SUMMARY
Fibroblast heterogeneity has long been recognized in mouse and human lungs, homeostasis, and disease states. However, there is no common consensus on fibroblast subtypes, lineages, biological properties, signaling, and plasticity, which severely hampers our understanding of the mechanisms of fibrosis. To comprehensively classify fibro-blast populations in the lung using an unbiased approach, single-cell RNA sequencing was performed with mesenchymal preparations from either uninjured or bleomycin-treated mouse lungs. Single-cell transcriptome analyses classified and defined six mesenchymal cell types in normal lung and seven in fibrotic lung. Furthermore, delineation of their differentiation trajectory was achieved by a machine learning method. This collection of single-cell transcriptomes and the distinct classification of fibroblast subsets provide a new resource for understanding the fibroblast landscape and the roles of fibroblasts in fibrotic diseases.
In Brief
Xie et al. have analyzed mesenchymal cell subpopulations at single-cell resolution and have demonstrated known subtypes and a newly emerging subtype during pulmonary fibrosis in mouse lung.
INTRODUCTION
Fibrosis is an evolutionary body strategy to rapidly close and repair wounds (Bochaton-Piallat et al., 2016; Gurtner et al., 2008). In the lung, fibrosis occurs when there is an ongoing epithelial injury (Liang et al., 2016; Thomas et al., 2002). Fibrosis in patients with idiopathic pulmonary fibrosis (IPF) results in persistent and relentlessly progressive lung scarring (Thannickal et al., 2014; Thum, 2014; Tzouvelekis and Kaminski, 2015), which leads to ~40,000 deaths every year in the US. The major effector cells in this process are the mesenchymal cells (MCs) (Li et al., 2011). MCs are believed to consist of multiple subtypes that are being intensively investigated (Kumar et al., 2014; Lee et al., 2017; Xie et al., 2016; Zepp et al., 2017), but it is unclear how many mesenchymal subtypes exist and how they differ from or are related to one another, and their cellular biology is poorly defined. Thus, these limitations hinder severely our ability to understand the cellular events and the molecular signaling pathways in the distinct subsets of fibroblasts in fibrogenesis, and to develop precise cellular models and animal models of lung fibrosis.
Pulmonary MCs are suggested to be extremely heterogeneous in IPF (Jordana et al., 1988) and in mouse models (Rock et al., 2011), suggesting that they could be derived from different cell types, represent different stages of activation, or may be influenced by the surrounding milieu. MC clones separated by Thy1 seem to have different morphology, growth characteristics, display of antigens, and collagen and fibronectin production (Derdak et al., 1992). Subsets of MCs distinguished by Pdgfrα expression were reported to express different levels of α-smooth muscle actin (α SMA) (Kimani et al., 2009). The regional airway MCs were suspected to be distinct from the distal lung MCs in terms of morphology, collagen and α SMA expression, and proliferation (Kotaru et al., 2006). Using genetic lineage tools to characterize lung MCs has provided some insights into subtypes. Fgf10 lineage MCs (El Agha et al., 2012); pericytes trace labeled with NG2, FoxJ1, or Foxd1 (Hung et al., 2013; Rock et al., 2011); or Plin2-traced lipofibroblasts (El Agha et al., 2017) were suggested to contribute to α SMA-expressing myofibroblasts and various MC subsets. We recently reported that Tbx4-lineage cells compose a large fibroblast population within the lung, including α SMA+, Col1α 1+, NG2+, vimentin+, desmin+, Pdgfrα +, and Pdgfrβ+ fibroblasts (Xie et al., 2016). These data suggest the existence of cellular subpopulations of fibroblasts, which vary with anatomical locations, gene expression, and cell surface markers. However, the enumeration of cell types and their definition can be controversial based on restricted markers available to identify, isolate, and manipulate. Biased morphology, physical properties, localization, molecular markers, functions, and developmental origins would alter the assignment of diversification and cellular differentiation for mesenchymal subtypes. Therefore, a systematic map of evolutionary pulmonary mesenchymal heterogeneity in both steady-state and pathological conditions remains unexposed.
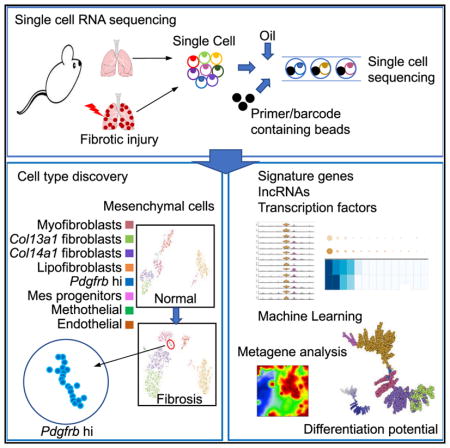
To overcome these challenges, efforts have been made to systematically classify lung MCs. The Lung Gene Expression in Single-Cell (LungGENS) program separated MCs into proliferative mesenchymal progenitor, myofibroblast/smooth muscle, pericyte, intermediate fibroblast 1 and 2 and matrix fibroblast on embryonic (E) 16.5, and FB (LipoFibroblast/Matrix Fibroblast) and myofibroblast/smooth muscle on E18.5 and postnatal mouse lungs by single-cell RNA sequencing (scRNA-seq) analysis using the Fluidigm C1 platform (Du et al., 2015). However, the numbers of fibroblasts included in the studies were small because of the limitation of the C1 platform. To conquer some of these obstacles, we used an unbiased approach: Drop-Seq single-cell RNA-seq (scRNA-seq; 10x Genomics) with much larger numbers of MCs to better assess the diversity of pulmonary MCs, leading to the identification of new subtypes of fibroblasts, and refine their existing classifications. We further assessed the signature genes, enriched extracellular and soluble protein coded genes, key transcription factors, and, notably, expressed long non-coding RNAs (lncRNAs) for each subtype. In addition, pseudo-time analysis was used to delineate the mesenchymal cellular paths of differentiation. Overall, our analysis provides a comprehensive map of the subtypes of the stromal taxonomy in steady-state in adult mice and fibrotic lung.
RESULTS
Classification of Mesenchymal Heterogeneity by scRNA-Seq in Normal and Fibrotic Mouse Lung Tissues
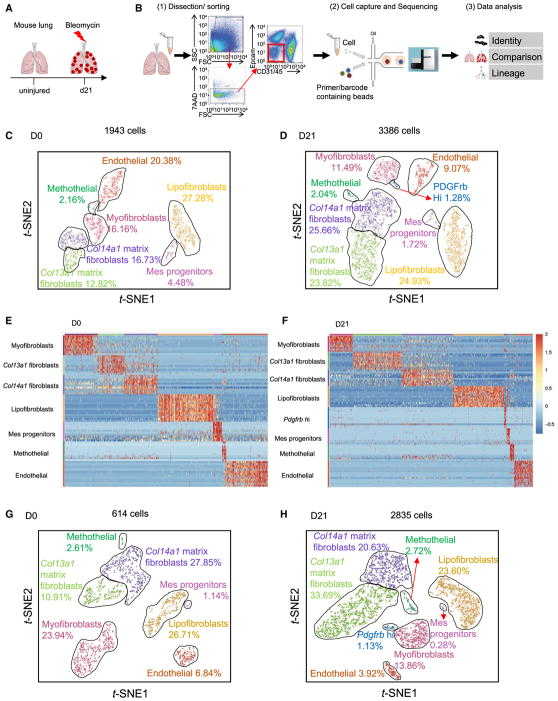
We set out to comprehensively identify and define subpopulations of the MCs between normal and fibrotic lung tissues. We treated αSMA-GFP;Tbx4-Cre;Rosa26-tdTomato mice with bleo-mycin and harvested the lungs after injury (Figure 1A). We obtained enriched MCs by fluorescence-activated cell sorting (FACS) Epcam−CD31−45− cells from single lung homogenates and performed scRNA-seq using the 10x Genomics Chromium platform (Figure 1B). We profiled 1,943 cells from normal mouse lung and 3,386 cells from fibrotic αSMA-GFP;Tbx4-Cre;Rosa26-tdTomato mouse lung. We visualized the cells in two dimensions according to their expression profiles by t-distributed stochastic neighborhood embedding (t-SNE) projections. Six subtypes as MCs in normal lung and seven subtypes in fibrotic lung were well segregated (Figures 1C and 1D). Endothelial cells also were included in the analysis. The other cell types such as epithelial cells contaminated during flow sorting were minimal and easily identifiable, and were eliminated from further analysis. We tentatively classified mesenchymal populations based on their preferential or distinctive marker expression and relations to known cell types. The compositions of these clusters were myofibroblasts, 16% in normal and 11% in fibrotic lung; Col13a1 matrix fibroblasts, 13% in normal and 24% in fibrotic lung; Col14a1 matrix fibroblasts, 17% in normal and 26% in fibrotic lung; lipofibroblasts, 27% in normal and 25% in fibrotic lung; mesenchymal progenitors, 5% in normal and 2% in fibrotic lung; mesothelial cells, 2% in normal and 2% in fibrotic lung; and endothelial cells, 20% in normal and 9% in fibrotic lung. A new Pdgfrb high (hi) subpopulation appeared only in the fibrotic lung, which comprised ~1% of all MCs (Figures 1C and 1D). Heatmaps of normalized MC profiles revealed normalized expression of the top variable genes in each MC subtype (Figures 1E and 1F). We further analyzed tdTomato (tdT)+GFP+ cells from αSMA-GFP;Tbx4-Cre;Rosa26-tdTomato mice to confirm the MC subtypes (Figures 1G and 1H). As expected, these subtypes and the patterns of composition were consistently reproduced in the analyses of 614 MCs in normal and 2,835 MCs in fibrotic tdT+GFP+ cells. Together, these unbiased analyses delineated the fibroblast heterogeneity in the adult mouse lungs and further identified mesenchymal subpopulation changes during lung fibrogenesis.
