SUMMARY
The Mycoplasma pneumoniae terminal organelle functions in adherence and gliding motility and is comprised of at least eleven substructures. We used electron cryotomography to correlate impaired gliding and adherence function with changes in architecture in diverse terminal organelle mutants. All eleven substructures were accounted for in the prkC, prpC, and P200 mutants, and variably so for the HMW3 mutant. Conversely, no terminal organelle substructures were evident in HMW1 and HMW2 mutants. The P41 mutant exhibits a terminal organelle detachment phenotype and lacked the bowl element normally present at the terminal organelle base. Complementation restored this substructure, establishing P41 as either a component of the bowl element or required for its assembly or stability, and that this bowl element is essential to anchor the terminal organelle but not for leverage in gliding. Mutants II-3, III-4, and topJ exhibited a visibly lower density of protein knobs on the terminal organelle surface. Mutants II-3 and III-4 lack accessory proteins required for a functional adhesin complex, while the topJ mutant lacks a DnaJ-like co-chaperone essential for its assembly. Taken together, these observations expand our understanding of the roles of certain terminal organelle proteins in the architecture and function of this complex structure.
Keywords: Mycoplasma pneumoniae, terminal organelle, electron cryotomography, mutants, bacterial ultrastructure
ABBREVIATED SUMMARY
Mycoplasma pneumoniae mutants defective in adherence and gliding were analyzed by electron cryotomography for corresponding changes in terminal organelle architecture. Ultrastructural changes ranged from complete loss of all eleven terminal organelle substructures in some mutants, to changes in specific substructures such as the protein knobs on the terminal organelle surface (green) or the bowl substructure (purple) at its base. Somewhat surprisingly, for other mutants no differences in terminal organelle architecture were evident.

INTRODUCTION
Mycoplasma pneumoniae is a common human respiratory tract pathogen causing community-acquired tracheobronchitis and atypical pneumonia (Esposito et al., 2004; Waites and Talkington, 2004). This novel, cell wall-less prokaryote is the leading cause of pneumonia in older children and young adults, but incidence is increasing in the very young and elderly, probably due to expanding, closely housed populations in these age groups. Symptoms are often flu-like, though characteristically persistent, and can lead to serious sequelae including extra-pulmonary spread, diverse immunopathology, and chronic pulmonary damage associated with onset and exacerbation of asthma.
Mycoplasmas are Gram-positive phylogenetically, but degenerative evolution has led to significant genome reduction. As a result, mycoplasmas lack genes for cell wall synthesis, many catabolic and biosynthetic pathways, and other capabilities common to model bacteria. Despite its minimal genome, M. pneumoniae possesses a unique and remarkably complex terminal organelle (Biberfeld and Biberfeld, 1970). This membrane-bound extension of the cell body mediates adherence to host epithelium, is the motor for gliding motility, and likely plays a role in the cell cycle, where its duplication is coordinated with chromosome replication and cell division (Hasselbring et al., 2006; Seto et al., 2001.). The terminal organelle is defined at the ultrastructural level by an electron-dense core that is often oriented along the long axis of the cell and is comprised of at least eleven distinct substructures, as revealed by electron cryotomography (ECT; Fig. 1) (Henderson and Jensen, 2006). These substructures include tightly packed protein knobs lining the inner and outer surface of the terminal organelle membrane at its distal end (Fig. 1, elements A and B, respectively); a terminal button comprised of three distinct subunits abutting the protein complexes along the inner surface of the terminal organelle membrane (Fig. 1, elements C–E); two parallel, angled, segmented rods of different length and thickness and often oriented longitudinally (Fig. 1, elements F and G); and a bowl complex at the base, or proximal end, of the terminal organelle (Fig. 1, elements H–K).
Fig. 1.
Schematic of M. pneumoniae terminal organelle architecture. Panel A: Substructural elements distinguishable by electron cryotomography are designated A–K according to Henderson and Jensen (2006). Subcellular localization of specific terminal organelle proteins to the bowl complex (elements H–K; red), paired segmented rods (elements F and G; blue), terminal button (elements C–E; green), or membrane (elements A and B; salmon) are indicated. Panel B: Tomogram slice of terminal organelle (sagittal view) with substructures false-colored according to the corresponding elements in Panel A.
Our understanding of the identity and function of proteins that comprise the terminal organelle comes largely from mutant analysis (Table 1), cross-linking and other biochemical approaches (Balish et al., 2001; Layh-Schmitt et al., 2000; Willby et al., 2004), and sub-cellular localization by antibody labeling and fluorescent protein fusions (Balish et al., 2003; Baseman et al., 1982; Hu et al., 1982; Seto et al., 2001; Stevens and Krause 1991; Stevens and Krause, 1992). Protein P1 functions directly in receptor binding and cell gliding and forms a complex with protein P90 and probably proteins P30 and P40 (Layh-Schmitt et al., 2000; Miyata and Hamaguchi, 2016; Nakane et al., 2011; Nakane et al., 2015; Seto et al., 2005). These P1 adhesin complexes are thought to correspond both to the prominent knobs on the terminal organelle surface (Fig. 1, elements A and B) (Henderson and Jensen, 2006; Kawamoto et al., 2016; Nakane et al., 2015; Seybert et al., 2006), as well as the nap seen in standard electron micrographs (EMs) (Baseman et al., 1982; Nakane et al., 2015). Proteins HMW1 and HMW2 are required to form or stabilize the paired segmented rods (Willby et al., 2004), with HMW2 a component specifically of the thicker rod (Balish et al., 2003; Bose et al., 2009; Nakane et al., 2015). Proteins P24, P41, P200, MPN387, and TopJ localize to the base of the terminal organelle, in the vicinity of the bowl complex (Cloward and Krause, 2009; Hasselbring and Krause, 2007a; Jordan et al., 2007; Nakane et al., 2015). While mutant analysis and protein localization studies complement the detailed terminal organelle architecture apparent by ECT, these approaches have not been previously combined in a comprehensive manner. Here we compared tomograms of wild-type M. pneumoniae and several terminal organelle mutants to correlate specific proteins with terminal organelle substructures, and in so doing, further elucidate their roles in terminal organelle architecture and function.
