Abstract
Flavonoids are secondary metabolites derived from the general phenylpropanoid pathway and are widespread throughout the plant kingdom. The functions of flavonoids are diverse, including defense against phytopathogens, protection against UV light damage and oxidative stress, regulation of auxin transport and allelopathy. One of the most conspicuous functions of flavonoids has long attracted the attention of pollinators and scientist alike: the vivid shades of red, pink, orange, blue and purple on display in the flowers of angiosperms. Thus, flavonoid pigments have perhaps been the most intensely studied phenylpropanoids. From Mendel to McClintock and up to the present, studies centered on flavonoid pigments have resulted in some of the most important scientific discoveries of the last 150 years, including the first examples of transcriptional regulation in plants. Here we focus on the highly conserved MYB–bHLH–WD repeat (MBW) transcriptional complex model for the regulation of the flavonoid pigment pathway. We will survey the history of the MBW model spanning the last three decades, highlighting the major findings that have contributed to our current understanding. In particular, recent discoveries regarding WRKY protein control of the flavonoid pigment pathway and its relationship to the MBW complex will be emphasized. In addition, we will discuss recent findings about the regulation of the beet betalain pigment pathway, and how a MYB member of the MBW complex was co-opted to regulate this chemically unrelated but functionally equivalent pathway.
Keywords: Anthocyanins, Betalains, Flavonoids, MBW, Proanthocyanidins, WRKY
Introduction to MYBs, bHLHs and WD Repeat Proteins
The myeloblastosis protein (MYB) DNA-binding domain defines a class of transcriptional regulators found in all eukaryotic organisms. This domain was first identified in the v-myb oncogene of the avian myeloblastosis virus and in its cellular homolog c-myb. Animal MYB transcription factors typically contain three imperfect MYB repeats (R1R2R3 repeats) and comprise a small family of two or three proteins with roles in cell proliferation (Lipsick 1996). Each repeat is about 52 amino acids long, containing regularly spaced tryptophan residues and folding into a helix–turn–helix variant related to those of prokaryotic repressors.
The MYB gene family expanded dramatically in higher plants, with Arabidopsis containing approximately 339 MYB genes and rice containing about 230 MYB genes (Feller et al. 2011). Accordingly, MYB proteins function in a diverse array of processes including the regulation of plant form, the cell cycle, cell differentiation, metabolism and stress response (for a general review of plant MYB transcription factors, see Dubos et al. 2010, Feller et al. 2011).
The majority of plant MYB proteins contain two imperfect MYB repeats corresponding to the R2R3 Myb repeats of the three repeat R1R2R3 animal MYB proteins. Sequence-specific DNA binding has been demonstrated for several R2R3 MYB transcription factors, with these Mybs binding to one or more DNA sequence types (Romeo et al. 1998, Feller et al. 2011). Additionally, R2R3 plant MYB proteins may contain an acidic transcriptional activation domain. In the context of the MYB–bHLH–WD repeat protein (MBW) complex considered in this review, a notable exception to the R2R3 organization is a small group of single MYB (R3) repeat proteins of Arabidopsis (Schellmann et al. 2002). These proteins have only one MYB domain, lack a transcriptional activation domain and function as negative regulators of transcription.
The basic helix–loop–helix (bHLH) domain was originally identified as a similar region shared among a group of DNA-binding proteins from animals (Murre et al. 1989). At approximately the same time, the R locus of Zea mays (maize) was shown to encode a bHLH transcriptional regulator of biosynthetic genes necessary for pigment production (Ludwig et al. 1989). A typical bHLH domain contains about 18 hydrophilic and basic amino acids at its N-terminal side followed by two amphipathic α-helices separated by a loop. Functionally, the basic region is important for making DNA contacts while the HLH region allows dimerization between bHLH proteins (Massari and Murre 2000). Animal bHLH proteins have broad functions in regulating cell proliferation and differentiation.
