Significance
The lamin proteins are an important component of the nuclear scaffold. Abnormal lamins can cause a variety of disorders called laminopathies, one of which is the premature aging disorder Hutchinson-Gilford progeria syndrome (HGPS). Cells from HGPS patients show improvement when treated with rapamycin and everolimus. These drugs stimulate the cellular system that disposes of abnormal proteins. Several other laminopathies also produce abnormal lamin proteins, so we hypothesized that everolimus would also be beneficial. We treated cells from six different laminopathy patients with everolimus and saw improved nuclear shape in most cells. Treatment also increased the cells’ ability to multiply and delayed senescence. These results suggest that everolimus might have clinical benefit for multiple laminopathy syndromes.
Keywords: everolimus, laminopathies, lamin, mTOR, progeria
Abstract
LMNA encodes the A-type lamins that are part of the nuclear scaffold. Mutations in LMNA can cause a variety of disorders called laminopathies, including Hutchinson-Gilford progeria syndrome (HGPS), atypical Werner syndrome, and Emery-Dreifuss muscular dystrophy. Previous work has shown that treatment of HGPS cells with the mTOR inhibitor rapamycin or with the rapamycin analog everolimus corrects several of the phenotypes seen at the cellular level—at least in part by increasing autophagy and reducing the amount of progerin, the toxic form of lamin A that is overproduced in HGPS patients. Since other laminopathies also result in production of abnormal and potentially toxic lamin proteins, we hypothesized that everolimus would also be beneficial in those disorders. To test this, we applied everolimus to fibroblast cell lines from six laminopathy patients, each with a different mutation in LMNA. Everolimus treatment increased proliferative ability and delayed senescence in all cell lines. In several cell lines, we observed that with treatment, there is a significant improvement in nuclear blebbing, which is a cellular hallmark of HGPS and other lamin disorders. These preclinical results suggest that everolimus might have clinical benefit for multiple laminopathy syndromes.
Laminopathies are a diverse group of disorders caused by a defect in the proteins that make up the nuclear lamina, which is composed of intermediate filament proteins called lamins. In humans, the major lamins are lamin A, lamin B1, lamin B2, and lamin C. All of the A-type lamins, including lamins A and C, are alternatively spliced from the LMNA gene (1). There are many laminopathies caused by defects in LMNA, including Emery-Dreifuss muscular dystrophy, atypical Werner syndrome, limb-girdle muscular dystrophy type 1B, Charcot-Marie-Tooth disease type 2, dilated cardiomyopathy, Dunnigan-type familial partial lipodystrophy, mandibuloacral dysplasia with type A lipodystrophy, and Hutchinson-Gilford progeria syndrome (HGPS) (2, 3). The relationship between LMNA genotype and phenotype is highly complex. Several LMNA mutations that cause the same or similar disorders do not cluster in the same functional region of the protein (3). Loss or gain of tissue-specific lamin A/C–protein interactions may explain how LMNA mutations cause such different, though at times overlapping, phenotypes.
Two of the disorders that can be caused by LMNA mutations, atypical Werner syndrome and HGPS, have some features of premature aging. A majority of classic Werner syndrome cases are caused by mutations in WRN, a helicase; however, there are several mutations in LMNA that have been observed in patients with atypical Werner syndrome (4). In most cases of HGPS, a de novo G to T mutation in exon 11 of LMNA activates a cryptic splice donor. This results in an increase in a form of lamin A, called progerin, with an internal, in-frame deletion of 50 amino acids removing a cleavage site important for protein maturation (5–7). HGPS is a rare premature-aging syndrome that causes an aged appearance, sclerotic skin, small size, alopecia, bone dysplasia, hearing loss, and impaired mobility as a result of joint contractures (8). Children with HGPS exhibit accelerated atherosclerosis leading to death at a mean age of 14.6 y due to myocardial infarction or stroke (9).
Cells from patients with HGPS exhibit an irregular nuclear shape or “blebbing” of the nucleus (5, 10), altered histone modification patterns, altered transcription (11), impaired DNA damage response (12, 13), early senescence, and decreased replicative ability at later passages (14, 15). These changes at the cellular level are also seen in cells from persons of advanced age (16). Irregular nuclear shape and chromatin disorganization have also been reported in cells from patients with other LMNA mutations (17–21).
Rapamycin and its analogs are of interest as potential new treatments for disorders associated with deposition of insoluble protein aggregates (22). Rapamycin inhibits TOR (target of rapamycin), a serine/threonine kinase that responds to environmental conditions and growth factors by regulating cell growth and metabolism (23). In mammals, the single TOR gene encodes mTOR (mammalian target of rapamycin), which functions as a component of two complexes, mTORC1 and mTORC2. Rapamycin inhibits mTORC1 but can also inhibit mTORC2 with prolonged treatment (24, 25). When cellular environmental conditions are favorable, mTORC1 signaling promotes ribosome biogenesis, transcription, and protein synthesis. If conditions are unfavorable (hypoxia, stress, low nutrient levels) or if cells are exposed to rapamycin, mTORC1 is inhibited and autophagy is promoted (23). Life span extension has been observed with rapamycin treatment in Drosophila, yeast, nematodes, and most recently, mouse (26).