Table 1.
Summary of defects and phenotypes in M. pneumoniae terminal organelle mutants examined here.
| Mutant | Defect | P1 Localization | Phenotype | Altered Substructures Noted Here | References |
|---|---|---|---|---|---|
| P200 | No P200 | Not tested | Reduced gliding speed | None | Jordan et al., 2007 |
| prkC | Truncated PrkC protein kinase | Not tested | Reduced gliding frequency | None | Page and Krause, 2013 |
| prpC | No PrpC protein phosphatase | Not tested | Elevated gliding frequency | None | Page and Krause, 2013 |
| HMW3 | No HMW3 | Variable | Reduced adherence | Variable: None, or terminal button / bowl complex absent | Willby and Krause, 2002 |
| P41 P41-C |
No P41 or P24 P41 restored / no P24 |
Polar Not tested |
Detached terminal organelle; reduced gliding speed and frequency Like wild-type |
No bowl substructure (K in Fig. 1) None |
Hasselbring and Krause, 2007a; 2007b |
| II-3 | No P30; reduced P65 | Not polar | No adherence / gliding | Reduced density of protein knobs (A in Fig. 1) | Baseman et al., 1987; Krause et al., 1982; Romero-Arroyo et al., 1999 |
| III-4 | No P40/P90 | Not polar | No adherence/gliding | Reduced density of protein knobs (A in Fig. 1) | Krause et al., 1982; Waldo et al., 2005 |
|
topJ topJ-C |
No DnaJ-like co-chaperone TopJ restored |
Not polar Not tested |
No adherence / gliding Like wild-type |
Reduced density of protein knobs (A in Fig. 1) None |
Cloward and Krause, 2009 |
| C1 | No HMW2; reduced HMW1, HMW3, P65, P41, P30, P24 | Not polar | No adherence / gliding | No core substructures evident | Krause et al., 1997 |
| H9 | Truncated HMW2; reduced HMW1, HMW3, P65, P41, P30, P24 | Not polar | No adherence / gliding | No core substructures evident | Krause et al., 1997 |
| M6 | No HMW1; truncated P30; reduced HMW2, HMW3, P65 | Not polar | No adherence / gliding | No core substructures evident | Layh-Schmitt et al., 1995 |
RESULTS AND DISCUSSION
The range of ultrastructural defects impacting terminal organelle function varied
The M. pneumoniae mutants examined here share in common the loss and/or truncation of one or more terminal organelle proteins, resulting in altered adherence, gliding, or both. Tomograms for most of these mutants revealed clear changes to terminal organelle architecture that involved specific substructures, as detailed below. However, all eleven substructures were consistently present in the prkC, prpC, and P200 mutants, and variably present in the HMW3 mutant, based upon examination of multiple tomograms for each. This was not surprising for the prkC mutant (Fig. 2 and supplemental movie 2; see supplemental movie 1 for comparison with wild-type M. pneumoniae), for which transposon insertion results in a truncated protein kinase that is predicted to retain activity but lack a localization domain, or for the prpC mutant (Fig. 2 and supplemental movie 3), lacking the cognate protein phosphatase (Page and Krause, 2013). Truncation of PrkC has a subtle impact on terminal organelle function, with no effect on cytadherence but a significantly lower gliding frequency. Conversely, loss of PrpC results in a significantly elevated gliding frequency. All known terminal organelle proteins are present at wild-type levels in both mutants (Page and Krause, 2013), consistent with identification of all core substructures in tomograms here. As likely regulatory rather than structural proteins, both may be present in low abundance, and attempts at their detection by using antibodies raised against multiple antigenic peptides for each were unsuccessful (Page and Krause, 2013).
Fig. 2.
Tomograms of prkC and prpC mutants, having all terminal organelle substructures accounted for and indistinguishable from wild-type M. pneumoniae (WT), based on the examination of multiple tomograms for each mutant. The upper-left panel is a dorsal/ventral view, while the remaining panels are sagittal views. Scale bars: 100nm.
The P200 mutant adheres at wild-type levels but glides at a reduced speed and lacks a large protein of undefined function that localizes to the region of the bowl complex (Jordan et al., 2007; Nakane et al., 2015). Nevertheless, all terminal organelle substructures, including bowl elements H-K, were accounted for in the P200 mutant (Fig. 3 and supplemental movie 4). P200 lacks homology to proteins of known function but has six “enriched in aromatic and glycine residues” (EAGR) boxes, which were identified as protein interaction domains in the closely related Mycoplasma genitalium (Calisto et al., 2012), and might mediate assembly into a higher-order complex. Thus it came as a surprise here that loss of P200 did not visibly affect the major substructures at the base of the terminal organelle.
Fig. 3.
