Abstract
Development of cell replacement therapies in diabetes requires understanding of the molecular underpinnings of β-cell maturation. The circadian clock regulates diverse cellular functions important for regulation of β-cell function and turnover. However, postnatal ontogenesis of the islet circadian clock and its potential role in β-cell maturation remain unknown. To address this, we studied wild-type Sprague-Dawley as well as Period1 luciferase transgenic (Per1:LUC) rats to determine circadian clock function, clock protein expression, and diurnal insulin secretion during islet development and maturation process. We additionally studied β-cell–specific Bmal1-deficient mice to elucidate a potential role of this key circadian transcription factor in β-cell functional and transcriptional maturation. We report that emergence of the islet circadian clock 1) occurs during the early postnatal period, 2) depends on the establishment of global behavioral circadian rhythms, and 3) leads to the induction of diurnal insulin secretion and gene expression. Islet cell maturation was also characterized by induction in the expression of circadian transcription factor BMAL1, deletion of which altered postnatal development of glucose-stimulated insulin secretion and the associated transcriptional network. Postnatal development of the islet circadian clock contributes to early-life β-cell maturation and should be considered for optimal design of future β-cell replacement strategies in diabetes.
Introduction
Cell replacement strategies using stem cell–derived β-cells hold therapeutic potential for patients with diabetes. Multiple groups have developed differentiation protocols designed to produce glucose-responsive β-cell–like products (1–3). Although significant progress has been made, the ultimate goal to recapitulate an adult β-cell functional and transcriptional framework remains elusive (4). In order for β-cell replacement therapy to be successful, it is imperative to elucidate the physiological and molecular underpinnings of early-life islet cell functional and transcriptional maturation.
The circadian clock is an intrinsic timekeeping system comprised of transcription-translation feedback loops designed to optimize cellular function to changes in environment, generated by behavioral circadian cycles (e.g., sleep/wake, fast/feeding). A substantial portion of the mammalian genome displays regulatory oversight by the circadian clock, which drives temporal regulation of diverse cellular and molecular functions (5). The core of the circadian clock is comprised of transcriptional activators CLOCK and its heterodimer BMAL1 and repressor genes that encode Period (PER1/2) and Cryptochrome (CRY1/2) proteins (6). Secondary regulatory loops involving nuclear receptors Rev-Erbα/β and RORα/β provide additional molecular control by acting as respective transcriptional repressors and activators of Bmal1 (7). In β-cells, circadian clocks regulate varied cellular processes such as glucose-stimulated insulin secretion, cell turnover, response to oxidative stress, and mitochondrial function (8–11). Circadian regulation of insulin secretion is particularly critical for normal β-cell physiology in humans, where the clock functions to restrain insulin secretion during the inactive/sleep phase and optimize insulin secretion during the active/feeding phase of the circadian cycle (10,12). Thus, circadian regulation of glucose tolerance and insulin secretion is a characteristic feature of glucose homeostatic control, and disruption of normal circadian rhythms is associated with increased prevalence of glucose intolerance, metabolic syndrome, and diabetes (8–10,13–15).
There is a paucity of data on the role of circadian clocks in cell development, differentiation, and maturation (16). However, studies suggest that circadian clock proteins are expressed during the embryonic, neonatal, and early postnatal periods in mammalian cell types and contribute to cell differentiation and maturation processes (16–18). Most notably, characteristic phenotypes of premature aging, glucose intolerance, and shortened life span in mice with embryonic germ line deletion of Bmal1 (key circadian clock transcription factor) are strikingly attenuated in mice in which Bmal1 expression is conditionally preserved during embryogenesis (19).
To date, however, temporal emergence of the islet circadian clock during the neonatal and postnatal period remains largely unexplored. It is also unknown whether circadian clocks play a contributory role to early-life islet cell development as well as functional and transcriptional maturation. The primary objectives of the current study were to 1) examine temporal ontogenesis of islet circadian clock function, core clock protein expression, and diurnal regulation of insulin secretion during early-life islet cell development and maturation and to 2) test whether postnatal ontogenesis of the islet circadian clock contributes to morphological, functional, and transcriptional maturation of β-cells.
Research Design and Methods
Animals
A total of 167 rat pups (Sprague-Dawley and Wistar background, equal proportions male and female) and 60 mouse pups (C57B6 background, equal proportions male and female) aged 2–30 days were used in the current study. Specifically, for longitudinal assessment of islet circadian clock bioluminescence, 30 pups of transgenic rats were used in which mouse Period1 promoter was linked to a luciferase reporter (Per1:LUC) (20). An additional 137 rat pups (Charles River Laboratories, Wilmington, MA) were used for circadian gene and protein expression, metabolic and circadian behavior profiling, and islet perifusion studies. Furthermore, 60 β-cell–specific Bmal1 knockout (β-Bmal1−/−) and respective control (β-Bmal1+/+) mouse pups were generated by breeding mice with floxed Bmal1 gene (B6.129S4 [Cg]-Arntltm1Weit/J; The Jackson Laboratory, Bar Harbor, ME) (21) crossed with mice expressing a Cre-mediated recombination system driven by the rat Insulin2 promoter (Tg [Ins2-cre] 1Dam/J; The Jackson Laboratory) (22). All animals were housed under either a standard 12-h light, 12-h dark (LD) cycle or 24-h constant light exposure (LL) (>100 Lux light intensity) and provided with ad libitum chow diet (14% fat, 32% protein, and 54% carbohydrates; Harlan Laboratories, Indianapolis, IN). By convention, the time of lights on (0600 h) is denoted as zeitgeber/circadian time (ZT/CT) 0 and time of lights off (1800 h) as ZT/CT 12. All experimental procedures involving animals were approved by the Mayo Clinic and University of California, Los Angeles, institutional animal care committees.
Assessment of Circadian Clock Function in Islets Isolated From Per1:LUC Rats
After euthanasia, pancreatic islets were isolated from Per1:LUC rat pups at 2, 7, 15, 24, and 30 days postbirth using the standard collagenase method (23). The validation of Per1:LUC transgenic rats for study of islet circadian clock function has previously been described (13,24). Batches of 50 islets matched by size and diameter were placed on polytetrafluoroethylene membranes (Millipore, Billerica, MA) in culture dishes containing culture medium (serum-free, no sodium bicarbonate, no phenol red DMEM [D5030; Sigma-Aldrich, St. Louis, MO]) supplemented with 10 mmol/L HEPES, pH 7.2; 2 mmol/L glutamine; 2% B27 serum-free supplement (17504; Gibco, Carlsbad, CA); and 0.1 mmol/L d-luciferin (L-8220; Biosynth, Itaska, IL). Culture dishes containing islets were immediately placed into a LumiCycle luminometer (Actimetrics, Wilmette, IL) inside a light-resistant 37°C chamber with bioluminescence recorder photomultiplier tube detector. The bioluminescence signal was counted in 1-min bins every 10 min for at least 5 days. Data were normalized by subtraction of the 24-h running average from raw data and then smoothed with a 2-h running average (LumiCycle Data Analysis; Actimetrics).
