Significance
The majority of cellular proteins undergo rapid degradation and synthesis to minimize the toxic effect to cells and tissues and to guarantee normal cellular functions. It has been appreciated that proteins with longer half-lives exist in certain cells and tissues. Here we identify synaptic long-lived proteins by high-resolution mass spectrometry. In general, synaptic proteins exhibit slower turnover than cytosolic proteins, and synaptic protein turnover from mouse brain is enhanced by enriched environment exposure. Moreover, protein half-lives are dynamically regulated during changes in neuronal activity. These findings demonstrate the existence of long-lived proteins in synapses in the brain and support a potential role for them in synaptic plasticity and learning and memory.
Keywords: long-lived proteins, protein turnover, enriched environment, neuronal activity, mass spectrometry
Abstract
Memory formation is believed to result from changes in synapse strength and structure. While memories may persist for the lifetime of an organism, the proteins and lipids that make up synapses undergo constant turnover with lifetimes from minutes to days. The molecular basis for memory maintenance may rely on a subset of long-lived proteins (LLPs). While it is known that LLPs exist, whether such proteins are present at synapses is unknown. We performed an unbiased screen using metabolic pulse-chase labeling in vivo in mice and in vitro in cultured neurons combined with quantitative proteomics. We identified synaptic LLPs with half-lives of several months or longer. Proteins in synaptic fractions generally exhibited longer lifetimes than proteins in cytosolic fractions. Protein turnover was sensitive to pharmacological manipulations of activity in neuronal cultures or in mice exposed to an enriched environment. We show that synapses contain LLPs that may underlie stabile long-lasting changes in synaptic structure and function.
Synapses have the ability to change in strength and structure, a process referred to as synaptic plasticity. Certain forms of synaptic plasticity, such as long-term potentiation (LTP) or depression (LTD), can be maintained for many months to years (1). It is widely believed that LTP and LTD are the cellular correlates of learning and memory. Many of the key cellular signaling events and molecular rearrangements underlying LTP and LTD have been identified and characterized (2, 3). The earliest events leading to LTP and LTD initiation involve activation of cellular signaling machinery, which can last on the order of seconds to minutes. Later phases of LTP and LTD, which can be examined experimentally over several hours, involve the synthesis, recruitment, and capture of specific proteins that produce changes in the structure and strength of the synapse. Inhibition of protein synthesis during this period is sufficient to impair synaptic plasticity and memory formation (4). Importantly, several hours following establishment of LTP/LTD and learning, the plasticity and memory are transformed into a relatively stable form that is resistant to protein synthesis inhibitors. However, during this maintenance phase of synaptic plasticity the majority of proteins known to constitute these synapses will turn over, being degraded and replaced with a time course typically of hours to a few days (5–8). While a great deal is known about the molecular mechanisms that initiate synaptic plasticity events, very little is known of the molecular processes that enable synaptic strength and memories to persist for extended periods. It is widely believed that some aspects of synapse structure will remain stable for extended periods to promote the retention of memories after their formation (9–11). One possibility is that extremely long-lived proteins (LLPs) may reside at synapses and form part of the molecular basis for long-term memory maintenance.
Previous studies have shown the existence of LLPs in the body. For example, crystallin and collagen, which make up the lens of the eye and cartilage, respectively, last for decades. Components of the nuclear pore complex of nondividing cells and some histones have also been shown to last for years (12–15). The extreme stability of some of these proteins is obviously critical for the structural integrity of relatively metabolically inactive regions of the body, such as the lens and cartilage. However, it is less clear why proteins in the nuclear pore complex and certain histones need to be stable. Presumably, the stability of these proteins in these dynamic subcellular domains is important for their structural and functional role in cells. The potential presence of LLPs in synapses may indicate that they play critical roles in long-lasting synaptic processes, such as long-term plasticity and memory. Unlike other long-lived molecules, such as DNA, that employ several mechanisms to repair damage encountered over the lifetime of the molecule, little is known regarding such mechanisms that act on LLPs (16, 17). It is known that proteins will suffer oxidative damage over time, and these insults to LLPs may contribute to the aging process. Such has been shown to be the case for crystallin and collagen, where age-related damage contributes to the formation of cataracts and cartilage stiffening, respectively (18–20). In metabolically active neurons, it is possible that damage to LLPs associated with memory storage may contribute to cognitive decline during aging.
Earlier studies have suggested that synaptic kinases can mediate the long-term maintenance of plasticity and memory. For example, activation of Ca2+/calmodulin-dependent protein kinase type II (CaMKII) is required for the induction of LTP and increased phosphorylation and functionality of AMPA receptors (AMPARs) (3, 21–25), and autophosphorylation of CaMKII has been proposed to maintain LTP and long-term memory (3, 25). Studies have also found that a constitutively active brain-specific variant of PKCζ, called PKMζ, could be generated following memory formation and maintain the memory through sustained kinase activity (26). However, recent studies have shown that memory formation and maintenance can occur in the absence of PKMζ (27, 28), although a recent study reports that another atypical isoform, PKCi, may compensate for PKMζ (29). Studies in aplysia have suggested that during facilitation (memory formation) the translation regulator CPEB protein (cytoplasmic polyadenylation element binding protein) can undergo a conformational shift resulting in prion-like properties, in which the prion conformation can be transmitted to newly made copies, enabling persistent memory storage (30–32). This mechanism has since been reported to occur in drosophila and mice (33, 34).
While it has long been observed that the formation of memory requires the synthesis of new proteins, it is unknown how these proteins become recruited to or captured by the synapses specifically associated with a memory or plasticity event and contribute to stable memory formation. The synaptic tag hypothesis posits that a molecular rearrangement occurs at active synapses that enable those synapses to recruit the necessary factors for long-term memory formation (35, 36). One possibility is that during the formation of the synaptic tag or other structural changes to the synapse, certain proteins will undergo a conformational shift resulting in a highly stable protein/complex that may be extremely long-lived. Another possibility is that LLPs can become recruited or captured by activated synapses and then reside at these synapses for long periods, forming part of the molecular basis for the synaptic tag and long-term memory storage.
Although many synaptic proteins have been studied and shown to play critical roles in brain function, their turnover kinetics have not yet been explored systemically. In the current study, we have performed SILAM (stable isotope labeling in mammals) (37, 38) in combination with subcellular fractionation to quantify protein turnover rates during control and experience-dependent plasticity conditions (i.e., enriched environment). With this approach, we have characterized the turnover kinetics for a large population of the synaptic proteome and have identified populations of synaptic LLPs. We also have performed SILAC (stable isotope labeling by amino acids in cell culture) (39) using primary neuronal cultures to characterize protein turnover kinetics during persistent neuronal activity changes and have identified populations of proteins that are stabilized or destabilized by activity. In summary, the identification and characterization of synaptic LLPs may provide insight into new molecular mechanisms contributing to synaptic function and learning and memory, as well as new avenues to understand and combat aging-related diseases of the nervous system.
Results
Measurement of Protein Turnover in the Synaptic Proteome in Mice.
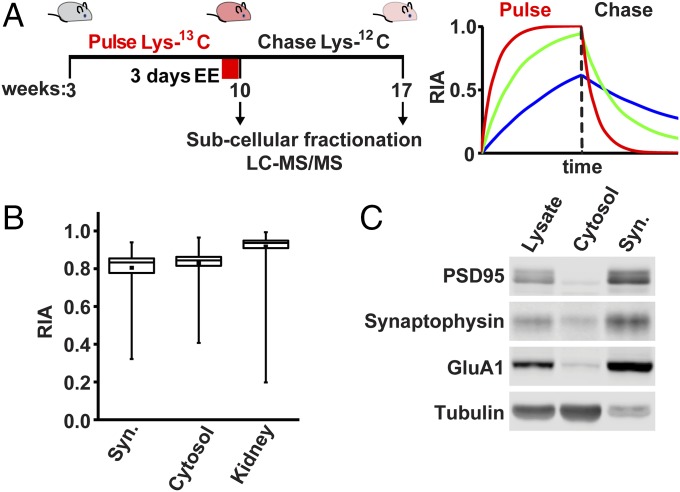
To systematically measure synaptic protein turnover in vivo and identify synaptic LLPs, we performed metabolic labeling in mice using heavy/light isotope-containing rodent chow. Following weaning, 3-wk-old mice were fed with a diet enriched in stable heavy-isotope-labeled amino acids (Lys-13C6) for a pulse period of 7 wk, during which time the majority of newly synthesized proteins in the mouse incorporate the heavy isotope label (discussed below). The mice were then switched to feed containing unlabeled (light) amino acids (Lys-12C6) for a chase period of 7 wk, during which time the majority of heavy-labeled proteins are replaced with newly made unlabeled proteins (Fig. 1 A and B). Our in vivo metabolic labeling strategy resulted in >80% labeling efficiency of synaptosomal and cytosolic fractions of mouse forebrain during the pulse (Fig. 1B). The kidney proteome showed labeling efficiency at >90% (Fig. 1B), a significantly higher relative isotope abundance (RIA) [heavy/(heavy + light)] compared with forebrain proteins, likely due to the higher rates of cellular renewal and proliferation in the kidney. Stable proteins with slow turnover incorporate heavy isotope label at a slower rate during the pulse but also retain a greater portion of heavy isotope label at the end of the chase (Fig. 1A). Just before chase, one cohort of mice remained in their home cages, while another underwent 3 d of enriched environment (EE), a condition expected to drive neuronal activity and plasticity across a large portion of the brain (40–42) (Fig. 1A). Mice brains were harvested at the end of the pulse or chase periods followed by dissection of the forebrain (cortex and hippocampus) and subcellular fractionation to isolate synaptosomes. This region of the brain was chosen for analysis as it is known to be critical for learning and memory. The quality of synaptosomes used for proteomic analyses was monitored by Western blot of synaptic proteins in sequential fractions (Fig. 1C). Proteins were digested with Lys-C protease to generate peptides and analyzed by high-resolution MS to identify peptides and quantify heavy/light ratios. First, the peak intensities of the heavy and light isotope-containing peptides from different tissues or subcellular fractions were obtained and labeling efficiency was validated by calculating RIA. Our dataset consists of four groups of two mice each, control pulse 7 wk, control chase 7 wk, EE pulse 7 wk, and EE chase 7 wk. MS analysis identified over 5,500 proteins from the synaptosome fractions, with 2,272 proteins quantified for RIA values obtained in all eight mice (Fig. S1).
