Abstract
Plant genomes encode a variety of short peptides acting as signaling molecules. Since the discovery of tomato systemin, a myriad of peptide signals, ranging in size, structure and modifications, have been found in plants. Moreover, new peptides are still being identified. Surprisingly, non-plant organisms, especially pathogens, also produce peptides which exert hormonal activities against host plants by hijacking their endogenous reception systems. In this review, we focus on short secretory peptides ranging from five to 20 amino acids. We first summarize recent advances in understanding relationships between the bioactivities and structures of plant peptide hormones. Subsequently, we introduce the topic of peptides produced by non-plant organisms. Lastly, we describe artificial peptides synthesized in laboratories, which possess intriguing bioactive properties beyond those of natural peptide hormones.
Keywords: CLE, Ligand, LRR-RK, Pathogens, Peptide hormone, Receptor
Introduction
In this mini-review, we aim to discuss how molecular structures of plant peptide hormones have been shaped and how one can design artificial peptide hormones with novel biological functions. Since the discovery of tomato systemin (Pearce et al. 1991), a number of different classes of peptide signals have been identified. They act extracellularly through recognition by their receptors on the plasma membrane of target cells. Based on (i) mature peptide structures and (ii) modes of trafficking into the extracellular space, these peptide signals, or plant peptide hormones, are classified into three groups: secreted small peptides, non-secreted small peptides and secreted cysteine-rich peptides (CRPs) (Matsubayashi 2014). The first two are both approximately 5–20 amino acids in length and do not undergo intramolecular disulfide bonding while the CRPs consist of 50–100 amino acids and have a relatively fixed structure due to intracellular disulfide bridges (Ohki et al. 2011). In this mini-review, we focus on the short peptides, all of which are perceived by leucine-rich repeat receptor kinases (LRR-RKs). Interestingly, such peptide signals are also made by phytopathogens to hijack functions of host plants. In the first and second sections, we will describe variations found in plants and parasitic phytopathogens, respectively. Since this review focuses on molecular structures of peptides, especially in dicots, please refer to other literature for the latest information and discussion on biological functions of peptide signals in diverse plant species including monocots (Grienenberger and Fletcher 2015, Higashiyama and Takeuchi 2015, Je et al. 2016, Okamoto et al. 2016, Somssich et al. 2016, Zipfel and Oldroyd 2017, Stegmann et al. 2017). In addition to the naturally occurring mechanisms shaping these peptide hormones, we will introduce synthetic approaches to design novel bioactive peptides in the last section.
Made in Plants
Peptide classes, receptors and structural insights
Small peptide signals of plants include systemin, PSK (phytosulfokine), HypSys (hydroxyproline-rich glycopeptide systemin), Pep1, CLE (CLAVATA3/EMBRYO SURROUNDING REGION-related)/TDIF (tracheary element differentiation inhibitory factor), PSY (plant peptide containing sulfated tyrosine), CEP (C-terminally encoded peptide), RGF/CLEL/GLV (root meristem growth factor/CLE-like/GOLVEN), PIP (PAMP-INDUCED PEPTIDE), IDA (INFLORESCENCE DEFICIENT IN ABSCISSION) and CIF (Casparian strip integrity factor) subclasses (Pearce et al. 1991, Matsubayashi and Sakagami 1996, Pearce et al. 2001, Huffaker et al. 2006, Ito et al. 2006, Ohyama et al. 2008, Ohyama et al. 2009, Matsuzaki et al. 2010, Okamoto et al. 2013, Hou et al. 2014, Schardon et al. 2016, Doblas et al. 2017, Nakayama et al. 2017). They are encoded in the genome as precursor proteins and mature into active forms via post-translational processing including proteolytic cleavage by proteases (Tamaki et al. 2013, Engineer et al. 2014, Schardon et al. 2016) and modifications of specific residues by modifying enzymes (Hieta and Myllyharju 2002, Tiainen et al. 2005, Yuasa et al. 2005, Komori et al. 2009, Ogawa-Ohnishi et al. 2013) such as tyrosine sulfation, proline hydroxylation and hydroxyproline arabinosylation.