Tomograms of terminal organelle mutant P200. The P200 protein localizes to the vicinity of the bowl complex (Jordan et al., 2007; Nakane et al., 2015), substructures H–K of which were all accounted for here (red arrowheads). The lower-left panel is a dorsal/ventral view, while the other panels are sagittal views. Scale bars: 100nm.
HMW3 localizes to the distal end of the terminal organelle core, including the region corresponding to the terminal button (Fig. 1, elements C-E) (Nakane et al., 2015; Stevens and Krause, 1992). HMW3 has two large acidic and proline-rich (APR) domains (Krause and Balish, 2004; Ogle and Krause, 1991), which have been shown for the TopJ protein to be required for association with the Triton X-100-insoluble, cytoskeletal fraction, or triton shell (Cloward and Krause, 2010). HMW3 is relatively abundant and appears to exist as polymerized filaments that surround the core at its distal end (Stevens and Krause, 1992). Loss of HMW3 is accompanied by reduced levels of terminal button protein P65 and a pronounced impact on terminal organelle function, including poor cytadherence, no gliding motility, and inconsistent localization of P1 to the terminal organelle (Willby and Krause, 2002). Given these features, we expected the substructures of the terminal button would be consistently altered or missing in the absence of HMW3. While we did indeed observe incomplete cores in this mutant, where paired rods were clearly present but elements of the terminal button and/or the bowl complex were not distinguishable (Fig. 4A–B), we also noted that all core substructures, including the terminal button elements, were present for many of the terminal organelles examined (Fig. 4C–4F, and supplemental movie 5). This variability in the presence of complete terminal organelle cores was unique to the HMW3 mutant but consistent with variability previously noted in P1 localization to the terminal organelle in this mutant (Willby and Krause, 2002). The variability observed here included the protein knobs on the terminal organelle surface (see Fig. 1, element A) which were only occasionally present at wild-type densities (e.g. Fig. 4F, salmon arrowheads). An explanation we favor to account for the inconsistent core architecture and P1 localization in this mutant is that HMW3 occupies the low-density region that separates the core substructures from the cell membrane and perhaps facilitates bridging the terminal button and the P1 adhesin complexes in a manner necessary for the latter to be fully functional. By this model, HMW3 surrounds the substructures near the distal end of the core, perhaps as the slender filaments not resolved by ECT but evident in some images of fixed and stained thin sections of whole cells by standard EM (Wilson and Collier, 1976). This model is consistent with the previous observation that HMW3 may be associated with the inner surface of the terminal organelle membrane and collapses onto the core when cells are extracted with Triton X-100 (Stevens and Krause, 1992).
Fig. 4.
Tomograms illustrating the presence of complete and incomplete terminal organelle cores in the HMW3 mutant. Panels A–C are dorsal/ventral views, while panels D and E are sagittal views. In panel F, the lower-right cell is a dorsal/ventral view and the upper left cell is a sagittal view. Panels A and B show incomplete cores, where the rod structure was present but the bowl complex and terminal button were not evident. Panels C–F show complete cores. HMW3 localizes to the distal region of the terminal organelle, including the region of the terminal button (Nakane et al., 2015; Stevens and Krause, 1992). Nevertheless, we accounted for substructures C–E of the terminal button (green arrowheads) in many terminal organelles of the HMW3 mutant. Panel D shows two cores linked at the terminal button (white arrow) in a chevron-like arrangement, as only observed with the HMW3 mutant, with black arrowheads indicating the bowl elements at the base of each core. The presence of the protein knobs on the terminal organelle was variable, with panel F illustrating cells where these structures were clearly abundant (salmon arrowheads). Scale bars: 100nm.
The terminal button and bowl complex are believed to assemble after the paired rods of the terminal organelle core (Krause and Balish, 2004), and the absence of terminal button and bowl elements despite the presence of the paired rods in some terminal organelles of the HMW3 mutant is consistent with that assembly sequence. HMW3 is thus not required for rod assembly or assembly of the terminal button or bowl complex, but appears to facilitate the latter. Finally, adjacent cores linked at the terminal button and separated at their bases in a chevron arrangement were also seen in some cells for the HMW3 mutant (Fig. 4, white arrow), confirming previous observations from standard EMs of thin sections (Willby and Krause, 2002). Core duplication precedes cell division (Hasselbring et al., 2006; Seto et al., 2001), where wild-type cores normally separate at the terminal button after duplication (Kawamoto et al., 2016), by a process that likewise seems impaired in the absence of HMW3. Collectively these observations suggest a role in spatial organization role for HMW3 in terminal organelle assembly and function.
