Abstract
Living systems employ protein pattern formation to regulate important life processes in space and time. Although pattern-forming protein networks have been identified in various prokaryotes and eukaryotes, their systematic experimental characterization is challenging owing to the complex environment of living cells. In turn, cell-free systems are ideally suited for this goal, as they offer defined molecular environments that can be precisely controlled and manipulated. Towards revealing the molecular basis of protein pattern formation, we outline two complementary approaches: the biochemical reverse engineering of reconstituted networks and the de novo design, or forward engineering, of artificial self-organizing systems. We first illustrate the reverse engineering approach by the example of the Escherichia coli Min system, a model system for protein self-organization based on the reversible and energy-dependent interaction of the ATPase MinD and its activating protein MinE with a lipid membrane. By reconstituting MinE mutants impaired in ATPase stimulation, we demonstrate how large-scale Min protein patterns are modulated by MinE activity and concentration. We then provide a perspective on the de novo design of self-organizing protein networks. Tightly integrated reverse and forward engineering approaches will be key to understanding and engineering the intriguing phenomenon of protein pattern formation.
This article is part of the theme issue ‘Self-organization in cell biology’.
Keywords: pattern formation, self-organization, reaction–diffusion systems, synthetic biology, in vitro reconstitution, Min system
1. Introduction
Living systems accomplish remarkable feats when regulating themselves in space and time. Across all scales, this exquisite regulation relies on biological self-organization for spatiotemporal order to emerge against the thermodynamic drive towards equilibrium. In self-organizing systems, nonlinear interactions between components under out-of-equilibrium conditions give rise to emergent phenomena that can exceed the spatial and temporal dimensions of individual interactions by several orders of magnitude. In single cells, nanometre-scale proteins self-organize via reaction–diffusion mechanisms into micrometre-scale patterns to regulate fundamental cellular processes. For example, in eukaryotes the Cdc42 network establishes polarity in budding yeast [1], Par proteins maintain polarity in the Caenorhabditis elegans zygote [2] and F-actin/RhoA waves regulate animal cell cytokinesis [3]. Similarly, bacteria rely on pattern-forming protein networks to organize their intracellular space. Many of these systems are based on a P-loop NTPase of the ParA/MinD-family, which can switch between a nucleoside diphosphate (NDP)-bound ‘inactive’ and a nucleoside triphosphate (NTP)-bound ‘active’ state, allowing reversible binding to an intracellular surface such as a lipid membrane or nucleoid DNA [4]. Such networks are vital for diverse intracellular processes, as exemplified by the MinCDE and PomXYZ systems regulating cell division [5,6], the ParABS system regulating DNA and plasmid segregation [7], or the FlhF/FlhG system determining the localization of flagella [8]. With a small number of functional elements, which are often encoded in a modular fashion on the same genetic operon, bacterial systems present an intriguing opportunity to study protein pattern formation.
Despite the identification of self-organizing protein networks and pioneering theoretical studies on the generic principles of protein pattern formation [9,10], experimental characterization in live cells remains challenging, as perturbations are limited by the networks' vital roles in essential cellular processes. Furthermore, owing to their inherent nonlinearity, reaction–diffusion networks are highly sensitive to experimental conditions to a level that can be difficult to control in vivo. By contrast, well-defined chemical self-organizing networks, such as those of the Belousov–Zhabotinsky reaction [11] or heterogeneous CO oxidation on platinum surfaces [12], are now relatively well understood because the conditions are readily amenable to systematic manipulation. Drawing inspiration from such chemical systems suggests a practical and complementary approach to investigate pattern formation: the reconstitution and design of minimal self-organizing systems in controllable in vitro environments.
With highly defined, adjustable and reproducible experimental conditions, in vitro systems are ideally suited to address fundamental questions regarding protein pattern formation. On a general level, what are the minimal molecular requirements for self-organization and by which mechanisms do small-scale interactions of biomolecules give rise to large-scale spatiotemporal patterns? Although mathematical approaches have established theoretical principles, it is largely unclear how they are implemented on a molecular level in real-life systems. Moreover, nature could have evolved additional or even alternative strategies for pattern formation. A related question is how the features of a specific system's components, such as the biochemical and physical properties of self-organizing proteins, quantitatively determine the system's emergent properties. Here, systematically characterizing how a system responds to controlled perturbations, e.g. changes in the identity or concentrations of the molecular players, can indicate why reaction rates or diffusion coefficients are tuned in a certain way. This could also shed light on how homologous systems function in vastly different contexts. For example, the Min system is found in both longitudinally and transversally dividing bacteria [13,14] and it would be intriguing to understand the molecular differences allowing the system to function under its respective geometric confines.
Two complementary approaches can be conceived to address such questions. First, an identified self-organizing network can be ‘reverse engineered’, whereby different parts of the system are independently perturbed to infer their function, from which a picture of the system's underlying mechanisms may be pieced together. Alternatively, a self-organizing system can be designed, or ‘forward engineered’, from first principles. This more challenging approach is capable of identifying truly general principles, but still benefits greatly from prior information, from either theoretical studies or well-understood natural systems. In this article, we discuss both the reconstitution and design of self-organizing networks. First, in the context of the Escherichia coli Min system, we illustrate the reverse engineering approach by characterizing the effects of altered protein activity and concentration in a simple in vitro system [15]. We then conclude by providing a perspective on the de novo design of artificial pattern-forming protein networks.