Fig. 1.
Metabolic labeling in mice and identification of synaptic LLPs. (A, Left) Schematic of metabolic labeling in mice. At 3 wk old, male mice were fed with chow containing heavy-isotope-labeled Lys-13C6 for 7 wk, followed by unlabeled chow for 7 wk. Mice were exposed to either control or EE for 3 d before chasing with unlabeled chow. Mice were killed at the end of the pulse or chase periods followed by tissue isolation, subcellular fractionation, and analysis by LC-MS/MS. (A, Right) Stable proteins incorporate less label during pulse but retain more during chase. Unstable proteins rapidly acquire and then lose heavy label. (B) RIA of heavy lysine incorporated in the proteome during the pulse of forebrain synaptosomes (Syn.), cytosol, and or whole kidney. Box plot shows high levels of heavy labeling were achieved on average; individual protein species show variable labeling according to their turnover. Labeling efficiency was significantly higher in the kidney compared with brain, indicating higher rates of protein metabolism (P < 0.05, Student’s t test). Data obtained from eight mice. Error bars indicate the full distribution of data points. (C) Subcellular fractionation of mouse forebrain was monitored by Western blot analyses of synaptic and nonsynaptic proteins. Whole forebrain lysate, cytosol, and synaptosomal fractions (Syn.) were analyzed.
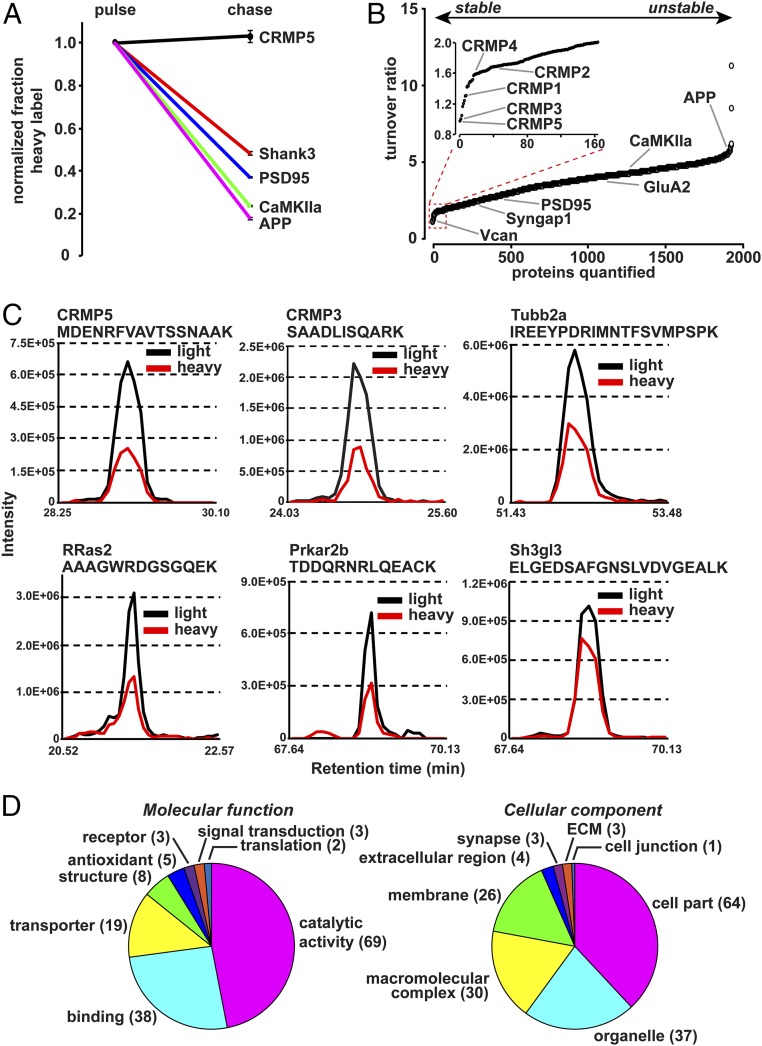
During the chase period, RIA values decrease as heavy isotope labels are replaced by light ones (Fig. 2A). The ratio of RIA during pulse to RIA during chase allows for a measure of protein turnover with a value of 1 showing no turnover and greater values indicating higher turnover rates (Fig. 2B). Synaptosome protein turnover measured from all four control mice spanned an order of magnitude (Fig. 2B). We defined LLPs as those proteins which retained at least 50% of the heavy label obtained during the pulse after the 7-wk chase period, corresponding to an RIA turnover ratio of 2.0 or less, indicating a half-life of several weeks or months, discovering 164 synaptosomal LLPs from control mice (Table 1). In comparison, the average RIA turnover ratio was 3.60 and the median was 3.74. The core synaptic scaffold protein PSD95 (Dlg4) showed a turnover ratio of 2.69, while amyloid beta A4 protein (App), which is known to undergo proteolytic cleavage, showed a high RIA turnover ratio of 5.61 (Fig. 2B). Reconstructed MS1 extracted ion chromatograms of proteins with slow turnover ratio showed a large amount of remaining heavy lysine after 7 wk of chase period (Fig. 2C and Fig. S2). Data obtained from the two replicates of control or EE conditions were highly correlated (R2con. pulse = 0.95, R2con. chase = 0.92, R2EE pulse = 0.89, R2EE chase = 0.93) (Fig. S3). Gene Ontology analysis revealed that LLPs showed a broad spectrum of molecular function and cellular components including cytoskeletal proteins, mitochondrial proteins, scaffold and signaling molecules, enzymes, and ECM proteins (Fig. 2D).
Fig. 2.
Characterization of synaptic LLPs. (A) Examples of decline in heavy label during chase, normalized to pulse. The decrease in heavy label-containing peptides during the chase period is used to indicate protein turnover, with greater decline indicating higher turnover. Data obtained from four mice from control group. (B) Plot indicates ranked turnover ratio for 2,272 proteins. The turnover ratio is calculated as the RIApulse/RIAchase. A value close to 1 indicates slow turnover, and larger values indicate greater protein turnover. (Inset) The 164 proteins defined as LLPs with a turnover ratio of 2 or less. Data obtained from four mice from control group. (C) MS1 extracted-ion chromatogram of representative peptides of LLPs from the chase period. Intensities of isotopes from eluted peptides were aligned according to their retention time and plotted for representative LLP peptides for CRMP5, CRMP3, Tubb2a, RRas2, Prkar2b, and Sh3gl3. Light isotope signal is plotted in black and heavy signal in red. (D) Chart indicates molecular functions and cellular components of 164 synaptosomal LLPs defined by at least 50% heavy label accumulated during pulse remaining after chase.
Table 1.