HGPS cells treated with rapamycin show a reduction in nuclear blebbing, enhanced cell proliferation, and a delay in the onset of cellular senescence (27). A similar effect was observed after treatment with the rapamycin analog, everolimus (28). The proposed mechanism for these improvements with rapamycin is that treatment promotes the removal of toxic, insoluble aggregates of progerin by means of the enhanced autophagic-lysosomal pathway (27). Autophagy normally occurs in the cytoplasm and would not be expected to remove a nuclear protein like progerin; however, progerin aggregates become mislocalized during mitosis and can be found in the cytoplasm (29). Furthermore, there is increasing evidence that misfolded and aggregated proteins may be disposed of within the nucleus (30).
Everolimus is generally better tolerated clinically than rapamycin (31), so we chose this mTOR inhibitor for further study. We hypothesized that treatment with everolimus would improve the phenotype of cells derived from patients with other laminopathies, since many of these involve the production of abnormal proteins that may also form aggregates and contribute to cell death.
Results
Primary fibroblast cell lines with mutations in LMNA were selected for treatment with everolimus (Fig. 1 and Table 1). Such cell lines are not widely available, but we were able to obtain six: two lines from patients with atypical HGPS (PSADFN414 and PSADFN425); two atypical Werner syndrome lines (NORWAY1010 and AG04110); one line from an Emery-Dreifuss muscular dystrophy patient (GM23780); and one line from an HGPS patient with the most common causative mutation (HGADFN167). We also included the normal cell line HGFDFN168 (father of HGADFN167). For each experiment, cells were treated with either 0.1 μM everolimus (dissolved in ethanol) or the same volume of ethanol (vehicle control).
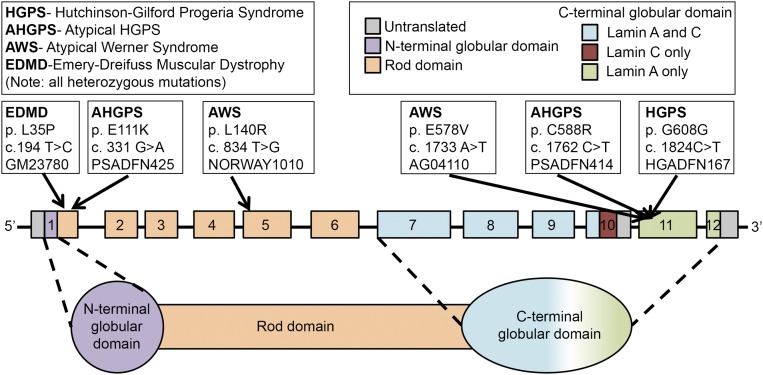
Fig. 1.
Diagram of the locations of LMNA mutations in the cell lines used in this study. Each putative causative mutation is indicated along with the name of the cell line that harbors it and the disease it causes. The top left key shows the name of each disorder, and the top right key shows the color code for the domains of lamin A and/or C. The LMNA gene is not drawn to scale. Adapted with permission from ref. 7.
Table 1.
