Significance
There has been increasing interest in hippocampal subfield morphometry in aging and disease using in vivo MRI. However, research on in vivo morphometry is hampered by the lack of a definitive reference model describing regional effects of aging and disease pathology on the hippocampus. To address this limitation, we built a 3D probabilistic atlas of the hippocampus combining postmortem MRI with histology, allowing us to investigate Alzheimer’s disease (AD)-related effects on hippocampal subfield morphometry, derived from histology. Our results support the hypothesis of differential involvement of hippocampal subfields in AD, providing further impetus for more granular study of the hippocampus in aging and disease during life. Furthermore, this atlas provides an important anatomical reference for hippocampal subfield research.
Keywords: hippocampal subfields, Alzheimer’s disease, ex vivo MRI, histology, computational anatomy
Abstract
Although the hippocampus is one of the most studied structures in the human brain, limited quantitative data exist on its 3D organization, anatomical variability, and effects of disease on its subregions. Histological studies provide restricted reference information due to their 2D nature. In this paper, high-resolution (∼200 × 200 × 200 μm3) ex vivo MRI scans of 31 human hippocampal specimens are combined using a groupwise diffeomorphic registration approach into a 3D probabilistic atlas that captures average anatomy and anatomic variability of hippocampal subfields. Serial histological imaging in 9 of the 31 specimens was used to label hippocampal subfields in the atlas based on cytoarchitecture. Specimens were obtained from autopsies in patients with a clinical diagnosis of Alzheimer's disease (AD; 9 subjects, 13 hemispheres), of other dementia (nine subjects, nine hemispheres), and in subjects without dementia (seven subjects, nine hemispheres), and morphometric analysis was performed in atlas space to measure effects of age and AD on hippocampal subfields. Disproportional involvement of the cornu ammonis (CA) 1 subfield and stratum radiatum lacunosum moleculare was found in AD, with lesser involvement of the dentate gyrus and CA2/3 subfields. An association with age was found for the dentate gyrus and, to a lesser extent, for CA1. Three-dimensional patterns of variability and disease and aging effects discovered via the ex vivo hippocampus atlas provide information highly relevant to the active field of in vivo hippocampal subfield imaging.
The hippocampus is a complex structure formed by multiple layers that interlock in an intricate geometric pattern. The neuroanatomy literature defines cytoarchitecturally and connectomically distinct subfields of the hippocampus (1–3), which are believed to be differentially involved in memory functions (4) and to be selectively affected by Alzheimer’s disease (AD), aging, epilepsy, and other disorders (5). Recently, a growing number of in vivo human MRI studies have focused their attention on hippocampal subfields (6–8). Numerous manual protocols (9) and several automated tools (10–12) have been developed for labeling hippocampal subfields on in vivo MRI scans.
However, despite intense interest in the recent literature, existing knowledge of the anatomical variability of the hippocampal region and the effects of development, aging, and disease on hippocampal subfields is limited. Neuroanatomy studies of the hippocampus offer meticulous descriptions of hippocampal anatomy based on cytoarchitecture (1–3, 13, 14), but they typically do so with sparse sampling in small numbers of subjects and hence offer very limited 3D information with regard to the distribution and variability of hippocampal subfields. Neuropathological studies have characterized the differential effects of aging and dementia on hippocampal subfields (e.g., refs. 15 and 16), but they provide limited reference information for MRI research because of the 2D nature of histology, difficulty of extrapolating quantitative measures such as neuronal counts to 3D, and inconsistencies between studies. While new 3D histology techniques such as CLARITY (17) may soon address these limitations, in today’s literature there is little commonality between information derived from in vivo MRI studies (volume, thickness, and shape) and information derived from neuroanatomy and neuropathology studies (cell counts, cytoarchitecture, and molecular pathology, all 2D).
The current work addresses this gap in knowledge by applying the techniques of computational morphometry to a unique dataset of 200 × 200 × 200 µm3-resolution ex vivo MRI of 31 specimens from 25 donors and serial histology of the hippocampal region with 200-µm–slice spacing in nine specimens. The donors were older adults with and without dementia (nine AD, nine other dementia, and seven no dementia). To our knowledge, this is the largest ex vivo 3D-imaging dataset of the hippocampal region ever described in the literature, for which highly customized computational morphometry pipelines are developed to coregister histology and MRI for each specimen with histology and to compute a diffeomorphic mapping between the MRI scans of the 31 specimens and a common anatomical “atlas.” Leveraging this atlas, we carry out 3D morphometric analysis of the hippocampal subfields [cornu ammonis (CA) 1, 2 and 3, stratum radiatum lacunosum moleculare (SRLM), dentate gyrus (DG), and the lateral portion of the subiculum (SUB)] and characterize hippocampal anatomy and variability and evaluate the effects of AD and aging on subfield volume and thickness and other anatomical features.
Results
Computational Atlas of the Hippocampus.
