Introduction
Diabetic retinopathy (DR) is the leading cause of blindness among U.S. working-aged adults aged 20–74 years.1 The Diabetes Control and Complications Trial (DCCT) showed the strongest factors (duration of diabetes and hemoglobin A1c) explained 11% of the risk of developing retinopathy.2 Similarly, the Wisconsin Epidemiologic Study of Diabetic Retinopathy (WESDR),3 a large population based study, showed that hemoglobin A1c, cholesterol and blood pressure only accounted for 10% of the risk of developing retinopathy, which suggests that other factors may influence the variation of DR. Twin studies and family studies have implicated strong genetic components in DR with heritability scores ranging from 25% to 52% for proliferative diabetic retinopathy (PDR) in either type 1 or type 2 diabetes (DM).4 However, previous analyses, including candidate gene and genome wide association studies (GWAS), have failed to identify genes that are reproducibly associated with DR.5–16 This failure has been attributed to small sample size, incomplete phenotyping of patients, and lack of data on rare variants in such studies.
Whole exome sequencing (WES) of individuals at the phenotypic extremes of disease has previously been successful in identifying genetic factors in conditions for which genetic analyses of common variants have failed.17 In contrast to GWAS, which employs SNPs mostly in non-coding regions to identify common markers that are in linkage disequilibrium with the functional or causal variants, studies in individuals with extreme phenotypes have often detected rare variants in coding regions with large functional effects. WES is a highly effective approach in discovering genes underlying multifactorial diseases. Only one study employing this approach has been published thus far. It found three genes that were associated with protection from DR using a gene-based approach.11
WES of extreme phenotypes is a study design conceived and used successfully by Emond et al. in identifying DCTN4 gene as a modifier of chronic Pseudomonas aeruginosa infection in patients with cystic fibrosis.18 In their study, they performed WES on 41 patients and 48 controls. The same strategy was successfully applied by the same group in discovering another two genes, CAV2 and TMC6, as modifiers of cystic fibrosis.19
Here, we hypothesized that rare or novel variants, especially the null alleles, are enriched in patients with PDR and may be involved in the pathogenesis of diabetic retinopathy. We used WES to identify rare variants of large effect in individuals at the extremes of the phenotypic spectrum of diabetic retinopathy: no DR with at least 10 years of diabetes mellitus (controls) and PDR (cases). We focused especially on frameshift, nonsense, and spicing variants at the canonical splice sites as these are expected to have severe consequences on gene expression and would have a larger functional impact on the pathogenesis of DR. After candidate genes were identified, we performed functional validation studies and investigated the RNA expression of these genes in human retinal endothelial cells (hRECs) under high glucose conditions.
Methods
Study population
The Massachusetts Eye and Ear Infirmary (MEEI), University of Mississippi Medical Center (UMMC), Dean McGee Eye Institute and the University of Oklahoma (DMEI) Institutional Review Boards approved all research involving human subjects. Written informed consent was obtained from all patients. Patients were recruited from two sources. The first is the African American Proliferative Diabetic Retinopathy Study (AAPDR), which has been previously described.20–22 All patients from the AAPDR study had a known diagnosis of type 2 diabetes mellitus by the 2003 American Diabetes Association criteria and/or by being on anti-diabetic medication. All patients had bilateral, dilated wide-field fundus photography. Level of retinopathy was scored using the Early Treatment Diabetic Retinopathy Study (ETDRS) adaptation of the modified Airlie House classification23 and determined by masked ophthalmologist graders. The second source of patients was from MEEI and DMEI. These patients had type 1 or type 2 diabetes mellitus and consisted of mixed ethnicities. All patients had PDR and surgical treatment with vitrectomy. For the analyses, cases were defined as patients with PDR in at least one eye. Controls were patients with no diabetic retinopathy in either eye and with at least 10 years of diagnosed diabetes.20 All controls were from the AAPDR study and were all African American.
After consent was obtained, blood samples were obtained from PDR patients (n=57) and patients with no DR (n=13). DNA was extracted from whole blood and stored at −80 degrees Celsius until the sequencing was ready to be performed. Thirty-one out of the 57 cases and all 13 controls were from the AAPDR Study.20 The 26 remaining cases recruited from the MEEI and DMEI were of different ethnicities. For the analyses, cases were divided into two groups: one group consisting of individuals from the AAPDR Study (AA cases)20 and one group with participants from MEEI and DMEI of mixed ethnicities (ME cases). Analyses included AA cases versus controls (AA group) and ME cases versus controls (AA group). Demographic and clinical information was obtained directly as part of the AAPDR study and from the electronic medical record for MEEI and DMEI patients.
Whole-exome sequencing
Exome capture was performed using Agilent SureSelect Human All Exon V5 kit (Agilent Technologies, Santa Clara, CA) as per the manufacturer’s instructions. Paired-end sequencing (2 × 101 base pair) was performed on an Illumina (San Diego, CA) HiSeq 2000 Next-Generation Sequencing system using v3.0 SBS chemistry with flow cell lane cluster densities of ~700 – 800 K/mm2 on average. One sample was loaded per flow cell lane to obtain a minimum 10× read depth across ~96% of the target regions.
Exome Data Analyses
WES data was analyzed with the MEEI Bioinformatics Center standard pipeline (based on human reference genome GRCh37), as previously described,24 and updated using BWA (version 0.6.2), Samtools (version 0.1.16 and 0.1.18) and latest version of ENSEMBL, (http://www.ensembl.org/index.html), 1000 Genomes Project (http://www.1000genomes.org), Exome Variant Server (EVS) (http://evs.gs.washington.edu/EVS/), SIFT (http://sift.jcvi.org/), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), UK10K Project (http://www.uk10k.org) and Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org; release 0.3). A coverage depth cutoff of 10× was then applied. Heterozygous was defined as a fraction of a variant base between 0.25–0.75 and homozygous was defined as above 0.75.
Only variants likely to alter protein function, such as missense and loss-of-function mutations, were kept for subsequent analysis. Annotations such as the phastCons score and the GERP scores were extracted from batch downloaded data files for human reference genome build hg19 from the UCSC Genome Browser. Variants were filtered further to include only those with a minor allele frequency less than 0.1% in data from the 1000 Genomes Project, EVS, and ExAC.
Cell culture and experimental protocol
hRECs (Cell Systems) were grown in endothelial cell growth medium EBM-2 and singlequots (Lonza), antibiotics (penicillin and streptomycin- Lonza), and 4% FBS (Atlanta Biologicals). Media was changed every 48 hours until cells reached 80 to 90% confluency. Cells were divided and treated under normal glucose (5 mM of D-glucose – Sigma-Aldrich) and high glucose (30 mM of D-glucose) conditions for 72 hours. After 72 hours, cells were harvested for RNA extraction.