Description of cell lines used including disease, LMNA mutation and clinical phenotype
| Cell line | Disease | Mutation in LMNA | Clinical phenotype |
| AG04110 | Atypical Werner syndrome | Exon 11, heterozygous c.1733 A>T (p.E578V) | Short stature, dysmorphic features, large coarse brown freckles over the entire body, thin skin on hands and feet, poor dentition, scoliosis, atrophic skin changes, beak nose, and high-pitched voice |
| NORWAY1010 | Atypical Werner syndrome | Exon 5, heterozygous c.834 T>G (p.L140R) | Died at age 36 y, cataracts, premature atherosclerosis, gray/thin hair, scleroderma-like skin, osteoporosis, soft tissue calcification, hypogonadism, aortic stenosis/insufficiency |
| GM23780 | Emery-Dreifuss muscular dystrophy | Exon 1, heterozygous c.104 T>C (p.L35P) | Achieved most motor function milestones and then lost them, sample collected at age 14 y, clinically affected, diagnostic muscle biopsy |
| PSADFN414 | Atypical HGPS | Exon 11, heterozygous c.1762T>C (p.C588R) | Small stature, mandibular hypoplasia, dental overcrowding, slow-growing coarse scalp hair, high narrow palate, thin and atrophic skin, little s.c. fat, joint pain, osteoarthritis, osteolysis, mild tricuspid valve regurgitation, sinus arrhythmia |
| PSADFN425 | Atypical HGPS | Exon 1, heterozygous c.331 G>A (p.E111K) | Small stature, mandibular hypoplasia, reduced s.c. fat and muscle mass, thinned hair, decreased bone density, dental overcrowding, stiff joints, elevated blood pressure, tricuspid valve regurgitation, systolic murmur, aortic stenosis at 20.5 y |
| HGADFN167 | Classical HGPS | Exon 11, heterozygous c.1824 C>T (p.G608G) | Classical HGPS phenotype with low body weight, short stature, muscle contractures, hearing loss, osteoporosis, dysplastic skeletal changes, dental crowding, narrow tented palate, s.c. paucity of fat, insulin resistance, scleroderma-like areas, skin fragility, paucity of scalp hair, and progressive cardiovascular disease; death at 17.2 y due to cardiac arrest |
| HGFDFN168 | Normal control | None (exon 11 sequenced) | None (father of HGADFN167, normal control) |
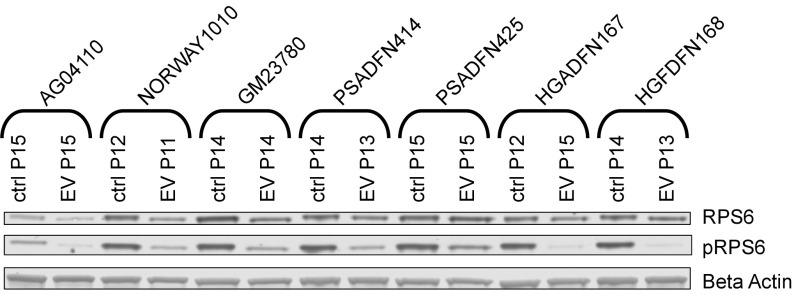
To assess biological effect of the drug treatment, we measured reduction in the level of phosphorylated ribosomal protein S6 (pRPS6), which is commonly used as a biomarker to reflect mTOR inhibition. We observed a reduction in pRPS6 levels in all cell lines after 2 wk of everolimus treatment (Fig. 2).
Fig. 2.
Reduction in pRPS6 levels with everolimus treatment. Western blots of RPS6 and pRPS6 are shown for everolimus-treated and vehicle control-treated cell lines. All cell lines were treated three times per week for 2 wk with 0.1 μM everolimus (EV) or vehicle control (ctrl). Passage (P) number at collection is indicated.
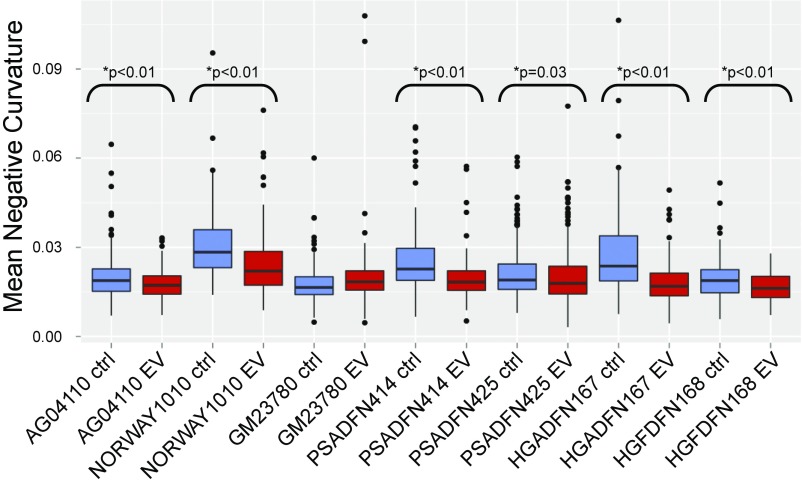
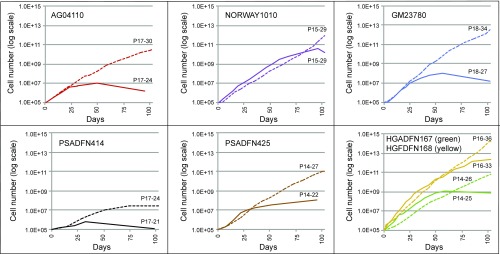
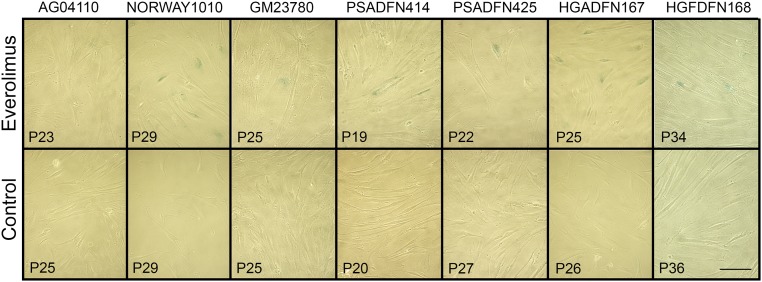