Identified LLPs from mouse forebrain synaptosomes
| Gene symbol | Average turnover ratio | Gene symbol | Average turnover ratio | Gene symbol | Average turnover ratio | Gene symbol | Average turnover ratio | Gene symbol | Average turnover ratio |
| Crmp5 | 0.970 | Cycs | 1.638 | Exog | 1.721 | Coro2a | 1.842 | Tamm41 | 1.920 |
| Crmp3 | 0.992 | Ncan | 1.638 | Got2 | 1.722 | Serac1 | 1.842 | Cltc | 1.926 |
| Vcan | 1.039 | Gap43 | 1.649 | Slc44a2 | 1.723 | Idh3a | 1.846 | Aldh5a1 | 1.928 |
| Hapln1 | 1.159 | Slc13a5 | 1.660 | Nipsnap1 | 1.727 | Clta | 1.847 | Slc25a5 | 1.929 |
| Mbp | 1.199 | Lactb | 1.668 | Samm50 | 1.735 | Tuba1b | 1.851 | C1qbp | 1.940 |
| Plp1 | 1.238 | Gnb2 | 1.670 | Phb | 1.736 | Hras | 1.853 | Sept3 | 1.944 |
| Crmp1 | 1.293 | Sh3gl1 | 1.674 | Gng4 | 1.740 | Hspd1 | 1.858 | Aifm1 | 1.944 |
| Tubb2a | 1.304 | Cyc1 | 1.676 | Sod2 | 1.741 | Acat1 | 1.859 | Aco2 | 1.948 |
| Sh3gl3 | 1.408 | Minos1 | 1.678 | Mcu | 1.745 | Cltb | 1.862 | Ckmt1 | 1.949 |
| Rras2 | 1.422 | Rala | 1.680 | Sh3bgrl2 | 1.748 | Atp5j | 1.866 | Ndufb7 | 1.950 |
| Cnp | 1.425 | Crmp2 | 1.681 | Nt5m | 1.765 | Lin7a | 1.868 | Gng3 | 1.951 |
| Dlat | 1.461 | Nipsnap3b | 1.682 | Rgs6 | 1.767 | Atp5a1 | 1.870 | Amph | 1.957 |
| Gnaz | 1.484 | Traf3 | 1.683 | Me3 | 1.769 | Mrps36 | 1.874 | Snca | 1.957 |
| Dlst | 1.486 | Dld | 1.685 | Pdha1 | 1.775 | Snap91 | 1.875 | Tubb3 | 1.958 |
| Nefh | 1.495 | Tnr | 1.689 | Sfxn3 | 1.777 | Epb41l2 | 1.876 | Ndufb10 | 1.958 |
| Gnao1 | 1.510 | Prkar2a | 1.691 | Uqcrc1 | 1.778 | Slc25a11 | 1.878 | Marcks | 1.969 |
| Ina | 1.558 | Atp5c1 | 1.694 | Uqcrb | 1.781 | Fh1 | 1.883 | Uqcrfs1 | 1.969 |
| Gnao1 | 1.574 | Phb2 | 1.695 | Uqcrc2 | 1.787 | 2310061I04Rik | 1.884 | Atad3a | 1.971 |
| Crmp4 | 1.580 | Txnrd2 | 1.700 | Ptprz1 | 1.790 | Abcb8 | 1.885 | Ndufb3 | 1.975 |
| Pdhx | 1.580 | Palm | 1.704 | Atp5b | 1.795 | Hexa | 1.885 | Ndufb9 | 1.976 |
| Nfasc | 1.589 | Atp5h | 1.705 | Gbas | 1.800 | Rac3 | 1.886 | Fahd2a | 1.978 |
| Pdhb | 1.589 | Stx1a | 1.706 | Abcg2 | 1.801 | Mmab | 1.889 | Suclg1 | 1.978 |
| Basp1 | 1.592 | Atp5f1 | 1.707 | Cs | 1.806 | Slc25a12 | 1.890 | Actn1 | 1.979 |
| Tubb4b | 1.596 | Atp5l | 1.707 | Tuba4a | 1.806 | Prdx3 | 1.895 | D10Jhu81e | 1.982 |
| Sirt2 | 1.609 | Tuba1a | 1.707 | Scai | 1.814 | Slc25a4 | 1.895 | Ndufb5 | 1.983 |
| Nefl | 1.609 | Atp5k | 1.708 | ATP8 | 1.817 | Ndufb8 | 1.897 | Cisd1 | 1.985 |
| Tpm4 | 1.611 | Atp5o | 1.710 | Timm9 | 1.819 | Gk | 1.898 | Ndufa8 | 1.986 |
| Prkar2b | 1.614 | Atp5d | 1.712 | Apoo | 1.823 | Tufm | 1.902 | Ndufb11 | 1.989 |
| Gng2 | 1.618 | Aldh1b1 | 1.713 | Acot9 | 1.825 | Vdac1 | 1.902 | Prdx5 | 1.989 |
| Gnb1 | 1.619 | Mtch2 | 1.716 | Vdac3 | 1.828 | Mecr | 1.905 | Vdac2 | 1.991 |
| Bdh1 | 1.619 | Epb41l3 | 1.716 | Hdhd3 | 1.829 | Syn1 | 1.912 | Slc2a3 | 1.993 |
| Nefm | 1.625 | Map6 | 1.716 | Sept11 | 1.838 | Nrn1 | 1.914 | Chchd3 | 1.996 |
| Sept6 | 1.630 | Mdh2 | 1.719 | Sfxn1 | 1.839 | Sgip1 | 1.919 |
Listed are the gene symbols (columns 1, 3, 5, 7, and 9) and average turnover ratio (columns 2, 4, 6, 8, and 10) from control group of SILAM mice. Same gene symbols represent different isoforms.
For example, we identified myelin sheath proteins such as Plp1 (myelin proteolipid protein), proteoglycans or proteoglycan-related proteins such as Vcan (versican core protein), and Hapln1 (hyaluronan and proteoglycan link protein) as long-lived, in agreement with earlier studies (Table 1) (8, 43). We particularly noted a family of five microtubule-associated proteins, collapsin response mediator protein (CRMP1–5), also called dihydropyrimidinase-related proteins (Dpysl1-5), which were all found to have extremely slow turnover: CRMP1 (1.29), CRMP2 (1.68), CRMP3 (0.99), CRMP4 (1.58), and CRMP5 with a turnover ratio of 0.97 showed no turnover over in 7 wk (Fig. 2 A and B and Table 1). CRMP proteins have been previously characterized as playing a role in axon guidance and development of neuronal circuits as well as synaptic plasticity (44) and have not previously been described as extremely stable proteins. Other notable LLPs include small GTPase proteins of the Ras family (Rras2), BAR domain proteins (Sh3gl1 and Sh3gl3), which have prominent functions in synaptic vesicle endocytosis and receptor trafficking, and the type-II regulatory subunits of PKA (Table 1). We found that other identified proteins with higher turnover were grouped in the same cellular components, molecular functions, or protein classes as LLPs (Fig. S4 A and B).
Protein Turnover Is Accelerated in Cytosol Compared with Synaptosomes.
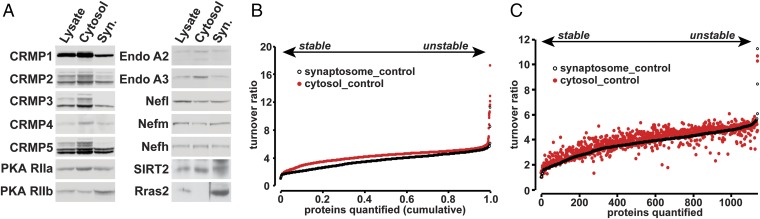
Several LLPs identified in the synaptosomes are also present in the cytoplasm, including CRMP isoforms and PKA subunits (Fig. 3A). We also examined protein turnover from the cytoplasmic fraction of mouse forebrains and were able to quantify 2,119 proteins. Cytosolic proteins showed an almost uniform shift toward higher turnover compared with synaptosomes (Fig. 3B). The median protein turnover ratio from control group was 4.35 in cytosol compared with 3.71 in synaptosomes (P < 0.001, Student’s t test, Fig. 3B). Although proteins from cytosolic fraction seemed to have higher turnover ratio under both control conditions, certain proteins were disproportionally affected, as shown by the paired plot in Fig. 3C. These findings suggest that some proteins may form relatively stable complexes in synapses that reduce their turnover or that proteins in the cytoplasm are more readily accessible to protein degradation machinery, accelerating their turnover.
Fig. 3.
Synaptosome proteins are stabilized relative to cytosolic proteins. (A) Western blot of forebrain subcellular fractions. Many LLPs are localized in both cytosol and synaptosomal fractions (Syn.). (B) Plot indicates the ranked protein turnover ratios of synaptosome and cytosol proteomes plotted against the fraction of the total number of proteins identified. Cytosolic proteins showed a statistically significant increase in protein turnover under control conditions (P < 0.001, Student’s t test). (C) Plot indicates the ranked protein turnover from synaptosomal fraction and the paired values of proteins from cytosolic fraction. The spread indicates that turnover ratio of certain proteins was disproportionally affected by their subcellular localization. Data obtained from four control mice.
Protein Turnover Kinetics Are Accelerated by Experience-Dependent Synaptic Plasticity.
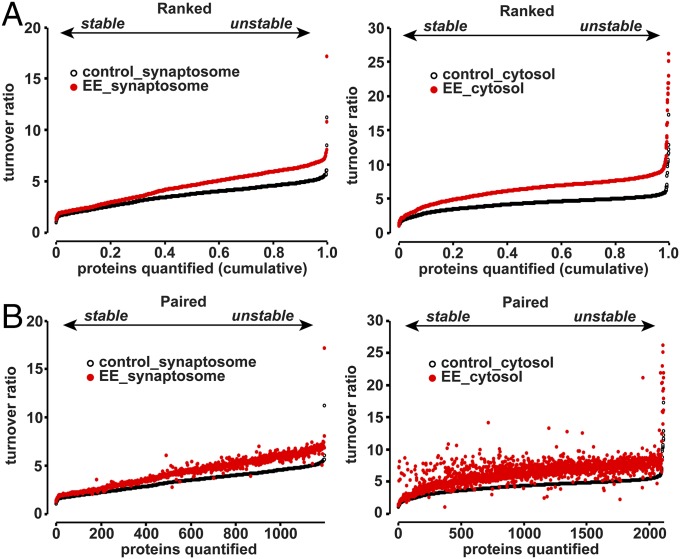
Previous studies have shown that EE exposure can result in altered gene expression, proliferation of neural progenitor cells, and altered threshold for synaptic plasticity (40, 41, 45). We have found that acute EE exposure can induce expression of plasticity-related genes, such as Arc and Homer1a, and enhance phosphorylation of AMPARs throughout the cortex and hippocampus (46). It is clear that protein synthesis and degradation play critical roles in synapse maintenance and plasticity and the cross-talk between these two distinct protein pathways must be properly orchestrated. However, how changes in global neuronal activity affect protein turnover are not clear. In our experiment, mice underwent 3-d EE exposure, with novel objects changed daily, just before the chase period. Our expectation was that EE would induce synaptic plasticity across large parts of the forebrain and certain heavy-labeled proteins would show altered stability 7 wk later at the end of the chase. Surprisingly, we found that synaptosome proteins from EE-exposed mice showed an almost uniform shift toward increased protein turnover when comparing the ranked turnover ratios from control mice (Fig. 4A; median turnover ratio 3.80 in control mice and 4.91 in EE exposed mice, P < 0.001, Student’s t test). Exposure to EE also resulted in increase of turnover ratio of proteins from cytosolic fractions (Fig. 4A; median turnover ratio 4.35 in control mice and 6.52 in EE exposed mice, P < 0.001, Student’s t test). This finding suggests that enhanced neuronal activity during exposure to an enriched environment accelerated the turnover of the majority of proteins, consistent with results showing that synaptic plasticity controls both protein synthesis and degradation (5–7, 47–53). Although there seemed to be an overall shift toward higher turnover in EE mice, certain proteins were disproportionally affected, as shown by the paired plot in Fig. 4B. We identified Casein kinase Iα as the only protein stabilized by greater than 40% in EE-exposed mice compared with control and 247 proteins destabilized by more than 40% compared with control (Table S1). Amyloid precursor protein APP showed a disproportionate destabilization in EE-exposed mice, consistent with findings that proteolytic processing of APP is accelerated by neuronal activity (Table S1) (54).