Receptors for these peptides have been identified genetically and biochemically (Butenko et al. 2014). The major receptor class is the LRR-RKs (Shiu and Bleecker 2001). LRR-RKs are single transmembrane domain kinases containing extracellular LRRs which can participate in versatile molecular recognition. The receptors for CLE/TDIF (Hirakawa et al. 2008, Ogawa et al. 2008), IDA (Santiago et al. 2016), CEP (Tabata et al. 2014), Pep1 (Yamaguchi et al. 2006), RGF (Shinohara et al. 2016), PIP1 (Hou et al. 2014) and CIF (Doblas et al. 2017, Nakayama et al. 2017) are in the subclass XI of the LRR-RK family, while the PSK receptor PSKR is in the subclass X (Matsubayashi et al. 2002). Binding of the peptide hormones to their receptors is thought to recruit additional co-receptors for the activation of downstream signaling events by transphosphorylation between kinase domains in proximity (Fig. 1A). Co-crystal structures of several peptide–receptor pairs have been solved recently (Song et al. 2016a). The extracellular region of the receptors contains an LRR, which forms a superhelix structure providing the structural backbone to form an interaction surface for a corresponding peptide ligand. In each peptide–receptor complex, a peptide molecule is stretched along the inner surface of the superhelix (Fig. 1B). Generally, peptide ligands act as a molecular glue to stabilize the interaction between each corresponding receptor and its co-receptor (Fig. 1A, B) (Tang et al. 2015, Morita et al. 2016, Santiago et al. 2016, Song et al. 2016b, Zhang et al. 2016). Interestingly, in the case of PSK perception by PSKR, the peptide binding to the receptor triggers its allosteric change, which allows the binding of the co-receptor SERK (SOMATIC EMBRYOGENESIS RECEPTOR KINASE) to PSKR (Wang et al. 2015). In some cases, LRR-RKs may work with other classes of proteins to perceive peptide signals and/or trigger intracellular signaling, such as single transmembrane LRR proteins harboring only extracellular LRRs without intracellular kinase domains (Jeong et al 1999, Nadeau and Sack 2002) and transmembrane kinase proteins containing only intracellular kinase domains without extracellular LRRs (Müller et al. 2008) (Fig. 1C). However, the mechanisms of direct peptide recognition and signal transduction by these proteins remain to be understood precisely (Bleckmann et al. 2010, Kinoshita et al. 2010, Zhu et al. 2010, Nimchuk et al. 2011, Bommert et al. 2013, Stahl et al. 2013, Ishida et al. 2014).
Fig. 1.
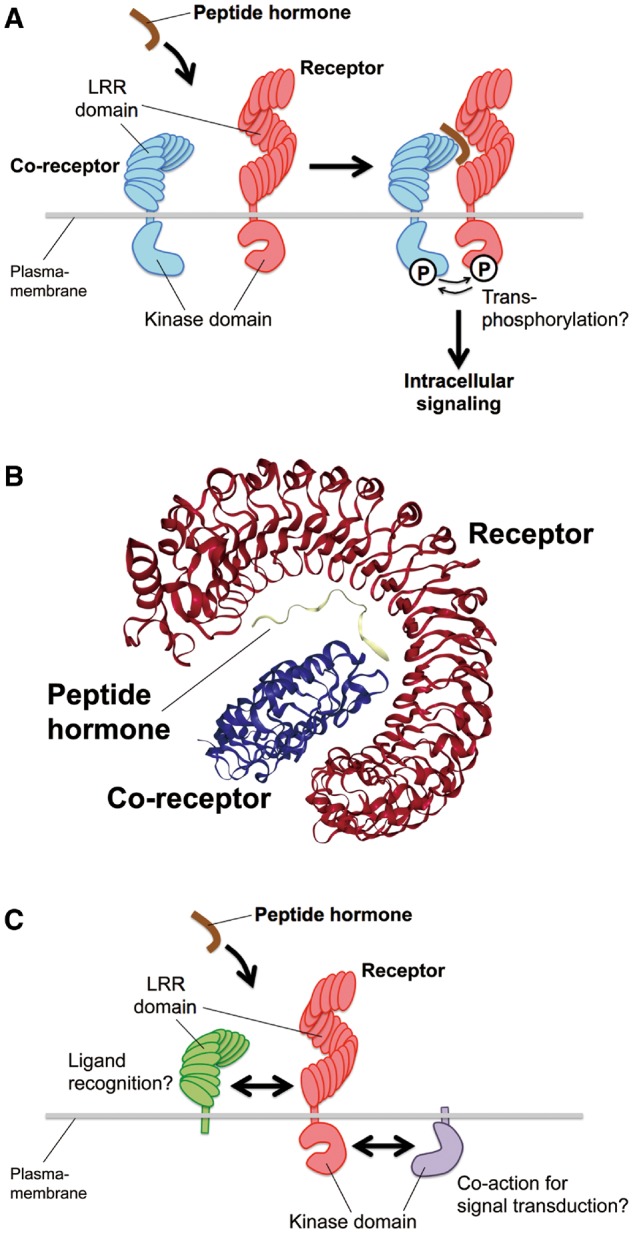
A proposed action of a peptide hormone and its receptor–co-receptor pair in the ‘molecular glue’ model. (A) A receptor (red) interacts with its co-receptor (blue) only in the presence of a peptide hormone (brown). Upon binding of the peptide, it is considered that the receptor and co-receptor can phosphorylate each other, triggering the intracellular signaling. (B) The co-crystal structure of the IDA peptide (yellow) and LRR domains of its receptor HAESA (red) and co-receptor SERK1 (blue) (PBD accession number: 5IYX) (Santiago et al. 2016) is shown as an example of the peptide–receptor–co-receptor complex. The structure was illustrated using the NGL viewer in the Protein Data Bank website. (C) LRR proteins without kinase domains (green) may participate in direst recognition of ligands with LRR-RK receptors (red). Also, transmembrane kinases without extracellular domains (purple) may act to trigger intracellular signal transduction co-ordinately with LRR-RK receptors (red).
