Abstract
Deficits in Schwann cell–mediated remyelination impair functional restoration after nerve damage, contributing to peripheral neuropathies. The mechanisms mediating block of remyelination remain elusive. Here, through small-molecule screening focusing on epigenetic modulators, we identified histone deacetylase 3 (HDAC3; a histone-modifying enzyme) as a potent inhibitor of peripheral myelinogenesis. Inhibition of HDAC3 enhanced myelin growth and regeneration and improved functional recovery after peripheral nerve injury in mice. HDAC3 antagonizes the myelinogenic neuregulin–PI3K–AKT signaling axis. Moreover, genome-wide profiling analyses revealed that HDAC3 represses promyelinating programs through epigenetic silencing while coordinating with p300 histone acetyltransferase to activate myelination-inhibitory programs that include the HIPPO signaling effector TEAD4 to inhibit myelin growth. Schwann cell–specific deletion of either Hdac3 or Tead4 in mice resulted in an elevation of myelin thickness in sciatic nerves. Thus, our findings identify the HDAC3–TEAD4 network as a dual-function switch of cell-intrinsic inhibitory machinery that counters myelinogenic signals and maintains peripheral myelin homeostasis, highlighting the therapeutic potential of transient HDAC3 inhibition for improving peripheral myelin repair.
Myelination around axons via Schwann cells (SCs) is essential for saltatory conduction of action potentials and for nerve function in the peripheral nervous system. Deficient myelination or failure to properly remyelinate contributes to functional deficits in peripheral nerves after injury and various forms of peripheral neuropathies1–3. Myelin sheaths are often the primary site of damage during the initial stages of pathological insults4. Despite the regenerative capacity retained in peripheral nerves after injury and disease in humans, remyelination occurs at a slow rate and is ultimately compromised with a thinner myelin sheath, resulting in incapacitating defects in functional reinnervation of the target tissue and sensorimotor function recovery and in neuropathic pain3,5. At present, the mechanisms underlying the block of remyelination after nerve trauma and peripheral neuropathies remain poorly understood.
The timing of myelinogenesis depends upon a balance between extrinsic signals and intracellular regulatory networks. Reciprocal axo–glial communication is critical for establishing the proper myelin thickness for saltatory conduction in nerve fibers. The growth factor neuregulin (NRG1) type III, which is produced by axons, induces myelin sheath formation through PI3K–AKT–ERK activation in SCs6,7. Receptor-mediated pathways, including those mediated by receptor tyrosine protein kinase erbB-2 (ERBB2)–ERBB3, integrin– laminin, and GPR126, in SCs can sense axonal signals and regulate myelination accordingly6–9. In addition, a series of promyelinating regulators, including octamer-binding transcription factor-6 (OCT6; also called POU3F1); Sry-related HMG-box gene 10 (SOX10); early growth response 2 (EGR2; KROX20); Yin Yang 1 (YY1); Yes-associated protein (YAP); Transcriptional coactivator with PDZ-binding motif (TAZ); and zinc finger E-box-binding homeobox 2 (ZEB2; also called SIP1), and negative regulatory cues, such as NOTCH-; SOX2-; and c-JUN-mediated signaling and the PI3K–AKT inhibitors phosphatase and tensin homolog (PTEN) and discs large homolog 1 (DLG1), function at different stages to coordinate SC lineage progression and myelination10–17. Currently, an explanation as to how SCs interpret the key axonal signal NRG1 to control peripheral myelin thickness remains elusive.
Recent studies indicate that epigenetic and chromatin modifiers play important roles in myelination in vertebrate nervous systems10,18,19. Histone-modifying enzymes, such as histone deacetylases 1 and 2, SWI–SNF chromatin-remodeling complexes, nucleosome-remodeling-and-deacetylase complexes, and polycomb repressive complexes, are crucial for SC differentiation, myelin formation, and myelin maintenance10,19,20. Critical questions remain as to how the epigenetic modifiers integrate multiple pathways to ensure that the appropriate amount of myelin is wrapped around axons during myelination and remyelination after injury. As many of the epigenetic modifiers act as repressors of gene expression, we hypothesized that deficient or limited SC remyelination after injury or pathological insult is caused by epigenetic silencing of the regulators required for myelinogenesis and myelin regeneration.
RESULTS
Identification of HDAC3 as an inhibitor of SC myelination by epigenetic screening
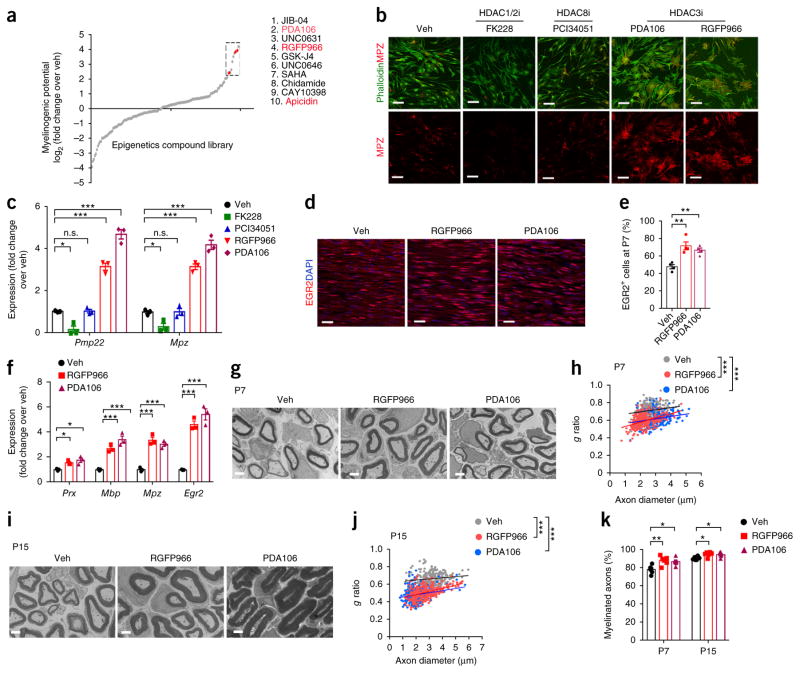
EGR2 is a promyelinating regulator that serves as an indicator of myelinogenic potential. We developed an unbiased gene-expression-based assay using real-time qPCR indexing of Egr2 to identify inhibitors of epigenetic enzymes that blocked SC maturation. Cultured rat SCs were treated with a library of epigenetic compounds targeting these enzymes, including methyltransferases, demethylases, histone acetyl-transferases, histone deacetylases (HDACs), and acetylated lysine–reader proteins. We identified a set of compounds that enhanced Egr2 expression over vehicle treatment by fivefold or more (Fig. 1a and Supplementary Fig. 1). Among these lead compounds were three inhibitors that preferentially targeted HDAC3: pimelic diphenylamide 106 (PDA106), RGFP966, and apicidin21,22. The convergent effects of chemically dissimilar compounds suggest that effects were exerted through HDAC3 inhibition.
Figure 1.
Small-molecule epigenetics compound library screen for inhibitors of SC myelination. (a) Relative expression of Egr2 in rat SC cultures treated with epigenetic drugs (gray dots). Data are presented as the mean from three independent experiments. The top ten compounds (boxed) that induced expression of Egr2 by more than fivefold over that induced by vehicle (veh) treatment are listed at the right. Red dots, compounds that preferentially target HDAC3. (b) Representative images of immunostaining for MPZ (red) and phalloidin (green, marking F-actin) in rat SC cultures treated with vehicle, HDAC1/2 inhibitor (HDAC1/2i; FK228), HDAC8i (PCI34051), or HDAC3i (PDA106 or RGFP966) for 24 h. n = 3 independent experiments with 15 images for each experiment. Scale bars, 50 μm. (c) Transcript levels of myelin-related genes Pmp22 and Mpz in rat SC cultures treated with the indicated HDAC inhibitors. Transcript levels were determined by qRT–PCR and were normalized to those in vehicle-treated cells. Data are presented as mean ± s.e.m.; n = 3 independent experiments; one-way ANOVA with Tukey’s multiple-comparisons test (Pmp22: F(4, 10) = 154.7, Pveh versus FK228 = 0.0144, Pveh versus PCI34051 > 0.9999, Pveh versus RGFP966 < 0.0001, Pveh versus PDA106 < 0.0001; Mpz: F(4, 10) = 122.3, Pveh versus FK228 = 0.0399, Pveh versus PCI34051 > 0.9999, Pveh versus RGFP966 < 0.0001, Pveh versus PDA106 < 0.0001). (d) Representative images of longitudinal cryosections of sciatic nerves from mice treated with vehicle, RGFP966, or PDA106. Immunostaining was performed at P7 for EGR2 (red) and DAPI (blue, marking nuclei). n = 4 mice per group with 5 images for each mouse. Scale bars, 20 μm. (e) The percentage of EGR2+ cells among total DAPI+ cells at P7 in sciatic nerves from mice treated with vehicle or an HDAC3 inhibitor. Data are presented as mean ± s.e.m.; n = 4 mice per group; one-way ANOVA with Tukey’s multiple-comparisons test (F(2, 9) = 14.7, Pveh versus RGFP966 = 0.0016, Pveh versus PDA106 = 0.0071). (f) Results from qRT–PCR showing expression of myelination-associated genes in sciatic nerves from mice treated with vehicle, RGFP966, or PDA106. Data are presented as mean ± s.e.m.; n = 3 independent experiments; one-way ANOVA with Tukey’s multiple-comparisons test (Prx: F(2, 6) = 10.56, Pveh versus RGFP966 = 0.0418, Pveh versus PDA106 = 0.0103; Mbp: F(2, 6) = 41.28, Pveh versus RGFP966 = 0.0019, Pveh versus PDA106 = 0.0003; Mpz: F(2, 6) = 52.8, Pveh versus RGFP966 = 0.0002, Pveh versus PDA106 = 0.0008; Egr2: F(2, 6) = 60.53, Pveh versus RGFP966 = 0.0004, Pveh versus PDA106 = 0.0001). (g) Representative electron micrographs of sciatic nerve cross-sections at P7 from mice treated with vehicle, RGFP966, or PDA106. n = 3 mice per group with 10 images for each mouse. Scale bars, 2 μm. (h) Quantification of g ratios for axons in sciatic nerves at P7 from mice treated with vehicle, RGFP966, or PDA106. n = 301 axons from 3 vehicle-treated mice, 300 axons from 3 RGFP966-treated mice, and 240 axons from 3 PDA106-treated mice; one-way ANOVA with Tukey’s multiple-comparisons test (F(2, 838) = 213.9, Pveh versus RGFP966 < 0.0001, Pveh versus PDA106 < 0.0001). (i) Representative electron micrographs of sciatic nerve cross-sections taken at P15 from mice treated with vehicle, RGFP966, or PDA106. n = 3 mice per group with 10 images for each mouse. Scale bars, 2 μm. (j) Quantification of g ratios for axons in sciatic nerves at P15 from mice treated with vehicle, RGFP966, or PDA106. n = 300 axons from 3 mice for each group; one-way ANOVA with Tukey’s multiple-comparisons test (F(2, 897) = 540.5, Pveh versus RGFP966 < 0.0001, Pveh versus PDA106 < 0.0001). (k) The percentage of myelinated axons among total axons in sciatic nerves at P7 and P15 from mice treated with vehicle, RGFP966, or PDA106. Data are presented as mean ± s.e.m.; n = 6 mice per group; one-way ANOVA with Tukey’s multiple-comparisons test (P7: F(2, 15) = 8.807, Pveh versus RGFP966 = 0.0042, Pveh versus PDA106 = 0.0111; P15: F(2, 15) = 6.541, Pveh versus RGFP966 = 0.0136, Pveh versus PDA106 = 0.0239). n.s., not significant; *P < 0.05, **P < 0.01, ***P < 0.001.
To investigate the effects of HDAC inhibition on SC maturation, we treated SCs isolated from neonatal rat sciatic nerve with inhibitors of HDAC1 and 2 (HDAC1/2; FK228), HDAC8 (PCI34051), and HDAC3 (RGFP966 and PDA106). Treatment with the HDAC1/2 inhibitor reduced extension of SC processes and expression of the mature SC markers myelin protein zero (MPZ) and peripheral myelin protein 22 (PMP22; Fig. 1b,c), corroborating an essential role for HDAC1/2 in SC maturation23,24. HDAC8 inhibition had a minimal effect on SC morphology (Fig. 1b). In contrast, treatment with HDAC3 inhibitors resulted in an expansion of cellular processes; actin cytoskeleton augmentation, as shown by phalloidin staining; and induction of MPZ and PMP22 expression, as assessed by immunocytochemistry and qRT–PCR (Fig. 1b,c), suggesting that inhibition of HDAC3 activity promotes the maturation process of SCs in vitro.
To evaluate the effects of HDAC3 inhibition in vivo, we administered RGFP966 or PDA106 via intraperitoneal (i.p.) injection into neonatal mice daily beginning at postnatal day (P) 1. Treatment continued until sciatic nerves were excised for analysis at P7 and P15. Both HDAC3 inhibitors caused an elevation in the number of EGR2-expressing mature SCs in sciatic nerves, consistent with our in vitro data (Fig. 1d,e). Treatment also significantly elevated expression of myelination-associated genes, including Prx, Mbp, Mpz, and Egr2, as compared to vehicle-treated control mice at P7 (Fig. 1f). Ultrastructural analysis of sciatic nerves by electron microscopy (EM) revealed an increase in myelin thickness (assessed through analysis of g ratios, defined as the inner axon diameter divided by the outer diameter of the myelinated sheath; Fig. 1g–j) and the percentage of myelinated axons (Fig. 1k) in HDAC3-inhibitor-treated mice relative to controls at P7 and P15. Although treatment initiated at either P7 or P14 enhanced myelination, treatment starting at P7 produced a more profound enhancement of myelin thickness than that beginning at P14; however, the numbers of EGR2+ SCs in the groups were comparable at P24, indicating that growth of myelin sheath in SCs is more responsive to HDAC3 inhibitors at earlier developmental stages (Supplementary Fig. 2).
