Abstract
Objectives
Circular RNAs (circRNAs) have been proven to function as competing endogenous RNAs to interact with microRNAs (miRNAs) and influence the expression of miRNA target mRNAs. In this study, we investigated whether circRNAs could act as competing endogenous RNAs to regulate the pathological process of intervertebral disc degeneration (IVDD).
Methods
The role and mechanism of a circRNA, circVMA21, in IVDD were explored in nucleus pulposus (NP) cells and degenerative NP tissues from patients and rat models. The interaction between circVMA21 and miR-200c as well as the target mRNA, X linked inhibitor-of-apoptosis protein (XIAP), was examined.
Results
The decreased expression of XIAP in the inflammatory cytokines-treated NP cells and the degenerative NP tissues was directly associated with excessive apoptosis and imbalance between anabolic and catabolic factors of extracellular matrix. miR-200c regulated NP cell viability and functions through inhibiting XIAP. circVMA21 acted as a sponge of miR-200c and functioned in NP cells through targeting miR-200c and XIAP. Intradiscal injection of circVMA21 alleviated IVDD in the rat model.
Conclusions
CircVMA21 could alleviate inflammatory cytokines-induced NP cell apoptosis and imbalance between anabolism and catabolism of extracellular matrix through miR-200c-XIAP pathway. It provides a potentially effective therapeutic strategy for IVDD.
Keywords: cytokines, osteoarthritis, tnf-alpha, low back pain, inflammation
Introduction
It is well documented that low back pain is a common condition and the leading cause of disability globally.1 A widely recognised contributor to low back pain is intervertebral disc (IVD) degeneration, which is the major cause of a series of degenerative disc diseases. A disc is composed of the inner nucleus pulposus (NP) and surrounded by annulus fibrosus. Nucleus pulposus cells (NPCs) are the main type of cells residing in the NP and responsible for synthesising components of extracellular matrix (ECM) such as type II collagen (collagen II) and aggrecan. These proteins are the major functional compositions of the IVD to maintain disc height and confront diverse external mechanical compression. Multiple abnormal stimuli can increase the levels of inflammatory cytokines (eg, interleukin -1β (IL-1β) and tumour necrosis factor-α (TNF-α)) in the NP, and then induce an imbalance between anabolic and catabolic activities of NPCs, such as increased generation of catabolic factors (eg, matrix metalloproteinases (MMP) and a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS)) and inhibited expression of anabolic factors (eg, collagen II and aggrecan), as well as excessive NPC apoptosis. These adverse factors initiate or accelerate intervertebral disc degeneration (IVDD).2–6 Thus, it is necessary to find an effective way to inhibit NPC apoptosis, attenuate inflammatory response, and reverse the imbalance between the anabolism and catabolism within the NP microenvironment.
MicroRNAs (miRNAs) are small, single-stranded, non-coding RNA molecules that impede protein production by directly interacting with the 3’untranslated region (UTR)of the target mRNAs. miR-200c is often associated with various cancers because it exhibits tumour suppressive behaviour by blocking epithelial to mesenchymal transition of cancer cells.7 Moreover, recent studies have revealed its ability to induce cell apoptosis by targeting different mRNAs.8 9 In addition, miRNAs have gained considerable attention as regulators of gene expression and play important roles in the prevention and treatment of IVDD.10 11
Circular RNAs (circRNAs) are another type of RNAs that form loop structures without 5′-3′ polarities and polyadenylated tails. Most of the circRNAs are endogenous non-coding RNAs, conserved between different species and showed a higher degree of stability than linear mRNAs.12 13 They mainly arise from one or multiple exons by a backsplice mechanism. Although the specific biogenesis of circRNAs is not completely clear, many studies have found a reliable model of circularisation driven by paring between flanking intron sequences. For example, complementary Alu repetitive elements within the flanking introns prompt the formation of exon-derived circRNAs.14 15 Lately, some circRNAs have been proven to be enriched with miRNA binding sites, and function as competing endogenous RNAs (ceRNAs) to interact with miRNAs and influence the expression of target mRNAs.16–20 Until now, it remains unclear whether circRNAs can act as ceRNAs to regulate the viability and functions of NPCs, and the pathological process of IVDD. In this study, we identified a circRNA derived from vacuolar ATPase assembly factor (VMA21) gene (named circVMA21; also termed hsa_circ_0091702 in CircBase (http://www.circbase.org)) in the NP and systemically investigated its role in cell and animal model of IVDD.
Materials and methods
Construction of miRNA and circRNA vector
Vectors were constructed by amplified DNA fragments including the sequence of pre-miR-200c or third exon of VMA21 gene with flanking introns containing complementary Alu elements.
CircRNA inhibition
CircVMA21 was knocked down using specific small interfering RNAs (siRNAs) targeting the backsplice region.
Luciferase reporter assay
The 3'-UTR of X linked inhibitor-of-apoptosis protein (XIAP) gene or circVMA21 fragments were inserted into the luciferase vector. Cells were transfected with the vectors and miR-200c.
A rat model of IVDD
The circVMA21 vectors were injected into the IVDs of rat models. Radiographic and histological examinations were performed to evaluate the change in severity of IVDD (see online supplementary methods for details). All sequences are listed in online supplementary table S1.
annrheumdis-2017-212056supp001.docx (45.9KB, docx)
annrheumdis-2017-212056supp002.docx (18.7KB, docx)
Results
miR-200c was upregulated in degenerative NP tissues and involved in the regulation of NPC viability and functions
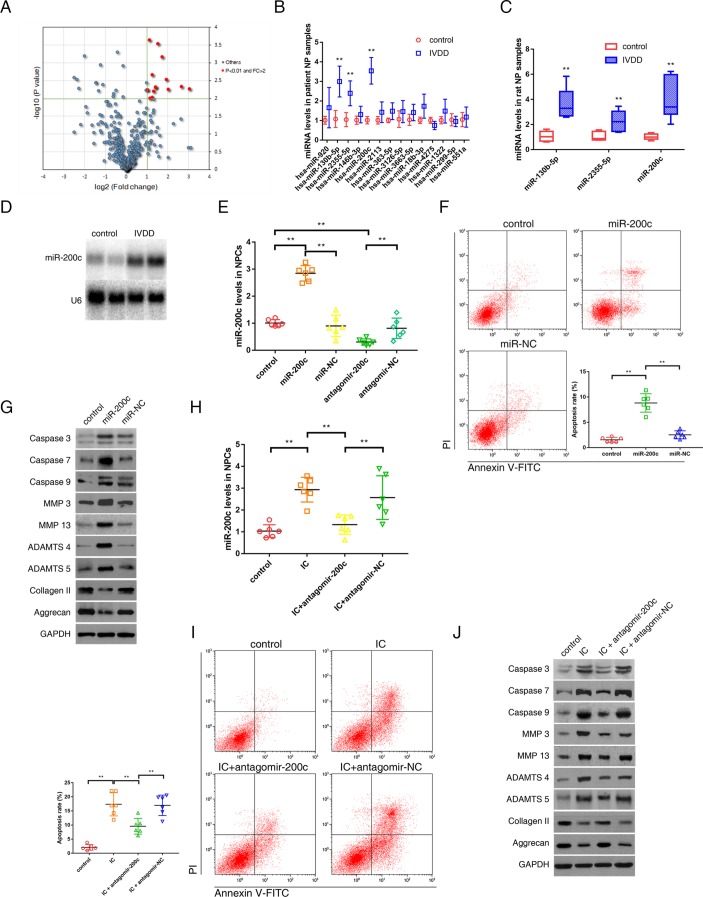
Differential expression of miRNAs in degenerative NP tissues was investigated using microarray data obtained from National Center for Biotechnology Information Gene Expression Omnibus database (GSE45856). Of 2672 miRNAs detected by miRNA microarray, 14 miRNAs were upregulated in the degenerative NP tissues compared with the controls when using the criteria of mean fold change >2.0 and P values <0.01 (figure 1A). These candidate miRNAs were chosen to analyse the validation using quantitative real-time reverse transcription-PCR (qRT-PCR) assay. Using the above-mentioned criteria, miR-200c, miR-130b-5p and miR-2355–5 p were observed to be significantly upregulated in the degenerative NP tissues compared with the controls, and miR-200c had the highest level of upregulation (figure 1B). The expression pattern of these miRNAs in the rat model of IVDD was consistent with that in the patient samples (figure 1C). Northern blot also confirmed the increase in miR-200c levels in the degenerative NP tissues (figure 1D). Therefore, miR-200c was selected for further analysis. The effects of miR-200c in NPCs were investigated. miR-200c-overexpressing cells (figure 1E) demonstrated increased percentage of apoptotic cells (figure 1F), elevated caspase activation, increased expression of MMP-3, MMP-13, ADAMTS-4 and ADAMTS-5, and decreased expression of collagen II and aggrecan (figure 1G). These results indicated the proapoptotic and procatabolic effect of miR-200c in NPCs. The treatment of TNF-α and IL-1β increased miR-200c levels in NPCs, which could be suppressed by miR-200c antagonist (figure 1H). The enhanced apoptosis and changes in ECM metabolism response to the treatment of TNF-α and IL-1β were markedly suppressed after miR-200c knockdown (figure 1I,J). Thus, the loss-of-function and gain-of-function experiments supported a critical role of miR-200c in negatively regulating NPC survival and functions.
