Abstract
The deterministic force of natural selection and stochastic influence of drift shape RNA virus evolution. New deep-sequencing and microfluidics technologies allow us to quantify the effect of mutations and trace the evolution of viral populations with single-genome and single-nucleotide resolution. Such experiments can reveal the topography of the genotype-fitness landscapes that shape the path of viral evolution. By combining historical analyses, like phylogenetic approaches, with high-throughput and high-resolution evolutionary experiments, we can observe parallel patterns of evolution that drive important phenotypic transitions. These developments provide a framework for quantifying and anticipating potential evolutionary events. Here, we examine emerging technologies that can map the selective landscapes of viruses, focusing on their application to pathogenic viruses. We identify areas where these technologies can bolster our ability to study the evolution of viruses and to anticipate and possibly intervene in evolutionary events and prevent viral disease.
Graphical Abstract
Dolan et al review experimental approaches to the study of evolutionary landscapes of viruses and the forces driving the dynamics of evolving virus populations. Combing various approaches can elucidate evolutionary mechanisms and pathways underlying phenotypic transitions, allowing us to anticipate and possibly intervene in evolutionary events to prevent viral disease.
INTRODUCTION
Predicting evolution is a key goal in virology, especially for pathogenic viruses (Holmes, 2013). Accurately forecasting viral evolutionary dynamics has implications for vaccine design and the maintenance, drug design, and surveillance of viral pathogens. Ultimately, the evolutionary path of a viral population is shaped by selection. The deterministic influence of natural selection tends to increase the fitness of the population over time, through positive selection of beneficial mutations. Natural selection constrains the evolutionary path by removing lethal and deleterious mutations from the population through negative, or purifying, selection. As a result, certain evolutionary trajectories are more likely than others. Therefore, understanding the selective forces that drive and constrain the evolutionary trajectories of specific viruses is critical to evolutionary prediction. New approaches and technologies to study specific phenotypic transitions that underlie host transmission or pathogenesis, could inform future prediction, surveillance, and intervention strategies.
Although RNA viruses have very limited genetic capacity, and a correspondingly limited repertoire of evolutionary solutions, they present unique challenges to evolutionary prediction. Stochastic influences of genetic drift, mutation, and recombination all limit our ability to anticipate evolutionary events. RNA viruses display high mutation rates (as high as 1 mutation in 1000 bases) and many engage in frequent recombination and reassortment, creating novel genotypes from co-circulating strains. RNA viruses also undergo frequent population bottlenecks as they pass from host to host, and encounter changes in their selective environment (Grubaugh et al., 2015, 2017; Varble et al., 2014). These bottlenecks are especially important in the evolution of viruses that transmit between highly divergent host species (Recently reviewed in (McCrone and Lauring, 2017)).
The feasibility of predicting viral evolution depends on the breadth and scale of the question being asked, and requires careful consideration of the relevant selective pressures and evolutionary parameters. The life histories of individual viruses, and the biological complexity associated with specific evolutionary challenges, influence the predictability of their evolutionary trajectories. Predicting the evolutionary response to a strong and specific selective pressure, such as treatment with a single direct-acting antiviral, is a more tractable question than predicting the next zoonotic spillover event. In the latter case, prediction is impeded by several ecological and biological factors influencing the likelihood of a specific outcome. It follows that there is significant disagreement about the utility and feasibility of approaches to predict virus evolution, and the resources that should be devoted to surveillance initiatives (Geoghegan and Holmes, 2017).
Despite the challenges in anticipating evolutionary events, there are on-going efforts to predict virus evolution in response to specific selection. One of the most notable examples is the forecasting of seasonal influenza dynamics to guide vaccine strain selection. Current methods of vaccine formulation, based on the observation of currently circulating strains, have been moderately, but variably, successful in maintaining efficacious vaccine stocks year-to-year, with efficacy normally around 30–50% against Influenza A virus (IAV) strains (Flannery et al., 2017; Jackson et al., 2017). Predictive models of RNA virus evolution could increase these rates as they are adopted into strain selection procedures (Du et al., 2017; Luksza and Lässig, 2014; Neher et al., 2016). Moreover, predictive methods could be used to anticipate and monitor for potential egg-adaptive mutations in the influenza glycoprotein hemagglutinin, which arise during some vaccine production procedures, leading to reduced efficacy (Zost et al., 2017). Efforts to anticipate and predict other viral evolutionary transitions, such as the emergence of drug resistance or increased pathogenesis, would also benefit from improved predictive models of virus evolution.
Here, we provide a perspective on current experimental approaches to the study of evolutionary landscapes of viruses and the forces that drive the dynamics of evolving virus populations. We highlight how combining empirical approaches to study the effect of individual mutations with historical analyses of virus evolution can inform our efforts to anticipate the evolution and emergence of new viral genotypes. We also draw examples from experimental and theoretical work outside of virology that define the limits of prediction and inform future study of viral evolutionary biology. We argue that, although prediction at certain scales is currently out of the question, mapping the biological and physical constraints on evolutionary trajectories can inform our understanding of the evolutionary potential of RNA viruses. These data can also leverage historical studies by testing mechanistic hypotheses they raise and their improving underlying evolutionary models. Together these approaches can elucidate evolutionary mechanisms and pathways underlying important phenotypic transitions, and better anticipate, monitor, and respond to evolutionary events in nature.
Virus evolution and predictability
As with all genetic organisms, the forces of mutation, drift, and natural selection shape the course of virus evolution and determine its predictability (Kryazhimskiy et al., 2014; Lenski et al., 1991). The sequence space of an RNA virus constitutes a vast network of genotypes connected by single mutational steps (Figure 1A). Individuals in a population sample adjacent genotypes through mutation. Due to their high mutation rates and often large population sizes, RNA viruses rapidly sample adjacent genotypes in sequence space, enabling rapid adaptation to changes in the selective environment. Each mutation has a characteristic effect on the reproductive fitness of the virus, or a mutational fitness effect (MFE), and selection acts on individual alleles based on their MFE. Beneficial alleles, those with positive fitness effects, are driven to fixation by positive selection. Deleterious and lethal alleles are removed from the population by negative selection. Together, these selective forces shape the evolutionary trajectories of virus populations across sequence space.
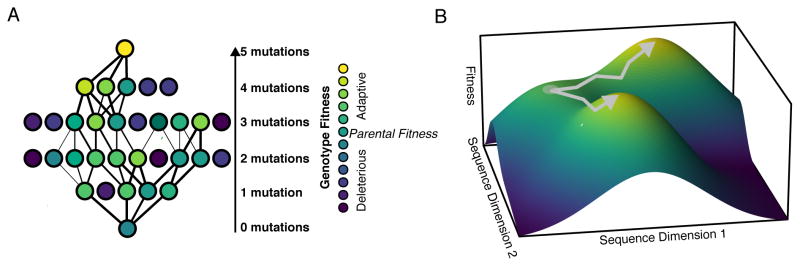
Figure 1. Example genotype-fitness landscapes.
A.) The network of genotypes corresponding to a simple 5-locus, 2-mutation system, showing all 32 possible genotype combinations (25) connected by single nucleotide substitutions. The likelihood of traversing a given link (acquiring a mutation) is shown as the line weight. Interactions between mutations (epistasis) shapes the complexity of the network. B.) From a starting point (grey dot), populations explore the topography of the genotype-fitness landscape by acquiring mutations. The slope of the landscape represents the change in fitness associated with genotypic change. Natural selection drives populations (grey lines) up the fitness gradient to local fitness maxima.
Sewall Wright described genotype-fitness landscapes (Figure 1B) in 1932 as a representation of phenotype, or fitness, as a function of genotype of a population (Wright, 1932). The topography of the landscape represents the fitness effects associated with movement (evolutionary change) through sequence space (McCandlish, 2011). The trajectories of viral populations across these landscapes are shaped by stochastic and deterministic forces. The deterministic force of natural selection tends to drive populations towards fitness maxima, or peaks in the fitness landscape, by fixing adaptive mutations. Conversely, genetic drift tends to shift the population to lower fitness by primarily fixing deleterious mutations.
Understanding why populations evolve along one trajectory, or a set of trajectories, and not others requires an understanding of how positive and negative selection act on viral genomes and their encoded proteins. The maintenance of viral fitness, enzyme function and protein stability constrains the repertoire of viable mutations (Acevedo et al., 2014; Wylie and Shakhnovich, 2011). Protein stability is a particularly important determinant of mutational fitness effects, and mutations which destabilize protein structures are rapidly removed by negative selection (Bloom et al., 2007). Because their genomes are dense with coding regions, models of mutational fitness based on calculations of the difference in thermodynamic stability between WT and mutant residues can correlate surprisingly well with empirically determined mutational fitness effects on viral genomes (Wylie and Shakhnovich, 2011). Environmental influences, such as host immunity or drug pressure, also shape the fitness landscape. In contrast to negative selection, which limits diversity, these extrinsic selective pressures are not static, and their fluctuations drive positive selection of new adaptive alleles in the viral population.