HDAC3 inhibition enhances SC remyelination and functional recovery after sciatic nerve transection
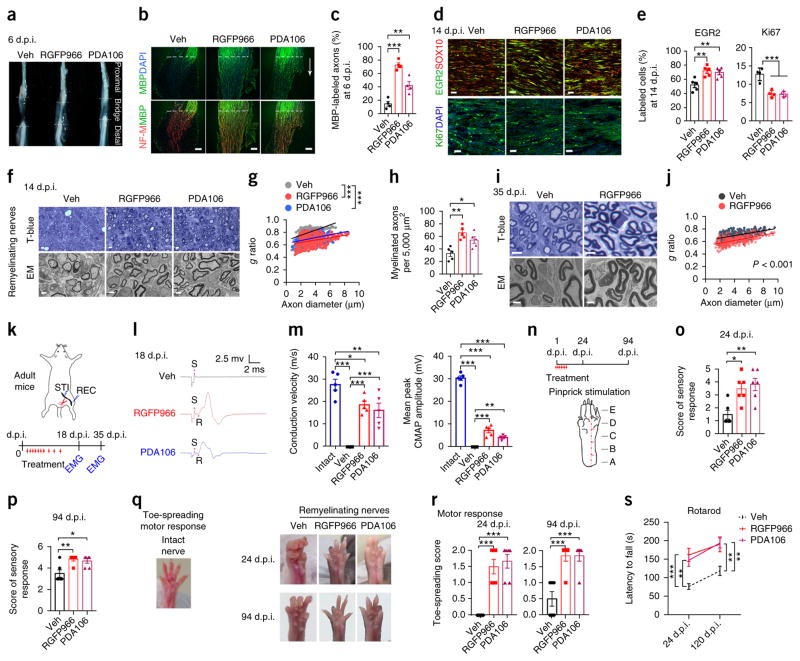
We next evaluated whether treatment with HDAC3 inhibitors after nerve injury would enhance remyelination and functional recovery of nerve conduction, which depends on the myelination state5,25. We performed sciatic nerve transection of the right hindlimb in a group of 8-week-old adult mice and then treated these mice with an HDAC3 inhibitor, either RGFP966 or PDA106 at a dose that resulted in effective inhibition of HDAC3 activity without adverse effects daily over the first week and every 2 d during the second week for a total of 3 d21,26,27. After peripheral nerve transection in mice, denervated SCs resume proliferation, which is followed by differentiation and remyelination of regenerated axons11. We focused our analysis on the regenerating site, defined as a tissue bridge consisting of the SC–axon growth tip proximal to the injury11,12. At 6 days post injury (d.p.i.), when SCs had extensively migrated into bridge tissue5,28, the bridge appeared transparent despite considerable axonal regrowth in the vehicle-treated mice, suggesting that little myelin regeneration occurred spontaneously within this time period (Fig. 2a). In contrast, opaque regeneration tracks were observed within the bridges of RGFP966- and PDA106-treated mice (Fig. 2a), suggesting an accelerated regeneration of myelin sheath. The percentage of myelin basic protein (MBP)+ myelinating axons entering the bridge of regenerating sciatic nerves was significantly higher in RGFP966- and in PDA106-treated mice than in vehicle-treated mice (Fig. 2b,c). Following treatment with an HDAC3 inhibitor, the proportion of EGR2+ maturing SCs in the tissue bridge increased (Fig. 2d,e) and the percentage of Ki67+ cells decreased, suggesting increased SC differentiation accompanied by reduced proliferation (Fig. 2d,e). To further assess the extent of remyelination, we performed EM analysis on the regenerating sciatic nerves. At 14 d.p.i., both RGFP966- and PDA106-treated mice exhibited significantly thicker myelin sheaths in axons of different calibers in the regenerating bridge (Fig. 2f,g and Supplementary Fig. 3a) and an increased number of myelinated axons as compared to vehicle-treated mice (Fig. 2h). Similarly, at 18 and 35 d.p.i., there was a significant increase in myelin thickness after HDAC3 inhibitor treatment relative to that following vehicle treatment (Fig. 2i,j and Supplementary Fig. 3b,c).
Figure 2.
Treatment with an HDAC3 inhibitor enhances SC remyelination and functional recovery after sciatic nerve transection. (a) Representative bright-field images of injured sciatic nerves at 6 d.p.i. Mice (~8 weeks old) were treated i.p. with vehicle, RGFP966, or PDA106 beginning on the day after sciatic nerve transection. Bridge tissues are present between the two dashed lines. n = 4 mice per group. (b) Representative images of longitudinal sections of tissue bridges (regions under the dashed lines) of injured sciatic nerves at 7 d.p.i. after the indicated treatment. Sections were immunostained for neurofilament-M (NF-M; red), MBP (green), and counterstained with DAPI (blue, marking nuclei). The arrow indicates proximal to distal direction. n = 4 mice per group. Scale bars, 50 μm. (c) The percentage of MBP-labelled axons among total axons within the tissue bridge after the indicated treatment. Data are presented as mean ± s.e.m.; n = 4 mice per group; one-way ANOVA with Tukey’s multiple-comparisons test (F(2, 9) = 40.64, Pveh versus RGFP966 < 0.0001, Pveh versus PDA106 = 0.0053). (d) Representative images of longitudinal cryosections of injured sciatic nerves at 14 d.p.i. immunostained for EGR2 (green) and SOX10 (red; top) or for Ki67 (green) and DAPI (blue; bottom). n = 4 or 5 mice per group with 5 images for each mouse. Scale bar, 20 μm. (e) Percentage of EGR2+ (left) or Ki67+ (right) cells among SOX10+ cells and DAPI+ cells, respectively, at 14 d.p.i. in the injured sciatic nerves of mice treated with vehicle or an HDAC3 inhibitor. Data are presented as mean ± s.e.m.; n = 4 or 5 mice per group; one-way ANOVA with Tukey’s multiple-comparisons test (EGR2: F(2, 12) = 9.912, Pveh versus RGFP966 = 0.0042, Pveh versus PDA106 = 0.0091; Ki67: F(2, 9) = 25.95, Pveh versus RGFP966 = 0.0004, Pveh versus PDA106 = 0.0004). (f) Representative toluidine blue–stained (t-blue) images (top) and electron micrographs (EM; bottom) of cross-sections of regenerating tissue bridge in sciatic nerves at 14 d.p.i. from mice treated with vehicle or the indicated compound. n = 5 mice per group with 10 images for each mouse. Scale bars, 5 μm (top) and 2 μm (bottom). (g) Quantification of g ratios for axons of regenerated sciatic nerves at 14 d.p.i. from mice treated with vehicle, RGFP966, or PDA106. n = 308 axons from 3 vehicle-treated mice, 300 axons from 3 RGFP966-treated mice, and 322 axons from 3 PDA106-treated mice; one-way ANOVA with Tukey’s multiple-comparisons test (F(2, 927) = 294.3, Pveh versus RGFP966 < 0.0001, Pveh versus PDA106 < 0.0001). (h) Quantification of the number of myelinated axons per 5,000 μm2 of area of injured sciatic nerves at 14 d.p.i. from mice treated with vehicle, RGFP966, or PDA106. Data are presented as mean ± s.e.m.; n = 5 mice per group; one-way ANOVA with Tukey’s multiple-comparisons test (F(2, 12) = 12.05, Pveh versus RGFP966 = 0.0011, Pveh versus PDA106 = 0.022). (i) Representative toluidine blue–stained images (top) and electron micrographs (bottom) of regenerated sciatic nerve cross-sections at 35 d.p.i. from mice treated with vehicle or RGFP966. n = 5 mice per group with 10 images for each mouse. Scale bars, 5 μm (top) and 2 μm (bottom). (j) Quantification of g ratios for the axons of regenerated sciatic nerves at 35 d.p.i. from mice treated with vehicle or RGFP966. n = 303 axons from 3 mice for each group; two-tailed unpaired Student’s t-test (P < 0.0001, t = 15.29, degrees of freedom (d.f.) = 604). (k) Schematic depicting sciatic nerve transection and electrophysiological analysis. Mice were treated daily (marked with red arrows) for the first week and then every 2 d until 14 d.p.i. Electrophysiological analyses were performed at 18 d.p.i. and 35 d.p.i. EMG, electromyography; STI, stimulus; REC, recording. (l) Representative recordings of CMAPs for regenerated sciatic nerves at 18 d.p.i. from mice treated with vehicle, RGFP966, or PDA106. S, stimulus; R, initiation of CMAP response. n = 5 mice per group. (m) Quantification of the conduction velocity and mean peak of CMAP amplitude of intact or injured sciatic nerves from mice treated with vehicle or RGFP966. Data are presented as mean ± s.e.m.; n = 5 mice per group; one-way ANOVA with Tukey’s multiple-comparisons test (conduction velocity: F(3, 16) = 27.75, Pintact versus veh < 0.0001, Pintact versus RGFP966 = 0.046, Pintact versus PDA106 = 0.0096, Pveh versus RGFP966 < 0.0001, Pveh versus PDA106 = 0.0004; mean peak of CMAP amplitude: F(3, 16) = 335.2, Pintact versus veh < 0.0001, Pintact versus RGFP966 < 0.0001, Pintact versus PDA106 < 0.0001, Pveh versus RGFP966 < 0.0001, Pveh versus PDA106 = 0.0055). (n) Schematic of HDAC3 inhibitor treatment (red arrows) for 10 d (top) and pinprick stimulation to the plantar surface of the paw (bottom); each letter marks a site at which stimulation was applied. (o,p) Quantification of pinprick stimulation assays for sensory recovery at 24 d.p.i. (o) and 94 d.p.i. (p) in mice treated with vehicle or an HDAC3 inhibitor. Data are presented as mean ± s.e.m.; n = 6 mice per group; one-way ANOVA with Tukey’s multiple-comparisons test (24 d.p.i.: F(2, 15) = 9.053, Pveh versus RGFP966 = 0.011, Pveh versus PDA106 = 0.0036; 94 d.p.i.: F(2, 15) = 8.382, Pveh versus RGFP966 = 0.0051, Pveh versus PDA106 = 0.013). (q) Photographs of the paw of an uninjured mouse and of the paws of mice with nerve transection treated with vehicle or an HDAC3 inhibitor for 10 d. (r) Quantification of toe-spreading reflex at 24 d.p.i. and 94 d.p.i. in mice treated with vehicle or an HDAC3 inhibitor. Data are presented as mean ± s.e.m.; n = 6 mice per group; one-way ANOVA with Tukey’s multiple-comparisons test (24 d.p.i.: F(2, 15) = 26.76, Pveh versus RGFP966 < 0.0001, Pveh versus PDA106 < 0.0001; 94 d.p.i.: F(2, 15) = 16.84, Pveh versus RGFP966 = 0.0004, Pveh versus PDA106 = 0.0004). (s) Latency (s) to fall off the accelerating rotarod at 24 d.p.i. and 120 d.p.i. for mice treated with vehicle or an HDAC3 inhibitor. Data are presented as mean ± s.e.m.; n = 8 mice per group; one-way ANOVA with Tukey’s multiple-comparisons test (24 d.p.i.: F(2, 21) = 10.04, Pveh versus RGFP966 = 0.0008, Pveh versus PDA106 = 0.0043; 120 d.p.i.: F(2, 21) = 7.392, Pveh versus RGFP966 = 0.0073, Pveh versus PDA106 = 0.0051). *P < 0.05, **P < 0.01, ***P < 0.001.
To investigate whether the increase in remyelination after HDAC3 inhibition improved motor function in injured nerves, we measured in vivo conduction velocity and compound muscle action potentials (CMAPs) in injured sciatic nerves at different stages (18 and 35 d.p.i.) in adult mice treated with RGFP966 or PDA106 for 2 weeks (Fig. 2k). At 18 d.p.i., CMAP activity was barely detectable after electrical stimulation of injured nerves in vehicle-treated mice. In contrast, an increase in conduction velocity and CMAP amplitude was observed in the injured nerves of inhibitor-treated mice (Fig. 2l,m). At 35 d.p.i., electrophysiological recordings indicated weak CMAP activity in the injured nerves of vehicle-treated mice, whereas treatment with RGFP966 substantially improved motor function in nerves (Supplementary Fig. 4).
Furthermore, we found that short-term 10-d treatment of injured mice with an HDAC3 inhibitor starting at 1 d.p.i. enhanced recovery of both sensory and motor functions, as measured by response to pinprick stimulation (Fig. 2n–p) and toe-spreading reflex (Fig. 2q,r), respectively, at 24 d.p.i. This result was observed 3 months after treatment (94 d.p.i.; Fig. 2p,r), when nerve function remained compromised in vehicle-treated control mice; this is in keeping with the increased myelin thickness in HDAC3-inhibitor-treated mice at this stage (Supplementary Fig. 3de,). Moreover, the rotarod test confirmed functional improvement in motor performance after inhibitor treatment (Fig. 2s).
Myelin morphology was unaltered in uninjured intact nerves of the left hindlimb during the treatment period (Supplementary Fig. 5), indicating that transient administration of an HDAC3 inhibitor did not elicit hypermyelination in uninjured nerves of adult mice at 2 months of age. There was an age-related decline in the level of HDAC3 expression in SCs of adult peripheral nerves as compared to developing peripheral nerves (Supplementary Fig. 6). Considering the age- associated deficits in functional recovery after nerve injury in aged mice compared with young adult mice at 2 months of age29, we assessed the efficacy of HDAC3 inhibition in 10- to 12-month-old mice and found that this treatment enhanced nerve conductivity after sciatic nerve transection (Supplementary Fig. 7). These data show that pharmacological attenuation of HDAC3 activity after sciatic nerve transection accelerates SC remyelination and improves functional recovery.
Ablation of Hdac3 in SCs leads to hypermyelination in sciatic nerves
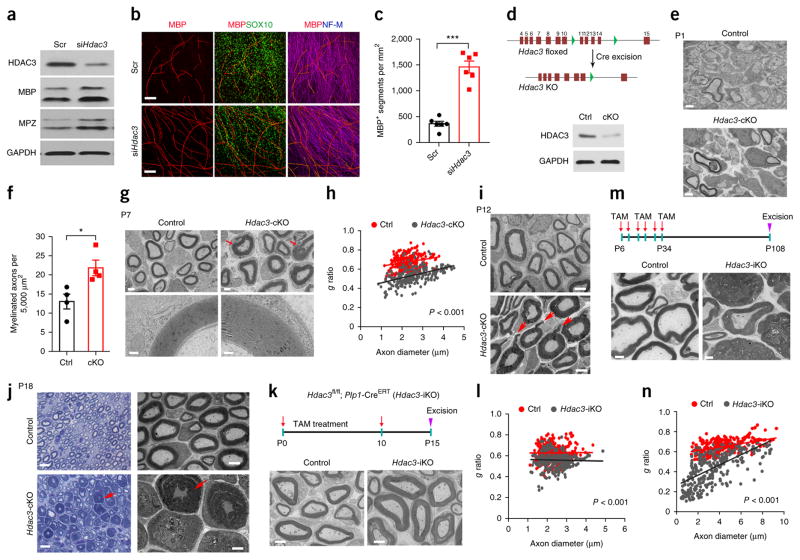
To determine whether the effect of HDAC3 inhibitors in promoting myelination is intrinsic to SCs, we transfected rat SC cultures with an siRNA designed to silence Hdac3 expression (siHdac3; Fig. 3a). Hdac3 silencing resulted in increased amounts of the myelin proteins MBP and MPZ (Fig. 3a). To assess the myelination capacity of SCs, we performed an SC–neuron co-culture assay in which primary rat SCs (SOX10+) treated with or without siHdac3 were seeded onto dorsal root ganglion (DRG) cultures30. After 10 d of culture in pro-myelinating medium containing 50 μg/ml ascorbic acid, the extent of myelination, as determined through immunolabeling for MBP, was substantially greater in Hdac3-deficient SC cultures than the control-transfected SCs (Fig. 3b). Further, the number of myelinated segments was increased by approximately fourfold in Hdac3-deficient SC cultures relative to controls (Fig. 3c).
