Abstract
Endo-β-mannanase (EC 3.2.1.78) is involved in hydrolysis of the mannan-rich cell walls of the tomato (Lycopersicon esculentum Mill.) endosperm during germination and post-germinative seedling growth. Different electrophoretic isoforms of endo-β-mannanase are expressed sequentially in different parts of the endosperm, initially in the micropylar endosperm cap covering the radicle tip and subsequently in the remaining lateral endosperm surrounding the rest of the embryo. We have isolated a cDNA from imbibed tomato seeds (LeMAN2) that shares 77% deduced amino acid sequence similarity with a post-germinative tomato mannanase (LeMAN1). When expressed in Escherichia coli, the protein encoded by LeMAN2 cDNA was recognized by anti-mannanase antibody and exhibited endo-β-mannanase activity, confirming the identity of the gene. LeMAN2 was expressed exclusively in the endosperm cap tissue of tomato seeds prior to radicle emergence, whereas LeMAN1 was expressed only in the lateral endosperm after radicle emergence. LeMAN2 mRNA accumulation and mannanase activity were induced by gibberellin in gibberellin-deficient gib-1 mutant seeds but were not inhibited by abscisic acid in wild-type seeds. Distinct mannanases are involved in germination and post-germinative growth, with LeMAN2 being associated with endosperm cap weakening prior to radicle emergence, whereas LeMAN1 mobilizes galactomannan reserves in the lateral endosperm.
Tomato (Lycopersicon esculentum Mill.) seeds have become a favored model system to analyze the physiological mechanisms and molecular and cell biology of seed germination (Hilhorst et al., 1998; Welbaum et al., 1998; Bradford et al., 2000). The tomato embryo is surrounded by a rigid endosperm that forms a mechanical restraint to embryo expansion. The region of the endosperm enclosing the radicle tip, termed the endosperm cap, weakens to allow radicle emergence (Groot and Karssen, 1987). The endosperm cell walls contain approximately 60% Man (Groot et al., 1988; Dahal et al., 1997), probably in the form of galactomannan or galactoglucomannan polymers that constitute the major carbohydrate reserves of the endosperm and contribute to its rigidity. Endo-(1,4)-β-mannanase (EC 3.2.1.78), which can hydrolyze internal bonds within mannan polymers, has therefore been investigated with respect to its potential role in degradation of the endosperm cell walls associated with tissue weakening and reserve mobilization (Groot et al., 1988; Nonogaki et al., 1992, 1995, 1998a, 1998b; Nonogaki and Morohashi, 1996; Toorop et al., 1996; Voigt and Bewley, 1996; Dahal et al., 1997; Still and Bradford, 1997; Still et al., 1997). Mannanase may also be involved in the mechanism of germination in seeds of other plant species (Watkins et al., 1985; Dutta et al., 1994, 1997; Downie et al., 1997; Sánchez and de Miguel, 1997).
Mannanase activity appears initially in the endosperm cap of tomato seeds prior to radicle emergence and subsequently increases markedly in the remaining lateral endosperm following radicle emergence (Groot et al., 1988; Nonogaki et al., 1992; Nomaguchi et al., 1995; Nonogaki and Morohashi, 1996). In a physiological and biochemical sense, seed germination sensu stricto encompasses only the events occurring in imbibed seeds prior to radicle emergence (Bewley and Black, 1994). After radicle emergence, subsequent post-germinative development is more properly designated as seedling growth. To distinguish germination-specific biochemical processes from post-germinative events, we will refer to the period of germination from imbibition to radicle emergence from the seed as “germinative” development and to the period after radicle protrusion as “post-germinative” development.
A single germinative mannanase isoform (Mα) and three post-germinative mannanase isoforms (M1, M2, M3) that can be distinguished by different electrophoretic mobilities are expressed in tomato endosperm (Nonogaki and Morohashi, 1996; Toorop et al., 1996; Voigt and Bewley, 1996). The germinative Mα isoform is localized to the endosperm cap and is thought to be involved in weakening of this tissue prior to radicle emergence, whereas the post-germinative isoforms are associated with mobilization of cell wall mannan reserves in the lateral endosperm during seedling growth (Nonogaki and Morohashi, 1996; Bewley, 1997). In addition at least two mannanase isoforms are present in the embryo (Toorop et al., 1996; Voigt and Bewley, 1996; Nonogaki et al., 1998a), and mannanase activity is also detected in ripening tomato fruits (Pressey, 1989). Bewley et al. (1997) isolated and partially characterized a cDNA encoding one of the post-germinative mannanases (M1). Southern hybridization with this cDNA suggested that a family of four or more mannanase genes was present in the tomato genome. Whether the different electrophoretic isoforms present in germinating tomato seeds represented different genes or post-translational modifications of a single protein was not known.
We report here the cloning and characterization of a cDNA encoding an endo-β-mannanase that is expressed specifically in the endosperm cap of tomato seeds prior to radicle emergence. Expression of the gene is induced by gibberellin (GA) but is not repressed by abscisic acid (ABA), consistent with the effects of these hormones on germinative mannanase activity (Groot et al., 1987; Toorop et al., 1996; Dahal et al., 1997; Still and Bradford, 1997). Both the amino acid sequence and the spatial and temporal expression patterns of this gene differ from that of the post-germinative mannanase reported previously (Bewley et al., 1997). Thus, at least two genes with different tissue-specific expression patterns are responsible for mannanase activity in the endosperm of germinating and germinated tomato seeds.
RESULTS
Isolation of the Germinative Mannanase cDNA
Because the expression of mannanase in tomato seeds before radicle protrusion is induced by GA (Groot et al., 1988), a cDNA library prepared from gib-1 seeds imbibed in GA for 24 h (prior to radicle emergence) was screened with a partial-length (0.9-kb) cDNA of the post-germinative mannanase LeMAN1 (Bewley et al., 1997). Four positive clones isolated from this screen had sequences similar to that of the post-germinative mannanase cDNA. The longest cDNA insert was rescued into pBK-CMV vector, subcloned into pBluescript II KS (Stratagene, La Jolla, CA), and designated LeMAN2 (Lycopersicon esculentum mannanase 2).