Assessment of Circadian Activity
For behavioral activity monitoring, dams and pups were housed in cages outfitted with an optical beam sensor system (Respironics, Murrysville, PA) to monitor circadian behavioral rhythms in activity in either LD or LL circadian cycle. To quantitatively assess dominant circadian period in activity, we used χ2 periodogram algorithm analysis (ClockLab; Actimetrics, Evanston, IL).
Assessment of Circadian Metabolic Profiles
Diurnal plasma samples were collected on alternate days at 1000 h (ZT 4 [day]) and 2200 h (ZT 16 [night]) via lateral saphenous venipuncture into chilled, EDTA-treated microcentrifuge tubes and immediately centrifuged at 4°C for subsequent analysis. Blood glucose was measured with the FreeStyle Lite blood glucose measuring system (Abbott Laboratories, Abbott Park, IL). Plasma insulin was measured using rat ELISA (Alpco Diagnostics, Salem, NH).
Immunofluorescence Analysis of Clock Protein Expression, Islet Cell Mass and Turnover
Paraffin-embedded pancreatic sections were stained for insulin (ab7842, 1:100; Abcam, Cambridge, MA), glucagon (G2654, 1:500; Sigma-Aldrich), BMAL1 (ab3350, 1:500; Abcam), and PER1 (AB2201, 1:100; Millipore). In addition, adjacent deparaffinized sections were coimmunostained for TUNEL (In Situ Cell Death Detection Kit, TMR Red, 12156792910; Roche Diagnostics, Nutley, NJ) and insulin (as above) for quantification of β-cell apoptosis and for insulin and Ki-67 (mouse anti Ki-67; BD Pharmingen, San Jose, CA) for determination of β-cell replication. All slides were viewed, imaged, and analyzed using a Zeiss Axio Observer Z1 microscope (Carl Zeiss Microscopy, LLC, Thornwood, NY) and ZenPro software (Carl Zeiss Microscopy, LLC). Human pancreas was procured from the Mayo Clinic autopsy archives with approval from the Institutional Research Biosafety Board.
Islet Perifusion Analysis
Upon isolation, islets were recovered overnight in standard RPMI 1640 medium containing 10% FBS. Insulin secretion was assessed by an islet perifusion method (Biorep Technologies, Miami, FL) as previously described (14). In short, groups of 20 islets were perifused at basal glucose (4 mmol/L) and stimulated glucose (16 mmol/L), and effluent was collected at 2-min intervals. Insulin concentration was measured using insulin ELISA (Alpco Diagnostics).
Islet RNA Extraction, Quantitative Real-time PCR, and Affymetrix GeneChip
Isolated islets were washed with PBS to remove RPMI medium and immediately transferred to RNeasy Lysis Buffer RLT (Qiagen, Valencia, CA) and stored at −80°C for subsequent RNA isolation. Total RNA was extracted using an RNeasy Mini Kit (Qiagen), quantified by NanoDrop ND-100 (BioTek, Winooski, VT), and assessed for overall integrity using agarose gel electrophoresis. Microarray analysis was conducted according to the manufacturer’s instructions for the GeneChip WT Pico Reagent Kit (Affymetrix, Santa Clara, CA). Briefly, the first round of cDNA was generated from 2 ng total RNA using primers that contain a T7 promoter sequence and synthesized to single-stranded cDNA using RNase H and DNA polymerase with the addition of a 3′ adaptor. The products were in vitro transcribed to generate complimentary RNA using T7 RNA polymerase and then bead purified (Affymetrix). Complimentary RNA was then reverse transcribed to single-stranded cDNA, and 5.5 µg single-stranded cDNA was fragmented and hybridized onto Affymetrix mouse transcriptome arrays at 45°C for 16 h. Subsequent to hybridization, the arrays were washed and stained with streptavidin-phycoerythrin and then scanned in a GeneChip Scanner 3000 (Affymetrix). Control parameters were confirmed to be within normal ranges before normalization, and data reduction was initiated. For quantitative real-time PCR (qRT-PCR), mRNA was converted into cDNA using an iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). qRT-PCR was performed on the StepOnePlus machine (Applied Biosystems, Carlsbad, CA). Data were analyzed using the 2−ΔΔCT method with mRNA levels normalized to Actin.
Statistical Analysis and Calculations
Parameters of Per1-driven luciferase rhythms emanating from islets were calculated using the Lumicycle Analysis software (Actimetrics) as previously described (24). β-Cell function was estimated by using HOMA of β-cell function (HOMA-β) (HOMA-β = [20 × insulin]/[glucose − 3.5], with insulin in milliunits per liter and glucose in millimoles per liter) (25). Circadian qRT-PCR data were statistically analyzed by two-way ANOVA with Sidak multiple comparisons test to assess effects of time and age (GraphPad Prism, version 6.0). Gene-level normalization for whole transcriptome analysis and signal summarization was done using the Affymetrix Expression Consol. A fold regulation cutoff set to 2 was used to identify upregulated or downregulated transcripts. Differentially expressed genes were subjected to Gene Ontology (GO) enrichment analysis using DAVID (26). GO terms with a false discovery rate <0.05 were considered to be significantly enriched. Data in graphs are presented as means ± SEM and assumed to be statistically significant at P < 0.05.
Results
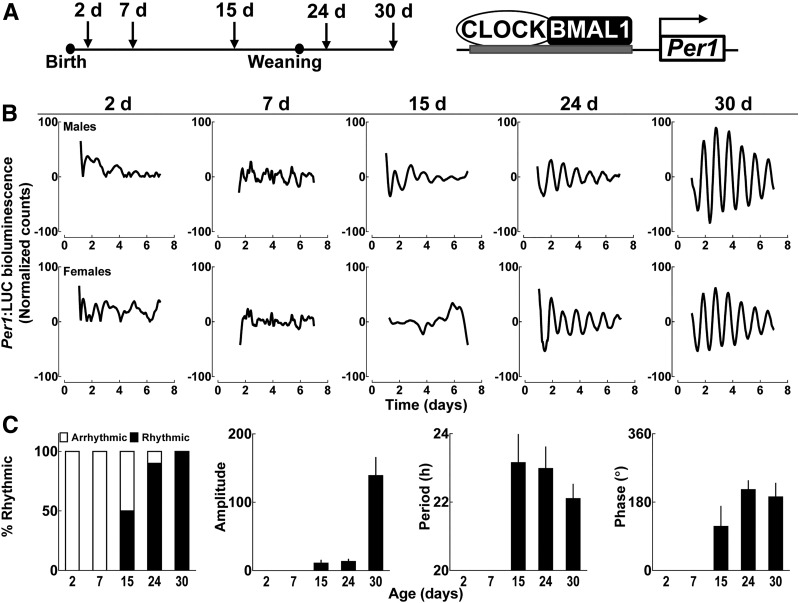
We first used tracking of islet cell bioluminescence with Per1 luciferase fusion construct in Per1:LUC transgenic rats for assessment of islet circadian clock function ex vivo (27), which was examined at multiple time points during the postnatal islet maturation period (e.g., postnatal days 2, 7, 15, 24, and 30) (Fig. 1A). Despite detection of a Per1-driven bioluminescence signal from isolated rat islets of all ages, no detectable circadian rhythmicity was observed in neonatal islets at 2 and 7 days postbirth (Fig. 1B and C). Circadian rhythmicity in Per1 bioluminescence gradually emerged during postnatal day 15 and was fully established by postnatal day 30, characterized by a robust oscillatory period (22.1 ± 0.5 h), amplitude, and phase angle of circadian oscillations, consistent with previous studies in adult islets (Fig. 1B and C) (24). Notably, the process of weaning per se did not appear to significantly impact establishment of amplitude, period, or phase of islet circadian oscillations in Per1:LUC transgenic rats (Supplementary Fig. 1).
