Abstract
The bacterial type IV secretion systems (T4SSs) are a highly functionally and structurally diverse superfamily of secretion systems found in many species of gram-negative and -positive bacteria. Collectively, the T4SSs can translocate DNA and monomeric and multimeric protein substrates to a variety of bacterial and eukaryotic cell types. Detailed phylogenomics analyses have established that the T4SSs evolved from ancient conjugation machines whose original functions were to disseminate mobile DNA elements within and between bacterial species. How members of the T4SS superfamily evolved to recognize and translocate specific substrate repertoires to prokaryotic or eukaryotic target cells is a fascinating question from evolutionary, biological, and structural perspectives. In this chapter, we will summarize recent findings that have shaped our current view of the biological diversity of the T4SSs. We focus mainly on two subtypes, designated as the types IVA (T4ASS) and IVB (T4BSS) systems that respectively are represented by the paradigmatic Agrobacterium tumefaciens VirB/VirD4 and Legionella pneumophila Dot/Icm T4SSs. We present current information about the composition and architectures of these representative systems. We also describe how these and a few related T4ASS and T4BSS members evolved as specialized nanomachines through acquisition of novel domains or subunits, a process that ultimately generated extensive genetic and structural mosaicism among this secretion superfamily. Finally, we present new phylogenomics information establishing that the T4BSSs are much more broadly distributed than initially envisioned.
Keywords: Type IV secretion, Conjugation, DNA transfer, Pathogenesis, Effector translocation, Legionella Dot/Icm, Coupling protein, Traffic ATPases
1. Introduction: The ABC’s of T4SS Classification
The T4SSs are widely distributed among gram-negative (Gram−) and gram-positive (Gram+) bacteria, and they mediate a broad range of functions to the benefit of their bacterial hosts (Grohmann et al. 2018). Various schemes have emerged to classify T4SSs, an undertaking complicated by the extreme genetic and functional heterogeneity of this secretion superfamily. For example, T4SSs have been classified on the basis of function as: (i) conjugation systems, (ii) effector translocators, or (iii) contact-independent DNA/protein exchange systems (Cascales and Christie 2003). The conjugation systems are the largest subfamily, present in nearly all bacterial species and some archaeal species (Guglielmini et al. 2013). These systems are specifically employed for dissemination of associated mobile genetic elements, although they also deliver a small number of protein substrates independently of DNA. The “effector translocators” deliver effector proteins to prokaryotic or eukaryotic cells (Asrat et al. 2015; Bhatty et al. 2013; Kubori and Nagai 2015; Souza et al. 2015). These substrates enable the competitive outgrowth of T4SS-carrying donor cells in polymicrobial communities, or bacterial colonization and spread in pathogenic settings through the disruption of eukaryotic host cell physiological processes. The contact-independent exchange systems, currently consisting of only a few members, function in release of DNA or protein substrates to the milieu or, alternatively, uptake of exogenous DNA (Locht et al. 2011; Ramsey et al. 2011; Stingl et al. 2010).
Alternative classification schemes are derived from phylogenetic analyses. One scheme, originating from studies of Escherichia coli conjugation systems, grouped the conjugative systems according to the conjugative pilus elaborated as F-, P-, or I-type. The F conjugation system was the earliest characterized T4SS; these systems are present in many species of Enterobacteriaceae as well as other members of gamma- and alphaproteobacteria (Arutyunov and Frost 2013). The F-type plasmids code for long, flexible pili that dynamically extend and retract, a property enabling highly efficient transfer in both solid surface and liquid matings (Clarke et al. 2008; Silverman and Clarke 2010). The P-type systems in contrast elaborate shorter, more rigid pili. These types of pili are produced by well-characterized conjugation systems encoded by E. coli plasmids RP4, R388, and pKM101, as well as the Agrobacterium tumefaciens VirB/VirD4 system (Arutyunov and Frost 2013; Christie et al. 2005; Backert and Meyer 2006). The I-type plasmids typically encode two types of pili, one similar to P-type pili and a second similar to type IV pili. Type IV pili are ancestrally unrelated to conjugative pili, but reminiscent of F-pili; they extend and retract, and this property enables efficient transfer of I-type plasmids in liquid media (Nagai and Kubori 2011; Sampei et al. 2010; Thanassi et al. 2012).
By far, the most detailed T4SS classifications to date are derived from phylogenetic analyses of the highly conserved ATPases associated with these systems. Nearly all T4SSs have two signature ATPases, which according to the unifying nomenclature of the A. tumefaciens VirB/VirD4 T4SS are homologs of the VirD4 and VirB4 subunits. By tracing the evolutionary history of the VirB4 ATPases, and using VirD4 to root the tree, Guglielmini et al. identified eight distinct clades into which all presently identified T4SSs can be assigned (Guglielmini et al. 2013). This work also supported a model for how the T4SSs evolved. The VirD4 and VirB4 ATPases are related to DNA motor proteins SpoIIIE and FtsK, which use the energy of ATP hydrolysis to translocate along DNA (Gomis-Ruth et al. 2004; Middleton et al. 2005). The VirD4 and VirB4 ATPases thus might originally have functioned as DNA motors, carrying out activities associated with DNA metabolism. Eventually, both ATPases were coupled with an envelope-spanning channel, which itself was probably an ancient protein translocation system. The resulting conjugation machines appear to have originated in diderm species (cell envelopes with inner and outer membranes) and then diversified to function in monoderm species (single membrane cell envelopes). Finally, and only recently in the evolutionary scale, further diversification led to the extant conjugation systems and dedicated effector translocator and exchange systems (Guglielmini et al. 2013).
A simple classification scheme, we will use in this chapter parses T4SSs into T4ASS and T4BSS types; these are represented respectively by the paradigmatic A. tumefaciens VirB/VirD4 and Legionella pneumophila Dot/Icm systems (see Fig. 1) (Christie and Vogel 2000). The T4ASS transporters encompass the P- and F-type systems and share a conserved set of approximately 12 subunits related to the 11 VirB proteins and VirD4 subunit comprising the A. tumefaciens VirB/VirD4 T4SS (Chandran Darbari and Waksman 2015; Christie et al. 2005). The T4BSS transporters evolved from I-type conjugation systems. The representative L. pneumophila Dot/Icm system, so named because the Isberg and Shuman groups independently discovered it and respectively named it Dot and Icm (defective in organelle trafficking/intracellular multiplication system), requires over >25 proteins of which only a few are VirB homologs (Marra et al. 1992; Sadosky et al. 1993; Berger and Isberg 1993; Berger et al. 1994; Brand et al. 1994; Nagai and Kubori 2011; Voth et al. 2012). The T4ASS and T4BSS classification scheme does not encompass all T4SSs, but suffices here as we explore the evolutionary diversification of T4SSs.
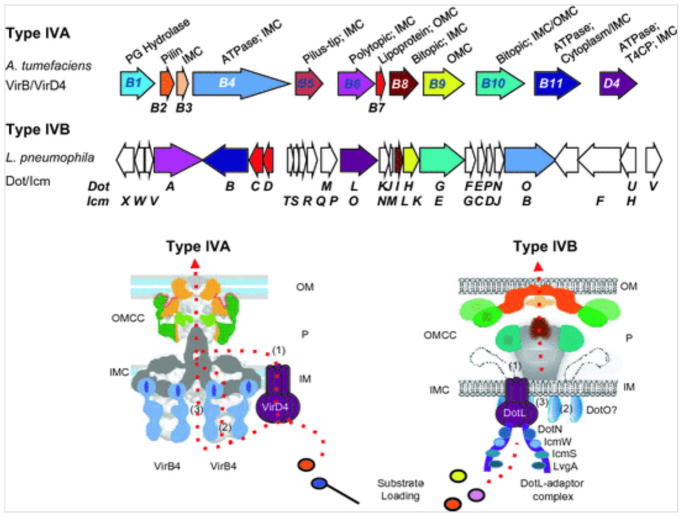
Fig. 1.
Gene arrangements and architectures of the A. tumefaciens VirB/VirD4 type IVA and L. pneumophila Dot/Icm type IVB secretion systems. Upper: gene arrangements of the two systems with color-coding of the genes encoding homologous subunits; unshaded genes are unique for the T4BSS transporters. The VirB/VirD4 subunit enzymatic functions and associations with inner membrane complex (IMC), outer membrane core complex (OMCC), or pilus are listed. PG Hydrolase, peptidoglycan hydrolase; T4CP, type IV coupling protein. Lower: architectures of the T4ASS and T4BSS machineries based on the R388-encoded VirB3–10 structure (Low et al. 20124) and the L. pneumophila Dot/Icm system (Ghosal et al. 2017) adapted with permission from the publishers. OM, outer membrane; P, periplasm; IM, inner membrane; OMCC, outer membrane core complex; IMC, inner membrane complex. For both systems, three different routes for substrate passage across the inner membrane are presented: (1) through the central channel formed by the VirD4/DotL hexamer, (2) through the channel formed by the VirB4/DotO hexamer, or (3) through a channel formed by other IMC subunits, e.g., VirB6/DotA and VirB8/DotI. The IMC of the Dot/Icm system has not been structurally analyzed. For both systems, substrates are delivered through an OMCC channel to the cell surface. For the Dot/Icm system, the DotL—adaptor complex involved in substrate recruitment is shown (Kwak et al. 2017)
2. Function, Structure, and Diversification of T4ASS and T4BSSs
There is now ample genetic, biochemical, and structural evidence that T4SSs evolved as supramolecular structures composed of modules of functionally distinct subassemblies (Fig. 1). At the base of the T4SS, the highly conserved VirD4 subunit functions as a receptor to mediate recruitment of substrates. Because of their functions in linking substrates with the T4SS channel, members of the VirD4 superfamily also have been termed type IV coupling proteins or T4CPs (Cabezon et al. 1997). VirD4 coordinates its ATPase functions with one or two other ATPases represented by the A. tumefaciens VirB4 and VirB11 subunits (Atmakuri et al. 2004; Cascales and Christie 2004; Pena et al. 2012; Ripoll-Rozada et al. 2013; Savvides et al. 2003). The VirD4/VirB4/VirB11 energy center localizes at the cytoplasmic entrance to the T4SS channel where it processes substrates for translocation and might also energize translocation through the channel. This energy center associates with a second-large subassembly that in Gram− bacterial systems is termed the inner membrane complex (IMC) (Low et al. 2014). The IMC directs the translocation of substrates across the inner membrane (IM). It physically interacts with another large subassembly termed the outer membrane core complex (OMCC), which is responsible for conveying the substrate through the periplasm and across the outer membrane (Christie et al. 2005; Low et al. 2014). In the Gram− bacterial systems, the T4SSs additionally elaborate extracellular structures such as conjugative pili that are important for establishing productive contacts with target cells (Lawley et al. 2003). VirB4 and VirB11, when present (see below), but not VirD4, coordinate with the IMC and OMCC subassemblies for pilus assembly. Conversely, the entire ATPase energy center plus the IMC and OMCC subassemblies, but not the extended pilus, are required for substrate transfer. In the following sections, we summarize information about the various modules comprising the T4ASS and T4BSS transporters and about adaptations acquired by these modules throughout evolution that have enabled functional diversification.
2.1. The VirD4 Receptor and Its Role in Substrate Selection
VirD4-like ATPases are associated with nearly all T4SSs, and the presence of a VirD4-like gene in sequenced bacterial genomes can serve to identify new T4SS gene clusters (Bhatty et al. 2013). A role for VirD4 subunits in substrate reception was suggested by early genetic studies, which showed that VirD4 subunits can sometimes be exchanged, resulting in a switch in the substrate specificity of the chimeric system (Cabezon et al. 1997). VirD4 receptor function was then firmly established using a formaldehyde (FA) crosslinking assay termed transfer DNA immunoprecipitation (TrIP) (Cascales and Christie 2004). In this assay, a DNA substrate was subjected to formaldehyde crosslinking during transit through the A. tumefaciens VirB/VirD4 T4SS, and crosslinking of the DNA to individual machine subunits was detected by immunoprecipitation and PCR amplification. DNA substrates were crosslinked with VirD4, even independently of other VirB machine subunits, and a virD4 mutation abolished all detectable substrate crosslinks with the VirB subunits. These findings confirm VirD4’s role in initiating the docking of DNA substrates with the T4SS (Atmakuri et al. 2004).
