Abstract
In this study, we report the gut microbial composition and predictive functional profiles of zebrafish, Danio rerio, fed with a control formulated diet (CFD), and a gluten formulated diet (GFD) using metagenomics approach and bioinformatics tools. The microbial communities of the GFD-fed D. rerio displayed heightened abundances of Legionellales, Rhizobiaceae, and Rhodobacter, as compared to the CFD-fed counterparts. Predicted metagenomics of microbial communities (PICRUSt) in GFD-fed D. rerio showed KEGG functional categories corresponding to bile secretion, secondary bile acid biosynthesis, and the metabolism of glycine, serine, and threonine. The CFD-fed D. rerio exhibited KEGG functional categories of bacteria-mediated cobalamin biosynthesis, which was supported by the presence of cobalamin synthesizers such as Bacteroides and Lactobacillus. Though these bacteria were absent in GFD-fed D. rerio, a comparable level of the cobalamin biosynthesis KEGG functional category was observed, which could be contributed by the compensatory enrichment of Cetobacterium. Based on these results, we conclude D. rerio to be a suitable alternative animal model for the use of targeted metagenomics approach along with bioinformatics tools to further investigate the relationship between the gluten diet and microbiome profile in the gut ecosystem leading to the gastrointestinal diseases and other undesired adverse health effects.
Keywords: NextGen Sequencing, 16S rRNA, QIIME, PICRUSt, Pathogens
1. Introduction
The Neolithic Revolution from hunter-gatherer survival into agricultural strategies redefined diet (Freeman, 2013), incorporating larger quantities of wheat products, barley, and rye – of which the prolamin fraction is known to contain wheat protein gluten (Béres et al., 2014; Thompson et al., 2005). Wheat gluten is comprised of the water soluble monomeric protein gliadin, and insoluble multimeric glutenin, and is immunostimulatory to patients with genetically predisposed celiac disease (CD), and the etiologic agent of non-celiac gluten sensitivity (NCGS) (Howdle, 2006; Narrowe et al., 2015; Sanz, 2015). The pathophysiology of CD has been extensively probed, of which gluten induced autoimmune responses manifest aberrant intestinal mucosa conditions, characterized by inflammation and lesions, leading to malabsorption at the site of immune response followed by diarrhea and steatorrhea (Murray, 1999; Van Kessel et al., 2011). For NCGS, despite a reported ~6% of the human population affected, little is known of the factors underlying and/or progressing enteropathy, beyond the involvement of gluten (Elli et al., 2015; Sapone et al., 2012). Both CD and NCGS have received attention regarding population shifts and perturbations in intestinal bacteria (microbiota) that may be directly or indirectly correlated with disease states (A Daulatzai, 2015; Lotta et al., 2016; Samsel and Seneff, 2013; Sanz, 2015). Examinations of the microbiota of patients with CD and NCGS have concluded similar increases of harmful gram-negative bacteria, pathobionts and overall microbial dysbiosis (A Daulatzai, 2015; Béres et al., 2014 ).
To explore the effect of a gluten formulated diet (GFD) on both the composition and the metabolic profile of the microbiota, an examination of a model animal lacking a pre-existing aversion to gluten is warranted. Recently, the zebrafish Danio rerio has been established as a model organism to study for various genetic and environmental aspects of health, disease, and embryological development (Barut and Zon, 2000; Elli, and Roncoroni et al,. 2015; Sadler et al., 2013; Siccardi III et al., 2009; Watts et al., 2016). Many anatomical and physiological characteristics of the D. rerio digestive system, including disease states, are shared with the mammalian digestive system (Sadler, and Rawls et al., 2013; Wang et al., 2010). Also, the usefulness of this organism is emphasized by the ability to formulate feeds to promote growth, health, or disease in the laboratory setting (Siccardi III et al., 2009; Watts et al., 2016; Watts et al., 2012). Because of this, it is possible to formulate a gluten diet to elucidate the effect of the microbial composition in the gut ecosystem of laboratory raised D. rerio.
In recent years, NextGen sequencing (NGS) technologies have made it possible to describe the microbiota occurring in the gastrointestinal tracts of host organisms, by targeting the 16S rRNA genes of the collective bacterial genomes (microbiome) as they change in response to diet (Chen et al., 2014; Savarese et al., 2014; Turnbaugh et al., 2009; Umu et al., 2015). It is proposed that dietary gluten will alter the microbiome in the D. rerio gut ecosystem, resulting in microbial populations that are pathobionts and/or metabolically unfavorable to maintain and promote host health and physiology. In this study, we determined the microbial compositions in the gut ecosystem of D. rerio fed with a gluten formulated diet (GFD) or a control formulated diet (CFD), using NextGen Illumina Miseq targeting the V4 region within 16S rRNA gene. In addition, we have compared the gut microbiota-driven predictive metabolic profiles of the GFD- and CFD-fed D. rerio using the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) (ver.1.0.0), based on the16S rRNA gene sequence datasets.
2. Materials and methods
2.1. Culture of D. rerio and Sample Preparation
D. rerio (wild type strain AB) (Zebrafish International Resource Center, Eugene, OR), were cultivated from embryos in the NORC Aquatic Animal Research Core at the University of Alabama at Birmingham (UAB). D. rerio (28 days old) were maintained at 28 °C in 2.8 L tanks on a recirculating rack system (Aquaneering, Inc., San Diego, CA) and at a density of five fish per Liter. The aquaculture water was purified through a 5 μm sediment filter, and subsequently passed through reverse-osmosis mediated by charcoal, followed by ion exchange resin (Kent Marine, Franklin, WI). Synthetic sea salt (Instant Ocean, Blacksburg, VA) was added to maintain a conductivity of 1500 IS/cm. The water pH of 7.4 was sustained through periodic adjustment using sodium bicarbonate. One group of D. rerio was fed CFD, containing marine fish protein and other ingredients (fish meal is the standard protein source in most lab animal diets including zebrafish), and another group of D. rerio was fed GFD, containing wheat gluten as the primary source of protein (Table 2). All other ingredients were included at identical concentrations in both diets (note that the fish meal protein source contains slightly more fat than the wheat gluten protein source). The animals were fed three times per day. Excess food particles in the culture were removed from the tank via siphoning every other day. After 12 weeks, both the CFD (n=2) and the GFD (n=2) samples (biological replicates) of D. rerio were euthanized and guts were removed intact, flash frozen in liquid nitrogen, and preserved at −80°C until used for DNA extraction and preparation for NGS. All experiments with the vertebrate animal (D. rerio) were conducted according to the guideline elaborated in “Guide for the Care and Use of Laboratory Animals,” which was published by the National Research Council of the National Academies. The protocols used in this study were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Alabama at Birmingham under the Animal Project Number (APN) IACUC-08196.
