by postponing toxin expression, which is the main driver of symptoms and morbidity. Given the established links between nutrient availability and toxin expression in C. difficile, we further examined this phenomenon in vitro by assessing toxin gene transcription when ethanolamine was added to the growth medium (Figure 7). We observed no change in tcdA or tcdB transcription or protein production, suggesting that unlike other nutrients, there is no clear link between ethanolamine utilization and the regulation of toxin expression. In addition, ethanolamine supplementation did not impact sporulation frequency or motility gene regulation, as other nutrient sources do (Antunes et al., 2011; Antunes et al., 2012; El Meouche et al., 2013; McKee et al., 2013; Edwards et al., 2014; Edwards et al., 2016a; Nawrocki et al., 2016). The lack of ethanolamine influence on both sporulation and toxin expression strongly suggest that the major global nutritional regulators of sporulation, CodY and CcpA, do not respond directly or indirectly to ethanolamine. The increased virulence of the eutA mutant may be due to an overall lower nutrient availability in vivo, leading to earlier activation of toxin gene expression and virulence, but more work is needed to understand how ethanolamine specifically impacts C. difficile in the host.
It was anticipated that eutA and the downstream eut gene products would be required for ethanolamine metabolism, which was validated by growth assessments with ethanolamine (Figure 4). Less evident was the dispensability of EutG, which was not needed for the growth of C. difficile in ethanolamine. EutG is a predicted alcohol dehydrogenase that is involved in the enzymatic conversion of acetaldehyde to ethanol and is important for growth in ethanolamine for other species (Stojiljkovic et al., 1995). The acetaldehyde generated by the initial steps of ethanolamine metabolism is highly toxic to cells and must be detoxified, though the alternative pathway involving the EutE aldehyde dehydrogenase and the EutD phosphotransacetylase is still available in the eutG mutant for acetaldehyde disposal. The EutE-EutD path generates acetylphosphate, which could then be used to generate acetate, or shunted to either the TCA cycle or biosynthetic pathways (Stojiljkovic et al., 1995; Brinsmade et al., 2005; Garsin, 2010). Additionally, C. difficile strain 630 encodes another five predicted alcohol dehydrogenases that could potentially compensate for the loss of EutG: CD0274, CD0334, CD2966, CD3006 and CD3105. However, the expression and functions of these factors in ethanolamine metabolism have not been investigated.
As observed in other species that encode extensive ethanolamine utilization mechanisms, the entire C. difficile eut gene cluster is induced when ethanolamine is present (Del Papa and Perego, 2008; Garsin, 2010; Anderson et al., 2015). eut gene induction in C. difficile is predicted to be controlled by the EutV-EutW two-component system, which was characterized previously in E. faecalis (Del Papa and Perego, 2008; Fox et al., 2009). In E. faecalis, EutW acts as the ethanolamine sensor histidine kinase that acts on the RNA-binding response regulator, EutV (Fox et al., 2009). Many Gram-negative species encode a positive regulator, EutR, an AraC-like DNA-binding protein that is needed for activation of eut gene expression (Tsoy et al., 2009). Although C. difficile encodes eutV-eutW and not the prototype Gram-negative EutR regulator, most C. difficile strains do encode a MarR-family regulator downstream from the eut gene cluster (CDR20291_1847 in strain R20291; Figure 2A). However, this MarR-family regulator is not present in the 630 genome. No changes in expression of this regulator were observed in the R20291 strain under any of the conditions tested (Figure 6C). Further studies would be needed to determine if this putative regulator contributes to the differences in ethanolamine-dependent growth and gene expression observed in these strains.
The data also demonstrated that the eut genes were constitutively expressed at low levels in MMM during logarithmic phase, and subsequently repressed as cells entered stationary phase growth (Figure 3). This repression is consistent with the previous observation that the early sporulation sigma factor, SigF, represses eut gene expression in C. difficile (Saujet et al., 2013). Since SigF is a transcriptional activator, SigF-dependent repression of the eut genes is likely indirect. This stationary phase repression was overcome when ethanolamine was present (Figure 3), but no ethanolamine-dependent changes in spore formation or germination were observed (Figure S10 and S11). Stationary phase repression of ethanolamine genes has also been observed in E. faecalis, wherein the FsrABC quorum sensing regulatory system represses eut expression in post-exponential phase cells (Bourgogne et al., 2006). Sequenced C. difficile genomes do not encode apparent FsrABC proteins, but similar Agr-like systems have been identified in some strains (Martin et al., 2013). However, examination of transcription in an agrA mutant of strain R20291 did not uncover an impact of the Agr system on eut gene expression (Martin et al., 2013).
These results demonstrate both the function of the ethanolamine gene cluster and the importance of ethanolamine utilization on pathogenesis in an animal model of infection. Complementation studies or in frame deletions of individual ethanolamine utilization genes and regulators could help to clarify the precise regulatory mechanisms and functions of these factors, which we were unable to attain. Further study is needed to determine the precise regulatory mechanisms that govern eut gene expression in C. difficile, in particular to understand what function, if any, the putative MarR-like regulator plays in this process. Moreover, understanding the roles of individual metabolites, such as ethanolamine, in CDI, could help inform ways to circumvent pathogenesis and potentially guide treatment strategies for the prevention of fulminant disease.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
Table 2 provides a list of the strains and plasmids used in this study. C. difficile strains were cultured in an anaerobic chamber (Coy Laboratory Products) at 37°C with an atmosphere of 10% H2, 5% CO2, and 85% N2, as previously described (Bouillaut et al., 2011; Edwards et al., 2013). C. difficile strains were routinely cultured in brain heart infusion-supplemented (BHIS) broth and agar plates (Smith et al., 1981) or TY medium (adjusted to pH 7.4 prior to autoclaving)(Garnier and Cole, 1986). Two to ten µg of thiamphenicol ml−1, or 5 µg of erythromycin ml−1 (Sigma-Aldrich) was used to supplement BHIS medium, as needed. Escherichia coli strains were cultured at 37°C in LB medium supplemented with 20 µg chloramphenicol ml−1 and 100 µg ampicillin ml−1. Following conjugations, 50 µg kanamycin ml−1 was used to counter-select against E. coli (Purcell et al., 2012). To induce the germination of C. difficile spores, BHIS medium was supplemented with 0.1% taurocholate (Sigma-Aldrich) (Sorg and Sonenshein, 2008). d-fructose (0.2%) was added to overnight cultures to prevent sporulation, as needed (Putnam et al., 2013). Minimal medium growth curves were performed using a modified minimal medium (MMM) based on previously described complete defined media (Karlsson et al., 1999; Cartman et al., 2012). This modified minimal medium contained lower final concentrations of amino acids and d-glucose than that used by Cartman et al. (Supplementary File S1). The base salts and amino acids of the minimal medium were supplemented with 5 mM d-glucose and/or 15 mM ethanolamine hydrochloride or liquid ethanolamine (Sigma-Aldrich), as noted. Growth curves in modified minimal medium (MMM) were performed as follows: log-phase cultures were grown to an OD600 of approximately 0.6 and back-diluted 6-fold to an OD600 of 0.1; the resulting dilution (OD600 0.1) was then used to inoculate minimal medium at a further 1:10 dilution (2.5 ml into 22.5 ml of MMM). A minimum of three biological replicates were performed for each growth curve shown. The pH of cultures and solutions was measured as previously described (Edwards et al., 2016b).
