Abstract
Ecological networks are composed of interacting communities that influence ecosystem structure and function. Fungi are the driving force for ecosystem processes such as decomposition and carbon sequestration in terrestrial habitats, and are strongly influenced by interactions with invertebrates. Yet, interactions in detritivore communities have rarely been considered from a network perspective. In the present study, we analyse the interaction networks between three functional guilds of fungi and insects sampled from dead wood. Using DNA metabarcoding to identify fungi, we reveal a diversity of interactions differing in specificity in the detritivore networks, involving three guilds of fungi. Plant pathogenic fungi were relatively unspecialized in their interactions with insects inhabiting dead wood, while interactions between the insects and wood-decay fungi exhibited the highest degree of specialization, which was similar to estimates for animal-mediated seed dispersal networks in previous studies. The low degree of specialization for insect symbiont fungi was unexpected. In general, the pooled insect–fungus networks were significantly more specialized, more modular and less nested than randomized networks. Thus, the detritivore networks had an unusual anti-nested structure. Future studies might corroborate whether this is a common aspect of networks based on interactions with fungi, possibly owing to their often intense competition for substrate.
Keywords: weighted networks, nestedness, spore dispersal, dead wood, plant pathogenic fungi, symbiont fungi
1. Introduction
Interactions between species shape ecological communities and networks, and drive evolution. Ecosystems therefore consist of complex networks that vary in structure depending on the specificity and frequency of the interacting species. Highly specific species interactions often result in very specialized networks with low robustness to species loss [1], where extinction of one species also leads to the loss of connected species from the network. As species are currently going extinct at an alarmingly high rate [2], knowledge of ecological networks and interactions is becoming increasingly important in order to understand and hopefully prevent extinction cascades.
Several studies have underlined the importance of pollination and other well-known interactions such as predation, herbivory and animal-mediated seed dispersal for ecosystem structure and function (e.g. [3–5]). However, our knowledge of biotic interactions is highly skewed towards macroscopic organisms [6], and network studies have largely focused on well-known interactions such as pollination [7,8]. There are few studies of interactions between bacteria, fungi or invertebrates at the community level, despite their overwhelming abundance and species diversity [9–12]. Bacteria and fungi are integral to terrestrial and freshwater ecosystems through their roles as pathogens, symbionts and decomposers [13–17]. Up to 90% of terrestrial plant production enters the detrital food chain [18], where the microbiota of bacteria, fungi and invertebrates determine rate of decomposition and carbon sequestration [16,17].
Invertebrates can have a significant influence on ecosystem processes through interactions with bacteria or fungi, as demonstrated for rate of decomposition, nutrient cycling and mycorrhizal symbiosis in laboratory experiments [19–21]. However, the role of invertebrates in the detritivore community is rarely considered from a network perspective, in contrast with the intensively studied functional roles of invertebrates as pollinators or herbivores [7,8]. In the present study, we show that network analysis of understudied species groups such as insects and fungi can reveal hidden interactions and elucidate the structure of detritivore communities.
Ecological networks are shaped by the frequency of interactions between species, which in turn is partly determined by abundance of the species and their interaction specialization. The tendency of species in a network to exhibit specialized interactions can be summed up at the network level as degree of specialization [22,23]. For instance, as pollinators are generally more specialized in their resource use than seed-feeding animals, pollination networks in general have a higher degree of specialization than networks based on animal-mediated seed dispersal [22].
If specialist species mainly interact with a proper subset of the interaction partners of generalist species, this results in a nested network structure. Nested networks are generally robust against random species loss [24], while networks with a high degree of specialization are more vulnerable [25]. Networks can also be organized into compartments called modules, in which species interact frequently within the modules and infrequently between modules. If within-module interactions are dominant in number, the network is said to have high modularity [26]. Modules might be the product of spatial or temporal variability in interactions, for instance if interaction frequency depends on overlap in phenology, or they might consist of closely related species or species with similar trait syndromes owing to convergent evolution [27,28]. Thus, the structure of an interaction network can reveal selective pressures shaping the interactions and the robustness of networks to species loss.
In the present study, we analyse specialization, nestedness and modularity of insect–fungus networks sampled from dead wood in boreal forests. These networks are vital for the functioning of forest ecosystems, as they are the driving force for decomposition and nutrient cycling in these habitats [29–31]. Understanding how these networks are structured is therefore integral to understanding the basis for ecosystem processes in forests. We used DNA metabarcoding to identify fungi extracted from individual insects, which enabled us to include interactions involving microscopic fungal structures such as spores, hyphae or yeast. We compiled quantitative (i.e. weighted) networks for interactions between insects inhabiting dead wood and three functional groups of fungi; insect symbiont fungi, wood-decay fungi and plant pathogenic fungi.
As we do not have replicates of each network, this study is not a test of differences between these groups, but rather an exploratory first step into largely unchartered territory for network analysis in terms of both methodology [6] (i.e. the combination of DNA metabarcoding and quantitative networks) and study organisms (i.e. detritivorous insects and fungi). In line with the few comparable previous studies [32,33], we demonstrate that such novel network analysis might reveal network structures differing from those of more well-known interactions, underlining the necessity of expanding the scope of network studies.
2. Methods
This study is based on data from Jacobsen et al. [34], where a more detailed description of insect sampling, DNA-analysis and bioinformatics can be found.
We sampled beetles from recently cut logs of aspen (Populus tremula L.) that had been placed at eight sites in two production forests in south-eastern Norway; Losby forest holdings (Latitude 55.98, Longitude 10.68, 150–300 m above sea level (a.s.l.) and Løvenskiold-Vækerø (LV) forest holdings (Latitude 54.49, Longitude 21.24, 200–500 m.a.s.l.). Both forest landscapes lie within the southern boreal vegetation zone [35] and consist mainly of spruce (Picea abies (L.) H.Karst.), with pine (Pinus sylvestris L.), birch (Betula pubescens Ehrh.) and aspen as subdominants.
Beetles were sampled individually with tweezers directly from the logs or from sticky traps on the logs, on 11 occasions during May to August in 2014 and 2015. The sticky traps were exposed for one or two days prior to insect sampling. The tweezers were sterilized with ethanol and fire between handling of each insect. The insects were killed by freezing at −80°C and identified to species or genus in a sterile environment using sterilized equipment. Insects that could not be confidently identified at least to genus by the first author (R.M.J.) were not analysed further (13 of 654 individuals). We selected 343 wood-inhabiting beetle individuals, i.e. species or genera with larval development either in dead wood or in fungal fruit bodies on dead wood [36,37], for analysis of fungal DNA.
