Abstract
During neurogenesis, dynamic developmental cues, transcription factors and histone modifying enzymes regulate the gene expression programs by modulating the activity of neural-specific enhancers. How transient developmental signals coordinate transcription factor recruitment to enhancers and to which extent chromatin modifiers contribute to enhancer activity is starting to be uncovered. Here, we take advantage of neural stem cells as a model to unravel the mechanisms underlying neural enhancer activation in response to the TGFβ signaling. Genome-wide experiments demonstrate that the proneural factor ASCL1 assists SMAD3 in the binding to a subset of enhancers. Once located at the enhancers, SMAD3 recruits the histone demethylase JMJD3 and the remodeling factor CHD8, creating the appropriate chromatin landscape to allow enhancer transcription and posterior gene activation. Finally, to analyze the phenotypical traits owed to cis-regulatory regions, we use CRISPR–Cas9 technology to demonstrate that the TGFβ-responsive Neurog2 enhancer is essential for proper neuronal polarization.
INTRODUCTION
During central nervous system development, multipotent neuroepithelial precursor cells originate specialized neurons and glial cells (1,2). The identity of the cells generated along neurogenesis is determined by the transcriptional programs operating on each cell type. The different gene expression outputs are the result of the interplay between developmental cues and epigenetic factors that control the activity of specific neural promoters and enhancers. Enhancers are distal cis-regulatory elements essential to control gene expression programs in a spatial-temporal manner during development (3). To do that, they integrate extrinsic and intrinsic signaling cues by containing clusters of recognition motifs for either lineage-specific transcription factors (TFs) or effectors of developmental signaling pathways (4). Moreover, TFs are dependent on the recruitment of coactivator proteins in order to activate transcription in the chromatin context (5). Historically, enhancers have been difficult to investigate due to the lack of identification criteria; however, recent epigenomic approaches to identify enhancer sequences at a genome-wide level have facilitated their study. Chromatin and TF signatures have allowed not only the unbiased detection of enhancers but also the classification of enhancers into active, primed and poised (6–8).
Despite the massive identification of enhancers that has been carried out in neural progenitors (9–11), the field still lacks data on how signaling cascades govern transient enhancer activations that lead to neural-fate changes. To address this question we investigate the mechanisms by which neural enhancers become activated by transforming growth factor β (TGFβ) pathway. As vertebrate neurogenesis is an asynchronized series of events, the study of molecular mechanisms involved in enhancer activation is very difficult in the developing embryo. As an alternative, we have used neural stem cells (NSCs) extracted from cerebral cortices of mouse embryos from E12.5. These NSCs adherent cultures are a very reliable model to study neurogenesis in comparison to the heterogeneity of the embryo (12). In response to TGFβ, NSCs lose multipotency and commit to the neuronal lineage (13–15), although TGFβ alone is not sufficient to differentiate NSCs into mature neurons. Mechanistically, TGFβ transduces signals from the plasma membrane to the nucleus by interacting with the serine/threonine kinases-type I and type II receptors. TGFβ binding leads to phosphorylation and activation of the effectors SMAD2 and SMAD3, that next interact with SMAD4 to enter the nucleus and regulate gene expression (16,17). At the cellular level, TGFβ controls growth, differentiation, migration and adhesion, in a cell context-dependent manner (18). Thus, the biological output of TGFβ action depends on the subset of genes and/or enhancers that are regulated (19), and this relies on the particular combination of co-factors participating in each cellular context. Indeed, several chromatin modifier enzymes have been identified to be associated with activated SMAD proteins (histone acetyltransferases CBP/p300, P/CAF or the remodeling factor BRG1) (17,20,21). Particularly, the TGFβ effectors interact with the lysine demethylase (KDM) JMJD3 in embryonic stem cells (ESCs) (22,23) and in neural progenitors to facilitate neuronal differentiation induction (15).
Here, we show that TGFβ signaling pathway activates a specific set of SMAD-responsive enhancers involved in neuronal commitment. Using genome-wide data, we identify that SMAD3 binding to neural enhancers coincides with the proneural achaete-scute family bHLH transcription factor 1 (ASCL1). SMAD3 binding to enhancers is associated with an increase in eRNA transcription that in turns correlates with gene activation. We uncover that this process is dependent on the action of the SMAD3 cofactor JMJD3 and the previously unknown partner CHD8 (chromodomain helicase DNA-binding protein 8). Importantly, we unequivocally identify a TGFβ responsive enhancer that drives Neurogenin 2 (Neurog2) gene expression and proper neuronal differentiation of NSCs.
MATERIALS AND METHODS
Cell culture and differentiation
Mouse NSCs were dissected from cerebral cortices of C57BL/6J mouse fetal brains (E12.5) and cultured in poly-d-lysine (5 μg/ml, 2 h 37°C) and laminin (5 μg/ml 37°C, 4 h 37°C) precoated dishes following the previous published procedures (24). NSCs were grown with a medium prepared by mixing equal parts of DMEM F12 (without Phenol Red, Gibco) and Neural Basal Media (Gibco) containing Penicillin/Streptomycin, Glutamax (1%), N2 and B27 supplements (Gibco), non essential aminoacids (0.1 mM), sodium pyruvat (1 mM), HEPES (5 mM), Heparin (0.2 mg/l), bovine serum albumin (0.8 mg/l) and β-mercaptoethanol (0.01 mM) as previously described (15). Fresh recombinant human Epidermal Growth Factor (EGF) (R&D systems) and Fibroblast Growth Factor (FGF) (Invitrogen) to 20 and 10 ng/ml final concentrations respectively were added to the growing media. Medium, supplements, EGF and FGF form the so-called expansion medium. Under these conditions, NSCs maintain the ability to self-renew and to generate a wide range of differentiated neural cell types (24–26). For NSCs differentiation experiments Parental and ΔNeurog2 enh cells were plated in 24-well plates pre-coated with poly-d-lysine (5 μg/ml, 2 h 37°C) and laminin (5 μg/ml 37°C, 4 h 37°C) at a seeding density of 0.1 × 106 cells per well in NSCs expansion medium. After 24 h, expansion medium was replaced by differentiating medium, consisting in the same components of the expansion medium but without EGF and FGF (27–29). Fresh differentiating medium was supplied every 2 days and after 3, 6 or 8 days, cells were fixed and stained for indirect immunofluorescence. Under these conditions, NSCs differentiate toward neurons, astrocytes and oligodendrocytes (30). TGFβ (Millipore) was used at a final concentration of 5 ηg/ml. Human HEK293T cells were cultured in DMEM supplemented with 10% of fetal bovine serum (Gibco) and 1% of Penicillin/Streptomycin (31).
Antibodies and reagents
TGFβ was acquired from Millipore (GF111). Antibodies used were anti: H3K27me3 (Millipore, 07449), H3K4me1 (Millipore, 07436), H3K4me2 (Millipore, 07030), H3K27me2 (Cell signaling, 9728S), H3K27ac (Abcam, ab4729), H3K4me3 (Abcam, ab8580), SMAD3 (Abcam, ab28379), phospho-SMAD3 (Cell Signaling, mAb9520), SMAD2/3 (BD Bioscience, 610842), ASCL1 (BD Pharmingen, 556604), JMJD3 (raised in the laboratory using amino acids 798–1095), CHD8 was raised in Dr José Carlos Reyes laboratory (32), β-TUBULIN III (TUJ1, Covance, MMS-435P), GFAP (Dako, z0334), NESTIN (Abcam, ab5968), HuC/D (MP, A21271), DAPI (ThermoFisher, D1306), β-TUBULIN (Millipore, MAB3408), VINCULIN (Sigma, V9131), HA tag (Abcam, ab20084) and MYC tag (Abcam, ab9132).
Plasmids and recombinant proteins
Previously published specific lentiviral vectors were either purchased from Sigma or cloned in pLKO.1 puro vector using AgeI and EcoRI sites, brackets indicate target sequence: pLKO-random (CAACAAGATGAAGAGCACC), pLKO-mSMAD3 (CCTTACCACTATCAGAGAGTA), pLKO-mASCL1 (CCACGGTCTTTGCTTCTGTTT), pLKO-mJMJD3 (CCTCTGTTCTTGAGGGACAAA), and pLKO-mCHD8 (TGCCTGGAAGAAATTGGAG). pCIG-HA-ASCL1, pCIG, pCIG-FLAG-SMAD3-S/D and pCIG-FLAG-SMAD3-S/A were kindly provided by Dr. Elisa Martí (33). pCIG-MYC-JMJD3 was described in (34). Luciferase pGL3-promoter and renilla pRL-TK vector were purchased in Promega. Ctgf(-102), Nrip3(-3,5) and Neurog2(-6) enhancer regions were extracted by PCR from mouse genomic DNA and cloned into luciferase reporter pGL3-promoter by using MluI and BglII restriction sites. Primers sequences can be found in Supplementary Table S1. Empty backbone for CRISPR–Cas9 constructs was obtained from Addgene (#42230).
Chick in ovo electroporation
Eggs from White-Leghorn chickens were incubated at 38.5°C and 70% humidity. Embryos were staged following Hamburger and Hamilton (HH) (35). Chick embryos were electroporated with purified plasmid DNA at 1 μg/μl in H2O with 50 ηg/ml of Fast Green. Plasmid DNA was injected into the lumen of HH11–HH12 neural tubes, electrodes were placed at both sides of the neural tube and embryos were electroporated by an IntracelDual Pulse (TSS-100) electroporator delivering five 50 ms square pulses of 20–25 V.