2. Reverse engineering protein pattern formation with the reconstituted E. coli Min system
(a). The E. coli Min system—an archetypal model system for protein self-organization
Among intracellular self-organizing systems, the E. coli MinCDE system has been particularly fruitful for understanding the molecular basis of protein pattern formation and is therefore a major model system for experimentalists [15–19] and theoreticians [20–23] alike. In E. coli, the Min system, together with the nucleoid occlusion mechanism [24], positions the cytokinetic machinery at mid-cell to ensure division of the bacterial mother cell into two equally sized daughters [13]. MinC directly antagonizes assembly of the early divisome protein FtsZ into a higher-order ring structure that is central to cytokinesis [25–27]. In order to specifically inhibit cell division at the poles but not at mid-cell, MinC interacts with MinD, which together with MinE oscillates from cell pole to cell pole, generating a time-averaged inhibitory gradient with a minimum at mid-cell [19]. In vitro reconstitution of purified Min proteins on model membrane systems confirmed that, in the presence of ATP, MinD and MinE are sufficient for Min protein self-organization [15]. Furthermore, reconstitution experiments revealed a rich variety of qualitatively different dynamics dependent on the experimental conditions [15,28–32]. Whereas the biologically relevant pole-to-pole oscillations and other geometry-dependent dynamics emerge in microchambers with cell-like geometries [30,32], a variety of different patterns emerge on flat supported lipid bilayers (SLBs) [15,29]. Notably, while the reconstituted Min system displays many of the properties of the in vivo patterns, the patterns' length scale is roughly tenfold higher in vitro [15]. One of the most prominent patterns is that of travelling waves, which emerge in the most basic experimental setup conceivable to reconstitute Min protein dynamics: a flat supported lipid membrane and a bulk reservoir containing purified MinD, MinE and ATP [15]. As the protein dynamics in this system are not influenced by additional factors such as geometric constraints [30–32] or hydrodynamic flow in the bulk phase [29], this system presents an ideal starting point for reverse engineering the Min system and thereby elucidating essential requirements and multi-scale relationships of protein pattern formation.
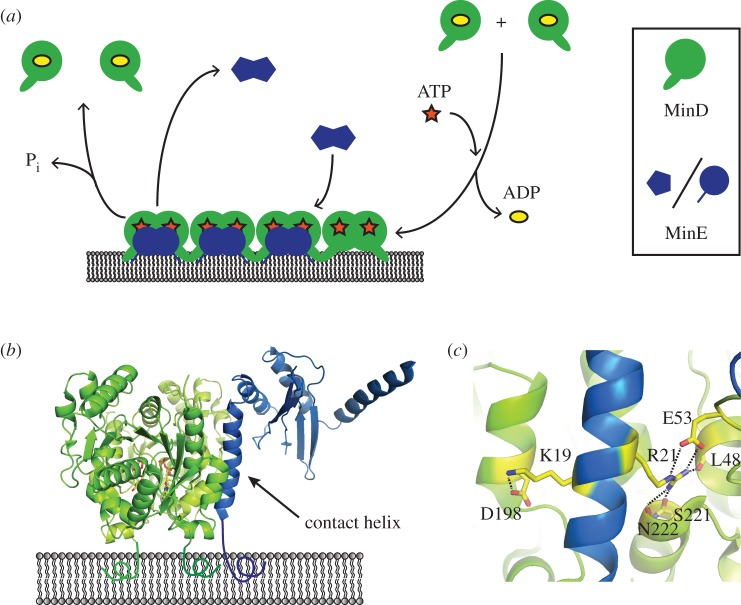
At the core of Min protein dynamics is the ATP-dependent cycling of MinD and MinE between a lipid membrane and a bulk phase (figure 1a). Upon exchanging ATP for ADP, MinD dimerizes and binds cooperatively to the membrane via a C-terminal amphipathic membrane targeting sequence (MTS) [34–37]. As the affinity of a MinD monomer's single MTS is insufficient for membrane binding, ATP-dependent dimerization acts as a molecular switch for membrane localization by increasing the local concentration of the MTS [34,35,37]. Whereas this switch occurs spontaneously upon ATP binding, the opposing process—ATP hydrolysis and subsequent membrane detachment—is facilitated by MinE, which acts as an ATPase activating protein [35,38]. Upon sensing membrane-bound MinD dimers, MinE undergoes a conformational change that stabilizes its interaction with MinD via release of a ‘contact helix’ [33,39]. Importantly, MinE can also interact with the lipid membrane through an N-terminal MTS [33]. In the membrane-bound MinDE complex, MinE then stimulates MinD's ATPase activity, resulting in detachment of MinD and eventually MinE from the membrane [33]. Thus, MinD and MinE have opposing roles in pattern formation: while membrane-bound MinD recruits MinE as well as—through its cooperative binding—further MinD to the membrane, MinE acts as an antagonist to MinD by promoting its depletion from the membrane.
Figure 1.
Molecular interactions underlying Min protein dynamics. (a) Simplified scheme of MinD and MinE cycling between a bulk phase and a lipid membrane. MinD binds to the membrane after exchanging ATP for ADP. It then recruits MinE, which, after undergoing a conformational change, forms a membrane-bound complex with MinD. In this complex, MinE triggers detachment of MinD via stimulation of its ATPase activity. (b) Structure of the membrane-bound MinD–MinE complex with MinD in green and MinE in blue (PDB: 3R9J), based on reference [33]. (c) Residues K19 and R21 in MinE's contact helix interact with MinD via hydrogen bonds, depicted as dashed lines.
It is well established that the opposing roles of MinD and MinE are important for self-organization. However, as reaction–diffusion systems are, in general, sensitive to even small parameter changes, it is intriguing to go one step further and ask how exactly large-scale Min protein patterns are influenced by the molecular-scale properties of the proteins. Recently, it was demonstrated that MinE's membrane affinity is an important modulatory parameter for large-scale Min protein patterns [29,40]. However, the effects of the Min proteins' other biochemical features, such as enzymatic rates, interaction affinities and conformational dynamics on pattern formation remain mysterious. By reverse engineering the Min system via mutagenesis, the contributions of such factors can be systematically dissected. In the long term, this approach promises to answer the following questions. First, which biochemical activities are strictly required for pattern formation? Second, which activities are non-essential but serve as important modulatory parameters? Last, how exactly do such activities regulate patterns, both individually and coupled with other factors such as protein concentration?