RNA Extraction and Quantitative (Real-Time) Reverse-Transcription Polymerase Chain Reaction
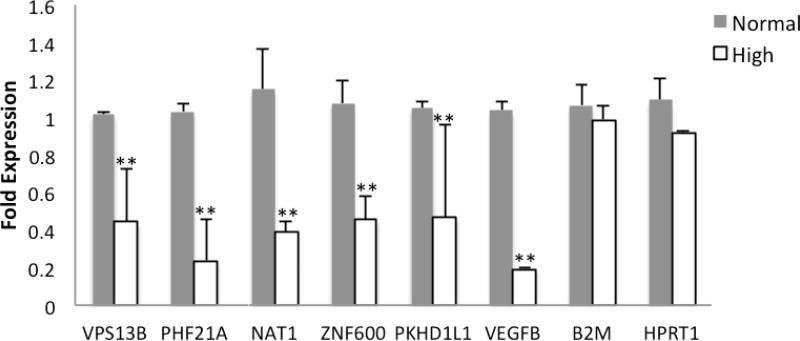
Total RNA was extracted from samples using the RNeasy Mini Kit (Qiagen) following the manufacture instructions. cDNA was prepared with 900 ng RNA using the iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc., Hercules, CA). This was then probed for quantitative (real-time) reverse-transcription polymerase chain reaction using Faststart Universal SYBR Green Master (Hoffmann-La Roche, Basel, Switzerland). Primers were designed using either the NCBI Gene website, or Ensembl Genome website and were purchased from Integrated DNA Technologies. Fold changes were normalized using two different housekeeping genes: HPRT1 and B2M. Forty-four candidate genes and two calibrator genes were screened under high glucose conditions. Genes regulated under high glucose conditions are listed in figure 1.
Figure 1. Expression levels of candidate genes in human retinal endothelial cells (hRECs) after 72 hours of high glucose exposure.
All candidate genes were screened under high glucose conditions and genes regulated under high glucose conditions are listed. Gene expression of VPS13B, PHF21A, NAT1, ZNF600, PKHD1L1, VEGFB were measured in hRECs following 72 hours of treatment with 30mM D-glucose. Fold changes were calculated by 2−ΔΔCT method using B2M and HPRT1 as housekeeping genes and cells grown in 4% FBS media with 5 mM D-glucose as control. Error bars, ±1 standard deviation. **p ≤ 0.05 significantly different than control.
Results
Table 1 shows the characteristics of study participants with PDR compared to those with no DR. Participants with PDR in both AA and ME case groups were more likely to have a shorter duration of diabetes (p < 0.001) and higher HbA1c (p = 0.001) than controls.
Table 1.
Demographics and clinical characteristics
| Cases | Controls | AA cohort | ME cohort |
||
|---|---|---|---|---|---|
| AA cohort (n=31) |
ME cohort (n=26) |
(n=13) | p-value | p-value | |
| Gender | |||||
| Male (%) | 15 (48.4) | 13 (50) | 5 (38.5) | 0.4478 | 0.7342 |
| Female (%) | 16 (51.6) | 13 (50) | 8 (61.5) | ||
| Race | |||||
| African American (%) | 31 (100) | 7 (26.9) | 13 (100) | ||
| Caucasian (%) | 0 | 11 (42.3) | 0 | ||
| Hispanic (%) | 0 | 4 (15.4) | 0 | ||
| Other (%) | 0 | 4 (15.4) | 0 | ||
| Diabetes | |||||
| Type 1 | 0 | 8 (30.8) | 0 | ||
| Type 2 | 31 (100) | 18 (69.2) | 13 (100) | ||
| Age at study entry (mean, years) | 54.8 | 50.2 | 62.1 | 0.145 | 0.08516 |
| Duration of diabetes | 6.4 | 10.4 | 23.9 | <0.001 | <0.001 |
| Hb A1c (%) | 8.6 | 8.3 | 6.8 | <0.001 | <0.001 |
WES identified 721 candidate genes with rare or novel variants in cases with PDR, which were not present in the control samples. These genes were filtered further to include only genes with greater than 2 cases in the ME cohort or greater than 3 cases in the AA cohort. Table 2 lists the variants in 16 candidate genes identified in our AA cohort using these criteria. All variants had an allele frequency of less than 1% in the 1000 genomes project, UK10K Project, and ExAC.
Table 2.
List of 16 candidate genes in African American cases of severe proliferative diabetic retinopathy
| Gene | chr | pos | ref | alt | Nature | Transcript | Protein | dbSNP |
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| HGVSc | HGVSp | |||||||
| AKR1C3 | 10 | 5141639 | C | T | stop_gained,splice_region_variant | NM_003739.5:c.568C>T | NP_003730.4:p.Gln190Ter | rs140580498 |
| KIAA1751 | 1 | 1888057 | A | G | splice_donor_variant | NM_001080484.1:c.2016+2T>C | - | rs191790164 |
| CD96 | 3 | 111342600 | G | A | splice_acceptor_variant | NM_005816.4:c.1181−1G>A | - | rs77738677,COSM4917117 |
| CRIPAK | 4 | 1388651 | T | -CA | frameshift_variant | NM_175918.3:c.352_353delTC | NP_787114.2:p.Ser118ThrfsTer289 | Novel |
| CRIPAK | 4 | 1389433 | C | A | stop_gained | NM_175918.3:c.1134C>A | NP_787114.2:p.Cys378Ter | rs145208075 |
| RGMA | 15 | 93616946 | A | G | splice_donor_variant | NM_001166283.1:c.38+2T>C | - | rs3942115 |
| ZNF77 | 19 | 2933649 | G | C | stop_gained | NM_021217.2:c.1476C>G | NP_067040.1:p.Tyr492Ter | rs34789013,COSM4076533 |
| MPZL3 | 11 | 118104210 | T | -AC | frameshift_variant | NM_198275.1:c.645_646delTA | NP_938016.1:p.Met216GlyfsTer9 | rs144871575 |
| NLRP12 | 19 | 54299165 | G | A | stop_gained | NM_001277126.1:c.3049C>T | NP_001264055.1:p.Arg1017Ter | rs35064500 |
| FAM92A1 | 8 | 94740519 | T | -AAGTA | frameshift_variant | NM_145269.3:c.866_870delAGTAA | NP_660312.2:p.Lys289AsnfsTer9 | Novel |
| EFCAB3 | 17 | 60472551 | T | C | splice_donor_variant | NM_173503.3:c.488+2T>C | - | rs73329490 |
| HNRNPCL1 | 1 | 12907683 | G | A | stop_gained | NM_001013631.1:c.460C>T | NP_001013653.1:p.Arg154Ter | rs142211889 |
| HNRNPCL1 | 1 | 12907352 | T | -C | frameshift_variant | NM_001013631.1:c.791delA | NP_001013653.1:p.Asp264ValfsTer6 | rs545031916 |