Figure 1. Clustering of Mesenchymal Cells by Single-Cell RNA Sequencing.
(A) Sketch of bleomycin-induced pulmonary fibrosis mouse model.
(B) Workflow depicts rapid dissociation and sorting of MCs from lung tissue for generating scRNA transcriptome profiles.
(C and D) 2D visualization of single-cell clustering of MC profiles inferred from RNA-seq data for all MCs in normal (C) and fibrotic (D) αSMA-GFP;Tbx4-Cre;Rosa26-tdTomato lung samples. Six major classes of MCs in normal lung and seven major classes of MCs in fibrotic lung were detected. Endothelial cells also were included in the analysis. The percentage of each cell population was indicated. Colored bar coded as indicated.
(E and F) Heat maps of MC normalized signal show MC subtypes changes by top genes (columns) for individual MC subtype cells (rows) in normal (E) and fibrotic
(F) αSMA-GFP;Tbx4-Cre;Rosa26-tdTomato lung samples.
(G and H) Clustering plots depicting single-cell RNA-seq datasets for normal (G) and fibrotic (H) Tbx4-lineage+α SMA+ MCs acquired from marker-based fluorescence-activated cell sorting (FACS).
Single-Cell Profiling of Myofibroblasts
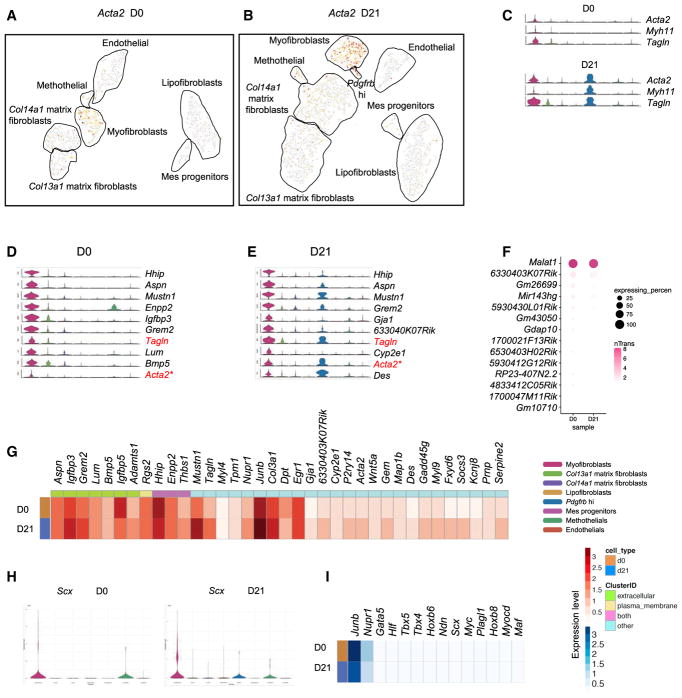
Using known marker genes for myofibroblasts, including Acta2 (Hinz et al., 2007), Myh11 (Hsia et al., 2016), and Tagln (Du et al., 2015; Robin et al., 2013) (Figures 2A–2C), a myofibroblast cluster was readily identified. Top 10 genes were highly and specifically expressed in myofibroblasts and absent or much less expressed in other MC subtypes, providing a series of novel markers that can distinguish myofibroblasts from other fibroblasts under both normal and fibrotic conditions (Figures 2D and 2E). The expression levels of the top highly expressed lncRNAs in myofibroblasts; the top 36 most abundant expressed genes, including extracellular and plasma membrane coding genes; and the most abundant transcription factors were analyzed and compared between normal and fibrotic myofibroblast subtypes (Figures 2F–2I). Among the newly identified putative myofibroblast markers, several are of particular interest. Hhip, which has been reported to be involved in maintaining normal lung function and alveolar structures (Lao et al., 2016), is the highest specifically expressing gene in the myofibroblast subtype. Aspn has been reported to be significantly expressed in IPF lung samples (Leng et al., 2013). Mustn1, which has been reported to be expressed in skeletal muscle, is believed to be an essential regulator of myogenic differentiation and myofusion (Liu et al., 2010). All three of these were better markers of myofibroblasts than was Acta2, which is consistent with the results of a recent report (Sun et al., 2016). Junb is a component of transcription complex AP1 (Andreucci et al., 2002). AP1, which has been shown to play a key role in fibrotic diseases, and Scx, a transcription factor reported to be a critical regulator of the cardiac fibroblast/myofibroblast phenotype (Bagchi et al., 2016), are showing up as the most distinctive transcription factors of myofibroblast subtypes in both normal and fibrotic conditions. These analyses identified novel markers and transcription networks in the myofibroblast subtype.
Figure 2. Transcriptional Profile of Myofibroblasts.
(A and B) Expression patterns of Acta2 (D0, A; D21, B) in representation as in Figures 1C and 1D.
(C) Violin plots showing known myofibroblast markers Acta2, Myh11, and Tagln gene expression across all MC clusters.
(D and E) Representative markers were distinct in normal (D) and fibrotic (E) myofibroblast clusters predicted in the scRNA-seq data.
(F) Top lncRNAs enriched in myofibroblasts. The size of each circle depicts the percentage of cells in the subtype in which the marker was detected, and its color depicts the average transcript count in expressing cells (nTrans).
(G) Heatmap of top significant genes, including extracellular and plasma membrane genes in this subtype. Rows correspond to normal (D0) and fibrotic (D21) myofibroblast subtypes, and columns correspond to the mean of the single-cell gene expression signature arranged by expression locations.
(H) Signature transcription factor Scx expression across all MC subtypes.
(I) Enrichment patterns of transcription factors in the myofibroblast subtype.
Transcriptional Signature Associated With Col13a1 Matrix Fibroblasts
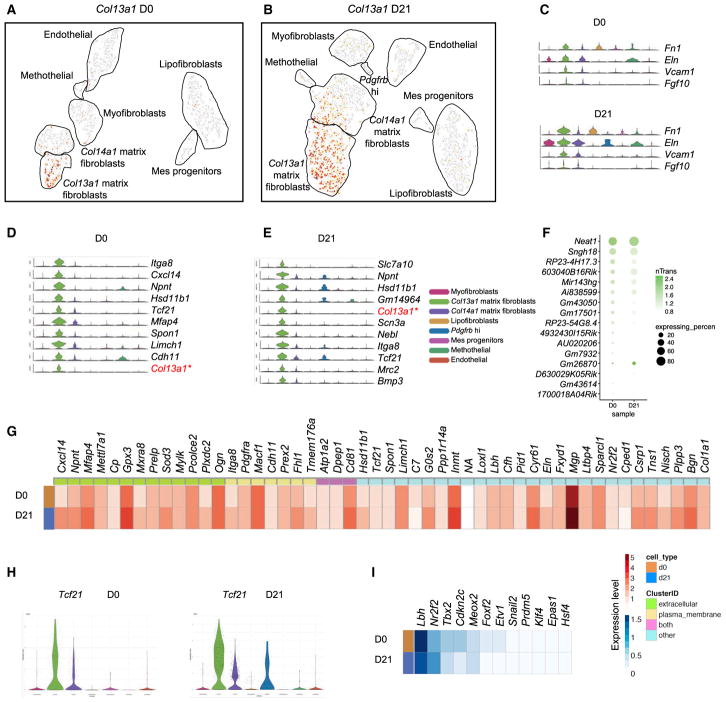
Matrix fibroblasts express signature genes associated with extracellular matrix and cell adhesion. We found that these Col1a1 highly expressing matrix fibroblasts clustered together (Figures S1A and S1B). Col13a1 and Col14a1 are highly discriminative markers within the matrix fibroblast clusters, segregating them into two distinct subtypes. We refer to these subsets henceforth as Col13a1 and Col14a1 matrix fibroblasts (Figures 3A and 3B). Previously reported matrix fibroblast markers are dominantly expressed in both Col13a1 and Col14a1 matrix fibro-blasts (Figure 3C). Col13a1 matrix fibroblasts were accurately delineated by Itga8, Cxcl14, Npnt, and other top signature genes in both normal and fibrotic lung (Figures 3D and 3E). Most of the highly expressed lncRNAs were analyzed and ordered by the range of their expression (Figure 3F), with Neat1 being the most abundantly expressed lncRNA. The top 50 significantly expressed genes were listed with discrimination of the extracellular and plasma membrane expressing genes, and their expression levels were depicted as heatmaps (Figure 3G). Chemokine Cxcl14 is the most distinct extracellular expressed gene in Col13a1 matrix fibroblasts. Plasma membrane associated gene Itga8 can uniquely delineate Col13a1 matrix fibroblasts. Both Cxcl14 and Igta8 are increased in fibrotic Col13a1 matrix fibroblasts. Transcription factor Tcf21 strongly marked the Col13a1 matrix fibroblasts, with more abundant expression at the fibrotic phase (Figure 3H). Tcf21 has been used to lineage trace resident cardiac fibroblasts during pathologic remodeling (Xiang et al., 2017). Other top transcription factors include Lbh, Nr2f2, Tbx2, and Meox2 (Figure 3I).
Figure 3. Matrix Fibroblast Subtype Enriched in Col13a1.
(A and B) Visualization of normal (A) and fibrotic (B) Col13a1 gene expression using a t-SNE plot.
(C) Expression patterns of known matrix fibroblast marker genes across MC subtypes.
(D and E) Core distinct expressed genes in Col13a1 subtype from both normal (D) and fibrotic (E) lungs were indicated by violin plots.
(F) lncRNAs identified in the Col13a1 subtype.
(G) Heatmap of significantly expressed genes with the indication of their cellular locations was compared between normal and fibrotic Col13a1 subtypes.
