Abstract
Gene expression in Gram-negative bacteria is regulated at many levels, including transcription initiation, RNA processing, RNA/RNA interactions, mRNA decay, and translational controls involving enzymes that alter translational efficiency. In this chapter we discuss the various enzymes that control transcription, translation and RNA stability through RNA processing and degradation. RNA processing is essential to generate functional RNAs, while degradation helps control the steady-state level of each individual transcript. For example, all the pre-tRNAs are transcribed with extra nucleotides at both their 5′ and 3′ termini, which are subsequently processed to produce mature tRNAs that can be aminoacylated. Similarly, rRNAs that are transcribed as part of a 30S polycistronic transcript, are matured to individual 16S, 23S and 5S rRNAs. Decay of mRNAs plays a key role in gene regulation through controlling the steady-state level of each transcript, which is essential for maintaining appropriate protein levels. In addition, degradation of both translated and non-translated RNAs recycles nucleotides to facilitate new RNA synthesis. To carry out all these reactions Gram-negative bacteria employ a large number of endonucleases, exonucleases, RNA helicases, and poly(A) polymerase as well as proteins that regulate the catalytic activity of particular ribonucleases. Under certain stress conditions an additional group of specialized endonucleases facilitate the cell’s ability to adapt and survive. Many of the enzymes, such as RNase E, RNase III, polynucleotide phosphorylase, RNase R, and poly(A) polymerase I participate in multiple RNA processing and decay pathways.
INTRODUCTION
All living organisms, including the Gram-negative bacteria, have two major classes of RNA molecules. Messenger RNAs (mRNAs) contain the information for the synthesis of the various proteins that are required for a living cell. The so-called non-translated RNAs that include transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), and small regulatory RNAs (sRNAs) provide the RNA components for ribosome assembly, protein synthesis and the regulation of mRNA functionality based on RNA/RNA interactions. The highly diverse functions that these RNAs perform within the cell are possible due to numerous enzymes that are involved in post-transcriptional RNA metabolism. However, many of these enzymes have overlapping activities. Besides the normal cellular complement of enzymes that carry out the above functions, there are ribonucleases which are specifically associated with particular stress conditions as part of toxin/antitoxin systems.
Based on our current knowledge of bacterial RNases, there are no dedicated RNases for either RNA degradation or processing. Many of the ribonucleases and RNA helicases participate in more than one pathway, which is visually described in Fig. 1. For example, endoribonuclease E (RNase E) is involved in almost all aspects of RNA metabolism. RNase III is very important for initiation of rRNA processing, but it also participates in mRNA decay and sRNA degradation. Similarly, although RNase P is essential for tRNA 5′ end maturation, it also participates in tRNA processing and mRNA decay. The ribonuclease activities of most of the exonucleases are highly redundant, since they can complement each other. Furthermore, as single-stranded RNAs can rapidly fold into more complex forms containing secondary and tertiary structures, RNA helicases play an important role in various aspects of post-transcriptional processing and decay. In this chapter ribonucleases and RNA helicases found in various Gram-negative bacteria are discussed in the context of their in vivo biological functions. The basic properties of all the ribonucleases and RNA helicases are outlined in Table 1. Readers are encouraged to review a complementary chapter in this book entitled “RNases and helicases in Gram-positive bacteria” by Durand and Condon.
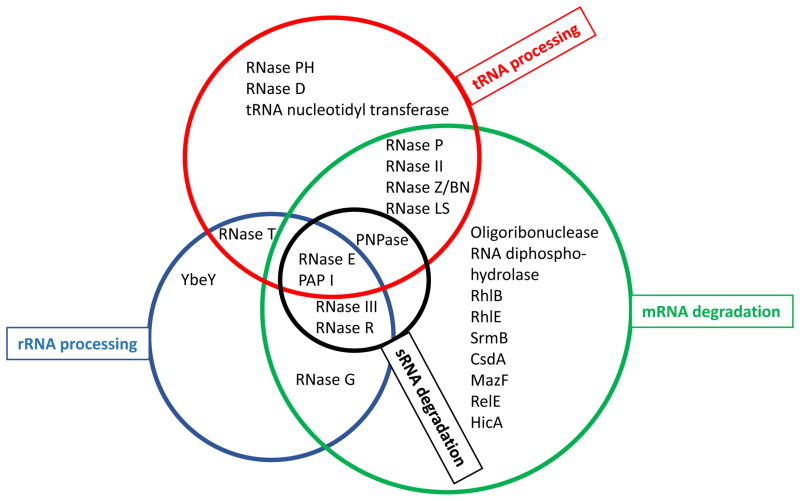
Fig. 1.
Venn diagram of ribonucleases in E. coli showing their involvement in the four major RNA metabolic pathways in Gram-negative bacteria. The participation of the various proteins is only included in pathways where it has been established that they play a significant role. In addition, it is possible that some proteins, such as YbeY, are involved in additional pathways.
Table 1.
Proteins involved in post-transcriptional RNA metabolism in E. coli.
| Name | Gene | Mode of Action | Substrate Preference | Functions* |
|---|---|---|---|---|
| RNase E | rne | Endonuclease | Single-stranded RNA | mRNA decay, tRNA processing, rRNA processing, sRNA degradation |
| RNase III | rnc | Endonuclease | Double-stranded RNA | rRNA processing, mRNA decay, sRNA degradation |
| RNase P | rnpA, rnpB | Endonuclease | Single-stranded RNA | tRNA processing, mRNA decay, rRNA processing |
| RNase G | rng | Endonuclease | Single-stranded RNA | rRNA processing, mRNA decay, tRNA processing |
| YbeY | ybeY | Endonuclease | Single-stranded RNA | rRNA processing |
| RNase Z/BN | rnz | Endonuclease/3′ → 5′ exonuclease | Single-stranded RNA | mRNA decay, tRNA processing |
| RNase LS | rnlA | Endonuclease | Single-stranded RNA | mRNA decay |
| RNase T | rnt | 3′ → 5′ exonuclease | Single-stranded RNA | tRNA processing, rRNA processing |
| RNase PH | rph | 3′ → 5′ exonuclease | Single-stranded RNA | tRNA processing |
| RNase D | rnd | 3′ → 5′ exonuclease | Single-stranded RNA | tRNA processing |
| PNPase | pnp | 3′ → 5′ exonuclease | Single-stranded RNA | mRNA decay, sRNA degradation, tRNA processing |
| RNase II | rnb | 3′ → 5′ exonuclease | Single-stranded RNA | mRNA decay, tRNA processing |
| RNase R | rnr | 3′ → 5′ exonuclease | Single-stranded RNA | rRNA processing, sRNA degradation |
| Oligoribonuclease | orn | 3′ → 5′ exonuclease | Single-stranded RNA | mRNA decay |
| Poly(A) polymerase | pcnB | polymerase | Single-stranded RNA | mRNA decay, tRNA maturation |
| Hfq | hfq | RNA binding protein | Single-stranded RNA | mRNA decay, polyadenylation, sRNA metabolism |
| tRNA nucleotidyl transferase | cca | polymerase | tRNAs | tRNA maturation |
| RNA diphosphohydrolase | rppH | phosphatase | 5′ triphosphate or 5′ diphosphate | mRNA decay |
| RhlB | rhlB | Helicase | Double-stranded RNA | mRNA decay |
| RhlE | rhlE | Helicase | Double-stranded RNA | mRNA decay |
| SrmB | srmB | Helicase | Double-stranded RNA | mRNA decay |
| CsdA | csdA | Helicase | Double-stranded RNA | mRNA decay under low temperature |
| DbpA | dbpA | Helicase | Double-stranded RNA | Ribosome biogenesis |
| Enolase | eno | Enolase | ? | RNA metabolism ? |
| MazF | mazF | mRNA interferase | Single-stranded RNA | mRNA decay, stress management |
| RelE | relE | mRNA interferase | Single-stranded RNA | mRNA decay, stress management |
| HicA | hicA | mRNA interferase | Single-stranded RNA | mRNA decay, stress management |
| Colicin E5 | tRNA targeting enzyme | Single-stranded RNA | Stress management | |
| Colicin D | tRNA targeting enzyme | Single-stranded RNA | Stress management | |
| Colicin E3 | tRNA targeting enzyme | Single-stranded RNA | Stress management | |
| Cas6A | cas6 | Endonuclease | Single-stranded RNA | Immune defense |
Listed in order of the importance of the enzyme
?: Role of enolase in RNA metabolism is yet to be defined.
GENERAL MESSENGER RNA DECAY
Initiation of mRNA decay by endonucleases
The decay of mRNAs plays a major role in the post-transcriptional regulation of gene expression, since it helps to control the steady-state level of each individual mRNA and in turn the protein level. As a result, considerable effort has been devoted to document the mechanisms of mRNA decay in E. coli, which still serves as the model organism for Gram-negative bacteria. Apirion was the first to propose a model for mRNA decay in 1973 (1), which involved a combination of endoribonucleases and exoribonucleases. However, at that time the evidence supporting his hypothesis was not particularly strong. Subsequently, RNase E was initially identified based on its role in rRNA processing as discussed later in this chapter (2). At about the same time as RNase E was discovered, the ams-1 allele (altered mRNA stability) was isolated and shown to affect cell viability. Strains carrying the ams-1 mutation demonstrated an increase in the half-life of bulk mRNA at the nonpermissive temperature (3, 4). Subsequently, the ams and rne loci were shown to encoded the same protein, which is now called RNase E (5).
