Abstract
In mammals, two integral membrane proteins, sperm IZUMO1 and egg CD9, regulate sperm–egg fusion, and their roles are critical, but yet unclear. Recent studies, however, indicate interesting connections between the sperm–egg fusion and virus‐induced cell–cell fusion. First, CD9‐containing exosome‐like vesicles, which are released from wild‐type eggs, can induce the fusion between sperm and CD9‐deficient egg, even though CD9‐deficient eggs are highly refractory to the fusion with sperm. This finding provides strong evidence for the involvement of CD9‐containing, fusion‐facilitating vesicles in the sperm–egg fusion. Secondly, there are similarities between the generation of retroviruses in the host cells and the formation of small cellular vesicles, termed exosomes, in mammalian cells. The exosomes are involved in intercellular communication through transfer of proteins and ribonucleic acids (RNAs) including mRNAs and microRNAs. These collective studies provide an insight into the molecular mechanism of membrane fusion events.
Keywords: CD9, Exosome, Fertilization, Membrane fusion, Tetraspanin
Introduction
Fertilization is a sequential event that includes cell–cell adhesion, cell–cell fusion, and activation of cellular signaling, which allows the resumption of the egg cell cycle arrested at the stage of meiotic metaphase II (Fig. 1). In mammals, two kinds of membrane protein families, the cell adhesion molecule “integrin” [1, 2] and the membrane‐anchored protease “a disintegrin and metalloprotease (ADAM)” [3, 4, 5, 6, 7, 8, 9], were biochemically identified in mammalian eggs and sperm, respectively, and immunocytochemically confirmed to localize on their outer cell membranes. The integrin family, which is expressed in many types of cells in animals, mediates cell–cell and cell–matrix interaction and intercellular communication, including cell adhesion and cell–cell fusion [10, 11, 12, 13, 14]. On the other hand, the ADAM family has a characteristic domain that is homologous to an extracellular region of the integrin family [4, 15]. The presence of the domain conserved between the integrin and ADAM families indicated that these two protein families play a role in sperm–egg adhesion and/or fusion [2, 15, 16, 17, 18]. As expected, antibodies against the extracellular regions of these protein families were shown to significantly reduce the rate of sperm–egg binding and fusion in mice [1]. However, when genetically manipulated mice were produced, both male and female mice displayed no overt anomalies in both sperm–egg membrane adhesion and fusion [18, 19, 20, 21, 22] (Table 1), suggesting that the integrin and ADAM families are not essential for these events.
Figure 1.
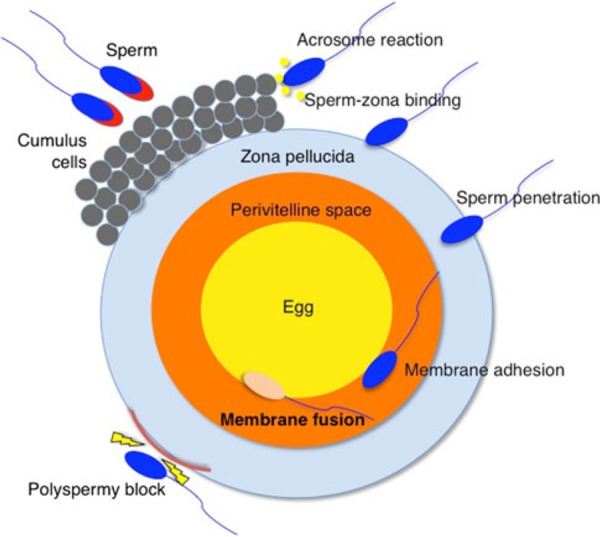
Series of steps from sperm–egg interaction to fusion during mammalian fertilization. This is an overview of mammalian fertilization. Fertilization is divided into multiple steps: interaction of sperm–somatic cells (termed cumulus cells), binding of sperm to the extracellular matrix (termed zona pellucida), and penetration of the egg. After the sperm penetrates the zona pellucida, it can bind and fuse to the egg cell membrane. Successful fertilization requires not only that a sperm and egg fuse, but also that polyspermy block occurs
Table 1.
Predicted players in sperm–egg fusion in mammals
| Gene name | Category of coding protein | Expression | Phenotypes of gametes in KO mice | References |
|---|---|---|---|---|
| ACE, testis‐specific | Secreted protein, angiotensin‐converting enzyme | Sperm | Failure of sperm transport into the oviduct and zona‐binding | [30] |
| ACE3, testis‐specific | Secreted protein, angiotensin‐converting enzyme | Sperm | No fertilizing defect | [34] |
| Acrin1, MN7 | Intra‐acrosomal protein | Sperm | Unknown | [35] |
| Acrin2, MC41 | Intra‐acrosomal protein | Sperm | Unknown | [35] |
| ADAM1 | Membrane protein | Sperm | Failure of sperm transport into the oviduct and zona‐binding | [31] |
| ADAM2 | Membrane protein | Sperm | Impaired binding of oviduct and zona with sperm | [32] |
| ADAM3 | Membrane protein | Sperm | Impaired binding of oviduct and zona with sperm | [33] |
| Basigin, MC31, CE9, CD147 | Membrane protein | Sperm | Unknown | [37] |
| E‐cadherin | Membrane protein | Egg and sperm | Impaired membrane adhesion, but normal fusion | [42] |
| N‐cadherin | Membrane protein | Egg and sperm | Unknown | [44] |
| Calmegin | ER chaperone | Sperm | Failure of sperm transport into the oviduct and zona‐binding | [25, 26] |
| Calreticulin3 | ER chaperone | Sperm | Failure of sperm transport into the oviduct and zona‐binding | [27] |
| β‐catenin | Cytoplasmic and nuclear protein | Egg and sperm | Impaired membrane adhesion, but normal fusion | [42] |
| CD81 | Membrane protein, tetraspanin | Egg | Impaired fusion with sperm | [75, 76, 77] |
| CD9 | Membrane protein, tetraspanin | Egg and sperm | Defective fusion with sperm | [57, 58, 59] |
| CD98 | Membrane protein | Egg | Unknown | [46] |
| CrispI | Secreted protein | Sperm | Normal fertility, but impaired fertilization in vitro | [47] |
| Crisp2 | Secreted protein | Sperm | Unknown | [48] |
| Equatorin, MN9 | Membrane‐anchored protein | Sperm | Unknown | [49, 50] |
| IGSF8 | Membrane protein, IgSF | Egg | No fertilizing defect | [51] |
| Integrin α3 | Membrane protein | Egg | Normal membrane adhesion and fusion | [21] |
| Integrin α6 | Membrane protein | Egg | Normal membrane adhesion and fusion | [20] |
| Integrin α9 | Membrane protein | Egg | Reduced fertilizing ability | [22] |
| Integrin αV | Membrane protein | Egg | Unknown | [21] |
| Integrin β1 | Membrane protein | Egg | Normal membrane adhesion and fusion | [20] |
| Integrin β3 | Membrane protein | Egg | Unknown | [21] |
| Izumo1 | Membrane protein, IgSF | Sperm | Defective fusion with egg | [60] |
| Izumo2 | Membrane protein, IgSF | Sperm | Unknown | [52] |
| Izumo3 | Membrane protein, IgSF | Sperm | Unknown | [52] |
| Izumo4 | Membrane protein, IgSF | Sperm | Unknown | [52] |
| PDILT | Protein disulfide isomerase | Sperm | Failure of sperm transport into the oviduct and zona‐binding | [28] |
| PMIS2 | Unidentified | Sperm | Failure of sperm transport into the oviduct and zona‐binding | [29] |
| SPESP1 | Equatorial segment protein | Sperm | Morphological defects of sperm | [53] |
| TMEM190 | Membrane protein | Sperm | No fertilizing defect | [54] |
| Tpst2 | Tyrosylprotein sulfotransferase | Sperm | Failure of sperm transport into the oviduct and zona‐binding | [29] |
| Tssk6 | Serine kinase | Sperm | Morphological defects of sperm | [55, 56] |
In the past, many genes predicted to participate in the sperm–egg fusion have emerged in mammals, but contrary to the expectations, most were found to be dispensable [23, 24] (Table 1). The sperm are required to migrate to the oviduct, where the ovulated eggs and sperm meet. Gene disruption experiments have been used to produce at least eight mouse lines with gene disruption for two testis‐specific chaperones expressed in the endoplasmic reticulum (ER) (Calmegin and Calreticulin 3) [25, 26, 27], a protein disulfide isomerase (PDILT) [28], a tyrosylprotein sulfotransferase (TPST2) [29], a testis‐specific angiotensin‐converting enzyme (ACE) [30], and three members of the ADAM family (ADAM1A, ADAM2, and ADAM3) [31, 32, 33]. In these mice, the sperm showed impaired migration into the oviduct and lost their zona pellucida‐binding (zona‐binding) ability when mixed with cumulus‐free eggs in vitro. As reported for each of the ADAM1a‐deficient, ADAM3‐deficient, and PDILT‐deficient lines, these mutant sperm could fertilize cumulus‐surrounded eggs. These results indicate that the primary cause for infertility in these mouse strains is not a defect in sperm–zona binding, but the inability of sperm to migrate from the uterus into the oviduct.