| SIGLEC11 | 19 | 50463539 | A | -G | frameshift_variant | NM_001135163.1:c.600delT | NP_001128635.1:p.Arg201GlufsTer37 | rs547387871 |
| ATP12A | 13 | 25266666 | T | -CGGA | frameshift_variant | NM_001185085.1:c.1186_1189delTCGG | NP_001172014.1:p.Ser396ThrfsTer6 | rs557563746 |
| TMEM217 | 6 | 37182972 | A | G | splice_donor_variant | NM_145316.3:c.*2T>C | - | rs116076202 |
| FAM132A | 1 | 1178848 | G | A | stop_gained | NM_001014980.2:c.616C>T | NP_001014980.1:p.Gln206Ter | rs115005664 |
| SLC5A9 | 1 | 48694627 | G | A | splice_donor_variant | NM_001135181.1:c.339+1G>A | - | rs149485404 |
| SLC5A9 | 1 | 48703520 | G | A | splice_donor_variant | NM_001135181.1:c.1536+1G>A | - | rs775853981 |
| SLC5A9 | 1 | 48697766 | G | A | stop_gained | NM_001135181.1:c.915G>A | NP_001128653.1:p.Trp305Ter | rs61997217 |
| Variant Frequency | Conservation scores | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| HGVSc | 1KG | UK10K | ExAC | EVS | phastCons | GERP (range: −12.3–6.17) |
Max | Samples |
| NM_003739.5:c.568C>T | 0.002396 | 0.000132 | 0.000608 | T=2/C=8598;T=29/C=4377;T=31/C=12975 | 1 | 2.67 | 73.2392 | 3 |
| NM_001080484.1:c.2016+2T>C | 0.000998 | 0 | 0.00043 | G=0/A=8454;G=22/A=4134;G=22/A=12588 | 0.747 | 3.2 | - | 3 |
| NM_005816.4:c.1181−1G>A | 0.003594 | 0 | 0.001302 | A=1/G=8597;A=61/G=4345;A=62/G=12942 | 0.94 | 4.02 | - | 3 |
| NM_175918.3:c.352_353delTC | 0 | 0 | 0.001209 | - | 0 | −1.9 | 1.5807 | 2 |
| NM_175918.3:c.1134C>A | 0.003594 | 0 | 0.00075 | A=0/C=8600;A=27/C=4379;A=27/C=12979 | 0.646 | 0.757 | 1.5807 | 1 |
| NM_001166283.1:c.38+2T>C | 0.004593 | 0 | 0.001184 | G=0/A=3178;G=13/A=1359;G=13/A=4537 | 0.23 | 1.42 | - | 3 |
| NM_021217.2:c.1476C>G | 0.003994 | 0 | 0.001326 | C=0/G=8600;C=62/G=4344;C=62/G=12944 | 0.039 | −1.5 | 2.2823 | 3 |
| NM_198275.1:c.645_646delTA | 0.004593 | 0 | 0.001279 | - | 0.988 | 1.94 | 20.2794 | 3 |
| NM_001277126.1:c.3049C>T | 0.004193 | 0 | 0.001665 | A=0/G=8600;A=79/G=4327;A=79/G=12927 | 0.117 | −1.31 | 0.1836 | 3 |
| NM_145269.3:c.866_870delAGTAA | 0 | 0 | 0 | - | 0.13 | 3.04 | 18.0953 | 3 |
| NM_173503.3:c.488+2T>C | 0.003594 | 0 | 0.001634 | C=0/T=8600;C=68/T=4338;C=68/T=12938 | 1 | 5.99 | - | 3 |
| NM_001013631.1:c.460C>T | 0.004593 | 0 | 0.001035 | A=0/G=8594;A=17/G=4387;A=17/G=12981 | 0.926 | −2.02 | 0.5411 | 1 |
| NM_001013631.1:c.791delA | 0.004393 | 0 | 0.000837 | - | 0.011 | 1.09 | 0.5411 | 2 |
| NM_001135163.1:c.600delT | 0.001997 | 0 | 0.000169 | - | 0 | −6.23 | 0.5998 | 3 |
| NM_001185085.1:c.1186_1189delTCGG | 0.003794 | 0 | 0.000908 | - | 1 | 5.63 | 0.5259 | 3 |
| NM_145316.3:c.*2T>C | 0.003794 | 0 | 0.001279 | G=0/A=8600;G=65/A=4341;G=65/A=12941 | 0.083 | 1.73 | - | 3 |
| NM_001014980.2:c.616C>T | 0.003195 | 0 | 0.000832 | A=1/G=8423;A=56/G=4222;A=57/G=12645 | 1 | 3.8 | 0.2202 | 3 |
| NM_001135181.1:c.339+1G>A | 0.002796 | 0 | 0.000474 | A=0/G=8600;A=20/G=4386;A=20/G=12986 | 1 | 5.51 | 0.8064 | 2 |
| NM_001135181.1:c.1536+1G>A | 0 | 0 | 3.16E-05 | - | 1 | 5.02 | - | 1 |
| NM_001135181.1:c.915G>A | 0.003395 | 0 | 0.001073 | A=0/G=8600;A=55/G=4351;A=55/G=12951 | 1 | 6.04 | 0.6774 | 2 |
1KG, 1000 genomes; UK10K, UK10K Project; ExAC, Exome Aggregation Consortium; EVS, exome variant server
The 1000 Genomes database included large cohorts from various ethnicities: African, Latino, East Asian, European and South Asian. The ExAC database included sequences from African American, Latino, East Asian, Finnish, Non-Finnish European, and South Asian ethnicities. The EVS exome database included two ethnically distinct U.S. populations: African-Americans and European-Americans. The UK10K database had a majority of sequences from samples of European-descent.
One candidate gene in particular, SLC5A9, a sodium-dependent glucose transporter, had null allele variants in 5 cases in the AA cohort: two samples with rs149485404 (chr1:48694627 G>A) resulting in a +1bp 5' splice site, two samples with rs61997217 (chr1:48697766 G>A) resulting in a p.W305 protein change and one with rs775853981 (chr1:48703520 G>A) causing a +1bp 5' splice site mutation. All three variants occurred at evolutionally conserved positions (phastCons scores of 1, 1, and 1, respectively). The phastCons score, ranging from 0–1, represents the posterior probability that the base position is at its most conserved state according to phastCons's phylogenetic hidden Markov model.25
In our ME cohort, we identified 28 candidate genes on the basis of their carrying null alleles in at least two subjects with PDR and none in the controls (Table 3). Twenty-three variants from 17 genes were novel or have been reported at very low frequencies by the EVS, 1000 genomes project, UK10K Project or ExAC. One variant was found in 4 cases in the ME cohort. This variant was a frameshift deletion in the ZNF600 gene (c.1934_1935del), resulting in a NP_940859.2:p.Lys645AsnfsTer9 protein change. This variant had a low allele frequency in the commonly referenced exome databases: 0.0018 (1000 genomes), 0 (UK10K), and 0.00068 (ExAC).
Table 3.
