Abstract
During the demographic history of the Pan clade, there has been gene-flow between species, likely >200,000 years ago. Bonobo haplotypes in three subspecies of chimpanzee have been identified to be segregating in modern-day chimpanzee populations, suggesting that these haplotypes, with increased differentiation, may be a target of natural selection. Here, we investigate signatures of adaptive introgression within the bonobo-like haplotypes in chimpanzees using site frequency spectrum-based tests. We find evidence for subspecies-specific adaptations in introgressed regions involved with male reproduction in central chimpanzees, the immune system in eastern chimpanzees, female reproduction and the nervous system in Nigeria-Cameroon chimpanzees. Furthermore, our results indicate signatures of balancing selection in some of the putatively introgressed regions. This might be the product of long-term balancing selection resulting in a similar genomic signature as introgression, or possibly balancing selection acting on alleles reintroduced through gene flow.
Keywords: chimpanzee, introgression, natural selection, balancing selection
Introduction
Introgression events, or the admixture between two closely related species has been described previously in plants and animals (Arnold and Martin 2009), between humans and Neanderthals (Smith et al. 2005; Green et al. 2010), Denisovans (Reich et al. 2011), and an unknown hominin (Mondal et al. 2016), and most recently between chimpanzees and bonobos (de Manuel et al. 2016). After an introgression event occurs, two scenarios, outside of neutrality, are possible. Either the new haplotype is not beneficial to the new species or the new variants may allow for some adaptive advantage. The first case, which is more common, tends to be removed through purifying selection or drift. This was described both for introgressed segments in the human genome and for introgressed segments in the chimpanzee genome (Sankararaman et al. 2014; de Manuel et al. 2016; Juric et al. 2016; Kuhlwilm et al. 2016). The second, rare case, describes adaptive introgression, where variants introduced through introgression will increase in frequency through adaptive selection.
Exploring the role that introgression plays in the evolution of a species has been limited (Arnold and Martin 2009; Hedrick 2013). However, some examples of adaptive advantage exist; for instance, the increased resistance to herbivores in the sunflower (Whitney et al. 2006), or warfarin resistance in mice (Song et al. 2011). Within humans, introgression from Denisovans allowed for adaptation to high altitudes in Tibetans (Huerta-Sanchez et al. 2014) and Neanderthal admixture benefits to the immune response (Dannemann et al. 2016; Deschamps et al. 2016).
Here, we present the first study of selection specifically in introgressed regions of the chimpanzee genome, using the results from de Manuel et al. (2016). After a selection scan in the introgressed regions, an enrichment test is used in order to investigate possible phenotypes selected, identifying candidate genes that may have an adaptive advantage. Conversely, we also discuss the possibility that a few of the introgressed regions may indicate the presence of long-term balancing selection. Overall, we discuss the possibility that introgression allows for natural selection to act on a highly differentiated fraction of the genome.
Materials and Methods
Full genome sequences from 18 P.t. troglodytes, 19 P.t. schweinfurthii, 10 P.t. ellioti, and 10 bonobos were obtained from a previous study (Prado-Martinez et al. 2013); as well as the coordinates of introgressed regions (de Manuel et al. 2016). We calculated Tajima’s D (Tajima 1989), Fu and Li’s D and F (Fu and Li 1993) for the whole genome of each population. All statistics were calculated in 30-kb windows with a 3-kb sliding window. Windows were masked if less than five SNPs were present, following Pybus et al. (2014, 2015). The windows were deemed as introgressed if any part was within an introgressed region.
Full methods available in the Supplementary Material online.
Results
Following de Manuel et al. (2016), 54.7 Mb of the chimpanzee genome are detected with a signature of bonobo introgression in any non-P.t. verus individual. These segments are significantly depleted of genic content (P < 2.2 × 10−16, Wilcoxon rank test), that is, they contain fewer protein-coding bases than found in random regions, similar to patterns of gene flow in the human lineage (Sankararaman et al. 2014). However, some protein coding substitutions are present (supplementary table S1, supplementary information 1.1, Supplementary Material online).