Figure 3.
Hdac3 ablation leads to hypermyelination during peripheral nerve development. (a) Western blots for HDAC3, MPZ, and MBP in co-cultures of DRGs and SCs treated with siHdac3 or scrambled control siRNA (Scr). n = 2 independent experiments. GAPDH served as a loading control. (b) Rat SCs treated with siHdac3 or control siRNA were seeded onto rat DRGs. After 10 d, co-cultures were immunostained for MBP (red), SOX10 (green), and neurofilament-M (blue). n = 6 independent experiments with 10 images for each experiment. Scale bars, 100 μm. (c) Quantification of the number of MBP+ segments per mm2 of area in myelinating co-cultures of DRGs and SCs treated with siHdac3 or control siRNA. Data are presented as mean ± s.e.m.; n = 6 independent experiments; two-tailed unpaired Student’s t-test (P < 0.0001, t = 8.79, d.f. = 10). (d) Top, a schematic showing Cre-mediated excision of floxed Hdac3 exons 11–14. Bottom, a western blot of sciatic nerves showing a marked decrease in HDAC3 in Hdac3-cKO mice relative to controls (ctrl) at P21. n = 3 independent experiments. GAPDH served as a loading control. (e) EM analysis of cross-sections of sciatic nerves from control and Hdac3-cKO mice at P1. n = 4 mice per group with 10 images for each mouse. Scale bars, 1 μm. (f) Quantification of the number of myelinated axons per 5,000 μm2 of sciatic nerve from control and Hdac3-cKO mice at P1. Data are presented as mean ± s.e.m.; n = 4 mice per group; two-tailed unpaired Student’s t-test (P = 0.0207, t = 3.114, d.f. = 6). (g) Top, EM analysis of cross-sections of control and Hdac3-cKO sciatic nerves at P7. Arrows indicate regions of thick myelin sheaths in Hdac3-cKO mice. Scale bars, 2 μm. Bottom, high-magnification images of myelin wrapped around axons of similar diameter. n = 3 mice per group with 10 images for each mouse. Scale bars, 0.2 μm. (h) Quantification of g ratios of axons at P7 from control and Hdac3-cKO mice. n = 250 axons from 3 control mice and 252 axons from 3 Hdac3-cKO mice; two-tailed unpaired Student’s t-test (P < 0.0001, t = 19.91, d.f. = 500). (i) Representative electron micrographs of cross-sections of sciatic nerves from control and Hdac3-cKO mice at P12. n = 4 mice per group with 10 images for each mouse. Arrows indicate myelin outfoldings. Scale bars, 2 μm. (j) Representative light microscopy images and electron micrographs of sciatic nerve cross-sections from control and Hdac3-cKO mice at P18. Arrows indicate excessive myelin. n = 4 mice per group with 10 images for each mouse. Scale bars, 10 μm (left) and 4 μm (right). (k) Top, a schematic of tamoxifen (TAM) administration. Tamoxifen was injected i.p. into lactating dams once per day until P10. Sciatic nerves were excised at P15. Bottom, representative electron micrographs of cross-sections of sciatic nerves from control heterozygotes and Hdac3-iKO mice. n = 4 mice per group with 10 images for each mouse. Scale bars, 2 μm. (l) Quantification of g ratios of axons at P15 from control and Hdac3-iKO mice. n = 303 axons from 3 control mice and 305 axons from 3 Hdac3-iKO mice; two-tailed unpaired Student’s t-test (P < 0.0001, t = 11.09, d.f. = 606). (m) Top, a schematic of tamoxifen administration. Mice were treated with tamoxifen from P6 to P10, from P18 to P22, and from P30 to P34. Sciatic nerves were excised at P108. Bottom, representative electron micrographs of cross-sections of sciatic nerves from control and Hdac3-iKO mice. n = 4 mice per group with 10 images for each mouse. Scale bars, 2 μm. (n) Quantification of g ratios for axons at P108 from control and Hdac3-iKO mice. n = 302 axons from 3 control mice and 304 axons from 3Hdac3-iKO mice; two-tailed unpaired Student’s t-test (P < 0.0001, t = 21.79, d.f. = 604). *P < 0.05, ***P < 0.001.
To assess the cell-intrinsic role for HDAC3 in SC myelination in vivo, we generated mice lacking Hdac3 in SCs by crossing mice in which Hdac3 was flanked by loxP sites (floxed; Hdac3fl/fl)31 with a mouse line in which cells of SC lineage expressed Cre recombinase under the control of the 2′,3′-cyclic nucleotide 3′ phosphodiesterase (Cnp) promoter (Cnp-Cre)32 (Fig. 3d). HDAC3 expression was lower in the sciatic nerves of Hdac3fl/fl; Cnp-Cre+/− (referred to here as Hdac3-cKO) mice than in controls (Fig. 3d and Supplementary Fig. 8). Hdac3+/fl; Cnp-Cre+/− or Hdac3fl/fl littermates were phenotypically normal during SC development and were used as controls.
The number of SOX10-expressing cells of SC lineage was comparable between the sciatic nerves of control mice and Hdac3-cKO mutants, and the proportion of differentiated EGR2+ SCs was higher in Hdac3-cKO mutants than in controls at P7 (Supplementary Fig. 9). Ultrastructural analysis of sciatic nerves from Hdac3-cKO neonates at P1 showed an increase in myelinated axons relative to controls, indicating an accelerated onset of myelination in the absence of Hdac3 (Fig. 3e,f). By P7, an active phase of myelinogenesis, Hdac3-cKO axons were hypermyelinated, with significantly more layers of myelin lamellae than observed in controls (Fig. 3g,h). At P12, the formation of localized myelin protrusions or outfoldings appeared in hypermyelinated fibers (Fig. 3i). At P18, massive myelin lamellar wrapping was observed in toluidine blue–stained semithin sections and EM graphs of cross-sections (Fig. 3j) and longitudinal sections (Supplementary Fig. 10a) from sciatic nerves of Hdac3-cKO mice; this wrapping eventually led to demyelination in adulthood (Supplementary Fig. 10b). Similar phenotypes were also detected in another mouse line with Hdac3 deletion, in which cells of SC lineage expressed Cre recombinase under the control of the desert hedgehog (Dhh) promoter (Hdac3fl/fl; Dhh-Cre+/− (referred to as Hdac3-cKODhh)33; Supplementary Fig. 11).
To exclude effects of Hdac3 deletion on embryonic SC development, we inactivated Hdac3 in postnatal SCs in a mouse line with expression of tamoxifen-inducible Cre recombinase driven by the proteolipid protein (myelin) 1 (Plp1) promoter (Plp1-CreERT)34 through tamoxifen administration from P0 to P10 (Fig. 3k). At P15, Hdac3fl/fl; Plp1-CreERT (referred to as Hdac3-iKO) mice exhibited hypermyelination in sciatic nerves as compared to controls (Fig. 3k,l).
To further examine effects of Hdac3 deletion on premyelinating SCs, we ablated Hdac3 through treating Hdac3-iKO mice with tamoxifen from P6 to P34 (Fig. 3m) and excised their sciatic nerves at P108. The redundant myelin sheaths in SCs from tamoxifen-treated Hdac3-iKO mice were dramatically thicker than in controls, to the point of causing axonal resection (Fig. 3m,n). Ablation of Hdac3 induced at the early postnatal stage P11 exerted a more robust effect on the enhancement of myelin thickness than ablation at P30 (Supplementary Fig. 12). In contrast, tamoxifen administration after mice had reached adulthood did not alter myelin morphology substantially, despite a modest increase of myelin thickness in axons with a small diameter (Supplementary Fig. 13); this result is consistent with the low expression levels of HDAC3 in adult nerves.
Hdac3 deletion enhances SC remyelination and myelin thickness after injury
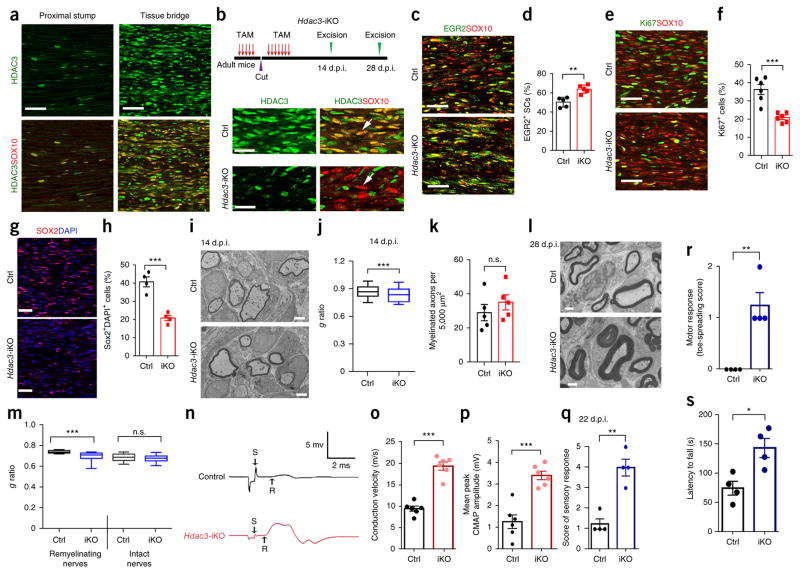
To test whether the effect of HDAC3 inhibitors on remyelination was due to inhibition of HDAC3 in SCs and not activity in other cell types, we used a cell-type-specific strategy, in which we ablated HDAC3 in SCs of Hdac3-iKO mice using tamoxifen after nerve transection injury. HDAC3 expression was detected in denervated SCs marked by SOX10, and the number of HDAC3+ SCs increased substantially in the regenerating bridge tissues after nerve transection at 14 d.p.i. as compared to uninjured adult sciatic nerves (Fig. 4a). We ablated HDAC3 by treating 8-week-old Hdac3-iKO and control mice with tamoxifen before and after sciatic nerve transection. This treatment efficiently removed HDAC3 specifically from the SCs of Hdac3-iKO mice (Fig. 4b). At 14 d.p.i., a time point at which remyelination of the distal stump occurs11,28, we observed an increase in differentiating EGR2+ SCs in the tissue bridge of Hdac3-iKO mice as compared to controls (Fig. 4c,d), and the SC proliferation rate and number of immature SOX2+ SCs were reduced in Hdac3-iKO mice (Fig. 4e–h).
Figure 4.
Ablation of Hdac3 promotes remyelination after nerve injury. (a) Immunofluorescence staining for HDAC3 (green) and SOX10 (red) in sciatic nerves from wild-type mice at 14 d.p.i. showing re-expression of HDAC3 in SCs in the lesion area following nerve cut. SCs in the intact proximal stump nerves show minimal HDAC3 immunoreactivity. n = 5 mice per group with 5 images for each mouse. Scale bars, 50 μm. (b) Top, schematic of nerve transection in control (Hdac3fl/+; Plp1-CreERT) and Hdac3-iKO mice. Tamoxifen was injected i.p. into 8-week-old mice for 5 d, followed by a 7-d rest before nerve cut; mice were then treated daily with tamoxifen for 7 d after injury. Bottom, immunofluorescence staining for SOX10 (red) and HDAC3 (green) in sciatic nerves from control and Hdac3-iKO mice in the regenerating site at 14 d.p.i. n = 5 mice per group with 5 images for each mouse. Arrows indicate the absence of HDAC3 in SOX10+ SCs. Scale bars, 20 μm. (c) Immunofluorescence staining for EGR2 (green) and SOX10 (red) in control and Hdac3-iKO sciatic nerves in the regenerating region at 14 d.p.i. n = 5 mice per group with 5 images for each mouse. Scale bars, 50 μm. (d) The percentage of EGR2+ and SOX10+ colabelled cells among total SOX10+ cells in the regenerating area in sciatic nerves from control and Hdac3-iKO mice at 14 d.p.i. Data are presented as mean ± s.e.m.; n = 5 mice per group; two-tailed unpaired Student’s t-test (P < 0.0019, t = 4.554, d.f. = 8). (e) Immunofluorescence staining for Ki67 (green) and SOX10 (red) in sciatic nerves from control and Hdac3-iKO mice in the regenerating region at 14 d.p.i. n = 6 mice per group with 5 images for each mouse. Scale bars, 50 μm. (f) Quantification of Ki67+ SCs in the regenerating area in sciatic nerves from control and Hdac3-iKO mice at 14 d.p.i. Data are presented as mean ± s.e.m.; n = 6 mice per group; two-tailed unpaired Student’s t-test (P < 0.0004, t = 5.222, d.f. = 10). (g) Immunolabeling for SOX2 (red) and counterstaining with DAPI (blue, marking nuclei) in the regenerating region of sciatic nerves from control and Hdac3-iKO mice at 14 d.p.i. Scale bars, 50 μm. (h) Quantification of the percentage of SOX2+ cells in g. Data are presented as mean ± s.e.m.; n = 4 mice per group with 5 images for each mouse; two-tailed unpaired Student’s t-test (P < 0.0007, t = 6.43, d.f. = 6). (i) Electron micrographs of transverse sections of tissue bridges from control and Hdac3-iKO mice at 14 d.p.i. n = 5 mice per group with 5 images for each mouse. Scale bars, 2 μm. (j) Box plots of g ratios for sciatic nerves from control and Hdac3-iKO mice at 14 d.p.i. in remyelinated regions. Whiskers show the minimum and maximum, and boxes extend from the first to the third quartiles with midlines at the medians; n = 102 remyelinated axons from 3 control mice and 104 remyelinated axons from 3 Hdac3-iKO mice; two-tailed unpaired Student’s t-test (P = 0.0002, t = 3.8, d.f. = 204). (k) Quantification of the number of myelinated axons per μm2 in the cross sections of tissue bridges from control and Hdac3-iKO mice at 14 d.p.i. Data are presented as mean ± s.e.m; n = 5 mice per group; two-tailed unpaired Student’s t-test (P < 0.3799, t = 0.9294, d.f. = 8). (l) Electron micrographs of transverse sections of tissue bridges from control and Hdac3-iKO mice at 28 d.p.i. n = 5 mice per group with 5 images for each mouse. Scale bars, 2 μm. (m) Box plots of g ratios for remyelinating sciatic nerves from control and Hdac3-icKO mice at 28 d.p.i. Whiskers show the minimum and maximum, boxes extend from the first to the third quartiles with cross lines at the medians; n = 150 remyelinated axons from 3 mice for each group; one-way ANOVA with Tukey’s multiple-comparisons test (F(3, 596) = 108.1; remyelinating nerves: Pctrl versus iKO < 0.0001; intact nerves: Pctrl versus iKO = 0.0675). (n) Representative recordings of CMAPs for regenerated sciatic nerves from control and Hdac3-iKO mice awt 20 d.p.i. n = 6 mice per group. (o,p) Quantification of the conduction velocity (o) and mean peak of CMAP amplitude (p) of injured sciatic nerves from control and Hdac3-icKO mice. Data are presented as mean ± s.e.m.; n = 6 mice per group; two-tailed unpaired Student’s t-test (o: P < 0.0001, t = 8.856, d.f. = 10; p: P = 0.0002, t = 5.754, d.f. = 10). (q,r) Results from pinprick stimulation assays for sensory function (q) and toe-spreading reflex for motor function (r) in control and Hdac3-iKO mice at 22 d.p.i. Data are presented as mean ± s.e.m.; n = 4 mice per group; two-tailed unpaired Student’s t-test (q: P = 0.0012, t = 5.745, d.f. = 6; r: P = 0.0025, t = 5, d.f. = 6). (s) Latency to fall (s) off the accelerating rotarod for control and Hdac3-iKO mice at 22 d.p.i. Data are presented as mean ± s.e.m.; n = 4 mice per group; two-tailed unpaired Student’s t-test (P = 0.016, t = 3.42, d.f. = 6). *P < 0.05, **P < 0.01, ***P < 0.001.
