Abstract
Type IV collagen is a major component of the basement membrane and interacts with numerous other basement membrane proteins. Many of these interactions are poorly characterized. Type IV collagen is abundantly post-translationally modified with 3-hydroxyproline (3-Hyp), but 3-Hyp's biochemical role in type IV collagen's interactions with other proteins is not well established. In this work, we present binding data consistent with a major role of 3-Hyp in interactions of collagen IV with glycoprotein VI and nidogens 1 and 2. The increased binding interaction between type IV collagen without 3-Hyp and glycoprotein VI has been the subject of some controversy, which we sought to explore, whereas the lack of binding of nidogens to type IV collagen without 3-Hyp is novel. Using tandem MS, we show that the putative glycoprotein VI-binding site is 3-Hyp–modified in WT PFHR-9 type IV collagen, but not in PFHR-9 cells in which prolyl-3-hydroxylase 2 (P3H2) has been knocked out (KO). Moreover, we observed altered 3-Hyp occupancy across many other sites. Using amino acid analysis of type IV collagen from the WT and P3H2 KO cell lines, we confirm that P3H2 is the major, but not the only 3-Hyp–modifying enzyme of type IV collagen. These findings underscore the importance of post-translational modifications of type IV collagen for interactions with other proteins.
Keywords: post-translational modification (PTM), basement membrane, collagen, mass spectrometry (MS), protein chemistry, collagen IV, protein interactions, tandem mass spectrometry
Introduction
The biochemical role of 3-hydroxyproline (3-Hyp)2 has been the subject of considerable interest because it was shown that the loss of 3-hydroxyproline in fibrillar collagens is associated with severe osteogenesis imperfecta (1). In contrast to the more common 4-hydroxyproline (2), the presence of 3-hydroxyproline does not appear to directly alter overall collagen stability (3). The loss of 3-Hyp modification of single residue homologous to Pro986 of human type I collagen may result from loss of any of the components of the P3H1 (1)–CRTAP (4)–CypB (5) complex and is associated with severe osteogenesis imperfecta. It was subsequently shown that high myopia and severe lens phenotypes are associated with loss of P3H2 activity in humans (6, 7). Type IV collagen is likely the major substrate of P3H2 and is highly modified in 3-hydroxyproline, and it is unknown whether there is a direct correlation between modification of individual 3-hydroxyproline sites and the observed phenotypes. The 3-Hyp occupancy of Pro986 in collagen I is normally nearly 100% (4); however, the 3-Hyp occupancy of only a single site of type IV collagen is known (8) and may vary from site to site. Characterizing the occupancy of potential 3-Hyp sites is important because, in analogy to type I collagen, the highly modified sites could be expected to have the greatest functional significance in type IV collagen.
Pokidysheva et al. (9) observed early embryonic lethality from platelet aggregation in P3H2-null mice and suggested that this lethality occurs through platelet glycoprotein VI (GP6) binding to type IV collagen rich Reichert's membrane only in the P3H2-null mice. Using model collagen peptides, Gly–Pro–4-Hyp repeat peptides had previously been shown to be a substrate for GP6 (10); however, Pokidysheva et al. (9) showed that Gly–3-Hyp–4-Hyp repeats fail to bind GP6 and are ineffective as a platelet agonist. Thus, the similar loss of 3-Hyp from Gly–3-Hyp–4-Hyp repeat sequences in type IV collagen was predicted to turn type IV collagen into a platelet agonist through its interaction with GP6 in these mice. The physiological relevance of this interaction has been questioned because disorders of blood clotting or embryonic lethality were not observed in a different P3H2-null mouse line (8) nor in humans lacking P3H2 activity (6, 7). However, these latter observations leave open the question of whether type IV collagen lacking 3-hydroxyproline is capable of binding GP6, with more minor clotting defects. Type IV collagen forms a wide array of different interactions in the basement membrane (11), but post-translational modifications of type IV collagen have never been shown to be required for these interactions.
Post-translational modifications, including prolyl-hydroxylation of hypoxia-inducible factor 1-α (HIF-1α) to enable interaction with pVHL (12, 13), have a major role in altering protein–protein interactions. The following work presents evidence supporting a role for 3-hydroxyproline in binding GP6 to type IV collagen. We show that the putative GP6-binding domain(s) consisting of tandem GPP repeats are modified in type IV collagen from WT PFHR-9 cells, but not from PFHR-9 cells lacking P3H2. We also demonstrate that type IV collagen with reduced 3-Hyp content produced by the latter cells has greatly enhanced binding to GP6. Conversely, 3-hydroxyproline modification is shown here to be required for binding of nidogen 1 and 2 to type IV collagen. Thus, 3-hydroxyproline may either increase or decrease binding of proteins that interact with type IV collagen, suggesting that alteration of protein–protein interactions may be a major role for 3-hydroxyproline at other sites within type IV collagen and other collagens. The 3-hydroxyproline occupancy of many sites on WT PFHR-9 type IV collagen is also examined using tandem MS to understand how highly modified sites may play a role in other protein–protein interactions.
Results
PFHR-9 is a murine cell line chosen for this work because it has been previously used to purify type IV collagen (α12α2) (14, 15). We wished to produce a cell line with a homozygous knockout of P3H2 and used CRISPR/Cas9 because of its ease of targeting and high efficiency of DNA double-strand cleavage. Type IV collagen from these cells is used to compare the properties of type IV collagen with or without 3-Hyp. To examine the feasibility of a homozygous knockout, the ploidy of this teratocarcinoma cell line was determined by fixation and karyotyping (Fig. S1). Karyotyping revealed that PFHR-9 cells were aneuploid with 37 chromosomes on average. Chromosome 16, which carries LEPREL1 (coding for P3H2) is diploid, whereas chromosome 8 is amplified by a duplication and Robertsonian translocation, making it effectively triploid, and this chromosome carries the COL4A1 and COL4A2 genes, coding for the most abundant type IV collagen isotype produced by PFHR-9. A full summary of the karyotyping results is presented in Fig. S1 and its associated annotation.
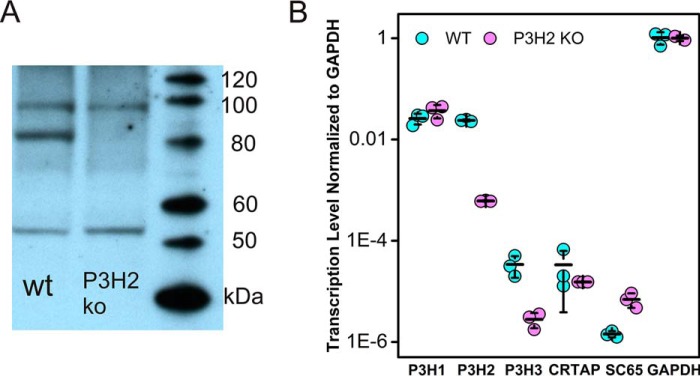
Knockouts of P3H2 from PFHR-9 cells were produced by random introduction of premature stop codons using CRISPR/Cas9 within the fourth exon of LEPREL1. Cell lysates were screened for P3H2 protein using immunoblot against the enzymatic domain of P3H2 (Fig. 1A), and clones lacking an 80-kDa band corresponding to P3H2 were subsequently lysed to isolate mRNA, which was reverse-transcribed to produce cDNAs that were subsequently quantitated through qRT-PCR. The level of LEPREL1 mRNA was decreased (Fig. 1B) for the 6A6 clone relative to WT PFHR-9 consistent with induction of nonsense-mediated decay by the stalled ribosomes. These experiments showed higher expression of P3H1 and P3H2 than for P3H3, the P3H1 complex partner CRTAP, or SC65, a similar protein to CRTAP. In addition to a nearly 40-fold decrease in P3H2 transcription in 6A6 clone (p < 2.0 × 10−5), a 12-fold decrease in P3H3 transcription was observed (p < 2.0 × 10−3), as well as a nearly 5-fold increase in SC65 transcription (p < 0.05). A cDNA of the 6A6 clone containing exon 4 of LEPREL1 was sequenced and revealed to contain a two-base deletion, resulting in a premature stop codon in exon 4 (Fig. S2). For the remainder of this article, clone 6A6 will be referred to as P3H2 KO PFHR-9.
Figure 1.
Immunoblot of P3H2 and transcription levels of P3Hs and related genes. A, chemiluminescent blot with an antibody against P3H2. P3H2 shows up as an 80-kDa band present in the WT but absent in 6A6 (P3H2 KO). B, the level of transcription of several genes related to P3Hs, and a GAPDH control transcript was examined by qRT-PCR of oligo(dT) generated first strand cDNAs of total RNA from PFHR-9 cells. Transcription levels are normalized to GAPDH transcription through absolute difference in threshold PCR cycle number and plotted logarithmically, assuming a one-cycle difference represents an exact 2-fold difference in transcription. The error bars indicate the standard deviation of cycle number.