Figure 1.
miR-200c was upregulated in the degenerative NP tissues and involved in the regulation of NPC viability and functions. (A) Differential upregulation of miRNAs detected by microarray in degenerative NP tissues compared with controls. Volcano plots were constructed using fold-change values and P values. The vertical green line corresponds to 2.0-fold upregulation between degenerative samples and controls, and the horizontal green line represents a P value of 0.01. The red point in the plot represents the differentially upregulated miRNAs with statistical significance. (B) qRT-PCR analysis confirmed the upregulated miRNAs in the degenerative NP samples from patients with IVDD. n=12; **P<0.01 compared with the controls. (C) qRT-PCR analysis confirmed the upregulated miRNAs in the degenerative NP samples from the rat model of IVDD. n=8; **P<0.01 compared with the controls. (D) Representative northern blots showing miR-200c levels in the NP samples from patients with or without IVDD. (E) NPCs were transfected with miR-200c, miR-negative control (NC), antagomir-200c or antagomir-NC. miR-200c levels in NPCs were analysed by qRT-PCR. **P<0.01. (F) Representative dot plots of apoptosis flow cytometry detection were shown after Annexin V-FITC/propidium iodide (PI) dual staining. The transfection of miR-200c increased apoptosis rate of NPCs. **P<0.01. (G) Western blot analysed protein expression of apoptotic effector caspases (caspase-3, caspase-7 and caspase-9), catabolic enzymes (MMP-3, MMP-13, ADAMTS-4 and ADAMTS-5) and extracellular matrix (ECM) compositions (collagen II, aggrecan) in NPCs after transfection of miR-200c. (H) NPCs were transfected with miR-200c antagonist or its NC, and then treated with inflammatory cytokines (IC; interleukin 1β plus tumour necrosis factor α). qRT-PCR showed increased miR-200c levels in NPCs treated with IC, which could be converted by transfection of miR-200c antagonist. **P<0.01. (I) Representative dot plots of apoptosis flow cytometry detection were shown after Annexin V-FITC/PI dual staining. The knockdown of miR-200c inhibited apoptosis induced by IC in NPCs. **P<0.01. (J) Western blot analysis showed that miR-200c knockdown attenuated the apoptotic and catabolic response and reversed the decreased expression of ECM compositions induced by IC treatment in NPCs. ADAMTS, a disintegrin and metalloproteinase with thrombospondin motifs; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IVDD, intervertebral disc degeneration; miRNA, microRNAs; MMP, matrix metalloproteinases; NP, nucleus pulposus; NPC, nucleus pulposus cells; qRT-PCR, quantitative real-time reverse transcription-PCR.
miR-200c regulated NPC viability and functions through inhibiting its target, XIAP
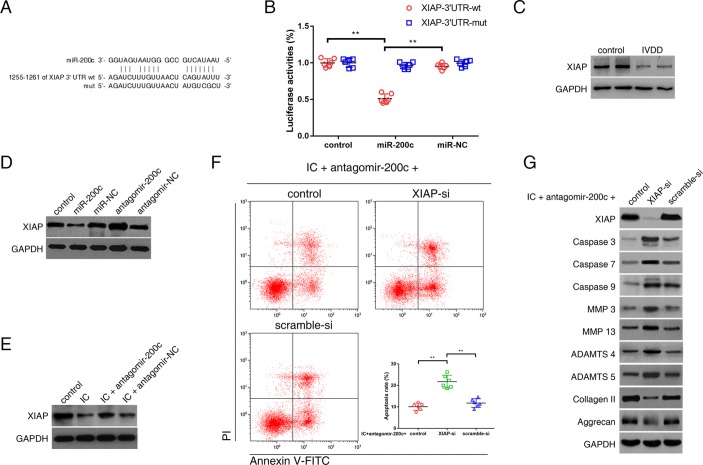
As predicted by bioinformatic programs, XIAP, a well-known regulator of apoptotic pathway, is a potential target of miR-200c (figure 2A). Gene ontology (GO) analysis revealed a correlation between the upregulated miRNAs in the degenerative NP tissues and the regulation of apoptotic signalling pathway. The luciferase signal of the wild-type XIAP reporter was suppressed by miR-200c, whereas introduction of mutations abolished the inhibitory effect of miR-200c (figure 2B). A decrease in XIAP expression was observed in the degenerative NP tissues compared with the controls (figure 2C). Elevated levels of miR-200c reduced XIAP expression, whereas knockdown of endogenous miR-200c increased XIAP expression in NPCs (figure 2D, online supplementary figure S1A). The levels of XIAP pre-mRNA were not changed in the degenerative NP tissues or miR-200c-overexpressing NPCs (online supplementary figure S1B,C). The treatment of TNF-α and IL-1β induced a decrease in the levels of XIAP protein in NPCs, while the miR-200c antagonist attenuated this decrease (figure 2E). Further, we investigated whether miR-200c and XIAP were functionally related in NPCs. The results showed that XIAP knockdown induced NPC apoptosis, exacerbated catabolic response and reduced expression of ECM compositions (online supplementary figure S1D,E). Furthermore, the absence of XIAP markedly counteracted the effects of miR-200c antagonist in the NPCs treated with TNF-α and IL-1β (figure 2F,G), indicating that miR-200c exerted its functions through XIAP.