In plants, bHLH proteins comprise the second largest transcription factor class. They have very diverse roles in processes ranging from regulation of secondary metabolism, cellular differentiation and patterning, plant growth and development via the regulation of brassinosteroid and ABA hormone signaling pathways, iron uptake in roots and phytochrome-mediated light signaling. bHLH proteins can regulate transcription by recognizing E-boxes, DNA sequences with the consensus of CANNTG (for a general review on plant bHLH proteins, see Feller et al. 2011).
Unlike the Myb and bHLH domains, it is difficult to define a distinct family of proteins based solely on the presence of a WD repeat motif (WDR; Smith et al. 1999, van Nocker and Ludwig, 2003). The WD was defined as an approximately 40 amino acid structural repeat ending with tryptophan–aspartic acid (W–D). Indeed, there can be uncertainty in defining and identifying WDRs due to large variations in the position and length of structural elements composing WDR motifs found in different proteins. For example, Arabidopsis contains 237 proteins with at least four WDRs (269 total proteins with at least one WDR). These 237 proteins were classified into 143 distinct families with about 113 of these families showing clear homology to WDR proteins from yeast, fly and/or human. Thus, WDRs are found in a diverse array of proteins spanning a broad spectrum of functions (for a review of WDR protein functions in plants, see van Nocker and Ludwig 2003, Miller et al. 2016). The WDRs generally serve as a protein–protein interaction platform for the formation of complexes and as mediators of transient connections between other proteins. The WDR motif was first identified in a Gβ subunit of heterotrimeric G proteins (Fong et al. 1986). The crystal structure of this protein shows that the WDRs adopt a β-propeller fold. The plant WDR proteins considered in this review are small proteins consisting only of four or five WDRs such as Transparent Testa Glabra 1 (TTG1) of Arabidopsis and its homologs controlling plant pigment production in other species (de Vetten et al. 1997, Walker et al. 1999, Miller et al. 2016).
The Research History of the MBW complex
The early years: foundational contributions from maize genetics
Because plants must regulate flavonoid pigment biosynthesis developmentally and in response to various biotic and abiotic stresses, this pathway has historically been an excellent model for the study of transcriptional regulation. Indeed, pioneering work performed 30 years ago in maize led to the discovery of transcription factors in plants. The first plant transcription factor gene cloned, via a transposon tagging approach, was Colorless1 (C1) from maize (Cone et al. 1986, Paz-Ares et al. 1987). c1 mutants lack anthocyanins in the aleurone layer of maize kernels and are transcriptionally down-regulated for a set of flavonoid biosynthetic genes. C1 encodes a MYB regulator of the anthocyanin biosynthetic pathway and, coincidentally, was the locus disrupted in Barbara McClintock’s breeding experiments that led to the discovery of transposable elements in the 1950s. This discovery represents the earliest milestone in the study of transcriptional regulation in plants and the first clue leading to the now canonical MBW model for the control of the flavonoid pigment pathway in all plants studied to date.
Just 2 years later, Ludwig et al. (1989) reported another landmark discovery in the field of plant transcriptional regulation that ultimately put the ‘B’ in the MBW complex: these researchers identified in maize the first plant bHLH gene encoded by the Red (R) locus required for pigment synthesis in the seed. Later in the same year, Chandler et al. (1989) reported the cloning of an additional regulatory locus, Booster (B), encoding a bHLH protein homologous to R. Depending on the allele, the B locus controls the anthocyanin pathway only in the plant (B-l) or in both the plant and the seed (B-Peru). Before the R and B loci were cloned, genetic and biochemical studies demonstrated the necessity of these loci for the activity of pigment production in the seed and plant, and for activating the enzymatic activity encoded by known pigment biosynthetic genes. Moreover, breeding experiments indicated that a functional R or B allele was not sufficient to activate the expression of anthocyanin structural genes, and thus pigment synthesis, in the absence of a functional C1 allele in the kernel (Chandler et al. 1989). Later, Cone et al. (1993) showed that the Purple Plant (Pl) locus required for anthocyanin pigments in the plant body encodes a MYB protein and close homolog to the C1 transcription factor. Similar to the kernel requiring C1 and R, activation of the anthocyanin pathway in the plant body requires both the Pl MYB and the B bHLH proteins. In addition, transactivation of anthocyanin structural gene promoter–reporter contructs bombarded into maize tissues also demonstrated a requirement for both a MYB and a bHLH protein (Goff et al. 1990). However, Goff et al. (1992) reported a novel biochemical observation that helped clarify the genetic interaction data and defined a cornerstone of the MBW transcriptional model as we understand it today: a physical interaction between B and C1 was demonstrated in yeast two-hybrid assays suggesting that these regulators function together in a transcriptional complex.