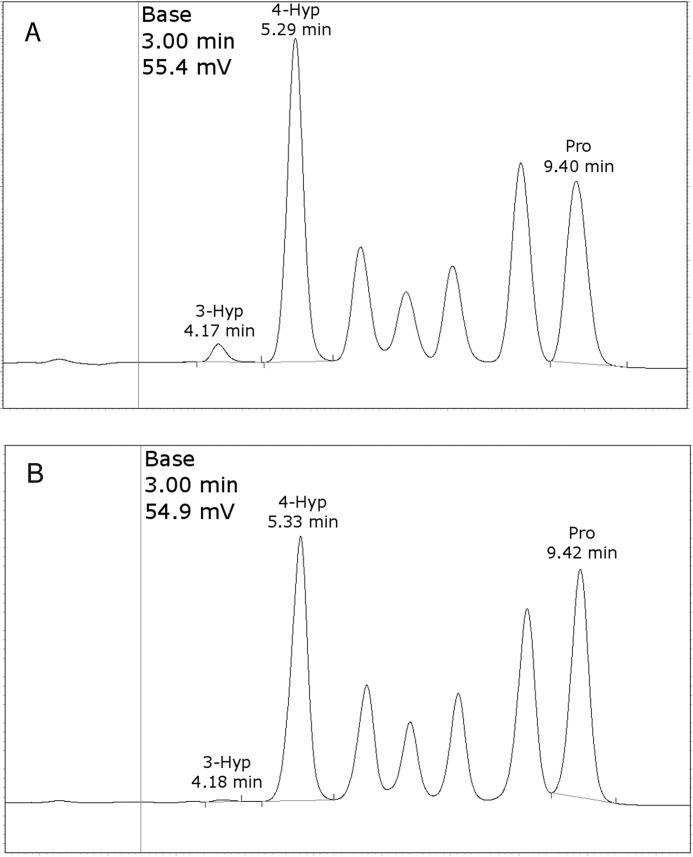
Type IV collagen was purified from the media of both PFHR-9 WT and P3H2 KO cells (Fig. S3). Purified type IV collagen was acid hydrolyzed, and free amino acids from this hydrolysate were separated by cation exchange column. Separated amino acids were quantitated by absorbance detection of post-column (ninhydrin) derivatized amino acids on a Hitachi L-8800A amino acid analyzer, using isoleucine as an internal standard. Chromatographic resolution and sensitivity was sufficient to achieve separation and quantitation of 4-Hyp and 3-Hyp (Fig. 2 and Table 1). Both WT and P3H2 KO type IV collagen showed similar compositions for most amino acids as would be expected for hydrolysates of pure type IV collagen (Table S1). The level of 4-Hyp and Lys-OH was similar between material isolated from the different cell lines but with slightly decreased hydroxylation observed in the material isolated from P3H2 KO cells (Table 1). Both samples had over 80% 4-Hyp occupancy and over 90% Lys-OH occupancy; however, 3-Hyp was almost completely absent from P3H2 KO type IV collagen, with on average of 4.2 modified residues per triple helix for the P3H2 KO, versus 30.6 3-Hyp residues for material isolated from the WT cells (Table 1 and Fig. 2). These results are consistent with P3H2 being the major, although not the sole, 3-Hyp–modifying enzyme for type IV collagen, despite the presence of P3H1 transcription.
Figure 2.
Amino acid analysis of WT (A) and P3H2 KO-derived (B) PFHR-9 type IV collagen. Overall quantitation is based on integration of peaks that had been annotated according to elution time, as indicated. Chromatograms were scaled to normalize the combined height of 4-Hyp and Pro peaks above baseline for concentration differences between the samples.
Table 1.
Amino acid analysis quantitation of hydroxyproline and hydroxylysine
Experimental concentration was derived from peak integration and the average of three runs. Experimental concentrations for each individual amino acid were normalized to the total Pro + 3-Hyp + 4-Hyp or Lys + Lys-OH content. The average occupancy is expressed at a fraction of the normalized experimental concentration divided by the number of corresponding motifs (i.e. GPP for 3-Hyp) present in a type IV collagen α1 α1 α2 trimer lacking the N-terminal signaling peptide.
| Amino acid | WT |
P3H2 KO |
||
|---|---|---|---|---|
| Experimental composition | Normalized composition | Experimental composition | Normalized composition | |
| 3-Hyp | 31.5 (3.35%) | 30.6/109 (GPP) | 4.0 (0.46%) | 4.2/109 (GPP) |
| 4-Hyp | 587.1 (62.4%) | 571.0/591 (GXP) | 505.1 (58.5%) | 535.3/591 (GXP) |
| Pro | 321.9 (34.2%) | 313.0/915 (P) | 354.8 (41.1%) | 376.1/915 (P) |
| Pro + 3-Hyp + 4-Hyp | 940.5 (100%) | 915/915 (P) | 863.8 (100%) | 915/915 (P) |
| Lys | 46.5 (17.6%) | 45.2/257 (K) | 68.6 (25.5%) | 65.5/257 (K) |
| Lys-OH | 217.6 (82.4%) | 211.8/202 (GXK) | 200.8 (74.5%) | 191.5/202 (GXK) |
| Lys + Lys-OH | 264.1 (100%) | 257/257 (K) | 269.4 (100%) | 257/257 (K) |
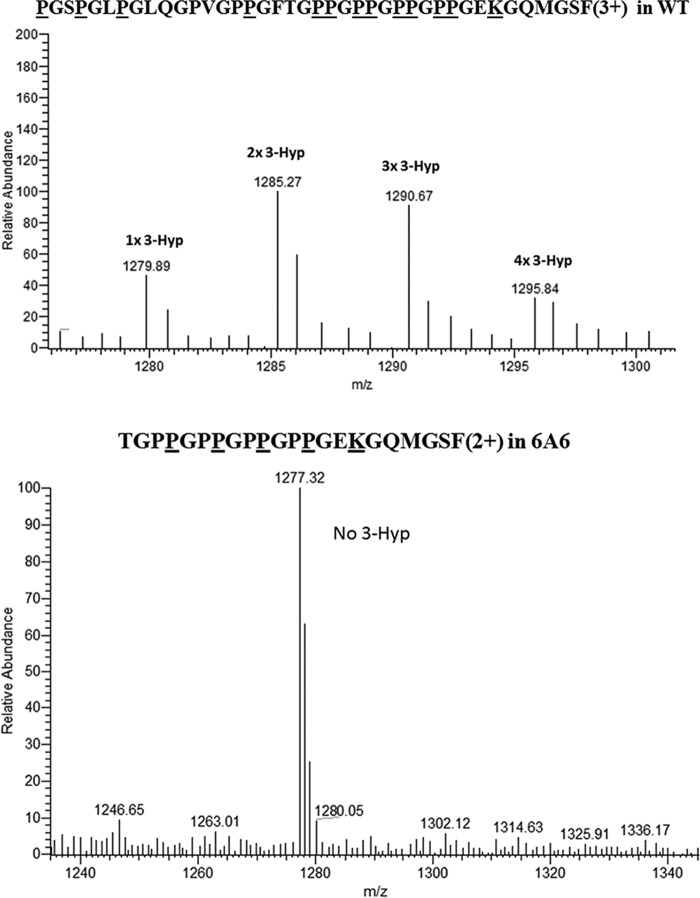
PFHR-9 WT and P3H2 KO type IV collagen proteolytic digests using trypsin alone, chymotrypsin then trypsin, or CNBr then trypsin were analyzed separately by Micro quadrupole time-of-flight mass spectrometer (Waters) and by VELOS electrospray linear ion trap (Thermo) mass spectrometers. Many fragmentation spectra from these experiments could be assigned to modified Gly-Pro-Pro sequences with variable 3-Hyp occupancy (Table 2, Figs. 3 and 4, and Figs. S9–S40) from WT and P3H2 KO type IV collagen. Mass spectrometry data from P3H2 KO type IV collagen suggested a loss of 3-Hyp from the longest tandem Gly-Pro-Pro repeat fragmentation spectrum that could be observed (Fig. 3), a (GPP)4 sequence in the type IV collagen α1 chain, and a variable level of modification is observed in WT type IV collagen, with one to four 3-Hyps present in the (GPP)4 sequence (Fig. 3). Additionally, 3-Hyp–modified GPPGPP sequences could be found in the MS data from WT type IV collagen. Each repeat sequence contained at least one residue with greater than 50% 3-Hyp occupancy (see Table 2 and accompanying supporting data for Pro1348 of the type IV collagen α1 chain, and Pro386, Pro644, Pro970 and Pro1419 (Fig. 4) of α2 chain). A recent publication failed to observe 3-Hyp modification at several tandem GPP sites in type IV collagen from two different sources (15). Unlike this work, however, the degree of modification of 3-Hyp sites across was not made clear, and so it is difficult to determine whether this previous work observed a complete lack of modification at these sites or whether the modified species were simply never observed in the data sets examined. Chemically synthesized Gly–Pro–4-Hyp repeat sequences are capable of being modified by heterologously expressed P3H2 (16). This suggests that GPP repeats are a common 3-Hyp modification motif for P3H2. Because 3-Hyp is isobaric to 4-Hyp (with mass 16 Da), these modified residues cannot be distinguished by using MS alone. The enzymatic activity of 4-hydroxylases and 3-hydroxylases within Gly-Pro-Pro have been previously characterized and showed that generally 4-Hyp modification must occur before 3-Hyp activity can occur within individual sites (17, 18). Therefore, in the case of ambiguous spectral assignments, we have chosen to assume that in collagenous Gly–X–Y sequences, all Y position prolines are 4-hydroxyproline, all X position prolines are 3-hydroxyproline, and all potential 4-hydroxyproline sites are modified before any 3-hydroxyproline sites are modified within any particular precursor peptide, in the absence of contradicting evidence.
Table 2.
