Summary
Host–microbiota mutualism has been established during long‐term co‐evolution. A diverse and rich gut microbiota plays an essential role in the development and maturation of the host immune system. Education of the adaptive immune compartment by gut microbiota antigens is important in establishing immune balance. In particular, a critical time frame immediately after birth provides a ‘window of opportunity’ for the development of lymphoid structures, differentiation and maturation of T and B cells and, most importantly, establishment of immune tolerance to gut commensals. Depending on the colonization niche, antigen type and metabolic property of different gut microbes, CD4 T‐cell responses vary greatly, which results in differentiation into distinct subsets. As a consequence, certain bacteria elicit effector‐like immune responses by promoting the production of pro‐inflammatory cytokines such as interferon‐γ and interleukin‐17A, whereas other bacteria favour the generation of regulatory CD4 T cells and provide help with gut homeostasis. The microbiota have profound effects on B cells also. Gut microbial exposure leads to a continuous diversification of B‐cell repertoire and the production of T‐dependent and ‐independent antibodies, especially IgA. These combined effects of the gut microbes provide an elegant educational process to the adaptive immune network. Contrariwise, failure of this process results in a reduced homeostasis with the gut microbiota, and an increased susceptibility to various immune disorders, both inside and outside the gut. With more definitive microbial–immune relations waiting to be discovered, modulation of the host gut microbiota has a promising future for disease intervention.
Keywords: Microbiota, T cell, B cell, immune homeostasis, autoimmunity
Introduction
When we think about the function of biological organs today, it is impossible to neglect the critical role of microbiota, especially the microbiota inhabiting the gastrointestinal tract, due to its enormous size, its diversity and, most importantly, its effects on the biological functions of the host. There are about 3·8 × 1013 bacteria, belonging to up to 1000 species,1, 2 colonizing the gut of a healthy human adult, plus a variety of fungi, viruses and archaea bacteria, which have been less studied.3, 4 Homeostasis between the host and its microbiota is the result of long‐term co‐evolution: gut microbiota relies on the host environment and nutrients for its survival, and in return provides the host with essential metabolites that promote a proper functioning of multiple organ systems. Generally, in a healthy adult gut, Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria and Verrucomicrobia are the five dominant bacterial phyla, with the first two being the most relatively abundant.5 Descending from the duodenum to the distal colon, due to physiological and immunological differences along the intestine, the number and diversity of bacteria increase, and the predominant bacterial families change.6 The continuous mutual selection between the host and the microbes during evolution has resulted in a set of host‐specific microbiota. For instance, although the phyla are similar between human and mouse gut microbiota, there are significant differences in the bacterial species in different hosts.7, 8 The full maturation of the host immune system is dependent on their natural harbouring of host‐specific microbiota, because colonization of germ‐free (GF) mice with human or rat microbiota fails to induce an effective immune response and fails to provide protection against enteric infection.8 Segmented filamentous bacteria (SFB, also known as Candidatus Savagella),9 which are potent inducers of the murine adaptive immune response including gut T helper type 17 (Th17) cell differentiation10, 11, 12 and IgA production,13, 14 are genetically different in mice and rats.15 And interestingly, although it has been intensively studied in mouse models, a species that is genetically related to SFB has not been identified as yet in humans.16
In the past decade, many studies have focused on the interactions between the host immune system and the gut microbiota. The innate immune system rapidly responds to the gut microbiota in an antigen‐nonspecific manner through the activation of pattern recognition receptors, and releases cytokines such as interferon‐α, interleukin‐18 (IL‐18) and IL‐22 to promote epithelial antimicrobial responses such as the production of antimicrobial peptides.17 The recently defined innate lymphoid cells mimic the cytokine production of T cells in an antigen‐nonspecific way.18 Although mice lacking adaptive immunity survive well with their microbiota in a specific pathogen‐free (SPF) environment, they have a greater vulnerability to opportunistic infections, such as Pneumocystis carinii pneumonia.19 Similarly, HIV‐infected individuals are very susceptible to infections due to their greatly decreased CD4 T cells.20 Different from the innate immune system, the adaptive immune compartment recognizes specific microbial antigens through its highly mutated cell surface receptors21 and depending on the type of bacteria it encounters, naive T cells can differentiate into either effector T cells to fight against the bacteria, or into regulatory T (Treg) cells to tolerate their presence and promote mutualism. Although it takes time for the adaptive immune system to differentiate and proliferate to respond to microbial antigens after the first encounter, some of the antigen‐experienced memory cells survive long‐term and provide a strong and timely response in a recall encounter.22
In this review, we will focus on the interactions between the host adaptive immune system and the gut microbiota, in particular how the adaptive immune compartment recognizes microbiota antigens and regulates microbiota composition to maintain gut homeostasis, and reciprocally how an abnormal composition of the microbiota or dysbiosis affects the host immune system and may result in mucosal or systemic immune disorders.