RNase E, an essential enzyme in E. coli, is one of its largest proteins. It is divided into an N-terminal catalytic region and a C-terminal scaffold region. Experiments have shown that the catalytic region of RNase E is located in the first 500 amino acids of the protein (6) and that it is associated with the inner membrane of the cell through a short amino sequence that is immediately downstream of the catalytic region (7). A very important advance in the analysis of mRNA decay pathways was the finding that RNase E was associated with several other enzymes involved in RNA degradation. First it was shown that RNase E forms a complex with polynucleotide phosphorylase (PNPase), a 3′ → 5′ exonuclease (8). Subsequently, it was determined that the RhlB RNA helicase and the glycolytic enzyme enolase were also included in this multiprotein complex, which was called the “degradosome” (8, 9). However, mRNA decay is not significantly affected in the absence of either degradosome assembly and membrane attachment, as shown by deletions of the carboxy terminal region that lack either the degradosome scaffold region (10) or both the degradosome scaffold region and the membrane attachment site (11). The degradosome is discussed in more detail in the Chapter by Bandyra and Luisi.
It appears that RNase E can recognize its substrates in more than one way. Mackie demonstrated that the enzyme was inhibited by the triphosphate moiety found at the 5′ terminus of RNA transcripts (12). Thus, RNase E processing is significantly enhanced by the removal of the pyrophosphate from the 5′ terminal triphosphate of many primary transcripts. While conversion of the 5′ triphosphate to a 5′ monophosphate was believed to be catalyzed by the RNA pyrophosphohydrolase (RppH) encoded by rppH (13), a recent report suggests that an unidentified enzyme converts the triphosphate to a diphosphate, which is the substrate of choice for the RppH protein (14). Although RNase E prefers to bind to substrates containing a 5′ monophosphate, a number of experiments have demonstrated that it can also cleave both mRNAs and tRNAs using a direct entry mechanism (15–19).
Several laboratories attempted to determine if there was sequence specificity associated with RNase E cleavages by comparing the sequences of known cleavage sites (20–22). These studies revealed that the enzyme prefers to cleave RNA in single-stranded A/U rich regions. Further work has shown that the cleavage sites are usually found either upstream or downstream of secondary structures. A recent RNAseq study suggests a minimal 5 nt RNase E consensus cleavage site as “RN↓WUU” (with R as G/A, W as A/U and N as any nucleotide) with a strong preference for uridine at +2 position (23).
High density tiling arrays as well as RNAseq experiments have demonstrated that RNase E is responsible for the initiation of mRNA decay for over 50% of all the transcripts generated during exponential growth in E. coli (18, 24). However, besides RNase E, there are a significant number of other endonucleases that initiate the decay of mRNAs as minor players. For example, RNase III, which will be discussed in more detail later in the context of rRNA processing, has been shown to affect the steady-state levels of up to 10% of the mRNAs in exponentially growing cells of E. coli resulting in either destabilization or stabilization of the transcripts (24, 25). RNase P, which is a ribozyme that contains an RNA catalytic subunit (26), specifically cleaves a small number of polycistronic mRNAs in intercistronic regions (27).
In addition, many Gram-negative bacteria have an RNase E ortholog called RNase G. A major distinction between the two enzymes is that RNase G lacks the degradosome scaffold region (28, 29). RNase G has been most studied in E. coli. It has been shown to target specific mRNAs and work in tandem with RNase E (30). Similar to RNase E, the enzyme prefers single stranded AU-rich sequences and is 5′ end dependent (31), but is present in much lower amounts than RNase E (32). In vitro, both RNase E and RNase G have similar substrate specificity. However, RNase G cleaves 5′ terminus of pre-16S rRNA precursors at different site than RNase E (28, 29). Although it was originally thought that RNase G could complement the conditional lethality associated with RNase E mutants (32, 33), it has now been shown that complementation only takes place in the presence of a mutationally altered RNase G protein (34).
RNase Z, another endonuclease found in E. coli and other Gram-negative bacteria was initially identified in eukaryotes based on its ability to cleave tRNA precursors endonucleolytically to generate a 3′ terminus that was a substrate for tRNA nucleotidyl transferase, which adds the CCA determinant (35). In E. coli the enzyme seems to function primarily in mRNA decay (36), but can also serve in a backup role in some aspects to tRNA processing (see section on tRNA processing). Interestingly RNase Z, which was originally identified as RNase BN (37), also has 3′ → 5′ exonuclease activity and will be discussed later in the section on tRNA processing.
RNase LS, another endonuclease encoded by rnlA, has been shown to play a very limited role in the decay in both bacterial mRNAs and phage encoded mRNAs in E. coli (38) and is also part of the rnlAB toxin/antitoxin module (39).
The role of 3′ → 5′ exonucleases in mRNA decay
RNase II, RNase R and PNPase are the three major 3′ → 5′ exonucleases affecting mRNAs and rRNAs in most Gram-negative bacteria. RNase II and RNase R degrade single-stranded RNA employing a hydrolytic mechanism, releasing mononucleotides (40, 41). It has been shown that RNase II accounts for ~95% of the hydrolytic RNase activity in E. coli (42). RNase II is strongly inhibited by secondary structures (43), while RNase R can easily degrade RNA molecules containing secondary structures (44), but requires a single-stranded region of at least 7 nt in order to bind (45). Interestingly, RNase R is more important in stationary phase cells than RNase II (46).
In contrast, PNPase degrades RNA using a phosphorolytic mechanism that requires inorganic phosphate, releasing nucleoside diphosphates (47). Since the equilibrium constant for this reaction is one, the enzyme can also synthesize RNA in an untemplated reaction employing nucleoside diphosphates to generate single-stranded RNA containing all four nucleotides (47). In fact, the enzyme can work both biosynthetically and degradatively in E. coli (48) and other Gram-negative bacteria. PNPase exists in at least two multiprotein complexes. The degradosome, which contains RNase E, PNPase, the RhlB RNA helicase and enolase and the polyadenylation complex, which contains poly(A) polymerase (PAP I), PNPase and the RNA binding protein Hfq (49). RhlB is also reported to be associated with PNPase independent of the degradosome assembly helping its exonucleolytic activity in vitro (50), although such an effect has yet to be observed in vivo (51).
While none of these three ribonucleases are essential for cell viability by themselves, inactivation of both RNase II and PNPase results in synthetic lethality (52). Of most significance is that at the nonpermissive temperature (44°C) there is a large accumulation of partially degraded mRNAs in a pnp-7 rnb-500 double mutant, demonstrating a significant role for these two enzymes in the mRNA turnover (52). It has also been shown that a pnp-7 rnr double mutant is a synthetic lethal, but a rnb rnr strain is viable (41).
While the decay of the majority of mRNAs is initiated via endonucleolytic cleavages employing a combination of RNase E, RNase III, RNase G, RNase P, RNase Z and RNase LS, the degradation of a significant number of transcripts is also initiated by exonucleases PNPase, RNase II and RNase R. A series of experiments employing either macroarrays (53) or RNAseq (54) have demonstrated that inactivation of any of the three exonucleases leads to significant changes in the steady-state levels of between 5–10% of the mRNAs. It is also thought that RNase R is more important in ribosome quality control by degrading nonfunctional rRNAs and tRNAs rather than mRNAs. In contrast, PNPase and RNase II are more involved in mRNA decay (53) and some aspects of tRNA processing (51) and rRNA degradation (55). It should also be noted that unlike Gram-positive bacteria (56), there do not appear to be any 5′ → 3′ exonucleases in Gram-negative species.
A common feature of all three of these exonucleases is that they cannot degrade a RNA substrate completely, leaving short oligonucleotides of 2–4 in length (57–59). Many bacteria contain another 3′ → 5′ exoribonuclease called oligoribonuclease, which specifically degrades these short oligonucleotides using a hydrolytic mechanism (60, 61). The complete degradation of the short oligonucleotides that remain after the action of RNase II, RNase R and PNPase seems to be essential, since there is evidence that oligoribonuclease is essential for cell viability (62). It is not clear at this time if an enzyme such as RNase T can also function on short oligoribonucleotides. Fig. 2 presents a model for general mRNA decay in E. coli initiated by RNase E and RNase G.
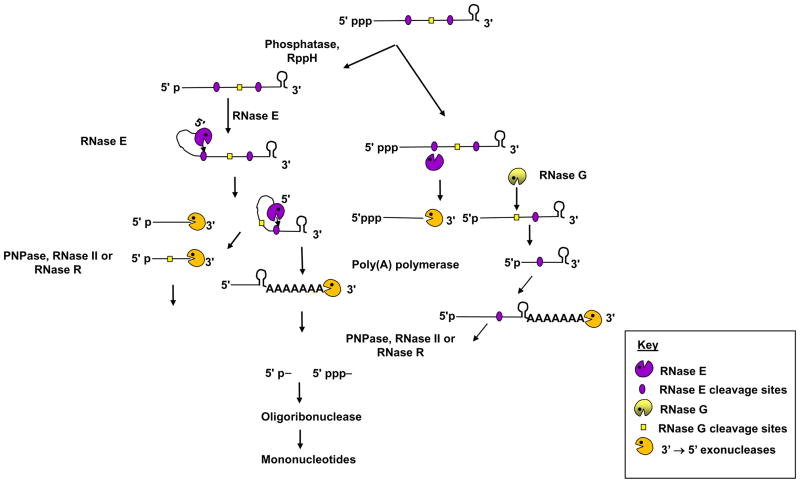
Fig. 2.
Model for the initiation of mRNA decay by RNase E. For the sake of simplicity, the other proteins associated with the RNase E-based degradosome are not shown. In addition, this model is independent of whether RNase E is associated with the inner membrane of E. coli. 5′ monophosphate RNA, a preferred substrate for RNase E is degraded via 5′ end dependent pathway. In contrast, 5′ triphosphate RNA is degraded via RNase E internal entry mechanism. Any endonucleolytically cleaved fragments with strong secondary structures, such as one containing a Rho-independent transcription terminator shown here, undergoes polyadenylation by PAP I. Subsequently, all decay intermediates are degraded by 3′ → 5′ exonucleases (PNPase, RNase II and RNase R) followed by oligoribonuclease to mononucleotides. Figure is not drawn to scale.