(b). Modulation of Min protein patterns by MinE activity and concentration
Here, we exemplify the reverse engineering approach to protein self-organization by investigating how the degree of MinD ATPase stimulation regulates large-scale Min protein patterns. It is generally established that MinE's stimulation of MinD's ATPase rate is a key step for Min protein pattern formation [38]. Furthermore, the wavelength and velocity of Min waves were shown to depend on the MinE/MinD concentration ratio [15,17,29]. However, it is unknown how exactly Min protein patterns are affected if the level of ATPase stimulation is decreased.
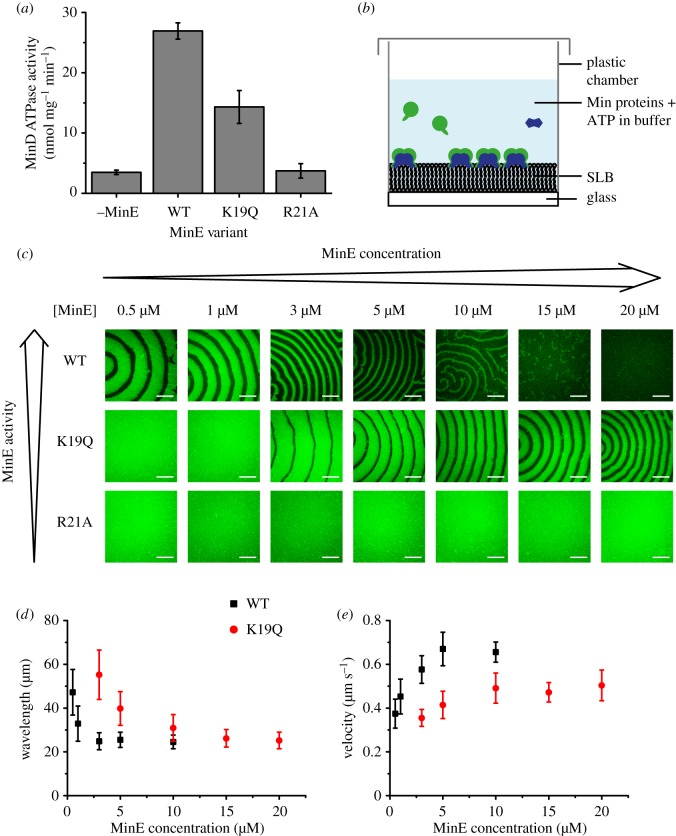
MinE interacts with MinD via a contact helix formed by residues 13–26 (figure 1b) [33]. Several residues, including K19 and the highly conserved R21, form hydrogen bonds with MinD (figure 1c) and mutations in these residues can compromise the MinD–MinE interaction [33,38,41]. To dissect the effects of reduced MinD ATPase stimulation by MinE, we investigated pattern formation by two mutant proteins, MinE K19Q and MinE R21A, that have been shown to be impaired in MinD ATPase stimulation [38,41]. While MinE R21A was incapable of significant ATPase stimulation, MinE K19Q was reported to increase MinD's ATPase rate, albeit at lower levels than the wild-type (WT) [38,41]. We confirmed the effects of these mutations by assaying MinD's ATPase activity in the presence of MinE and liposomes (figure 2a). We then reconstituted MinE WT or mutant proteins together with MinD on flat SLBs (figure 2b) and investigated pattern formation by confocal microscopy (figure 2c).
Figure 2.
Modulation of large-scale Min protein patterns by MinE activity and concentration. (a) ATPase stimulation assay with WT MinE and MinE K19Q and R21A using 4 µM MinD, 4 µM MinE and 0.2 mg ml−1 small unilamellar vesicles made of E. coli polar lipids. Error bars represent standard deviations (N = 3). (b) Schematic of the self-organization assay on flat SLBs. (c) Confocal images of the self-organization assay at different MinE concentrations with MinD constant at 1 µM with 20% eGFP-MinD. Scale bars, 50 µm. Dependence of the mean wavelength (d) and velocity (e) of WT and K19Q waves on MinE concentration (MinD at 1 µM). Error bars represent standard deviations (N ≥ 7 waves from three independent experiments).
At low MinE/MinD ratios, where WT MinE supported Min protein self-organization, both MinE K19Q and R21A were incapable of symmetry breaking and pattern formation (figure 2c). Instead, MinD homogeneously covered the membrane in a protein ‘carpet’, similar to when MinE is absent in the assay [15]. We then tested whether pattern formation could be rescued at higher MinE mutant levels by increasing the MinE concentration while keeping MinD constant at 1 µM (figure 2c). MinE R21A was unable to generate Min protein patterns even at high MinE/MinD ratios, consistent with its reported inability to stimulate MinD's enzymatic activity even at elevated concentrations [41]. This confirms that MinD ATPase stimulation by MinE is an essential requirement for pattern formation and explains the high conservation of the R21 residue [41]. In contrast to MinE R21A, Min protein patterns emerged at elevated concentrations of MinE K19Q (figure 2c), consistent with the reported rescue of WT-like ATPase stimulation at higher mutant concentrations [38]. Strikingly, while the mutant protein patterns required a higher MinE/MinD ratio, they also tolerated a higher excess of MinE relative to MinD. This emergence of Min protein patterns in a limited concentration range can be understood by considering MinE's functional role of antagonizing MinD accumulation on the membrane. When MinE's activity or concentration is too low, MinE's antagonism towards MinD is too weak to allow symmetry breaking, which results in a homogeneous distribution of MinD on the membrane. In turn, if MinE's antagonistic activity is too strong, MinD cannot accumulate effectively on the bilayer, resulting in uniform depletion of MinD from the membrane.