Early‐life host–microbiota interactions and ‘window of opportunity’
It is widely accepted that the first burst of microbial encounter occurs at the moment of birth.23 Although evidence of prenatal microbiota in the placenta has emerged, the numbers and effects are small compared with microbial colonization after birth.24, 25, 26 Several factors, including mode of delivery, breastfeeding,23, 27 antibiotics28 and environmental exposure,29, 30 have been shown to greatly modulate the dominant bacteria of the neonate's early gut colonizers, which can exert long‐term health effects in the offspring.31 Therefore, restoring the gut microbiota of newborns delivered by caesarean section with exposure of maternal vaginal fluids or addition of probiotics into formula may lower disease susceptibility in childhood, and even into adulthood.32 Besides microbial antigens,33 breast milk contains a considerable amount of maternal antibodies that not only help to establish the microbial composition, but also dampen excessive follicular T‐cell and germinal B‐cell responses to gut microbes in neonatal mice.34 Maternal IgA has been shown to provide protection of the newborn from epithelial translocation of opportunistic bacteria such as Ochrobactrum anthropi, and to provide long‐term benefits by preventing intestinal inflammation.35 The transition from breast milk/formula to solid food triggers the first major wave of the gut microbiota expansion,36 and depending on the diet and lifestyles, the composition of the gut microbiota continues to change with age towards adulthood.37
The gut microbiota remains unstable during neonatal life, which creates a crucial ‘window of opportunity’ for the development of the host immune system. This process generally takes about 4–6 weeks in mice, and 2–3 years in children,38, 39, 40, 41 until the microbial community reaches a relatively stable status, which is then maintained into adulthood. Because of the close proximity between the enteric microbes and the gut mucosal barrier, the most dramatic impact that the microbiota brings to host immune development is on the mucosal compartment. Although cryptopatches, Peyer's patches (PPs) and mesenteric lymph nodes form prenatally,42 postnatal microbial stimulation further promotes their development, including the formation of isolated lymphoid tissues from cryptopatches,43 further recruitment of T and B cells into the lamina propria (LP) and PPs,44 and T‐cell differentiation and B‐cell maturation (discussed below). Depending on the variations of gut microbial introduction and colonization postnatally, host immune development can differ dramatically and result in altered immune reactivity that could affect the host's susceptibility to diseases such as autoimmune and allergic diseases in later life. Insufficient microbial stimulation in neonatal mice results in increased IgE class switching of mucosal B cells and elevated serum IgE level in a CD4 T‐cell‐dependent manner, a process reversed by colonization with conventional gut microbiota immediately after birth, but not in adulthood.45 Similarly, invariant natural killer T‐cell accumulation is observed in GF mice, which is associated with increased susceptibility to inflammatory bowel disease compared with SPF mice. However, microbiota conventionalization of neonatal GF mice reduced the increase of invariant natural killer T cells in the gut and lung, whereas colonization in adulthood had no such beneficial effect.46 This time‐sensitive perturbation of invariant natural killer T‐cell homeostasis was regulated by bacterial sphingolipids produced by Bacteroides fragilis.47 Early‐life exposure to antibiotics also has a potentially detrimental impact on the host due to the instability of gut microbiota and hence an altered differentiation of immune cells.48 Gut microbiota disruption introduced by early‐life antibiotics can remain altered during adulthood even with normal environmental exposure.49 Oral administration of antibiotics in adult mice ameliorated psoriasis severity, whereas neonatal exposure to antibiotics exaggerated psoriasis progression, an effect mediated by IL‐22‐producing T cells in the skin.49 Neonatal antibiotic exposure in both mice and humans enhanced the sensitization to various allergens due to an altered gut microbiota, resulting in increased risk of childhood atopy, asthma and food allergies.50, 51, 52 Treg cells play an important role for establishment of immune tolerance in this early life. Compared with adult mice, neonatal mice harbour a higher fraction of Treg cells in their gut CD4 compartment as well as a stronger suppressive function.53 Maternal antibodies help to prevent the translocation of luminal antigens present in the dam.35 As a consequence, effector CD4 cell functions are actively repressed before weaning.53 However, at weaning, shifts in the gut microbiota are associated with induction of peripheral Treg cells (pTreg), and a change in thymic Treg (tTreg)/pTreg ratio.54 There is a critical time window before weaning for the establishment of commensal‐specific pTreg cells in mice via goblet‐cell‐associated passages, and disruption of these goblet‐cell‐associated passages during this critical time leads to an impaired control of gut homeostasis later in adulthood.55
As the host ages, the gut microbiota remains relatively stable with minor fluctuations. However, antibiotics and dietary changes can still bring compositional and functional changes to the gut microbiota.37, 56, 57, 58
T cell–gut microbiota interactions
Conventionally, T‐cell education refers to thymic selection, by which immature T cells with high self‐reactivity are eliminated or converted to Treg cells to prevent autoimmunity.59 More recent studies have included peripheral education by gut microbial antigens as a form of T‐cell education, which also maintains homeostasis.60, 61 In addition to physical separation of the microbiota from the intestinal immune cells by the mucus, the epithelial layer, antimicrobial peptides and secretory antibodies, a major component maintaining gut homeostasis are Treg cells, including canonical Foxp3‐expressing Treg cells,62 IL‐10‐expressing Foxp3− Treg type 1 cells,63 and the newly defined regulatory intraepithelial CD4+ CD8αα + T cells.64 Although Treg cells are present in the gut mucosa postnatally regardless of microbial colonization,54 in adult mice, certain endogenous bacteria (Clostridium clusters IV, XIVa and XVIII),65, 66 bacterial products (B. fragilis polysaccharide),67 or bacterial metabolites (short‐chain fatty acids including acetate, butyrate and propionate),68, 69, 70 can induce functional Treg cells in the colonic LP (Fig. 1) and provide protection to immune‐related diseases locally or systemically.71, 72 Further analyses of transcription factors and T‐cell receptor (TCR) repertoire suggest that gut Treg cells that are present before weaning are mainly of thymus origin (tTreg), because they express the tTreg‐specific transcription factor Helios and surface marker Neuropilin‐1.73, 74, 75 In contrast, Treg cells induced by microbiota colonization express low levels of Helios,54 and they may use a different TCR repertoire,60, 61 indicating that they are a result of pTreg induction instead of expansion of tTreg cells. Induction of pTreg cells was shown to occur primarily in the mesenteric lymph nodes with robust Foxp3+ cell proliferation.54 As stated earlier, gut Treg cells are required to help establish oral tolerance to food antigens as well as to the enteric microbiota. Co‐transfer of Treg cells with CD45RBhi T cells abrogates colitis in immunodeficient mice.76 It has also been suggested that conversion from Th17 cells into Treg cells in late‐phase colitis helps to resolve inflammation.77 MyD88‐Stat3‐dependent sensing of gut microbiota in Treg cells is indispensable for the induction of intestinal IgA and the restraint of pro‐inflammatory T‐cell responses in the gut.78 Using the CBir1 flagellin TCR transgenic mouse model, of which > 85% CD4 T cells recognize the epitope expressed on the flagellin of the Lachnospiraceae family of Clostridiales, Treg cells provide tolerance to commensal bacteria by promoting the survival of antigen‐specific IgA+ B cells,79 because depletion of Treg cells with anti‐CD25 resulted in the loss of intestinal IgA B cells. Similarly, using adoptive transfer models, it was shown that failure of pTreg induction for selected microbiota antigens results in T effector cell differentiation and increased susceptibility to intestinal inflammation.80 Nonetheless, Treg cells can accomplish their suppressive function both in antigen‐specific and bystander ways through the secretion of anti‐inflammatory cytokines transforming growth factor‐β and IL‐10,81 and the induction of IgA, either directly or through the promotion of T follicular helper (Tfh) cells78, 82 and T follicular regulatory cells,78, 83 to maintain gut homeostasis.
Figure 1.
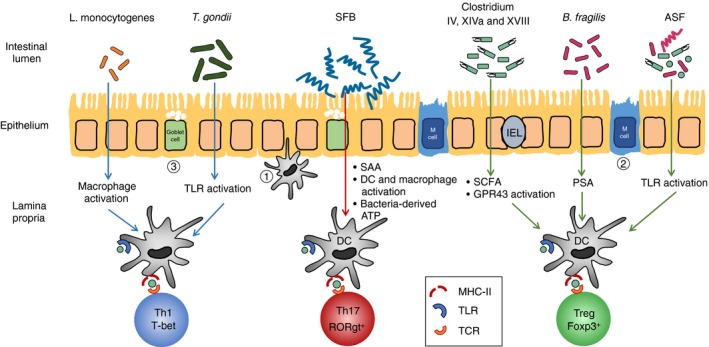
Microbiota induction of intestinal regulatory T (Treg)/ effector T (Teff) cell differentiation. Commensal bacteria such as Clostridium cluster IV, XIVa and XVIII, Bacteroides fragilis and altered Schaedler flora (ASF) promote the differentiation and expansion of Treg cells in the gut through various mechanisms, whereas microbially derived ATP and epithelium‐adhering bacteria such as segmented filamentous bacteria (SFB) stimulate the induction of intestinal Th17 cells. Antigen‐specific Th1 cell differentiation can be promoted by intracellular pathogens such as Listeria monocytogenes and Toxoplasma gondii. Microbiota antigens are sampled via (1) transepithelial dendrites of dendritic cells (DC), (2) transcytosis through microfold cells (M cell), or (3) goblet cell‐associated antigen passages (GAP) and then induce T‐cell differentiation in the mesenteric lymph nodes or de novo in the lamina propria. Teff cells and Treg cells are both plastic and can convert into each other and into other T‐cell subsets under certain conditions.