Fig. 4.
Protein turnover is accelerated by EE experience. (A) Plot indicates the ranked turnover ratios of synaptosomal and cytosolic proteins from control mice and mice that underwent 3 d of EE exposure. EE exposure resulted in an almost uniform increase in protein turnover that was statistically significant (P < 0.001, Student’s t test). Data obtained from four control mice and four EE mice. (B) Plot indicates the ranked protein turnover from control mice and the paired values obtained from EE mice. The spread indicates that certain proteins from synaptosomal and cytosolic fractions were disproportionally affected by EE exposure. Data obtained from four control mice and four EE mice.
Measurement of Protein Half-Lives in Cultured Neurons with Pharmacological Manipulations of Neuronal Activity.
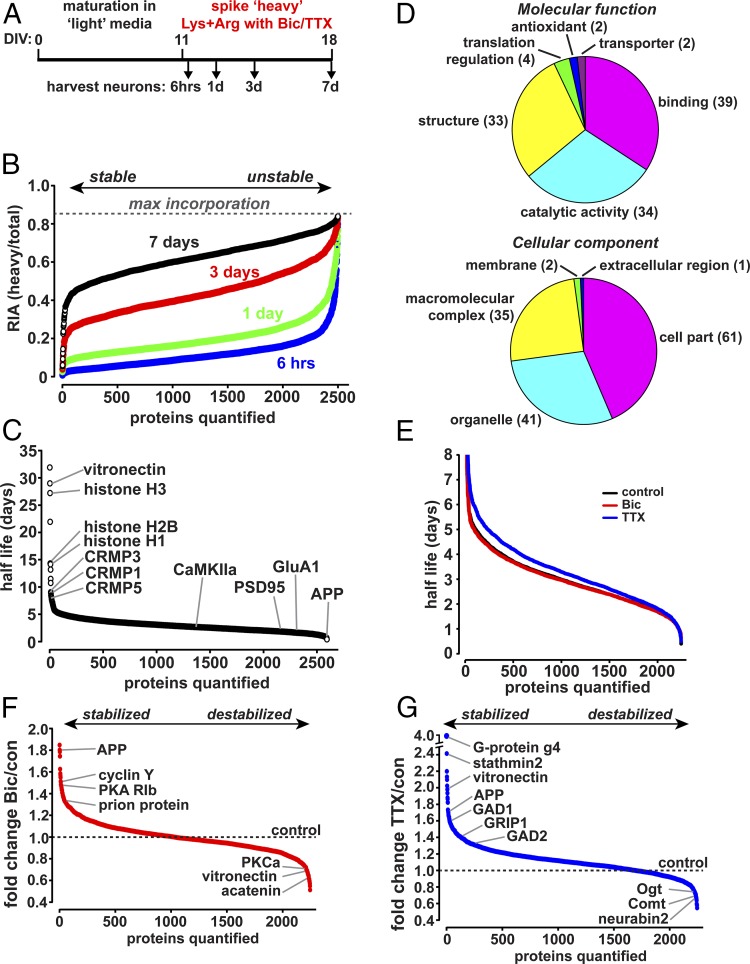
Exposure to EE accelerated protein turnover in mice, suggesting that protein lifetime kinetics may be sensitive to neuronal activity. Neuronal activity changes have also been found to lead to changes in protein expression in vitro (55, 56). To further address the effect of neuronal activity on protein turnover, we adapted metabolic labeling using stable isotope-labeled amino acids in primary cultured rat cortical neurons (Fig. 5A). Rat cortical neurons were grown in culture under normal growth conditions until synapses began to form at day in vitro (DIV) 11 and treated thereafter with a fivefold excess of heavy isotope-labeled lysine and arginine. These heavy amino acids are incorporated into newly synthesized proteins, which can then be detected using MS(Fig. 5B and Table 2). Heavy isotope labeled proteins accumulated as new proteins are synthesized, while older unlabeled proteins decrease in abundance as they are degraded. With a fivefold excess of heavy amino acids the theoretical maximum RIA will be 0.833 of the total protein. Proteins with the highest turnover approach the maximum incorporation within a few hours of isotope labeling, while proteins with the lowest turnover incorporate much less isotope label, even after 7 d of labeling (Fig. 5B). Thus, at different times after the addition of labeled amino acids the RIA can be determined and the rate at which proteins exchange light for heavy amino acids can be used to calculate the half-life of individual protein species (57) (Fig. 5 B and C). Coincident with the addition of heavy-labeled amino acids, neuronal network activity was increased by the addition of bicuculline (Bic), an antagonist of inhibitory GABAA receptors, or suppressed by the addition of TTX, an inhibitor of voltage-gated sodium channels, to induce global increases or decreases in neuronal activity, respectively. Total neuronal lysates were then collected 6 h and 1, 3, or 7 d after the addition of heavy amino acids together with Bic or TTX (Fig. 5A); proteins were digested with trypsin to generate peptides and then subjected to high-resolution MSanalysis.
Fig. 5.
Protein turnover in cultured neurons is regulated by neuronal activity. (A) Schematic of metabolic labeling in cultured neurons. Rat cortical neurons were grown for 11 d in vitro (DIV11) under normal conditions. The media was then spiked with a 5× excess of heavy isotope labeled lysine and arginine with or without the addition of Bic (20 μM) or TTX (1 μM). Whole-cell lysate was obtained at indicated times following treatment and analyzed by MS. (B) Plot indicates the ranked RIA for 2,593 proteins identified/quantified in all four time points as indicated. RIA increases steadily with time after labeling, approaching the maximum RIA value of 0.833. Proteins with slower turnover incorporate less isotope label while proteins with higher turnover rapidly acquire the isotope label. The rate of isotope incorporation is used to calculate half-life. (C) Plot indicates ranked half-lives in days for 2,593 proteins from control-treated neurons. Half-lives calculated from four separate time points. (D) Plot indicates the molecular functions and cellular components for 117 proteins with a half-life of 5 d or longer. (E) Plot indicates ranked half-lives for 2,245 proteins from control-, Bic-, and TTX-treated neurons. Half-lives calculated from four separate time points. TTX resulted in a statistically significant shift toward longer protein half-lives (P < 0.001, Student’s t test), while Bic treatment caused a slight decrease in overall protein half-lives that was not significant (Student’s t test). (F) Plot indicates the fold change in protein half-life from Bic-treated neurons over control ranked from proteins stabilized by Bic (Bic/con > 1) to those destabilized (Bic/con < 1). The dashed line indicates the control protein half-life. (G) Plot indicates the fold change in protein half-life from TTX-treated neurons over control ranked from proteins stabilized by TTX (TTX/con > 1) to those destabilized (TTX/con < 1). The dashed line indicates the control protein half-life.
Table 2.
Identified LLPs from cultured rat cortical neurons
| Gene symbol | Average half-life, d | Gene symbol | Average half-life, d | Gene symbol | Average half-life, d | Gene symbol | Average half-life, d |
| LOC680498 | 31.89 | Rpsa | 6.72 | Eef1b2 | 5.51 | Coro1a | 5.19 |
| Vtn | 28.98 | Tubb2a | 6.71 | Rps4x | 5.51 | LOC103694877 | 5.19 |
| LOC684762 | 27.21 | Rps6 | 6.54 | Eef1g | 5.48 | Arsa | 5.19 |
| LOC102551184 | 21.95 | Tmsb4x | 6.52 | Acat2 | 5.47 | Rbmx | 5.18 |
| LOC100910200 | 14.31 | Eef1d | 6.39 | LOC103689992 | 5.45 | Naxe | 5.18 |
| Hist1h1b | 14.21 | Pebp1 | 6.30 | Acat1 | 5.45 | Rpl5 | 5.18 |
| LOC680322 | 14.16 | Crmp2 | 6.27 | LOC100911402 | 5.42 | Rpl27a | 5.15 |
| Hist2h2aa3 | 13.19 | Atp5j | 6.22 | Nup93 | 5.41 | Tubb6 | 5.14 |
| Mapt | 11.47 | H2afy2 | 6.09 | Rpl11 | 5.39 | Rps15 | 5.12 |
| H3f3c | 10.73 | Ddah2 | 6.02 | Srsf7 | 5.39 | Dpp7 | 5.12 |
| Ncam1 | 9.04 | Ndufb5 | 5.91 | Rps2 | 5.39 | Snrpf | 5.12 |
| Tubb2b | 8.83 | Pafah1b3 | 5.89 | Rpl14 | 5.35 | Tpm4 | 5.12 |
| Crmp3 | 8.45 | Ap2a2 | 5.87 | Alg9 | 5.34 | Dhodh | 5.09 |
| Lmnb1 | 8.40 | Hist3h2ba | 5.79 | Actc1 | 5.33 | Rpl7 | 5.09 |
| Basp1 | 8.21 | Sfxn3 | 5.74 | Cotl1 | 5.32 | H2afy | 5.09 |
| Crmp1 | 8.20 | Sod1 | 5.74 | Prkar2a | 5.32 | Ppa1 | 5.09 |
| Tubb5 | 8.01 | Hist1h1d | 5.72 | Rps3a | 5.30 | Tuba1b | 5.09 |
| Tubb4b | 7.75 | Hnrnpl | 5.68 | Hnrnpa3 | 5.29 | Gdi1 | 5.09 |
| Tubb3 | 7.71 | Rps8 | 5.68 | Hnrnpa1 | 5.27 | Idh1 | 5.08 |
| Prkar2b | 7.52 | LOC102555453 | 5.65 | Cct4 | 5.27 | Stip1 | 5.07 |
| Nop10 | 7.51 | C1qbp | 5.64 | Rps20 | 5.26 | Ranbp1 | 5.07 |
| Hnrnpa3 | 7.40 | Rpl18 | 5.64 | Fdxr | 5.25 | Katnal2 | 5.06 |
| Rac3 | 7.29 | Nme1 | 5.62 | Rplp1 | 5.25 | Hnrnpd | 5.06 |
| Crmp1 | 7.22 | Ncam1 | 5.62 | LOC100362830 | 5.25 | Tpm3 | 5.03 |
| Tuba1a | 7.20 | Gap43 | 5.61 | Rpl7a | 5.25 | Rpl4 | 5.03 |
| Nme2 | 7.19 | Arhgdia | 5.59 | Fabp5 | 5.23 | Psme2 | 5.02 |
| Crmp5 | 7.13 | Rpl23 | 5.57 | Nipsnap1 | 5.22 | Mtch2 | 5.02 |
| Mapt | 6.89 | Rpl10a | 5.56 | Tufm | 5.21 | Ybx1 | 5.02 |
| Snrpa | 6.78 | Atp5h | 5.54 | Prdx2 | 5.21 | Cct5 | 5.00 |
Listed are the gene symbols (columns 1, 3, 5, and 7) and calculated average half-life (days) (columns 2, 4, 6, and 8) obtained from control group of SILAC experiment using cultured rat cortical neurons. Same gene symbols represent different isoforms.