Level of modification of individual 3-Hyp sites in type IV collagen from WT or P3H2 KO (6A6) PFHR-9 cells
Modification levels are based on MS1 spectra, which can be found with the corresponding figure (i.e. 197 COL4A2 MS1 and MS2 data are found in Fig. S29). Peak occupancies are estimated to the nearest 5% based on MS1 peak height. F, figure; SF, supplemental figure; NA, not applicable.
| Residue no. | Occupancy |
Figure |
||
|---|---|---|---|---|
| WT | P3H2 KO | WT | P3H2 KO | |
| % | % | |||
| 137 COL4A1 | 0 | 0 | SF9 | SF23 |
| 149 COL4A1 | 20 | 0 | SF9 | SF23 |
| 198 COL4A1 | 100 | No data | F3/ (MS2 SF36) | NA |
| 204 COL4A1 | 60 | 0 | F3/ (MS2 SF36) | F3/ (MS2 SF35) |
| 207 COL4A1 | 60 | 0 | F3/ (MS2 SF36) | F3/ (MS2 SF35) |
| 210 COL4A1 | 60 | 0 | F3/ (MS2 SF36) | F3/ (MS2 SF35) |
| 213 COL4A1 | 60 | 0 | F3/ (MS2 SF36) | F3/ (MS2 SF35) |
| 239 COL4A1 | No data | 0 | NA | SF37 |
| 330 COL4A1 | 0 | 0 | SF10 | SF24 |
| 372 COL4A1 | No data | 0 | NA | SF25 |
| 478 COL4A1 | 60 | 0 | SF11 | SF26 |
| 484 COL4A1 | 0 | 0 | SF11 | SF26 |
| 587 COL4A1 | 0 | No data | SF12 | NA |
| 647 COL4A1 | 15 | 0 | SF13 | SF27 |
| 1214 COL4A1 | 8–35 | No data | SF40 | NA |
| 1345 COL4A1 | 25 | No data | SF14 | NA |
| 1348 COL4A1 | 55 | No data | SF14 | NA |
| 1367 COL4A1 | 0 | No data | SF15 | NA |
| 1424 COL4A1 | 80 | No data | SF16a/b | NA |
| 1436 COL4A1 | 15 | 0 | SF16a/b | SF28 |
| 74 COL4A2 | 80 | 15 | SF38 | SF38 |
| 197 COL4A2 | No data | 0 | NA | SF29 |
| 343 COL4A2 | 60 | No data | SF17 | NA |
| 386 COL4A2 | 50 | 0 | SF18 | SF30 |
| 389 COL4A2 | 5 | No data | SF18 | NA |
| 586 COL4A2 | 0 | 0 | SF19 | SF31 |
| 594 COL4A2 | 0 | No data | SF19 | NA |
| 638 COL4A2 | 40 | 0 | SF20 | SF32 |
| 644 COL4A2 | 70 | 0 | SF20 | SF32 |
| 647 COL4A2 | 15 | 0 | SF20 | SF32 |
| 728 COL4A2 | 65 | No data | SF39 | NA |
| 970 COL4A2 | 60 | 0 | SF21 | SF33 |
| 973 COL4A2 | 0 | 0 | SF21 | SF33 |
| 1419 COL4A2 | 60 | 0 | F4/ (MS2 SF22) | F4/ (MS2 SF34) |
Figure 3.
Extent of hydroxylation in WT and P3H2 KO type IV collagen GPPGPPGPPGPP peptide. Top panel, averaged MS scans from m/z 1275 to 1300 in chromatograms where CNBr + tryptic type IV collagen α1 hydroxylated peptide from WT cells elute. Underlined residues in the amino acid sequence show predicted 3-and 4-hydroxproline modification sites, as well as a modified glucosylgalacotosyl hydroxylysine and oxidized methionine. Bottom panel, averaged MS scans from m/z 1250 to 1300 where chymotryptic type IV collagen α1 peptide from P3H2 KO cells elute. Underlined residues in the amino acid sequence show predicted 4-hydroxyproline modification sites and modified glucosylgalacotosyl hydroxylysine. Residues Pro204, Pro207, Pro210, and Pro213 in the WT peptide were at least partially hydroxylated, as the indicated masses corresponding to a mixture of peptides containing 2–5 extra hydroxylation sites, whereas these putative 3-hydroxproline sites were not observed in P3H2 KO peptides.
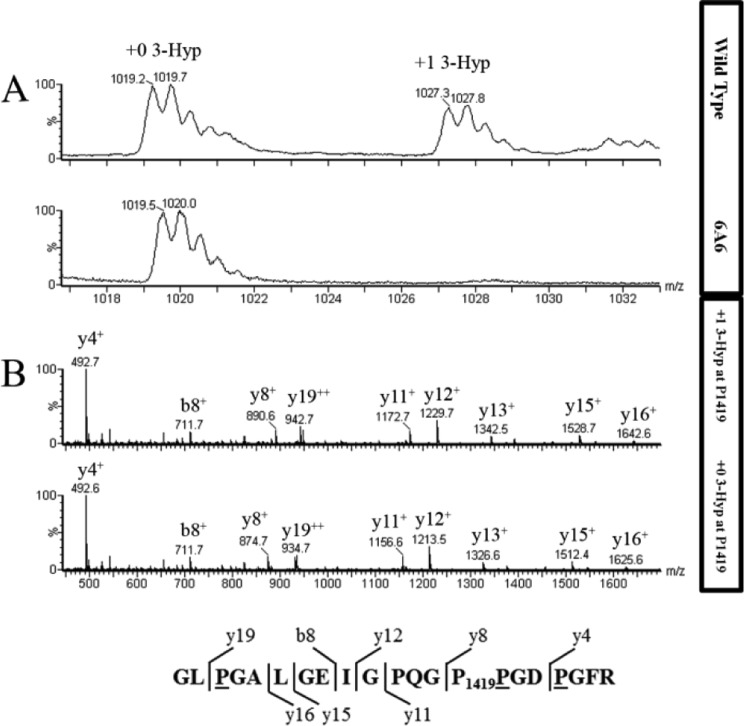
Figure 4.
Extent of hydroxylation in type IV collagen α-2 residue P1419 from WT and P3H2 KO cells. A, averaged MS scans from m/z 1016 to 1034 in chromatograms where tryptic type IV collagen α2 peptide from WT (top panel) and P3H2 KO (bottom panel) cells elute. Amino acid sequence is shown with observed b and y ions. B, fragment spectrum for this peptide from m/z 450 to 1700 for WT (top panel) and P3H2 KO (bottom panel) cells. The b and y ions and their m/z values are indicated for each labeled peak. Underlined residues in the amino acid sequence at the bottom are predicted to be hydroxylated, and Pro1419 is predicted to be 3-hydroxylated, based on the +16 unit mass shifts of the y8 fragment ion and larger y ions in the peptide from WT cells. As expected, peptides from both WT and KO cells contain three 4-Hyp residues.
One tryptic peptide sequence, GQPGAVGPQGYNGP74PGLQGFPGLQGR, which was found in both WT and P3H2 KO material, has a likely 3-Hyp modification at Pro74 in the α2-chain, which is within the N-terminal 7S domain of this protein. It is modified in type IV collagen from both WT and P3H2 KO cells (Fig. S38, a and b), although to a much greater extent in collagen IV from PFHR-9 WT cells. We were unable to observe additional 3-Hyp–modified sites in P3H2 KO cells (Table 2, Figs. 3 and 4, and Figs. S9–S37, S39, and S40). These observations support the hypothesis that P3H2 is the major but not sole prolyl-3-hydroxylase–modifying collagen IV, earlier suggested by the amino acid analysis results.
A sequence comparison between mouse and human type IV collagen α1- and α2-chains is shown in Figs. S4 and S5. The location of the identified 3-Hyp sites is shown in the mouse sequences. Interestingly, all modified proline residues in the mouse sequences are conserved in the human sequences with one exception in the α2-chain.
The results of Pokidysheva et al. (9) suggested that Gly–Pro–4-Hyp repeats in type IV collagen from knockouts of P3H2 would bind GP6, accounting for the differences in clotting phenotype in embryonic mice. To attempt to answer this question, label-free analysis of type IV collagen/GP6 binding was examined using surface plasmon resonance with the GP6–collagen-binding domain (GP6–CBD), used in (9). To ensure that well folded and stable collagen would be coupled to surface plasmon resonance substrates, the secondary structure of type IV collagen was examined using CD. The wavelength scan showed a positive peak at ∼222 nm for both WT and P3H2 KO type IV collagen, consistent with collagenous secondary structure (Fig. S6). The CD temperature scan monitored at 220 nm showed two melting transitions for both WT (Fig. S7a) and P3H2 KO material (Fig. S7b): a large transition at 40 °C and a lesser transition at 52 °C, consistent with previous optical rotary dispersion results monitored at 365 nm from PFHR-9 type IV collagen (14). Altogether this shows that type IV collagen obtained from either WT or P3H2 KO cells is folded and that there is very little difference in overall secondary structure or stability for the differently 3-Hyp–modified type IV collagen.