ENZYMES INVOLVED IN TRANSFER RNA PROCESSING
In E. coli and many other Gram-negative bacteria, all of the tRNAs are encoded with extra nucleotides at both their 5′ and 3′ ends. They occur as either monocistronic or polycistronic transcripts that contain only tRNAs, tRNAs and mRNAs, or tRNAs and rRNAs. Fig. 3 presents some of the pathways involved in tRNA processing. Many of these transcripts are terminated in a Rho-independent manner, which generates a stem-loop at the 3′ terminus. The processing of the majority of these tRNA transcripts is initiated by RNase E, which either removes the Rho-independent transcription terminators (63–65) or cleaves in the intercistronic regions (11, 66). In some cases, RNase G and RNase Z can inefficiently substitute for RNase E (65) (Fig. 3A). In at least one well-documented case, the Rho-independent transcription terminator on the leuX primary transcript is removed exonucleolytically by PNPase (51) (Fig. 3B).
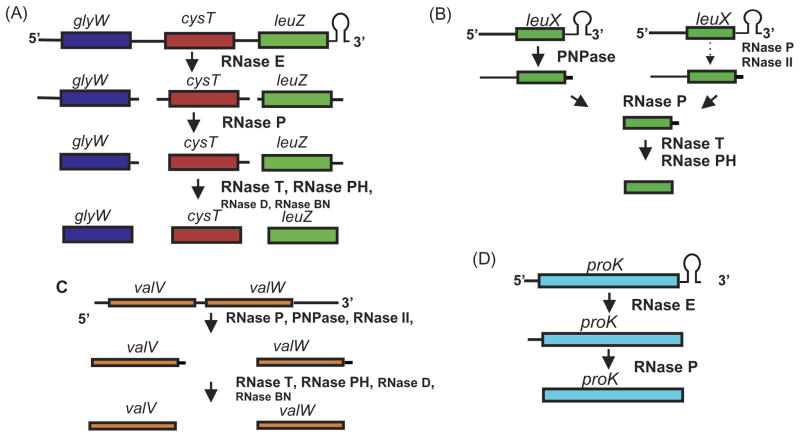
Fig. 3.
Diagrammatic representation of four independent pathways of tRNA processing. (A). Processing of the glyW cyst leuZ polycistronic operon. RNase E initiates processing by cleaving the polycistronic transcript to release pre-tRNAs (11). Processing at the 5′ termini is carried out by RNase P. Maturation of the 3′ termini is usually carried out by RNase T and/or RNase PH. If these two enzymes are not present, RNase D and/or RNase BN can complete the process. (B) Processing of the monocistronic leuX transcript (51). The Rho-independent transcription terminator is removed exonucleolytically by PNPase. In the absence of PNPase, a combination of RNase P and RNase II can digest the terminator. Subsequently, RNase P matures the 5′ terminus, while RNase T and RNase PH complete the process at the 3′ terminus. (C) Processing of the valV valW polycistronic operon (67). RNase P separates valV and valW pre-tRNAs by cleaving at their respective mature 5′ ends while PNPase and RNase II shorten the 3′ Rho-depended terminator. Subsequently, 3′ → 5′ exonucleases (RNase T/RNase PH/RNase D/RNase BN) matures the 3′ ends. (D). Processing of the monocistronic proK transcript (63). RNase E removes the Rho-independent transcription terminator to generate the mature 3′ terminus without the need of any of the 3′ → 5′ exonucleases. RNase P cleaves at the mature 5′ end. Figure is not drawn to scale.
Interestingly, many polycistronic tRNA transcripts do not utilize RNase E, but rather are dependent on RNase P for their initial processing (67) (Fig. 3C). Some require the initial removal of the Rho-independent transcription terminator by RNase E before RNase P can process the rest of the transcript (64, 65, 67). The RNase P cleavages occur at the mature 5′ termini (65), while RNase E cleavages, with the exception of the three proline tRNAs (63), leave extra nucleotides downstream of the CCA determinant (11, 66). A surprising observation from the RNase P processed transcripts was that the enzyme cleaves the polycistronic transcripts starting from the 3′ terminus and not the 5′ terminus (64).
An interesting feature of tRNA maturation in Gram-negative bacteria is that there is only one ribonuclease, RNase P, that generates the mature 5′ terminus of all the tRNA species (26). In contrast, maturation of the 3′ terminus can be carried out by a variety of 3′ → 5′ exonucleases including RNase T, RNase PH, RNase D, RNase BN, and RNase II (42). PNPase and RNase II can also remove extra nucleotides from the 3′ terminus of a pre-tRNA but they cannot complete the final maturation process to expose the CCA determinant (51, 65).
The bulk (79/86) of the pre-tRNAs in E. coli employ a combination of RNase T and RNase PH for their final 3′ end maturation (68). However, RNase T appears to be the most important enzyme of the two based on its unique substrate specificity. Specifically, unlike other ribonucleases, it specifically stops at the terminal CCA because of its inhibition by the presence of C ribonucleotides within its catalytic site (69). Thus, tRNAs containing C residues downstream of CCA are more dependent on RNase PH, which is not inhibited by C residues (64). Inactivation of both RNase T and RNase PH leads to rapid accumulation of 3′ immature tRNAs which becomes substrates for PAP I (68). Contrary to mRNAs, PAP I adds short poly(A) tails (generally <5 nucleotides) to the immature tRNAs (68). However, instead of undergoing degradation as defective tRNAs (70), a majority of the polyadenylated tRNAs are matured slowly by inefficient exonucleases, such as RNase D and RNase BN/Z (68). These results suggest that PAP I helps regulate functional tRNA levels in E. coli (68). However, excess PAP I polyadenylates mature tRNAs inhibiting aminoacylation and protein synthesis resulting in rapid cell death (71).
Recently, it has been shown that the three proline tRNAs do not require exonucleolytic processing at their 3′ termini (63) (Fig. 3D). Rather, RNase E removes the Rho-independent transcription terminator by cleaving immediately downstream of the CCA determinant (63). It is not at this time what accounts for the altered substrate specificity of RNase E in the case of the proline tRNA transcripts.
It has now been shown that the 3′ → 5′ exonuclease RNase BN and that the endoribonuclease RNase Z are encoded by the same gene (72) and the enzyme does not remove the CCA determinant from pre-tRNAs (73). Although it has been shown that RNase D and RNase BN can participate in 3′ end maturation (37, 74), it is not clear at this time how significant a role they play in cells that contain RNase T and RNase PH. As described above, RNase Z has been shown to play a role in mRNA decay (36).
With the possible exception of RNase T and RNase BN, the other 3′ → 5′ exonucleases can conceivably partially degrade into the CCA determinant that is required for aminoacylation. The repair of such termini can be carried out by the enzyme tRNA nucleotidyl transferase, which can add C, CC or CCA in an untemplated reaction (75). Interestingly, this enzyme is not essential for cell viability in E. coli (76), most likely due to its overlapping biosynthetic activity with PNPase and PAP I (77). Recently, it has been suggested that the enzyme plays a role in tRNA quality control by adding CCACCA tag to defective tRNA molecules which are preferentially degraded by RNase R(78).
ENZYMES INVOLVED IN RIBOSOMAL RNA PROCESSING
All functional rRNAs in Gram-negative bacteria are matured from large (over 3 Kb) 30S rRNA polycistronic transcripts that contain the coding sequences for the 16S, 23S and 5S rRNAs as well as at least one tRNA and a significant amount of spacer sequences in between (Fig. 4). Inverted repeats lead to two large stem-loop structures containing the 16S and 23S species as part of large loops (Fig. 4). Initial processing by RNase III, an enzyme that is specific for double-stranded RNA (79, 80), within the double-stranded stems releases 17S (pre-16S), 25S (pre-23S), and 9S (pre-5S) rRNA precursors (81–83) as well as the embedded tRNAs.
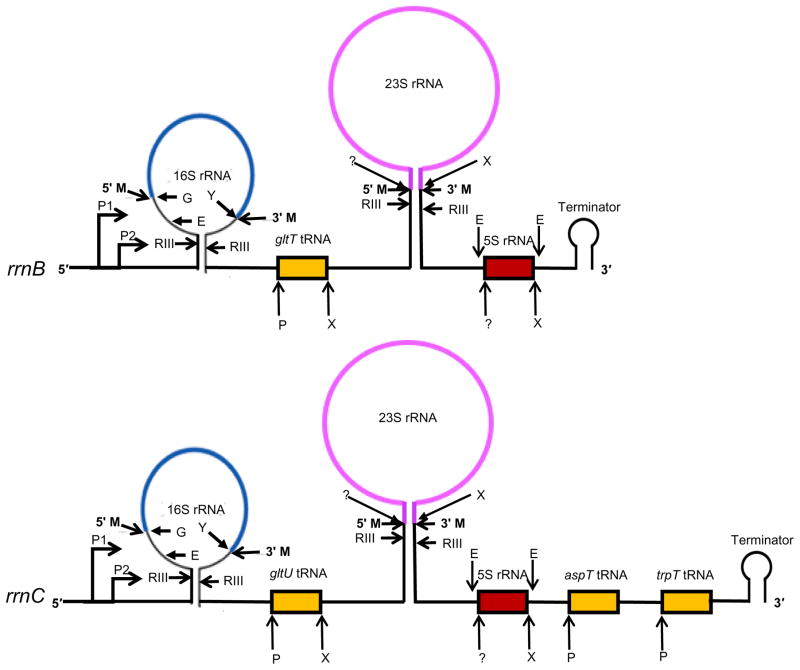
Fig. 4.