(H) Violin plot of Tcf21 gene expression, which is highlighted in the Col13a1 subtype.
(I) Heatmap showing changes in top transcription factors between normal and fibrotic Col13a1 subtypes.
Delineation of Col14a1 Matrix Fibroblasts
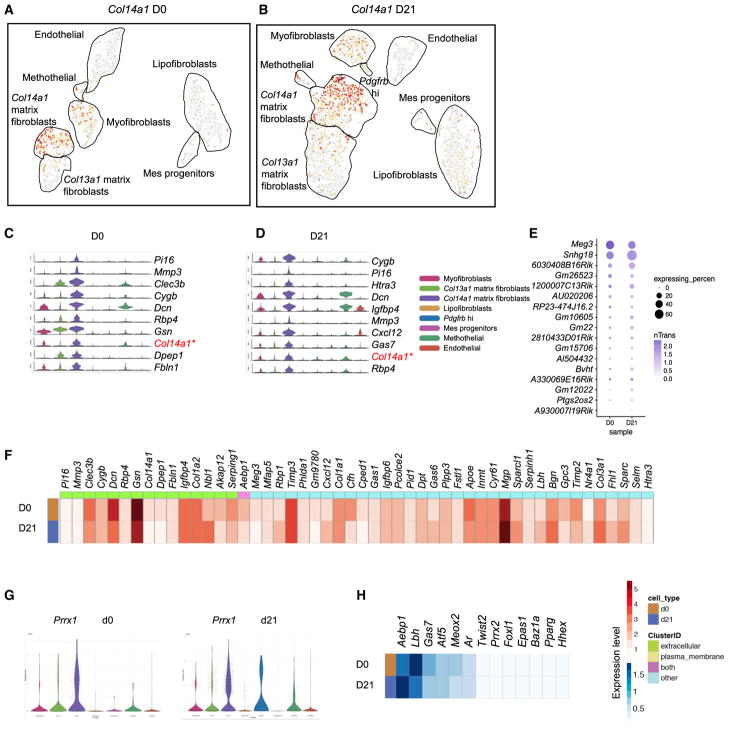
Unlike Col13a1 matrix fibroblasts, Col14a1 matrix fibroblasts distinguish themselves by harboring a unique set of significant genes, including Pi16, Mmp3, Cygb, and Rtp4 (Figures 4A–4D). Meg3 and Snhg18 are the most abundant lncRNAs expressed in the Col14a1 matrix fibroblasts. The percentage of the cells expressing Meg3 in Col14a1 matrix fibroblasts and the average transcript count of Meg3 are decreased, whereas the percentage of cells expressing Snhg18 is increased in fibrotic Col14a1 matrix fibroblasts (Figure 4E). Meg3 lncRNA has been suggested to be expressed in tissue fibrosis (He et al., 2014; Piccoli et al., 2017). The top discriminative extracellular expressing genes are Pi16 and Mmp3 for Col14a1 matrix fibroblasts, but with low transcript levels. Clec3b and Dcn are more significantly expressed, but with less distinction (Figure 4F). For transcription factors, mesoderm homeobox gene Prrx1 is the top factor in Col14a1 matrix fibroblasts, with Aebp1 and Lbh highly expressed as well (Figures 4G and 4H).
Figure 4. Characterization of Col14a1 Matrix Fibroblasts.
(A and B) Col14a1 expression in normal (A) and fibrotic (B) MC subtypes projected by t-SNE
(C and D) Enriched signature genes within the normal (C) and fibrotic (D) Col14a1 MC subtype.
(E) Subtype significantly expressed lncRNAs.
(F) Significantly differentiated genes with the indication of cellular location enriched in the Col14a1 subtype.
(G) Prrx1 as the master transcription factor in the Col14a1 subtype.
(H) Heatmap showing the highly distinct transcription factors.
Elucidation of a Lipofibroblast Gene Signature
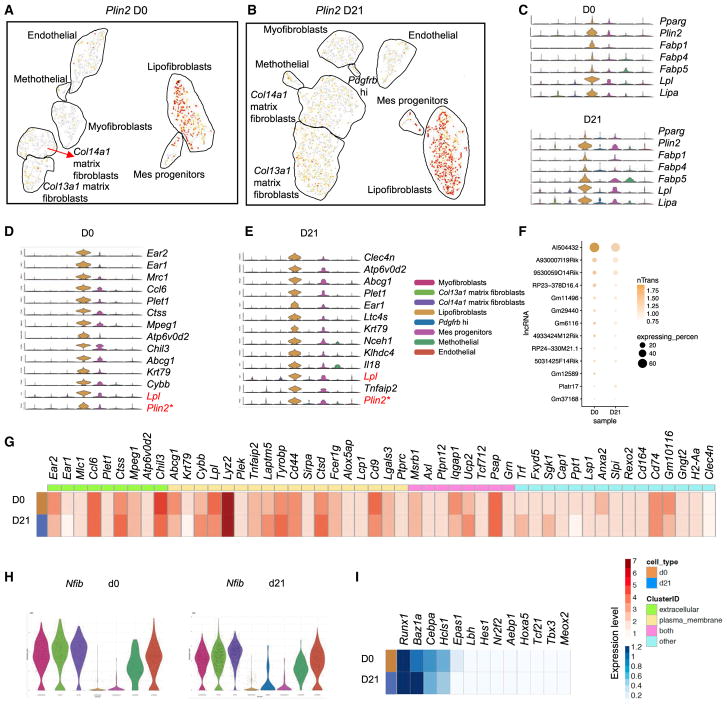
Lipofibroblasts are lipid-containing interstitial fibroblasts (McGowan and Torday, 1997; Torday and Rehan, 2016). The previously suggested markers for lipofibroblasts include Adrp (El Agha et al., 2017; Schultz et al., 2002), Pparg (Varisco et al., 2012), Fabp1, Fabp 4, Fabp5 (Chen et al., 1998, 2012; Li et al., 2016), and Lpl (Imamura et al., 2002). We found that these Adrp and Pparg highly expressing MCs were clustered together to form a distinct subpopulation, and at the same time, this cluster also expresses considerable levels of Fabp1,4,5, Lpl, and Lipa. Therefore, we referred to this cluster as lipofibroblasts (Figures 5A–5C). We further examined the cluster and found that this subtype expresses immune function-related genes along with mesenchymal genes. The top signature genes include Ear1 and Ear2, Mrc1, Ccl6, Plet1, Abcg1, and Krt79 (Figures 5D and 5E). Furthermore, the cluster also expresses common fibro-blast genes such as Pdgfra, Vim, Col4a1, and Fn1 (Figure S2A). Therefore, the expression patterns of the lipofibroblast subtype possessed a lipid synthesis and transport gene signature, as well as a mesenchymal feature. The top expressing lncRNA is AI504432, which is located on the opposite strand of the Kcna3 gene, with expression level and percentage decreased in fibrotic lipofibroblasts (Figure 5F). We also compared the expression levels for the top 50 highest expressed genes, including extracellular and plasma membrane genes, between normal and fibrotic lipofibroblasts (Figure 5G). It is interesting that M2 macrophage-like signature genes (Chil3, Mrc1, IL18, and CD9) (Lechner et al., 2017), together with mesenchymal genes (Mlc1, Plek, CD44, Ptpn12, and Slpi), are expressed in the lipofibroblast subtype (Figures S2B and 5G). The most distinctive transcription factor for the lipofibroblast is Nfib, and its expression level is the lowest within the MC subtypes (Figure 5H). Other top expressed transcription factors include Runx1, Baz1a, Cebpa, and Hcls1 (Figure 5I).
Figure 5. Molecular Census of Lipofibroblasts.
(A and B) Signature gene Plin2 expression was visualized in t-SNE plots of normal (A) and fibrotic (B) MCs.
(C) Violin plots of single-cell expression levels of known lipofibroblast genes across the MC subtypes.
(D and E) Top unique expressed genes in the normal (D) and fibrotic (E) lipofibroblast subtype.
(F) Expression patterns of lncRNAs in normal and fibrotic lipofibroblasts.
(G) Averaged expression of lipofibroblast significant genes in heatmap view. Genes were labeled with the cellular location, as indicated.
(H) Nfib as the most significantly lower expressed transcription factor across the MC subtypes.
(I) Top transcription factors shown in the lipofibroblast subtype.
Identification of a Subgroup of Potential Mesenchymal Progenitors
Mesenchymal progenitors are characterized by their self-renewal capacity and a signature enriched with proliferative genes. The term ‘‘mesenchymal progenitor’’ was adapted from the nomenclature by the LungGENS project ‘‘Proliferative Mesenchymal Progenitors’’ (Du et al., 2015). We observed a cluster of cells expressing high levels of Top2a and Mki67. We henceforth hypothesized that these cells could act as mesenchymal progenitors (Figures S3A–S3C). We further characterized the predominantly expressed genes within this cluster and found that Hist1h2ap, Ube2c, H2afx, Cks2, Hmgb2, and Ccnb2 were distinct markers. These genes are related to cell cycle, cell proliferation, DNA metabolism, nuclear division, and mitotic cell cycle (Figures S3D and S3E). They are variably expressed during normal and fibrotic status but typically present in the mesenchymal progenitor cluster. Top expressed lncRNAs in this cluster were Malat1 and Lockd (Figure S3F). The most significant extra-cellular expressed gene is Hmgb2. The distinct plasma membrane genes are S100a8 and Cd52. Given the insufficiency of S100a8 surface expression, CD52 would be a better cell surface marker for the mesenchymal progenitors (Figure S3G). In addition, Hmgb2 is the most highly expressed transcription factor in the mesenchymal progenitor subtype (Figure S3H), together with Ezh2, Uhrf1, Mcm6, Hmgb3, and Mcm5 as the specifically expressed transcription factors in mesenchymal progenitors (Figure S3I). The LungGENS project identified 453 signature genes for the proliferative mesenchymal progenitors (PMPs). These 453 genes showed up in our differentiated gene list for mesenchymal progenitors (both D0 and D21) (Table S1), with various p value ranks. Within the 23 genes that were upregulated and log2 fold change >2 in the D0 mesenchymal progenitors, 13 genes were identical as PMP signature genes. Forty-six genes are upregulated and log2 fold change >2 in the D21 mesenchymal progenitors, and 23 genes are the same as PMP signature genes (Table S2). The gene signatures of PMPs and mesenchymal progenitors are similar, regardless that PMPs were sampled from E16.5 mouse lung.