At the binding surface of the peptide hormone and receptor, both the main and side chains of the peptide form multiple hydrogen bonds and/or hydrophobic contacts with the receptor. In some cases, side chain modifications of peptide ligands directly interact with receptor residues such as the PSK–PSKR pair that involves two sulfate groups of PSK in the interaction surface (Wang et al. 2015). The sulfate group of RGF1 is recognized by the RxGG motif that is conserved among RGF receptors (Song et al. 2016b). The hydroxyproline residue of IDA peptide forms hydrogen bonds with the receptor (Santiago et al. 2016). In contrast, hydroxyprolines of CLE peptides do not directly interact to their receptors (Morita et al. 2016, Zhang et al. 2016). Further arabinosylation of hydroxyprolines is found in some CLEs, and the arabinosylation is important for bioactivity (Ohyama et al. 2009, Okamoto et al. 2013, Xu et al. 2015). A proposed role for the arabinosylation is to force a conformational distortion on the peptide backbone in a highly directional manner, conferring a significant increase in affinity for the corresponding receptors (Shinohara and Matsubayashi 2013).
The reported co-crystal structures of subclass XI LRR-RKs and their peptide ligands also show that the conserved RxR motifs of the receptors are involved in the interaction with the free carboxyl group of the last residue of TDIF/CLE41, Pep1, RGF1 or IDA (Song et al. 2016a), which is in agreement with the report on the SOL1 (SUPPRESSOR OF LLP1 1) protease required for the maturation of functional CLE19 peptide by cleaving off the C-terminal extension in its precursor (Tamaki et al. 2013).
Collectively, shapes of peptide hormones and their recognition by corresponding receptors have been co-ordinately elaborated during their molecular evolution.
Diversification under evolutionary constraints
All plant peptide hormone genes identified so far belong to gene families. Each family contains small variations in the mature ligand sequences. Such minor variations could be formed under certain evolutionary pressures in their molecular evolution. The contribution of each amino acid residue in peptide hormones can be examined by analysis of structure–activity relationship using mutated peptides. A typical method is alanine scanning in which every residue of a peptide hormone is substituted one by one with alanine. If the alanine substitution of a certain residue affects the bioactivity, the side chain of the residue must play an important role in exerting the specific bioactivity. For example, the sixth glycine of TDIF/CLE41 peptide is important for its kinked structure that is recognized by its receptor TDR/PXY, and indeed the substitution of the glycine by alanine abolishes the bioactivity (Ito et al. 2006, Morita et al. 2016).