The 1,481-bp LeMAN2 cDNA (GenBank accession no. AF184238) contained an open reading frame encoding a protein of 415 amino acids (Fig. 1). A putative signal peptide sequence of 22 amino acids was identified at the amino terminus of the protein (underlined in Fig. 1). The mature protein of 393 amino acids was encoded from the Cys residue at nucleotide 83 to Ile at nucleotide 1,259, with a predicted Mr of 44,379 and pI of pH 5.7. The predicted amino acid sequence of the protein encoded by LeMAN2 was compared with the post-germinative mannanase protein encoded by LeMAN1, with expressed sequence tag (AtMAN1) and genomic sequences (AtMAN2) from Arabidopsis and with two fungal mannanases (Fig. 1). Overall amino acid sequence homology (identity plus similarity) between LeMAN1 and LeMAN2 was 78% (72% nucleotide sequence identity). LeMAN2 contained additional amino acids compared with LeMAN1 protein (e.g. amino acids 26–35 and 131–158; Fig. 1), accounting for the greater predicted size of LeMAN2 (44 versus 39 kD for LeMAN1). Potential catalytic sites (asterisks in Fig. 1) and a potential N-glycosylation site (Asn-Gly-Ser; amino acids 50–52, double underlined in Fig. 1) that have been identified in the post-germinative mannanase (Bewley et al., 1997) were also present in LeMAN2 protein. The Arabidopsis sequences were about 40% (AtMAN2) and 50% (AtMAN1), similar to either of the two tomato cDNAs. Both predicted amino acid sequences from Arabidopsis shared the most highly conserved regions found in tomato, but AtMAN2 contained a region (amino acids 126–150) that was absent from AtMAN1 and corresponded to the same additional region in LeMAN2 noted above (Fig. 1). The amino acid sequence similarity between the tomato mannanases and fungal mannanases (Aspergillus aculeatus [Christgau et al., 1994] and Trichoderma reeseii [accession no. AAA34208]) was approximately 30%. It is interesting that the amino acid sequences in LeMAN2 and AtMAN2 that were absent from the LeMAN1 and AtMAN1 proteins showed high homology to the fungal mannanase proteins in this region (Fig. 1).
Figure 1.
Alignment of amino acid sequences of A. aculeatus (asp; accession no. AAA67426), T. reeseii (trr; accession no. AAA34208), Arabidopsis (AtMAN1, expressed sequence tag accession no. AAD20927; and AtMAN2, translated protein sequence from genomic clone K1F13.9 [www.kazusa.or.jp/kaos/]), and tomato post-germinative (LeMAN1, accession no. AAB87859) and germinative (LeMAN2, cDNA accession no. AF184238) mannanases. The putative signal peptide (22 amino acids) in LeMAN2 is underlined. The double-underlined amino acids indicate a possible N-glycosylation site, and putative catalytic sites conserved across fungal and plant mannanases are indicated by asterisks (Bewley et al., 1997). Amino acids identical in three or more of the sequences are highlighted by reverse shading.
Expression of the Protein Encoded by LeMAN2 cDNA
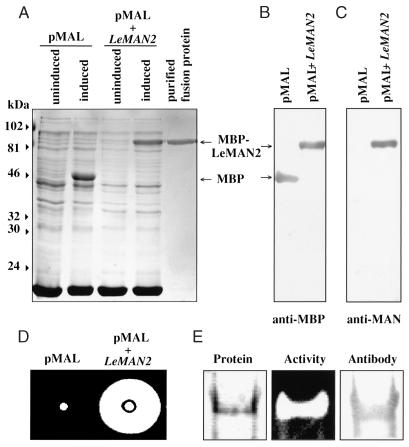
To confirm that the LeMAN2 cDNA encodes endo-β-mannanase protein, the cDNA was inserted into a maltose-binding protein overexpression vector and transformed into Escherichia coli. When the transformed cells were induced for protein expression by adding isopropylthio-β-d-galactoside (IPTG), a strong intensity band with a molecular mass of 87 kD was observed in the bacterial lysates, matching the predicted size of the fusion protein (maltose-binding protein [43 kD] plus LeMAN2 mannanase [44 kD]; Fig. 2A, pMAL + LeMAN2). This protein band was absent in the uninduced cells and in both induced and uninduced cells containing the empty vector (Fig. 2A). The putative fusion protein band was recognized by both anti-maltose-binding protein antibody (Fig. 2B) and antibody to one of the post-germinative mannanases (anti-M3 mannanase antibody; Nonogaki et al., 1995; Fig. 2C). These results confirm that the 87-kD overexpressed protein contains the maltose-binding::mannanase fusion protein. Extracts of the induced bacterial cells containing the LeMAN2 insert showed endo-β-mannanase activity, which could not be detected in extracts of bacterial cells that contained the empty pMAL vector, indicating that the overexpressed recombinant protein was an active form of mannanase (Fig. 2D). When the fusion protein was purified to homogeneity using a maltose-binding protein affinity resin (Fig. 2A), the fractions containing the fusion protein showed high mannanase activity and were recognized by the antimannanase antibody (Fig. 2E). The gel diffusion assay method for mannohydrolase activity is specific for endo-type enzymes (Downie et al., 1994), so LeMAN2 clearly encodes an endo-β-mannanase. In addition, the purified fusion protein was able to degrade endosperm cap cell walls. When fusion protein was added to 500 μg of isolated cell walls, 250 μg of reducing sugars was released. To test whether LeMAN2 mannanase alone can weaken endosperm cap tissue, we vacuum infiltrated the active recombinant fusion protein into isolated endosperm caps. However, no change in the strength (puncture force) of the endosperm cap tissue was detected after 2 d of incubation at 25°C (data not shown).
Figure 2.
Overexpression of the protein encoded by LeMAN2 cDNA. A, Protein profiles visualized by Coomassie Blue staining of the lysates of induced or uninduced bacterial cells containing the empty vector (pMAL) or the vector containing LeMAN2 cDNA insert (pMAL + LeMAN2) and the purified fusion protein. The positions of maltose-binding protein (MBP) and the recombinant fusion protein (MBP-LeMAN2) are indicated by the arrows on the right. Molecular masses (kD) are shown on the left. B and C, Immunoblots of the lysates of induced bacterial cells without (pMAL) or with (pMAL + LeMAN2) the cDNA insert, probed with anti-maltose-binding protein antibody (anti-MBP) and anti-M3 mannanase antibody (anti-MAN; Nonogaki et al., 1995), respectively. D, Gel diffusion assays for mannanase activity of the lysates from induced bacterial cells without (pMAL) or with (pMAL + LeMAN2) the cDNA insert. The activity is logarithmically proportional to the size of the cleared area on the gel plate (see Fig. 4). E, Native PAGE of purified maltose-binding protein-LeMAN2 fusion protein. Left, Coomassie-stained protein; center, gel assay for endo-β-mannanase activity; right, immunoblot probed with anti-M3 mannanase antibody.