Seto and Miyata (2003) previously reported that no electron-dense core is apparent in standard EMs of mutants lacking HMW1 or HMW2. Here we confirmed and extended that observation, noting that no core rods or any other core substructures were evident in mutants H9, C1, and M6 (Fig. 5 and supplemental movies 6–8, respectively). H9 and C1 have a transposon insertion at different sites in the gene encoding HMW2 (Krause et al., 1997), a major component of the larger segmented rod of the terminal organelle core (Nakane et al., 2015). Loss or truncation of HMW2 impacts the synthesis or stability of proteins HMW1, HMW3, P65, P41, P30, and P24 (Jordan et al., 2001; Krause et al., 1982; Popham et al., 1997), which likely accounts for the lack of substructures in tomograms of these mutants. Conversely, mutant M6 lacks HMW1 (Layh-Schmitt et al., 1995), which is required for HMW2 stability (Willby et al., 2004), and thus the absence of core substructures in the M6 mutant likewise was not unexpected. It has been suggested that HMW1 is a component of the smaller plate of the core (Fig. 1, element G; Nakane et al., 2015), although the data are ambiguous and may reflect a more peripheral position (Balish et al., 2001). Finally, tomograms of the HMW1 and HMW2 mutants occasionally revealed long, narrow, segmented ribbons (Fig. 5, arrowheads) not previously seen in standard EMs. These ribbons exhibited no pattern with respect to location within the cell and often appeared twisted, in some cases in helical bundles (Fig. 5, arrows). Such ribbons were absent in wild-type or other mutant tomograms and may represent a terminal organelle substructure that has failed to fold or localize correctly in the absence of the paired rods of the core, which may serve to orient normal core assembly. Perhaps significantly, segmented filamentous structures, sometimes bundled, were also recently described in other bacteria (Dobro et al., 2017).
Fig. 5.
Tomograms of mutants for which no terminal organelle substructures were evident. From the top: mutants C1, H9, and M6. Arrowheads, ribbon structures seen in some tomograms of these mutants; arrows, bundled ribbons. Scale bars: 100nm.
P41 is required for the bowl element
The P41 mutant exhibits a striking phenotype when gliding, where the terminal organelle pulls away from the cell body, eventually detaches, and continues to glide independently for about an hour (Hasselbring and Krause, 2007a). Tomograms of this mutant revealed the presence of all core substructures except the bowl element (see Fig. 1, element K) that is normally found at the base of the terminal organelle in wild-type M. pneumoniae (Fig. 6, circles, and supplemental movie 9). All other elements of the bowl complex were accounted for in the P41 mutant (Fig. 6, red arrowheads). Tomograms also frequently revealed detached terminal organelles of varying sizes, their membranes having re-sealed following separation from the cell body (Fig. 6, white arrows). In addition, we noted cells with no terminal organelle (not shown), and membrane vesicles of various sizes (white arrowheads), likely generated during the forceful detachment of terminal organelles, were much more common with the P41 mutant than for all other strains examined. Finally, the absence of the bowl element did not affect the protein knobs on the terminal organelle surface (salmon arrowheads), which is consistent with the ability of the P41 mutant to attach normally to surfaces and retain some gliding capacity.
Fig. 6.
Tomograms of the P41mutant (top panels), which exhibits a terminal organelle detachment phenotype, and the P41 mutant complemented with recombinant P41 (rP41) and P24 (rP24) (middle panels), or with recombinant P24 alone (bottom panels). Panel A, dorsal/ventral view of terminal organelle cores; panels B and C, sagittal views of cores. Circles, terminal organelle lacking bowl element K; white arrows, detached terminal organelles; white arrowheads, membrane vesicles; red arrowheads, bowl complex substructures; salmon arrowhead, protein knobs on terminal organelle surface. Panels D and E are sagittal views of terminal organelle cores, with arrowheads the same as above. Panel F is a dorsal/ventral view of the terminal organelle core in the cell in lower center, while the cell in the upper center has two cores, one a dorsal/ventral view and the second a sagittal view. Panel G is a dorsal/ventral view of the terminal organelle core. Circles, terminal organelle lacking bowl element K. Scale bars: 100nm.
Transposon disruption of the P41 gene has a polar effect on the downstream protein P24. Complementation of this mutant by using a recombinant transposon to deliver the wild-type alleles for P41 and P24 restores a wild-type phenotype (Hasselbring and Krause, 2007a), and here tomograms of the complemented mutant revealed no detached terminal organelles, with an intact bowl complex evident (Fig. 6, middle panels). In contrast, the bowl element was consistently absent in tomograms of the P41 mutant complemented with a transposon carrying only the recombinant wild-type P24 allele (Fig. 6, bottom panels; circles). Thus, we conclude that P41 is either a component of the bowl structure itself or is required for its assembly or stability. Finally, it has been suggested that M. pneumoniae motility is achieved in part by conformational changes in the rod structure, utilizing the bowl for leverage (Henderson and Jensen, 2006; Kawamoto et al., 2016). However, the P41 mutant and its detached terminal organelles are motile despite the absence of the bowl element K, suggesting that this substructure specifically is not essential for such leverage in the model proposed by Henderson and Jensen (2006). Filaments can be seen radiating from the base of the terminal organelle into the cell body in M. pneumoniae triton shells (Göbel et al., 1981), and bowl element K may serve to anchor those filaments. The nature of any linkage between the cell membrane and the bowl complex is not known, but the variety in detached terminal organelle size in this mutant suggests that the terminal organelle separates imprecisely from the cell body in the absence of P41.