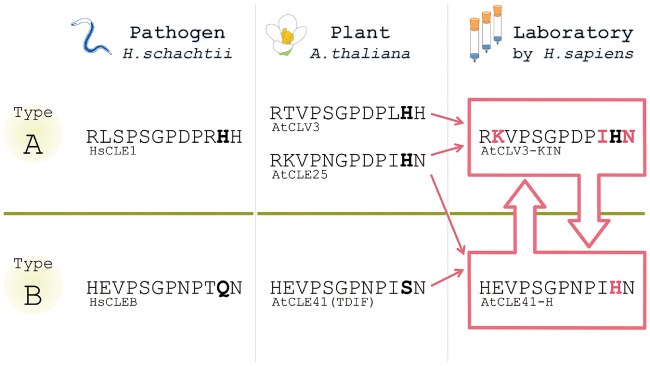
On the other hand, some residues can play a role to avoid activating unwanted signaling. Very recently, an intriguing example of this case was reported (Hirakawa et al. 2017). The CLE-family peptides are classified into two subfamilies; one group (A-type) that can affect the shoot and root meristems and the other (B-type) that affects the vascular meristem (Cock and McCormick 2001, Ito et al. 2006, Whitford et al. 2008). B-type CLEs have the characteristic serine residue at the 11th position in the mature form (Fig. 2) that is conserved only among the B-type peptides within the CLE family (Oelkers et al. 2008, Hirakawa and Bowman 2015). Surprisingly, the mutation of the 11th serine into histidine results in the acquisition of the A-type activity without losing the original B-type activity (Fig. 2). Such a striking property has been overlooked by the previous alanine scanning, which classified the 11th serine as a ‘dispensable’ residue for bioactivity (Ito et al. 2006). These suggest that the 11th serine may be kept unchanged to avoid unwanted signaling which disrupts the well-organized signaling network for growth and development.
Fig. 2.
CLE peptide hormones made in plants, pathogens and laboratories. Amino acid sequences of representative CLE peptides are shown. CLE peptides produced by plants and pathogens are classified into A and B types depending on their activities. CLE peptides in one group do not exert the activity of the other group, indicating that there exists a strict specificity barrier (green) between the two groups. However, it was recently reported that some artificial CLE peptides synthesized in laboratories show both activities beyond the specificity barrier (Hirakawa et al. 2017), as indicated by open pink arrows. Solid pink arrows indicate the flows to design the synthetic bi-functional peptides. Black bold font indicates the characteristic 11th residues. Pink bold font indicates swapped residues to create the bi-functional peptides. See the main text for a detailed explanation.
Made in Pthogens
Homologs of plant peptide hormones are found in phytopathogen genomes, which may be acquired either via convergent evolution or by horizontal gene transfer from host plants (Olsen and Skriver 2003). Parasitic nematodes enter the plant root and alter its tissue structure to form feeding cells/tissues such as syncytia and giant cells (Mitchum et al. 2012). For this purpose, parasitic nematodes secrete effector proteins to hijack developmental systems of host plants. The first example of secretory peptide mimics produced by parasitic nematodes is HgCLE1/syv46 of the soybean cyst nematode Heterodera glycines, which shows similarity to the A-type CLE peptides of host plants (Wang et al. 2001, Olsen and Skriver 2003). HgCLE1 is expressed mainly in the esophageal gland and released into plant cells via the stylet (Wang et al. 2005). The precursor protein of HgCLE1 peptide contains a domain essential for its subcellular trafficking into the apoplast, allowing the nematode-derived CLE peptide to interact with the extracellular domains of target receptors in host plants (Wang et al. 2010, Replogle et al. 2011). In addition to the A-type CLE peptides, B-type CLE homologs were also reported recently in Heterodera schachtii (Guo et al. 2017). Since A-type and B-type CLEs synergistically promote vascular thickening in plants (Whitford et al. 2008), the nematodes may have exploited this synergistic effect for maximizing their successful parasitism. Interestingly, the CLE peptide sequences in nematodes are slightly different from those of plant CLE peptides (Fig. 2; Yamaguchi et al. 2016), which may reflect differences in maturation processes between nematode and plant CLE peptides. In addition to short CLE peptides, functional homologs of the CRP-type peptide hormone RALF (rapid alkalinization factor) are also found in fungal pathogens (Masachis et al. 2016, Thynne et al. 2016). The significance of differences in peptide sequences between homologs derived from plants and pathogens has not been well understood. As yet uncovered constraints may have existed in the evolution of the plant–pathogen interaction.