In total, greater than 6,700 proteins were identified. Using this approach, we have been able to obtain half-life estimates for 2,593 proteins in control neurons (calculated from all four time points), and 2,245 neuronal proteins identified/quantified in all four time points from control, Bic, and TTX treatment, including stable and short-lived proteins, demonstrating the feasibility of this approach (Fig. 5C, Fig. S5, and Table S2). Reconstructed MS1 elution profile of proteins with longer half-life showed a small amount of incorporated heavy arginine or lysine after 7 d of pulse period while proteins with shorter half-life showed higher portion of incorporated heavy arginine or lysine (Fig. S6); 117 proteins (4.5% of the total) showed a half-life of 5 d or longer, including cytoplasmic, nuclear, cytoskeletal, ribosomal, mitochondrial, and cell-surface proteins with various molecular functions such as binding, catalytic, and structure (Fig. 5D). Several LLPs identified in the in vivo labeling studies in mice, such as CRMPs and type-II PKA regulatory subunits, were also found to be relatively long-lived in cultured cells in vitro (Fig. 5C), suggesting that metabolic stability may be an inherent property of these proteins. Histone subunits also showed notable longevity consistent with previous studies (43). Suppression of neuronal activity with TTX resulted in significantly slower protein turnover and greater protein stability, whereas increased neuronal activity with Bic treatment caused a slight acceleration of protein turnover that was not statistically significant (median half-life: 2.83 d control; 3.13 d TTX; 2.80 d Bic) (Fig. 5E). While TTX treatment caused an overall decrease in protein turnover and Bic treatment caused a slight increase, specific proteins showed stabilization or destabilization (Fig. 5 F and G), perhaps as part of the homeostatic adaptation to altered neuronal activity (58, 59). We defined Bic or TTX treatment as having a specific effect on protein turnover if the half-life was stabilized or destabilized by 40% or greater compared with control treatment. With these criteria, we found that Bic treatment caused the stabilization of 28 proteins and destabilization of 37 proteins; TTX treatment caused the stabilization of 135 proteins and destabilization of 17 proteins (Table S2). Intriguingly, the overall reduction in protein turnover in cultured neurons upon suppression of neuronal activity was similar to the accelerated protein turnover observed following EE exposure in mice, suggesting a strong relationship between protein turnover and neuronal activity. Since EE and neuronal activity are known to regulate protein synthesis rates this could complicate the determination of the turnover rates of some proteins. Further studies of individual proteins will be required to address this question.
Discussion
Previous studies have shown that the human body contains many LLPs, such as collagen in cartilage, that can have half-lives longer than 100 y (14, 60). LLPs have also been found to form stable structures within the metabolically active environment of the cell, such as histone octamers and nuclear pore complexes. The extreme stability of these proteins is likely important for the structure and function of chromatin and the nuclear pore complex. LLPs may be especially important in neurons in the brain, which have a very limited capacity for renewal. We hypothesized that a subset of LLPs may reside in neuronal synapses, and this extreme stability would be important in long-lasting structural and functional properties of synapses, such as the maintenance of long-term plasticity and memory. Interestingly, LLPs would be expected to accumulate oxidative damage over time, potentially forming a source of vulnerability during aging. Synaptic LLPs that maintain memories could therefore play a critical role in age-related cognitive decline.
We used unbiased and systematic metabolic labeling and high-resolution MSin mouse brain synaptosomes and cultured neurons to characterize protein turnover and identify novel neuronal LLPs. We found that the synaptosomal proteome from adult mice does indeed contain LLPs, many of which have not been previously described for their longevity, including type-II regulatory subunits of PKA, several components of the ECM, and all five members of the CRMP family of microtubule binding proteins. CRMP5 in particular showed almost no turnover during the 7-wk chase period. CRMPs and others were also found to be among the most stable proteins in cultured neurons in vitro, suggesting that longevity is not unique to the adult brain and may be an inherent property of these proteins. Furthermore, we provide evidence both in mouse brain and cultured neurons that global protein turnover is sensitive to neuronal activity.
Having identified many LLPs within neuronal synapses, major questions yet to be addressed are what mechanism(s) allows for this exceptional longevity and what functional role this extreme stability conveys to the proteins and the synapse. Moreover, why would neurons maintain these synaptic LLPs that in the metabolically active environment of the synapse would be potentially exposed to oxidative damage? Protein turnover rates span a very wide range, from minutes to decades, and also vary between tissues and cell types (43). The turnover of a protein is determined by multiple factors including the (i) primary sequence, (ii) cellular localization, and (iii) incorporation in larger protein complexes or cellular structures. The primary sequence can confer inherent stability when proteins adopt a particularly stable 3D conformation such as the β-can structure of GFP, or instability through inherently disordered regions or motifs that target the protein for proteolytic processing, such as the amyloid precursor protein APP, which was found to be among the most unstable proteins in mouse brain and cultured neurons. Longevity can also be conferred by targeting proteins to metabolically inactive environments such as the ECM, where proteins such as collagen can reside for very long periods. This likely explains the longevity of several ECM LLPs identified in our study, such as Vcan and Hapln1. Longevity may be conferred as proteins are incorporated in larger protein complexes or cellular structures where the individual proteins cannot be replaced without disruption of the complex and consequences to cellular function. The latter was proposed as the mechanism that allows for the exceptional longevity of histones and nuclear pore complex components (43), where renewal of these proteins may require transient disruption of the nuclear envelope or chromatin structures, causing loss of nuclear/cytoplasmic compartmentalization and altered gene expression. Interestingly, we found that protein turnover was significantly higher in the cytoplasm than in synaptosomes, even for the same protein species found in both fractions, suggesting that these proteins may be incorporated into stable complexes or structures upon targeting to the synapse.
Some of the synaptic LLPs discovered in the current study may achieve longevity through particularly stable 3D folded structures. However, there is currently no evidence to suggest this is the case. Indeed, many of the LLPs we have identified have related proteins that were not long-lived. For example, RRas2 and type-II PKA regulatory subunits were found to be highly stable, while members of the structurally related Rab family or type-I PKA regulatory subunits were not. Additional structural or bioinformatic analyses may reveal certain sequence elements in the primary structure of LLPs that confer longevity.
In the second case, we identified several components of the ECM, such as Vcan and Hapln1, whose longevity could be explained by localization to the metabolically inactive extracellular space. It has been proposed that part of the long-term maintenance of memories may come from the perineuronal net (PNN), a lattice-like structure of ECM that wraps certain neurons and dendritic processes. The PNN remains flexible during development and fully crystalizes as the brain matures, remaining largely stable for the remainder of life (11). By wrapping certain synapses in an inflexible structure, the PNN may constrain and maintain the structure of dendritic spines and axonal boutons. During synaptic plasticity, extracellular proteases have been reported to be transiently activated to remodel the PNN (11). As an extracellular structure, the proteins that make up the PNN may be highly resistant to typical cellular protein degradative machinery and damage from intracellular metabolites, offering a substantial advantage for the maintenance of memories. A recent study showed that not only was the PNN of the adult mouse sensory cortex temporarily regulated by fear learning, but it was also necessary for fear learning (61). Since the synaptic long-lived proteome includes both intracellular and extracellular proteins, we suggest that long-term maintenance of synapses and memories will require the coordinated organization of intracellular and extracellular LLP complexes.
Longevity may be conferred by incorporation of proteins into larger complexes or structures (62). CRMPs and type-II PKA regulatory subunits are known to form protein complexes, which we propose are required to maintain certain functional or structural properties of synapses. Similar to histones and nuclear pore complexes, replacement of synaptic LLPs may transiently disrupt synapse structure, leading to loss of information storage or cognitive function. Type-II PKA regulatory subunits are important to position the PKA holoenzyme by interaction with anchoring scaffold proteins and are known to play a role in synaptic plasticity and synaptic tagging (63, 64). It has been recently shown that PKA catalytic subunits remain tethered to their type-II regulatory subunits upon activation by cAMP, limiting the activity of the kinase to within 25 nm of the site of anchoring (65). As synaptic LLPs, type-II PKA regulatory subunits could be important to maintain and limit the activity of the PKA holoenzyme within synaptic nanodomains to maintain some aspect of synapse structure or functionality for long periods.