Lobulations, or blebs, of the nuclear membrane have been observed at a much higher rate in HGPS cells compared to normal cells, especially after multiple passages in culture. Blebs have also been observed in cells with other LMNA mutations. We explored whether everolimus could improve the nuclear morphology of cells from patients with different laminopathies. Given the limitation of subjective assessments, we developed a machine vision approach to identify nuclei in images of immunofluorescence-labeled cells, define the borders of the nuclei, and measure aspects of the nuclei. The metric mean negative curvature (MNC) was used to assess the severity of the blebbing (Fig. 3). Cells were treated three times per week for 2 wk with everolimus or vehicle control, and then nuclei were immunofluorescently labeled with a lamin A/C antibody and imaged. First, we assessed whether nuclear blebbing is a feature of the six laminopathy cell lines in this study. Two of the vehicle-treated lines, AG04110 and PSADFN425, did not demonstrate a difference in blebbing compared with the vehicle-treated normal control (P = 0.58 and P = 0.25, respectively, Wilcoxon rank sum test, two sided), so increased nuclear blebbing does not appear to be associated with these mutations. In most of the cell lines, the median MNC of the everolimus-treated cells was less than the median MNC of the control-treated cells, including the normal control (Fig. 4). The one exception was GM23780, which shows a higher median MNC with everolimus treatment. GM23780 nuclei have an unusually elongated shape, which may make MNC a less informative metric for measuring improvement in this line.
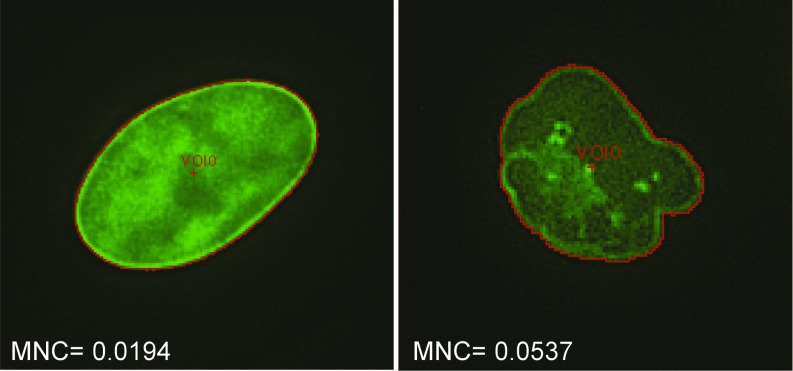
Fig. 3.
MNC is an objective measurement of nuclear blebbing that can be determined by machine vision. Shown are two example nuclei stained with a lamin A/C antibody, one unblebbed and one blebbed. The perimeter of each nucleus, which is used to determine MNC, is shown in red.
Fig. 4.
MNC values for everolimus-treated and control-treated cell lines. All cell lines were treated three times per week for 2 wk with 0.1 μM everolimus or vehicle control. Cells were fixed and labeled with lamin A/C antibody, and nuclei were photographed with an immunofluorescent microscope. All P values were calculated using a one-sided Wilcoxon rank sum test.
The proliferative ability of HGPS cells in culture is reduced compared with normal cells, and it has been previously shown that rapamycin treatment can improve this defect. We observed the proliferation rates of the laminopathy fibroblasts with everolimus or control treatment and found that with everolimus treatment, several of the lines were able proliferate better than their control-treated counterparts (Fig. 5). The everolimus-treated cells did show a decrease in proliferation initially, which has been noted elsewhere (27, 28), but later surpassed the control-treated cells in most of the lines. Several of the everolimus-treated cell lines (AG04110, PSADFN414, and PSADFN425) continued growing for 30 d or more past the time when control-treated cells of the same line stopped dividing. Normal fibroblasts (cell line HGFDFN168) showed increased proliferation with everolimus treatment as well, which has been observed previously with rapamycin treatment (27).
Fig. 5.
Proliferation is improved with everolimus treatment. Cell numbers over time of everolimus-treated (dashed lines) and vehicle control-treated (solid lines) cell lines are shown. All cell lines were treated three times per week with 0.1 μM everolimus or vehicle control. The cells were treated for 102 d, or until they ceased reaching confluence for at least 30 d. At each division, the cells were counted. Passage (P) number at the start and end of treatment for everolimus-treated and vehicle control-treated cells is indicated.