When viral population sizes are very large, they exist as swarms of related genotypes, often referred to as a quasispecies (Andino and Domingo, 2015; Eigen, 1971, 1996; Lauring and Andino, 2010). Although driven by the same evolutionary forces, selection in these populations acts on the entire mutational distribution due to intense mutational pressure (Wilke, 2005). Although not a universal model for virus evolution across all spatial and temporal scales (Holmes and Moya, 2002), quasispecies theory conveys the nature of large RNA virus populations as distributions of genotypes across sequence space. It also makes important predictions about the evolution of viral populations. First, the consensus sequence of the population does not necessarily represent the most common sequences in the population. Instead, the population exists as a dynamic equilibrium of variants surrounding a master sequence, or master sequences, that emerge in response to selection on the population. Second, the master genotypes are positioned in sequence space by past selection to maximize accessibility to neutral and adaptive neighbors and buffer mutation by increasing the probability of neutral mutations (i.e. mutational robustness) (Forster et al., 2006; Lauring et al., 2012; Montville et al., 2005). Third, under given selective conditions, specific quasispecies distributions emerge reproducibly as a function of the fitness landscape. These complex population and dynamic population structures appear to be relevant to evolution within and between hosts (Korboukh et al., 2014; Stern et al., 2014; Vignuzzi et al., 2006; Xiao et al., 2017). Quantifying the dynamics of viral quasispecies and understanding their relevance to viral biology and pathogenesis represents a major technical challenge in experimental RNA virus evolution.
Identifying patterns of positive selection
Determining the diversity of adaptive paths available to a population in response to selection is key to predicting viral evolution (Figure 2A). The traditional methods for identifying potential evolutionary trajectories are historical methods, such as phylogenetics, and experimental selection and clonal sequencing. These tools have been used for decades to map the patterns of mutations that escape antibody recognition or confer resistance to drug pressure. Although evolution is largely unpredictable at some scales (Kryazhimskiy et al., 2014; Nelson et al., 2006; Sailer and Harms, 2017a), the existence of parallel evolution in phenotypic or genotypic evolution has emerged as a common theme in evolutionary studies (Sackman et al., 2017; Stewart et al., 1988; Xiao et al., 2017; Xue et al., 2017). In cases where viruses are subject to similar selective pressures in parallel or repeatedly over time, for example within individual cells, hosts, or host populations, common adaptive mutations are often selected (Morley and Turner, 2017; Xue et al., 2017). In large populations, selection drives the emergence of reproducible quasispecies structures under common selective pressures. These repeated patterns often reflect the key selective pressures underlying important phenotypic transitions, such as escape from immune selection, changes in virulence, or shifts in host tropism. As such, these common signatures could potentially serve as indicators for future surveillance or intervention.
Figure 2. Experimental techniques to study viral evolutionary paths and aid in prediction.
A. A 2D genotype-fitness landscape underlying a hypothetical phenotypic transition (from purple to gold). B. Traversal of a viral population through such a landscape can be examined in a number of experimental ways. Phylogenetic reconstruction examines the evolutionary history of a viral population. Experimental evolution techniques measure the precise trajectories a given viral population takes to navigate a particular selective environment. Library-based screening allows further manipulation of the viral population, by exploring the local topography of the genotype-fitness landscape. C.) High resolution sequencing, such as Circular Sequencing (CirSeq), allows for the tracking of most single nucleotide mutations in a viral population, even at low frequencies. The error correction of CirSeq relies on the generation of head-to-tail cDNA repeats from circularized, fragmented RNA. Only mutations occurring in a majority of these repeats are identified as true mutations. If combined with serial passaging, such high resolution sequencing experiments can be used to estimate fitness effects across the genome and describe the evolutionary landscapes of viruses. D.) Deep mutational scanning (DMS) uses successive rounds of mutagenic PCR to generate a library of all possible codon or nucleotide substitutions of a given sequence. This library is then cloned back into the viral sequence for rescue and creation of mutant viruses. This population undergoes some form of selection (e.g. antibody selection), and the frequency of codon mutants postselection is determined by deep sequencing. A variety of methods can be used to determine amino acid preference at individual site across the gene of interest. Such preferences can be used to better inform phylogenetic reconstruction of viral evolution. E.) Microfluidics allow for massively parallel evolutionary experimentation and fine control of evolutionary and selective parameters influencing the evolution of viral populations. The ability to observe individual host cells, and individual virus particles and genomes, enables the quantification of important biological heterogeneity and dynamics underlying the infection process.
Experimental interrogation of historical parallelism
The repeated emergence of circulating vaccine-derived poliovirus (cVDPVs) provides another notable example of parallelism in virus evolution and emergence. Live, attenuated oral poliovirus vaccines (OPV) were developed as alternatives to the inactivated poliovirus vaccine (IPV). These vaccines, attenuated by adaptation to low temperatures, provide many benefits, including the development of more effective mucosal immunity (Sabin and Boulger, 1973). However, because the virus is active, vaccinated individuals shed live virus in their stool for 3–6 weeks post-vaccination (Alexander et al., 1997). In populations of low vaccine coverage, this attenuated virus can be spread, accumulating mutations along the way that reverse its attenuated phenotype, leading to outbreaks of poliomyelitis. Outbreaks of virulent cVDPV have thwarted global poliovirus eradication efforts (Kew et al., 2005).
Recent analysis of the patterns of selection across independent vaccine-derived virus populations revealed common adaptive mutations across the virus genome, including modifications to the viral internal ribosome entry site (IRES) and coding sequences (Stern et al., 2017). These mutations often occurred sequentially, suggesting that the path to OPV virulence is contingent on initial gatekeeper mutations in the 5′ UTR and capsid protein, VP1, that reverse the originally engineered attenuating mutations. These key mutational steps toward cVDPV emergence occurred in nearly all sequenced isolates, and likely reverse the temperature sensitivity of the vaccine strain (Bouchard et al., 1995).
In addition, cVDPV emergence is associated with recombination between attenuated OPV strains and circulating Coxsackieviruses (Guillot et al., 2000), related viruses that are also members of the Picornaviridae family. Recombination between co-circulating viruses serves as a shortcut to circumvent the initial gatekeeper mutations, by replacing the entire 5′ of the OPV virus with that of another virus. After these mutations sweep the population, they are followed by further sweeps of adaptive mutations that enhance the fitness of the virus (Stern et al., 2017). Notably, these observations were subsequently verified, demonstrating that the gatekeeper mutations enhance the ability of the temperature-sensitive, attenuated virus to replicate at higher temperatures.
This study provides a framework for combining historical and experimental methods to map the evolutionary pathway of a virus, and reconstituting the selective conditions in a more controlled experimental setting within the laboratory. The observed path to poliovirus virulence has informed new strategies to prevent the emergence of cVDPV in future polio vaccines. Genetically engineering the vaccine strain with mutations that reduce its adaptive capacity, by preventing its ability to accumulate adaptive mutations or to recombine with other viruses, could prevent the reversion of attenuating mutations (Vignuzzi et al., 2008; Xiao et al., 2016). Moreover, this study emphasizes the importance of evolutionary parallelism to the prediction of future adaptation.
Microfluidic approaches for high-throughput analysis of parallel evolution
The repeated deployment of OPV provides a rather unique opportunity for mapping the adaptive trajectories of a viral population. Historical methods typically only identify a small number of mutations that are fixed in the population, but do not identify the complete repertoire of adaptive mutations available to a viral population. Recently, microfluidic technologies have emerged as a powerful tool to miniaturize traditional laboratory experiments, enabling massively parallel evolutionary experiments. These tools provide remarkable utility in the study of virus evolution. Microfluidic technologies can be used to sort, infect and monitor the phenotype of individual cells, thus creating the potential for parallel experiments that can identify common evolutionary trajectories from independent experiments. They also allow fine control of many experimental variables simultaneously, and improved resolution of viral and host cell dynamics during infection.
Although their potential for understanding dynamic biological processes is clear from recent in vitro and in vivo studies of cellular transcription (Dixit et al., 2016; Macosko et al., 2015; Zilionis et al., 2017) and protein translation (Bjornson et al., 2013), these technologies have not yet been widely adopted for the study of viral infection. However, early results have already demonstrated the profound heterogeneity in viral and cellular phenotypes during infection, even in monocultured cell lines (Guo et al., 2017; Russell et al., 2018).