ACE3, a testis‐specific ACE homologue, is identified as an IZUMO1‐interacting protein in sperm [34]. Ace3‐deficient mice showed that the localization of IZUMO1 spread in a little wider area on sperm, but the elimination of ACE3 did not result in a loss of sperm fertilizing ability [34].
Two monoclonal antibodies (mAb), mMN7 and mMC41, against intra‐acrosomal proteins, ACRIN1/MN7 and ACRIN2/MC41, respectively, significantly inhibited fertilization of zona‐intact eggs in a dose‐dependent manner, but did not influence the fertilization of zona‐free eggs [35]. This result suggests that ACRIN1 and ACRIN2 are involved in the sperm–zona pellucida interaction before or during penetration of the zona pellucida.
BASIGIN/MC31/CE9/CD147 is a transmembrane glycoprotein and also acts as a receptor essential for erythrocyte invasion by Plasmodium falciparum [36]. A mAb against this protein significantly inhibited fertilization of cumulus‐intact, zona‐intact, and zona‐free rat eggs in a dose‐dependent manner. By contrast, sperm–egg membrane binding was not affected [37]. These findings suggest that this protein facilitates sperm–egg fusion, but the gene‐disrupted sperm have been not analyzed.
Before membrane adhesion, both sperm and egg retain the cell–cell adhesion complex comprising β‐catenin and E‐cadherin [38, 39, 40, 41, 42, 43]. Once membrane adhesion occurs, β‐catenin is immediately ubiquitinated and probably degraded in both the sperm and egg, thereby initiating membrane fusion between these two cells. The absence of β‐catenin results in a reduction in the ability of sperm to adhere to an egg, but sperm–egg fusion occurs normally [42]. This result indicates that β‐catenin contributes partly to sperm–egg membrane adhesion, but does not play an essential role in this event. N‐cadherin is also expressed in the human gonads and gametes [44], but the gene‐disrupted sperm have not been investigated.
CD98 is a glycoprotein composed of SLC3A2 and SLC7A5 that forms the large neutral amino acid transporter [45], and is expressed on mouse eggs [46]. MAbs against CD9 and CD98 cooperatively inhibit in vitro fertilization [46], but the gene‐disrupted eggs have not been analyzed.
Epididymal protein CRISPI is a member of the cysteine‐rich secretory proteins family. The CRISP1‐deficient sperm presented a decreased level of protein tyrosine phosphorylation during capacitation, and an impaired ability to fertilize both zona‐intact and zona‐free eggs in vitro, but they exhibited normal fertility [47]. Testicular CRISP2 is also expected to be involved in sperm–egg fusion [48], but the gene‐disrupted mice have not been investigated.
EQUATORIN/MN9 is a sperm‐specific type I transmembrane protein and a widely distributed acrosomal protein in mammalian sperm [49, 50]. During the acrosome reaction, some amount of this protein translocates to the plasma membrane, covering the equatorial region. The mAb against this protein inhibited both in vitro and in vivo fertilization. In addition, the gamete interaction‐related domain recognized by the MN9 antibody is post‐translationally modified. The modified domain was identified near threonine 138, which was most likely to be O‐glycosylated when analyzed by amino acid substitution, dephosphorylation, and O‐glycosylation inhibitor assays. Immunoelectron‐microscopic analysis showed EQUATORIN on the hybrid vesicles surrounded by amorphous substances at advanced stage of acrosome reaction. Thus, the established EQUATORIN‐based progression model will be useful for analyzing not only the behavior of EQUATORIN, but also of other molecules of interest involved in the acrosome reaction.
An immunoglobulin superfamily member, IGSF8, tightly associates with CD9 on the egg surface and is undetectable on the surface of CD9‐deficient eggs [51]. However, the IGSF8‐deficient female mice showed no fertilization defect in vitro or in vivo [51], indicating that IGSF8 is dispensable in fertility.
A family of four genes (Izumo1, 2, 3, and 4) is mostly expressed in the sperm with known and potential roles in sperm–egg fusion [52], but the gene‐disrupted mice have not been investigated.
It is widely accepted that the equatorial segment of the acrosome‐reacted sperm is important in initiating fusion with the egg plasma membrane during fertilization. A mouse line lacking sperm equatorial segment protein 1 (SPESP1) was generated [53]. The average number of pups that were fathered by Spesp1 +/− and Spesp1 −/− male mice was significantly lower than that of wild‐type fathers. Fewer sperm were found to migrate into oviducts and fewer eggs were fertilized. The sperm produced in Spesp1 +/− and Spesp1 −/− male mice showed a lower fusing ability compared with the wild‐type sperm. Moreover, scanning electron microscopy revealed that the membrane in the equatorial segment area, which usually forms an acrosomal sheath, disappears after acrosome reaction in Spesp1‐deficient mice, suggesting that SPESP1 is necessary to produce the fully fusioncompetent sperm.
TMEM190, a small transmembrane protein containing the trefoil domain, was identified by proteomic analysis of mouse sperm [54]. Two structural features of TMEM190, trefoil domain and small transmembrane protein, are predicted to form a protein–protein complex required during fertilization. TMEM190 is an inner‐acrosomal membrane protein of cauda epididymal sperm. During the acrosome reaction, TMEM190 partly relocated onto the surface of the equatorial segment, on which sperm–egg fusion occurs. Moreover, TMEM190 and IZUMO1 were co‐localized in mouse sperm both before and after the acrosome reaction. Its role in fertilization is probably dispensable since TMEM190‐deficient male mice were normally fertile.
TSSK6 is a member of the testis‐specific serine kinase family of proteins and is expressed post‐meiotically in male germ cells [55, 56]. The sperm produced by Tssk6‐deficient mice present defects that prevent the successful fertilization of eggs in vitro and the fusion to zona‐free eggs. Tssk6‐deficient sperm fails to relocate IZUMO1 during the acrosome reaction. TSSK6 is involved in sperm–egg fusion through the regulation of actin polymerization and changes in IZUMO1 localization.