Finally, we compared the wavelength and velocity of the wave patterns formed by WT MinE and MinE K19Q (figure 2d,e). At relatively low MinE concentrations, the K19Q mutant displayed a significantly higher wavelength and lower velocity than WT MinE (figure 2d,e), consistent with a slower oscillation period observed for this mutant in vivo [38]. With increasing MinE concentration, the wavelength decreased and the velocity increased for both proteins (figure 2d,e), in agreement with earlier studies of WT MinE [15]. In this way, the wave properties displayed by WT MinE at low concentrations could be rescued by elevating the mutant's concentration. While the K19Q mutant stimulated MinD's ATPase activity to around 50% of the WT's level at the tested concentrations (figure 2a), the mutant concentration had to be increased by roughly one order of magnitude to rescue the behaviour observed with lower concentrations of WT MinE on SLBs (figure 2c–e). This can be explained with the observation that MinE's stimulation of MinD's ATPase activity follows a higher-order concentration dependency [29].
In summary, Min protein patterns form in a characteristic concentration range that can be modulated by mutation. Furthermore, the spatiotemporal properties of Min protein patterns are determined by both MinE concentration and activity. This interplay also suggests two complementary ways in which Min protein patterns may be tuneable in vivo. On the time scale of cell growth and division, bacteria could transiently adjust the MinE concentration ratio to change the spatiotemporal properties of Min patterns. In turn, on the evolutionary time scale, a similar but inheritable effect on the patterns is possible by adjusting the effective ATPase rate via mutation.
Of course, elucidating the complete set of biochemical determinants of Min protein pattern formation requires the comprehensive characterization of all molecular processes implicated in self-organization, and here we have merely illustrated our approach towards this end. Besides reaction and binding rates, the mobility of proteins in solution and on the membrane could be another important factor for modulating the patterns. A first step towards characterizing this parameter was taken by Martos et al., who compared Min waves on supported and free-standing membranes [42]. This revealed that the patterns' wavelength and velocity are increased on free-standing membranes, whose higher fluidity is associated with a higher protein diffusivity on the membrane [42]. Future protein engineering efforts could address the mobility of MinD and MinE individually, which may reveal further insights into the effect of protein mobility. Besides protein features, the geometric boundary conditions also influence Min protein patterns [18,30,32]. Thus, it is intriguing to speculate that changes in primary sequence can adapt the Min system to function in different geometries. In the long term, reverse engineering the Min system could yield a multi-dimensional ‘phase diagram’ of the various factors influencing pattern formation. This would uncover important multi-scale dependencies between the properties of nanometre-scale proteins and micrometre-scale patterns. Finally, such relationships would also allow ‘educated guesses’ on how to design a biochemical pattern-forming system with desirable properties.
3. Forward engineering: towards protein pattern formation by design
Forward engineering comprises the establishment of a high-level design upon which complexity can be built. In biology, this approach has become increasingly adopted through the emergence of ‘synthetic’ biology, which seeks to establish frameworks for designing biological systems with predefined properties [43]. Such strategies require sets of parts or modules that can be assembled into new systems at some level of abstraction from their underlying properties. Ideally, there should exist mechanisms or models whereby any assembly of components can achieve a predictable outcome.
While reverse engineering—perturbing existing complex systems to pick apart their underlying mechanisms—is the predominant investigational approach in biology, forward engineering offers unique benefits. First, by identifying key principles that are broadly applicable owing to abstraction from specific species, contexts or evolutionary outcomes, theories can be definitively tested through experiment. Second, it offers great potential in terms of engineering non-natural systems and subsequent industrial applications. Key characteristics for both strategies are summarized in table 1.
Table 1.
Key feature comparison between reverse and forward engineering strategies.
| reverse engineering | forward engineering |
|---|---|
| findings may be limited to specific context (e.g. model organism) | abstraction enables broader context, testing of key principles |
| dissect: molecular interactions enzymatic activity kinetics |
design: modular functions network topologies connectivity |
| natural systems | artificial systems (de novo) |
| perturb complex systems (top-down) | build-up complexity (bottom-up) |
| outcome of historical evolution | explore alternative outcomes |
| problem of cross-talk | bio-orthogonal systems |
The forward engineering approach is exemplified by one of synthetic biology's most ambitious goals: the bottom-up synthesis of an ‘artificial cell’ or ‘proto-cell’ capable of basic life functions such as metabolism, division and evolution [44]. Systems capable of spatiotemporal self-organization will play important roles in the realization of these goals. At a more fundamental level, the application of forward engineering to pattern formation is highly attractive, as complex spatiotemporal behaviours can arise from the interaction of only a few components. However, it is also challenging owing to the necessary nonlinear dependencies that make such systems highly sensitive to their parameters. In the remainder of this paper, we consider the prospects for forward engineering complex pattern formation in the context of examples where similar approaches have successfully been applied in vitro and in vivo.
(a). Prospects for forward engineering biomolecular pattern formation
It is important to note that with forward engineering, there is significant latitude in the manner in which mimicking systems can be constructed. For example, there are now multiple examples of membrane-spanning pores constructed of DNA [45–47] that mimic natural ion channel and pore-forming toxin proteins. Despite their radically different construction, they nonetheless successfully mimic key biological phenomena. Heavily artificial systems can still offer significant mechanistic insight, aside from their inherent potential for non-natural applications. We therefore begin with a few examples of more ‘unnatural’ systems before concluding with a consideration of protein-based systems operating in vivo.