Hdac3-iKO mice had thicker myelin in transected sites than controls, as determined through EM analysis at 14 d.p.i. (Fig. 4i,j), although the number of myelinating axons was comparable between the groups (Fig. 4k). At 28 d.p.i., myelin sheaths were much thicker in the regenerating region of nerves in Hdac3-iKO mice than in controls (Fig. 4l), with significantly lower g ratios than in control lesion sites or contralateral regions of intact nerves (Fig. 4m). Tamoxifen treatment during this period did not alter myelin thickness in the uninjured nerves (Fig. 4m).
At 20 d.p.i., in vivo CMAP activity was barely detectable after electrical stimulation of injured nerves in control mice (Fig. 4n). In contrast, we observed significantly greater CMAP signals in the injured nerves of Hdac3-iKO mice, as reflected by elevated conduction velocities and mean peak amplitudes (Fig. 4o,p). Furthermore, Hdac3-iKO mice exhibited enhanced sensory function, as measured through pinprick stimulation (Fig. 4q), and motor function, as determined through toe-spreading reflexes and a rotarod test (Fig. 4r,s). These observations suggest that HDAC3 inhibition in denervated SCs accelerates SC maturation, enhances remyelination, and improves functional recovery.
HDAC3 negatively regulates NRG1–PI3K–AKT signaling to repress SC myelination
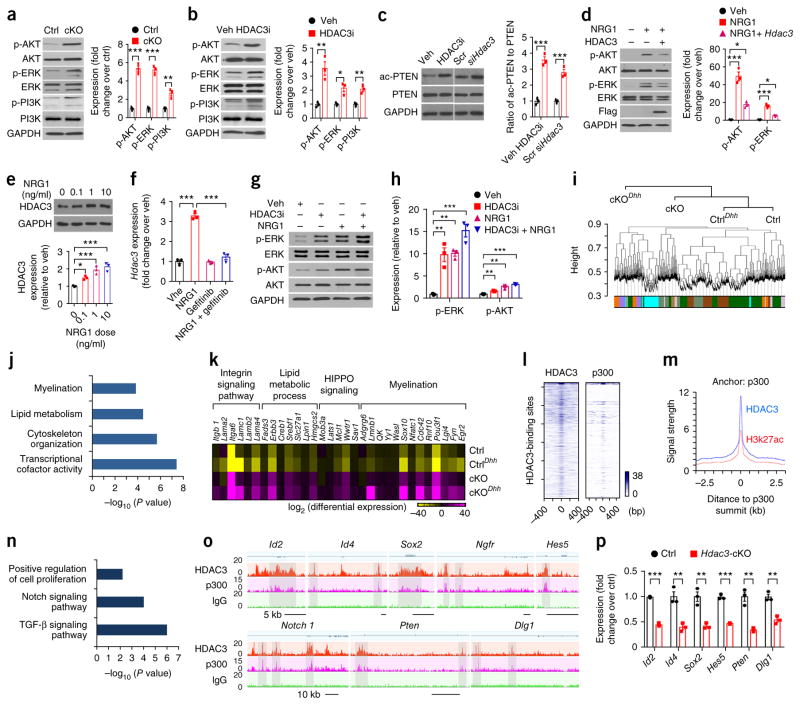
The hypermyelinating phenotype observed in Hdac3-cKO mice resembles that of mice overexpressing NRG1 type III (ref. 35). As we did not detect significant differences in activation of the NRG1 receptors ErbB2 or ErbB3, as determined through the ratio of phosphorylated ErbB (p-ErbB) to ErbB, between the nerves of control and Hdac3-cKO mice at P13 (Supplementary Fig. 14), we hypothesized that the effect of HDAC3 on SC myelination was due to activation of downstream pathways, such as PI3K–AKT and MAPK–ERK36–38. Indeed, we detected an increase in the levels of p-AKT, p-PI3K, and p-ERK in the nerves of Hdac3-cKO mice at P13 as compared to controls (Fig. 5a). Similarly, inhibition of HDAC3 by PDA106 in purified rat SCs led to an elevation in p-AKT, p-PI3K, and p-ERK levels (Fig. 5b). In addition, treatment with HDAC3 inhibitor or siHdac3 significantly increased the level of acetylated PTEN (Fig. 5c), which resulted in a reduction of PTEN activity39, leading to activation of PI3K–AKT signaling.
Figure 5.
Activation of the PI3K–AKT pathway promotes myelination in the sciatic nerves of Hdac3-mutant mice. (a,b) Representative western blots (left) and quantification (right) of p-AKT, p-ERK, and p-PI3K in sciatic nerves from control and Hdac3-cKO mice at P13 (a) and rat SC cultures treated with vehicle or PDA106 (b). The band densities of phosphorylated forms were normalized to the respective levels of total AKT, ERK, or PI3K. GAPDH served as a loading control. Data are presented as mean ± s.e.m.; n = 3 independent western blot experiments; two-tailed unpaired Student’s t-test (a: p-AKT: P = 0.0002, t = 12.91, d.f. = 4; p-ERK: P = 0.0001, t = 14.17, d.f. = 4; p-PI3K: P = 0.005, t = 5.583, d.f. = 4; b: p-AKT: P = 0.006, t = 5.222, d.f. = 4; p-ERK: P = 0.0118, t = 4.392, d.f. = 4; p-PI3K: P = 0.005, t = 5.708, d.f. = 4). (c) Representative western blots (left) and quantification (right) of acetylated PTEN (ac-PTEN) and PTEN in rat SC cultures treated with PDA106 or transfected with siHdac3. GAPDH served as a loading control. Data are presented as mean ± s.e.m.; n = 3 independent western blot experiments; two-tailed unpaired Student’s t-test (PHDAC3i versus veh = 0.0004, tHDAC3i versus veh = 10.63, d.f. = 4; PsiHDAC3 versus scr = 0.00097, tsiHDAC3 versus scr = 8.673, d.f. = 4). (d) Representative western blots (left) and quantification (right) of p-AKT, AKT, p-ERK, and ERK in rat SC cultures treated with NRG1 with or without Hdac3 overexpression (+ Hdac3). p-AKT and p-ERK were normalized to AKT and ERK, respectively. Data are presented as mean ± s.e.m.; n = 3 independent western blot experiments; one-way ANOVA with Tukey’s multiple-comparisons test (p-AKT: F(2, 6) = 66.62, Pveh versus NRG1 < 0.0001, Pveh versus NRG1 + Hdac3 = 0.017; p-ERK: F(2, 6) = 109.2, Pveh versus NRG1 < 0.0001, Pveh versus NRG1 + Hdac3 = 0.0106). (e) Top, representative western blot of HDAC3 levels in SC cultures treated with the indicated concentrations of NRG1 for 1 h. GAPDH served as a loading control. Bottom, NRG1-induced HDAC3 expression relative to vehicle treatment. Data are presented as mean ± s.e.m.; n = 3 independent western blot experiments; one-way ANOVA with Tukey’s multiple-comparisons test (F(3, 8) = 23.72, P0 versus 0.1 = 0.0415, P0 versus 1 = 0.0009, P0 versus 10 = 0.0003). (f) Results from qPCR showing expression of Hdac3 in primary SCs treated with vehicle, NRG1, gefitinib, or combined NRG1 and gefitinib. Data are presented as mean ± s.e.m.; n = 3 independent experiments; one-way ANOVA with Tukey’s multiple-comparisons test (F(3, 8) = 125.8; PNRG1 versus veh < 0.0001, PNRG1 versus gefitinib < 0.0001, PNRG1 versus NRG1 + gefitinib < 0.0001). (g) Western blots of rat SCs showing p-AKT, p-ERK, and p-PI3K levels after treatment with vehicle, PDA106, NRG1, or combined PDA106 and NRG1; GAPDH served as a loading control. (h) Quantification of relative expression of the indicated protein following the indicated treatment over expression induced by vehicle; p-AKT and p-ERK were normalized to AKT and ERK, respectively. Data are presented as mean ± s.e.m.; n = 3 independent western blot experiments; one-way ANOVA with Tukey’s multiple-comparisons test (p-ERK: F(3, 8) = 29.83, Pveh versus HDAC3i = 0.0018, Pveh versus NRG1 = 0.0015, Pveh versus HDAC3i + NRG1 < 0.0001; p-AKT: F(3, 8) = 41.62, Pveh versus HDAC3i = 0.0494, Pveh versus NRG1 = 0.0002, Pveh versus HDAC3i + NRG1 < 0.0001). (i) Top, gene dendrogram based on RNA-seq data sets for sciatic nerves from Hdac3-cKO (cKO), Hdac3fl/+; Cnp-Cre (Ctrl), Hdac3-cKODhh (cKODhh), and Hdac3fl/+; Dhh-Cre (CtrlDhh) mice at P6. Bottom, coexpression modules are color-coded among Hdac3-mutant sciatic nerves and corresponding controls. The module colors represent coexpressed gene modules. (j) Results from gene ontology analysis showing that genes associated with myelination, lipid metabolism, cytoskeleton organization, and transcription cofactor activity were upregulated in sciatic nerves from Hdac3-cKO mice relative to controls at P6. (k) Heat map showing representative upregulated genes in each category of interest from RNA-seq analysis of sciatic nerves from cKO, Ctrl, cKODhh, and CtrlDhh at P6. Each RNA sample was pooled from 6 mice per genotype and sequenced once. (l) Heat maps of HDAC3 and p300 ChIP–seq signals in rat SCs grown under the differentiation condition. Each line on the y axis represents a genomic region that is within ± 0.4 kb of the corresponding HDAC3 summit. (m) Results from ChIP–seq enrichment analysis of the profiles of HDAC3 and H3K27ac around p300 peak summits in rat SCs under the differentiation condition. (n) Results from gene ontology analysis showing that the downregulated genes targeted by both HDAC3 and p300 (HDAC3 peaks within 5 kb of the transcription start site) were significantly enriched for genes related to regulation of cell proliferation, components of the Notch signaling pathway, and components of the TGF-β signaling pathway. (o) ChIP signals over representative gene loci of negative regulators of myelination that are targeted by both HDAC3 and p300. n = 2 independent experiments. The y axis marks the ChIP-seq enrichment values. (p) Results from qPCR showing reduced expression of genes related to myelination inhibition in sciatic nerves from Hdac3-cKO mice relative to control mice at P7. Data are presented as mean ± s.e.m.; n = 3 independent experiments; two-tailed unpaired Student’s t-test (Id2: P < 0.0001, t = 23.59, d.f. = 4; Id4: P = 0.0051, t = 5.558, d.f. = 4; Sox2: P = 0.0044, t = 5.791, d.f. = 4; Hes5: P < 0.0001, t = 16.51, d.f. = 4; Pten: P = 0.0022, t = 7.007, d.f. = 4; Dlg1: P = 0.0029, t = 6.485, d.f. = 4). *P < 0.05, **P < 0.01, ***P < 0.001.
Treatment of primary rat SCs with recombinant NRG1 type III for 1 h led to upregulation of HDAC3 expression (Fig. 5e). This effect was occluded by the NRG1 signaling inhibitor gefitinib (Fig. 5f), which blocks the signaling of the ErbB2 and ErbB3 receptors activated by NRG1 engagement40. Conversely, HDAC3 overexpression inhibited the phosphorylation of AKT and ERK that was induced by NRG1 in SCs (Fig. 5d). In addition, treatment with NRG1 or the HDAC3 inhibitor PDA106 increased the p-ERK and p-AKT levels in SCs (Fig. 5g,h), and combined treatment of NRG1 together with the HDAC3 inhibitor further enhanced p-ERK levels (Fig. 5g,h). These observations suggest that NRG1 may induce expression of HDAC3, which in turn acts as a negative regulator of the NRG1 signaling pathway.