CRMP1–5, some of the most stable proteins from the current study, are involved in neuronal development, axonal outgrowth, cell migration, and protein trafficking, through interaction with microtubules and regulation of microtubule dynamics (44, 66). Microtubules have been recognized as stable structures that support neuronal structures such as dendritic shafts and axons in mature neurons. Imaging studies focused on microtubule-associated proteins such as EB3 and Tau show that microtubules, which are typically absent from dendritic spines, transiently invade spines during synaptic plasticity, potentially delivering cargo required for synapse remodeling (67–70). Both tubulin and CRMPs undergo extensive reversible posttranslational modifications that regulate microtubule dynamics (71, 72). Phosphorylation of CRMP isoforms by Cdk5 and GSK3β at conserved sites causes the dissociation of CRMPs from the microtubule network (44, 66, 73, 74). We hypothesize that CRMPs may form a long-lasting complex with microtubules generating particularly stable conformations of the microtubule network that can maintain structural or functional properties of synapses. Combinations of modifications on CRMPs and tubulin may form a localized molecular code to direct protein and vesicular traffic or other cytoskeletal dynamics. Since the modifications on tubulin and CRMPs are reversible, the formation and maintenance of new memories during synaptic plasticity could require the local recruitment or remodeling of stable CRMP complexes.
To investigate the changes of protein turnover during global and persistent neuronal activity, we exposed mice to an EE or treated cultured neurons in vitro with Bic or TTX. EE is known to induce behavioral, structural, and molecular changes throughout much of the brain (41, 42, 75). Our expectation was that robust memory formation during EE exposure would lead to the stabilization of memory-associated proteins. Surprisingly, EE induced an almost uniformly increased turnover of the synaptosomal proteome. In cultured cells, we found that blocking neuronal activity with TTX caused the opposite effect of almost uniform protein stabilization. The lack of overall effect from Bic treatment on protein turnover suggests that the basal activity of cultured neurons may already be quite high. Indeed, it is known that neuronal activity of neurons in culture is much higher than that typically observed in vivo. Together these findings suggest that ongoing neuronal activity plays an important role in global protein metabolism. However, in these experiments we also observed stabilization or destabilization of certain proteins, which may form part of the homeostatic adaptation to changes in neuronal activity (58, 59) and the long-term maintenance of memory.
Memories can last a lifetime, but many of the proteins that make up the synapse are degraded and replaced regularly. Here, we have used unbiased and systematic methods to examine the turnover kinetics of the neuronal and synaptic proteome. We found that overall protein metabolism is highly sensitive to neuronal activity and experience and that synaptic proteins are relatively stable compared with the cytosol. Importantly, we have identified a population of extremely LLPs in synapses which could potentially open new avenues of research into the long-term maintenance of plasticity and memory and age-related cognitive decline.
Materials and Methods
Animal Use and EE.
All animals were treated in accordance with the Johns Hopkins University Animal Care and Use Committee guidelines. For all experiments mice were age 8–10 wk. Mice were either left in their home cage or allowed to explore an enriched environment, a physiologically relevant condition expected to drive neuronal activity and synaptic plasticity (40–42), for 2 h (a large cage containing novel objects, tubes, and strings of beads suspended from the cage lid). Mice were then anesthetized by inhalation of isoflurane for 15 s followed immediately by cervical dislocation; brains were removed and forebrains (cerebral cortex and hippocampus) were dissected in ice-cold PBS and immediately frozen on dry ice. Samples were kept at −80 °C until used for subcellular fractionation.
Neuronal Cultures.
Cortical neurons obtained from Sprague-Dawley rats at embryonic day 18 were initially prepared in Neurobasal media (Invitrogen) supplemented with 2% B-27, 2 mM GlutaMax, 50 U/mL penicillin, 50 mg/mL streptomycin, and 5% horse serum (Invitrogen) and plated onto poly-l-lysine–coated tissue culture dishes. Cells were then transferred and maintained in a humidified tissue culture incubator at 37 °C in a 95% air and 5% CO2 mixture; 5 mM FDU (5-fluoro-2′-deoxyuridine; Sigma) was added at DIV 4 and cells were thereafter maintained in glia-conditioned NM1 (Neurobasal media with 2% B-27, 2 mM GlutaMax, 50 U/mL penicillin, 50 mg/mL streptomycin, and 1% horse serum). Cultured cortical neurons were fed twice per week. For all experiments, cortical neurons (grown for 13–14 d in vitro) were plated at a density of 800,000 cells per well into standard six-well tissue culture plates.
SILAM.
Metabolic labeling of mice with heavy isotope was accomplished as previously described with minor modifications (37, 76, 77). Briefly, following weaning at 3 wk old (P21), young male mice were fed with a diet enriched in stable heavy-isotope-labeled amino acids [MouseExpress l-Lysine (13C6, 99%) MOUSE FEED, Lys-13C6] for a pulse period of 7 wk. The mice were then switched to feed containing unlabeled (light) amino acids (MouseExpress Unlabeled MOUSE FEED, Lys-12C6) for a chase period of 7 wk, during which time the majority of heavy-labeled proteins are replaced with newly made unlabeled proteins.
At the end of the pulse period, just before chase, one cohort of mice remained in their home cages, while another cohort of mice was transferred to an EE cage for 3 d as described above. Mice from control and EE, pulse and pulse-chase labeling were then killed, tissues were harvested, and cortex and hippocampus were dissected and flash-frozen for further subcellular fractionation to yield synaptosomes and cytosol fractions.
Subcellular Fractionation and Western Blotting.
Tissues (cortex and hippocampus) collected after pulse- or pulse-chase labeling were homogenized using a Tenbroeck tissue grinder in homogenization buffer [320 mM sucrose, 5 mM sodium pyrophosphate, 1 mM EDTA, 10 mM Hepes, pH 7.4, 200 nM okadaic acid, and protease inhibitor mixture (Roche)]. The homogenate was then centrifuged at 800 × g for 10 min at 4 °C to yield P1 (nuclear fraction) and S1 fractions. S1 fraction was further centrifuged at 17,000 × g for 20 min at 4 °C to yield P2 (membrane/crude synaptosome) and S2 (cytosol) fractions. P2 was resuspended in homogenization buffer and layered onto nonlinear sucrose gradient cushion (1.2 M, 1.0 M, and 0.8 M sucrose from bottom to top) then centrifuged at 82.500 × g for 2 h at 4 °C. Synaptosomes were collected at the interface of 1.0 M and 1.2 M sucrose cushion. Collected synaptosomes were diluted with 10 mM Hepes, pH 7.4, to reach a final concentration to 320 mM sucrose and then centrifuged at 150,000 × g for 30 min at 4 °C. The final synaptosome pellet was resuspended in 50 mM Hepes, pH 7.4, followed by protein quantification and biochemical analysis.
For Western blotting analysis, samples were quantified using BCA protein assay kit and loaded onto 9 or 12% SDS/PAGE (depending on the molecular weights of the protein of interest). Proteins were transferred to PVDF membrane, and the membranes were blocked with Odyssey blocking buffer for fluorescent detection or 3% BSA in Tris-buffered saline with Tween (TBST) for chemiluminescent detection for 1 h at room temperature. Primary antibodies were resuspended in Odyssey blocker or 3% BSA in TBST for overnight at 4 °C with gentle rocking. Antibodies were used as follows: mouse anti-GluA1 (homemade clone 4.9D, 1:5,000), rabbit anti-CRMP1 (1:2,000; Abcam), rabbit anti-CRMP2 (1:300,000; Abcam), rabbit anti-CRMP3 (1:2,000; Abcam), rabbit anti-CRMP4 (1:2,000; Abcam), rabbit anti-CRMP5 (1:2,000; Abcam), rabbit anti-RRas2 (1:1,000; Abcam), mouse anti-endophilin 2 (1:1,000; Santa Cruz), mouse anti-endophilin 3 (1:1,000; Santa Cruz), rabbit anti-PKA2 beta regulatory subunit (1:10,000; Abcam), rabbit anti-PKA R2 regulatory subunit 2 alpha (1:5,000; Abcam), mouse anti-SIRT2 (1:500; Santa Cruz), mouse anti-neurofilament-H (1:1,000; Cell Signaling), mouse anti-neurofilament-M (1:1,000; Cell Signaling), mouse anti-neurofilament-L (1:1,000; Cell Signaling), mouse anti-PSD95 (1:1,000,000; NeuroMab), rabbit anti-synaptophysin (1:5,000; Abcam), and mouse anti-alpha tubulin (1:400,000; Sigma). Primary antibodies were removed and membranes were washed followed by secondary antibodies in blocking solutions as follows: donkey anti-rabbit IgG IRDye 680 conjugate (1:100,000; LI-COR), donkey anti-mouse IgG IRDye 800 conjugate (1:100,000; LI-COR), donkey anti-rabbit IgG HRP conjugate (1:6,000; GE Healthcare Life Sciences), and sheep anti-mouse IgG HRP conjugate (1:6,000; GE Healthcare Life Sciences). Blots were developed using either LI-COR Odyssey CLx Imaging system (LI-COR) or Luminata Forte Western HRP substrate (EMD Millipore) and imaged using manual film exposure.
SILAC and Drug Treatment.