Fungal DNA was extracted from the beetles following a modified CTAB protocol [38] and amplified by polymerase chain reaction (PCR) on an Eppendorf Thermal Cycler (VWR, Radnor, USA) using primers internal transcribed spacer (ITS) 4 (ITS4) [39] and fITS7 [40]. The PCR products were cleaned using Wizard® SV Gel and PCR Clean-Up System (Promega, Madison, USA) and pooled according to strength of the bands in gel electrophoresis. Pooled samples were cleaned with the ChargeSwitch® kit (Invitrogen, California, USA), DNA-concentration was measured with the Qubit® BR DNA kit (Invitrogen, California, USA), and the sample quality was confirmed by Nanodrop™ (Thermo Fisher Scientific, Madison, USA). The samples were submitted to GATC Biotech for adaptor-ligation and Illumina HiSeq Rapid Run 300 bp paired-end sequencing. Quality control and clustering of the resulting sequences was conducted with the SCATA pipeline (https://scata.mykopat.slu.se/, accessed 5 July 2016). The sequences were subsampled to 10 000 per beetle sample prior to clustering. Taxonomy was assigned to the representative sequences of each operational taxonomic unit (OTU) taking the top hit of a Basic Local Alignment Search Tool (BLASTn [41]) search against the NCBI (National Centre for Biotechnology Information) and UNITE [42] databases. OTUs with e-values <e-10 and bit-scores >100 were annotated to species level if ITS homology was 100–98%, genus for 97.9–95%, family or order for 94.9–80%, phylum for 79.9–70% and ‘fungus’ for lower homology or e-values >e-10 and bit-scores <100. Taxonomy was updated according to the taxonomic database Dyntaxa (https://www.dyntaxa.se/, accessed 24 February 2017) and MycoBank (http://www.mycobank.org/, export date 26 October 2017). For further statistical analysis only OTUs represented by at least 20 reads were included, since we wanted to focus on widespread fungi more likely to be important in interactions.
(a). Classification of fungal functional groups
Fungal OTUs annotated to species or genus level were analysed further, in networks including all OTUs (electronic supplementary material, table S1) and in networks including OTUs classified into functional groups based on the FUNGuild database [43] and various literature (see the electronic supplementary material, tables S2–S4). Groups were non-overlapping. We analysed networks with the three most abundant (in terms of number of sequences) functional groups:
(i) insect symbionts (electronic supplementary material, table S5); this group included known insect symbionts such as Ophiostoma spp. or Phialophoropsis spp., and yeast species isolated from insect guts in previous studies such as Candida spp. and Cryptococcus spp., that were assumed to be endosymbionts. Fungal parasites or pathogens of insects were not included (only eight OTUs matched these functional groups according to FUNGuild);
(ii) wood-decayers (electronic supplementary material, table S6); this group included fungi in the class Agaricomycetes known to inhabit dead wood, in which the majority of species produce large fruit bodies and large quantities of spores that attract spore-feeding insects during sporulation (e.g. [44,45]); and
(iii) plant pathogens (electronic supplementary material, table S7); this group included pathogens of living plants. Plant pathogenic fungi known to be insect symbionts such as Ophiostoma spp. were excluded, since these functional groups were meant to mainly reflect the relationship between the insects and the fungi.
(b). Statistics
All analyses were conducted in R v. 3.3.2 [46].
The number of beetle individuals in which each fungal OTU occurred was used as a basis for quantitative/weighted networks. Excluding insect species represented by single individuals did not change the results and these species were therefore included in the network analysis. Network specialization was estimated by the standardized two-dimensional Shannon entropy H2′ [47] using the package bipartite v. 2.07 [48]. This index defines the degree of specialization in a network as the deviation from the expected probability distribution of interactions, which assumes that a species interacts with another species in proportion to its total frequency of occurrence in the network (i.e. terminal row or column sums). We estimated the species-level specialization by the standardized Kullback–Leibler distance d′ [47]. The species-level specialization index is defined as a species' deviation of the utilization of potential partners that is expected based on their terminal row or column sums, i.e. link numbers in the fungus x insect interaction matrix. Both H2′ and d′ range from 0 for most generalized to 1 for most specialized.
Modularity of the networks was estimated with the QuanBiMo algorithm developed by Dormann & Strauss [26] and implemented as function ‘computeModules’ in the bipartite package. Modularity Q ranges from 0, meaning that there are no more links between species in a module than expected by chance, to 1 which signifies maximum modularity for the network. As the QuanBiMo algorithm is based on a stochastic process, we estimated modularity 10 times for each network and reported the mean value. To estimate nestedness of the network, we used the weighted version of the nestedness metric based on overlap and decreasing fill, abbreviated WNODF [49]. This metric ranges from 0 for networks without nested structure, to 100 for perfectly nested networks.
We tested the statistical significance of the metrics for each network by simulating null models (n = 1000). Null model P followed Patefield's algorithm [50] as implemented in the function ‘r2dtable’ in R, which randomizes network interactions with the restriction of fixed marginal sums (i.e. the sum of interactions for each species was kept constant). We also tested null model V, which in addition to fixed marginal sums also keeps connectance (i.e. proportion of realized links in the interaction matrix) of the network constant as proposed by Vazquez et al. [51] and implemented in function ‘quasiswap_count’ in the vegan package v. 2.4–2. We performed two-sided tests of the network metric value against the distribution of the null model metric values. Finally, we repeated all network analyses for subsets of the insect–fungus networks with species numbers standardized to those of the smallest network.