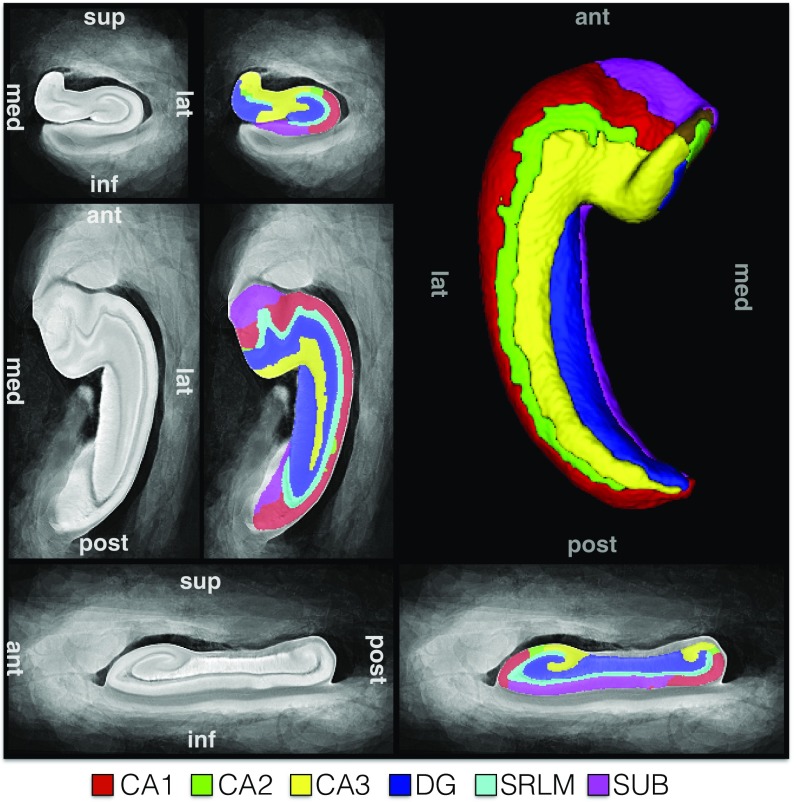
Fig. 1 shows the “final” MRI atlas constructed from the 31 specimens (Table 1) as a synthetic average MR image and a consensus segmentation of hippocampal subfields derived from histology. Each specimen’s MRI has a one-to-one correspondence to this atlas (SI Appendix, Fig. S10). Quantitative and visual evaluation of atlas quality at different stages of atlas construction and comparisons with an alternative atlas-building strategy are presented in SI Appendix, section S2.1 and Figs. S9–S12. Overall, the atlas-generation strategy achieves excellent groupwise alignment between ex vivo MRI scans and captures the average shape and anatomical configuration of hippocampal subfields.
Fig. 1.
The hippocampus atlas obtained by groupwise registration of the MRI scans of 31 ex vivo specimens and its hippocampal subfield segmentation derived from serial histology in nine specimens. The atlas represents the average hippocampal anatomy.
Table 1.
Demographic data for the three patient groups
| Group | N subjects | Age (y) | Sex | N specimens | Side | Fixation time (d) |
| Alzheimer’s disease (AD) | 7 | 71 ± 13 | 4 male, 3 female | 9 | 5 left, 4 right | 283 ± 236 |
| Other dementia (OD) | 9 | 74 ± 9 | 5 male, 4 female | 9 | 3 left, 6 right | 54 ± 43 |
| Nondementia control (NC) | 9 | 79 ± 5 | 3 male, 6 female | 13 | 7 left, 6 right | 266 ± 158 |
Other dementia diagnoses include: corticobasal degeneration (2), dementia with lewy bodies (1), frontotemporal lobar degeneration – TDP43 (2), progressive supranuclear palsy (1), progressive supranuclear palsy and a low probability of AD (1), cerebrovascular disease and a low probability of AD (1) from CNDR, and dementia not otherwise specified (1) from NDRI. Demographics per subject are shown in SI Appendix, Table S1.
Anatomy and Variability in Normal Controls.
The first two columns in Table 2 show anatomical features and variability in nondementia controls (NC; note that subiculum volume is not reported, as its full extent was not incorporated into the atlas). CA1 is the largest subfield at 1.6–8.6 times the size of other subfields. There is a considerable variability in subfield volume, with minima and maxima differing on the order of 1.7–2.5-fold across subject. On average, the hippocampus has ∼14 folds and digitations (see SI Appendix, section S1.4.8 for methods), of which four are in the hippocampal head and ∼10 in the body and tail. Interestingly, in some subjects a clear transition from the head digitations superiorly to the more inferiorly located body/tail folds could be observed, often bridged by a laterally located digitation at the transition from head to body. See SI Appendix, Fig. S13 for an example. A positive association at a trend level was found for the total number of digitations and folds with hippocampal volume (P = 0.067), but no evidence was found for a specific association with one of the subfields (all P values between 0.12–0.20).
Table 2.
Description of hippocampal anatomy in the NC, AD, and OD groups with statistical analyses for NC vs. AD and the associations with age
| Measure | NC | AD | OD | NC vs. AD | Age correlation in NC | |||||||
| Mean (SD) | Range | Mean (SD) | Range | Mean (SD) | Range | %* | t | P | b (%)* | t | P | |
| CA1 volume, mm3 | 762 (161) | 401–1,010 | 451 (130) | 338–743 | 707 (118) | 545–856 | −46.0 | −4.8 | 0.00045 | −2.4 | −2.5 | 0.045 |
| CA2 volume, mm3 | 89 (17) | 63–112 | 66 (21) | 42–101 | 96 (28) | 71–159 | −32.2 | −4.2 | 0.0011 | −0.5 | −0.5 | 0.62 |
| CA3 volume, mm3 | 271 (47) | 193–339 | 239 (32) | 194–288 | 259 (48) | 187–354 | −18.1 | −2.7 | 0.018 | −1.5 | −1.6 | 0.16 |
| SRLM volume, mm3 | 423 (68) | 311–546 | 254 (98) | 155–457 | 455 (103) | 269–622 | −44.9 | −5.1 | 0.00026 | −1.7 | −2.2 | 0.07 |
| DG volume, mm3 | 496 (117) | 341–680 | 381 (43) | 307–459 | 454 (106) | 303–620 | −28.7 | −3.1 | 0.0099 | −3.2 | −3.3 | 0.017 |
| HF volume, mm3 | 2,553 (414) | 1,664–3,039 | 1,717 (366) | 1,326–2,529 | 2,429 (479) | 1,880–3,156 | −37.6 | −5.0 | 0.00031 | −2.0 | −3.4 | 0.015 |
| DG/CA1 ratio | 0.66 (0.12) | 0.51–0.92 | 0.88 (0.15) | 0.62–1.12 | 0.64 (0.09) | 0.53–0.8 | 37.3 | 3.4 | 0.0055 | −0.4 | −0.4 | 0.71 |
| CA1 thickness, mm | 1.61 (0.19) | 1.15–1.89 | 1.30 (0.15) | 1.11–1.56 | 1.54 (0.10) | 1.40–1.70 | −22.2 | −4.2 | 0.0013 | −0.9 | −1.3 | 0.25 |
| CA2 thickness, mm | 1.25 (0.17) | 0.95–1.56 | 1.13 (0.17) | 0.81–1.42 | 1.25 (0.17) | 1.00–1.53 | −14.4 | −2.0 | 0.065 | 0.3 | 0.3 | 0.76 |