Figure 2.
miR-200c-regulated NPC viability and functions through inhibiting its target mRNA, X chromosome-linked inhibitor-of-apoptosis protein (XIAP). (A) 3′-UTR region of XIAP mRNA was found to harbour a binding site for miR-200c. (B) NPCs were transfected with miR-200c or miR-negative control (NC), and then transfected with the luciferase constructs of the wild-type XIAP-3′UTR (3′UTR-wt) or the mutated XIAP-3′UTR (3′UTR-mut). Luciferase reporter assay found that miR-200c exclusively decreased luciferase activity of the wild-type reporter plasmids. n=6; **P<0.01. (C) Western blot analysis revealed lower expression of XIAP in the degenerative NP tissues compared with the controls. (D) NPCs were transfected with miR-200c, miR-NC, antagomir-200c or antagomir-NC. Western blot analysis showed that the expression of XIAP was suppressed by miR-200c upregulation and elevated by miR-200c knockdown. (E) NPCs were transfected with antagomir-200c or its NC, and then treated with inflammatory cytokines (IC; interleukin 1β and tumour necrosis factor-α). Western blot analysis showed decreased XIAP expression in NPCs treated with IC, which could be alleviated by transfection of miR-200c antagonist. (F, G) NPCs were cotransfected with antagomir-200c and XIAP siRNA (XIAP-si) or scramble siRNA (scramble-si), and then exposed to IC. (F) Representative dot plots of apoptosis flow cytometry detection were shown after Annexin V-FITC/propidium iodide (PI) dual staining. The knockdown of XIAP attenuated the inhibitory effects of miR-200c antagonist on apoptosis induced by IC in NPCs. **P<0.01. (G) Western blot analysed protein expression of apoptotic effector caspases (caspase-3, caspase-7 and caspase-9), catabolic enzymes (MMP-3, MMP-13, ADAMTS-4 and ADAMTS-5) and extracellular matrix compositions (collagen II, aggrecan) in NPCs. The knockdown of XIAP interfered with the effects of miR-200c antagonist on the expression of these functional proteins in IC-treated NPCs. ADAMTS, a disintegrin and metalloproteinase with thrombospondin motifs; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IVDD, intervertebral disc degeneration; MMP, matrix metalloproteinases; NP, nucleus pulposus; NPC, nucleus pulposus cell; UTR, untranslated region.
annrheumdis-2017-212056supp003.jpg (259.3KB, jpg)
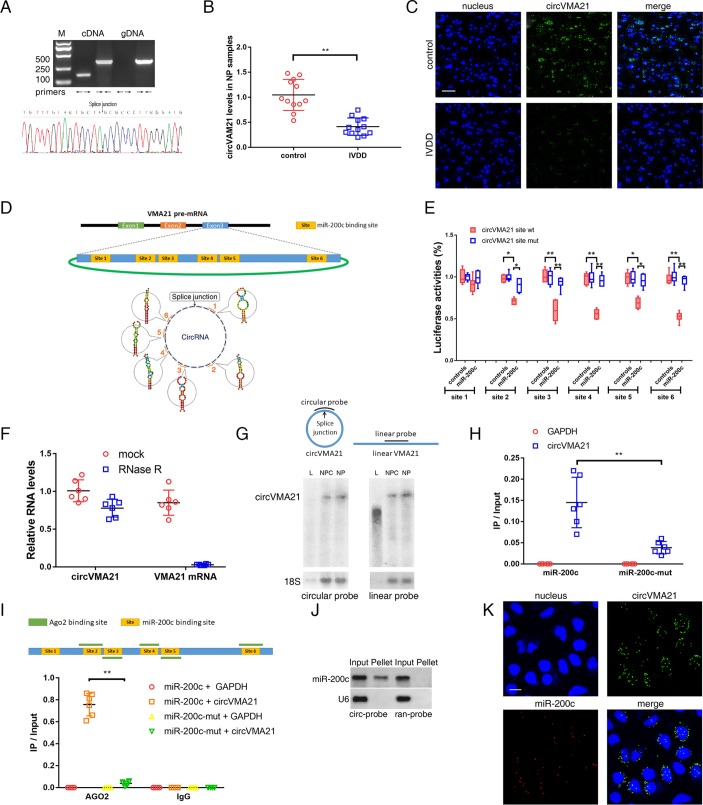
CCircVMA21 acted as a sponge of miR-200c. miR-200c is predicted to have binding sites for several circRNAs by starBase. We screened the top 20 predicted circRNAs according to their clipreadNum scores. The existence of the selected circRNAs in the NP samples was detected using specific divergent primers by qRT-PCR analysis. Predicted splice junction of circVMA21 (online supplementary table S2) was validated in the NP tissues (figure 3A, upper). The amplified product using divergent primers was confirmed in accordance with the sequence of circVMA21 by sequencing (figure 3A, lower). CircVMA21 levels were markedly reduced in the degenerative NP tissues compared with the controls by qRT-PCR analysis (figure 3B). This result was also confirmed by RNA fluorescence in situ hybridisation (FISH) (figure 3C, online supplementary figure S2A). The level of circVMA21 was positively correlated with XIAP level in the degenerative NP tissues (online supplementary figure S2B). To test whether circVMA21 can be bounded by miR-200c, we compared the sequence of circVMA21 with that of miR-200c using RNAhybrid and noticed that circVMA21 contains six putative target sites of miR-200c (figure 3D, online supplementary table S3). Of them, five sites were validated by the luciferase assay (figure 3E). This circRNA was abundant and resistant to RNase R treatment in contrast to mRNA (figure 3F). Northern blot analysis showed that linear VMA21 was detectable by a linear probe but not a circular probe within the splice sites (figure 3G). Pull-down assay revealed a more enrichment of circVMA21 in the miR-200c-captured fraction compared with the introduction of miR-200c mutation that disrupted the binding site of miR-200c for circVMA21 (figure 3H). CircVMA21 is derived from the last exon of VMA21 gene mainly transcribing the 3’UTR of mRNA. It has been found to own 69 binding sites of Argonaute 2 (AGO2) protein as shown in the CircInteractome (online supplementary table S4). Crosslinking and immunoprecipitation (CLIP) sequence revealed at least 30 AGO2-bound regions located in the 3’UTR of VMA21 mRNA (online supplementary table S5). Of them, five regions are overlapped with the binding sites of miR-200c (figure 3I, upper). AGO2 immunoprecipitation found that endogenous circVMA21 pulled down with AGO2 was enriched in NPCs transfected with miR-200c but not its mutant (figure 3I, lower), indicating that miR-200c facilitated the association between AGO2 and circVMA21. Northern blot analysis revealed that circVMA21 could in reverse pull down miR-200c (figure 3J). RNA FISH found colocalisation of circVMA21 and miR-200c in the cytoplasm of NPCs (figure 3K, online supplementary figure S2C). All things considered, our results indicated that circVMA21 was able to directly bind to miR-200c in NPCs.