Table 2.
A description of the diet compositions used in this study. Diet composition and proximate analysis of the control formulated diet (CFD. containing fish meal as the primary protein source. The Scoular Company-C.P.S.P.90) and the gluten formulated diet (GFD. containing wheat gluten as the primary protein source. MP Biomedicals) are elaborated in this table.
| Ingredienta | Amount included (g/100 g total) | |
|---|---|---|
| CFD | GFD | |
| Fish protein hydrolysate (82%) | 59.00 | – |
| Wheat gluten (80%) | – | 60.00 |
| Dextrin | 13.85 | 13.85 |
| Soy lecithin | 4.00 | 4.00 |
| Canthaxanthin | 2.31 | 2.31 |
| Ascorbylpalmitate | 0.04 | 0.04 |
| Vitamin premix BML-2 | 4.00 | 4.00 |
| Mineral mix BTm | 3.00 | 3.00 |
| Betaine | 0.15 | 0.15 |
| Potassium phosphate monobasic | 1.15 | 1.15 |
| Alginate | 5.38 | 5.38 |
| Cholesterol | 0.12 | 0.12 |
| Menhaden oil | 4.67 | 4.67 |
| Corn oil | 2.33 | 2.33 |
| Diet proximate analysis | CFD | GFD |
| Moisture (%)b,c | 10.97, 10.72 | 9.58, 9.45 |
| Fat(%)c,d | 15.86, 15.65 | 11.10, 10.90 |
| Fiber (%)c | 1.77, 1.67 | 1.73, 1.85 |
| Protein (%)c,e | 45.30, 45.19 | 49.50, 49.09 |
| Ash (%)c | 8.37, 8.34 | 5.95, 5.98 |
Content by percentage of dry matter.
Content by percentage, as fed.
Duplicate measures (analysis by MVTL Laboratories, Minnesota).
Fat by ethyl ether extraction.
Protein = N × 6.25.
2.2. DNA Extraction and 16S rRNA Amplicon Library Preparation
Metacommunity DNA was extracted in triplicates (technical replicates) from each CFD and GFD sample using the Zymo Research Fecal DNA isolation kit (Irvine, CA; catalog #D6010) and then combined into single samples to perform NGS. For amplicon library preparation from the purified DNA, uniquely barcoded oligonucleotide primers (Forward primer V4: 5′-AATGATACGGCGACCACCGAGATCTACACTATGGTAATTGTGTGCCAGCMGCCGCGGTAA-3′; and Reverse primer V4: 5′-CAAGAGAAGACGGCATACGAGATNNNNNNAGTCAGTCAGCCGGACTACHVGGGTWTCTAAT-3′) (Eurofins Genomics, Inc., Huntsville, AL) were used for PCR to amplify the V4 region of the 16S rRNA gene (Kumar et al., 2014). All PCR reagents and cycling parameters were used as described previously (Kumar et al., 2014).
2.3. Illumina Miseq, Sequence Processing, and Bioinformatics Workflow
The PCR amplified V4 segment of the16S rRNA were subjected to NGS (250 bp, paired end) using Illumina Miseq™ platform (Kozich et al., 2013; Kumar et al., 2014). The raw FASTQ sequence files were used for quality checking using FastQC (Andrews, 2010), and ambiguous sequences were filtered or trimmed by using FASTX toolkit (Gordon and Hannon, 2010). Then, overlapping regions of the paired-end reads were merged, and then the chimera sequences were removed by USEARCH (Edgar, 2010).
The processed sequences were then clustered into Operational Taxonomic Units (OTUs) at 97% sequence similarity using UCLUST (Edgar, 2010), and then representative sequences were aligned by PyNAST (Caporaso et al., 2010). The taxonomic identifications were assigned to each representative sequence through the Ribosomal Database Project (RDP) classifier (Wang et al., 2007), trained using the Greengenes (ver. 13.8) (McDonald et al., 2012) at 60% confidence threshold. Stack column bar graphs representing the relative abundances of the bacterial taxa in CFD and GFD samples were generated from the filtered OTU table (>0.0005% abundance).
2.4 PICRUSt Analyses for Predictive functions
PICRUSt (ver 1.0.0) (Langille et al., 2013) was used to reveal the predictive functions of the microbial communities from each sample. To determine the predicted functions of each sample, the seq.fasta files were used to assign OTUs by closed reference OTU approach against the GreenGenes database (ver 13.5) at a 97% identity. The resultant OTU table was then normalized, and “predict_metagenomes.py” and “categorize_by_functions.py” commands were used to predict functions by referencing the assigned GreenGenes Ids of the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology (KO) database (Kanehisa and Goto, 2000; Kanehisa et al., 2014). The predicted functions were merged into hierarchical categories (Level 1, Level 2, and Level 3) in all samples.
2.5. Statistical Analysis of the Bacterial Diversity and their Predicted Functions
The OTU tables were used to calculate alpha (rarefaction curve and Shannon diversity (Shannon et al., 1964) and beta diversity (principle coordinate analysis (PCoA) plot through QIIME (ver. 1.8.0) (Caporaso et al., 2010). In order to calculate beta diversity, microbial communities between samples were compared using UniFrac metrics (Lozupone et al., 2006). The PCoA plot and a jackknife were created based on the weighted UniFrac phylogenetic distances through QIIME (ver. 1.8.0). A similarity percentages procedure (SIMPER) analysis was also conducted to estimate the contribution of each taxon to the contrast between CFD and GFD samples using the SIMPER function in the vegan package of R statistical software (Oksanen et al., 2013). Then, the “heatmap.2” function in R package (Oksanen et al., 2013) was used to visualize a top 25 most highly abundant taxa, found from the SIMPER analysis, into a heatmap. For the predicted function analysis (PICRUSt, ver. 1.0.0), the output from “categorize_by_functions.py” command was then used for two-group box-plot analysis implemented in Statistical Analysis of Metagenomic Profiles (STAMP, ver. 2.1.3) (Parks et al., 2014). Welch’s t-test was used for two-group comparison of the CFD and GFD samples with confidence intervals set to 95% (0.95).