Made in Laboratories
As mentioned above, natural peptide hormones are made in living organisms and have been optimally shaped under evolutionary pressures. In contrast, chemical synthesis in laboratories does not have such restrictions and thus could enable new design principles for functional peptides. In theory, engineering of artificial bioactive molecules could be accomplished for any type of hormones. For example, one may imagine a molecule which exerts both auxin and cytokinin activities by simply coupling IAA and kinetin. However, considering the structural information on the ligand-binding pockets of the auxin and cytokinin receptors (Tan et al. 2007, Hothorn et al. 2011), this imaginary bi-functional molecule is difficult to design. On the other hand, synthesis of bi-functional peptides that bind and activate two distinct CLE receptors was reported recently (Hirakawa et al. 2017). CLV3 and CLE25 both belong to the A-type CLE peptides and affect the shoot and root meristems. They have four amino acid substitutions compared with each other (Fig. 2). Surprisingly, systematic swapping of these residues led to the discovery of a synthetic peptide that exerts the B-type activity in addition to the original A-type activity (Fig. 2: CLV3-KIN that has the CLV3 backbone with K, I and N substitutions derived from CLE25) by interacting with both A-type and B-type CLE receptors. As mentioned above, TDIF/CLE41 can also acquire bi-functionality by an amino acid substitution (Fig. 2: CLE41-H). These studies suggest that building blocks for designing unnatural bi-functional peptides (such as CLV3-KIN and CLE41-H) exist in the natural diversity in the genome. Further identification of such cryptic bioactivities will be a future challenge toward engineering cell–cell signaling in plants.
Peptides are chains of amino acids linked by amide bonds (peptide bonds). Peptide-like molecules with different main chain structures are collectively called peptidomimetics. Peptidomimetics have been developed especially in the field of drug discovery, pursuing enhanced in vivo stability and activity (Vagner et al. 2008). A previous study adopted this approach to understand the structure–activity relationship of CLE peptides, and the ninth proline residue was substituted with a series of N-modified peptoids, such as sarcosine (N-methylglycine), to control the bioactivity (Kondo et al. 2011). Besides peptoids, synthetic routes for new molecular designs of peptidomimetics have been explored not only for pure chemistry but also for development of bioengineering approaches. By harnessing diversity in molecular structures of peptides/peptidomimetics, which may also include unnatural side chains, we may be able to expand toolkits for peptide hormone studies toward creating unprecedented bioactivities.
Funding
This research was supported by Ministry of Education, Culture, Sports, Science and Technology (MEXT)/Japan Society for the Promotion of Science (JSPS) KAKENHI [grant Nos. 14J08452 to Y.H.; JP26291057 and JP16H01237 to K.U.T.; JP16H01462 and JP17H03695 to N.U.] and the Howard Hughes Medical Institute (HHMI) and Gordon and Betty Moore Foundation (GBMF) [grant No. GBMF3035 to K.U.T]. Y.H. was a JSPS Postdoctoral Fellow; K.U.T. is an HHMI–GBMF Investigator.
Disclosures
The authors have no conflicts of interest to declare.
Glossary
Abbreviations
- CEP
C-terminally encoded peptide
- CIF
Casparian strip integrity factor
- CLE
CLAVATA3/EMBRYO SURROUNDING REGION-related
- CRP
cysteine-rich peptide
- IDA
INFLORESCENCE DEFICIENT IN ABSCISSION
- LRR-RK
leucine-rich repeat receptor kinase
- PIP
PAMP-INDUCED PEPTIDE
- PSK
phytosulfokine
- RGF
root meristem growth factor
- SERK
SOMATIC EMBRYOGENESIS RECEPTOR KINASE
- TDIF
tracheary element differentiation inhibitory factor
References
- Bleckmann A., Weidtkamp-Peters S., Seidel C.A., Simon R. (2010) Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 152: 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P., Je B.I., Goldshmidt A., Jackson D. (2013) The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 502: 555–558. [DOI] [PubMed] [Google Scholar]
- Butenko M.A., Wildhagen M., Albert M., Jehle A., Kalbacher H., Aalen R.B. et al. (2014) Tools and strategies to match peptide–ligand receptor pairs. Plant Cell 26: 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock J.M., McCormick S. (2001) A large family of genes that share homology with CLAVATA3. Plant Physiol. 126: 939–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblas V.G., Smakowska-Luzan E., Fujita S., Alassimone J., Barberon M., Madalinski M. et al. (2017) Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science 355: 280–284. [DOI] [PubMed] [Google Scholar]