Figure 3.
circVMA21 acted as a sponge of miR-200c. (A) Agarose gel electrophoresis found that divergent primers (←→) amplified circVMA21 in complementary DNA (cDNA) but not genomic DNA (gDNA) (upper). The amplified product of specific divergent primers was confirmed in accordance with the sequence of circVMA21 by sequencing (lower). (B) qRT-PCR analysis detected circVMA21 levels in the NP samples from patients with or without IVDD. n=12; **P<0.01. (C) The expression of circVMA21 was detected in the NP samples from patients with or without IVDD by RNA fluorescence in situ hybridisation (FISH). circVMA21 probe was labelled with Alexa 488. Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). Scale bar=50 µm. (D) circVMA21 is transcribed from the third exon of the VMA21 gene and contains six putative binding sites complementary to miR-200c. (E) NPCs were transfected with miR-200c and luciferase constructs of circVMA21 containing wild-type putative miR-200c binding sites (circVMA21 site wt) or mutated sites (circVMA21 site mut). n=6; *P<0.05, **P<0.01. (F) qRT-PCR analysis for the abundance of circVMA21 and VMA21 mRNA in NPCs with or without RNase R treatment. The amounts were normalised to the value of circVMA21 measured in the mock treatment. n=6. (G) Northern blot analysis showed that linear VMA21 was detectable by a linear but not circular probe. L, linear VMA21 transcribed in vitro; NPC, total RNAs extracted from NPCs; NP, total RNAs extracted from NP tissue samples; circular probe, probe within splice site; linear probe, head-to-tail probe. (H) The biotinylated miR-200c or its mutant (miR-200c-mut) was transfected into NPCs. The RNA levels of circVMA21 and GAPDH were quantified by qRT-PCR analysis, and the relative ratios of immunoprecipitate (IP) to input were plotted. **P<0.01. (I) CLIP sequence revealed five AGO2-bound regions overlapped with the binding sites of miR-200c within the circVMA21 sequence (upper). AGO2 RNA immunoprecipitation in NPCs transfected with miR-200c or its mutant. The levels of circVMA21 and GAPDH were quantified by qRT-PCR analysis, and the relative ratios of IP to input were plotted. **P<0.01 (lower). (J) miR-200c was pulled down by the circular probe for circVMA21 (circ-probe) but not random probe (ran-probe). The levels of miR-200c were detected by northern blot. Input, 20% samples were loaded; Pellet, all samples were loaded. (K) RNA FISH for colocalisation of circVMA21 and miR-200c in cytoplasm of NPCs. circVMA21 and miR-200c probes were labelled with Alexa 488 and Cy-5, respectively. Nuclei were stained with DAPI. Scale bar=10 µm. AGO2, Argonaute 2; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IVDD, intervertebral disc degeneration; NP, nucleus pulposus; NPC, nucleus pulposus cells; qRT-PCR, quantitative real-time reverse transcription-PCR.
annrheumdis-2017-212056supp004.docx (15.1KB, docx)
annrheumdis-2017-212056supp005.jpg (757.6KB, jpg)
annrheumdis-2017-212056supp006.docx (16.5KB, docx)
annrheumdis-2017-212056supp007.docx (19.4KB, docx)
annrheumdis-2017-212056supp008.docx (15.1KB, docx)
circVMA21 functioned in NPCs through targeting miR-200c and XIAP
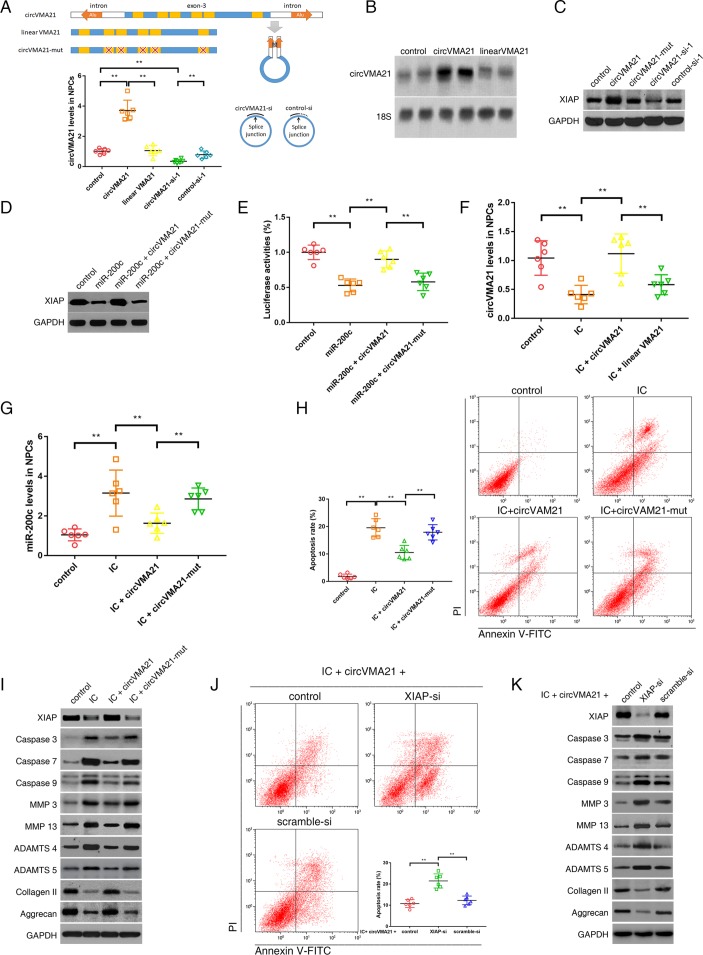
NPCs were infected by constructed adenovirus harbouring circVMA21, linear VMA21, circVMA21 siRNA or control siRNA. The results of qRT-PCR showed that the infection of adenovirus circVMA21 resulted in an overexpression of circVMA21 in the NPCs and that siRNA depressed the expression of circVMA21 (figure 4A). Knockdown of VMA21 mRNA did not affect circVMA21 levels (online supplementary figure S3A). Likewise, circVMA21 siRNA had no effect on mRNA levels (online supplementary figure S3B). Northern blot analysis also confirmed the elevated amount of circVMA21 after the intake of exogenous circVMA21 (figure 4B). Then, the effect of circVMA21 on XIAP expression was detected by western blot assay. Upregulation of circVMA21 increased the expression of XIAP, whereas circVMA21 knockdown induced a decrease in the XIAP expression (figure 4C). Similar changes were observed from a well-known target of miR-200c, zinc finger E-box binding homeobox transcription factor 1 (ZEB1; online supplementary figure S3C). Moreover, the upregulation of circVMA21 counteracted the inhibitory effect of miR-200c on XIAP expression (figure 4D) and activity (figure 4E). Taken together, these results suggested that circVMA21 acted as a functional sponge of miR-200c to regulate XIAP expression and activity. Next, we investigated the function of circVMA21 in the TNF-α-treated and IL-1β-treated NPCs. After the treatment of the inflammatory cytokines, qRT-PCR analysis found a decrease in circVMA21 levels and an increase in miR-200c levels in NPCs, both of which could be reversed by the upregulation of circVMA21 (figure 4F,G). As a consequence, the enforced expression of circVMA21 restrained the apoptotic and catabolic effects of these cytokines, and promoted the expression of collagen II and aggrecan (figure 4H,I). Additionally, we explored whether XIAP was the downstream mediator of circVMA21 in the inflammatory cytokines-treated NPCs. We cotransfected circVMA21 and XIAP siRNA into NPCs, and observed that the positive effects of circVMA21 on NPC vitality and functions were attenuated in the absence of XIAP (figure 4J,K). Collectively, these data indicated that circVMA21 functioned in NPCs through modulating miR-200c and XIAP.