| CA3 thickness, mm | 1.35 (0.15) | 1.17–1.54 | 1.32 (0.07) | 1.25–1.44 | 1.32 (0.10) | 1.12–1.43 | −3.7 | −0.8 | 0.42 | 0 | 0 | 0.99 |
| SRLM thickness, mm | 0.87 (0.08) | 0.77–1.05 | 0.65 (0.13) | 0.52–0.88 | 0.94 (0.10) | 0.74–1.04 | −26.2 | −4.7 | 0.00052 | 0 | 0 | 0.99 |
| DG thickness, mm | 1.65 (0.33) | 1.26–2.37 | 1.47 (0.07) | 1.37–1.60 | 1.61 (0.20) | 1.29–1.95 | −13.2 | −1.5 | 0.15 | −1.6 | −1.3 | 0.23 |
| SUB thickness, mm | 1.52 (0.10) | 1.40–1.74 | 1.27 (0.15) | 1.02–1.49 | 1.40 (0.20) | 1.18–1.78 | −14.3 | −3.0 | 0.011 | 0.1 | 0.3 | 0.78 |
| A. HF volume, mm3 | 1,422 (204) | 947–1,747 | 1,015 (242) | 737–1,514 | 1,386 (281) | 1,033–1,932 | −35 | −5.3 | 0.00019 | −1.2 | −2.0 | 0.097 |
| P. HF volume, mm3 | 1,131 (259) | 656–1,498 | 701 (178) | 464–1,015 | 1,043 (245) | 765–1,538 | −41 | −3.6 | 0.0036 | −3.0 | −2.9 | 0.029 |
| Head dig (n) | 4 (0.82) | 3–5 | 3.22 (0.44) | 3–4 | 3.78 (0.83) | 3–5 | −19.0 | 2.3 | 0.039 | 1.7 | 1.8 | 0.11 |
| Body/tail folds (n) | 9.92 (1.19) | 8–12 | 8.88 (0.83) | 8–10 | 11.14 (2.04) | 9–15 | −8.4 | −1.3 | 0.24 | −1.4 | −2.1 | 0.078 |
| Total dig and folds (n) | 13.92 (1.04) | 12–16 | 12 (0.76) | 11–13 | 14.86 (2.04) | 12–18 | −12.8 | 3.7 | 0.0032 | −0.5 | −1.8 | 0.13 |
| HF extent, mm | 40 (5) | 31–51 | 40 (3) | 35–43 | 41 (4) | 35–48 | −5.5 | −1.0 | 0.36 | −1.5 | −2.3 | 0.062 |
The reported percent change is adjusted for covariates, NC, normal controls; AD, Alzheimer’s disease; OD, other dementia; CA, cornu ammonis; SRLM, stratum radiatum lacunosum moleculare. DG, dentate gyrus; dig, digitation; HF, hippocampal formation; SUB, subiculum; A, anterior; P, posterior. The means and ranges for the OD group are also listed. The OD group is highly heterogeneous diagnostically, but overall, their hippocampi are relatively spared compared with AD. The primary rationale for including the OD group in the atlas was not for statistical comparison with the NC and AD groups, but to increase the number of samples used to capture the anatomical variability of the hippocampus in the atlas.
The hippocampal tail shows great anatomical variability in the coronal plane, and our hypothesis is that this is primarily attributable to its angulation. We therefore resliced the posterior third of the hippocampus, as a working definition of the hippocampal tail to follow the curve in a consistent manner, in three places orthogonal to the long axis of the hippocampus, often at a clockwise rotation (see SI Appendix, section S1.4.9). We observed that the appearance of the posterior hippocampus is “body-like” in all resliced sections. In a small number of specimens the third resliced section in the very posterior portion of the hippocampus contained little DG and slightly deviated from the common body-like appearance. See SI Appendix, Fig. S14 for examples.
Alzheimer’s Disease.
Table 2 shows that subfield volumes are considerably smaller in AD compared with NC subjects, especially CA1 and SRLM. Importantly, the CA1/DG volume ratio is significantly smaller in AD than in NC, indicating a larger effect of AD on CA1 than on DG. Besides volume, measures of SRLM and CA1 average thickness are also significantly decreased in AD compared with NC, indicating that the observed volume loss is partly due to thinning of these structures. SUB thickness (the lateral portion) was also significantly decreased in AD compared with NC. Moreover, a significant decrease in volume of both the anterior and posterior hippocampus could be observed, as well as a decrease in head digitations in AD compared with NC.
Aging.
DG volume was significantly associated with age (Table 2; −3.3%/y) and, to a lesser extent, CA1 (−2.5%/y). Interestingly, whereas SRLM was one of the most affected regions in AD, the effect of aging in SRLM seems to be much smaller and only reached a trend level. Interestingly, none of the analyses with the thickness measures reached significance, indicating that thinning of these regions in the context of aging is less pronounced. Hippocampal extent trended to be negatively associated with age, decreasing 1.6% per year. Finally, posterior hippocampal volume was significantly and negatively associated with age, but anterior hippocampal volume only at a trend level. This was reflected in the digitation/fold analyses, where fold number in the body/tail, but not in the head, is related to age at a trend level.