Figure 1.
Postnatal emergence of the islet circadian clock function elucidated through real-time bioluminescence monitoring of islets isolated from Per1:LUC transgenic rats. A: Diagrammatic representation of study design indicating that pancreatic islets were isolated from Per1:LUC rat pups at 2, 7, 15, 24, and 30 days postbirth to assess Per1-driven bioluminescence. This is indicative of Per1 promoter activation mediated through binding of core clock transcription factors CLOCK and BMAL1 to upstream regulatory sequences. B: Representative examples of circadian Per1-driven bioluminescence rhythms in batches of 50 islets isolated from Per1:LUC male and female rat pups at postnatal days 2, 7, 15, 24, and 30. Note the appearance of rhythms on day 15 and robust circadian rhythmicity by day 30. C: Bar graphs represent (from left to right) percentage of pups exhibiting statistically significant circadian bioluminescence rhythms, mean amplitude, period, and phase angle of Per1 bioluminescence rhythms at 2, 7, 15, 24, and 30 day postbirth. Values are mean ± SEM (n = 4–10 per data point). The bioluminescence signal was counted in 1-min bins every 10 min for at least 5 days. Data were normalized by subtraction of the 24-h running average from raw data and then smoothed with a 2-h running average (Lumicycle Data Analysis; Actimetrics). d, days.
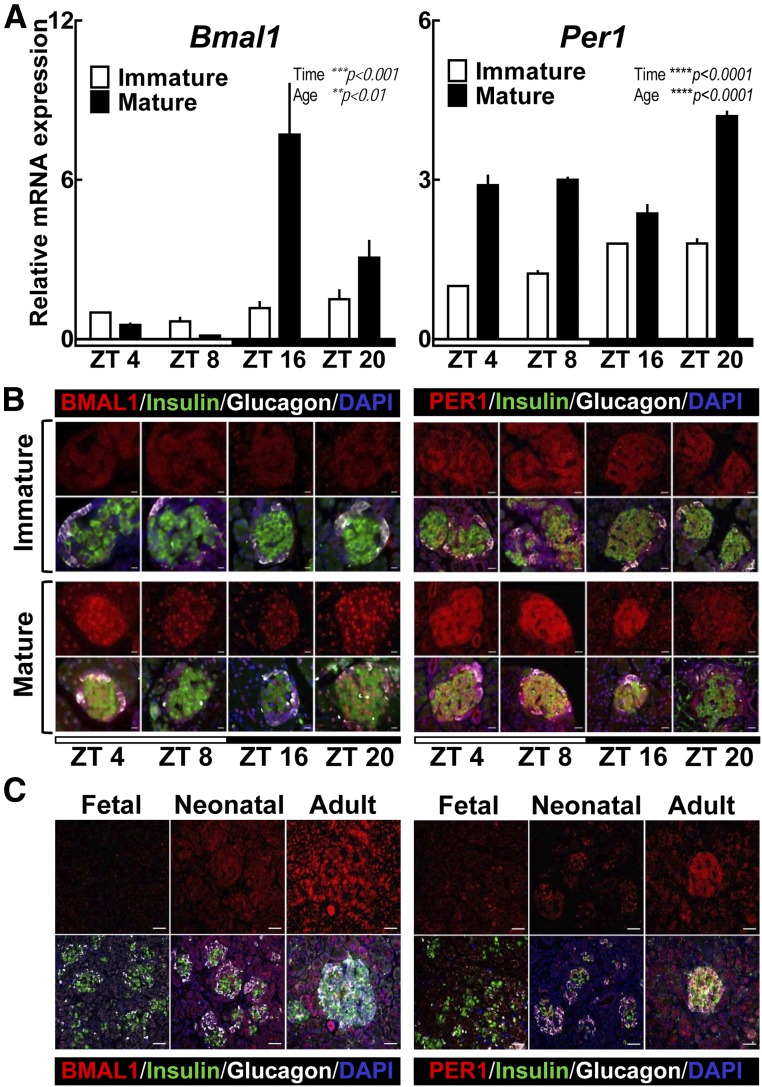
Development of the mammalian circadian clock is associated with induction of a core circadian clock transcription factor Bmal1 (28). Indeed, absence of Bmal1 expression corresponds with complete loss of circadian rhythms, a property unique to Bmal1 among all canonical clock genes (29). Consistent with data obtained in Per1:LUC rats, qRT-PCR analysis performed at multiple time points throughout the circadian cycle revealed a clear quantitative suppression and lack of circadian rhythmicity in Bmal1 (and its core clock gene target Per1) expression in immature (day 2) compared with mature (day 30) rat islets (Fig. 2A) (P < 0.0001 for age and time variables). Consistent with mRNA data, BMAL1 and PER1 protein expression was repressed in immature rat islets but demonstrated robust expression in mature (day 30) pancreatic β- and α-cells (Fig. 2B). Furthermore, BMAL1 expression was also repressed in neonatal and fetal endocrine human pancreas, implying a common mechanism mediating maturation of the islet circadian clock in rodents and humans (Fig. 2C).
Figure 2.
Postnatal emergence of circadian expression patterns of BMAL1 and PER1 in rat and human pancreatic islets. A: Normalized Bmal1 and Per1 mRNA expression obtained from whole islet lysates of immature (2–5 days old) and mature (30 days old) rat pups collected during ZT 4 (1000 h), 8 (1400 h), 16 (2200 h), and 20 (0200 h). Values are mean ± SEM (n = 3–5) fold change with immature pups at ZT 4 set as 1. Statistical significance was determined by two-way ANOVA with Sidak multiple comparisons test to assess effects of time and age (GraphPad Prism, version 6.0). **P < 0.01; ***P < 0.001; ****P < 0.0001. B: Representative examples of pancreatic islets stained by immunofluorescence for BMAL1 or PER1 (red), insulin (green), and glucagon (white) and counterstained with nuclear marker DAPI (blue) imaged at ×63 (BMAL1 [scale bars, 10 µm]) and ×43 (PER1 [scale bars, 20 µm]) magnification, respectively, in immature (2–5 days old) and mature (30 days old) rat pups collected during day (light cycle) (ZT 4 and 8) and night (dark cycle) (ZT 16 and 20) (representative of n = 3 per time point). C: Representative examples of human pancreatic islets stained by immunofluorescence for BMAL1 or PER1 (red), insulin (green), and glucagon (white) and counterstained with nuclear marker DAPI (blue) imaged at ×20 magnification (scale bars, 50 µm) in fetal (31 weeks’ gestation), neonatal (1 day old), and adult (74 years old) islets (all without diabetes).