Protein knob density was reduced on some mutants
All internal substructures were observed in the II-3, III-4, and topJ mutants. However, the abundance of the protein knobs on the terminal organelle surface (element A in Fig. 1) was visually lower for these mutants compared to wild-type M. pneumoniae (Fig. 7 and 8 and supplemental movies 10–12). These protein knobs are of particular interest, given their likely direct involvement in receptor binding and cell gliding, and their reduced density correlates with the loss of adherence and gliding motility for these three mutants. Furthermore, for mutants II-3 and III-4 their reduced density also correlates with the absence of the terminal organelle nap normally seen by negative staining in standard EMs of wild-type M. pneumoniae (Baseman et al., 1982). Kawamoto et al. (2016) recently reported the markedly reduced abundance of these terminal organelle knobs on a mutant that lacks both the P1 adhesin and accessory proteins P40 and P90, concluding that these knobs correspond to the P1 adhesin complex. Moreover, isolated complexes of the P1/P90 homologs in the closely related Mycoplasma genitalium are structurally similar by ECT to the nap complexes seen on intact cells (Scheffer et al., 2017). Our results here support and extend those findings, as tomograms of mutant III-4, lacking P40 and P90 but not P1, likewise revealed visually lower numbers of protein knobs on the terminal organelle surface. P1 surface exposure on this mutant is comparable to wild-type, based on accessibility to lactoperoxidase-mediated radioiodination (Baseman et al., 1982) and antibody labeling (Seto and Miyata, 2003), but it remains non-functional in the absence of accessory proteins P40/P90 (Waldo et al., 2005). In this regard, Baseman et al. (1982) noted that cooperation between P1-associated proteins may permit essential lateral mobility or activation of the membrane for formation of the nap. The same probably applies to the reduced density of protein knobs on the P30 mutant II-3, where P1 is also present at wild-type levels on the mycoplasma surface (Baseman et al., 1982; Seto and Miyata, 2003) but likewise fails to form functional adhesin complexes. Thus P1, P40, and P90 are necessary but not sufficient for the presence of these terminal organelle knobs at wild-type densities. Interestingly, Kawamoto et al. (2016) noted a high degree of heterogeneity in the particles on wild-type M. pneumoniae and suggested that they may be conformationally dynamic, while Scheffer et al. (2017) described significant variation in the degree of tilt of nap complexes with respect to the membrane in M. genitalium.
Fig. 7.
Tomograms of mutants II-3 and III-4, revealing reduced abundance of protein complexes on the terminal organelle surface compared to wild-type M. pneumoniae (WT; salmon arrowheads). Scale bars: 100nm.
Fig. 8.
Tomograms of the topJ mutant revealing reduced abundance of protein complexes on the terminal organelle surface compared to wild-type M. pneumoniae (WT; salmon arrowheads). Scale bars: 100nm.
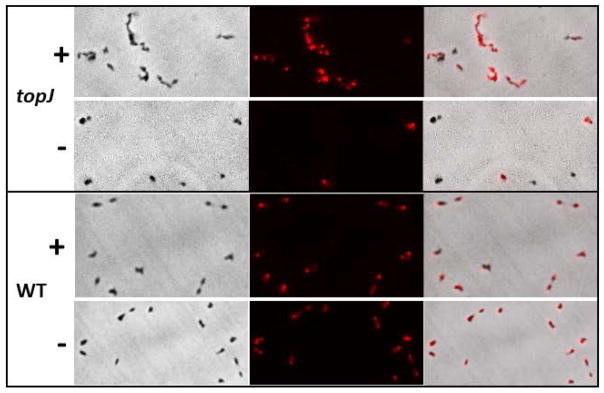
The low abundance of protein knobs was most pronounced for the topJ mutant (Fig. 8), which has normal levels of P1, P30, and P40/P90 but lacks the DnaJ-like co-chaperone TopJ (Cloward and Krause, 2009; 2011). Complementation of the topJ mutant restored normal protein knob density (data not shown). P1 exhibits a strikingly lower accessibility to protease with the topJ mutant than with wild-type M. pneumoniae or other terminal organelle mutants examined (Cloward and Krause, 2011). This inaccessibility appears to reflect poor processing of P1 to the mycoplasma membrane, as P1 accessibility to immunofluorescence labeling differed here with and without membrane permeabilization (Fig. 9). In the absence of membrane permeabilization approximately 40% of the cells examined exhibited P1 labeling, but that percentage nearly doubled with membrane permeabilization. In contrast, no difference in P1 labeling with and without membrane permeabilization was seen with wild-type M. pneumoniae. J-domain co-chaperones partner with DnaK to stabilize protein complexes for protein translocation and macromolecular assembly. The reduced P1 accessibility to trypsin or antibodies with the topJ mutant, relative to wild-type M. pneumoniae, is consistent with the low density of protein knobs on the terminal organelle surface, and like mutants II-3 and III-4, reflects an inability to assemble a functional adhesin complex. Attempts to quantify differences in protein knob abundance noted visually were impeded by the missing wedge effect on tomograms. We anticipate that such quantitation will be required to further elucidate the dynamic relationship between the protein knobs and terminal organelle function in M. pneumoniae.
Fig. 9.
Immunofluorescence analysis of the topJ mutant (upper panels) and wild-type M. pneumoniae (WT; lower panels) with P1-specific monoclonal antibodies with (+) and without (-) membrane permeabilization. Panels in the left column are phase contrast images, in the center column immunofluorescence images, and in the right column merged images.
EXPERIMENTAL PROCEDURES
Mycoplasma strains and culture conditions
These studies utilized wild-type M. pneumoniae strain M129 (Lipman et al., 1969), broth-passage 18, which retains virulence in experimentally infected hamsters, as well as several previously characterized terminal organelle mutants of M129 (Table 1). Spontaneously arising mutants originating from M129 included II-3, III-4, and M6. Mutants generated by transposon insertion in M129 included C1, H9, HMW3, P41, P200, prpC, prkC, and topJ. We also examined complemented derivatives of the P41 and topJ mutants. Mycoplasmas were grown in tissue culture flasks in SP-4 medium (Tully et al., 1977) at 37°C until mid-log phase and harvested as described previously (Cloward and Krause, 2009). Gentamicin (18 μg/ml) was included for transformants with the transposon Tn4001mod, while chloramphenicol (24 μg/ml) was included for transformants with the transposon Tn4001cat, which was used for delivery of recombinant alleles for complementation.