Southern Hybridization
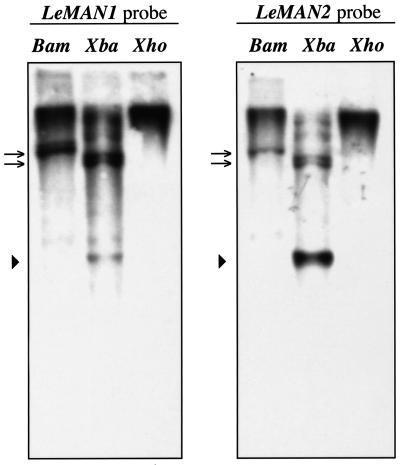
The hybridization patterns of the LeMAN1 and LeMAN2 cDNAs with tomato genomic DNA were compared using Southern hybridization (Fig. 3). Both cDNAs hybridized to the same sets of DNA fragments, confirming that multiple mannanase genes are present in the tomato genome (Fig. 3; Bewley et al., 1997). However, some DNA fragments hybridized more strongly to the LeMAN1 cDNA (Fig. 3, arrows), whereas other bands showed a stronger signal with the LeMAN2 cDNA (Fig. 3, arrowhead). This supports the sequence data indicating that different genes encode the germinative and post-germinative mannanases.
Figure 3.
Southern blots of tomato genomic DNA hybridized with full-length cDNAs of LeMAN1 (left) and LeMAN2 (right). Genomic DNA (10 μg) isolated from tomato leaves was digested with BamHI (Bam), XbaI (Xba), and XhoI (Xho). Bands marked with arrows hybridized more strongly with LeMAN1, whereas the band marked with an arrowhead hybridized more strongly to LeMAN2 (see text for details).
Expression of LeMAN2 and LeMAN1 mRNA in Tomato Seeds
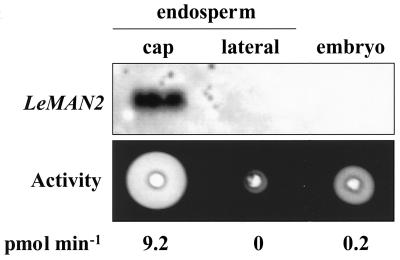
Since LeMAN2 was isolated from a cDNA library prepared from tomato seeds prior to radicle emergence, the LeMAN2 protein is likely to be a germinative mannanase. Only the Mα germinative mannanase specific to the endosperm cap is present in the endosperm at this time (Nonogaki and Morohashi, 1996), but two embryo-specific mannanases are also present in germinating tomato seeds (Nonogaki et al., 1998a). To investigate in which tissue(s) of imbibed seeds the LeMAN2 mRNA is expressed, RNA gel-blot analyses were performed. When total RNA from dissected seed parts (endosperm cap, lateral endosperm, and embryo) from wild-type tomato seeds imbibed in water for 24 h was hybridized with a full-length LeMAN2 RNA probe, the transcript was detected only from the endosperm cap, indicating that LeMAN2 mRNA is specifically expressed in this tissue (Fig. 4). The endosperm cap tissue also contained high mannanase activity, whereas little or no activity was detected from the lateral endosperm at this time (Fig. 4). Although some mannanase activity was present in the embryo as well, no hybridization between the LeMAN2 and embryonic mRNA was detected (Fig. 4). Since the northern hybridization was performed under relatively high stringency conditions (70°C washing), hybridization was also conducted at low stringency (55°C). However, even at low stringency, no hybridization could be detected with embryo RNA (data not shown).
Figure 4.
Northern blot of total RNA and gel diffusion assay for mannanase activity of protein extracts from the endosperm caps, the lateral endosperms, and whole embryos of wild-type tomato seeds imbibed in water for 24 h. The northern blot was hybridized with a LeMAN2 riboprobe. Endo-β-mannanase activity is indicated beneath each lane.
To compare the tissue specificity of expression of LeMAN2 (see above) with that of LeMAN1 (Bewley et al., 1997), total RNA was extracted from the endosperm caps of seeds prior to radicle emergence and from the lateral endosperms of germinated seedlings at different stages of development. To directly compare the hybridization patterns, the same sets of RNA samples were loaded on the same gel, transferred to the same membrane, processed using the same anti-digoxigenin (DIG) antibody solution following hybridization to the different probes, and exposed to the same x-ray film. The patterns of hybridization by LeMAN1 and LeMAN2 riboprobes to these RNA samples were completely different (Fig. 5). When LeMAN2 was used as a probe, a strong signal was detected in the RNA sample from the endosperm cap of seeds prior to radicle emergence, and only faint signals were detected at postemergence stages (Fig. 5). On the other hand, when the LeMAN1 probe was used, hybridization was detected specifically in the RNA samples from post-germinative lateral endosperms after radicle growth had begun (Fig. 5), although a faint band could also be seen in the endosperm caps prior to radicle emergence after a longer exposure to the x-ray film (data not shown). Thus, under the conditions used, there is little cross-hybridization of riboprobes prepared from each cDNA.
Figure 5.
Northern blots of total RNA from the endosperm caps of 24-h-imbibed tomato seeds prior to radicle emergence (0 mm) and from the lateral endosperms of germinated seeds (2–20 mm in radicle length), hybridized with LeMAN1 and LeMAN2 riboprobes, respectively. A representative seed or seedling at each stage is shown. Ethidium bromide-stained ribosomal RNA bands are shown under the blots to indicate RNA loading of each lane.
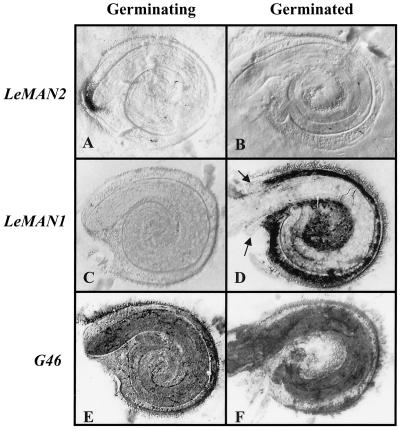
We used tissue printing to determine more precisely the tissue and germination stage specificity of LeMAN2 and LeMAN1 expression (Fig. 6). LeMAN2 mRNA was detected exclusively in the endosperm cap of germinating seeds (Fig. 6A) and was absent from germinated seeds (Fig. 6B). The LeMAN1 probe, on the other hand, did not hybridize to prints of germinating seeds (Fig. 6C), but hybridized strongly to the lateral endosperm of germinated seeds (Fig. 6D). Neither of these probes hybridized to the embryo at either stage of development (Fig. 6, A–D). Tissues corresponding to the endosperm cap in germinated seeds (arrows in Fig. 6D) did not hybridize to the LeMAN1 probe. Sense probes prepared from LeMAN2 and LeMAN1 did not hybridize with tissue prints from either germinating or germinated seeds (data not shown). A constitutively expressed mRNA encoding a ribosomal protein (termed G46; Cooley et al., 1999) was used to indicate mRNA binding to the tissue print membranes. G46 mRNA was present in all parts of the seed, with the greatest abundance in the radicle tip of germinating seeds (Fig. 6E) and in the embryo of germinated seeds (Fig. 6F). The hybridization with G46 demonstrates that RNA is relatively uniformly transferred to the membrane by tissue printing and confirms the specificity of the mannanase probes, consistent with the northern blots of RNA from the dissected tissues (Figs. 4 and 5). Thus, expression of LeMAN2 is specific to the endosperm cap prior to radicle emergence, whereas LeMAN1 expression is localized to the lateral endosperm after radicle emergence.