3. RESULTS
3.1. Total sequence reads, quality trimming, and OTUs information
A total of 563,879 raw sequence reads from four samples (2 samples from CFD and 2 samples from GFD) of D. rerio were listed in Table 1. After quality-based trimming and filtering processes, a total 383,220 sequences were used for further bioinformatics analyses. Within these reads, 114,804 sequences clustered into 471 OTUs from CFD1 sample, and 114,916 sequences clustered into 439 OTUs from the CFD2 sample. Similarly, 79,906 sequences clustered into 331 OTUs from the GFD1 sample; and 73,594 sequences clustered into 322 OTUs from the GFD2 sample (Table 1). The observed species (OTUs) rarefaction curves (Supplementary Fig. 1) were generated for each sample, to show that the sequencing depth was sufficient to produce stable and unbiased estimates of species richness.
Table 1.
Raw and trimmed sequence reads following NextGen sequencing of the V4 segment of the 16S rRNA gene. The number of OTUs and calculated alpha diversity of four gut microbiome samples of D. rerio fed with control formulated diet (CFD, n = 2) and gluten formulated diet (GFD, n = 2) are listed.
| CFD1 | CFD2 | GFD1 | GFD2 | Total | |
|---|---|---|---|---|---|
| Number of raw sequences | 146,253 | 153,866 | 138,447 | 125,313 | 563,879 |
| Number of sequences after trimming and filtering processes | 114,804 | 114,916 | 79,906 | 73,594 | 383,220 |
| Number of OTUs | 471 | 439 | 331 | 322 | 1563 |
| Shannon diversity | 5.880 | 6.252 | 5.382 | 5.496 | – |
3.2. Microbial diversity in CFD and GFD samples
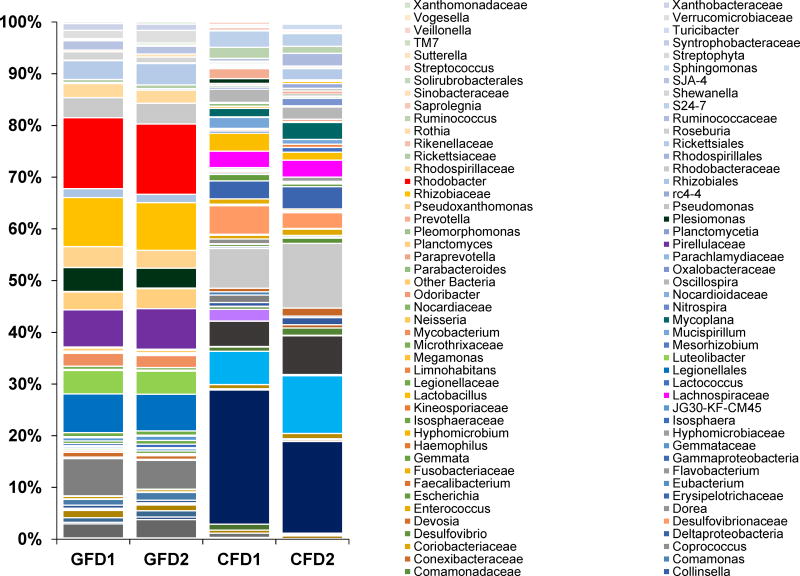
The relative abundances of microbial taxa found to the most resolvable level (up to family or genus) in four samples elaborated in Fig. 1. Proteobacteria and Actinobacteria were commonly found in both CFD and GFD samples. However, Firmicutes and Bacteroidetes were present at higher abundances in the CFD samples, whereas Planctomycetes, Fusobacteria, and Verrucomicrobia appeared as more abundant in the GFD samples (Fig. 1). A comparative analysis of the bacterial distribution at the lower taxonomic levels in CFD and GFD samples showed that, Bacteroides, Desulfovibrionaceae, Bifidobacterium, Lachnospiraceae, Oscillospira, and Ruminococcus were found only in CFD samples, whereas Rhodobacter, Legionellales, Pirellulaceae, Luteolibacter, and Pseudoxanthomonas in GFD samples (Fig. 1). Interestingly, BLAST analysis (Altschul et al., 1990; Morgulis et al., 2008) of most of sequences representing the Legionellales identified as Legionella anisa (99% identity with E-value of 1e –126), Legionella norrlandica (99% identity with E-value of 1e –126) and uncultured Legionella sp. (Identity: 97–99%, E-value: 1e–126 – 6e–115). A detailed list of the distribution of taxonomic groups at the genus and the family levels for CFD and GFD sample have been elaborated in Fig. 1.
Fig. 1. Stacked column bar graph showing the distribution and abundances of bacterial communities in D. rerio fed with control formulated diet (CFD) and gluten formulated diet (GFD).
The figure shows the relative abundances that were analyzed using QIIME (v1.8.0), and the graph was generated using Microsoft Excel software (Microsoft, Seattle, WA) (GFD1 and GFD2 = Gluten formulated diet sample 1 and 2; CFD1 and CFD2 = Control formulated diet sample 1 and 2).