Figure 4.
circVMA21 functioned in NPCs through targeting miR-200c and XIAP. (A) The third exon of VMA21 gene along with approximate 1 kb flanking intron sequences containing complementary Alu elements was amplified to construct circVMA21 vector. The exon with and without mutation was used as controls. NPCs were transfected with circVMA21, linear VMA21, circVMA21 siRNA-1 (circVMA21-si-1) or control siRNA-1 (control-si-1), and circVMA21 levels were analysed by qRT-PCR. **P<0.01 (lower). (B) NPCs were transfected with circVMA21 or linear VMA21 for northern blot analysis, and the blots were probed against circVMA21 with 18S ribosomal RNA as an internal control. (C) NPCs were transfected with circVMA21, its mutation (circVMA21-mut), circVMA21-si or scramble-si. XIAP expression was analysed by western blot assay. The expression of XIAP was enhanced after circVMA21 upregulation and reduced after circVMA21 knockdown. (D) NPCs were cotransfected with miR-200c and circVMA21 or circVMA21-mut. Western blot assay showed that circVMA21 blocked the inhibitory effect of miR-200c on XIAP expression. (E) NPCs were cotransfected with XIAP 3′UTR luciferase construct, miR-200c and circVMA21 or circVMA21-mut. Luciferase assay showed that circVMA21 blocked the inhibitory effects of miR-200c on XIAP activity. **P<0.01. (F,G,H,I) NPCs were transfected with circVMA21 or a control (linear VMA21 or circVMA21-mut), and then treated with inflammatory cytokines (IC; interleukin 1β and tumour necrosis factor-α). (F) qRT-PCR showed a decrease in circVMA21 expression in NPCs treated with IC, which could be converted by transfection of circVMA21. **P<0.01. (G) qRT-PCR showed an increase in miR-200c levels in NPCs treated with IC, which could be downregulated by transfection of circVMA21. **P<0.01. (H) Representative dot plots of apoptosis flow cytometry detection were shown after Annexin V-FITC/propidium iodide (PI) dual staining. The transfection of circVMA21 inhibited apoptosis induced by IC in NPCs. **P<0.01. (I) Western blot analysed expression of XIAP, apoptotic effector caspases (caspase-3, caspase-7 and caspase-9), catabolic enzymes (MMP-3, MMP-13, ADAMTS-4 and ADAMTS-5) and extracellular matrix (ECM) compositions (collagen II, aggrecan) in NPCs. The transfection of circVMA21 attenuated the apoptotic and catabolic response, and rescued the reduced expression of ECM compositions induced by IC treatment. (J,K) NPCs were cotransfected with circVMA21 and XIAP siRNA (XIAP-si) or scramble siRNA (scramble-si), and then exposed to IC. (J) Representative dot plots of apoptosis flow cytometry detection were shown after Annexin V-FITC/PI dual staining. The inhibitory effect of circVMA21 on NPC apoptosis was attenuated after the knockdown of XIAP. **P<0.01. (K) Western blot analysed the expression of XIAP, apoptotic effector caspases (caspase-3, caspase-7 and caspase-9), catabolic enzymes (MMP-3, MMP-13, ADAMTS-4 and ADAMTS-5) and ECM compositions (collagen II, aggrecan) in NPCs. The knockdown of XIAP impaired the protective effect of circVMA21 on NPC functions. ADAMTS, a disintegrin and metalloproteinase with thrombospondin motifs; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; MMP, matrix metalloproteinases; NPC, nucleus pulposus cells; qRT-PCR, quantitative real-time reverse transcription-PCR; XIAP, X linked inhibitor-of-apoptosis protein.
annrheumdis-2017-212056supp009.jpg (186.6KB, jpg)
Intradiscal injection of circVMA21 alleviated IVDD in a rat model
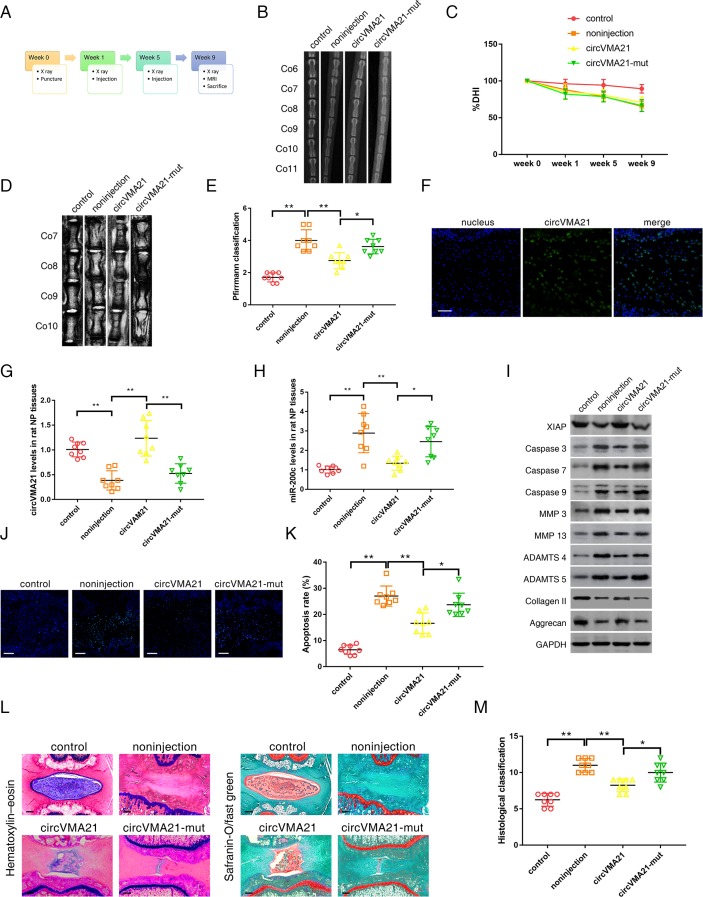
We successfully established a rat model of IVDD by the needle puncture (figure 5A). At 1 and 5 weeks after the puncture, adenoviral human circVMA21 were injected into the punctured IVDs using a 33-gauge fine needle. X-rays obtained at time 1, 5 and 9 weeks demonstrated progressive disc space narrowing over time in all IVD punctured groups. At each time point after injection, no significant difference in the percentage of disc height index (%) was noted between the circVMA21 group and the non-injection or circVMA21-mut group (figure 5B,C, online supplementary figure S4A). At 9 weeks after injection, the MRI degeneration score of the IVDs was significantly lower in the circVMA21 group than in the non-injection group (figure 5D,E, online supplementary figure S4B). In vivo RNA FISH found circVMA21 located in the NP region (figure 5F, online supplementary figure S4C). After the injection of adenoviral circVMA21, the levels of circVMA21 in the degenerative NP tissues were remarkably elevated, while the levels of miR-200c were decreased (figure 5G,H). The injection of adenoviral circVMA21 alleviated the degenerative changes of the NP, such as enhanced apoptotic and catabolic response, and reduced ECM compositions in the rat model of IVDD (figure 5I,J,K, online supplementary figure S4D,E). The histological score was significantly higher in the non-injection group than in the circVMA21 group at 9 weeks (figure 5L,M). Taken together, these results revealed the positive effects of elevated circVMA21 levels on attenuating NPC apoptosis, inhibiting ECM catabolism, promoting anabolism in the NP and resultantly alleviating IVDD in vivo.
Figure 5.