Classification of Mesothelial Cells
Mesothelial cells provide a slippery, non-adhesive, and protective surface that wrap the internal organs and the body’s cavities. WT1 lineage mesothelial cells were shown to give rise to desmin+CD34+ fibroblasts, as well as bronchial and vascular smooth muscle cells (Cano et al., 2013; Dixit et al., 2013). The previously reported markers for mesothelial cells are Wt1, Upk3b, Lrrn4, Msln, and Calb2 (Du et al., 2015; Kanamori-Katayama et al., 2011; Que et al., 2008; Rinkevich et al., 2012). Notably, cells enriched for these common genes are clustered tightly, and we subsequently identified them as the mesothelial cell subtype (Figures S4A–S4C). Identification of the subtype distinguished genes revealed Lgals2, Cxcl13, Gpm6a, Rspo1, and Nkain4 as novel putative markers for mesothelial cells (Figures S4D and S4E). The top lncRNA for the mesothelial subtype is Gm12840 (Figure S4F). The 50 most significant genes were highly and specifically expressed in mesothelial cells (Figure S4G). Zinc finger gene Bnc1 is the best transcription factor uniquely identified in this cluster (Figure S4H). Other highly distinctive transcription factors include Aebp1, Wt1, Gata6, and pdlim4 (Figure S4I).
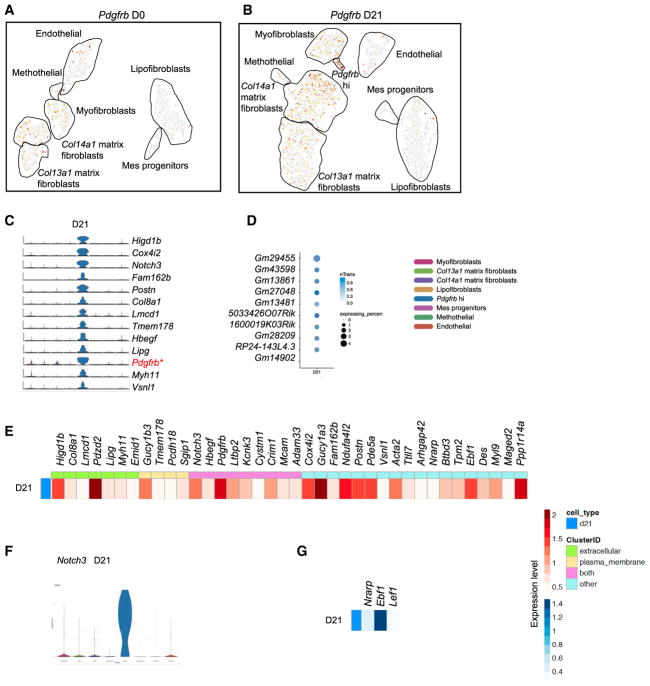
Discovering the Newly Emerging Pdgfrb High Fibroblasts in Fibrotic Lung
The comparison of normal and fibrotic MCs led us to uncover a newly emerging MC subtype expressing high levels of Pdgfrb. The Pdgfrb expressing cells were scattered in the myofibro-blast, matrix fibroblast, mesothelial, and endothelial subtypes in normal MCs; the same pattern was found in fibrotic MCs, but the proportion of Pdgfrb expressing cells was increased. When using Pdgfrβ antibody to stain fibrotic lungs, the labeled cells were expanded as compared to normal lungs (Barron et al., 2016; Xie et al., 2016), which is consistent with our scRNA-seq data (Figure S5A). In addition, the Pdgfrb highly expressing MCs were clustered together and can be distinguished collectively from the nearby myofibroblast subtype (Figures 6A and 6B). Thus, despite co-expressing genes such as Acta2 and Kcnk3, Pdgfrb hi and myofibroblast subtypes were separated and represent distinct clusters. Pdgfrb hi fibroblasts expressed perfectly discriminating markers, including Higd1b, Cox4i2, Notch3, Fam162b, Postn, Col8a1, Lmcd1, Tmem178, Hbegf, and Lipg (Figure 6C). lncRNAs found in the Pdgfrb hi subtype were not remarkable (Figure 6D). Postn, Higd1b, and Col8a1 were highly distinctive extracellular expressing genes. Plasma membrane expressing genes other than Pdgfrb were Gucy1b3, Tmem178, Pcdh18, and Sgip1 (Figure 6E). The Pdgfrb hi subtype was best delineated by transcription factor Notch3 (Figure 6F) and Notch downstream effector Nrarp. Other transcription factors such as Ebf1 and Lef1 were discriminatively expressed in this subtype (Figure 6G). It is interesting that Nrarp also is involved in stabilizing LEF1 in regulating Wnt signaling (Ishitani et al., 2005), suggesting a role of the Notch-Wnt signaling pathways in this Pdgfrb hi subtype. Because Notch3 and Pdgfrb have been suggested to be pericyte markers, we checked the other previously suggested pericyte markers in the current scRNA-seq data: Mcam (Cd146) (Barron et al., 2016), labeled Pdgfrb hi, endothelial, lipofibroblast subtypes (Figure S5B); Cspg4 (Ng2) (Barron et al., 2016; Hung et al., 2013), labeled few cells scattered in the Pdgfrb hi, myofibroblast, matrix fibroblast; and lipofibroblast MC subtypes (Figure S5C). These data suggested that Pdgfrb hi fibroblasts are newly emerging MCs in response to the fibrotic injury, but there was no distinct pericyte cluster in the analysis. The potential overlaps with pericytes anatomically and functionally remain to be determined.
Figure 6. Newly Emerging Fibrotic MC Subtype Expressing a High Level of Pdgfrb.
(A and B) Pdgfrb expression in normal (A) and fibrotic (B) MC subtypes.
(C–G) Highly unique genes (C), lncRNAs (D), significantly expressed extracellular and plasma membrane expressing genes (E), top transcription factor (F), and enriched transcription factors (G) expressed in fibrotic Pdgfrb hi subtype.
Uncovering the Transcriptional Program of Endothelial Cells
Given the fact that a close relation exists between endothelial cells and fibroblasts (Kumar et al., 2014; Xie et al., 2016), we included endothelial cells in the analysis. We detected a cluster of cells enriched for endothelial cell markers, including Pecam1, Cdh5, Edn1, Kdr, Ets1, and Gata2 (Figures S6A–S6D). A set of strong, unique signature genes, including Cldn5, Cyyr1, Clic5, Clec14a, and Tspan7, were found to be highly and specifically expressed in endothelial cells from both normal and fibrotic lungs (Figures S6E and S6F). Bvht is the top endothelial expressed lncRNA and the prevalence of this lncRNA in endothelial cluster is ~30% (Figure S6G). Bvht has been shown to have a key role in cardiac differentiation, including vascular endothelium (Klattenhoff et al., 2013). The top 50 most distinct signature genes in endothelial cells were plotted as a heatmap and analyzed for the differences between normal and fibrotic lungs (Figure S6H). The most uniquely discriminative transcription factor is Sox18, which is slightly increased in the endothelial cluster in fibrotic lungs (Figure S6I). Transcription factors, including Epas1, Klf2, Ppp1r16, Gata2, Sox17, and Ahr, also are significantly expressed in endothelial cells (Figure S6J).
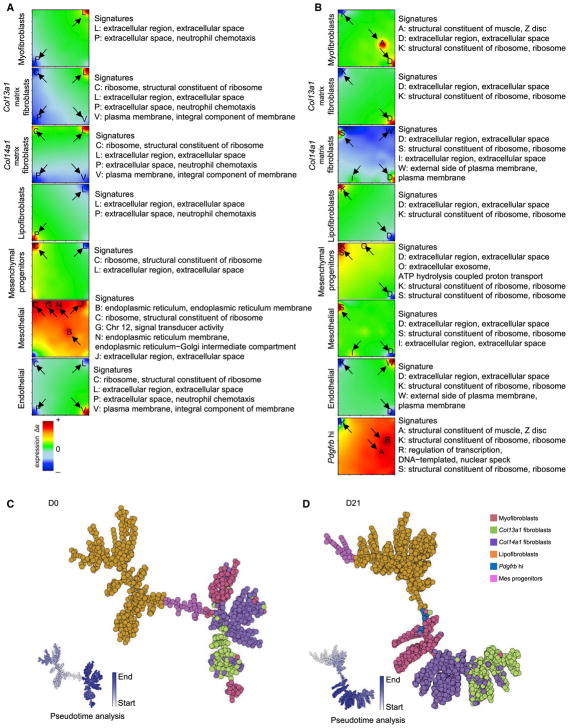
Differentiation Potential of the MC Subtypes
We used self-organizing maps (SOMs) by single-cell R analysis tool based on SOM machine learning (SCRAT) (Camp et al., 2017) to determine and envision coincidental gene sets exhibited in each population of MCs during fibrogenesis formation. The analysis revealed that MC subtypes expressed variable signatures under normal and fibrotic conditions (Figures 7A and 7B). We demonstrated multiple subtype-specific gene signatures, including extracellular region, extracellular space, neutrophil chemotaxis, ribosome, structural constituent of ribosomes, plasma membrane, integral component of membrane, endoplasmic reticulum, endoplasmic reticulum membrane, chromosome 12, signal transducer activity, structural constituent of muscle, ATP hydrolysis-coupled proton transport, regulation of transcription, and DNA-templated nuclear speck (Figures 7A and 7B). Notably, at the fibrotic stage, the myofibroblast subtype acquired a gene signature involving the structural constituent of muscle. Col13a1 and Col14a1 matrix fibroblasts as well as lipofibroblasts lost the signature of neutrophil chemotaxis. Mesothelial cells displayed gene signatures similar to Col14a1 matrix fibroblasts following fibrotic injury.