The other main T‐cell subset that has been studied the most regarding microbiota education comprises Th17 cells. The CD4+ Th17 subset is characterized by expression of master transcription factor Rorγt, and the production of cytokines including IL‐17F, IL‐17A, IL‐21 and IL‐22.84 Th17 cells are considered to be pathogenic because of their ubiquitous presence in a variety of inflammatory disorders. However, more recent studies have revealed a Th17 homeostatic role in the gut.85 In SPF mice, Th17 cells are among the most abundant effector CD4 T cells in the healthy gut LP. Unlike the gene expression signature of pathogenic Th17 cells isolated from an inflammatory milieu, homeostatic Th17 cells have a significantly lower level of pro‐inflammatory transcripts including Tnf, Ifng and Il23a, and an up‐regulated expression of anti‐inflammatory genes, such as Ctla4, Icos and Il22.86 The majority of Th17 cells in the murine intestinal LP are reactive against microbiota antigens, in particular to SFB.87, 88 GF mice or mice provided from the Jackson Laboratory (Bar Harbor, ME), which do not harbour SFB, have low numbers of Th17 cells in their gut LP.10, 11 Unlike other commensals that are mainly present in the mucus layer or intestinal lumen, SFB attach and penetrate into the epithelium of terminal ileum.11, 16 It is postulated that this unique property results in the induction of Th17 cells. In agreement with this hypothesis, epithelial cell adhesive bacteria Citrobacter rodentium, Escherichia coli O157, and a mixture of 20 bacterial strains isolated from human gut with properties of epithelial adherence were shown to induce gut Th17 cells in mice.89 Although it is still not entirely clear where and how the induction of Th17 cells happens, several groups have shown that this process primarily happens de novo in the small intestine and requires MHC‐II expressing intestinal dendritic cells and macrophages.88, 90 Innate lymphoid 3 cells are also suggested to participate in the induction of SFB‐specific Th17 cells through secretion of IL‐22.91 SFB‐specific Th17 cells are found in the peripheral lymphoid organs beyond the intestine, which could potentially contribute to the vulnerability to some autoimmune diseases in genetically susceptible hosts.92, 93 In addition to previously proposed molecular mimicry and bystander activation theories as to the mechanism for microbiota‐reactive T cells to trigger autoimmunity,94, 95 a recent study in a lung autoimmune mouse model showed that SFB selectively expanded Th17 cells that expressed dual TCR, one of which recognized SFB peptide and the other directed at a self‐antigen.96
Compared with the induction of intestinal Treg and Th17 cells, much less is known about the role of specific commensal microbiota on Th1 and Th2 cell differentiation. Reduced abundance and diversity of Bacteroidetes were associated with a decreased Th1 response in infants delivered by caesarean section.97 Colonization of GF mice with the polysaccharide‐expressing bacteria B. fragilis corrected an imbalance between Th1 and Th2 cells.98 Listeria monocytogenes 99 and Toxoplasma gondii 100 infections drove an antigen‐specific Th1 response in the host. Colon‐resident Helicobacter spp. have dual roles for T‐cell responses, including pTreg induction at homeostasis, and driving differentiation of Foxp3 to effector T cells during colitis.101 The microbiota has effects on Tfh cell number and function, and conversely Tfh in turn can modulate the microbiota. Compared with SPF mice, GF mice have a substantial reduction of Tfh cells in the PPs.102 Programmed cell death‐1‐deficient mice, which lack Tfh cells, showed a significant reduction of anaerobic bacteria in the gut.103 Also, Tfh cells are able to sense bacterial ATP through the receptor P2X7, and in turn shape gut microbiota composition.104 SFB promotes PPs Tfh cell differentiation and dissemination into systemic sites, leading to an increased production of autoantibodies and worsened arthritis.93
B‐cell–gut microbiota interactions
T‐cell‐dependent and ‐independent B cells complete the other half of the adaptive immune system via production of antibodies that protect the host against microbial invasions.105 T‐cell‐dependent B‐cell production of antibodies has been linked to microbial antigen exposure. Although B cells are present in the gut‐associated lymphoid tissues, including PPs and mesenteric lymph nodes, before birth,42 microbiota antigens and microbial metabolites, such as short‐chain fatty acids, strongly promote plasma cell differentiation in both mucosal and systemic sites.106 IgA serves as the major form of secretory antibody present at the mucosal surface and so plays a critical role in maintaining gut homeostasis.107 Potential mechanisms include binding and prevention of uptake of microbial antigens in the lumen,79 bacterial disruption and agglutination,108, 109 enchaining of growing bacteria,110 and neutralization of pathogenic bacterial toxins.111, 112 Multiple mechanisms have been advanced to explain the establishment of mutualism between secretory IgA and gut microbiota. Secretory IgA can induce members of the microbiota such as Bacteroides thetaiotaomicron to down‐regulate the expression of pro‐inflammatory surface epitopes.113 Coating of some luminal bacteria by secretory IgA guides bacterial entry into the PPs, where a germinal centre response is induced and a positive loop of antigen‐specific IgA production is established.114 Microbial antigen recognition mediated by different MHC repertoires also contributes to altered IgA repertoires, which in turn modulates microbiota composition in the gut.115
Due to physical proximity, the gut microbiota greatly influence the production of intestinal IgA.13 Lack of intestinal microbial stimulation results in fewer numbers of IgA+ plasma cells in the gut and reduced abundance of IgA.14, 116, 117 This is possibly because of a compromised development of isolated lymphoid tissues – a major site for T‐cell‐independent IgA production.118 SFB potently promotes T‐cell‐independent IgA production through stimulation of postnatal development of isolated lymphoid tissues and tertiary lymphoid tissue in the gut.13, 14 A fraction of anti‐microbial IgA in the gut is polyreactive and generated from this high‐capacity low‐affinity pathway.119, 120, 121 However, the majority of intestinal IgA is T‐cell‐dependent, particularly that directed at bacterial protein antigens, and is part of a low‐capacity and high‐affinity pathway. This T‐dependent IgA mainly occurs in the PPs by B cells interacting with antigen‐loaded dendritic cells in a CCR6‐dependent manner.122, 123 Bacteria such as SFB and Mucispirillum sp. able to adhere to epithelial cells are potent inducers of T‐cell‐dependent IgA,120 probably by enhanced uptake of their antigens into dendritic cells. IgA‐producing B cells home to the intestinal LP, where IgA is produced and then transported across the epithelium into the gut lumen through polymeric immunoglobulin receptor expressed on the basolateral side of epithelial cells.124 Polymeric immunoglobulin receptor deficiency leads to the abrogation of IgA and IgM transcytosis, resulting in increased serum IgG antibodies against gut commensals and pathogens,125 demonstrating the important role of secretory antibodies in limiting systemic exposure to microbiota antigens.