Selection Scans of the Introgressed Regions
As background selection will remove the majority of introgressed haplotypes, in order to test the strength of background selection, we compared the B scores (McVicker et al. 2009) of the introgressed segments as compared with random regions across the genome. In introgressed fragments, these scores are significantly higher (P < 2.2 × 10−16, Wilcoxon rank test), showing weaker background selection. This suggests that bonobo alleles were more often tolerated in neutrally evolving regions of the genome, analogous to Neanderthal haplotypes in modern humans (Sankararaman et al. 2016; Vernot et al. 2016).
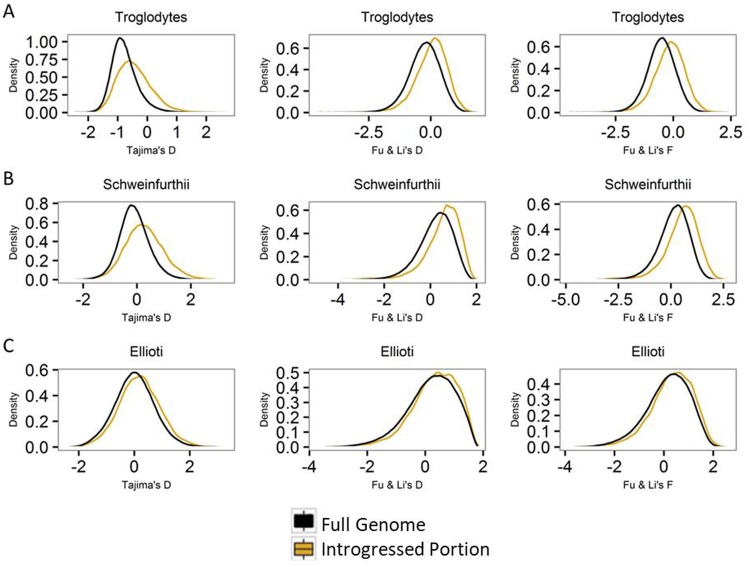
To detect adaptive evolution, three selection tests (Tajima’s D, Tajima 1989 and Fu and Li’s D and F, Fu and Li 1993) based on site frequency spectrum (SFS) analysis were applied. When comparing the genome-wide distribution of the SFS-based statistics with the introgressed portion, in all cases, the three selection tests are shifted toward positive values within the introgressed regions (fig. 1). A deviation in the positive direction indicates a higher proportion of intermediate frequency variants, as seen in the genome-wide distribution of the SFS (supplementary fig. S1, Supplementary Material online). This shift toward positive values cannot be explained by sampling (supplementary fig. S2, supplementary information 1.2, Supplementary Material online). The shift in SFS values may be due either to balancing selection or what is expected under introgression, and may be difficult to discern between the two without additional information (see below).
Fig. 1.
—Statistical distribution of Tajima’s D, Fu and Li’s D and F. Black represents the whole genome and yellow represents the introgressed regions. (A) Pan troglodytes troglodytes. (B) Pan troglodytes schweinfurthii. (C) Pan troglodytes ellioti.
In order to detect the genes (and the possible underlying functions) that have been under selection, we use an outlier approach in the introgressed regions of each of the subspecies, selecting the 25 minimum (regions likely under a selective sweep) and maximum (regions under adaptive introgression or balancing selection) regions for each statistic (supplementary tables S2–S7, Supplementary Material online). It is important to note here, that when compared with the whole genome, the extreme introgressed regions fall into the top 0.7% for the positive tail and the bottom 0.4% for the negative tail.
Positively Selected Introgression—Negative Tail and Tissue Enrichment
For each of the subspecies, we have a list of genes in or closest to the introgressed regions (supplementary tables S2–S4, Supplementary Material online) that have extreme negative values for the three selection tests and thus are very likely to have evolved under positive selection. We first performed an enrichment analysis with DEPICT (Pers et al. 2015) in order to relate the set of genes to specific tissues and likely, to a phenotype. We then explore the genes in or closest to each region through a literature search to identify genes with a known association either by function, by expression (among top three median expression when comparing across all tissues as obtained by GTEx; GTEx Consortium 2017), or likely inclusion in functional pathways as obtained by the PathCards database (Belinky et al. 2015).