From these studies, in order to ensure the success of fertilization in mammals, overlapping functions of multiple proteins seem to be needed. In other words, there will be more than one way for a sperm and an egg to fuse, which may reduce the malfunction that occurs in sperm or eggs lacking a single gene. Exceptionally, CD9 on the egg membrane [57, 58, 59] and IZUMO1 on the sperm membrane [60] are factors proved to be essential for the sperm–egg fusion in gene disruption experiments (Fig. 2).
Figure 2.
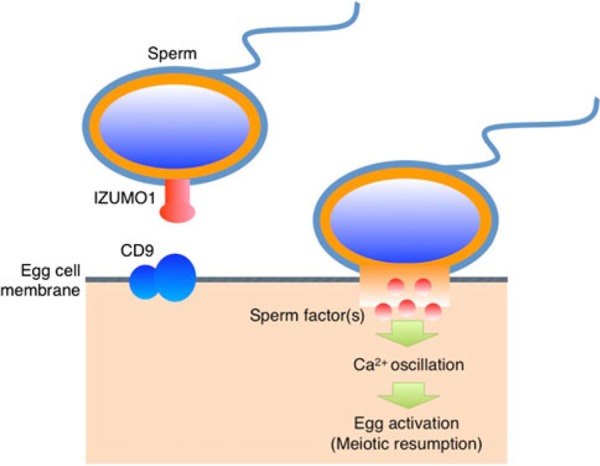
Players identified in sperm–egg fusion. IZUMO1 is expressed on the sperm membrane, and Izumo1‐deficient sperm show a defect in fusion with the egg cell membrane. CD9 is expressed on the egg cell membrane and functions in fusion with the sperm. Two membrane proteins, IZUMO1 and CD9, are essential for sperm–egg fusion in mice. Direct interaction between CD9 and IZUMO1 has not been identified, andunidentified sperm and egg factors may be involved in sperm–egg fusion. After sperm–egg membrane fusion occurs, a sperm factor triggers Ca2+ oscillations [95] and initiates egg activation in mammals
CD9 and its role in cellular function
CD9 gene encoding a 24‐kDa protein is transcribed in all types of mammalian cells [61]. This protein is localized on the cell membranes and partly on endosomes, and it is expected to be involved in cell–cell adhesion, because CD9 associates with the integrin family [61]. CD9 is also known as a motility‐related protein 1 (MRP‐1), which plays a role in suppressing tumor metastasis [62]. As depicted in Fig. 3, CD9 has two extracellular loops, four transmembrane domains, and two short cytoplasmic domains. Its functional domain is expected to be positioned in a large extracellular loop (LEL), because CD9 associates with other membrane proteins via LEL in vitro [61]. In addition, due to its significantly higher levels in mesenchymal and embryonic stem cells versus fibroblastic cells, CD9 is useful as one of the cell surface markers for isolating undifferentiated cells from mixed cell populations in mice and humans [63].
Figure 3.
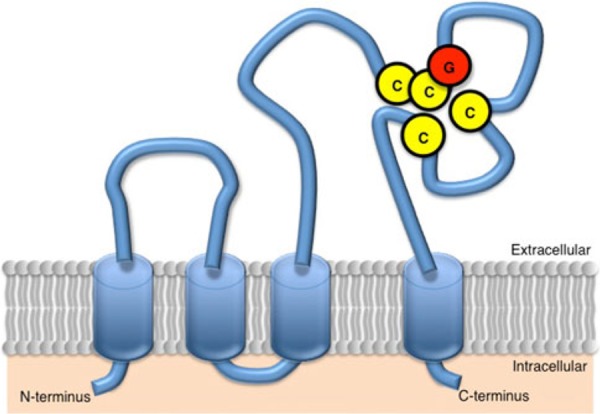
Structural features of tetraspanin CD9. CD9 is a member of the tetraspan‐membrane protein family, termed tetraspanin, and its molecular mass is 24 kDa. The structural features of CD9 include four transmembrane domains, two extracellular loops, short and large extracellular loops (SEL and LEL), and two short cytoplasmic tails. CD9 has cysteine–cysteine–glycine (CCG) residues (amino acids 152–154) as a tetraspanin‐specific motif and two other cysteines within LEL
In order to clarify in vivo roles of CD9, three laboratories independently generated CD9 −/− mice [57, 58, 59]. All strains of the CD9 −/− mice consistently showed severe female subfertility, whereas the CD9 −/− male mice were fertile. Moreover, the CD9‐deficient eggs exhibited severely reduced fusion ability with sperm. Since these findings, CD9 has been studied as one of the crucial factors in sperm–egg fusion in mammals. A functionally essential domain of CD9 was predicted to be located within the LEL in sperm–egg fusion [64, 65]; however, even though CD9‐binding proteins have been identified in non‐gamete cells, LEL‐binding, potentially fusion‐related proteins have not been found yet.
Tetraspanin family
CD9 belongs to a membrane protein family, collectively termed “tetraspanin”, which encompasses 35 members in mammals, such as CD9, CD37, CD53, CD63, CD81, CD82, and CD151 [61], 30 in nematodes [66, 67], and 30 in flies [68, 69]. The tetraspanin family is often thought to act as scaffolding proteins, anchoring multiple proteins to one area of the cell membrane in tissue formation [61] and even in infectious diseases [70]. Generally, pathogenic microorganisms, such as bacteria, viruses, parasites, or fungi, cause infectious diseases; the diseases can be spread, directly or indirectly, from one organism to another [71]. Members of this family are related to the onset of infectious and parasitic diseases; for instance, they are involved in cell–cell transmission of HIV‐1 virus [72, 73]. After its primary infection into host cells, CD9, CD63, CD81, and CD82 are enriched at HIV‐1 budding sites of HIV‐1 virions. When tetraspanin‐containing HIV‐1 particles are next formed and released from host cells, they become 10‐fold more infectious than the cell‐free virus particles [73]. In mice, CD81 is also required for malarial parasites to commit to infection of the hepatocyte [74]. Malarial sporozoites, the cell form that infects new hosts, are transmitted into livers of the mammalian hosts through bites from infected mosquitoes, but the sporozoites failed to infect hepatocytes of CD81 −/− mice, suggesting that CD81 is involved in the sporozoite entry into hepatocytes as a host factor. Furthermore, CD81 is involved in mammalian reproductive capacity. In other words, CD81 −/− female mice were subfertile, because CD81‐deficient eggs exhibited the impaired sperm fusion ability [75, 76]. In addition, CD81 is expressed on CD9‐deficient eggs, and CD9 is also expressed on CD81‐deficient eggs at the expression rate comparable with that of wild‐type eggs, indicating that CD9 and CD81 independently work in sperm–egg fusion [77]. On the other hand, plants have more than 60 members of tetraspanin [78, 79], but their roles in membrane fusion‐related events are unclear.