Table 2.
Bacterial Strains and plasmids
| Plasmid or Strain | Relevant genotype or features | Source, construction or reference |
|
|---|---|---|---|
| Strains | |||
| E. coli | |||
| HB101 | F− mcrB mrr hsdS20(rB− mB−) recA13 leuB6 ara-14 proA2 lacY1 galK2 xyl-5 mtl-1 rpsL20 | B. Dupuy | |
| MC101 | HB101 pRK24 | B. Dupuy | |
| DH5α Max Efficiency | F− Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rk−, mk+) phoA supE44 λ−thi− 1 gyrA96 relA1 | Invitrogen | |
| MC341 | HB101 pRK24 pMC263 | This study | |
| MC344 | HB101 pRK24 pMC266 | This study | |
| C. difficile | |||
| R20291 | Clinical isolate; ribotype 027 | (Stabler et al., 2009) | |
| 630Δerm | ErmS derivative of strain 630 | N. Minton (Hussain et al., 2005) | |
| MC310 | 630Δerm spo0A::ermB | (Edwards et al., 2014) | |
| MC346 | 630Δerm eutG::ermB | This study | |
| MC394 | 630Δerm eutA::ermB | This study | |
| Plasmids | pCE240 | C. difficile Targetron construct based on pJIR750ai (group II intron, ermB::RAM ltrA) catP | C. Ellermeier (Ho and Ellermeier, 2011) |
| pRK24 | Tra+, Mob+; bla, tet | (Thomas and Smith, 1987) | |
| pCR2.1 | bla kan | Invitrogen | |
| pMC123 | E. coli-C. difficile shuttle vector; bla, catP | (McBride and Sonenshein, 2011b) | |
| pMC253 | pCR2.1 Group II intron targeting region for eutA | This study | |
| pMC256 | pCR2.1 Group II intron targeting region for eutG | This study | |
| pMC257 | pCE240 Group II intron targeted to eutA | This study | |
| pMC260 | pCE240 Group II intron targeted to eutG | This study | |
| pMC263 | pMC123 Group II intron targeted to eutA, ermB::RAM ltrA catP | This study | |
| pMC266 | pMC123 Group II intron targeted to eutG, ermB::RAM ltrA catP | This study | |
Cloning and strain construction
Oligonucleotides used in this study are listed in Table 3. Construction details of plasmids used in this study can be found in Supplementary File S2. 630 genomic sequence (NC_009089.1) was used for primer design and null mutations were generated in 630Δerm using a Targetron-based intron, as previously described (McBride and Sonenshein, 2011a; Purcell et al., 2012). C. difficile colonies were screened by PCR to verify intron insertion into the target gene. C. difficile DNA was isolated as described previously (Harju et al., 2004; Edwards et al., 2016a). Plasmids and cloned sequences were verified by standard sequencing (Eurofins MWG Operon). Conjugations of pMC263 and pMC266 into C. difficile strain 630Δerm were performed as previously described (McBride and Sonenshein, 2011a). Given the very large eut genomic cluster (~17 kB), standard complementation of the Targetron-based mutant was unattainable in our hands. In lieu of complementation, whole-genome resequencing of the eutA mutant was performed to verify that the eutA phenotype was a direct result of the insertional mutation, and not unintended second-site mutations (detailed below).
Table 3.
Oligonucleotides
| Primer | Sequence (5’→3’) | Use/locationa | Source or reference |
|---|---|---|---|
| EBS universal | 5’-CGAAATTAGAAACTTGCGTTCAGTAAAC-3’ | Targetron cloning | Sigma-Aldrich |
| oMC44 | 5’-CTAGCTGCTCCTATGTCTCACATC-3’ | rpoC PCR (CD0067) | (McBride and Sonenshein, 2011a) |
| oMC45 | 5'-CCAGTCTCTCCTGGATCAACTA-3’ | rpoC PCR (CD0067) | (McBride and Sonenshein, 2011a) |
| oMC112 | 5'-GGCAAATGTAAGATTTCGTACTCA-3' | tcdB PCR (CD0660) | (Edwards et al., 2014) |
| oMC113 | 5'-TCGACTACAGTATTCTCTGAC-3' | tcdB PCR (CD0660) | (Edwards et al., 2014) |
| oMC339 | 5’-GGGCAAATATACTTCCTCCTCCAT-3’ | sigE PCR (CD2643) | (Edwards et al., 2014) |
| oMC340 | 5’-TGACTTTACACTTTCATCTGTTTCTAGC-3’ | sigE PCR (CD2643) | (Edwards et al., 2014) |
| oMC547 | 5’-TGGATAGGTGGAGAAGTCAGT-3’ | tcdA PCR (CD0663) | (Edwards et al., 2014) |
| oMC548 | 5’-GCTGTAATGCTTCAGTGGTAGA-3’ | tcdA PCR (CD0663) | (Edwards et al., 2014) |
| oMC655 | 5’-GAAATAGTACCAGACCCACCAATA-3’ | eutG PCR (CD1907) | This study |
| oMC656 | 5’-TATTGCAGAACCACCACCTAAT-3’ | eutG PCR (CD1907) | This study |
| oMC657 | 5’-CTAAAGAAGATATACATACAGGAGCAGT-3’ | eutA PCR (CD1912) | This study |
| oMC658 | 5’-TAAATCAGGTCCTGCTGTTGC-3’ | eutA PCR (CD1912) | This study |
| oMC659 | 5’-ATCAGGAGATAAATTAGCAGGTCTT-3’ | eutB PCR (CD1913) | This study |
| oMC660 | 5’-GCCACTGCAGGATTATTTCTTATATC-3’ | eutB PCR (CD1913) | This study |
| oMC661 | 5’-GTTGATGATGAACCACTGTTAAGG-3’ | eutV PCR (CD1910) | This study |
| oMC662 | 5’-TTCAACAGCTTCAAATCCATCAC-3’ | eutV PCR (CD1910) | This study |
| oMC663 | 5’-TGTACTAAGTAAATTTGATGATGAAGCA-3’ | eutW PCR (CD1911) | This study |
| oMC664 | 5’-TGACCAGTTTATATTCCTGTGTTAAAG-3’ | eutW PCR (CD1911) | This study |