Although microfluidics encompasses a broad range of tools, two applications are especially useful in the study of evolution of RNA viruses. First, the sequestration and manipulation of individual viral particles for genotypic and phenotypic characterization is a fundamentally new way to perform virology (Guo et al., 2017). Recent studies have quantified such phenotypes as viability (Tao et al., 2015a), replication kinetics (Guo et al., 2017), and antibody binding and neutralization (Rotem et al., 2016). Depending on the platform, deep-sequencing technologies are limited by read length and accuracy and therefore require probabilistic reconstruction of the haplotype diversity of the population which limits resolution (Di Giallonardo et al., 2014). Single-genome sequencing with droplet-based barcoding approaches overcomes this limitation of deep-sequencing haplotype diversity to be determined at single-virus resolution (Lan et al., 2016; Tao et al., 2015b). The ability to determine the structure and dynamics of viral populations in vivo will open long-standing questions to investigation, including the nature of viral population structures and quasispecies dynamics associated with virus infection.
The second major benefit of microfluidics is the ability to analyze individual host cells. Infection is a spatially and temporally heterogeneous process. Viral populations encounter heterogeneous selective environments across many scales, from individual host cell types and cellular immune states within the host, to variability in host genotypes. Therefore, quantifying genotypic heterogeneity in viral populations as well as phenotypic heterogeneity in the host environment is critical to understanding the selective forces that drive virus evolution within a host. However, most experiments are performed using bulk preparations of virus or cellular RNA from cultured cells or complex tissues, masking much of the heterogeneity present and obscuring a more realistic view of the infection process. Single-cell transcriptome sequencing studies of cells infected with Zika virus (Zanini et al., 2018) and Influenza A virus (IAV) (Russell et al., 2018) have already demonstrated the profound heterogeneity present even in simple models of infection.
The ability to finely control host cells and viruses has also made microfluidics a useful tool for miniaturizing and parallelizing viral experimental evolution to explore the influence of evolutionary parameters on the evolution of viral populations (Fischer et al., 2015; Guo et al., 2017; Rotem et al., 2016; Tao et al., 2015a) (Figure 2C). Microfluidic technologies allow simultaneous observation of the genotypes and phenotypes of host and virus at single-cell and single-virus resolution, providing a holistic view of the infection. By enabling parallel evolutionary experiments in conditions that mimic specific selective pressures, these technologies can, in some cases, enumerate the limited number of evolutionary solutions available to overcome specific challenges.
Defining genome-wide negative selection with deep-sequencing
Negative selection, or purifying selection, constrains the evolutionary trajectory by removing deleterious genotypes from populations. Mapping the regions of the genome that evolve under strong negative selection provides insights into the structural and functional constraints on the viral genome and encoded proteins. Sites evolving under strong purifying selection can also suggest targets for vaccine or antiviral development with improved resistance profiles. However, identifying sites evolving under negative selection has historically been a challenge experimentally and phylogenetically, as it is often impossible to distinguish from sampling bias (Zhai et al., 2009). In other words, distinguishing those mutations which cannot occur from those that have not occurred. Recent advances in deep sequencing and library-based mutation screening allow the robust measurement of site-specific negative selection through the observation of dynamics of low frequency alleles in naturally evolving populations (Acevedo et al., 2014; Jabara et al., 2011; Kennedy et al., 2014; Kosik et al., 2018) and the high-resolution screening of mutant libraries before and after selection.
Mapping mutational fitness with population sequencing
Ultra-deep sequencing technologies are necessary to accurately assess the swarm of mutations normally hidden below the limits of conventional next generation sequencing (NGS) approaches. Although traditional NGS approaches can readily detect the accumulation of adaptive alleles to high frequencies, deleterious or lethal alleles are held to low frequencies by negative selection and therefore require increased depth to accurately estimate their frequencies. New techniques have recently been developed to counteract the error rate associated with NGS, allowing accurate detection of low frequency variants in viral populations. Circular Sequencing (Figure 2A), or CirSeq, is a library preparation and analysis technique that relies on circularization of fragmented viral RNA and subsequent rolling-circle reverse transcription to generate tandem cDNA repeats from template RNAs (Acevedo et al., 2014). A consensus sequence is generated from each tandem repeat, where only errors appearing on the majority of repeats are considered true mutations. Another high resolution sequencing technique, Primer ID, relies on the incorporation of random barcodes to the primers used for initial cDNA synthesis (Zhou et al., 2015). These barcodes are then used to computationally infer the frequency and consensus sequences of the template RNAs.
Recently, CirSeq, Primer ID, and Luria-Delbrück fluctuation tests have all been used to measure the rates of individual nucleotide substitutions of RNA viruses (e.g. A to G, T to C, G to A) (Acevedo et al., 2014; Korboukh et al., 2014; Pauly et al., 2017; Xiao et al., 2016). In serial passage experiments of large viral populations, the MFE of individual mutations can be estimated by comparison of an allele’s frequency trajectory to its respective mutation rate. Neutral mutations will tend to accumulate in the population as they are produced through mutation, while deleterious mutations will accumulate at a rate slower than the respective mutation rate due to their removal from the population by negative selection (Acevedo et al., 2014; Whitfield and Andino, 2016). With the coverage and error correction afforded by ultra-deep sequencing, fitness values can be assigned to even low frequency mutations, thus revealing the complete spectrum of evolutionary dynamics in viral populations (Figure 2A,C).
Mutagenesis-based screening
Deep Mutational Scanning (DMS) (Araya and Fowler, 2011; Fowler et al., 2010) uses error-prone PCR or random mutagenesis to create libraries of all possible nucleotide or codon substitutions of a target locus (Jiang et al., 2016; Roscoe et al., 2013; Thyagarajan and Bloom, 2014; Wu et al., 2013, 2014a, 2014b) (Figure 2D). These mutant libraries are used to generate, or ‘rescue’, populations of mutant virus. After generation, the pool of viable mutant viruses may be sequenced and compared to the mutagenized library to examine effects on viral fitness. Alternatively, the viable virus population may be subjected to some form of selection, and the effect of each mutation is assessed by comparison to the variant frequency before and after selection. The site-specific preference for individual residues is generally quantified in terms of the enrichment of individual mutations or the overall preference of each site for each amino acid (Ashenberg et al., 2017; Thyagarajan and Bloom, 2014; Wu et al., 2014a). Some saturating mutagenesis approaches can specifically avoid clones with multiple mutations (EMPIRIC; (Hietpas et al., 2011)), while others permit a fraction of the resulting clones to incorporate mutations at multiple sites (Thyagarajan and Bloom, 2014). In the latter case, most codon mutations are represented more than once in the generated libraries, meaning a given mutant’s enrichment in the library represents an average of all genetic backgrounds.
In a recent study in IAV, DMS was used to identify residues in the IAV nucleoprotein (NP), which affect sensitivity to the cellular restriction factor, myxovirus-resistance protein A (MxA) that binds to NP and prevents viral replication (Ashenberg et al., 2017). The authors identified a number of mutations that increased NP resistance or sensitivity to MxA binding. Importantly, this approach identified not only residues previously unknown to affect MxA resistance, but also those identified in previous epidemics, demonstrating the efficacy of this approach in identifying established, causative mutations. Most identified mutants conferred higher sensitivity to MxA, illustrating negative selection and detection of additional constraints host-virus interactions place on influenza evolution. Due to their small genome sizes, RNA viruses are especially amenable and experimentally tractable to mutagenesis approaches. In fact, a recent study reported mutagenesis of the entire influenza genome to identify determinants of interferon-sensitivity (Du et al., 2018).
Future challenges and opportunities
Dynamic Landscapes
The concept of a single, static fitness landscape is useful conceptually, but in reality RNA virus evolution takes place across dynamic, responsive genotype-fitness landscapes (Fig 3A). Determining the fitness effects of individual mutations experimentally provides a picture of the selective pressures that influence the evolutionary trajectory contingent on a given genotype, and a given selective environment. However, any change in selective pressure will rapidly reshape the viral fitness landscape (Figure 3A–B). In this way, fitness landscapes can instead be considered as time-dependent fitness seascapes (Merrell, 1994; Mustonen and Lässig, 2009). A changing fitness landscape can render previously beneficial mutations neutral, or even detrimental (Figure 3C). Depending on the timescale and frequency of selective change, mutations can behave as neutral over time if the fitness seascape continuously shifts during the time of fixation (Mustonen and Lässig, 2009).
Figure 3. Challenges in predicting viral evolution.