3. Results
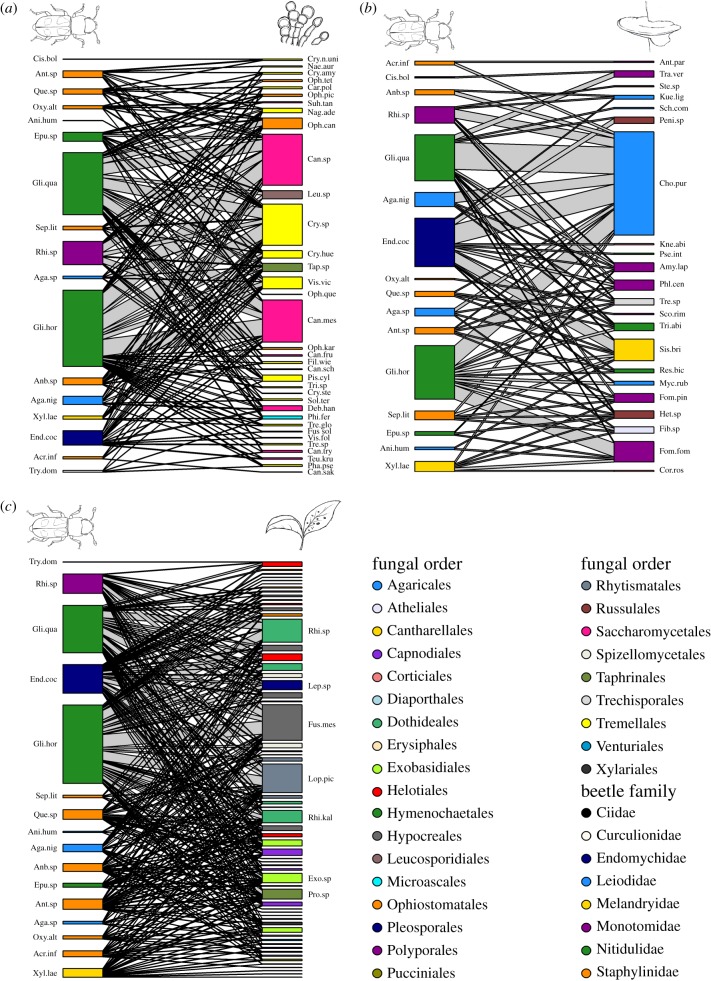
Fungal DNA was obtained from 187 wood-inhabiting beetle individuals of 17 species or genera (electronic supplementary material, table S8). The DNA metabarcoding analyses resulted in 1069 fungal OTUs represented by more than 20 sequences and distributed on a total of 1 714 063 sequences. Of these OTUs, 449 were annotated to species or genus and analysed further in networks with the insects, either including all fungi or separated into functional groups; 35 species or genera of fungi (356 279 sequences) were classified as insect symbionts, 22 (48 196 sequences) were classified as wood-decayers in the class Agaricomycetes and 61 (158 133 sequences) were classified as plant pathogens (figure 1).
Figure 1.
Networks of wood-inhabiting beetles and fungi classified as (a) insect symbionts, (b) wood-decayers or (c) plant pathogens. Sizes of boxes and interaction lines represent number of occurrences of the fungi in the insect samples. Colours denote taxonomic grouping; order for fungi and family for insects (all in the order Coleoptera). See the electronic supplementary material, tables S2–S7 for full names of abbreviations. (Online version in colour.)
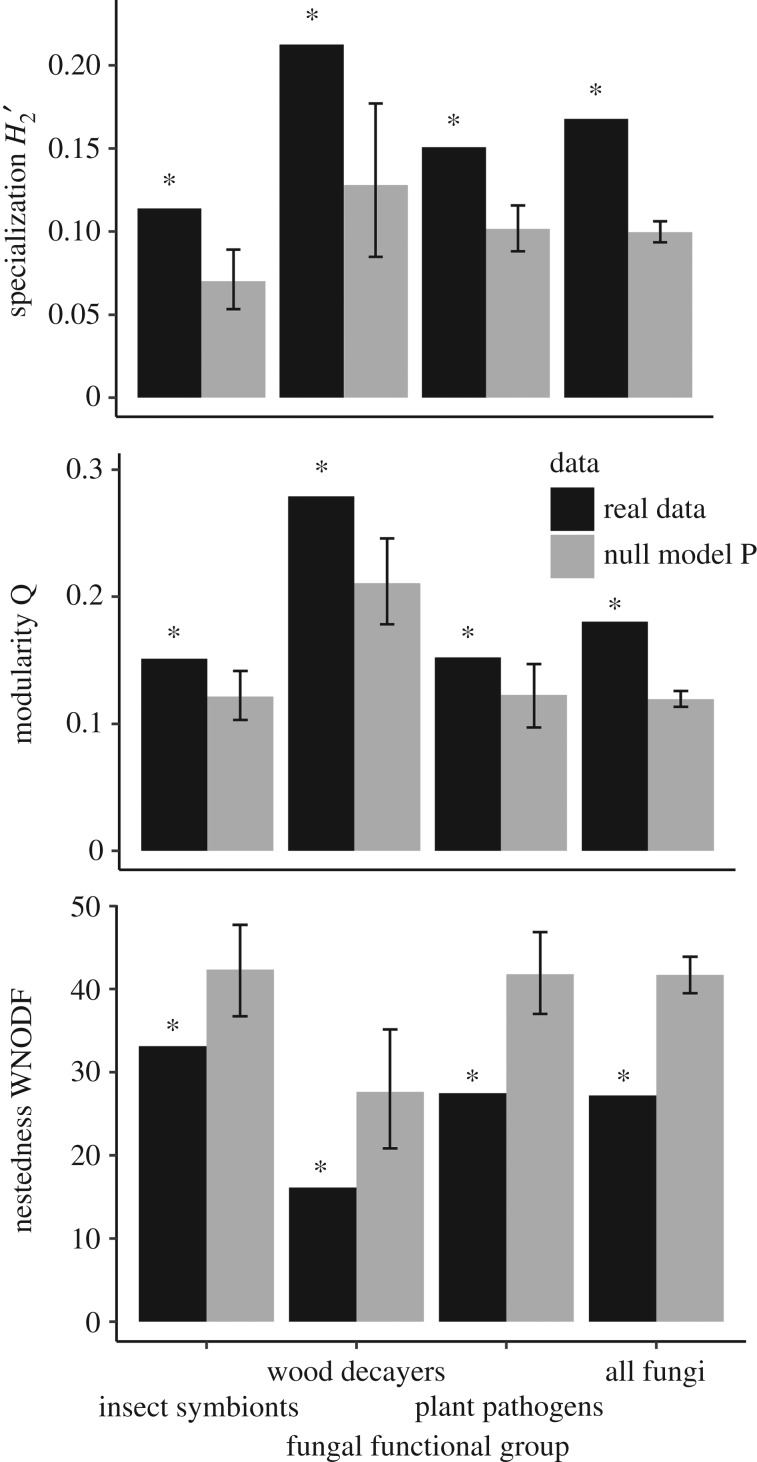
All insect–fungus networks were significantly more specialized, more modular and less nested than the null model with randomized interactions (figure 2). The results were relatively similar when compared with the null model which also had constant connectance (electronic supplementary material, figure S1), and for the subsampled networks with standardized species numbers (electronic supplementary material, figure S2). The network with wood-decay fungi had the highest degree of specialization and modularity (H2′ = 0.21, Q = 0.28; figure 2). Correspondingly, it also had the lowest nestedness (WNODF = 16.14; figure 2). However, when comparing standardized values of nestedness (real value − mean value of randomization/s.e. of randomizations), the network with plant pathogenic fungi had the lowest values (−5.68 standardized WNODF compared with −3.14 for wood-decay fungi and −3.29 for insect symbiont fungi).
Figure 2.