| oMC665 | 5’-AAAAGCTTTTGCAACCCACGTCGATCGTGAAGGAAGGCTTGTAGTGCGCCCAGATAGGGT-3’ | eutA intron retargeting (CD1912) | This study |
| oMC666 | 5’-CAGATTGTACAAATGTGGTGATAACAGATAAGTCCTTGTAAATAACTTACCTTTCTTTGT-3’ | eutA intron retargeting (CD1912) | This study |
| oMC667 | 5’-CGCAAGTTTCTAATTTCGGTTCTTCCTCGATAGAGGAAAGTGTCT-3’ | eutA intron retargeting (CD1912) | This study |
| oMC674 | 5’-AAAAGCTTTTGCAACCCACGTCGATCGTGAATCTACCTCGTTAGTGCGCCCAGATAGGGTG-3’ | eutG intron retargeting (CD1907) | This study |
| oMC675 | 5’-CAGATTGTACAAATGTGGTGATAACAGATAAGTCTCGTTAATTAACTTACCTTTCTTTGT-3’ | eutG intron retargeting (CD1907) | This study |
| oMC676 | 5’-CGCAAGTTTCTAATTTCGGTTGTAGATCGATAGAGGAAAGTGTCT-3’ | eutG intron retargeting (CD1907) | This study |
| oMC714 | 5’-GGAGAAGGCTCACTAAAAGC-3’ | eutG verification | This study |
| oMC715 | 5’-GCCTCTTTGCTTGTTTAAGAC-3’ | eutA verification | This study |
| oMC716 | 5’-GTAGCCATTGTACCTACTTGAAG-3’ | eutA verification | This study |
| oMC855 | 5’-TCATAGCTAACCCTAATGAAGACAT-3’ | eutS PCR (CD1908) | This study |
| oMC856 | 5’-GCAACATCTGCTGCTATTATTGA-3’ | eutS PCR (CD1908) | This study |
| oMC857 | 5’-GACACTCCTGGTGAGTATATGG-3’ | eutP PCR (CD1909) | This study |
| oMC858 | 5’-GTGTGGCATCATACACTATTGC-3’ | eutP PCR (CD1909) | This study |
| oMC859 | 5’-TCCTAAGGATGCAGAAGCTTATTT-3’ | eutC PCR (CD1914) | This study |
| oMC860 | 5’-CAGCATGGTCTGCTCTAAATCT-3’ | eutC PCR (CD1914) | This study |
| oMC861 | 5’-ACTGCAGACTGTGATGATGTT-3’ | eutL PCR (CD1915) | This study |
| oMC862 | 5’-CAGCACCTGCATACATTGATTT-3’ | eutL PCR (CD1915) | This study |
| oMC863 | 5’-GGAGATGTCGGTGCTGTAAA-3’ | eutM PCR (CD1916) | This study |
| oMC864 | 5’-TGTTCTTAATGTACCAACTCTGGA-3’ | eutM PCR (CD1916) | This study |
| oMC865 | 5’-TGGTGAAGATAAGGAAGCCAATA-3’ | eutE PCR (CD1917) | This study |
| oMC866 | 5’-GATGTTGGGTTTGTTGAAGGTATAA-3’ | eutE PCR (CD1917) | This study |
| oMC867 | 5’-GCAGCAGATGCAATGGTTAAA-3’ | eutK PCR (CD1918) | This study |
| oMC868 | 5’-CAGCTCCAACATCTCCTCTTAC-3’ | eutK PCR (CD1918) | This study |
| oMC869 | 5’-TTTGGCGCTAAATTGGATGAAA-3’ | eutT PCR (CD1919) | This study |
| oMC870 | 5’-TTCCTTAACAAGCTTTGGCATATC-3’ | eutT PCR (CD1919) | This study |
| oMC871 | 5’-ATAGAAGTTGAAGCATCAGGAAGAC-3’ | eutD PCR (CD1920) | This study |
| oMC872 | 5’-CATGCATATTGACCTGGTTGAGATA-3’ | eutD PCR (CD1920) | This study |
| oMC873 | 5’-CCTAAAGCTGTGATTGTATTTGAACAAG-3’ | eut hyp PCR (CD1921) | This study |
| oMC874 | 5’-ATTTCCAAGTGCTATATTCGATAGTCC-3’ | eut hyp PCR (CD1921) | This study |
| oMC875 | 5’-AACTAAAGACACATACAAGGAAGGC-3’ | eutN PCR (CD1922) | This study |
| oMC876 | 5’-TCTAGCTGAGGAACCTTTGGATATAA-3’ | eutN PCR (CD1922) | This study |
| oMC877 | 5’-AGGCGTTGTTGAAAGTAGTAAAGT-3’ | pduT PCR (CD1923) | This study |
| oMC878 | 5’-ACTGTCTACTGATGGTATTACAGTCT-3’ | pduT PCR (CD1923) | This study |
| oMC879 | 5’-TGGTCTTGGAGAACAATTTGAAGA-3’ | eutH PCR (CD1924) | This study |
| oMC880 | 5’-ATCAGCAAGAACTGGTGCTAAA-3’ | eutH PCR (CD1924) | This study |
| oMC881 | 5’-AAGTGGAACTAAAGACACAGTAGAT-3’ | eutQ PCR (CD1925) | This study |
| oMC882 | 5’-CCTAATCTTGGGCTTTCCTCTAAT-3’ | eutQ PCR (CD1925) | This study |
| oMC972 | 5’-ATAAGAGCCACAGTGGTTGG-3’ | eutA PCR (CD1912) | This study |
| oMC973 | 5’-CTGGCAAGTTCTTAAGTGGAAAC-3’ | eutA PCR (CD1912) | This study |
| oMC1588 | 5’-GGGATTGCATATGGAAATCAATGTTATAG-3’ | CD1906 PCR | This study |
| oMC1589 | 5’-TTTACAAACCAACTAGCTTTATCCATTTC-3’ | CD1906 PCR | This study |
| oMC1593 | 5’-GGCTTTAGGAATGGTTGGTATAGTT-3’ | CD0742 PCR | This study |
| oMC1594 | 5’-CTCCACAGAGATTGAAGATTGGTATT-3’ | CD0742 PCR | This study |
| oMC1595 | 5’-AAACTAGGAGGTCTTGACAGATTAGA-3’ | CD3167 PCR | This study |
| oMC1596 | 5’-GGTCAAGGATAGTATCAAGTGCAAATA-3’ | CD3167 PCR | This study |
| oMC1634 | 5’-CTTGTTTAGCAGGTATGGCATTTAAC-3’ | eutG (CD1907; 3’) PCR | This study |
| oMC1635 | 5’-TTCCACCTAATACATGTGCAATAGAA-3’ | eutG (CD1907; 3’) PCR | This study |
| oMC1687 | 5’-GAAGTACATATGCTAACAGCGATTG-3’ | CDR20291_1847 | This study |
| oMC1688 | 5’-GAGATATTGCACCTTTAGTTCTGTTC-3’ | CDR20291_1847 | This study |
| fliCqF | 5’-TACAAGTTGGAGCAAGTTATGGAAC-3’ | fliC PCR (CD0239) | (McKee et al., 2013) |
| fliCqR | 5’-GTTGTTATACCAGCTGAAGCCATTA-3’ | fliC PCR (CD0239) | (McKee et al., 2013) |
| qRTpilA1F | 5’-TGGCAGTTCCAGCTTTATTTAGTAAT-3’ | pilA1 PCR (CD3513) | (Purcell et al., 2016) |
| qRTpilA1R | 5’-AAGATAATGCTGCACTCTTAACTGAA-3’ | pilA1 PCR (CD3513) | (Purcell et al., 2016) |
Single-nucleotide polymorphisms (SNP) analysis