VirD4 receptors typically are composed of an N-terminal transmembrane domain (NTD) and a cytoplasmic moiety that functions in substrate reception. The receptor moiety consists of a conserved nucleotide binding domain (NBD) and a sequence-variable α-helical bundle termed the all-alpha-domain (AAD) (Alvarez-Martinez and Christie 2009; Gomis-Ruth et al. 2001). The AAD is located at the cytoplasmic pole of the VirD4 hexamer in an optimal position for recruitment of substrates from the cytoplasm (Gomis-Ruth et al. 2001). A combination of in vivo mutational studies evaluating effects of AAD point or deletion mutations or domain swaps and in vitro binding studies with purified AADs firmly support a role for the AAD in the engagement of cognate substrates (de Paz et al. 2010; Schroder et al. 2002; Whitaker et al. 2015, 2016).
Many VirD4 subunits also possess sequence-variable C-terminal domains (CTDs) that are typically enriched in acidic residues (Alvarez-Martinez and Christie 2009; Kwak et al. 2017). When present, these CTDs also play important roles in substrate recruitment, as shown by studies of the F and pKM101 conjugation systems (see Figs. 1 and 2). In the F transfer system, the TraD T4CP possesses a CTD that strongly enhances the efficiency of F plasmid transfer through the F-encoded T4SS (Sastre et al. 1998). This is achieved through formation of a specific contact between an acidic motif at the extreme C terminus of TraD with TraM, an accessory component of the relaxosome required for nicking at the F plasmid’s origin of transfer (oriT) (Beranek et al. 2004; Lu and Frost 2005; Lu et al. 2008). F-encoded T4SS also is capable of mobilizing the transfer of the non-self-transmissible plasmid RSF1010; however, TraDF ’s CTD inhibits RSF1010 transfer. These findings show that TraD’s CTD functions as a specificity checkpoint by ensuring efficient F plasmid transfer while blocking transfer of the parasitic RSF1010 plasmid. The E. coli pKM101-encoded T4SS has long been considered a conjugation system dedicated to the transfer of the pKM101 transfer intermediate. However, recently it was shown that the pKM101 T4SS could be reconfigured to translocate heterologous effector proteins to E. coli recipients. This was achieved by swapping the receptor domain of the TraJ T4CP with receptor domains of VirD4 homologs associated with effector translocator systems functioning in alpha-proteobacterial species, including A. tumefaciens, Anaplasma phagocytophilum, and Wolbachia pipientis (Whitaker et al. 2016). The VirD4 homologs from these alpha-proteobacterial species possess long acidic CTDs (Alvarez-Martinez and Christie 2009), and deletions of these domains from the corresponding chimeric receptors had the interesting phenotypes of enhancing transfer of certain effectors while diminishing transfer of other effectors (Whitaker et al. 2016). Based on these findings, it was proposed that the VirD4 CTD contributes not only to substrate selection but also coordinates the presentation of substrates—either in abundance or temporally—to the T4SS channel.
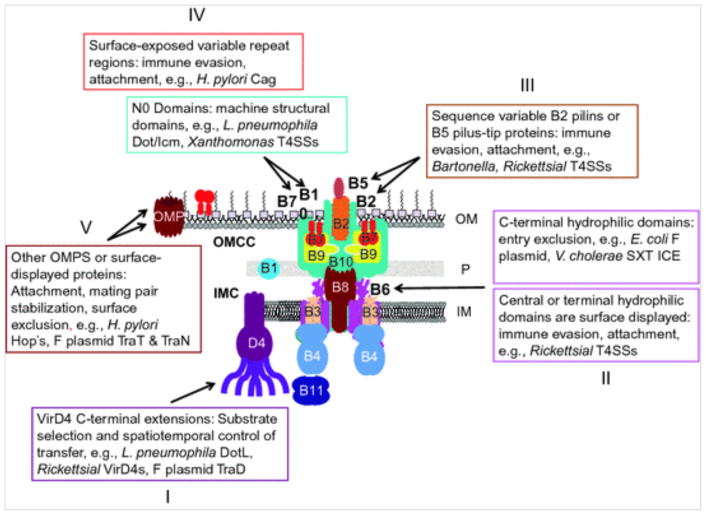
Fig. 2.
Evolutionary adaptations of T4SS subunits for machine diversification. An archetypal T4ASS type with the VirB subunits is shown. Five different types of subunit adaptations with known or postulated functions are depicted: (I) VirD4 subunits with C-terminal extensions involved in substrate recruitment, (II) “extended-VirB6” subunits with C-terminal or central hydrophilic domains that extend across the outer membrane, (III) sequence-variable VirB2 and VirB5 pilus-associated subunits, (IV) VirB7 and VirB10 subunits with variable repeat regions or N0 structural domains, (V) other outer membrane proteins (OMPs) or surface-displayed lipoproteins co-opted by T4SSs for novel functions. See text for details
Studies of VirD4-like DotL functioning in the L. pneumophila Dot/Icm system have identified several interesting structural and mechanistic features. First, DotL also has a long, C-terminal CTD for which there is also now strong experimental and structural evidence for a role in substrate recruitment. DotL was earlier shown through genetic and biochemical approaches to interact with several chaperones (here termed adaptors) that are required for translocation of effectors through the Dot/Icm channel (Buscher et al. 2005; Vincent et al. 2012; Sutherland et al. 2012) (see Sect. 2.2). Recently, an X-ray structure showed that DotL’s C-terminal domain (CTD) interacts with the stabilizing subunit DotN and three adaptors IcmS, IcmW, and LvgA sequentially along its length (Kwak et al. 2017; Xu et al. 2017). The findings underscore the importance of the CTDs of T4CPs for effector diversification and spatiotemporal control of effector presentation to the T4SS channel. Additionally, DotL interacts with and is stabilized by two IM-associated proteins DotM (IcmP) and DotN (IcmJ) (Vincent et al. 2012). Interestingly, mutations in genes for DotL, DotM, or DotN confer lethality when the mutant strains are grown in axenic media (Buscher et al. 2005). Thus, DotL assembles as a large complex with membrane-associated DotM and DotN and at least three cytosolic adaptors IcmS, IcmW, and LvA at the base of the Dot/Icm machine where it functions not only as a substrate loading platform but also to regulate channel activity (see Fig. 1).
2.2. The Role of Accessory Factors in Substrate Recruitment
Translocation of DNA and protein substrates through the T4SS often relies on association with cognate accessory factors, also termed chaperones or adaptors. These accessory factors can contribute to substrate processing as in the case of DNA substrates, or in maintenance of substrates in an unfolded, translocation-competent form as in the case of effector proteins (see Alvarez-Martinez and Christie 2009). In early studies of A. tumefaciens, the secretion chaperone VirE1 was shown to be required for translocation of the VirE2 effector through the VirB/VirD4 T4SS (Atmakuri et al. 2003). Several other T4ASSs are now known to rely on chaperones or adaptors, which typically are small, acidic cytoplasmic proteins, for recruitment of substrates (see Alvarez-Martinez and Christie 2009). Recent studies of the Dot/Icm T4BSS, however, have established the capacity of the T4BSS transporters to deploy multiple adaptors for recruitment of effectors (Fig. 1). Intriguingly, these adaptors act in pairwise fashion to promote translocation of distinct subsets of effectors. For example, IcmS and IcmW independently promote translocation of certain substrates, while coordinating with each other or with the adaptor LvgA to mediate transfer of other effectors (Zuckman et al. 1999; Coers et al. 2000; Ninio et al. 2005; Luo and Isberg 2004; Vincent and Vogel 2006; Xu et al. 2017). The evolved ability of DotL to bind different adaptors, which in different combinations recruit distinct subsets of effectors, accounts at least in part for the significant—and to date unprecedented—expansion of the Dot/Icm repertoire (see Sect. 5 and Chapter “ Subversion of Host Membrane Dynamics by the Legionella Dot/Icm Type IV Secretion System ”).
IcmR also is a small, acidic adaptor-like protein but appears to regulate the function of its partner subunit, IcmQ, by a distinct mechanism. IcmR interacts with IcmQ (Dumenil and Isberg 2001), and both IcmQ and IcmR are essential for growth of L. pneumophila in macrophages, a Dot/Icm-dependent salt-sensitivity phenotype, and evasion of lysosomes (Coers et al. 2000). Interestingly, IcmQ localizes on the surface of the bacterium shortly after infection and also inserts into lipid membranes to form pores by a mechanism regulated by IcmR (VanRheenen et al. 2004). The crystal structures of IcmR and IcmQ interacting domains confirmed that the interaction is mediated through the N-terminal part of IcmQ and the middle region of IcmR (Raychaudhury et al. 2009). Recently, the structure of full-length IcmQ in complex with IcmR was solved, revealing that the C-terminal domain of IcmQ contains an NAD+ binding module. The presence of this module suggests that the IcmR–IcmQ complex binds to membranes, where the NAD(+)-bound form of the complex might promote stabilizing interactions with, or modification of, a protein in the Dot/Icm machine (Farelli et al. 2013).
2.3. The Inner Membrane Complex (IMC)
Once VirD4 binds a substrate, it delivers the substrate to the VirB11 and VirB4 ATPase presumably for further processing prior to delivery through the T4SS (Atmakuri et al. 2004; Cascales and Christie 2004; Li et al. 2012). The VirB4 ATPases are signatures of all T4SSs characterized to date. These subunits are phylogenetically related to VirD4 subunits, and they also assemble as homohexamers. In the only high-resolution structure of a T4SS generated to date, two hexamers of VirB4 are situated side-by-side at the base of the IMC (Fig. 1) (see Chapter “ Structural and Molecular Biology of Type IV Secretion Systems ”). By contrast, homologs of VirB11 are associated with only ~20% of known T4SSs (Cabezon et al. 2014). For example, VirB11 subunits are associated with the A. tumefaciens VirB/VirD4 and L. pneumophila Dot/Icm T4SSs, but not with the E. coli F plasmid-encoded T4SSs or systems elaborated in Gram+ species (Berger and Christie 1994; Lawley et al. 2003; Sexton et al. 2004b; Bhatty et al. 2013). VirB11 ATPases are members of the AAA+ ATPase superfamily, but partition predominantly with the cytosolic fraction and in contrast to VirB4 might interact dynamically with the T4SS in response to substrate binding or another signal (Sexton et al. 2004b).
The ATP energy complex interacts with integral membrane components of the IMC, and the importance of these interactions is evidenced by results in the A. tumefaciens system showing that catalytic activities of the three ATPases are required for formation of formaldehyde-crosslinks between DNA substrates and two IMC subunits, VirB6 and VirB8 (Atmakuri et al. 2004). In the T4ASSs, the IMC consists minimally of the VirB-like subunits: (i) VirB3, a small, two-pass membrane protein that interacts with and stabilizes the VirB4 ATPase, (ii) VirB6, a multi-pass subunit that forms several stabilizing contacts with other IMC constituents, and (iii) VirB8 and VirB10, both typically consisting of a short N-terminal cytoplasmic domain, a TM domain, and structurally-conserved periplasmic domains (Low et al. 2014).