In vivo luciferase assay
Enhancer activation by the TGFβ-pathway was assayed in chicken neural tubes. Chick embryos were electroporated at HH11–HH12 with pCIG-SMAD3-S/D, pCIG-SMAD3-S/A or empty pCIG, together with the luciferase reporter constructs and renilla for ovoelectroporation efficiency normalization. Embryos were harvested after 48 h incubation in ovo and Dual Luciferase Reporter Assay System (Promega) was utilized to lyse neural tubes and measure luciferase and renilla activities.
Lentiviral transduction
Lentiviral transduction was carried out as previously described (36). Extended protocol is provided in Supplementary Materials and Methods.
CoIP and ChIP assays
Coimmunoprecipitation (CoIP) experiments were performed as previously described (34). Chromatin immunoprecipitation (ChIP) assays were essentially performed as described (37,38) with modifications: 1 × 106 NSCs untreated or treated with TGFβ (5 ng/ml, for the indicated times) were fixed with formaldehyde 1% 10 min. Fixation was stopped by addition of 0.125 M glycine diluted in H2O. Cells were lysed in 1% SDS lysis buffer (1% SDS; 10 mM EDTA pH 8.0; 50 mM Tris–HCl pH 8.1). Sonication step was performed in a Bioruptor sonicator and shredded chromatin was used for each immunoprecipitation. ChIP DNA was analyzed by qPCR with SYBR Green (Roche) in a LightCycler 480 PCR system (Roche) using specific primers (see Supplementary Table S1). Percentage of input was used for the quantification of the immunoprecipitated material with respect to the total starting chromatin. See expanded protocol in Supplementary Materials and Methods.
RNA extraction and qPCR
TRIZOL reagent (Invitrogen) was used to extract RNA, following the manufacturer instructions. Reverse transcription was performed with 2 μg of RNA using High Capacity cDNA reverse transcription kit (Invitrogen) and qPCR was performed with SYBR Green (Roche) in a LightCycler 480 (Roche) using specific primer pairs (see Supplementary Table S1). Extended protocol is provided in Supplementary Materials and Methods.
Indirect immunofluorescence and cell counting
Cells were fixed for 20 min in 4% paraformaldehyde (diluted in phosphate buffer 0.1 M, pH 7.4) and permeabilized with PBS-Triton X-100 (0.1%) before blocking at room temperature for 1 h in 1% BSA (in PBS with 0.1% Triton X-100) before overnight incubation at 4°C with primary antibodies. Finally, cells were incubated for 2 h at room temperature with Alexa-conjugated secondary IgG antibodies (Jackson ImmunoResearch) and 0.1 ng/μl DAPI (ThermoFisher, D1306). Images were captured by Leica SP5 confocal microscope using LAS-AF software.
Number of neurites per cell and percentage of uni/bipolar or multipolar neurons were quantified by direct counting of 10 randomly selected fields considering as multipolar the neurons with more than three neurites. Data show mean of n = 60 cells. Measure of the average length of the longest neurite in Parental and ΔNeurog2 enh neurons was performed on representative fields using LAS AF Leica Microsystems Version: 1.8.2 build 1465 software.
Western blot
Immunoblotting was performed using standard procedures and visualized by means of an ECL kit (Amersham).
Size-exclusion chromatography
Size exclusion chromatography was performed with whole NSCs extracts in a Superose-6 10/300 gel filtration column (GE Healthcare) on AKTA purifier system (GE Healthcare). Proteins were detected by Western Blot.
ChIP-seq data acquisition and analysis
ChIP-seq data were downloaded from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) (Accessions used in this paper are specified in Supplementary Table S2). For all of the accessions excepting for ASCL1, SMAD3 and JMJD3, bed files were already available and were used for enhancer identification and Venn diagram construction. For enhancer identification we used -intersectbed command from BEDTools with a minimum of overlapping base pairs of 50 (39). ChIP-seq captions were obtained from IGV genome browser (40). Extended protocol is provided in Supplementary Materials and Methods.
CRISPR–Cas9
In order to delete Neurog2(-6) enhancer, primer pairs of gRNA (Supplementary Table S1) were designed flanking the mm9 coordinates chr3:127 326 051–127 334 232 using the online tool http://crispr.mit.edu/. Selected pair of primers has a score of 88 (left) and 87 (right) and specificity was assessed observing that the highest off-target score for each pair of primers was theoretically low (1.4 left, 1.3 right). gRNA were cloned in pX330-U6-Chimeric_BB-CBh-hSpCas9 vector using BbsI sites. Left and right-cutting plasmids were nucleofected in NSCs with an Amaxa Nucleofector (Lonza) following manufacturer instructions. After puromycin selection (2μg/ml) and detection analysis with conventional PCR, heterogeneous population carrying a majority of homozygotic deletions was used for experiments. Adequate CRISPR–Cas9 deletion was additionally assessed by Sanger sequencing and SnapGene viewer was used to generate sequence pictures.
Statistical analysis
Quantitative data were expressed as mean and standard deviation (SD) (for immunofluoresce countings and RNA transcription experiments) and as mean and standard error of the mean (SEM) (for ChIPs). At least two or three biologically independent experiments were performed. The significance of differences between groups was assessed using the Student's t-test (*P < 0.05; **P < 0.01; ***P < 0.001).
MIQE guidelines
This manuscript provides all the information recommended by MIQE Guidelines (Supplementary Dataset 1). Details on MIQE Guidelines for qPCR experiments are mainly provided in the RNA isolation and qPCR methods section and figure legends. MIQE check-list and primer list has been included as Supplementary Dataset 1 and Supplementary Table S1 respectively. RF is the lab member who performed all the qPCR assays.
RESULTS
Epigenetic identification of neural enhancers
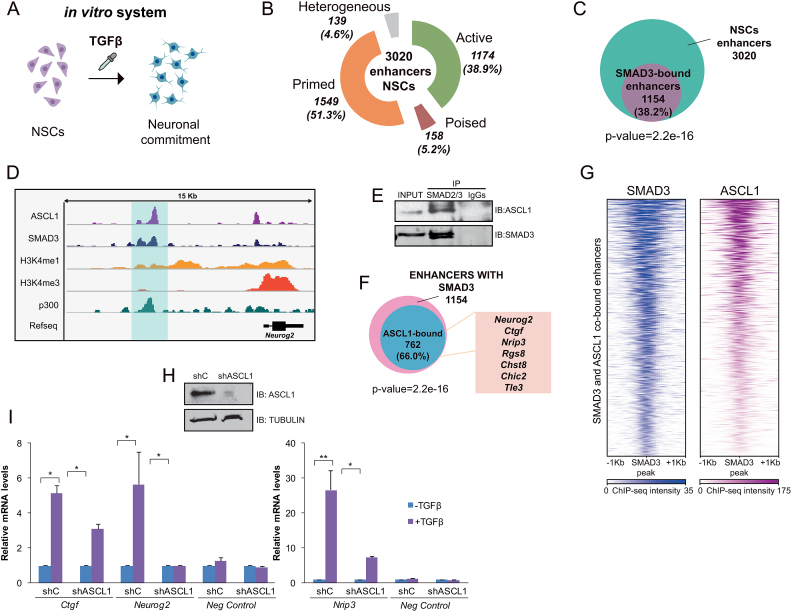
To evaluate the functional relevance of enhancers during neural differentiation, we utilized NSCs from cortices of mouse embryos (E12.5) to create neural progenitor cells as a model (Figure 1A). Using previously published epigenomic data and following the well-established enhancer identification criteria (8) we analyzed the enhancer landscape in NSCs. For that purpose, we combined ChIP-seq of p300, H3K4me1, H3K4me3, H3K27ac and H3K27me3 generated in cortex progenitors (11) to classify the enhancers in active enhancers: presence of p300, H3K4me1 and H3K27ac; poised enhancers: binding of p300 and presence of H3K4me1 and H3K27me3 modifications; primed enhancers: marked with H3K4me1 and p300 and heterogeneous enhancers: presence of p300, H3K4me1 and both H3K27ac and H3K27me3. We identified 3020 putative enhancers in NSCs (defined as presence of p300, H3K4me1 and low levels of H3K4me3) (Figure 1B and D, and Supplementary Dataset 2). Among these, we found 1174 (38.9%) active enhancers, 158 (5.2%) poised enhancers, 1549 (51.3%) primed enhancers and 139 (4.6%) heterogeneous enhancers (Figure 1B). The lower number of poised enhancers in NSCs compared to ESCs is in agreement with previous reports indicating that the poised enhancers seem to be especially abundant in pluripotent cells (41). Nevertheless, in order to understand how relevant the poised enhancers are at this stage we analyzed how many of them become activated in terminally differentiated neurons. By comparing previously published datasets of active enhancers in neurons from E16.5 embryos (42), we found that 74 out of the 158 poised enhancers in progenitors (46.8%) became active after the stage-transition (Supplementary Figure S1A). Notably, some classical neuronal genes associated to these enhancers are included in this list (Camk2n1, Kcnd3, Tle3, Amigo1…). As H3K4me2 histone mark has also been associated to enhancers (43), we also cataloged the putative neural enhancers using this modification instead of H3K4me1 (together with p300 binding and low levels of H3K4me3). A similar set of data, although reduced in number and enriched in active enhancers was generated (Supplementary Figure S1B and Supplementary Dataset 2). Moreover, the number of mouse promoters co-localizing with the enhancers identified with H3K4me1/2 was very low (8 and 9, respectively), (Supplementary Figure S1C), validating the enhancer analysis.
Figure 1.