MC394 (eutA::ermB) and its parent strain, 630Δerm, underwent SNP analysis to determine if any changes occurred in the eutA mutant other than the intended insertional disruption. Sample preparation and analyses were performed by the Yerkes Nonhuman Primate Genomics Core (Emory University). Libraries were generated from genomic DNA using the Illumina NexteraXT DNA kit, and dual barcoding and sequencing primers were used according to the manufacturer protocol. Libraries were validated by microelectrophoresis on an Agilent 2100 Bioanalyzer, quantified, pooled, and clustered on an Illumina miSeq V3 flowcell. The clustered flowcell was sequenced on an Illumina miSeq in 275-base paired-read reactions. Per sample reads were mapped to the C. difficile 630 reference genome (GenBank no. AM180355). CLC Genomics Workbench v9.0 was used for all variant analysis, as previously described (Childress et al., 2016). No nucleotide changes were observed between the mutant and parent strain genomes.
RNA isolation and quantitative reverse transcription PCR analysis (qRT-PCR)
qRT-PCR was performed as described previously with minor modifications (Edwards et al., 2014). C. difficile cultures were started in BHIS broth supplemented with 0.1% taurocholate and 0.2% fructose until they reached mid-logarthmic growth (OD600 of ~0.5). Cultures were then diluted 1:10 into 70:30 broth (70% SMC, 30% BHIS medium)(Putnam et al., 2013), with and without the addition of 15 mM ethanolamine hydrochloride (Sigma-Aldrich). Samples for RNA were collected at logarithmic growth (OD600 of ~0.5), mid-stationary phase (two hours after reaching an OD600 of ~1.0, T2), and at late stationary phase (four hours after reaching an OD600 of ~1.0, T4). Samples for RNA isolation were diluted in 1:1 acetone-ethanol and stored at −80°C. RNA isolation was performed as previously described (Dineen et al., 2010; McBride and Sonenshein, 2011a). Samples were treated with the Ambion DNAse Turbo Kit to remove contaminating genomic DNA, and cDNA synthesis was performed using a Bioline Tetro cDNA synthesis kit. qRT-PCR was performed with 50 ng of cDNA as template, using the Bioline Sensi-Fast SYBR and Fluroescein kit on a Roche LightCycler 96. qRT-PCR reactions were performed in technical triplicates and reactions containing no reverse transcriptase were included as negative controls. The PrimerQuest tool provided by Integrated DNA Technologies was used to generate qRT-PCR primers. The primer efficiencies for each primer set were determined using genomic DNA standards. qRT-PCR results were calculated by the comparative cycle threshold method (Schmittgen and Livak, 2008), with the expression of the amplified transcript being normalized to the internal control transcript, rpoC. A minimum of three biological replicates were performed and analyzed for each experimental set.
Hamster studies
Hamster studies were conducted as previously described with minor modifications (Woods et al., 2016). Male and female Syrian golden hamsters (Mesocricetus auratus) weighing between 70 to 110 grams were purchased from Charles River Laboratories. Hamsters were kept in an animal biosafety level 2 room within the Emory University Division of Animal Resources. Hamsters were housed individually with sterile rodent pellets and sterile water provided ad libitum. Hamsters were orally gavaged with clindamycin (30 mg/kg) to induce susceptibility to C. difficile seven days prior to inoculation (Day −7). Hamsters were inoculated by oral gavage with 5000 spores of 630Δerm or MC394 (eutA::ermB) (Day 0). Hamsters treated with clindamycin, but not inoculated with C. difficile spores, served as negative controls. Spores were prepared as previously described (Edwards et al., 2014; Edwards and McBride, 2016; Woods et al., 2016). Following inoculation, hamsters were monitored for signs of disease, such as diarrhea or wet tail, weight loss, and lethargy. Hamsters were weighed and fecal pellets were collected once daily, if possible. Hamsters were considered moribund if they lost ≥ 15% of their body weight and/or exhibited severe disease symptoms. Hamsters meeting these criteria were euthanized in accordance with the American Veterinary Medical Association guidelines. Cecal contents were collected from animals post-mortem. Fecal and cecal contents were diluted in 1× PBS and plated onto taurocholate cycloserine cefoxitin fructose agar (TCCFA) plates to determine the number of C. difficile CFU (George et al., 1979; Wilson et al., 1981). Inoculated TCCFA plates were incubated anaerobically for 48 hours prior to CFU enumeration. Cecal content enumerations were analyzed using the Student’s two-tailed t test and animal survival data were analyzed using the log rank test. Three independent hamster experiments were conducted with cohorts of 5 to 6 hamsters per C. difficile strain; a total of 17 hamsters per strain were used.