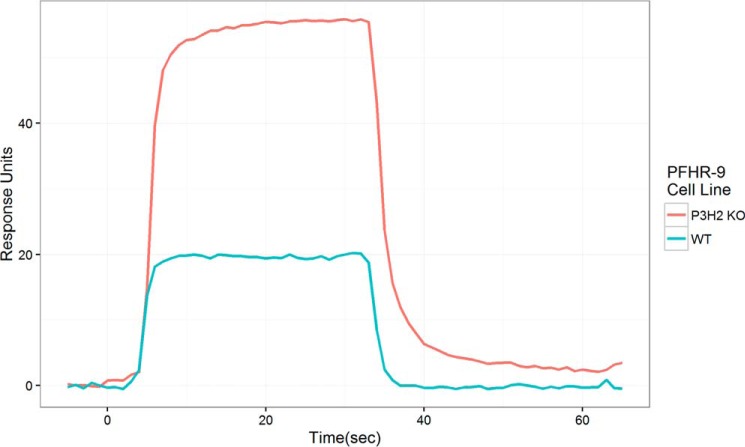
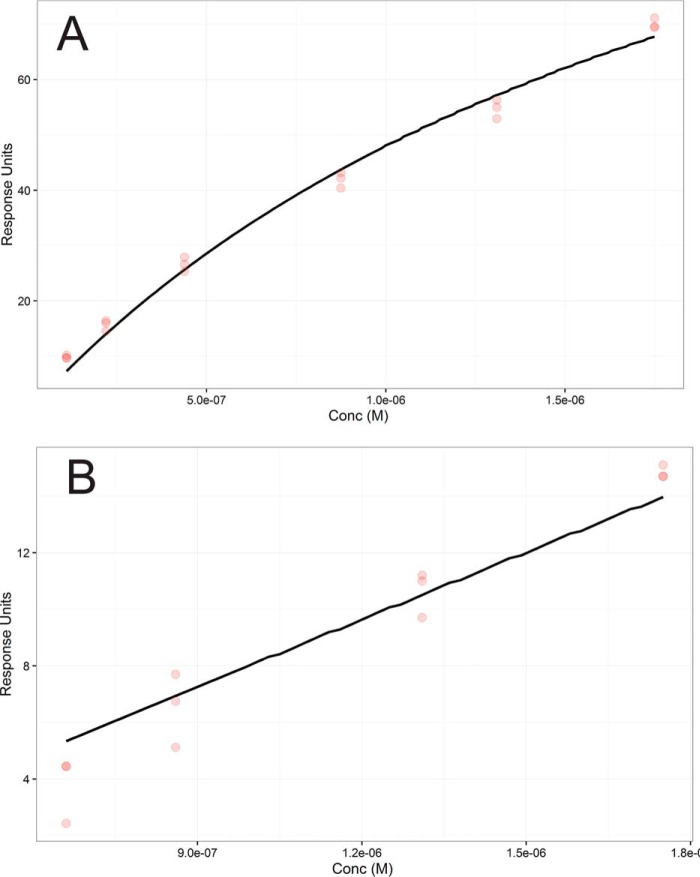
The GP6–CBD was obtained by heterologous expression and flowed over a surface plasmon resonance chips coupled with either 2000 RU of P3H2 KO type IV collagen or 2600 RU of WT type IV collagen. Normalized for this difference in innate response, P3H2 KO type IV collagen showed a greater response than WT type IV collagen for each tested concentration. Both substrates showed rapid binding and dissociation kinetics; however, dissociation kinetics was slower (though still too rapid for quantitative estimation) for binding to P3H2 KO type IV collagen (Fig. 5). Fitting to a 1:1 steady-state binding model was used to estimate the affinity of GP6–CBD to collagen IV having or lacking 3-Hyp within Gly–Pro–4-Hyp sequences. Because multiple repeat sequences substrates for GP6–CBD binding are modified in the WT type IV collagen but unmodified in the P3H2 KO, the 1:1 binding model is only an estimate resulting in an effective Kd. This fit revealed that binding to P3H2 KO type IV collagen (effective Kd = 2.1 μm) is much stronger than to WT type IV collagen (effective Kd = 84 μm) (Fig. 6).
Figure 5.
Surface plasmon resonance binding of the collagen-binding domain of GP6–CBD to type IV collagen from WT or P3H2 koPFHR-9 cells. Injection was from 5 to 35 s, and dissociation was from 35 to 65 s. Concentration for all injections was 35 μg/ml (1.75 μm). Each curve is an average of three injections. Response was normalized for the amount of bound collagen IV.
Figure 6.
Steady-state affinities of GP6–CBD for type IV collagen from surface plasmon resonance data. A, data from GP6–CBD binding to P3H2 KO PFHR-9 type IV collagen. B, data from GP6–CBD binding to WT PFHR-9 type IV collagen. Three replicate responses are shown for each of four concentrations as red dots. Steady-state 1:1 model fit is shown as a black line. Response units for each data point are an average of the response from t = 20 s to t = 30 s for each 30-s injection. The modeled Kd for P3H2 KO type IV collagen was 2.1 and 84 μm for the WT.
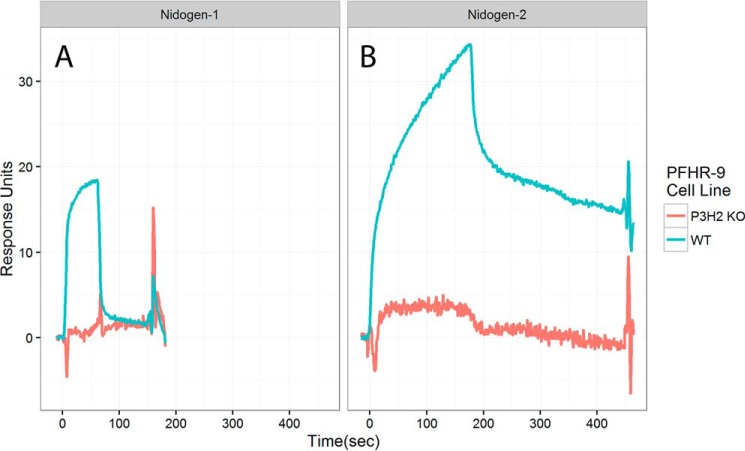
Nidogen-1 and nidogen-2 are basement membrane proteins that may link type IV collagen and laminin networks (16), as well as binding to other basement membrane proteins. To determine whether nidogen-1 and nidogen-2 binding is affected by the presence of 3-Hyp, these proteins were flowed over type IV collagen coupled chips. Binding of nidogen-1 to P3H2 KO type IV collagen was absent, whereas WT type IV collagen bound nidogen-1 as expected. Nidogen-2 binding to P3H2 KO type IV collagen was nearly absent, whereas binding to WT type IV collagen showed a greater response and dissociated less rapidly than nidogen-1 (Fig. 7). This suggests a requirement for 3-Hyp in nidogen 1 and 2 binding to type IV collagen. Additionally, binding of SPARC was tested with collagen binding SPARC construct BM40ΔIαC (17). Binding of this construct was also decreased for type IV collagen from the P3H2 KO, with a roughly 50% decrease in overall binding response (Fig. S8).
Figure 7.
Surface plasmon resonance binding of nidogens to type IV collagen from WT or P3H2 KO PFHR-9 cells. For nidogen-1 (A), the concentration was 100 μg/ml. For nidogen-2 (B), the concentration was 80 μg/ml. Each curve is an average of three injections. Response was normalized for the amount of bound type IV collagen.
Discussion
The precise binding site for nidogen on type IV collagen is unknown, but EM data support binding nearest to the modified site at Pro1214 of the type IV collagen α1 chain (19). It is interesting to note that disruptions in nidogen-1 (20) (as well as other basement membrane proteins, such as collagen XVIII (21) and SPARC (22)) are associated with abnormal lens phenotypes in mice. Thus, one could speculate that loss of nidogen binding to type IV collagen in lens capsule at least partially underlies the lens phenotype observed in humans and mice with disruptions in P3H2 activity. The initial discovery of the type IV collagen–nidogen–laminin complex suggested (19) a strong interaction in material isolated from Engelbreth–Holm–Swarm rat chondrosarcoma tumor, having a similar overall 3-Hyp composition to material derived from PFHR-9 cells. However, a recent report using magnetic bead pulldown from epidermis suggested that collagen IV/laminin networks cross-linked with nidogen were separable, but those with perlecan were not (23), implying that perlecan cross-links are more prevalent and therefore more functionally important in basement membrane stability in this tissue. The results presented here suggest that 3-hydroxyproline in type IV collagen could alter the composition of type IV collagen/laminin cross-links to favor nidogen. Lens capsule type IV collagen is known to be highly enriched in 3-hydroxyproline (24), and lens capsule is impacted in nidogen-1 knockout mice, where lens fiber cells infiltrate the posterior lens capsule (20). On the other hand, epidermal P3H2 expression is very low (25). Nidogen knockout in Caenorhabditis elegans (26), which lacks a prolyl-3-hydroxylase homolog (27), does not grossly alter type IV collagen or laminin immunolocalization. Thus, the importance of nidogen cross-links to basement membrane structure may depend upon the presence and occupancy of 3-hydroxyproline in basement membrane type IV collagen, although the precise site(s) involved remain to be determined. In this way, 3-hydroxyproline could be used to tune the properties of specialized basement membranes, such as the lens capsule.
The substrate specificity of P3H1 is highly constrained, capable of 3-Hyp modifying the sequence GLPGPIGPPGPR in type I collagen but not the highly similar sequence GHPGPIGPPGPR in type III collagen (28). In contrast, this work shows that many sites are 3-Hyp–modified in type IV collagen in diverse sequence contexts. These sequences also differ greatly from the clade A2, A3, and B2 sequences thought to be modified by P3H2 in fibrillar collagens (28). This suggests that P3H2 is a much less selective enzyme than P3H1. The amino acid analysis results presented suggest that P3H2 activity is responsible for only ∼85% of 3-Hyp, and tandem MS permitted the assignment of one site (Pro74) that is clearly modified in the P3H2 KO cells. This GPQGYNGPPGFP sequence differs from the canonical P3H1 3-Hyp sequence, suggesting that another enzyme, perhaps P3H3, may be responsible for modification here. The sequence determinants of 3-Hyp modification by P3H2 are unknown. The variable level of modification of sites throughout type IV collagen suggests that most sites are capable of being modified, and a large number might be fully modified in 3-Hyp repeats of type IV collagen from lens capsule simply because of the level of P3H2 activity present in lens epithelial cells. The highly variable 3-Hyp occupancy from site to site underscores the importance in quantitating the degree of 3-Hyp, beyond the mere presence or absence of modification at different sites. A recent report (15) observed a distinct pattern of modification from that presented here. These differences may arise from the source of type IV collagen (Engelbreth–Holm–Swarm rat chondrosarcoma tumor and bovine lens type IV collagen versus PFHR-9 collagen) but might also be resolved if the level of modification was known. The average 3-Hyp occupancy estimated from the tandem MS data were ∼36% (Table 2) for the observed sites, similar to that estimated from amino acid analysis results for the overall protein at 28% (30.9 modified residues/109 GPP motifs; Table 1). Thus, the MS data presented here provide a substantially more detailed examination of 3-Hyp modification of type IV collagen than previously available, and the occupancy data are even more credible because of its consistency with the amino acid analysis results.