Processing of rRNA operons in E. coli. The rrnB and rrnC operons are shown as model operons. RNase III (RIII) cleaves the 30S rRNA transcript first within the double-stranded stems formed by the spacer sequences adjacent to the mature 16S and 23S rRNAs, generating 17S, 25S, and 9S pre-rRNAs. The functional mature 16S rRNA is generated from 17S pre-rRNA after initial RNase E (E) cleavage followed by RNase G (G) at the mature 5′ end and removal of extra 33 nts at the 3′ ends by YbeY (Y) along with multiple exoribonucleases (not shown). A p5S precursor is generated from the the 9S precursor by initial RNase E cleavage at three nt upstream (E) and downstream (E) of the mature termini of the mature 5S rRNA. The mature 5′ end of the tRNAs are generated by RNase P (P) cleavage. Exoribonucleases (X) (primarily RNase T) are responsible for the 3′ end maturation of the tRNAs, 23S rRNA, and 5S rRNA, but the ribonuclease(s) (?) responsible for the maturation of 5′ ends of 23S and 5S rRNAs remain unidentified. The model is not drawn to scale.
The pre-16S species containing 115 extra nucleotides at its 5′ terminus is initially cleaved endonucleolytically by RNase E removing 60 nucleotides. Subsequently, RNase G endonucleolytically removes the remaining 55 nt to generate the mature 5′ terminus of the 16S rRNA (28, 29). The extra 33 nt found at the 3′ terminus of the pre-16S rRNA are removed either by the YbeY endonuclease or a combination of RNase II, RNase PH, PNPase, and RNase R (84–87).
The pre-5S species is cleaved by RNase E to within 3 nt on each side of the mature sequence (81, 88–91). The three extra nucleotides at the 3′ end of the pre-5S rRNA are removed by the 3′ → 5′ exonuclease RNase T. Nothing is currently known about how the three extra nucleotides at the 5′ end of 5S rRNA species are processed.
In the case of the pre-23S species, the RNase III cleavages leave 7–9 extra nucleotides at the 3′ terminus and either three or seven extra nucleotides at the 5′ terminus (89, 92). The final maturation of the 3′ terminus can be carried out either by RNase T alone or by a combination of PAP I, RNase II, RNase PH, and RNase T (91). In fact, the pre-23S rRNA species is an excellent target for PAP I in the wild type cells (49, 93). Although it is not clear at this time how the 5′ end is matured, since Gram-negative bacterium do not contain 5′ → 3′ exonucleases, this step most likely is carried out by an endonuclease. Recent experiments suggest that RNase III might in fact carry out this reaction (Chardhuri et al. 2018, submitted to Molecular Microbiology).
The tRNAs embedded in the rRNA operons are released as pre-tRNAs following RNase III cleavage of the primary transcript and RNase E action on the 9S precursor (Fig. 4). Presumably, these pre-tRNAs employ the various enzymes described in the section on tRNA processing for final maturation (See above).
The ribosome, an essential component of protein synthetic machinery, is assembled using mature 23S, 16S rRNA and 5S rRNAs. While it has been shown that pre-23S rRNAs can be incorporated to form functional ribosomes (81), incorporation of pre-16S species do not result in functional ribosomes (94). Thus, it was predicted that RNase III would be an essential enzyme because of the need to separate the 16S, 23S and 5S species from the larger polycistronic transcript. However, it has been shown that rnc deletion mutants are viable and show only small defects in their growth rates (90, 95). Clearly there is an alternative mechanism for generating rRNAs that can be incorporated into functional ribosomes in the absence of RNase III.
PROCESSING AND DECAY OF SMALL REGULATORY RNAS
Recent experiments have shown that in a wide range of organisms small RNAs (sRNA) play more widespread regulatory roles than previously envisioned. In prokaryotes, sRNAs generally range in size from ~ 50 to 400 nt in length. Gram-negative bacteria contain > 100 sRNAs that are encoded on plasmids or the genome (96–99). In fact, high density tiling array analysis of RNase E and RNase III mutants have suggested that there may in fact be as many as 300 sRNAs in E. coli (24, 100). While sRNAs that do not undergo any processing to be functional cannot be ruled out, all sRNAs identified to date require some initial processing by RNase E processing of many sRNAs for them to be functional (101–103). In case of MicC, conversion of 5′ triphosphate to 5′ monophosphate enhanced RNase E mediated decay of ompD (104).
Based on their interactions with their target mRNAs, sRNAs can inhibit translation by masking the ribosome binding site (RBS) (105) or in some cases binding outside of the RBS (106) followed by rapid turnover of the transcripts. Other sRNAs control Rho-dependent termination at the 5′ UTR of the transcripts by blocking the action of Rho and resulting in increased translation of the transcripts (107). The association of many sRNAs with a functionally important cognate RNA-binding protein (Hfq) is critical for its functionality. For example, RyhB directly promotes mRNA instability by forming a RNA/protein complex containing Hfq that attracts RNase E (108). A recent study suggests that a large number of sRNAs in Salmonella enterica are associated with a conserved RNA-binding protein ProQ (109). In some cases, RNase III is responsible for the decay of an mRNA targeted by a sRNA (110).
An interesting question relates to the degradation of sRNAs. Since these are relatively small molecules and highly structured, it was not expected that their degradation would be initiated by endonucleolytic attack. However, both RNase E and RNase III play important roles in degrading sRNAs when they are bound to their target mRNAs (111, 112). The association of Hfq with sRNA-mRNA complexes facilitates this degradation process (113), but the experiments of Andrade et al. (114) have shown that sRNAs not associated with Hfq were not substrates for either RNase E or RNase III. Rather, PNPase seemed to be required for their degradation (114), a surprising result since these sRNAs are highly structured. However, it has previously been shown that PNPase can degrade the Rho-independent transcription terminator associated with the leuX primary transcript (51). The recent demonstration of that PNPase can both protect and degrade sRNAs is consistent with these observations (115). However, since PNPase has been shown to be inhibited by strong secondary structures (43), it is likely that either polyadenylation by PAP I or unwinding of the secondary structures by one of the DEAD-box RNA helicases (see below) facilitate degradation of sRNAs by PNPase (114, 116). In contrast, sRNAs, such as ryhB, sgrS and cyaR, are destabilized in the absence of PNPase, presumably by RNase E (117).
RNA HELICASES
Although RNAs are transcribed as single-stranded molecules, both nontranslated and translated transcripts readily form secondary and tertiary structures, which are critical for their proper functioning and stability. For example, many mRNAs are terminated with Rho-independent transcription terminators, a double-stranded stem-loop structure, which also serve as stability elements for mRNAs. All tRNAs assume their tertiary cloverleaf structures as soon as they are transcribed, a step that is crucial for their maturation by various ribonucleases, post-transcriptional modification, and their functionality as amino acid carriers. The formation of large stem-loops in rRNA precursors generate cleavage sites for RNase III. The mature 16S and 23S rRNAs form complex arrays of secondary and tertiary structures in the generation of functional ribosomes. In addition, changes in growth temperatures, such as cold and heat shock, result in altered mRNA structure, which has a significant effect on translation efficiency. It is thus not surprising that bacteria contain helicases that use the energy derived from the hydrolysis of ATP to alter the structure of various RNA molecules. This class of enzyme is characterized by the DEXD/H (usually referred to as the DEAD-box) amino acid motif.
The best characterized of the DEAD-box RNA helicases are the five paralogs (rhlB, rhlE, srmB, dbpA and csdA, formerly called deaD) found in E. coli. A strain lacking all the helicases is still viable (118). Only loss of DeaD or SrmB causes significant growth defects and alterations in ribosomal RNA processing and maturation at both 30°C and 37°C (118–120).
RhlB is involved in mRNA decay through its association with the RNase E-based degradosome (9, 121) as well as separately with PNPase (50) and is discussed in more detail in the chapter by Bandryra and Luisi. It has also been shown that RhlE can also associate with the RNase E-based degradosome under certain conditions (122). It aids in the degradation of mRNAs and seems to regulate the roles of other RNA helicases associated with ribosome maturation (122, 123). The SrmB and CsdA helicases function in the process of ribosome biogenesis (119, 120, 124). CsdA has also been associated with a “cold shock degradosome” (125). The DbpA helicase activity is dependent on 23S rRNA (126–128).
SPECIALIZED RIBONUCLEASES
Toxin/antitoxin systems
The phenomenon of bacterial persistence was originally discovered by Bigger in 1944 (129). Simply put in any bacterial population some cells grow considerably slower than others. These slow growing cells are inherently more resistant to antibiotics, which are more effective in killing rapidly growing bacteria. In 1983 Moyed et al. (130) showed that mutations in the hipA locus led to increased levels of persister cells. These mutations were part of the hipBA toxin-antitoxin (TA) locus. Since then multiple TA systems have been identified which are broadly classified into five types. Generally, most toxins target an mRNA to inhibit translation either in a ribosome dependent or independent manner. These are sequence specific endonucleases and have been called “mRNA interferases.” Readers are encouraged to study the chapter: “Leaderless mRNA: Novel Aspects of Ancestral transcripts” in this book for more details.
E. coli contains a large number of TA loci including mazEF (2 copies), relBE (6 copies), hipAB, rnlAB, and hicAB (131). In the case of RelE, the protein cleaves an mRNA positioned at the ribosomal A site, between the second and third base of the A-site codon (132–134) leading to translation inhibition. The MazF endonuclease probably has the most sequence specificity of any E. coli endonuclease, with the possible exception of the ribonucleases (bacteriocins) that target tRNAs (see next section). It cleaves mRNAs site-specifically at ACA base motifs independent of ribosomes (135). In contrast, the HicA ribonuclease, a protein of only 58 amino acids, cleaves mRNAs without any ribosomal involvement in an apparent random fashion (136).
RNase LS, which was discussed in the section of mRNA decay, is part of the rnlAB TA module (39). It is not clear whether there is any cleavage specificity associated with this enzyme or whether it has specific targets or whether it plays any role in persistence.
Bacteriocins function as ribonucleases
Bacteria can produce bacteriocins that can kill other bacteria that are living in close proximity. Colicin, the earliest discovered bacteriocin is encoded by the Col-plasmids. E. coli cells not carrying the same or cognate plasmid are killed by the released colicin (137). Colicin production is induced by nutrient starvation, the stringent response, the SOS response and various other stress responses (137). Many of the colicins use the BtuB receptor for cell entry, which is involved in vitamin B12 uptake. There are several types of Colicins, but here we only focus on those encoding an RNase activity.