A) The fitness landscapes encountered by viruses are dynamic, making the maximal fitness a moving target. B.) Contour plot showing the movement of the optimal genotype in response to the shifting landscape in A. C.) Changes in the selective environment change the fitness effects of specific mutations. D.) Bottleneck effect. In transmitting a population of mutants, smaller samples will not accurately represent the distribution of mutations in the parental population and will result in the loss of rare beneficial mutations in the most-fit class. E.) When small populations are transmitted, the mean fitness of the transmitted population is highly variable. F.) Epistatic interactions create historical contingency in the path of evolution and constrain populations evolution (0 = wild-type locus; 1 = mutant). Mutation at the second position (mutant B) is lethal on its own, but viable when epistatic with a mutation at the third position (mutants C and D).
In the case of RNA viruses, selective pressures change especially rapidly. Whether from host transmission, intrahost spread, or host antiviral responses, the influence of selection on specific genomic loci changes over time. The evolutionary responses to particular transient selective pressures often lead to fitness tradeoffs by destabilizing proteins or perturbing viral metabolic processes, thus requiring compensatory mutations (Gong et al., 2013; Wu et al., 2016). In this way, a shifting landscape creates a moving target for the virus, forcing the virus to constantly adapt while balancing adaptive mutations with those that keep core viral functions intact (Lässig et al., 2017). Accurate prediction must not only take into account a virus’ long-term response to a fixed landscape, but also its evolutionary response as the landscape itself shifts.
Identifying and recapitulating the selective landscapes a virus will face constitutes a major challenge to the experimental study of RNA virus evolution in vivo and in vitro. In vivo, host tissues constitute distinct selective microenvironments, each presenting a unique repertoire of selective pressures to the virus population and, therefore, defining a unique fitness landscape. In the case of poliovirus (PV), subpopulations of virus isolated from distinct tissues of transgenic mice expressing the human poliovirus receptor (hPVR) exhibited unique patterns of genetic diversity (Xiao et al., 2017). By analyzing the host gene expression and viral evolution over the course of infection, the authors found the ability of the virus to rapidly evolve is required to overcome the distinct selective challenges within individual tissues. Rapid evolution is only required for spread in immune-competent mice, suggesting the ability to evolve rapidly is required in response to changes in the selective environment due to immune activation.
These results are corroborated by recent studies that demonstrate the dynamics of RNA virus evolution are altered depending on the rate at which the environment of a virus population changes (Morley and Turner, 2017; Morley et al., 2015). When sindbis virus (SINV) populations were introduced to a novel host cell type, a majority of replicates exhibited rapid fixation of the same initial point mutation (Morley and Turner, 2017). Conversely, a more gradual introduction of environmental changes did not lead to such rapid fixation of single mutations, but rather more frequent cohorts of multiple mutations rising together, suggesting increased clonal interference when environmental factors change more slowly. A sudden change of environment also induced rapid and consistent adaptation in influenza. When cell culture- and egg-passaged influenza was inoculated into healthy subjects, strong purifying selection removed egg-adapted mutations (Sobel Leonard et al., 2016).
These studies highlight not only the role changing fitness landscapes have on viral evolution, but that the dynamics of the landscape itself can affect viral population dynamics. Host characteristics like the kinetics of the immune response, or the rate at which a new potential host enters a virus’ environment, will alter the evolutionary solutions available to a virus population and the likelihood of important phenotypic transitions. The resolution afforded by techniques such as DMS and ultra-deep sequencing will enable experimenters much greater control of important selective parameters in order to mimic realistic fitness landscapes. Future studies that apply these methods to interrogate the shifting landscapes characteristic of intrahost and interhost transmission, for example (Stern et al., 2014; Xiao et al., 2017), could further clarify the key barriers to transmission, and the potential adaptive mutations that facilitate it.
Predicting host jumps
Over half of recent human outbreaks have been zoonotic in nature (Smith et al., 2014); that is, they were passed from animals to humans. Such outbreaks represent extreme examples of viruses traversing shifts in their selective landscape. New hosts can exert radically different selection pressures on a viral population (Bordería et al., 2015). During host shifts, the virus may encounter changes in temperature, antiviral immune systems, or host factor repertoires. Due to their epidemiological importance, the host and viral factors that influence transmission are a major focus of historical and experimental studies in viral systems (Grubaugh et al., 2017; Liu et al., 2016; Varble et al., 2014). Phylogenetic and reverse-genetic approaches have implicated a number of mutations in the recent high profile outbreaks of Ebola virus (EBOV) and Zika virus (ZIKV) (Diehl et al., 2016; Liu et al., 2017; Urbanowicz et al., 2016; Yuan et al., 2017). In the case of chikungunya virus (CHIKV), which is typically vectored by the mosquito Aedes aegypti, a single mutation in the envelope protein, E1, of CHIKV (A226V) adapted the virus for increased transmission by the Aedes albopictus mosquito and fueled an outbreak on Réunion island (Tsetsarkin et al., 2007). Analysis of CHIKV diversity in the salivary glands of adult Ae. albopictus mosquitoes revealed that A226V also emerged reproducibly under laboratory conditions (Stapleford et al., 2014).
Although accurately predicting specific novel host transmission events is highly unlikely, the example of CHIKV-A226V illustrates that, given the right scope and approach, an outbreak-associated gain-of-function mutation could be anticipated by experimental means. Selection experiments that focus on relevant animal models and modes of transmission provide a powerful tool to identify such mutations. However, studies to select for these gain-of-function mutations experimentally in animals are practically difficult and ethically problematic. Using the technologies described here to assay populations of viral variants in parallel selection experiments for specific phenotypes could aid in the identification of mutations with outbreak potential and inform surveillance efforts. Reconstituting the key selective pressures and barriers associated with specific modes of transmission will require combining these technologies with relevant cell lines and organoid culture systems (Qian et al., 2016) to more confidently extrapolate to real world host populations. Developing this level of experimental complexity constitutes a major challenge for the field but one accelerated by the technologies outlined here. In the future, it will be important to weigh the safety of such approaches against their advantages and their potential to provide reliable predictive information (Imperiale and Casadevall, 2015).
Population size fluctuation and stochasticity
The dynamic population size of viruses is especially problematic for predicting evolutionary trajectories. The influence of drift and selection depends on population size (Szendro et al., 2013). In sufficiently large populations, selection acts deterministically on mutant genomes, where they rise to fixation at a probability relative to the selection coefficient. In very large populations, evolution is slow because only mutations with very high fitness will be able to fix. When the population is small, during so-called fitness population bottlenecks, stochastic drift dominates. This is because in small samples of the viral population, such as those transmitted between hosts, the allele fitness distribution of the transmitted population deviates from the distribution the parental population (Figure 3D and E).
Fluctuations in population size, punctuated by stochastic bottlenecks such as transmission between hosts or during in vivo spread, are a common feature of the life history of many viruses and thought to drive their evolutionary dynamics (Gutiérrez et al., 2012; Stapleford et al., 2014; Tully and Fares, 2009; Varble et al., 2014). Although virus population sizes fluctuate widely during infection and transmission, evolutionary histories tend to reflect the population size during past bottlenecks, where mutations were most likely to fix, known as the effective population size. RNA viruses tend to have small effective population sizes due to the frequent selective bottlenecks, leading to an outsized influence of stochastic drift on the evolutionary trajectories of many virus populations, potentially influencing their robustness to mutation(Bennett et al., 2010; McCrone et al., 2017). In the future, combining approaches to modulate evolutionary parameters under controlled conditions provide a means to investigate the influence of various of specific parameters on virus evolution, such as genetic or chemical approaches to modulate mutation and recombination rates, or microfluidic technologies to modulate population sizes and coinfection rates.
Epistasis and contingent evolution
Complexity arises in genotype-fitness landscapes when interactions occur between mutations that yield unexpected phenotypes compared to their phenotypes in isolation. These epistatic interactions between mutations create so-called ruggedness, fitness ridges and valleys that render many potentially adaptive paths through sequence space inaccessible (Gong et al., 2013; Weinreich et al., 2006). These interactions determine the complexity of the genotype-fitness landscape and, therefore, influence the path of evolution and its predictability. Epistasis can influence the order in which mutations can occur (Shah et al., 2015; Starr et al., 2017) and the accessibility of certain evolutionary paths can depend on preceding substitutions (Shah et al., 2015; Starr et al., 2017). High-order epistasis, interactions between more than two mutations, has recently been appreciated as an important influence on evolutionary trajectories (Sailer and Harms, 2017b; Weinreich et al., 2013). Although the influence of any one interaction is small, because of their combinatorial nature, high-order epistatic interactions constitute a powerful influence in aggregate, determining the long-term evolutionary trajectory of evolving populations.