Glycoprotein VI is a platelet protein involved in the initiation of the collagen-dependent clotting response (29). The major binding motif for GP6 is a Gly–Pro–4-Hyp sequence repeated at least twice, with the highest binding observed for four or more such repeats in tandem (10). Every Gly–Pro–4-Hyp repeat observed in this work in WT collagen has over 50% occupancy for at least one 3-Hyp, consistent with these repeats being a motif for P3H2 modification. Additionally the surface plasmon resonance results show a corresponding decrease in affinity when the Gly–Pro–4-Hyp repeats are converted to 3-Hyp–containing repeats. These results are consistent with previous comparisons between GP6 binding to Gly–3-Hyp–4-Hyp and Gly–Pro–4-Hyp repeat peptides (9). Gly–Pro–4-Hyp, but not Gly–3-Hyp–4-Hyp repeat peptides were also shown there to be an agonist for platelet aggregation, and the physiological relevance of binding to type IV collagen could be examined analogously by testing sufficient quantities of type IV collagen obtained from these or similar PFHR-9 cells. GP6 has been scrutinized as a target for antithrombotics (30), and common human GP6 variants may vary in thrombotic efficiency (31). Thus, understanding the determinants of GP6 interaction with collagen may be important to understanding GP6 related thrombotic risk factors. Integrin α2β1 is also involved in the collagen-dependent clotting response, although less essential than GP6 (32). Like GP6, integrin α2β1 binds with greater affinity to type I collagen than to type IV collagen (33). Thus, multiple mechanisms could be used to inhibit platelet activation at endothelial type IV collagen, of which prolyl-3-hydroxylation may be one.
LAIR-1 (leukocyte-associated immunoglobulin-like receptor 1) and GP6 are both expressed from the chromosome 19 immunoreceptor cluster in humans (34), and LAIR-1 contains an immunoglobulin-like collagen-binding domain, similar to GP6. Also like GP6, LAIR-1 binds to Gly–Pro–4-Hyp repeats but not Gly-Pro-Pro repeats (35). Whereas LAIR-1 has been shown to bind to collagen types I, II, III, VII, and XVII (35), its competence at binding type IV collagen has not been reported. It would be interesting to test whether the collagen-binding domain of LAIR-1 is capable of binding type IV collagen and to Gly–3-Hyp–4-Hyp repeats. LAIR-1 is an inhibitory immune receptor (36); therefore, a putative inability to bind endothelial type IV collagen because of its 3-Hyp content could be functionally related to leukocyte immune response to occur in the vasculature.
When P3H1 was originally isolated, it was shown to form a complex with CypB and CRTAP (37). Subsequent studies have shown that loss of 3-Hyp modification of P986 of type I collagen in vivo is associated with loss of any of these three complex members, which also causes severe osteogenesis imperfecta (1, 4, 5). To investigate whether P3H2 has similar complex members, transcription of CRTAP and the SC65 genes, both of which have much lower transcription than P3H2, was quantified by qPCR. Both SC65 and CRTAP have similar N-terminal domains to those found in P3Hs, consisting of four TPR domains. The much greater transcription level of P3H2 compared with other P3Hs, or to SC65 or CRTAP suggests that P3H2 may form a very different complex than P3H1. P3H1 is transcribed in these cells at a similar level as P3H2, but CRTAP is transcribed at a much lower level. Because P3H1 requires CRTAP for activity, P3H1 is likely to be largely catalytically inactive in these cells. Intriguingly, the prolyl-4-hydroxylase (P4HA2) has recently been reported as a recessive nonsyndromic high-myopia gene, like P3H2 (38). It would be interesting to test whether P4HA2 can form a complex with P3H2 and whether prolyl-3-hydroxylation is decreased in tissues from P4HA2-null mice (39).
Experimental procedures
RNA extraction
RNA was extracted from lysates of 5 × 106 PFHR-9 cells from T-75 flasks that had reached confluence using a QIAGen RNeasy mini kit using manufacturer's instructions. The cells were lysed by multiple aspiration cycles through DNase/RNase free 18-gauge needle and syringe. Work was performed in sterile cell culture hood, with work area cleaned using RNAZap® solution.
cDNA first-strand synthesis
The Superscript® III first-strand synthesis system was used to synthesis cDNAs for qRT-PCR using the manufacturer's instructions. Synthesis of cDNAs was primed using a final concentration of 2.5 μm oligo(dT)20, with 1 μg of total PFHR-9 RNA from QIAGen RNeasy prep per 20-μl reaction. RNA concentration and quality was quantitated using a Nanodrop spectrophotometer before addition to reaction mix. The reaction was terminated by incubation at 85 °C for 5 min and then incubation on ice. Following termination the reaction mixture was collected by centrifugation, and 1 μl of RNase H was added to digest total DNA for 20 min at 37 °C and then stored at −20 °C.
qRT-PCR of PFHR-9 cDNA
Half of each (21 μl) cDNA reaction from the previous step was combined with 2× iQ SYBR Green Supermix to a volume of 200 μl. Next this mixture was combined with forward and reverse primer sets to a final concentration of 1× iQ SYBR Green Supermix, and 200 nm of each forward and reverse primer and a final volume of 16 μl was added to each well of 96-well iCycler iQ® PCR plates with three replicates for each combination of cDNA and primers, as well as primer-only controls against primer self-amplification. The samples were analyzed on a Bio-Rad iQ5 multicolor RT-PCR detection system and associated manufacturer's software. Amplification and analysis of cDNAs used the following temperature sequence: the samples were denatured at 95 °C for 10 min and then 40 two-step cycles alternated 95 °C for 15 s and 60 °C for 1 min, and real-time fluorescence acquisition occurred during the 60 °C steps. Following this analysis, the samples were heated for 95 °C to fully denature for 1 min and then cooled to 55 °C for 1 min. Melting curves were then acquired by incrementing temperature 0.5 °C every 10 s from 55 to 95 °C, whereas fluorescence was monitored in real time.
The primers used in these reactions were as follows: P3H1 forward, CCCCCAGCCTACACGTTCCG; P3H1 reverse, TGCCCCCTGGTGACAGCCTTC; P3H2 forward, ATGCTAAAACCGTGACTGCC; P3H2 reverse, CGCTCCAGTTCTCGGTAAAG; P3H3 forward, AGAGGGCCTACTACCAGTTG; P3H3 reverse, GTGTTGGGTTTGCCACAAAG; CRTAP forward, CGCCTCCCTTCCTAGGCCTGC; CRTAP reverse,GCGGAAGCTGTAGCGCTCGTA; SC65 forward, GGTCGCCGGCTGCTCCGTAA; SC65 reverse, GCGCCAGCTTTCGCCTTCGT; GAPDH forward, CCACCCAGAAGACTGTGGAT; and GAPDH reverse, TTCAGCTCTGGGATGACCTT. The primer products had the following sizes: P3H1, 194 bp; P3H2, 163 bp; P3H3, 78 bp; CRTAP, 168 bp; SC65, 200 bp; and GAPDH, 127 bp. Primers had been synthesized as DNA oligonucleotides purified by standard desalting and diluted to 100 μm by Integrated DNA Technologies into 10 mm Tris-HCl, pH 8.0, 1 mm EDTA.
Disruption of LEPREL1 gene in PFHR-9 cells
DNA oligonucleotides were annealed and ligated into GeneArt® CRISPR nuclease vector with OFP Reporter according to the manufacturer's instructions. The oligonucleotide sequences used for ligation and targeting of LEPREL1 exon 4 were TGGAGGTAGTCGTAATGCAAGTTTT and TTGCATTACGACTACCTCCACGGTG. Ligated vector was used to transform One Shot® TOP10 chemically competent Escherichia coli using heat shock using manufacturer's instructions, and the cultures were prepared in LB containing 50 μg/ml ampicillin from individual clones selected from LB agar + 50 μg/ml ampicillin plates. The plasmid was isolated from 100-ml cultures using a Qiagen Plasmid Plus Maxi kit using the manufacturer's instructions. Vector was incubated with Lipofectamine 2000 and Opti-MEM® using manufacturer's instructions. PFHR-9 cells were grown to confluence in T-175 flasks and transfected with 25 μg of vector in Lipofectamine 2000/Opti-MEM solution. The cells were incubated overnight, washed repeatedly with PBS, trypsinized, collected by centrifugation, and washed repeatedly with PBS + 1 mm EDTA + 25 mm HEPES + 1% FBS. The cells were resuspended in the previous buffer to a density of 107 cells/ml. The cells were stained with 1 μl/ml Live/Dead® Aqua Fixable cell stain that had been freshly reconstituted in DMSO for at least an hour before FACS sorting. Resuspended cells were filtered using a sterile 35-μm mesh-size FalconTM cell-strainer cap and loaded onto a BD InfluxTM cell sorter. Forward and side-scatter and doublet discriminator gating were used to isolate monodisperse cell populations. Selection of a viable and CRISPR/Cas9 active cell population was guided by Live/Dead® Aqua Fixable cell stain and OFP fluorescence, respectively, with a vehicle-only (Opti-MEM + Lipofectamine transfected) PFHR-9 control for OFP detection of autofluorescence. OFP fluorescence was excited with a 488-nm solid-state laser and observed at 670 nm through a 670/30-nm bandpass filter. Live/Dead Aqua fluorescence was excited with a 405-nm DPSS laser, and emission was observed at 520 nm through a 520/35-nm bandpass filter. All instrumental operations of flow cytometry were performed by technicians of the Oregon Health & Science University Flow Cytometry Shared Resource. The clones were sorted individually into tissue-culture treated 96-well plates containing DMEM + 10% FBS and subsequently passaged into 24-well and then 6-well plates upon attaining confluence.