Although studied for several decades, the role of gut microbiota in IgA induction was intensely investigated in the past few years. The application of flow cytometry with 16S rDNA sequencing, a technology referred as IgA‐Seq, has enabled the identification of IgA‐bound versus IgA‐unbound bacteria isolated from the gut. The IgA‐bound bacteria vary according to the study and the composition of the microbiota at different laboratories. The bacterial surface antigens bound by IgA in this approach are largely carbohydrates.121 Enrichment of E. coli and other Enterobacteriaceae was identified with high IgA coating in patients with Crohn's disease‐associated spondyloarthritis and diet‐dependent enteropathy, respectively.126, 127 Another study showed that colonization of GF mice with IgA‐coated bacteria from patients with inflammatory bowel disease, exacerbated dextran sulphate sodium‐induced colitis.128 However, the characterization of all IgA‐coated gut bacteria as ‘pathobionts’ is not warranted given the complexity of the microbiota and of bacteria–host interactions within the gut.
Gut IgA repertoires are highly diverse and distinct to individuals, which is even true in bacterial mono‐colonized mice.129 Using a reversible microbial colonization system in vivo, the production of antigen‐specific IgA did not require persistent bacterial colonization, and once an antigen‐specific B‐cell response was established in the gut, the magnitude of that IgA response was long‐lived unless new microbial species were encountered.117 As mice age, their IgA repertoires become more complex with new B‐cell clones continuously generated against new microbiota antigens; however, B‐cell clones induced in early life are also maintained, indicating a long‐lived memory B‐cell response.116 SPF mice have a much greater diversity of gut IgA repertoires than bacterial mono‐colonized mice or GF mice.129 These IgA‐switched memory B cells recirculate between multiple lymphoid tissues inside and outside the gut, and can differentiate to plasma cells in the mammary glands, which contributes to antibody production in the breast milk that helps protect and establish the microbiota in offspring gut.129
In addition to intestinal IgA, IgM and some IgG subclasses also bind gut microbiota, the majority of which are elicited through the T‐cell‐independent pathway.34, 130 In mouse, B1 cells serve as the major source for polyclonal low‐affinity anti‐commensal IgM responses as a primitive natural antibody response.130 However, in contrast to mice, humans have more abundant IgM+ plasma cells in the gut, which secrete IgM antibodies that help retain a diverse community of commensals in the mucus layer in synergy with IgA.131 Surprisingly, a considerable amount of IgG2b and IgG3 has been identified in the secretory compartment in the gut as well. The generation of these antibodies is dependent on Toll‐like receptor signalling through B cells but independent of T‐cell help, in that TCR‐βδ −/− mice have levels comparable to those of wild‐type mice.34 The IgD isotype is rare compared with other antibody isotypes, but recently it was shown that IgD class switch recombination happens preferentially in mucosal sites and is dependent on a diversified gut microbiota.132
Gut microbiota‐adaptive immunity interplay in immunological disorders, and potential therapeutic applications of microbiota modulation in disease interventions
Immune recognition of gut microbiota is shown to trigger multiple inflammatory disorders. For example, both mucosal and serum antibodies targeting gut commensal antigens are elevated in individuals with Crohn's disease and those with ulcerative colitis.133, 134, 135, 136 Infiltrating T cells in the gut of Crohn's disease patients react to luminal microbial antigens and produce pro‐inflammatory cytokines including tumour necrosis factor‐α, interferon‐γ and IL‐17A.137, 138, 139 Studies from our group and others have shown that flagellin of gut commensal bacteria, specifically bacteria belonging to Lachnospiraceae family of the Clostridiales, serve as immunodominant antigens in both experimental mouse colitis and in Crohn's patients.134, 140 CD4 T cells reactive to these flagellins can be activated during enteric pathogenic infections and form memory cells that persists in the host,141 then serve as a potential reservoir for pathogenic effector cells or regulatory cells when they are reactivated later. Adoptive transfer of naive CD4 cells from CBir1 TCR transgenic mice into immunodeficient mice induces severe colitis, which is dependent on the colonization of CBir1 flagellin‐expressing bacteria in the gut of the recipient mice.142 Lachnospiraceae A4, which has flagellins similar to CBir1, was shown to induce transforming growth factor‐β production in dendritic cells and so inhibit Th2 cell differentiation.143 In agreement with this, CBir1 TCR transgenic CD4 T cells isolated from inflamed colon of adoptive recipients have a strong profile of interferon‐γ and IL‐17A expression, indicating that this colitis is mainly driven by flagellin‐reactive Th1 and Th17 cells.142
Autoimmune diseases that are outside the gut can be triggered by the enteric microbiota. It has been shown that Prevotella copri colonization activates autoreactive T cells in the gut and correlates with increased susceptibility of rheumatoid arthritis.144, 145 Prevotella copri‐specific IgG and IgA were found in subgroups of individuals with new‐onset rheumatoid arthritis but not in disease or healthy controls. Gut microbiota is also needed for the induction and acceleration of experimental autoimmune encephalomyelitis (EAE) in the relapsing–remitting mouse model, via activation of myelin oligodendrocyte glycoprotein‐specific T cells and recruitment of autoantibody‐producing B cells into the brain‐draining lymph nodes.146 Human studies found that Akkermansia muciniphila and Acinetobacter calcoaceticus were expanded in untreated multiple sclerosis (MS) patients, and colonization of germ‐free myelin oligodendrocyte glycoprotein T‐cell transgenic mice or wild‐type mice with MS patient gut microbiota increased the incidence and disease severity of EAE, with a compromised Treg differentiation in the mesenteric lymph nodes.147
Gut dysbiosis is commonly associated with autoimmune disorders and metabolic syndromes in both mice and humans. Microbiome dysbiosis is defined as an altered gut microbiota composition together with a functional change in the microbial transcriptome, proteome or metabolome. However, a causal relationship between dysbiosis and the change of host immune response remains difficult to determine and awaits further definitive studies. Garrett et al.148 provided the first solid evidence that a disrupted consortium of gut microbiota in T‐bet‐deficient Rag−/− mice could be transmitted to wild‐type mice, and induce colitis in the latter. Studies have been performed to seek the relationship between dysbiosis and inflammatory bowel disease in humans as well. In a large cohort, an alteration of gut microbiota in treatment‐naive new‐onset Crohn's disease patients was identified; a set of taxa was identified with strong correlation with disease status, and antibiotic treatment further amplified the dysbiosis.149 Similarly, dysbiosis is observed in patients with ulcerative colitis, with a reduction of Roseburia hominis and the anti‐inflammatory bacteria Faecalibacterium prausnitzii 150 compared with healthy controls.151
Immune cells primed in the gut under the influence of an altered gut microbiota can contribute to the pathogenesis of systemic autoimmune diseases and can alter the outcome of cancer immunotherapy. Certain bacterial colonization, such as SFB in EAE and autoimmune arthritis, and Prevotella copri in murine autoimmune arthritis, are linked to increased disease susceptibility.92, 145, 152 Gut microbiota was shown to participate in the modulation of anti‐tumour immunotherapies as well, through the regulation of responses of different T‐cell subsets.153, 154, 155, 156 Therefore, normalization of the gut microbiota of patients with inflammatory bowel disease or autoimmune diseases is increasingly suggested as an approach of disease intervention. So far, the treatment of Clostridium difficile infection with faecal microbiota transplantation has been remarkably effective.157 However, the human gut microbiota is such a complex entity that it is under the influence of various factors, including host genetics, diet, environmental effects, antibiotics and competitions/interactions among members of the microbiota. With a small number of defined bacteria that are able to regulate the host immune responses, and countless unknown participants to be identified, further definitive evidence is required for a broader utilization of faecal microbiota transplantation and/or other proposed microbial modulation approaches such as probiotics and prebiotics in the treatment of dysbiosis‐associated diseases.