Epistasis and historical contingency determine the evolutionary potential of a given genotype and shape the path of evolution in complex and confounding ways (Harms and Thornton, 2014; Sailer and Harms, 2017b; Shah et al., 2015) (Figure 3F). Unlike single mutations, which can be explored completely on a given genomic background in the laboratory, the combinatorial nature of epistatic interactions renders an exhaustive exploration in the laboratory impossible and thus constitutes a major limitation on our ability to predict long-term evolution (Sailer and Harms, 2017a). However, high-throughput technologies like DMS provide new opportunities to characterize the influence of epistatic interactions on virus evolution through combinatorial studies of mutational fitness, allowing targeted exploration of low-order epistatic interactions (Araya et al., 2012; Bank et al., 2015; Olson et al., 2014)
Combining DMS approaches with phylogenetic analyses is emerging as an especially powerful approach for addressing historical contingency in past protein evolution and interrogating the epistatic networks that exist in and between proteins. A recent series of papers examining the evolution of DNA sequence specificity of the family of cellular steroid hormone receptor (SR) provides an example of the insights provided by this synergy. The authors inferred the ancestral sequences of the SR DNA-binding domain using a phylogenetic technique known as ancestral state reconstruction (McKeown et al., 2014). Analysis of the historical path of SR evolution identified signatures of epistasis and historical contingency between mutations as the family expanded its range of sequence binding specificities (Anderson et al., 2015). To explore the local mutational neighborhoods along the path of evolution, Starr and colleagues used DMS to assess the functionality and sequence specificity of 11 ancestral genotypes along the evolutionary transition between two distinct sequence specificities, revealing the profound effect of historical contingency on the path of evolution (Starr et al., 2017). The ancestral evolutionary states exhibited vastly different numbers of functional neighbors and thus highly variable access to potential evolutionary trajectories. Many of the historical mutations during the transition were neutral, but were required to access the current extant diversity in the protein family, emphasizing the importance of stochastic influence of drift and neutral evolution in the emergence of new phenotypes.
Defining common evolutionary strategies
As discussed previously, each genotype has unique evolutionary parameters and biological constraints that determine its evolutionary trajectory, thus the data gathered by new high-throughput and high-resolution approaches are largely genotype-specific. Viruses can vary dramatically in sequence and genomic organization, as well as their modes of transmission, their individual mutation, recombination, and reassortment rates, limiting the immediate application of insights of a given study to other systems. Anticipating the common potential mechanisms of adaptation across viruses requires an understanding of the broader principles that drive the evolution of viral populations. Therefore, a major challenge is divining the commonalities between evolutionary trajectories across viral genotypes and selective environments.
The high-throughput techniques described here yield large volumes of data on simultaneous dynamics of virus and host, and map the global evolutionary constraints acting on viral genomes and protein sequences. Analysis of these high dimensional data sets will increasingly depend on statistical modeling and learning techniques, such as machine learning or neural networks, to identify important features within the data. Applications of these approaches are becoming more common in biology (Pak and Kasarskis, 2015), including efforts toward predicting the spread of influenza (Luksza and Lässig, 2014), identifying determinants of virus-host transmission (Geoghegan et al., 2016; Olival et al., 2017), identifying the likely hosts of unknown viruses (Raj et al., 2011), and predicting protein structure (Raj et al. 2011; Jo et al. 2015; Wang et al. 2016), but are limited by the availability of adequate training data. As we collect more data such analytical techniques should identify additional commonalities and differences between viral species to elucidate the molecular and phenotypic mechanisms that lead to important evolutionary transitions.
CONCLUSIONS
New technologies are allowing us to probe the dynamics of viral populations with unprecedented depth, making it possible to directly measure key evolutionary parameters in evolving populations. Massively-parallel evolutionary experiments allow us to play the ‘evolutionary tape’ (Gould, 2011) thousands of times and measure the fitness effect of every mutation in across a viral genome. Combining these approaches with high-throughput and high-content phenotypic screening enables the correlation of fitness with specific phenotypic characteristics, such as replication kinetics or translation efficiency.
Library-based screening approaches like DMS can quantify the site-specific evolutionary constraints across the viral genome. Traditional evolutionary models used in phylogenetic inference apply substitution probabilities uniformly across a sequence. However, the fitness of specific substitutions across the genome is heterogeneous, due to the unique functional and chemical context. Integration of these site-specific data from library-based screening and evolutionary experiments has the potential to greatly improve the sensitivity and quality of phylogenetic inference by informing each evolutionary model with the specific evolutionary constraints of a given molecule (Bloom, 2014; Hilton et al., 2017).
In addition to the physical and biological constraints assessed by high-throughput experimental methods, transient and dynamic selective forces, such as intrahost immune states (Ben-Shachar et al., 2016; Xiao et al., 2017), seasonal weather changes (Bedford et al., 2010; Mordecai et al., 2017), and transmission dynamics (McCrone et al., 2017) drive the evolution of pathogenic RNA virus populations in nature. The influence of these factors depends on the viral population’s life history and, therefore, careful consideration is needed to design and interpret evolutionary experiments to study the evolutionary potential of viruses and inform prediction. In the context of strong selection on specific phenotypes, the evolutionary response to selection is often repeatable and involves only a few mutations (Longdon et al., 2017; Morley and Turner, 2017; Stern et al., 2017; Weinreich et al., 2006; Xiao et al., 2017; Xue et al., 2017). However, when considering more complex outcomes, such as spillover events, environmental and epidemiological factors add layers of additional complexity that must be considered (Campbell et al., 2018; Geoghegan and Holmes, 2017). Although predictive efforts will always be probabilistic, these improved experimental techniques enable us to probe the evolutionary forces and selective pressures that lead to viral populations and novel methods for modeling and detecting outbreaks should aid in efforts to better anticipate viral evolutionary trajectories.
Acknowledgments
The authors thank Dr. Edward Holmes and Dr. Jesse Bloom for helpful comments on this manuscript. This work is in part supported by NIH (R01 AI36178 and P01 AI091575) and the University of California (CCADD), DARPA Prophecy, and the Bill and Melinda Gates Foundation. PTD is supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number F32GM113483. ZJW is supported by the National Institutes of Health Microbial Pathogenesis and Host Defense Award Number NIH/NIAID T32 AI060537.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acevedo A, Brodsky L, Andino R. Mutational and fitness landscapes of an RNA virus revealed through population sequencing. Nature. 2014;505:686–690. doi: 10.1038/nature12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander JP, Jr, Gary HE, Jr, Pallansch MA. Duration of poliovirus excretion and its implications for acute flaccid paralysis surveillance: a review of the literature. J Infect Dis. 1997;175(Suppl 1):S176–S182. doi: 10.1093/infdis/175.supplement_1.s176. [DOI] [PubMed] [Google Scholar]