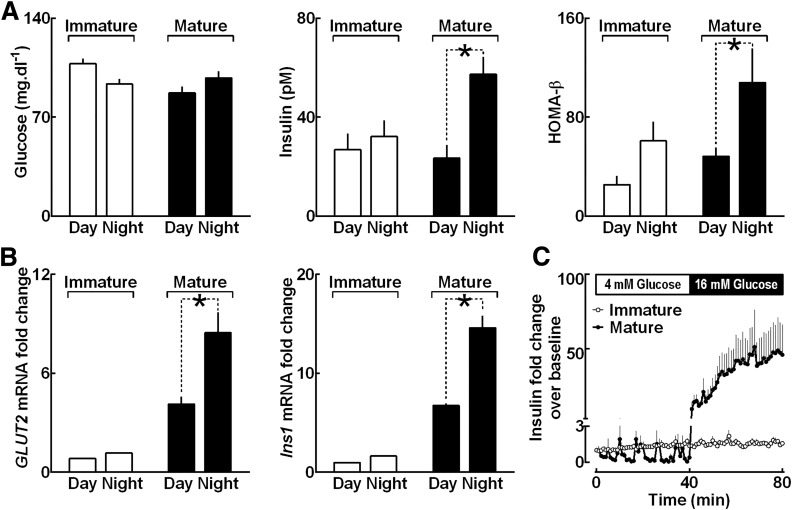
Next, we examined temporal development of circadian variations of in vivo insulin secretion and BMAL1-targeted mRNA expression in rat islets (Fig. 3). Diurnal glucose levels were comparable between immature (day 10) and mature (day 30) rats in vivo (Fig. 3A). However, immature (day 10) rats failed to exhibit characteristic circadian differences in plasma insulin concentrations as well as in the index of β-cell function derived from HOMA-β (P > 0.05 for ZT 4 vs. ZT 16 for both parameters) (Fig. 3A). In contrast, establishment of the islet circadian clock in mature 30-day-old rats was associated with emergence of circadian variations in plasma insulin (23.5 ± 6 vs. 57.4 ± 7 pmol/L for ZT 4 vs. ZT 16; P < 0.001) (Fig. 3A) and HOMA-β index (48.5 ± 7 vs. 108.0 ± 29 for ZT 4 vs. ZT 16; P < 0.05) (Fig. 3A). Consistent with these observations, islets isolated from 30-day-old pups displayed circadian expression patterns (P < 0.05 for ZT 4 vs. ZT 16) (Fig. 3B) of key BMAL1 target genes Glut2 and Ins1 (10), which are known regulators of glucose-stimulated insulin secretion. Consequently, immature rat islets also lacked glucose-stimulated insulin secretion when assessed in vitro by dynamic islet perifusion assay (Fig. 3C).
Figure 3.
Postnatal development of circadian variations of in vivo insulin secretion and clock-dependent mRNA expression in rat islets (A). Graphs showing diurnal differences in blood glucose, plasma insulin, and HOMA-β assessed in immature (10 days old) and mature (30 days old) rat pups. Each data point represents mean ± SEM (n = 6–10) (assumed to be statically significant at *P < 0.05). Day represents light phase 1000 h (ZT 4) and night 2200 h (ZT 16). B: Graphs showing Glut2 and Ins1 mRNA expression levels from whole islet lysates of immature (2–5 days old) and mature (30 days old) rat pups collected during the day (ZT 4–8) and at night (ZT 16–20). mRNA values from immature pups at daytime set as 1. Each data point represents mean ± SEM (n = 3–5) (assumed to be statically significant at P < 0.05). C: Graph showing data from in vitro islet perifusion in islets isolated from immature (2 days old) and mature (30 days old) rat pups expressed as mean ± SEM (n = 4–5 per group) insulin fold change over baseline profiles sampled at basal (4 mmol/L, 0–40 min) and stimulated (16 mmol/L, 40–80 min) glucose.
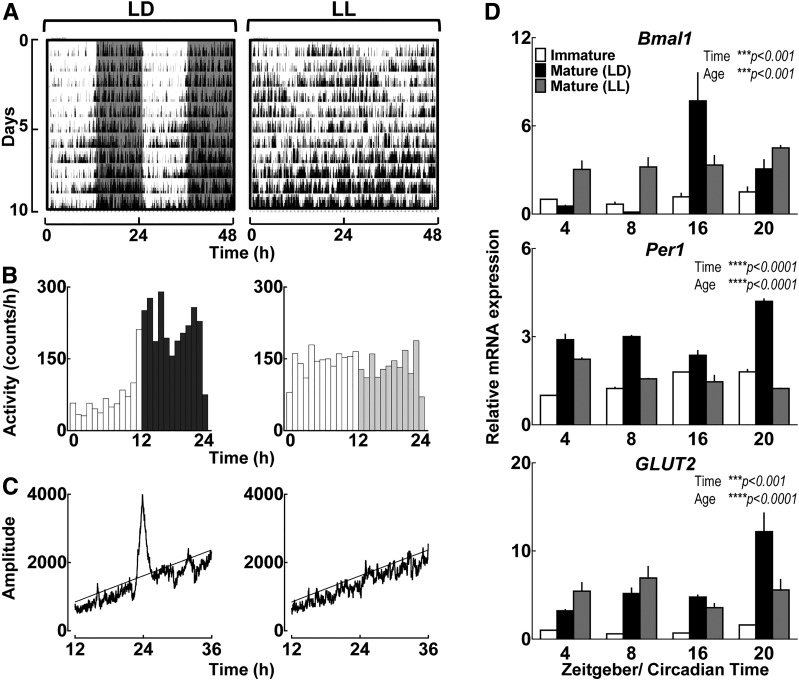
To gain insights into potential physiological mechanisms responsible for postnatal emergence of the circadian clock in pancreatic islets, we performed experiments to test whether early-life establishment of global behavioral circadian rhythms is required for maturation of the islet circadian clock. To accomplish this, we exposed rat dams and pups after parturition to either the normal light/dark (LD) cycle or constant light (LL), a modality previously shown to disrupt development of the suprachiasmatic nucleus of the hypothalamus molecular clock and impair establishment of global behavioral circadian rhythms (30,31). As expected, postnatal exposure to LL led to complete disruption in circadian organization of behavioral activity rhythms during the postnatal period in dams (Fig. 4A–C). Importantly, islets isolated from mature (day 30) LL-exposed rats displayed immature circadian genotype characterized by failed postnatal induction in circadian expression patterns for Bmal1 and BMAL1 target genes Per1 and Glut2 (P < 0.001 for LL vs. LD) (Fig. 4D).
Figure 4.