Western blots and pH assessment
C. difficile strains were grown in BHIS medium containing 0.2% fructose and 0.1% taurocholate, as previously described. Cultures were diluted into BHIS medium and grown OD600 of ~0.5, then diluted 1:10 into TY medium with or without 15 mM ethanolamine, and grown for 24 h at 37°C. Cells were harvested by centrifugation, resuspended in SDS-PAGE loading buffer (without dye) and mechanically disrupted as previously described (Edwards et al., 2016a; Nawrocki et al., 2016). The pH of the cultures were also measured at the time of harvest, as previously described (Edwards et al., 2016b). Protein concentrations were assessed using a micro BCA assay (Thermo Scientific) and 8 µg of whole cell protein loaded onto a 12% polyacrylamide gel. Proteins were subsequently transferred from the SDS-PAGE gel onto nitrocellulose membranes (0.45 µM; Bio-Rad), and probed with mouse anti-TcdA antibody (Novus Biologicals). Membranes were then washed and probed with goat anti-mouse secondary Alexa Fluor 488 antibody (Life Technologies). Imaging and densitometry analyses were performed using a Bio-Rad ChemiDoc MP and Image Lab Software (Bio-Rad). Three biological replicates were analyzed for each strain and condition. Statistical analyses of the data were performed using a one-way ANOVA.
Phase contrast microscopy and direct count sporulation frequency
Samples for phase contrast microscopy were prepared as previously described (Edwards et al., 2014; Edwards and McBride, 2017). Briefly, samples were taken from 70:30 agar plates, resuspended in BHIS broth, and pipetted on to a 0.7% agarose pad on a glass slide. Micrographs were taken with a DS-Fi2 camera on a Nikon Eclipse Ci-L microscope with an X100 Ph3 oil-immersion objective. Direct count sporulation frequency was performed as previously described (Edwards et al., 2016a; Edwards and McBride, 2017). A minimum of 1000 cells were counted per strain and condition. Sporulation frequency was calculated by dividing the number of spores by the total number of cells present (spores and vegetative cells). MC310 (spo0A::ermB), a sporulation-defective mutant, was used as a negative control (Edwards et al., 2014). The mean sporulation frequencies and standard deviations were calculated from four independent experiments. Phase contrast sporulation frequency data were analyzed using a two-way repeated measures ANOVA.
Germination assays
C. difficile strains were grown on 70:30 sporulation agar and spores purified for germination studies as previously described, with some modifications (Nawrocki et al., 2016). Cultures on sporulation agar were removed from the anaerobic chamber after a minimum of 48 h of anaerobic growth and kept outside of the chamber in atmospheric oxygen overnight. Spore cultures were then scraped from the plates, resuspended in water, and frozen briefly at −80°C. After thawing, spore suspensions were centrifuged for 15 min at ~3200 × g in a swing bucket rotor, and the supernatant discarded. Spore pellets were then resuspended in 1 ml of a 1× PBS + 1% BSA solution, applied to a 12 ml 50% sucrose bed volume, and centrifuged at ~3200 × g for 20 min. The supernatant was then decanted and the spores were checked by phase-contrast microscopy for purity. Sucrose gradients were repeated until the preparations reached a purity of greater than 95%. Spore pellets were then washed three times with 1× PBS + 1% BSA and resuspended to an OD600 = 3.0. Germination assays were carried out as previously described (Sorg and Sonenshein, 2008, 2009).
Accession numbers
C. difficile strain 630, GenBank accession NC_009089.1; strain R20291, GenBank accession NC_0133161.1.
Statistical analysis
Statistical analyses were performed using Excel and GraphPad Prism version 7.0 for Windows.
Supplementary Material
Originality-Significance Statement.
This work represents the first investigation of ethanolamine utilization in C. difficile and our data demonstrate that ethanolamine represents more than simply an alternative nutrient source for this important pathogen. These results represent a significant advance in understanding how nutrition impacts C. difficile virulence.
Acknowledgments
We thank members of the McBride lab, Charles Moran, Joanna Goldberg, David Weiss, and Philip Rather for their helpful suggestions and discussions during the course of this work.
FUNDING INFORMATION
This research was supported by the U.S. National Institutes of Health through research grants DK087763, DK101870, AI109526 and AI116933 to S.M.M., and T32 AI106699 to K.L.N. The content of this manuscript is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
Footnotes
The authors have no conflicts of interest to declare.
References
- Anderson CJ, Clark DE, Adli M, Kendall MM. Ethanolamine signaling promotes Salmonella niche recognition and adaptation during infection. PLoS Pathog. 2015;11:e1005278. doi: 10.1371/journal.ppat.1005278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes A, Martin-Verstraete I, Dupuy B. CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol Microbiol. 2011;79:882–899. doi: 10.1111/j.1365-2958.2010.07495.x. [DOI] [PubMed] [Google Scholar]
- Antunes A, Camiade E, Monot M, Courtois E, Barbut F, Sernova NV, et al. Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile. Nucleic Acids Res. 2012;40:10701–10718. doi: 10.1093/nar/gks864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batah J, Deneve-Larrazet C, Jolivot PA, Kuehne S, Collignon A, Marvaud JC, Kansau I. Clostridium difficile flagella predominantly activate TLR5-linked NF-kappaB pathway in epithelial cells. Anaerobe. 2016;38:116–124. doi: 10.1016/j.anaerobe.2016.01.002. [DOI] [PubMed] [Google Scholar]
- Bouillaut L, McBride SM, Sorg JA. Genetic manipulation of Clostridium difficile. Curr Protoc Microbiol. 2011;Chapter 9(Unit 9A 2) doi: 10.1002/9780471729259.mc09a02s20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillaut L, Self WT, Sonenshein AL. Proline-dependent regulation of Clostridium difficile Stickland metabolism. J Bacteriol. 2013;195:844–854. doi: 10.1128/JB.01492-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillaut L, Dubois T, Sonenshein AL, Dupuy B. Integration of metabolism and virulence in Clostridium difficile. Res Microbiol. 2015;166:375–383. doi: 10.1016/j.resmic.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgogne A, Hilsenbeck SG, Dunny GM, Murray BE. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J Bacteriol. 2006;188:2875–2884. doi: 10.1128/JB.188.8.2875-2884.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinsmade SR, Paldon T, Escalante-Semerena JC. Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J Bacteriol. 2005;187:8039–8046. doi: 10.1128/JB.187.23.8039-8046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartman ST, Kelly ML, Heeg D, Heap JT, Minton NP. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Appl Environ Microbiol. 2012;78:4683–4690. doi: 10.1128/AEM.00249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, McClane BA, Fisher DJ, Rood JI, Gupta P. Construction of an alpha toxin gene knockout mutant of Clostridium perfringens type A by use of a mobile group II intron. Appl Environ Microbiol. 2005;71:7542–7547. doi: 10.1128/AEM.71.11.7542-7547.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childress KO, Edwards AN, Nawrocki KL, Woods EC, Anderson SE, McBride SM. The phosphotransfer protein CD1492 represses sporulation initiation in Clostridium difficile. Infect Immun. 2016 doi: 10.1128/IAI.00735-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Papa MF, Perego M. Ethanolamine activates a sensor histidine kinase regulating its utilization in Enterococcus faecalis. J Bacteriol. 2008;190:7147–7156. doi: 10.1128/JB.00952-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dineen SS, McBride SM, Sonenshein AL. Integration of metabolism and virulence by Clostridium difficile CodY. J Bacteriol. 2010;192:5350–5362. doi: 10.1128/JB.00341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dineen SS, Villapakkam AC, Nordman JT, Sonenshein AL. Repression of Clostridium difficile toxin gene expression by CodY. Mol Microbiol. 2007;66:206–219. doi: 10.1111/j.1365-2958.2007.05906.x. [DOI] [PubMed] [Google Scholar]
- Dowhan W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- Dubois T, Dancer-Thibonnier M, Monot M, Hamiot A, Bouillaut L, Soutourina O, et al. Control of Clostridium difficile physiopathology in response to cysteine availability. Infect Immun. 2016;84:2389–2405. doi: 10.1128/IAI.00121-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy B, Sonenshein AL. Regulated transcription of Clostridium difficile toxin genes. Mol Microbiol. 1998;27:107–120. doi: 10.1046/j.1365-2958.1998.00663.x. [DOI] [PubMed] [Google Scholar]
- Edwards AN, McBride SM. Initiation of sporulation in Clostridium difficile: a twist on the classic model. FEMS Microbiol Lett. 2014;358:110–118. doi: 10.1111/1574-6968.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AN, McBride SM. Isolating and Purifying Clostridium difficile Spores. Methods Mol Biol. 2016;1476:117–128. doi: 10.1007/978-1-4939-6361-4_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AN, McBride SM. Determination of the in vitro Sporulation Frequency of Clostridium difficile. Bio-protocol. 2017;7:1–8. doi: 10.21769/BioProtoc.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AN, Suarez JM, McBride SM. Culturing and maintaining Clostridium difficile in an anaerobic environment. J Vis Exp. 2013:e50787. doi: 10.3791/50787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AN, Nawrocki KL, McBride SM. Conserved oligopeptide permeases modulate sporulation initiation in Clostridium difficile. Infect Immun. 2014;82:4276–4291. doi: 10.1128/IAI.02323-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AN, Tamayo R, McBride SM. A novel regulator controls Clostridium difficile sporulation, motility and toxin production. Mol Microbiol. 2016a;100:954–971. doi: 10.1111/mmi.13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AN, Karim ST, Pascual RA, Jowhar LM, Anderson SE, McBride SM. Chemical and Stress Resistances of Clostridium difficile Spores and Vegetative Cells. Front Microbiol. 2016b;7:1698. doi: 10.3389/fmicb.2016.01698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Meouche I, Peltier J, Monot M, Soutourina O, Pestel-Caron M, Dupuy B, Pons JL. Characterization of the SigD regulon of C. difficile and its positive control of toxin production through the regulation of tcdR. PLoS One. 2013;8:e83748. doi: 10.1371/journal.pone.0083748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust LR, Connor JA, Roof DM, Hoch JA, Babior BM. Cloning, sequencing, and expression of the genes encoding the adenosylcobalamin-dependent ethanolamine ammonia-lyase of Salmonella Typhimurium. J Biol Chem. 1990;265:12462–12466. [PubMed] [Google Scholar]
- Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe. 2014;16:770–777. doi: 10.1016/j.chom.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimlaid KA, Bond JP, Schutz KC, Putnam EE, Leung JM, Lawley TD, Shen A. Global Analysis of the Sporulation Pathway of Clostridium difficile. PLoS Genet. 2013;9:e1003660. doi: 10.1371/journal.pgen.1003660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox KA, Ramesh A, Stearns JE, Bourgogne A, Reyes-Jara A, Winkler WC, Garsin DA. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc Natl Acad Sci U S A. 2009;106:4435–4440. doi: 10.1073/pnas.0812194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier T, Cole ST. Characterization of a bacteriocinogenic plasmid from Clostridium perfringens and molecular genetic analysis of the bacteriocin-encoding gene. J Bacteriol. 1986;168:1189–1196. doi: 10.1128/jb.168.3.1189-1196.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin DA. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat Rev Micro. 2010;8:290–295. doi: 10.1038/nrmicro2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin DA. Ethanolamine: a signal to commence a host-associated lifestyle? MBio. 2012;3:e00172–00112. doi: 10.1128/mBio.00172-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George WL, Sutter VL, Citron D, Finegold SM. Selective and differential medium for isolation of Clostridium difficile. J Clin Microbiol. 1979;9:214–219. doi: 10.1128/jcm.9.2.214-219.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose C, Eugenis I, Sun X, Edwards AN, McBride SM, Pride DT, et al. Immunogenicity and protective efficacy of recombinant Clostridium difficile flagellar protein FliC. Emerg Microbes Infect. 2016;5:e8. doi: 10.1038/emi.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girinathan BP, Braun S, Sirigireddy AR, Lopez JE, Govind R. Importance of Glutamate Dehydrogenase (GDH) in Clostridium difficile Colonization In Vivo. PLoS One. 2016;11:e0160107. doi: 10.1371/journal.pone.0160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonyar LA, Kendall MM. Ethanolamine and choline promote expression of putative and characterized fimbriae in enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2014;82:193–201. doi: 10.1128/IAI.00980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harju S, Fedosyuk H, Peterson KR. Rapid isolation of yeast genomic DNA: Bust n' Grab. BMC Biotechnol. 2004;4:8. doi: 10.1186/1472-6750-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TD, Ellermeier CD. PrsW is required for colonization, resistance to antimicrobial peptides, and expression of extracytoplasmic function sigma factors in Clostridium difficile. Infect Immun. 2011;79:3229–3238. doi: 10.1128/IAI.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain HA, Roberts AP, Mullany P. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Δerm) and demonstration that the conjugative transposon Tn916ΔE enters the genome of this strain at multiple sites. J Med Microbiol. 2005;54:137–141. doi: 10.1099/jmm.0.45790-0. [DOI] [PubMed] [Google Scholar]