Other plants join the fray: contributions from Petunia, Antirrhinum and Arabidopsis
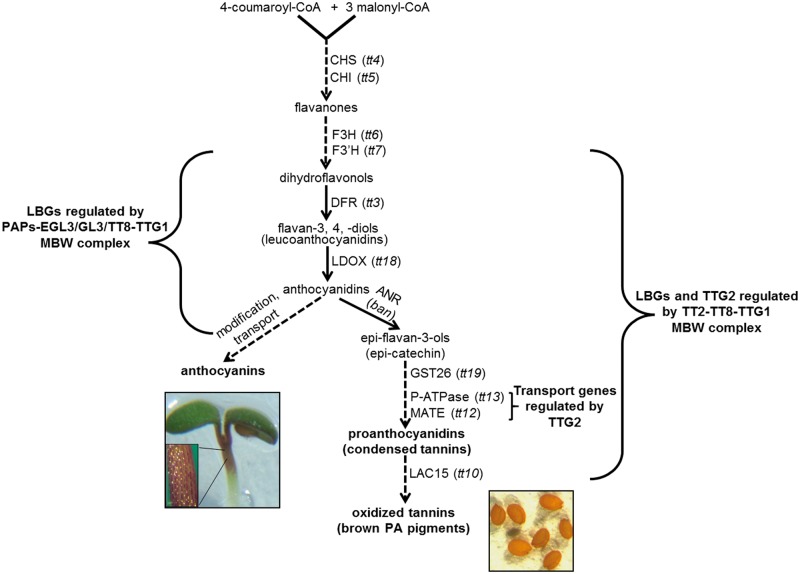
In addition to the early groundbreaking genetic work in maize, studies of the anthocyanin pathway in other species made important contributions to the emerging MBW model (Table 1). Petunia (Petunia hybrida) and snapdragon (Antirrhinum majus) were also major models for the study of anthocyanin biosynthesis, particularly in floral tissues. Arabidopsis thaliana was another key species for studying the synthesis and regulation of flavonoid pigments. Arabidopsis flowers, however, are naturally anthocyaninless but flavonoid-based proanthocyanidin pigments (PA; commonly known as tannins) are produced in the seed coat (testa), giving Arabidopsis seeds their characteristic brownish orange color (Fig. 1). Hence, the identification of pigment mutants in Arabidopsis primarily focused on convenient screens for yellow or lighter colored seed, otherwise known as transparent testa (tt) mutants (Koornneef 1990).
Table 1.
Composition of foundational MBW complexes and biological functions in the major plant pigment models
| Species | M–B–W | Function |
|---|---|---|
| Zea mays | C1–R/B–PAC1 | Anthocyanins in kernels. |
| P1–R/B–PAC1 | Anthocyanins in the plant body. | |
| Antirrhinum majus | Ros1/Ros2/Ve–Del–W? | Anthocyanin synthesis and patterning in flowers and the plant body. |
| Petunia hybrida | AN2–AN1–AN11 | Anthocyanins in flowers and the plant body. |
| PH4–AN1–AN11 | Flower color modification. | |
| Arabidopsis thaliana | PAPs–EGL3/GL3/TT8–TTG1 | Anthocyanins in the plant body. |
| TT2–TT8–TTG1 | PAs in seed coats. |
Fig. 1.