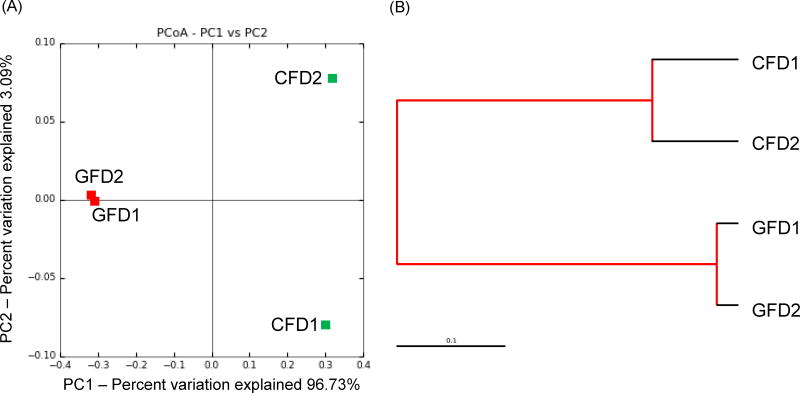
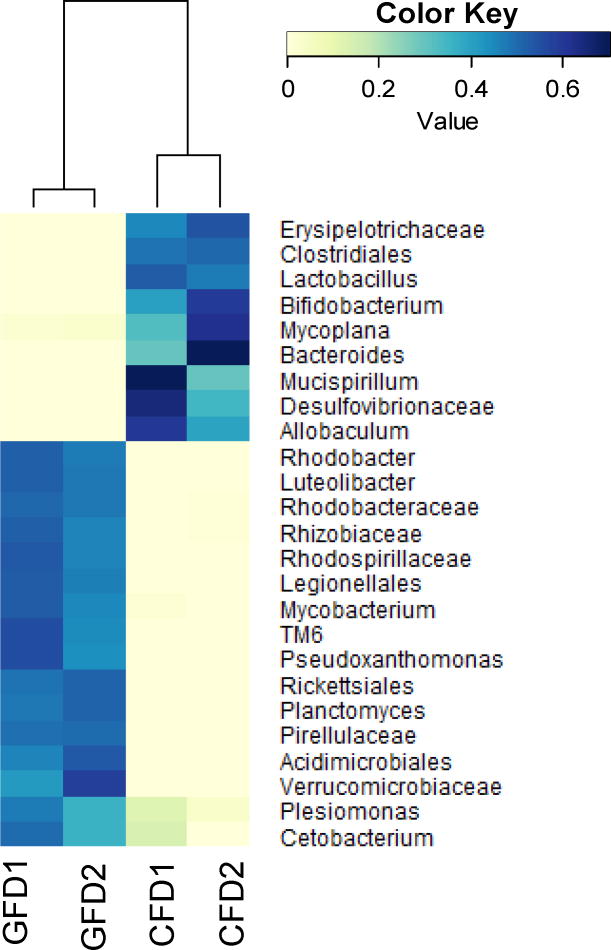
The Shannon diversity index revealed a low microbial diversity in GFD samples compared to the CFD samples (Table 1). The PCoA plot (Fig. 2a) and Jackknife (Fig. 2b) showed a closely related microbial community within each sample. The jackknife analysis also supported the PCoA plot clustering by showing the robustness of the differences between the CFD and the GFD samples. A SIMPER analyses showed that changes in the abundance of top 25 taxa contributed to 62% of the contrast between CFD and GFD samples, suggesting a distinct microbial community in GFD-fed D. rerio samples. In addition, a heatmap representing the SIMPER results showed low intra-sample variation (Fig. 3). The relative abundance for each bacterial taxon was shown by the color intensity with the legend elaborated in Fig. 3.
Fig. 2.
(A) Principle Coordinate Analysis (PCoA) plot, and (B) Jackknife, generated by QIIME (v1.8.0) representing the relationship between the composition of the gut bacterial communities in D. rerio fed with control formulated diet (CFD) and gluten formulated diet (GFD). (GFD1 and GFD2 = Gluten formulated diet sample 1 and 2; CFD1 and CFD2 = Control formulated diet sample 1 and 2).
Fig. 3. Heatmap showing the gut microbial compositions at the family level of D. rerio fed with control formulated diet (CFD) and gluten formulated diet (GFD).
The heatmap was generated based on the SIMPER (similarity percentages procedure) result, which represents the contribution of each taxon of the D. rerio gut samples. The SIMPER analysis and heatmap visualization were performed using the vegan package of R statistical software. (GFD1 and GFD2 = Gluten formulated diet sample 1 and 2; CFD1 and CFD2 = Control formulated diet sample 1 and 2).
3.3. Predicted metabolic functions using PICRUSt
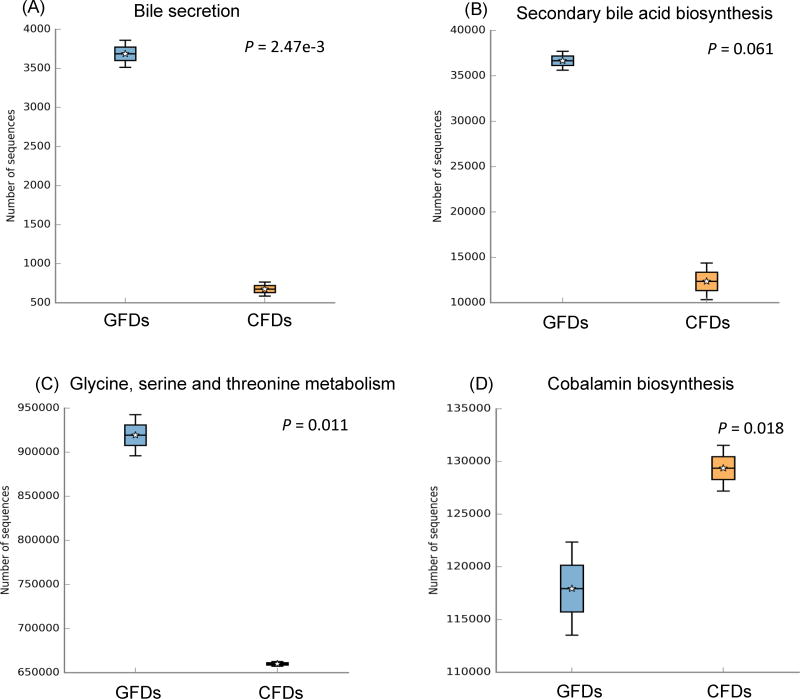
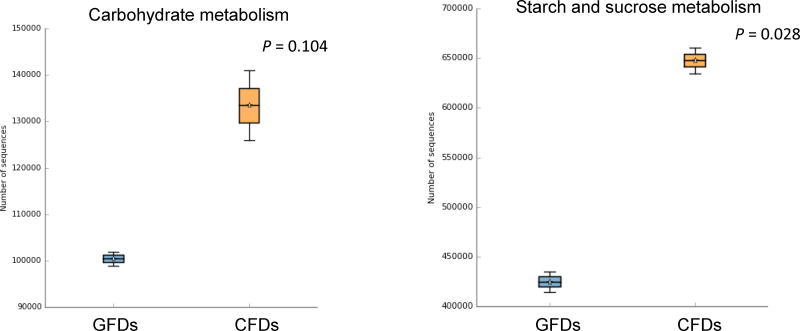
The results of the predicted metabolic functions of the microbial communities visualized by STAMP represented at Level 3 are as follows: bile secretion, secondary bile acid biosynthesis, glycine, serine and threonine metabolism, cobalamin biosynthesis, carbohydrate metabolism, and starch and sucrose metabolism (Fig. 4). In general, GFD samples showed a heightened relative abundance of the KEGG Level 3 categories corresponding to bile secretion (P-value = 2.47e–3), secondary bile acid biosynthesis (P-value = 0.061), and glycine, serine and threonine metabolism (P-value = 0.011) (Figs. 4a–c). In contrast, CFD samples showed a higher abundance of pathways related to cobalamin biosynthesis (P-value = 0.018), carbohydrate metabolism (P-value = 0.104), and starch and sucrose metabolism (P-value = 0.028) (Figs. 4d–f). Detail information of each KEGG metagenomics category, as well as other KEGG categories identified using PICRUSt (ver. 1.0.0) were listed in Supplementary Table 1.