- Jackson S, Calos M, Myers A, Self WT. Analysis of proline reduction in the nosocomial pathogen Clostridium difficile. J Bacteriol. 2006;188:8487–8495. doi: 10.1128/JB.01370-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoir C, Deneve C, Bouttier S, Barbut F, Hoys S, Caleechum L, et al. Adaptive strategies and pathogenesis of Clostridium difficile from in vivo transcriptomics. Infect Immun. 2013;81:3757–3769. doi: 10.1128/IAI.00515-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Przybilla K, Stuhler C, Schauer K, Slaghuis J, Fuchs TM, Goebel W. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol. 2006;188:556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansau I, Barketi-Klai A, Monot M, Hoys S, Dupuy B, Janoir C, Collignon A. Deciphering Adaptation Strategies of the Epidemic Clostridium difficile 027 Strain during Infection through In Vivo Transcriptional Analysis. PLoS One. 2016;11:e0158204. doi: 10.1371/journal.pone.0158204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasawa T, Ikoma S, Yamakawa K, Nakamura S. A defined growth medium for Clostridium difficile. Microbiology. 1995;141:371–375. doi: 10.1099/13500872-141-2-371. [DOI] [PubMed] [Google Scholar]
- Karberg M, Guo H, Zhong J, Coon R, Perutka J, Lambowitz AM. Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat Biotechnol. 2001;19:1162–1167. doi: 10.1038/nbt1201-1162. [DOI] [PubMed] [Google Scholar]
- Karlsson S, Burman LG, Akerlund T. Suppression of toxin production in Clostridium difficile VPI 10463 by amino acids. Microbiology. 1999;145(Pt 7):1683–1693. doi: 10.1099/13500872-145-7-1683. [DOI] [PubMed] [Google Scholar]
- Karlsson S, Burman LG, Akerlund T. Induction of toxins in Clostridium difficile is associated with dramatic changes of its metabolism. Microbiology. 2008;154:3430–3436. doi: 10.1099/mic.0.2008/019778-0. [DOI] [PubMed] [Google Scholar]
- Karlsson S, Lindberg A, Norin E, Burman LG, Akerlund T. Toxins, butyric acid, and other short-chain fatty acids are coordinately expressed and down-regulated by cysteine in Clostridium difficile. Infect Immun. 2000;68:5881–5888. doi: 10.1128/iai.68.10.5881-5888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall MM, Gruber CC, Parker CT, Sperandio V. Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. MBio. 2012;3 doi: 10.1128/mBio.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopke M, Straub M, Durre P. Clostridium difficile is an autotrophic bacterial pathogen. PLoS One. 2013;8:e62157. doi: 10.1371/journal.pone.0062157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. The role of toxin A and toxin B in Clostridium difficile infection. Nature. 2010;467:711–713. doi: 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- Larocque M, Chenard T, Najmanovich R. A curated C. difficile strain 630 metabolic network: prediction of essential targets and inhibitors. BMC Syst Biol. 2014;8:117. doi: 10.1186/s12918-014-0117-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson TJ, Ehrmann M, Boos W. Periplasmic glycerophosphodiester phosphodiesterase of Escherichia coli a new enzyme of the glp regulon. J Biol Chem. 1983;258:5428–5432. [PubMed] [Google Scholar]
- Liu Y, Leal NA, Sampson EM, Johnson CL, Havemann GD, Bobik TA. PduL is an evolutionarily distinct phosphotransacylase involved in B12-dependent 1,2-propanediol degradation by Salmonella enterica serovar typhimurium LT2. J Bacteriol. 2007;189:1589–1596. doi: 10.1128/JB.01151-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzader DH, Clark DE, Gonyar LA, Kendall MM. EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157: H7. J Bacteriol. 2013;195:4947–4953. doi: 10.1128/JB.00937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyerly DM, Saum KE, MacDonald DK, Wilkins TD. Effects of Clostridium difficile toxins given intragastrically to animals. Infect Immun. 1985;47:349–352. doi: 10.1128/iai.47.2.349-352.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyras D, O'Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, et al. Toxin B is essential for virulence of Clostridium difficile. Nature. 2009;458:1176–1179. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maadani A, Fox KA, Mylonakis E, Garsin DA. Enterococcus faecalis mutations affecting virulence in the Caenorhabditis elegans model host. Infect Immun. 2007;75:2634–2637. doi: 10.1128/IAI.01372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli GA, De Masi L, Rosenvinge EC, Carter M, Donnenberg MS. Identification, immunogenicity, and cross-reactivity of type IV pilin and pilin-like proteins from Clostridium difficile. Pathog dis. 2014;71:302–314. doi: 10.1111/2049-632X.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MJ, Clare S, Goulding D, Faulds-Pain A, Barquist L, Browne HP. The agr locus regulates virulence and colonization genes in Clostridium difficile 027. J Bacteriol. 2013;195:3672–3681. doi: 10.1128/JB.00473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Verstraete I, Peltier J, Dupuy B. The regulatory networks that control Clostridium difficile toxin synthesis. Toxins (Basel) 2016;8 doi: 10.3390/toxins8050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride SM, Sonenshein AL. The dlt operon confers resistance to cationic antimicrobial peptides in Clostridium difficile. Microbiology. 2011a;157:1457–1465. doi: 10.1099/mic.0.045997-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride SM, Sonenshein AL. Identification of a genetic locus responsible for antimicrobial peptide resistance in Clostridium difficile. Infect Immun. 2011b;79:167–176. doi: 10.1128/IAI.00731-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee RW, Mangalea MR, Purcell EB, Borchardt EK, Tamayo R. The second messenger cyclic Di-GMP regulates Clostridium difficile toxin production by controlling expression of sigD. J Bacteriol. 2013;195:5174–5185. doi: 10.1128/JB.00501-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore TC, Escalante-Semerena JC. The EutQ and EutP proteins are novel acetate kinases involved in ethanolamine catabolism: physiological implications for the function of the ethanolamine metabolosome in Salmonella enterica. Mol Microbiol. 2016;99:497–511. doi: 10.1111/mmi.13243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki KL, Edwards AN, Daou N, Bouillaut L, McBride SM. CodY-dependent regulation of sporulation in Clostridium difficile. J Bacteriol. 2016;198:2113–2130. doi: 10.1128/JB.00220-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechine S, Collignon A. Immune responses induced by Clostridium difficile. Anaerobe. 2016;41:68–78. doi: 10.1016/j.anaerobe.2016.04.014. [DOI] [PubMed] [Google Scholar]