HDAC3 inhibits promyelinating programs and recruits p300 to activate a myelination-inhibitory network
Although the Hdac3-deficient phenotype resembles that of NRG1-overexpressing mice, unique features, such as myelin outfoldings that are not induced by NRG1 overexpression, suggest that HDAC3 might regulate additional regulatory networks independent of NRG1 that block peripheral myelin overgrowth. To define the HDAC3-regulated genetic program that controls formation of myelin sheaths by SCs, we performed unbiased transcriptome profiling of control, Hdac3-cKO, and Hdac3-cKODhh sciatic nerves at P6, an active phase of SC differentiation. We applied weighted gene coexpression network analysis (WGCNA) to the data41 to identify coexpressed gene modules with the most significant changes in both Hdac3-deficient models (Fig. 5i). Our data indicate an upregulation of genes associated with myelination, lipid metabolic and biosynthetic processes, HIPPO signaling, and integrin signaling processes in Hdac3 mutants (Fig. 5j,k) all are processes known to be critical for SC myelination.
To further identify HDAC3 target genes with altered expression in Hdac3-cKO mutants, we performed chromatin immunoprecipitation and sequencing (ChIP–seq) analysis of HDAC3 genomic occupancy in SCs cultured under conditions fostering proliferation or differentiation. The intensity of HDAC3-binding sites was higher in differentiating SCs than in proliferating SCs (Supplementary Fig. 15a,b). Recent studies indicate that, in the absence of the histone acetyltransferase (HAT) coactivator p300, HDAC3 occupancy represses target gene expression, while HDAC3 and p300, when both present, coordinately target enhancers marked by H3K27ac to promote expression of target genes, producing an overall increase in net HAT activity42,43. HDAC3 targeted a set of genes different from those targeted by p300 in SCs (Fig. 5l). These HDAC3-targeted genes with minimal p300 co-occupancy included the myelin-related genes Pmp22, Mpz, Mbp, Erbb2, and Gjb1 and promyelinating transcriptional regulatory genes such as Egr2, Zeb2, and Srebf1 (Supplementary Fig. 15c). In accordance, expression of these genes was increased in the sciatic nerves of Hdac3-cKO mice (Fig. 5i–k), whereas overexpression of HDAC3 inhibited expression of these myelination-related genes (Supplementary Fig. 15d). Furthermore, knockdown of HDAC3 in SCs led to an increased level of the activating histone mark H3K27ac on the enhancers of myelination-associated gene loci (Supplementary Fig. 16).
These data reveal that HDAC3 and p300 targeted the same set of genes in differentiating SCs that coincide with enhancer elements marked by H3K27ac (Fig. 5l,m). The gene loci bound by both HDAC3 and p300 were significantly enriched for genes encoding cell-growth-related factors and negative regulators of myelination (Fig. 5n). These included Notch1, Hes5, Id2, Id4, Sox2, and Ngfr (p75; Fig. 5o), as well as Pten and Dlg1, which are negative regulators of PI3K–AKT signaling (Fig. 5o). Accordingly, expression of these genes was downregulated in the nerves of Hdac3-cKO mutants (Fig. 5p). Similarly, at 14 d.p.i., expression of HDAC3–p300-targeted genes encoding negative regulators of myelination was upregulated in the regenerating tissue bridge, whereas myelination-associated genes and promyelination regulators were downregulated (Supplementary Fig. 17). Our data position coordination between HDAC3 and p300 at the apex of a gene regulatory network that controls both activation and inhibition of SC myelination via distinct mechanisms.
HIPPO effector TEAD4, an HDAC3–p300 target, is an inhibitor of SC myelin growth
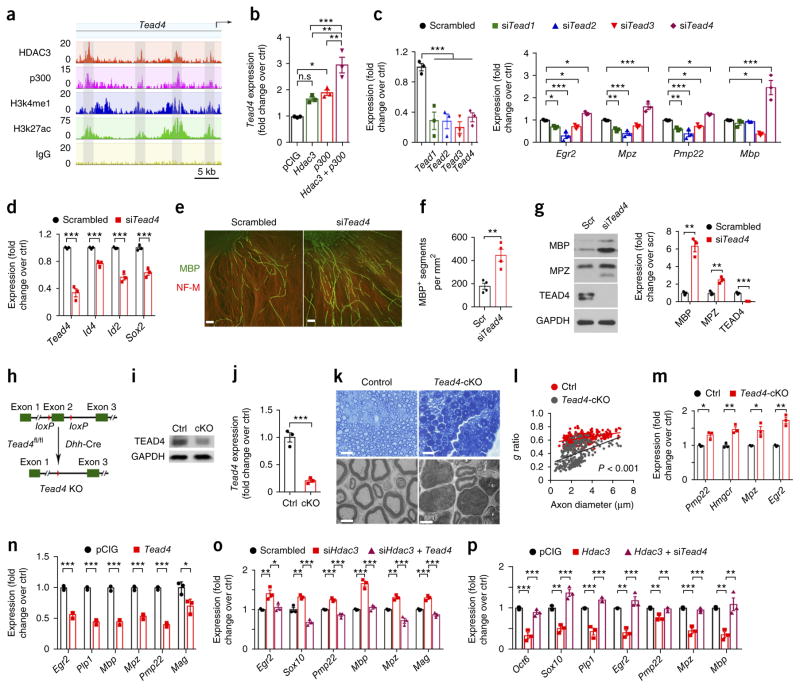
To identify new regulatory factors that inhibit myelin overgrowth, we next examined potential transcription factors regulated through the coordinated action of HDAC3 and p300. Tead4 was identified among the transcription factor gene loci co-occupied by HDAC3 and p300 (Supplementary Table 1). The TEA-domain transcription factor 4 (Tead4) locus was enriched for the H3K27ac and H3K4me1 histone modifications, which mark active and poised enhancers44, respectively, on HDAC3–p300 co-targeted elements (Fig. 6a), suggesting that HDAC3 and p300 co-occupancy may activate Tead4 expression. In contrast, the loci of TEAD family members Tead1–Tead3 were mainly targeted by HDAC3 but not p300 (Supplementary Fig. 18a). Overexpression of HDAC3 and p300 in SCs promoted Tead4 expression to a significantly greater extent than either HDAC3 or p300 overexpression alone (Fig. 6b). Conversely, Tead4 expression was significantly downregulated in siHdac3-treated SCs and the sciatic nerves of Hdac3-cKO mice (Supplementary Fig. 18b).
Figure 6.
HDAC3 inhibits myelinogenesis by activating the inhibitory factor TEAD4. (a) Genome browser view of the distribution of HDAC3, p300, and the H3K4me1 and H3K27ac marks on the Tead4 locus. n = 2 independent experiments for ChIP–seq of HDAC3, p300, and IgG; n = 1 experiment for histone marks. (b) Results from qRT–PCR analysis of Tead4 expression in SCs transfected with control vector pCMV-IRES-GFP (pCIG), a vector for expression of Hdac3, or a vector for expression of both Hdac3 and p300. Data are presented as mean ± s.e.m.; n = 3 independent experiments; one-way ANOVA with Tukey’s multiple-comparisons test (F(3, 8) = 25.48; PpCIG versus Hdac3 = 0.0705, PpCIG versus p300 = 0.0164, PpCIG versus Hdac3 + p300 = 0.0001, PHdac3 versus Hdac3 + p300 = 0.0022, Pp300 versus Hdac3 + p300 = 0.0077). (c) Left, results from qRT–PCR validation of inhibition of Tead1–Tead4 expression with the indicated siRNAs. Right, results from qRT–PCR analyses for expression of Egr2, Mpz, Pmp22, and Mbp in SCs treated with the indicated siRNAs. Data are presented as mean ± s.e.m.; n = 3 independent experiments; one-way ANOVA with Tukey’s multiple-comparisons test (left: F(4, 10) = 15.72, Pscr versus Tead1 = 0.0009, Pscr versus Tead2 = 0.0009, Pscr versus Tead3 = 0.0004, Pscr versus Tead4 = 0.001; right: Egr2: F(4, 10) = 31.07, Pscr versus Tead1 = 0.0333, Pscr versus Tead2 = 0.0001, Pscr versus Tead3 = 0.0398, Pscr versus Tead4 = 0.0281; Mpz: F(4, 10) = 40.63, Pscr versus Tead1 = 0.0076, Pscr versus Tead2 = 0.0007, Pscr versus Tead4 = 0.0006; Pmp22: F(4, 10) = 28.81, Pscr versus Tead1 = 0.0048, Pscr versus Tead2 = 0.0002, Pscr versus Tead3 = 0.041, Pscr versus Tead4 = 0.0399; Mbp: F(4, 10) = 27.26, Pscr versus Tead3 = 0.0499, Pscr versus Tead4 = 0.0002). (d) qRT–PCR analyses of Id4, Id2, and Sox2 in siTead4-treated SCs. Data are presented as mean ± s.e.m.; n = 3 independent experiments; two-tailed unpaired Student’s t-test (Tead4: P = 0.0004, t = 11.12, d.f. = 4; Id4: P = 0.0011, t = 8.44, d.f. = 4; Id2: P = 0.0005, t = 10.09, d.f. = 4; Sox2: P = 0.0013, t = 8.069, d.f. = 4). (e) Rat SCs transfected with siTead4 or a scrambled control siRNA were seeded onto rat DRGs. After 10 d, co-cultures were stained for MBP (green) and neurofilament-M (red). n = 3 independent experiments with 10 images for each experiment. Scale bars, 100 μm. (f) Quantification of the number of MBP+ segments per mm2 of area formed in myelinating co-cultures of DRGs and SCs transfected with siTead4 or control siRNA. Data are presented as mean ± s.e.m.; n = 4 independent experiments; two-tailed unpaired Student’s t-test (P = 0.0037, t = 4.591, d.f. = 6). (g) Left, western blots of TEAD4, MPZ, and MBP in myelinating co-cultures of DRGs and rat SCs transfected with siTead4 or control siRNA. GAPDH served as a loading control. Right, relative siTead4-induced expression of the indicated protein over the expression induced by the scrambled control. The corresponding band densities were normalized to GAPDH. Data are presented as mean ± s.e.m.; n = 3 independent western blot experiments; two-tailed unpaired Student’s t-test (MBP: P = 0.0011, t = 8.412, d.f. = 4; MPZ: P = 0.0042, t = 5.885, d.f. = 4; TEAD4: P = 0.0002, t = 13.2, d.f. = 4). (h) A schematic showing excision of the floxed exon 2 of the Tead4 allele upon recombination mediated by Dhh-Cre in SCs. (i) Western blot analysis of sciatic nerves showing a marked decrease in TEAD4 in Tead4-cKO mice as compared to controls at P120. GAPDH served as a loading control. n = 3 independent western blot experiments. (j) Results from qRT–PCR analysis of Tead4 expression in sciatic nerves from control or Tead4-cKO mice. Data are presented as mean ± s.e.m.; n = 3 independent experiments; two-tailed unpaired Student’s t-test (P = 0.0009, t = 8.774, d.f. = 4). (k) Cross-sections from control and Tead4-cKO sciatic nerves at P120 imaged with light microscopy after toluidine blue staining (top) and electron microscopy (bottom). n = 3 mice per group with 5 images for each mouse. Scale bars, 10 μm (top) and 4 μm (bottom). (l) Quantification of g ratios of myelinated axons from control and Tead4-cKO mice at P120. n = 303 axons from 3 mice for each group; two-tailed unpaired Student’s t-test (P < 0.0001, t = 25.51, d.f. = 604). (m) Results from qRT–PCR analysis for expression of Pmp22, Hmgcr, Mpz, and Egr2 in sciatic nerves from Tead4-cKO and control mice at P120. Data are presented as mean ± s.e.m.; n = 3 mice per group; two-tailed unpaired Student’s t-test (Pmp22: P = 0.0106, t = 4.532, d.f. = 4; Hmgcr: P = 0.0049, t = 5.619, d.f. = 4; Mpz: P = 0.0191, t = 3.798, d.f. = 4; Egr2: P = 0.0014, t = 7.916, d.f. = 4). (n) Results from qRT–PCR analysis for expression of Egr2, Plp1, Mbp, Mpz, Pmp22, and Mag in SCs transfected with a vector to overexpress Tead4. Data are presented as mean ± s.e.m.; n = 3 independent experiments; two-tailed unpaired Student’s t-test (Egr2: P = 0.0003, t = 12, d.f. = 4; Plp1: P < 0.0001, t = 19.58, d.f. = 4; Mbp: P < 0.0001, t = 15.65, d.f. = 4; Mpz: P = 0.0001, t = 14.88, d.f. = 4; Pmp22: P < 0.0001, t = 24.77, d.f. = 4; Mag: P = 0.0264, t = 3.435, d.f. = 4). (o) Results from qRT–PCR analysis for expression of myelination-associated genes in SCs treated with siHdac3 and transfected with control vector or a vector for Tead4 expression. Data are presented as mean ± s.e.m.; n = 3 independent experiments; one-way ANOVA with Tukey’s multiple-comparisons test (Egr2: F(2, 6) = 11.52, Pscr versus siHdac3 = 0.0079, PsiHdac3 versus siHdac3 + pTead4 = 0.0159; Sox10: F(2, 6) = 52.54, Pscr versus siHdac3 = 0.004, PsiHdac3 versus siHdac3 + pTead4 = 0.0001; Pmp22: F(2, 6) = 57.59, Pscr versus siHdac3 = 0.0009, PsiHdac3 versus siHdac3 + pTead4 = 0.0001; Mbp: F(2, 6) = 63.97, Pscr versus siHdac3 = 0.0001, PsiHdac3 versus siHdac3 + pTead4 = 0.0002; Mpz: F(2, 6) = 47.15, Pscr versus siHdac3 = 0.004, PsiHdac3 versus siHdac3 + pTead4 = 0.0001; Mag: F(2, 6) = 56.23, Pscr versus siHdac3 = 0.0007, PsiHdac3 versus siHdac3 + pTead4 = 0.0001). (p) Results from qRT–PCR analysis of expression of myelination-associated genes in SCs that overexpress HDAC3 treated with either control siRNA or siTead4. Data are presented as mean ± s.e.m.; n = 3 independent experiments; one-way ANOVA with Tukey’s multiple-comparisons test (Oct6: F(2, 6) = 46.18, Pscr versus pHdac3 = 0.0002, PpHdac3 versus pHdac3 + siTead4 = 0.0005; Sox10: F(2, 6) = 48.37, Pscr versus pHdac3 = 0.0025, PpHdac3 versus pHdac3 + siTead4 = 0.0001; Plp1: F(2, 6) = 58.43, Pscr versus pHdac3 = 0.0005, PpHdac3 versus pHdac3 + siTead4 = 0.0001; Egr2: F(2, 6) = 34.41, Pscr versus pHdac3 = 0.0016, PpHdac3 versus pHdac3 + siTead4 = 0.0004; Pmp22: F(2, 6) = 15.21, Pscr versus pHdac3 = 0.0046, PpHdac3 versus pHdac3 + siTead4 = 0.0067; Mpz: F(2, 6) = 50.16, Pscr versus pHdac3 = 0.0002, PpHdac3 versus pHdac3 + siTead4 = 0.0003; Mbp: F(2, 6) = 16.23, Pscr versus pHdac3 = 0.0068, PpHdac3 versus pHdac3 + siTead4 = 0.0035). *P < 0.05, **P < 0.01, ***P < 0.001.