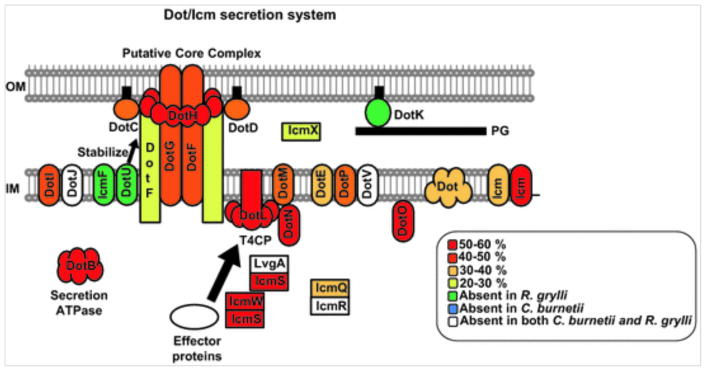
The Dot/Icm systems rely on homologs of VirD4, VirB4, and VirB11 for substrate recruitment and processing, but only two IMC subunits bear relatedness to VirB proteins. DotI is a bitopic IM protein whose periplasmic region has a VirB8-like structural fold despite weak primary sequence relatedness to VirB8 subunits. DotI interacts with DotJ, which appears to be a truncated form of DotI lacking the VirB8-fold (Nagai and Kubori 2011). DotA is a polytopic membrane-spanning protein reminiscent of the VirB6 signature subunits of the T4ASS transporters and is required for intracellular growth in macrophages and in ameba (Roy and Isberg 1997; Berger et al. 1994). Very intriguingly, however, DotA also is secreted by the Dot/Icm T4SS (Nagai and Roy 2001). The secreted form of DotA is truncated due to a proteolytic processing event and assembles as a hollow ring. These findings led to a proposal that DotA forms a channel in target cell membranes as a prerequisite for delivery of Dot/Icm substrates into the eukaryotic cell host (Nagai and Roy 2001). Phylogenetic analysis of different dotA segments from clinical and environmental strains showed that recombination and frequent non-synonymous mutations have played an important role in dotA evolution (Costa et al. 2010; Ko et al. 2003). Rapid evolution of DotA thus may have contributed to the enhancement of bacterial fitness in certain environmental niches. The Dot/Icm IMC consists of other IM-associated proteins including IcmF and DotU(IcmH). IcmF and DotU interact, and icmF and dotU mutations affect the stability of the DotF, DotG, and DotH OMCC subunits, suggesting that the IcmF/DotU complex stabilizes the Dot/Icm T4SS (Segal et al. 1998; Sexton et al. 2004a). Interestingly, the icmF/dotU gene pair is present in a wide variety of Gram− bacteria, not in association with T4SS loci but rather with loci encoding type VI secretion systems (T6SSs) (Cascales 2008; Bingle et al. 2008). Other small IM-spanning Dot/Icm proteins including IcmT, IcmV, IcmC, DotV, and DotP(IcmD) contribute to Dot/Icm function and might be part of the IMC (see Fig. 4).
Fig. 4.
Conservation of the type IVB Dot/Icm secretion system among Legionella pneumophila, Coxiella burnetii, and Rickettsiella grylli. Strains Paris, RSA493, and NZ_AAQJ, respectively are taken as representatives for their species. The Dot/Icm secretion complex proteins have been colored according to the percentage of amino acid identity among the three corresponding orthologous for each Dot/Icm component (red for the most conserved ones to clear yellow for the less conserved ones). Green and blue, proteins absent in R. grylli and C. burnetii, respectively; white, proteins of L. pneumophila absent in both other bacteria (modified from Nagai and Kubori 2011)
Perhaps one of the most intriguing mysteries surrounding type IV secretion is the route(s) by which substrates are conveyed across the cytoplasmic membrane (Grohmann et al. 2018). Based on available structure—function information summarized above for the T4ASS and T4BSS transporters, three possible translocation routes can be envisioned: (1) substrates pass directly through the lumen of the VirD4 hexamer, (2) substrates are transferred to the VirB4 hexamer for delivery through its central channel, or (3) after engaging with the ATPases for processing/unfolding, substrates pass through a channel composed of IMC subunits, e.g., VirB6 and VirB8 (Fig. 1). Whether the translocation route(s) is conserved among all T4ASSs and T4BSSs, and whether different substrates, e.g., DNA versus protein, are routed through the same pathway(s) remain fascinating questions for future studies.
2.4. Evolutionary Adaptations of IMC Subunits
Many T4SSs have evolved adapted forms of IMC components for novel functions. VirB6 in particular offers a remarkable example of an IMC subunit that has been extensively modified throughout evolution. The signature feature of VirB6 subunits is their five to seven membrane-spanning configurations. However, many larger variants termed “extended VirB6” subunits carry one or more large hydrophilic domains. Such variants are widely distributed among conjugation and effector translocator systems and in members of both the T4ASS and T4BSS groups (Alvarez-Martinez and Christie 2009). These domains appear to play important roles in specifying donor cell interactions with other bacteria or eukaryotic host cells (Fig. 2). In the F-type systems, for example, TraG subunits carry an N-terminal polytopic motif and a large ~600 residue C-terminal domain (Arutyunov and Frost 2013; Lawley et al. 2003). TraGF is involved in entry exclusion, a process that blocks redundant DNA transfer between identical donor cells. In such donor–donor contacts, TraGF ’s C-terminal domain of one donor cell establishes contact with TraSF, an inner membrane protein present in the paired donor cell (Audette et al. 2007). This contact is achieved either by extension of TraGF across the outer membranes of both donor cells, or by proteolytic cleavage of the C-terminal domain of TraGF followed by active translocation into the paired donor cell. The TraGF –TraSF interaction signals a nonproductive donor–donor cell junction and blocks DNA transfer (Anthony et al. 1999). Similar findings were reported for homologs of TraG and TraS encoded by the SXT ICE (integrative and conjugative element) of Vibrio cholerae (Marrero and Waldor 2007).
In Rickettsia spp., the T4SSs encode multiple copies of “extended-VirB6” subunits with sizes ranging from 600 to over 1400 residues (Gillespie et al. 2009, 2010). The large hydrophilic domains are positioned centrally or at one or both terminal regions, they vary considerably in sequence composition, and many contain multiple repeat regions. Interestingly, VirB6 domains were identified on the surfaces of Wolbachia, Ehrlichia, and Rickettsia cells, which supports the notion that these domains are somehow conveyed to the cell surface where they contribute to establishment of endosymbiotic or pathogenic relationships (Rances et al. 2008).
The T4BSS transporters also encode “extended-VirB6” subunits. E. coli plasmid R64 encodes TraY, a 745-residue protein with an unusual hydropathy profile (Sampei et al. 2010). The N- and C-terminal thirds of the protein each possess between four and six predicted TM motifs, whereas the central third is hydrophilic and predicted to reside in the periplasm. In the L. pneumophila Dot/Icm system, VirB6-like DotA is ~300 residues larger than TraY and possesses the same general hydropathy profile with multiple N- and C-terminal TM domains flanking a central hydrophilic domain. As mentioned above, however, DotA localizes both in the inner membrane and is secreted to the milieu in a Dot/Icm T4SS-dependent manner where it forms ring-like oligomers (Nagai and Roy 2001). How DotA is partitioned to these different locations where it contributes to effector translocation across the cytoplasmic membrane or into target cells remains unknown.
2.5. The Outer Membrane Core Complex (OMCC)
The IMCs are connected to OMCCs in ways that are not well structurally defined to mediate substrate passage across the periplasm and OM. Among well-characterized T4ASSs, the OMCCs are composed of homologs or orthologs of the lipoprotein VirB7, outer membrane-associated VirB9, and the C-terminal half of VirB10. The OMCC is intrinsically stable and stabilizing for most of the other VirB subunits, and structures of several OMCC’s from E. coli plasmids (R388, pKM101) and the A. tumefaciens VirB/VirD4 T4SSs have now been solved by transmission- or cryo-electron microscopy (Fronzes et al. 2009; Low et al. 2014; Gordon et al. 2017). These are structurally-conserved, large ~1 MDa barrel-shaped structures composed of 14 copies of each of the 3 VirB-like subunits. They are envisioned to form a structural scaffold for the translocation channel, although the architecture and composition of the channel remain undefined (Low et al. 2014) (see Chapter “ Structural and Molecular Biology of Type IV Secretion Systems ”). Very recently, a low-resolution structure was obtained for the OMCC of a T4ASS distantly related to the A. tumefaciens VirB/VirD4 system, namely the Helicobacter pylori Cag T4SS (Frick-Cheng et al. 2016). Interestingly, the OMCC is considerably larger (~41 nm as opposed to ~18 nm for the VirB/VirD4-like T4SSs) and composed of at least five subunits (VirB7-like CagT, VirB9-like CagX, VirB10-like CagY, Cag3, CagM). Nevertheless, the Cag OMCC adopts a ring-shaped architecture that generally resembles OMCCs of the VirB/VirD4-like T4ASSs (see also Chapter “ The Helicobacter pylori Type IV Secretion System Encoded by the cag Pathogenicity Island: Architecture, Function and Signaling ”).
In the L. pneumophila Dot/Icm, the OMCC also is composed of five subunits, DotG(IcmE), DotF(IcmG), DotC, DotD, and DotH (Figs. 3 and 4). DotG is required for Dot/Icm function. It is configured as an IM-spanning subunit that extends into the periplasm where it constitutes part of the OMCC. DotG subunits are large (~1000 to 1500-kDa), sequence-variable subunits of which only the extreme C termini bears sequence similarities to the VirB10 subunits of the T4ASSs. DotF similarly spans the IM and has a large periplasmic domain, but it is important for machine function in the ameba Acanthamoeba castellanii and not in human macrophages (Purcell and Shuman 1998; Segal and Shuman 1999a; Luo and Isberg 2004; Sutherland et al. 2013). DotC and DotD are outer membrane lipoproteins required for machine assembly (Yerushalmi et al. 2005). DotD possesses a disordered N-terminal domain and a globular C-terminal domain with an N0 structural fold that might connect the OMCC with the IMC or play a more dynamic role in regulating substrate passage (Nagai and Kubori 2011; Nakano et al. 2010; Souza et al. 2011). DotH also is a critical OMCC subunit and is dependent on lipoproteins DotC and DotD for delivery to the OM protein (Andrews et al. 1998; Nakano et al. 2010). Although DotH has features similar to VirB9 (Watarai et al. 2001; Nakano et al. 2010), it also was reported to comprise part of a fibrous structure that covers the entire bacterial surface that enhances internalization of bacteria (Watarai et al. 2001). Finally, two subunits, DotK(IcmN) and IcmX, are localized in the OM or periplasm, but are not predicted to form part of the OMCC. DotK is a predicted lipoprotein that carries an OmpA peptidoglycan-binding domain, but is not required for Dot/Icm function (Segal et al. 1998; Segal and Shuman 1999a; Yerushalmi et al. 2005). IcmX is a 50-kDa, periplasmic protein shown to be required for pore formation in the membrane of the eukaryotic cell and might participate in regulating the trafficking of the Legionella containing vacuole (Edelstein et al. 1999; Matthews and Roy 2000; Sadosky et al. 1993).
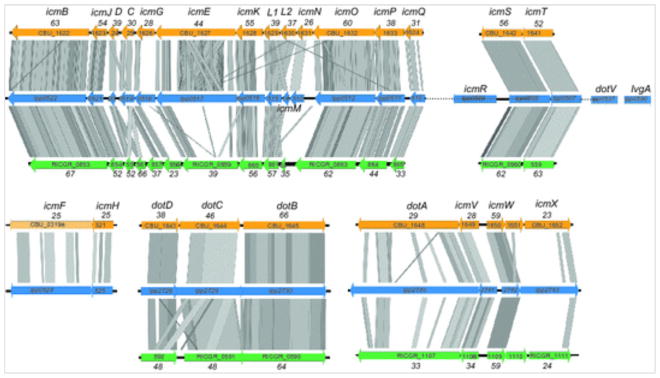
Fig. 3.
Syntenic regions of the Dot/Icm system encoding genes among three bacterial species. L. pneumophila strain Paris, C. burnetii strain RSA493, and R. grylli strain NZ_AAQJ are represented. The genomic organization and Blastx comparisons of the regions encoding for the T4BSS machinery in these three species are depicted. The gray color code represents the blast matches; the darker the gray the better the blast match
The Dot/Icm OMCC also adopts a ring-shaped structure, whose assembly requires DotC, DotD, DotH, and DotG, but not DotF (Kubori et al. 2014; Vincent et al. 2006). These findings were recently extended with presentation of the first in-situ structure of a T4SS, solved by cryo-electron tomography (Ghosal et al. 2017). These studies determined that the Dot/Icm T4SS assembles as a large cone-shaped structure predominantly at L. pneumophila cell poles. Indeed, secretion of effectors from the polar-localized Dot/Icm machine recently was shown to be essential for virulence (Jeong et al. 2017). The polar-localized OMCC presents as several densities that were envisioned as a “WiFi” structure that extends from the OM across the periplasm to the IM where it presumably interacts with an IMC subassembly whose structure has not yet been defined (Fig. 1).