Figure 6.
Tissue printing and hybridization of germinating (24-h-imbibed) and germinated (5-mm radicle length) tomato seeds. Seeds were bisected, pressed onto a membrane, and hybridized with antisense riboprobes for LeMAN2 (A and B), LeMAN1 (C and D), and G46 (constitutively expressed ribosomal protein mRNA used as a printing control; E and F). The dark areas indicate hybridization. The arrows in D indicate the remaining endosperm cap tissue in the germinated seeds, which does not hybridize with the LeMAN1 riboprobe. Representative images are shown of more than 20 individual seeds printed and hybridized for each condition.
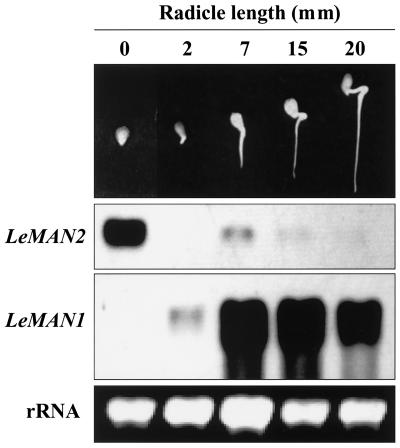
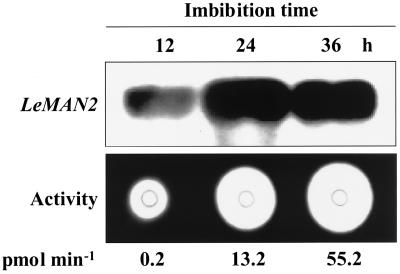
Given the timing and location of its expression, it is likely that LeMAN2 is involved in cell wall hydrolysis associated with endosperm cap weakening prior to radicle protrusion. There is no detectable mannanase activity in dry tomato seeds, and activity begins to increase 6 to 12 h after imbibition (Groot et al., 1988). LeMAN2 transcript was present in the endosperm cap within 12 h of imbibition and increased markedly by 24 h before declining slightly by 36 h (Fig. 7). The timing of LeMAN2 expression corresponded with the appearance of mannanase activity in the same tissue, although the peak of mRNA accumulation occurred earlier than the maximum enzyme activity, as would be expected (Fig. 7). Radicle protrusion of wild-type seeds was first observed 40 to 48 h after imbibition (data not shown), well after the increase in LeMAN2 message and mannanase activity.
Figure 7.
Northern blot of total RNA and gel diffusion assay for mannanase activity of the protein extracts from the endosperm caps of wild-type tomato seeds imbibed in water for 12, 24, and 36 h. The northern blot was hybridized with a LeMAN2 riboprobe. Endo-β-mannanase activity is indicated beneath each lane.
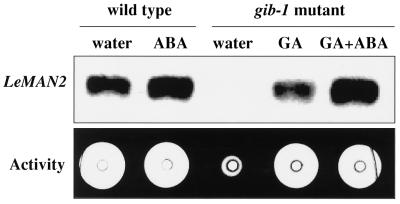
Hormonal regulation of the expression of LeMAN2 in wild-type and gib-1 tomato seeds was also examined. Although wild-type seeds can germinate in water, germination of gib-1 mutant tomato seeds is dependent on application of exogenous GA. LeMAN2 mRNA could not be detected in the endosperm caps of gib-1 mutant seeds incubated in water, and mannanase activity was barely detectable (Fig. 8). In contrast, both LeMAN2 mRNA expression and mannanase activity were induced in the endosperm caps of gib-1 seeds imbibed in GA4+7 (Fig. 8). The expression of LeMAN2 mRNA and mannanase activity in the endosperm caps of wild-type seeds in water or gib-1 seeds in GA was not inhibited by 100 μm ABA (Fig. 8), although radicle protrusion was prevented in both cases (data not shown).
Figure 8.
Northern blot of total RNA and gel diffusion assay for mannanase activity of protein extracts from the endosperm caps of wild-type tomato seeds imbibed for 24 h in water or 100 μm ABA, and gib-1 mutant seeds imbibed for 24 h in water, 100 μm GA, or 100 μm GA plus 100 μm ABA. The northern blot was hybridized with a LeMAN2 riboprobe. Endo-β-mannanase activity was 31.6 pmol min−1 in all cases except gib-1 seeds incubated in water, where activity was 0 pmol min−1.
DISCUSSION
Nonogaki and Morohashi (1996) showed that the mannanase protein found in the endosperm caps of germinating tomato seeds prior to radicle emergence was a different isoform from the mannanase proteins found in the lateral endosperm after radicle emergence. Since a polyclonal antibody raised against one of the post-germinative mannanases recognized the polypeptides of the germinative mannanase (Nonogaki et al., 1995; Nonogaki and Morohashi, 1996), it was expected that the amino acid sequences of the germinative and post-germinative mannanase proteins would be relatively similar, at least at certain epitopes. However, it was not clear whether these different mannanase isoforms were the products of post-translational modifications of a single protein or whether different genes encoded those proteins. We show here that a cDNA isolated from a cDNA library prepared from imbibed seeds, termed LeMAN2, has a sequence similar to but different from the sequence of a post-germinative mannanase cDNA LeMAN1 (Fig. 1). This suggested that the germinative and post-germinative mannanases are encoded by different genes, which was confirmed by Southern hybridization where the full-length DNA probes of LeMAN1 and LeMAN2 hybridized to specific fragments of tomato genomic DNA with different affinities (Fig. 3).
The deduced amino acid sequence of the protein encoded by LeMAN2 showed common features of mannanase proteins. Like the post-germinative LeMAN1 mannanase (Bewley et al., 1997), the LeMAN2 protein has a predicted signal peptide (Fig. 1) that is expected to be involved in targeting of this enzyme to the cell walls. Glu residues that are hypothesized to be catalytic sites and are conserved across fungal and plant mannanases (Bewley et al., 1997) were present in LeMAN2 protein as well (Fig. 1, asterisks; Glu-212 and Glu-329). The predicted amino acid sequence of mature LeMAN2 was longer than that for mature LeMAN1 (393 versus 346 amino acids), with predicted Mr of 44,000 for LeMAN2 and 39,000 for LeMAN1. This agrees with previous studies where the germinative mannanase isoform was larger than the post-germinative isoform, although the predicted Mrs of the mature proteins are approximately 10% larger than would be expected based on their mobilities on SDS-PAGE (Nonogaki and Morohashi, 1996). Overexpressed recombinant protein of LeMAN2 hydrolyzed locust bean galactomannan in a gel diffusion assay and was identified by anti-mannanase antibody (Fig. 2E). These results show conclusively that LeMAN2 encodes an endo-β-mannanase protein.