- Anderson DW, McKeown AN, Thornton JW. Intermolecular epistasis shaped the function and evolution of an ancient transcription factor and its DNA binding sites. Elife. 2015;4:e07864. doi: 10.7554/eLife.07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino R, Domingo E. Viral quasispecies. Virology. 2015;479–480:46–51. doi: 10.1016/j.virol.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya CL, Fowler DM. Deep mutational scanning: assessing protein function on a massive scale. Trends Biotechnol. 2011;29:435–442. doi: 10.1016/j.tibtech.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya CL, Fowler DM, Chen W, Muniez I, Kelly JW, Fields S. A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function. Proc Natl Acad Sci U S A. 2012;109:16858–16863. doi: 10.1073/pnas.1209751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashenberg O, Padmakumar J, Doud MB, Bloom JD. Deep mutational scanning identifies sites in influenza nucleoprotein that affect viral inhibition by MxA. PLoS Pathog. 2017;13:e1006288. doi: 10.1371/journal.ppat.1006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bank C, Hietpas RT, Jensen JD, Bolon DNA. A systematic survey of an intragenic epistatic landscape. Mol Biol Evol. 2015;32:229–238. doi: 10.1093/molbev/msu301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford T, Cobey S, Beerli P, Pascual M. Global Migration Dynamics Underlie Evolution and Persistence of Human Influenza A (H3N2) PLoS Pathog. 2010;6:e1000918. doi: 10.1371/journal.ppat.1000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett SN, Drummond AJ, Kapan DD, Suchard MA, Muñoz-Jordán JL, Pybus OG, Holmes EC, Gubler DJ. Epidemic dynamics revealed in dengue evolution. Mol Biol Evol. 2010;27:811–818. doi: 10.1093/molbev/msp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shachar R, Schmidler S, Koelle K. Drivers of Inter-individual Variation in Dengue Viral Load Dynamics. PLoS Comput Biol. 2016;12:e1005194. doi: 10.1371/journal.pcbi.1005194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornson ZB, Nolan GP, Fantl WJ. Single-cell mass cytometry for analysis of immune system functional states. Curr Opin Immunol. 2013;25:484–494. doi: 10.1016/j.coi.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JD. An experimentally determined evolutionary model dramatically improves phylogenetic fit. Mol Biol Evol. 2014;31:1956–1978. doi: 10.1093/molbev/msu173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JD, Raval A, Wilke CO. Thermodynamics of Neutral Protein Evolution. Genetics. 2007;175:255–266. doi: 10.1534/genetics.106.061754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordería AV, Isakov O, Moratorio G, Henningsson R, Agüera-González S, Organtini L, Gnädig NF, Blanc H, Alcover A, Hafenstein S, et al. Group selection and contribution of minority variants during virus adaptation determines virus fitness and phenotype. PLoS Pathog. 2015;11:e1004838. doi: 10.1371/journal.ppat.1004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard MJ, Lam DH, Racaniello VR. Determinants of attenuation and temperature sensitivity in the type 1 poliovirus Sabin vaccine. J Virol. 1995;69:4972–4978. doi: 10.1128/jvi.69.8.4972-4978.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell F, Strang C, Ferguson N, Cori A, Jombart T. When are pathogen genome sequences informative of transmission events? PLoS Pathog. 2018;14:e1006885. doi: 10.1371/journal.ppat.1006885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl WE, Lin AE, Grubaugh ND, Carvalho LM, Kim K, Kyawe PP, McCauley SM, Donnard E, Kucukural A, McDonel P, et al. Ebola Virus Glycoprotein with Increased Infectivity Dominated the 2013–2016 Epidemic. Cell. 2016;167:1088–1098e6. doi: 10.1016/j.cell.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giallonardo F, Töpfer A, Rey M, Prabhakaran S, Duport Y, Leemann C, Schmutz S, Campbell NK, Joos B, Lecca MR, et al. Full-length haplotype reconstruction to infer the structure of heterogeneous virus populations. Nucleic Acids Res. 2014;42:e115. doi: 10.1093/nar/gku537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell. 2016;167:1853–1866. e17. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, King AA, Woods RJ, Pascual M. Evolution-informed forecasting of seasonal influenza A (H3N2) Sci Transl Med. 2017:9. doi: 10.1126/scitranslmed.aan5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Xin L, Shi Y, Zhang TH, Wu NC, Dai L, Gong D, Brar G, Shu S, Luo J, et al. Genome-wide identification of interferon-sensitive mutations enables influenza vaccine design. Science. 2018;359:290–296. doi: 10.1126/science.aan8806. [DOI] [PubMed] [Google Scholar]
- Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971;58:465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- Eigen M. On the nature of virus quasispecies. Trends Microbiol. 1996;4:216–218. doi: 10.1016/0966-842X(96)20011-3. [DOI] [PubMed] [Google Scholar]
- Fischer AE, Wu SK, Proescher JBG, Rotem A, Chang CB, Zhang H, Tao Y, Mehoke TS, Thielen PM, Kolawole AO, et al. A high-throughput drop microfluidic system for virus culture and analysis. J Virol Methods. 2015;213:111–117. doi: 10.1016/j.jviromet.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Flannery B, Chung JR, Thaker SN, Monto AS, Martin ET, Belongia EA, McLean HQ, Gaglani M, Murthy K, Zimmerman RK, et al. Interim Estimates of 2016–17 Seasonal Influenza Vaccine Effectiveness - United States, February 2017. MMWR Morb Mortal Wkly Rep. 2017;66:167–171. doi: 10.15585/mmwr.mm6606a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster R, Adami C, Wilke CO. Selection for mutational robustness in finite populations. J Theor Biol. 2006;243:181–190. doi: 10.1016/j.jtbi.2006.06.020. [DOI] [PubMed] [Google Scholar]
- Fowler DM, Araya CL, Fleishman SJ, Kellogg EH, Stephany JJ, Baker D, Fields S. High-resolution mapping of protein sequence-function relationships. Nat Methods. 2010;7:nmeth.1492. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan JL, Holmes EC. Predicting virus emergence amid evolutionary noise. Open Biol. 2017:7. doi: 10.1098/rsob.170189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan JL, Senior AM, Di Giallonardo F, Holmes EC. Virological factors that increase the transmissibility of emerging human viruses. Proceedings of the National Academy of Sciences. 2016;113:4170–4175. doi: 10.1073/pnas.1521582113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong LI, Suchard MA, Bloom JD. Stability-mediated epistasis constrains the evolution of an influenza protein. Elife. 2013;2:e00631. doi: 10.7554/eLife.00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ. Full House. Harvard University Press; 2011. [Google Scholar]
- Grubaugh ND, Smith DR, Brackney DE, Bosco-Lauth AM, Fauver JR, Campbell CL, Felix TA, Romo H, Duggal NK, Dietrich EA, et al. Experimental evolution of an RNA virus in wild birds: evidence for host-dependent impacts on population structure and competitive fitness. PLoS Pathog. 2015;11:e1004874. doi: 10.1371/journal.ppat.1004874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh ND, Fauver JR, Rückert C, Weger-Lucarelli J, Garcia-Luna S, Murrieta RA, Gendernalik A, Smith DR, Brackney DE, Ebel GD. Mosquitoes Transmit Unique West Nile Virus Populations during Each Feeding Episode. Cell Rep. 2017;19:709–718. doi: 10.1016/j.celrep.2017.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot S, Caro V, Cuervo N, Korotkova E, Combiescu M, Persu A, Aubert-Combiescu A, Delpeyroux F, Crainic R. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J Virol. 2000;74:8434–8443. doi: 10.1128/jvi.74.18.8434-8443.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Li S, Caglar MU, Mao Z, Liu W, Woodman A, Arnold JJ, Wilke CO, Huang TJ, Cameron CE. Single-Cell Virology: On-Chip Investigation of Viral Infection Dynamics. Cell Rep. 2017;21:1692–1704. doi: 10.1016/j.celrep.2017.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez S, Michalakis Y, Blanc S. Virus population bottlenecks during within-host progression and host-to-host transmission. Curr Opin Virol. 2012;2:546–555. doi: 10.1016/j.coviro.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Harms MJ, Thornton JW. Historical contingency and its biophysical basis in glucocorticoid receptor evolution. Nature. 2014;512:203–207. doi: 10.1038/nature13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietpas RT, Jensen JD, Bolon DNA. Experimental illumination of a fitness landscape. Proc Natl Acad Sci U S A. 2011;108:7896–7901. doi: 10.1073/pnas.1016024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton SK, Doud MB, Bloom JD. phydms: Software for phylogenetic analyses informed by deep mutational scanning. 2017 doi: 10.7717/peerj.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC. What can we predict about viral evolution and emergence? Curr. Opin Virol. 2013;3:180–184. doi: 10.1016/j.coviro.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC, Moya A. Is the quasispecies concept relevant to RNA viruses? J Virol. 2002;76:460–465. doi: 10.1128/JVI.76.1.460-462.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperiale MJ, Casadevall A. A new synthesis for dual use research of concern. PLoS Med. 2015;12:e1001813. doi: 10.1371/journal.pmed.1001813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc Natl Acad Sci U S A. 2011;108:20166–20171. doi: 10.1073/pnas.1110064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson ML, Chung JR, Jackson LA, Phillips CH, Benoit J, Monto AS, Martin ET, Belongia EA, McLean HQ, Gaglani M, et al. Influenza Vaccine Effectiveness in the United States during the 2015–2016 Season. N Engl J Med. 2017;377:534–543. doi: 10.1056/NEJMoa1700153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Liu P, Bank C, Renzette N, Prachanronarong K, Yilmaz LS, Caffrey DR, Zeldovich KB, Schiffer CA, Kowalik TF, et al. A Balance between Inhibitor Binding and Substrate Processing Confers Influenza Drug Resistance. J Mol Biol. 2016;428:538–553. doi: 10.1016/j.jmb.2015.11.027. [DOI] [PubMed] [Google Scholar]