Electron cryotomography and image analysis
Mycoplasma cultures grown on Quantifoil grids (EMS, Hatfield, PA) previously treated with 10-nm gold fiducial markers were plunge-frozen in liquid ethane or ethane/propane mixture using a Vitrobot (FEI Co., Hillsboro, OR). Samples were maintained at a temperature < −150°C during transfer to the electron microscope. Images were acquired with an FEI Polara 300 kV field emission gun, transmission electron microscope equipped with a Gatan energy filter and a lens-coupled 4k × 4k UltraCam, later upgraded to a direct detection K2 Summit camera (Gatan, Inc., Pleasanton, CA). Data were collected with Leginon (Suloway et al., 2009) and UCSFTomo (Zheng et al., 2007) using 27.5Kx nominal magnification, 8–10 μm defocus, 120–200 electrons per square Å total dose, through a range of ± 65° at 1° increments. Images were aligned using fiducial markers and merged by weighted back-projection or SIRT with IMOD software to produce 3-D reconstructions (Mastronarde, 1997). We examined a minimum of 10 tomograms for each strain, and >20 tomograms for most strains, in order to determine the presence or absence of specific substructures. As individual 2-D slices do not always capture all elements of a 3-D structure, representative tomogram series in movie format for each strain are included in Supporting Information.
Immunofluorescence microscopy
Mycoplasmas were cultured in borosilicate glass chamber slides (Thermo Fisher Scientific, Waltham, MA) or glass coverslips bound to molds of polydimethylsiloxane (Ellsworth Adhesives, Germantown, WI). For the topJ mutant the glass was pre-coated in 0.01% poly-L-lysine. Samples were prepared as described (Cloward and Krause, 2011) except that cells were fixed with 4.0% formaldehyde-2.0% glutaraldehyde in SP-4 lacking fetal bovine serum (pH adjusted to 7.0), and 5% bovine serum albumin was used in place of 5% dry milk. Post-fixation extraction with 0.5% Tween was omitted for samples analyzed without membrane permeabilization. Images captured as described (Hasselbring et al., 2005) were processed with OpenLab Imaging Software (version 5.5.0; Perkin Elmer).
Supplementary Material
Acknowledgments
This work was supported by Public Health Service research grants AI49194 and AI110098 from the National Institute of Allergy and Infectious Diseases to D.C.K. We acknowledge Mitch Balish for helpful discussions. We declare no competing interests.
Footnotes
AUTHOR CONTRIBUTIONS
Study conception and design: D.C.K. and G.J.J.; data acquisition and analysis: D.C.K., S.C., J.S., A.J., E.S.S., and G.J.J.; writing the manuscript: D.C.K., S.C., and G.J.J.
References
- Balish MF, Hahn TW, Popham PL, Krause DC. Stability of Mycoplasma pneumoniae cytadherence-accessory protein HMW1 correlates with its association with the triton shell. J Bacteriol. 2001;183:3680–3688. doi: 10.1128/JB.183.12.3680-3688.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balish MF, Santurri RA, Ricci AM, Lee KK, Krause DC. Localization of Mycoplasma pneumoniae cytadherence-associated protein HMW2 by fusion with green fluorescent protein: Implications for attachment organelle structure. Mol Microbiol. 2003;47:49–60. doi: 10.1046/j.1365-2958.2003.03282.x. [DOI] [PubMed] [Google Scholar]
- Baseman JB, Cole RM, Krause DC, Leith DK. Molecular basis for cytadsorption of Mycoplasma pneumoniae. J Bacteriol. 1982;151:1514–1522. doi: 10.1128/jb.151.3.1514-1522.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biberfeld G, Biberfeld P. Ultrastructural features of Mycoplasma pneumoniae. J Bacteriol. 1970;102:855–861. doi: 10.1128/jb.102.3.855-861.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose SR, Balish MF, Krause DC. Mycoplasma pneumoniae cytoskeletal protein HMW2 and the architecture of the terminal organelle. J Bacteriol. 2009;191:6741–6748. doi: 10.1128/JB.01486-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisto BM, Broto A, Martinelli L, Querol E, Piñol J, Fita I. The EAGR box structure: a motif involved in mycoplasma motility. Mol Microbiol. 2012;86:382–393. doi: 10.1111/j.1365-2958.2012.08200.x. [DOI] [PubMed] [Google Scholar]
- Cloward JM, Krause DC. Mycoplasma pneumoniae J-domain protein required for terminal organelle function. Mol Microbiol. 2009;71:1296–1307. doi: 10.1111/j.1365-2958.2009.06602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloward JM, Krause DC. Functional domain analysis of the Mycoplasma pneumoniae co-chaperone TopJ. Mol Microbiol. 2010;77:158–169. doi: 10.1111/j.1365-2958.2010.07196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloward JM, Krause DC. Loss of co-chaperone TopJ impacts adhesin P1 presentation and terminal organelle maturation in Mycoplasma pneumoniae. Mol Microbiol. 2011;81:528–539. doi: 10.1111/j.1365-2958.2011.07712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobro MJ, Oikonomou CM, Piper A, Cohen J, Guo K, Jensen T, Tadayon J, Donermeyer J, Park Y, Solis BA, Kjaer A, Jewett AI, McDowall AW, Chen S, Chang YW, Shi J, Subramanian P, Iancu CV, Li Z, Briegel A, Tocheva EI, Pilhofer M, Jensen GJ. Uncharacterized bacterial structures revealed by electron cryotomography. J Bacteriol. 2017;199:e00100–17. doi: 10.1128/JB.00100-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito S, Blasi F, Bosis S, Droghetti R, Faelli N, Lastrico A, Principi N. Aetiology of acute pharyngitis: the role of atypical bacteria. J Med Microbiol. 2004;53:645–651. doi: 10.1099/jmm.0.05487-0. [DOI] [PubMed] [Google Scholar]