Depletion of Tead4, but not Tead1, Tead2, or Tead3, significantly increased expression of myelination-associated genes Egr2, Pmp22, Mbp, and Mpz (Fig. 6c) while repressing expression of genes associated with inhibition of SC differentiation, such as Id2, Id4, and Sox2 (Fig. 6d). The change in gene expression patterns upon depleting Tead4 correlated with increased myelination in the DRG–SC co-culture paradigm: the knockdown of Tead4 in SCs produced an increased number of myelin segments as compared to control-siRNA-treated SCs (Fig. 6e,f), which was confirmed by an elevated level of the myelin proteins MBP and MPZ in SCs (Fig. 6g). These results identify TEAD4 as a direct target of HDAC3–p300 regulation, which negatively regulates SC myelination.
To determine whether genetic ablation of Tead4 in the SC lineage produces hypermyelination phenotypes similar to Hdac3 deletion in vivo, we selectively ablated the expression of Tead4 in cells of SC lineage through breeding mice harboring a floxed Tead4 allele45 with the mouse line in which SC lineage cells expressed Cre recombinase from the Dhh promoter (Fig. 6h–j).Tead4fl/fl; Dhh-Cre+/− (referred to as Tead4-cKO) mice were born at the expected Mendelian frequency. At early postnatal stages, morphometric analysis did not reveal substantial alterations in expression of mature SC markers (Supplementary Fig. 19a–d), whereas myelin thickness was slightly increased relative to controls (Supplementary Fig. 19e,f). The myelin internodal length was not significantly altered in Tead4-cKO mice at P14 (Supplementary Fig. 20). In contrast, in adult sciatic nerves, Tead4-cKO mutants developed profound hypermyelination and excessive myelin wraps, as shown by toluidine blue staining, ultrastructure, and g ratio analyses (Fig. 6k,l), suggesting that sustained absence of TEAD4 leads to myelin overgrowth. Accordingly, we also observed an increase in expression of the major myelination-related genes in Tead4-cKO nerves through qRT–PCR (Fig. 6m), whereas overexpression of TEAD4 in SCs inhibited expression of myelination-related genes (Fig. 6n).
To test whether Tead4 loss is responsible for upregulation of myelin genes in Hdac3-deficient SCs, we inhibited Hdac3 expression using siRNA and then overexpressed Tead4. Silencing Hdac3 led to an upregulation in expression of myelination-associated genes, whereas Tead4 overexpression repressed their expression (Fig. 6o). Conversely, Hdac3 overexpression inhibited the expression of myelination-related genes and Tead4 inhibition partially restored their expression (Fig. 6p).
DISCUSSION
Functional regeneration and recovery after nerve injury requires not only axonal regrowth but also remyelination of the regenerated axons5,25. Despite the remarkable axonal regeneration capacity of peripheral nerves, myelin sheaths regenerated after injury are substantially thinner than myelin formed during development4,5. Slow and compromised remyelination could contribute to the limited restoration of sensory and motor functions observed after proximal nerve injury in humans29,46.
In a small-molecule epigenetic screen, we found that pharmacological inhibitors of HDAC3 markedly elevated myelinogenic potential. Further, transient inhibition of HDAC3 activity robustly accelerated myelination and promoted functional recovery after peripheral nerve injury in mice. As peripheral nerve regeneration involves multiple processes, including axonal regeneration, SC migration, macrophage infiltration, and endothelial cell growth5,11,47,48, HDAC3 inhibitors may also influence remyelination in a non-cell-autonomous manner, as pharmacological inhibition of HDAC3 enhanced axonal regrowth and blood vessel formation after nerve injury (Supplementary Fig. 21). Nonetheless, cell-type-specific genetic ablation demonstrated that Hdac3 loss in cells of SC lineage sufficed to increase SC maturation and myelinogenesis during nerve development and promoted denervated SC redifferentiation and myelination.
Inhibition of HDAC3 enhanced nerve conductivity after nerve injury not only in young adult mice but also in aging mice, suggesting that targeting HDAC3 may rejuvenate the age-related decline in functional recovery after nerve injury. As the axonal regeneration rate is comparable between young and aged mice (>20 months age) after nerve injury49, enhancement of SC myelination by HDAC3 inhibition likely contributes to conductivity restoration in aging mice, suggesting a potential therapeutic strategy for improving myelin repair in elderly individuals who often suffer from idiopathic peripheral neuropathy50.
Although nuclear export of HDAC5 in DRG neurons is critical for nerve regeneration51, we did not detect nuclear translocation of HDAC5 in SCs during nerve regeneration (Supplementary Fig. 22). HDAC3 exerts unique functions distinct from those of other class I HDACs, as inhibition of HDAC1/2 suppresses, rather than promotes, myelin sheath growth23,24 and blocks SC remyelination52, although transient pharmacological inhibition of HDAC1/2 increases the number of repair cells to promote axonal regrowth17,52. This suggests distinct functions among individual HDAC family members in SC myelination and remyelination. In contrast to its later function as an inhibitor of SC maturation and myelinogenesis, HDAC3 promotes oligodendrocyte specification early in development42. Thus, HDAC3 may have a distinct role in development of myelinating cells in the central and peripheral nervous systems, although the potential HDAC3 functions in oligodendrocyte myelin growth and repair remain to be determined. Both the in vitro and in vivo pharmacological and genetic approaches described here provide evidence that HDAC3 acts in a cell-intrinsic manner on immature or premyelinating SCs to delimit myelin growth and regeneration.
Our results suggest that HDAC3 functions in a negative regulatory loop that restrains the activity of NRG1–PI3K–AKT signaling to ensure proper myelination and myelin growth. We found that HDAC3 antagonizes NRG1 signaling and its downstream myelinogenic program through blocking the activation of PI3K–AKT and ERK at the cellular level. It is possible that HDAC3, which can shuttle between the cytoplasm and nucleus53, may regulate the acetylation state of PTEN or PI3K–AKT to control kinase activity or turnover as in other contexts54. Alternatively, HDAC3 may indirectly regulate factors that inhibit NRG1–PI3K–AKT signaling. Our genomic occupancy and expression profiling analyses revealed that HDAC3, in coordination with p300, directly activates expression of Pten and Dlg1. In contrast to the loss of Pten-associated Dlg1, which leads to a transient increase of myelin thickness during development55, deletion of Hdac3 in SCs resulted in profound, sustained myelin sheath growth.
The HDAC3 expression pattern parallels that of NRG1 type III, for which the expression of genes increases during perinatal stages and then decreases in adulthood56. Downregulation of HDAC3 in adult nerves may correlate with a low level of NRG1; therefore, a minimal level of inhibition exerted by HDAC3 is sufficient to maintain myelin homeostasis in adult nerves. Alternatively, the low HDAC3 levels in adulthood are related to the previous formation of deacetylated chromatin or heterochromatin over myelination-related gene loci, obviating the need for histone deacetylation in adulthood. Thus, a developmental window of opportunity exists for modulating HDAC3 activity in immature or pre-myelinating SCs. Our observations suggest a potential role for a balance between NRG1–PI3K–AKT signaling and HDAC3 activity in fine-tuning myelin sheath thickness for optimal conduction velocity and nerve functions during development and recovery from injury.
Our genome-wide analysis of HDAC3 occupancy and transcriptome profiling revealed that HDAC3 inhibits the transcription of genes encoding promyelinating regulators in SCs. Inhibition of HDAC3 increased transcription of these genes and the deposition of activating H3K27ac marks on their enhancers and promoters. Thus, our observations suggest that HDAC3 exerts an inhibitory effect on SC myelination at least in part through epigenetic silencing of the promyelination program. Strikingly, we found that HDAC3 and p300 co-occupy the regulatory elements of genes associated with myelination inhibition, including Notch, Id2, Id4, Pten, and Dlg1. Coordination between HDAC3 and p300, which have similar temporal expression patterns (Supplementary Fig. 23), exerts net activating effects to activate a myelination-inhibitory network. Thus, HDAC3 may act as a dual-function switch via exerting an inhibitory effect through the epigenetic silencing of genes encoding proteins with promyelinating functions and through the recruitment of p300 to activate genes with products that inhibit SC myelination. The genome-wide HDAC3–p300 targeting analyses revealed the epigenetic landscape that underlies regulation of myelin growth; these will be an instructive reference for identifying regulatory networks that block peripheral myelin regrowth post-injury.
The genomic occupancy analyses further identified an HDAC3–p300 target, TEAD4, as a previously unrecognized inhibitor of SC myelin growth. In contrast to TEAD1, which cooperates with YAP–TAZ to activate myelination programs14–16, TEAD4 can also inhibit expression of myelination-associated genes in SCs, indicating a function divergent from those of other TEAD family members in SC myelination. Overexpressing YAP with TEAD4 led to net stimulation of Egr2 promoter activity in vitro57, whereas TEAD4 overexpression alone in the absence of YAP inhibited Egr2 expression, suggesting that there are context-dependent TEAD4 functions in gene regulation. Nonetheless, deletion of Tead4 in mice led to elevation of Egr2 and hypermyelination, consistent with the notion that TEAD4 functions as a default repressor and that YAP promotes tissue growth by relieving TEAD-mediated repression58.
The present study uncovered a previously unrecognized HDAC3-dependent pathway and epigenetic silencing that impact SC myelin growth and regeneration, providing insight into the epigenetic mechanisms underlying myelinopathy-associated diseases and nerve injury. Why functional recovery after trauma is slow and compromised remains unclear. Although it is possible that there is an insufficient supply of NRG1 after peripheral nerve injury, we found that the number of HDAC3-expressing SCs increased substantially after nerve injury. Thus, HDAC3 likely counters the activity of promyelinating signals, such as PI3K–AKT–ERK signaling. Inactivation of HDAC3 increased myelin thickness of SCs after nerve damage, suggesting that HDAC3 may limit full restoration of normal myelin thickness following injury.
Although the excessive hypermyelination seen with sustained Hdac3 ablation would not be a desirable endpoint, the potent activity of the druggable enzyme HDAC3 in the regulation of myelin sheath growth nevertheless highlights a therapeutic potential for time- or dose-delimited and controlled HDAC3 attenuation, which might allow SC re-entry into active myelinogenesis, overcoming the remyelination block in individuals with demyelinating neuropathies or nerve damage. In the mouse models described here, acceleration of myelin repair by measured and limited treatment with HDAC3 inhibitors facilitated timely recovery of conduction velocity as well as sensory and motor functions. Importantly, the myelin morphology in uninjured nerves was unaltered by treatment with HDAC3 inhibitors, indicating that transient HDAC3 inhibition is not detrimental to these nerves. Treatment with pharmacological HDAC3 inhibitors activated expression of myelination-associated genes in human SC-derived cell lines (Supplementary Fig. 24), suggesting that HDAC3 inhibition will promote the myelination program in human SCs. Given that HDAC inhibitors are generally well tolerated and have been clinically approved for treating certain cancers59,60, pharmacological attenuation of HDAC3 activity and its regulatory circuits, including its downstream effector TEAD4, may represent a therapeutic avenue for enhancement of myelin repair in patients suffering from peripheral neuropathies and nerve trauma.
ONLINE METHODS
Mice
Mice homozygous for floxed alleles of Hdac3 (Hdac3fl/fl)31 and Tead4 (Tead4fl/fl)45 were crossed with mice carrying Dhh-Cre33 or Cnp-Cre32 to generate Hdac3- and Tead4-mutant mice, respectively. Mice with tamoxifen-inducible Hdac3 knockout were generated through crossing Hdac3fl/fl mice with Plp1-CreERT mice34, followed by tamoxifen injection. Mice of either sex were used in the study, and littermates were used as controls unless otherwise indicated. The mouse strains used in this study were generated and maintained on a mixed C57Bl/6; 129Sv background and were housed in a vivarium with a 12-h light–12-h dark cycle. All studies complied with all relevant animal use guidelines and ethical regulations. All animal use and study protocols were approved by the Institutional Animal Care and Use Committee at the Cincinnati Children’s Hospital Medical Center, Ohio, USA.
Rat primary Schwann cell culture
SCs from the sciatic nerves of newborn rats (1–2 d old) were isolated as described previously12. SCs were grown in DMEM with 10% FBS (Life Technologies), supplemented with 10 ng/ml neuregulin 1 (NRG1 type III; 396-HB-050, R&D Systems) and 5 μM forskolin (Sigma, F6886) plus -glutamine and penicillin–streptomycin; this is hereafter denoted as SC proliferation medium. Cells between passages two and five were used in all experiments. >95% SC purity was achieved, as assessed by positive SOX10 and S100β immunoreactivity. To initiate differentiation, SCs were cultured in differentiation medium consisting of DMEM supplemented with 0.5% FBS and 1 mM dibutyl cyclic AMP (Sigma, D0627) along with -glutamine and penicillin–streptomycin for 3 d. Human neurofibroma-derived SC lines SNF02.2 (ATCC, CRL-2885) and SNF96.2 (ATCC, CRL-2884) were propagated in DMEM supplemented with 10% FBS plus -glutamine and penicillin–streptomycin. All tissue culture containers and coverslips were coated with 50 μg/ml poly(-lysine) (Sigma, P7890) in PBS for at least 30 min at room temperature and then rinsed with distilled water.
Small-molecule compound screening
96-well plates were coated with poly(-lysine) (50 μg/ml) for 30 min at room temperature and then rinsed with distilled water and air dried. In each well, 5,000 primary SCs isolated from rat neonates were seeded and maintained in chemically defined medium consisting of DMEM with 10% FBS (Life Technologies) supplemented with 10 ng/ml NRG1 (R&D Systems, 396-HB-050) and 5 μM forskolin (Sigma, F6886) plus -glutamine and penicillin–streptomycin for 2 d before screening. Each compound in the Epigenetics compound library (Cayman, 11076) was diluted to a final concentration of 5 μM in medium consisting of DMEM supplemented with 0.5% FBS, -glutamine, and penicillin–streptomycin. SCs were treated with the diluted compounds for 8 h. These SCs were washed with PBS and then treated with the SingleShot Cell Lysis Kit (172-5080, Bio-Rad), following the manufacturer’s instructions. The cell lysates from each well were collected for qRT–PCR using the iTaq Universal SYBR Green One-Step Kit (Bio-Rad, 172-5150), following the manufacturer’s instructions. Each compound was tested in triplicate.