List of 28 candidate genes in Mixed Ethnicity cases of severe proliferative diabetic retinopathy
| Gene | chr | pos | ref | alt | Nature | Transcript HGVSc |
Protein HGVSp |
dbSNP |
|---|---|---|---|---|---|---|---|---|
| ABCA7 | 19 | 1049426 | A | -C | frameshift_variant | NM_019112.3:c.2544delC | NP_061985.2:p.Thr849ProfsTer6 | Novel |
| ABCA7 | 19 | 1044707 | G | -GGGCACCTGGT | frameshift_variant | NM_019112.3:c.1179_1189delGGGGCACCTGG | NP_061985.2:p.Leu396AlafsTer45 | Novel |
| ABCA7 | 19 | 1063762 | G | T | stop_gained | NM_019112.3:c.5851G>T | NP_061985.2:p.Glu1951Ter | Novel |
| ABHD17A | 19 | 1881528 | A | -AG | frameshift_variant | NM_031213.3:c.39_40delCT | NP_112490.3:p.Phe13LeufsTer27 | Novel |
| ANO2 | 12 | 5941617 | C | T | splice_donor_variant | NM_001278596.1:c.788+1G>A | - | rs200258741 |
| ANO2 | 12 | 5842031 | C | G | splice_donor_variant | NM_001278596.1:c.1449+1G>C | - | rs200963904 |
| BPIFB6 | 20 | 31631146 | C | A | stop_gained | NM_174897.2:c.1302C>A | NP_777557.1:p.Tyr434Ter | rs140595029 |
| BPIFB6 | 20 | 31619550 | C | T | stop_gained, splice_region_variant | NM_174897.2:c.97C>T | NP_777557.1:p.Gln33Ter | rs774063482 |
| C15orf32 | 15 | 93015466 | A | T | stop_gained | NM_153040.2:c.88A>T | NP_694585.1:p.Lys30Ter | rs115999940 |
| CCDC105 | 19 | 15121689 | G | -CC | frameshift_variant | NM_173482.2:c.55_56delCC | NP_775753.2:p.Pro19SerfsTer129 | Novel |
| CDKL1 | 14 | 50799034 | T | -GTTG | frameshift_variant | NM_004196.3:c.912_915delAACA | NP_004187.2:p.Thr305GlyfsTer4 | Novel |
| CDKL1 | 14 | 50862418 | C | G | splice_donor_variant | NM_004196.3:c.171+1G>C | - | rs200559651 |
| CEP192 | 18 | 13056620 | C | T | missense_variant | NM_032142.3:c.4031C>T | NP_115518.3:p.Thr1344Ile | rs144622986 |
| COL6A5 | 3 | 130095079 | G | A | splice_acceptor_variant | NM_153264.6:c.68−1G>A | - | rs142846354 |
| CRIPAK | 4 | 1389424 | T | -CA | frameshift_variant | NM_175918.3:c.1125_1126delTC | NP_787114.2:p.Thr377ValfsTer30 | Novel |
| CRIPAK | 4 | 1388466 | T | -CA | frameshift_variant | NM_175918.3:c.167_168delTC | NP_787114.2:p.Leu56HisfsTer351 | Novel |
| DNHD1 | 11 | 6592054 | C | T | stop_gained | NM_144666.2:c.13312C>T | NP_653267.2:p.Arg4438Ter | rs80197979 |
| DNHD1 | 11 | 6566679 | C | T | stop_gained | NM_144666.2:c.4510C>T | NP_653267.2:p.Gln1504Ter | rs536843662 |
| DNHD1 | 11 | 6587805 | T | -GTTACCCCAA | splice_region_variant,intron_variant | NM_144666.2:c.11207−12_11207−3delTGTTACCCCA | - | Novel |
| DNHD1 | 11 | 6579307 | C | T | stop_gained | NM_144666.2:c.8782C>T | NP_653267.2:p.Arg2928Ter | rs199752008 |
| GPATCH1 | 19 | 33579035 | T | -TTA | splice_region_variant,intron_variant | NM_018025.2:c.74−5_74−3delTTT | - | Novel |
| HMCN1 | 1 | 186060001 | G | A | stop_gained | NM_031935.2:c.9839G>A | NP_114141.2:p.Trp3280Ter | rs375502689 |
| KIF24 | 9 | 34306386 | A | -CT | frameshift_variant | NM_194313.2:c.676_677delGT | NP_919289.2:p.Val226LeufsTer18 | Novel |
| KIF24 | 9 | 34269293 | T | -TGTC | frameshift_variant | NM_194313.2:c.1402_1405delACAA | NP_919289.2:p.Thr468ArgfsTer7 | Novel |
| LRBA | 4 | 151936623 | A | T | splice_donor_variant | NM_006726.4:c.−220+2T>A | - | Novel |
| LRBA | 4 | 151765814 | G | A | missense_variant | NM_006726.4:c.4457C>T | NP_006717.2:p.Ala1486Val | rs149639181 |
| LRP8 | 1 | 53793510 | C | -TG | frameshift_variant | NM_001018054.2:c.75_76delGC | NP_001018064.1:p.Gln25HisfsTer10 | Novel |
| MSH2 | 2 | 47641558 | G | -TAAA | splice_donor_variant,intron_variant | NM_000251.2:c.942+1_942+4delGTAA | - | Novel |
| NAT1 | 8 | 18080115 | C | T | stop_gained | NM_001160175.1:c.745C>T | NP_001153647.1:p.Arg249Ter | rs5030839,CM981375 |
| PHF21A | 11 | 45957289 | T | -G | frameshift_variant,splice_region_variant | NM_016621.3:c.1545delA | NP_057705.3:p.Glu517LysfsTer14 | Novel |
| PKHD1L1 | 8 | 110477066 | C | T | stop_gained | NM_177531.4:c.8005C>T | NP_803875.2:p.Gln2669Ter | rs72687034 |
| PKHD1L1 | 8 | 110460558 | C | G | stop_gained | NM_177531.4:c.5963C>G | NP_803875.2:p.Ser1988Ter | Novel |
| SLC6A13 | 12 | 368999 | C | -A | frameshift_variant | NM_001243392.1:c.221delT | NP_001230321.1:p.Val74GlyfsTer32 | Novel |
| SLURP1 | 8 | 143822563 | A | G | stop_lost | NM_020427.2:c.310T>C | NP_065160.1:p.Ter104ArgextTer16 | rs62636565 |
| TTC22 | 1 | 55247197 | C | A | stop_gained | NM_001114108.1:c.1429G>T | NP_001107580.1:p.Glu477Ter | rs61733131 |
| TTC22 | 1 | 55251251 | G | A | stop_gained | NM_017904.3:c.1087C>T | NP_060374.2:p.Arg363Ter | rs112886857 |
| UPK3A | 22 | 45683240 | C | A | stop_gained | NM_006953.3:c.396C>A | NP_008884.1:p.Tyr132Ter | rs138640270 |
| UPK3A | 22 | 45685025 | G | A | splice_donor_variant | NM_006953.3:c.571+1G>A | - | rs145723454,COSM5859557 |
| VPS13B | 8 | 100844595 | A | -G | splice_acceptor_variant | NM_017890.4:c.9406−1delG | - | Novel |
| ZDHHC11B | 5 | 710709 | C | -TG | 3_prime_UTR_variant | XM_003118532.5:c.*1695_*1696delAG | - | Novel |
| ZDHHC11 | 5 | 840725 | C | -AG | frameshift_variant | NM_024786.2:c.668_669delTG | NP_079062.1:p.Phe224ProfsTer46 | Novel |
| ZNF600 | 19 | 53269073 | C | -TT | frameshift_variant | NM_198457.2:c.1935_1936delAG | NP_940859.2:p.Lys645AsnfsTer9 | Novel |
| Variant Frequency | Conservation scores | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| HGVSc | 1KG | UK10K | ExAC | EVS | phastCons | GERP (range: −12.3–6.17) |
Max | Samples |
| NM_019112.3:c.2544delC | 0 | 0 | 5.62E-05 | 0 | 1 | 2.99 | 25.0107 | 1 |