Tetraspanin‐like proteins have also been identified in fungi, and their molecular masses (more than 200 kDa) are greater than those of tetraspanin identified in animals and plants (20–30 kDa) [80]. An appressorium is a specialized cell typical of fungal plant pathogens that is used to infect host plants. By analyzing a non‐pathogenic mutant, punchless, isolated from the rice blast fungus Magnaporthe grisea, tetraspanin‐like PLS1 (MgPLS1) has been shown to control the appressorrial function, which is essential for the fungal penetration into host leaves [81]. Similarly, Colletotrichum lindemuthianum PLS1 (ClPLS1) is a functional homologue of MgPLS1, and the non‐pathogenic ClPLS1‐deficient mutant to bean leaves exhibits a defect in the formation and positioning of the penetration pore [82]. On the other hand, MgPLS1 and PaPls1 genes are functional orthologues, since MgPLS1 fully complements the germination defect of the PaPls1‐null mutant in Podospora anserine [83]. Yet, MgPLS1 is required for the formation of the penetration peg originating at the pore of M. grisea melanized appressorium, while it is required for ascospore germination in P. anserine [83]. The phenotypes in both fungal species strongly differ. However, the appressorium and the ascospore share similar physiological features [83]. First, P. anserina ascospores and M. grisea appressoria both germinate through the differentiation of, respectively, a germination peg and a penetration peg after an induction. Second, they are heavily melanized cells. In both species, the melanin is deposited between the membrane and the cell wall as a dense continuous layer, and acts as a semi‐permeable membrane‐retaining substance affecting osmosis. Besides these shared characteristics, P. anserina ascospore germination and M. grisea appressorium peg formation rely on similar physiological processes such as the catabolism of lipids. This evidence suggests that the appressorium and the ascospore are physiologically comparable organs. Consequently, since the invasion of pathogenic fungi into leaves is an event closely related to membrane fusion events, these studies indicate that tetraspanin‐like PLS1s are involved in the membrane fusion‐related event between fungus and plant.
Taken together, these results suggest that members of the tetraspanin family are closely related to membrane fusion‐related events in multicellular organisms. Nonetheless, their fusogenic activity corresponding to fusogenic transmembrane proteins, such as syncytin, identified in human placenta [84], and virus envelope proteins [85], has not been identified, and their physiological activities are still unclear.
Tetraspanin as a major component of exosomes
In mammals, cell‐cultured media contain nano‐sized membrane vesicles, but these are not attractive to researchers, because they cannot be structurally distinguished from the debris of dead cells [86]. Recent studies have shown that the vesicles, termed exosomes, are derived from living cells, but not dead cells [87]. Furthermore, they have been proven to play a significant role in the mediation of adaptive immune reactions to pathogens and tumors through the enhancement of antigen‐specific T cell responses [86, 88]. Besides immune cells, the exosomes are released from a wide range of normal and malignant mammalian cell types, and their diameter is estimated to range from 50 to 90 nm [88]. The protein composition of exosomes varies with the origin of cells, yet the exosomes commonly contain a ganglioside GM3, two kinds of heat shock proteins (HSP70 and HSP90), and tetraspanin [88]. The exosomes also contain transcripts, mRNA, and microRNA, which are thought to be shuttled from one cell to another, thereby influencing protein synthesis in recipient cells [89].
Exosome‐like vesicles are released from mouse eggs
At least two reports suggest that CD9 contributes to the organization of the cell membrane in eggs. First, CD9 is transferred from the egg to the fertilizing sperm present in the perivitelline space, implying the involvement of a process similar to trogocytosis, which is a mechanism for the cell‐to‐cell contact‐dependent transfer of membrane fragments from antigen‐presenting cells to lymphocytes in immune responses to pathogens [90]. Secondly, CD9 deficiency alters the length and density of microvilli on the egg cell membrane [91]. However, treatment with fixatives often disturbs membrane organization and modifies the localization of membrane proteins. The above two studies exhibited the membrane localization of CD9 in eggs treated with paraformaldehyde.
On the other hand, the potential of enhanced green fluorescent protein‐tagged CD9 (CD9‐EGFP) was used as a reporter protein for studying sperm–egg fusion in living mouse eggs [92]. Interestingly, in eggs just before fertilization, CD9‐EGFP was significantly accumulated within the perivitelline space that completely surrounded the eggs and lay between the egg cell membrane and the zona pellucida. In addition, when the eggs were carefully treated with fixatives, immunoelectron‐microscopic analysis of wild‐type eggs also revealed that CD9 was not only present in the perivitelline space, but also incorporated into vesicles of varying size (50–200 nm in diameter) without a sectional profile of a typical lipid bilayer [92]. Membrane vesicles were also previously detected via electron microscope within the perivitelline space of their eggs in opossums [93] and humans [94]. Moreover, the above study demonstrates that the vesicles identified in mouse eggs share CD9, GM3, and HSP90 with exosomes, and these components are absent in eggs lacking CD9 and are reproduced by CD9‐EGFP, expression restricted to the eggs [92]. These results provide evidence as to the nature of CD9 in mouse eggs. First, CD9‐incorporated exosome‐like vesicles are produced in mouse eggs and are released outside the egg cell membrane just before fertilization. Secondly, CD9 is essential for the formation of the exosome‐like vesicles (hereafter referred to as egg exosomes) in mouse eggs.
The finding of egg exosomes gives us the idea that they facilitate the sperm–egg fusion. As expected, CD9‐containing egg exosomes rendered sperm capable of fusing with CD9‐deficient eggs [92] (Fig. 4). Concretely, CD9‐deficient eggs could not fuse with sperm, but the co‐existence of wild‐type eggs resulted in 60–70% of the CD9‐deficient eggs fusing with at least one sperm [92]. Thus, sperm can fuse with CD9‐deficient eggs with impaired microvilli via the egg exosomes released from wild‐type eggs, which means that the egg exosomes, but not the egg microvilli, are essential for sperm–egg fusion.
Figure 4.
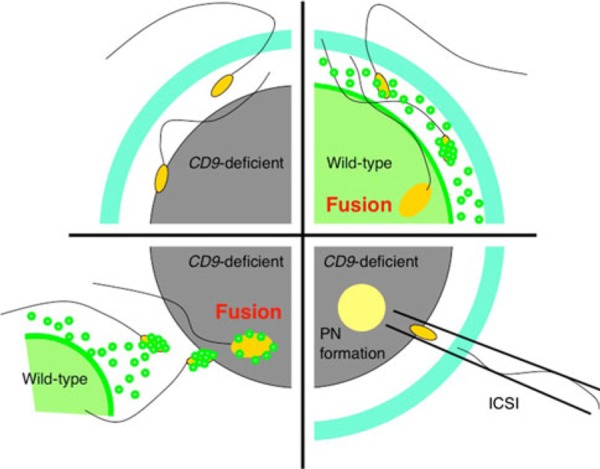
Overview of the studies of CD9‐deficient eggs. In wild‐type eggs, CD9‐containing egg exosomes are released from wild‐type eggs before any interaction with the sperm (upper right diagram). Shortly after the sperm penetrates the perivitelline space, the egg exosomes are transferred on the acrosome‐reacted sperm head. Then, a sperm fuses with the egg cell membrane. Interaction between the sperm and the exosomes is an essential step for sperm‐fusing ability. In contrast, CD9‐deficient eggs cannot release the egg exosomes, which are correlated with the formation of microvilli on the egg cell membrane (upper left diagram). The sperm cannot fuse to the cell membrane of the CD9‐deficient egg. On the other hand, when the zona pellucida is removed from the eggs, the sperm is able to interact with the egg exosomes released from wild‐type eggs and can fuse with the CD9‐deficient egg (lower left diagram). By co‐incubation with wild‐type eggs, the sperm can fuse with a similar number of CD9‐deficient and wild‐type eggs. Intracytoplasmic sperm injection (ICSI) is an in vitro fertilization procedure in which a single sperm head is injected directly into an egg (lower right diagram). This procedure is most commonly used to overcome male infertility and fusion defects in CD9‐deficient eggs
Conclusions
The close relation between egg exosomes and sperm–egg fusion raises the question of how egg exosomes facilitate such fusion. According to a previous report, exosomes contain both functional mRNA and microRNA, which are shuttled from one cell to another, affecting the recipient cell's ability to produce protein [89]. Moreover, HIV‐1 utilizes the exosome biogenesis pathway for the formation of infectious particles, and in macrophages, HIV‐1 assembles into an intracellular plasma membrane domain‐containing tetraspanin (i.e., CD9, CD81, CD53, or CD63) [73]. Thus, the exosomes play at least two roles in regulating cell function: first, shuttling proteins and RNAs (mRNAs and microRNAs) from one cell to another, and secondly, forming infectious particles. These two roles may be required for sperm–egg fusion in mammals. In conclusion, the studies of exosomes will present a useful strategy for regulating the cell‐to‐cell spread of specific viruses and fertilization ability.