Epigenomic identification of neural enhancers. (A) Schematic view of the model used to study neural enhancer activation upon the TGFβ differentiation signal. (B) Number and type of identified enhancers using ChIP-seq data for p300, H3K4me1, H3K4me3, H3K27ac and H3K27me3. The percentage related to the total number of enhancers is displayed in the graphic. (C) Venn diagram showing the number of neural enhancers bound by SMAD3 in NSCs treated with TGFβ for 30 min. P-value is the result of an equal proportions test performed between SMAD3-bound enhancers and a random set. (D) IGV capture showing the chromatin landscape around the Neurog2 gene. Neurog2(-6) enhancer is highlighted in blue. Tracks display ChIP-seq in NSCs treated for 30 min with TGFβ (SMAD3) or untreated cells (ASCL1, p300, H3K4me1 and H3K4me3). (E) Endogenous SMAD2/3 was immunoprecipitated from NSCs and the presence of SMAD3 and ASCL1 in the immunopellet was determined by immunoblot with the antibodies indicated on the right part of the figure. Figure is representative of at least two biological independent experiments. (F) Venn diagram showing the number of neural SMAD3-bound enhancers upon TGFβ treatment for 30 min that contain ASCL1 TF in untreated NSCs. The names of some genes putatively regulated by these enhancers are indicated. (G) Heatmap representation of SMAD3 (left) and ASCL1 (right) binding on the SMAD3 and ASCL1 co-occupied enhancers. Scales indicate ChIP-seq intensities. (H) NSCs were infected with lentivirus expressing shRNA control (shC) or shRNA specific for ASCL1 (shASCL1) cloned into pLKO vector. Forty eight hours later, total protein extracts were prepared and the ASCL1 and TUBULIN levels were determined by immunoblot. (I) shC and shASCL1 cells were treated with TGFβ for 6 h (3 h for Ctgf) and mRNA levels of TGFβ-responsive genes associated to the enhancers bound by SMAD3 and ASCL1 were analyzed by qPCR. Data were normalized to Rps23 housekeeping gene and figure shows values relative to shC samples. Ccne3 gene, a non TGFβ-responsive gene was used as a control. Results are the mean of three biological independent experiments. Error bars indicate SD. *P < 0.05; **P < 0.01 (Student's t-test).
Once identified the putative enhancers in NSCs, we sought to understand the mechanism underlying the dynamic activation of the enhancers in response to a developmental signal involved in neuronal fate. To do that, we studied the response of the enhancers to the well-characterized TGFβ pathway. This signaling cascade is already active under basal conditions to allow progenitor proliferation (14,15). Further TGFβ stimulation leads to the full gene activation that drives neuronal commitment in vitro and in vivo (14,15,44,45) (Figure 1A) (Supplementary Figure S1D and Supplementary Dataset 3). To decipher whether TGFβ pathway is directly implicated in NSCs enhancer activation, we analyzed the SMAD3 genomic distribution in NSCs upon TGFβ signaling with our previously published SMAD3 ChIP-seq (15). After combining the data, we identified 1154 (38.2%) putative NSCs enhancers that were bound by SMAD3 (P-value 2.2e–16, equal proportions test against a random sample) (Figure 1C), reinforcing the potential role of TGFβ-signaling in NSCs.
ASCL1 interacts with SMAD3 at NSCs enhancers
It has been described that SMAD genome-binding pattern shares many targets with cell-type specific TFs important for cell identity (46,47). We then, questioned about the neural specific factor/s that could cooperate with SMAD3 to target neural enhancers. Basic helix-loop-helix (bHLH) proteins have largely been demonstrated to be crucial players in chromatin regulation (48). Concretely, the bHLH protein ASCL1 has been shown to exert a critical role regulating neural gene expression by binding and opening the chromatin structure at enhancers (49). Thus, we tested whether ASCL1 and SMAD3 physically interact by CoIP assays. Figure 1E shows that endogenous ASCL1 and SMAD3 interact together. Next, we analyzed the colocalization of SMAD3 and ASCL1 at the genome wide level using previously published ChIP-seq data from NSCs (15,49). Doing that, we identified 762 (66%) SMAD3-bound enhancers that also contained ASCL1 (P-value 2.2e-16, equal proportion test against a random sample), (Figure 1F and G). Following these findings we decided to test whether TGFβ and ASCL1 had any functional association. To do that, ASCL1 protein levels were transiently depleted by transduction of lentivirus containing specific ASCL1 shRNA into the NSCs (Figure 1H and Supplementary Figure S1E). The transient removal of ASCL1 did not affect SMAD3 levels, proliferation rate or differentiation status of the NSCs (Supplementary Figure S1E, F and G). After viral transduction, the expression of some TGFβ-responsive genes associated to enhancers bound by ASCL1 and SMAD3 (Figure 1F) was tested by qPCR in the shASCL1 and control cell lines. We chose Ctgf, Nrip3 and Neurog2 genes that cover the spectrum of transcriptional levels in our previously published microarray data (15,50). Ccne3 gene, which does not respond to TGFβ, was used as a negative control. Results in Figure 1I demonstrate that ASCL1 is essential to fully activate TGFβ-targets in NSCs. In ASCL1 depleted cells, the response to TGFβ of Neurog2 was totally abolished and Nrip3 and Ctgf induction was severely decreased. All things considered, these data point to the proneural factor ASCL1 as a new partner of SMAD3 in forebrain enhancer recognition.
Neural enhancers are dynamically activated by TGFβ
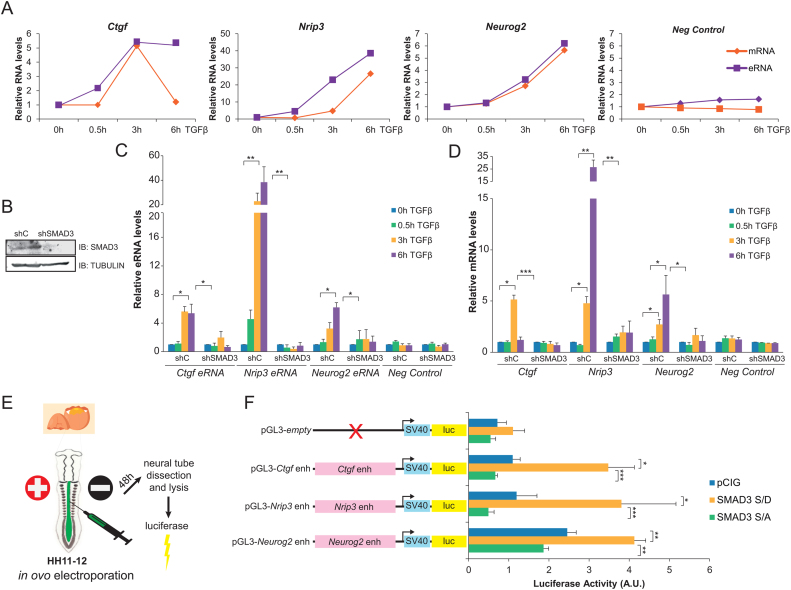
Once identified the SMAD3-ASCL1 bound neural enhancers, we investigated whether they became fully active upon TGFβ signaling. To this end, we evaluated the transcription from the enhancers, measuring the enhancer RNAs (eRNAs) which serve as readout of enhancer activation (51). We chose for posterior analysis the Nrip3(–3.5), Ctgf(–102) and Neurog2(–6) enhancers. These enhancers are associated to the previously analyzed genes (Figure 1I) and represent the two major functional enhancer categories: active (Nrip3 and Ctgf) and poised (Neurog2). In NSCs, eRNAs were detected as early as 30 min after TGFβ stimulation (Figure 2A). Interestingly, we observed a strong correlation on both the magnitude of the transcription and the stability among the eRNAs and their corresponding mRNAs (Figure 2A). To further confirm that enhancer activation was dependent on TGFβ pathway and thus, of SMAD3 binding, NSCs depleted of SMAD3 were generated (Figure 2B) and eRNA synthesis was tested. In concordance with the aforementioned results, eRNAs were hardly induced in shSMAD3 cells compared to the control cell line (Figure 2C). Similarly, mRNA of the associated genes was not produced upon TGFβ addition in the shSMAD3 NSCs (Figure 2D).
Figure 2.
SMAD3-bound enhancers respond to TGFβ. (A) NSCs cells were treated with TGFβ. eRNA from the indicated enhancers was determined by qPCR and the mRNA of the associated genes was analyzed and represented together with the eRNA levels. Transcription values were normalized to the housekeeping gene Rps23 and figure shows values relative to shC samples. Progesterone-responsive Fabp4 gene and eRNA were used as negative controls. Results are the mean of three biological independent experiments. Errors bars represent SD. *P < 0.05; **P < 0.01 (Student's t-test). (B) NSCs were infected with lentivirus expressing shRNA control (shC) or shRNA specific for SMAD3 (shSMAD3). Forty eight hours later, total protein extracts were prepared and the SMAD3 and TUBULIN levels were determined by immunoblot. (C and D) Control (shC) or SMAD3 depleted (shSMAD3) NSCs cells were treated for the indicated time with TGFβ. Then, total RNA was prepared and the levels of the indicated eRNA (C) or mRNA (D) were determined by qPCR. Transcription values were normalized to the housekeeping gene Rps23 and figure shows values relative to time 0h. Progesterone-responsive Fabp4 eRNA or mRNA were used as negative controls. Results are the mean of three biological independent experiments. Errors bars represent SD. *P < 0.05; **P < 0.01. (E) Schematic representation of the in vivo ovoelectroporation and luciferase experiment. (F) HH11–12 embryos were electroporated in ovo with pGL3-empty or the pGL3-enhancer fusions together with pCIG (empty vector), SMAD3-S/D (pseudo-phosphorylated mutant) or SMAD3-S/A (mutant that cannot be phosphorylated). 48h-PE neural tubes were dissected, tissue was disaggregated and the luciferase activity was measured using the Promega dual kit. Data represent ratios between luciferase and renilla in arbitrary units. Values are the mean of three experiments from four to six embryos. Error bars indicate SD. *P < 0.05; **P < 0.01; ***P < 0.001 (Student's t-test).