| NM_019112.3:c.1179_1189delGGGGCACCTGG | 0.002196 | 0 | 0.000437 | 0 | 0.043 | 0.601 | 47.8482 | 1 |
| NM_019112.3:c.5851G>T | 0 | 0 | 0 | 0 | 1 | 3.52 | 45.96 | 1 |
| NM_031213.3:c.39_40delCT | 0 | 0 | 4.09E-05 | 0 | 1 | 3.69 | 6.9166 | 2 |
| NM_001278596.1:c.788+1G>A | 0.000799 | 0 | 0.000223 | T=0/C=8366;T=13/C=3889;T=13/C=12255 | 1 | 5.72 | - | 1 |
| NM_001278596.1:c.1449+1G>C | 0.0002 | 0 | 0.00012 | G=0/C=8394;G=8/C=4088;G=8/C=12482 | 1 | 4.76 | - | 1 |
| NM_174897.2:c.1302C>A | 0.002396 | 0 | 0.000852 | A=1/C=8599;A=36/C=4370;A=37/C=12969 | 0.923 | −1.93 | 0 | 1 |
| NM_174897.2:c.97C>T | 0 | 0 | 2.37E-05 | 0 | 0.199 | 4.33 | 0 | 1 |
| NM_153040.2:c.88A>T | 0.001997 | 0 | 0.000616 | T=0/A=8596;T=25/A=4371;T=25/A=12967 | 0.004 | −0.477 | 0 | 2 |
| NM_173482.2:c.55_56delCC | 0.000799 | 0.001851 | 0.000449 | 0 | 0.053 | 2.77 | 0.0052 | 2 |
| NM_004196.3:c.912_915delAACA | 0 | 0.000264 | 8.68E-05 | 0 | 0 | −2.4 | 6.1617 | 1 |
| NM_004196.3:c.171+1G>C | 0.0002 | 0.000264 | 0.000245 | G=2/C=8598;G=1/C=4405;G=3/C=13003 | 1 | 4.21 | - | 1 |
| NM_032142.3:c.4031C>T | 0.000998 | 0 | 0.000395 | T=0/C=8600;T=17/C=4389;T=17/C=12989 | 0.292 | 3.22 | 4.438 | 2 |
| NM_153264.6:c.68−1G>A | 0.001398 | 0 | 0.000385 | 0 | 0.017 | 4.13 | - | 1 |
| NM_175918.3:c.1125_1126delTC | 0.004593 | 0 | 0 | 0 | 0.185 | −1.1 | 1.5807 | 1 |
| NM_175918.3:c.167_168delTC | 0 | 0 | 0.000428 | 0 | 0.001 | −2.22 | 1.5807 | 1 |
| NM_014675.3:c.3618C>T | 0.004393 | 0 | 0.001302 | T=0/C=8552;T=60/C=4284;T=60/C=12836 | 0.97 | −5.31 | 22.5101 | 1 |
| NM_144666.2:c.4510C>T | 0.0002 | 0 | 6.26E-05 | 0 | 0 | −3.18 | 0.6055 | 1 |
| NM_144666.2:c.11207−12_11207−3delTGTTACCCCA | 0 | 0 | 0.000088 | 0 | 0.001 | 3.07 | 1.0815 | 1 |
| NM_144666.2:c.8782C>T | 0.000799 | 0.000661 | 0.000655 | T=2/C=3180;T=1/C=1383;T=3/C=4563 | 0.015 | −2.28 | 0.7669 | 1 |
| NM_018025.2:c.74−5_74−3delTTT | 0 | 0 | 0 | 0 | 0.814 | 0.368 | - | 2 |
| NM_031935.2:c.9839G>A | 0 | 0 | 2.37E-05 | A=1/G=8597;A=0/G=4406;A=1/G=13003 | 1 | 5.98 | 2.3114 | 2 |
| NM_194313.2:c.676_677delGT | 0 | 0 | 0.000032 | 0 | 0.996 | 4.58 | 0.5141 | 1 |
| NM_194313.2:c.1402_1405delACAA | 0.000799 | 0 | 0.001889 | 0 | 1 | 5.42 | 1.2178 | 1 |
| NM_006726.4:c.−220+2T>A | 0 | 0 | 0 | 0 | 1 | 3.77 | - | 1 |
| NM_006726.4:c.4457C>T | 0.000399 | 0 | 8.68E-05 | A=1/G=8599;A=0/G=4406;A=1/G=13005 | 1 | 5.19 | 19.9326 | 1 |
| NM_001018054.2:c.75_76delGC | 0 | 0 | 0.003842 | 0 | 0.298 | 2.53 | 0.257 | 2 |
| NM_000251.2:c.942+1_942+4delGTAA | 0 | 0 | 0 | 0 | 1 | 4.65 | - | 2 |
| NM_001160175.1:c.745C>T | 0.002796 | 0.002116 | 0.00266 | T=27/C=8573;T=9/C=4397;T=36/C=12970 | 0 | −0.582 | 1.1163 | 2 |
| NM_016621.3:c.1545delA | 0 | 0 | 0 | 0 | 0.973 | −4.14 | 6.0199 | 2 |
| NM_177531.4:c.8005C>T | 0.000799 | 0.001984 | 0.001353 | T=17/C=8201;T=0/C=3728;T=17/C=11929 | 1 | 5.78 | 0.21 | 1 |
| NM_177531.4:c.5963C>G | 0 | 0 | 0 | 0 | 0.995 | 2.49 | 0.1821 | 1 |
| NM_001243392.1:c.221delT | 0.001797 | 0.003174 | 0.003709 | 0 | 0.136 | 4.33 | 6.1782 | 2 |
| NM_020427.2:c.310T>C | 0.001198 | 0 | 0.000499 | G=0/A=8598;G=22/A=4384;G=22/A=12982 | 0 | −1.35 | 1.4982 | 3 |
| NM_001114108.1:c.1429G>T | 0.001597 | 0 | 0.00024 | 0 | 0.995 | 3.93 | 5.2207 | 1 |
| NM_017904.3:c.1087C>T | 0.002796 | 0.000132 | 0.00027 | 0 | 0.016 | −0.397 | 0.3699 | 1 |
| NM_006953.3:c.396C>A | 0 | 0 | 7.90E-06 | A=0/C=8600;A=1/C=4405;A=1/C=13005 | 1 | 2.26 | 0 | 1 |
| NM_006953.3:c.571+1G>A | 0.003594 | 0 | 0.001294 | A=1/G=8599;A=50/G=4356;A=51/G=12955 | 0.999 | 4.68 | - | 1 |
| NM_017890.4:c.9406−1delG | 0 | 0 | 0 | 0 | 0.998 | 5.55 | - | 2 |
| XM_003118532.5:c.*1695_*1696delAG | 0 | 0 | 0 | 0 | 0.061 | 0 | 7.7216 | 1 |
| NM_024786.2:c.668_669delTG | 0 | 0 | 0.000111 | 0 | 0 | −1.99 | 5.5912 | 1 |
| NM_198457.2:c.1935_1936delAG | 0.001797 | 0 | 0.000679 | 0 | 0.008 | −2.23 | 1.9314 | 4 |
1KG, 1000 genomes; UK10K, UK10K Project; ExAC, Exome Aggregation Consortium; EVS, exome variant server
We also sought to determine if there were rare or novel variants in two well-studied genes believed to be important in the pathogenesis of diabetes or DR: vascular endothelial growth factor (VEGFB) and apolipoprotein B (apoB). VEGF has several members including VEGF-A, VEGF-B, VEGF-C, VEGF-D and placental growth factor (PlGF). Decreased VEGF-B signaling in rodent models of type 2 diabetes has been found to restore insulin sensitivity. In addition, VEGF-B levels have been found to be significantly correlated with diabetic retinopathy.26–28 Three nonsynonymous variants in VEGFB were detected in 4 PDR patients (3 AA cases, 1 ME case) as shown in Table 4. One variant was novel, (chr11:64002973 G>A). ApoB is the main component of low-density lipoprotein cholesterol (LDL-C) and apoB100 has been found to be associated with diabetic retinopathy.29, 30 Eight non-synonymous variants were identified with four variants that were novel (table 5).
Table 4.
VEGFB variants in severe PDR cases
| Gene | chr | pos | ref | alt | RS ID | Amino acid (protein) |
|---|---|---|---|---|---|---|
| VEGFB | 11 | 64004924 | G | A | rs138325963 | p.R148H |
| 11 | 64005108 | C | T | rs61384522 | p.L176F | |
| 11 | 64002973 | G | A | novel | +1bp 5' splice site |
| Variant Frequency | Samples | |||
|---|---|---|---|---|
|
| ||||
| 1KG | UK10K | ExAC | EVS | |
| 0.00479233 | 0 | 0.001383 | A=1/G=8585;A=61/G=4337;A=62/G=12922 | 2 |
| 0.00359425 | 0.0006746 | T=0/C=8226;T=27/C=4185;T=27/C=12411 | 1 | |
| 0 | 0.000793 | 1.58E-05 | none | 1 |
Table 5.