Postnatal establishment of global behavioral circadian rhythms is required for maturation of circadian clock function in pancreatic islets. Representative behavioral profiles (actograms) of circadian activity (A) and mean circadian activity (expressed as counts/60 min) (B) shown across the 24-h day in rat pups monitored for 10 days under either LD cycle or LL exposure during early postnatal period (days 2–12). Shaded areas represent periods of dark. C: Representative χ2 periodograms of activity recordings in LD-exposed (left) vs. LL-exposed (right) pups. Horizontal lines represent statistical significant threshold in determination of dominant circadian period. Note the absence of circadian period in LL pups. D: Normalized Bmal1, Per1, and Glut2 mRNA expression obtained from whole islet lysates of immature (2–5 days old) and mature (30 days old) rat pups exposed to either LD or disrupted LL circadian cycle during the postnatal period (days 0–30) and collected during ZT 4 (1000 h), 8 (1400 h), 16 (2200 h), and 20 (0200 h). Values are mean ± SEM (n = 3–5) fold change with immature pups at ZT 4 set as 1. Statistical significance was determined by two-way ANOVA with Sidak multiple comparisons test to assess effects of time and age (GraphPad, version 6.0). ***P < 0.001; ****P < 0.0001. Data for “Immature” and “Mature (LD)” islets are replotted from Fig. 2A.
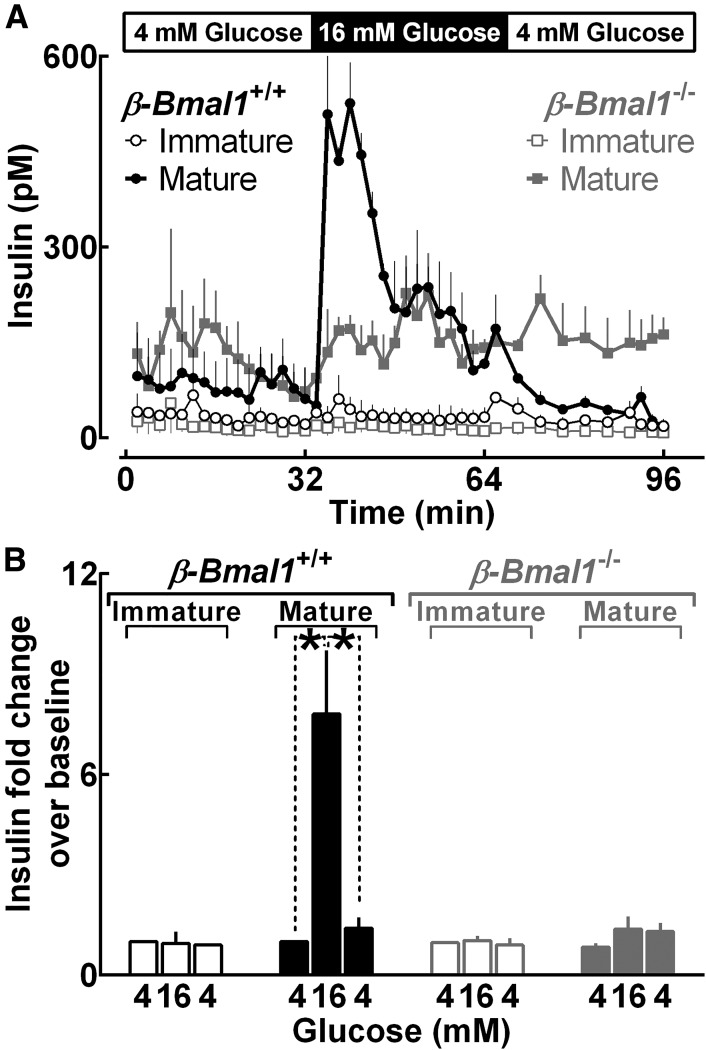
Next, to determine whether postnatal induction of Bmal1 expression contributes to morphological, functional, and/or transcriptional β-cell maturation, we generated β-cell–specific Bmal1 knockout mice (β-Bmal1−/−) by crossing Bmal1fl/fl mice with rat Insulin2 promoter Cre (RIP-Cre) mice. As expected, BMAL1 expression was suppressed in mature (25 days old) β-Bmal1−/− (Bmal1fl/fl;RIPCre/+) mice compared with control β-Bmal1+/+ (Bmal1fl/fl) (Fig. 5A). Interestingly, measures of β- and α-cell mass, β-cell proliferation, and β-cell apoptosis were comparable between immature (5 days old) and mature (25 days old) β-Bmal1−/− and β-Bmal1+/+ mice (P > 0.05 for all parameters) (Fig. 5B), suggesting that postnatal induction of BMAL1 does not contribute to early-life expansion of islet cell mass. In contrast, whereas transition of islets from the immature (5 days old) to mature (25 days old) stage in control (β-Bmal1+/+) mice resulted in expected maturation of glucose-stimulated insulin secretion (Fig. 6), islets isolated from mature (25 days old) β-Bmal1−/− mice phenocopied glucose-stimulated insulin response in immature (5 days old) islets (Fig. 6).
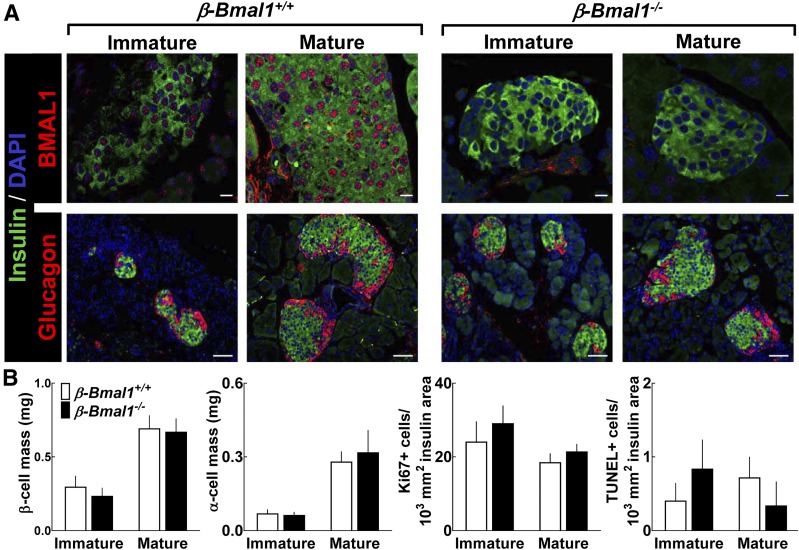
Figure 5.
Embryonic deletion of Bmal1 in pancreatic β-cells does not alter postnatal development of islet morphology, postnatal islet cell expansion, or β-cell turnover. A: Representative examples of pancreatic islets stained by immunofluorescence for BMAL1 or glucagon (red) and insulin (green) and counterstained with nuclear marker DAPI (blue) imaged at ×63 (BMAL1 [scale bars, 10 µm]) and ×20 (glucagon [scale bars, 50 µm]) magnification in immature (5 days old) and mature (25 days old) β-Bmal1−/− and control β-Bmal1+/+ mice. B: Graphs showing β-cell mass, α-cell mass, proliferation, and apoptosis in immature (5 days old) and mature (25 days old) β-Bmal1+/+ and β-Bmal1−/− mouse pups. Data are expressed as mean ± SEM (n = 3–7 per group).