Figure 7. Metagene Analysis and Differential Potential of MCs.
(A and B) Metagene profile for each MC subtype in normal status (A) and fibrotic status (B). Arrows mark overexpressed and underexpressed metagene signatures. Red shows overexpression and blue shows underexpression.
(C and D) Lineage bifurcation of five MC subtypes in normal (C) and fibrotic lung (D). Cells on the same or neighboring branches are expected to be more hierarchically related. Color coding indicates pseudo-time scores of the cells.
We then projected MCs onto the SCRAT for sample similarity and pseudo-time analysis, which provides information inferring lineage trajectories from single-cell expression data in the form of 2D bifurcation. We assigned major MC subtypes onto SCRAT, including myofibroblasts, Col13a1 matrix fibroblasts, Col14a1 matrix fibroblasts, lipofibroblasts, mesenchymal progenitors, and Pdgfrb hi MCs. We found that the correlation-spanning tree and trajectory report displayed a directed hierarchical relation of the various subgroups, starting from mesenchymal progenitors and bifurcated to other MC subtypes (Figures 7C and 7D). It is interesting that our SCRAT analysis demonstrated a different lineage hierarchy among the MC subpopulations between normal and fibrotic stages. In the normal lung, mesenchymal progenitors bifurcated to lipofibroblasts and Col14a1 matrix fibroblasts, whereas Col14a1 matrix fibroblasts diverged to myofibroblasts and Col13a1 matrix fibroblasts. In the fibrotic lung, mesenchymal progenitors branched to lipofibroblasts and then lineage differentiated to the Pdgfrb hi subtype, myofibroblasts, Col14a1 matrix fibroblasts, and Col13a1 matrix fibroblasts sequentially.
Comparison between the Present Study and Recent Reports of Single-Cell Sequencing of Mesenchymal Cells
Recent single-cell studies reported by Zepp et al. (2017) showed that distinct Axin2+Pdgfrα + mesenchymal alveolar niche cell (MANC) and Axin2+ mesenchymal progenitor (AMP) subpopulations are found by scRNA-seq. We extracted the significant genes of MANCs and AMPs from the reports of Zepp et al. (2017) and compared the transcriptional programs of these two subgroups with our study (Figures S7A and S7B). AMPs, which have higher Acta2 expression levels, show correlation with our myofibroblast subgroup on D0 and Pdgfrb hi subgroup on D21. MANCs, which are expanded post-injury and mainly Pdgfrα +, are similar to the Col13a1/Col14a1 matrix fibroblasts. Our analysis revealed two more mesenchymal cell types in the normal lung compared to the Zepp et al. (2017) study. This may be because the cells included in our study are a larger population. Our analysis included all of the Epcam−CD31−CD45− cells, and Tbx4 lineage cells represent ~90% of the total Epcam− CD31− CD45− cells (Xie et al., 2016). All of the Axin2+, Axin2+Pdgfrα +, Pdgfrα +, and other cells compose ~50% of the Epcam− CD31− CD45− cells, when interpreted from Figures S1H and S1I in Zepp et al. (2017). Wnt2+ cells may constitute only a portion of the rest of the mesenchymal cells because Wnt2+ cells have ~85% overlap with Pdgfrα + cells and ~30% overlap with Axin2+ cells. It is unexpected to see that the scRNA-seq analysis segregated Wnt2+ cells so well from the other populations when they are supposed to have large overlaps. All of these in turn may lead this previous analysis to reveal limited mesenchymal subgroups.
A recent single-cell study by Lee et al. (2017) showed that Lgr5 and Lgr6 lineage cells are epithelial niche-promoting MCs located in alveolar and airway compartments, respectively. In our study, most of the Lgr5 and Lgr6 expressing cells are found within Acta2 hi expressing myofibroblasts (Figure S7C). In the D0 single-cell analysis, there are 93 Lgr5+ cells, 44 Lgr6+ cells, 13 Lgr5+/Lgr6+ cells, and 19 Lgr6+/Acta2+ cells. By D21, there are 124 Lgr5+ cells, 120 Lgr6+ cells, 31 Lgr5+/Lgr6+ cells, and 71 Lgr6+/Acta2+ cells. Lgr5 and Lgr6 may be the two subclusters of the myofibroblast subgroup with distinct locations. The Lgr5+/Lgr6+ cells found in our analysis are consistent with the claim by Lee et al. (2017) that Lgr6 marks cell populations expressing Lgr5.
DISCUSSION
In this study, we used unbiased single-cell transcriptome analyses to comprehensively classify the MC subtypes and cell lineage potential of individual MCs in the normal and fibrotic mouse lung. The analyses identified adult pulmonary MCs, including myofibroblasts, Col13a1 matrix fibroblasts, Col14a1 matrix fibroblasts, lipofibroblasts, mesenchymal progenitors, and mesothelial cells, as heterogeneous populations. In addition, the Pdgfrb hi fibroblast subpopulation was found to emerge in fibrogenesis. Our data provided combinatorial information of the signature genes, lncRNAs, extracellular and plasma membrane genes, and transcription factors for each of the MC subtypes. The fibroblast differentiation potential analyses identified different cell lineage trajectories between normal homeostasis and fibrotic conditions.
Are Myofibroblasts the Major Expanded MCs in Fibrotic Lung?
Myofibroblasts express α SMA with features reminiscent of both fibroblasts and smooth muscle cells (Hinz et al., 2007), and are the vital players in fibrotic diseases (Wynn and Ramalingam, 2012). When performing α SMA antibody staining or using αSMA-GFP-reporting mice, α SMA+ cells are dramatically expanded during fibrogenesis (Xie et al., 2016). We observed that Acta2 highly expressing cells are within the myofibroblast subtype; the low Acta2-expressing cells that express Col1a1 are matrix fibroblast subtypes. The percentage of matrix fibroblasts is ~30% in normal MCs and increases to 50% in the fibrotic MCs. These scRNA-seq analyses suggest that α SMA is not specific enough to discriminate myofibroblasts from matrix fibro-blasts. Therefore, many previously reported data that α SMA-expressing cells expand in lung fibrosis may be in fact mainly the result of matrix fibroblast amplification.
Lipofibroblasts Are Further Delineated under Homeostatic and Fibrotic Conditions
Lipofibroblasts contain lipid in the form of large cytoplasmic lipid droplets without a limiting biomembrane or lipid vacuoles (McGowan and Torday, 1997; Tahedl et al., 2014). Lipofibroblasts are involved in alveolar development and regeneration associated with alveolar epithelial type II cells (AECII) surfactant synthesis and vitamin A (retinoic acid) storage (Tahedl et al., 2014), and contribute to the AECII stem cell niche in the adult mouse lung (Barkauskas et al., 2013). Adipose differentiation-related protein (ADRP, encoded by Plin2) is believed to be the major component that mediates the consumption of lipid inclusions in lipofibroblasts and their subsequent transport to AECII cells (Friedmacher et al., 2014). Common adipocyte genes, including Pparg, Plin2, Fabp1, Fabp4, Fabp5, Lpl, and Lipa, are featured in lipofibroblasts (Chen et al., 1998, 2012; El Agha et al., 2017; Imamura et al., 2002; Li et al., 2016; Schultz et al., 2002; Varisco et al., 2012). On the basis of these previously suggested lipofibroblast markers, we identified the lipofibroblast subtype in which these markers are substantially highly expressed in our scRNA-seq data. These cells also express common fibroblast markers such as Vim, Col4a1, and Fn1. Concurrently, this subtype may exhibit signature genes related to immune responses. Specifically, an M2-like macrophage gene signature also was found in the subtype. However, lipofibroblasts do not express classical macrophage markers such as Ccr2, Lst1, Ms4a6c, plac8, and Ifitm3. These M2-like signature genes include Chil3, Mrc1(CD206), IL18, and CD9. A recent study showed that a subset of M2-like macrophages was defined by using scRNA-seq of FACS-sorted 68 CD45+, CSF1R− GFP+, F4/80+, and Ly6G−individual cells from mouse lung 7 days post-partial pneumonectomy and was a component of the regenerative AECII niche (Lechner et al., 2017). The relation between lipofibroblasts and M2-like macrophages warrants further investigation.
Are Lipofibroblasts Pdgfra+ Cells?
It is reported that Pdgfrα was expressed in a population of stem cell antigen-1 (Sca1)+ and CD34+, CD45−, CD31−, and Thy-1+ cells, which also were lipid-staining positive (McQualter et al., 2009), Pdgfra+ cells from Pdgfra;H2B-GFP mice contained lipid droplets and were supportive of the expansion and differentiation of epithelial cells in vitro (Barkauskas et al., 2013). A subpopulation of Pdgfrα+ fibroblasts are α SMA+ peribronchiolar smooth muscle and myofibroblasts in alveolar development (Chen et al., 2012; Endale et al., 2017). By using Pdgfra-GFP mice, Pdgfrα expressing precursor cells differentiate into myofibroblasts as well as lipofibroblasts, while the constitutive Pdgfracre mice revealed that the Pdgfrα signaling is restricted to bronchial smooth muscle cells and alveolar fibroblasts. Therefore, the overlap and the differences between Pdgfrα + fibroblasts and lipofibroblasts can be delineated. It is interesting that our scRNA-seq data showed that Pdgfra+ cells were mainly within Col13a1 and Col14a1 matrix fibroblasts. Pdgfra expression was low in Acta2+ myofibroblasts and lipofibroblasts (Figure S2B). Some Pdgfra+ matrix fibroblasts expressed Adrp, Lpl, and Lipa, but not Pparg and Fabp1,4,5. These results provide new insights into Pdgfra+ cells and suggest that they are mainly matrix fibro-blasts and can be transdifferentiated from lipofibroblasts.