The Arabidopsis flavonoid pathway for the synthesis of anthocyanins and oxidized tannins, or proanthocyanidins (PAs). The large brackets indicate the late biosynthetic genes (LBGs) directly regulated by the MBW complex: on the left is the anthocyanin-specific MBW complex; on the right is the PA-specific MBW complex. The small bracket on the right indicates the narrow subset of LBGs encoding the transport steps regulated by TTG2. Dashed lines indicate multiple steps. Depicted on the left is a wild-type seedling expressing purplish red anthocyanins in the hypocotyl and parts of the cotyledons early in Arabidopsis development. Depicted on the right are Arabidopsis wild-type dry seed showing the brownish orange PA pigments expressed in the seed coat.
In Petunia, Antirrhinum and Arabidopsis, genes encoding MYB and bHLH proteins were identified as regulators of anthocyanin structural genes as in maize, demonstrating broad conservation of this regulatory mechanism in the plant kingdom. Delila, encoding a bHLH factor homologous to the product of R that controls pigment accumulation in Antirrhinum flowers, was the first dicot anthocyanin regulatory locus cloned (Goodrich et al. 1992). Later, it was shown that three Antirrhinum MYB genes, Rosea1, Rosea2 and Venosa, homologous to maize C1, differentially regulate structural genes of the anthocyanin pathway (Schwinn et al. 2006). In Petunia, Anthocyanin 1 and 2 (AN1 and AN2) were shown to encode a MYB and a bHLH transcription factor, respectively, necessary for pigmentation of floral organs (Quattrocchio et al. 1999, Spelt et al. 2000). Interestingly, the first anthocyanin regulatory locus cloned from Petunia, ANTHOCYANIN 11 (AN11), encoded neither a MYB nor a bHLH but a novel regulator protein containing five WDRs (de Vetten et al. 1997), a motif originally noted in a bovine G protein β subunit (Fong et al. 1986). Almost 20 years after its discovery (Koornneef 1981), the TTG1 locus of Arabidopsis was also shown to encode a WDR protein (Walker et al. 1999) homologous to that encoded by AN11. However, the relationship between the WDR protein and the MYB/bHLH transcriptional regulatory model was not immediately clear.
An epidermal cell fate digression in the development of the anthocyanin MBW model
Ironically, the now well-established placement of the WDR protein in the MBW complex did not come from anthocyanin studies in maize or the Petunia or Antirrhinum floral pigment model species, but instead from trichome cell fate studies in Arabidopsis. Besides lacking both anthocyanins and proanthocyanidin (PA) pigments, the transparent testa glabra1 (ttg1) mutant of Arabidopsis is deficient for trichome (plant hairs) initiation, root hair patterning and differentiation of the mucilage-producing outer seed coat cells. Although it had been known for some time that the maize R bHLH gene could rescue the entire suite of ttg1 mutant phenotypes (Lloyd et al. 1992), the first MBW complex bHLH genes identified in Arabidopsis were the PA pigment regulator TT8 (Nesi et al. 2000) and the trichome regulator GLABRA 3 (GL3). In yeast two-hybrid analysis, Payne et al. (2000) reported that GL3 could physically interact (via different domains) with both TTG1 and the Arabidopsis GL1 trichome regulator, a homolog of MYB anthocyanin regulators (Oppenheimer et al. 1991). Moreover, GL1 and TTG1 were not capable of direct physical interaction but could presumably form a complex together with GL3. Thus, the now canonical MBW transcriptional complex that regulates anthocyanin production throughout the plant kingdom (Table 1; Fig. 1) was first proposed in the context of Arabidopsis trichome production (Payne et al. 2000). A specific combination of MYB and bHLH proteins in Arabidopsis has since been identified for the regulation of specific TTG1-dependent epidermal cell fate pathways (Nesi et al. 2001, Bernhardt et al. 2003, Zhang et al. 2003, Bernhardt et al. 2005, Gonzalez et al. 2008, Gonzalez et al. 2009). Also, WDR proteins orthologous to AN11 and TTG1 have been identified as anthocyanin regulators in other species, including Pale Aleurone Color1 (PAC1) of maize (Carey et al. 2004).