Regional Thickness Maps.
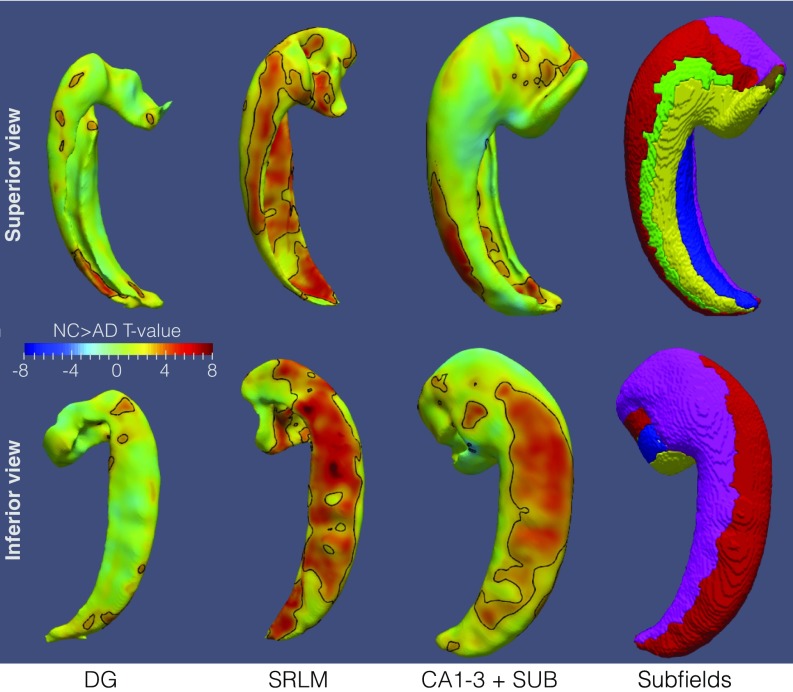
To examine more localized effects of AD and aging on the hippocampus, we performed regional thickness analyses on hippocampal layers, DG, SRLM, and combined CA1-3 and SUB (SI Appendix, section S1.4.6). Fig. 2 plots differences in thickness between AD and NC. The set of locations that survive false discovery rate (FDR) correction (18) at the α = 0.05 threshold includes most of the SRLM and large extents of CA1 and SUB, particularly on the inferior and lateral aspects. A much more sparse set of locations on the DG survives FDR correction, mostly posteriorly. This is also shown in Movie S1. No regions in the regional thickness analysis for aging survived FDR correction.
Fig. 2.
Statistical maps of regional thickness difference between AD and NC subjects, covarying for age and sex. Separate maps are generated for the dentate gyrus, stratum radiatum lacunosum moleculare (SRLM), and outer gray matter layers of the hippocampus. The maps are in atlas space, viewed from the superior (Top) and inferior (Bottom). Locations that survive the false discovery rate correction at the α = 0.05 threshold are demarcated by a black outline.
Effects of AD on Hippocampal Shape.
SI Appendix, Fig. S15 and Movie S2 visualize shape changes that correspond to the transition between NC and AD groups in high-dimensional atlas space. The thinning of the CA1 and SRLM are clearly visible in the coronal views, whereas the change in the DG is less pronounced. Overall, the shape change associated with AD is more complex than global shrinking, likely explained by different degrees of shrinking in different subregions. Subtle straightening of the posterior hippocampus along the long axis can be observed in AD compared with NC.
Discussion
Our probabilistic 3D atlas of the human hippocampus uniquely combines ultra–high-resolution postmortem MRI and histologically derived information on subfield boundaries using specimens from older controls and patients with AD and other dementias. This atlas allowed us to investigate detailed hippocampal anatomy and variability in 3D in older controls and investigate AD- and age-related effects on hippocampal morphometry.
AD- and Age-Related Effects on Hippocampal Subfields.
We used the atlas to investigate AD- and age-related effects on hippocampal subfields, ultimately to inform in vivo research on this topic. Findings from in vivo MRI studies have been very inconsistent, especially with regard to the aging literature (19). The AD literature has been more consistent (19), but the relative contribution of CA1 and DG has also been unclear (e.g., refs. 20 and 21). Although histological studies have provided the best possible ground truth so far, even results from these studies have been inconsistent both for aging (e.g., refs. 15 and 22) and for AD (e.g., refs. 15 and 23). Additionally, as noted above, it is unclear how well measures like cell count translate to volumes as commonly used in vivo, how well definitions of subfields in the two modalities compare, and how much selection of histology sections, sometimes only sampled from the hippocampal body (23), affects the results and may have caused inconsistencies between studies. In this atlas we were able to combine the “best of both worlds” and use histology to derive subfield boundaries, but perform analyses in 3D MRI space, incorporating the full length of the hippocampus and obtaining volume measures, similar to those in in vivo MRI.
AD.
For AD, we found that all subfields were affected but that the largest decreases could be observed in CA1 and SRLM, providing evidence for a relatively selective effect of AD on hippocampal subfields. The size of the decrease in CA1 and SRLM (∼45%) suggests that differential effects might potentially be observed in earlier stages of the disease. Interestingly, the CA1/DG ratio was also significantly different in AD compared with normal controls. This supports the notion that CA1 is affected more than the DG in AD, consistent with the greater neurofibrillary tangle burden in the former described by Braak and Braak (24). While we were not able to investigate SUB volume, we did observe a significant decrease in thickness in its lateral extent, potentially reflecting neurofibrillary pathology in the later stages of the disease.
SRLM.