2.6. Evolutionary Adaptations of OMCC Subunits
Like the IMC-associated VirB6 subunits, the OMCC components have undergone diversification during evolution through acquisition of novel domains or motifs. This is mainly observed with the VirB7-like lipoproteins and the VirB10 scaffold proteins. In A. tumefaciens, VirB7 is a small ~4.5 kDa lipoprotein tethered to the inner leaflet of the outer membrane and required for stabilization of other OMCC subunits (Fernandez et al. 1996). However, in many systems, the VirB7-like lipoproteins have acquired surface-variable regions as shown for H. pylori CagT (Terradot and Waksman 2011; Backert et al. 2015), or N0 domains as shown for Xanthomonas citri VirB7 and L. pneumophila DotD (Nakano et al. 2010; Souza et al. 2011). Surface-variable CagT is required for CagA translocation and pilus biogenesis (Ding et al. 2012; Johnson et al. 2014) and might also contribute to immune evasion by H. pylori (Fischer 2011). The N0 domains of VirB7Xac and DotDLp are envisioned to form additional rings within or at the base of the OMC of possible importance for channel gating or communication between the IMC and the OMCC (Nakano et al. 2010; Souza et al. 2011) (Fig. 2).
The VirB10-like subunits are among the most sequence- and structurally-variable subunits of the T4SSs (Fig. 2). Only a small C-terminal region of H. pylori CagY is similar to VirB10 and a large central region is composed of multiple repeats (Aras et al. 2003). This central region is surface-displayed and associates with a pilus structure (Barrozo et al. 2013; Rohde et al. 2003), and during infection this region undergoes extensive rearrangements that disrupt or activate the Cag T4SS (Aras et al. 2003; Barrozo et al. 2013). Through host immune-driven recombination, CagY is postulated to function as a sensor of the host immune response and, in turn, regulate Cag T4SS function to maximize persistent infection (Barrozo et al. 2013). In E. coli, plasmid R64 encodes a T4BSS and TraO closely resembles VirB10 in size and predicted overall structure (Sampei et al. 2010). As mentioned above, however, in the closely related L. pneumophila Dot/Icm system, DotG is over 1000 residues and only the C-terminal region resembles VirB10 (Segal et al. 1998; Vogel et al. 1998). Like CagY, DotG possesses central variable repeats consisting in part of multiple sets of pentapeptide repeats between the N-terminal transmembrane domain and the C-terminal conserved region (Segal et al. 1998). Furthermore, N-proximal regions of DotG subunits from different L. pneumophila species are highly variable. At this time, however, there is no evidence that the sequence-variable regions of DotG are surface-displayed.
3. T4SS-Mediated Modulation of Target Cell Attachment
T4SSs elaborate conjugative pili or other types of surface adhesins to establish contacts with potential recipients (Bhatty et al. 2013). T4SSs also have evolved other functions to block nonproductive or deleterious cell–cell contacts or to evade immune surveillance. These modulatory functions appear to have arisen by three mechanisms (i) expression of variant forms of pilin subunits that may or may not assemble as pili, (ii) acquisition of surface-exposed domains by signature IMC or OMCC subunits, and (iii) linkage of genes specifying surface-associated functions with T4SS loci (Fig. 2). These adaptations either enhance productive or inhibit nonproductive cell–cell contacts.
3.1. T4ASS P- and F-pili
Gram− bacterial conjugation machines elaborate conjugative pili to initiate contacts with target cells to facilitate formation of direct cell–cell contacts termed mating junctions. There are two well-characterized groups of conjugative pili, P-pili produced by various E. coli plasmids and the A. tumefaciens VirB/VirD4 T4SS and F-pili elaborated by the E. coli F plasmid (Lawley et al. 2003; Schroder and Lanka 2005). The P-type pili are thick (90–110 Å), rigid, and short although length measurements are complicated by the fact that isolated pili are typically broken (Bradley 1980; Bradley et al. 1980; Paranchych and Frost 1988). These pili do not appear to undergo cycles of extension/retraction, but instead accumulate in the milieu, either through breakage or an active sloughing mechanism. Donors elaborating these pili typically mate efficiently only on solid surfaces. By contrast, F-type pili are typically ~90 Å in width and flexible, and range in length up to 1 micron (see Chapter “ Structural and Molecular Biology of Type IV Secretion Systems ”). These pili dynamically extend and retract, enabling donor cells to bind and draw recipient cells into physical contact for establishment of the mating junction. Although genetic requirements for production of P-pili are nearly the same as for elaboration of the mating channel, assembly of F-pili additionally requires several F-specific proteins (TraF, -H, -U, -W, and TrbI) that are required for pilus extension and retraction (Arutyunov and Frost 2013; Clarke et al. 2008).
Interestingly, F-type systems also elaborate other surface-exposed proteins or domains to promote or block F plasmid transfer. For example, once the F-pilus retracts, bringing donor and recipient cells into juxtaposition, F-systems also encode OM-associated TraN, which binds OmpA and possibly LPS on the recipient cell surface to stabilize the mating junction (Fig. 2) (Klimke et al. 2005). Additionally, as noted above, the C-terminal region of VirB6-like TraG blocks redundant DNA transfer in donor–donor contacts through interactions with TraS (Audette et al. 2007). In matings with F-minus recipients, however, TraG’s C-terminal domain coordinates with TraN to stabilize the mating junction (Audette et al. 2007; Firth and Skurray 1992). Finally, the F-type and other conjugation systems employ surface or entry exclusion systems to block redundant DNA transfer among populations of donor cells (Fig. 2) (Garcillan-Barcia and de la Cruz 2008; Lawley et al. 2003). Besides the TraG/TraS entry exclusion system, F-systems encode a lipoprotein, TraT, that is, exported to the E. coli cell surface. TraT forms higher-order oligomers and appears to block initiating or stabilizing contacts of donor cells with each other, possibly by impeding the binding of the F-pilus or of TraN to OmpA in donor-donor cell contacts.
3.2. Evolutionary Adaptations of T4ASS-Associated Surface Structures
Surprisingly, at this time only two systems functioning in the delivery of effectors to eukaryotic cells have been shown to produce pili, the A. tumefaciens VirB/VirD4 system and the H. pylori Cag system (Aly and Baron 2007; Fullner et al. 1996; Kwok et al. 2007; Johnson et al. 2014; Tegtmeyer et al. 2017). Assembly of pili by the latter system is more complex than the former in its requirement for VirB-like subunits (VirB9-like CagX, VirB7-like CagT, VirB8-like CagV) as well as several Cag-specific proteins (Cag3, CagM). By contrast, VirB-like subunits such as VirB2-like CagC and VirB10-like CagY that are required for pilus production in the A. tumefaciens and closely related systems are not required for Cag pilus production (Noto et al. 2015). Furthermore, in addition to VirB5-like CagL, other subunits including CagI, CagH, a domain of CagY and the CagA substrate itself associate with the pilus tip. The surface display of CagL, CagI, CagY, and CagA appears to be biologically relevant in view of evidence that these subunits bind integrin receptors on host epithelial cell surfaces (Backert and Tegtmeyer 2017; Conradi et al. 2012). Comparative genomic studies also have supplied evidence for the diversification of surface-localized Cag subunits, presumably under evolutionary selective pressures in the human host (see Chapter “ The Helicobacter pylori Type IV Secretion System Encoded by the cag Pathogenicity Island: Architecture, Function, and Signaling ”).
Diversification of T4SS-associated pili or pilins during the evolution of pathogen–host relationships is potentially widespread (Fig. 2). Bartonella spp. carry two general types of T4SS loci, VirB/VirD4-like systems responsible for effector translocation and Trw systems that lack associated VirD4-like receptors (Eicher and Dehio 2012). The Trw systems have the coding capacity for multiple variant forms of VirB2- and VirB5-like subunits, suggestive of a function not in substrate transfer but rather in production of variant forms of surface-exposed pilins or pili. The Trw system is essential for erythrocyte invasion, and it is postulated that the variant pili/pilins might facilitate interactions with different erythrocyte receptors, either within the reservoir host population (e.g., different blood group antigens) or among different reservoir hosts (Dehio 2008). Rickettsia spp. genomes, particularly among members of family Anaplasmataceae, also show a proliferation of many VirB2 variants (Gillespie et al. 2010). These T4SSs also might elaborate surface-variable pilins or pili to modulate attachment to different host cell types or for immune evasion.
Various effector translocators lack genes for VirB5 subunits, which are required for pilus assembly. The best-characterized example is the Bordetella pertussis Ptl system, which possesses a VirB2-like pilin but not a VirB5 homolog or a detectable pilus (Locht et al. 2011). This might be attributed to the evolution of the Ptl system for export of its PT cargo into the milieu without a requirement for host cell binding. Rickettsia spp. T4SSs also lack discernible VirB5 homologs, raising the possibility that the infection cycle of these obligate intracellular pathogens also might not require elaboration of a pilus for binding of the mammalian host membrane (Gillespie et al. 2010). In fact, it is interesting to note that substrate transfer by the well-characterized A. tumefaciens VirB/VirD4 T4SS does not require production of extended pili, as shown by the isolation of “uncoupling” mutations that block pilus assembly without affecting substrate transfer (Jakubowski et al. 2003, 2005; Sagulenko et al. 2001). These observations suggest that in certain environmental or infection niches, the production of adhesive pili might benefit the bacterial host by enhancing the efficiency of substrate transfer. In other settings, pilus production might impose a fitness cost or another disadvantage to cell viability, resulting in the evolution of T4SSs dependent on other cell surface proteins for target cell binding and mating junction formation.
3.3. T4BSS DNA Transfer and Dot/Icm Systems
T4BSS (IncI) conjugation systems have evolved to deliver their DNA substrates efficiently both on solid surface and in liquid matings (Komano et al. 2000). Efficient transfer on solid surfaces is attributed to the capacity of these systems to elaborate a thick rigid pilus, although this pilus has not been extensively investigated. Transfer in liquid, however, is mediated by an associated type IV pilus, which despite the nomenclature is phylogenetically unrelated to the T4SS-encoded pili (Yoshida et al. 1999). Interestingly, the type IV pilus is subject to sequence variation in the pilus tip protein, PilV, due to the presence of a plasmid-encoded site-specific recombination system termed the “shufflon” (Gyohda and Komano 2000; Yoshida et al. 1999). PilV promotes efficient binding of lipopolysaccharides on the surfaces of recipient bacterial cells, but the shufflon introduces variation in the C-terminal region of the pilus tip protein PilV, which in turn determines recipient specificity during liquid matings. Thus, the T4BSS conjugation systems have co-opted a type IV pilus gene cluster and an associated shufflon to ensure efficiency as well as specificity to the mating reaction.
The T4BSS Dot/Icm system evolved from an ancestral IncI conjugation system, but it did not retain the type IV pilus gene cluster and also apparently does not produce a thick rigid pilus. Rather, L. pneumophila with an intact dot/icm locus elaborate a fibrous material covering the surface of cells. This fibrous mesh was proposed to facilitate specific stages of the L. pneumophila infection cycle (Watarai et al. 2000), and might also account for the capacity of the Dot/Icm T4SS to conjugatively transfer a DNA substrate to recipient bacterial cells (Vogel and Isberg 1999).
4. Evolution of Dot/Icm Secretion System
The availability of many bacterial genome sequences has enabled detailed phylogenomics studies exploring the distribution of T4BSS transporters both within Legionella and among other species of Gammaproteobacteria (Figs. 3 and 4). The T4BSS apparatus was first identified in Legionella, but since was shown function in Coxiella burnetii (Segal and Shuman 1999b; Seshadri et al. 2003; Sexton and Vogel 2002) and Rickettsiella grylli (Leclerque and Kleespies 2008; Nagai and Kubori 2011). C. burnetii is an intracellular pathogen responsible for Q fever in humans (Larson et al. 2016), whereas bacteria of the genus Rickettsiella are obligate intracellular pathogens of a wide variety of arthropods. The genera Legionella, Coxiella, and Rickettsiella belong to the same order, Legionellales, within the group of Gammaproteobacteria. In accordance with their common ancestries, the dot/icm loci from these genera are highly similar in sequence and gene organization (Fig. 3). In line with predicted architectural and functional similarities between these systems, several C. burnetii dot/icm genes were shown to complement the corresponding dot/icm mutations in L. pneumophila (Zusman et al. 2003; Zamboni et al. 2003). The T4SSs from the three genera, do however, exhibit a few differences: (a) lvgA and icmR are absent from Coxiella and Rickettsiella, although icmR functional homologs have been found in both organisms (b) dotJ(icmM) and dotV are apparently absent from Coxiella and Rickettsiella (c) icmF and icmH are not present in Rickettsiella and icmF is fragmented in Coxiella, and (d) icmL is duplicated in Coxiella and Rickettsiella although in the latter the duplication is shorter (Fig. 4) (Segal et al. 2005).