Fig. 4. Box-plot showing the predictive functions of bacterial communities in D. rerio fed with control formulated diet (CFD) and gluten formulated diet (GFD).
The plot was generated from the STAMP analysis based on the PICRUSt results. For group comparison, Welch’s t-test was used to calculate variance between the two groups (i.e. CFD and GFD). The P-value for the total variance of the two groups is listed in the Box-plots. (GFDs = Gluten formulated diet sample 1 and 2; CFDs = Control formulated diet sample 1 and 2).
4. Discussion
This study has elucidated the distinct community composition and predictive KEGG metagenomic functional profiles of GFD-fed D. rerio gut microbiome, as compared to D. rerio fed with CFD. Since this study performed NGS approach, the plateau in a rarefaction curve (Supplementary Fig. 1) was used to suggest adequate sample replicates to generate a meaningful experimental outcome for this study. In general, the PCoA plot (Fig. 2a), Jackknife (Fig. 2b) and heatmap (Fig. 3) that accounts for changes in the relative abundances between communities revealed a low intra-sample variation.
The CFD samples displayed a generally beneficial gram-positive microbial profile (Figs. 1 and 3), representing Clostridiales, Allobaculum, Ruminococcus, Lactobacillus, and Bifidobacterium (Duncan et al., 2007; Stefka et al., 2014; Tojo et al., 2014; Zhang et al., 2013), along with Bacteroides – a gram-negative taxon that is known to produce cobalamin as a metabolite at high efficiency, and has been observed in the gut of goldfish, common carp, Nile tilapia, and ayu (Tsuchiya et al., 2008). In contrast, the resultant microbiota following a GFD showed a microbial profile with elevated gram-negative taxa (Figs. 1 and 3) including Cetobacterium and Rhodobacter. In addition, bacteria such as Legionella and Mycobacterium were also identified and the BLAST analysis of the representative OTU sequences and showed sequence identities with previously reported pathogenic L. anisa and L. norrlandica, and Mycobacterium fortuitum (98% identity with E-value of 1e –121) sequences, respectively (Newton et al., 2010; Sanderson and Hermon-Taylor, 1992). These results corroborated previously reported microbiota profiles in children with CD, which have shown to contribute to decrease beneficial gram-positive taxa such as Bifidobacterium, and elevated occurrences of gram-negative pathogenic bacteria, including Legionella (Kopečný et al., 2008; Morgulis et al., 2008; Sanderson and Hermon-Taylor, 1992).
Interestingly, the GFD-fed D. rerio gut microbial profile showed a heightened abundance of Rhodobacter, members of which are known to assimilate bile to cholesterol, creating bile acid (Afrose et al., 2010; Salma et al., 2007). Such functional categories were supported in our study by PICRUSt analysis, which showed heightened bile secretion functional category in KEGG of the GFD samples, presumably contributing to a high gut bile acid environment (Salma et al., 2007). As a potential response to the increased primary bile acid, pathways related to an elevated secondary bile acid biosynthesis functional KEGG category were also found in the GFD-fed D. rerio microbiota samples. This supports previously reported study where the Rhodobacter bacteria-exclusive pathways deconjugate primary bile acids (Ridlon and Hylemon, 2006; Ridlon et al., 2014), are implicated in damaging intestinal mucosa (Vuoristo et al., 1988), and are considered co-mutagenic and co-carcinogenic in the onset of colorectal cancer (Hill, 1990; Nagengast et al., 1995; Valko et al., 2001). In addition, the KEGG categories of secondary bile acid biosynthesis by members of Rhodobacter, Legionella, and Mycobacterium found in our GFD-fed D. rerio samples were in accordance with results reported elsewhere (Kanehisa and Goto, 2000; Kimura et al., 2014).
Conversely, the CFD-fed D. rerio microbiota displayed a high abundance of Bacteroides, Bifidobacterium, and Lactobacillus, which were absent in the GFD-fed D. rerio, members of which have been previously reported to suppress secondary bile acid through the assimilation of cholic acid, and reversion to primary bile acid (Ridlon and Hylemon, 2006). Additionally, the GFD-fed D. rerio gut microbial community displayed relatively heightened metabolic functional KEGG categories related to glycine, serine, and threonine metabolisms. Under specific circumstances, serine and threonine precede glycine in the one-carbon metabolic pathway, and hyperactivity of these pathways has been implicated in oncogenesis (Amelio et al., 2014; Locasale, 2013; McKnight, 2014). Interestingly, dietary restrictions of the amino acids serine and glycine have been shown to impede tumor development (Maddocks et al., 2013; Tavana and Gu, 2013). Thus increased productions of these amino acids by gut bacteria observed in this study provide a predictive outlook of the potentially developing pathogenic profile of the GFD induced gut microbiota.
As compared to the CFD-fed D. rerio, a marked increase in Cetobacterium was observed in the GFD-fed D. rerio, a genus that has been identified in the gut microbiota of sea mammals (Foster et al., 1995), fathead minnows Pimephales promelas (Narrowe et al., 2015), carp Cyprinus carpio (Van Kessel et al., 2011), and in previous culture independent microbiome investigations of D. rerio (Roeselers et al., 2011). Cetobacterium, and particularly C. somerae, are known to synthesize cobalamin in fish without dietary sources of the vitamin (Van Kessel et al., 2011), signifying a compensatory enrichment of the taxa in response to either malabsorption of the vitamin, or its deficiency in the diet (Dahele and Ghosh, 2001). Importantly, members of Cetobacterium are bile resistant (Duncan, and Louis, 2007; Finegold et al., 2003), which may explain their elevated appearance in a high bile gut environment of GFD samples. Nevertheless, cobalamin biosynthesis KEGG pathways were observed to be heightened in the CFD-fed D. rerio, suggesting an overall decrease in beneficial cobalamin producing bacteria in GFD-fed fish.