For P.t. troglodytes (supplementary table S2, Supplementary Material online), 25 extreme negative regions for each statistic were selected, and due to overlaps between statistics, 38 total regions were considered with 35 unique genes. We find two (medical subject heading) MeSH terms (table 1) from the tissue enrichment analysis that are significant (P < 0.05). These tissues are testis and the retina. For the first group (testis), there are a total of 13 genes of interest. For eye-related sense organs there are a total of two regions with confirmed function in eye-related traits (fig. 2A and supplementary table S2, supplementary information 2.1, Supplementary Material online). Thus, among the regions that were introgressed from bonobo into P.t. troglodytes, there is an enrichment of genes whose phenotypes are related to maleness, with interesting genes that are highly expressed in testis and may have had a special male reproductive function.
Table 1.
DEPICT Algorithm Results for Tissue Expression of Introgressed Regions in Each Subspecies
| MeSH First Level | MeSH Second Level | MeSH Number | P Value |
|---|---|---|---|
| Pan troglodytes troglodytes | |||
| Testis | Endocrine system | A06.407.312.782 | 0.028 |
| Retina | Sense organs | A09.371.729 | 0.04 |
| Pan troglodytes schweinfurthii | |||
| Synovial fluid | Musculoskeletal system | A02.835.583.443.800.800 | 7.487×10−5 |
| Monocytes | Hemic and immune systems | A15.378.316.580 | 0.007 |
| Myeloid cells | Cells | A11.627 | 0.028 |
| Mononuclear phagocyte system | Hemic and immune systems | A15.382.812 | 0.031 |
| Umbilical veins | Cardiovascular system | A07.231.908.670.874 | 0.032 |
| Portal system | Cardiovascular system | A07.231.908.670 | 0.032 |
| Bone marrow cells | Hemic and immune systems | A15.378.316 | 0.036 |
| Hematopoietic system | Hemic and immune systems | A15.378 | 0.036 |
| Endothelial cells | Cells | A11.436.275 | 0.037 |
| Phagocytes | Hemic and immune systems | A15.382.680 | 0.042 |
| Blood | Hemic and immune systems | A15.145 | 0.043 |
| Veins | Cardiovascular system | A07.231.908 | 0.047 |
| Pan troglodytes ellioti | |||
| Oocytes | Cells | A11.497.497.600 | 0.042 |
| Ovum | Urogenital System | A05.360.490.690 | 0.042 |
| Neural stem cells | Cells | A11.872.653 | 0.046 |
| Retina | Sense Organs | A09.371.729 | 0.046 |
Fig. 2.
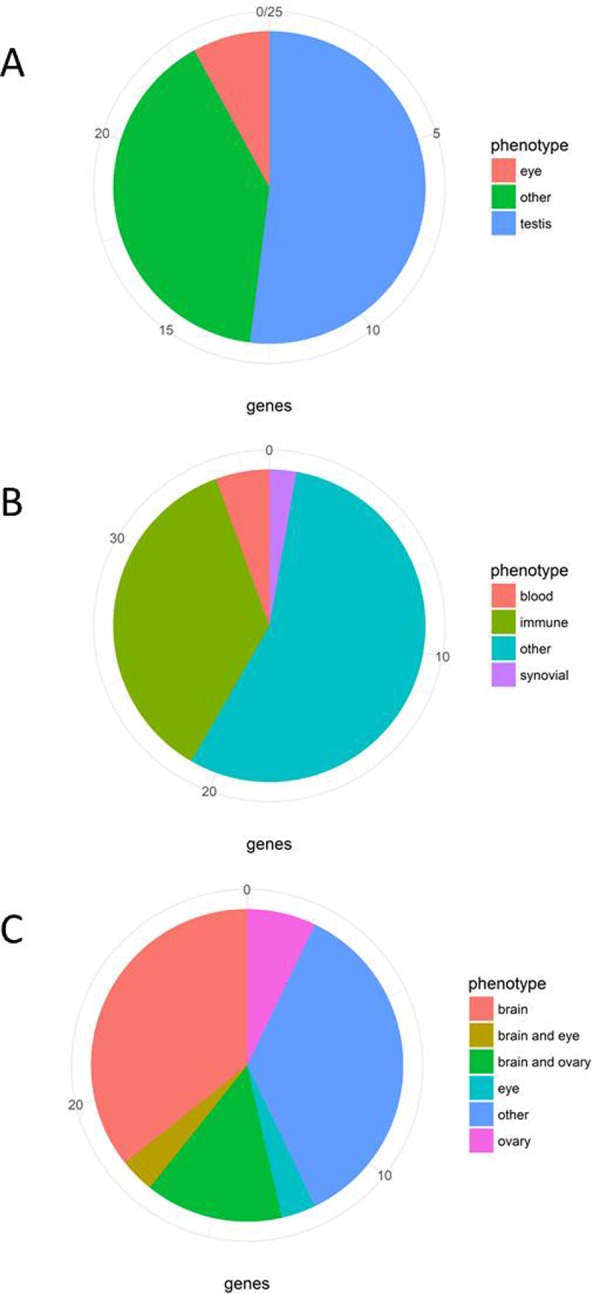
—Proportion of genes in or nearest to each introgressed region with DEPICT tissue enrichment analysis through a literature search. (A) Pan troglodytes troglodytes. (B) Pan troglodytes schweinfurthii. (C) Pan troglodytes ellioti.