Disclosures
The authors have no competing interests.
References
- 1. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 2016; 14:e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C et al A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010; 464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol 2014; 14:405–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Virgin HW. The virome in mammalian physiology and disease. Cell 2014; 157:142–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Human Microbiome Project Consortium . Structure, function and diversity of the healthy human microbiome. Nature 2012; 486:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 2016; 14:20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xiao L, Feng Q, Liang S, Sonne SB, Xia Z, Qiu X et al A catalog of the mouse gut metagenome. Nat Biotechnol 2015; 33:1103–8. [DOI] [PubMed] [Google Scholar]
- 8. Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB et al Gut immune maturation depends on colonization with a host‐specific microbiota. Cell 2012; 149:1578–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thompson CL, Vier R, Mikaelyan A, Wienemann T, Brune A. ‘Candidatus Arthromitus’ revised: segmented filamentous bacteria in arthropod guts are members of Lachnospiraceae . Environ Microbiol 2012; 14:1454–65. [DOI] [PubMed] [Google Scholar]
- 10. Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB et al Specific microbiota direct the differentiation of IL‐17‐producing T‐helper cells in the mucosa of the small intestine. Cell Host Microbe 2008; 4:337–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U et al Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009; 139:485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gaboriau‐Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C et al The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 2009; 31:677–89. [DOI] [PubMed] [Google Scholar]
- 13. Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun 1999; 67:1992–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lecuyer E, Rakotobe S, Lengline‐Garnier H, Lebreton C, Picard M, Juste C et al Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity 2014; 40:608–20. [DOI] [PubMed] [Google Scholar]
- 15. Prakash T, Oshima K, Morita H, Fukuda S, Imaoka A, Kumar N et al Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of Th17 cell differentiation. Cell Host Microbe 2011; 10:273–84. [DOI] [PubMed] [Google Scholar]
- 16. Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C et al The genome of th17 cell‐inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe 2011; 10:260–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature 2016; 535:65–74. [DOI] [PubMed] [Google Scholar]
- 18. Artis D, Spits H. The biology of innate lymphoid cells. Nature 2015; 517:293–301. [DOI] [PubMed] [Google Scholar]
- 19. Roths JB, Marshall JD, Allen RD, Carlson GA, Sidman CL. Spontaneous Pneumocystis carinii pneumonia in immunodeficient mutant SCID mice. Natural history and pathobiology. Am J Pathol 1990; 136:1173–86. [PMC free article] [PubMed] [Google Scholar]
- 20. Okoye AA, Picker LJ. CD4+ T‐cell depletion in HIV infection: mechanisms of immunological failure. Immunol Rev 2013; 254:54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature 2007; 449:819–26. [DOI] [PubMed] [Google Scholar]
- 22. Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science 1996; 272:54–60. [DOI] [PubMed] [Google Scholar]
- 23. Dominguez‐Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N et al Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA 2010; 107:11971–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med 2014; 6:237ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep 2016; 6:23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lauder AP, Roche AM, Sherrill‐Mix S, Bailey A, Laughlin AL, Bittinger K et al Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome 2016; 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yoshioka H, Iseki K, Fujita K. Development and differences of intestinal flora in the neonatal period in breast‐fed and bottle‐fed infants. Pediatrics 1983; 72:317–21. [PubMed] [Google Scholar]
- 28. Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I et al Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 2006; 118:511–21. [DOI] [PubMed] [Google Scholar]
- 29. Gupta VK, Paul S, Dutta C. Geography, ethnicity or subsistence‐specific variations in human microbiome composition and diversity. Front Microbiol 2017; 8:1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg‐Lyons D et al Cohabiting family members share microbiota with one another and with their dogs. Elife 2013; 2:e00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science 2016; 352:539–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dominguez‐Bello MG, De Jesus‐Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A et al Partial restoration of the microbiota of cesarean‐born infants via vaginal microbial transfer. Nat Med 2016; 22:250–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A et al Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr 2017; 171:647–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koch MA, Reiner GL, Lugo KA, Kreuk LS, Stanbery AG, Ansaldo E et al Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell 2016; 165:827–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rogier EW, Frantz AL, Bruno ME, Wedlund L, Cohen DA, Stromberg AJ et al Secretory antibodies in breast milk promote long‐term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc Natl Acad Sci USA 2014; 111:3074–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Laursen MF, Bahl MI, Michaelsen KF, Licht TR. First foods and gut microbes. Front Microbiol 2017; 8:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S et al Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 2010; 107:14691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature 2012; 489:220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R et al Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA 2011; 108(Suppl 1):4578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez‐Bello MG, Contreras M et al Human gut microbiome viewed across age and geography. Nature 2012; 486:222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schloss PD, Schubert AM, Zackular JP, Iverson KD, Young VB, Petrosino JF. Stabilization of the murine gut microbiome following weaning. Gut Microbes 2012; 3:383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van de Pavert SA, Mebius RE. New insights into the development of lymphoid tissues. Nat Rev Immunol 2010; 10:664–74. [DOI] [PubMed] [Google Scholar]
- 43. Eberl G, Lochner M. The development of intestinal lymphoid tissues at the interface of self and microbiota. Mucosal Immunol 2009; 2:478–85. [DOI] [PubMed] [Google Scholar]
- 44. Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012; 489:231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early‐life colonization shapes long‐term IgE levels. Cell Host Microbe 2013; 14:559–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A et al Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012; 336:489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk‐Hasdemir D et al Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 2014; 156:123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Keeney KM, Yurist‐Doutsch S, Arrieta MC, Finlay BB. Effects of antibiotics on human microbiota and subsequent disease. Annu Rev Microbiol 2014; 68:217–35. [DOI] [PubMed] [Google Scholar]