In the case of anthocyanin pigment regulation in Arabidopsis, clear identification of the MBW complex was complicated by the redundancy of bHLH and MYB loci, and by the fact that Arabidopsis evolved different MBW complexes for anthocyanin and PA pigment regulation (Table 1; Fig. 1). The tt mutant screen approach described earlier yielded a pair of TTG1-dependent regulators, TT8 and TT2 (Nesi et al. 2000, Nesi et al. 2001). TT8 and TT2 encode a bHLH and MYB protein, respectively, that together with TTG1 regulate the biosynthesis of PA pigments in the Arabidopsis seed coat (Baudry et al. 2004). However, tt8 and tt2 mutants show no other ttg1-like mutant phenotypes, even displaying normal pigmentation in young Arabidopsis seedlings where anthocyanins are developmentally produced and TT8 is also expressed. This suggested a distinct, as yet unknown, set of TTG1-dependent MYB and bHLH regulators controlling anthocyanin biosynthesis in the plant body. As hypothesized, the characterization of a third bHLH regulatory gene, ENHANCER of GLABRA 3 (EGL3), uncovered the partially redundant roles of the three Arabidopsis bHLH proteins in regulating anthocyanin biosynthesis (Zhang et al. 2003). In addition, ectopic gene expression and RNA interference (RNAi) approaches helped identify four partially redundant Arabidopsis MYB genes (PAP1, PAP2, MYB113 and MYB114, collectively known as PAP MYBs for production of anthocyanin pigment) dedicated to the regulation of the anthocyanin pathway in the shoot (Borevitz et al. 2000, Gonzalez et al. 2008). Thus, in retrospect, it is clear why the Arabidopsis anthocyanin-specific MBW complex long evaded detection in screens focused on tt phenotypes.
Distillation of universal features of MBW regulation
After 30 years of investigating the regulation of anthocyanin biosynthesis via the MBW complex, some general trends may be noted across the numerous plant species studied to date. Based in part on the timing of structural gene expression and by the subset of structural genes regulated by the MBW complex, the flavonoid biosynthetic pathway is subdivided into ‘early’ steps and ‘late’ steps. The MBW complex is predominantly required for the regulation of late flavonoid biosynthetic genes over early genes (Fig. 1). However, the particular pathway steps comprising the late and early sets and the degree of regulation of the sets differ between species and even tissues (Taylor and Briggs 1991, Martin et al. 1991, Quattrocchio et al. 1993, Deboo et al. 1995, Winkel-Shirley et al. 1995, Pelletier and Winkel Shirley 1996, Pelletier et al. 1997, Zhang et al. 2003, Morita et al. 2006, Gonzalez et al. 2008). Moreover, the MBW complex is necessary and sufficient for the direct transcriptional regulation of late flavonoid biosynthetic genes (Baudry et al. 2004, Gonzalez et al. 2008, Xu et al. 2014). In addition, just as different MBW combinations can discriminate between different TTG1-dependent epidermal pathways in Arabidopsis, so too can the exact composition of pigment MBW complexes influence which type of flavonoid pigment is produced, which biosynthetic pathway genes are activated and the strength of target gene expression (Table 1; Fig. 1; Zhang et al. 2003, Baudry et al. 2004, Schwinn et al. 2006, Lea et al. 2007, Feyissa et al. 2009). Thus, different MYB and bHLH members of pigment MBW complexes within and across species have functionally diversified for more subtle regulation of the flavonoid pathway, with the MYBs contributing more of the fine-tuning of pigment production in terms of pigment type and patterns in the plant (see Xu et al. 2015 for a thorough review on the structure, function and regulation of the flavonoid MBW complex; see Davies et al. 2012 for a review on how diversification of MBW complex members accomplish fine control of the flavonoid pigment pathway leading to pattern formation).