In the case of P.t. schweinfurthii (supplementary table S3, Supplementary Material online), the 25 most extreme regions for each statistic are combined in 38 regions and the DEPICT algorithm of enrichment gives 12 MeSH terms (table 1) from the tissue enrichment analysis that are significant. These 12 tissues are involved with the Synovial Compartment, Immune System, Cardiovascular System, and Blood. We find, for phenotypes related to blood and immune function as well as the synovial compartment, 16 interesting genes among the 36 considered (fig. 2B and supplementary table S3, supplementary information 2.2, Supplementary Material online). The overall result of the genes under positive selection in the introgressed regions points to immunity, specifically of genes expressed in white blood cells. Many different genes have been preserved and driven, by selection, into an immunity response that may have benefited from the past exposure in another species.
In the P.t. ellioti subspecies (supplementary table S4, Supplementary Material online), the 25 most extreme regions for each statistic overlap resulting in 30 total regions and 28 genes. When analyzed by DEPICT, four MeSH terms (table 1) are significant in the tissue enrichment analysis. They include Oocyte and Ovum, the Nervous System, and the Retina. For the first two tissue categories, oocyte and ovum, we find eight genes of note. For the second phenotypic category, neural stem cells, 15 genes have some relationship to brain phenotypes in the literature. Lastly, for eye-related sense organs, two genes have been shown to play a role in the development of the retina (fig. 2C and supplementary table S4, supplementary information 2.3, Supplementary Material online). Thus, for P.t. ellioti, we see enrichment in genes related to femaleness and neural functions.
Adaptive Introgression and Balancing Selection—Positive Tail
The segments identified in the positive tail consist of regions both under long-term balancing selection and adaptive introgression, that is, regions that are considered as introgressed because they are in excess of heterozygotes. Because these two biological processes are distinct it is not logical to apply any enrichment test to these regions as a cohesive group.
We discuss here genes in the extreme positive tail with the unique combination that their coding region falls within the introgressed region (supplementary tables S5–S7, column 6, Supplementary Material online) and where the corresponding region in bonobo (supplementary tables S8–S10, supplementary information 3.1–3.3, Supplementary Material online) has statistical values near zero (indicating no significant selection of any kind). These regions can be considered as adaptive introgression because they are segregating at intermediate frequencies and have not been removed by purifying selection nor been subject to long-term balancing selection during the history of the Pan clade. Their evolutionary significance may be either recent balancing selection or a complex situation leading to an excess of heterozygotes. We discuss the specific candidate genes under an adaptive advantage in the supplementary information 3.1–3.3, Supplementary Material online. Overall, regions with signatures of adaptive introgression and segregating at intermediate frequencies in chimpanzees but not in bonobos tend to lie within protein-coding regions, and several of these genes contain protein-coding substitutions (supplementary table S1, Supplementary Material online). This underscores the interesting evolutionary benefit that may arise through introgression between species.