The expression of LeMAN2 mRNA was localized specifically to the endosperm cap tissue prior to radicle emergence (Figs. 4–6), whereas LeMAN1 was expressed only in the lateral endosperm after radicle emergence (Figs. 5 and 6). Only one isoform of endo-β-mannanase (Mα) is expressed exclusively in the endosperm cap of tomato seeds prior to radicle emergence (Nonogaki and Morohashi, 1996), consistent with the fact that no cDNAs coding for LeMAN1 were isolated from our cDNA library prepared from seeds prior to radicle emergence and screened using the LeMAN1 cDNA. Moreover, the timing of the accumulation of LeMAN2 message during germination corresponds to the increase in mannanase activity in the endosperm cap prior to radicle emergence (Fig. 7), and the pattern of hormonal regulation of LeMAN2 gene expression corresponded with mannanase activity (Fig. 8). Therefore, we conclude that LeMAN2 encodes the Mα endo-β-mannanase that is endosperm cap specific, GA responsive, and expressed only prior to radicle emergence.
In addition to the germinative and post-germinative endosperm mannanases, embryo-specific mannanase isoforms are also present in germinating tomato seeds (Fig. 4; Nonogaki et al., 1998a). It is likely that de novo synthesis of these proteins occurs, in that immunoblots showed that the amounts of the embryo-specific mannanase proteins increase during germination (Nonogaki et al., 1998a). One would therefore expect mRNA of embryonic mannanases to be present as well. However, we could not detect hybridization of LeMAN2 probes to embryonic mRNA even at low stringency (Fig. 4; data not shown). Neither LeMAN2 nor LeMAN1 probes hybridized to tissue prints of embryonic tissues (Fig. 6). It is possible that the mannanase activity in the embryo is due to the activation of pre-existing precursor protein(s), although this seems unlikely given the measured increase in total immunoreactive mannanase protein during germination (Nonogaki et al., 1998a). Alternatively, the abundance of the embryonic mannanase mRNAs might be very low, as mannanase activity in the embryo was only 2% of that in the endosperm cap prior to radicle emergence (Fig. 4). In addition, the sequence homology between the endospermic and the embryonic mannanase gene(s) might be insufficient to allow cross-hybridization, since little cross-hybridization occurred between the full-length LeMAN2 and LeMAN1 riboprobes (Figs. 5 and 6), despite considerable sequence homology (Fig. 1). Thus, we anticipate that additional divergent mannanase gene(s) that are expressed in the embryos of germinating tomato seeds remain to be identified.
In addition to the tissue specificity (Figs. 4–6) and timing (Fig. 7) of its expression, considerable circumstantial evidence supports a key role during germination for the endo-β-mannanase encoded by LeMAN2. For example, the expression of LeMAN2 was induced in gib-1 seeds by application of GA (Fig. 8), which also is required for radicle protrusion. On the other hand, ABA delays or prevents radicle emergence but had no effect on expression of LeMAN2 in either wild-type seeds or gib-1 seeds in the presence of GA (Fig. 8). However, contrary to results with excised endosperm caps treated with GA and ABA (Groot and Karssen, 1992), endosperm caps of intact seeds continue to weaken in the presence of ABA (Toorop, 1998; F. Chen, C.-T. Wu, and K.J. Bradford, unpublished results), consistent with the presence of LeMAN2 mannanase activity (Fig. 8). Changes in tomato endosperm cap cell wall structure during germination have been observed by electron microscopy (Nonogaki et al., 1998b; Toorop, 1998), and in Datura ferox seeds, which also express mannanase prior to radicle emergence (Sánchez and de Miguel, 1997), the endosperm cap tissue showed marked degradation of the galactomannan-rich cell walls prior to radicle emergence (Sánchez et al., 1990; Mella et al., 1995). However, infiltration of recombinant LeMAN2 fusion protein into isolated endosperm caps did not reduce the force required to penetrate the endosperm cap tissue (puncture force; see Groot and Karssen, 1987). This might be due to inability of the exogenously applied protein to reach active sites inside the cell walls or to the presence of the maltose-binding component in the fusion protein. On the other hand, the enzyme is active on tomato cell wall components, as the recombinant protein released 50% of the weight of isolated endosperm cap cell walls as reducing sugars.
Another possibility is that mannanase alone is not sufficient to degrade intact cell walls and that some cofactor(s), for example, other cell wall proteins like expansins (McQueen-Mason et al., 1992; Cosgrove, 1997), are needed for tissue weakening. A specific expansin gene (LeEXP4) is expressed in the endosperm cap tissue at the same time as LeMAN2 (Bradford et al., 2000), along with a number of other cell wall hydrolases, including polygalacturonase, cellulase, arabinosidase, and xyloglucan endotransglycosylase (Leviatov et al., 1995; Sitrit et al., 1999; Bradford et al., 2000). Thus, the LeMAN2 germinative mannanase may be one component among several that are required for cell wall disassembly and tissue weakening in the endosperm cap to allow radicle emergence.
We have reported here the cloning and characterization of the endo-β-mannanase (LeMAN2) that is expressed specifically in the endosperm cap tissue of tomato seeds prior to radicle emergence and in response to GA. Despite much correlative evidence supporting its role in germination, it has yet to be demonstrated directly that this enzyme is responsible for the endosperm weakening required for radicle emergence. Now that the gene has been identified, we are constructing LeMAN2 antisense transgenic wild-type plants and LeMAN2-overexpressing gib-1 plants to critically test whether this endo-β-mannanase is necessary or sufficient for endosperm cap weakening and therefore whether it is a key component in the mechanism of tomato seed germination.
MATERIALS AND METHODS
Plant Material and Seed Germination
Tomato (Lycopersicon esculentum Mill.) seeds, either from wild-type (cv Moneymaker) plants or homozygous GA-deficient mutant plants (gib-1) were used in this study. The gib-1 mutant and its isogenic parent line were originally obtained from Dr. Cees Karssen (Wageningen Agricultural University, The Netherlands). Mutant plants were sprayed three times per week with 100 μm GA4+7 to revert the dwarf habit and to allow more vigorous growth and fertility. After fruits were harvested, seeds were collected, treated with 0.25 m HCl, dried to 6% moisture content (fresh-weight basis), and stored at −20°C until they were used (Ni and Bradford, 1993). For germination, 100 or 200 tomato seeds were placed in Petri dishes on two layers of filter papers moistened with 12 mL of water or test solutions and incubated at 25°C in the dark. For hormone treatments, seeds were imbibed in the presence of 100 μm GA4+7 and/or 100 μm ABA.