The large decrease in SRLM volume, but also thickness, supports work on this region in in vivo MRI studies (e.g., refs. 25 and 26). Especially notable is the work from Kerchner et al. (25, 27), reporting a decrease in SRLM thickness in mild AD compared with controls and even in APOE-ε4 carriers compared with noncarriers using 7T MRI. In fact, the current study, being the ex vivo counterpart of these in vivo studies, provides validation of SRLM atrophy in AD using measures similar to in vivo studies. It also matches histopathology studies, where indeed SRLM of CA1 is one of the first regions where tau pathology accumulates (28, 29), making SRLM an interesting potential marker of early disease pathology.
Age.
According to our data, age was only associated with DG and with CA1, but, interestingly, in contrast to AD, only at a trend level with the SRLM. The age-related decreases found in the current work were larger than reported in the literature; 2.0% decrease per year in hippocampal volume in our data compared with a reported decrease of 1.41% (30). This difference is potentially due to a difference in population, e.g., difference in age range, but could also be due to a difference in methodology.
Anterior and posterior hippocampal volume and digitations/folds.
A significant decrease in anterior hippocampal volume and, to a lesser extent, in posterior hippocampal volume was observed in AD compared with controls. Interestingly, the opposite pattern was observed for aging where only the posterior hippocampus showed a significant decrease. These findings match reports from in vivo studies in mild cognitive impairment (31) and aging (ref. 32, but note ref. 33). Similarly, we found a negative association between the number of folds and digitations and both age (trend) and AD, occurring in anterior regions in AD and in posterior regions in aging, matching our results on volume loss in these regions in AD and aging.
In general, these results support the idea of differential and potentially even diverging effects of aging and AD on hippocampal subfields and other anatomical features, providing further impetus for research on these regions during life. Using the regional thickness maps and shape analysis, we were able to explore AD-related atrophy of the hippocampus in 3D in the postmortem domain and found that atrophy in the subregions showed a more complex pattern than global shrinking. This further indicates potential usefulness of more regional hippocampal analyses.
Characterization of Hippocampal Anatomy and Variability.
This atlas and dataset also allowed us to characterize hippocampal anatomy and anatomical variability in more detail than would be possible with only histology or in vivo MRI data. This characterization together with the atlas, which will be made publicly available, can be used to both validate and inform protocols for segmentation of hippocampal subfields on in vivo MRI, an area of active methodological research, including an ongoing effort to harmonize segmentation protocols across the scientific community (9, 34). Additionally, the data on the typical ranges of subfield volumes and subfield volume ratios may be used as a validity check for new and existing in vivo MRI segmentation protocols. Interestingly, we found that variability observed in the appearance of the hippocampal tail in the coronal plane is really due to the bending of the tail. When sectioning the tail in the direction of the tail bend, the tail has a body-like structure in all subjects. This information could potentially benefit in vivo segmentation of the tail, which is generally omitted from segmentation protocols due to perceived anatomical complexity.
Atlases of the Hippocampus in Prior Work.
Our ex vivo hippocampus atlas is distinct from previous related efforts. Iglesias et al. (10) constructed a probabilistic atlas of the hippocampal formation using high-resolution ex vivo MRI of 15 specimens from older adults. However, this atlas was developed without histology, and subfields were traced on the basis of MRI signal alone. Furthermore, the authors did not extensively discuss hippocampal anatomic variability and effects of disease on hippocampal subfields. Goubran et al. (35) coanalyzed ex vivo MRI and serial histology in 15 specimens extracted during epilepsy surgery and quantified relationships between MRI and histology measures, as well as to presurgical in vivo MRI. However, that study did not employ computational anatomy techniques to build a probabilistic atlas. Our earlier work developed a probabilistic atlas from a handful of ex vivo MRI scans (36) and described methods for coregistration of ex vivo MRI and serial histology of the hippocampus (37). The current atlas expands on this work and combines these efforts within a single large-scale atlas and uses them to examine the effects of AD and aging on hippocampal subfields.
Limitations.
A significant limitation is that in subjects from the National Disease Research Interchange (NDRI) brain bank we only had access to clinical diagnoses. This means that some older controls may already harbor AD pathology, and some AD subjects may have received an incorrect diagnosis. However, the fact that we observed very strong differences in hippocampal subfield volumes in this potentially “dirty” sample is very encouraging, as we would expect even stronger effects to be observed in a “clean” sample with pathologically confirmed diagnoses.
Another limitation is the limited inclusion of the subiculum. During atlas construction, registration is guided by semiautomatic segmentations of the hippocampus and SRLM (SI Appendix, section S1.2) from a previous iteration of our technique (38), which made assumptions about hippocampus/SRLM topology that are not valid in the medial portion of the subiculum. Subsequently, the semiautomatic segmentations included only the lateral subiculum, which was analyzed for thickness but not volume. We are planning to label and include the full subiculum along with several parahippocampal gyrus subregions in a future version of the atlas. Another limitation is that the measures of thickness (average and pointwise) reported in this paper are based on a mathematical definition (distance from the boundary of a subfield to its Voronoi skeleton) and do not directly reflect the thickness of the anatomical lamina that compose hippocampal subfields, particularly in the label DG, where the structure is composed of two substructures (DG and hilus).
Additionally, the study is limited by the fact that subfield measurements were derived from a template-based segmentation of the 31 specimens, which inevitably introduced errors relative to the underlying “true” anatomical subfield boundaries (as seen in SI Appendix, Fig. S12). This approach was taken because histological processing was performed in only 9 of the 31 specimens, which was highly labor- and resource-intensive, with each specimen requiring 3–4 mo of combined time for staining, scanning, reconstruction, and annotation. As the histology annotation and histology-guided subfield segmentation in MRI space took more than 20 h per specimen, we were also not able to perform a reliability analysis. Indeed, small segmentation and registration errors may have occurred, affecting the final atlas set. However, it should be noted that this is a probabilistic atlas, and small nonsystematic errors in the placement of the subfield boundaries are likely to cancel out in the final atlas segmentation. Moreover, volumes of the subfields could have been affected by slight nonlinear distortions due to the MRI scanner gradient nonlinearity. Finally, it is unclear whether extraction of the brain from the skull has a localized effect on shape or whether there is a differential effect of formalin fixation on different cell types. However, it seems highly unlikely that such factors alone would explain the very strong observed aging and AD effects.