Given the facile ‘programmability’ of synthetic nucleic acids and the advantages of cell-free systems, as discussed above, it is not surprising that many forward engineering approaches towards biomolecular pattern formation have been pursued in vitro using polynucleotides rather than proteins. Building on the utility of inorganic chemical reaction networks (CRNs) capable of spatiotemporal self-organization, CRNs using biochemical components have recently begun to be developed (biochemical reaction networks, BRNs) [48]. These often use DNA in combination with DNA-processing enzymes such as polymerases, nickases and exonucleases. Most examples exhibit temporal behaviours such as bistability or oscillations under batch or stirred conditions [49] and are well reviewed by van Roekel et al. [48].
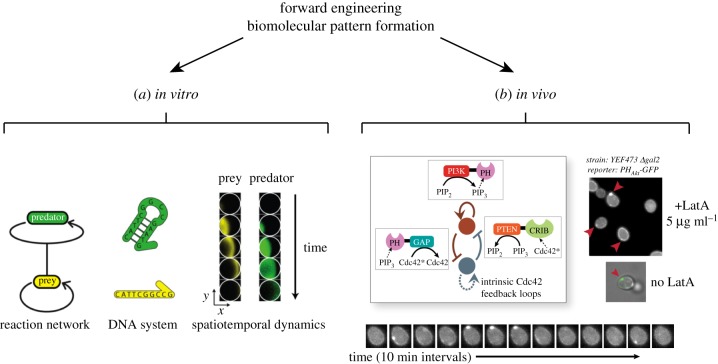
Of particular relevance to our comparison of the Min system is the work of Padirac et al. [50]. The authors used a DNA-based system of oligonucleotides, polymerase, exonuclease and nickase. The system was designed to exhibit a ‘predator–prey’ mechanism that has been theoretically proposed to exhibit oscillations. Remarkably, when reconstituted into unstirred closed reactors, this reaction network produced travelling waves and spirals (figure 3a) [50]. Similarly, cell-free transcription–translation systems, in combination with immobilization or compartmentalization techniques, have been used to produce spatial phenomena, such as gradients and moving fronts of gene expression, or spatial patterns emulating those occurring during Drosophila development [52–55].
Figure 3.
Two examples of forward engineering approaches to biomolecular pattern formation. (a) In vitro: Padirac et al. engineered a DNA-based biochemical reaction network of the ‘predator–prey’ type [50]. Reproduced with permission from Padirac et al. [50]. Copyright © 2013 American Chemical Society. (b) In vivo: Chau et al. designed regulatory circuits for self-organized polarization in Saccharomyces cerevisiae [51]. Reproduced with permission from Chau et al. [51]. Copyright © 2012 Elsevier.
These studies demonstrate the possibility of de novo systems capable of complex spatiotemporal pattern formation. Pleasingly, they have also shown reasonable agreement with theoretical models, or have been successfully guided by modelling and simulation [50,51,53–55]. It seems likely that DNA-based reaction networks will remain attractive for further studies, owing to their greater programmability compared with the inherent difficulties of protein engineering. Nevertheless, the Min system suggests that spatiotemporal patterns can emerge from only two proteins that reversibly associate with a lipid membrane in an energy-dependent fashion [15]. Synthetic protein pattern formation might therefore be built with significantly fewer components and without the requirement of synthesis and degradation reactions, as compared with most approaches based on gene expression or DNA circuits.
The most promising example of forward engineering applied to the self-organization of proteins so far focuses on the more elementary case of cellular polarization. Chau et al. constructed and tested synthetic regulatory networks of different topologies in yeast to systematically investigate and engineer the symmetry breaking process underlying cellular polarization [51]. Symmetry breaking is a basic requirement for both self-organized pattern formation and polarization. Theoretical work has established the role of local positive feedback and global inhibition as key to such processes [10,56,57]. Chau et al. began by using a coarse-grained computational model to explore all possible networks formed by positive feedback, mutual inhibition and inhibition with positive feedback motifs. They then constructed and tested these networks in Saccharomyces cerevisiae using chimeric proteins that regulated the density of phosphatidylinositol(3,4,5)-trisphosphate (PIP3) in the cell membrane (figure 3b). While each motif on its own was capable of weakly polarizing PIP3 under a narrow range of conditions, topologies that combined motifs produced significantly more robust and long-lasting polarization [51].
One of the most attractive aspects of the study of Chau et al. is the complementary use of computational modelling, albeit at quite a high level of abstraction. With further development, one could imagine a virtuous engineering cycle where the outcomes of systematic reverse engineering studies could be used to parameterize increasingly detailed models that could then be built and tested. In vitro reconstitution experiments, as exemplified earlier, would be a rich source of parametric data. Such a strategy would hopefully lead to a convergence of the two engineering approaches, arriving at, for example, a clearly elucidated connection between primary amino acid sequence and its physical outcome on spatial pattern formation.
In summary, self-organization is essential for many cellular processes such as cell division and motility, and so a detailed mechanistic understanding will be essential both in basic science and in engineered applications such as the already discussed proto-cells. Further, bio-orthogonal self-organizing systems could be useful for the engineering of natural cells to enhance industrial applications, such as the production of non-natural products, or for medicine, such as in immunotherapy [58,59]. To achieve such applications, de novo systems will need to be highly tuneable, and capable of organizing and interfacing with down-stream processes, cytoskeletal elements and organelles. It follows that, while it can be argued that a complex system will not be fully understood until it has been systematically built up from its component parts, bottom-up strategies still benefit greatly from knowledge derived by dissecting natural systems in a top-down fashion. We therefore believe that tight integration between reverse and forward engineering will be essential for realizing these goals.
4. Material and methods
(a). Plasmids
Mutations were introduced by site-directed mutagenesis into our previously described His-MinE expression vector [15] using the GeneArt® Site–Directed Mutagenesis System (Invitrogen, Carlsbad, CA, USA) with the forward and reverse primer pairs ACAGCCAACATTGCACAGGAACGGCTGCAGATT and AAT-CTGCAGCCGTTCCTGTGCAATGTTGGCTGT for the K19Q mutation, and AACATTGCAAAAGAAGCGCTGCAGATTATT-GT and ACAATAATCTGCAGCGCTTCTTTTGCAATGTT for the R21A mutation. The presence of the mutations was verified by sequencing.