- Kennedy SR, Schmitt MW, Fox EJ, Kohrn BF, Salk JJ, Ahn EH, Prindle MJ, Kuong KJ, Shen J-C, Risques R-A, et al. Detecting ultralow-frequency mutations by Duplex Sequencing. Nat Protoc. 2014;9:2586–2606. doi: 10.1038/nprot.2014.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew OM, Sutter RW, de Gourville EM, Dowdle WR, Pallansch MA. VACCINE-DERIVED POLIOVIRUSES AND THE ENDGAME STRATEGY FOR GLOBAL POLIO ERADICATION*. Annu Rev Microbiol. 2005;59:587–635. doi: 10.1146/annurev.micro.58.030603.123625. [DOI] [PubMed] [Google Scholar]
- Korboukh VK, Lee CA, Acevedo A, Vignuzzi M, Xiao Y, Arnold JJ, Hemperly S, Graci JD, August A, Andino R, et al. RNA Virus Population Diversity, an Optimum for Maximal Fitness and Virulence. J Biol Chem. 2014;289:29531–29544. doi: 10.1074/jbc.M114.592303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik I, Ince WL, Gentles LE, Oler AJ, Kosikova M, Angel M, Magadán JG, Xie H, Brooke CB, Yewdell JW. Influenza A virus hemagglutinin glycosylation compensates for antibody escape fitness costs. PLoS Pathog. 2018;14:e1006796. doi: 10.1371/journal.ppat.1006796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S, Rice DP, Jerison ER, Desai MM. Microbial evolution. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science. 2014;344:1519–1522. doi: 10.1126/science.1250939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Haliburton JR, Yuan A, Abate AR. Droplet barcoding for massively parallel single-molecule deep sequencing. Nat Commun. 2016;7:ncomms11784. doi: 10.1038/ncomms11784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lässig M, Mustonen V, Walczak AM. Predicting evolution. Nature Ecology & Evolution. 2017;1 doi: 10.1038/s41559-017-0077. s41559-017-0077-017. [DOI] [PubMed] [Google Scholar]
- Lauring AS, Andino R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010;6:e1001005. doi: 10.1371/journal.ppat.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring AS, Acevedo A, Cooper SB, Andino R. Codon usage determines the mutational robustness, evolutionary capacity, and virulence of an RNA virus. Cell Host Microbe. 2012;12:623–632. doi: 10.1016/j.chom.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-Term Experimental Evolution in Escherichia coli. I. Adaptation and Divergence During 2,000 Generations. Am Nat. 1991;138:1315–1341. [Google Scholar]
- Liu J, Liu Y, Nie K, Du S, Qiu J, Pang X, Wang P, Cheng G. Flavivirus NS1 protein in infected host sera enhances viral acquisition by mosquitoes. Nature Microbiology. 2016;1:nmicrobiol201687. doi: 10.1038/nmicrobiol.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liu J, Du S, Shan C, Nie K, Zhang R, Li X-F, Zhang R, Wang T, Qin C-F, et al. Evolutionary enhancement of Zika virus infectivity in Aedes aegypti mosquitoes. Nature. 2017;545:nature22365. doi: 10.1038/nature22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longdon B, Day JP, Alves JM, Smith SCL, Houslay TM, McGonigle JE, Tagliaferri L, Jiggins FM. Host shifts result in parallel genetic changes when viruses adapt to closely related species. 2017 doi: 10.1371/journal.ppat.1006951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luksza M, Lässig M. A predictive fitness model for influenza. Nature. 2014;507:57–61. doi: 10.1038/nature13087. [DOI] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCandlish DM. Visualizing fitness landscapes. Evolution. 2011;65:1544–1558. doi: 10.1111/j.1558-5646.2011.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrone JT, Lauring AS. Genetic bottlenecks in intraspecies virus transmission. Curr Opin Virol. 2017;28:20–25. doi: 10.1016/j.coviro.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrone JT, Woods RJ, Martin ET, Malosh RE, Monto AS, Lauring AS. The evolutionary dynamics of influenza A virus within and between human hosts 2017 [Google Scholar]
- McKeown AN, Bridgham JT, Anderson DW, Murphy MN, Ortlund EA, Thornton JW. Evolution of DNA specificity in a transcription factor family produced a new gene regulatory module. Cell. 2014;159:58–68. doi: 10.1016/j.cell.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell DJ. The Adaptive Seascape: The Mechanism of Evolution. U of Minnesota Press; 1994. [Google Scholar]
- Montville R, Froissart R, Remold SK, Tenaillon O, Turner PE. Evolution of Mutational Robustness in an RNA Virus. PLoS Biol. 2005;3:e381. doi: 10.1371/journal.pbio.0030381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordecai EA, Cohen JM, Evans MV, Gudapati P, Johnson LR, Lippi CA, Miazgowicz K, Murdock CC, Rohr JR, Ryan SJ, et al. Detecting the impact of temperature on transmission of Zika, dengue, and chikungunya using mechanistic models. PLoS Negl Trop Dis. 2017;11:e0005568. doi: 10.1371/journal.pntd.0005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley VJ, Turner PE. Dynamics of molecular evolution in RNA virus populations depend on sudden versus gradual environmental change. Evolution. 2017;71:872–883. doi: 10.1111/evo.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley VJ, Mendiola SY, Turner PE. Rate of novel host invasion affects adaptability of evolving RNA virus lineages. Proc Biol Sci. 2015;282:20150801. doi: 10.1098/rspb.2015.0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustonen V, Lässig M. From fitness landscapes to seascapes: non-equilibrium dynamics of selection and adaptation. Trends Genet. 2009;25:111–119. doi: 10.1016/j.tig.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Neher RA, Bedford T, Daniels RS, Russell CA, Shraiman BI. Prediction, dynamics, and visualization of antigenic phenotypes of seasonal influenza viruses. Proc Natl Acad Sci U S A. 2016;113:E1701–E1709. doi: 10.1073/pnas.1525578113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Simonsen L, Viboud C, Miller MA, Taylor J, George KS, Griesemer SB, Ghedin E, Ghedi E, Sengamalay NA, et al. Stochastic processes are key determinants of short-term evolution in influenza a virus. PLoS Pathog. 2006;2:e125. doi: 10.1371/journal.ppat.0020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olival KJ, Hosseini PR, Zambrana-Torrelio C, Ross N, Bogich TL, Daszak P. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546:646. doi: 10.1038/nature22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson CA, Wu NC, Sun R. A comprehensive biophysical description of pairwise epistasis throughout an entire protein domain. Curr Biol. 2014;24:2643–2651. doi: 10.1016/j.cub.2014.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak TR, Kasarskis A. How next-generation sequencing and multiscale data analysis will transform infectious disease management. Clin Infect Dis. 2015;61:1695–1702. doi: 10.1093/cid/civ670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly MD, Procario MC, Lauring AS. A novel twelve class fluctuation test reveals higher than expected mutation rates for influenza A viruses. Elife. 2017:6. doi: 10.7554/eLife.26437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Dewar M, Palacios G, Rabadan R, Wiggins CH. Identifying hosts of families of viruses: a machine learning approach. PLoS One. 2011;6:e27631. doi: 10.1371/journal.pone.0027631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscoe BP, Thayer KM, Zeldovich KB, Fushman D, Bolon DNA. Analyses of the effects of all ubiquitin point mutants on yeast growth rate. J Mol Biol. 2013;425:1363–1377. doi: 10.1016/j.jmb.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem A, Serohijos A, Chang C, Wolfe J, Fischer A, Mehoke T, Zhang H, Tao Y, Ung L, Choi J-M, et al. Tuning the course of evolution on the biophysical fitness landscape of an RNA virus. 2016 doi: 10.1093/molbev/msy131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Trapnell C, Bloom JD. Extreme heterogeneity of influenza virus infection in single cells. Elife. 2018:7. doi: 10.7554/eLife.32303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin AB, Boulger LR. History of Sabin attenuated poliovirus oral live vaccine strains. J Biol Stand. 1973;1:115–118. [Google Scholar]