For SILAC experiments, cortical neurons were grown for 11 d and then glia-conditioned NM1 containing heavy isotope-labeled l-Lysine:HCl [Lys8, (13C6, 99%; 15N2, 99%)] and l-Arginine:HCl [Arg10, (13C6, 99%; 15N4, 99%)] (Cambridge Isotope Laboratories) were added to culture dishes to achieve a heavy isotope spiked-in condition at 5:1 (heavy:light) ratio. Neurons were simultaneously treated with 20 μM Bic or 1 μM TTX to induce homeostatic scaling-up or -down and harvested after 6 h and 1, 3, and 7 d. Neurons were fed with glia-conditioned NM1 containing heavy lysine and arginine and additional Bic or TTX at concentrations that preserved the 5:1 heavy:light ratio. Bic and TTX were supplemented on day 3 of homeostatic scaling to compensate the loss of initial Bic and TTX. At each harvesting time point, neurons were gently washed twice with ice-cold PBS then scraped out using cell harvesting buffer [10 mM Hepes, pH 7.4, 320 mM sucrose, 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 1 mM EDTA/EGTA, 200 nM okadaic acid, and protease inhibitor mixtures (Roche)] and centrifuged at 1,000 × g for 5 min at 4 °C. Pelleted neurons were snap-frozen and kept at −80 °C until use. Pelleted neurons were lysed in 100 mM Tris⋅HCl, pH 8.0, containing 4% SDS (wt/vol) and 100 mM DTT, sonicated, and centrifuged at 16,000 × g at 15 °C.
Liquid Chromatography–MS/MS Analysis.
An Orbitrap Elite mass spectrometer or Orbitrap Fusion Tribrid or Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific) interfaced with Easy nLC 1000 liquid chromatography (LC) system (Thermo Fisher Scientific) or Easy nLC 1200 LC systems (Thermo Fisher Scientific) were used for LC-MS/MS analysis (see SI Materials and Methods for details).
Additional detailed materials and methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank all members of the R.L.H. laboratory for helpful comments, discussion, and critical reading of the manuscript. We also thank Drs. Min-Sik Kim and Gajanan J. Sathe for helping design experimental approach and proteomic data acquisition. This work was supported by NIH Grants P50MH100024 and R01NS036715 (to R.L.H.) and P50NS038377 and R01CA184165 (to A.P.), NIH shared instrumentation Grant S10OD021844 (to A.P.), and the Center for Proteomics Discovery at Johns Hopkins University and the Canadian Institute for Health Research (G.H.D.).
Footnotes
The authors declare no conflict of interest.
Data deposition: All MS data and search results have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD007156 and project name Identification of Long Lived Synaptic Proteins by Comprehensive Proteomic Analysis of Synaptosome Protein Turnover; the data are directly accessible via https://www.ebi.ac.uk/pride/archive/projects/PXD007156.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720956115/-/DCSupplemental.
References
- 1.Gale GD, et al. Role of the basolateral amygdala in the storage of fear memories across the adult lifetime of rats. J Neurosci. 2004;24:3810–3815. doi: 10.1523/JNEUROSCI.4100-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huganir RL, Nicoll RA. AMPARs and synaptic plasticity: The last 25 years. Neuron. 2013;80:704–717. doi: 10.1016/j.neuron.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lisman J. The CaM kinase II hypothesis for the storage of synaptic memory. Trends Neurosci. 1994;17:406–412. doi: 10.1016/0166-2236(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 4.Frey U, Krug M, Reymann KG, Matthies H. Anisomycin, an inhibitor of protein synthesis, blocks late phases of LTP phenomena in the hippocampal CA1 region in vitro. Brain Res. 1988;452:57–65. doi: 10.1016/0006-8993(88)90008-x. [DOI] [PubMed] [Google Scholar]
- 5.Fonseca R, Vabulas RM, Hartl FU, Bonhoeffer T, Nägerl UV. A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron. 2006;52:239–245. doi: 10.1016/j.neuron.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 6.Hanus C, Schuman EM. Proteostasis in complex dendrites. Nat Rev Neurosci. 2013;14:638–648. doi: 10.1038/nrn3546. [DOI] [PubMed] [Google Scholar]
- 7.Cajigas IJ, Will T, Schuman EM. Protein homeostasis and synaptic plasticity. EMBO J. 2010;29:2746–2752. doi: 10.1038/emboj.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price JC, Guan S, Burlingame A, Prusiner SB, Ghaemmaghami S. Analysis of proteome dynamics in the mouse brain. Proc Natl Acad Sci USA. 2010;107:14508–14513. doi: 10.1073/pnas.1006551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gogolla N, Caroni P, Lüthi A, Herry C. Perineuronal nets protect fear memories from erasure. Science. 2009;325:1258–1261. doi: 10.1126/science.1174146. [DOI] [PubMed] [Google Scholar]
- 10.Happel MF, et al. Enhanced cognitive flexibility in reversal learning induced by removal of the extracellular matrix in auditory cortex. Proc Natl Acad Sci USA. 2014;111:2800–2805. doi: 10.1073/pnas.1310272111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsien RY. Very long-term memories may be stored in the pattern of holes in the perineuronal net. Proc Natl Acad Sci USA. 2013;110:12456–12461. doi: 10.1073/pnas.1310158110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savas JN, Toyama BH, Xu T, Yates JR, 3rd, Hetzer MW. Extremely long-lived nuclear pore proteins in the rat brain. Science. 2012;335:942. doi: 10.1126/science.1217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verzijl N, et al. Effect of collagen turnover on the accumulation of advanced glycation end products. J Biol Chem. 2000;275:39027–39031. doi: 10.1074/jbc.M006700200. [DOI] [PubMed] [Google Scholar]
- 15.Truscott RJW, Schey KL, Friedrich MG. Old proteins in man: A field in its infancy. Trends Biochem Sci. 2016;41:654–664. doi: 10.1016/j.tibs.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin Z, Dimitrijevic A, Aswad DW. Accelerated protein damage in brains of PIMT+/− mice; a possible model for the variability of cognitive decline in human aging. Neurobiol Aging. 2015;36:1029–1036. doi: 10.1016/j.neurobiolaging.2014.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu JX, Doyle HA, Mamula MJ, Aswad DW. Protein repair in the brain, proteomic analysis of endogenous substrates for protein L-isoaspartyl methyltransferase in mouse brain. J Biol Chem. 2006;281:33802–33813. doi: 10.1074/jbc.M606958200. [DOI] [PubMed] [Google Scholar]
- 18.Bloemendal H, et al. Ageing and vision: Structure, stability and function of lens crystallins. Prog Biophys Mol Biol. 2004;86:407–485. doi: 10.1016/j.pbiomolbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 19.Haus JM, Carrithers JA, Trappe SW, Trappe TA. Collagen, cross-linking, and advanced glycation end products in aging human skeletal muscle. J Appl Physiol (1985) 2007;103:2068–2076. doi: 10.1152/japplphysiol.00670.2007. [DOI] [PubMed] [Google Scholar]
- 20.Sharma KK, Santhoshkumar P. Lens aging: Effects of crystallins. Biochim Biophys Acta. 2009;1790:1095–1108. doi: 10.1016/j.bbagen.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barria A, Muller D, Derkach V, Griffith LC, Soderling TR. Regulatory phosphorylation of AMPA-type glutamate receptors by CaM-KII during long-term potentiation. Science. 1997;276:2042–2045. doi: 10.1126/science.276.5321.2042. [DOI] [PubMed] [Google Scholar]
- 22.Lisman JE, Goldring MA. Feasibility of long-term storage of graded information by the Ca2+/calmodulin-dependent protein kinase molecules of the postsynaptic density. Proc Natl Acad Sci USA. 1988;85:5320–5324. doi: 10.1073/pnas.85.14.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malinow R, Schulman H, Tsien RW. Inhibition of postsynaptic PKC or CaMKII blocks induction but not expression of LTP. Science. 1989;245:862–866. doi: 10.1126/science.2549638. [DOI] [PubMed] [Google Scholar]
- 24.Lee HK, Barbarosie M, Kameyama K, Bear MF, Huganir RL. Regulation of distinct AMPA receptor phosphorylation sites during bidirectional synaptic plasticity. Nature. 2000;405:955–959. doi: 10.1038/35016089. [DOI] [PubMed] [Google Scholar]
- 25.Rossetti T, et al. Memory erasure experiments indicate a critical role of CaMKII in memory storage. Neuron. 2017;96:207–216.e2. doi: 10.1016/j.neuron.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sacktor TC. How does PKMζ maintain long-term memory? Nat Rev Neurosci. 2011;12:9–15. doi: 10.1038/nrn2949. [DOI] [PubMed] [Google Scholar]
- 27.Lee AM, et al. Prkcz null mice show normal learning and memory. Nature. 2013;493:416–419. doi: 10.1038/nature11803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volk LJ, Bachman JL, Johnson R, Yu Y, Huganir RL. PKM-ζ is not required for hippocampal synaptic plasticity, learning and memory. Nature. 2013;493:420–423. doi: 10.1038/nature11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsokas P, et al. Compensation for PKMζ in long-term potentiation and spatial long-term memory in mutant mice. eLife. 2016;5:e14846. doi: 10.7554/eLife.14846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raveendra BL, et al. Characterization of prion-like conformational changes of the neuronal isoform of Aplysia CPEB. Nat Struct Mol Biol. 2013;20:495–501. doi: 10.1038/nsmb.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- 32.Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 33.Fioriti L, et al. The persistence of hippocampal-based memory requires protein synthesis mediated by the prion-like protein CPEB3. Neuron. 2015;86:1433–1448. doi: 10.1016/j.neuron.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 34.Majumdar A, et al. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell. 2012;148:515–529. doi: 10.1016/j.cell.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 35.Frey U, Morris RG. Synaptic tagging: Implications for late maintenance of hippocampal long-term potentiation. Trends Neurosci. 1998;21:181–188. doi: 10.1016/s0166-2236(97)01189-2. [DOI] [PubMed] [Google Scholar]
