Abstract
Background
Carbapenems are considered “drugs of last resort” in many life-threatening infections. Advent of carbapenemases like KPC, OXA-48, VIM, IMP, and NDM have greatly affected the efficacy of these drugs, posing serious threat to global health and infection control. NDM bears special significance to the India subcontinent, labeled as place of origin and reservoir. NDM tends to escape detection by routine phenotypic methods, requiring molecular confirmation. This study utilizes nested, multiplex polymerase chain reaction (PCR) for reliable detection of blaNDM-1 in nosocomial Enterobacteriaceae isolates.
Methods
This study was conducted to detect prevalence of blaNDM-1, blaIMP, blaVIMand blaKPC genes by multiplex PCR among multidrug/carbapenem-resistant nosocomial Enterobacteriaceae isolates. From March 2013 to April 2014, 100 consecutive non-repeat isolates of Enterobacteriaceae from various inpatient clinical samples were analyzed. Imipenem-resistant isolates identified by Kirby Bauer disk diffusion method with Clinical and Laboratory Standards Institute guidelines were further subjected to nested, multiplex PCR to simultaneously detect blaNDM-1, blaIMP, blaVIMand blaKPC genes.
Results
Out of 100 isolates, 17 (17%) were found to be imipenem-resistant. blaNDM-1 was detected in all 17 isolates by nested, multiplex PCR. blaVIM was co-carried in 4 isolates while one isolate co-harbored blaIMP with blaNDM-1. Imipenem resistance and NDM-1 carriage was predominant amongst Klebsiella isolates. Maximum NDM-1 producers were isolated from the intensive care unit (70.6%).
Conclusion
NDM-1 prevalence in nosocomial Enterobacteriaceae isolates in our hospital was found to be 17%. A nested, multiplex PCR was used for rapid detection of various carbapenemase genes with high sensitivity and specificity which is essential not only for favorable patient outcome but also for timely implementation of appropriate infection control practices to prevent further spread of such organisms.
Keywords: Carbapenemases, New Delhi Metallobetalactamase (NDM), Multiplex PCR (blaNDM-1, blaIMP, blaVIM and blaKPC), ICU
Introduction
β-Lactam resistance in Enterobacteriaceae has shown an alarming increase in frequency and spread, posing an ominous threat to public health. Among the most important issues in the antimicrobial resistance today, resistance to carbapenems, colloquially considered the ‘drugs of last resort’ for life-threatening infections in Enterobacteriaceae is mediated primarily by carbapenemases, functionally divided into serine-dependent (e.g., KPC, OXA-48) and metallo-β-lactamase (e.g., VIM, IMP, and NDM). New Delhi Metallobetalactamase-1 (NDM-1) bears special significance to India, labeled as the country of origin of this deadly enzyme, first identified in a Klebsiella pneumoniae isolate from a Swedish patient of Indian origin hospitalised in New Delhi in 2007.1 Since then, studies worldwide have reported the emergence of NDM-1, most linking it to a possible reservoir in the Indian subcontinent.2
Earlier restricted largely to the Enterobacteriaceae, the plasmid-mediated blaNDM-1 gene3 has since spread rapidly to non-clonally related isolates. The same plasmid often carries other drug-resistance genes as well, giving rise to multidrug resistant organisms (MDROs). Non-plasmid-mediated resistances may confer complete resistance to all antibiotics, including tigecycline and colistin3 severely limiting treatment options and increasing risk of nosocomial spread.
These often escape detection by routine laboratory methods, giving a false susceptibility report which in turn leads to ineffective and unnecessary antibiotic use, further aggravating the problem of resistance. Hence, rapid detection of these organisms is of utmost importance in limiting their emergence as well as spread, improving patient outcome and enhancing infection control. This has gained great interest amongst researchers in these past few years. Absence of phenotypic manifestation of gene, susceptible albeit elevated carbapenem minimum inhibitory concentrations (MICs) and inability of automated systems in distinguishing from other resistance mechanisms (e.g., extended spectrum beta-lactamases (ESBLs) and/or porin loss)3 necessitate molecular detection of the gene as the gold standard in diagnosis.12, 13, 14, 15, 16, 19, 20, 21, 22
This prospective study was undertaken at a tertiary care hospital in New Delhi over a period of one year (March 2013 to April 2014) to detect prevalence of blaNDM-1, blaIMP, blaVIM and blaKPC genes by multiplex polymerase chain reaction (PCR) among MDR and carbapenem-resistant nosocomial isolates of the Enterobacteriaceae family.