Screening and verification of clones with disruption of LEPREL1
NP-40 lysates of PFHR-9 were immunoblotted to detect loss of P3H2, the protein product of LEPREL1 gene. Briefly, trypsinized and then PBS-washed cells were lysed by resuspended in NP-40 and passed repeatedly through an 18-gauge needle. Clarified lysate was reduced by the addition of BOLT® LDS sample buffer containing BOLT® sample reducing agent and separated on 4–12% BOLTTM Bis-Tris gels using the manufacturer's instructions. MagicMarkTM XP Western Protein Standard was used as a protein size ladder. The gel was blotted onto nitrocellulose membrane for 16 h at 10 V in 4.5 mm sodium tetraborate decahydrate at 4 °C. The membrane was blocked with 5% (w/v) dry skim milk in TBS for 1 h at room temperature and then incubated overnight at 4 °C with rabbit anti-mouse P3H2 (Proteintech 15723-1-AP) with 1:500 dilution in TBS with 0.5% dry skim milk. The membrane was then repeatedly rinsed in room temperature TBS and subsequently incubated for 1 h at room temperature in goat anti-rabbit HRP conjugate (Sigma–Aldrich A0545) with 1:1000 dilution in TBS with 0.5% dry skim milk. The membrane was again repeatedly rinsed in TBS, then blotted nearly dry, and incubated with a PierceTM ECL Western blotting substrate kit according to the manufacturer's instructions and exposed for 1–30 min with Denville Scientific HyBlot ES® high sensitivity autoradiography film in Kodak X-Omatic regular intensifying screen in a dark room and then developed in a Kodak X_OMAT 2000A Film Processor. Under these conditions, it was possible to observe a 95-kDa band present in both WT PFHR-9 and CRISPR/Cas9 transfected clones, as well as a 80-kDa band present in the WT cells but absent from some of the clones. Clones lacking the 80-kDa band and with decreased LEPREL1 transcription (see above qRT-PCR procedure) were sequenced at exon 4 to verify disruption of exon 4. Briefly, cDNAs of exon 4 were generated by first-strand synthesis using oligo(dT) as a template, then amplified, and quantitated using the above qRT-PCR procedure with exon 4 spanning primers with a concentration of 250 nm. The primers used in the amplification were CGTGCTCTATGTGAGGGGGC and GGCCCTCTGCAGCTAACTT and were purified by Integrated DNA Technologies and then purified by standard desalting. The cDNAs produced by this amplification were run on a 1.2% TAE agarose gel, and then cDNAs were purified from reaction mixture using a QIAquick PCR purification kit, using the manufacturer's instructions. Sequencing was accomplished by Sequetech, using 50 ng of purified cDNA and 10 μm sequencing primer. The sequencing primers used here were CGTGCTCTATGTGAGGGGGC and GGCCCTCTGCAGCTAACTT (identical to amplification primers), as well as TAAATCCTCCCGGGCCTCAA.
Purification of type IV collagen
Type IV collagen was purified from PFHR-9 medium using a previously published procedure with several modifications (14). Here, equal volumes of media were harvested from confluent cells grown in T-175 flasks and CELLBIND 850-cm2 roller bottles rather than being grown in dishes. Ascorbic acid phosphate solutions (Wako, 013-12061, 50 μg/ml) were made fresh and added to the cells every 2 days when medium was collected rather than adding ascorbic acid daily. The purification step using a 1-ml Mono S 5/5 column instead used a 1-ml HiTrap SP HP Sephorose column (GE Healthcare 17-1151-010). Additionally, the final gel filtration purification step using Superose 6 medium was omitted. All other procedures were performed similarly to the published protocol.
Digestion of type IV collagen
To digest type IV collagen in trypsin, lyophilized samples were resuspended in equal volume as the pre-lyophilisate of 1 m Tris-HCl, pH 8.0, with 20 ng/μl trypsin and digested for 18 h at 37 °C. To digest type IV collagen in chymotrypsin, lyophilized samples were resuspended in equal volume as the pre-lyophilasate of 100 mm Tris-HCl, pH 8.0, 10 mm CaCl2 with 20 ng/μl chymotrypsin and digested for 18 h at room temperature. To digest type IV collagen in CNBr, sample was dried from acetic acid (50 mm) by centrifugal evaporation and resuspended in 70% formic acid containing CNBr having a molar ratio of 1:100 methionine residues:CNBr molecules, sealed, incubated overnight, and then lyophilized to remove the reaction mixture.
MS analysis by Q-TOF spectrometer
Identification of peptides produced by proteolytic digest was performed on a Q-TOF Micromass spectrometer (Waters, Billerica, MA) equipped with an electrospray ionization source. The data were collected with the MassLynx (version 4.1) data acquisition software (Waters) and processed using Mascot Distiller (Matrix Software, London, UK). HPLC was performed with nanoACQUITY (Waters) system using a 75-μm × 100-mm 3-μm Atlantis dC18 column as the analytical column and a 180-μm × 20-mm 5-μm Symmetry C18 column as the trapping column. Chromatographic mobile phases consisted of solvents A (0.1% formic acid and 99.9% water (v/v)) and B (0.1% formic acid and 99.9% acetonitrile (v/v)). Peptide samples were loaded onto the trapping column and equilibrated 4 min in 99% solvent A followed by a 180-min gradient to 60% solvent A, 40% solvent B at a constant flow rate of 0.8 μl/min. Analysis was performed in survey scan mode. Tryptic peptides were identified from MS/MS spectra by a Mascot search against the National Center for Biotechnology Information nr database (peptide tolerance 1.0 Da, MS/MS tolerance 1.0 Da).
MS analysis by LTQ Velos
Protein digests were separated using LC with an Agilent 1100 series capillary LC system (Agilent Technologies), then delivered to an LTQ Velos dual pressure linear ion trap mass spectrometer (Thermo Fisher) using electrospray ionization with an Ion Max source (Thermo Fisher) fitted with a 34-gauge metal needle (Thermo Fisher) and 2.7-kV source voltage. Xcalibur version 2.0 was used to control the system. The samples were applied at 20 μl/min to a trap cartridge (Michrom BioResources) and then switched onto a 0.5 × 250 mm Zorbax SB-C18 column with 5-mm particles (Agilent Technologies) using a mobile phase containing 0.1% formic acid, 7–30% acetonitrile gradient over 60 min, and 10 μl/min flow rate. A normalized collision energy of 35 was used. Data-dependent collection of MS/MS spectra used the dynamic exclusion feature of the instrument's control software (repeat count equal to 1, exclusion list size of 50, exclusion duration of 30 s, and exclusion mass width of −1 to +4) to obtain MS/MS spectra of the three most abundant parent ions (minimum signal of 5000) following each survey scan from m/z 400 to 2000. The tune file was configured with no averaging of microscans, a maximum inject time of 200 ms, and automatic gain control targets of 3 × 104 in MS1 mode and 1 × 104 in MS2 mode.
Amino acid analysis of type IV collagen
Acid hydrolysis was performed in six 50-mm Pyrex culture tubes placed in Pico Tag reaction vessels fitted with a sealable cap (Eldex Laboratories, Inc., Napa, CA). Samples were placed in culture tubes, dried in a SpeedVac (GMI, Inc. Albertsville, MN), and then placed into a reaction vessel that contained 250 ml of 6 m HCl (Pierce) containing 2% phenol (Sigma–Aldrich). The vessel was then purged with argon gas and evacuated using the automated evacuation work station Eldex hydrolysis/derivatization work station (Eldex Laboratories, Inc.). Closing the valve on the Pico Tag cap maintained the vacuum during hydrolysis at 105 °C for 24 h. The hydrolyzed samples were then dried in a Savant SpeedVac. The dried samples were dissolved in 100 ml of 0.02 m HCl containing an internal standard (100 μm norvaline; Sigma). Appropriate further dilutions were made using the same dilution solvent for concentrated samples. Analysis was performed by ion-exchange chromatography with postcolumn ninhydrin derivatization and visible detection (440 nm/570 nm) with a Hitachi L-8800A amino acid analyzer (Hitachi High Technologies America, Inc., San Jose, CA) running the EZChrom Elite software (Scientific Software, Inc., Pleasanton, CA).
Coupling of type IV collagen to CM5 chip
Type IV collagen was coupled to Biacore CM5 chips using the manufacturer's instructions for amine coupling. The surface was activated by exposure to freshly made 0.2 m (1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide, 50 mm N-hydroxysuccinimide. Following activation, one channel was coupled with collagen IV in 50 mm sodium acetate, pH 5.0, and the other was blocked using 1 m ethanolamine HCl, pH 8.5, as a protein binding control. The amount of coupled collagen differed between chips (2000 RU for WT type IV collagen versus 2600 RU for P3H2 KO type IV collagen) and was used to normalize the overall response.
Surface plasmon resonance of type IV collagen-binding proteins
In all cases, identical running conditions were used for both WT PFHR-9 type IV collagen and P3H2 KO type IV collagen on a Biacore X (GE Healthcare) instrument. BIACORE X Control Software Version 2.3 was used for all instrument control operations, and data were analyzed using BIAevaluation version 4.1. GP6–CBD was purified as described below and dialyzed into 0.01 m Hepes buffer, pH 7.4, containing 0.15 m NaCl, 3 mm EDTA, and 0.005 % (v/v) P20 (HBS-EP) buffer. The same buffer was filtered using a 0.22-μm filter degassed and reused as running buffer for binding studies. The temperature was set to 25 °C, flow rate was set to 10 μl/min, and 5 μl of GP6–CBD sample was injected, with a 30-s dissociation step. Purified human nidogen-1 and nidogen-2 were a kind gift of Dr. Takako Sakaki and were prepared as described previously (40). Both samples were dialyzed into HBS-EP, which was filtered with a 0.22-μm filter, degassed, and reused as running buffer for binding studies. For nidogen-1, 10 μl were injected at 10 μl/min with a 90-s dissociation period followed by a standard wash step. For nidogen-2, 30 μl were injected at 10 μl/min with a 270-s dissociation period followed by a standard wash step. Purified BM40-ΔI-αC sample was a kind gift of Dr. Takako Sakaki. BM40-ΔI-αC was dialyzed into HBS-P + 5 mm CaCl2, which was filtered with a 0.22-μm filter, degassed, and reused as running buffer for binding studies. For this sample, 50 μl was injected at 5 μl/min with a 600-s dissociation period followed by a standard wash step.