Conversely, we classify regions as under long-term balancing selection when they are shared among all subspecies of chimpanzee and bonobo. To find the most interesting candidates, first, we selected the most extreme 25 positive regions for one of the three statistics in each subspecies of chimpanzee (Top 0.4% tail genome-wide; supplementary tables S5–S7, Supplementary Material online) and kept regions only when the corresponding region in the bonobo genome has a value for one of the statistics in the top 1% tail of its genome-wide distribution (supplementary tables S11, Supplementary Material online); indicated in the last column of supplementary tables S5–S7, Supplementary Material online. Out of the 21 unique regions with these conditions, the majority (17/21) are also in the top 5% genome-wide tail for at least another subspecies (supplementary fig. S3 and table S11, last two columns, Supplementary Material online) and some are in the top fraction for all: STEAP1, UNC5D, RIOK2, ZWINT, PCDH9, COL11A1, and SLC16A7. Based on this combination, it is likely that these regions have been undergoing long-term balancing selection during the evolutionary history of the Pan clade. Due to this type of selective force, these regions have an increased genetic diversity as well as unusually old lineages (Charlesworth 2006). Interestingly, some regions under long-term balancing selection in the Pan clade (supplementary information 4, supplementary tables S5–S7, Supplementary Material online) are falling outside of coding regions, implicating the possibility of the importance of balancing selection in regulatory regions.
Discussion
We have established that the introgressed regions of bonobo genome into chimpanzee are depleted of purifying selection indicating that the haplotypes that have survived do not confer a negative fitness within the chimpanzee genome. Previous studies (Arnold and Martin 2009) have identified reproductive traits as candidates for adaptive introgression in different species. This mechanism may increase the viability of hybrid offspring and counter reproductive isolation.
There have been well-established differences between the reproductive traits of bonobos and chimpanzees, which may make gene flow difficult. For instance, the estrous cycle of the two species is understood to be quite different, with prolonged sexual swellings in bonobo females (Furuichi 1987), discrepancies in ovulation timing (Ryu et al. 2015), and the age of menarche differing by 3 years (Behringer et al. 2014). Two MeSH terms for P.t. ellioti are involved with ovulation, six genes are highly expressed in the female reproductive organs, and two genes have a known function within the female cycle. The most striking enrichment for adaptive introgression in P.t. troglodytes falls within male-related tissues and genes. When comparing chimpanzees and bonobos, some evidence indicates a differentiation between the ratio of body size and testis size and with midpiece volume (the part of a spermatozoon between the head and the tail; Anderson and Dixson 2002). Furthermore, testosterone levels are known to fluctuate in chimpanzee males while remaining consistent in bonobos (Wobber et al. 2012). Although no evidence is available for subspecies-specific phenotypes related to male reproduction, selection within these introgressed regions in P.t. troglodytes may indicate a benefit gained from bonobo introgression. We indeed find 15 genes with implication in testis phenotypes, specifically four genes having functional importance. We also find two protein-coding substitutions in genes involved in fertility, which might be strong candidates for adaptation. This evidence together indicates that selection may have acted to facilitate the admixture of these two differentiated species.
In general, the majority of evidence for differences between the two species is in behavior (as reviewed by Gruber and Clay 2016). Behavioral traits are of course extremely complex and difficult to research, however, we do want to highlight that one of the MeSH terms from enrichment in P.t. ellioti is neural stem cells; and that nine of the closest genes are implicated in some brain function, specifically with seven of those functionally involved in the development of the nervous system. Behaviorally bonobos are viewed as “delayed” because they exhibit playful behavior throughout life (Hare et al. 2012) and due to slower development of endocranial volume in juveniles (Durrleman et al. 2012). These two species are well-established to be social animals and the selection of bonobo haplotypes may also have had an impact on allowing their gene flow.
Interesting, P.t. schweinfurthii has MeSH terms involved in immune function. This, on the surface, is surprising as it is well-established that P.t. schweinfurthii and bonobos live in adjacent habitats; and would therefore come in contact with extremely similar pathogens. However, evidence suggests that this subspecies specifically is a reservoir for Simian Immunodeficiency Virus (SIV, the Pan equivalent to HIV) while no wild infections have been found in bonobos (Li et al. 2012), indicating a difference in their immune response to viral infection. Intriguingly, four genes closest to introgressed regions of P.t. schweinfurthii were placed in the HIV life cycle according to PathCards (Belinky et al. 2015); with the addition that six closest genes have known function within the immune system, indicating a likely boost to the immune system in this subspecies.