Material and methods
100 consecutive non-repeat isolates of Enterobacteriaceae isolated from various clinical specimens such as urine, pus, blood, sputum and body fluids from various medical and surgical wards and the intensive care unit (ICU) were included in the study.
Identification of Enterobacteriaceae: The isolates were identified till species level based upon standard bio-chemical methods.4
Antibiotic susceptibility testing: The antimicrobial susceptibility testing was done by the Kirby Bauer disk diffusion method on Mueller Hinton agar and commercially available antimicrobial discs (HiMedia Laboratories Pvt. Ltd., 23, Vadhani Ind. Est., LBS Marg, Mumbai, India) were used. Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 25923 and Pseudomonas aeruginosa ATCC 27853 were used as controls. Results were interpreted according to Clinical and Laboratory Standards Institute (CLSI) guidelines (M100-S20-U Jun 2010). For isolates with imipenem zone size <13 mm, MICs of carbapenems were determined by Vitek 2 Compact automated system (bioMérieux, Marcy-l′Etoile, France) in accordance with the CLSI recommendations and manufacturers’ instructions. MIC values of colistin and tigecycline were not carried out. All Enterobacteriaceae isolates resistant to imipenem were directly subjected to molecular detection of blaNDM-1, blaIMP, blaVIM and blaKPC genes by multiplex PCR. No phenotypic methods of carbapenemase detection were undertaken.
Molecular methods
All carbapenem resistant isolates were subjected to an automated multiplex PCR technique provided by Aus Diagnostics Pty Ltd for rapid detection of resistance genes blaNDM-1, blaVIM, blaIMP and blaKPC by following steps:
-
1.
DNA extraction – DNA extraction was done from all isolates with QIAamp DNA mini kits (QIAGEN, Str. 1, 40724 Hilden, Germany) as per manufacturer's instructions for DNA extraction from fresh cultures. Filtrate containing the DNA was taken up for PCR
-
2.
Multiplex PCR – Multiplex Tandem PCR kits provided by Aus Diagnostics Pty Ltd.
(205 Victoria Street Beaconsfield NSW 2015 Australia) were used for two sequential PCR steps.
Step 1 consisted of 15 cycles of multiplexed pre-amplification using primers homologous to all targets in the panel. Step 2 utilized the product from Step 1 diluted into individual wells for real time PCR reactions. Primers for Step 2 were ‘nested inside’ the primers used for Step 1. Till this point, the procedure was automated by the Mark II Easy-Plex™ 72 liquid handling robot system (AusDiagnostics Pty Ltd.)
Step 2 real time PCR reaction was performed in the Rotor-Gene of the Mark II Easy-Plex™ 72 system. Eva-Green™ fluorescent dye was incorporated into the DNA being formed in the specific amplification reaction. DNA amplification was then measured by the increase in fluorescence produced by the dye.
Step 1 was carried out in a 7-well tube strip (for seven samples), amplifying the five targets (including SPIKE, an internal control). A 72-well Easy-Plex™ ring is provided for performing Step 2.
Internal control – An artificial DNA target called SPIKE acted as an internal control for integrity of reagents (polymerase, primers, etc.), equipment function (thermal cyclers) and the presence of inhibitors in the sample and was contained in each Step 1 tube, in addition to target primers.
Reaction mixture for Step 1
Cycling conditions for Step 1
Reaction mixture for Step 2
Cycling conditions for Step 2
Primers for both steps were patented by AusDiagnostics Pty Ltd and not revealed.
Interpretation
Results were read based on fluorescent signal produced by detection and amplification of any of the target genes, blaNDM-1, blaVIM, blaIMP and blaKPC. Results were generated both in tabular form as well as in the form of amplification curves.
Control group
A total of 30 nosocomial E. coli and K. pneumoniae isolates from similar clinical samples but sensitive to imipenem were also collected and tested during the same study period for detection of MBL genes (blaNDM-1, blaVIM, blaIMP and blaKPC) which might be harbored without phenotypic expression of the resistance mechanism.