Network specialization, modularity and weighted nestedness for networks between wood-inhabiting beetles and the fungal functional groups insect symbionts, wood-decayers and plant pathogens, or all fungi annotated to species or genus. Black bars represent the original networks, while grey bars represent networks randomized with constant marginal sums according to null model P [50] with 95% confidence intervals (CI). Asterisks (*) above the black bars signify significant differences between the original and the randomized networks.
We re-calculated the network metrics with OTUs annotated as Chondrostereum purpureum (Pers. : Fr.) Pouzar excluded from the network of wood-decayers, since this species was visibly fruiting on the logs during insect sampling and could have occurred in all samples indiscriminately. Indeed, DNA from C. purpureum was isolated from 43% of the insect samples, including 12 of 17 taxa. Excluding C. purpureum from the wood-decayer network resulted in even higher specialization (H2′ = 0.29, null model P 95% confidence interval (CI) = 0.13–0.23), higher modularity (Q = 0.40, null model P 95% CI = 0.28–0.36) and lower nestedness (WNODF = 9.38, null model P 95% CI = 12.00–23.23, albeit higher standardized WNODF = −2.9). Without C. purpureum, the network between wood-inhabiting beetles and wood-decay fungi was organized in six modules (figure 3).
Figure 3.
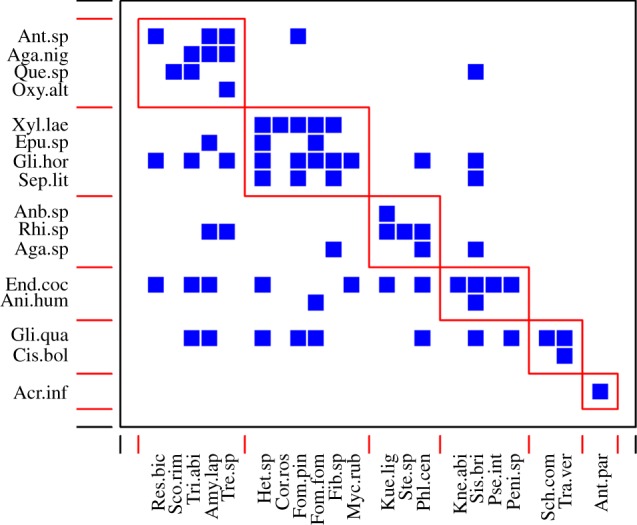
Modules in the network between wood-inhabiting beetles and wood-decay fungi with C. purpureum excluded, as organized by the QuanBiMo algorithm [26]. Lines demarcate modules, squares indicate interactions between insects and fungi. See the electronic supplementary material, tables S3 and S6 for full names of abbreviations. (Online version in colour.)
We estimated specialization at the species level for all networks (electronic supplementary material, tables S9–S14), but focus here on interactions with the more well-known wood-decay fungi. In interactions with wood-decay fungi, the insect species Endomychus coccineus (Linnaeus, 1758) was significantly (p-value = 0.005) more specialized and Glischrochilus hortensis (Geoffroy, 1785) was nearly significantly (p-value = 0.053) more specialized than expected from the null model (electronic supplementary material, table S11), with index values (d′) of 0.25 and 0.18, respectively. Among the wood-decay fungi, OTUs annotated as Trametes versicolor (L. : Fr.) Pilát., Fomes fomentarius (L. : Fr.) Fr. and Sistotrema brinkmannii (Bres.) J. Erikss. were significantly specialized with index values of 0.45, 0.38 and 0.24 (p-values < 0.05), respectively (electronic supplementary material, table S12).
4. Discussion
This study shows that species of two very diverse eukaryotic kingdoms, insects and fungi, interact in structured networks. The networks had an anti-nested structure (i.e. they were less nested than randomized networks), they were specialized, though not to a high degree, and interacting species were compartmentalized in modules. The lack of a nested network structure might indicate a relatively low species redundancy, which could mean that the insect–fungus networks are vulnerable to species loss [7], although the relatively low degree of specialization (H2′ = 0.21 or less) might increase robustness [1] and species within modules might fulfil similar interaction functions.
Although non-nested structures have been demonstrated more often for quantitative, weighted networks than for qualitative, binary networks, anti-nested structures do not seem to be common for either network type [52]. However, previous studies using molecular methods to identify mycorrhizal fungi interacting with plants have also documented anti-nested networks [32,33,53]. Toju et al. [33] found that this anti-nested structure seemed to be explained by reduced fungal host range overlap, causing a checkerboard pattern of interactions. They suggested that this pattern might be caused by competitive exclusion by the fungi, preventing other species of fungi from interacting with their plant host. Although the insects in the present study are not presumed to function as a substrate and thus a site of competition for the fungi (possibly with the exception of the symbiont fungi), their interactions with the fungi might reflect competitive exclusion structuring fungal communities at shared habitats such as dead wood, where competition for substrate can be fierce [54]. Future studies might confirm whether anti-nestedness is a common aspect of interaction networks involving fungi.
Both degree of specialization (H2′ = 0.15) and modularity (Q = 0.15) were relatively low for the network between plant-pathogenic fungi and insects. Although there are examples of plant-pathogenic fungi being dispersed by insects in species-specific interactions [55], the insects analysed in the present study only included species inhabiting dead wood. Thus, it is not unexpected that their interactions with pathogens of living plants were relatively unspecific, perhaps only based on shared forest habitats. Furthermore, plant pathogenic fungi known to be symbionts of insects, such as Ophiostoma spp., were classified as insect symbionts rather than plant pathogens, as we considered this to be the aspect of their ecology most likely to affect their interaction networks with insects. The versatile ecology of fungi, where trophic mode might vary depending on context, complicates classification into functional groups [43]. Fungi have been documented to shift between an endophytic and a plant pathogenic lifestyle, or between a mycorrhizal and a saprotrophic lifestyle, to mention a few of the examples summarized by Selosse et al. [56]. Thus, the classification of fungi into functional groups in the present study is likely to be highly simplified and relatively uncertain for some taxa, especially the insect symbionts. Nevertheless, this tentative classification allows us to explore differences in the structure of networks involving different groups of fungi and build hypotheses for further studies.
The network with fungi annotated as insect symbionts had a surprisingly low degree of specialization (H2′ = 0.11). This group included fungi that might live in mutualistic or commensalistic symbiosis with insects, as insect parasites and pathogens were not included. Most of these species were classified as insect symbionts based on previous isolation from beetle guts (references in electronic supplementary material, table S2). In comparison, in a study by Shukla et al. [57] bacterial endosymbionts had a relatively high degree of specialization (H2′ = 0.35) even in an intraspecific network with males, females and larvae of one dung beetle species. Modularity was also relatively low (Q = 0.15), considering that intimate interactions tend to result in more modular networks [58]. Our results indicate that many of the fungal species found in insect guts might be unspecific symbionts, or simply contaminants from food or habitat that do not function as symbionts. Certainly, yeast fungi like Candida spp. and Cryptococcus spp. can occur in several different environments such as soil or dead wood [59–62], where insects are also abundant. Some of the fungi isolated from beetle guts do seem to be more closely associated with the habitat than with the beetle species [63]. However, endosymbionts can be relatively unspecific with regard to insect host species, especially if they are transmitted horizontally [15]. Further in-depth studies, including microscopy and experimentation, are required to clarify to what extent fungi such as Candida mesenterica and the other taxa tentatively classified as insect symbionts in the present study spend part of their life living as symbionts on or in insects, and whether this affects the insects.