APOB variants in severe PDR cases
| Gene | Variant | Amino acid (protein) |
|---|---|---|
| APOB | rs12713540 | p.S3801T |
| rs61741974 | p.F3753L | |
| Novel | p.E3382Q | |
| rs72653099 | p.L3076M | |
| Novel | p.G2540V | |
| Novel | p.K2110fs | |
| Novel | p.L1568M | |
| rs140877474 | p.S1459G |
To determine whether the candidate genes may play a role in proliferative diabetic retinopathy, we assessed 44 candidate genes for changes in expression in human retinal endothelial cells (hRECs) cultured in high or normal glucose conditions using RT-PCR. We found that the expression of six of our candidate genes including VEGFB, VPS13B, PHF21A, NAT1, ZNF600, PKHD1L1 was reduced in hRECs cultured in high glucose (Figure 1).
Discussion
We performed a WES study using an extreme phenotype design on 57 PDR patients and 13 controls without retinopathy to detect genes contributing to disease risk. We selected for rare or novel protein-truncating variants and identified a total of 44 candidate genes: 16 genes in our AA case group and 28 genes in ME case group. The final variants identified were not observed or were observed at very low frequencies in the 1000 genomes, UK10K, ExAC and EVS exome databases. In both combined cohorts, 25 novel variants in 19 genes were identified. To our knowledge, these genes have not been previously identified in other candidate gene studies or from genome-wide association studies in population cohorts in DR. Furthermore, our functional study suggests a potential role of six candidate genes in the pathogenesis of PDR.
Prior studies have identified several candidate genes in association with diabetic retinopathy.31, 32 In our AA cases with type 2 diabetes, one gene related to angiogenesis, TMEM217 on chromosome 6, was identified as a candidate gene. TMEM217 is a transmembrane protein involved in the MAPK pathway. One GWAS study suggested an association of two polymorphisms with DR, but none of the variants were statistically significant after Bonferroni correction.33 One study found a 2-fold increase in gene expression of TMEM217 in vascular endothelial cells treated with DMU-212, a derivative of resveratrol that possesses potent pro-apoptotic and anti-angiogenic effects.34 This suggests a potential role for TMEM217 in the signal transduction of inflammation and apoptosis pathways, which are involved in the pathogenesis of DR.35
We also discovered a 2 bp frameshift mutation in ZNF600 with an allele frequency of 4/26 or 0.154 within our ME cohort. This mutation was absent in the controls and its allele frequency was very low in the commonly referenced exome databases: 0.0018 (1000 genomes), 0 (UK10K), and 0.00068 (ExAC). It is statistically unlikely (p < 0.01), therefore, to see 4/26 unrelated PDR patients to have this particular variant even after applying Bonferroni multiple testing correction for the number of candidate variants after filtering, as listed in Table 3. ZNF600 is a zinc finger protein that may be involved in transcriptional regulation. In a recent GWAS study, ZNF600 was found to be significantly associated with novel phospholipid loci.36 Phospholipids are key regulators of intracellular processes and have been implicated in the pathology of type 2 diabetes.37 Disruption of their metabolism has diverse metabolic consequences and may be associated with diabetic retinopathy.37–39
Another interesting candidate gene was SLC5A9, where three rare variants were identified in five patients in the AA cohort. SLC5A9 (SGLT4) is found on chromosome 1 and is a sodium-dependent glucose transporter.40 Hypoxia, growth factors and cytokines upregulate glucose transport in endothelial cells in diabetes.41 Although usually found in the intestine and kidney, this gene was also found to be expressed in hREC in our study (Table 2). Solute carrier family (SLC) proteins play a role in insulin secretion and several studies have looked at the association of SLC proteins with DR.42–44 Some have found nominal associations45 and others have found no association to DR.42
One interesting candidate variant, rs115005664, found in three cases in the AA cohort was a non-synonymous mutation in the FAM132A gene (CTRP12/Adipolin). FAM132A is an adipose-derived insulin-sensitizing factor and functions as an adipokine, which are cytokines that are secreted by adipose tissue. Acute and chronic hyperinsulinemic states were found to have significantly increased circulating levels of adipolin.46 Adipolin also has anti-inflammatory effects that exert beneficial actions on glucose metabolism.47 Some adipokines are known to regulate (repress) endothelial angiogenesis, e.g. adiponectin48, and may play a role in diabetic cardiovascular and metabolic complications such as DR.
We also examined two well-studied genes of biological relevance in diabetic retinopathy: VEGFB and apoB. VEGF is considered a primary promoter of the neovascularization in PDR. A number of polymorphisms in VEGFB have been analyzed either with DR or severe DR.16, 49–51 Our study identified three non-synonymous variants in 4 PDR patients: rs138325963, rs61384522 and one novel variant. These variants have not been associated in prior genetic studies and warrant further investigation. In addition, ApoB was also analyzed with eight non-synonymous variants identified including four novel ones. ApoB is the main component of LDL-C and is correlated with atherogenicity.52,53
To date, only one other study has used extreme phenotype design with WES to identify candidate genes associated with diabetic retinopathy. In their study, Shtir et al. performed whole exome sequencing on 64 diabetics with diabetic retinopathy (controls) and 43 diabetics without retinopathy (cases), all of Saudi descent.11 They identified three genes, NME3, LOC728699, and FASTK, whose increased rare variant burden appeared to protect against DR. We did not identify these genes in our study and this may be due to the difference in patient populations. While Shtir et al. described a cohort of Saudi descent, our study’s population were primarily Caucasian and African American.
In this study, we also examined the expression of 44 genes and found that six of these candidate genes (VEGFB, VPS13B, PHF21A, NAT1, ZNF600, PKHD1L1) are regulated by culture conditions resembling the diabetic milieu. We demonstrate that high glucose challenge significantly decreases the expression of all six genes in human retinal endothelial cells. These results suggest an important role of hyperglycemia-induced suppression of these genes in diabetic retinopathy. Although additional functional analyses are necessary, our data allow us to postulate a model whereby a diabetic milieu interacts with rare gene variation to modify the susceptibility to PDR.
We recognize several limitations of this study. Our cases of mixed ethnicities were compared to controls consisting of only African American patients. This may not provide optimal or adequately comparable results because of racial differences in allele frequencies and genetic risks for the disease. Non-Hispanic blacks, for example, are known to have significantly higher risk for diabetic retinopathy than the non-Hispanic whites.54 A candidate rare variant might have different allele frequencies in different ethnic groups. However, we divided our cases into two cohorts in order to minimize this problem, with a threshold difference of greater than two cases between either cohort compared to controls. Many of the identified candidate rare variants this way do have different allele frequencies in the European Americans comparing to African Americans, as indicated by the EVS column in the tables. If such a candidate gene or variant is indeed associated with increased risk for diabetic retinopathy, it would therefore contribute to different amount of genetic risk in each ethnic group. It is also possible that these variants are associated with ethnicity and not disease itself. Further validation in other cohorts will be necessary to determine this. The number of controls was limited because we chose a very stringent definition of the extreme of phenotype – no diabetic retinopathy despite at least 10 years of type 2 diabetes. The limited sample size could have led to false positive results, underscoring the need for further follow-up functional characterization of these changes.
We have identified numerous candidate genes that warrant further investigation. This mode of analysis allows us to begin to understand the complex genomics of PDR, and may help to identify the pathways that contribute to the disease process in this population. The global burden of diabetic retinopathy is significant, and efforts in precision health have galvanized new ways to identify at-risk patients and spurred clinical discoveries in targeted treatments. Future in vitro and in vivo studies involving knockdown of gene targets are warranted. The variants we identified highlight the spectrum of defects potentially relevant in this population. Identifying genes that may be responsible for severity of disease may allow for the development of new therapies, alleviating significant morbidity worldwide.