Figure 6.
Embryonic deletion of Bmal1 in pancreatic β-cells compromises postnatal functional maturation. Insulin concentration profiles during islet perifusion expressed as mean ± SEM (A) and corresponding mean fold change over baseline (B) sampled at 4 mmol/L glucose (0–32 min), 16 mmol/L glucose (32–64 min), and 4 mmol/L glucose (64–96 min) in isolated islets from immature (5 days old [n = 3]) and mature (25 days old [n = 5]) β-Bmal1−/− and control β-Bmal1+/+ mouse pups. *P < 0.05.
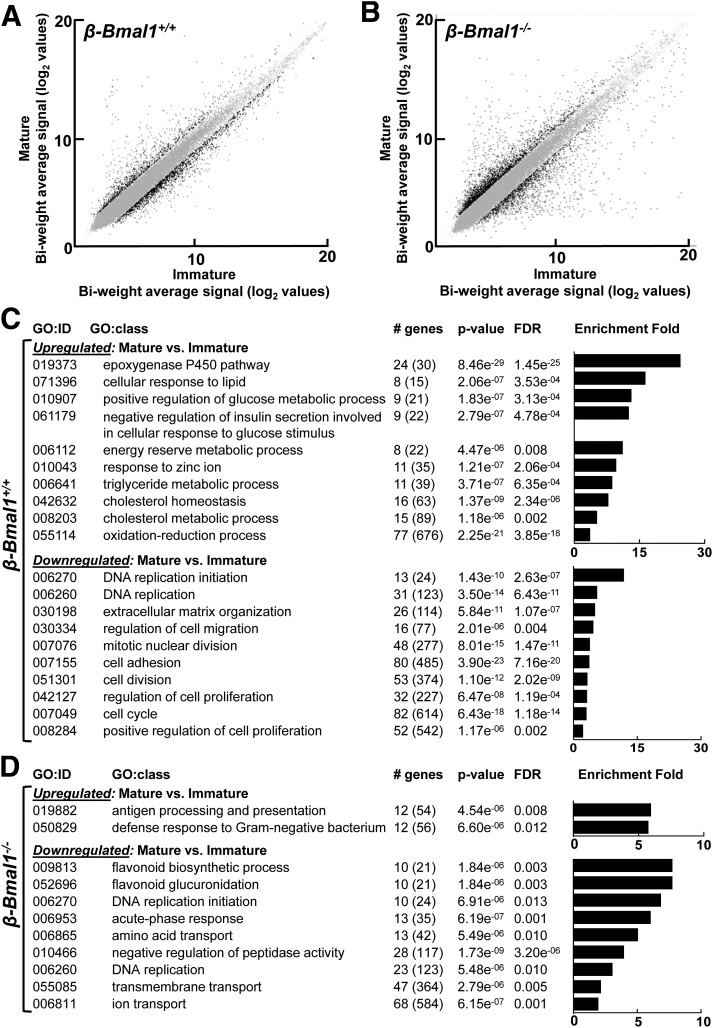
Consistent with these observations, transition of islets from the immature (5 days old) to mature (25 days old) stage was associated with enrichment in transcriptional pathways shown to be critical for regulation of β-cell function (e.g., GO:0055114 [oxidation-reduction process], GO:010043 [response to zinc ion], GO:0010907 [regulation of glucose metabolic process], etc.) (Fig. 7), which was observed only in control β-Bmal1+/+, but not in Bmal1-deficient, islets (Fig. 7). In contrast, in both β-Bmal1+/+ and β-Bmal1−/−, transition of islets from the immature (5 days old) to mature (25 days old) stage was associated with downregulation of genes enriched for pathways regulating cell proliferation and cell cycle (e.g., GO:0006260 [DNA replication]), an observation consistent with previous findings (32). Also, Bmal1 deletion in β-cells did not affect transcriptional expression of major islet hormones (e.g., Iapp), key β-cell development transcription factors (e.g., Mafa, Ucn3, Pdx1), or disallowed genes (e.g., Ldha, Slc16a1) (Supplementary Fig. 2).
Figure 7.
Embryonic deletion of Bmal1 in pancreatic β-cells modulates postnatal transcriptional maturation. Scatterplots showing biweight log2 average expression values of all genes in mature (day 25) vs. immature (day 5) pancreatic islets of β-Bmal1+/+ (A) and β-Bmal1−/− (B) mice. Gray dots indicate unchanged expression, while black dots represent up- and downregulated genes with fold regulation cutoff set at 2. Tables showing significantly up- and downregulated GO biological processes with a false discovery rate (FDR) <0.05 in islets from 25-day-old mature compared with 5-day-old immature β-Bmal1+/+ (C) and β-Bmal1−/− (D) mouse pups. Differentially expressed genes were subjected to GO enrichment analysis using DAVID (26).
Discussion
Cell replacement therapy is a promising therapeutic option to restore β-cell functional mass in diabetes (33). However, the ultimate success of cell replacement therapy in diabetes relies on the ability to fully recapitulate adult β-cell phenotype and transcriptional networks in stem cell–generated β-cells (4). Pancreatic islets undergo a complex functional maturation process throughout the early neonatal and postnatal period during which they acquire phenotypical characteristics of adult endocrine cells, the most important of which is glucose-stimulated insulin secretion (34–37). The current study demonstrates that development of the islet circadian oscillator also occurs during the early postnatal period and is associated with the establishment of circadian control of insulin secretion and gene expression. Furthermore, our data also indicate that postnatal islet cell maturation is characterized by induction in the expression of a core circadian clock transcription factor, BMAL1, an observation shown to be consistent between rodent and human islets.
Molecular and physiological mechanisms underlying embryonic development and postnatal maturation of the circadian oscillator are incompletely understood (16). In general, studies suggest that circadian clocks develop gradually during the embryonic and neonatal period beginning with 1) emergence of clock gene expression, 2) start of low-amplitude transcriptional and translational oscillations, and 3) eventual development of high-amplitude circadian oscillations attributed to the induction of core transcriptional activators (e.g., BMAL1) (16). Ontogeny of circadian rhythms in various tissues is complex but is dictated in part by anticipation of diurnal changes influencing particular cellular function (e.g., light/dark, fast/feeding cycles). For example, circadian pacemaker neurons of the suprachiasmatic nucleus of the hypothalamus start to express clock genes as early as embryonic day 19 but do not fully mature until postnatal days 3–10 (P3–P10), which coincides with the time an animal begins to respond to changes in the photoperiod (38). On the other hand, hepatic clocks do not exhibit complete circadian oscillations until P20–P30, coincident with ontogenesis of circadian rhythms in feeding (39).