The network between wood-inhabiting beetles and wood-decay fungi had the highest degree of specialization in this study (H2′ = 0.21). However, this is still much lower than the specialization of pollinator–plant networks (H2′ = 0.60 [22]), ant–myrmecophyte networks (H2′ = 0.80 [22,64]) or legume–rhizobium bacteria networks (H2′ = 0.85 [65]). Instead, it was closer to that of networks based on more opportunistic interactions, such as ants harvesting honeydew from true bugs (H2′ = 0.43 [23]) or nectar from plants (H2′ = 0.25 [22]), or animal-mediated seed dispersal (H2′ = 0.18–0.47 [22,66,67]). This indicates that the network between wood-inhabiting beetles and wood-decay fungi was based upon similarly opportunistic yet reciprocal interactions that would result in a moderate degree of specialization. Spore feeding and subsequent spore dispersal by the beetles could represent such an interaction [34]. In line with this hypothesis, the nitidulid beetle G. hortensis has frequently been registered on sporulating fruit bodies of wood-decay fungi such as the polypore F. fomentarius [44,45], although its habitat is fresh dead wood [36]. In the present study, this beetle species was found to be significantly more specialized on wood-decay fungi than expected by chance, and F. fomentarius was isolated from 11 individuals of G. hortensis. This beetle species might therefore function as a moderately specific propagule vector for F. fomentarius, providing targeted dispersal to fresh dead wood [34]. Although the network between wood-living beetles and wood-decay fungi might be a food web without dispersal benefits to the fungi, the beetles were sampled from dead wood that had recently been cut and placed in these forests, without any other visible fungal fruit bodies than those of C. purpureum.
If the network between wood-inhabiting beetles and wood-decay fungi was based on spore feeding and dispersal, its degree of specialization might be constrained by the same factors that limit specialization of animal-mediated seed dispersal networks [68]. Optimal dispersal of both spores and seeds requires the propagule vector to move away from the source and deliver the propagule not to a conspecific, but to a suitable habitat. The propagule source has no means to direct the vector, its only chance is to attract vectors that share its habitat. Fungal odour has been shown to attract several different species of beetles inhabiting dead wood [69–71], and odour release increases during sporulation [72]. Fomes fomentarius and certain other polypore species also aggregate spores on top of their fruit bodies, which are visited by several wood-inhabiting insects [44]. Aggregation of spores and increased odour emission during sporulation thus seem to function as attractants to wood-inhabiting insects, in much the same way as brightly coloured fruits attract seed dispersing animals. As such, there is a basis for selection favouring a certain degree of reciprocity and specialization between wood-decay fungi and insects. However, spore dispersal effectiveness would be low if the insects were highly specialized spore-feeders that only moved between sporulating fruit bodies, without dispersing the spores to unoccupied substrates. For seed dispersal, it has been shown that generalist frugivores can be very effective seed dispersers [73,74] and that species in highly diverse frugivore assemblages fulfil complementary roles [75,76]. These mechanisms promote diversified interactions and generalized dispersal systems [77], restraining the degree of specialization in seed dispersal networks [22,66,67] and possibly in the potential spore dispersal network in the present study.
It should be noted that certain aspects of network structure can be subject to strong spatial and temporal variability [67,78,79]. Our networks were based on pooled datasets of beetles sampled over two seasons in two different landscapes, but the necessity of sampling beetles individually resulted in a sample size that was too low to explore spatial and temporal variability in network structure. However, the distribution of sampled individuals was relatively even between landscapes, and the majority of individuals were sampled during the second year. Furthermore, network level measures tend to exhibit a lower temporal and spatial variability than species level measures [79]. In any case, our study is but an exploratory first step into novel methodology and understudied interactions, which can hopefully provide future research with a foundation for important working hypotheses regarding detritivore networks and the use of DNA metabarcoding for discerning microscopic interactions.
In conclusion, our results demonstrate that there is a diversity of hidden interactions in detritivore networks. These interactions could have significant influence on fungal communities in dead wood [62,80], and thereby affect important ecosystem functions such as carbon sequestration and decomposition [31]. We encourage the use of molecular methods to include microscopic organisms in future network studies [6], as the unusual network structures demonstrated in this and previous studies [32,33,53] underline the importance of expanding the scope of network analysis to understudied and functionally important organisms such as fungi.
Supplementary Material
Supplementary Material
Acknowledgements
We would like to thank Sindre Ligaard for advice during insect identification, Synnøve Botnen for tutoring in DNA analysis, Sundy Maurice and Janina Fuss for advice and help during laboratory work, Marit M. Bjorbækmo for indispensable help with bioinformatics, Sebastian Seibold for sharing data on fungal guilds, Marie Davey and Elisabet Ottosson for advice and information about fungal ecology, Jens Mogens Olesen for advice on network analysis, and Markus A.K. Sydenham for inspiration.
Data accessibility
Raw data (fastq-files), barcode and primer mapping file, OTU table and representative sequence files have been accessioned in Dryad with http://dx.doi.org/10.5061/dryad.3t2d4 [81].
Authors' contributions
R.M.J., T.B., H.K. and A.S.-T. conceived the idea and designed the methodology. R.M.J. did the fieldwork, laboratory work, analyses and led the writing of the manuscript. All authors contributed critically to the drafts and gave final approval for publication.
Competing interests
The authors declare no competing interests.
Funding
We received no funding for this study.