- 49. Zanvit P, Konkel JE, Jiao X, Kasagi S, Zhang D, Wu R et al Antibiotics in neonatal life increase murine susceptibility to experimental psoriasis. Nat Commun 2015; 6:8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK et al Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci USA 2014; 111:13145–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D et al Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 2016; 22:1187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Murk W, Risnes KR, Bracken MB. Prenatal or early‐life exposure to antibiotics and risk of childhood asthma: a systematic review. Pediatrics 2011; 127:1125–38. [DOI] [PubMed] [Google Scholar]
- 53. Torow N, Yu K, Hassani K, Freitag J, Schulz O, Basic M et al Active suppression of intestinal CD4+TCRαβ + T‐lymphocyte maturation during the postnatal period. Nat Commun 2015; 6:7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nutsch K, Chai JN, Ai TL, Russler‐Germain E, Feehley T, Nagler CR et al Rapid and efficient generation of regulatory T cells to commensal antigens in the periphery. Cell Rep 2016; 17:206–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Knoop KA, Gustafsson JK, McDonald KG, Kulkarni DH, Coughlin PE, McCrate S et al Microbial antigen encounter during a preweaning interval is critical for tolerance to gut bacteria. Sci Immunol 2017; 2:eaao1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Smits SA, Leach J, Sonnenburg ED, Gonzalez CG, Lichtman JS, Reid G et al Seasonal cycling in the gut microbiome of the Hadza hunter‐gatherers of Tanzania. Science 2017; 357:802–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Suez J, Korem T, Zeevi D, Zilberman‐Schapira G, Thaiss CA, Maza O et al Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature 2014; 514:181–6. [DOI] [PubMed] [Google Scholar]
- 58. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA et al Linking long‐term dietary patterns with gut microbial enterotypes. Science 2011; 334:105–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see). Nat Rev Immunol 2014; 14:377–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N et al Peripheral education of the immune system by colonic commensal microbiota. Nature 2011; 478:250–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Haribhai D, Williams JB, Jia S, Nickerson D, Schmitt EG, Edwards B et al A requisite role for induced regulatory T cells in tolerance based on expanding antigen receptor diversity. Immunity 2011; 35:109–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol 2012; 30:531–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vieira PL, Christensen JR, Minaee S, O'Neill EJ, Barrat FJ, Boonstra A et al IL‐10‐secreting regulatory T cells do not express Foxp3 but have comparable regulatory function to naturally occurring CD4+CD25+ regulatory T cells. J Immunol 2004; 172:5986–93. [DOI] [PubMed] [Google Scholar]
- 64. Cervantes‐Barragan L, Chai JN, Tianero MD, Di Luccia B, Ahern PP, Merriman J et al Lactobacillus reuteri induces gut intraepithelial CD4+CD8αα + T cells. Science 2017; 357:806–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H et al Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013; 500:232–6. [DOI] [PubMed] [Google Scholar]
- 66. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y et al Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011; 331:337–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T‐cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA 2010; 107:12204–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM et al The microbial metabolites, short‐chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013; 341:569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D et al Commensal microbe‐derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013; 504:446–50. [DOI] [PubMed] [Google Scholar]
- 70. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P et al Metabolites produced by commensal bacteria promote peripheral regulatory T‐cell generation. Nature 2013; 504:451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chu H, Khosravi A, Kusumawardhani IP, Kwon AH, Vasconcelos AC, Cunha LD et al Gene‐microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 2016; 352:1116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ochoa‐Reparaz J, Mielcarz DW, Wang Y, Begum‐Haque S, Dasgupta S, Kasper DL et al A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol 2010; 3:487–95. [DOI] [PubMed] [Google Scholar]
- 73. Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y et al Expression of Helios, an Ikaros transcription factor family member, differentiates thymic‐derived from peripherally induced Foxp3+ T regulatory cells. J Immunol 2010; 184:3433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Weiss JM, Bilate AM, Gobert M, Ding Y, Curotto de Lafaille MA, Parkhurst CN et al Neuropilin 1 is expressed on thymus‐derived natural regulatory T cells, but not mucosa‐generated induced Foxp3+ T reg cells. J Exp Med 2012; 209:1723–42, S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yadav M, Louvet C, Davini D, Gardner JM, Martinez‐Llordella M, Bailey‐Bucktrout S et al Neuropilin‐1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo . J Exp Med 2012; 209:1713–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Singh B, Read S, Asseman C, Malmstrom V, Mottet C, Stephens LA et al Control of intestinal inflammation by regulatory T cells. Immunol Rev 2001; 182:190–200. [DOI] [PubMed] [Google Scholar]
- 77. Gagliani N, Amezcua Vesely MC, Iseppon A, Brockmann L, Xu H, Palm NW et al Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature 2015; 523:221–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang S, Charbonnier LM, Noval Rivas M, Georgiev P, Li N, Gerber G et al MyD88 adaptor‐dependent microbial sensing by regulatory T cells promotes mucosal tolerance and enforces commensalism. Immunity 2015; 43:289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell‐IgA response to the intestinal microbiota. Proc Natl Acad Sci USA 2009; 106:19256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y et al Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature 2012; 482:395–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Schmidt A, Oberle N, Krammer PH. Molecular mechanisms of Treg‐mediated T cell suppression. Front Immunol 2012; 3:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tsuji M, Komatsu N, Kawamoto S, Suzuki K, Kanagawa O, Honjo T et al Preferential generation of follicular B helper T cells from Foxp3+ T cells in gut Peyer's patches. Science 2009; 323:1488–92. [DOI] [PubMed] [Google Scholar]
- 83. Kawamoto S, Maruya M, Kato LM, Suda W, Atarashi K, Doi Y et al Foxp3+ T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 2014; 41:152–65. [DOI] [PubMed] [Google Scholar]
- 84. Korn T, Bettelli E, Oukka M, Kuchroo VK. IL‐17 and Th17 cells. Annu Rev Immunol 2009; 27:485–517. [DOI] [PubMed] [Google Scholar]