It is also worth noting that several other transcription factor proteins can directly interact with the flavonoid pigment MBW complex either to repress or to promote pigment production. For example, hormone-regulated anthocyanin biosynthesis in Arabidopsis is repressed by jasmonate ZO-1 interaction motif (ZIM) domain proteins interacting with bHLH regulators of the pigment pathway (Qi et al. 2011). Similarly, Squamous Promoter Binding Protein-Like 9 (SPL9) represses pigment accumulation by directly binding to Arabidopsis PAP MYBs, thus disrupting the MBW complex (Gou et al. 2011). In contrast, the TCP3 bHLH protein enhances the transcriptional activity of the flavonoid pigment MBW complex in Arabidopsis by direct interaction with the R2R3 MYBs, thus promoting pigment biosynthesis (Li and Zachgo 2013). Similarly, TT1 (a WIP type zinc finger transcription factor) can interact with the TT2 MYB member of the PA MBW complex and enhance its transcriptional activity (Appelhagen et al. 2011). (For a more thorough review on the post-translational regulation of the flavonoid pigment MBW complex, see Xu et al. 2015.)
Elucidation of WRKY Factor Control Mechanisms Reveals a New Twist on the MBW Pigment Regulatory Model
For about the last 15 years, a wrinkle in the flavonoid pigment MBW regulatory paradigm existed in the form of the TTG2 locus of Arabidopsis. In 2002, Smyth and co-workers (Johnson et al. 2002) reported that TTG2 encoded a WRKY class transcription factor (containing the WRKY amino acid motif) that is necessary for trichome and seed coat development. As the ‘tt’ designation implies, among the phenotypes in the ttg2-1 mutant is a lack of PA flavonoid pigments in the seed coat. In addition, it was shown that the TTG2 gene is regulated by the MBW complex in the context of trichome and seed coat development (Johnson et al. 2002, Lepiniec et al. 2006, Ishida et al. 2007, Gonzalez et al. 2008, Zhao et al, 2008). Unlike other TT regulators such as TT8 and TT2, TTG2 is not required for the expression of Banyuls (BAN), the first committed step in the PA branch of the flavonoid pigment pathway (Debeaujon et al. 2003). However, like tt2 and tt8 mutants, the ttg2 mutant pigment phenotype is restricted to the seed coat; no lack of anthocyanins in the plant body has been observed in ttg2 mutants (Johnson et al. 2002, Lepiniec et al. 2006, Appelhagen et al. 2014, Gonzalez et al. 2016). These observations raised some interesting questions: if the MBW complex (specifically TT2/TT8/TTG1) is necessary and sufficient for the direct regulation of pigment biosynthetic genes (especially of BAN and other PA pathway-specific genes), how exactly does TTG2 regulate the pathway? Aside from being a direct transcriptional target, what exactly is the nature of the relationship, particularly at the protein level, between TTG2 and the MBW complex?
Another interesting question relates to how taxonomically widespread the WRKY strategy is for the control of the pigment pathway in plants. Among all of the flavonoid pigment model species studied, it appeared that the evolution of a WRKY protein specifically and narrowly controlling the PA branch of the pigment pathway may be unique to Arabidopsis, or unique to instances of PA pathway regulation and perhaps not anthocyanin regulation. Again, considering that Arabidopsis pigment mutant screens focused on tt phenotypes while screens in other plant models focused on anthocyanin phenotypes in floral organs, it is not surprising (perhaps even expected) that genes and regulatory mechanisms unique to PA biosynthesis would be uncovered.
However, a recent discovery revealed the conservation of the WRKY-based regulatory mechanism operating beyond the Arabidopsis PA pathway and in the anthocyanin pathway in Petunia (Verweij et al. 2016). This and other findings reported in three recent publications have uncovered some fascinating insights regarding the novel mechanisms by which WRKY factors together with the MBW complex regulate the anthocyanin and PA pigment pathways, and the trichome pathway in Arabidopsis (Pesch et al. 2014, Gonzalez et al. 2016, Verweij et al. 2016).
Some recent answers to long-standing questions: reconciliation between the WRKY and MBW control mechanisms suggests new regulatory models.