CircVMA21 alleviated IVDD in vivo. (A) A flow diagram of the experiments in vivo. A total of 32 rats were randomly divided into four groups: non-puncture group (control), non-injection with puncture group (non-injection), circVMA21 injection with puncture group (circVMA21), and circVMA21 mutant injection with puncture group (circVMA21-mut). (B) Radiographs of the indicated groups were obtained 9 weeks after needle puncture. Co6/7, Co8/9 and Co10/11 were punctured with Co7/8 and Co9/10 left intact. (C) Changes in disc height index (DHI) of the indicated groups after needle puncture. The DHI was measured at weeks 0, 1, 5 and 9 time point. A significant decrease of the %DHI was observed in all puncture groups at 1 week after surgery (P<0.01). At each time point after puncture, a significant decrease of %DHI was noted in all puncture groups compared with the control group (P<0.01). No significant difference was found in the %DHI between all puncture groups. (D) MRIs of the indicated groups were obtained 9 weeks after needle puncture. Co6/7, Co8/9 and Co10/11 were punctured with Co7/8 and Co9/10 left intact. (E) The MRI grade in the indicated groups at 9 weeks after needle puncture. The degree of disc degeneration by MRI grade was significantly lower in the circVMA21 group than in the non-injection group. *P<0.05, **P<0.01. (F) In vivo RNA fluorescence in situ hybridisation found circVMA21 located in the NP region. Blue fluorescence (4,6-diamidino-2-phenylindole, DAPI) indicating cell nucleus; green fluorescence (Alexa 488) indicating circVMA21. Scale bar=100 µm. (G) qRT-PCR showed that the decreased levels of circVMA21 in the punctured IVDs were rescued by the injection of circVMA21. **P<0.01. (H) qRT-PCR showed that the increased levels of miR-200c in the punctured IVDs were depressed by the injection of circVMA21. *P<0.05, **P<0.01. (I) Western blot analysed the expression of XIAP, apoptotic effector caspases (caspase-3, caspase-7 and caspase-9), catabolic enzymes (MMP-3, MMP-13, ADAMTS-4 and ADAMTS-5) and extracellular matrix (ECM) compositions (collagen II, aggrecan) in the rat NP tissues. The injection of circVMA21 alleviated the degenerative changes of the NP such as enhanced apoptotic and catabolic response, and reduced the expression of ECM compositions in the rat model of IVDD. (J) Terminal deoxynucleotidyl transferase-mediated dUTP nick end labelling (TUNEL) staining of the IVDs in the indicated groups at 9 weeks after needle puncture. Blue fluorescence (DAPI) indicating total cells; green fluorescence (fluorescein isothiocyanate) indicating TUNEL positive cells. Scale bar=100 µm. (K) A significant decrease in the apoptosis rate was noted in the circVMA21 group compared with the non-injection group. *P<0.05, **P<0.01. (L) H&E (left) and safranin-O/fast green (right) staining of the IVDs in the indicated groups at 9 weeks after needle puncture. Scale bar=100 µm. (M) A significant decrease in the grade of IVDD was noted in the circVMA21 group compared with the non-injection group. *P<0.05, **P<0.01. ADAMTS, a disintegrin and metalloproteinase with thrombospondin motifs; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IVD, intervertebral disc; IVDD, intervertebral disc degeneration; MMP, matrix metalloproteinases; NP, nucleus pulposus; qRT-PCR, quantitative real-time reverse transcription-PCR; XIAP, X linked inhibitor-of-apoptosis protein.
annrheumdis-2017-212056supp010.jpg (2.1MB, jpg)
Discussion
Multiple lines of evidence have shown that certain miRNAs could target distinct genes related to the development and progression of IVDD, and play roles in regulating the vitality and functions of NPCs.10 11 In this study, we first identified miR-200c as a key miRNA involved in IVDD. miR-200c participated in the proapoptotic response and imbalanced expression of anabolic and catabolic factors. We then investigated the potential effects of circRNAs on the regulatory functions of miR-200c in the NPCs. The results revealed that circVMA21 markedly decreased the activity of miR-200c by capturing it and suppressed its functions. Therefore, a mechanism was proposed in which circVMA21 sponged miR-200c to inhibit NPC apoptosis, promote ECM anabolism and suppress ECM catabolism, and consequently delayed the progression of IVDD.
CircRNAs are a type of widespread, tissue-specific and conserved endogenous non-coding RNAs in mammalian cells. Although the specific functions of most circRNAs still remain unclear, accumulating evidence has revealed a role of circRNAs as miRNA sponges.16–20 No free ends can render circRNAs to evade destabilisation and degradation mediated by miRNAs. Several recent studies have indicated the availability of circRNAs as miRNA sponges to take part in the occurrence and progression of various diseases.21–23 Nevertheless, there are still arguments against miRNA sponges being a major function of circRNAs because of their low amount or lack of reiterated miRNA binding sites.24 25 In the present study, qRT-PCR, northern blot and FISH assay revealed abundant circVMA21 in NPCs. VMA21 gene is located on the X chromosome and encodes an integral membrane protein to assist in the assembly of the V-ATPase.26 circVMA21 is generated by back splicing of the third exon of VMA21 gene. It mainly comprises 3’UTR of VMA21 mRNA, so its sequence is relatively conserved between human and rat (online supplementary table S6). The bioinformatics analysis found circVMA21 containing multiple target sites of miR-200c, which was validated by luciferase, pull-down, RNA binding protein immunoprecipitation (RIP and FISH analyses. Furthermore, the expression of miR-200c target mRNA, XIAP, was positively modulated by circVMA21. Thus, the binding sites of circVMA21 for miR-200c were proven effective.
annrheumdis-2017-212056supp011.docx (14.4KB, docx)
In this study, we selected TNF-α and IL-1β as the agents to induce a range of pathogenic responses in NPCs, because they have vital roles in the pathological process of IVDD.2 6 27 Our results were consistent with previous findings that the stimulation of NPCs with these cytokines caused a similar pattern of changes observed in patients with IVDD or animal models, including excessive NPC apoptosis, enhanced expression of the ECM-degrading enzymes (MMP-3, MMP-13, ADAMTS-4 and ADAMTS-5), and inhibited expression of the ECM proteins (collagen II and aggrecan).28–30 Deletion of XIAP remarkably impaired the antiapoptotic and anti-inflammatory ability of circVMA21 or miR-200c antagonist, confirming that XIAP was the direct target of circVMA21 and miR-200c to suppress the effects of TNF-α and IL-1β in NPCs. XIAP belongs to the inhibitor-of-apoptosis proteins (IAP) that represent a family of endogenous caspase inhibitors. Accumulated TNF-α and IL-1β are thought to activate the effector caspases to exert apoptotic effects in NPCs via the extrinsic and intrinsic pathways.4 5 Of note, XIAP is the only IAP that can bind and directly inhibit the activity of the three most important apoptosis effector caspases, caspase-3, caspase-7 and caspase-9.31 32 The absence or inhibition of XIAP increases the formation and subsequent activation of the death inducing signalling complex, and renders the majority of cells to be sensitised to death receptor-induced apoptosis, such as with TNF-α.33 34 Mehrkens et al 35 recently reported that notochordal cell-derived conditioned medium upregulates the genomic expression of XIAP and thus inhibits inflammatory cytokines-induced NPC apoptosis. Moreover, the blockage of caspase signalling attenuates IVDD by inhibiting NPC apoptosis and by regulating the expression of matrix metabolism enzymes. Yamada et al 36 find that caspase-3 knockdown reduces the production of matrix-degrading enzymes (MMP-3, MMP-13, ADAMTS-4 and ADAMTS-5) and increases the expression of proanabolic proteins (tissue inhibitor of metalloproteinases-1, collagen II and aggrecan). XIAP knockdown-induced activation of caspases could lead to consequent matrix metabolism imbalance as shown in the present study.