Another defect observed in HGPS cells is premature senescence. To assess senescence, everolimus- and control-treated cells from each line were stained for β-galactosidase activity. In all of the lines, we observed β-galactosidase–positive cells in the control-treated cells, but none in the everolimus-treated cells (Fig. 6 and Fig. S1).
Fig. 6.
Senescence-associated β-galactosidase staining of everolimus-treated and control-treated cells. Cells were treated three times per week with 0.1 μM everolimus or vehicle control. Passage (P) number of each cell line is indicated in the lower left corner. (Scale bar: 100 μm). All control lines have β-galactosidase–positive cells (indicated by blue staining), but everolimus-treated lines are negative.
Lastly, we assessed whether everolimus treatment results in a quantitative reduction in lamin proteins. We did not observe an overall reduction in lamin A or C in the fibroblast lines after everolimus treatment for 2 wk (Fig. S2).
Discussion
Fibroblast cells from laminopathy patients with six different LMNA mutations showed improvement in at least two key phenotypic measures of cellular health upon everolimus treatment. First, all everolimus-treated laminopathy cultures showed an increase in proliferative ability, with three of five lines continuing to proliferate after control cultures had ceased to grow (AG04110, PSADFN414, and PSADFN425). Second, all treated cell lines displayed a reduction in senescence by β-galactosidase staining. In addition, of the four laminopathy cell lines that exhibited nuclear blebbing, three demonstrated significant improvement in nuclear morphology (a reduction in nuclear MNC) after everolimus treatment.
A previous study showed that the benefit of rapamycin treatment of HGPS fibroblasts is based, at least in part, on increased autophagy of the mutant progerin protein (27). Although most of the laminopathy fibroblast lines studied here do not produce progerin, they each produce some form of abnormal lamin A proteins that might well accumulate as insoluble complexes in the nucleus and cytoplasm after multiple mitoses. We therefore hypothesized that mTOR inhibition could provide benefit for these cell lines through a similar mechanism, by increasing autophagy and eliminating aggregated abnormal lamin protein. Unfortunately, our experiments with autophagy inhibitors in concert with everolimus were unsuccessful due to the extreme cell toxicity of the autophagy inhibitors. The autophagy inhibitors were not tolerated for the length of time needed for everolimus treatment.
mTOR inhibitors have shown benefit in mouse models with a Lmna deletion or mutation. Rapamycin treatment increases survival in Lmna-knockout homozygous mice, a model that develops dilated cardiomyopathy and skeletal muscle dystrophy (32). Ramos et al. demonstrated that mTORC1 signaling is increased in tissues linked to the pathology and that rapamycin treatment reduces this signaling and, thus, increases autophagy (32). Unlike the laminopathy cell lines studied here that have an aberrant form of lamin A and/or C, these mice have a lack of lamin A and C altogether. Clearly, rapamycin is not improving the phenotype of these mice through clearing lamin A and/or C, but perhaps clearance of other deleterious proteins is causing the improvement. In LmnaH222P/H222P mice, a model of Emery-Dreifuss muscular dystrophy, treatment with the rapamycin analog tesirolimus has been shown to improve heart function (33, 34). Again, an increase in mTORC1 signal was present in disease-relevant tissue, and treatment increased autophagy, although lamin A/C aggregates or a reduction in lamin A/C were not observed. It is possible that improvement in overall autophagy is responsible for the amelioration of the phenotype observed in both of these models and in the laminopathy cell lines treated in this study with everolimus as well.
Previous treatment strategies for HGPS have focused on farnesyltransferase inhibitors, since the toxic protein in this disorder is permanently farnesylated as a result of loss of a cleavage site used in the final posttranslational step (7). Inhibiting farnesylation (and thus quantitatively reducing the amount of progerin) has shown benefit in HGPS cell models (35), mouse models (36), and patients in a clinical trial (37). The molecular defect in some of the other laminopathies is not the overproduction of a permanently farnesylated prelamin, but rather the production of a mature lamin containing a missense mutation in the protein. Although farnesyltransferase inhibitors have proved useful in treating HGPS, their use in other laminopathies does not have the same rationale. Alternative therapeutic approaches are needed.
Everolimus is approved for the management of transplant rejection and for cancer treatment and has been administered to children over long periods of time with a good safety record. It is being tested for treatment of HGPS in an ongoing clinical trial (https://clinicaltrials.gov/). Cellular data presented here, including improvements in the growth, senescence, and nuclear morphology of cell lines from several different laminopathies, indicate that everolimus might also be a viable treatment option for other disorders in this category of rare diseases.
Materials and Methods
Laminopathy Fibroblast Cell Lines and Patient Phenotypes.