In a previous study (Bonder et al., 2016), the effect of a gluten-free diet on the gut microbiome in humans with no preexisting gastrointestinal disorders showed changes in the gut microbiome structure and microbial metabolic pathways involved in carbohydrate and starch metabolisms. In support of their observations, the CFD-fed zebrafish in our study showed a noteworthy occurrence of Clostridiaceaes, Clostridiales, Coriobacteriaceae, and Collinsella, as well as a low abundance of Veillonella, Ruminococcus, and Roseburia. To further support the predicted functional attributes performed by Bonder et al. (2016) on the gluten free microbiome, our PICRUSt results showed a heightened relative abundance of functional categories related to sugar metabolisms (Figs. 4e–f) in the CFD samples. Interestingly, Clostridiales was found to be the most dominant of the aforementioned bacteria in our CFD samples, members of which are known to perform a variety of anaerobic carbohydrate metabolisms including Alpha- and Beta-linked saccharides hydrolysis (Xia et al., 2015), thereby implicating this taxon in the increased abundance of carbohydrate metabolisms in CFD-fed zebrafish samples.
Both GFD- and CFD-fed D. rerio in our study showed gut microbial taxa that were conserved between both sample types, a result which corroborates aspects of the previously suggested core gut microbiota of domesticated and recently caught wild D. rerio (Roeselers et al., 2011). Of the described core phyla, Proteobacteria and Firmicutes were observed in both sample groups in this study, though to varying degrees of relative abundances. Specifically, Proteobacteria was more heightened in the GFD samples as compared to the CFD-fed D. rerio, which showed a higher observance of Firmicutes. Another described core phylum, Fusobacteria, although commonly observed between GFD and CFD samples, was more heightened in GFD samples. Lastly, members of the phylum Actinobacteria, as well as family Aeromonadaceae and genus Shewanella were shared among both sample types in this study. Overall, the GFD and CFD samples shared core microbial taxa that seem to be persistent with the previously described core microbiota of D. rerio (Roeselers et al., 2011).
Diet-induced variations in gut microbial population have been observed in human (David et al., 2014) as well as proxy mammalian (Turnbaugh et al., 2009), and other vertebrate and invertebrate model organisms (Newton et al., 2013), including D. rerio (Oka et al., 2010; Watts et al., 2016) to predict circumstantial deviations from healthy reference microbiome communities. Such investigations have also been conducted to understand the changes in the gut microbiome influenced by gluten in context to pre-existing diseases such as CD and NCGS. (A Daulatzai, 2015; Béres et al., 2014; Lotta, and Katri, 2016; Samsel and Seneff, 2013).
Although fecal samples are generally accepted for microbiome investigations in humans, recent reports have indicated that by including tissue biopsy techniques from multiple regions of the gastrointestinal tract, it is possible to achieve a more comprehensive and appropriate representation of the microbial communities contributing to gut tissue health (Huse et al., 2014, Bashir et al., 2016 ; Momozawa et al., 2011). In accordance with these previous reports, we have sampled the entire zebrafish gut tissue that included the internal digesta in our study.
This study for the first time demonstrated a uniform and distinct gut microbial profile in D. rerio fed with GFD, which contrasted their counterparts fed with CFD. By using a 16S rRNA gene-based metagenomics approach, we have determined the differences in the bacterial community compositions between GFD- and CFD-fed D. rerio using bioinformatics software such as QIIME and R, and most importantly were able to use this taxonomic information to predict KEGG functional profiles of those bacterial communities through PICRUSt and STAMP. From this approach, we have corroborated previous studies of gluten-related aberrant gut microbiome in humans with disease profiles (CD and NCGS), and thereby conclude D. rerio to be a suitable alternative vertebrate model organism for the investigations of diet-induced variations of gut microbiome, particularly as it relates to diseases or pathophysiology influenced by gluten enriched diet regimen.
Supplementary Material
Acknowledgments
We thank the UAB Institutional Animal Care and Use Committee for their support in this research. We thank Edward Partridge, MD (Comprehensive Cancer Center; grant P30AR050948), Robert Kimberly, MD (Center for Clinical Translational Science; grant UL1TR001417), Michael Saag, MD (Center for AIDS Research; grant 5P30AI027767), and David Allison, PhD (UAB NORC; grant NIH P30DK056336) for providing support with the center grants and core facilities for this work. We also thank Matthew Pace and T.D. Todd of CAS IT for computer support; Cheaha UABgrid by UAB ITRC for providing the HPC support necessary for bioinformatics analyses of the NGS data.
Footnotes
Conflict of interests
The authors declare no conflicts and financial interest with this study.
Accession codes: Sequence data have been deposited on the publicly available database located at the SRA of NCBI, under accession number SRP077792.
Supplementary data: Available at the journal online.