(b). Protein purification
His-MinD, His-eGFP-MinD as well as WT and mutant His-MinE were expressed and purified as described previously [15,40].
(c). Preparation of small unilamellar vesicles and supported lipid bilayers
Small unilamellar vesicles (SUVs) and SLBs composed of E. coli polar lipids (Avanti Polar Lipids, Alabaster, AL, USA) were prepared, as described previously [40].
(d). ATPase assay
Measurement of MinD's basal and MinE-stimulated ATPase activity was performed with an ATP/NADH-coupled assay, as described previously [40].
(e). Self-organization assay
Self-organization assays were performed on SLBs, essentially as described previously [15,40]. First, SLBs on glass were generated by vesicle fusion of E. coli polar lipid SUVs [40]. Then, 1 µM MinD incorporating 20% eGFP-MinD, 2.5 mM ATP (F. Hoffmann-La Roche, Basel, Switzerland) and MinE at varying concentrations were added to Min buffer (25 mM Tris-HCl pH 7.5, 150 mM KCl, 5 mM MgCl2) (200 µl total volume) on top of SLBs. Samples were incubated for several hours to provide abundant time for pattern formation.
(f). Microscopy and image processing
Confocal imaging was performed using a Zeiss LSM780 confocal laser scanning microscope with a Zeiss C-Apochromat 40×/1.20 water-immersion objective (Carl Zeiss, Oberkochen, Germany). Image processing was carried out using Fiji [60]. As intensities varied substantially with MinE concentration, image brightness and contrast were adjusted for better visibility. As adjustments were performed equally for all conditions, some images are depicted outside the dynamic range. Thus, image intensities in figure 2 are not comparable. Any adjustments were applied uniformly to the entire image field.
Acknowledgements
We thank the Biochemistry Core Facility at MPI-B for help with protein purification and members of the Schwille laboratory for useful discussions.
Data accessibility
All relevant data are within the manuscript. Materials are available upon request.
Authors' contributions
S.K. and P.S. designed the research. S.K. performed experiments and analysed the data. S.K, L.H. and P.S. wrote the manuscript.
Competing interests
We declare no competing interests.
Funding
S.K. and P.S. acknowledge financial support from the DFG via project A09 of the SFB1032 ‘Nanoagents for the spatiotemporal control of molecular and cellular reactions'. S.K. was also supported by a DFG fellowship from the Graduate School of Quantitative Biosciences Munich (QBM). L.H. is the recipient of a Research Fellowship from the Alexander von Humboldt Foundation. This work is part of the MaxSynBio consortium, which is jointly funded by the Federal Ministry of Education and Research of Germany and the Max Planck Society.
References
- 1.Kozubowski L, Saito K, Johnson JM, Howell AS, Zyla TR, Lew DJ. 2008. Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr. Biol. 18, 1719–1726. ( 10.1016/j.cub.2008.09.060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goehring NW, Trong PK, Bois JS, Chowdhury D, Nicola EM, Hyman AA, Grill SW. 2011. Polarization of PAR proteins by advective triggering of a pattern-forming system. Science 334, 1137–1141. ( 10.1126/science.1208619) [DOI] [PubMed] [Google Scholar]
- 3.Bement WM, et al. 2015. Activator–inhibitor coupling between Rho signalling and actin assembly makes the cell cortex an excitable medium. Nat. Cell Biol. 17, 1471–1483. ( 10.1038/ncb3251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lutkenhaus J. 2012. The ParA/MinD family puts things in their place. Trends Microbiol. 20, 411–418. ( 10.1016/j.tim.2012.05.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kretschmer S, Schwille P. 2016. Pattern formation on membranes and its role in bacterial cell division. Curr. Opin. Cell Biol. 38, 52–59. ( 10.1016/j.ceb.2016.02.005) [DOI] [PubMed] [Google Scholar]
- 6.Schumacher D, Bergeler S, Harms A, Vonck J, Huneke-Vogt S, Frey E, Sogaard-Andersen L. 2017. The PomXYZ proteins self-organize on the bacterial nucleoid to stimulate cell division. Dev. Cell 41, 299–314.e213. ( 10.1016/j.devcel.2017.04.011) [DOI] [PubMed] [Google Scholar]
- 7.Hu L, Vecchiarelli AG, Mizuuchi K, Neuman KC, Liu J. 2017. Brownian ratchet mechanisms of ParA-mediated partitioning. Plasmid 92, 12–16. ( 10.1016/j.plasmid.2017.05.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bange G, Sinning I. 2013. SIMIBI twins in protein targeting and localization. Nat. Struct. Mol. Biol. 20, 776–780. ( 10.1038/nsmb.2605) [DOI] [PubMed] [Google Scholar]
- 9.Turing AM. 1952. The chemical basis of morphogenesis. Phil. Trans. R. Soc. Lond. B 237, 37–72. ( 10.1098/rstb.1952.0012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gierer A, Meinhard H. 1972. Theory of biological pattern formation. Kybernetik 12, 30–39. ( 10.1007/Bf00289234) [DOI] [PubMed] [Google Scholar]
- 11.Zaikin AN, Zhabotinsky AM. 1970. Concentration wave propagation in two-dimensional liquid-phase self-oscillating system. Nature 225, 535–537. ( 10.1038/225535b0) [DOI] [PubMed] [Google Scholar]
- 12.Jakubith S, Rotermund HH, Engel W, Vonoertzen A, Ertl G. 1990. Spatiotemporal concentration patterns in a surface-reaction: propagating and standing waves, rotating spirals, and turbulence. Phys. Rev. Lett. 65, 3013–3016. ( 10.1103/Physrevlett.65.3013) [DOI] [PubMed] [Google Scholar]