More recently, evidence has been presented for the existence of T4BSSs in other species. For example, a Dot/Icm system was identified in the fish pathogen Piscirickettsia salmonis, and reminiscent of the L. pneumophila infection process, the phagosome-lysosome fusion event is inhibited during Piscirickettsia infection (Gomez et al. 2013). Phylogenetic studies place the Piscirickettsia as a member of the order Thiotrichales (Mauel et al. 1999). The presence of a Dot/Icm system in this order thus pushes back the origin of the T4BSSs to the common ancestor of the orders Legionellales and Thiotrichales. Genome sequence studies also have revealed the presence of T4BSS gene clusters in other proteobacteria including Marinobacter aquaeolei, Xanthomonas campestris, and Burkholderia vietnamiensis (Nagai and Kubori 2011). In these organisms, genes for core subunits of the T4SS are often present in several clusters around the genome, yet ancillary adaptors and other subunits, e.g., icmS, icmR, icmX, icmV, seem to be found only in the order Legionellales (Nagai and Kubori 2011).
The availability of genome sequences of different Legionella species allows us now to study the evolution of the Dot/Icm system in greater depth. Comparison of the different Dot/Icm encoding genes among the more than 50 available Legionella genomes shows that this secretion system is highly conserved despite the large phylogenetic distance between some of the Legionella species. Indeed, all dot/icm genes known from L. pneumophila are also present in all other Legionella species sequenced so far (Fig. 4). The only exception is icmR, although one or two non-homologous genes appear to be functional homologs in other Legionella species; these genes were termed FIR for functional homologs of IcmR (Feldman and Segal 2004; Feldman et al. 2005). Our recent analysis comprising 80 Legionella strains belonging to 58 different Legionella species confirms this observation and further suggests that FIR proteins are extremely fast-evolving (Gomez et al., submitted). Interestingly, the order and orientation of the genes encoding the Dot/Icm are completely conserved among the different species comprising the genus Legionella (Burstein et al. 2016). The only differences are a few insertions between some of the dot/icm genes in some species that apparently are not related to the T4BSS group. These insertions are conserved between phylogenetically closely related species, suggesting that the subregions encoding Dot/Icm components are tightly regulated (Burstein et al. 2016).
5. Comparisons of Effectors Secreted Through Different Dot/Icm Secretion Systems
In 2002, Nagai and collaborators demonstrated that the protein RalF is secreted by the Dot/Icm secretion system (Nagai et al. 2002). Since these early studies, a panoply of experimental and bioinformatics techniques has uncovered in excess of 300 Dot/Icm-translocated substrates (Finsel and Hilbi 2015; Hubber and Roy 2010). These effectors, which represent about 10% of the L. pneumophila genome, are unprecedented in their total number in just one strain. Only a subset of these effectors have been characterized, and unfortunately in many cases mutations of candidate effectors have not yielded discernible phenotypes possibly as a result of functional redundancy (Luo and Isberg 2004; O’Connor et al. 2012; Finsel and Hilbi 2015). The evolution of multiple mechanisms to subvert the eukaryotic host likely can be attributed to the molecular arms race that evolved between individual strains of L. pneumophila and the broad spectrum of protozoan hosts encountered in its natural environment.
Dot/Icm substrates have been identified through distinct signatures. Most notably, they often carry conserved eukaryotic protein domains, e.g., serine–threonine kinases, ubiquitin ligases, Sel-1, Sec7, U-box, F-box, ankyrin repeats (Cazalet et al. 2004; de Felipe et al. 2005; Cazalet et al. 2010; Gomez-Valero et al. 2011). These domains were likely acquired by horizontal gene transfer and their presence reflects the long-standing co-evolution of Legionella spp. with their protozoan hosts (de Felipe et al. 2005; Gomez-Valero et al. 2011; Gomez-Valero and Buchrieser 2013; Gomez-Valero et al. 2014; Lurie-Weinberger et al. 2010). Searches for homologs of the many L. pneumophila Dot/Icm substrates in other Legionella species identified only seven that are common among 40 species analyzed (Burstein et al. 2016; Gomez-Valero, submitted). However, when applying in-silico approaches, many putative effectors are identified in the newly sequenced Legionella genomes. This suggests that Legionella spp., typically have large substrate repertoires, although the effector set can be quite distinct from species to species (Gomez-Valero, submitted). The functions of effectors that have been revealed up to now target many cellular pathways and different eukaryotic organelles, including cell uptake and exit, endocytosis, vesicle trafficking, autophagy, mitochondria, cytoskeleton, ubiquitination/proteasome, ribosome, transcription factors, and the nucleus (Escoll et al. 2016; Finsel and Hilbi 2015; Qiu and Luo 2017; Sherwood and Roy 2016). (see Chapter “ Subversion of Host Membrane Dynamics by the Legionella Dot/Icm Type IV Secretion System ”).
The effectors translocated through the C. burnetii Dot/Icm system have not yet been as extensively studied, due to earlier limitations in genetic manipulations and axenic growth of this bacterium (Beare et al. 2009). Currently, 133 protein substrates have been identified in C. burnetii representing about 6% of the open reading frames of its genome (Chen et al. 2010; Carey et al. 2011; Qiu and Luo 2017). These are involved in the subversion of vesicle trafficking, lipid metabolism of the Coxiella containing vacuole, host gene expression, autophagy, cell death, and immunity (Moffatt et al. 2015; Qiu and Luo 2017). Strikingly, only six of these effectors have homologs in L. pneumophila (Qiu and Luo 2017). Additionally, in contrast to the redundancy we find among Legionella effectors, most C. burnetii effector mutants fail to grow inside host cells (Moffatt et al. 2015; Weber et al. 2013). The apparent lack of redundancy among effectors in this species might be due to the comparatively narrow host range of C. burnetii compared with L. pneumophila (Qiu and Luo 2017) (see Chapter “ Subversion of Host Membrane Dynamics by the Legionella Dot/Icm Type IV Secretion System ”).
Recently, studies have begun to identify Dot/Icm effectors in Rickettsiella and Piscirickettsia. Using bioinformatics approaches, 18 putative Dot/Icm substrates were predicted for R. grylli, of which six were homologs of effectors in L. pneumophila strain Philadelphia (Lifshitz et al. 2013). In Piscirickettsia, four effectors have been identified through genetic screening (Labra et al. 2016) that exhibit clear matches with effectors in either L. pneumophila or C. burnetii. These effectors all have eukaryotic-like protein domains, revealing a possible common function upon translocation to the eukaryotic host during the establishment of pathogen–host relationships.
6. Concluding Remarks
Structural, functional, and phylogenetic studies continue to shape our understanding of the fascinating and complex T4SSs. The structural advances are accelerating and promise in the near future to generate structures of several paradigmatic systems at or near atomic resolution. Continued work in this area also should allow for more detailed comparisons of systems from different ancestries, e.g., types T4ASS and T4BSS. In conjunction, in vivo functional tests and mutational studies are critically important lines of study for assigning biological relevance of structures and for defining dynamic processes relating to machine biogenesis, mating junction formation, and substrate transfer. In the future, the implementation of high-resolution imaging techniques, including single-cell analyses, will provide a detailed understanding of early steps in machine biogenesis and substrate recruitment and trafficking. These and other approaches also need to address the challenging questions of how the T4SSs are activated by extracellular, e.g., target cell contact, or intracellular, e.g., substrate docking, signals. Finally, recent studies have shed light on the fact that T4SSs can function as mediators of antagonistic as well as cooperative interbacterial interactions. Further work along this line will generate a broader picture of the role of type IV secretion in the shaping and maintenance of polymicrobial communities in different environmental and host niches.
The work of Guglielmini and colleagues (Guglielmini et al. 2013) is a compelling example of how the increasing number of available genome sequences provides a wider view of the studied paradigmatic systems. These types of phylogenomics studies will lead to further refinements of the T4SS classification schemes and contribute to a better understanding of the evolution of these systems.
To fully decipher how a conserved machinery like the T4SS is able to adapt to so many different bacteria, hosts, and sets of effectors will remain a fascinating challenge for many years to come. Importantly, the information we have and will continue to acquire remains an invaluable resource for the translational goals of inhibiting T4SS machine functions or killing bacterial hosts that deploy these machines in clinical settings. Indeed, several important advances along these lines have been recently reported (see Chapters “ Mechanisms of Conjugative Transfer and Type IV Secretion-Mediated Effector Transport in Gram-Positive Bacteria ” and “ Coupling Proteins in Type IV Secretion ”).
Acknowledgments
Work in the Christie laboratory was supported by NIH grants R01GM48476 and R21AI105454. Work in the CB laboratory is financed by the Institut Pasteur, the grants N°ANR-10-LABX-62-IBEID, the Fondation pour la Recherche Médicale (FRM) grant N° DEQ 20120323697, and the Infect-ERA project EUGENPATH (ANR-13-IFEC-0003-02).