In some cases, seeds were dissected into the micropylar tip and the remainder of the seed as previously described (Nonogaki et al., 1992), and the embryonic tissues were removed from each part using forceps. The embryoless micropylar tip and the embryoless remainder of the seed were denoted as the endosperm cap and lateral endosperm, respectively.
Isolation of cDNAs
A cDNA library was constructed using mRNA from gib-1 seeds imbibed in 100 μm GA4+7 for 24 h using a λZAP Express cDNA Synthesis Kit (Stratagene) according to the manufacturer's instructions. The cDNA library was screened by hybridization of nitrocellulose filter plaque replicas with a partial-length (0.9-kb) cDNA of the LeMAN1 post-germinative mannanase cDNA (provided by J.D. Bewley; Bewley et al., 1997; accession no. AF017144) labeled with enhanced chemiluminescence (ECL)-labeling reagents (Amersham Pharmacia Biotech, Piscataway, NJ). Hybridization was at 42°C overnight using ECL gold buffer including 5% (w/v) blocking reagent (Amersham Pharmacia Biotech) and 0.5 m NaCl after 1 h of prehybridization at the same temperature. Following hybridization, the membranes were washed twice for 20 min each at 42°C with 6 m urea, 0.5% (w/v) SDS, and 0.5× SSC and twice for 5 min each at room temperature with 2× SSC. Independent inserts in the library vector pBK-CMV were sequenced by the Advanced Plant Genetics Facility at the University of California, Davis. Sequence comparisons were made using DNASTAR software (DNASTAR, Madison, WI). Signal peptide prediction was performed using the Signal IP version 1.1 server (www.cbs.dtu.dk/services/SignalIP; Nielsen et al., 1997).
Expression of Recombinant Protein in Escherichia coli
The coding region (without the signal peptide) of the mannanase cDNA (amino acids 23–415; Fig. 1) was amplified by PCR using a BamHI site-linked forward primer (5′-CGGGATCCTGTGAAGCTAGGGTT-3′) and a XbaI site-linked reverse primer (5′-CGTCTAGACTAAATCTTAACC- AAATG-3′). The product was digested with BamHI and XbaI and ligated into the BamHI and XbaI sites of the maltose-binding protein expression vector pMAL-c2 (New England Biolabs, Beverly, MA). The empty vector and the vector containing insertion were transformed into competent cells of a proteinase-deficient strain (BL21) of E. coli, and the resulting transformant cells were selected using blue-white screening with IPTG-Xgal (5-bromo-4-chloro-3-indolyl-β-d-galactoside) plates. After incubation of a 1% (v/v) overnight culture for 4 h at 37°C, protein expression was induced by addition of IPTG to a final concentration of 2 mm and further incubation at 37°C for 2 h. The bacterial cells were harvested by centrifugation at 6,000g and the pellet was dissolved in sonication buffer (50 mm sodium phosphate buffer, pH 8.0, containing 0.3 m NaCl and 1 mg/mL lysozyme [Boehringer Mannheim, Indianapolis]). After overnight freezing at −20°C, the bacterial lysates were thawed and centrifuged at 10,000g for 10 min, and the supernatants were collected. Expressed proteins were examined by SDS-PAGE of the supernatants (crude extracts) of induced or uninduced bacterial cultures with or without the insertion. For purification of the fusion protein, the supernatant of an induced bacterial culture with the insertion was applied to a maltose-binding protein affinity column (amylose resin, New England Biolabs). The fusion protein was eluted from the column with 10 mm maltose. The fractions containing the fusion protein were examined by SDS-PAGE, mixed, and dialyzed against 10 mm Tris [tris(hydroxymethyl)aminomethane]-HCl, pH 7.5, overnight at 4°C. The dialysate was stored at −80°C.
PAGE and Immunoblotting
Proteins were separated by SDS-PAGE using 10% (w/v) acrylamide gels according to Laemmli (1970). Native PAGE was performed in 7.5% (w/v) gels according to Davis (1964), except that ammonium peroxydisulfate was used in place of riboflavin in the stacking gel. After electrophoresis, proteins were transferred to polyvinylidine difluoride membranes using a semidry blotter (TRAN-BLOT SD, Bio-Rad Laboratories, Hercules, CA) and were blocked with 5% (w/v) non-fat milk in phosphate-buffered saline containing 0.5% (v/v) Tween 20. Antimaltose-binding protein antibody (New England Biolabs) or anti-M3 mannanase antibody (Nonogaki et al., 1995) was used for immunoblotting at 1:5,000 dilution. Bound antibody was detected using horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin (Sigma Immunochemicals, St. Louis). The bands were detected on x-ray film after the reaction with the chemiluminescence reagent Renaissance (DuPont-NEN Products, Boston).
Endo-β-Mannanase Extraction and Assays
Endo-β-mannanase activity was extracted from tomato seeds or seed parts by homogenizing the tissues in 50 mm potassium phosphate buffer, pH 6.8, with a mortar and pestle. The homogenate was centrifuged at 10,000g for 5 min, and the supernatant was used as the enzyme solution. The endo-β-mannanase activity was assayed by the modified gel diffusion method (Still et al., 1997). Agarose (0.8% [w/v]) plates containing 0.05% (w/v) locust bean galactomannan (Sigma, St. Louis) were solidified, and wells were formed on the plates by scoring with a 3-mm cork borer and removing the plug by suction. The extracts (10 μL) from tomato seed parts or purified recombinant protein solution (10 μL) were applied to the wells, and the plates were incubated at 25°C for 24 h. After incubation, the agarose gel plates were stained by 0.5% (w/v) Congo red dye (Sigma) as described previously (Still et al., 1997). The hydrolyzed areas were visible as clear circles on a dark background. The diameter of the hydrolyzed area is logarithmically related to the enzyme activity and was quantified by comparison with authentic endo-β-mannanase standards as described previously (Still et al., 1997). This agarose gel method was also used for activity staining of native PAGE gels. After electrophoresis, a native gel was overlaid on top of the substrate-containing agarose gel and incubated at 25°C for 1 h. The activity band was detected as a transparent zone on the substrate gel after staining as described above.
Isolation of Endosperm Cap Cell Walls
Endosperm cap cell walls were prepared essentially according to Groot et al. (1988). Briefly, 100 endosperm caps were dissected from the seeds, and the testas were removed. The tissue was homogenized in 1.5 mL water and centrifuged at 10,000g for 5 min. The supernatant was removed, and the pellet was washed three times each with 1 m NaCl, 70% (v/v) ethanol, and chloroform-methanol (2:1) and then was dried at room temperature. The dried cell wall material (0.5 mg) was suspended in 50 mm sodium acetate buffer, pH 4.5, and subjected to enzyme digestion with the recombinant maltose-binding::mannanase fusion protein (about 15 μg) at 25°C for 20 h. After removing the insoluble cell wall fraction by centrifugation, reducing sugars released into the supernatant were assayed by the phenol-sulfuric acid method (Dubois et al., 1956).