- 13.de Boer PA, Crossley RE, Rothfield LI. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56, 641–649. ( 10.1016/0092-8674(89)90586-2) [DOI] [PubMed] [Google Scholar]
- 14.Leisch N, Verheul J, Heindl NR, Gruber-Vodicka HR, Pende N, den Blaauwen T, Bulgheresi S. 2012. Growth in width and FtsZ ring longitudinal positioning in a gammaproteobacterial symbiont. Curr. Biol. 22, R831–R832. ( 10.1016/j.cub.2012.08.033) [DOI] [PubMed] [Google Scholar]
- 15.Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P. 2008. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science 320, 789–792. ( 10.1126/science.1154413) [DOI] [PubMed] [Google Scholar]
- 16.Raskin DM, de Boer PA. 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl Acad. Sci. USA 96, 4971–4976. ( 10.1073/pnas.96.9.4971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vecchiarelli AG, Li M, Mizuuchi M, Mizuuchi K. 2014. Differential affinities of MinD and MinE to anionic phospholipid influence Min patterning dynamics in vitro. Mol. Microbiol. 93, 453–463. ( 10.1111/mmi.12669) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu F, van Schie BG, Keymer JE, Dekker C. 2015. Symmetry and scale orient Min protein patterns in shaped bacterial sculptures. Nat. Nanotechnol. 10, 719–726. ( 10.1038/nnano.2015.126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Z, Lutkenhaus J. 1999. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34, 82–90. ( 10.1046/j.1365-2958.1999.01575.x) [DOI] [PubMed] [Google Scholar]
- 20.Halatek J, Frey E. 2012. Highly canalized MinD transfer and MinE sequestration explain the origin of robust MinCDE-protein dynamics. Cell Rep. 1, 741–752. ( 10.1016/j.celrep.2012.04.005) [DOI] [PubMed] [Google Scholar]
- 21.Kruse K. 2002. A dynamic model for determining the middle of Escherichia coli. Biophys. J. 82, 618–627. ( 10.1016/S0006-3495(02)75426-X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meinhardt H, de Boer PA. 2001. Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc. Natl Acad. Sci. USA 98, 14 202–14 207. ( 10.1073/pnas.251216598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang KC, Meir Y, Wingreen NS. 2003. Dynamic structures in Escherichia coli: spontaneous formation of MinE rings and MinD polar zones. Proc. Natl Acad. Sci. USA 100, 12 724–12 728. ( 10.1073/pnas.2135445100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu LJ, Errington J. 2012. Nucleoid occlusion and bacterial cell division. Nat. Rev. Microbiol. 10, 8–12. ( 10.1038/nrmicro2671) [DOI] [PubMed] [Google Scholar]
- 25.Bisson-Filho AW, et al. 2017. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739–743. ( 10.1126/science.aak9973) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl Acad. Sci. USA 96, 14 819–14 824. ( 10.1073/pnas.96.26.14819) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, Xiao J. 2017. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355, 744–747. ( 10.1126/science.aak9995) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivanov V, Mizuuchi K. 2010. Multiple modes of interconverting dynamic pattern formation by bacterial cell division proteins. Proc. Natl Acad. Sci. USA 107, 8071–8078. ( 10.1073/pnas.0911036107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vecchiarelli AG, Li M, Mizuuchi M, Hwang LC, Seol Y, Neuman KC, Mizuuchi K. 2016. Membrane-bound MinDE complex acts as a toggle switch that drives Min oscillation coupled to cytoplasmic depletion of MinD. Proc. Natl Acad. Sci. USA 113, E1479–E1488. ( 10.1073/pnas.1600644113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caspi Y, Dekker C. 2016. Mapping out Min protein patterns in fully confined fluidic chambers. eLife 5, 35 ( 10.7554/eLife.19271) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zieske K, Chwastek G, Schwille P. 2016. Protein patterns and oscillations on lipid monolayers and in microdroplets. Angew. Chem. Int. Edn 55, 13 455–13 459. ( 10.1002/anie.201606069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zieske K, Schwille P. 2014. Reconstitution of self-organizing protein gradients as spatial cues in cell-free systems. eLife 3, 11858 ( 10.7554/eLife.03949) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park KT, Wu W, Battaile KP, Lovell S, Holyoak T, Lutkenhaus J. 2011. The Min oscillator uses MinD-dependent conformational changes in MinE to spatially regulate cytokinesis. Cell 146, 396–407. ( 10.1016/j.cell.2011.06.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Z, Lutkenhaus J. 2003. A conserved sequence at the C-terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Mol. Microbiol. 47, 345–355. ( 10.1046/j.1365-2958.2003.03321.x) [DOI] [PubMed] [Google Scholar]
- 35.Lackner LL, Raskin DM, de Boer PAJ. 2003. ATP-dependent interactions between Escherichia coli Min proteins and the phospholipid membrane in vitro. J. Bacteriol. 185, 735–749. ( 10.1128/jb.185.3.735-749.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szeto TH, Rowland SL, Rothfield LI, King GF. 2002. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc. Natl Acad. Sci. USA 99, 15 693–15 698. ( 10.1073/pnas.232590599) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szeto TH, Rowland SL, Habrukowich CL, King GF. 2003. The MinD membrane targeting sequence is a transplantable lipid-binding helix. J. Biol. Chem. 278, 40 050–40 056. ( 10.1074/jbc.M306876200) [DOI] [PubMed] [Google Scholar]