- Engineer C.B., Ghassemian M., Anderson J.C., Peck S.C., Hu H., Schroeder J.I. (2014) Carbonic anhydrases, EPF2 and a novel protease mediate CO2 control of stomatal development. Nature 513: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger E., Fletcher J.C. (2015) Polypeptide signaling molecules in plant development. Curr. Opin. Plant Biol. 23: 8–14. [DOI] [PubMed] [Google Scholar]
- Guo X., Wang J., Gardner M., Fukuda H., Kondo Y., Etchells J.P. et al. (2017) Identification of cyst nematode B-type CLE peptides and modulation of the vascular stem cell pathway for feeding cell formation. PLoS Pathog. 13: e1006142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieta R., Myllyharju J. (2002) Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. Effective hydroxylation of proline-rich, collagen-like, and hypoxia-inducible transcription factor alpha-like peptides. J. Biol. Chem. 277: 23965–23971. [DOI] [PubMed] [Google Scholar]
- Higashiyama T., Takeuchi H. (2015) The mechanism and key molecules involved in pollen tube guidance. Annu. Rev. Plant Biol. 66: 393–413. [DOI] [PubMed] [Google Scholar]
- Hirakawa Y., Bowman J.L. (2015) A role of TDIF peptide signaling in vascular cell differentiation is conserved among euphyllophytes. Front. Plant Sci. 6: 1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y., Shinohara H., Kondo Y., Inoue A., Nakanomyo I., Ogawa M. et al. (2008) Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 105: 15208–15213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y., Shinohara H., Welke K., Irle S., Matsubayashi Y., Torii K.U. et al. (2017) Cryptic bioactivity capacitated by synthetic hybrid plant peptides. Nat. Commun. 8: 14318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M., Dabi T., Chory J. (2011) Structural basis for cytokinin recognition by Arabidopsis thaliana histidine kinase 4. Nat. Chem. Biol. 7: 766–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S., Wang X., Chen D., Yang X., Wang M., Turrá D. et al. (2014) The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7. PLoS Pathog. 10: e1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A., Pearce G., Ryan C.A. (2006) An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA 103: 10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Tabata R., Yamada M., Aida M., Mitsumasu K., Fujiwara M. et al. (2014) Heterotrimeric G proteins control stem cell proliferation through CLAVATA signaling in Arabidopsis. EMBO Rep 15: 1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., Dohmae N. et al. (2006) Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313: 842–845. [DOI] [PubMed] [Google Scholar]
- Je B.I., Gruel J., Lee Y.K., Bommert P., Arevalo E.D., Eveland A.L. et al. (2016) Signaling from maize organ primordia via FASCIATED EAR3 regulates stem cell proliferation and yield traits. Nat. Genet. 48: 785–791. [DOI] [PubMed] [Google Scholar]
- Jeong S., Trotochaud A.E., Clark S.E. (1999) The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11: 1925–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A., Betsuyaku S., Osakabe Y., Mizuno S., Nagawa S., Stahl Y. et al. (2010) RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137: 3911–3920. [DOI] [PubMed] [Google Scholar]
- Komori R., Amano Y., Ogawa-Ohnishi M., Matsubayashi Y. (2009) Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 15067–15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Yokomine K., Nakagawa A., Sakagami Y. (2011) Analogs of the CLV3 peptide: synthesis and structure–activity relationships focused on proline residues. Plant Cell Physiol. 52: 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masachis S., Segorbe D., Turrá D., Leon-Ruiz M., Fürst U., El Ghalid M. et al. (2016) A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat. Microbiol. 1: 16043. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y. (2014) Posttranslationally modified small-peptide signals in plants. Annu. Rev. Plant Biol. 65: 385–413. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y., Ogawa M., Morita A., Sakagami Y. (2002) An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296: 1470–1472. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y., Sakagami Y. (1996) Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc. Natl. Acad. Sci. USA 93: 7623–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y., Ogawa-Ohnishi M., Mori A., Matsubayashi Y. (2010) Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329: 1065–1067. [DOI] [PubMed] [Google Scholar]