Cells and information (genotype and patient phenotype) for the HGFDFN168, HGADFN167, PSADFN414, and PSADFN425 cell lines were obtained from the Cell and Tissue Bank and the Medical and Research Database, respectively, of the Progeria Research Foundation (www.progeriaresearch.org). PSADFN414 and PSADFN425 have been described previously (identified as APS 400.5 and APS 500.3, respectively) (38).
PSADFN425 is a compound heterozygote for the mutation listed in Table 1 and for LMNA intron 6, 1158-44 C>T. The mutation found in intron 6 is present in the 1000 Genomes Project database (39) and in one of the patient’s unaffected parents and is unlikely to be causative. AG04110 and GM23780 were purchased from the Coriell Institute for Medical Research (Coriell Cell Repositories, https://ccr.coriell.org/) and genotype and patient phenotype information was obtained from the online listing for those lines. NORWAY1010 was a gift from Junko Oshima, University of Washington, Seattle, and genotype and patient phenotype information was previously published (18).
Control and Experimental Treatments.
Cells were treated with media containing 0.1 μM everolimus (S1120; Selleck Chemicals) in ethanol or with media containing vehicle control (the same volume of ethanol) three times per week (Monday, Wednesday, and Friday). The media used was Minimum Essential Media (10370021; Life Technologies) supplemented with 13% heat-inactivated FBS (10438018; Life Technologies), 1× l-Glutamine (25030081; Life Technologies), and 1× Antibiotic-Antimycotic (15240096; Life Technologies).
Proliferation Studies.
Each cell line was trypsinized and counted, and two 25-cm2 culture-treated vented flasks were seeded with 1 × 105 cells each. The media on the cells was changed as described above, and split 1:3 when approaching 90% confluency. One-third of the cells were seeded into a new 25-cm2 flask. Cells were counted on a Z2 COULTER COUNTER Cell and Particle Counter (Beckman Coulter), and the media/cell volume was recorded. The total number of cells was calculated and then multiplied by 3 for each split the line had undergone between the start of the assay and the count to adjust for cells that were removed at each split. If the cells were less than 90% confluent 30 d after the last split, the cells were counted and not cultured further. The data are available in Table S1.
Senescence-Associated β-Galactosidase Assay.
Cells from 0.1 μM everolimus-treated and control-treated flasks that had been growing for the same amount of time (not necessarily the same passage due to differences between the proliferation rates of the drug-treated and vehicle control-treated cells) were trypsinized and seeded onto a six-well plate. HeLa cells transfected with a β-galactosidase expression plasmid were used as a positive control, and nontransfected HeLa cells were used as a negative control. Cells were stained using a senescence-associated β-galactosidase cell staining kit according to the manufacturer’s protocol (9860; Cell Signaling Technologies). Photographs were taken on a light microscope at 20× magnification.
Western Blots.
For RPS6/pRPS6 and lamin A/C detection in laminopathy and control fibroblasts (Fig. 2 and Fig. S2), cells were treated with 0.1 μM everolimus or vehicle control three times per week for 2 wk, and the cells were last treated ∼4 to 5 h before collection. Cells were suspended in 1× LI-COR protein loading buffer diluted in ddH2O (928-40004; LI-COR). Cells were boiled for 3 min and cooled at room temperature for 5 min. Ten micrograms of protein was used per sample, and NuPAGE Sample Reducing Agent was added before loading (NP0004; ThermoFisher Scientific). Protein was loaded into each well of a 10% Bis-Tris gel and electrophoresed in 1× 3-(N-morpholino) propanesulfonic acid buffer.
Protein was transferred from the gel to a nitrocellulose membrane (wet transfer) using an iBlot gel transfer device (Invitrogen), and the membrane was blocked with shaking over night at 4 °C in 1:1 1× PBS:Odyssey Blocking Solution (927-40000; LI-COR). The membrane was incubated with two primary antibodies (one for the protein of interest and one for the control protein) and two secondary antibodies (each with shaking at room temperature for 3 and 1 h, respectively, in 1:1 1× PBS:Odyssey Blocking Solution with 0.1% Tween 20 and antibody).
The following primary antibodies were used: S6 Ribosomal Protein (5G10) Rabbit mAb (2217; Cell Signaling Technology), Phospho-S6 Ribosomal Protein (Ser235/236) (D57.2.2E) XP Rabbit mAb (4858; Cell Signaling Technology), Anti-beta Actin antibody (mouse, ab8226; Abcam; used for Fig. 2 and Fig. S2), and Lamin A/C (N-18) Antibody (goat, sc-6215; Santa Cruz Biotechnology, Inc.; used for Fig. S2). The following secondary antibodies were used as appropriate: IRDye 680RD Donkey (polyclonal) Anti-Mouse IgG (926-68072; LI-COR), IRDye 680RD Donkey (polyclonal) Anti-Rabbit IgG (926-68073; LI-COR), IRDye 800CW Donkey Anti-Mouse IgG (926-32212; LI-COR), IRDye 800CW Donkey Anti-Rabbit IgG (926-32213; LI-COR), and IRDye 800CW Donkey Anti-goat IgG (925-32214; LI-COR). Membranes were scanned using a LI-COR Odyssey CLx.