In vivo administration of HDAC3 inhibitors
The HDAC3 inhibitors RGFP966 (MedKoo, 510205; a selective HDAC3 inhibitor with an IC50 of 80 nM; half-life, ~1 h) and PDA106 (MedKoo, 406720; preference toward HDAC3 inhibition with a Ki of 14 nM; half-life, ~8 h) can pass through the blood–brain barrier and are benzamide-type HDAC3-selective inhibitors21,26,27. They were dissolved in DMSO (calculated to be 10% of the final volume) and diluted with 30% hydroxypropyl-β-cyclodextrin and 0.1 M acetate, pH 5.4. PDA106 and RGFP966 were administered at doses of 100 mg per kg body weight and 10 mg per kg body weight, respectively and separately, as previously described21,26. Pups were given subcutaneous injections of drugs daily over the first week and i.p. injections every 2 d during the second week for a total of 3 d. Sciatic nerves were then collected and analyzed at the time points indicated in Figure 1d–k. For treatment of adult mice, drugs were i.p. injected daily after sciatic nerve transection injury in the first week and every 2 d after the first week for 3 d; they were analyzed at corresponding time points in Figure 2. Administrations of the HDAC3 inhibitors were well tolerated, and no harmful side effects were observed in the mice throughout the entire period of experiments.
SC–DRG co-culture
DRG neurons were isolated from embryonic day (E) 16.5 rat spinal cords and plated as explants on collagen-coated coverslips. Cultures were maintained in serum-free neurobasal medium (NB medium; 2% B27 supplement, 2 mM -glutamine, 0.4% glucose, and 50 ng/ml 2.5 S nerve growth factor (NGF) (Harlan, 005017)). Non-neuronal cells were removed via feeding the cultures with NB medium containing 5-fluorodeoxyuridine and uridine. SCs were isolated from sciatic nerves taken at P2 and expanded in SC proliferation medium. SC–DRG co-cultures were established through seeding purified DRG neuron cultures with 100,000 SCs in culture medium (MEM, 10% FBS, 2 mM -glutamine, 0.4% glucose, and 50 ng/ml 2.5 S NGF). 3 d after SC plating, C medium was supplemented with 50 μg/ml ascorbic acid (Sigma, A0278) to initiate myelination. SC–DRG co-cultures were allowed to myelinate for 10 d, with fresh media provided every 2 d. To determine the extent of myelination in SC–DRG co-cultures, the total number of MBP+ segments was counted in micrographs from 10–12 random fields per coverslip.
Tamoxifen induction of floxed allele deletion
Tamoxifen (Sigma, T5648) was dissolved to a stock concentration of 20 mg/ml in a vehicle of ethanol and sunflower seed oil (1:9 vol/vol). For perinatal tamoxifen injections, tamoxifen stock was injected i.p. at 2 mg/100 μl to lactating mice, thus administering tamoxifen to neonatal pups, beginning at P0. Tamoxifen was injected i.p. into pups after P5. Control mice were treated identically with tamoxifen. Sciatic nerves of pups were analyzed at the dates indicated in Figure 1g,i by immunostaining and EM.
Sciatic nerve transection injury
Young or aging adult mice were under general anesthesia with injection of a mixture of ketamine (90 mg per kg body weight) and xylazine (10 mg per kg body weight). Right sciatic nerves were exposed and transected at midthigh level. Exposed left sciatic nerves were used as uncut controls. Mice were treated with HDAC3 inhibitors or tamoxifen to delete floxed Hdac3 alleles following injury. Nerves were collected at the time points indicated in Figures 2 and 4 following surgery and processed for immunohistochemistry or EM.
Assessment of sensory and motor recovery
After nerve transection injury, mice were tested for sensory recovery through pinprick assays. An austerlitz insect pin (size 000; Fine Science Tools, Inc.) was gently applied to the plantar surface of the paw without moving the paw or penetrating the skin. The most lateral part of the plantar surface of the hind paw, corresponding to the sensory field of the sciatic nerve, was divided into five areas, A–E. The pinprick was applied twice from the most lateral toe to the heel. A response was considered positive when the mouse briskly removed its paw, and the mouse was graded 1 for this area. Then, the next area was tested. Experimenters assigning scores were blinded to mouse genotype.
To assess motor recovery after nerve transection injury, the movement of toes was evaluated. The reappearance of the toe-spreading reflex results from reinnervation of the small muscles of the foot and was scored as follows: 0, no spreading; 1, intermediate spreading with all toes; and 2, full spreading. Full spreading was defined as spreading of the toes that was complete, wide, and sustained for at least 2 s. Mice were scored when a full response was observed on the side contralateral to injury. Mice were evaluated twice in each experimental session, with at least a 45-min interval between sessions. Experimenters assigning scores were blinded to mouse genotype. For rotarod analysis, mice were tested on a rotarod apparatus specifically designed for mouse usage (Med Associates) to evaluate motor function. Mice were trained for 3 d and tested three times with 1-h intervals between testing trials. An acceleration setting was used to test motor function: the initial speed was 4 r.p.m. and there was gradual acceleration to a maximum speed of 40 r.p.m. The mean latency to fall (in seconds) of three separate trials on the rotarod apparatus is reported. Experiments performing recordings were blinded to treatment groups and mouse genotypes.
Immunofluorescence staining
The sciatic nerves of mice at defined ages were dissected and fixed for 15 min in 4% PFA in 0.1 M sodium phosphate buffer (pH 7.4), embedded in optimal cutting temperature (OCT) compound, cryoprotected in 25% sucrose, and sectioned at 12 μm as longitudinal or cross-sections using a cryostat or at 30 μm using a vibratome. Tissue sections or cells were permeabilized and blocked in blocking buffer (0.3% Triton X-100 and 5% normal donkey serum in PBS) for 1 h at 25 °C, followed by incubation with primary antibodies overnight at 4 °C. Antibodies against the following proteins were used: HDAC3 (rabbit; Santa Cruz Biotechnology, sc-11417), MPZ (rabbit; Abcam, ab31851), MBP (goat; Santa Cruz Biotechnology, sc-13914 and mouse; BioLegend, 836501), NF-M (rabbit; Millipore, AB1987), SOX10 (goat; Santa Cruz Biotechnology, sc-17342), EGR2 (rabbit; Santa Cruz, sc-20690), Ki67 (rabbit; Thermo Scientific, RM-9106), SOX2 (goat; Santa Cruz Biotechnology, sc-17320), HDAC5 (mouse; Sigma, H4538), CD31 (rat; BD Pharmingen, 553370), and CASPR (mouse; NeuroMab, 75-001). Secondary antibodies conjugated to Cy2, Cy3, or Cy5 were from Jackson ImmunoResearch Laboratories (catalog numbers 705-165-147, 705-225-147, 711-225-152, 711-165-152, 711-175-152, 715-165-150, and 712-165-150). All images were acquired using a Nikon C2+ confocal microscope.
Electron microscopy and morphometric analysis
Mice were perfused with 4% PFA with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.2. Sciatic nerves were dissected and fixed in the same fixative solution overnight. Nerves were rinsed in PBS, postfixed in 1% OsO4 in PBS for 1 h, dehydrated in a graded ethanol series, infiltrated with propylene oxide, and embedded in Epon. Semithin sections were stained with toluidine blue, and thin sections were stained with lead citrate. The morphometric measurements were performed in toluidine blue–stained semithin sections. The number of myelinated axons per nerve was analyzed in ultrathin sections using a JEOL 1200 EXII electron microscope, as previously described12.
Transient transfections
For plasmid transfections, rat SCs were transfected with expression vectors using Lipofectamine 3000 (Life Technologies) per the manufacturer’s protocol and assayed by immunocytochemistry, western blotting, or qRT–PCR analysis. Differentiation was assessed through immunostaining for EGR2. Multiple images were taken from each coverslip to obtain representative images from all areas of the coverslip, and at least 400 cells per coverslip were counted using ImageJ (https://imagej.nih.gov/ij/).
For siRNA knockdown in SCs, we used Lipofectamine RNAiMAX (Life Technologies) per the manufacturer’s instructions; cells were then subjected to qRT–PCR. siHdac3: SASI_Rn01_00031908 and SASI_Rn01_00031910; siTead1: SASI_Rn02_00327154; siTead2: SASI_Rn02_00229556; siTead3: SASI_Rn02_ 00209091; siTead4: SASI_Rn02_00317135 and SASI_Rn02_00317137.
Western blotting
For western blotting, the perineurium and epineurium were removed from sciatic nerves before snap freezing and storage at −80 °C. Sciatic nerves and rat SCs were lysed in RIPA buffer containing protease and phosphatase inhibitors. Western blot analysis was performed as described previously42. GAPDH (Millipore, MAB374) was used as an input control. Antibodies against the following proteins were used: HDAC3 (rabbit; Santa Cruz Biotechnology, sc-11417), MPZ (rabbit; Abcam, ab31851), MBP (goat; Santa Cruz Biotechnology, sc-13914), AKT (rabbit; Cell Signaling Technology, 9272), p-AKT (rabbit; Cell Signaling Technology, 9271), ERK (rabbit; Cell Signaling Technology, 4695), p-ERK (rabbit; Cell Signaling Technology, 4730), p-PI3K (rabbit; Cell Signaling Technology, 4228), PI3K (rabbit; Millipore, ABS233), TEAD4 (rabbit; Santa Cruz Biotechnology, sc-101184), Flag (rabbit; Cell Signaling Technology, 2368), PTEN (rabbit; Cell Signaling Technology, 9559), and acetylated PTEN (rabbit; Signalway Antibody, HW139). Corresponding secondary antibodies conjugated to HRP were from Jackson ImmunoResearch Laboratories (catalog numbers 111-035-144 and 111-035-003).
RNA isolation and real-time qPCR
RNA from purified SCs or sciatic nerves was extracted using TRIzol (Life Technologies). cDNA was synthesized from 1 μg RNA using iScript Reverse Transcription Supermix (Bio-Rad) according to the manufacturer’s instructions. Real-time qPCR was performed using the StepOnePlus Real-time PCR System (Applied Biosystems) with quantitative SYBR green PCR mix (Bio-Rad, 170-8880), as previously described12. PCR primer sequences are available upon request.
RNA sequencing and data analysis
RNA from the sciatic nerves of controls and Hdac3 mutants was extracted using TRIzol (Life Technologies), followed by purification using an RNeasy Mini Kit (Qiagen). RNA-seq libraries were prepared using the Illumina RNA-Seq Preparation Kit and sequenced by a HiSeq 2000 sequencer. RNA-seq reads were aligned to mm10 using TopHat with default settings (http://tophat.cbcb.umd.edu/). We used Cuff-diff to (1) estimate fragments per kilobase of transcript per million mapped reads (FPKM) values for known transcripts and to (2) analyze differentially expressed transcripts. In all differential expression tests, a difference was considered significant if the q value was less than 0.05 (Cuff-diff default). A heat map of gene expression was generated using the R language (version 3.2.1) and was generated on the basis of log2 (FPKM). Gene ontology analysis of gene expression changes was performed using Gene Set Enrichment Analysis (GSEA; http://www.broadinstitute.org/gsea/index.jsp). We used ToppCluster (https://toppcluster.cchmc.org/) to construct the network of genes belonging to over-represented GO-term categories.
ChIP–seq assays
ChIP assays were performed as described previously with minor modifications12. Briefly, purified rat SCs (~20 million cells) grown in the proliferation or differentiation (9 h in 1 mM cAMP-containing medium) condition were fixed for 10 min at room temperature with 1% formaldehyde–containing medium. Nuclei were isolated and sonicated in sonication buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA; 0.5 mM EGTA; and protease inhibitor cocktail). Sonicated chromatin (~300 μg) was used for immunoprecipitation via incubation with antibodies overnight at 4 °C. Prerinsed protein A/G PLUS-agarose beads (50 μl) were added to each ChIP reaction and incubated for 1 h at 4 °C. The beads were then incubated in 200 μl elution buffer at 65 °C for 20 min to elute immunoprecipitated materials. The ChIP–seq libraries were prepared using the NEBNext ChIP-seq Library Prep Master Mix Set for Illumina (NEB, E6240L) and then were run on the Illumina sequencer HiSeq 2000. Two ChIP–seq replicates were performed for diff_HDAC3, p300, and IgG, and one replicate was performed for Pro_HDAC3, siHdac3_ H3K27ac, Scr_H3K27ac, H3k27ac, and H3K4me1. Antibodies against the following proteins were used: HDAC3 (Santa Cruz Biotechnology, sc-11417), p300 (rabbit; Santa Cruz, sc-585), H3K27ac (rabbit; Abcam, ab4729), and H3K4me1 (rabbit; Abcam, ab8895).
ChIP–seq peak-calling and data analysis
All sequencing data were mapped to rat genome assembly rn5, and ChIP–seq peak calling was performed as previously described using model-based analysis of ChIP-seq (MACS, version 1.4.2; http://liulab.dfci.harvard.edu/MACS) with default parameters to get primary binding regions. To ensure that our data were of high quality and reproducibility, we called peaks with enrichment ≥10-fold over control (P ≤ 10−9) and compared the peak sets using the ENCODE overlap rules. The identified primary regions were further filtered using the following criteria, to define a more stringent protein–DNA interactome: (1) the P value cutoff was set to ≤10−9; and (2) we required an enrichment of sixfold and peak height >5.
The genome-wide distribution of protein-binding regions was determined by HOMER (http://homer.salk.edu/homer/index.html) in reference to Ensembl RGSC3.4.61 release. For all ChIP–seq data sets, WIG files were generated with MACS, which were subsequently visualized using Mochiview v1.46. HDAC3–p300 co-occupancy was defined as at least a 25% overlap of the HDAC3 peak by the p300 peak in the genome regions determined as co-occupied by HDAC3 and p300 through ChIP–seq, and occupancy was analyzed with Pearson’s correlation and ToppCluster (https://toppcluster.cchmc.org/). ChIP–seq heat maps were ordered by strength of binding. The heat maps were drawn using the Heatmap tools provided by Cistrome (http://cistrome.org/ap).