- 85. Hueber W, Sands BE, Lewitzky S, Vandemeulebroecke M, Reinisch W, Higgins PD et al Secukinumab, a human anti‐IL‐17A monoclonal antibody, for moderate to severe Crohn's disease: unexpected results of a randomised, double‐blind placebo‐controlled trial. Gut 2012; 61:1693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhao Q, Harbour SN, Kolde R, Latorre IJ, Tun HM, Schoeb TR et al Selective induction of homeostatic Th17 cells in the murine intestine by cholera toxin interacting with the microbiota. J Immunol 2017; 199:312–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL et al Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 2014; 510:152–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG et al Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 2014; 40:594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S et al Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 2015; 163:367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Panea C, Farkas AM, Goto Y, Abdollahi‐Roodsaz S, Lee C, Koscso B et al Intestinal monocyte‐derived macrophages control commensal‐specific Th17 responses. Cell Rep 2015; 12:1314–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ et al An IL‐23R/IL‐22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell 2015; 163:381–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T‐cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA 2011; 108(Suppl 1):4615–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Teng F, Klinger CN, Felix KM, Bradley CP, Wu E, Tran NL et al Gut microbiota drive autoimmune arthritis by promoting differentiation and migration of Peyer's patch T follicular helper cells. Immunity 2016; 44:875–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Albert LJ, Inman RD. Molecular mimicry and autoimmunity. N Engl J Med 1999; 341:2068–74. [DOI] [PubMed] [Google Scholar]
- 95. Munz C, Lunemann JD, Getts MT, Miller SD. Antiviral immune responses: triggers of or triggered by autoimmunity? Nat Rev Immunol 2009; 9:246–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Bradley CP, Teng F, Felix KM, Sano T, Naskar D, Block KE et al Segmented filamentous bacteria provoke lung autoimmunity by inducing gut‐lung axis Th17 cells expressing dual TCRs. Cell Host Microbe 2017; 22:697–704 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C et al Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 2014; 63:559–66. [DOI] [PubMed] [Google Scholar]
- 98. Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005; 122:107–18. [DOI] [PubMed] [Google Scholar]
- 99. Hsieh CS, Macatonia SE, Tripp CS, Wolf SF, O'Garra A, Murphy KM. Development of Th1 CD4+ T cells through IL‐12 produced by Listeria‐induced macrophages. Science 1993; 260:547–9. [DOI] [PubMed] [Google Scholar]
- 100. Benson A, Pifer R, Behrendt CL, Hooper LV, Yarovinsky F. Gut commensal bacteria direct a protective immune response against Toxoplasma gondii . Cell Host Microbe 2009; 6:187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Chai JN, Peng Y, Rengarajan S, Solomon BD, Ai TL, Shen Z et al Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Sci Immunol 2017; 2:eaal5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kubinak JL, Petersen C, Stephens WZ, Soto R, Bake E, O'Connell RM et al MyD88 signaling in T cells directs IgA‐mediated control of the microbiota to promote health. Cell Host Microbe 2015; 17:153–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kawamoto S, Tran TH, Maruya M, Suzuki K, Doi Y, Tsutsui Y et al The inhibitory receptor PD‐1 regulates IgA selection and bacterial composition in the gut. Science 2012; 336:485–9. [DOI] [PubMed] [Google Scholar]
- 104. Perruzza L, Gargari G, Proietti M, Fosso B, D'Erchia AM, Faliti CE et al T follicular helper cells promote a beneficial gut ecosystem for host metabolic homeostasis by sensing microbiota‐derived extracellular ATP. Cell Rep 2017; 18:2566–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Hou L, Sasakj H, Stashenko P. B‐cell deficiency predisposes mice to disseminating anaerobic infections: protection by passive antibody transfer. Infect Immun 2000; 68:5645–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kim M, Qie Y, Park J, Kim CH. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe 2016; 20:202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Mantis NJ, Rol N, Corthesy B. Secretory IgA's complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol 2011; 4:603–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Hendrickx AP, Top J, Bayjanov JR, Kemperman H, Rogers MR, Paganelli FL et al Antibiotic‐driven dysbiosis mediates intraluminal agglutination and alternative segregation of Enterococcus faecium from the intestinal epithelium. MBio 2015; 6:e01346–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Levinson KJ, De Jesus M, Mantis NJ. Rapid effects of a protective O‐polysaccharide‐specific monoclonal IgA on Vibrio cholerae agglutination, motility, and surface morphology. Infect Immun 2015; 83:1674–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Moor K, Diard M, Sellin ME, Felmy B, Wotzka SY, Toska A et al High‐avidity IgA protects the intestine by enchaining growing bacteria. Nature 2017; 544:498–502. [DOI] [PubMed] [Google Scholar]
- 111. Johnson S, Sypura WD, Gerding DN, Ewing SL, Janoff EN. Selective neutralization of a bacterial enterotoxin by serum immunoglobulin A in response to mucosal disease. Infect Immun 1995; 63:3166–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Tokuhara D, Yuki Y, Nochi T, Kodama T, Mejima M, Kurokawa S et al Secretory IgA‐mediated protection against V. cholerae and heat‐labile enterotoxin‐producing enterotoxigenic Escherichia coli by rice‐based vaccine. Proc Natl Acad Sci USA 2010; 107:8794–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2007; 2:328–39. [DOI] [PubMed] [Google Scholar]
- 114. Fransen F, Zagato E, Mazzini E, Fosso B, Manzari C, El Aidy S et al BALB/c and C57BL/6 mice differ in polyreactive IgA abundance, which impacts the generation of antigen‐specific iga and microbiota diversity. Immunity 2015; 43:527–40. [DOI] [PubMed] [Google Scholar]
- 115. Kubinak JL, Stephens WZ, Soto R, Petersen C, Chiaro T, Gogokhia L et al MHC variation sculpts individualized microbial communities that control susceptibility to enteric infection. Nat Commun 2015; 6:8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Lindner C, Wahl B, Fohse L, Suerbaum S, Macpherson AJ, Prinz I et al Age, microbiota, and T cells shape diverse individual IgA repertoires in the intestine. J Exp Med 2012; 209:365–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M et al Reversible microbial colonization of germ‐free mice reveals the dynamics of IgA immune responses. Science 2010; 328:1705–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Tsuji M, Suzuki K, Kitamura H, Maruya M, Kinoshita K, Ivanov II et al Requirement for lymphoid tissue‐inducer cells in isolated follicle formation and T cell‐independent immunoglobulin A generation in the gut. Immunity 2008; 29:261–71. [DOI] [PubMed] [Google Scholar]
- 119. Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell‐independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 2000; 288:2222–6. [DOI] [PubMed] [Google Scholar]
- 120. Bunker JJ, Flynn TM, Koval JC, Shaw DG, Meisel M, McDonald BD et al Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity 2015; 43:541–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Bunker JJ, Erickson SA, Flynn TM, Henry C, Koval JC, Meisel M et al Natural polyreactive IgA antibodies coat the intestinal microbiota. Science 2017; 358:eaan6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 2004; 303:1662–5. [DOI] [PubMed] [Google Scholar]