Purification of GP6–CBD
Plasmid pET23_Trx-GP6–CBD (ampicillin resistance) was transduced into OneShot® BL21(DE3) chemically competent E. coli cells (ThermoFisher, C600003) using the manufacturer's instructions, plated on LB-agar plates with 50 μg/ml ampicillin sodium salt, and grown at 37 °C overnight. A single colony was selected from the plate and grown for 16 h in 10 ml of LB medium with ampicillin at 37 °C. This growth culture was diluted 1:100 in LB with ampicillin, shaken for 5 h at 37 °C, and then induced using 1 mm isopropyl β-d-thiogalactopyranoside overnight at 25 °C with shaking. The cells were collected by centrifugation and lysed by sonication in chilled B-PER buffer. Cell debris was collected at 7,000 rpm for 30 min on an SS-34 rotor, and the supernatant was discarded. The pellet was resuspended in unfolding buffer consisting of 0.1 m Tris-HCl, pH 8.2, 10 mm DTT, 5 mm EDTA, 8 m guanidinium HCl. The resuspended sample was centrifuged for 30 min at 15,000 rpm in an SS-34 rotor, and this pellet was discarded. The unfolded supernatant was passed through a 0.22-μm filter and diluted 1:10 into refolding buffer consisting of 50 mm Tris-HCl, 50 mm sodium phosphate, 5 mm EDTA, 10 mm reduced GSH, 1 mm oxidized GSH, 1 m arginine, pH 8.8, and incubated overnight with gentle stirring at room temperature. The sample was then dialyzed into nickel column loading buffer (50 mm sodium phosphate, pH 7.0, 500 mm NaCl) and loaded onto a nickel-bound HiTrapTM chelating column, and the sample that eluted in loading buffer plus 500 mm imidazole was collected. The sample was cleaved by dialysis into thrombin cleavage buffer (50 mm Tris-HCl, 150 mm NaCl, pH 8.0) and then overnight incubation with 1 unit/ml high purity bovine thrombin (MP Biomedicals, 02154163). The cleaved sample was loaded onto a nickel-bound HiTrapTM chelating column that had been equilibrated with thrombin cleavage buffer, and the sample that flowed through was collected. This sample was dialyzed into 20 mm HEPES pH 7.0 buffer, and loaded onto an HiTrapTM SP HP column equilibrated with the same buffer. The sample eluting from the column at equilibration buffer + 200 mm NaCl was collected and stored at 4 °C. The amino acid sequence of the Trx-GP6–CBD construct is: MGHHHHHHGSGMSDKIIHLTDDSFDTDVLKADGAILVDFWAEWCGPCKMIAPILDEIADEYQGKLTVAKLNIDQNPGTAPKYGIRGIPTLLLFKNGEVAATKVGALSKGQLKEFLDANLAGSGSGLVPR↓GSGPLPKPSLQALPSSLVPLEKPVTLRCQGPPGVDLYRLEKLSSSRYQDQAVLFIPAMKRSLAGRYRCSYQNGSLWSLPSDQLELVATGVFAKPSLSAQPGPAVSSGGDVTLQCQTRYGFDQFALYKEGDPAPYKNPERWYRASFPIITVTAAHSGTYRCYSFSSRDPYLWSAPSDPLELVVTGAS. The thrombin cleavage site is indicated by a downward arrow. The final purified product, GP6–CBD, is the portion of the protein C-terminal to the thrombin cleavage site and has a mass of 20.1 kDa.
Author contributions
N. T. M., K. D. Z., and H. P. B. data curation; N. T. M. and K. D. Z. formal analysis; N. T. M. and K. D. Z. investigation; N. T. M. and E. N. P. methodology; N. T. M., K. D. Z., and E. N. P. writing-original draft; N. T. M., K. D. Z., E. N. P., and H. P. B. writing-review and editing; E. N. P. and H. P. B. conceptualization; H. P. B. resources; H. P. B. funding acquisition; H. P. B. validation; H. P. B. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Takako Sakaki for the gift of recombinant proteins and Dr. Sergey Boudko for the gift of the Trx-GP6–CBD vector. We thank the Oregon Health and Science University Research Cytogenetics Core for the karyotyping of PFHR-9 and the Oregon Health and Science University Flow Cytometry Shared Resource for FACS analysis and sorting of CRISPR/Cas9 clones. We also thank the Analytical Core Facility of Shriners Hospitals for Children in Portland for amino acid analysis and MS and the Oregon Health & Science University Proteomics shared resource for additional MS.
This work was supported by Shriners Hospital for Children Grants 85100 and 85500 (to H. P. B.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains text, Table S1, and Figs. S1–S40.
- 3-Hyp
- 3(S)-hydroxy-l-proline
- 4-Hyp
- 4(R)-hydroxy-l-proline
- GP6
- glycoprotein VI
- P3H2
- prolyl-3-hydroxylase 2
- qRT-PCR
- quantitative RT-PCR
- CBD
- collagen-binding domain
- P4HA2
- prolyl-4-hydroxylase
- CRTAP
- cartilage associated protein
- GPP
- Gly-Pro-Pro
- P3H1
- prolyl-3-hydroxylase 1
- P3H3
- prolyl-3-hydroxylase 3
- SC65
- endoplasmic reticulum protein 65
- RU
- resonance units.
References
- 1. Cabral W. A., Chang W., Barnes A. M., Weis M., Scott M. A., Leikin S., Makareeva E., Kuznetsova N. V., Rosenbaum K. N., Tifft C. J., Bulas D. I., Kozma C., Smith P. A., Eyre D. R., and Marini J. C. (2007) Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat. Genet. 39, 359–365 10.1038/ng1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gorres K. L., and Raines R. T. (2010) Prolyl 4-hydroxylase. Crit. Rev. Biochem. Mol. Biol. 45, 106–124 10.3109/10409231003627991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mizuno K., Peyton D. H., Hayashi T., Engel J., and Bächinger H. P. (2008) Effect of the -Gly-3(S)-hydroxyprolyl-4(R)-hydroxyprolyl-tripeptide unit on the stability of collagen model peptides. FEBS J. 275, 5830–5840 10.1111/j.1742-4658.2008.06704.x [DOI] [PubMed] [Google Scholar]
- 4. Morello R., Bertin T. K., Chen Y., Hicks J., Tonachini L., Monticone M., Castagnola P., Rauch F., Glorieux F. H., Vranka J., Bächinger H. P., Pace J. M., Schwarze U., Byers P. H., Weis M., et al. (2006) CRTAP is required for prolyl 3-hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell 127, 291–304 10.1016/j.cell.2006.08.039 [DOI] [PubMed] [Google Scholar]
- 5. van Dijk F. S., Nesbitt I. M., Zwikstra E. H., Nikkels P. G., Piersma S. R., Fratantoni S. A., Jimenez C. R., Huizer M., Morsman A. C., Cobben J. M., van Roij M. H., Elting M. W., Verbeke J. I., Wijnaendts L. C., Shaw N. J., et al. (2009) PPIB mutations cause severe osteogenesis imperfecta. Am. J. Hum. Genet. 85, 521–527 10.1016/j.ajhg.2009.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mordechai S., Gradstein L., Pasanen A., Ofir R., El Amour K., Levy J., Belfair N., Lifshitz T., Joshua S., Narkis G., Elbedour K., Myllyharju J., and Birk O. S. (2011) High myopia caused by a mutation in LEPREL1, encoding prolyl 3-hydroxylase 2. Am. J. Hum. Genet. 89, 438–445 10.1016/j.ajhg.2011.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo H., Tong P., Peng Y., Wang T., Liu Y., Chen J., Li Y., Tian Q., Hu Y., Zheng Y., Xiao L., Xiong W., Pan Q., Hu Z., and Xia K. (2014) Homozygous loss-of-function mutation of the LEPREL1 gene causes severe non-syndromic high myopia with early-onset cataract. Clin. Genet. 86, 575–579 10.1111/cge.12309 [DOI] [PubMed] [Google Scholar]
- 8. Hudson D. M., Joeng K. S., Werther R., Rajagopal A., Weis M., Lee B. H., and Eyre D. R. (2015) Post-translationally abnormal collagens of prolyl 3-hydroxylase-2 null mice offer a pathobiological mechanism for the high myopia linked to human LEPREL1 mutations. J. Biol. Chem. 290, 8613–8622 10.1074/jbc.M114.634915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pokidysheva E., Boudko S., Vranka J., Zientek K., Maddox K., Moser M., Fässler R., Ware J., and Bächinger H. P. (2014) Biological role of prolyl 3-hydroxylation in type IV collagen. Proc. Natl. Acad. Sci. U.S.A. 111, 161–166 10.1073/pnas.1307597111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smethurst P. A., Onley D. J., Jarvis G. E., O'Connor M. N., Knight C. G., Herr A. B., Ouwehand W. H., and Farndale R. W. (2007) Structural basis for the platelet-collagen interaction: the smallest motif within collagen that recognizes and activates platelet glycoprotein VI contains two glycine-proline-hydroxyproline triplets. J. Biol. Chem. 282, 1296–1304 10.1074/jbc.M606479200 [DOI] [PubMed] [Google Scholar]
- 11. Parkin J. D., San Antonio J. D., Pedchenko V., Hudson B., Jensen S. T., and Savige J. (2011) Mapping structural landmarks, ligand binding sites, and missense mutations to the collagen IV heterotrimers predicts major functional domains, novel interactions, and variation in phenotypes in inherited diseases affecting basement membranes. Hum. Mutat. 32, 127–143 10.1002/humu.21401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kurban G., Duplan E., Ramlal N., Hudon V., Sado Y., Ninomiya Y., and Pause A. (2008) Collagen matrix assembly is driven by the interaction of von Hippel-Lindau tumor suppressor protein with hydroxylated collagen IV α2. Oncogene 27, 1004–1012 10.1038/sj.onc.1210709 [DOI] [PubMed] [Google Scholar]