Of the RNase type, colicin E3 is the most studied. It interacts with intact ribosomes, cleaving the 3′ region of the 16S rRNA between A1493 and G1494 in the decoding A-site, leading to functional inactivation (138–140). In contrast, colicin E5 and colicin D use different cell surface receptors and specifically cleave selected tRNAs. For example, colicin E5 cleaves between the Q and U residues (positions 34 and 35) in the anticodon arm of tRNAHis, tRNAAsn, tRNAAsp, and tRNATyr (141). Colicin D cleaves four of the six tRNAArg species between positions 38 and 39 at the 3′ end of the anticodon loop.
Additional tRNA cleaving ribonucleases have been identified in Shigella and Salmonella as part of the vapBC toxin/antitoxin system (142). In this case, translation is inhibited by cleavage of the initiator tRNAMet at the 3′ end of the anticodon loop (143). While it had been suggested that the tRNA cleavages lead to cell death, recent work has shown that cells are not immediately killed (141). Rather the tRNA cleavages serve to help regulate cell growth in order to survive a variety of stresses conditions (141).
CRISPR/Cas systems
Unlike the well-characterized restriction/modification systems that provide bacteria broad range protection from phage attack (144), CRISPR/Cas systems provide adaptive immunity to the bacterial strains that carry them. The term CRISPR was first used by Jansen et al. (145). These systems are found in almost all archaea and about half of the bacteria that have been examined (146). The pathway found in E. coli K12 falls into the Type IE system. Simply put, bacteria carrying a CRISPR/Cas system have one or more CRISPR arrays, which contain short spacer sequences derived from previous bacteriophage infections or exposure to foreign bacterial plasmids, that are separated by identical repeat sequences (147). For any of the spacers to function properly in terms of targeting an invading bacteriophage, the CRISPR array must be transcribed and processed. In E. coli, five Cas proteins form a multiprotein complex called Cascade (148). The cas6a gene encodes a metal independent endoribonuclease that cleaves in the repeat sequences to yield 61–62 nt crRNAs that have extra nucleotides at both their 5′ and 3′ ends. These species remain bound to the Cascade complex to facilitate its interaction with the target DNA (147). Interestingly, the Cas6a ribonuclease is normally not expressed in E. coli (147). Readers are encouraged to read the excellent recent review by Mohanraju et al. (149) on diverse CRSPR/Cas systems for more details.
CONTROL OF RIBONUCLEASES
There are many different ways that bacteria respond to the ever changing environments in which they live. The regulation of various ribonuclease activities is one of them. The levels of RNase E, RNase III and PNPase are autoregulated based on specific circumstances by modulating the stability of their respective mRNAs (150–152). Under certain stress conditions the activity of RNase E is also controlled by global protein regulators, such as RraA and RraB (153, 154). RraA has also been shown to interact with RhlB helicase, a component of the degradosome, to inhibit its RNA-binding and helicase activities (155). The protein YmdB, which is expressed upon cold shock or entry into stationary phase, has been shown to inhibit RNase III activity by preventing its dimerization (156). The level of the 3′ → 5′ exoribonuclease RNase R increases significantly upon entry into stationary phase or cold shock compared to the exponential phase cells due to reduced proteolysis (157).
CONCLUSIONS
Although there has been tremendous progress in our understanding of the mechanisms of post-transcriptional RNA processing and decay over the past 25 years, many questions remain to be answered. With the discovery of new enzymes some aspects of RNA processing have gotten closer attention and some conventional models have had to be significantly revised. However, even after the complete sequencing of multiple E. coli genomes, the functions for over one third of the genes are still unknown. Are there additional ribonucleases present in the cell that have not yet been identified?
Perhaps all of the enzymes involved in mRNA decay have been identified, but the regulation of the process is still not well-understood (158) and all the targets of the various enzymes have yet to be identified. For example, RNase III initiates the processing of the 30S rRNA, but the enzyme is not essential for cell viability. Clearly there must be an alternative processing pathway to generate the 16S, 23S and 5S species that are required to produce functional ribosomes. In addition, the ribonuclease(s) required for the 5′ end maturation of both 23S and 5S rRNA are still a mystery.
Our understanding of the role of PAP I and PNPase as polyadenylating enzymes in Gram-negative bacteria is also still limited, especially the function of the polynucleotide tails synthesized by PNPase. First of all, it is not yet clear how these two enzymes select their substrates. The available data (49, 159, 160) suggest that PAP I can add poly(A) tails to both the full-length and decay intermediates. However, in the case of full-length transcripts, those terminated with a Rho-independent transcription terminator are the preferred substrates and the poly(A) tails are mostly added downstream of the terminator. In contrast, PNPase adds polynucleotide tails to both full-length transcripts terminating in a Rho-dependent fashion and decay intermediates. While poly(A) tail addition clearly targets transcripts for decay (161), no such data is available for polynucleotide tails. However, the extremely long unstructured polynucleotide tails added by PNPase (49, 160) suggest that they also facilitate degradation of its substrates by exonucleases by providing them unstructured substrates.
The notion of cellular compartmentalization in bacteria (162), similar to what occurs in eukaryotes, has further complicated our understanding of the process of post-transcriptional regulation by various enzymes. Since the degradosome is associated with the inner membrane, does that mean that all mRNA decay has to occur at this location? What about the transcripts that are decayed by enzymes other than RNase E? In fact, limited dispersion of some transcripts from the site of transcription was observed in a recent study (163). Clearly, identification of new player(s) and their regulatory processes will help us develop a better understanding of RNA processing and degradation.
Acknowledgments
This work was supported by a NIH grant (GM081554) to S.R.K.
References
- 1.Apirion D. Degradation of RNA in Escherichia coli: A hypothesis. Mol Gen Genet. 1973;122:313–322. doi: 10.1007/BF00269431. [DOI] [PubMed] [Google Scholar]
- 2.Misra TK, Apirion D. RNase E, an RNA processing enzyme from Escherichia coli. J Biol Chem. 1979;254:11154–11159. [PubMed] [Google Scholar]
- 3.Kuwano M, Ono M, Endo H, Hori K, Nakamura K, Hirota Y, Ohnishi Y. Gene affecting longevity of messenger RNA: a mutant of Escherichia coli with altered mRNA stability. Mol Gen Genet. 1977;154:279–285. doi: 10.1007/BF00571283. [DOI] [PubMed] [Google Scholar]
- 4.Ono M, Kuwano M. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of mRNA. J Mol Biol. 1979;129:343–357. doi: 10.1016/0022-2836(79)90500-x. [DOI] [PubMed] [Google Scholar]
- 5.Babitzke P, Kushner SR. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc Natl Acad Sci U S A. 1991;88:1–5. doi: 10.1073/pnas.88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDowall KJ, Cohen SN. The N-terminal domain of the rne gene product has RNase E activity and is non-overlapping with the arginine-rich RNA-binding motif. J Mol Biol. 1996;255:349–355. doi: 10.1006/jmbi.1996.0027. [DOI] [PubMed] [Google Scholar]
- 7.Khemici V, Poljak L, Luisi BF, Carpousis AJ. The RNase E of Escherichia coli is a membrane-binding protein. Mol Microbiol. 2008;70:799–813. doi: 10.1111/j.1365-2958.2008.06454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 9.Py B, Higgins CF, Krisch HM, Carpousis AJ. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 10.Ow MC, Liu Q, Kushner SR. Analysis of mRNA decay and rRNA processing in Escherichia coli in the absence of RNase E-based degradosome assembly. Mol Microbiol. 2000;38:854–66. doi: 10.1046/j.1365-2958.2000.02186.x. [DOI] [PubMed] [Google Scholar]
- 11.Ow MC, Kushner SR. Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli. Genes Dev. 2002;16:1102–15. doi: 10.1101/gad.983502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mackie GA. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- 13.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 14.Luciano DJ, Vasilyev N, Richards J, Serganov A, Belasco JG. A novel RNA phosphorylation state enables 5′ end-dependent degradation in Escherichia coli. Mol Cell. 2017;67:44–54. e6. doi: 10.1016/j.molcel.2017.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrey SM, Mackie GA. Roles of the 5′-phosphate sensor domain in RNase E. Mol Microbiol. 2011;80:1613–24. doi: 10.1111/j.1365-2958.2011.07670.x. [DOI] [PubMed] [Google Scholar]
- 16.Baker KE, Mackie GA. Ectopic RNase E sites promote bypass of 5′-end-dependent mRNA decay in Escherichia coli. Molecular Microbiol. 2003;47:75–88. doi: 10.1046/j.1365-2958.2003.03292.x. [DOI] [PubMed] [Google Scholar]
- 17.Kime L, Clarke JE, Romero AD, Grasby JA, McDowall KJ. Adjacent single-stranded regions mediate processing of tRNA precursors by RNase E direct entry. Nucleic Acids Res. 2014;42:4577–89. doi: 10.1093/nar/gkt1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke JE, Kime L, Romero AD, McDowall KJ. Direct entry by RNase E is a major pathway for the degradation and processing of RNA in Escherichia coli. Nucleic Acids Res. 2014;42:11733–51. doi: 10.1093/nar/gku808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kime L, Jourdan SS, Stead JA, Hidalgo-Sastre A, McDowall KJ. Rapid cleavage of RNA by RNase E in the absence of 5′ monophosphate stimulation. Mol Microbiol. 2010;76:590–604. doi: 10.1111/j.1365-2958.2009.06935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ehretsmann CP, Carpousis AJ, Krisch HM. Specificity of Escherichia coli endoribonuclease RNase E: In vivo and in vitro analysis of mutants in a bacteriophage T4 mRNA processing site. Genes & Develop. 1992;6:149–159. doi: 10.1101/gad.6.1.149. [DOI] [PubMed] [Google Scholar]