In addition to caspases inhibition, a growing body of evidence exists to support the modulatory role for XIAP in inflammation. Loss of XIAP facilitates the proinflammatory effect of TNF-α and causes severe sterile inflammation, which can be reduced by anti-TNF therapy.34 37 38 Meanwhile, XIAP suppresses inappropriate or excess IL-1β activity, while absence of XIAP promotes excessive IL-1β secretion in different cell types.39 40 During the process of IVDD, TNF-α and IL-1β are produced by both leucocytes and NPCs themselves.41 Therefore, XIAP seemed to function in NPCs by blocking the effects of exogenous TNF-α and IL-1β, and by inhibiting endogenous generation of these cytokines.
The age gap between the patients with and without IVDD may lead to a bias in this study. Nevertheless, it is an inevitable confounding factor when detecting the samples from human NP tissues because of the clinicopathological characteristics of IVDD. The results of in vitro and in vivo experiments would help eliminate the influence of age. In addition to ceRNA, there may exist other potential mechanisms of circVMA21 to regulate the process of IVDD, for example, through interacting with RNA binding proteins (RBPs) other than AGO2. RBPs CLIP shows that the 3’UTR of VMA21 mRNA can bind to abundant human antigen R (HuR), an extensively studied RBP with substantial regulatory effects on the stability and translation of multiple mRNAs. The binding of some circRNAs to HuR has been identified to prevent HuR binding to mRNAs and thus lower their translation.42 In addition, the mechanism of the decrease in circVMA21 levels during the degenerative process remains unclear. Further investigation is needed to completely understand the role of circVMA21 in IVDD.
In summary, circVMA21 could alleviate inflammatory cytokines-induced NPC apoptosis and imbalance between anabolism and catabolism of ECM through the miR-200c-XIAP pathway (figure 6). It provides a potentially effective therapeutic strategy for IVDD.
Figure 6.
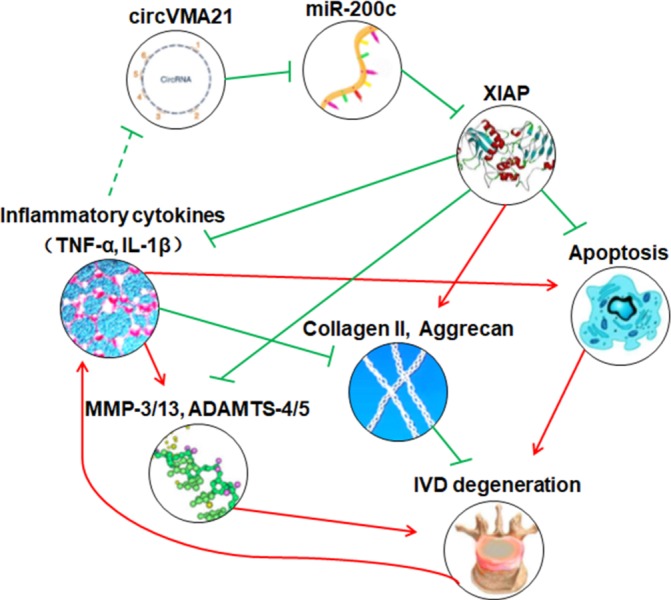
Schematic of the working hypothesis. The decreased expression of XIAP in the inflammatory cytokines-treated NPCs and the degenerative NP tissues is directly associated with excessive NPC apoptosis and imbalance between anabolism and catabolism of extracellular matrix. The treatment of circVMA21 could inhibit these adverse factors through binding miR-200c, and thus delay the progression of intervertebral disc degeneration. ADAMTS, a disintegrin and metalloproteinase with thrombospondin motifs; IL-1β, interleukin-1β; IVD, intervertebral disc; MMP, matrix metalloproteinases; NP, nucleus pulposus; NPC, nucleus pulposus cells; TNF-α, tumour necrosis factor-α; XIAP, X linked inhibitor-of-apoptosis protein.
Footnotes
XC and LZ contributed equally.
Handling editor: Tore K Kvien
Contributors: XC and JZ designed the experiments. XC, LZ, KZ, GZ, YH and XS performed the experiments and acquired the data. XC, LZ, CZ and HL analysed the data. XC, YML and JZ supervised the project and wrote the manuscript.
Funding: This work was supported by grants from the National Natural Science Foundation of China (81572168, 81401830), and the Industry-Academy-Research Cooperation Project of Shanghai Science and Technology Committee (13DZ1940504, 13DZ1940505), the Natural Science Foundation of Shanghai, the Department Integration Foundation (160065), and the Fundamental Research Program Funding of the Ninth People’s Hospital Affiliated to Shanghai Jiao Tong University School of Medicine.
Competing interests: None declared.
Patient consent: Obtained.