Results
Phenotypic studies
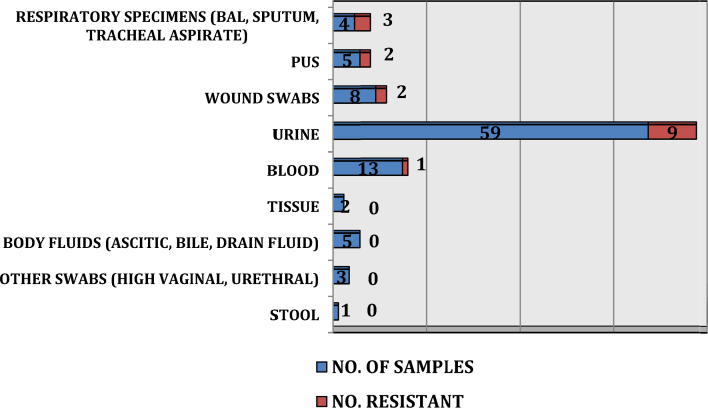
Maximum samples were urine (59) followed by blood (13), wound swabs (8), pus (5), body fluids (ascitic, bile, drain fluid) (5), respiratory specimens (BAL, sputum, tracheal aspirate) (4), high vaginal swabs (2), tissue specimens (2), urethral swab (1) and stool (1). These were mainly from Medical (45) and Surgical wards (40) while 15 samples were from ICU (adult and neonatal). E. coli (57%) and Klebsiella spp. (22%) were the commonest isolates. Distribution of isolates is shown in Fig. 1.
Fig. 1.
Distribution of isolates amongst Enterobacteriaceae.
MICs of carbapenems determined by Vitek 2 Compact automated system were 8–64 μg/ml. Among the 100 isolates studied, 17 isolates were found to be imipenem resistant of which 12 (70.6%) belonged to Klebsiella spp. and 5 (29.4%) were E coli. Rest all Enterobacteriaceae isolates were susceptible to imipenem (Fig. 2).
Fig. 2.
Comparison of distribution of isolates vs NDM-1 producers.
Imipenem resistance was maximum among the respiratory isolates (75%) followed by pus (40%) and wound swabs (25%). Urine isolates showed a low prevalence of imipenem resistance (15.25%) followed by blood (7.69%) (Fig. 3).
Fig. 3.
Distribution of imipenem resistance in various specimens.
Molecular methods
All 17 imipenem-resistant isolates were subjected to nested, multiplex PCR for rapid detection of resistance genes blaNDM-1, blaVIM, blaIMP and blaKPC as described above.
All imipenem-resistant isolates in our study were found to be carrying the blaNDM-1 gene (100%). blaVIM was detected in 4 isolates (23.5%) but all these isolates were positive for blaNDM-1 as well. One isolate co-harbored blaIMP with blaNDM-1. blaKPC was not detected in any of the isolates (Table 1).
Table 1.
Distribution of genes among carbapenem-resistant isolates.
| Gene | Total no. of imipenem resistant isolates | No. of positive by multiplex PCR | % |
|---|---|---|---|
| blaNDM-1 | 17 | 17 | 100% |
| blaVIM | 17 | 4 | 23.5% |
| blaIMP | 17 | 1 | 5.88% |
| blaKPC | 17 | 0 | 0 |
Thus, imipenem resistance as well as NDM-1 production was found to be predominant amongst Klebsiella isolates at our hospital (Table 2).
Table 2.
Distribution of NDM-1 producing organisms.
| Organism | Total no. of imipenem resistant isolates | MBL producers |
||
|---|---|---|---|---|
| Number | % | Gene | ||
| Klebsiella pneumoniae | 12 | 12 | 100% | blaNDM-1 |
| E. coli | 5 | 5 | 100% | blaNDM-1 |
Out of the 100 nosocomial Enterobacteriaceae isolates studied, majority (85/100) were from medical and surgical wards while 15 came from the ICU. However, 12 of the 15 ICU isolates (70.6%) turned out to be carbapenem-resistant and NDM producers whereas the wards had only 5 such isolates (5.9%) (Fig. 1).
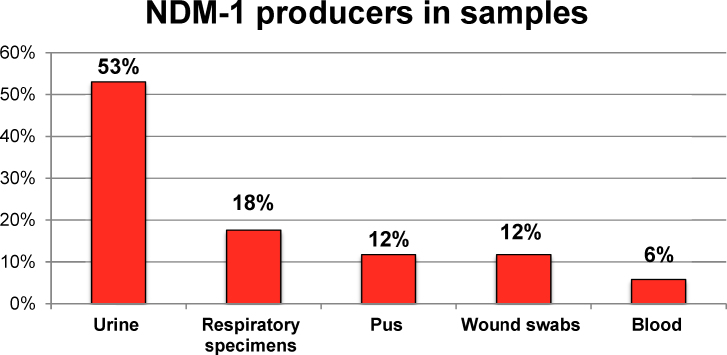
Imipenem-resistant urinary isolates harbored majority of NDM producers (53%) followed by respiratory specimens (17.6%), pus and wound swabs (11.76% each) and blood cultures (5.8%). These results are depicted in relevant tables/figures (Fig. 4, Fig. 5).