- Göbel U, Speth V, Bredt W. Filamentous structures in adherent Mycoplasma pneumoniae cells treated with nonionic detergents. J Cell Biol. 1981;91:537–543. doi: 10.1083/jcb.91.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselbring BM, Jordan JL, Krause DC. Mutant analysis reveals a specific requirement for protein P30 in Mycoplasma pneumoniae gliding motility. J Bacteriol. 2005;187:6281–6289. doi: 10.1128/JB.187.18.6281-6289.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselbring BM, Jordan JL, Krause RW, Krause DC. Terminal organelle development in the cell wall-less bacterium Mycoplasma pneumoniae. Proc Natl Acad Sci U S A. 2006;103:16478–16483. doi: 10.1073/pnas.0608051103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselbring BM, Krause DC. Cytoskeletal protein P41 is required to anchor the terminal organelle of the wall-less prokaryote Mycoplasma pneumoniae. Mol Microbiol. 2007a;63:44–53. doi: 10.1111/j.1365-2958.2006.05507.x. [DOI] [PubMed] [Google Scholar]
- Hasselbring BM, Krause DC. Proteins P24 and P41 function in the regulation of terminal-organelle development and gliding motility in Mycoplasma pneumoniae. J Bacteriol. 2007b;189:7442–7449. doi: 10.1128/JB.00867-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson GP, Jensen GJ. Three-dimensional structure of Mycoplasma pneumoniae’s attachment organelle and a model for its role in gliding motility. Mol Microbiol. 2006;60:376–385. doi: 10.1111/j.1365-2958.2006.05113.x. [DOI] [PubMed] [Google Scholar]
- Hu PC, Cole RM, Huang YS, Graham JA, Gardner DE, Collier AM, Clyde WA., Jr Mycoplasma pneumoniae infection: role of a surface protein in the attachment organelle. Science. 1982;216:313–315. doi: 10.1126/science.6801766. [DOI] [PubMed] [Google Scholar]
- Jordan JL, Berry KM, Balish MF, Krause DC. Stability and subcellular localization of cytadherence-associated protein P65 in Mycoplasma pneumoniae. J Bacteriol. 2001;183:7387–7391. doi: 10.1128/JB.183.24.7387-7391.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan JL, Chang HY, Balish MF, Holt LS, Bose SR, Hasselbring BM, Waldo RH, 3rd, Krunkosky TM, Krause DC. Protein P200 is dispensable for Mycoplasma pneumoniae hemadsorption but not gliding motility or colonization of differentiated bronchial epithelium. Infect Immun. 2007;75:518–522. doi: 10.1128/IAI.01344-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto A, Matsuo L, Kato T, Yamamoto H, Namba K, Miyata M. Periodicity in attachment organelle revealed by electron cryotomography suggests conformational changes in gliding mechanism of Mycoplasma pneumoniae. mBio. 2016;7:e00243–16. doi: 10.1128/mBio.00243-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause DC, Balish MF. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol Microbiol. 2004;51:917–924. doi: 10.1046/j.1365-2958.2003.03899.x. [DOI] [PubMed] [Google Scholar]
- Krause DC, Leith DK, Wilson RM, Baseman JB. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect Immun. 1982;35:809–817. doi: 10.1128/iai.35.3.809-817.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause DC, Proft T, Hedreyda CT, Hilbert H, Plagens H, Herrmann R. Transposon mutagenesis reinforces the correlation between Mycoplasma pneumoniae cytoskeletal protein HMW2 and cytadherence. J Bacteriol. 1997;179:2668–2677. doi: 10.1128/jb.179.8.2668-2677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layh-Schmitt G, Hilbert H, Pirkl E. A spontaneous hemadsorption-negative mutant of Mycoplasma pneumoniae exhibits a truncated adhesin-related 30-kilodalton protein and lacks the cytadherence-accessory protein HMW1. J Bacteriol. 1995;177:843–846. doi: 10.1128/jb.177.3.843-846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layh-Schmitt G, Podtelejnikov A, Mann M. Proteins complexed to the P1 adhesin of Mycoplasma pneumoniae. Microbiol. 2000;146:741–747. doi: 10.1099/00221287-146-3-741. [DOI] [PubMed] [Google Scholar]
- Lipman RP, Clyde WA, Jr, Denny FW. Characteristics of virulent, attenuated, and avirulent Mycoplasma pneumoniae strains. J Bacteriol. 1969;100:1037–1043. doi: 10.1128/jb.100.2.1037-1043.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- Miyata M, Hamaguchi T. Integrated information and prospects for gliding mechanism of the pathogenic bacterium Mycoplasma pneumoniae. Front Microbiol. 2016;7:960. doi: 10.3389/fmicb.2016.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane D, Adan-Kubo J, Kenri T, Miyata M. Isolation and characterization of P1 adhesin, a leg protein of the gliding bacterium Mycoplasma pneumoniae. J Bacteriol. 2011;193:715–722. doi: 10.1128/JB.00796-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane D, Kenri T, Matsuo L, Miyata M. Systematic structural analyses of attachment organelle in Mycoplasma pneumoniae. PLoS Pathog. 2015;11:e1005299. doi: 10.1371/journal.ppat.1005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle KE, Krause DC. Cloning and analysis of the gene encoding the cytadherence phase-variable protein HMW3 from Mycoplasma pneumoniae. Gene. 1991;97:69–75. doi: 10.1016/0378-1119(91)90011-y. [DOI] [PubMed] [Google Scholar]