We also analyzed the levels of H3K27ac and H3K4me2 chromatin marks by ChIP assays. Data in Supplementary Figure S2A and B shows that the analyzed enhancers display H3K27ac and H3K4me2. After TGFβ addition, a time-dependent increase on the H3K27ac and H3K4me2 levels is observed at Ctgf(–102) and Nrip3(–3.5) enhancers but not at the intergenic region used as a negative control. No changes on H3K4me1 and H3K4me3 levels were detected (Supplementary Figure S2C and D), yet a clear increase on H3K4me3 was observed at the promoter of the analyzed genes (Supplementary Figure S2E) correlating with full activation in response to TGFβ (Figure 2D). Altogether, these data demonstrate that TGFβ signaling pathway stimulation results in enhancer activation and subsequent transcription of associated genes in vitro.
The above described findings support the idea of TGFβ pathway activating relevant enhancers for neural development. Therefore, we tested whether SMAD3 regulates the analyzed enhancers in an in vivo model of neurogenesis, the chick embryo neural tube (Figure 2E). This model was chosen because it has been previously shown that activation of TGFβ pathway by overexpression of a constitutively active pseudo-phosphorylated form of SMAD3 (SMAD3-S/D) promotes neuronal differentiation (15,33). With this goal, Ctgf(–102), Nrip3(–3.5) and Neurog2(–6) enhancers were cloned into the pGL3 promoter vector that contains the SV40 promoter fused to the luciferase reporter gene (Figure 2F). Then, chick embryos were electroporated in ovo (EP) at HH11–12 stage with the luciferase reporter constructs together with either the empty vector (pCIG) or SMAD3-S/D expressing plasmid (Figure 2E). Figure 2F shows that the expression of the luciferase downstream the enhancers was increased by co-EP of the SMAD3-S/D constitutively active mutant, while the expression of the luciferase with the promoter alone (pGL3-empty) was not affected. On the contrary, co-EP of a SMAD3 mutant that cannot be phosphorylated (SMAD3-S/A) and acts as a dominant negative form of SMAD3 (33), blocked the TGFβ induction of the analyzed enhancers. The results demonstrate that the investigated enhancers are activated in vivo in response to TGFβ signaling pathway.
JMJD3 is recruited to neural enhancers in a TGFβ-dependent manner
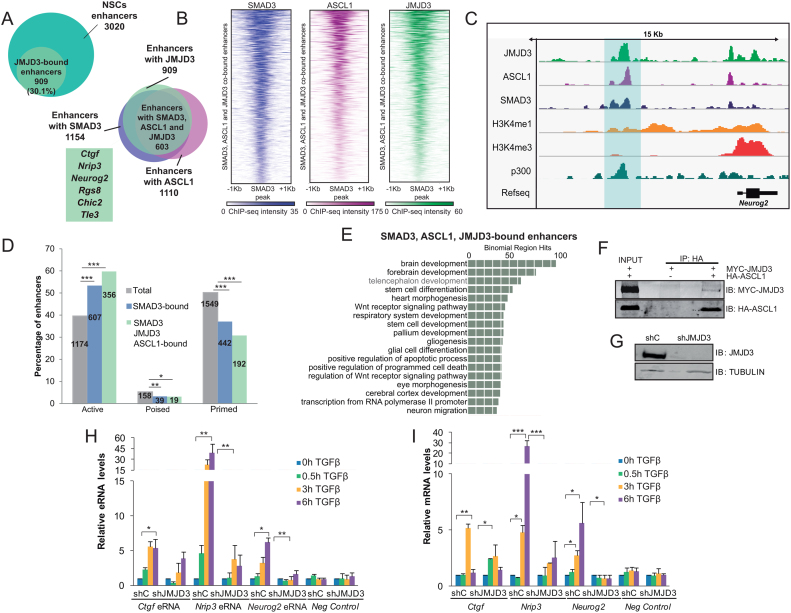
Next, we sought to identify the molecular machinery that collaborates with ASCL1/SMAD3 for full enhancer activation after TGFβ signaling. Previous work in our laboratory has demonstrated that JMJD3 interacts and cooperates with SMAD3 at promoters to induce the TGFβ-neurogenic program in vitro and in vivo (15,50). Thus, we wondered whether JMJD3 also affects TGFβ transcriptional response by contributing to enhancer activation. To address this hypothesis, we first identified the JMJD3 bound enhancers upon TGFβ using our reported ChIP-seq data (15). Results in Figure 3A and C shows that JMJD3 was found in 30.1% of total forebrain enhancers. Interestingly, among these, 66.3% contained also ASCL1 and SMAD3 (20% of total enhancers), showing a strong colocalization of the three proteins at enhancers (Figure 3A–C). SMAD3 and JMJD3 were found at the three types of enhancers (active, primed and poised). However, there is an increase in the percentage of active enhancers when the three partners, ASCL1/SMAD3/JMJD3, are occupying the regions (P-value 1.1e–15, equal proportions test between the ASCL1/SMAD3/JMJD3-bound enhancers and the total enhancers), suggesting that ASCL1 and JMJD3 contribute to enhancer activation in response to TGFβ (Figure 3D). Moreover, GREAT analysis (52) of the ASCL1/SMAD3/JMJD3-bound enhancers returned categories involved in neurogenesis (Figure 3E). The fact that JMJD3 co-occupies enhancers together with ASCL1 prompted us to test whether JMJD3 interacts with ASCL1 in NSCs. CoIP experiments showed a clear interaction between these proteins (Figure 3F), supporting the idea that JMJD3, SMAD3 and ASCL1 form a functional complex at neural enhancers.
Figure 3.
JMJD3 interacts with SMAD3 and ASCL1 at neural enhancers. (A) Venn diagrams showing the number and percentage of JMJD3-bound enhancers after 0.5 h of TGFβ treatment in NSCs (upper panel) and number of enhancers with overlapping ASCL1 (before TGFβ), SMAD3 and JMJD3 peaks (upon 30 min of TGFβ addition) in NSCs. Genes associated to the identified enhancers are indicated. (B) Heatmap showing JMJD3 binding to neural enhancers co-bound by ASCL1, SMAD3 and JMJD3. Scales depict ChIP-seq intensities. (C) IGV capture showing the chromatin landscape around the Neurog2 gene. Enhancer is highlighted in blue. Tracks display ChIP-seq in NSCs treated for 30 min with TGFβ (SMAD3 and JMJD3) or untreated cells (ASCL1, p300, H3K4me1 and H3K4me3). (D) The percentage of active, poised and primed total enhancers, and those bound by SMAD3 or SMAD3/ASCL1/JMJD3 are depicted. Numbers inside the bars indicate absolute number of enhancers. An equal proportions test was performed and asterisks show P-values: *P < 0.05; **P < 0.01; ***P < 0.001. (E) GREAT analysis showing GO Biological Process of the enhancers co-occupied by ASCL1/SMAD3/JMJD3, analysis was performed using as a background the whole Mus musculus genome. (F) 293T cells were transfected with HA-ASCL1 and MYC-JMJD3 as indicated. ASCL was precipitated using the HA tag antibody and the presence of JMJD3 and ASCL1 in the immunopellet was determined by immunoblot with MYC and HA antibodies respectively. Figure is representative of at least three biological independent experiments. (G) NSCs were infected with lentivirus expressing shRNA control (shC) or shRNA specific for JMJD3 (shJMJD3). Forty eight hours later, total protein extracts were prepared and the JMJD3 and TUBULIN levels were determined by immunoblot. (H and I) shC or shJMJD3 NSCs were treated for the indicated times with TGFβ. Levels of the indicated eRNA (J) or mRNA (K) were determined by qPCR. Transcription values were normalized to the housekeeping gene Rps23 and figure shows values relative to time 0 h. Progesterone-responsive Fabp4 eRNA or mRNA were used as negative controls. Results are the mean of three biological independent experiments. Errors bars represent SD. *P < 0.05; **P < 0.01 (Student's t-test).
JMJD3 contributes to full enhancer activation in response to TGFβ
Once identified the neural enhancers bound by ASCL1, SMAD3 and JMJD3 we evaluated the contribution of this KDM to full neural enhancer activation upon TGFβ signaling. To do that, we efficiently depleted JMJD3 from NSCs using lentivirus expressing specific shRNAs (Figure 3G) and the activity of Ctgf(–102), Nrip3(–3.5) and Neurog2(–6) enhancers in response to TGFβ was analyzed. Figure 3H shows that the synthesis of eRNAs was profoundly diminished in shJMJD3 NSCs upon TGFβ stimulation, correlating with the lack of induction of the associated genes (Figure 3I). Notably, transcriptional level of Utx, another member of the JMJD3 KDM family, was unchanged in the tested conditions (Supplementary Figure S3A). Within the chromatin context, TGFβ treatment was unable to increase neither H3K27ac nor H3K4me2 levels in the Ctgf(–102) enhancer and H3K27ac in the Nrip3(–3.5) enhancer in JMJD3 depleted cells (Supplementary Figure S3B and C). These data strongly suggest that JMJD3 is required for full enhancer activation upon TGFβ stimulation.