This study identifies 13 genes as under adaptive introgression or balancing selection that were previously identified as under selection in chimpanzees; specifically, ADAM22, FAM162B, VPS41, INPP4B, NCAM2, SORCS1, ANO2, CCSER1, ZPLD1, HS6ST3, and GALNTL6 (positive selection) and HLA-DQA1 and AKR1E2 (balancing selection; Cagan et al. 2016). On the whole, the regions which contain introgression tend to fall into the extreme tails genome-wide (well within the extreme 1%). It is therefore likely for these specific genes to be classified as under selection.
Finally, long-term balancing selection can be rare and difficult to identify (Charlesworth 2006) and here we have found regions that may have interesting features. There is evidence for balancing selection occurring outside coding regions; specifically in a promoter (Wilson et al. 2006), in the 5′ regulatory region ∼2 kb outside the coding region (Bamshad 2002), and upstream ∼4 kb outside the coding region (Sun et al. 2011). However, no study has satisfactorily attributed this evolutionary force this far outside of genes and for several regions, while here, we identify 15 regions in the extreme tails which are between 30 and 600 kb to the closest gene. These data, however, are uniquely rich and show similar patterns of balancing selection between two species (and, in most cases, across the three subspecies), making a strong case for the importance of long-term long-distance balancing selection.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This study has been possible thanks to grant BFU2016-77961-P (AEI/FEDER, UE) awarded by the Agencia Estatal de Investigación (Spain) and with the support of Secretaria d’Universitats i Recerca del Departament d’Economia i Coneixement de la Generalitat de Catalunya (GRC 2014 SGR 866). J.N. was supported by an FI PhD fellowship by Agaur (FI-DGR 2015). M.K. was supported by a DFG fellowship (KU 3467/1-1). T.M.-B. is supported by MINECO BFU2014-55090-P (FEDER), U01 MH106874 grant, Howard Hughes International Early Career and Secretaria d’Universitats i Recerca del Departament d’Economia i Coneixement de la Generalitat de Catalunya.
Literature Cited
- Anderson MJ, Dixson AF. 2002. Sperm competition-Motility and the midpiece in primates. Nature, 416(6880):496–496. [DOI] [PubMed] [Google Scholar]
- Arnold ML, Martin NH.. 2009. Adaptation by introgression. J Biol. 8(9):82.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ. 2002. A strong signature of balancing selection in the 5′ cis-regulatory region of CCR5. Proc Natl Acad Sci U S A. 99(16):10539–10544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer V, Deschner T, Deimel C, Stevens JMG, Hohmann G.. 2014. Age-related changes in urinary testosterone levels suggest differences in puberty onset and divergent life history strategies in bonobos and chimpanzees. Horm Behav. 66(3):525–533. [DOI] [PubMed] [Google Scholar]
- Belinky F, et al. , 2015. PathCards: multi-source consolidation of human biological pathways. Database 2015(0):bav006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagan A, et al. , 2016. Natural selection in the great apes. Mol Biol Evol. 33(12):3268–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. 2006. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2(4):e64–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannemann M, Andres AM, Kelso J.. 2016. Introgression of Neandertal- and Denisovan-like haplotypes contributes to adaptive variation in human Toll-like receptors. Am J Hum Genet. 98(2):399.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Manuel M, et al. , 2016. Chimpanzee genomic diversity reveals ancient admixture with bonobos. Science 354(6311):477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps M, et al. , 2016. Genomic signatures of selective pressures and introgression from Archaic hominins at human innate immunity genes. Am J Hum Genet. 98(1):5–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrleman S, Pennec X, Trouvé A, Ayache N, Braga J.. 2012. Comparison of the endocranial ontogenies between chimpanzees and bonobos via temporal regression and spatiotemporal registration. J Hum Evol. 62(1):74–88. [DOI] [PubMed] [Google Scholar]
- Fu YX, Li WH.. 1993. Statistical tests of neutrality of mutations. Genetics 133(3):693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi T. 1987. Sexual swelling, receptivity, and grouping of wild pygmy chimpanzee females at Wamba, Zaire. Primates 28(3):309–318. [Google Scholar]