Compound muscle action potential recording
To analyze nerve conduction and motor unit function, single CMAPs were recorded in vivo from the lateral gastrocnemius muscles of injured or noninjured mice during electrical stimulation of the sciatic nerve under sodium pentobarbital anesthesia (50 mg per kg body weight, i.p.) as described previously12. CMAPs were amplified and obtained using a Micro 1401 data acquisition unit and were analyzed using Spike2 software (http://ced.co.uk/products/spkovin). Three successive electrical stimulations of the sciatic nerve at 2 mA (0.25–0.5 Hz, 0.1-ms duration) were initiated immediately proximal to the tibial, sural, and common peroneal branches via a stimulus isolator connected to the Micro 1401. Conduction velocity was calculated after determining the latency of CMAP onset relative to the stimulus artifact induced by electrical stimulation of the sciatic nerve, and the distance between the recording and stimulating electrodes was measured directly on the nerve. Peak CMAP amplitude and CMAP duration were calculated from each stimulation paradigm. The average of the stimulations of the sciatic nerve for each paradigm were obtained and then averaged across animals.
Statistical analyses
All analyses were done using GraphPad Prism 6.00 (San Diego, CA; http://www.graphpad.com/). Data are shown in dot plots or bar graphs as mean ± s.e.m. P < 0.05 is deemed statistically significant. Data distribution was assumed to be normal, but this was not formally tested. Statistical analysis was performed using two-tailed unpaired Student’s t-tests between two samples and one-way ANOVA with Tukey’s post hoc analysis for multiple comparisons or as indicated. Quantifications were performed from at least three experimental groups in a blinded fashion. The n value was defined as the number of experiments that were repeated independently with similar results. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those generally employed in the field. No randomization was used to collect all the data, but data were quantified with blinding. No animals or data points were excluded from analyses.
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data availability
All high-throughput data mentioned in the paper are publicly available from GEO under accession GSE93161. All data that support the findings of this study are available from the corresponding author upon reasonable request. Uncropped images of blots can be found in Supplementary Figures 25–27.
Supplementary Material
Supplementary Table 1. The loci of genes associcated with negative regulation targeted by HDAC 3 and p300 co-occupancy
Acknowledgments
We thank M. Wegner, J. Wells, and E. Hurlock for critical reading of the manuscript. We are grateful to E. Olson (University of Texas Southwestern Medical Center), D. Meijer (University of Edinburgh), and M. Wegner (Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU)) for Hdac3-floxed mice, Dhh-Cre mice, and antibodies against EGR2 and KROX20, respectively, and to N. Wu and L. Xu for technical support. This study was funded in part by the US National Institutes of Health (NIH; grant no. R37NS096359 and R01NS075243 to Q.R.L.; R35NS097303 to B.D.T.; and R01AR064551-01A1 to M.P.J.) and the National Multiple Sclerosis Society (grant no. NMSS-RG1507 to Q.R.L.).
Footnotes
AUTHOR CONTRIBUTIONS
X.H., L.Z., and Q.R.L. designed the experiments, analyzed the data, and wrote the manuscript with input from all authors. L.F.Q. and M.P.J. carried out CMAP analysis. X.H., L.Z., X.L., A.L., G.K., and X.D. performed the in vitro, in vivo, gene profiling, ChIP–seq, and in silico analyses. R.R.W., W.Z., S.-O.Y., J.B.R., M.X., K.-A.N., and B.D.T. provided resources and inputs. A.B. and K.N. provided floxed Tead4 and Cnp-Cre animals, respectively. Q.R.L. supervised the project.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Suter U, Scherer SS. Disease mechanisms in inherited neuropathies. Nat Rev Neurosci. 2003;4:714–726. doi: 10.1038/nrn1196. [DOI] [PubMed] [Google Scholar]
- 2.Scherer SS, Wrabetz L. Molecular mechanisms of inherited demyelinating neuropathies. Glia. 2008;56:1578–1589. doi: 10.1002/glia.20751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nave KA, Sereda MW, Ehrenreich H. Mechanisms of disease: inherited demyelinating neuropathies—from basic to clinical research. Nat Clin Pract Neurol. 2007;3:453–464. doi: 10.1038/ncpneuro0583. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Y, Notterpek L. Promoting peripheral myelin repair. Exp Neurol. 2016;283(Pt. B):573–580. doi: 10.1016/j.expneurol.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen ZL, Yu WM, Strickland S. Peripheral regeneration. Annu Rev Neurosci. 2007;30:209–233. doi: 10.1146/annurev.neuro.30.051606.094337. [DOI] [PubMed] [Google Scholar]
- 6.Nave KA, Werner HB. Myelination of the nervous system: mechanisms and functions. Annu Rev Cell Dev Biol. 2014;30:503–533. doi: 10.1146/annurev-cellbio-100913-013101. [DOI] [PubMed] [Google Scholar]
- 7.Salzer JL. Schwann cell myelination. Cold Spring Harb Perspect Biol. 2015;7:a020529. doi: 10.1101/cshperspect.a020529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monk KR, Feltri ML, Taveggia C. New insights on Schwann cell development. Glia. 2015;63:1376–1393. doi: 10.1002/glia.22852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feltri ML, Poitelon Y, Previtali SC. How Schwann cells sort axons: new concepts. Neuroscientist. 2016;22:252–265. doi: 10.1177/1073858415572361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stolt CC, Wegner M. Schwann cells and their transcriptional network: evolution of key regulators of peripheral myelination. Brain Res. 2016;1641:101–110. doi: 10.1016/j.brainres.2015.09.025. [DOI] [PubMed] [Google Scholar]
- 11.Jessen KR, Mirsky R, Lloyd AC. Schwann cells: development and role in nerve repair. Cold Spring Harb Perspect Biol. 2015;7:a020487. doi: 10.1101/cshperspect.a020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu LM, et al. Zeb2 recruits HDAC-NuRD to inhibit Notch and controls Schwann cell differentiation and remyelination. Nat Neurosci. 2016;19:1060–1072. doi: 10.1038/nn.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quintes S, et al. Zeb2 is essential for Schwann cell differentiation, myelination and nerve repair. Nat Neurosci. 2016;19:1050–1059. doi: 10.1038/nn.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poitelon Y, et al. YAP and TAZ control peripheral myelination and the expression of laminin receptors in Schwann cells. Nat Neurosci. 2016;19:879–887. doi: 10.1038/nn.4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng Y, et al. A reciprocal regulatory loop between TAZ/YAP and G-protein Gαs regulates Schwann cell proliferation and myelination. Nat Commun. 2017;8:15161. doi: 10.1038/ncomms15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grove M, et al. YAP/TAZ initiate and maintain Schwann cell myelination. eLife. 2017;6:e20982. doi: 10.7554/eLife.20982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arthur-Farraj PJ, et al. c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron. 2012;75:633–647. doi: 10.1016/j.neuron.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emery B, Lu QR. Transcriptional and epigenetic regulation of oligodendrocyte development and myelination in the central nervous system. Cold Spring Harb Perspect Biol. 2015;7:a020461. doi: 10.1101/cshperspect.a020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pereira JA, Lebrun-Julien F, Suter U. Molecular mechanisms regulating myelination in the peripheral nervous system. Trends Neurosci. 2012;35:123–134. doi: 10.1016/j.tins.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Ma KH, Svaren J. Epigenomic reprogramming in peripheral nerve injury. Neural Regen Res. 2016;11:1930–1931. doi: 10.4103/1673-5374.197133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malvaez M, et al. HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. Proc Natl Acad Sci USA. 2013;110:2647–2652. doi: 10.1073/pnas.1213364110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Y, Hsieh J. HDAC3 controls gap 2/mitosis progression in adult neural stem/progenitor cells by regulating CDK1 levels. Proc Natl Acad Sci USA. 2014;111:13541–13546. doi: 10.1073/pnas.1411939111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacob C, et al. HDAC1 and HDAC2 control the transcriptional program of myelination and the survival of Schwann cells. Nat Neurosci. 2011;14:429–436. doi: 10.1038/nn.2762. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, et al. HDAC-mediated deacetylation of NF-κB is critical for Schwann cell myelination. Nat Neurosci. 2011;14:437–441. doi: 10.1038/nn.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bei F, et al. Restoration of visual function by enhancing conduction in regenerated axons. Cell. 2016;164:219–232. doi: 10.1016/j.cell.2015.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rai M, et al. HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PLoS One. 2008;3:e1958. doi: 10.1371/journal.pone.0001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia H, et al. The effects of pharmacological inhibition of histone deacetylase 3 (HDAC3) in Huntington’s disease mice. PLoS One. 2016;11:e0152498. doi: 10.1371/journal.pone.0152498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parrinello S, et al. EphB signaling directs peripheral nerve regeneration through Sox2-dependent Schwann cell sorting. Cell. 2010;143:145–155. doi: 10.1016/j.cell.2010.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Painter MW, et al. Diminished Schwann cell repair responses underlie age-associated impaired axonal regeneration. Neuron. 2014;83:331–343. doi: 10.1016/j.neuron.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watkins TA, Emery B, Mulinyawe S, Barres BA. Distinct stages of myelination regulated by γ-secretase and astrocytes in a rapidly myelinating CNS coculture system. Neuron. 2008;60:555–569. doi: 10.1016/j.neuron.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montgomery RL, et al. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lappe-Siefke C, et al. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet. 2003;33:366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- 33.Jaegle M, et al. The POU proteins Brn-2 and Oct-6 share important functions in Schwann cell development. Genes Dev. 2003;17:1380–1391. doi: 10.1101/gad.258203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doerflinger NH, Macklin WB, Popko B. Inducible site-specific recombination in myelinating cells. Genesis. 2003;35:63–72. doi: 10.1002/gene.10154. [DOI] [PubMed] [Google Scholar]
- 35.Michailov GV, et al. Axonal neuregulin-1 regulates myelin sheath thickness. Science. 2004;304:700–703. doi: 10.1126/science.1095862. [DOI] [PubMed] [Google Scholar]
- 36.Sheean ME, et al. Activation of MAPK overrides the termination of myelin growth and replaces Nrg1/ErbB3 signals during Schwann cell development and myelination. Genes Dev. 2014;28:290–303. doi: 10.1101/gad.230045.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cotter L, et al. Dlg1–PTEN interaction regulates myelin thickness to prevent damaging peripheral nerve overmyelination. Science. 2010;328:1415–1418. doi: 10.1126/science.1187735. [DOI] [PubMed] [Google Scholar]
- 38.Norrmén C, Suter U. Akt/mTOR signalling in myelination. Biochem Soc Trans. 2013;41:944–950. doi: 10.1042/BST20130046. [DOI] [PubMed] [Google Scholar]
- 39.Okumura K, et al. PCAF modulates PTEN activity. J Biol Chem. 2006;281:26562–26568. doi: 10.1074/jbc.M605391200. [DOI] [PubMed] [Google Scholar]
- 40.Erjala K, et al. Signaling via ErbB2 and ErbB3 associates with resistance and epidermal growth factor receptor (EGFR) amplification with sensitivity to EGFR inhibitor gefitinib in head and neck squamous cell carcinoma cells. Clin Cancer Res. 2006;12:4103–4111. doi: 10.1158/1078-0432.CCR-05-2404. [DOI] [PubMed] [Google Scholar]
- 41.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L, et al. Hdac3 interaction with p300 histone acetyltransferase regulates the oligodendrocyte and astrocyte lineage fate switch. Dev Cell. 2016;36:316–330. doi: 10.1016/j.devcel.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yagi R, et al. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134:3827–3836. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- 46.Lundborg G. A 25-year perspective of peripheral nerve surgery: evolving neuroscientific concepts and clinical significance. J Hand Surg Am. 2000;25:391–414. doi: 10.1053/jhsu.2000.4165. [DOI] [PubMed] [Google Scholar]
- 47.Brosius Lutz A, Barres BA. Contrasting the glial response to axon injury in the central and peripheral nervous systems. Dev Cell. 2014;28:7–17. doi: 10.1016/j.devcel.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 48.Cattin AL, et al. Macrophage-induced blood vessels guide Schwann cell–mediated regeneration of peripheral nerves. Cell. 2015;162:1127–1139. doi: 10.1016/j.cell.2015.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang H, Lichtman JW. Motor axon regeneration and muscle reinnervation in young adult and aged animals. J Neurosci. 2013;33:19480–19491. doi: 10.1523/JNEUROSCI.4067-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki M. Peripheral neuropathy in the elderly. Handb Clin Neurol. 2013;115:803–813. doi: 10.1016/B978-0-444-52902-2.00046-1. [DOI] [PubMed] [Google Scholar]
- 51.Cho Y, Sloutsky R, Naegle KM, Cavalli V. Injury-induced HDAC5 nuclear export is essential for axon regeneration. Cell. 2013;155:894–908. doi: 10.1016/j.cell.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brügger V, et al. Delaying histone deacetylase response to injury accelerates conversion into repair Schwann cells and nerve regeneration. Nat Commun. 2017;8:14272. doi: 10.1038/ncomms14272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sadoul K, Boyault C, Pabion M, Khochbin S. Regulation of protein turnover by acetyltransferases and deacetylases. Biochimie. 2008;90:306–312. doi: 10.1016/j.biochi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 55.Noseda R, et al. DDIT4/REDD1/RTP801 is a novel negative regulator of Schwann cell myelination. J Neurosci. 2013;33:15295–15305. doi: 10.1523/JNEUROSCI.2408-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang LL, Liu ZY, Huang JH, Luo ZJ. Expression pattern of neuregulin-1 type III during the development of the peripheral nervous system. Neural Regen Res. 2015;10:65–70. doi: 10.4103/1673-5374.150708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fernando RN, et al. Optimal myelin elongation relies on YAP activation by axonal growth and inhibition by Crb3/Hippo pathway. Nat Commun. 2016;7:12186. doi: 10.1038/ncomms12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koontz LM, et al. The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev Cell. 2013;25:388–401. doi: 10.1016/j.devcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mottamal M, Zheng S, Huang TL, Wang G. Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents. Molecules. 2015;20:3898–3941. doi: 10.3390/molecules20033898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, Seto E. HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harb Perspect Med. 2016;6:a026831. doi: 10.1101/cshperspect.a026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. The loci of genes associcated with negative regulation targeted by HDAC 3 and p300 co-occupancy
Data Availability Statement
All high-throughput data mentioned in the paper are publicly available from GEO under accession GSE93161. All data that support the findings of this study are available from the corresponding author upon reasonable request. Uncropped images of blots can be found in Supplementary Figures 25–27.