As JMJD3 is a KDM responsible for H3K27me2/3 demethylation (53,54), we wondered whether JMJD3-bound enhancers were demethylated during TGFβ induced activation. To answer this question, we selected three H3K27me2/3-marked poised enhancers from genes that are activated upon TGFβ-stimulation (Supplementary Figure S4F) (15): Neurog2(–6), Chic2(–26), Tle3(–114) and Ctgf(–102) as an active enhancer devoid of H3K27me2/3 marks. Then, we determined the levels of H3K27me2/3 in control and JMJD3 depleted NSCs before and upon TGFβ stimulation by ChIP-qPCR. Our results did not show any significant change on the H3K27me3 levels associated to Neurog2(–6) enhancer. Only an increase of H3K27me3 was observed in JMJD3 depleted cell line upon TGFβ addition (Supplementary Figure S4A and B). In the case of Chic2(–26) and Tle3(–114) H3K27me3 decreased upon stimulation; however, these changes were independent of JMJD3 (Supplementary Figure S4A and B). The active enhancer Ctgf(-102) did not suffered any methylation change in any of the conditions tested (Supplementary Figure S4A and B). Interestingly, the promoters associated to these poised enhancers were efficiently demethylated upon TGFβ (Supplementary Figure S4C); accordingly, levels of the PRC2 complex subunit EZH2 decreased and the H3K4me3 levels resulted incremented (Supplementary Figure S4D and E). Correspondingly, at the mRNA level, treatment with TGFβ boosted gene activation (Supplementary Figure S4F). These results suggest that H3K27 methylated enhancers exhibit different behaviour upon TGFβ-stimulation and that JMJD3 main role at enhancers is neither the demethylation of H3K27me3 nor the anti-H3K27me2 accumulation effect, suggesting a co-existence of catalytic-dependent and independent roles.
CHD8, a novel SMAD/JMJD3 partner
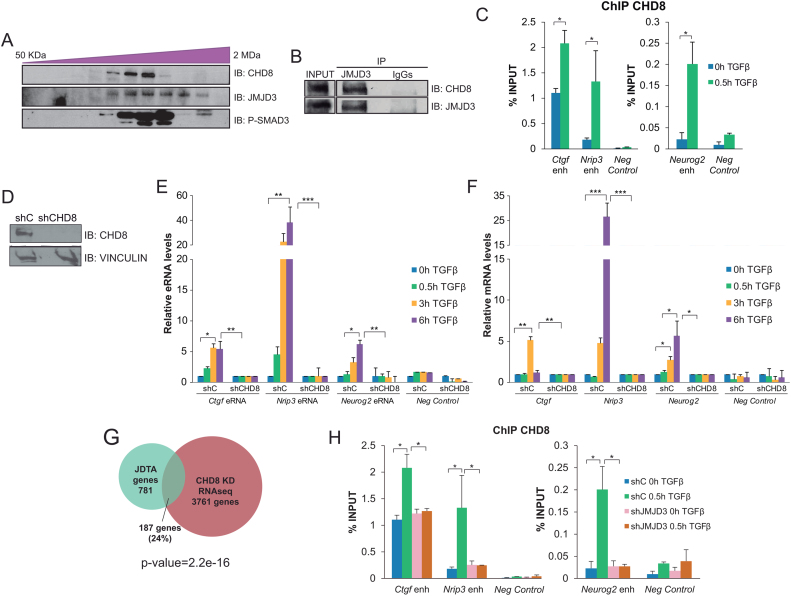
CHD8 belongs to a wide family of ATP-dependent remodelers that bind and open chromatin at enhancer regions (55,56). Moreover, CHD8 is essential for early neurogenesis (57,58). Thus, we considered the possibility that CHD8 could contribute to full neural enhancer activation upon TGFβ signaling. To investigate this hypothesis, we first checked whether CHD8 could be physically contacting SMAD3 and JMJD3. For that purpose, we used size-exclusion chromatography on whole NSCs extracts and we observed that CHD8 co-eluted with the phosphorylated form of SMAD3. Consistently, JMJD3 co-eluted in the same fractions (Figure 4A). Additionally, by CoIP experiments we demonstrated that endogenous CHD8 interacted with JMJD3 (Figure 4B). Then, we investigated whether CHD8 recruitment to the neural enhancer was dependent on TGFβ. Using ChIP-qPCR assays we observed that CHD8 was already bound to these active enhancers (due to the basal activity of TGFβ pathway) and TGFβ stimulation significantly increased at the analyzed enhancers upon TGFβ-stimulation (Figure 4C). Next, we tested whether CHD8 was required for the full enhancer activation induced by TGFβ. Depletion of CHD8 protein levels using specific shRNAs (Figure 4D) blocked the enhancer activity (Figure 4E), and the full activation of the associated genes (Figure 4F). Additionally, genome-wide analysis showed that 24% of the genes regulated by TGFβ that depend on JMJD3 (JDTA) (15) were also regulated by CHD8 (P-value 2.2e–16, equal proportions test against a random sample of genes) (59). (Figure 4G). Then, we speculate that JMJD3 could be contributing to CHD8 high-affinity binding. To test this hypothesis we investigated the role of JMJD3 in CHD8 binding to neural enhancers upon TGFβ. To do that, we analyzed the chromatin association of CHD8 in JMJD3 depleted cells. The results showed a clear decrease on CHD8 recruitment to the analyzed enhancers upon TGFβ stimulation in shJMJD3 cells (Figure 4H). Altogether, these data demonstrate that CHD8 is a new SMAD3/JMJD3 partner at neural enhancers that is essential to full enhancer activation upon TGFβ signaling.
Figure 4.
CHD8 facilitates enhancer activation. (A) Size-exclusion chromatography immunoblots depicting the co-elution of CHD8, JMJD3 and phospho-SMAD3. (B) Endogenous JMJD3 was precipitated from NSCs using JMJD3 antibody and the presence of CHD8 in the immunopellet was determined by immunoblot with the antibodies indicated on the right part of the figure. IgGs were used as negative control. Figure is representative of at least three biological independent experiments. (C) ChIP of CHD8 in NSCs treated for 0 and 0.5 h with TGFβ and analyzed by qPCR at the indicated enhancers. An intergenic region devoid of histone marks was used as negative control. Results are the mean of two biological independent experiments. Errors bars represent SEM. *P<0.05 (Student's t-test). (D) Immunoblot showing the CHD8 levels in NSCs infected with lentivirus expressing shRNA control (shC) or shRNA specific for CHD8 (shCHD8). Total protein extracts were prepared from NSCs 48 h after infection and CHD8 and VINCULIN levels were detected by immunoblot. (E and F) shC or shCHD8 NSCs cells were treated for the indicated time with TGFβ. Then, total RNA was purified and the levels of the indicated eRNA (E) and mRNA (F) were determined by qPCR. Expression values were normalized to the housekeeping gene Rps23, and figure shows values relative to time 0h. Progesterone-responsive Fabp4 eRNA or mRNA respectively were used as negative controls. Results are the mean of three biological independent experiments. Errors bars represent SD. *P < 0.05; **P < 0.01; ***P < 0.001 (Student's t-test). (G) Venn diagram showing the number and percentage of genes that are regulated by TGFβ pathway in a JMJD3 dependent manner (JDTA genes) in NSCs that are also regulated by CHD8 in the RNA-seq of cortical progenitors. (H) ChIP of CHD8 in control (shC) or JMJD3 depleted (shJMJD3) NSCs cells treated for 0 and 0.5 h with TGFβ and analyzed by qPCR at the indicated enhancers. An intergenic region was used as negative control. Results are the mean of two biological independent experiments. Errors bars represent SEM. *P < 0.05 (Student's t-test).
The TGFβ-responsive enhancer Neurog2(–6) is essential for proper neuronal differentiation
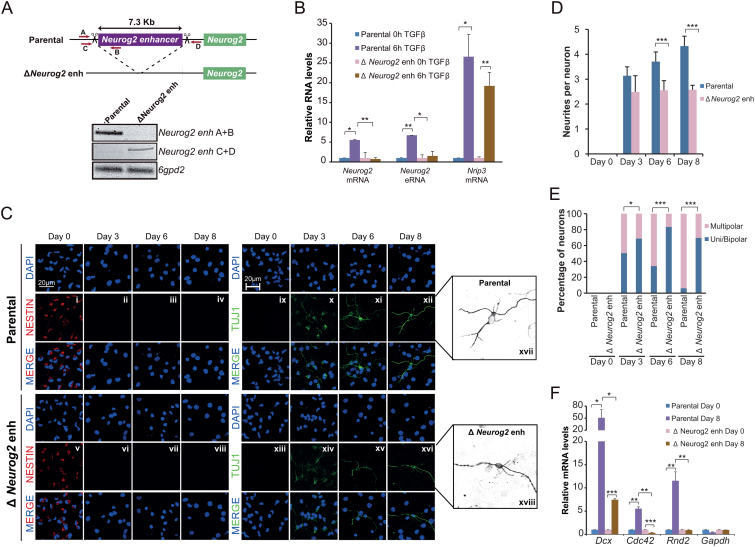
To demonstrate the physiological contribution of TGFβ-responsive enhancers to neuronal commitment, we used CRISPR–Cas9 technology to delete an enhancer associated to a relevant gene for neurogenesis, Neurog2(–6) (Figure 5A and Supplementary Figure S5A). This regulatory region has been functionally tested in vivo by transgenic mouse assays that were positive for enhancer activity in forebrain and midbrain (60). Importantly, Neurog2 is an essential bHLH TF that promotes neuronal differentiation, blocks glial differentiation and is essential for proper neuronal morphogenesis and migration (61–64). Thus, by using CRISPR–Cas9 technology we deleted Neurog2(-6) enhancer (ΔNeurog2 enh) as demonstrated by multiplex PCR (Figure 5A) and genomic sequencing (Supplementary Figure S5B). Next, the parental and the ΔNeurog2 enh NSCs were treated with TGFβ and the expression of Neurog2 was evaluated by RT-qPCR. Results show that Neurog2 gene remained silenced in non- treated parental and ΔNeurog2 enh NSCs (Figure 5B), suggesting that this poised enhancer is not responsible of maintaining Neurog2 gene in an inactive state before differentiation induction, and, hence, does not act as a silencer. After TGFβ addition, a clear Neurog2 activation was observed in the parental line but not in ΔNeurog2 enh cells (Figure 5B). Accordingly, synthesis of Neurog2 eRNA was not observed in ΔNeurog2 enh cell line (Figure 5B). These effects on Neurog2 gene induction were not due to a lack of TGFβ response, because other TGFβ targets, such as Nrip3, were clearly upregulated in these cells upon TGFβ treatment (Figure 5B). These data demonstrate that the Neurog2(-6) enhancer plays an essential and non-redundant role during the induction of its target gene upon TGFβ. To further investigate the developmental relevance of this enhancer, we evaluated the consequences of lacking Neurog2(–6) enhancer during neuronal differentiation and morphogenesis in vitro. To do that, we tested the differentiation capacity of parental and ΔNeurog2 enh NSCs by immunostaining using the neural progenitor marker NESTIN, (present in dividing cells and downregulated upon differentiation) and specific markers for neurons (TUBB3, known as TUJ1, and HuC/D) (Figure 5C and Supplementary Figure S5C). After three days in differentiating medium, both cell lines stopped proliferating (Figure 5Cii and vi and Supplementary Figure S5D) and the number of cells that expressed neuronal markers were similar in parental and ΔNeurog2 enh cells (Figure 5Cx and xiv and Supplementary Figure S5E). Nevertheless, ΔNeurog2 enh cells showed lower TUJ1 intensity than parental NSCs (Figure 5C, e.g. x versus xiv). Interestingly, ΔNeurog2 enh cells failed to properly differentiate into neurons (Figure 5C, e.g. xii versus xvi). After 6 days in differentiating medium, the number of neurites per cell was clearly reduced when compared with neurons from the parental cell line, going from 4–5 in control to 2–3 in ΔNeurog2 enh TUJ1+ cells (Figure 5C e.g. xvii versus xviii; Supplementary Figure S5C e.g. i versus ii, quantified in Figure 5D). In addition, the neurite length was also reduced (Figure 5C e.g. xvii versus xviii; quantified in Supplementary Figure S5F) and interestingly, in ΔNeurog2 enh pseudoneurons, a high number of cells presented only one, two (uni/bipolar neurons) or none neurites (Figure 5C e.g. xvii versus xviii; quantified in Figure 5E). These morphological alterations could be pointing to a misregulation of the transcription of cytoskeleton genes such as Rnd2, Cdc42 or Dcx. For that reason, we tested their expression in parental and ΔNeurog2 enh cell lines and we observed that the latter showed a clear misinduction of these genes upon differentiation (Figure 5F). These results are in agreement with previous reports indicating that NEUROG2 protein controls the expression of these cytoskeleton regulators (24,33,62).