Acknowledgments
This review is based on our previous studies, which were supported by a grant from The Ministry of Health, Labor and Welfare, and a grant‐in‐aid for Scientific Research, The Ministry of Education, Culture, Sports, and Technology of Japan.
Conflict of interest
The author declares that they have no conflict of interest.
References
- 1. Almeida EA, Huovila AP, Sutherland AE, Stephens LE, Calarco PG, Shaw LM, Mercurio AM, Sonnenberg A, Primakoff P, Myles DG, White JM. Mouse egg integrin alpha 6 beta 1 functions as a sperm receptor. Cell, 1995, 81, 1095–1104 10.1016/S0092-8674(05)80014-5 [DOI] [PubMed] [Google Scholar]
- 2. Chen H, Sampson NS. Mediation of sperm–egg fusion: evidence that mouse egg alpha6beta1 integrin is the receptor for sperm fertilinbeta. Chem Biol, 1999, 6, 1–10 10.1016/S1074-5521(99)80015-5 [DOI] [PubMed] [Google Scholar]
- 3. Blobel CP, Wolfsberg TG, Turck CW, Myles DG, Primakoff P, White JM. A potential fusion peptide and an integrin ligand domain in a protein active in sperm–egg fusion. Nature, 1992, 356, 248–252 10.1038/356248a0 [DOI] [PubMed] [Google Scholar]
- 4. Wolfsberg TG, Bazan JF, Blobel CP, Myles DG, Primakoff P, White JM. The precursor region of a protein active in sperm–egg fusion contains a metalloprotease and a disintegrin domain: structural, functional, and evolutionary implications. Proc Natl Acad Sci USA, 1993, 90, 10783–10787 10.1073/pnas.90.22.10783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weskamp G, Blobel CP. A family of cellular proteins related to snake venom disintegrins. Proc Natl Acad Sci USA, 1994, 91, 2748–2751 10.1073/pnas.91.7.2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Myles DG, Kimmel LH, Blobel CP, White JM, Primakoff P. Identification of a binding site in the disintegrin domain of fertilin required for sperm–egg fusion. Proc Natl Acad Sci USA, 1994, 91, 4195–4198 10.1073/pnas.91.10.4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Evans JP, Schultz RM, Kopf GS. Characterization of the binding of recombinant mouse sperm fertilin alpha subunit to mouse eggs: evidence for function as a cell adhesion molecule in sperm–egg binding. Dev Biol, 1997, 187, 94–106 10.1006/dbio.1997.8612 [DOI] [PubMed] [Google Scholar]
- 8. Yuan R, Primakoff P, Myles DG. A role for the disintegrin domain of cyritestin, a sperm surface protein belonging to the ADAM family, in mouse sperm–egg plasma membrane adhesion and fusion. J Cell Biol, 1997, 137, 105–112 10.1083/jcb.137.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Waters SI, White JM. Biochemical and molecular characterization of bovine fertilin alpha and beta (ADAM 1 and ADAM 2): a candidate sperm–egg binding/fusion complex. Biol Reprod, 1997, 56, 1245–1254 10.1095/biolreprod56.5.1245 [DOI] [PubMed] [Google Scholar]
- 10. Elices MJ, Osborn L, Takada Y, Crouse C, Luhowskyj S, Hemler ME, Lobb RR. VCAM‐1 on activated endothelium interacts with the leukocyte integrin VLA‐4 at a site distinct from the VLA‐4/fibronectin binding site. Cell, 1990, 60, 577–584 10.1016/0092-8674(90)90661-W [DOI] [PubMed] [Google Scholar]
- 11. Menko AS, Boettiger D. Occupation of the extracellular matrix receptor, integrin, is a control point for myogenic differentiation. Cell, 1987, 51, 51–57 10.1016/0092-8674(87)90009-2 [DOI] [PubMed] [Google Scholar]
- 12. Rosen GD, Sanes JR, LaChance R, Cunningham JM, Roman J, Dean DC. Roles for the integrin VLA‐4 and its counter receptor VCAM‐1 in myogenesis. Cell, 1992, 69, 1107–1119 10.1016/0092-8674(92)90633-N [DOI] [PubMed] [Google Scholar]
- 13. Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell, 1996, 84, 345–357 10.1016/S0092-8674(00)81279-9 [DOI] [PubMed] [Google Scholar]
- 14. Schwander M, Leu M, Stumm M, Dorchies OM, Ruegg UT, Schittny J, Muller U. Beta1 integrins regulate myoblast fusion and sarcomere assembly. Dev Cell, 2003, 4, 673–685 10.1016/S1534-5807(03)00118-7 [DOI] [PubMed] [Google Scholar]
- 15. Evans JP. Fertilin beta and other ADAMs as integrin ligands: insights into cell adhesion and fertilization. Bioessays, 2001, 23, 628–639 10.1002/bies.1088 [DOI] [PubMed] [Google Scholar]
- 16. Bronson RA, Fusi FM, Calzi F, Doldi N, Ferrari A. Evidence that a functional fertilin‐like ADAM plays a role in human sperm–oolemmal interactions. Mol Hum Reprod, 1999, 5, 433–440 10.1093/molehr/5.5.433 [DOI] [PubMed] [Google Scholar]
- 17. Zhu X, Bansal NP, Evans JP. Identification of key functional amino acids of the mouse fertilin beta (ADAM2) disintegrin loop for cell–cell adhesion during fertilization. J Biol Chem, 2000, 275, 7677–7683 10.1074/jbc.275.11.7677 [DOI] [PubMed] [Google Scholar]
- 18. McLaughlin EA, Frayne J, Bloomerg G, Hall L. Do fertilin beta and cyritestin play a major role in mammalian sperm–oolemma interactions? A critical re‐evaluation of the use of peptide mimics in identifying specific oocyte recognition proteins. Mol Hum Reprod, 2001, 7, 313–317 10.1093/molehr/7.4.313 [DOI] [PubMed] [Google Scholar]
- 19. Cho C, Ge H, Branciforte D, Primakoff P, Myles DG. Analysis of mouse fertilin in wild‐type and fertilin beta(−/−) sperm: evidence for C‐terminal modification, alpha/beta dimerization, and lack of essential role of fertilin alpha in sperm–egg fusion. Dev Biol, 2000, 222, 289–295 10.1006/dbio.2000.9703 [DOI] [PubMed] [Google Scholar]
- 20. Miller BJ, Georges‐Labouesse E, Primakoff P, Myles DG. Normal fertilization occurs with eggs lacking the integrin alpha6beta1 and is CD9‐dependent. J Cell Biol, 2000, 149, 1289–1296 10.1083/jcb.149.6.1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. He ZY, Brakebusch C, Fassler R, Kreidberg JA, Primakoff P, Myles DG. None of the integrins known to be present on the mouse egg or to be ADAM receptors are essential for sperm–egg binding and fusion. Dev Biol, 2003, 254, 226–237 10.1016/S0012-1606(02)00043-X [DOI] [PubMed] [Google Scholar]