- Mitchum M.G., Wang X., Wang J., Davis E.L. (2012) Role of nematode peptides and other small molecules in plant parasitism. Annu. Rev. Phytopathol. 50: 175–195. [DOI] [PubMed] [Google Scholar]
- Morita J., Kato K., Nakane T., Kondo Y., Fukuda H., Nishimasu H. et al. (2016) Crystal structure of the plant receptor-like kinase TDR in complex with the TDIF peptide. Nat. Commun. 7: 12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R., Bleckmann A., Simon R. (2008) The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell. 20: 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau J.A., Sack F.D. (2002) Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700. [DOI] [PubMed] [Google Scholar]
- Nakayama T., Shinohara H., Tanaka M., Baba K., Ogawa-Ohnishi M., Matsubayashi Y. (2017) A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science 355: 284–286. [DOI] [PubMed] [Google Scholar]
- Nimchuk Z.L., Tarr P.T., Ohno C., Qu X., Meyerowitz E.M. (2011) Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr. Biol. 21: 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelkers K., Goffard N., Weiller G.F., Gresshoff P.M., Mathesius U., Frickey T. (2008) Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol. 8: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M., Matsushita W., Matsubayashi Y. (2013) Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nat. Chem. Biol. 9: 726–730. [DOI] [PubMed] [Google Scholar]
- Ohki S., Takeuchi M., Mori M. (2011) The NMR structure of stomagen reveals the basis of stomatal density regulation by plant peptide hormones. Nat. Commun. 2: 512. [DOI] [PubMed] [Google Scholar]
- Ohyama K., Ogawa M., Matsubayashi Y. (2008) Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 55: 152–160. [DOI] [PubMed] [Google Scholar]
- Ohyama K., Shinohara H., Ogawa-Ohnishi M., Matsubayashi Y. (2009) A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5: 578–580. [DOI] [PubMed] [Google Scholar]
- Okamoto S., Shinohara H., Mori T., Matsubayashi Y., Kawaguchi M. (2013) Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat. Commun. 4: 2191. [DOI] [PubMed] [Google Scholar]
- Okamoto S., Tabata R., Matsubayashi Y. (2016) Long-distance peptide signaling essential for nutrient homeostasis in plants. Curr. Opin. Plant Biol. 34: 35–40. [DOI] [PubMed] [Google Scholar]
- Olsen A.N., Skriver K. (2003) Ligand mimicry? Plant-parasitic nematode polypeptide with similarity to CLAVATA3. Trends Plant Sci. 8: 55–57. [DOI] [PubMed] [Google Scholar]
- Pearce G., Moura D.S., Stratmann J., Ryan C.A. (2001) Production of multiple plant hormones from a single polyprotein precursor. Nature 411: 817–820. [DOI] [PubMed] [Google Scholar]
- Pearce G., Strydom D., Johnson S., Ryan C.A. (1991) A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253: 895–897. [DOI] [PubMed] [Google Scholar]
- Replogle A., Wang J., Bleckmann A., Hussey R.S., Baum T.J., Sawa S. et al. (2011) Nematode CLE signaling in Arabidopsis requires CLAVATA2 and CORYNE. Plant J. 65: 430–440. [DOI] [PubMed] [Google Scholar]
- Santiago J., Brandt B., Wildhagen M., Hohmann U., Hothorn L.A., Butenko M.A. et al. (2016) Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife 5: e15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardon K., Hohl M., Graff L., Pfannstiel J., Schulze W., Stintzi A. et al. (2016) Precursor processing for plant peptide hormone maturation by subtilisin-like serine proteinases. Science 354: 1594–1597. [DOI] [PubMed] [Google Scholar]
- Shinohara H., Matsubayashi Y. (2013) Chemical synthesis of Arabidopsis CLV3 glycopeptide reveals the impact of hydroxyproline arabinosylation on peptide conformation and activity. Plant Cell Physiol. 54: 369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H., Mori A., Yasue N., Sumida K., Matsubayashi Y. (2016) Identification of three LRR-RKs involved in perception of root meristem growth factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 113: 3897–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B. (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98: 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich M., Je B.I., Simon R., Jackson D. (2016) CLAVATA–WUSCHEL signaling in the shoot meristem. Development 143: 3238–3248. [DOI] [PubMed] [Google Scholar]