- 36.Martin KC, Kosik KS. Synaptic tagging–Who’s it? Nat Rev Neurosci. 2002;3:813–820. doi: 10.1038/nrn942. [DOI] [PubMed] [Google Scholar]
- 37.McClatchy DB, Dong MQ, Wu CC, Venable JD, Yates JR., 3rd 15N metabolic labeling of mammalian tissue with slow protein turnover. J Proteome Res. 2007;6:2005–2010. doi: 10.1021/pr060599n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu CC, MacCoss MJ, Howell KE, Matthews DE, Yates JR., 3rd Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal Chem. 2004;76:4951–4959. doi: 10.1021/ac049208j. [DOI] [PubMed] [Google Scholar]
- 39.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 40.Kempermann G, Gast D, Gage FH. Neuroplasticity in old age: Sustained fivefold induction of hippocampal neurogenesis by long-term environmental enrichment. Ann Neurol. 2002;52:135–143. doi: 10.1002/ana.10262. [DOI] [PubMed] [Google Scholar]
- 41.Rampon C, et al. Effects of environmental enrichment on gene expression in the brain. Proc Natl Acad Sci USA. 2000;97:12880–12884. doi: 10.1073/pnas.97.23.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nithianantharajah J, Hannan AJ. Enriched environments, experience-dependent plasticity and disorders of the nervous system. Nat Rev Neurosci. 2006;7:697–709. doi: 10.1038/nrn1970. [DOI] [PubMed] [Google Scholar]
- 43.Toyama BH, et al. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013;154:971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamashita N, Goshima Y. Collapsin response mediator proteins regulate neuronal development and plasticity by switching their phosphorylation status. Mol Neurobiol. 2012;45:234–246. doi: 10.1007/s12035-012-8242-4. [DOI] [PubMed] [Google Scholar]
- 45.Ramírez-Amaya V, Balderas I, Sandoval J, Escobar ML, Bermúdez-Rattoni F. Spatial long-term memory is related to mossy fiber synaptogenesis. J Neurosci. 2001;21:7340–7348. doi: 10.1523/JNEUROSCI.21-18-07340.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diering GH, Heo S, Hussain NK, Liu B, Huganir RL. Extensive phosphorylation of AMPA receptors in neurons. Proc Natl Acad Sci USA. 2016;113:E4920–E4927. doi: 10.1073/pnas.1610631113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yi JJ, Ehlers MD. Ubiquitin and protein turnover in synapse function. Neuron. 2005;47:629–632. doi: 10.1016/j.neuron.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 48.Colledge M, et al. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40:595–607. doi: 10.1016/s0896-6273(03)00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan X, Li D, Lichti CF, Green TA. Dynamic proteomics of nucleus accumbens in response to acute psychological stress in environmentally enriched and isolated rats. PLoS One. 2013;8:e73689. doi: 10.1371/journal.pone.0073689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan X, Li D, Zhang Y, Green TA. Differential phosphoproteome regulation of nucleus accumbens in environmentally enriched and isolated rats in response to acute stress. PLoS One. 2013;8:e79893. doi: 10.1371/journal.pone.0079893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li C, Niu W, Jiang CH, Hu Y. Effects of enriched environment on gene expression and signal pathways in cortex of hippocampal CA1 specific NMDAR1 knockout mice. Brain Res Bull. 2007;71:568–577. doi: 10.1016/j.brainresbull.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 52.Lichti CF, et al. Environmental enrichment alters protein expression as well as the proteomic response to cocaine in rat nucleus accumbens. Front Behav Neurosci. 2014;8:246. doi: 10.3389/fnbeh.2014.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Restivo L, et al. Enriched environment promotes behavioral and morphological recovery in a mouse model for the fragile X syndrome. Proc Natl Acad Sci USA. 2005;102:11557–11562. doi: 10.1073/pnas.0504984102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu J, et al. Arc/Arg3.1 regulates an endosomal pathway essential for activity-dependent β-amyloid generation. Cell. 2011;147:615–628. doi: 10.1016/j.cell.2011.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piccoli G, et al. Proteomic analysis of activity-dependent synaptic plasticity in hippocampal neurons. J Proteome Res. 2007;6:3203–3215. doi: 10.1021/pr0701308. [DOI] [PubMed] [Google Scholar]
- 56.Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- 57.Cohen LD, et al. Metabolic turnover of synaptic proteins: Kinetics, interdependencies and implications for synaptic maintenance. PLoS One. 2013;8:e63191. doi: 10.1371/journal.pone.0063191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature. 1998;391:892–896. doi: 10.1038/36103. [DOI] [PubMed] [Google Scholar]
- 59.O’Brien RJ, et al. Activity-dependent modulation of synaptic AMPA receptor accumulation. Neuron. 1998;21:1067–1078. doi: 10.1016/s0896-6273(00)80624-8. [DOI] [PubMed] [Google Scholar]
- 60.Sivan SS, et al. Collagen turnover in normal and degenerate human intervertebral discs as determined by the racemization of aspartic acid. J Biol Chem. 2008;283:8796–8801. doi: 10.1074/jbc.M709885200. [DOI] [PubMed] [Google Scholar]
- 61.Banerjee SB, et al. Perineuronal nets in the adult sensory cortex are necessary for fear learning. Neuron. 2017;95:169–179.e3. doi: 10.1016/j.neuron.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McShane E, et al. Kinetic analysis of protein stability reveals age-dependent degradation. Cell. 2016;167:803–815.e21. doi: 10.1016/j.cell.2016.09.015. [DOI] [PubMed] [Google Scholar]
- 63.Huang T, McDonough CB, Abel T. Compartmentalized PKA signaling events are required for synaptic tagging and capture during hippocampal late-phase long-term potentiation. Eur J Cell Biol. 2006;85:635–642. doi: 10.1016/j.ejcb.2006.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhong H, et al. Subcellular dynamics of type II PKA in neurons. Neuron. 2009;62:363–374. doi: 10.1016/j.neuron.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith FD, et al. Local protein kinase A action proceeds through intact holoenzymes. Science. 2017;356:1288–1293. doi: 10.1126/science.aaj1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Charrier E, et al. Collapsin response mediator proteins (CRMPs): Involvement in nervous system development and adult neurodegenerative disorders. Mol Neurobiol. 2003;28:51–64. doi: 10.1385/MN:28:1:51. [DOI] [PubMed] [Google Scholar]
- 67.Hu X, Viesselmann C, Nam S, Merriam E, Dent EW. Activity-dependent dynamic microtubule invasion of dendritic spines. J Neurosci. 2008;28:13094–13105. doi: 10.1523/JNEUROSCI.3074-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jaworski J, et al. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron. 2009;61:85–100. doi: 10.1016/j.neuron.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 69.Dent EW, Merriam EB, Hu X. The dynamic cytoskeleton: Backbone of dendritic spine plasticity. Curr Opin Neurobiol. 2011;21:175–181. doi: 10.1016/j.conb.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kapitein LC, Yau KW, Hoogenraad CC. Microtubule dynamics in dendritic spines. Methods Cell Biol. 2010;97:111–132. doi: 10.1016/S0091-679X(10)97007-6. [DOI] [PubMed] [Google Scholar]
- 71.Janke C, Kneussel M. Tubulin post-translational modifications: Encoding functions on the neuronal microtubule cytoskeleton. Trends Neurosci. 2010;33:362–372. doi: 10.1016/j.tins.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 72.Song Y, Brady ST. Post-translational modifications of tubulin: Pathways to functional diversity of microtubules. Trends Cell Biol. 2015;25:125–136. doi: 10.1016/j.tcb.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cole AR, et al. GSK-3 phosphorylation of the Alzheimer epitope within collapsin response mediator proteins regulates axon elongation in primary neurons. J Biol Chem. 2004;279:50176–50180. doi: 10.1074/jbc.C400412200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Uchida Y, et al. Semaphorin3A signalling is mediated via sequential Cdk5 and GSK3beta phosphorylation of CRMP2: Implication of common phosphorylating mechanism underlying axon guidance and Alzheimer’s disease. Genes Cells. 2005;10:165–179. doi: 10.1111/j.1365-2443.2005.00827.x. [DOI] [PubMed] [Google Scholar]
- 75.Kempermann G, Kuhn HG, Gage FH. More hippocampal neurons in adult mice living in an enriched environment. Nature. 1997;386:493–495. doi: 10.1038/386493a0. [DOI] [PubMed] [Google Scholar]
- 76.McClatchy DB, Yates JR., 3rd Stable isotope labeling in mammals (SILAM) Methods Mol Biol. 2014;1156:133–146. doi: 10.1007/978-1-4939-0685-7_8. [DOI] [PubMed] [Google Scholar]
- 77.Rauniyar N, McClatchy DB, Yates JR., 3rd Stable isotope labeling of mammals (SILAM) for in vivo quantitative proteomic analysis. Methods. 2013;61:260–268. doi: 10.1016/j.ymeth.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 78.Kim MS, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 80.Olsen JV, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 81.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 82.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 83.Tyanova S, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 84.Mi H, et al. PANTHER version 11: Expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pratt JM, et al. Dynamics of protein turnover, a missing dimension in proteomics. Mol Cell Proteomics. 2002;1:579–591. doi: 10.1074/mcp.m200046-mcp200. [DOI] [PubMed] [Google Scholar]
- 87.Vizcaíno JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.