Are the Newly Emerging Pdgfrb Hi Cells during Fibrogenesis Pericytes?
Our scRNA-seq observations suggested that the Pdgfrb hi subtype is a newly emerging MC population in response to the fibrotic injury. Pericytes were reported to be marked by Pdgfrβ protein expression and have established their interactions with endothelial cells (Barron et al., 2016). When we used Pdgfrβ antibody to stain fibrotic lungs, the labeled cells were expanded compared to normal lungs (Barron et al., 2016; Xie et al., 2016). By analyzing the expression of pericyte markers (Pdgfrb, Mcam, and Cspg4), our scRNA-seq data did not indicate a unique cluster for pericytes but suggested a phenomenon that pericytes are heterogeneous and plastic populations (Barron et al., 2016) and that they are overlapping with myofibroblasts and matrix fibroblasts. The newly emerging Pdgfrb hi MC subtype has uniquely expressed genes that can be cleanly separated from other MCs.
Mesothelial Cells Contribute to Mesenchyme Expansion
Mesothelium contributes to lung mesenchyme during lung development (Que et al., 2008). Wt-1 lineage-traced mesothelium gives rise to interstitial fibroblasts and myofibroblasts, which reside outside the blood vessels and alveoli in the embryonic lung (Que et al., 2008). The contribution of mesothelium to mesenchyme also was detected in disease contexts, including peritoneal, liver, and lung fibrosis (Li et al., 2013; Lua et al., 2015; von Gise et al., 2016). In addition, mesothelial cells can be lineage traced by mesenchymal transcription factor Tbx4 in the lung (Kumar et al., 2014; Xie et al., 2016). Our scRNA-seq data demonstrated well-clustered mesothelial cells in total MCs and in tdT+GFP+ cells from αSMA-GFP;Tbx4-Cre;Rosa26-tdTomato mice. Analyses of SOM gene sets revealed that the mesothelial cells displayed gene signatures similar to Col14a1 matrix fibroblasts in fibrotic lung. Significant gene and transcription factor analyses confirmed previously identified mesothelial markers. Thus, our data confirmed that mesothelial cells contribute to mesenchyme expansion.
Regulation of Fibroblast Subtypes by lncRNAs
lncRNAs are emerging as valuable mediators for fibrotic disease (Thum, 2014; Tzouvelekis and Kaminski, 2015). Notably, Malat1 is highly expressed in the myofibroblast and mesenchymal progenitor subtype. MALAT1 is one of the top expressed lncRNAs in patients with nonalcoholic steatohepatitis fibrosis. Its expression was increased in activated hepatic stellate cells (Leti et al., 2017). Neat1 is the most abundantly expressed lncRNA in Col13a1 matrix fibroblasts. It was reported that Neat1 expression was significantly elevated in mouse liver fibrosis and activated hepatic stellate cells. Suppression of Neat1 decreased liver fibrosis (Yu et al., 2017). Furthermore, Meg3 is the most distinctly expressed lncRNA in Col14a1 matrix fibroblasts, and its expression was decreased upon fibrotic injury. It is interesting that Meg3 has been reported to be the highest expressed lncRNA found in cardiac fibroblasts by global lncRNA profiling, and its inhibition in vivo decreased cardiac fibrosis. Silencing of Meg3 in cardiac fibroblasts decreased cardiac fibrosis (Piccoli et al., 2017). Thus, exploration of these distinguished lncRNAs in different mesenchymal subtypes will provide new insights into fibrogenesis.
Conceptual Exploration of Subgroup Trajectory
The trajectory analysis has been used to delineate the cell lineage differentiation in developmental or disease conditions (Savage et al., 2017; Yan et al., 2017). Pseudo-time analysis implies the state of the lineage initiation subgroup and the potential capacity of transdifferentiation of each subgroup. Cells located on the same or adjacent branches are expected to be more hierarchically related compared to cells on the neighboring branches in a given trajectory tree.
The mesenchymal progenitors are the lineage-initiation MC subgroup in both normal and fibrotic lung. The demonstration that mesenchymal progenitors have the capacity to reconstitute an entire mesenchymal trajectory tree suggested a similarity between the mesenchymal progenitors and previously reported mesenchymal stem cells, although the differences between the two cell types cannot be ignored. Mesenchymal stem cells were reported to be able to differentiate into multiple cell types, including fibroblasts, adipocytes, osteoblasts, chondrocytes, myocytes, and neurons. Our scRNA-seq analysis defined mesenchymal progenitors as resident lung cells. A trajectory analysis found that these progenitors could directly differentiate into lipofibroblasts and Col14a1 matrix fibroblasts. Our trajectory analysis also suggested that lipofibroblasts are poised for commitment to myofibroblasts through Pdgfrb hi subgroup in fibrotic lungs. This prediction is well correlated with a recent report that lipogenic fibroblasts or lipofibroblasts are a source of activated myofibroblasts in lung fibrosis (El Agha et al., 2017).
Experimental and biological characterizations such as precise lineage tracing and transcriptional conversion of mesenchymal progenitors (and mesenchymal subgroups) and the potential hierarchical differentiation in normal and in disease conditions warrant further investigation. Nevertheless, our trajectory analysis is inconsistent with previous reports and is our attempt to provide a conceptual framework to unmask the hierarchical relations between the mesenchymal subgroups.
In summary, the single-cell transcriptomic analyses dissected heterogeneous MC subtypes in both normal adult and fibrotic mouse lung. These comprehensive analyses provide transcription profiles for delineating mesenchymal taxonomy and add significantly to our understanding of fibroblast subpopulations in lung health and disease by providing a new toolbox to explore effector functions in disease. The study revealed distinctive molecular signatures for mesenchymal subsets in the lung, providing a foundation to augment our understanding of fibro-blast subpopulations and to identify cell markers at the protein level, localization in the lung, signaling programs, and future functional significance. Further identification of ‘‘pathogenic’’ fibroblast subpopulations in lung fibrosis will enable us to develop therapeutic targets, as well as more precise cellular and animal models for patients with progressive pulmonary fibrosis.
EXPERIMENTAL PROCEDURES
Mice
Triple-heterozygous αSMA-GFP;Tbx4-Cre;Rosa26-tdTomato mice were used. All of the mice were on a C57BL/6 background. The mouse studies were approved by the Institutional Animal Care and Use Committee at Cedars-Sinai Medical Center (protocols IACUC004722 and IACUC004751).
Sequencing Library Construction Using the 10x Genomics Chromium Platform
scRNA-seq libraries were prepared per the Single Cell 3′ Reagent Kit User Guide v2 (10x Genomics). Cellular suspensions were loaded on a Chromium Controller instrument (10x Genomics) to generate single-cell gel bead-in-emulsions (GEMs). GEM-reverse transcriptions (GEM-RTs) were performed in a Veriti 96-well thermal cycler (Thermo Fisher Scientific). After RT, GEMs were harvested and the cDNAs were amplified and cleaned up with the SPRIselect Reagent Kit (Beckman Coulter). Indexed sequencing libraries were constructed using the Chromium Single-Cell 3′ Library Kit (10x Genomics) for enzymatic fragmentation, end-repair, A-tailing, adaptor ligation, ligation cleanup, sample index PCR, and PCR cleanup. The barcoded sequencing libraries were quantified by quantitative PCR using the KAPA Library Quantification Kit (KAPA Biosystems). Sequencing libraries were loaded on a NextSeq500 (Illumina) with a custom sequencing setting (26 bp for read 1 and 98 bp for read 2) to obtain a sequencing depth of ~80,000 reads per cell.
Statistical Method
We used Cell Ranger version 1.3.1 (10x Genomics) to process raw sequencing data and Cell Ranger R kit version 2.0.0 and Seurat suite version 2.0.0 (Butler and Satija, 2017; Macosko et al., 2015) for downstream analysis. For clustering, principal-component analysis was performed for dimension reduction. Top 10 principal components (PCs) were selected by using a permutation-based test implemented in Seurat and passed to t-SNE for clustering visualization. sSeq version 1.0.0 integrated in the Cell Ranger R kit was used for modeling the gene expression with negative binomial distribution to identify genes whose expression was enriched in specific clusters. The Benjamini-Hochberg procedure was used for correcting errors of multiple testing.
Supplementary Material
Highlights.
Distinct MC subtypes were defined by single-cell transcriptome analysis
Lipofibroblasts were identified
Fibrotic Pdgfrb high MC subtype emerges post-injury
Integrative analysis of MC trajectories was constructed by machine learning
Acknowledgments
This study was supported by NIH grants P01 HL108793, R01 HL060539 (to P.W.N.), and R01 HL122068 (to D.J.). The authors would like to acknowledge Nargess Hassanzadeh-Kiabi and Andres Lopez at the Flow Cytometry Core of Cedars-Sinai Medical Center in Los Angeles for their assistance with FACS. The authors also are grateful for the assistance provided by Chintda Santiskulvong at Genomics Core of Cedars-Sinai Medical Center for single-cell RNA sequencing.
Footnotes
DATA AND SOFTWARE AVAILABILITY
The accession number for the raw data files of the RNA-seq analyses reported in this paper is GEO: GSE104154.
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.03.010.