Figure 5.
TGFβ-responsive enhancers are essential for neuronal differentiation. (A) Schematic representation of the CRISPR/Cas9 experimental approach used to delete Neurog2(–6) enhancer in NSCs. Two gRNAs flanking Neurog2(-6) enhancer region were used to create the deletion. Red arrows represent primers to test the deletion. PCR using Neurog2 deletion and 6gpd2 pairs of primers is shown in the top of the figure in parental and ΔNeurog2 enh NSC lines. (B) Parental and ΔNeurog2 enh cell lines were treated with TGFβ for 6 h. Total RNA was prepared and the expression levels of Neurog2 mRNA or eRNA were determined by qPCR. mRNA level of Nrip3 was used as a TGFβ response control. Transcription values were normalized to the housekeeping gene Rps23, and figure shows values relative to time 0 h. Errors bars represent SD. Results are representatives of two biological independent experiments. *P < 0.05; **P < 0.01 (Student's t-test). (C–E) Parental and ΔNeurog2 enh cell lines were cultured in differentiating medium. After 3, 6 or 8 days cells were fixed and stained with NESTIN, TUJ1 antibody and DAPI (C). Number of neurites per cell (D) and the percentage of uni/bipolar or multipolar pseudoneurons (E) were quantified by direct counting of 10 randomly selected fields. Data show mean of n = 60 cells. Error bars indicate SD. *P < 0.05; ***P < 0.001 (Student's t-test). (F) Parental and ΔNeurog2 enh cell lines were maintained in differentiating medium for 8 days. Total RNA was purified and the expression levels of the indicated genes were determined by qPCR. Transcription values were normalized to the housekeeping gene Ubc, and figure shows values relative to Day 0 samples. Errors bars represent SD. Results are representatives of two biological independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 (Student's t-test).
These results demonstrate that the Neurog2(–6) enhancer is sufficient to modulate the acquisition of phenotypical traits during neuronal commitment. Altogether, these data show that TGFβ-responsive enhancers play an essential role for the induction of a major regulator of neuronal differentiation of NSCs.
DISCUSSION
In this paper we provide a detailed molecular description of enhancer activation in response to the TGFβ signaling pathway during neurogenesis. Our data uncover an unforeseen interplay between the TFs SMAD3 and ASCL1 and the chromatin modifiers JMJD3 and CHD8 that modulates neural enhancer activity towards the exit of the stem state upon TGFβ signaling. We demonstrate that neural enhancers work as platforms to integrate developmental signals, cell-type specific TFs and epigenetic regulators to fine-tune neuronal commitment. A major caveat in the field is the understanding of the specificity of SMAD3 transcriptional response. In particular, the identity of the SMAD3 partners in different lineages is an intriguing issue. Recent studies have shown that master TFs, such as OCT4 in ESCs, MYOD1 in myotubes and PU.1 in pro-B cells select cell-type-specific response to TGFβ signaling providing specificity to the TGFβ response in these particular cellular contexts (46). Our work describes for the first time an interplay between TGFβ-pathway and the bHLH ASCL1 (Figure 1). Our results demonstrate that the proneural TF, ASCL1, assists SMAD3 in chromatin binding at enhancers and their ulterior activation in NSCs. ASCL1, as well as other bHLH, is a main regulator of neurogenesis (65,66). Interestingly, ASCL1 works as a pioneer factor in a neurogenic context and its binding to enhancers results in disruptions in the chromatin landscape, allowing the posterior binding of signaling factors and cofactors (49). The functional cooperation between ASCL1 and SMAD3 provides specificity to TGFβ response in neural context and opens new avenues to understand the functional interplay between intrinsic factors and extrinsic signals during development.
In addition to the factors that help SMAD3 recruitment, TGFβ plasticity is dependent on the coactivator proteins, mainly chromatin acting factors, which regulate transcription in the genomic context. Although a large number of SMAD3 cofactors have been previously described, how they provide specificity to TGFβ response is still unknown. Our studies extent the list of SMAD3 cofactors, by showing that JMJD3 and CHD8 are essential to activate TGFβ-responsive enhancers in NSCs.
JMJD3 is an essential cofactor during neural fate establishment (15,21). Our lab has previously demonstrated that JMJD3 interacts with SMAD3 at promoters in NSCs (15,50,67). In this study, we went further on that cooperation demonstrating that JMJD3 is essential to fully activate TGFβ-responsive enhancers (Figure 3). The molecular link between our previously published SMAD3/JMJD3 interaction at promoters and the one now described at enhancers remains unclear. However, we hypothesize that the recruitment of TGFβ cofactors is dependent on the three-dimensional structure of the chromatin and that the chromatin topology might constrain or facilitate the TF and cofactor interplay and the potential enhancer-promoter contacts. Another critical player for neural enhancer activation in response to TGFβ is CHD8 (Figure 4). This protein is an ATP-dependent chromatin remodeler factor in vitro and in vivo. In concordance, CHD8 binds enhancers and facilitated their remodeling in response to progesterone (55). Markedly, CHD8 plays an essential role in neurogenesis (57,58). Recently, CHD8 has called much attention due to its relevance in autism spectrum disorder (ASD). Functional analysis demonstrate that CHD8 regulates many ASD risk genes involved in neurodevelopment, synaptic function and WNT and p53 signaling pathways (58,59,68–72). Our data suggest that in addition to WNT and p53 pathways, TGFβ signaling might be also orchestrating CHD8 targeting during neurogenesis. This new discovered partnership could be related to ASD or other developmental or neurodegenerative disorders in which TGFβ is involved. Although more experiments are required to fully understand the interaction between TGFβ and CHD8, the discovery of the functional axis SMAD3/JMJD3/CHD8 provides insights into how histone modifier enzymes and chromatin remodelers are coordinated to fully activate SMAD3 targeted neural enhancers in a temporal specific manner during neural development.
Using CRISPR–Cas9 genetic deletions, we show that Neurog2(–6) poised enhancer is necessary for the induction of its target gene upon TGFβ signaling activation (Figure 5). Interestingly, Neurog2(–6) enhancer deletion did not result in the activation of the gene in NSCs, supporting the idea that the poised enhancers have not repressive activity (73) as it has been previously proposed (74–76). Major developmental and cell-identity genes are frequently regulated by multiple and sometimes redundant enhancers (77,78). However, our results clearly demonstrate that the deletion of a single poised enhancer totally blocks the expression of Neurog2 after TGFβ and compromise normal neuronal differentiation. This extends previous observations indicating that certain enhancers can control gene expression in a non-redundant manner in different cellular contexts (73,79,80).