Acknowledgments
Funding: We gratefully acknowledge support from the following organizations for this research: Research to Prevent Blindness William and Mary Greve Special Scholar Award (LS) and Career Development Award (LS); National Eye Institute (K12EY16335 (LAK); EY22302 (LS); R21EY037061 (LAK)); NIH grants (R00EY021624; UH2NS100121-01 (JFAV); E. Matilda Ziegler Foundation for the Blind (LAK); Karl Kirchgessner Foundation (LAK); Massachusetts Lions Eye Research Fund (JFAV); Alcon Research Institute; American Diabetes Association (1-11-CT-51); Harvard Catalyst; Department of Ophthalmology, Harvard Medical School (LAK and JFAV).
References
- 1.Ruta LM, Magliano DJ, Lemesurier R, et al. Prevalence of diabetic retinopathy in Type 2 diabetes in developing and developed countries. Diabet Med. 2013;30:387–98. doi: 10.1111/dme.12119. [DOI] [PubMed] [Google Scholar]
- 2.Lachin JM, Genuth S, Nathan DM, et al. Effect of glycemic exposure on the risk of microvascular complications in the diabetes control and complications trial--revisited. Diabetes. 2008;57:995–1001. doi: 10.2337/db07-1618. [DOI] [PubMed] [Google Scholar]
- 3.Klein R, Klein BE, Moss SE, et al. The Wisconsin Epidemiologic Study of Diabetic Retinopathy: XVII. The 14-year incidence and progression of diabetic retinopathy and associated risk factors in type 1 diabetes. Ophthalmology. 1998;105:1801–15. doi: 10.1016/S0161-6420(98)91020-X. [DOI] [PubMed] [Google Scholar]
- 4.Hietala K, Forsblom C, Summanen P, et al. Heritability of proliferative diabetic retinopathy. Diabetes. 2008;57:2176–80. doi: 10.2337/db07-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheu WH, Kuo JZ, Lee IT, et al. Genome-wide association study in a Chinese population with diabetic retinopathy. Hum Mol Genet. 2013;22:3165–73. doi: 10.1093/hmg/ddt161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Awata T, Yamashita H, Kurihara S, et al. A genome-wide association study for diabetic retinopathy in a Japanese population: potential association with a long intergenic non-coding RNA. PLoS One. 2014;9:e111715. doi: 10.1371/journal.pone.0111715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu YP, Hallman DM, Gonzalez VH, et al. Identification of Diabetic Retinopathy Genes through a Genome-Wide Association Study among Mexican-Americans from Starr County, Texas. J Ophthalmol. 2010;2010 doi: 10.1155/2010/861291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grassi MA, Tikhomirov A, Ramalingam S, et al. Genome-wide meta-analysis for severe diabetic retinopathy. Hum Mol Genet. 2011;20:2472–81. doi: 10.1093/hmg/ddr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang YC, Lin JM, Lin HJ, et al. Genome-wide association study of diabetic retinopathy in a Taiwanese population. Ophthalmology. 2011;118:642–8. doi: 10.1016/j.ophtha.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 10.Burdon KP, Fogarty RD, Shen W, et al. Genome-wide association study for sight-threatening diabetic retinopathy reveals association with genetic variation near the GRB2 gene. Diabetologia. 2015;58:2288–97. doi: 10.1007/s00125-015-3697-2. [DOI] [PubMed] [Google Scholar]
- 11.Shtir C, Aldahmesh MA, Al-Dahmash S, et al. Exome-based case-control association study using extreme phenotype design reveals novel candidates with protective effect in diabetic retinopathy. Hum Genet. 2016;135:193–200. doi: 10.1007/s00439-015-1624-8. [DOI] [PubMed] [Google Scholar]
- 12.Hosseini SM, Boright AP, Sun L, et al. The association of previously reported polymorphisms for microvascular complications in a meta-analysis of diabetic retinopathy. Hum Genet. 2015;134:247–57. doi: 10.1007/s00439-014-1517-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grassi MA, Tikhomirov A, Ramalingam S, et al. Replication analysis for severe diabetic retinopathy. Invest Ophthalmol Vis Sci. 2012;53:2377–81. doi: 10.1167/iovs.11-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng D, Wang J, Zhang R, et al. Common variants in or near ZNRF1, COLEC12, SCYL1BP1 and API5 are associated with diabetic retinopathy in Chinese patients with type 2 diabetes. Diabetologia. 2015;58:1231–8. doi: 10.1007/s00125-015-3569-9. [DOI] [PubMed] [Google Scholar]
- 15.Cheung CY, Hui EY, Lee CH, et al. Impact of Genetic Loci Identified in Genome-Wide Association Studies on Diabetic Retinopathy in Chinese Patients With Type 2 Diabetes. Invest Ophthalmol Vis Sci. 2016;57:5518–5524. doi: 10.1167/iovs.16-20094. [DOI] [PubMed] [Google Scholar]
- 16.Abhary S, Hewitt AW, Burdon KP, et al. A systematic meta-analysis of genetic association studies for diabetic retinopathy. Diabetes. 2009;58:2137–47. doi: 10.2337/db09-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuen RK, Thiruvahindrapuram B, Merico D, et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat Med. 2015;21:185–91. doi: 10.1038/nm.3792. [DOI] [PubMed] [Google Scholar]
- 18.Emond MJ, Louie T, Emerson J, et al. Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat Genet. 2012;44:886–9. doi: 10.1038/ng.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emond MJ, Louie T, Emerson J, et al. Exome Sequencing of Phenotypic Extremes Identifies CAV2 and TMC6 as Interacting Modifiers of Chronic Pseudomonas aeruginosa Infection in Cystic Fibrosis. PLoS Genet. 2015;11:e1005273. doi: 10.1371/journal.pgen.1005273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penman A, Hancock H, Papavasileiou E, et al. Risk Factors for Proliferative Diabetic Retinopathy in African Americans with Type 2 Diabetes. Ophthalmic Epidemiol. 2016;23:88–93. doi: 10.3109/09286586.2015.1119287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davoudi S, Papavasileiou E, Roohipoor R, et al. Optical Coherence Tomography Characteristics of Macular Edema and Hard Exudates and Their Association with Lipid Serum Levels in Type 2 Diabetes. Retina. 2016;36:1622–9. doi: 10.1097/IAE.0000000000001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papavasileiou E, Davoudi S, Roohipoor R, et al. Association of serum lipid levels with retinal hard exudate area in African Americans with type 2 diabetes. Graefes Arch Clin Exp Ophthalmol. 2016 doi: 10.1007/s00417-016-3493-9. [DOI] [PubMed] [Google Scholar]
- 23.Grading diabetic retinopathy from stereoscopic color fundus photographs--an extension of the modified Airlie House classification. ETDRS report number 10. Early Treatment Diabetic Retinopathy Study Research Group. Ophthalmology. 1991;98:786–806. [PubMed] [Google Scholar]
- 24.Bujakowska KM, Consugar M, Place E, et al. Targeted exon sequencing in Usher syndrome type I. Invest Ophthalmol Vis Sci. 2014;55:8488–96. doi: 10.1167/iovs.14-15169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siepel A, Bejerano G, Pedersen JS, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aiello LP. Angiogenic pathways in diabetic retinopathy. N Engl J Med. 2005;353:839–41. doi: 10.1056/NEJMe058142. [DOI] [PubMed] [Google Scholar]