- 22. Vjugina U, Zhu X, Oh E, Bracero NJ, Evans JP. Reduction of mouse egg surface integrin alpha9 subunit (ITGA9) reduces the egg's ability to support sperm–egg binding and fusion. Biol Reprod, 2009, 80, 833–841 10.1095/biolreprod.108.075275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Okabe M, Cummins JM. Mechanisms of sperm–egg interactions emerging from gene‐manipulated animals. Cell Mol Life Sci, 2007, 64, 1945–1958 10.1007/s00018-007-7037-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ikawa M, Inoue N, Benham AM, Okabe M. Fertilization: a sperm's journey to and interaction with the oocyte. J Clin Invest, 2010, 120, 984–994 10.1172/JCI41585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ikawa M, Wada I, Kominami K, Watanabe D, Toshimori K, Nishimune Y, Okabe M. The putative chaperone calmegin is required for sperm fertility. Nature, 1997, 387, 607–611 10.1038/42484 [DOI] [PubMed] [Google Scholar]
- 26. Yamagata K, Nakanishi T, Ikawa M, Yamaguchi R, Moss SB, Okabe M. Sperm from the calmegin‐deficient mouse have normal abilities for binding and fusion to the egg plasma membrane. Dev Biol, 2002, 250, 348–357 10.1006/dbio.2002.0803 [DOI] [PubMed] [Google Scholar]
- 27. Ikawa M, Tokuhiro K, Yamaguchi R, Benham AM, Tamura T, Wada I, Satouh Y, Inoue N, Okabe M. Calsperin is a testis‐specific chaperone required for sperm fertility. J Biol Chem, 2011, 286, 5639–5646 10.1074/jbc.M110.140152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tokuhiro K, Ikawa M, Benham AM, Okabe M. Protein disulfide isomerase homolog PDILT is required for quality control of sperm membrane protein ADAM3 and male fertility [corrected]. Proc Natl Acad Sci USA, 2012, 109, 3850–3855 10.1073/pnas.1117963109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marcello MR, Jia W, Leary JA, Moore KL, Evans JP. Lack of tyrosylprotein sulfotransferase‐2 activity results in altered sperm–egg interactions and loss of ADAM3 and ADAM6 in epididymal sperm. J Biol Chem, 2011, 286, 13060–13070 10.1074/jbc.M110.175463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kondoh G, Tojo H, Nakatani Y, Komazawa N, Murata C, Yamagata K, Maeda Y, Kinoshita T, Okabe M, Taguchi R, Takeda J. Angiotensin‐converting enzyme is a GPI‐anchored protein releasing factor crucial for fertilization. Nat Med, 2005, 11, 160–166 10.1038/nm1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cho C, Bunch DO, Faure JE, Goulding EH, Eddy EM, Primakoff P, Myles DG. Fertilization defects in sperm from mice lacking fertilin beta. Science, 1998, 281, 1857–1859 10.1126/science.281.5384.1857 [DOI] [PubMed] [Google Scholar]
- 32. Kim E, Yamashita M, Nakanishi T, Park KE, Kimura M, Kashiwabara S, Baba T. Mouse sperm lacking ADAM1b/ADAM2 fertilin can fuse with the egg plasma membrane and effect fertilization. J Biol Chem, 2006, 281, 5634–5639 10.1074/jbc.M510558200 [DOI] [PubMed] [Google Scholar]
- 33. Nishimura H, Cho C, Branciforte DR, Myles DG, Primakoff P. Analysis of loss of adhesive function in sperm lacking cyritestin or fertilin beta. Dev Biol, 2001, 233, 204–213 10.1006/dbio.2001.0166 [DOI] [PubMed] [Google Scholar]
- 34. Inoue N, Kasahara T, Ikawa M, Okabe M. Identification and disruption of sperm‐specific angiotensin converting enzyme‐3 (ACE3) in mouse. PLoS ONE, 2010, 5, e10301 10.1371/journal.pone.0010301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saxena DK, Tanii I, Yoshinaga K, Toshimori K. Role of intra‐acrosomal antigenic molecules acrin 1 (MN7) and acrin 2 (MC41) in penetration of the zona pellucida in fertilization in mice. J Reprod Fertil, 1999, 117, 17–25 10.1530/jrf.0.1170017 [DOI] [PubMed] [Google Scholar]
- 36. Crosnier C, Bustamante LY, Bartholdson SJ, Bei AK, Theron M, Uchikawa M, Mboup S, Ndir O, Kwiatkowski DP, Duraisingh MT, Rayner JC, Wright GJ. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum . Nature, 2011, 480, 534–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saxena DK, Toshimori K. Molecular modifications of MC31/CE9, a sperm surface molecule, during sperm capacitation and the acrosome reaction in the rat: is MC31/CE9 required for fertilization?. Biol Reprod, 2004, 70, 993–1000 10.1095/biolreprod.103.021667 [DOI] [PubMed] [Google Scholar]
- 38. Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. β‐Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell, 2001, 105, 533–545 10.1016/S0092-8674(01)00336-1 [DOI] [PubMed] [Google Scholar]
- 39. Nagafuchi A, Takeichi M, Tsukita S. The 102 kD cadherin‐associated protein: similarity to vinculin and posttranscriptional regulation of expression. Cell, 1991, 65, 849–857 10.1016/0092-8674(91)90392-C [DOI] [PubMed] [Google Scholar]
- 40. Nagafuchi A, Takeichi M. Cell binding function of E‐cadherin is regulated by the cytoplasmic domain. EMBO J, 1988, 7, 3679–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vries WN, Evsikov AV, Haac BE, Fancher KS, Holbrook AE, Kemler R, Solter D, Knowles BB. Maternal beta‐catenin and E‐cadherin in mouse development. Development, 2004, 131, 4435–4445 10.1242/dev.01316 [DOI] [PubMed] [Google Scholar]
- 42. Takezawa Y, Yoshida K, Miyado K, Sato M, Nakamura A, Kawano N, Sakakibara K, Kondo T, Harada Y, Ohnami N, Kanai S, Miyado M et al. β‐catenin is a molecular switch that regulates transition of cell–cell adhesion to fusion. Sci Rep, 2011, 1, 68 10.1038/srep00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Valenta T, Hausmann G, Basler K. The many faces and functions of beta‐catenin. EMBO J, 2012, 31, 2714–2736 10.1038/emboj.2012.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marin‐Briggiler CI, Lapyckyj L, Gonzalez Echeverria MF, Rawe VY, Alvarez Sedo C, Vazquez‐Levin MH. Neural cadherin is expressed in human gametes and participates in sperm–oocyte interaction events. Int J Androl, 2010, 33, e228–e239 10.1111/j.1365-2605.2009.00999.x [DOI] [PubMed] [Google Scholar]
- 45. Ohgimoto S, Tabata N, Suga S, Nishio M, Ohta H, Tsurudome M, Komada H, Kawano M, Watanabe N, Ito Y. Molecular characterization of fusion regulatory protein‐1 (FRP‐1) that induces multinucleated giant cell formation of monocytes and HIV gp160‐mediated cell fusion. FRP‐1 and 4F2/CD98 are identical molecules. J Immunol, 1995, 155, 3585–3592 [PubMed] [Google Scholar]