- 21.McDowall KJ, Kaberdin VR, Wu S-W, Cohen SN, Lin-Chao S. Site-specific RNase E cleavage of oligonucleotides and inhibition by stem-loops. Nature. 1995;374:287–290. doi: 10.1038/374287a0. [DOI] [PubMed] [Google Scholar]
- 22.McDowall KJ, Lin-Chao S, Cohen SN. A + U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J Biol Chem. 1994;269:10790–10796. [PubMed] [Google Scholar]
- 23.Chao Y, Li L, Girodat D, Forstner KU, Said N, Corcoran C, Smiga M, Papenfort K, Reinhardt R, Wieden HJ, Luisi BF, Vogel J. In vivo cleavage mapilluminates the central role of RNase E in coding and non-coding RNA pathways. Mol Cell. 2017;65:39–51. doi: 10.1016/j.molcel.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stead MB, Marshburn S, Mohanty BK, Mitra J, PCL, Ray D, Hughes T, Kushner SR. Analysis of E. coli RNase E and RNase III activity in vivo using tiling microarrays. Nucleic Acids Res. 2010;39:3188–3203. doi: 10.1093/nar/gkq1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon GC, Cameron JC, Pfleger BF. RNA sequencing identifies new RNase III cleavage sites in Escherichia coli and reveals increased regulation of mRNA. MBio. 2017:8. doi: 10.1128/mBio.00128-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altman S. Ribonuclease P: an enzyme with a catalytic RNA subunit. Advances in enzymology and related areas of molecular biology. 1989;62:1–36. doi: 10.1002/9780470123089.ch1. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Altman S. A specific endoribonuclease, RNase P, affects gene expression of polycistronic operon mRNAs. Proc Natl Acad Sci USA. 2003;100:13213–13218. doi: 10.1073/pnas.2235589100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wachi M, Umitsuki G, Shimizu M, Takada A, Nagai K. Escherichia coli cafA gene encodes a novel RNase, designated as RNase G, involved in processing of the 5′ end of 16S rRNA. Biochem Biophys Res Comm. 1999;259:483–488. doi: 10.1006/bbrc.1999.0806. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Pandit S, Deutscher MP. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 1999;18:2878–85. doi: 10.1093/emboj/18.10.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ow MC, Perwez T, Kushner SR. RNase G of Escherichia coli exhibits only limited functional overlap with its essential homologue, RNase E. Mol Microbiol. 2003;49:607–22. doi: 10.1046/j.1365-2958.2003.03587.x. [DOI] [PubMed] [Google Scholar]
- 31.Tock MR, Walsh AP, Carroll G, McDowall KJ. The CafA protein required for the 5′-maturation of 16 S rRNA is a 5′-end-dependent ribonuclease that has context-dependent broad sequence specificity. J Biol Chem. 2000;275:8726–32. doi: 10.1074/jbc.275.12.8726. [DOI] [PubMed] [Google Scholar]
- 32.Lee K, Bernstein JA, Cohen SN. RNase G complementation of rne null mutation identified functional interrelationships with RNase E in Escherichia coli. Mol Microbiol. 2002;43:1445–1456. doi: 10.1046/j.1365-2958.2002.02848.x. [DOI] [PubMed] [Google Scholar]
- 33.Deana A, Belasco JG. The function of RNase G in Escherichia coli is constrained by its amino and carboxyl termini. Molecular Microbiol. 2004;51:1205–1217. doi: 10.1046/j.1365-2958.2003.03905.x. [DOI] [PubMed] [Google Scholar]
- 34.Chung D-H, Min Z, Wang B-C, Kushner SR. Single amino acid changes in the predicted RNase H domain of E. coli RNase G lead to the complementation of RNase E mutants. RNA. 2010;16:1371–1385. doi: 10.1261/rna.2104810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schiffer S, Rosch S, Marchfelder A. Assigning a function to a conserved group of proteins: the tRNA 3′ processing enzymes. EMBO J. 2002;21:2769–2677. doi: 10.1093/emboj/21.11.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perwez T, Kushner SR. RNase Z in Escherichia coli plays a significant role in mRNA decay. Mol Microbiol. 2006;60:723–37. doi: 10.1111/j.1365-2958.2006.05124.x. [DOI] [PubMed] [Google Scholar]
- 37.Asha PK, Blouin RT, Zaniewski R, Deutscher MP. Ribonuclease BN: Identification and partial characterization of a new tRNA processing enzyme. Proc Natl Acad Sci USA. 1983;80:3301–3304. doi: 10.1073/pnas.80.11.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otsuka Y, Yonesaki T. A novel endoribonuclease, RNase LS, in Escherichia coli. Genetics. 2005;169:13–20. doi: 10.1534/genetics.104.033290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koga M, Otsuka Y, Lemire S, Yonesaki T. Escherichia coli rnlA and rnlB compose a novel toxin-antitoxin system. Genetics. 2011;187:123–30. doi: 10.1534/genetics.110.121798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nossal NG, Singer MF. The processive degradation of individual polynucleotide chains. J Biol Chem. 1968;243:913–922. [PubMed] [Google Scholar]
- 41.Cheng ZF, Zuo Y, Li Z, Rudd KE, Deutscher MP. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J Biol Chem. 1998;273:14077–14080. doi: 10.1074/jbc.273.23.14077. [DOI] [PubMed] [Google Scholar]
- 42.Kelly KO, Deutscher MP. The presence of only one of five exoribonucleases is sufficient to support the growth of Escherichia coli. J Bacteriol. 1992;174:6682–6684. doi: 10.1128/jb.174.20.6682-6684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spickler C, Mackie GA. Action of RNase II and polynucleotide phosphorylase against RNAs containing stem-loops of defined structure. J Bacteriol. 2000;182:2422–2427. doi: 10.1128/jb.182.9.2422-2427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hossain ST, Malhotra A, Deutscher MP. How RNase R degrades structured RNA: Role of the helicase activity and the S1 domain. J Biol Chem. 2016;291:7877–87. doi: 10.1074/jbc.M116.717991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem. 2006;281:29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- 46.Andrade JM, Cairrao F, Arraiano CM. RNase R affects gene expression in stationary phase: regulation of ompA. Molecular Microbiol. 2006;60:219–228. doi: 10.1111/j.1365-2958.2006.05092.x. [DOI] [PubMed] [Google Scholar]
- 47.Grunberg-Manago M. Polynucleotide phosphorylase. Prog Nucl Acids Res. 1963;1:93–133. [Google Scholar]
- 48.Mohanty BK, Kushner SR. Polynucleotide phosphorylase functions both as a 3′ – 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:11966–71. doi: 10.1073/pnas.220295997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol Microbiol. 2004;54:905–20. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 50.Lin P-H, Lin-Chao S. RhlB helicase rather than enolase is the B-subunit of the Escherichia coli polynucleotide phosphorylase (PNPase)-exoribonucleolytic complex. Proc Natl Acad Sci USA. 2005;102:16590–16595. doi: 10.1073/pnas.0500994102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohanty BK, Kushner SR. Processing of the Escherichia coli leuX tRNA transcript, encoding tRNAleu5, requires either the 3′–5′ exoribonuclease polynucleotide phosphorylase or RNase P to remove the Rho-independent transcription terminator. Nucleic Acids Res. 2010;38:597–607. doi: 10.1093/nar/gkp997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Donovan WP, Kushner SR. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1986;83:120–4. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohanty BK, Kushner SR. Genomic analysis in Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol Microbiol. 2003;50:645–58. doi: 10.1046/j.1365-2958.2003.03724.x. [DOI] [PubMed] [Google Scholar]
- 54.Pobre V, Arraiano CM. Next generation sequencing analysis reveals that the ribonucleases RNase II, RNase R and PNPase affect bacterial motility and biofilm formation in E. coli. BMC Genomics. 2015;16:72. doi: 10.1186/s12864-015-1237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Basturea GN, Zundel MA, Deutscher MP. Degradation of ribosomal RNA during starvation: comparison to quality control during steady-state growth and a role for RNase PH. RNA. 2011;17:338–45. doi: 10.1261/rna.2448911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Durand S, Tomasini A, Braun F, Condon C, Romby P. sRNA and mRNA turnover in Gram-positive bacteria. FEMS Microbiol Rev. 2015;39:316–30. doi: 10.1093/femsre/fuv007. [DOI] [PubMed] [Google Scholar]
- 57.Amblar M, Barbas A, Gomez-Peurtas P, Arraiano CM. The role of the S1 domain in exoribonucleolytic activity: Substrate specificity and multimerization. RNA. 2007;13:317–327. doi: 10.1261/rna.220407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singer MF. Phosphorolysis of oligoribonucleotides by polynucleotide phosphorylase. J Biol Chem. 1958;232:211–28. [PubMed] [Google Scholar]
- 59.Cannistraro VJ, Kennell D. The processive reaction mechanism of ribonuclease II. J Mol Biol. 1994;243:930–43. doi: 10.1006/jmbi.1994.1693. [DOI] [PubMed] [Google Scholar]
- 60.Datta AK, Niyogi K. A novel oligoribonuclease of Escherichia coli. II. Mechanism of action. J Biol Chem. 1975;250:7313–9. [PubMed] [Google Scholar]
- 61.Niyogi SK, Datta AK. A novel oligoribonuclease of Escherichia coli I. Isolation and properties. J Biol Chem. 1975;250:7307–7312. [PubMed] [Google Scholar]