- 123. Reboldi A, Arnon TI, Rodda LB, Atakilit A, Sheppard D, Cyster JG. IgA production requires B cell interaction with subepithelial dendritic cells in Peyer's patches. Science 2016; 352:aaf4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Song W, Bomsel M, Casanova J, Vaerman JP, Mostov K. Stimulation of transcytosis of the polymeric immunoglobulin receptor by dimeric IgA. Proc Natl Acad Sci USA 1994; 91:163–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Johansen FE, Pekna M, Norderhaug IN, Haneberg B, Hietala MA, Krajci P et al Absence of epithelial immunoglobulin A transport, with increased mucosal leakiness, in polymeric immunoglobulin receptor/secretory component‐deficient mice. J Exp Med 1999; 190:915–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Viladomiu M, Kivolowitz C, Abdulhamid A, Dogan B, Victorio D, Castellanos JG et al IgA‐coated E. coli enriched in Crohn's disease spondyloarthritis promote TH17‐dependent inflammation. Sci Transl Med 2017; 9:eaaf9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kau AL, Planer JD, Liu J, Rao S, Yatsunenko T, Trehan I et al Functional characterization of IgA‐targeted bacterial taxa from undernourished Malawian children that produce diet‐dependent enteropathy. Sci Transl Med 2015; 7:276ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L et al Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014; 158:1000–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Lindner C, Thomsen I, Wahl B, Ugur M, Sethi MK, Friedrichsen M et al Diversification of memory B cells drives the continuous adaptation of secretory antibodies to gut microbiota. Nat Immunol 2015; 16:880–8. [DOI] [PubMed] [Google Scholar]
- 130. Ehrenstein MR, Notley CA. The importance of natural IgM: scavenger, protector and regulator. Nat Rev Immunol 2010; 10:778–86. [DOI] [PubMed] [Google Scholar]
- 131. Magri G, Comerma L, Pybus M, Sintes J, Llige D, Segura‐Garzon D et al Human secretory IgM emerges from plasma cells clonally related to gut memory B cells and targets highly diverse commensals. Immunity 2017; 47:118–34 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Choi JH, Wang KW, Zhang D, Zhan X, Wang T, Bu CH et al IgD class switching is initiated by microbiota and limited to mucosa‐associated lymphoid tissue in mice. Proc Natl Acad Sci USA 2017; 114:E1196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Macpherson A, Khoo UY, Forgacs I, Philpott‐Howard J, Bjarnason I. Mucosal antibodies in inflammatory bowel disease are directed against intestinal bacteria. Gut 1996; 38:365–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Lodes MJ, Cong Y, Elson CO, Mohamath R, Landers CJ, Targan SR et al Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest 2004; 113:1296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Christmann BS, Abrahamsson TR, Bernstein CN, Duck LW, Mannon PJ, Berg G et al Human seroreactivity to gut microbiota antigens. J Allergy Clin Immunol 2015; 136:1378–86 e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Saxon A, Shanahan F, Landers C, Ganz T, Targan S. A distinct subset of antineutrophil cytoplasmic antibodies is associated with inflammatory bowel disease. J Allergy Clin Immunol. 1990; 86:202–10. [DOI] [PubMed] [Google Scholar]
- 137. Calderon‐Gomez E, Bassolas‐Molina H, Mora‐Buch R, Dotti I, Planell N, Esteller M et al Commensal‐specific CD4+ cells from patients with Crohn's disease have a T‐helper 17 inflammatory profile. Gastroenterology 2016; 151:489–500 e3. [DOI] [PubMed] [Google Scholar]
- 138. Pirzer U, Schonhaar A, Fleischer B, Hermann E, Meyer zum Buschenfelde KH. Reactivity of infiltrating T lymphocytes with microbial antigens in Crohn's disease. Lancet 1991; 338:1238–9. [DOI] [PubMed] [Google Scholar]
- 139. Hegazy AN, West NR, Stubbington MJT, Wendt E, Suijker KIM, Datsi A et al Circulating and tissue‐resident CD4+ T cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology 2017; 153:1320–37 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Duck LW, Walter MR, Novak J, Kelly D, Tomasi M, Cong Y et al Isolation of flagellated bacteria implicated in Crohn's disease. Inflamm Bowel Dis 2007; 13:1191–201. [DOI] [PubMed] [Google Scholar]
- 141. Hand TW, Dos Santos LM, Bouladoux N, Molloy MJ, Pagan AJ, Pepper M et al Acute gastrointestinal infection induces long‐lived microbiota‐specific T cell responses. Science 2012; 337:1553–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Feng T, Wang L, Schoeb TR, Elson CO, Cong Y. Microbiota innate stimulation is a prerequisite for T cell spontaneous proliferation and induction of experimental colitis. J Exp Med 2010; 207:1321–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Wu W, Liu HP, Chen F, Liu H, Cao AT, Yao S et al Commensal A4 bacteria inhibit intestinal Th2‐cell responses through induction of dendritic cell TGF‐β production. Eur J Immunol 2016; 46:1162–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Pianta A, Arvikar S, Strle K, Drouin EE, Wang Q, Costello CE et al Evidence of the immune relevance of Prevotella copri, a gut microbe, in patients with rheumatoid arthritis. Arthritis Rheumatol 2017; 69:964–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C et al Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013; 2:e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C et al Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 2011; 479:538–41. [DOI] [PubMed] [Google Scholar]
- 147. Berer K, Gerdes LA, Cekanaviciute E, Jia X, Xiao L, Xia Z et al Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Natl Acad Sci USA 2017; 114:10719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Garrett WS, Lord GM, Punit S, Lugo‐Villarino G, Mazmanian SK, Ito S et al Communicable ulcerative colitis induced by T‐bet deficiency in the innate immune system. Cell 2007; 131:33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Gevers D, Kugathasan S, Denson LA, Vazquez‐Baeza Y, Van Treuren W, Ren B et al The treatment‐naive microbiome in new‐onset Crohn's disease. Cell Host Microbe 2014; 15:382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez‐Humaran LG, Gratadoux JJ et al Faecalibacterium prausnitzii is an anti‐inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 2008; 105:16731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V et al A decrease of the butyrate‐producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014; 63:1275–83. [DOI] [PubMed] [Google Scholar]
- 152. Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y et al Gut‐residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 2010; 32:815–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Sivan A, Corrales L, Hubert N, Williams JB, Aquino‐Michaels K, Earley ZM et al Commensal Bifidobacterium promotes antitumor immunity and facilitates anti‐PD‐L1 efficacy. Science 2015; 350:1084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C et al Anticancer immunotherapy by CTLA‐4 blockade relies on the gut microbiota. Science 2015; 350:1079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV et al Gut microbiome modulates response to anti‐PD‐1 immunotherapy in melanoma patients. Science 2017; 359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R et al Gut microbiome influences efficacy of PD‐1‐based immunotherapy against epithelial tumors. Science 2017; 359:91–7. [DOI] [PubMed] [Google Scholar]
- 157. van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM et al Duodenal infusion of donor feces for recurrent Clostridium difficile . N Engl J Med 2013; 368:407–15. [DOI] [PubMed] [Google Scholar]