References
- 1.Pocock MJ, Evans DM, Memmott J. 2012. The robustness and restoration of a network of ecological networks. Science 335, 973–977. ( 10.1126/science.1214915) [DOI] [PubMed] [Google Scholar]
- 2.Barnosky AD, et al. 2011. Has the Earth's sixth mass extinction already arrived? Nature 471, 51–57. ( 10.1038/nature09678) [DOI] [PubMed] [Google Scholar]
- 3.Biesmeijer JC, et al. 2006. Parallel declines in pollinators and insect-pollinated plants in Britain and the Netherlands. Science 313, 351–354. ( 10.1126/science.1127863) [DOI] [PubMed] [Google Scholar]
- 4.Peres CA, Emilio T, Schietti J, Desmoulière SJ, Levi T. 2016. Dispersal limitation induces long-term biomass collapse in overhunted Amazonian forests. Proc. Natl Acad. Sci. USA 113, 892–897. ( 10.1073/pnas.1516525113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ripple WJ, Beschta RL. 2012. Trophic cascades in Yellowstone: the first 15 years after wolf reintroduction. Biol. Conserv. 145, 205–213. ( 10.1016/j.biocon.2011.11.005) [DOI] [Google Scholar]
- 6.Toju H. 2015. High-throughput DNA barcoding for ecological network studies. Popul. Ecol. 57, 37–51. ( 10.1007/s10144-014-0472-z) [DOI] [Google Scholar]
- 7.Bascompte J, Jordano P. 2007. Plant-animal mutualistic networks: the architecture of biodiversity. Annu. Rev. Ecol. Evol. Syst. 38, 567–593. ( 10.1146/annurev.ecolsys.38.091206.095818) [DOI] [Google Scholar]
- 8.Ings TC, et al. 2009. Ecological networks: beyond food webs. J. Anim. Ecol. 78, 253–269. ( 10.1111/j.1365-2656.2008.01460.x) [DOI] [PubMed] [Google Scholar]
- 9.Hamilton AJ, et al. 2010. Quantifying uncertainty in estimation of tropical arthropod species richness. Am. Nat. 176, 90–95. ( 10.1086/652998) [DOI] [PubMed] [Google Scholar]
- 10.Hamilton AJ, et al. 2011. Correction. Am. Nat. 177, 544–545. ( 10.1086/659643) [DOI] [Google Scholar]
- 11.Hawksworth D. 2012. Global species numbers of fungi: are tropical studies and molecular approaches contributing to a more robust estimate? Biodivers. Conserv. 21, 2425–2433. ( 10.1007/s10531-012-0335-x) [DOI] [Google Scholar]
- 12.Locey KJ, Lennon JT. 2016. Scaling laws predict global microbial diversity. Proc. Natl Acad. Sci. USA 113, 5970–5975. ( 10.1073/pnas.1521291113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benítez M-S, Hersh MH, Vilgalys R, Clark JS. 2013. Pathogen regulation of plant diversity via effective specialization. Trends Ecol. Evol. 28, 705–711. ( 10.1016/j.tree.2013.09.005) [DOI] [PubMed] [Google Scholar]
- 14.Clemmensen K, et al. 2013. Roots and associated fungi drive long-term carbon sequestration in boreal forest. Science 339, 1615–1618. ( 10.1126/science.1231923) [DOI] [PubMed] [Google Scholar]
- 15.Engel P, Moran NA. 2013. The gut microbiota of insects: diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. ( 10.1111/1574-6976.12025) [DOI] [PubMed] [Google Scholar]
- 16.Gessner MO, Swan CM, Dang CK, McKie BG, Bardgett RD, Wall DH, Hättenschwiler S. 2010. Diversity meets decomposition. Trends Ecol. Evol. 25, 372–380. ( 10.1016/j.tree.2010.01.010) [DOI] [PubMed] [Google Scholar]
- 17.Nielsen UN, Ayres E, Wall DH, Bardgett RD. 2011. Soil biodiversity and carbon cycling: a review and synthesis of studies examining diversity–function relationships. Eur. J. Soil Sci. 62, 105–116. ( 10.1111/j.1365-2389.2010.01314.x) [DOI] [Google Scholar]
- 18.Cebrian J. 1999. Patterns in the fate of production in plant communities. Am. Nat. 154, 449–468. ( 10.1086/303244) [DOI] [PubMed] [Google Scholar]
- 19.A'Bear AD, Jones TH, Boddy L. 2014. Size matters: what have we learnt from microcosm studies of decomposer fungus–invertebrate interactions? Soil Biol. Biochem. 78, 274–283. ( 10.1016/j.soilbio.2014.08.009) [DOI] [Google Scholar]
- 20.De Meester N, Gingold R, Rigaux A, Derycke S, Moens T. 2016. Cryptic diversity and ecosystem functioning: a complex tale of differential effects on decomposition. Oecologia 182, 559–571. ( 10.1007/s00442-016-3677-3) [DOI] [PubMed] [Google Scholar]
- 21.Gange AC, Bower E, Brown VK. 2002. Differential effects of insect herbivory on arbuscular mycorrhizal colonization. Oecologia 131, 103–112. ( 10.1007/s00442-001-0863-7) [DOI] [PubMed] [Google Scholar]
- 22.Blüthgen N, Menzel F, Hovestadt T, Fiala B, Blüthgen N. 2007. Specialization, constraints, and conflicting interests in mutualistic networks. Curr. Biol. 17, 341–346. ( 10.1016/j.cub.2006.12.039) [DOI] [PubMed] [Google Scholar]
- 23.Ivens AB, von Beeren C, Blüthgen N, Kronauer DJ. 2016. Studying the complex communities of ants and their symbionts using ecological network analysis. Annu. Rev. Entomol. 61, 353–371. ( 10.1146/annurev-ento-010715-023719) [DOI] [PubMed] [Google Scholar]
- 24.Thébault E, Fontaine C. 2010. Stability of ecological communities and the architecture of mutualistic and trophic networks. Science 329, 853–856. ( 10.1126/science.1188321) [DOI] [PubMed] [Google Scholar]
- 25.Kaiser-Bunbury CN, Mougal J, Whittington AE, Valentin T, Gabriel R, Olesen JM, Blüthgen N. 2017. Ecosystem restoration strengthens pollination network resilience and function. Nature 542, 223–227. ( 10.1038/nature21071) [DOI] [PubMed] [Google Scholar]
- 26.Dormann CF, Strauss R. 2014. A method for detecting modules in quantitative bipartite networks. Methods Ecol. Evol. 5, 90–98. ( 10.1111/2041-210X.12139) [DOI] [Google Scholar]
- 27.Lewinsohn TM, Inácio Prado P, Jordano P, Bascompte J, Olesen JM. 2006. Structure in plant–animal interaction assemblages. Oikos 113, 174–184. ( 10.1111/j.0030-1299.2006.14583.x) [DOI] [Google Scholar]