In our study, we purposefully chose to assess the emergence of the islet circadian clock function during distinct phases: 1) when rat pups are fully reliant on the dam for nutrients and thus display 24-h suckling behaviors (e.g., P2–P10) (40), 2) start of the transition to independent chow feeding behavior coinciding with development of gastrointestinal adaptions for carbohydrates (e.g., P15) (40), and 3) weaned pups exhibiting independent circadian feeding patterns (e.g., P24–30) (40). Our results show gradual emergence of circadian oscillations in islets starting at postnatal day 15, which coincides with the development of gastrointestinal adaptions for independent feeding activity cycles (40). Notably, establishment of circadian feeding cycles has been demonstrated to entrain the phase of peripheral circadian clocks (e.g., liver) (41,42), an observation recently extended to pancreatic islets (43). Consistent with these observations, our studies show that postnatal establishment of global behavioral circadian rhythms in activity and feeding cycles is required for the emergence of circadian clock function in pancreatic islets. Furthermore, the process of weaning and specifically transition from lipid-rich maternal milk to carbohydrate-rich chow has also recently been reported to trigger functional β-cell maturation and enhanced mitogenic potential in rodents (36). However, our data show that failure to wean pups from a lipid-rich milk diet does not appear to impact establishment of amplitude, period, or phase of the islet circadian clock, suggesting that weaning per se is not the main mediator of islet clock maturation process.
Recent in vitro studies shed some light on molecular mechanisms underlying induction of circadian clock function during ES cell differentiation and maturation (28,44). Using mouse embryonic stem cells, investigators demonstrated that the circadian clock is absent in the population of pluripotent stem cells and develops during cellular terminal differentiation via an intrinsic cellular program (28,44). Importantly, when the same differentiated cells are reprogrammed back to pluripotency by forced expression of reprogramming factors (e.g., c-Myc), the circadian clock appears to be repressed (28,44). Interestingly, c-Myc–mediated repression of Bmal1 has recently been shown to characterize cancer cell phenotype and hypothesized to underlie the recapitulation of cell pluripotency and enhanced proliferative potential (45,46). This observation is particularly relevant for endocrine cell development in the pancreas, given that early-life differentiation and proliferation of pancreatic progenitors is critical for the establishment of proper postnatal β-cell mass (47). Thus, our studies suggest that the repression of the circadian clock during the early neonatal period may promote postnatal β-cell proliferation. Interestingly, recent studies suggest that transient accumulation of reactive oxygen species (ROS) serves as a mediator of early postnatal β-cell proliferation and β-cell mass expansion (32). Intriguingly, since BMAL1 provides transcriptional control over intracellular ROS scavenging network, BMAL1-deficient β-cells are characterized by enhanced ROS accumulation (8).
Our study also shows that postnatal induction of BMAL1 in β-cells is a contributory factor to β-cell functional and transcriptional maturation and, particularly, establishment of circadian control of glucose-stimulated insulin secretion. However, it is important to note that early-life Bmal1 deletion in β-cells did not significantly impact postnatal expression of established developmental maturation markers (e.g., Ucn3, Pdx1, and Mafa) or key β-cell disallowed genes (e.g., Ldha). Complex molecular mechanisms define maturation of glucose-stimulated insulin secretion. This process has been shown to be driven by cellular transition from predominantly glycolytic to mitochondrial metabolism (36,37), induction of key β-cell–specific transcription factors (34,37,48,49) (e.g., Ucn3, Mafa, Pdx1, ERRϒ), methylation-dependent repression of transcripts involved in anaerobic metabolism (35), and alterations in islet microRNA environment (50). This implies that interaction among diverse physiological and molecular mechanisms is required to achieve full functional maturity of β-cells. In this regard, our studies suggest that postnatal induction of BMAL1 and circadian clock mechanism should be considered another potential contributory factor to the β-cell maturation process. Indeed, recently elucidated BMAL1 transcriptional targets in β-cells encompass diverse cellular pathways critical for regulation of glucose-stimulated insulin secretion (e.g., glucose transport, SNARE interactions, mitochondrial function, protein transport, etc.) (10).
It is important to acknowledge some limitations and additional questions raised by our study. It remains unknown whether β-cell maturation per se, and specifically postnatal induction in key development transcription factors (e.g., Mafa, Ucn3, Pdx1, etc.), also contributes to early-life development of the β-cell circadian oscillator. Since PDX1 expression has been shown to colocalize with BMAL1/CLOCK within active β-cell enhancers (10), postnatal PDX-1 expression appears to be important for full maturation of the circadian clock function in islets. In addition, recent studies suggest that BMAL1 may exhibit clock-independent functions, particularly during embryonic development and the neonatal period in mice (19). In support of this premise, many pathologies typically associated with whole-body embryonic Bmal1 knockout mice are absent in mice in which deletion of Bmal1 was restricted to adult life (e.g., 3 months) (19). Additional studies will be required to address whether nonoscillatory or clock-independent function of Bmal1 contributes to regulation of β-cell function.
In conclusion, circadian clocks regulate many cellular and molecular functions required for proper islet function and turnover. The current study suggests that early-life development of the islet circadian clock plays a contributory role in the development of mature β-cell phenotype that should be taken into consideration in the optimal design of future β-cell replacement strategies in diabetes.
Supplementary Material
Article Information
Acknowledgments. The authors thank Dr. Peter C. Butler (University of California, Los Angeles) and Dr. Anil Bhushan (University of California, San Francisco) for helpful discussions and insightful comments.
Funding. The authors acknowledge funding support from the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (R01-DK-098468 to A.V.M.), and the Center for Regenerative Medicine (Mayo Clinic, Rochester, MN).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. K.R. contributed to study design, conducted experiments, and assisted with the data analysis and interpretation and preparation of the manuscript. J.Q. contributed to study design, conducted experiments, and reviewed the manuscript. K.S.G. performed bioinformatics data analysis and reviewed the manuscript. S.D. contributed to study design and data interpretation and reviewed the manuscript. C.S.C. contributed to study design and data interpretation and reviewed the manuscript. A.V.M. designed and interpreted the studies and wrote the manuscript. A.V.M. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db17-0850/-/DC1.