- 62.Ghosh S, Deutscher MP. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc Natl Acad Sci USA. 1999;96:4372–4377. doi: 10.1073/pnas.96.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mohanty BK, Petree JR, Kushner SR. Endonucleolytic cleavages by RNase E generate the mature 3′ termini of the three proline tRNAs in Escherichia coli. Nucleic Acids Res. 2016;44:6350–6362. doi: 10.1093/nar/gkw517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agrawal A, Mohanty BK, Kushner SR. Processing of the seven valine tRNAs in Escherichia coli involves novel features of RNase P. Nucleic Acids Res. 2014;42:11166–79. doi: 10.1093/nar/gku758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mohanty BK, Kushner SR. Rho-independent transcription terminators inhibit RNase P processing of the secG leuU and metT tRNA polycistronic transcripts in Escherichia coli. Nucleic Acids Res. 2008;36:364–75. doi: 10.1093/nar/gkm991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Z, Deutscher MP. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA. 2002;8:97–109. doi: 10.1017/s1355838202014929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mohanty BK, Kushner SR. Ribonuclease P processes polycistronic tRNA transcripts in Escherichia coli independent of ribonuclease E. Nucleic Acids Res. 2007;35:7614–25. doi: 10.1093/nar/gkm917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mohanty BK, Maples VF, Kushner SR. Polyadenylation helps regulate functional tRNA levels in Escherichia coli. Nucleic Acids Res. 2012;40:4589–4603. doi: 10.1093/nar/gks006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zuo Y, Deutscher MP. Mechanism of action of RNase T. I. Identification of residues required for catalysis, substrate binding, and dimerization. J Biol Chem. 2002;277:50155–9. doi: 10.1074/jbc.M207706200. [DOI] [PubMed] [Google Scholar]
- 70.Li Z, Reimers S, Pandit S, Deutscher MP. RNA quality control: degradation of defective transfer RNA. EMBO J. 2002;21:1132–1138. doi: 10.1093/emboj/21.5.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mohanty BK, Kushner SR. Deregulation of poly(A) polymerase I in Escherichia coli inhibits protein synthesis and leads to cell death. Nucleic Acids Res. 2013;41:1757–1766. doi: 10.1093/nar/gks1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ezraty B, Dahlgren B, Deutscher MP. The RNase Z homologue encoded by Escherichia coli elaC gene is RNase BN. J Biol Chem. 2005;280:16542–16545. doi: 10.1074/jbc.C500098200. [DOI] [PubMed] [Google Scholar]
- 73.Dutta T, Deutscher MP. Mode of action of RNase BN/RNase Z on tRNA precursors: RNase BN does not remove the CCA sequence from tRNA. J Biol Chem. 2010;285:22874–81. doi: 10.1074/jbc.M110.141101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang JR, Deutscher MP. Transfer RNA is a substrate for RNase D in vivo. J Biol Chem. 1988;263:17909–17912. [PubMed] [Google Scholar]
- 75.Deutscher MP, Evans JA. Transfer RNA nucleotidyltransferase repairs all transfer RNAs randomly. J Mol Biol. 1977;109:593–7. doi: 10.1016/s0022-2836(77)80093-4. [DOI] [PubMed] [Google Scholar]
- 76.Zhu L, Deutscher MP. tRNA nucleotidyltransferase is not essential for Escherichia coli viability. The EMBO journal. 1987;6:2473–7. doi: 10.1002/j.1460-2075.1987.tb02528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reuven NB, Zhou Z, Deutscher MP. Functional overlap of tRNA nucleotidyltransferases, poly(A) polymerase I, and polynucleotide phosphorylase. J Biol Chem. 1997;272:33255–33259. doi: 10.1074/jbc.272.52.33255. [DOI] [PubMed] [Google Scholar]
- 78.Wellner K, Czech A, Ignatova Z, Betat H, Morl M. Examining tRNA 3′-ends in Escherichia coli: A teamwork between CCA-adding enzyme, RNase T and RNase R. RNA. 2017 doi: 10.1261/rna.064436.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kindler P, Keil TV, Hofschneider PH. Isolation and characterization of an RNase III deficient mutant of Escherichia coli. Mol Gen Genet. 1973;126:53–69. doi: 10.1007/BF00333481. [DOI] [PubMed] [Google Scholar]
- 80.Robertson HD, Webster RE, Zinder ND. A nuclease specific for double-stranded RNA. Virology. 1967;12:718–719. doi: 10.1016/0042-6822(67)90048-7. [DOI] [PubMed] [Google Scholar]
- 81.King TC, Sirdeshmukh R, Schlessinger D. RNase III cleavage is obligate for maturation but not for function of Escherichia coli pre-23S rRNA. Proc Natl Acad Sci USA. 1984;81:185–188. doi: 10.1073/pnas.81.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Srivastava AK, Schlessinger D. Coregulation of processing and translation: mature 5′ termini of Escherichia coli 23S ribosomal RNA form in polysomes. Proc Natl Acad Sci U S A. 1988;85:7144–8. doi: 10.1073/pnas.85.19.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gutgsell NS, Jain C. Coordinated regulation of 23S rRNA maturation in Escherichia coli. J Bacteriol. 2010;192:1405–9. doi: 10.1128/JB.01314-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sulthana S, Deutscher MP. Multiple exoribonucleases catalyze maturation of the 3′ terminus of 16S ribosomal RNA (rRNA) J Biol Chem. 2013;288:12574–9. doi: 10.1074/jbc.C113.459172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rasouly A, Schonbrun M, Shenhar Y, Ron EZ. YbeY, a heat shock protein involved in translation in Escherichia coli. J Bacteriol. 2009;191:2649–55. doi: 10.1128/JB.01663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jacob AI, Kohrer C, Davies BW, RajBhandary UL, Walker GC. Conserved bacterial RNase YbeY plays key roles in 70S ribosome quality control and 16S rRNA maturation. Mol Cell. 2013;49:427–38. doi: 10.1016/j.molcel.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Davies BW, Kohrer C, Jacob AI, Simmons LA, Zhu J, Aleman LM, Rajbhandary UL, Walker GC. Role of Escherichia coli YbeY, a highly conserved protein, in rRNA processing. Mol Microbiol. 2010;78:506–18. doi: 10.1111/j.1365-2958.2010.07351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roy MK, Singh B, Ray BK, Apirion D. Maturation of 5S rRNA: Ribonuclease E cleavages and their dependence on precursor sequences. Eur J Biochem. 1983;131:119–127. doi: 10.1111/j.1432-1033.1983.tb07238.x. [DOI] [PubMed] [Google Scholar]
- 89.Sirdeshmukh R, Krych M, Schlessinger D. Escherichia coli 23S ribosomal RNA truncated at its 5′ terminus. Nucl Acid Res. 1985;13:1185–1192. doi: 10.1093/nar/13.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Babitzke P, Granger L, Olszewski J, Kushner SR. Analysis of mRNA decay and rRNA processing in Escherichia coli multiple mutants carrying a deletion in RNase III. J Bacteriol. 1993;175:229–39. doi: 10.1128/jb.175.1.229-239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li Z, Pandit S, Deutscher MP. Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T. RNA. 1999;5:139–146. doi: 10.1017/s1355838299981669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bram RJ, Young RA, Steitz JA. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of Escherichia coli. Cell. 1980;19:393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- 93.Mohanty BK, Kushner SR. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol Microbiol. 1999;34:1094–108. doi: 10.1046/j.1365-2958.1999.01673.x. [DOI] [PubMed] [Google Scholar]
- 94.Wireman JW, Sypherd PS. In vitro assembly of 30S ribosomal particles from precursor 16S RNA of Escherichia coli. Nature. 1974;247:552–4. doi: 10.1038/247552a0. [DOI] [PubMed] [Google Scholar]
- 95.Takiff HE, Baker T, Copeland T, Chen SM, Court DL. Locating essential Escherichia coli genes by using mini-Tn10 transposons: The pdxJ operon. J Bacteriol. 1992;174:1544–1553. doi: 10.1128/jb.174.5.1544-1553.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–91. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Raghavan R, Groisman EA, Ochman H. Genome-wide detection of novel regulatory RNAs in E. coli. Genome Res. 2011;21:1487–97. doi: 10.1101/gr.119370.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thomason MK, Bischler T, Eisenbart SK, Forstner KU, Zhang A, Herbig A, Nieselt K, Sharma CM, Storz G. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J Bacteriol. 2015;197:18–28. doi: 10.1128/JB.02096-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bilusic I, Popitsch N, Rescheneder P, Schroeder R, Lybecker M. Revisiting the coding potential of the E. coli genome through Hfq co-immunoprecipitation. RNA Biol. 2014;11:641–54. doi: 10.4161/rna.29299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lybecker M, Zimmermann B, Bilusic I, Tukhtubaeva N, Schroeder R. The double-stranded transcriptome of Escherichia coli. Proc Natl Acad Sci U S A. 2014;111:3134–9. doi: 10.1073/pnas.1315974111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci U S A. 2010;107:9602–7. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chao Y, Li L, Girodat D, Forstner KU, Said N, Corcoran C, Smiga M, Papenfort K, Reinhardt R, Wieden HJ, Luisi BF, Vogel J. In vivo cleavage map illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol Cell. 2017;65:39–51. doi: 10.1016/j.molcel.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guo MS, Updegrove TB, Gogol EB, Shabalina SA, Gross CA, Storz G. MicL, a new sigmaE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 2014;28:1620–34. doi: 10.1101/gad.243485.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bandyra KJ, Said N, Pfeiffer V, Gorna MW, Vogel J, Luisi BF. The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol Cell. 2012;47:943–53. doi: 10.1016/j.molcel.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol. 2004;186:6698–705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Desnoyers G, Bouchard MP, Masse E. New insights into small RNA-dependent translational regulation in prokaryotes. Trends Genet. 2013;29:92–8. doi: 10.1016/j.tig.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 107.Sedlyarova N, Shamovsky I, Bharati BK, Epshtein V, Chen J, Gottesman S, Schroeder R, Nudler E. sRNA-Mediated Control of Transcription Termination in E. coli. Cell. 2016;167:111–121. e13. doi: 10.1016/j.cell.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Masse R, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Smirnov A, Forstner KU, Holmqvist E, Otto A, Gunster R, Becher D, Reinhardt R, Vogel J. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc Natl Acad Sci U S A. 2016;113:11591–11596. doi: 10.1073/pnas.1609981113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Viegas SC, Silva IJ, Saramago M, Domingues S, Arraiano CM. Regulation of the small regulatory RNA MicA by ribonuclease III: a target-dependent pathway. Nucleic Acids Res. 2011;39:2918–30. doi: 10.1093/nar/gkq1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Afonyushkin T, Vecerek B, Moll I, Blasi U, Kaberdin VR. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucl Acid Res. 2005;33:1678–1689. doi: 10.1093/nar/gki313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes & Develop. 2005;19:2276–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr Opin Microbiol. 2007;10:134–9. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 114.Andrade JM, Pobre V, Matos AM, Arraiano CM. The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq. RNA. 2012;18:844–55. doi: 10.1261/rna.029413.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bandyra KJ, Sinha D, Syrjanen J, Luisi BF, De Lay NR. The ribonuclease polynucleotide phosphorylase can interact with small regulatory RNAs in both protective and degradative modes. RNA. 2016 doi: 10.1261/rna.052886.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Viegas SC, Pfeiffer V, Sittka A, Silva IJ, Vogel J, Arraiano CM. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 2007;35:7651–64. doi: 10.1093/nar/gkm916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.De Lay N, Gottesman S. Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA. 2011;17:1172–89. doi: 10.1261/rna.2531211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jagessar KL, Jain C. Functional and molecular analysis of Escherichia coli strains lacking multiple DEAD-box helicases. RNA. 2010;16:1386–92. doi: 10.1261/rna.2015610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Charollais J, Dreyfus M, Iost I. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucl Acid Res. 2004;32:2751–2759. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Molecular Microbiol. 2003;48:1253–1265. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- 121.Miczak A, Kaberdin VR, Wei C-L, Lin-Chao S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc Natl Acad Sci USA. 1996;93:3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Khemici V, Toesca I, Poljak L, Vanzo NF, Carpousis AJ. The RNase E of Escherichia coli has at least two binding sites for DEAD-box RNA helicases: functional replacement of RhlB by RhlE. Mol Microbiol. 2004;54:1422–30. doi: 10.1111/j.1365-2958.2004.04361.x. [DOI] [PubMed] [Google Scholar]
- 123.Jain C. The E. coli RhlE RNA helicase regulates the function of related RNA helicases during ribosome assembly. RNA. 2008;14:381–9. doi: 10.1261/rna.800308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Peil L, Virumae K, Remme J. Ribosome assembly in Escherichia coli strains lacking the RNA helicase DeaD/CsdA or DbpA. FEBS J. 2008;275:3772–82. doi: 10.1111/j.1742-4658.2008.06523.x. [DOI] [PubMed] [Google Scholar]
- 125.Prud’homme-Genereux A, Beran RK, Iost I, Ramey CS, Mackie GA, Simons RW. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a ‘cold shock degradosome’. Mol Microbiol. 2004;54:1409–21. doi: 10.1111/j.1365-2958.2004.04360.x. [DOI] [PubMed] [Google Scholar]
- 126.Fuller-Pace FV, Nicol SM, Reid AD, Lane DP. DbpA: a DEAD box protein specifically activated by 23s rRNA. EMBO J. 1993;12:3619–26. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nicol SM, Fuller-Pace FV. The “DEAD box” protein DbpA interacts specifically with the peptidyltransferase center in 23S rRNA. Proc Natl Acad Sci U S A. 1995;92:11681–5. doi: 10.1073/pnas.92.25.11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tsu CA, Uhlenbeck OC. Kinetic analysis of the RNA-dependent adenosinetriphosphatase activity of DbpA, an Escherichia coli DEAD protein specific for 23S ribosomal RNA. Biochemistry. 1998;37:16989–96. doi: 10.1021/bi981837y. [DOI] [PubMed] [Google Scholar]
- 129.Bigger JW. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet. 1944;294:497–500. [Google Scholar]
- 130.Moyed HS, Bertrand KP. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1983;155:768–75. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gerdes K, Maisonneuve E. Bacterial persistence and toxin-antitoxin loci. Annu Rev Microbiol. 2012;66:103–23. doi: 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- 132.Christensen SK, Gerdes K. RelE toxins from bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol Microbiol. 2003;48:1389–400. doi: 10.1046/j.1365-2958.2003.03512.x. [DOI] [PubMed] [Google Scholar]
- 133.Neubauer C, Gao YG, Andersen KR, Dunham CM, Kelley AC, Hentschel J, Gerdes K, Ramakrishnan V, Brodersen DE. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell. 2009;139:1084–95. doi: 10.1016/j.cell.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell. 2003;112:131–140. [Google Scholar]
- 135.Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Molec Cell. 2003;12:913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 136.Jorgensen MG, Pandey DP, Jaskolska M, Gerdes K. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J Bacteriol. 2009;191:1191–9. doi: 10.1128/JB.01013-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Senior BW, Holland IB. Effect of colicin E3 upon the 30S ribosomal subunit of Escherichia coli. Proc Natl Acad Sci U S A. 1971;68:959–63. doi: 10.1073/pnas.68.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bowman CM, Dahlberg JE, Ikemura T, Konisky J, Nomura M. Specific inactivation of 16S ribosomal RNA induced by colicin E3 in vivo. Proc Natl Acad Sci U S A. 1971;68:964–8. doi: 10.1073/pnas.68.5.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Boon T. Inactivation of ribosomes in vitro by colicin E 3 and its mechanism of action. Proc Natl Acad Sci U S A. 1972;69:549–52. doi: 10.1073/pnas.69.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ogawa T. tRNA-targeting ribonucleases: molecular mechanisms and insights into their physiological roles. Biosci Biotechnol Biochem. 2016;80:1037–45. doi: 10.1080/09168451.2016.1148579. [DOI] [PubMed] [Google Scholar]
- 142.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–76. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Winther KS, Gerdes K. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc Natl Acad Sci U S A. 2011;108:7403–7. doi: 10.1073/pnas.1019587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, Dybvig K, Firman K, Gromova ES, Gumport RI, Halford SE, Hattman S, Heitman J, Hornby DP, Janulaitis A, Jeltsch A, Josephsen J, Kiss A, Klaenhammer TR, Kobayashi I, Kong H, Kruger DH, Lacks S, Marinus MG, Miyahara M, Morgan RD, Murray NE, Nagaraja V, Piekarowicz A, Pingoud A, Raleigh E, Rao DN, Reich N, Repin VE, Selker EU, Shaw PC, Stein DC, Stoddard BL, Szybalski W, Trautner TA, Van Etten JL, Vitor JM, Wilson GG, Xu SY. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–12. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–75. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 146.Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu Rev Genet. 2012;46:311–39. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 148.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV, van der Oost J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353:aad5147. doi: 10.1126/science.aad5147. [DOI] [PubMed] [Google Scholar]
- 150.Jain C, Belasco JG. RNase E autoregulates its synthesis by controlling the degradation rate of its own mRNA in Escherichia coli: unusual sensitivity of the rne transcript to RNase E activity. Genes & Develop. 1995;9:84–96. doi: 10.1101/gad.9.1.84. [DOI] [PubMed] [Google Scholar]
- 151.Jarrige A-C, Mathy N, Portier C. PNPase autocontrols its expression by degrading a double-stranded structure in the pnp mRNA leader. EMBO J. 2001;20:6845–6855. doi: 10.1093/emboj/20.23.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Matsunaga J, Simons EL, Simons RW. RNase III autoregulation: Structure and function of rncO, the posttranscriptional “operator”. RNA. 1996;2:1228–1240. [PMC free article] [PubMed] [Google Scholar]
- 153.Lee K, Zhan X, Gao J, Feng Y, Meganathan R, Cohen SN, Georgiou G. RraA: a protein inhibitor of RNase E activity that globally modulates RNA abundance in E. coli. Cell. 2003;114:623–634. [PubMed] [Google Scholar]
- 154.Gao JJ, Lee K, Zhao M, Qiu J, Zhan XM, Saxena A, Moore CJ, Cohen SN, Georgiou G. Differential modulation of E. coli mRNA abundance by inhibitory proteins that alter the composition of the degradosome. Molecular Microbiol. 2006;61:394–406. doi: 10.1111/j.1365-2958.2006.05246.x. [DOI] [PubMed] [Google Scholar]
- 155.Gorna MW, Pietras Z, Tsai YC, Callaghan AJ, Hernandez H, Robinson CV, Luisi BF. The regulatory protein RraA modulates RNA-binding and helicase activities of the E. coli RNA degradosome. RNA. 2010;16:553–62. doi: 10.1261/rna.1858010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kim KS, Manasherob R, Cohen SN. YmdB: a stress-responsive ribonuclease-binding regulator of E. coli RNase III activity. Genes & Develop. 2008;22:3497–3508. doi: 10.1101/gad.1729508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liang W, Malhotra A, Deutscher MP. Acetylation regulates the stability of a bacterial protein: growth stage-dependent modification of RNase R. Mol Cell. 2011;44:160–6. doi: 10.1016/j.molcel.2011.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mohanty BK, Kushner SR. Regulation of mRNA decay in bacteria. Ann Rev Micbiol. 2016;70:25–44. doi: 10.1146/annurev-micro-091014-104515. [DOI] [PubMed] [Google Scholar]
- 159.Mohanty BK, Kushner SR. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 2006;34:5695–704. doi: 10.1093/nar/gkl684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mohanty BK, Kushner SR. Bacterial/archaeal/organellar polyadenylation. WIREs RNA. 2010;2:256–276. [Google Scholar]
- 161.Mohanty BK, Kushner SR. Residual polyadenylation in poly(A) polymerase I (pcnB ) mutants of Escherichia coli does not result from the activity encoded by the f310 gene. Mol Microbiol. 1999;34:1109–19. doi: 10.1046/j.1365-2958.1999.01674.x. [DOI] [PubMed] [Google Scholar]
- 162.Keiler KC. RNA localization in bacteria. Curr Opin Microbiol. 2011;14:155–9. doi: 10.1016/j.mib.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Montero Llopis P, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonet T, Jacobs-Wagner C. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]