Fig. 4.
Distribution of NDM-1 producers in various specimens.
Fig. 5.
Multiplex Tandem PCR report for isolate carrying both blaNDM-1 and blaVIM. (a) Tabulated version and (b) cycling curves.
None of the imipenem-susceptible control isolates harbored any of the target genes.
Discussion
Carbapenem resistance in clinical Enterobacteriaceae isolates has been reported worldwide. Imipenem resistance was detected in 17% of isolates in our study, 12 belonging to Klebsiella spp. (70.6%) while the remaining 5 isolates were E. coli (29.4%).
MIC values for imipenem, meropenem and ertapenem ranged from 8–64 μg/ml for all isolates.
Indian studies report a range of 15–29% of imipenem resistance from different specimens from ICU and various wards.5, 6, 7 The blaNDM-1 gene was detected in all imipenem-resistant isolates in our study (100%) and was most prevalent in Klebsiella spp. Recent studies from other Indian tertiary hospitals corroborate our findings of carbapenem resistance and carriage of MBLs, especially NDM (40–92%) amongst carbapenem-resistant Enterobacteriaceae (CRE)8, 9, 10, 11, 12, 13, 14, 15, 16 with researchers from various regions of the country utilizing conventional or multiplex, real time PCR formats similar to the one used in our study. Deshpande et al reported 91.67% carriage of blaNDM-1 gene among the carbapenem-resistant isolates in her study from Mumbai8 similar to studies from Pune9, 10, 11 including a recent study on NDM producing extraintestinal pathogenic E. coli (ExPEC) strains, where NDM-1 carriage was 69%. blaNDM-1 in studies from South Indian tertiary care hospitals ranged from 40% in Vellore, Tamil Nadu12, 13 to 81% in Mangalore, Karnataka.14 Wattal et al. in their study in a tertiary care hospital in North India found 100% NDM-1 carriage in CRE (Carbapenem Resistant Enterobacteriaceae) isolates from ICU blood cultures.15 Bora et al. in their study from the Northeast detected 71.42% of E coli isolates harboring blaNDM-1.16
These findings are also supported by the data from Europe and North America.17, 18 The SMART study7 detected NDM producers in 50% of all Enterobacteriaceae isolates with reduced susceptibility to carbapenems. Other studies also show a wide range of prevalence of NDM-1 producers amongst CRE isolates. A study of carbapenemase-producing Enterobacteriaceae from Russia and other Baltic countries detected blaNDM-1 in 15 out of 77 CRE isolates (19.48%).19 A study on an outbreak of NDM-1 producing Enterobacteriaceae in Italy in 2011 analyzed 44 isolates, out of which 6 isolates harbored the blaNDM-1 gene (13.63%).20 Also, maximum number of NDM-1 producers belonged to Klebsiella spp.
High prevalence of NDM-1 producers in our study can be attributed to limitation of sampling bias. Ours is a tertiary care hospital with superspeciality departments such as liver, renal and bone marrow/stem cell transplant, malignant diseases treatment center and departments dealing with conditions such as hematological malignancies, joint replacement and reconstructive surgery. Moreover, cases are referred from most of northern India, already subjected to various antimicrobial regimens in primary and secondary medical care. Thus, sample population is limited with known risk factors of multidrug resistance acquisition (general debility, immunosuppression, malignancy and antibiotic exposure). This is further corroborated by the finding of maximum NDM producers being isolated from the ICU.
This was a pilot study conducted to validate the utility of rapid and robust molecular detection of carbapenem resistance amongst hospital-acquired Enterobacteriaceae isolates at a premier tertiary care hospital. Early detection of such isolates not only facilitates patient care but also proves invaluable in infection control and prevention of spread/emergence of multidrug resistance.