- 28.Olesen JM, Bascompte J, Dupont YL, Jordano P. 2007. The modularity of pollination networks. Proc. Natl Acad. Sci. USA 104, 19 891–19 896. ( 10.1073/pnas.0706375104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fekete I, Kotroczó Z, Varga C, Nagy PT, Várbíró G, Bowden RD, Tóth JA, Lajtha K. 2014. Alterations in forest detritus inputs influence soil carbon concentration and soil respiration in a Central-European deciduous forest. Soil Biol. Biochem. 74, 106–114. ( 10.1016/j.soilbio.2014.03.006) [DOI] [Google Scholar]
- 30.Ulyshen MD. 2016. Wood decomposition as influenced by invertebrates. Biol. Rev. 91, 70–85. ( 10.1111/brv.12158) [DOI] [PubMed] [Google Scholar]
- 31.van der Wal A, Geydan TD, Kuyper TW, de Boer W. 2013. A thready affair: linking fungal diversity and community dynamics to terrestrial decomposition processes. FEMS Microbiol. Rev. 37, 477–494. ( 10.1111/1574-6976.12001) [DOI] [PubMed] [Google Scholar]
- 32.Toju H, Guimarães PR, Olesen JM, Thompson JN. 2014. Assembly of complex plant–fungus networks. Nat. Commun. 5, 5273 ( 10.1038/ncomms6273) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toju H, Guimarães PR, Olesen JM, Thompson N. 2015. Below-ground plant–fungus network topology is not congruent with above-ground plant–animal network topology. Sci. Adv. 1, e1500291 ( 10.1126/sciadv.1500291) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobsen RM, Kauserud H, Sverdrup-Thygeson A, Bjorbækmo MM, Birkemoe T. 2017. Wood-inhabiting insects can function as targeted vectors for decomposer fungi. Fungal Ecol. 29, 76–84. ( 10.1016/j.funeco.2017.06.006) [DOI] [Google Scholar]
- 35.Moen A. 1998. Nasjonalatlas for Norge: Vegetasjon (Norwegian national atlas: vegetation). Hønefoss, Norway: Norwegian Mapping Authority. [Google Scholar]
- 36.Dahlberg A, Stokland JN. 2004. Vedlevande arters krav på substrat. Skogsstyrelsen, Rapport 7, 1–74. [Google Scholar]
- 37.Wheeler Q, Blackwell M. 1984. Fungus-insect relationships: perspectives in ecology and evolution. New York, NY: Columbia University Press. [Google Scholar]
- 38.Murray M, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. ( 10.1093/nar/8.19.4321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR protocols: a guide to methods and applications (eds Innis M, Gelfland D, Sninsky J, White T), pp. 315–322. San Diego, CA: Academic Press. [Google Scholar]
- 40.Ihrmark K, et al. 2012. New primers to amplify the fungal ITS2 region—evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 82, 666–677. ( 10.1111/j.1574-6941.2012.01437.x) [DOI] [PubMed] [Google Scholar]
- 41.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215, 403–410. ( 10.1006/jmbi.1990.9999) [DOI] [PubMed] [Google Scholar]
- 42.Abarenkov K, et al. 2010. The UNITE database for molecular identification of fungi: recent updates and future perspectives. New Phytol. 186, 281–285. ( 10.1111/j.1469-8137.2009.03160.x) [DOI] [PubMed] [Google Scholar]
- 43.Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG. 2016. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. ( 10.1016/j.funeco.2015.06.006) [DOI] [Google Scholar]
- 44.Hågvar S. 1999. Saproxylic beetles visiting living sporocarps of Fomitopsis pinicola and Fomes fomentarius. Nor. J. Entomol. 46, 25–32. [Google Scholar]
- 45.Schigel DS. 2011. Polypore-beetle associations in Finland. Ann. Zool. Fenn. 48, 319–348. ( 10.5735/086.048.0601) [DOI] [Google Scholar]
- 46.R Core Team. 2016. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 47.Blüthgen N, Menzel F, Blüthgen N. 2006. Measuring specialization in species interaction networks. BMC Ecol. 6, 9 ( 10.1186/1472-6785-6-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dormann CF, Gruber B, Fründ J. 2008. Introducing the bipartite package: analysing ecological networks. R News 8, 8–11. [Google Scholar]
- 49.Almeida-Neto M, Ulrich W. 2011. A straightforward computational approach for measuring nestedness using quantitative matrices. Environ. Model. Softw. 26, 173–178. ( 10.1016/j.envsoft.2010.08.003) [DOI] [Google Scholar]
- 50.Patefield W. 1981. Algorithm AS 159: an efficient method of generating random R × C tables with given row and column totals. J. R. Stat. Soc. Ser. C Appl. Stat. 30, 91–97. ( 10.2307/2346669) [DOI] [Google Scholar]
- 51.Vázquez DP, Melián CJ, Williams NM, Blüthgen N, Krasnov BR, Poulin R. 2007. Species abundance and asymmetric interaction strength in ecological networks. Oikos 116, 1120–1127. ( 10.1111/j.0030-1299.2007.15828.x) [DOI] [Google Scholar]
- 52.Staniczenko PP, Kopp JC, Allesina S. 2013. The ghost of nestedness in ecological networks. Nat. Commun. 4, 1391 ( 10.1038/ncomms2422) [DOI] [PubMed] [Google Scholar]
- 53.Bahram M, Harend H, Tedersoo L. 2014. Network perspectives of ectomycorrhizal associations. Fungal Ecol. 7, 70–77. ( 10.1016/j.funeco.2013.10.003) [DOI] [Google Scholar]
- 54.Boddy L. 2000. Interspecific combative interactions between wood-decaying basidiomycetes. FEMS Microbiol. Ecol. 31, 185–194. ( 10.1111/j.1574-6941.2000.tb00683.x) [DOI] [PubMed] [Google Scholar]
- 55.Piepenbring M, Hagedorn G, Oberwinkler F. 1998. Spore liberation and dispersal in smut fungi. Botanica Acta 111, 444–460. ( 10.1111/j.1438-8677.1998.tb00732.x) [DOI] [Google Scholar]
- 56.Selosse MA, Schneider-Maunoury L, Martos F. 2018. Time to re-think fungal ecology? Fungal ecological niches are often prejudged. New Phytol. 217, 968–972. ( 10.1111/nph.14983) [DOI] [PubMed] [Google Scholar]
- 57.Shukla SP, Sanders JG, Byrne MJ, Pierce NE. 2016. Gut microbiota of dung beetles correspond to dietary specializations of adults and larvae. Mol. Ecol. 25, 6092–6106. ( 10.1111/mec.13901) [DOI] [PubMed] [Google Scholar]
- 58.Fontaine C, Guimarães PR, Kéfi S, Loeuille N, Memmott J, van Der Putten WH, van Veen FJ, Thébault E. 2011. The ecological and evolutionary implications of merging different types of networks. Ecol. Lett. 14, 1170–1181. ( 10.1111/j.1461-0248.2011.01688.x) [DOI] [PubMed] [Google Scholar]