Ethics approval: Ethics Committee of Shanghai Ninth People’s Hospital.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. GBD 2015 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016;388:1545–602. 10.1016/S0140-6736(16)31678-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Risbud MV, Shapiro IM. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat Rev Rheumatol 2014;10:44–56. 10.1038/nrrheum.2013.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kepler CK, Markova DZ, Hilibrand AS, et al. Substance P stimulates production of inflammatory cytokines in human disc cells. Spine 2013;38:E1291–E1299. 10.1097/BRS.0b013e3182a42bc2 [DOI] [PubMed] [Google Scholar]
- 4. Ding F, Shao ZW, Xiong LM. Cell death in intervertebral disc degeneration. Apoptosis 2013;18:777–85. 10.1007/s10495-013-0839-1 [DOI] [PubMed] [Google Scholar]
- 5. Johnson ZI, Schoepflin ZR, Choi H, et al. Disc in flames: roles of TNF-α and IL-1β in intervertebral disc degeneration. Eur Cell Mater 2015;30:104–17. 10.22203/eCM.v030a08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dudek M, Yang N, Ruckshanthi JP, et al. The intervertebral disc contains intrinsic circadian clocks that are regulated by age and cytokines and linked to degeneration. Ann Rheum Dis 2017;76 10.1136/annrheumdis-2016-209428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kumar S, Nag A, Mandal CC. A Comprehensive Review on miR-200c, a promising cancer biomarker with therapeutic potential. Curr Drug Targets 2015;16:1381–403. 10.2174/1389450116666150325231419 [DOI] [PubMed] [Google Scholar]
- 8. Magenta A, Cencioni C, Fasanaro P, et al. miR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition. Cell Death Differ 2011;18:1628–39. 10.1038/cdd.2011.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schickel R, Park SM, Murmann AE, et al. miR-200c regulates induction of apoptosis through CD95 by targeting FAP-1. Mol Cell 2010;38:908–15. 10.1016/j.molcel.2010.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang HQ, Yu XD, Liu ZH, et al. Deregulated miR-155 promotes Fas-mediated apoptosis in human intervertebral disc degeneration by targeting FADD and caspase-3. J Pathol 2011;225:232–42. 10.1002/path.2931 [DOI] [PubMed] [Google Scholar]
- 11. Ji ML, Lu J, Shi PL, et al. Dysregulated miR-98 contributes to extracellular matrix degradation by targeting IL-6/STAT3 signaling pathway in human intervertebral disc degeneration. J Bone Miner Res 2016;31:900–9. 10.1002/jbmr.2753 [DOI] [PubMed] [Google Scholar]
- 12. Meng X, Li X, Zhang P, et al. Circular RNA: an emerging key player in RNA world. Brief Bioinform 2016:bbw045 10.1093/bib/bbw045 [DOI] [PubMed] [Google Scholar]
- 13. Guo Y, Luo F, Liu Q, et al. Regulatory non-coding RNAs in acute myocardial infarction. J Cell Mol Med 2017;21:1013–23. 10.1111/jcmm.13032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141–57. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang XO, Wang HB, Zhang Y, et al. Complementary sequence-mediated exon circularization. Cell 2014;159:134–47. 10.1016/j.cell.2014.09.001 [DOI] [PubMed] [Google Scholar]
- 16. Piwecka M, Glažar P, Hernandez-Miranda LR, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017;357:eaam8526 10.1126/science.aam8526 [DOI] [PubMed] [Google Scholar]
- 17. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333–8. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 18. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384–8. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 19. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016;7:11215 10.1038/ncomms11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res 2013;73:5609–12. 10.1158/0008-5472.CAN-13-1568 [DOI] [PubMed] [Google Scholar]
- 21. Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 2017;66:1151–64. 10.1002/hep.29270 [DOI] [PubMed] [Google Scholar]
- 22. Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016;37:2602–11. 10.1093/eurheartj/ehv713 [DOI] [PubMed] [Google Scholar]
- 23. Chen L, Zhang S, Wu J, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene 2017;36:4551–61. 10.1038/onc.2017.89 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24. Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014;15:409 10.1186/s13059-014-0409-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015;160:1125–34. 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- 26. Ramachandran N, Munteanu I, Wang P, et al. VMA21 deficiency prevents vacuolar ATPase assembly and causes autophagic vacuolar myopathy. Acta Neuropathol 2013;125:439–57. 10.1007/s00401-012-1073-6 [DOI] [PubMed] [Google Scholar]
- 27. Phillips KL, Cullen K, Chiverton N, et al. Potential roles of cytokines and chemokines in human intervertebral disc degeneration: interleukin-1 is a master regulator of catabolic processes. Osteoarthritis Cartilage 2015;23:1165–77. 10.1016/j.joca.2015.02.017 [DOI] [PubMed] [Google Scholar]
- 28. Hiyama A, Skubutyte R, Markova D, et al. Hypoxia activates the notch signaling pathway in cells of the intervertebral disc: implications in degenerative disc disease. Arthritis Rheum 2011;63:1355–64. 10.1002/art.30246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang J, Tian Y, Phillips KL, et al. Tumor necrosis factor α- and interleukin-1β-dependent induction of CCL3 expression by nucleus pulposus cells promotes macrophage migration through CCR1. Arthritis Rheum 2013;65:832–42. 10.1002/art.37819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang J, Markova D, Anderson DG, et al. TNF-α and IL-1β promote a disintegrin-like and metalloprotease with thrombospondin type I motif-5-mediated aggrecan degradation through syndecan-4 in intervertebral disc. J Biol Chem 2011;286:39738–49. 10.1074/jbc.M111.264549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Deveraux QL, Takahashi R, Salvesen GS, et al. X-linked IAP is a direct inhibitor of cell-death proteases. Nature 1997;388:300–4. 10.1038/40901 [DOI] [PubMed] [Google Scholar]
- 32. Eckelman BP, Salvesen GS. The human anti-apoptotic proteins cIAP1 and cIAP2 bind but do not inhibit caspases. J Biol Chem 2006;281:3254–60. 10.1074/jbc.M510863200 [DOI] [PubMed] [Google Scholar]
- 33. Vasilikos L, Spilgies LM, Knop J, et al. Regulating the balance between necroptosis, apoptosis and inflammation by inhibitors of apoptosis proteins. Immunol Cell Biol 2017;95:160–5. 10.1038/icb.2016.118 [DOI] [PubMed] [Google Scholar]
- 34. Wicki S, Gurzeler U, Wei-Lynn Wong W, et al. Loss of XIAP facilitates switch to TNFα-induced necroptosis in mouse neutrophils. Cell Death Dis 2016;7:e2422 10.1038/cddis.2016.311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mehrkens A, Matta A, Karim MZ, et al. Notochordal cell-derived conditioned medium protects human nucleus pulposus cells from stress-induced apoptosis. Spine J 2017;17:579–88. 10.1016/j.spinee.2017.01.003 [DOI] [PubMed] [Google Scholar]
- 36. Yamada K, Sudo H, Iwasaki K, et al. Caspase 3 silencing inhibits biomechanical overload-induced intervertebral disk degeneration. Am J Pathol 2014;184:753–64. 10.1016/j.ajpath.2013.11.010 [DOI] [PubMed] [Google Scholar]
- 37. Wong WW, Vince JE, Lalaoui N, et al. cIAPs and XIAP regulate myelopoiesis through cytokine production in an RIPK1- and RIPK3-dependent manner. Blood 2014;123:2562–72. 10.1182/blood-2013-06-510743 [DOI] [PubMed] [Google Scholar]
- 38. Lawlor KE, Khan N, Mildenhall A, et al. RIPK3 promotes cell death and NLRP3 inflammasome activation in the absence of MLKL. Nat Commun 2015;6:6282 10.1038/ncomms7282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vince JE, Wong WW, Gentle I, et al. Inhibitor of apoptosis proteins limit RIP3 kinase-dependent interleukin-1 activation. Immunity 2012;36:215–27. 10.1016/j.immuni.2012.01.012 [DOI] [PubMed] [Google Scholar]
- 40. Yabal M, Müller N, Adler H, et al. XIAP restricts TNF- and RIP3-dependent cell death and inflammasome activation. Cell Rep 2014;7:1796–808. 10.1016/j.celrep.2014.05.008 [DOI] [PubMed] [Google Scholar]
- 41. Le Maitre CL, Hoyland JA, Freemont AJ. Catabolic cytokine expression in degenerate and herniated human intervertebral discs: IL-1beta and TNFalpha expression profile. Arthritis Res Ther 2007;9:R77 10.1186/ar2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 2017;14:361–9. 10.1080/15476286.2017.1279788 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
annrheumdis-2017-212056supp001.docx (45.9KB, docx)
annrheumdis-2017-212056supp002.docx (18.7KB, docx)
annrheumdis-2017-212056supp003.jpg (259.3KB, jpg)
annrheumdis-2017-212056supp004.docx (15.1KB, docx)
annrheumdis-2017-212056supp005.jpg (757.6KB, jpg)
annrheumdis-2017-212056supp006.docx (16.5KB, docx)
annrheumdis-2017-212056supp007.docx (19.4KB, docx)
annrheumdis-2017-212056supp008.docx (15.1KB, docx)
annrheumdis-2017-212056supp009.jpg (186.6KB, jpg)
annrheumdis-2017-212056supp010.jpg (2.1MB, jpg)
annrheumdis-2017-212056supp011.docx (14.4KB, docx)