- Song W., Han Z., Wang J., Lin G., Chai J. (2016a) Structural insights into ligand recognition and activation of plant receptor kinases. Curr. Opin. Struct. Biol. 43: 18–27. [DOI] [PubMed] [Google Scholar]
- Song W., Liu L., Wang J., Wu Z., Zhang H., Tang J. et al. (2016b) Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth. Cell Res. 26: 674–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y., Grabowski S., Bleckmann A., Kühnemuth R., Weidtkamp-Peters S., Pinto K.G. et al. (2013) Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr. Biol. 23: 362–371. [DOI] [PubMed] [Google Scholar]
- Stegmann M., Monaghan J., Smakowska-Luzan E., Rovenich H., Lehner A., Holton N. et al. (2017) The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355: 287–289. [DOI] [PubMed] [Google Scholar]
- Tabata R., Sumida K., Yoshii T., Ohyama K., Shinohara H., Matsubayashi Y. (2014) Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346: 343–346. [DOI] [PubMed] [Google Scholar]
- Tamaki T., Betsuyaku S., Fujiwara M., Fukao Y., Fukuda H., Sawa S. (2013) SUPPRESSOR OF LLP1 1-mediated C-terminal processing is critical for CLE19 peptide activity. Plant J. 76: 970–981. [DOI] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L.I., Sharon M., Zheng C., Robinson C.V., Estelle M. et al. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645. [DOI] [PubMed] [Google Scholar]
- Tang J., Han Z., Sun Y., Zhang H., Gong X., Chai J. (2015) Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1. Cell Res. 25: 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thynne E., Saur I.M., Simbaqueba J., Ogilvie H.A., Gonzalez-Cendales Y., Mead O. et al. (2016) Fungal phytopathogens encode functional homologues of plant rapid alkalinisation factor (RALF) peptides. Mol. Plant Pathol. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiainen P., Myllyharju J., Koivunen P. (2005) Characterization of a second Arabidopsis thaliana prolyl 4-hydroxylase with distinct substrate specificity. J. Biol. Chem. 280: 1142–1148. [DOI] [PubMed] [Google Scholar]
- Vagner J., Qu H., Hruby V.J. (2008) Peptidomimetics, a synthetic tool of drug discovery. Curr. Opin. Chem. Biol. 12: 292–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Lee C., Replogle A., Joshi S., Korkin D., Hussey R. et al. (2010) Dual roles for the variable domain in protein trafficking and host-specific recognition of Heterodera glycines CLE effector proteins. New Phytol. 187: 1003–1017. [DOI] [PubMed] [Google Scholar]
- Wang J., Li H., Han Z., Zhang H., Wang T., Lin G. et al. (2015) Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature 525: 265–268. [DOI] [PubMed] [Google Scholar]
- Wang X., Allen R., Ding X., Goellner M., Maier T., de Boer J.M. et al. (2001) Signal peptide-selection of cDNA cloned directly from the esophageal gland cells of the soybean cyst nematode Heterodera glycines. Mol. Plant Microbe Interact. 14: 536–544. [DOI] [PubMed] [Google Scholar]
- Wang X., Mitchum M.G., Gao B., Li C., Diab H., Baum T.J. et al. (2005) A parasitism gene from a plant-parasitic nematode with function similar to CLAVATA3/ESR (CLE) of Arabidopsis thaliana. Mol. Plant Pathol. 6: 187–191. [DOI] [PubMed] [Google Scholar]
- Whitford R., Fernandez A., De Groodt R., Ortega E., Hilson P. (2008) Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc. Natl. Acad. Sci. USA 105: 18625–18630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Liberatore K.L., MacAlister C.A., Huang Z., Chu Y.H., Jiang K. et al. (2015) A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 47: 784–792. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Pearce G., Ryan C.A. (2006) The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc. Natl. Acad. Sci. USA 103: 10104–10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y.L., Ishida T., Sawa S. (2016) CLE peptides and their signaling pathways in plant development. J. Exp. Bot. 67: 4813–4826. [DOI] [PubMed] [Google Scholar]
- Yuasa K., Toyooka K., Fukuda H., Matsuoka K. (2005) Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. Plant J. 41: 81–94. [DOI] [PubMed] [Google Scholar]
- Zhang H., Lin X., Han Z., Qu L.J., Chai J. (2016) Crystal structure of PXY–TDIF complex reveals a conserved recognition mechanism among CLE peptide–receptor pairs. Cell Res. 26: 543–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Wang Y., Li R., Song X., Wang Q., Huang S. et al. (2010) Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis. Plant J. 61: 223–233. [DOI] [PubMed] [Google Scholar]
- Zipfel C., Oldroyd G.E. (2017) Plant signalling in symbiosis and immunity. Nature 543: 328–336. [DOI] [PubMed] [Google Scholar]