The contribution of H3K27me3 demethylation by JMJD3 to enhancer activation is an intriguing question. Our results indicate that H3K27me2/3 levels are not altered in the analyzed enhancers during activation in a JMJD3-dependent manner. Studying the data, two mechanisms might be envisioned to explain the role of JMJD3 and H3K27me3 at the analyzed poised enhancers (Supplementary Figure S4A). In Chic2(–26) and Tle3(–114) enhancers, the H3K27me3 levels decreased upon TGFβ in a JMJD3-independent manner. At these enhancers, JMJD3 function might not be related to H3K27me3, and could be linked to demethylation of other locus like the promoters (Supplementary Figure S4C) or associated with RNA-Polymerase II release at transcriptional starting sites (50,81). In the case of Neurog2(-6), the increase observed in H3K27me3 upon TGFβ in shJMJD3 cells might be related to the ability of the PRC2 complex to bind this enhancer. It has been recently shown that PRC2 might be working as an activator of neural poised enhancers, by facilitating loop formation (73). Thus, it is possible that in the absence of JMJD3, EZH2 could be targeting more efficiently the enhancer increasing the levels of H3K27me3 in response to TGFβ signal. In addition to the poised enhancers, JMJD3 is actively recruited to some non-H3K27-methylated enhancers such as Ctgf(–102). This data suggest that in addition to demethylation, other JMJD3 catalytic-independent functions might be involved in TGFβ-responsive enhancer activation as it has been previously proposed at promoters and gene bodies in different cellular contexts (50,82–85). These results also open the possibility that other essential factors different than histone H3 might be targeted by JMJD3 KDM activity upon TGFβ to facilitate transcription activation. To fully clarify these issues more work will be required.
In summary, our results highlight enhancers as TF-binding platforms where different modifying enzymes coordinate their activities to induce faithful gene activation. This study uncovers the molecular mechanism responsible for full enhancer activation in response to TGFβ signaling in a neural stem cell context. This involves the action of JMJD3 and CHD8 cofactors, which, by remodeling enhancers, previously pre-marked by ASCL1, activate the neuronal commitment program. Due to the broad range of TGFβ functions in areas of cancer and neurodegenerative disorders, this work paves the way for investigating the ASCL1/SMAD/JMJD3/CHD8 contribution to transcriptional regulation in other cellular contexts and helps to move forward our understanding of the myriad of crosstalk between epigenetics and developmental programs.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Mariona Arbonès and Dr Sonia Najas for technical help dissecting NSCs. We also thank Dr Elisa Martí for reagents. This work has been performed within the framework of the doctoral program in Biochemistry and Molecular Biology of the Universitat Autònoma de Barcelona.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Education and Science [BFU2015-69248-P, BFU20012-34261 to M.M.B.]; Fundaciò La Marató de TV3 [090210 to M.M.B.]; Jérôme Lejeune Fondation; FPU fellowship and a travelling fellowship from the Company of Biologists (to R.F.). Funding for open access charge: Spanish Ministry of Education and Science [BFU2015-69248].
Conflict of interest statement. None declared.
REFERENCES
- 1. Temple S. The development of neural stem cells. Nature. 2001; 414:112–117. [DOI] [PubMed] [Google Scholar]
- 2. Tiberi L., Vanderhaeghen P., van den Ameele J.. Cortical neurogenesis and morphogens: diversity of cues, sources and functions. Curr. Opin. Cell Biol. 2012; 24:269–276. [DOI] [PubMed] [Google Scholar]
- 3. Schaffner W. Enhancers, enhancers - from their discovery to today's universe of transcription enhancers. Biol. Chem. 2015; 396:311–327. [DOI] [PubMed] [Google Scholar]
- 4. Buecker C., Wysocka J.. Enhancers as information integration hubs in development: lessons from genomics. Trends Genet. 2012; 28:276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weake V.M., Workman J.L.. Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 2010; 11:426–437. [DOI] [PubMed] [Google Scholar]
- 6. Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zentner G.E., Tesar P.J., Scacheri P.C.. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011; 21:1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J.. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011; 470:279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Visel A., Blow M.J., Li Z., Zhang T., Akiyama J.A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F. et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009; 457:854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Visel A., Taher L., Girgis H., May D., Golonzhka O., Hoch R.V., McKinsey G.L., Pattabiraman K., Silberberg S.N., Blow M.J. et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013; 152:895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun J., Rockowitz S., Xie Q., Ashery-Padan R., Zheng D., Cvekl A.. Identification of in vivo DNA-binding mechanisms of Pax6 and reconstruction of Pax6-dependent gene regulatory networks during forebrain and lens development. Nucleic Acids Res. 2015; 43:6827–6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conti L., Cattaneo E.. Neural stem cell systems: physiological players or in vitro entities. Nat. Rev. Neurosci. 2010; 11:176–187. [DOI] [PubMed] [Google Scholar]
- 13. Roussa E., Wiehle M., Dunker N., Becker-Katins S., Oehlke O., Krieglstein K.. Transforming growth factor beta is required for differentiation of mouse mesencephalic progenitors into dopaminergic neurons in vitro and in vivo: ectopic induction in dorsal mesencephalon. Stem Cells. 2006; 24:2120–2129. [DOI] [PubMed] [Google Scholar]
- 14. Vogel T., Ahrens S., Buttner N., Krieglstein K.. Transforming growth factor beta promotes neuronal cell fate of mouse cortical and hippocampal progenitors in vitro and in vivo: identification of Nedd9 as an essential signaling component. Cereb. Cortex. 2010; 20:661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Estaras C., Akizu N., Garcia A., Beltran S., de la Cruz X., Martinez-Balbas M.A.. Genome-wide analysis reveals that Smad3 and JMJD3 HDM co-activate the neural developmental program. Development. 2012; 139:2681–2691. [DOI] [PubMed] [Google Scholar]
- 16. Shi Y., Massague J.. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003; 113:685–700. [DOI] [PubMed] [Google Scholar]
- 17. Feng X.H., Derynck R.. Specificity and versatility in tgf-beta signaling through Smads. Annu. Rev. Cell Dev. Biol. 2005; 21:659–693. [DOI] [PubMed] [Google Scholar]
- 18. Yang L., Moses H.L.. Transforming growth factor beta: tumor suppressor or promoter? Are host immune cells the answer. Cancer Res. 2008; 68:9107–9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Massague J. How cells read TGF-beta signals. Nat. Rev. Mol. Cell Biol. 2000; 1:169–178. [DOI] [PubMed] [Google Scholar]
- 20. Massague J., Seoane J., Wotton D.. Smad transcription factors. Genes Dev. 2005; 19:2783–2810. [DOI] [PubMed] [Google Scholar]
- 21. Burgold T., Spreafico F., De Santa F., Totaro M.G., Prosperini E., Natoli G., Testa G.. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS One. 2008; 3:e3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dahle O., Kumar A., Kuehn M.R.. Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci. Signal. 2010; 3:ra48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim S.W., Yoon S.J., Chuong E., Oyolu C., Wills A.E., Gupta R., Baker J.. Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev. Biol. 2011; 357:492–504. [DOI] [PubMed] [Google Scholar]
- 24. Currle D.S., Hu J.S., Kolski-Andreaco A., Monuki E.S.. Culture of mouse neural stem cell precursors. J. Vis. Exp. 2007; 152, doi:10.3791/152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pollard S.M., Conti L., Sun Y., Goffredo D., Smith A.. Adherent neural stem (NS) cells from fetal and adult forebrain. Cereb. Cortex. 2006; 16(Suppl. 1):i112–i120. [DOI] [PubMed] [Google Scholar]
- 26. Theus M.H., Ricard J., Liebl D.J.. Reproducible expansion and characterization of mouse neural stem/progenitor cells in adherent cultures derived from the adult subventricular zone. Curr. Protoc. Stem Cell Biol. 2012; doi:10.1002/9780470151808.sc02d08s20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Galderisi U., Peluso G., Di Bernardo G., Calarco A., D’Apolito M., Petillo O., Cipollaro M., Fusco F.R., Melone M.A.. Efficient cultivation of neural stem cells with controlled delivery of FGF-2. Stem Cell Res. 2013; 10:85–94. [DOI] [PubMed] [Google Scholar]
- 28. Schwindt T.T., Motta F.L., Gabriela F.B., Cristina G.M., Guimaraes A.O., Calcagnotto M.E., Pesquero J.B., Mello L.E.. Effects of FGF-2 and EGF removal on the differentiation of mouse neural precursor cells. An. Acad. Bras. Cienc. 2009; 81:443–452. [DOI] [PubMed] [Google Scholar]
- 29. Sun D., Zhou X., Yu H.L., He X.X., Guo W.X., Xiong W.C., Zhu X.J.. Regulation of neural stem cell proliferation and differentiation by Kinesin family member 2a. PLoS One. 2017; 12:e0179047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jori F.P., Galderisi U., Napolitano M.A., Cipollaro M., Cascino A., Giordano A., Melone M.A.. RB and RB2/P130 genes cooperate with extrinsic signals to promote differentiation of rat neural stem cells. Mol. Cell. Neurosci. 2007; 34:299–309. [DOI] [PubMed] [Google Scholar]
- 31. Blanco-Garcia N., Asensio-Juan E., de la Cruz X., Martinez-Balbas M.A.. Autoacetylation regulates P/CAF nuclear localization. J. Biol. Chem. 2009; 284:1343–1352. [DOI] [PubMed] [Google Scholar]
- 32. Rodriguez-Paredes M., Ceballos-Chavez M., Esteller M., Garcia-Dominguez M., Reyes J.C.. The chromatin remodeling factor CHD8 interacts with elongating RNA polymerase II and controls expression of the cyclin E2 gene. Nucleic Acids Res. 2009; 37:2449–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garcia-Campmany L., Marti E.. The TGFbeta intracellular effector Smad3 regulates neuronal differentiation and cell fate specification in the developing spinal cord. Development. 2007; 134:65–75. [DOI] [PubMed] [Google Scholar]
- 34. Akizu N., Estaras C., Guerrero L., Marti E., Martinez-Balbas M.A.. H3K27me3 regulates BMP activity in developing spinal cord. Development. 2010; 137:2915–2925. [DOI] [PubMed] [Google Scholar]
- 35. Hamburger V., Hamilton H.L.. A series of normal stages in the development of the chick embryo. 1951. Dev. Dyn. 1992; 195:231–272. [DOI] [PubMed] [Google Scholar]