- 59.Baldrian P, et al. 2012. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 6, 248–258. ( 10.1038/ismej.2011.95) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O'Brien HE, Parrent JL, Jackson JA, Moncalvo J-M, Vilgalys R. 2005. Fungal community analysis by large-scale sequencing of environmental samples. Appl. Environ. Microbiol. 71, 5544–5550. ( 10.1128/AEM.71.9.5544-5550.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ottosson E, Kubartová A, Edman M, Jönsson M, Lindhe A, Stenlid J, Dahlberg A. 2015. Diverse ecological roles within fungal communities in decomposing logs of Picea abies. FEMS Microbiol. Ecol. 91, fiv012 ( 10.1093/femsec/fiv012) [DOI] [PubMed] [Google Scholar]
- 62.Strid Y, Schroeder M, Lindahl B, Ihrmark K, Stenlid J. 2014. Bark beetles have a decisive impact on fungal communities in Norway spruce stem sections. Fungal Ecol. 7, 47–58. ( 10.1016/j.funeco.2013.09.003) [DOI] [Google Scholar]
- 63.Suh S-O, Blackwell M. 2005. Four new yeasts in the Candida mesenterica clade associated with basidiocarp-feeding beetles. Mycologia 97, 167–177. ( 10.1080/15572536.2006.11832850) [DOI] [PubMed] [Google Scholar]
- 64.Barriga PA, Dormann CF, Gbur EE, Sagers CL. 2015. Community structure and ecological specialization in plant–ant interactions. J. Trop. Ecol. 31, 325–334. ( 10.1017/S0266467415000139) [DOI] [Google Scholar]
- 65.Le Roux JJ, Mavengere NR, Ellis AG. 2016. The structure of legume–rhizobium interaction networks and their response to tree invasions. AoB Plants 8, plw038 ( 10.1093/aobpla/plw038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Correa SB, Arujo JK, Penha J, da Cunha CN, Bobier KE, Anderson JT. 2016. Stability and generalization in seed dispersal networks: a case study of frugivorous fish in Neotropical wetlands. Proc. R. Soc. B 283, 20161267 ( 10.1098/rspb.2016.1267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schleuning M, Blüthgen N, Flörchinger M, Braun J, Schaefer HM, Böhning-Gaese K. 2011. Specialization and interaction strength in a tropical plant–frugivore network differ among forest strata. Ecology 92, 26–36. ( 10.1890/09-1842.1) [DOI] [PubMed] [Google Scholar]
- 68.Wheelwright NT, Orians GH. 1982. Seed dispersal by animals: contrasts with pollen dispersal, problems of terminology, and constraints on coevolution. Am. Nat. 119, 402–413. ( 10.1086/283918) [DOI] [Google Scholar]
- 69.Johansson T, Olsson J, Hjältén J, Jonsson BG, Ericson L. 2006. Beetle attraction to sporocarps and wood infected with mycelia of decay fungi in old-growth spruce forests of northern Sweden. For. Ecol. Manag. 237, 335–341. ( 10.1016/j.foreco.2006.09.056) [DOI] [Google Scholar]
- 70.Jonsell M, Nordlander G. 1995. Field attraction of Coleoptera to odours of the wood-decaying polypores Fomitopsis pinicola and Fomes fomentarius. Ann. Zool. Fenn. 32, 391–402. [Google Scholar]
- 71.Leather SR, Baumgart EA, Evans HF, Quicke DL. 2013. Seeing the trees for the wood—beech (Fagus sylvatica) decay fungal volatiles influence the structure of saproxylic beetle communities. Insect Conserv. Diver. 7, 314–326. ( 10.1111/icad.12055) [DOI] [Google Scholar]
- 72.Fäldt J, Jonsell M, Nordlander G, Borg-Karlson A-K. 1999. Volatiles of bracket fungi Fomitopsis pinicola and Fomes fomentarius and their functions as insect attractants. J. Chem. Ecol. 25, 567–590. ( 10.1023/A:1020958005023) [DOI] [Google Scholar]
- 73.Carlo TA, Morales JM. 2016. Generalist birds promote tropical forest regeneration and increase plant diversity via rare-biased seed dispersal. Ecology 97, 1819–1831. ( 10.1890/15-2147.1) [DOI] [PubMed] [Google Scholar]
- 74.Wehncke E, Hubbell S, Foster R, Dalling J. 2003. Seed dispersal patterns produced by white-faced monkeys: implications for the dispersal limitation of neotropical tree species. J. Ecol. 91, 677–685. ( 10.1046/j.1365-2745.2003.00798.x) [DOI] [Google Scholar]
- 75.Escribano-Avila G, Calviño-Cancela M, Pías B, Virgos E, Valladares F, Escudero A. 2014. Diverse guilds provide complementary dispersal services in a woodland expansion process after land abandonment. J. Appl. Ecol. 51, 1701–1711. ( 10.1111/1365-2664.12340) [DOI] [Google Scholar]
- 76.McConkey KR, Brockelman WY. 2011. Nonredundancy in the dispersal network of a generalist tropical forest tree. Ecology 92, 1492–1502. ( 10.1890/10-1255.1) [DOI] [PubMed] [Google Scholar]
- 77.Schupp EW, Jordano P, Gómez JM. 2010. Seed dispersal effectiveness revisited: a conceptual review. New Phytol. 188, 333–353. ( 10.1111/j.1469-8137.2010.03402.x) [DOI] [PubMed] [Google Scholar]
- 78.Morris RJ, Sinclair FH, Burwell CJ. 2015. Food web structure changes with elevation but not rainforest stratum. Ecography 38, 792–802. ( 10.1111/ecog.01078) [DOI] [Google Scholar]
- 79.Trøjelsgaard K, Olesen JM. 2016. Ecological networks in motion: micro-and macroscopic variability across scales. Funct. Ecol. 30, 1926–1935. ( 10.1111/1365-2435.12710) [DOI] [Google Scholar]
- 80.Jacobsen RM, Birkemoe T, Sverdrup-Thygeson A. 2015. Priority effects of early successional insects influence late successional fungi in dead wood. Ecol. Evol. 5, 4896–4905. ( 10.1002/ece3.1751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jacobsen RM, Sverdrup-Thygeson A, Kauserud H, Birkemoe T. 2017. Data from: Wood-inhabiting insects can function as targeted vectors for decomposer fungi Dryad Digital Repository. ( 10.5061/dryad.3t2d4) [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Jacobsen RM, Sverdrup-Thygeson A, Kauserud H, Birkemoe T. 2017. Data from: Wood-inhabiting insects can function as targeted vectors for decomposer fungi Dryad Digital Repository. ( 10.5061/dryad.3t2d4) [DOI]
Supplementary Materials
Data Availability Statement
Raw data (fastq-files), barcode and primer mapping file, OTU table and representative sequence files have been accessioned in Dryad with http://dx.doi.org/10.5061/dryad.3t2d4 [81].