Various phenotypic methods of carbapenemase/MBL detection are also available, namely the modified Hodge test, agar diffusion tests like E-test, disk enhancement tests utilizing Zn and EDTA or commercially available combined disk tests like the KPC + MBL confirm ID pack and Neo-Sensitabs (Rosco Diagnostica) as well as chromogenic tests such as the Carba NP described by Nordmann et al and Dortet et al21, 22 which has recently been introduced as a confirmatory test for carbapenemase producers by CLSI M100-S25.23 however, there are conflicting reports as regards sensitivity and specificity of these tests in detecting carbapenemases/MBLs, further varied by the specific resistance-determinant gene. In addition, the turnaround time of >24 h of these tests is an added deterrant.13, 23, 24, 25, 26, 27, 28
Conclusion
Our study demonstrated a high prevalence of NDM-1 in carbapenem-resistant nosocomial Enterobacteriaceae isolates. This shows that the emergence of this enzyme is a major threat in the field of microbial drug resistance. Early detection is desirable for patient benefit and to avoid spread of such isolates. However, routine detection is controversial and difficult. Molecular detection with multiplex PCR remains the gold standard with high sensitivity and specificity but techniques are demanding and not available at most laboratories. Hence, detection of pan-resistant organisms by phenotypic methods in smaller peripheral hospitals assumes extreme importance as such organisms are likely to harbor NDM-1 gene. A timely warning conveyed to treating clinicians in such cases would avert preventable calamities, especially in acute cases in high output areas such as the ICUs and Burns Center and Long-term care areas such as Orthopedic wards by timely institution of appropriate treatment and infection control practices such as isolation.
Our study also highlights the undeniable importance of strict infection control practices and judicious use of antibiotics especially in setting of ICU and critical care wards, not only in preventing emergence and spread of such organisms but also in extending longevity of carbapenems, the last resort antibiotics in many life-threatening infections.
Conflicts of interest
The authors have none to declare.
Acknowledgements
This paper is based on Armed Forces Medical Research Committee Project No 4401/2013 granted by the Office of the Directorate General Armed Forces Medical Services and Defence Research Development Organization, Government of India.
References
- 1.Yong D., Toleman M.A., Giske C.G. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumarasamy K.K., Toleman M.A., Walsh T.R. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10(9):597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nordmann P., Naas T., Poirel L. Global spread of carbapenemase producing Enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koneman E. 1st ed. Lippincott Williams & Wilkins; 2016. Koneman's Color Atlas and Textbook of Diagnostic Microbiology. [Google Scholar]
- 5.Wankhede S.V., Iyer V.S., Bharadwaj R.S. The study of MBL producers in gram negative isolates from ICUs and wards. Indian J Basic Appl Med Res. 2011;1(1):38–46. [Google Scholar]
- 6.Rawat V., Singhai M., Verma P.K. Detection of different β-lactamases and their co-existence by using various discs combination methods in clinical isolates of Enterobacteriaceae and Pseudomonas spp. J Lab Phys. 2013;5(1):21–25. doi: 10.4103/0974-2727.115918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lascols C., Hackel M., Marshall S.H. Increasing prevalence and dissemination of NDM-1 metallo-β-lactamase in India: data from the SMART study (2009) J Antimicrob Chemother. 2011;66(9):1992–1997. doi: 10.1093/jac/dkr240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshpande P., Rodrigues C., Shetty A., Kapadia F., Hedge A., Soman R. New Delhi Metallo-lactamase (NDM-1) in Enterobacteriaceae: treatment options with carbapenems compromised. J Assoc Phys India. 2010;58(3):147–149. [PubMed] [Google Scholar]
- 9.Khajuria A., Praharaj A.K., Kumar M., Grover N., Aggarwal A. Multidrug resistant NDM-1 metallo-beta-lactamase producing Klebsiella pneumoniae sepsis outbreak in a neonatal intensive care unit in a tertiary care center at central India. Indian J Pathol Microbiol. 2014;57:65–68. doi: 10.4103/0377-4929.130900. [DOI] [PubMed] [Google Scholar]