DNA Extraction and Southern Hybridization
Genomic DNA was isolated from young tomato leaves (cv Moneymaker) as described by Murray and Thompson (1980) and modified by Bernatzky and Tanksley (1986). Genomic DNA (10 μg) was digested with the restriction enzymes BamHI, XbaI, and XhoI (New England Biolabs), separated on a 1.0% (w/v) agarose gel, and transferred to positively charged membranes (Hybond-N+, Amersham Pharmacia Biotech). Prehybridization, hybridization, washing, and detection were performed as described for cDNA library screening. To prepare the DNA probes, vectors containing the full-length mannanase cDNAs were digested with BamHI and XhoI, and the gel-purified insertions were used to make ECL-labeled probes (Amersham Pharmacia Biotech).
RNA Extraction and Northern Hybridization
Total RNA was extracted from seed parts (endosperm cap, lateral endosperm, or whole embryo) of germinating or germinated tomato seeds using a standard phenol extraction method (Sambrook et al., 1989). Total RNA (2–10 μg) was subjected to electrophoresis on 1.3% (w/v) agarose gels containing 7% (v/v) formaldehyde, transferred to a neutral membrane (Hybond-N, Amersham Pharmacia Biotech), and UV-cross-linked. RNA probes were prepared using a DIG-labeled dNTP mixture (Boehringer Mannheim). Hybridization was routinely done overnight at 60°C with a hybridization buffer containing 50% (v/v) deionized formamide, 4% (w/v) blocking reagent (Boehringer Mannheim), 0.2% (w/v) SDS, 0.1% (w/v) N-lauroylsarcosine, 5× SSC, and approximately 100 ng/mL RNA probe after 1 h prehybridization at the same temperature. The membranes were washed once for 25 min with 2× SSC, 0.1% (w/v) SDS at 70°C and twice for 25 min with 0.2× SSC, 0.1% (w/v) SDS at 70°C. The membranes were then blocked for 1 h with 5% (w/v) non-fat milk in 0.1 m maleic acid buffer, pH 7.5, containing 0.15 m NaCl and 0.3% (v/v) Tween 20 (buffer A) and were incubated with alkaline phosphatase-conjugated anti-DIG antibody (1:15,000 dilution) for 1 h at 25°C. After washing with buffer A, the membranes were subjected to chemiluminescence detection. The signal was detected on x-ray film after 5- to 20-min exposures. When the signals on two different membranes hybridized with different probes were compared, those membranes were exposed together on the same x-ray film for the same duration.
Tissue Printing and Hybridization
Germinating (24-h-imbibed) and germinated (5-mm radicle length) seeds were bisected with a razor blade. The cut surfaces were pressed for approximately 15 s onto a positively charged membrane (Hybond-N+, Amersham Pharmacia Biotech) supported on six layers of filter paper. After removing the tissue, the membrane was UV-cross-linked and hybridized with the LeMAN2 or LeMAN1 probes that were used for northern blots. The same stringency conditions as were used in northern blots were used for hybridization and washing of the tissue print membranes. The DIG-labeled probes were detected using alkaline phosphatase-conjugated anti-DIG antibody as described above, except that the signal was colorimetrically detected with 0.18 m Tris-HCl buffer, pH 8.8, containing 0.025 mg/mL 5-bromo-4-chloro-3-indolyl-phosphate, 0.1 mg/mL nitroblue tetrazolium, and 2 mm MgCl.
ACKNOWLEDGMENTS
We thank Prof. J.D. Bewley (University of Guelph, Guelph, Ontario, Canada) for providing the LeMAN1 cDNA and Prof. Y. Morohashi (Saitama University, Urawa, Japan) for providing the anti-M3 mannanase antibody. We also thank Feng Chen of our laboratory for preparation of genomic DNA and assistance with the puncture force analysis.
Footnotes
This research was supported by the National Science Foundation (grant no. IBN–9722978 to K.J.B.) and by a University of California (Davis) Katherine Esau postdoctoral fellowship (to H.N.).
LITERATURE CITED
- Bernatzky R, Tanksley SD. Genetics of actin-related sequences in tomato. Theor Appl Genet. 1986;72:314–321. doi: 10.1007/BF00288567. [DOI] [PubMed] [Google Scholar]
- Bewley JD. Breaking down the walls: a role for endo-β-mannanase in release from seed dormancy? Trends Plant Sci. 1997;2:464–469. [Google Scholar]
- Bewley JD, Black M. Seeds: Physiology of Development and Germination. Ed 2. New York: Plenum Press; 1994. [Google Scholar]
- Bewley JD, Burton RA, Morohashi Y, Fincher GB. Molecular cloning of a cDNA encoding a (1→4)-β-mannan endohydrolase from the seeds of germinated tomato (Lycopersicon esculentum) Planta. 1997;203:454–459. doi: 10.1007/s004250050214. [DOI] [PubMed] [Google Scholar]
- Bradford KJ, Chen F, Cooley MB, Dahal P, Downie B, Fukunaga KK, Gee OH, Gurusinghe S, Mella RA, Nonogaki H, Wu C-T, Yim K-O. Gene expression prior to radicle emergence in imbibed tomato seeds. In: Black M, Bradford KJ, Vazquez-Ramos J, editors. Seed Biology: Advances and Applications. Wallingford, UK: CAB International; 2000. pp. 231–251. [Google Scholar]
- Chrispeels MJ, Varner JE. Hormonal control of enzyme synthesis: on the mode of action of gibberellic acid and abscisin in aleurone layers of barley. Plant Physiol. 1967;42:1008–1016. doi: 10.1104/pp.42.7.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christgau S, Kauppinen S, Vind J, Kofod LV, Dalboge H. Expression cloning, purification and characterization of a β-1,4-mannanase from Aspergillus aculeatus. Biochem Mol Biol Int. 1994;33:917–925. [PubMed] [Google Scholar]
- Cooley MB, Yang H, Dahal P, Mella RA, Downie B, Haigh AM, Bradford KJ. Vacuolar H+-ATPase is expressed in response to gibberellin during tomato seed germination. Plant Physiol. 1999;121:1339–1347. doi: 10.1104/pp.121.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Relaxation in a high-stress environment: the molecular bases of extensible cell walls and cell enlargement. Plant Cell. 1997;9:1031–1041. doi: 10.1105/tpc.9.7.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahal P, Nevins DJ, Bradford KJ. Relationship of endo-β-d-mannanase activity and cell wall hydrolysis in tomato endosperm to germination rates. Plant Physiol. 1997;113:1243–1252. doi: 10.1104/pp.113.4.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BJ. Disc electrophoresis: II. Method and application to human serum proteins. Ann N Y Acad Sci. 1964;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- Downie B, Hilhorst HWM, Bewley JD. A new assay for quantifying endo-β-mannanase activity using Congo Red dye. Phytochemistry. 1994;36:829–835. [Google Scholar]