- Sackman AM, McGee LW, Morrison AJ, Pierce J, Anisman J, Hamilton H, Sanderbeck S, Newman C, Rokyta DR. Mutation-Driven Parallel Evolution during Viral Adaptation. Mol Biol Evol. 2017;34:3243–3253. doi: 10.1093/molbev/msx257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailer ZR, Harms MJ. Molecular ensembles make evolution unpredictable. Proc Natl Acad Sci U S A. 2017a doi: 10.1073/pnas.1711927114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailer ZR, Harms MJ. High-order epistasis shapes evolutionary trajectories. PLoS Comput Biol. 2017b;13:e1005541. doi: 10.1371/journal.pcbi.1005541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, McCandlish DM, Plotkin JB. Contingency and entrenchment in protein evolution under purifying selection. Proc Natl Acad Sci U S A. 2015;112:E3226–E3235. doi: 10.1073/pnas.1412933112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KF, Goldberg M, Rosenthal S, Carlson L, Chen J, Chen C, Ramachandran S. Global rise in human infectious disease outbreaks. J R Soc Interface. 2014;11:20140950. doi: 10.1098/rsif.2014.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel Leonard A, McClain MT, Smith GJD, Wentworth DE, Halpin RA, Lin X, Ransier A, Stockwell TB, Das SR, Gilbert AS, et al. Deep Sequencing of Influenza A Virus from a Human Challenge Study Reveals a Selective Bottleneck and Only Limited Intrahost Genetic Diversification. J Virol. 2016;90:11247–11258. doi: 10.1128/JVI.01657-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleford KA, Coffey LL, Lay S, Bordería AV, Duong V, Isakov O, Rozen-Gagnon K, Arias-Goeta C, Blanc H, Beaucourt S, et al. Emergence and Transmission of Arbovirus Evolutionary Intermediates with Epidemic Potential. Cell Host Microbe. 2014;15:706–716. doi: 10.1016/j.chom.2014.05.008. [DOI] [PubMed] [Google Scholar]
- Starr TN, Picton LK, Thornton JW. Alternative evolutionary histories in the sequence space of an ancient protein. Nature. 2017;549:409–413. doi: 10.1038/nature23902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern A, Bianco S, Yeh MT, Wright C, Butcher K, Tang C, Nielsen R, Andino R. Costs and benefits of mutational robustness in RNA viruses. Cell Rep. 2014;8:1026–1036. doi: 10.1016/j.celrep.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern A, Yeh MT, Zinger T, Smith M, Wright C, Ling G, Nielsen R, Macadam A, Andino R. The Evolutionary Pathway to Virulence of an RNA Virus. Cell. 2017;169:35–46. e19. doi: 10.1016/j.cell.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CB, Schilling JW, Wilson AC. Convergent evolution of lysozyme sequences? Nature. 1988;332:788. doi: 10.1038/332787c0. [DOI] [PubMed] [Google Scholar]
- Szendro IG, Franke J, de Visser JAGM, Krug J. Predictability of evolution depends nonmonotonically on population size. Proc Natl Acad Sci U S A. 2013;110:571–576. doi: 10.1073/pnas.1213613110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Rotem A, Zhang H, Chang CB, Basu A, Kolawole AO, Koehler SA, Ren Y, Lin JS, Pipas JM, et al. Rapid, targeted and culture-free viral infectivity assay in drop-based microfluidics. Lab Chip. 2015a;15:3934–3940. doi: 10.1039/c5lc00556f. [DOI] [PubMed] [Google Scholar]
- Tao Y, Rotem A, Zhang H, Cockrell SK, Koehler SA, Chang CB, Ung LW, Cantalupo PG, Ren Y, Lin JS, et al. Artifact-Free Quantification and Sequencing of Rare Recombinant Viruses by Using Drop-Based Microfluidics. Chembiochem. 2015b;16:2167–2171. doi: 10.1002/cbic.201500384. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B, Bloom JD. The inherent mutational tolerance and antigenic evolvability of influenza hemagglutinin. eLife Sciences. 2014;3:e03300. doi: 10.7554/eLife.03300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3:e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully DC, Fares MA. Shifts in the selection-drift balance drive the evolution and epidemiology of foot-and-mouth disease virus. J Virol. 2009;83:781–790. doi: 10.1128/JVI.01500-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowicz RA, McClure CP, Sakuntabhai A, Sall AA, Kobinger G, Müller MA, Holmes EC, Rey FA, Simon-Loriere E, Ball JK. Human Adaptation of Ebola Virus during the West African Outbreak. Cell. 2016;167:1079–1087. e5. doi: 10.1016/j.cell.2016.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varble A, Albrecht RA, Backes S, Crumiller M, Bouvier NM, Sachs D, García-Sastre A, tenOever BR. Influenza A virus transmission bottlenecks are defined by infection route and recipient host. Cell Host Microbe. 2014;16:691–700. doi: 10.1016/j.chom.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignuzzi M, Wendt E, Andino R. Engineering attenuated virus vaccines by controlling replication fidelity. Nat Med. 2008;14:154–161. doi: 10.1038/nm1726. [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Delaney NF, DePristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Lan Y, Wylie CS, Heckendorn RB. Should evolutionary geneticists worry about higher-order epistasis? Curr. Opin Genet Dev. 2013;23:700–707. doi: 10.1016/j.gde.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ZJ, Andino R. Characterization of Viral Populations by Using Circular Sequencing. J Virol. 2016;90:8950–8953. doi: 10.1128/JVI.00804-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke CO. Quasispecies theory in the context of population genetics. BMC Evol Biol. 2005;5:44. doi: 10.1186/1471-2148-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The roles of mutation, inbreeding, crossbreeding, and selection in evolution (na) 1932 [Google Scholar]
- Wu NC, Young AP, Dandekar S, Wijersuriya H, Al-Mawsawi LQ, Wu T-T, Sun R. Systematic identification of H274Y compensatory mutations in influenza A virus neuraminidase by high-throughput screening. J Virol. 2013;87:1193–1199. doi: 10.1128/JVI.01658-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NC, Young AP, Al-Mawsawi LQ, Olson CA, Feng J, Qi H, Chen S-H, Lu I-H, Lin C-Y, Chin RG, et al. High-throughput profiling of influenza A virus hemagglutinin gene at single-nucleotide resolution. Sci Rep. 2014a;4:srep04942. doi: 10.1038/srep04942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NC, Young AP, Al-Mawsawi LQ, Olson CA, Feng J, Qi H, Luan HH, Li X, Wu T-T, Sun R. High-throughput identification of loss-of-function mutations for anti-interferon activity in the influenza A virus NS segment. J Virol. 2014b;88:10157–10164. doi: 10.1128/JVI.01494-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NC, Du Y, Le S, Young AP, Zhang TH, Wang Y, Zhou J, Yoshizawa JM, Dong L, Li X, et al. Coupling high-throughput genetics with phylogenetic information reveals an epistatic interaction on the influenza A virus M segment. BMC Genomics. 2016;17:46. doi: 10.1186/s12864-015-2358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie CS, Shakhnovich EI. A biophysical protein folding model accounts for most mutational fitness effects in viruses. Proc Natl Acad Sci U S A. 2011;108:9916–9921. doi: 10.1073/pnas.1017572108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Rouzine IM, Bianco S, Acevedo A, Goldstein EF, Farkov M, Brodsky L, Andino R. RNA Recombination Enhances Adaptability and Is Required for Virus Spread and Virulence. Cell Host Microbe. 2016;19:493–503. doi: 10.1016/j.chom.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Dolan PT, Goldstein EF, Li M, Farkov M, Brodsky L, Andino R. Poliovirus intrahost evolution is required to overcome tissue-specific innate immune responses. Nat Commun. 2017;8:375. doi: 10.1038/s41467-017-00354-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue KS, Stevens-Ayers T, Campbell AP, Englund JA, Pergam SA, Boeckh M, Bloom JD. Parallel evolution of influenza across multiple spatiotemporal scales. Elife. 2017:6. doi: 10.7554/eLife.26875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Huang X-Y, Liu Z-Y, Zhang F, Zhu X-L, Yu J-Y, Ji X, Xu Y-P, Li G, Li C, et al. A single mutation in the prM protein of Zika virus contributes to fetal microcephaly. Science. 2017:eaam7120. doi: 10.1126/science.aam7120. [DOI] [PubMed] [Google Scholar]
- Zanini F, Pu S-Y, Bekerman E, Einav S, Quake SR. Single-cell transcriptional dynamics of flavivirus infection. Elife. 2018:7. doi: 10.7554/eLife.32942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai W, Nielsen R, Slatkin M. An investigation of the statistical power of neutrality tests based on comparative and population genetic data. Mol Biol Evol. 2009;26:273–283. doi: 10.1093/molbev/msn231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Jones C, Mieczkowski P, Swanstrom R. Primer ID Validates Template Sampling Depth and Greatly Reduces the Error Rate of Next-Generation Sequencing of HIV-1 Genomic RNA Populations. J Virol. 2015;89:8540–8555. doi: 10.1128/JVI.00522-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilionis R, Nainys J, Veres A, Savova V, Zemmour D, Klein AM, Mazutis L. Single-cell barcoding and sequencing using droplet microfluidics. Nat Protoc. 2017;12:44–73. doi: 10.1038/nprot.2016.154. [DOI] [PubMed] [Google Scholar]
- Zost SJ, Parkhouse K, Gumina ME, Kim K, Perez SD, Wilson PC, Treanor JJ, Sant AJ, Cobey S, Hensley SE. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc Natl Acad Sci U S A. 2017;114:12578–12583. doi: 10.1073/pnas.1712377114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost SJ, Parkhouse K, Gumina ME, Kim K, Perez SD, Wilson PC, Treanor JJ, Sant AJ, Cobey S, Hensley SE. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc Natl Acad Sci U S A. 2017;114:12578–12583. doi: 10.1073/pnas.1712377114. [DOI] [PMC free article] [PubMed] [Google Scholar]