Immunofluorescence.
After 2 wk of 0.1 μM everolimus or vehicle control treatment, cells were trypsinized, counted, and seeded onto eight-chamber treated culture slides (354108; Corning) at 25,000 cells per well. The cells were allowed to attach overnight. The next day, the cells were fixed with 4% paraformaldehyde (15710; Electron Microscopy Science) in 1× PBS and permeablized with 0.5% Triton X-100 in 1× PBS. Slides were blocked overnight with 4% BSA in 1× TBS at 4 °C. Primary antibody to lamin A/C (MAB3211; Millipore) was diluted 1:50 in 4% BSA in 1× TBS and applied for 5 h at room temperature, and then washed. Secondary antibody, Alexa Fluor 594 Donkey Anti-Mouse IgG (H+L) (A21203; Invitrogen) was diluted 1:1,000 in 4% BSA in 1× TBS and applied for overnight at 4 °C, and then washed. VECTASHIELD Antifade Mounting Medium with DAPI (H1200; Vector Laboratories, Inc.) was mixed with VECTASHIELD Antifade Mounting Medium (H1000; Vector Laboratories, Inc.) at a 1:4 ratio and applied to the wells, and the coverslip was affixed.
Nuclear Imaging and Analysis.
Images were taken using a DeltaVision Elite (GE Healthcare Life Sciences) fluorescent microscope with a 63× oil objective. Images were run through the MIPAV (Medical Image Processing, Analysis, and Visualization) application with the Nuclei Segmentation plugin. Segmented nuclei were viewed, and any with improper segmentation were removed. Remaining segmented nuclei were analyzed with the Nuclei Statistics plugin to determine the MNC. The Nuclei Statistic plugin was used to measure the features of the nucleus, including MNC. To determine the MNC, the perimeter of the nucleus is divided into 100 segments, and all segments with a negative change in slope (concave) are averaged.
The data are available in Table S2. The cell lines used in this work can be requested from their respective sources. MIPAV and relevant plugins are available at https://mipav.cit.nih.gov/.
Supplementary Material
Acknowledgments
We thank Even McCreedy and the Biomedical Imaging Research Services Section of the NIH Center for Information Technology for creating the MIPAV plugin for nuclear analysis. We thank Junko Oshima for the gift of the NORWAY1010 cell line, and Kan Cao for scientific guidance and discussions. We thank the Progeria Research Foundation for providing several cell lines.
Footnotes
Conflict of interest statement: F.S.C. and T.W.G. are coauthors on a 2017 research article. They did not collaborate directly on this work.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802811115/-/DCSupplemental.
References
- 1.Dittmer TA, Misteli T. The lamin protein family. Genome Biol. 2011;12:222. doi: 10.1186/gb-2011-12-5-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broers JL, Hutchison CJ, Ramaekers FC. Laminopathies. J Pathol. 2004;204:478–488. doi: 10.1002/path.1655. [DOI] [PubMed] [Google Scholar]
- 3.Dittmer TA, et al. Systematic identification of pathological lamin A interactors. Mol Biol Cell. 2014;25:1493–1510. doi: 10.1091/mbc.E14-02-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oshima J, Hisama FM. Search and insights into novel genetic alterations leading to classical and atypical Werner syndrome. Gerontology. 2014;60:239–246. doi: 10.1159/000356030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eriksson M, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Sandre-Giovannoli A, et al. Lamin a truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 7.Capell BC, Collins FS. Human laminopathies: Nuclei gone genetically awry. Nat Rev Genet. 2006;7:940–952. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- 8.Merideth MA, et al. Phenotype and course of Hutchinson-Gilford progeria syndrome. N Engl J Med. 2008;358:592–604. doi: 10.1056/NEJMoa0706898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon LB, et al. Progeria Clinical Trials Collaborative Impact of farnesylation inhibitors on survival in Hutchinson-Gilford progeria syndrome. Circulation. 2014;130:27–34. doi: 10.1161/CIRCULATIONAHA.113.008285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldman RD, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Rusinol A, Sinensky M, Wang Y, Zou Y. DNA damage responses in progeroid syndromes arise from defective maturation of prelamin A. J Cell Sci. 2006;119:4644–4649. doi: 10.1242/jcs.03263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manju K, Muralikrishna B, Parnaik VK. Expression of disease-causing lamin A mutants impairs the formation of DNA repair foci. J Cell Sci. 2006;119:2704–2714. doi: 10.1242/jcs.03009. [DOI] [PubMed] [Google Scholar]