References
- Daulatzai AM. Non-celiac gluten sensitivity triggers gut dysbiosis, neuroinflammation, gut-brain axis dysfunction, and vulnerability for dementia. CNS & Neurological Disorders-Drug Targets (Formerly Current Drug Targets-CNS & Neurological Disorders) 2015;14:110–131. doi: 10.2174/1871527314666150202152436. [DOI] [PubMed] [Google Scholar]
- Afrose S, Hossain M, Salma U, Miah AG, Tsujii H. Dietary karaya saponin and Rhodobacter capsulatus exert hypocholesterolemic effects by suppression of hepatic cholesterol synthesis and promotion of bile acid synthesis in laying hens. Cholesterol. 2010 doi: 10.1155/2010/272731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amelio I, Cutruzzola F, Antonov A, Agostini M, Melino G. Serine and glycine metabolism in cancer. Trends Biochem Sci. 2014;39:191–198. doi: 10.1016/j.tibs.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. FastQC: A quality control tool for high throughput sequence data. Reference Source. 2010 available at http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Barut BA, Zon LI. Realizing the potential of zebrafish as a model for human disease. Physiol Genomics. 2000;2:49–51. doi: 10.1152/physiolgenomics.2000.2.2.49. [DOI] [PubMed] [Google Scholar]
- Bashir M, Prietl B, Tauschmann M, Mautner SI, Kump PK, Treiber G, Wurm P, Gorkiewicz G, Hogenauer C, Pieber TR. Effects of high doses of vitamin D3 on mucosa-associated gut microbiome vary between regions of the human gastrointestinal tract. Eur J Nutr. 2016;55:1479–1489. doi: 10.1007/s00394-015-0966-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béres NJ, Sziksz E, Vannay Á, Szabó D, Pap D, Veres-Székely A, Arató A, Szabó AJ, Veres G. Role of the microbiome in celiac disease. International Journal of Celiac Disease. 2014;2:150–153. [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, He X, Huang J. Diet effects in gut microbiome and obesity. J Food Sci. 2014;79:R442–R451. doi: 10.1111/1750-3841.12397. [DOI] [PubMed] [Google Scholar]
- Choo JM, Leong LE, Rogers GB. Sample storage conditions significantly influence faecal microbiome profiles. Sci Rep. 2015:5. doi: 10.1038/srep16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahele A, Ghosh S. Vitamin B12 deficiency in untreated celiac disease. The American journal of gastroenterology. 2001;96:745–750. doi: 10.1111/j.1572-0241.2001.03616.x. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S, Louis P, Flint H. Cultivable bacterial diversity from the human colon. Lett Appl Microbiol. 2007;44:343–350. doi: 10.1111/j.1472-765X.2007.02129.x. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Elli L, Roncoroni L, Bardella MT. Non-celiac gluten sensitivity: time for sifting the grain. World journal of gastroenterology: WJG. 2015;21:8221. doi: 10.3748/wjg.v21.i27.8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegold SM, Vaisanen ML, Molitoris DR, Tomzynski TJ, Song Y, Liu C, Collins MD, Lawson PA. Cetobacterium somerae sp nov from human feces and emended description of the genus. Cetobacterium Syst Appl Microbiol. 2003;26:177–181. doi: 10.1078/072320203322346010. [DOI] [PubMed] [Google Scholar]
- Foster G, Ross H, Naylor R, Collins M, Ramos CP, Garayzabal FF, Reid R. Cetobacterium ceti gen. nov., sp nov., a new Gram-negative obligate anaerobe from sea mammals. Lett Appl Microbiol. 1995;21:202–206. doi: 10.1111/j.1472-765x.1995.tb01041.x. [DOI] [PubMed] [Google Scholar]
- Freeman HJ. The Neolithic revolution and subsequent emergence of the celiac affection. International Journal of Celiac Disease. 2013;1:19–22. [Google Scholar]
- Gordon A, Hannon G. Fastx-toolkit. Computer program distributed by the author, website. 2010 http://hannonlab.cshl.edu/fastx_toolkit/
- Hill M. Bile flow and colon cancer. Mutation Research/Reviews in Genetic Toxicology. 1990;238:313–320. doi: 10.1016/0165-1110(90)90023-5. [DOI] [PubMed] [Google Scholar]
- Howdle PD. Gliadin, glutenin or both? The search for the Holy Grail in coeliac disease. Eur J Gastroenterol Hepatol. 2006;18:703–706. doi: 10.1097/01.meg.0000221847.09792.34. [DOI] [PubMed] [Google Scholar]
- Huse SM, Young VB, Morrison HG, Antonopoulos DA, Kwon J, Dalal S, Arrieta R, Hubert NA, Shen L, Vineis JH, Koval JC, Sogin ML, Chang EB, Raffals LE. Comparison of brush and biopsy sampling methods of the ileal pouch for assessment of mucosa-associated microbiota of human subjects. Microbiome. 2014;2:5. doi: 10.1186/2049-2618-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Hisano Y, Kawahara A, Higashijima S-i. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep. 2014:4. doi: 10.1038/srep06545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopečný J, Mrázek J, Fliegerová K, Frühauf P, Tučková L. The intestinal microflora of childhood patients with indicated celiac disease. Folia Microbiol. 2008;53:214–216. doi: 10.1007/s12223-008-0028-8. [DOI] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Eipers P, Little RB, Crowley M, Crossman DK, Lefkowitz EJ, Morrow CD. Getting started with microbiome analysis: sample acquisition to bioinformatics. Current Protocols in Human Genetics. 2014:18. doi: 10.1002/0471142905.hg1808s82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nature Reviews Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotta N, Katri K, Katri L. The microbiota as a component of the celiac disease and non-celiac gluten sensitivity. Clinical Nutrition Experimental 2016 [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. The ISME journal. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight SL. A hypothetical means of treating or preventing cancer. Cancer & metabolism. 2014;2:O7. [Google Scholar]
- Momozawa Y, Deffontaine V, Louis E, Medrano JF. Characterization of bacteria in biopsies of colon and stools by high throughput sequencing of the V2 region of bacterial 16S rRNA gene in human. PLoS One. 2011;6:e16952. doi: 10.1371/journal.pone.0016952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schäffer AA. Database indexing for production MegaBLAST searches. Bioinformatics. 2008;24:1757–1764. doi: 10.1093/bioinformatics/btn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JA. The widening spectrum of celiac disease. The American journal of clinical nutrition. 1999;69:354–365. doi: 10.1093/ajcn/69.3.354. [DOI] [PubMed] [Google Scholar]
- Nagengast F, Grubben M, Van Munster I. Role of bile acids in colorectal carcinogenesis. Eur J Cancer. 1995;31:1067–1070. doi: 10.1016/0959-8049(95)00216-6. [DOI] [PubMed] [Google Scholar]