References
- Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aly KA, Baron C. The VirB5 protein localizes to the T-pilus tips in Agrobacterium tumefaciens. Microbiology. 2007;153:3766–3775. doi: 10.1099/mic.0.2007/010462-0. [DOI] [PubMed] [Google Scholar]
- Andrews HL, Vogel JP, Isberg RR. Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway. Infect Immun. 1998;66:950–958. doi: 10.1128/iai.66.3.950-958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony KG, Klimke WA, Manchak J, Frost LA. Comparison of proteins involved in pilus synthesis and mating pair stabilization from the related plasmids F and R100-1: insights into the mechanism of conjugation. J Bacteriol. 1999;181:5149–5159. doi: 10.1128/jb.181.17.5149-5159.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aras RA, Fischer W, Perez-Perez GI, Crosatti M, Ando T, Haas R, Blaser MJ. Plasticity of repetitive DNA sequences within a bacterial (Type IV) secretion system component. J Exp Med. 2003;198:1349–1360. doi: 10.1084/jem.20030381. https://doi.org/10.1084/jem.20030381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arutyunov D, Frost LS. F conjugation: back to the beginning. Plasmid. 2013;70:18–32. doi: 10.1016/j.plasmid.2013.03.010. https://doi.org/10.1016/j.plasmid.2013.03.010. [DOI] [PubMed] [Google Scholar]
- Asrat S, Davis KM, Isberg RR. Modulation of the host innate immune and inflammatory response by translocated bacterial proteins. Cell Microbiol. 2015;17:785–795. doi: 10.1111/cmi.12445. https://doi.org/10.1111/cmi.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmakuri K, Ding Z, Christie PJ. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol Microbiol. 2003;49:1699–1713. doi: 10.1046/j.1365-2958.2003.03669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmakuri K, Cascales E, Christie PJ. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol. 2004;54:1199–1211. doi: 10.1111/j.1365-2958.2004.04345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audette GF, Manchak J, Beatty P, Klimke WA, Frost LS. Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology. 2007;153:442–451. doi: 10.1099/mic.0.2006/001917-0. [DOI] [PubMed] [Google Scholar]
- Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol. 2006;9(2):207–217. doi: 10.1016/j.mib.2006.02.008. https://doi.org/10.1016/j.mib.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Backert S, Tegtmeyer N. Type IV secretion and signal transduction of Helicobacter pylori CagA through interactions with host cell receptors. Toxins (Basel) 2017:9. doi: 10.3390/toxins9040115. https://doi.org/10.3390/toxins9040115. [DOI] [PMC free article] [PubMed]
- Backert S, Tegtmeyer N, Fischer W. Composition, structure and function of the Helicobacter pylori cag pathogenicity island encoded type IV secretion system. Future Microbiol. 2015;10(6):955–965. doi: 10.2217/fmb.15.32. https://doi.org/10.2217/fmb.15.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrozo RM, et al. Functional plasticity in the type IV secretion system of Helicobacter pylori. PLoS Pathog. 2013;9:e1003189. doi: 10.1371/journal.ppat.1003189. https://doi.org/10.1371/journal.ppat.1003189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare PA, Howe D, Cockrell DC, Omsland A, Hansen B, Heinzen RA. Characterization of a Coxiella burnetii ftsZ mutant generated by Himar1 transposon mutagenesis. J Bacteriol. 2009;191:1369–1381. doi: 10.1128/JB.01580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beranek A, Zettl M, Lorenzoni K, Schauer A, Manhart M, Koraimann G. Thirty-eight C-terminal amino acids of the coupling protein TraD of the F-like conjugative resistance plasmid R1 are required and sufficient to confer binding to the substrate selector protein TraM. J Bacteriol. 2004;186:6999–7006. doi: 10.1128/JB.186.20.6999-7006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger BR, Christie PJ. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- Berger KH, Merriam JJ, Isberg RR. Altered intracellular targeting properties associated with mutations in the Legionella pneumophila dotA gene. Mol Microbiol. 1994;14:809–822. doi: 10.1111/j.1365-2958.1994.tb01317.x. [DOI] [PubMed] [Google Scholar]
- Bhatty M, Laverde Gomez JA, Christie PJ. The expanding bacterial type IV secretion lexicon. Res Microbiol. 2013;164:620–639. doi: 10.1016/j.resmic.2013.03.012. https://doi.org/10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: a beginner’s guide. Curr Opin Microbiol. 2008;11:3–8. doi: 10.1016/j.mib.2008.01.006. https://doi.org/10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Bradley DE. Morphological and serological relationships of conjugative pili. Plasmid. 1980;4:155–169. doi: 10.1016/0147-619x(80)90005-0. [DOI] [PubMed] [Google Scholar]
- Bradley DE, Taylor DE, Cohen DR. Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J Bacteriol. 1980;143:1466–1470. doi: 10.1128/jb.143.3.1466-1470.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand BC, Sadosky AB, Shuman HA. The Legionella pneumophila icm locus: a set of genes required for intracellular multiplication in human macrophages. Mol Microbiol. 1994;14:797–808. doi: 10.1111/j.1365-2958.1994.tb01316.x. [DOI] [PubMed] [Google Scholar]
- Burstein D, et al. Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat Genet. 2016;48:167–175. doi: 10.1038/ng.3481. https://doi.org/10.1038/ng.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscher BA, Conover GM, Miller JL, Vogel SA, Meyers SN, Isberg RR, Vogel JP. The DotL protein, a member of the TraG-coupling protein family, is essential for viability of Legionella pneumophila strain Lp02. J Bacteriol. 2005;187:2927–2938. doi: 10.1128/JB.187.9.2927-2938.2005. https://doi.org/10.1128/JB.187.9.2927-2938.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezon E, Sastre JI, de la Cruz F. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol Gen Genet. 1997;254:400–406. doi: 10.1007/s004380050432. [DOI] [PubMed] [Google Scholar]
- Cabezon E, Ripoll-Rozada J, Pena A, de la Cruz F, Arechaga I. Towards an integrated model of bacterial conjugation. FEMS Microbiol Rev. 2014;39:81–95. doi: 10.1111/1574-6976.12085. https://doi.org/10.1111/1574-6976.12085. [DOI] [PubMed] [Google Scholar]
- Carey KL, Newton HJ, Luhrmann A, Roy CR. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Path. 2011;7:e1002056. doi: 10.1371/journal.ppat.1002056. https://doi.org/10.1371/journal.ppat.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E. The type VI secretion toolkit. EMBO Rep. 2008;9:735–741. doi: 10.1038/embor.2008.131. https://doi.org/10.1038/embor.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 2003;1:137–150. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalet C, Gomez-Valero L, Rusniok C, Lomma M, Dervins-Ravault D, Newton HJ, Sansom FM, Jarraud S, Zidane N, Ma L, Bouchier C, Etienne J, Hartland EL, Buchrieser C. Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires’ disease. PLoS Genet. 2010;6(2):e1000851. doi: 10.1371/journal.pgen.1000851. https://doi.org/10.1371/journal.pgen.1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalet C, et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36:1165–1173. doi: 10.1038/ng1447. https://doi.org/10.1038/ng1447 (doi:ng1447 [pii]) [DOI] [PubMed] [Google Scholar]
- Chandran Darbari V, Waksman G. Structural biology of bacterial type IV secretion systems. Annu Rev Biochem. 2015;84:603–629. doi: 10.1146/annurev-biochem-062911-102821. https://doi.org/10.1146/annurev-biochem-062911-102821. [DOI] [PubMed] [Google Scholar]
- Chen C, et al. Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc Natl Acad Sci U S A. 2010;107:21755–21760. doi: 10.1073/pnas.1010485107. https://doi.org/10.1073/pnas.1010485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Vogel JP. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 2000;8:354–360. doi: 10.1016/s0966-842x(00)01792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M, Maddera L, Harris RL, Silverman PM. F-pili dynamics by live-cell imaging. Proc Natl Acad Sci U S A. 2008;105:17978–17981. doi: 10.1073/pnas.0806786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coers J, Kagan JC, Matthews M, Nagai H, Zuckman DM, Roy CR. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol Microbiol. 2000;38:719–736. doi: 10.1046/j.1365-2958.2000.02176.x. [DOI] [PubMed] [Google Scholar]
- Conradi J, Huber S, Gaus K, Mertink F, Royo GS, Strijowski U, Backert S, Sewald N. Cyclic RGD peptides interfere with binding of the Helicobacter pylori protein CagL to integrins alphaVbeta3 and alpha5beta1. Amino Acids. 2012;43(1):219–232. doi: 10.1007/s00726-011-1066-0. https://doi.org/10.1007/s00726-011-1066-0. [DOI] [PubMed] [Google Scholar]
- Costa J, Tiago I, Da Costa MS, Verissimo A. Molecular evolution of Legionella pneumophila dotA gene, the contribution of natural environmental strains. Environ Microbiol. 2010;12:2711–2729. doi: 10.1111/j.1462-2920.2010.02240.x. https://doi.org/10.1111/j.1462-2920.2010.02240.x. [DOI] [PubMed] [Google Scholar]
- de Felipe KS, Pampou S, Jovanovic OS, Pericone CD, Ye SF, Kalachikov S, Shuman HA. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol. 2005;187:7716–7726. doi: 10.1128/JB.187.22.7716-7726.2005. https://doi.org/10.1128/jb.187.22.7716-7726.2005 (doi:187/22/7716 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paz HD, Larrea D, Zunzunegui S, Dehio C, de la Cruz F, Llosa M. Functional dissection of the conjugative coupling protein TrwB. J Bacteriol. 2010;192:2655–2669. doi: 10.1128/JB.01692-09. https://doi.org/10.1128/JB.01692-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehio C. Infection-associated type IV secretion systems of Bartonella and their diverse roles in host cell interaction. Cell Microbiol. 2008;10:1591–1598. doi: 10.1111/j.1462-5822.2008.01171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, et al. Helicobacter pylori chaperone-like protein CagT plays an essential role in the translocation of CagA into host cells. J Microbiol Biotechnol. 2012;22:1343–1349. doi: 10.4014/jmb.1202.02025. [DOI] [PubMed] [Google Scholar]
- Dumenil G, Isberg RR. The Legionella pneumophila IcmR protein exhibits chaperone activity for IcmQ by preventing its participation in high-molecular-weight complexes. Mol Microbiol. 2001;40:1113–1127. doi: 10.1046/j.1365-2958.2001.02454.x. [DOI] [PubMed] [Google Scholar]
- Edelstein PH, Edelstein MA, Higa F, Falkow S. Discovery of virulence genes of Legionella pneumophila by using signature tagged mutagenesis in a guinea pig pneumonia model. Proc Natl Acad Sci U S A. 1999;96:8190–8195. doi: 10.1073/pnas.96.14.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicher SC, Dehio C. Bartonella entry mechanisms into mammalian host cells. Cell Microbiol. 2012;14:1166–1173. doi: 10.1111/j.1462-5822.2012.01806.x. https://doi.org/10.1111/j.1462-5822.2012.01806.x. [DOI] [PubMed] [Google Scholar]
- Escoll P, Mondino S, Rolando M, Buchrieser C. Targeting of host organelles by pathogenic bacteria: a sophisticated subversion strategy. Nat Rev Microbiol. 2016;14:5–19. doi: 10.1038/nrmicro.2015.1. https://doi.org/10.1038/nrmicro.2015.1. [DOI] [PubMed] [Google Scholar]
- Farelli JD, et al. IcmQ in the Type 4b secretion system contains an NAD+ binding domain. Structure. 2013;21:1361–1373. doi: 10.1016/j.str.2013.05.017. https://doi.org/10.1016/j.str.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Segal G. A specific genomic location within the icm/dot pathogenesis region of different Legionella species encodes functionally similar but nonhomologous virulence proteins. Infect Immun. 2004;72:4503–4511. doi: 10.1128/IAI.72.8.4503-4511.2004. https://doi.org/10.1128/IAI.72.8.4503-4511.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Zusman T, Hagag S, Segal G. Coevolution between nonhomologous but functionally similar proteins and their conserved partners in the Legionella pathogenesis system. Proc Natl Acad Sci U S A. 2005;102:12206–12211. doi: 10.1073/pnas.0501850102. https://doi.org/10.1073/pnas.0501850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez D, Spudich GM, Zhou XR, Christie PJ. The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J Bacteriol. 1996;178:3168–3176. doi: 10.1128/jb.178.11.3168-3176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsel I, Hilbi H. Formation of a pathogen vacuole according to Legionella pneumophila: how to kill one bird with many stones. Cell Microbiol. 2015;17:935–950. doi: 10.1111/cmi.12450. https://doi.org/10.1111/cmi.12450. [DOI] [PubMed] [Google Scholar]
- Firth N, Skurray R. Characterization of the F plasmid bifunctional conjugation gene, traG. Mol Gen Genet. 1992;232:145–153. doi: 10.1007/BF00299147. [DOI] [PubMed] [Google Scholar]
- Fischer W. Assembly and molecular mode of action of the Helicobacter pylori Cag type IV secretion apparatus. FEBS J. 2011;278:1203–1212. doi: 10.1111/j.1742-4658.2011.08036.x. https://doi.org/10.1111/j.1742-4658.2011.08036.x. [DOI] [PubMed] [Google Scholar]