Conclusions
We present a probabilistic atlas of the human hippocampus combining postmortem MRI and histology generated by applying highly customized computational morphometry techniques to the largest ex vivo hippocampus 3D-imaging dataset. This unique probabilistic atlas allowed us to investigate anatomical variability in the subfields of the human hippocampus and to describe aging and AD effects on hippocampal subfield morphometry at an unprecedented level of detail. This atlas will be made publicly available and can be used as a reference for future studies on hippocampal anatomy in older populations. The observed differential aging and AD effects on hippocampal subfields provide further motivation to pursue this in vivo and lend support for the usefulness of hippocampal subfields as biomarkers.
Materials and Methods
Specimens and Postmortem MRI.
Human brain specimens were obtained in accordance with the University of Pennsylvania Institutional Review Board guidelines. Preconsent during life and/or next-of-kin consent at death was given for all cases. In total 32 ex vivo hippocampal specimens from 26 autopsies were obtained (as noted below, one specimen was ultimately excluded from the atlas); 20 hemispheres from 14 subjects from the brain bank operated by the NDRI and 12 hemispheres from autopsies performed at the University of Pennsylvania Center for Neurodegenerative Disease Research (CNDR) in 12 subjects who previously participated in MRI research studies of aging and dementia. For specimens from NDRI, only clinical designation as AD or control was available. For CNDR specimens, pathological diagnoses were available. Specimens, fixed in 10% formalin solution for a minimum of 21 d, were imaged on a Varian 9.4 T animal scanner at 200 × 200 × 200 μm3 (160 × 160 × 160 μm3 in one specimen) resolution. Details of the imaging protocol used for each specimen are given in SI Appendix, section S1.1 and Table S1.
Manual Segmentation of SRLM and Hippocampus Boundary.
Applying rigid, affine, or deformable image registration algorithms directly to ex vivo MRI scans results in very poor alignment between specimens, regardless of parameters. This is due to large variability in amount of tissue cut and imaged, collapsing of cerebrospinal fluid spaces in some samples, and imaging artifacts, as well as anatomical variability. To overcome this obstacle, our approach leverages semimanually generated segmentations of the outer hippocampal boundary and the myelinated SRLM layer, similar to other published ex vivo hippocampal MRI atlases (10, 36). For more details, see SI Appendix, section S1.2 and Fig. S1.
MRI Atlas Generation.
The algorithm for constructing an atlas from ex vivo MRI scans is detailed in SI Appendix, section 1.3. In brief, the algorithm first performs groupwise registration between the segmentations of the SRLM and hippocampus of all specimens, which yields a pointwise correspondence map and an average SRLM/hippocampus shape. The algorithm then maps the MRI scans of all specimens into the space of the average shape by extrapolating pointwise correspondences to the entire image domain using a “geodesic shooting” algorithm that ensures that the resulting maps are smooth and invertible. Last, groupwise diffeomorphic image registration is performed on the MRI intensities to resolve residual misalignments between specimens. The use of manual SRLM and hippocampus segmentations to initialize groupwise MRI intensity registration results in much better alignment than when groupwise MRI intensity registration is performed without such initialization, as showing in SI Appendix, section 2.1.
Histological Imaging and Reconstruction.
Nine specimens, each from a different individual, underwent serial histological processing. The histology protocol, approach for matching histology to MRI, and the approach for mapping cytoarchitectural subfield boundaries from histology space to MRI space followed the general framework described in Adler et al. (37) with a number of modifications that are detailed in SI Appendix, sections S1.4.1–S1.4.4 and Fig. S2. Specimens were cut into ∼1-cm thick blocks (46 in total, for nine specimens), which were imaged separately with 200 × 200 × 200 μm3-resolution MRI. Blocks were embedded in paraffin and sectioned on a vibratome with 5-μm thickness and ∼200-μm spacing. Sections were stained using the Kluver–Barrera method (39) and digitally scanned at 0.5 μm × 0.5 μm resolution. For each block, the scanned sections were reconstructed in 3D and aligned to the block MRI using the interactive software HistoloZee (https://www.nitrc.org/projects/historecon). The resulting alignment was used to initialize the graph-theoretic automated histology reconstruction algorithm described in Adler et al. (37), wherein each histology section undergoes linear and deformable registration to the neighboring sections and to the matched slice of the MRI scan. An example of a histology stack reconstructed to the MRI is shown for a block and a whole MRI in SI Appendix, Figs. S3 and S4.
Hippocampal Subfield Segmentation in Histology and MRI.
Boundaries between hippocampal subfields CA1, CA2, CA3, DG, and SUB were annotated in each histology image on the basis of cytoarchitectural features, following the anatomical protocol by Ding and Van Hoesen (14) (note that SUB was not included in the atlas to its full extent and was therefore not included in the statistical analyses of volume). The annotations were reviewed by Dr. Ding and modified based on his feedback. Annotations were mapped into the block-space MRI of each specimen and used to guide the manual segmentation of hippocampal subfields in MRI space. SI Appendix, sections S1.4.3 and S1.4.4 and Figs. S5–S7 provide more details on this process, and specifically, SI Appendix, Figs. S6 and S16–S20 illustrate the labeling of subfields in a specimen MRI on the basis of histological annotations.