- 14.Bridger JM, Kill IR. Aging of Hutchinson-Gilford progeria syndrome fibroblasts is characterised by hyperproliferation and increased apoptosis. Exp Gerontol. 2004;39:717–724. doi: 10.1016/j.exger.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, et al. Ankyrin G overexpression in Hutchinson-Gilford progeria syndrome fibroblasts identified through biological filtering of expression profiles. J Hum Genet. 2006;51:934–942. doi: 10.1007/s10038-006-0042-0. [DOI] [PubMed] [Google Scholar]
- 16.Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toth JI, et al. Blocking protein farnesyltransferase improves nuclear shape in fibroblasts from humans with progeroid syndromes. Proc Natl Acad Sci USA. 2005;102:12873–12878. doi: 10.1073/pnas.0505767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, et al. LMNA mutations in atypical Werner’s syndrome. Lancet. 2003;362:440–445. doi: 10.1016/S0140-6736(03)14069-X. [DOI] [PubMed] [Google Scholar]
- 19.Filesi I, et al. Alterations of nuclear envelope and chromatin organization in mandibuloacral dysplasia, a rare form of laminopathy. Physiol Genomics. 2005;23:150–158. doi: 10.1152/physiolgenomics.00060.2005. [DOI] [PubMed] [Google Scholar]
- 20.Vigouroux C, et al. Nuclear envelope disorganization in fibroblasts from lipodystrophic patients with heterozygous R482Q/W mutations in the lamin A/C gene. J Cell Sci. 2001;114:4459–4468. doi: 10.1242/jcs.114.24.4459. [DOI] [PubMed] [Google Scholar]
- 21.Taimen P, et al. A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc Natl Acad Sci USA. 2009;106:20788–20793. doi: 10.1073/pnas.0911895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendelsohn AR, Larrick JW. Rapamycin as an antiaging therapeutic?: Targeting mammalian target of rapamycin to treat Hutchinson-Gilford progeria and neurodegenerative diseases. Rejuvenation Res. 2011;14:437–441. doi: 10.1089/rej.2011.1238. [DOI] [PubMed] [Google Scholar]
- 23.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 24.Sarbassov DD, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 25.Lamming DW, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335:1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao K, et al. Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in Hutchinson-Gilford progeria syndrome cells. Sci Transl Med. 2011;3:89ra58. doi: 10.1126/scitranslmed.3002346. [DOI] [PubMed] [Google Scholar]
- 28.Driscoll MK, et al. Automated image analysis of nuclear shape: What can we learn from a prematurely aged cell? Aging (Albany NY) 2012;4:119–132. doi: 10.18632/aging.100434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao K, Capell BC, Erdos MR, Djabali K, Collins FS. A lamin A protein isoform overexpressed in Hutchinson-Gilford progeria syndrome interferes with mitosis in progeria and normal cells. Proc Natl Acad Sci USA. 2007;104:4949–4954. doi: 10.1073/pnas.0611640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen SV, Poulsen EG, Rebula CA, Hartmann-Petersen R. Protein quality control in the nucleus. Biomolecules. 2014;4:646–661. doi: 10.3390/biom4030646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan R, Kay A, Berg WJ, Lebwohl D. Targeting tumorigenesis: Development and use of mTOR inhibitors in cancer therapy. J Hematol Oncol. 2009;2:45. doi: 10.1186/1756-8722-2-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramos FJ, et al. Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Sci Transl Med. 2012;4:144ra103. doi: 10.1126/scitranslmed.3003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi JC, et al. Temsirolimus activates autophagy and ameliorates cardiomyopathy caused by lamin A/C gene mutation. Sci Transl Med. 2012;4:144ra102. doi: 10.1126/scitranslmed.3003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi JC, Worman HJ. Reactivation of autophagy ameliorates LMNA cardiomyopathy. Autophagy. 2013;9:110–111. doi: 10.4161/auto.22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Capell BC, et al. Inhibiting farnesylation of progerin prevents the characteristic nuclear blebbing of Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2005;102:12879–12884. doi: 10.1073/pnas.0506001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Capell BC, et al. A farnesyltransferase inhibitor prevents both the onset and late progression of cardiovascular disease in a progeria mouse model. Proc Natl Acad Sci USA. 2008;105:15902–15907. doi: 10.1073/pnas.0807840105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gordon LB, et al. Clinical trial of a farnesyltransferase inhibitor in children with Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2012;109:16666–16671. doi: 10.1073/pnas.1202529109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garg A, et al. Atypical progeroid syndrome due to heterozygous missense LMNA mutations. J Clin Endocrinol Metab. 2009;94:4971–4983. doi: 10.1210/jc.2009-0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Auton A, et al. 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.