- Frick-Cheng AE, Pyburn TM, Voss BJ, McDonald WH, Ohi MD, Cover TL. Molecular and structural analysis of the Helicobacter pylori cag type IV secretion system core complex. MBio. 2016;7:e02001–e02015. doi: 10.1128/mBio.02001-15. https://doi.org/10.1128/mBio.02001-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronzes R, Schafer E, Wang L, Saibil HR, Orlova EV, Waksman G. Structure of a type IV secretion system core complex. Science. 2009;323:266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullner KJ, Lara JC, Nester EW. Pilus assembly by Agrobacterium T-DNA transfer genes. Science. 1996;273:1107–1109. doi: 10.1126/science.273.5278.1107. [DOI] [PubMed] [Google Scholar]
- Garcillan-Barcia MP, de la Cruz F. Why is entry exclusion an essential feature of conjugative plasmids? Plasmid. 2008;60:1–18. doi: 10.1016/j.plasmid.2008.03.002. https://doi.org/10.1016/j.plasmid.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Ghosal D, Chang YW, Jeong KC, Vogel JP, Jensen GJ. In situ structure of the Legionella Dot/Icm type IV secretion system by electron cryotomography. EMBO Rep. 2017;18:726–732. doi: 10.15252/embr.201643598. https://doi.org/10.15252/embr.201643598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ, et al. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS ONE. 2009;4:e4833. doi: 10.1371/journal.pone.0004833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ, Brayton KA, Williams KP, Diaz MA, Brown WC, Azad AF, Sobral BW. Phylogenomics reveals a diverse Rickettsiales type IV secretion system. Infect Immun. 2010;78:1809–1823. doi: 10.1128/IAI.01384-09. https://doi.org/10.1128/IAI.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez FA, Tobar JA, Henriquez V, Sola M, Altamirano C, Marshall SH. Evidence of the presence of a functional Dot/Icm type IV-B secretion system in the fish bacterial pathogen Piscirickettsia salmonis. PLoS ONE. 2013;8:e54934. doi: 10.1371/journal.pone.0054934. https://doi.org/10.1371/journal.pone.0054934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Valero L, Buchrieser C. Genome dynamics in Legionella: the basis of versatility and adaptation to intracellular replication. Cold Spring Harb Perspect Med. 2013:3. doi: 10.1101/cshperspect.a009993. https://doi.org/10.1101/cshperspect.a009993. [DOI] [PMC free article] [PubMed]
- Gomez-Valero L, Rusniok C, Cazalet C, Buchrieser C. Comparative and functional genomics of legionella identified eukaryotic like proteins as key players in host-pathogen interactions. Front Microbiol. 2011;2:208. doi: 10.3389/fmicb.2011.00208. https://doi.org/10.3389/fmicb.2011.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Valero L, et al. Comparative analyses of Legionella species identifies genetic features of strains causing Legionnaires’ disease. Genome Biol. 2014;15:505. doi: 10.1186/s13059-014-0505-0. https://doi.org/10.1186/PREACCEPT-1086350395137407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Ruth FX, Moncalian G, Perez-Luque R, Gonzalez A, Cabezon E, de la Cruz F, Coll M. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature. 2001;409:637–641. doi: 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- Gomis-Ruth FX, Sola M, de la Cruz F, Coll M. Coupling factors in macromolecular type-IV secretion machineries. Curr Pharm Des. 2004;10:1551–1565. doi: 10.2174/1381612043384817. [DOI] [PubMed] [Google Scholar]
- Gordon JE, et al. Use of chimeric type IV secretion systems to define contributions of outer membrane subassemblies for contact-dependent translocation. Mol Microbiol. 2017;105:273–293. doi: 10.1111/mmi.13700. https://doi.org/10.1111/mmi.13700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann E, Christie PJ, Waksman G, Backert S. Type IV secretion in gram-negative and gram-positive bacteria. Mol Microbiol. 2018 doi: 10.1111/mmi.13896. https://doi.org/10.1111/mmi.13896. [DOI] [PMC free article] [PubMed]
- Guglielmini J, de la Cruz F, Rocha EP. Evolution of conjugation and type IV secretion systems. Mol Biol Evol. 2013;30:315–331. doi: 10.1093/molbev/mss221. https://doi.org/10.1093/molbev/mss221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyohda A, Komano T. Purification and characterization of the R64 shufflon-specific recombinase. J Bacteriol. 2000;182:2787–2792. doi: 10.1128/jb.182.10.2787-2792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. https://doi.org/10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- Jakubowski SJ, Krishnamoorthy V, Christie PJ. Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion. J Bacteriol. 2003;185:2867–2878. doi: 10.1128/JB.185.9.2867-2878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski SJ, Cascales E, Krishnamoorthy V, Christie PJ. Agrobacterium tumefaciens VirB9, an outer-membrane-associated component of a type IV secretion system, regulates substrate selection and T-pilus biogenesis. J Bacteriol. 2005;187:3486–3495. doi: 10.1128/JB.187.10.3486-3495.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong KC, Ghosal D, Chang YW, Jensen GJ, Vogel JP. Polar delivery of Legionella type IV secretion system substrates is essential for virulence. Proc Natl Acad Sci U S A. 2017;114:8077–8082. doi: 10.1073/pnas.1621438114. https://doi.org/10.1073/pnas.1621438114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EM, Gaddy JA, Voss BJ, Hennig EE, Cover TL. Genes required for assembly of pili associated with the Helicobacter pylori cag type IV secretion system. Infect Immun. 2014;82:3457–3470. doi: 10.1128/IAI.01640-14. https://doi.org/10.1128/IAI.01640-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimke WA, Rypien CD, Klinger B, Kennedy RA, Rodriguez-Maillard JM, Frost LS. The mating pair stabilization protein, TraN, of the F plasmid is an outer-membrane protein with two regions that are important for its function in conjugation. Microbiology. 2005;151:3527–3540. doi: 10.1099/mic.0.28025-0. https://doi.org/10.1099/mic.0.28025-0. [DOI] [PubMed] [Google Scholar]
- Ko KS, Hong SK, Lee HK, Park MY, Kook YH. Molecular evolution of the dotA gene in Legionella pneumophila. J Bacteriol. 2003;185:6269–6277. doi: 10.1128/JB.185.21.6269-6277.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komano T, Yoshida T, Narahara K, Furuya N. The transfer region of IncI1 plasmid R64: similarities between R64 tra and legionella icm/dot genes. Mol Microbiol. 2000;35:1348–1359. doi: 10.1046/j.1365-2958.2000.01769.x. [DOI] [PubMed] [Google Scholar]
- Kubori T, Nagai H. The Type IVB secretion system: an enigmatic chimera. Curr Opin Microbiol. 2015;29:22–29. doi: 10.1016/j.mib.2015.10.001. https://doi.org/10.1016/j.mib.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Kubori T, Koike M, Bui XT, Higaki S, Aizawa S, Nagai H. Native structure of a type IV secretion system core complex essential for Legionella pathogenesis. Proc Natl Acad Sci U S A. 2014;111:11804–11809. doi: 10.1073/pnas.1404506111. https://doi.org/10.1073/pnas.1404506111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak MJ, et al. Architecture of the type IV coupling protein complex of Legionella pneumophila. Nat Microbiol. 2017;2:17114. doi: 10.1038/nmicrobiol.2017.114. https://doi.org/10.1038/nmicrobiol.2017.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, Misselwitz R, Berger J, Sewald N, König W, Backert S. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449(7164):862–866. doi: 10.1038/nature06187. https://doi.org/10.1038/nature06187. [DOI] [PubMed] [Google Scholar]
- Labra A, Arredondo-Zelada O, Flores-Herrera P, Marshall SH, Gomez FA. In silico identification and characterization of putative Dot/Icm secreted virulence effectors in the fish pathogen Piscirickettsia salmonis. Microb Pathog. 2016;92:11–18. doi: 10.1016/j.micpath.2015.12.002. https://doi.org/10.1016/j.micpath.2015.12.002. [DOI] [PubMed] [Google Scholar]
- Larson CL, Martinez E, Beare PA, Jeffrey B, Heinzen RA, Bonazzi M. Right on Q: genetics begin to unravel Coxiella burnetii host cell interactions. Future Microbiol. 2016;11:919–939. doi: 10.2217/fmb-2016-0044. https://doi.org/10.2217/fmb-2016-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Klimke WA, Gubbins MJ, Frost LS. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. 2003;224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- Leclerque A, Kleespies RG. Type IV secretion system components as phylogenetic markers of entomopathogenic bacteria of the genus Rickettsiella. FEMS Microbiol Lett. 2008;279:167–173. doi: 10.1111/j.1574-6968.2007.01025.x. https://doi.org/10.1111/j.1574-6968.2007.01025.x. [DOI] [PubMed] [Google Scholar]
- Li F, Alvarez-Martinez C, Chen Y, Choi KJ, Yeo HJ, Christie PJ. Enterococcus faecalis PrgJ, a VirB4-like ATPase, mediates pCF10 conjugative transfer through substrate binding. J Bacteriol. 2012;194:404140–404151. doi: 10.1128/JB.00648-12. https://doi.org/10.1128/JB.00648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifshitz Z, et al. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc Natl Acad Sci U S A. 2013;110:E707–E715. doi: 10.1073/pnas.1215278110. https://doi.org/10.1073/pnas.1215278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locht C, Coutte L, Mielcarek N. The ins and outs of pertussis toxin. FEBS J. 2011;278(23):4668–4682. doi: 10.1111/j.1742-4658.2011.08237.x. https://doi.org/10.1111/j.1742-4658.2011.08237.x. [DOI] [PubMed] [Google Scholar]
- Low HH, et al. Structure of a type IV secretion system. Nature. 2014;508:550–553. doi: 10.1038/nature13081. https://doi.org/10.1038/nature13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Frost LS. Mutations in the C-terminal region of TraM provide evidence for in vivo TraM-TraD interactions during F-plasmid conjugation. J Bacteriol. 2005;187:4767–4773. doi: 10.1128/JB.187.14.4767-4773.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Wong JJ, Edwards RA, Manchak J, Frost LS, Glover JN. Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation. Mol Microbiol. 2008;70:89–99. doi: 10.1111/j.1365-2958.2008.06391.x. [DOI] [PubMed] [Google Scholar]
- Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurie-Weinberger MN, Gomez-Valero L, Merault N, Glockner G, Buchrieser C, Gophna U. The origins of eukaryotic-like proteins in Legionella pneumophila. Int J Med Microbiol. 2010;300:470–481. doi: 10.1016/j.ijmm.2010.04.016. https://doi.org/10.1016/j.ijmm.2010.04.016. [DOI] [PubMed] [Google Scholar]
- Marra A, Blander SJ, Horwitz MA, Shuman HA. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc Natl Acad Sci U S A. 1992;89:9607–9611. doi: 10.1073/pnas.89.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero J, Waldor MK. Determinants of entry exclusion within Eex and TraG are cytoplasmic. J Bacteriol. 2007;189:6469–6473. doi: 10.1128/JB.00522-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews M, Roy CR. Identification and subcellular localization of the Legionella pneumophila IcmX protein: a factor essential for establishment of a replicative organelle in eukaryotic host cells. Infect Immun. 2000;68:3971–3982. doi: 10.1128/iai.68.7.3971-3982.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauel MJ, Giovannoni SJ, Fryer JL. Phylogenetic analysis of Piscirickettsia salmonis by 16S, internal transcribed spacer (ITS) and 23S ribosomal DNA sequencing. Dis Aquat Organ. 1999;35:115–123. doi: 10.3354/dao035115. https://doi.org/10.3354/dao035115. [DOI] [PubMed] [Google Scholar]
- Middleton R, Sjolander K, Krishnamurthy N, Foley J, Zambryski P. Predicted hexameric structure of the Agrobacterium VirB4 C terminus suggests VirB4 acts as a docking site during type IV secretion. Proc Natl Acad Sci U S A. 2005;102:1685–1690. doi: 10.1073/pnas.0409399102. https://doi.org/10.1073/pnas.0409399102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt JH, Newton P, Newton HJ. Coxiella burnetii: turning hostility into a home. Cell Microbiol. 2015;17:621–631. doi: 10.1111/cmi.12432. https://doi.org/10.1111/cmi.12432. [DOI] [PubMed] [Google Scholar]
- Nagai H, Kubori T. Type IVB secretion systems of Legionella and other Gram-negative bacteria. Front Microbiol. 2011;2:136. doi: 10.3389/fmicb.2011.00136. https://doi.org/10.3389/fmicb.2011.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H, Roy CR. The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. EMBO J. 2001;20:5962–5970. doi: 10.1093/emboj/20.21.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295:679–682. doi: 10.1126/science.1067025. [DOI] [PubMed] [Google Scholar]
- Nakano N, Kubori T, Kinoshita M, Imada K, Nagai H. Crystal structure of Legionella DotD: insights into the relationship between type IVB and type II/III secretion systems. PLoS Pathog. 2010;6:e1001129. doi: 10.1371/journal.ppat.1001129. https://doi.org/10.1371/journal.ppat.1001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio S, Zuckman-Cholon DM, Cambronne ED, Roy CR. The Legionella IcmS-IcmW protein complex is important for Dot/Icm-mediated protein translocation. Mol Microbiol. 2005;55:912–926. doi: 10.1111/j.1365-2958.2004.04435.x. [DOI] [PubMed] [Google Scholar]
- Noto JM, Lee JY, Gaddy JA, Cover TL, Amieva MR, Peek RM., Jr Regulation of Helicobacter pylori virulence within the context of iron deficiency. J Infect Dis. 2015;211:1790–1794. doi: 10.1093/infdis/jiu805. https://doi.org/10.1093/infdis/jiu805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor TJ, Boyd D, Dorer MS, Isberg RR. Aggravating genetic interactions allow a solution to redundancy in a bacterial pathogen. Science. 2012;338:1440–1444. doi: 10.1126/science.1229556. https://doi.org/10.1126/science.1229556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranchych W, Frost LS. The physiology and biochemistry of pili. Adv Microb Physiol. 1988;29:53–114. doi: 10.1016/s0065-2911(08)60346-x. [DOI] [PubMed] [Google Scholar]