Pesch et al. (2014) provided a wealth of novel insights into regulatory mechanism of TTG2 and its relationship to the MBW complex in the context of trichome development. These authors showed that TTG2 primarily directly regulates the trichome patterning R3 MYB gene TRIPTYCHON (TRY). In addition, this regulation is dependent on the MBW complex, although TTG2 enhances the MBW activation of TRY. Consistent with these observations, TTG2 was shown to interact physically with TTG1. Moreover, yeast three-hybrid experiments showed that the TTG2 protein and the GL3 bHLH trichome regulator interact via TTG1 as a mediator. Yeast four-hybrid experiments indicate that the trichome R2R3 MYB regulator GLABRA 1 (GL1) does not interrupt the GL3–TTG1–TTG2 interaction. Based on these findings, Pesch et al. (2014) present a model in which, after activation of the TTG2 gene by the trichome MBW complex, the TTG2 protein joins the complex via interaction with TTG1 to enhance the direct regulation of TRY.
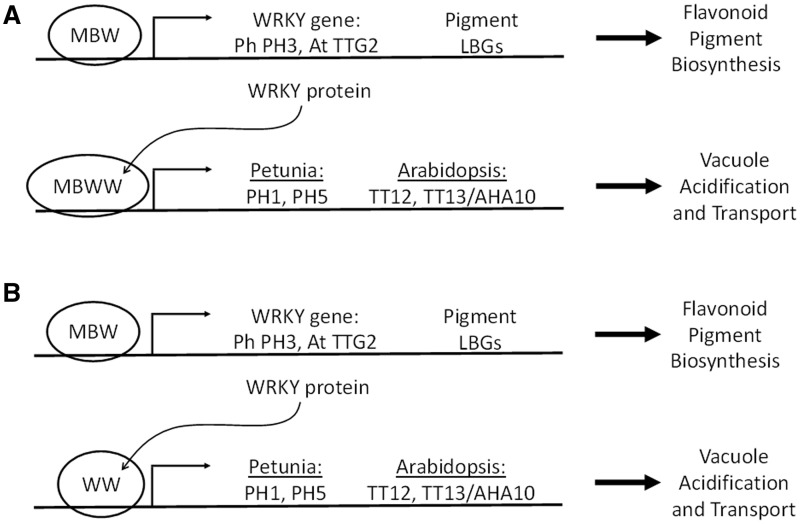
Verweij et al. (2016) recently reported the cloning of the PH3 locus in Petunia, which encodes a WRKY transcription factor homologous to TTG2. PH mutants of Petunia all show similar phenotypes consisting of bluish-hued flower petals and increased pH of petal homogenates (de Vlaming et al. 1983, van Houwelingen et al. 1998). Interestingly, ph6 was shown to be an allele of the AN1 bHLH gene that had retained the ability to activate anthocyanin biosynthesis but not the ability to acidify the vacuole (Spelt et al. 2002). Moreover, a distinct Petunia MYB regulator, PH4, can form a complex with AN1 and AN11 to activate a separate pathway that alters flower color (Table 1). This is accomplished by the PH4–AN1–AN11 complex targeting the expression of two genes, PH1 and PH5, both encoding tonoplast-localized P-ATPase transmembrane transporters. Besides vacuolar acidification, PH4 also regulates volatile floral emissions (Cna’ani et al. 2015). Thus, differential activation of distinct anthocyanin or vacuole acidification/volatile emission pathways is achieved by altering the MYB regulator (AN2 or PH4) or by altering the function of the AN1 bHLH factor (encoded by the AN1 or ph6 alleles). The PH3 WRKY factor was also shown to regulate PH1 and PH5. In addition, the PH4–AN1–AN11 MBW complex regulates PH3. Lastly, Verweij et al. (2016) showed that the PH3 WRKY factor physically interacts with the AN11 WDR in yeast two-hybrid analysis. Based on these findings, Verweij et al. (2016) present a model in which the Petunia MBW complex activates the expression of PH3. Then, the PH3 WRKY factor joins the MBW complex to target the expression of PH1 and PH5, ultimately resulting in the acidification of the vacuole as a means of flower color modification (Fig. 2A). The authors also speculate that a similar mechanism may be operating in Arabidopsis to acidify the vacuole of inner seed coat cells that produce PA pigments: the TT2–TT8–TTG1 MBW complex directly targets the expression of TTG2 and then a TT2–TT8–TTG1–TTG2 complex activates the expression of a tonoplast-localized P-ATPase encoded by TT13/AHA10. However, recent developments reported in Gonzalez et al. (2016) provide a more comprehensive mechanism of TTG2 control of the PA pathway through the specific regulation of genes involved in vacuolar transport of PA precursors (Fig. 1).