- Pechine S, Gleizes A, Janoir C, Gorges-Kergot R, Barc MC, Delmee M, Collignon A. Immunological properties of surface proteins of Clostridium difficile. J Med Microbiol. 2005;54:193–196. doi: 10.1099/jmm.0.45800-0. [DOI] [PubMed] [Google Scholar]
- Pechine S, Janoir C, Boureau H, Gleizes A, Tsapis N, Hoys S, et al. Diminished intestinal colonization by Clostridium difficile and immune response in mice after mucosal immunization with surface proteins of Clostridium difficile. Vaccine. 2007;25:3946–3954. doi: 10.1016/j.vaccine.2007.02.055. [DOI] [PubMed] [Google Scholar]
- Pitts AC, Tuck LR, Faulds-Pain A, Lewis RJ, Marles-Wright J. Structural insight into the Clostridium difficile ethanolamine utilisation microcompartment. PLoS One. 2012;7:e48360. doi: 10.1371/journal.pone.0048360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price-Carter M, Tingey J, Bobik TA, Roth JR. The Alternative Electron Acceptor Tetrathionate Supports B12-Dependent Anaerobic Growth of Salmonella enterica Serovar Typhimurium on Ethanolamine or 1,2-Propanediol. J Bacteriol. 2001;183:2463–2475. doi: 10.1128/JB.183.8.2463-2475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proulx P, Fung CK. Metabolism of phosphoglycerides in E. coli. IV. The positional specificity and properties of phospholipase A. Can J Biochem. 1969;47:1125–1128. doi: 10.1139/o69-181. [DOI] [PubMed] [Google Scholar]
- Purcell EB, McKee RW, McBride SM, Waters CM, Tamayo R. Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. J Bacteriol. 2012;194:3307–3316. doi: 10.1128/JB.00100-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell EB, McKee RW, Bordeleau E, Burrus V, Tamayo R. Regulation of Type IV Pili Contributes to Surface Behaviors of Historical and Epidemic Strains of Clostridium difficile. J Bacteriol. 2016;198:565–577. doi: 10.1128/JB.00816-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam EE, Nock AM, Lawley TD, Shen A. SpoIVA and SipL are Clostridium difficile spore morphogenetic proteins. J Bacteriol. 2013;195:1214–1225. doi: 10.1128/JB.02181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh A, DebRoy S, Goodson JR, Fox KA, Faz H, Garsin DA, Winkler WC. The mechanism for RNA recognition by ANTAR regulators of gene expression. PLoS Genet. 2012;8:e1002666. doi: 10.1371/journal.pgen.1002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saujet L, Monot M, Dupuy B, Soutourina O, Martin-Verstraete I. The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J Bacteriol. 2011;193:3186–3196. doi: 10.1128/JB.00272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saujet L, Pereira FC, Serrano M, Soutourina O, Monot M, Shelyakin PV, et al. Genome-Wide Analysis of Cell Type-Specific Gene Transcription during Spore Formation in Clostridium difficile. PLoS Genet. 2013;9:e1003756. doi: 10.1371/journal.pgen.1003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaria J, Chen JW, Useh N, He H, McDonough SP, Mao C, et al. Comparative nutritional and chemical phenome of Clostridium difficile isolates determined using phenotype microarrays. Int J Infect Dis. 2014;27C:20–25. doi: 10.1016/j.ijid.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- Smith CJ, Markowitz SM, Macrina FL. Transferable tetracycline resistance in Clostridium difficile. Antimicrob Agents Chemother. 1981;19:997–1003. doi: 10.1128/aac.19.6.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EA, Macfarlane GT. Enumeration of amino acid fermenting bacteria in the human large intestine: effects of pH and starch on peptide metabolism and dissimilation of amino acids. FEMS Microbiol Ecol. 1998;25:355–368. [Google Scholar]
- Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorg JA, Sonenshein AL. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J Bacteriol. 2009;191:1115–1117. doi: 10.1128/JB.01260-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikumar S, Fuchs TM. Ethanolamine utilization contributes to proliferation of Salmonella enterica serovar Typhimurium in food and in nematodes. Appl Environ Microbiol. 2011;77:281–290. doi: 10.1128/AEM.01403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, et al. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 2009;10:R102. doi: 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojiljkovic I, Baumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot CM, Young VB. Microbial and metabolic interactions between the gastrointestinal tract and Clostridium difficile infection. Gut Microbes. 2014;5:86–95. doi: 10.4161/gmic.27131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot CM, Young VB. Interactions Between the Gastrointestinal Microbiome and Clostridium difficile. Annu Rev Microbiol. 2015;69:445–461. doi: 10.1146/annurev-micro-091014-104115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot CM, Koenigsknecht MJ, Carlson PE, Jr, Hatton GE, Nelson AM, Li B, et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CM, Smith CA. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Ann Rev Microbiol. 1987;41:77–101. doi: 10.1146/annurev.mi.41.100187.000453. [DOI] [PubMed] [Google Scholar]
- Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- Tsoy O, Ravcheev D, Mushegian A. Comparative genomics of ethanolamine utilization. J Bacteriol. 2009;191:7157–7164. doi: 10.1128/JB.00838-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance JE. Phosphatidylserine and phosphatidylethanolamine in mammalian cells: two metabolically related aminophospholipids. J Lipid Res. 2008;49:1377–1387. doi: 10.1194/jlr.R700020-JLR200. [DOI] [PubMed] [Google Scholar]
- Wilson KH, Perini F. Role of competition for nutrients in suppression of Clostridium difficile by the colonic microflora. Infect Immun. 1988;56:2610–2614. doi: 10.1128/iai.56.10.2610-2614.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KH, Silva J, Fekety FR. Suppression of Clostridium difficile by normal hamster cecal flora and prevention of antibiotic-associated cecitis. Infect Immun. 1981;34:626–628. doi: 10.1128/iai.34.2.626-628.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods EC, Nawrocki KL, Suarez JM, McBride SM. The Clostridium difficile Dlt pathway is controlled by the ECF sigma factor, SigmaV, in response to lysozyme. Infect Immun. 2016 doi: 10.1128/IAI.00207-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa K, Kamiya S, Meng X, Karasawa T, Nakamura S. Toxin production by Clostridium difficile in a defined medium with limited amino acids. J Med Microbiol. 1994;41:319–323. doi: 10.1099/00222615-41-5-319. [DOI] [PubMed] [Google Scholar]
- Yoshino Y, Kitazawa T, Ikeda M, Tatsuno K, Yanagimoto S, Okugawa S, et al. Clostridium difficile flagellin stimulates toll-like receptor 5, and toxin B promotes flagellin-induced chemokine production via TLR5. Life Sci. 2013;92:211–217. doi: 10.1016/j.lfs.2012.11.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.