For each of the nine specimens with histology, subfield segmentations were warped into the MRI atlas space using the deformable transformations by groupwise registration. A consensus segmentation of the MRI atlas was obtained by application of voxel-wise majority voting among the nine segmentations with slight regularization by a Markov Random Field prior (SI Appendix, section S1.4.5). The histology-based subfield segmentation in atlas space was combined with the average MRI-based hippocampus and SRLM segmentations to yield the “final” atlas segmentation. The final atlas segmentation was warped back into the MRI space of each of the 32 specimens. The resulting atlas-based segmentations were inspected visually. A significant segmentation failure was detected in one specimen (NDRI12-R), likely due to an unusually large tail of the caudate nucleus that caused intensity-based normalization to fail along almost the entire length of the hippocampus. This specimen was excluded from the study, and the atlas was rebuilt using the remaining 31 specimens.
Statistical Analyses.
To test for the effect of diagnostic group or age on each summary measure (e.g., CA1 volume), we fitted a general linear model (GLM) with the summary measure as the dependent variable, diagnostic group (AD or NC; OD subjects were not included) or age in the NC subgroup as the independent variable, and sex and/or age as covariates. In subjects where both hemispheres were available, the left and right summary measures were averaged since there were no evident differences between the hemispheres. Similarly, for regional thickness measures, a GLM with thickness as the dependent variable, diagnostic group or age as independent variable, and sex and/or age as covariates was fitted at each point of the DG, SRLM, and SUB+CA1-3 surfaces. The FDR approach (18) was applied to obtain a corrected P value at each vertex. More details are given in SI Appendix, section S1.4.6. For establishing an association between the number of digitations/folds and volume, hemispheres were treated separately. We used a linear mixed effects model with digitations/folds as the dependent variable and subject as random effect. The Kenward–Roger approximation was applied at each vertex to obtain an uncorrected P value (40). The shape analysis approach used for visualizing the discriminant direction between AD and NC in atlas space is described in SI Appendix, section 1.4.7.
Supplementary Material
Acknowledgments
We gratefully acknowledge the tissue donors and their families. We also thank the staff at the NDRI brain bank and at CNDR for performing the autopsies and making tissue available for this project. This work was supported by the National Institute of Health (Grants R01 AG037376, R01 EB017255, R01 AG056014, R01 EB014146, P30 NS045839, P41 EB015893, R03 EB16923-01A1, AG038490, P30 AG010124, and P01 AG017586), the Wyncote Foundation, and the Newhouse Foundation. We acknowledge the donors of Alzheimer’s Disease Research, a program of the BrightFocus Foundation (to L.E.M.W.) for support of this research.
Footnotes
Conflict of interest statement: D.A.W. received consultation fees from Eli Lilly, Janssen, and Merck. D.A.W. receives grant support from Avid Radiopharmaceuticals/Eli Lilly, Biogen, Functional Neuromodulation, and Merck. J.B.T. has received research support from Eli Lilly. J.Q.T. may accrue revenue in the future on patents submitted by the University of Pennsylvania wherein he is co-inventor, and he received revenue from the sale of Avid to Eli Lilly as co-inventor on imaging-related patents submitted by the University of Pennsylvania.
This article is a PNAS Direct Submission.
Data deposition: The MRI data and reduced-resolution histology and intermediate results have been uploaded to https://www.nitrc.org/projects/pennhippoatlas/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801093115/-/DCSupplemental.
References
- 1.Duvernoy HM, et al. The Human Hippocampus. Springer; Berlin: 2005. pp. 1–232. [Google Scholar]
- 2.Insausti R, Amaral DG. In: The Human Nervous System. Mai JK, Paxinos G, editors. Elsevier Academic; San Diego, Calif.: 2012. [Google Scholar]
- 3.Ding SL, et al. Comprehensive cellular-resolution atlas of the adult human brain. J Comp Neurol. 2016;524:3127–3481. doi: 10.1002/cne.24080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yassa MA, Stark CE. Pattern separation in the hippocampus. Trends Neurosci. 2011;34:515–525. doi: 10.1016/j.tins.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Small SA, Schobel SA, Buxton RB, Witter MP, Barnes CA. A pathophysiological framework of hippocampal dysfunction in ageing and disease. Nat Rev Neurosci. 2011;12:585–601. doi: 10.1038/nrn3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mueller SG, et al. Measurement of hippocampal subfields and age-related changes with high resolution MRI at 4T. Neurobiol Aging. 2007;28:719–726. doi: 10.1016/j.neurobiolaging.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olsen RK, et al. Volumetric analysis of medial temporal lobe subregions in developmental amnesia using high-resolution magnetic resonance imaging. Hippocampus. 2013;23:855–860. doi: 10.1002/hipo.22153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanseeuw BJ, et al. Mild cognitive impairment: Differential atrophy in the hippocampal subfields. AJNR Am J Neuroradiol. 2011;32:1658–1661. doi: 10.3174/ajnr.A2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yushkevich PA, et al. Hippocampal Subfields Group (HSG) Quantitative comparison of 21 protocols for labeling hippocampal subfields and parahippocampal subregions in in vivo MRI: Towards a harmonized segmentation protocol. Neuroimage. 2015;111:526–541. doi: 10.1016/j.neuroimage.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iglesias JE, et al. Alzheimer’s Disease Neuroimaging Initiative A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. Neuroimage. 2015;115:117–137. doi: 10.1016/j.neuroimage.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pipitone J, et al. Alzheimer’s Disease Neuroimaging Initiative Multi-atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. Neuroimage. 2014;101:494–512. doi: 10.1016/j.neuroimage.2014.04.054. [DOI] [PubMed] [Google Scholar]