- Page CA, Krause DC. Protein kinase/phosphatase function correlates with gliding motility in Mycoplasma pneumoniae. J Bacteriol. 2013;195:1750–1757. doi: 10.1128/JB.02277-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham P, Hahn TW, Krebes K, Krause DC. Loss of HMW1 and HMW3 in noncytadhering mutants of Mycoplasma pneumoniae occurs post-translationally. Proc Natl Acad Sci USA. 1997;94:13979–13984. doi: 10.1073/pnas.94.25.13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Arroyo CE, Jordan J, Peacock SJ, Willby MJ, Farmer MA, Krause DC. Mycoplasma pneumoniae protein P30 is required for cytadherence and associated with proper cell development. J Bacteriol. 1999;181:1079–1087. doi: 10.1128/jb.181.4.1079-1087.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer MP, Gonzalez-Gonzalez L, Seybert A, Ratera M, Kunz M, Valpuesta JM, Fita I, Querol E, Piñol J, Martín-Benito J, Frangakis AS. Structural characterization of the NAP; the major adhesion complex of the human pathogen Mycoplasma genitalium. Mol Microbiol. 2017;105:869–879. doi: 10.1111/mmi.13743. [DOI] [PubMed] [Google Scholar]
- Seto S, Kenri T, Tomiyama T, Miyata M. Involvement of P1 adhesin in gliding motility of Mycoplasma pneumoniae as revealed by the inhibitory effects of antibody under optimized gliding conditions. J Bacteriol. 2005;187:1875–1877. doi: 10.1128/JB.187.5.1875-1877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto S, Layh-Schmitt G, Kenri T, Miyata M. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J Bacteriol. 2001;183:1621–1630. doi: 10.1128/JB.183.5.1621-1630.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto S, Miyata M. Attachment organelle formation represented by localization of cytadherence proteins and formation of the electron-dense core in wild-type and mutant strains of Mycoplasma pneumoniae. J Bacteriol. 2003;185:1082–1091. doi: 10.1128/JB.185.3.1082-1091.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A, Herrmann R, Frangakis AS. Structural analysis of Mycoplasma pneumoniae by cryo-electron tomography. J Struct Biol. 2006;156:342–354. doi: 10.1016/j.jsb.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Stevens MK, Krause DC. Localization of the Mycoplasma pneumoniae cytadherence-accessory proteins HMW1/4 in the cytoskeleton-like triton shell. J Bacteriol. 1991;173:1041–1050. doi: 10.1128/jb.173.3.1041-1050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens MK, Krause DC. Mycoplasma pneumoniae cytadherence phase-variable protein HMW3 is a component of the attachment organelle. J Bacteriol. 1992;174:4265–4274. doi: 10.1128/jb.174.13.4265-4274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Shi J, Cheng A, Pulokas J, Carragher B, Potter CS, Zheng SQ, Agard DA, Jensen GJ. Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol. 2009;167:11–18. doi: 10.1016/j.jsb.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully JG, Whitcomb RF, Clark HF, Williamson DL. Pathogenic mycoplasmas: cultivation and vertebrate pathogenicity of a new spiroplasma. Science. 1977;195:892–894. doi: 10.1126/science.841314. [DOI] [PubMed] [Google Scholar]
- Waites KB, Talkington DF. Mycoplasma pneumoniae and its role as a human pathogen. Clin Microbiol Rev. 2004;17:697–728. doi: 10.1128/CMR.17.4.697-728.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldo RH, III, Jordan JL, Krause DC. Identification and complementation of a mutation associated with loss of Mycoplasma pneumoniae virulence-specific proteins B and C. J Bacteriol. 2005;187:747–751. doi: 10.1128/JB.187.2.747-751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willby MJ, Balish MF, Ross SM, Lee KK, Jordan JL, Krause DC. HMW1 is required for stability and localization of HMW2 to the attachment organelle of Mycoplasma pneumoniae. J Bacteriol. 2004;186:8221–8228. doi: 10.1128/JB.186.24.8221-8228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willby MJ, Krause DC. Characterization of a Mycoplasma pneumoniae hmw3 mutant: implications for attachment organelle assembly. J Bacteriol. 2002;184:3061–3068. doi: 10.1128/JB.184.11.3061-3068.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MH, Collier AM. Ultrastructural study of Mycoplasma pneumoniae in organ culture. J Bacteriol. 1976;124:332–339. doi: 10.1128/jb.125.1.332-339.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Keszthelyi B, Branlund E, Lyle JM, Braunfeld MB, Sedat JW, Agard DA. UCSF tomography: an integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. J Struct Biol. 2007;157:138–147. doi: 10.1016/j.jsb.2006.06.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.