- Narrowe AB, Albuthi-Lantz M, Smith EP, Bower KJ, Roane TM, Vajda AM, Miller CS. Perturbation and restoration of the fathead minnow gut microbiome after low-level triclosan exposure. Microbiome. 2015;3:1. doi: 10.1186/s40168-015-0069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton HJ, Ang DK, van Driel IR, Hartland EL. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev. 2010;23:274–298. doi: 10.1128/CMR.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton IL, Sheehan KB, Lee FJ, Horton MA, Hicks RD. Invertebrate systems for hypothesis-driven microbiome research. Microbiome Science and Medicine. 2013:1. [Google Scholar]
- Oka T, Nishimura Y, Zang L, Hirano M, Shimada Y, Wang Z, Umemoto N, Kuroyanagi J, Nishimura N, Tanaka T. Diet-induced obesity in zebrafish shares common pathophysiological pathways with mammalian obesity. BMC Physiol. 2010;10:1. doi: 10.1186/1472-6793-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara R, Simpson GL, Solymos P, Stevens MHH, Wagner H. Community ecology package, version. 2. 2013. Package ‘vegan’. [Google Scholar]
- Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Hylemon PB. A potential role for resistant starch fermentation in modulating colonic bacterial metabolism and colon cancer risk. Cancer Biol Ther. 2006;5:273–274. doi: 10.4161/cbt.5.3.2728. [DOI] [PubMed] [Google Scholar]
- Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Current opinion in gastroenterology. 2014;30:332. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, Rawls JF. Evidence for a core gut microbiota in the zebrafish. The ISME journal. 2011;5:1595–1608. doi: 10.1038/ismej.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler KC, Rawls JF, Farber SA. Getting the inside tract: new frontiers in zebrafish digestive system biology. Zebrafish. 2013;10:129–131. doi: 10.1089/zeb.2013.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salma U, Miah A, Tareq K, Maki T, Tsujii H. Effect of dietary Rhodobacter capsulatus on egg-yolk cholesterol and laying hen performance. Poult Sci. 2007;86:714–719. doi: 10.1093/ps/86.4.714. [DOI] [PubMed] [Google Scholar]
- Samsel A, Seneff S. Glyphosate, pathways to modern diseases II: Celiac sprue and gluten intolerance. Interdiscip Toxicol. 2013;6:159–184. doi: 10.2478/intox-2013-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson J, Hermon-Taylor J. Mycobacterial diseases of the gut: some impact from molecular biology. Gut. 1992;33:145–147. doi: 10.1136/gut.33.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz Y. Microbiome and Gluten. Ann Nutr Metab. 2015;67:28–41. doi: 10.1159/000440991. [DOI] [PubMed] [Google Scholar]
- Sapone A, Bai JC, Ciacci C, Dolinsek J, Green PH, Hadjivassiliou M, Kaukinen K, Rostami K, Sanders DS, Schumann M. Spectrum of gluten-related disorders: consensus on new nomenclature and classification. BMC Med. 2012;10:13. doi: 10.1186/1741-7015-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarese M, Tousignant KD, Uno J. The effect of wheat products on the colonization of the microbiota in the intestines of zebrafish. The FASEB Journal. 2014;28:902. [Google Scholar]
- Shannon CE, Weaver W, Blahut RE, Hajek B. The mathematical theory of communication 1964 [Google Scholar]
- Siccardi AJ, III, Garris HW, Jones WT, Moseley DB, D’Abramo LR, Watts SA. Growth and survival of zebrafish (Danio rerio) fed different commercial and laboratory diets. Zebrafish. 2009;6:275–280. doi: 10.1089/zeb.2008.0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, Tjota MY, Seo GY, Cao S, Theriault BR. Commensal bacteria protect against food allergen sensitization. Proceedings of the National Academy of Sciences. 2014;111:13145–13150. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavana O, Gu W. The Hunger Games: p53 regulates metabolism upon serine starvation. Cell Metab. 2013;17:159–161. doi: 10.1016/j.cmet.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson T, Dennis M, Higgins L, Lee A, Sharrett M. Gluten-free diet survey: are Americans with coeliac disease consuming recommended amounts of fibre, iron, calcium and grain foods? J Hum Nutr Diet. 2005;18:163–169. doi: 10.1111/j.1365-277X.2005.00607.x. [DOI] [PubMed] [Google Scholar]
- Tojo R, Suárez A, Clemente MG, de los Reyes-Gavilán CG, Margolles A, Gueimonde M, Ruas-Madiedo P. Intestinal microbiota in health and disease: role of bifidobacteria in gut homeostasis. World J Gastroenterol. 2014;20:15163–15176. doi: 10.3748/wjg.v20.i41.15163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya C, Sakata T, Sugita H. Novel ecological niche of Cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Lett Appl Microbiol. 2008;46:43–48. doi: 10.1111/j.1472-765X.2007.02258.x. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umu ÖC, Frank JA, Fangel JU, Oostindjer M, da Silva CS, Bolhuis EJ, Bosch G, Willats WG, Pope PB, Diep DB. Resistant starch diet induces change in the swine microbiome and a predominance of beneficial bacterial populations. Microbiome. 2015;3:1. doi: 10.1186/s40168-015-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valko M, Morris H, Mazur M, Rapta P, Bilton RF. Oxygen free radical generating mechanisms in the colon: do the semiquinones of vitamin K play a role in the aetiology of colon cancer? Biochimica et Biophysica Acta (BBA)-General Subjects. 2001;1527:161–166. doi: 10.1016/s0304-4165(01)00163-5. [DOI] [PubMed] [Google Scholar]
- Van Kessel M, Dutilh BE, Neveling K, Kwint MP, Veltman JA, Flik G, Jetten MS, Klaren PH, Op den Camp H. Pyrosequencing of 16S rRNA gene amplicons to study the microbiota in the gastrointestinal tract of carp (Cyprinus carpio L.) AMB express. 2011;1:41. doi: 10.1186/2191-0855-1-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuoristo M, Tilvis R, Miettinen T. Serum plant sterols and lathosterol related to cholesterol absorption in coeliac disease. Clin Chim Acta. 1988;174:213–224. doi: 10.1016/0009-8981(88)90388-9. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Du J, Lam SH, Mathavan S, Matsudaira P, Gong Z. Morphological and molecular evidence for functional organization along the rostrocaudal axis of the adult zebrafish intestine. BMC Genomics. 2010;11:392. doi: 10.1186/1471-2164-11-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts SA, Lawrence C, Powell M, D’Abramo LR. The Vital Relationship Between Nutrition and Health in Zebrafish. Zebrafish. 2016 doi: 10.1089/zeb.2016.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts SA, Powell M, D’Abramo LR. Fundamental Approaches to the Study of Zebrafish Nutrition. ILAR J. 2012;53:144–160. doi: 10.1093/ilar.53.2.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Chin FY, Chao Y, Zhang T. Phylogeny-structured carbohydrate metabolism across microbiomes collected from different units in wastewater treatment process. Biotechnol Biofuels. 2015;8:172. doi: 10.1186/s13068-015-0348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Li S, Yang L, Huang P, Li W, Wang S, Zhao G, Zhang M, Pang X, Yan Z. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nature communications. 2013:4. doi: 10.1038/ncomms3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.