- 12.Yushkevich PA, et al. Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Hum Brain Mapp. 2015;36:258–287. doi: 10.1002/hbm.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mai JK, Paxinos G, Voss T. Atlas of the Human Brain. Academic; New York: 2008. pp. 1–271. [Google Scholar]
- 14.Ding SL, Van Hoesen GW. Organization and detailed parcellation of human hippocampal head and body regions based on a combined analysis of cyto- and chemoarchitecture. J Comp Neurol. 2015;523:2233–2253. doi: 10.1002/cne.23786. [DOI] [PubMed] [Google Scholar]
- 15.Simic G, Kostovic I, Winblad B, Bogdanovic N. Volume and number of neurons of the human hippocampal formation in normal aging and Alzheimer’s disease. J Comp Neurol. 1997;379:482–494. doi: 10.1002/(sici)1096-9861(19970324)379:4<482::aid-cne2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 16.West MJ, Kawas CH, Stewart WF, Rudow GL, Troncoso JC. Hippocampal neurons in pre-clinical Alzheimer’s disease. Neurobiol Aging. 2004;25:1205–1212. doi: 10.1016/j.neurobiolaging.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Chung K, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- 19.de Flores R, La Joie R, Chételat G. Structural imaging of hippocampal subfields in healthy aging and Alzheimer’s disease. Neuroscience. 2015;309:29–50. doi: 10.1016/j.neuroscience.2015.08.033. [DOI] [PubMed] [Google Scholar]
- 20.Mueller SG, et al. Hippocampal atrophy patterns in mild cognitive impairment and Alzheimer’s disease. Hum Brain Mapp. 2010;31:1339–1347. doi: 10.1002/hbm.20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.La Joie R, et al. Hippocampal subfield volumetry in mild cognitive impairment, Alzheimer’s disease and semantic dementia. Neuroimage Clin. 2013;3:155–162. doi: 10.1016/j.nicl.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.West MJ, Coleman PD, Flood DG, Troncoso JC. Differences in the pattern of hippocampal neuronal loss in normal ageing and Alzheimer’s disease. Lancet. 1994;344:769–772. doi: 10.1016/s0140-6736(94)92338-8. [DOI] [PubMed] [Google Scholar]
- 23.Rossler M, Zarski R, Bohl J, Ohm TG. Stage-dependent and sector-specific neuronal loss in hippocampus during Alzheimer’s disease. Acta Neuropathol. 2002;103:363–369. doi: 10.1007/s00401-001-0475-7. [DOI] [PubMed] [Google Scholar]
- 24.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 25.Kerchner GA, et al. Hippocampal CA1 apical neuropil atrophy in mild Alzheimer disease visualized with 7-T MRI. Neurology. 2010;75:1381–1387. doi: 10.1212/WNL.0b013e3181f736a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boutet C, et al. Detection of volume loss in hippocampal layers in Alzheimer’s disease using 7 T MRI: A feasibility study. Neuroimage Clin. 2014;5:341–348. doi: 10.1016/j.nicl.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerchner GA, et al. APOE ε4 worsens hippocampal CA1 apical neuropil atrophy and episodic memory. Neurology. 2014;82:691–697. doi: 10.1212/WNL.0000000000000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braak E, Braak H. Alzheimer’s disease: Transiently developing dendritic changes in pyramidal cells of sector CA1 of the Ammon’s horn. Acta Neuropathol. 1997;93:323–325. doi: 10.1007/s004010050622. [DOI] [PubMed] [Google Scholar]
- 29.Thal DR, et al. Alzheimer-related tau-pathology in the perforant path target zone and in the hippocampal stratum oriens and radiatum correlates with onset and degree of dementia. Exp Neurol. 2000;163:98–110. doi: 10.1006/exnr.2000.7380. [DOI] [PubMed] [Google Scholar]
- 30.Barnes J, et al. A meta-analysis of hippocampal atrophy rates in Alzheimer’s disease. Neurobiol Aging. 2009;30:1711–1723. doi: 10.1016/j.neurobiolaging.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin SB, Smith CD, Collins HR, Schmitt FA, Gold BT. Evidence that volume of anterior medial temporal lobe is reduced in seniors destined for mild cognitive impairment. Neurobiol Aging. 2010;31:1099–1106. doi: 10.1016/j.neurobiolaging.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malykhin NV, Bouchard TP, Camicioli R, Coupland NJ. Aging hippocampus and amygdala. Neuroreport. 2008;19:543–547. doi: 10.1097/WNR.0b013e3282f8b18c. [DOI] [PubMed] [Google Scholar]
- 33.Chen KH, Chuah LY, Sim SK, Chee MW. Hippocampal region-specific contributions to memory performance in normal elderly. Brain Cogn. 2010;72:400–407. doi: 10.1016/j.bandc.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Wisse LEM, et al. Hippocampal Subfields Group A harmonized segmentation protocol for hippocampal and parahippocampal subregions: Why do we need one and what are the key goals? Hippocampus. 2017;27:3–11. doi: 10.1002/hipo.22671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goubran M, et al. In vivo MRI signatures of hippocampal subfield pathology in intractable epilepsy. Hum Brain Mapp. 2016;37:1103–1119. doi: 10.1002/hbm.23090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yushkevich PA, et al. A high-resolution computational atlas of the human hippocampus from postmortem magnetic resonance imaging at 9.4 T. Neuroimage. 2009;44:385–398. doi: 10.1016/j.neuroimage.2008.08.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adler DH, et al. Histology-derived volumetric annotation of the human hippocampal subfields in postmortem MRI. Neuroimage. 2014;84:505–523. doi: 10.1016/j.neuroimage.2013.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adler DH, et al. Probabilistic atlas of the human hippcampus combining ex vivo MRI and histology. In: Ourselin S, Joskowicz L, Sabuncu M, Unal G, Wells W, editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science. Vol 9902 Springer, Cham; Switzerland: 2016. [Google Scholar]
- 39.Kluver H, Barrera E. A method for the combined staining of cells and fibers in the nervous system. J Neuropathol Exp Neurol. 1953;12:400–403. doi: 10.1097/00005072-195312040-00008. [DOI] [PubMed] [Google Scholar]
- 40.Halekoh U, Højsgaard S. A Kenward-Roger approximation and parametric bootstrap methods for tests in linear mixed models–The R package pbkrtest. J Stat Softw. 2014;59:1–30. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.