- Pena A, et al. The sexameric structure of a conjugative VirB4 protein ATPase provides new insights for a functional and phylogenetic relationship with DNA translocases. J Biol Chem. 2012;287:39925–39932. doi: 10.1074/jbc.M112.413849. https://doi.org/10.1074/jbc.M112.413849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell MW, Shuman HA. The Legionella pneumophila icmGCDJBF genes are required for killing of human macrophages. Infect Immun. 1998;66:2245–2255. doi: 10.1128/iai.66.5.2245-2255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Luo ZQ. Legionella and Coxiella effectors: strength in diversity and activity. Nat Rev Microbiol. 2017;15:591–605. doi: 10.1038/nrmicro.2017.67. https://doi.org/10.1038/nrmicro.2017.67. [DOI] [PubMed] [Google Scholar]
- Ramsey ME, Woodhams KL, Dillard JP. The gonococcal genetic island and type IV secretion in the pathogenic Neisseria. Front Microbiol. 2011;2:61. doi: 10.3389/fmicb.2011.00061. https://doi.org/10.3389/fmicb.2011.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rances E, Voronin D, Tran-Van V, Mavingui P. Genetic and functional characterization of the type IV secretion system in Wolbachia. J Bacteriol. 2008;190:5020–5030. doi: 10.1128/JB.00377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhury S, et al. Structure and function of interacting IcmR-IcmQ domains from a type IVb secretion system in Legionella pneumophila. Structure. 2009;17:590–601. doi: 10.1016/j.str.2009.02.011. https://doi.org/10.1016/j.str.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll-Rozada J, Zunzunegui S, de la Cruz F, Arechaga I, Cabezon E. Functional interactions of VirB11 traffic ATPases with VirB4 and VirD4 molecular motors in type IV secretion systems. J Bacteriol. 2013;195:4195–4201. doi: 10.1128/JB.00437-13. https://doi.org/10.1128/JB.00437-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde M, Puls J, Buhrdorf R, Fischer W, Haas R. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol. 2003;49:219–234. doi: 10.1046/j.1365-2958.2003.03549.x. [DOI] [PubMed] [Google Scholar]
- Roy CR, Isberg RR. Topology of Legionella pneumophila DotA: an inner membrane protein required for replication in macrophages. Infect Immun. 1997;65:571–578. doi: 10.1128/iai.65.2.571-578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadosky AB, Wiater LA, Shuman HA. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun. 1993;61:5361–5373. doi: 10.1128/iai.61.12.5361-5373.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagulenko E, Sagulenko V, Chen J, Christie PJ. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J Bacteriol. 2001;183:5813–5825. doi: 10.1128/JB.183.20.5813-5825.2001. https://doi.org/10.1128/JB.183.20.5813-5825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampei G, Furuya N, Tachibana K, Saitou Y, Suzuki T, Mizobuchi K, Komano T. Complete genome sequence of the incompatibility group I1 plasmid R64. Plasmid. 2010;64:92–103. doi: 10.1016/j.plasmid.2010.05.005. https://doi.org/10.1016/j.plasmid.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Sastre JI, Cabezon E, de la Cruz F. The carboxyl terminus of protein TraD adds specificity and efficiency to F-plasmid conjugative transfer. J Bacteriol. 1998;180:6039–6042. doi: 10.1128/jb.180.22.6039-6042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvides SN, et al. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 2003;22:1969–1980. doi: 10.1093/emboj/cdg223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder G, Lanka E. The mating pair formation system of conjugative plasmids—a versatile secretion machinery for transfer of proteins and DNA. Plasmid. 2005;54:1–25. doi: 10.1016/j.plasmid.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Schroder G, et al. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J Bacteriol. 2002;184:2767–2779. doi: 10.1128/JB.184.10.2767-2779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G, Shuman HA. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect Immun. 1999a;67:2117–2124. doi: 10.1128/iai.67.5.2117-2124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G, Shuman HA. Possible origin of the Legionella pneumophila virulence genes and their relation to Coxiella burnetii. Mol Microbiol. 1999b;33:669–667. 670. doi: 10.1046/j.1365-2958.1999.01511.x. [DOI] [PubMed] [Google Scholar]
- Segal G, Purcell M, Schuman HA. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc Natl Acad Sci U S A. 1998;95:1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G, Feldman M, Zusman T. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol Rev. 2005;29:65–81. doi: 10.1016/j.femsre.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Seshadri R, et al. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc Natl Acad Sci U S A. 2003;100:5455–5460. doi: 10.1073/pnas.0931379100. https://doi.org/10.1073/pnas.0931379100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton JA, Vogel JP. Type IVB secretion by intracellular pathogens. Traffic. 2002;3:178–185. doi: 10.1034/j.1600-0854.2002.030303.x. [DOI] [PubMed] [Google Scholar]
- Sexton JA, Miller JL, Yoneda A, Kehl-Fie TE, Vogel JP. Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex. Infect Immun. 2004a;72:5983–5992. doi: 10.1128/IAI.72.10.5983-5992.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton JA, Pinkner JS, Roth R, Heuser JE, Hultgren SJ, Vogel JP. The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J Bacteriol. 2004b;186:1658–1666. doi: 10.1128/JB.186.6.1658-1666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RK, Roy CR. Autophagy evasion and endoplasmic reticulum subversion: the yin and yang of Legionella intracellular infection. Annu Rev Microbiol. 2016;70:413–433. doi: 10.1146/annurev-micro-102215-095557. https://doi.org/10.1146/annurev-micro-102215-095557. [DOI] [PubMed] [Google Scholar]
- Silverman PM, Clarke MB. New insights into F-pilus structure, dynamics, and function. Integr Biol (Camb) 2010;2:25–31. doi: 10.1039/b917761b. https://doi.org/10.1039/b917761b. [DOI] [PubMed] [Google Scholar]
- Souza DP, Andrade MO, Alvarez-Martinez CE, Arantes GM, Farah CS, Salinas RK. A component of the Xanthomonadaceae type IV secretion system combines a VirB7 motif with a N0 domain found in outer membrane transport proteins. PLoS Pathog. 2011;7:e1002031. doi: 10.1371/journal.ppat.1002031. https://doi.org/10.1371/journal.ppat.1002031 (PPATHOGENS-D-10-00184 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza DP, et al. Bacterial killing via a type IV secretion system. Nat Commun. 2015;6:6453. doi: 10.1038/ncomms7453. https://doi.org/10.1038/ncomms7453. [DOI] [PubMed] [Google Scholar]
- Stingl K, Muller S, Scheidgen-Kleyboldt G, Clausen M, Maier B. Composite system mediates two-step DNA uptake into Helicobacter pylori. Proc Natl Acad Sci U S A. 2010;107:1184–1189. doi: 10.1073/pnas.0909955107. https://doi.org/10.1073/pnas.0909955107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland MC, Nguyen TL, Tseng V, Vogel JP. The Legionella IcmSW complex directly interacts with DotL to mediate translocation of adaptor-dependent substrates. PLoS Pathog. 2012;8:e1002910. doi: 10.1371/journal.ppat.1002910. https://doi.org/10.1371/journal.ppat.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland MC, Binder KA, Cualing PY, Vogel JP. Reassessing the role of DotF in the Legionella pneumophila type IV secretion system. PLoS ONE. 2013;8:e65529. doi: 10.1371/journal.pone.0065529. https://doi.org/10.1371/journal.pone.0065529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer N, Wessler S, Necchi V, Rohde M, Harrer A, Rau TT, Asche CI, Boehm M, Loessner M, Figueiredo C, Naumann M, Palmisano R, Solcia E, Ricci V, Backert S. A unique basolateral type IV secretion model for the CagA oncoprotein of Helicobacter pylori. Cell Host Microbe. 2017;22(552–560):e5. doi: 10.1016/j.chom.2017.09.005. https://doi.org/10.1016/j.chom.2017.09.005. [DOI] [PubMed] [Google Scholar]
- Terradot L, Waksman G. Architecture of the Helicobacter pylori Cag-type IV secretion system. FEBS J. 2011;278:1213–1222. doi: 10.1111/j.1742-4658.2011.08037.x. https://doi.org/10.1111/j.1742-4658.2011.08037.x. [DOI] [PubMed] [Google Scholar]
- Thanassi DG, Bliska JB, Christie PJ. Surface organelles assembled by secretion systems of Gram-negative bacteria: diversity in structure and function. FEMS Microbiol Rev. 2012;36:1046–1082. doi: 10.1111/j.1574-6976.2012.00342.x. https://doi.org/10.1111/j.1574-6976.2012.00342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRheenen SM, Dumenil G, Isberg RR. IcmF and DotU are required for optimal effector translocation and trafficking of the Legionella pneumophila vacuole. Infect Immun. 2004;72:5972–5982. doi: 10.1128/IAI.72.10.5972-5982.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent CD, Vogel JP. The Legionella pneumophila IcmS-LvgA protein complex is important for Dot/Icm-dependent intracellular growth. Mol Microbiol. 2006;61:596–613. doi: 10.1111/j.1365-2958.2006.05243.x. [DOI] [PubMed] [Google Scholar]
- Vincent CD, Friedman JR, Jeong KC, Buford EC, Miller JL, Vogel JP. Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol Microbiol. 2006;62:1278–1291. doi: 10.1111/j.1365-2958.2006.05446.x. [DOI] [PubMed] [Google Scholar]
- Vincent CD, Friedman JR, Jeong KC, Sutherland MC, Vogel JP. Identification of the DotL coupling protein subcomplex of the Legionella Dot/Icm type IV secretion system. Mol Microbiol. 2012;85:378–391. doi: 10.1111/j.1365-2958.2012.08118.x. https://doi.org/10.1111/j.1365-2958.2012.08118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Isberg RR. Cell biology of Legionella pneumophila. Curr Opin Microbiol. 1999;2:30–34. doi: 10.1016/s1369-5274(99)80005-8. [DOI] [PubMed] [Google Scholar]
- Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- Voth DE, Broederdorf LJ, Graham JG. Bacterial type IV secretion systems: versatile virulence machines. Future Microbiol. 2012;7:241–257. doi: 10.2217/fmb.11.150. https://doi.org/10.2217/fmb.11.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watarai M, Andrews HL, Isberg R. Formation of a fibrous structure on the surface of Legionella pneumophila associated with exposure of DotH and DotO proteins after intracellular growth. Mol Microbiol. 2000;39:313–329. doi: 10.1046/j.1365-2958.2001.02193.x. [DOI] [PubMed] [Google Scholar]
- Watarai M, Derre I, Kirby J, Growney JD, Dietrich WF, Isberg RR. Legionella pneumophila is internalized by a macropinocytotic uptake pathway controlled by the Dot/Icm system and the mouse Lgn1. locus. J Exp Med. 2001;194:1081–1096. doi: 10.1084/jem.194.8.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MM, et al. Identification of Coxiella burnetii type IV secretion substrates required for intracellular replication and Coxiella-containing vacuole formation. J Bacteriol. 2013;195:3914–3924. doi: 10.1128/JB.00071-13. https://doi.org/10.1128/JB.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker N, Chen Y, Jakubowski SJ, Sarkar MK, Li F, Christie PJ. The all-alpha domains of coupling proteins from the Agrobacterium tumefaciens VirB/VirD4 and Enterococcus faecalis pCF10-Encoded type IV secretion systems confer specificity to binding of cognate DNA substrates. J Bacteriol. 2015;197:2335–2349. doi: 10.1128/JB.00189-15. https://doi.org/10.1128/JB.00189-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker N, et al. Chimeric coupling proteins mediate transfer of heterologous type IV effectors through the Escherichia coli pKM101-encoded conjugation machine. J Bacteriol. 2016;198:2701–2718. doi: 10.1128/JB.00378-16. https://doi.org/10.1128/JB.00378-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, et al. Structural insights into the roles of the IcmS-IcmW complex in the type IVb secretion system of Legionella pneumophila. Proc Natl Acad Sci U S A. 2017 doi: 10.1073/pnas.1706883115. https://doi.org/10.1073/pnas.1706883115. [DOI] [PMC free article] [PubMed]
- Yerushalmi G, Zusman T, Segal G. Additive effect on intracellular growth by Legionella pneumophila Icm/Dot proteins containing a lipobox motif. Infect Immun. 2005;73:7578–7587. doi: 10.1128/IAI.73.11.7578-7587.2005. https://doi.org/10.1128/IAI.73.11.7578-7587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Kim SR, Komano T. Twelve pil genes are required for biogenesis of the R64 thin pilus. J Bacteriol. 1999;181:2038–2043. doi: 10.1128/jb.181.7.2038-2043.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni DS, McGrath S, Rabinovitch M, Roy CR. Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol Microbiol. 2003;49:965–976. doi: 10.1046/j.1365-2958.2003.03626.x. [DOI] [PubMed] [Google Scholar]
- Zuckman DM, Hung JB, Roy CR. Pore-forming activity is not sufficient for Legionella pneumophila phagosome trafficking and intracellular growth. Mol Microbiol. 1999;32:990–1001. doi: 10.1046/j.1365-2958.1999.01410.x. [DOI] [PubMed] [Google Scholar]
- Zusman T, Yerushalmi G, Segal G. Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. Infect Immun. 2003;71:3714–3723. doi: 10.1128/IAI.71.7.3714-3723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]