DECLARATION OF INTERESTS
The authors declare no competing interests
AUTHOR CONTRIBUTIONS
Conceptualization, T.X., P.W.N. and D.J.; Software, Y.W. and N.D.; Validation and Formal Analysis, T.X., Y.W., and N.D.; RNA-seq Experiments, T.X. and N.L.; Investigation, G.H., F.T., V.K., N.L., Y.G., Y.W., N.D., Z.L., C.Y., P.C., B.S., J.L., T.X., and D.J.; Writing – Original Draft, T.X.; Writing – Review & Editing, Y.W., J.T., D.J., and P.W.N.; Funding Acquisition, D.J. and P.W.N.
References
- Andreucci JJ, Grant D, Cox DM, Tomc LK, Prywes R, Goldhamer DJ, Rodrigues N, Bédard PA, McDermott JC. Composition and function of AP-1 transcription complexes during muscle cell differentiation. J Biol Chem. 2002;277:16426–16432. doi: 10.1074/jbc.M110891200. [DOI] [PubMed] [Google Scholar]
- Bagchi RA, Roche P, Aroutiounova N, Espira L, Abrenica B, Schweitzer R, Czubryt MP. The transcription factor scleraxis is a critical regulator of cardiac fibroblast phenotype. BMC Biol. 2016;14:21. doi: 10.1186/s12915-016-0243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, Hogan BL. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron L, Gharib SA, Duffield JS. Lung pericytes and resident fibroblasts: busy multitaskers. Am J Pathol. 2016;186:2519–2531. doi: 10.1016/j.ajpath.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochaton-Piallat ML, Gabbiani G, Hinz B. The myofibroblast in wound healing and fibrosis: answered and unanswered questions. F1000Res Published online April. 2016;26:2016. doi: 10.12688/f1000research.8190.1. https://doi.org/10.12688/f1000research.8190.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Satija R. Integrated analysis of single cell transcriptomic data across conditions, technologies, and species. bioRxiv. 2017 doi: 10.1038/nbt.4096. https://doi.org/10.1101/164889. [DOI] [PMC free article] [PubMed]
- Camp JG, Sekine K, Gerber T, Loeffler-Wirth H, Binder H, Gac M, Kanton S, Kageyama J, Damm G, Seehofer D, et al. Multilineage communication regulates human liver bud development from pluripotency. Nature. 2017;546:533–538. doi: 10.1038/nature22796. [DOI] [PubMed] [Google Scholar]
- Cano E, Carmona R, Muñoz-Chápuli R. Wt1-expressing progenitors contribute to multiple tissues in the developing lung. Am J Physiol Lung Cell Mol Physiol. 2013;305:L322–L332. doi: 10.1152/ajplung.00424.2012. [DOI] [PubMed] [Google Scholar]
- Chen H, Jackson S, Doro M, McGowan S. Perinatal expression of genes that may participate in lipid metabolism by lipid-laden lung fibro-blasts. J Lipid Res. 1998;39:2483–2492. [PubMed] [Google Scholar]
- Chen L, Acciani T, Le Cras T, Lutzko C, Perl AK. Dynamic regulation of platelet-derived growth factor receptor α expression in alveolar fibroblasts during realveolarization. Am J Respir Cell Mol Biol. 2012;47:517–527. doi: 10.1165/rcmb.2012-0030OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derdak S, Penney DP, Keng P, Felch ME, Brown D, Phipps RP. Differential collagen and fibronectin production by Thy 1+ and Thy 1-lung fibroblast subpopulations. Am J Physiol. 1992;263:L283–L290. doi: 10.1152/ajplung.1992.263.2.L283. [DOI] [PubMed] [Google Scholar]
- Dixit R, Ai X, Fine A. Derivation of lung mesenchymal lineages from the fetal mesothelium requires hedgehog signaling for mesothelial cell entry. Development. 2013;140:4398–4406. doi: 10.1242/dev.098079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Guo M, Whitsett JA, Xu Y. ‘LungGENS’: a web-based tool for mapping single-cell gene expression in the developing lung. Thorax. 2015;70:1092–1094. doi: 10.1136/thoraxjnl-2015-207035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Agha E, Al Alam D, Carraro G, MacKenzie B, Goth K, De Langhe SP, Voswinckel R, Hajihosseini MK, Rehan VK, Bellusci S. Characterization of a novel fibroblast growth factor 10 (Fgf10) knock-in mouse line to target mesenchymal progenitors during embryonic development. PLoS One. 2012;7:e38452. doi: 10.1371/journal.pone.0038452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Agha E, Moiseenko A, Kheirollahi V, De Langhe S, Crnkovic S, Kwapiszewska G, Kosanovic D, Schwind F, Schermuly RT, Henneke I, et al. Two-way conversion between lipogenic and myogenic fibroblastic phenotypes marks the progression and resolution of lung fibrosis. Cell Stem Cell. 2017;20:261–273e3. doi: 10.1016/j.stem.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endale M, Ahlfeld S, Bao E, Chen X, Green J, Bess Z, Weirauch MT, Xu Y, Perl AK. Temporal, spatial, and phenotypical changes of PDGFRα expressing fibroblasts during late lung development. Dev Biol. 2017;425:161–175. doi: 10.1016/j.ydbio.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmacher F, Fujiwara N, Hofmann AD, Takahashi H, Alvarez LA, Gosemann JH, Puri P. Prenatal retinoic acid increases lipofibroblast expression in hypoplastic rat lungs with experimental congenital diaphragmatic hernia. J Pediatr Surg. 2014;49:876–881. doi: 10.1016/j.jpedsurg.2014.01.017. [DOI] [PubMed] [Google Scholar]
- Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453:314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- He Y, Wu YT, Huang C, Meng XM, Ma TT, Wu BM, Xu FY, Zhang L, Lv XW, Li J. Inhibitory effects of long noncoding RNA MEG3 on hepatic stellate cells activation and liver fibrogenesis. Biochim Biophys Acta. 2014;1842:2204–2215. doi: 10.1016/j.bbadis.2014.08.015. [DOI] [PubMed] [Google Scholar]
- Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton-Piallat ML, Gabbiani G. The myofibroblast: one function, multiple origins. Am J Pathol. 2007;170:1807–1816. doi: 10.2353/ajpath.2007.070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia LT, Ashley N, Ouaret D, Wang LM, Wilding J, Bodmer WF. Myofibroblasts are distinguished from activated skin fibroblasts by the expression of AOC3 and other associated markers. Proc Natl Acad Sci USA. 2016;113:E2162–E2171. doi: 10.1073/pnas.1603534113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C, Linn G, Chow YH, Kobayashi A, Mittelsteadt K, Altemeier WA, Gharib SA, Schnapp LM, Duffield JS. Role of lung pericytes and resident fibroblasts in the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med. 2013;188:820–830. doi: 10.1164/rccm.201212-2297OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura M, Inoguchi T, Ikuyama S, Taniguchi S, Kobayashi K, Nakashima N, Nawata H. ADRP stimulates lipid accumulation and lipid droplet formation in murine fibroblasts. Am J Physiol Endocrinol Metab. 2002;283:E775–E783. doi: 10.1152/ajpendo.00040.2002. [DOI] [PubMed] [Google Scholar]
- Ishitani T, Matsumoto K, Chitnis AB, Itoh M. Nrarp functions to modulate neural-crest-cell differentiation by regulating LEF1 protein stability. Nat Cell Biol. 2005;7:1106–1112. doi: 10.1038/ncb1311. [DOI] [PubMed] [Google Scholar]
- Jordana M, Schulman J, McSharry C, Irving LB, Newhouse MT, Jordana G, Gauldie J. Heterogeneous proliferative characteristics of human adult lung fibroblast lines and clonally derived fibroblasts from control and fibrotic tissue. Am Rev Respir Dis. 1988;137:579–584. doi: 10.1164/ajrccm/137.3.579. [DOI] [PubMed] [Google Scholar]
- Kanamori-Katayama M, Kaiho A, Ishizu Y, Okamura-Oho Y, Hino O, Abe M, Kishimoto T, Sekihara H, Nakamura Y, Suzuki H, et al. LRRN4 and UPK3B are markers of primary mesothelial cells. PLoS ONE. 2011;6:e25391. doi: 10.1371/journal.pone.0025391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimani PW, Holmes AJ, Grossmann RE, McGowan SE. PDGF-Ralpha gene expression predicts proliferation, but PDGF-A suppresses transdifferentiation of neonatal mouse lung myofibroblasts. Respir Res. 2009;10:119. doi: 10.1186/1465-9921-10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaru C, Schoonover KJ, Trudeau JB, Huynh ML, Zhou X, Hu H, Wenzel SE. Regional fibroblast heterogeneity in the lung: implications for remodeling. Am J Respir Crit Care Med. 2006;173:1208–1215. doi: 10.1164/rccm.200508-1218OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar ME, Bogard PE, Espinoza FH, Menke DB, Kingsley DM, Krasnow MA. Mesenchymal cells. Defining a mesenchymal progenitor niche at single-cell resolution. Science. 2014;346:1258810. doi: 10.1126/science.1258810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao T, Jiang Z, Yun J, Qiu W, Guo F, Huang C, Mancini JD, Gupta K, Laucho-Contreras ME, Naing ZZ, et al. Hhip haploinsufficiency sensitizes mice to age-related emphysema. Proc Natl Acad Sci USA. 2016;113:E4681–E4687. doi: 10.1073/pnas.1602342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner AJ, Driver IH, Lee J, Conroy CM, Nagle A, Locksley RM, Rock JR. Recruited monocytes and type 2 immunity promote lung regeneration following pneumonectomy. Cell Stem Cell. 2017;21:120–134e7. doi: 10.1016/j.stem.2017.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Tammela T, Hofree M, Choi J, Marjanovic ND, Han S, Canner D, Wu K, Paschini M, Bhang DH, et al. Anatomically and functionally distinct lung mesenchymal populations marked by Lgr5 and Lgr6. Cell. 2017;170:1149–1163e12. doi: 10.1016/j.cell.2017.