- 27.Sun CY, Lee CC, Hsieh MF, et al. Clinical association of circulating VEGF-B levels with hyperlipidemia and target organ damage in type 2 diabetic patients. J Biol Regul Homeost Agents. 2014;28:225–36. [PubMed] [Google Scholar]
- 28.Hagberg CE, Mehlem A, Falkevall A, et al. Targeting VEGF-B as a novel treatment for insulin resistance and type 2 diabetes. Nature. 2012;490:426–30. doi: 10.1038/nature11464. [DOI] [PubMed] [Google Scholar]
- 29.Wu M, Chen Y, Wilson K, et al. Intraretinal leakage and oxidation of LDL in diabetic retinopathy. Invest Ophthalmol Vis Sci. 2008;49:2679–85. doi: 10.1167/iovs.07-1440. [DOI] [PubMed] [Google Scholar]
- 30.Yu JY, Lyons TJ. Modified Lipoproteins in Diabetic Retinopathy: A Local Action in the Retina. J Clin Exp Ophthalmol. 2013;4 doi: 10.4172/2155-9570.1000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liew G, Klein R, Wong TY. The role of genetics in susceptibility to diabetic retinopathy. Int Ophthalmol Clin. 2009;49:35–52. doi: 10.1097/IIO.0b013e31819fd5d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng DP. Human genetics of diabetic retinopathy: current perspectives. J Ophthalmol. 2010;2010 doi: 10.1155/2010/172593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin HJ, Huang YC, Lin JM, et al. Association of genes on chromosome 6, GRIK2, TMEM217 and TMEM63B (linked to MRPL14) with diabetic retinopathy. Ophthalmologica. 2013;229:54–60. doi: 10.1159/000342616. [DOI] [PubMed] [Google Scholar]
- 34.Miao Y, Cui L, Chen Z, et al. Gene expression profiling of DMU-212-induced apoptosis and anti-angiogenesis in vascular endothelial cells. Pharm Biol. 2016;54:660–6. doi: 10.3109/13880209.2015.1071414. [DOI] [PubMed] [Google Scholar]
- 35.Feenstra DJ, Yego EC, Mohr S. Modes of Retinal Cell Death in Diabetic Retinopathy. J Clin Exp Ophthalmol. 2013;4:298. doi: 10.4172/2155-9570.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demirkan A, van Duijn CM, Ugocsai P, et al. Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genet. 2012;8:e1002490. doi: 10.1371/journal.pgen.1002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Folsom AR, Zheng ZJ, et al. Plasma fatty acid composition and incidence of diabetes in middle-aged adults: the Atherosclerosis Risk in Communities (ARIC) Study. Am J Clin Nutr. 2003;78:91–8. doi: 10.1093/ajcn/78.1.91. [DOI] [PubMed] [Google Scholar]
- 38.Brugger B, Erben G, Sandhoff R, et al. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc Natl Acad Sci U S A. 1997;94:2339–44. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsumoto T, Kobayashi T, Kamata K. Role of lysophosphatidylcholine (LPC) in atherosclerosis. Curr Med Chem. 2007;14:3209–20. doi: 10.2174/092986707782793899. [DOI] [PubMed] [Google Scholar]
- 40.Tazawa S, Yamato T, Fujikura H, et al. SLC5A9/SGLT4, a new Na+-dependent glucose transporter, is an essential transporter for mannose, 1,5-anhydro-D-glucitol, and fructose. Life Sci. 2005;76:1039–50. doi: 10.1016/j.lfs.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 41.Takagi H, King GL, Aiello LP. Hypoxia upregulates glucose transport activity through an adenosine-mediated increase of GLUT1 expression in retinal capillary endothelial cells. Diabetes. 1998;47:1480–8. doi: 10.2337/diabetes.47.9.1480. [DOI] [PubMed] [Google Scholar]
- 42.Ng ZX, Kuppusamy UR, Tajunisah I, et al. Investigation of SLC2A1 26177A/G gene polymorphism via high resolution melting curve analysis in Malaysian patients with diabetic retinopathy. J Diabetes Complications. 2012;26:388–92. doi: 10.1016/j.jdiacomp.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 43.Fu LL, Lin Y, Yang ZL, et al. Association analysis of genetic polymorphisms of TCF7L2, CDKAL1, SLC30A8, HHEX genes and microvascular complications of type 2 diabetes mellitus. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2012;29:194–9. doi: 10.3760/cma.j.issn.1003-9406.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 44.Kirchhoff K, Machicao F, Haupt A, et al. Polymorphisms in the TCF7L2, CDKAL1 and SLC30A8 genes are associated with impaired proinsulin conversion. Diabetologia. 2008;51:597–601. doi: 10.1007/s00125-008-0926-y. [DOI] [PubMed] [Google Scholar]
- 45.Chen P, Ong RT, Tay WT, et al. A study assessing the association of glycated hemoglobin A1C (HbA1C) associated variants with HbA1C, chronic kidney disease and diabetic retinopathy in populations of Asian ancestry. PLoS One. 2013;8:e79767. doi: 10.1371/journal.pone.0079767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan BK, Lewandowski KC, O'Hare JP, et al. Insulin regulates the novel adipokine adipolin/CTRP12: in vivo and ex vivo effects. J Endocrinol. 2014;221:111–9. doi: 10.1530/JOE-13-0537. [DOI] [PubMed] [Google Scholar]
- 47.Enomoto T, Ohashi K, Shibata R, et al. Adipolin/C1qdc2/CTRP12 protein functions as an adipokine that improves glucose metabolism. J Biol Chem. 2011;286:34552–8. doi: 10.1074/jbc.M111.277319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brakenhielm E, Veitonmaki N, Cao R, et al. Adiponectin-induced antiangiogenesis and antitumor activity involve caspase-mediated endothelial cell apoptosis. Proc Natl Acad Sci U S A. 2004;101:2476–81. doi: 10.1073/pnas.0308671100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Awata T, Inoue K, Kurihara S, et al. A common polymorphism in the 5'-untranslated region of the VEGF gene is associated with diabetic retinopathy in type 2 diabetes. Diabetes. 2002;51:1635–9. doi: 10.2337/diabetes.51.5.1635. [DOI] [PubMed] [Google Scholar]
- 50.Zhao T, Zhao J. Association between the −634C/G polymorphisms of the vascular endothelial growth factor and retinopathy in type 2 diabetes: a meta-analysis. Diabetes Res Clin Pract. 2010;90:45–53. doi: 10.1016/j.diabres.2010.05.029. [DOI] [PubMed] [Google Scholar]
- 51.Abhary S, Burdon KP, Gupta A, et al. Common sequence variation in the VEGFA gene predicts risk of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2009;50:5552–8. doi: 10.1167/iovs.09-3694. [DOI] [PubMed] [Google Scholar]
- 52.Marcovina S, Packard CJ. Measurement and meaning of apolipoprotein AI and apolipoprotein B plasma levels. J Intern Med. 2006;259:437–46. doi: 10.1111/j.1365-2796.2006.01648.x. [DOI] [PubMed] [Google Scholar]
- 53.McQueen MJ, Hawken S, Wang X, et al. Lipids, lipoproteins, and apolipoproteins as risk markers of myocardial infarction in 52 countries (the INTERHEART study): a case-control study. Lancet. 2008;372:224–33. doi: 10.1016/S0140-6736(08)61076-4. [DOI] [PubMed] [Google Scholar]
- 54.Zhang X, Saaddine JB, Chou CF, et al. Prevalence of diabetic retinopathy in the United States, 2005–2008. JAMA. 2010;304:649–56. doi: 10.1001/jama.2010.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]