- 38.Hu Z, Lutkenhaus J. 2001. Topological regulation of cell division in E. coli: spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol. Cell 7, 1337–1343. ( 10.1016/S1097-2765(01)00273-8) [DOI] [PubMed] [Google Scholar]
- 39.Park KT, Villar MT, Artigues A, Lutkenhaus J. 2017. MinE conformational dynamics regulate membrane binding, MinD interaction, and Min oscillation. Proc. Natl Acad. Sci. USA 114, 7497–7504. ( 10.1073/pnas.1707385114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kretschmer S, Zieske K, Schwille P. 2017. Large-scale modulation of reconstituted Min protein patterns and gradients by defined mutations in MinE's membrane targeting sequence. PLoS ONE 12, e0179582 ( 10.1371/journal.pone.0179582) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park KT, Wu W, Lovell S, Lutkenhaus J. 2012. Mechanism of the asymmetric activation of the MinD ATPase by MinE. Mol. Microbiol. 85, 271–281. ( 10.1111/j.1365-2958.2012.08110.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martos A, Petrasek Z, Schwille P. 2013. Propagation of MinCDE waves on free-standing membranes. Environ. Microbiol. 15, 3319–3326. ( 10.1111/1462-2920.12295) [DOI] [PubMed] [Google Scholar]
- 43.Auslander S, Auslander D, Fussenegger M. 2017. Synthetic biology—the synthesis of biology. Angew. Chem. Int. Edn 56, 6396–6419. ( 10.1002/anie.201609229) [DOI] [PubMed] [Google Scholar]
- 44.Kretschmer S, Schwille P. 2014. Toward spatially regulated division of protocells: insights into the E. coli Min system from in vitro studies. Life 4, 915–928. ( 10.3390/life4040915) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langecker M, Arnaut V, Martin TG, List J, Renner S, Mayer M, Dietz H, Simmel FC. 2012. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 338, 932–936. ( 10.1126/science.1225624) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burns JR, Stulz E, Howorka S. 2013. Self-assembled DNA nanopores that span lipid bilayers. Nano Lett. 13, 2351–2356. ( 10.1021/nl304147f) [DOI] [PubMed] [Google Scholar]
- 47.Gopfrich K, Zettl T, Meijering AE, Hernandez-Ainsa S, Kocabey S, Liedl T, Keyser UF. 2015. DNA-tile structures induce ionic currents through lipid membranes. Nano Lett. 15, 3134–3138. ( 10.1021/acs.nanolett.5b00189) [DOI] [PubMed] [Google Scholar]
- 48.van Roekel HW, Rosier BJ, Meijer LH, Hilbers PA, Markvoort AJ, Huck WT, de Greef TF. 2015. Programmable chemical reaction networks: emulating regulatory functions in living cells using a bottom-up approach. Chem. Soc. Rev. 44, 7465–7483. ( 10.1039/c5cs00361j) [DOI] [PubMed] [Google Scholar]
- 49.Semenov SN, Wong ASY, van der Made RM, Postma SGJ, Groen J, van Roekel HWH, de Greef TFA, Huck WTS. 2015. Rational design of functional and tunable oscillating enzymatic networks. Nat. Chem. 7, 160–165. ( 10.1038/NCHEM.2142) [DOI] [PubMed] [Google Scholar]
- 50.Padirac A, Fujii T, Estevez-Torres A, Rondelez Y. 2013. Spatial waves in synthetic biochemical networks. J. Am. Chem. Soc. 135, 14 586–14 592. ( 10.1021/ja403584p) [DOI] [PubMed] [Google Scholar]
- 51.Chau AH, Walter JM, Gerardin J, Tang C, Lim WA. 2012. Designing synthetic regulatory networks capable of self-organizing cell polarization. Cell 151, 320–332. ( 10.1016/j.cell.2012.08.040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karzbrun E, Tayar AM, Noireaux V, Bar-Ziv RH. 2014. Synthetic biology. Programmable on-chip DNA compartments as artificial cells. Science 345, 829–832. ( 10.1126/science.1255550) [DOI] [PubMed] [Google Scholar]
- 53.Semenov SN, Markvoort AJ, Gevers WBL, Piruska A, de Greef TFA, Huck WTS. 2013. Ultrasensitivity by molecular titration in spatially propagating enzymatic reactions. Biophys. J. 105, 1057–1066. ( 10.1016/j.bpj.2013.07.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tayar AM, Karzbrun E, Noireaux V, Bar-Ziv RH. 2015. Propagating gene expression fronts in a one-dimensional coupled system of artificial cells. Nat. Phys. 11, 1037–1041. ( 10.1038/NPHYS3469) [DOI] [Google Scholar]
- 55.Isalan M, Lemerle C, Serrano L. 2005. Engineering gene networks to emulate Drosophila embryonic pattern formation. PLoS Biol. 3, e64 ( 10.1371/journal.pbio.0030064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wedlich-Soldner R, Altschuler S, Wu L, Li R. 2003. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299, 1231–1235. ( 10.1126/science.1080944) [DOI] [PubMed] [Google Scholar]
- 57.Mogilner A, Allard J, Wollman R. 2012. Cell polarity: quantitative modeling as a tool in cell biology. Science 336, 175–179. ( 10.1126/science.1216380) [DOI] [PubMed] [Google Scholar]
- 58.Roybal KT, Williams JZ, Morsut L, Rupp LJ, Kolinko I, Choe JH, Walker WJ, McNally KA, Lim WA. 2016. Engineering T cells with customized therapeutic response programs using synthetic Notch receptors. Cell 167, 419–432.e416. ( 10.1016/j.cell.2016.09.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morsut L, Roybal KT, Xiong X, Gordley RM, Coyle SM, Thomson M, Lim WA. 2016. Engineering customized cell sensing and response behaviors using synthetic Notch receptors. Cell 164, 780–791. ( 10.1016/j.cell.2016.01.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Meth. 9, 676–682. ( 10.1038/nmeth.2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript. Materials are available upon request.