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng D, Huan C, Xie T, Liang J, Wang J, Dai H, Wang C, Jiang D. Meta-analysis of genetic programs between idiopathic pulmonary fibrosis and sarcoidosis. PLoS One. 2013;8:e71059. doi: 10.1371/journal.pone.0071059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leti F, Legendre C, Still CD, Chu X, Petrick A, Gerhard GS, DiStefano JK. Altered expression of MALAT1 lncRNA in nonalcoholic steatohepatitis fibrosis regulates CXCL5 in hepatic stellate cells. Transl Res. 2017;190:25–39e21. doi: 10.1016/j.trsl.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jiang D, Liang J, Meltzer EB, Gray A, Miura R, Wogensen L, Yamaguchi Y, Noble PW. Severe lung fibrosis requires an invasive fibroblast phenotype regulated by hyaluronan and CD44. J Exp Med. 2011;208:1459–1471. doi: 10.1084/jem.20102510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang J, Asahina K. Mesothelial cells give rise to hepatic stellate cells and myofibroblasts via mesothelial-mesenchymal transition in liver injury. Proc Natl Acad Sci USA. 2013;110:2324–2329. doi: 10.1073/pnas.1214136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Ma S, Smith SM, Lee MK, Fischer A, Borok Z, Bellusci S, Li C, Minoo P. Mesodermal ALK5 controls lung myofibroblast versus lipofibroblast cell fate. BMC Biol. 2016;14:19. doi: 10.1186/s12915-016-0242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Zhang Y, Xie T, Liu N, Chen H, Geng Y, Kurkciyan A, Mena JM, Stripp BR, Jiang D, Noble PW. Hyaluronan and TLR4 promote surfactant-protein-C-positive alveolar progenitor cell renewal and prevent severe pulmonary fibrosis in mice. Nat Med. 2016;22:1285–1293. doi: 10.1038/nm.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Gersch RP, Hawke TJ, Hadjiargyrou M. Silencing of Mustn1 inhibits myogenic fusion and differentiation. Am J Physiol Cell Physiol. 2010;298:C1100–C1108. doi: 10.1152/ajpcell.00553.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lua I, Li Y, Pappoe LS, Asahina K. Myofibroblastic conversion and regeneration of mesothelial cells in peritoneal and liver fibrosis. Am J Pathol. 2015;185:3258–3273. doi: 10.1016/j.ajpath.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly parallel genome-wide expression proflifing of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan SE, Torday JS. The pulmonary lipofibroblast (lipid interstitial cell) and its contributions to alveolar development. Annu Rev Physiol. 1997;59:43–62. doi: 10.1146/annurev.physiol.59.1.43. [DOI] [PubMed] [Google Scholar]
- McQualter JL, Brouard N, Williams B, Baird BN, Sims-Lucas S, Yuen K, Nilsson SK, Simmons PJ, Bertoncello I. Endogenous fibroblastic progenitor cells in the adult mouse lung are highly enriched in the sca-1 positive cell fraction. Stem Cells. 2009;27:623–633. doi: 10.1634/stemcells.2008-0866. [DOI] [PubMed] [Google Scholar]
- Piccoli MT, Gupta SK, Viereck J, Foinquinos A, Samolovac S, Kramer FL, Garg A, Remke J, Zimmer K, Batkai S, Thum T. Inhibition of the cardiac fibroblast-enriched lncRNAMeg3 prevents cardiac fibrosis and diastolic dysfunction. Circ Res. 2017;121:575–583. doi: 10.1161/CIRCRESAHA.117.310624. [DOI] [PubMed] [Google Scholar]
- Que J, Wilm B, Hasegawa H, Wang F, Bader D, Hogan BL. Mesothelium contributes to vascular smooth muscle and mesenchyme during lung development. Proc Natl Acad Sci USA. 2008;105:16626–16630. doi: 10.1073/pnas.0808649105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkevich Y, Mori T, Sahoo D, Xu PX, Bermingham JR, Jr, Weissman IL. Identification and prospective isolation of a mesothelial precursor lineage giving rise to smooth muscle cells and fibroblasts for mammalian internal organs, and their vasculature. Nat Cell Biol. 2012;14:1251–1260. doi: 10.1038/ncb2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin YM, Penel N, Pérot G, Neuville A, Vélasco V, Ranchére-Vince D, Terrier P, Coindre JM. Transgelin is a novel marker of smooth muscle differentiation that improves diagnostic accuracy of leiomyosarcomas: a comparative immunohistochemical reappraisal of myogenic markers in 900 soft tissue tumors. Mod Pathol. 2013;26:502–510. doi: 10.1038/modpathol.2012.192. [DOI] [PubMed] [Google Scholar]
- Rock JR, Barkauskas CE, Cronce MJ, Xue Y, Harris JR, Liang J, Noble PW, Hogan BL. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci USA. 2011;108:E1475–E1483. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage P, Blanchet-Cohen A, Revil T, Badescu D, Saleh SMI, Wang YC, Zuo D, Liu L, Bertos NR, Munoz-Ramos V, et al. A targetable EGFR-dependent tumor-initiating program in breast cancer. Cell Rep. 2017;21:1140–1149. doi: 10.1016/j.celrep.2017.10.015. [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Torres E, Londos C, Torday JS. Role of adipocyte differentiation-related protein in surfactant phospholipid synthesis by type II cells. Am J Physiol Lung Cell Mol Physiol. 2002;283:L288–L296. doi: 10.1152/ajplung.00204.2001. [DOI] [PubMed] [Google Scholar]
- Sun KH, Chang Y, Reed NI, Sheppard D. α-Smooth muscle actin is an inconsistent marker of fibroblasts responsible for force-dependent TGFβ activation or collagen production across multiple models of organ fibrosis. Am J Physiol Lung Cell Mol Physiol. 2016;310:L824–L836. doi: 10.1152/ajplung.00350.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahedl D, Wirkes A, Tschanz SA, Ochs M, Muhlfeld C. How common is the lipid body-containing interstitial cell in the mammalian lung? Am J Physiol Lung Cell Mol Physiol. 2014;307:L386–L394. doi: 10.1152/ajplung.00131.2014. [DOI] [PubMed] [Google Scholar]
- Thannickal VJ, Zhou Y, Gaggar A, Duncan SR. Fibrosis: ultimate and proximate causes. J Clin Invest. 2014;124:4673–4677. doi: 10.1172/JCI74368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas AQ, Lane K, Phillips J, 3rd, Prince M, Markin C, Speer M, Schwartz DA, Gaddipati R, Marney A, Johnson J, et al. Heterozygosity for a surfactant protein C gene mutation associated with usual interstitial pneumonitis and cellular nonspecific interstitial pneumonitis in one kindred. Am J Respir Crit Care Med. 2002;165:1322–1328. doi: 10.1164/rccm.200112-123OC. [DOI] [PubMed] [Google Scholar]
- Thum T. Noncoding RNAs and myocardial fibrosis. Nat Rev Cardiol. 2014;11:655–663. doi: 10.1038/nrcardio.2014.125. [DOI] [PubMed] [Google Scholar]
- Torday JS, Rehan VK. On the evolution of the pulmonary alveolar lipofibroblast. Exp Cell Res. 2016;340:215–219. doi: 10.1016/j.yexcr.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzouvelekis A, Kaminski N. Epigenetics in idiopathic pulmonary fibrosis. Biochem Cell Biol. 2015;93:159–170. doi: 10.1139/bcb-2014-0126. [DOI] [PubMed] [Google Scholar]
- Varisco BM, Ambalavanan N, Whitsett JA, Hagood JS. Thy-1 signals through PPARγ to promote lipofibroblast differentiation in the developing lung. Am J Respir Cell Mol Biol. 2012;46:765–772. doi: 10.1165/rcmb.2011-0316OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gise A, Stevens SM, Honor LB, Oh JH, Gao C, Zhou B, Pu WT. Contribution of fetal, but not adult, pulmonary mesothelium to mesenchymal lineages in lung homeostasis and fibrosis. Am J Respir Cell Mol Biol. 2016;54:222–230. doi: 10.1165/rcmb.2014-0461OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA, Ramalingam TR. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med. 2012;18:1028–1040. doi: 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang FL, Fang M, Yutzey KE. Loss of β-catenin in resident cardiac fibroblasts attenuates fibrosis induced by pressure overload in mice. Nat Commun. 2017;8:712. doi: 10.1038/s41467-017-00840-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Liang J, Liu N, Huan C, Zhang Y, Liu W, Kumar M, Xiao R, D’Armiento J, Metzger D, et al. Transcription factor TBX4 regulates myofibroblast accumulation and lung fibrosis. J Clin Invest. 2016;126:3063–3079. doi: 10.1172/JCI85328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Gevaert O, Zheng GXY, Anchang B, Probert CS, Larkin KA, Davies PS, Cheng ZF, Kaddis JS, Han A, et al. Intestinal enter-oendocrine lineage cells possess homeostatic and injury-inducible stem cell activity. Cell Stem Cell. 2017;21:78–90e6. doi: 10.1016/j.stem.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Jiang Z, Chen B, Dong P, Zheng J. NEAT1 accelerates the progression of liver fibrosis via regulation of microRNA-122 and Kruppel-like factor 6. J Mol Med (Berl) 2017;95:1191–1202. doi: 10.1007/s00109-017-1586-5. [DOI] [PubMed] [Google Scholar]
- Zepp JA, Zacharias WJ, Frank DB, Cavanaugh CA, Zhou S, Morley MP, Morrisey EE. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell. 2017;170:1134–1148e10. doi: 10.1016/j.cell.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.