- Downie B, Hilhorst HWM, Bewley JD. Endo-β-mannanase activity during dormancy alleviation and germination of white spruce (Picea glauca) seeds. Physiol Plant. 1997;101:405–415. [Google Scholar]
- Dubois M, Giles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- Dutta S, Bradford KJ, Nevins DJ. Cell-wall autohydrolysis in isolated endosperms of lettuce (Lactuca sativa L.) Plant Physiol. 1994;104:623–628. doi: 10.1104/pp.104.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Nevins DJ, Bradford KJ. Endo-β-mannanase activity present in cell wall extracts of lettuce endosperm prior to radicle emergence. Plant Physiol. 1997;113:155–161. doi: 10.1104/pp.113.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot SPC, Karssen CM. Gibberellins regulate seed germination in tomato by endosperm weakening: a study with gibberellin-deficient mutants. Planta. 1987;171:525–531. doi: 10.1007/BF00392302. [DOI] [PubMed] [Google Scholar]
- Groot SPC, Karssen CM. Dormancy and germination of abscisic acid-deficient tomato seeds: studies with the sitiens mutant. Plant Physiol. 1992;99:952–958. doi: 10.1104/pp.99.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot SPC, Kieliszewska-Rokicka B, Vermeer E, Karssen CM. Gibberellin-induced hydrolysis of endosperm cell walls in gibberellin-deficient tomato seeds prior to radicle protrusion. Planta. 1988;174:500–504. doi: 10.1007/BF00634479. [DOI] [PubMed] [Google Scholar]
- Hilhorst HWM, Groot SPC, Bino RJ. The tomato seed as a model system to study seed development and germination. Acta Bot Neerl. 1998;47:169–183. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leviatov S, Shoseyev O, Wolf S. Involvement of endomannanase in the control of tomato seed germination under low temperature conditions. Ann Bot. 1995;76:1–6. [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ. Two endogenous proteins that induce cell wall expansion in plants. Plant Cell. 1992;4:1425–1433. doi: 10.1105/tpc.4.11.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mella RA, Maldonaldo S, Sánchez RA. Phytochrome-induced structural changes and protein degradation prior to radicle protrusion in Datura ferox seeds. Can J Bot. 1995;73:1371–1378. [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni B-R, Bradford KJ. Germination and dormancy of abscisic acid- and gibberellin-deficient mutant tomato (Lycopersicon esculentum) seeds: sensitivity of germination to abscisic acid, gibberellin and water potential. Plant Physiol. 1993;101:607–617. doi: 10.1104/pp.101.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Nomaguchi M, Nonogaki H, Morohashi Y. Development of galactomannan-hydrolyzing activity in the micropylar endosperm tip of tomato seed prior to germination. Physiol Plant. 1995;94:105–109. [Google Scholar]
- Nonogaki H, Matsushima H, Morohashi Y. Galactomannan hydrolyzing activity develops during priming in the micropylar endosperm tip of tomato seeds. Physiol Plant. 1992;85:167–172. [Google Scholar]
- Nonogaki H, Morohashi Y. An endo-β-mannanase develops exclusively in the micropylar endosperm of tomato seeds prior to radicle emergence. Plant Physiol. 1996;110:555–559. doi: 10.1104/pp.110.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H, Nomaguchi M, Morohashi Y. Endo-β-mannanases in the endosperm of germinated tomato seeds. Physiol Plant. 1995;94:328–334. [Google Scholar]
- Nonogaki H, Nomaguchi M, Morohashi Y, Matsushima H. Development and localization of endo-β-mannanase in the embryo of germinating tomato seeds. J Exp Bot. 1998a;49:1501–1507. [Google Scholar]
- Nonogaki H, Nomaguchi M, Okumoto N, Kaneko Y, Matsushima H, Morohashi Y. Temporal and spatial pattern of the biochemical activation of the endosperm during and following imbibition of tomato seeds. Physiol Plant. 1998b;102:236–242. [Google Scholar]
- Pressey R. Endo-β-mannanase in tomato fruit. Phytochemistry. 1989;28:3277–3280. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sánchez RA, de Miguel L. Phytochrome promotion of mannan-degrading enzyme activities in the micropylar endosperm of Datura ferox seeds requires the presence of embryo and gibberellin synthesis. Seed Sci Res. 1997;7:27–33. [Google Scholar]
- Sánchez RA, Sunell L, Labavitch JM, Bonner BA. Changes in endosperm cell walls of two Datura species before radicle protrusion. Plant Physiol. 1990;93:89–97. doi: 10.1104/pp.93.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitrit Y, Hadfield KA, Bennett AB, Bradford KJ, Downie B. Expression of a polygalacturonase associated with tomato seed germination. Plant Physiol. 1999;121:419–428. doi: 10.1104/pp.121.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Still DW, Bradford KJ. Endo-β-mannanase activity from individual tomato endosperm caps and radicle tips in relation to germination rates. Plant Physiol. 1997;113:21–29. doi: 10.1104/pp.113.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Still DW, Dahal P, Bradford KJ. A single-seed assay for endo-β-mannanase activity from tomato endosperm and radicle tissues. Plant Physiol. 1997;113:13–20. doi: 10.1104/pp.113.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toorop PE. The role of endo-β-mannanase activity in tomato seed germination. PhD thesis. The Netherlands: Wageningen Agricultural University; 1998. [Google Scholar]
- Toorop PE, Bewley JD, Hilhorst HWM. Endo-β-mannanase isoforms are present in the endosperm and embryo of tomato seeds, but are not essentially linked to the completion of germination. Planta. 1996;200:153–158. [Google Scholar]
- Voigt B, Bewley JD. Developing tomato seeds when removed from the fruit produce multiple forms of germinative and post-germinative endo-β-mannanase: responses to desiccation, abscisic acid and osmoticum. Planta. 1996;200:71–77. [Google Scholar]
- Watkins JT, Cantliffe DJ, Huber DJ, Nell TA. Gibberellic acid stimulated degradation of endosperm in pepper. J Am Soc Hortic Sci. 1985;110:61–65. [Google Scholar]
- Welbaum GE, Bradford KJ, Yim K-O, Booth DT, Oluoch MO. Biophysical, physiological and biochemical processes regulating seed germination. Seed Sci Res. 1998;8:161–172. [Google Scholar]