- 46. Takahashi Y, Bigler D, Ito Y, White JM. Sequence‐specific interaction between the disintegrin domain of mouse ADAM 3 and murine eggs: role of beta1 integrin‐associated proteins CD9, CD81, and CD98. Mol Biol Cell, 2001, 12, 809–820 10.1091/mbc.12.4.809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ros VG, Maldera JA, Willis WD, Cohen DJ, Goulding EH, Gelman DM, Rubinstein M, Eddy EM, Cuasnicu PS. Impaired sperm fertilizing ability in mice lacking cysteine‐rich secretory protein 1 (CRISP1). Dev Biol, 2008, 320, 12–18 10.1016/j.ydbio.2008.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Busso D, Goldweic NM, Hayashi M, Kasahara M, Cuasnicu PS. Evidence for the involvement of testicular protein CRISP2 in mouse sperm–egg fusion. Biol Reprod, 2007, 76, 701–708 10.1095/biolreprod.106.056770 [DOI] [PubMed] [Google Scholar]
- 49. Yamatoya K, Yoshida K, Ito C, Maekawa M, Yanagida M, Takamori K, Ogawa H, Araki Y, Miyado K, Toyama Y, Toshimori K. Equatorin: identification and characterization of the epitope of the MN9 antibody in the mouse. Biol Reprod, 2009, 81, 889–897 10.1095/biolreprod.109.077438 [DOI] [PubMed] [Google Scholar]
- 50. Toshimori K, Saxena DK, Tanii I, Yoshinaga K. An MN9 antigenic molecule, equatorin, is required for successful sperm‐oocyte fusion in mice. Biol Reprod, 1998, 59, 22–29 10.1095/biolreprod59.1.22 [DOI] [PubMed] [Google Scholar]
- 51. Inoue N, Nishikawa T, Ikawa M, Okabe M. Tetraspanin‐interacting protein IGSF8 is dispensable for mouse fertility. Fertil Steril, 2012, 98, 465–470 10.1016/j.fertnstert.2012.04.029 [DOI] [PubMed] [Google Scholar]
- 52. Grayson P, Civetta A. Positive selection and the evolution of izumo genes in mammals. Int J Evol Biol, 2012, 2012, 958164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fujihara Y, Murakami M, Inoue N, Satouh Y, Kaseda K, Ikawa M, Okabe M. Sperm equatorial segment protein 1, SPESP1, is required for fully fertile sperm in mouse. J Cell Sci, 2010, 123, 1531–1536 10.1242/jcs.067363 [DOI] [PubMed] [Google Scholar]
- 54. Nishimura H, Gupta S, Myles DG, Primakoff P. Characterization of mouse sperm TMEM190, a small transmembrane protein with the trefoil domain: evidence for co‐localization with IZUMO1 and complex formation with other sperm proteins. Reproduction, 2011, 141, 437–451 10.1530/REP-10-0391 [DOI] [PubMed] [Google Scholar]
- 55. Spiridonov NA, Wong L, Zerfas PM, Starost MF, Pack SD, Paweletz CP, Johnson GR. Identification and characterization of SSTK, a serine/threonine protein kinase essential for male fertility. Mol Cell Biol, 2005, 25, 4250–4261 10.1128/MCB.25.10.4250-4261.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sosnik J, Miranda PV, Spiridonov NA, Yoon SY, Fissore RA, Johnson GR, Visconti PE. Tssk6 is required for Izumo relocalization and gamete fusion in the mouse. J Cell Sci, 2009, 122, 2741–2749 10.1242/jcs.047225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Miyado K, Yamada G, Yamada S, Hasuwa H, Nakamura Y, Ryu F, Suzuki K, Kosai K, Inoue K, Ogura A, Okabe M, Mekada E. Requirement of CD9 on the egg plasma membrane for fertilization. Science, 2000, 287, 321–324 10.1126/science.287.5451.321 [DOI] [PubMed] [Google Scholar]
- 58. Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C. Severely reduced female fertility in CD9‐deficient mice. Science, 2000, 287, 319–321 10.1126/science.287.5451.319 [DOI] [PubMed] [Google Scholar]
- 59. Kaji K, Oda S, Shikano T, Ohnuki T, Uematsu Y, Sakagami J, Tada N, Miyazaki S, Kudo A. The gamete fusion process is defective in eggs of CD9‐deficient mice. Nat Genet, 2000, 24, 279–282 10.1038/73502 [DOI] [PubMed] [Google Scholar]
- 60. Inoue N, Ikawa M, Isotani A, Okabe M. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature, 2005, 434, 234–238 10.1038/nature03362 [DOI] [PubMed] [Google Scholar]
- 61. Hemler ME. Targeting of tetraspanin proteins—potential benefits and strategies. Nat Rev Drug Discov, 2008, 7, 747–758 10.1038/nrd2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Miyake M, Koyama M, Seno M, Ikeyama S. Identification of the motility‐related protein (MRP‐1), recognized by monoclonal antibody M31‐15, which inhibits cell motility. J Exp Med, 1991, 174, 1347–1354 10.1084/jem.174.6.1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Akutsu H, Miura T, Machida M, Birumachi J, Hamada A, Yamada M, Sullivan S, Miyado K, Umezawa A. Maintenance of pluripotency and self‐renewal ability of mouse embryonic stem cells in the absence of tetraspanin CD9. Differentiation, 2009, 78, 137–142 10.1016/j.diff.2009.08.005 [DOI] [PubMed] [Google Scholar]
- 64. Zhu GZ, Miller BJ, Boucheix C, Rubinstein E, Liu CC, Hynes RO, Myles DG, Primakoff P. Residues SFQ (173–175) in the large extracellular loop of CD9 are required for gamete fusion. Development, 2002, 129, 1995–2002 [DOI] [PubMed] [Google Scholar]
- 65. Kaji K, Oda S, Miyazaki S, Kudo A. Infertility of CD9‐deficient mouse eggs is reversed by mouse CD9, human CD9, or mouse CD81; polyadenylated mRNA injection developed for molecular analysis of sperm–egg fusion. Dev Biol, 2002, 247, 327–334 10.1006/dbio.2002.0694 [DOI] [PubMed] [Google Scholar]
- 66. Moribe H, Yochem J, Yamada H, Tabuse Y, Fujimoto T, Mekada E. Tetraspanin protein (TSP‐15) is required for epidermal integrity in Caenorhabditis elegans. J Cell Sci, 2004, 117, 5209–5220 10.1242/jcs.01403 [DOI] [PubMed] [Google Scholar]
- 67. Moribe H, Konakawa R, Koga D, Ushiki T, Nakamura K, Mekada E. Tetraspanin is required for generation of reactive oxygen species by the dual oxidase system in Caenorhabditis elegans . Plos Genet, 2012, 8, e1002957 10.1371/journal.pgen.1002957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kopczynski CC, Davis GW, Goodman CS. A neural tetraspanin, encoded by late bloomer, that facilitates synapse formation. Science, 1996, 271, 1867–1870 10.1126/science.271.5257.1867 [DOI] [PubMed] [Google Scholar]
- 69. Todres E, Nardi JB, Robertson HM. The tetraspanin superfamily in insects. Insect Mol Biol, 2000, 9, 581–590 10.1046/j.1365-2583.2000.00222.x [DOI] [PubMed] [Google Scholar]
- 70. Hassuna N, Monk PN, Moseley GW, Partridge LJ. Strategies for targeting tetraspanin proteins: potential therapeutic applications in microbial infections. BioDrugs, 2009, 23, 341–359 10.2165/11315650-000000000-00000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shiino T. Phylodynamic analysis of a viral infection network. Front Microbiol, 2012, 3, 278 10.3389/fmicb.2012.00278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Garcia E, Pion M, Pelchen‐Matthews A, Collinson L, Arrighi JF, Blot G, Leuba F, Escola JM, Demaurex N, Marsh M, Piguet V. HIV‐1 trafficking to the dendritic cell‐T‐cell infectious synapse uses a pathway of tetraspanin sorting to the immunological synapse. Traffic, 2005, 6, 488–501 10.1111/j.1600-0854.2005.00293.x [DOI] [PubMed] [Google Scholar]