- 36. Asensio-Juan E., Fueyo R., Pappa S., Iacobucci S., Badosa C., Lois S., Balada M., Bosch-Presegue L., Vaquero A., Gutierrez S. et al. The histone demethylase PHF8 is a molecular safeguard of the IFNgamma response. Nucleic Acids Res. 2017; 45:3800–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Frank S.R., Schroeder M., Fernandez P., Taubert S., Amati B.. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001; 15:2069–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Valls E., Blanco-Garcia N., Aquizu N., Piedra D., Estaras C., de la Cruz X., Martinez-Balbas M.A.. Involvement of chromatin and histone deacetylation in SV40 T antigen transcription regulation. Nucleic Acids Res. 2007; 35:1958–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rada-Iglesias A., Bajpai R., Prescott S., Brugmann S.A., Swigut T., Wysocka J.. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell. 2012; 11:633–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim T.K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D.A., Laptewicz M., Barbara-Haley K., Kuersten S. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010; 465:182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Luyten A., Zang C., Liu X.S., Shivdasani R.A.. Active enhancers are delineated de novo during hematopoiesis, with limited lineage fidelity among specified primary blood cells. Genes Dev. 2014; 28:1827–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z.. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009; 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Supek F., Bosnjak M., Skunca N., Smuc T.. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011; 6:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mullen A.C., Orlando D.A., Newman J.J., Loven J., Kumar R.M., Bilodeau S., Reddy J., Guenther M.G., DeKoter R.P., Young R.A.. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011; 147:565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Trompouki E., Bowman T.V., Lawton L.N., Fan Z.P., Wu D.C., DiBiase A., Martin C.S., Cech J.N., Sessa A.K., Leblanc J.L. et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011; 147:577–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Castro D.S., Guillemot F.. Old and new functions of proneural factors revealed by the genome-wide characterization of their transcriptional targets. Cell Cycle. 2011; 10:4026–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Raposo A.A., Vasconcelos F.F., Drechsel D., Marie C., Johnston C., Dolle D., Bithell A., Gillotin S., van den Berg D.L., Ettwiller L. et al. Ascl1 coordinately regulates gene expression and the chromatin landscape during neurogenesis. Cell Rep. 2015; 9:1544–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Estaras C., Fueyo R., Akizu N., Beltran S., Martinez-Balbas M.A.. RNA polymerase II progression through H3K27me3-enriched gene bodies requires JMJD3 histone demethylase. Mol. Biol. Cell. 2013; 24:351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li W., Notani D., Rosenfeld M.G.. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat. Rev. Genet. 2016; 17:207–223. [DOI] [PubMed] [Google Scholar]
- 52. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G.. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010; 28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. De Santa F., Totaro M.G., Prosperini E., Notarbartolo S., Testa G., Natoli G.. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007; 130:1083–1094. [DOI] [PubMed] [Google Scholar]
- 54. Agger K., Cloos P.A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K.. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007; 449:731–734. [DOI] [PubMed] [Google Scholar]
- 55. Ceballos-Chavez M., Subtil-Rodriguez A., Giannopoulou E.G., Soronellas D., Vazquez-Chavez E., Vicent G.P., Elemento O., Beato M., Reyes J.C.. The chromatin Remodeler CHD8 is required for activation of progesterone receptor-dependent enhancers. PLoS Genet. 2015; 11:e1005174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Marfella C.G., Imbalzano A.N.. The Chd family of chromatin remodelers. Mutat. Res. 2007; 618:30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nishiyama M., Nakayama K., Tsunematsu R., Tsukiyama T., Kikuchi A., Nakayama K.I.. Early embryonic death in mice lacking the beta-catenin-binding protein Duplin. Mol. Cell. Biol. 2004; 24:8386–8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Platt R.J., Zhou Y., Slaymaker I.M., Shetty A.S., Weisbach N.R., Kim J.A., Sharma J., Desai M., Sood S., Kempton H.R. et al. Chd8 mutation leads to autistic-like behaviors and impaired striatal circuits. Cell Rep. 2017; 19:335–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Durak O., Gao F., Kaeser-Woo Y.J., Rueda R., Martorell A.J., Nott A., Liu C.Y., Watson L.A., Tsai L.H.. Chd8 mediates cortical neurogenesis via transcriptional regulation of cell cycle and Wnt signaling. Nat. Neurosci. 2016; 19:1477–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Visel A., Minovitsky S., Dubchak I., Pennacchio L.A.. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 2007; 35:D88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sauvageot C.M., Stiles C.D.. Molecular mechanisms controlling cortical gliogenesis. Curr. Opin. Neurobiol. 2002; 12:244–249. [DOI] [PubMed] [Google Scholar]
- 62. Hand R., Bortone D., Mattar P., Nguyen L., Heng J.I., Guerrier S., Boutt E., Peters E., Barnes A.P., Parras C. et al. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005; 48:45–62. [DOI] [PubMed] [Google Scholar]
- 63. Ge W., He F., Kim K.J., Blanchi B., Coskun V., Nguyen L., Wu X., Zhao J., Heng J.I., Martinowich K. et al. Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:1319–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Heng J.I., Nguyen L., Castro D.S., Zimmer C., Wildner H., Armant O., Skowronska-Krawczyk D., Bedogni F., Matter J.M., Hevner R. et al. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature. 2008; 455:114–118. [DOI] [PubMed] [Google Scholar]
- 65. Bertrand N., Castro D.S., Guillemot F.. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002; 3:517–530. [DOI] [PubMed] [Google Scholar]
- 66. Wilkinson G., Dennis D., Schuurmans C.. Proneural genes in neocortical development. Neuroscience. 2013; 253:256–273. [DOI] [PubMed] [Google Scholar]
- 67. Fueyo R., Garcia M.A., Martinez-Balbas M.A.. Jumonji family histone demethylases in neural development. Cell Tissue Res. 2015; 359:87–98. [DOI] [PubMed] [Google Scholar]
- 68. De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S. et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014; 515:209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sugathan A., Biagioli M., Golzio C., Erdin S., Blumenthal I., Manavalan P., Ragavendran A., Brand H., Lucente D., Miles J. et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E4468–E4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cotney J., Muhle R.A., Sanders S.J., Liu L., Willsey A.J., Niu W., Liu W., Klei L., Lei J., Yin J. et al. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat. Commun. 2015; 6:6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wilkinson B., Grepo N., Thompson B.L., Kim J., Wang K., Evgrafov O.V., Lu W., Knowles J.A., Campbell D.B.. The autism-associated gene chromodomain helicase DNA-binding protein 8 (CHD8) regulates noncoding RNAs and autism-related genes. Transl. Psychiatry. 2015; 5:e568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Katayama Y., Nishiyama M., Shoji H., Ohkawa Y., Kawamura A., Sato T., Suyama M., Takumi T., Miyakawa T., Nakayama K.I.. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature. 2016; 537:675–679. [DOI] [PubMed] [Google Scholar]
- 73. Cruz-Molina S., Respuela P., Tebartz C., Kolovos P., Nikolic M., Fueyo R., van Ijcken W.F.J., Grosveld F., Frommolt P., Bazzi H. et al. PRC2 facilitates the regulatory topology required for poised enhancer function during pluripotent stem cell differentiation. Cell Stem Cell. 2017; 20:689–705. [DOI] [PubMed] [Google Scholar]
- 74. Spitz F., Furlong E.E.. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012; 13:613–626. [DOI] [PubMed] [Google Scholar]
- 75. Bonn S., Zinzen R.P., Girardot C., Gustafson E.H., Perez-Gonzalez A., Delhomme N., Ghavi-Helm Y., Wilczynski B., Riddell A., Furlong E.E.. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 2012; 44:148–156. [DOI] [PubMed] [Google Scholar]
- 76. Entrevan M., Schuettengruber B., Cavalli G.. Regulation of genome architecture and function by polycomb proteins. Trends Cell Biol. 2016; 26:511–525. [DOI] [PubMed] [Google Scholar]
- 77. Frankel N., Davis G.K., Vargas D., Wang S., Payre F., Stern D.L.. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010; 466:490–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A.. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Huang J., Liu X., Li D., Shao Z., Cao H., Zhang Y., Trompouki E., Bowman T.V., Zon L.I., Yuan G.C. et al. Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev. Cell. 2016; 36:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shin H.Y., Willi M., Yoo K.H., Zeng X., Wang C., Metser G., Hennighausen L.. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat. Genet. 2016; 48:904–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen S., Ma J., Wu F., Xiong L.J., Ma H., Xu W., Lv R., Li X., Villen J., Gygi S.P. et al. The histone H3 Lys 27 demethylase JMJD3 regulates gene expression by impacting transcriptional elongation. Genes Dev. 2012; 26:1364–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. De Santa F., Narang V., Yap Z.H., Tusi B.K., Burgold T., Austenaa L., Bucci G., Caganova M., Notarbartolo S., Casola S. et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 2009; 28:3341–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Miller S.A., Mohn S.E., Weinmann A.S.. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol. Cell. 2010; 40:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kang S.C., Kim S.K., Chai J.C., Kim S.H., Won K.J., Lee Y.S., Jung K.H., Chai Y.G.. Transcriptomic profiling and H3K27me3 distribution reveal both demethylase-dependent and independent regulation of developmental gene transcription in cell differentiation. PLoS One. 2015; 10:e0135276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Xun J., Wang D., Shen L., Gong J., Gao R., Du L., Chang A., Song X., Xiang R., Tan X.. JMJD3 suppresses stem cell-like characteristics in breast cancer cells by downregulation of Oct4 independently of its demethylase activity. Oncotarget. 2017; 8:21918–21929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.