- 13. Hon W. C., Wilson M. I., Harlos K., Claridge T. D., Schofield C. J., Pugh C. W., Maxwell P. H., Ratcliffe P. J., Stuart D. I., and Jones E. Y. (2002) Structural basis for the recognition of hydroxyproline in HIF-1α by pVHL. Nature 417, 975–978 10.1038/nature00767 [DOI] [PubMed] [Google Scholar]
- 14. Davis J. M., Boswell B. A., and Bächinger H. P. (1989) Thermal stability and folding of type IV procollagen and effect of peptidyl-prolyl cis-trans-isomerase on the folding of the triple helix. J. Biol. Chem. 264, 8956–8962 [PubMed] [Google Scholar]
- 15. Basak T., Vega-Montoto L., Zimmerman L. J., Tabb D. L., Hudson B. G., and Vanacore R. M. (2016) Comprehensive characterization of glycosylation and hydroxylation of basement membrane collagen IV by high-resolution mass spectrometry. J. Proteome Res. 15, 245–258 10.1021/acs.jproteome.5b00767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tiainen P., Pasanen A., Sormunen R., and Myllyharju J. (2008) Characterization of recombinant human prolyl 3-hydroxylase isoenzyme 2, an enzyme modifying the basement membrane collagen IV. J. Biol. Chem. 283, 19432–19439 10.1074/jbc.M802973200 [DOI] [PubMed] [Google Scholar]
- 17. Risteli J., Tryggvason K., and Kivirikko K. I. (1977) Prolyl 3-hydroxylase: partial characterization of the enzyme from rat kidney cortex. Eur. J. Biochem. 73, 485–492 10.1111/j.1432-1033.1977.tb11341.x [DOI] [PubMed] [Google Scholar]
- 18. Tryggvason K., Risteli J., and Kivirikko K. I. (1976) Separation of prolyl 3-hydroxylase and 4-hydroxylase activities and the 4-hydroxyproline requirement for synthesis of 3-hydroxyproline. Biochem. Biophys. Res. Commun. 76, 275–281 [DOI] [PubMed] [Google Scholar]
- 19. Aumailley M., Wiedemann H., Mann K., and Timpl R. (1989) Binding of nidogen and the laminin-nidogen complex to basement membrane collagen type IV. Eur J Biochem 184, 241–248 10.1111/j.1432-1033.1989.tb15013.x [DOI] [PubMed] [Google Scholar]
- 20. Dong L., Chen Y., Lewis M., Hsieh J. C., Reing J., Chaillet J. R., Howell C. Y., Melhem M., Inoue S., Kuszak J. R., DeGeest K., and Chung A. E. (2002) Neurologic defects and selective disruption of basement membranes in mice lacking entactin-1/nidogen-1. Lab. Invest. 82, 1617–1630 10.1097/01.LAB.0000042240.52093.0F [DOI] [PubMed] [Google Scholar]
- 21. Marneros A. G., and Olsen B. R. (2005) Physiological role of collagen XVIII and endostatin. FASEB J. 19, 716–728 10.1096/fj.04-2134rev [DOI] [PubMed] [Google Scholar]
- 22. Gilmour D. T., Lyon G. J., Carlton M. B., Sanes J. R., Cunningham J. M., Anderson J. R., Hogan B. L., Evans M. J., and Colledge W. H. (1998) Mice deficient for the secreted glycoprotein SPARC/osteonectin/BM40 develop normally but show severe age-onset cataract formation and disruption of the lens. EMBO J. 17, 1860–1870 10.1093/emboj/17.7.1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Behrens D. T., Villone D., Koch M., Brunner G., Sorokin L., Robenek H., Bruckner-Tuderman L., Bruckner P., and Hansen U. (2012) The epidermal basement membrane is a composite of separate laminin- or collagen IV-containing networks connected by aggregated perlecan, but not by nidogens. J. Biol. Chem. 287, 18700–18709 10.1074/jbc.M111.336073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schuppan D., Glanville R. W., Timpl R., Dixit S. N., and Kang A. H. (1984) Sequence comparison of pepsin-resistant segments of basement-membrane collagen α1(IV) chains from bovine lens capsule and mouse tumour. Biochem. J. 220, 227–233 10.1042/bj2200227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vranka J., Stadler H. S., and Bächinger H. P. (2009) Expression of prolyl 3-hydroxylase genes in embryonic and adult mouse tissues. Cell Struct. Funct. 34, 97–104 10.1247/csf.09002 [DOI] [PubMed] [Google Scholar]
- 26. Kang S. H., and Kramer J. M. (2000) Nidogen is nonessential and not required for normal type IV collagen localization in Caenorhabditis elegans. Mol. Biol. Cell 11, 3911–3923 10.1091/mbc.11.11.3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Capellini T. D., Dunn M. P., Passamaneck Y. J., Selleri L., and Di Gregorio A. (2008) Conservation of notochord gene expression across chordates: insights from the Leprecan gene family. Genesis 46, 683–696 10.1002/dvg.20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weis M. A., Hudson D. M., Kim L., Scott M., Wu J. J., and Eyre D. R. (2010) Location of 3-hydroxyproline residues in collagen types I, II, III and V/XI implies a role in fibril supramolecular assembly. J. Biol. Chem. 285, 2580–2590 10.1074/jbc.M109.068726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nieswandt B., and Watson S. P. (2003) Platelet-collagen interaction: is GPVI the central receptor? Blood 102, 449–461 10.1182/blood-2002-12-3882 [DOI] [PubMed] [Google Scholar]
- 30. Zahid M., Mangin P., Loyau S., Hechler B., Billiald P., Gachet C., and Jandrot-Perrus M. (2012) The future of glycoprotein VI as an antithrombotic target. J. Thromb. Haemost. 10, 2418–2427 10.1111/jth.12009 [DOI] [PubMed] [Google Scholar]
- 31. Watkins N. A., O'Connor M. N., Rankin A., Jennings N., Wilson E., Harmer I. J., Davies L., Smethurst P. A., Dudbridge F., Farndale R. W., and Ouwehand W. H. (2006) Definition of novel GP6 polymorphisms and major difference in haplotype frequencies between populations by a combination of in-depth exon resequencing and genotyping with tag single nucleotide polymorphisms. J. Thromb. Haemost. 4, 1197–1205 10.1111/j.1538-7836.2006.01937.x [DOI] [PubMed] [Google Scholar]
- 32. Holtkötter O., Nieswandt B., Smyth N., Müller W., Hafner M., Schulte V., Krieg T., and Eckes B. (2002) Integrin α2-deficient mice develop normally, are fertile, but display partially defective platelet interaction with collagen. J. Biol. Chem. 277, 10789–10794 10.1074/jbc.M112307200 [DOI] [PubMed] [Google Scholar]
- 33. Kern A., Eble J., Golbik R., and Kühn K. (1993) Interaction of type IV collagen with the isolated integrins α1β1 and α2β1. Eur J Biochem 215, 151–159 10.1111/j.1432-1033.1993.tb18017.x [DOI] [PubMed] [Google Scholar]
- 34. Lebbink R. J., de Ruiter T., Verbrugge A., Bril W. S., and Meyaard L. (2004) The mouse homologue of the leukocyte-associated Ig-like receptor-1 is an inhibitory receptor that recruits Src homology region 2-containing protein tyrosine phosphatase (SHP)-2, but not SHP-1. J. Immunol. 172, 5535–5543 10.4049/jimmunol.172.9.5535 [DOI] [PubMed] [Google Scholar]
- 35. Lebbink R. J., de Ruiter T., Adelmeijer J., Brenkman A. B., van Helvoort J. M., Koch M., Farndale R. W., Lisman T., Sonnenberg A., Lenting P. J., and Meyaard L. (2006) Collagens are functional, high affinity ligands for the inhibitory immune receptor LAIR-1. J. Exp. Med. 203, 1419–1425 10.1084/jem.20052554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meyaard L., Adema G. J., Chang C., Woollatt E., Sutherland G. R., Lanier L. L., and Phillips J. H. (1997) LAIR-1, a novel inhibitory receptor expressed on human mononuclear leukocytes. Immunity 7, 283–290 10.1016/S1074-7613(00)80530-0 [DOI] [PubMed] [Google Scholar]
- 37. Vranka J. A., Sakai L. Y., and Bächinger H. P. (2004) Prolyl 3-hydroxylase 1, enzyme characterization and identification of a novel family of enzymes. J. Biol. Chem. 279, 23615–23621 10.1074/jbc.M312807200 [DOI] [PubMed] [Google Scholar]
- 38. Guo H., Tong P., Liu Y., Xia L., Wang T., Tian Q., Li Y., Hu Y., Zheng Y., Jin X., Li Y., Xiong W., Tang B., Feng Y., Li J., et al. (2015) Mutations of P4HA2 encoding prolyl 4-hydroxylase 2 are associated with nonsyndromic high myopia. Genet. Med. 17, 300–306 10.1038/gim.2015.28 [DOI] [PubMed] [Google Scholar]
- 39. Aro E., Salo A. M., Khatri R., Finnilä M., Miinalainen I., Sormunen R., Pakkanen O., Holster T., Soininen R., Prein C., Clausen-Schaumann H., Aszódi A., Tuukkanen J., Kivirikko K. I., Schipani E., et al. (2015) Severe extracellular matrix abnormalities and chondrodysplasia in mice lacking collagen prolyl 4-hydroxylase isoenzyme II in combination with a reduced amount of isoenzyme I. J. Biol. Chem. 290, 16964–16978 10.1074/jbc.M115.662635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kohfeldt E., Sasaki T., Göhring W., and Timpl R. (1998) Nidogen-2: a new basement membrane protein with diverse binding properties. J. Mol. Biol. 282, 99–109 10.1006/jmbi.1998.2004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.