- 73. Wiley RD, Gummuluru S. Immature dendritic cell‐derived exosomes can mediate HIV‐1 trans infection. Proc Natl Acad Sci USA, 2006, 103, 738–743 10.1073/pnas.0507995103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Silvie O, Rubinstein E, Franetich JF, Prenant M, Belnoue E, Renia L, Hannoun L, Eling W, Levy S, Boucheix C, Mazier D. Hepatocyte CD81 is required for Plasmodium falciparum and Plasmodium yoelii sporozoite infectivity. Nat Med, 2003, 9, 93–96 10.1038/nm808 [DOI] [PubMed] [Google Scholar]
- 75. Rubinstein E, Ziyyat A, Prenant M, Wrobel E, Wolf JP, Levy S, Naour F, Boucheix C. Reduced fertility of female mice lacking CD81. Dev Biol, 2006, 290, 351–358 10.1016/j.ydbio.2005.11.031 [DOI] [PubMed] [Google Scholar]
- 76. Tanigawa M, Miyamoto K, Kobayashi S, Sato M, Akutsu H, Okabe M, Mekada E, Sakakibara K, Miyado M, Umezawa A, Miyado K. Possible involvement of CD81 in acrosome reaction of sperm in mice. Mol Reprod Dev, 2008, 75, 150–155 10.1002/mrd.20709 [DOI] [PubMed] [Google Scholar]
- 77. Ohnami N, Nakamura A, Miyado M, Sato M, Kawano N, Yoshida K, Harada Y, Takezawa Y, Kanai S, Ono C, Takahashi Y, Kimura K et al. CD81 and CD9 work independently as extracellular components upon fusion of sperm and oocyte. Biol Open, 2012, 1, 640–647 10.1242/bio.20121420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Huang S, Yuan S, Dong M, Su J, Yu C, Shen Y, Xie X, Yu Y, Yu X, Chen S, Zhang S, Pontarotti P et al. The phylogenetic analysis of tetraspanins projects the evolution of cell–cell interactions from unicellular to multicellular organisms. Genomics, 2005, 86, 674–684 10.1016/j.ygeno.2005.08.004 [DOI] [PubMed] [Google Scholar]
- 79. Chiu WH, Chandler J, Cnops G, Lijsebettens M, Werr W. Mutations in the TORNADO2 gene affect cellular decisions in the peripheral zone of the shoot apical meristem of Arabidopsis thaliana. Plant Mol Biol, 2007, 63, 731–744 10.1007/s11103-006-9105-z [DOI] [PubMed] [Google Scholar]
- 80. Lambou K, Tharreau D, Kohler A, Sirven C, Marguerettaz M, Barbisan C, Sexton AC, Kellner EM, Martin F, Howlett BJ, Orbach MJ, Lebrun MH. Fungi have three tetraspanin families with distinct functions. BMC Genomics, 2008, 9, 63 10.1186/1471-2164-9-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Clergeot PH, Gourgues M, Cots J, Laurans F, Latorse MP, Pepin R, Tharreau D, Notteghem JL, Lebrun MH. PLS1, a gene encoding a tetraspanin‐like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea . Proc Natl Acad Sci USA, 2001, 98, 6963–6968 10.1073/pnas.111132998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Veneault‐Fourrey C, Parisot D, Gourgues M, Lauge R, Lebrun MH, Langin T. The tetraspanin gene ClPLS1 is essential for appressorium‐mediated penetration of the fungal pathogen Colletotrichum lindemuthianum . Fungal Genet Biol, 2005, 42, 306–318 10.1016/j.fgb.2005.01.009 [DOI] [PubMed] [Google Scholar]
- 83. Lambou K, Malagnac F, Barbisan C, Tharreau D, Lebrun MH, Silar P. The crucial role of the Pls1 tetraspanin during ascospore germination in Podospora anserina provides an example of the convergent evolution of morphogenetic processes in fungal plant pathogens and saprobes. Eukaryot Cell, 2008, 7, 1809–1818 10.1128/EC.00149-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC Jr, McCoy JM. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature, 2000, 403, 785–789 10.1038/35001608 [DOI] [PubMed] [Google Scholar]
- 85. Hernandez LD, Hoffman LR, Wolfsberg TG, White JM. Virus‐cell and cell–cell fusion. Annu Rev Cell Dev Biol, 1996, 12, 627–661 10.1146/annurev.cellbio.12.1.627 [DOI] [PubMed] [Google Scholar]
- 86. Couzin J. Cell biology: the ins and outs of exosomes. Science, 2005, 308, 1862–1863 10.1126/science.308.5730.1862 [DOI] [PubMed] [Google Scholar]
- 87. Denzer K, Kleijmeer MJ, Heijnen HF, Stoorvogel W, Geuze HJ. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J Cell Sci, 2000, 113 (Pt 19) 3365–3374 [DOI] [PubMed] [Google Scholar]
- 88. Simons M, Raposo G. Exosomes–vesicular carriers for intercellular communication. Curr Opin Cell Biol, 2009, 21, 575–581 10.1016/j.ceb.2009.03.007 [DOI] [PubMed] [Google Scholar]
- 89. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol, 2007, 9, 654–659 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- 90. Barraud‐Lange V, Naud‐Barriant N, Bomsel M, Wolf JP, Ziyyat A. Transfer of oocyte membrane fragments to fertilizing spermatozoa. FASEB J, 2007, 21, 3446–3449 10.1096/fj.06-8035hyp [DOI] [PubMed] [Google Scholar]
- 91. Runge KE, Evans JE, He ZY, Gupta S, McDonald KL, Stahlberg H, Primakoff P, Myles DG. Oocyte CD9 is enriched on the microvillar membrane and required for normal microvillar shape and distribution. Dev Biol, 2007, 304, 317–325 10.1016/j.ydbio.2006.12.041 [DOI] [PubMed] [Google Scholar]
- 92. Miyado K, Yoshida K, Yamagata K, Sakakibara K, Okabe M, Wang X, Miyamoto K, Akutsu H, Kondo T, Takahashi Y, Ban T, Ito C et al. The fusing ability of sperm is bestowed by CD9‐containing vesicles released from eggs in mice. Proc Natl Acad Sci USA, 2008, 105, 12921–12926 10.1073/pnas.0710608105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Talbot P, DiCarlantonio G. Ultrastructure of opossum oocyte investing coats and their sensitivity to trypsin and hyaluronidase. Dev Biol, 1984, 103, 159–167 10.1016/0012-1606(84)90017-4 [DOI] [PubMed] [Google Scholar]
- 94. Dandekar P, Aggeler J, Talbot P. Structure, distribution and composition of the extracellular matrix of human oocytes and cumulus masses. Hum Reprod, 1992, 7, 391–398 [DOI] [PubMed] [Google Scholar]
- 95. Saunders CM, Larman MG, Parrington J, Cox LJ, Royse J, Blayney LM, Swann K, Lai FA. PLC zeta: a sperm‐specific trigger of Ca(2+) oscillations in eggs and embryo development. Development, 2002, 129, 3533–3544 [DOI] [PubMed] [Google Scholar]