- 10.Khajuria A., Praharaj A.K., Kumar M., Grover N. Emergence of Escherichia coli, co-producing NDM-1 and OXA-48 carbapenemases, in urinary isolates, at a tertiary care centre at Central India. J Clin Diagn Res. 2014;8(6):DC01–DC4. doi: 10.7860/JCDR/2014/7952.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ranjan A., Shaik S., Mondal A. Molecular epidemiology and genome dynamics of New Delhi metallo-beta-lactamase (NDM) producing extraintestinal pathogenic E. coli (ExPEC) strains from India. Antimicrob Agents Chemother. 2016 doi: 10.1128/AAC.01345-16. pii:AAC.01345-16 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anandan S., daModaran S., Gopi R., Bakthavatchala Y.D.M., Veeraraghavan B. Rapid screening for carbapenem resistant organisms: current results and future approaches. J Clin Diagn Res. 2015;9(9):DM01–DM03. doi: 10.7860/JCDR/2015/14246.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pragasam A.K., Sahni R.D., Anandan S. A pilot study on carbapenemase detection: do we see the same level of agreement as with the CLSI observations. J Clin Diagn Res. 2016;10(7):DC09–DC13. doi: 10.7860/JCDR/2016/16417.8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakraborty A., Adhikari P., Shenoy S. Molecular characterization and clinical significance of New Delhi metallo-beta-lactamases-1 producing Escherichia coli recovered from a South Indian tertiary care hospital. Indian J Pathol Microbiol. 2015;58:323–327. doi: 10.4103/0377-4929.162864. [DOI] [PubMed] [Google Scholar]
- 15.Wattal C., Raveendran R., Goel N., Oberoi J.K., Rao B.K. Ecology of blood stream infection and antibiotic resistance in intensive care unit at a tertiary care hospital in North India. Braz J Infect Dis. 2014;18(3):245–251. doi: 10.1016/j.bjid.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bora A., Ahmed G.U., Hazarika N.K. Incidence of blaNDM-1 gene in Escherichia coli isolates at a tertiary care referral hospital in Northeast India. Indian J Med Microbiol. 2013;31(3):250. doi: 10.4103/0255-0857.115628. [DOI] [PubMed] [Google Scholar]
- 17.Magiorakos A.P., Suetens C., Monnet D.L., Gagliotti C., Heuer O.E. The rise of carbapenem resistance in Europe: just the tip of the iceberg? Antimicrob Resist Infect Control. 2013;2(1):6. doi: 10.1186/2047-2994-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vital signs: carbapenem-resistant Enterobacteriaceae. Morbid Mortal Wkly Rep MMWR. 2013;62(9):165–170. [PMC free article] [PubMed] [Google Scholar]
- 19.Pavelkovich A., Balode A., Edquist P. Detection of carbapenemase-producing Enterobacteriaceae in the Baltic countries and St. Petersburg area. Biomed Res Int. 2014 doi: 10.1155/2014/548960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaibani P., Ambretti S., Berlingeri A. Outbreak of NDM-1-producing Enterobacteriaceae in northern Italy, July to August 2011. Euro Surveill. 2011;16(47):20–27. [PubMed] [Google Scholar]
- 21.Nordmann P., Poirel L., Dortet L. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2012;18(9):1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dortet L., Poirel L., Nordmann P. Rapid detection of carbapenemase-producing Pseudomonas spp. J Clin Microbiol. 2012;50(11):3773–3776. doi: 10.1128/JCM.01597-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clinical and Laboratory Standards Institute . Twenty fifth Informational Supplement. CLSI Document M100-25. Clinical and Laboratory Standards Institute; Wayne, PA: 2015. Performance Standards for Antimicrobial Susceptibility Testing. [Google Scholar]
- 24.Doyle D., Peirano G., Lascols C., Lloyd T., Church D.L., Pitout J.D. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J Clin Microbiol. 2012;50(12):3877–3880. doi: 10.1128/JCM.02117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasteran F., Veliz O., Rapoport M., Guerriero Corso A. Sensitive and specific modified Hodge test for KPC and metallo beta lactamase detection in Pseudomonas aeruginosa by use of a novel strain, Klebsiella pneumoniae ATCC 700603. J Clin Microbiol. 2011;49(12):4301–4303. doi: 10.1128/JCM.05602-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeremiah S.S., Balaji V., Anandan S., Sahni R.D. A possible alternative to the error prone modified Hodge test to correctly identify the carbapenemase producing Gram-negative bacteria. Indian J Med Microbiol. 2014;32:414–418. doi: 10.4103/0255-0857.142258. [DOI] [PubMed] [Google Scholar]
- 27.Tijet N., Boyd D., Patel S.N., Mulvey M.R., Melano R.G. Evaluation of the Carba NP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57(9):4578–4580. doi: 10.1128/AAC.00878-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dortet L., Bréchard L., Poirel L., Nordmann P. Impact of the isolation medium for detection of carbapenemase-producing Enterobacteriaceae using an updated version of the Carba NP test. J Med Microbiol. 2014;63(Pt 5):772–776. doi: 10.1099/jmm.0.071340-0. [DOI] [PubMed] [Google Scholar]