- Green RE, et al. , 2010. A draft sequence of the Neandertal genome. Science 328(5979):710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber T, Clay Z.. 2016. A comparison between bonobos and chimpanzees: a review and update. Evol Anthropol. 25(5):239–252. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium. 2017. Genetic effects on gene expression across human tissues. Nature 550(7675):204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare B, Wobber V, Wrangham R.. 2012. The self-domestication hypothesis: evolution of bonobo psychology is due to selection against aggression. Anim Behav. 83(3):573–585. [Google Scholar]
- Hedrick PW. 2013. Adaptive introgression in animals: examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol Ecol. 22(18):4606–4618. [DOI] [PubMed] [Google Scholar]
- Huerta-Sanchez E, et al. , 2014. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512(7513):194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric I, Aeschbacher S, Coop G.. 2016. The strength of selection against Neanderthal introgression. PLoS Genet. 12(11):e1006340.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlwilm M, et al. , 2016. Ancient gene flow from early modern humans into Eastern Neanderthals. Nature 530(7591):429–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. , 2012. Eastern chimpanzees, but not Bonobos, represent a Simian immunodeficiency virus reservoir. J Virol. 86(19):10776–10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G, Gordon D, Davis C, Green P.. 2009. Widespread genomic signatures of natural selection in Hominid evolution. PLoS Genet. 5(5):e1000471.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal M, et al. , 2016. Genomic analysis of Andamanese provides insights into ancient human migration into Asia and adaptation. Nat Genet. 48(9):1066–1070. [DOI] [PubMed] [Google Scholar]
- Pers TH, et al. , 2015. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 6:5890.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado-Martinez J, et al. , 2013. Great ape genetic diversity and population history. Nature 499(7459):471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus M, et al. 2014. 1000 Genomes Selection Browswer 1.0: a genome browser dedicated to signatures of natural selection in modern humans. Nucleic Acids Res. 42, D903–D909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus M, et al. 2015. Hierarchical boosting: a machine-learning framework to detect and classify hard selective sweeps in human populations. Bioinformatics, 31(24):3946–3952. [DOI] [PubMed] [Google Scholar]
- Reich D, et al. , 2011. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am J Hum Genet. 89(4):516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Hill DA, Furuichi T.. 2015. Prolonged maximal sexual swelling in wild bonobos facilitates affiliative interactions between females. Behaviour 152(3–4):285–311. [Google Scholar]
- Sankararaman S, et al. , 2014. The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507(7492):354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S, Mallick S, Patterson N, Reich D.. 2016. The combined landscape of Denisovan and Neanderthal ancestry in present-day humans. Curr Biol. 26(9):1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FH, Jankovic I, Karavanic I.. 2005. The assimilation model, modern human origins in Europe, and the extinction of Neandertals. Quant Int. 137(1):7–19. [Google Scholar]
- Song Y, et al. , 2011. Adaptive introgression of anticoagulant rodent poison resistance by hybridization between old world mice. Curr Biol. 21(15):1296–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, et al. , 2011. A signature of balancing selection in the region upstream to the human UGT2B4 gene and implications for breast cancer risk. Hum Genet. 130(6):767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical-method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123(3):585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B, et al. , 2016. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science 352(6282):235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney KD, Randell RA, Rieseberg LH.. 2006. Adaptive introgression of herbivore resistance traits in the weedy sunflower Helianthus annuus. Am Nat. 167(6):794–807. [DOI] [PubMed] [Google Scholar]
- Wilson JN, et al. , 2006. A hallmark of balancing selection is present at the promoter region of interleukin 10. Genes Immun. 7(8):680–683. [DOI] [PubMed] [Google Scholar]
- Wobber V, Lipson S, Hare B, Wrangham R, Ellison P.. 2012. Species differences in the ontogeny of testosterone production between chimpanzees and bonobos. Am J Phys Anthropol. 147:305–306. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.