References
- 1.Pagliuca FW, Millman JR, Gürtler M, et al. . Generation of functional human pancreatic β cells in vitro. Cell 2014;159:428–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rezania A, Bruin JE, Arora P, et al. . Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol 2014;32:1121–1133 [DOI] [PubMed] [Google Scholar]
- 3.Russ HA, Parent AV, Ringler JJ, et al. . Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. EMBO J 2015;34:1759–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson JD. The quest to make fully functional human pancreatic beta cells from embryonic stem cells: climbing a mountain in the clouds. Diabetologia 2016;59:2047–2057 [DOI] [PubMed] [Google Scholar]
- 5.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 2017;18:164–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet 2008;9:764–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Preitner N, Damiola F, Lopez-Molina L, et al. . The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 2002;110:251–260 [DOI] [PubMed] [Google Scholar]
- 8.Lee J, Moulik M, Fang Z, et al. . Bmal1 and β-cell clock are required for adaptation to circadian disruption, and their loss of function leads to oxidative stress-induced β-cell failure in mice. Mol Cell Biol 2013;33:2327–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcheva B, Ramsey KM, Buhr ED, et al. . Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 2010;466:627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perelis M, Marcheva B, Ramsey KM, et al. . Pancreatic β cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science 2015;350:aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rakshit K, Thomas AP, Matveyenko AV. Does disruption of circadian rhythms contribute to beta-cell failure in type 2 diabetes? Curr Diab Rep 2014;14:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boden G, Ruiz J, Urbain JL, Chen X. Evidence for a circadian rhythm of insulin secretion. Am J Physiol 1996;271:E246–E252 [DOI] [PubMed] [Google Scholar]
- 13.Qian J, Yeh B, Rakshit K, Colwell CS, Matveyenko AV. Circadian disruption and diet-induced obesity synergize to promote development of β-cell failure and diabetes in male rats. Endocrinology 2015;156:4426–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rakshit K, Hsu TW, Matveyenko AV. Bmal1 is required for beta cell compensatory expansion, survival and metabolic adaptation to diet-induced obesity in mice. Diabetologia 2016;59:734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A 2009;106:4453–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du Pré BC, Van Veen TA, Young ME, Vos MA, Doevendans PA, Van Laake LW. Circadian rhythms in cell maturation. Physiology (Bethesda) 2014;29:72–83 [DOI] [PubMed] [Google Scholar]
- 17.Brown SA. Circadian clock-mediated control of stem cell division and differentiation: beyond night and day. Development 2014;141:3105–3111 [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi Y, Ye Z, Hensch TK. Clock genes control cortical critical period timing. Neuron 2015;86:264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang G, Chen L, Grant GR, et al. . Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci Transl Med 2016;8:324ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamazaki S, Numano R, Abe M, et al. . Resetting central and peripheral circadian oscillators in transgenic rats. Science 2000;288:682–685 [DOI] [PubMed] [Google Scholar]
- 21.Storch KF, Paz C, Signorovitch J, et al. . Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell 2007;130:730–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Postic C, Shiota M, Niswender KD, et al. . Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem 1999;274:305–315 [DOI] [PubMed] [Google Scholar]
- 23.Rakshit K, Qian J, Ernst J, Matveyenko A. Circadian variation of the pancreatic islet transcriptome. Physiol Genomics 2016;48:677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian J, Block GD, Colwell CS, Matveyenko AV. Consequences of exposure to light at night on the pancreatic islet circadian clock and function in rats. Diabetes 2013;62:3469–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985;28:412–419 [DOI] [PubMed] [Google Scholar]
- 26.Huang DW, Sherman BT, Tan Q, et al. . The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 2007;8:R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welsh DK, Imaizumi T, Kay SA. Real-time reporting of circadian-regulated gene expression by luciferase imaging in plants and mammalian cells. Methods Enzymol 2005;393:269–288 [DOI] [PubMed] [Google Scholar]
- 28.Yagita K, Horie K, Koinuma S, et al. . Development of the circadian oscillator during differentiation of mouse embryonic stem cells in vitro. Proc Natl Acad Sci U S A 2010;107:3846–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bunger MK, Wilsbacher LD, Moran SM, et al. . Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 2000;103:1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohta H, Mitchell AC, McMahon DG. Constant light disrupts the developing mouse biological clock. Pediatr Res 2006;60:304–308 [DOI] [PubMed] [Google Scholar]
- 31.Ohta H, Yamazaki S, McMahon DG. Constant light desynchronizes mammalian clock neurons. Nat Neurosci 2005;8:267–269 [DOI] [PubMed] [Google Scholar]
- 32.Zeng C, Mulas F, Sui Y, et al. . Pseudotemporal ordering of single cells reveals metabolic control of postnatal β cell proliferation. Cell Metab 2017;25:1160–1175.e1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matveyenko A, Vella A. Regenerative medicine in diabetes. Mayo Clin Proc 2015;90:546–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blum B, Hrvatin S, Schuetz C, Bonal C, Rezania A, Melton DA. Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nat Biotechnol 2012;30:261–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dhawan S, Tschen SI, Zeng C, et al. . DNA methylation directs functional maturation of pancreatic β cells. J Clin Invest 2015;125:2851–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stolovich-Rain M, Enk J, Vikesa J, et al. . Weaning triggers a maturation step of pancreatic β cells. Dev Cell 2015;32:535–545 [DOI] [PubMed] [Google Scholar]
- 37.Yoshihara E, Wei Z, Lin CS, et al. . ERRγ is required for the metabolic maturation of therapeutically functional glucose-responsive β cells. Cell Metab 2016;23:622–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sládek M, Sumová A, Kováciková Z, Bendová Z, Laurinová K, Illnerová H. Insight into molecular core clock mechanism of embryonic and early postnatal rat suprachiasmatic nucleus. Proc Natl Acad Sci U S A 2004;101:6231–6236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sládek M, Jindráková Z, Bendová Z, Sumová A. Postnatal ontogenesis of the circadian clock within the rat liver. Am J Physiol Regul Integr Comp Physiol 2007;292:R1224–R1229 [DOI] [PubMed] [Google Scholar]
- 40.Henning SJ. Postnatal development: coordination of feeding, digestion, and metabolism. Am J Physiol 1981;241:G199–G214 [DOI] [PubMed] [Google Scholar]
- 41.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 2000;14:2950–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saini C, Suter DM, Liani A, Gos P, Schibler U. The mammalian circadian timing system: synchronization of peripheral clocks. Cold Spring Harb Symp Quant Biol 2011;76:39–47 [DOI] [PubMed] [Google Scholar]
- 43.Rakshit K, Qian J, Colwell CS, Matveyenko AV. The islet circadian clock: entrainment mechanisms, function and role in glucose homeostasis. Diabetes Obes Metab 2015;17(Suppl. 1):115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Umemura Y, Koike N, Matsumoto T, et al. . Transcriptional program of Kpna2/Importin-α2 regulates cellular differentiation-coupled circadian clock development in mammalian cells. Proc Natl Acad Sci U S A 2014;111:E5039–E5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altman BJ, Hsieh AL, Sengupta A, et al. . MYC disrupts the circadian clock and metabolism in cancer cells. Cell Metab 2015;22:1009–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shostak A, Diernfellner A, Brunner M. MYC inhibits the clock and supports proliferation. Cell Cycle 2016;15:3323–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dhawan S, Georgia S, Bhushan A. Formation and regeneration of the endocrine pancreas. Curr Opin Cell Biol 2007;19:634–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aguayo-Mazzucato C, Koh A, El Khattabi I, et al. . Mafa expression enhances glucose-responsive insulin secretion in neonatal rat beta cells. Diabetologia 2011;54:583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Meulen T, Xie R, Kelly OG, Vale WW, Sander M, Huising MO. Urocortin 3 marks mature human primary and embryonic stem cell-derived pancreatic alpha and beta cells. PLoS One 2012;7:e52181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacovetti C, Rodriguez-Trejo A, Guay C, et al. . MicroRNAs modulate core-clock gene expression in pancreatic islets during early postnatal life in rats. Diabetologia 2017;60:2011–2020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.