Fig. 2.
Possible MBW and WRKY transcriptional models for the regulation of the flavonoid pigment pathway. In (A), the MBW complex controls the expression of the LBGs and the WRKY gene (PH3 in Petunia, TTG2 in Arabidopsis). The WRKY protein then possibly enters the MBW complex, via physical interaction with the WDR protein (AN11 in Petunia and TTG1 in Arabidopsis), and narrows the range of LBG targets to the vacuole acidification and transport genes: PH1 and PH5 in Petunia, TT12 and TT13/AHA10 in Arabidopsis. In (B), the MBW complex regulates the LBGs and the WRKY gene but then the WRKY and WDR proteins physically interact possibly to form a separate transcriptional complex, without the MYB and bHLH proteins, that narrowly targets the vacuole acidification and transport genes. Ph: Petunia hybrida; At: Arabidopsis thaliana; LBGs: late biosynthetic genes of the flavonoid pigment pathway.
In Arabidopsis, a quantitative PCR approach comparing the expression of a broad set of pigment biosynthetic genes between wild-type and ttg2 developing seeds revealed only two obvious targets of regulation: TT12 and TT13/AHA10 (Gonzalez et al. 2016; Fig. 1). This is in contrast to the much broader regulation by the MBW complex of the late biosynthetic gene subset (which includes TT12 and TT13/AHA10). TT12 encodes a MATE (multidrug and toxin extrusion)-type vacuolar transporter necessary for the proton gradient-dependent pumping of epicatechin-3′-O-glucoside (E3′OG), a glycosylated PA pigment precursor (Marinova et al. 2007). TT13/AHA10 encodes a tonoplast-localized P3A-ATPase (Baxter et al. 2005, Appelhagen et al. 2015) that presumably establishes the H+ gradient necessary to power TT12. In addition, TTG2 is capable of binding a 1 kb fragment of DNA containing several putative WRKY boxes immediately upstream of the TT12 gene, suggesting direct regulation. A similar mechanism has been described in Medicago involving an epicatechin-specific glucosyltransferase (yet to be identified in Arabidopsis) and a MATE-type pump for the transport of E3′OG PA precursors into the vacuole of seed coat cells (Zhao and Dixon 2009, Pang et al. 2013). Similar to findings in Petunia and in the trichome pathway in Arabidopsis, yeast two-hybrid and bimolecular fluorescence complementation (BiFC) experiments demonstrated an interaction between TTG2 and TTG1, but not between TTG2 and other MBW complex members such as TT2 and TT8 (Gonzalez et al. 2016). Moreover, overexpressing TT12 in ttg2 mutants restored the seed coat color phenotype to almost wild-type levels, consistent with the findings that TTG2 has a narrow range of targets consisting of one or two genes involved specifically in vacuolar transport. These findings are consistent with at least two regulatory models presented in Gonzalez et al. (2016) and in Fig. 2. In one model, the TT2–TT8–TTG1 MBW complex directly activates TTG2. Similar to the models presented in Pesch et al. (2014) and Verweij et al. (2016), TTG2 then interacts with TTG1 to form a TT2–TT8–TTG1–TTG2 quartet (Fig. 2A). The consequence of this ‘MBWW quartet’ model is to narrow greatly the target range of the MBW complex from all the late flavonoid pigment biosynthetic genes to just two vacuolar transport genes (TT12 and TT13/AHA10) near the end of PA pathway (Fig. 1). Alternatively, the observations made thus far do not rule out the possibility of a second model, the ‘WW duo’ model, in which a transcriptional complex centered on the TTG1 WDR protein and the TTG2 WRKY factor, but omitting the TT2 MYB and TT8 bHLH factors, regulates only the vacuolar transport steps in the PA pathway (Fig. 2B). In this model, the TTG1–TTG2 complex would reinforce the MBW complex in the regulation of TT12. It is also possible that these two TTG2 regulatory models are not mutually exclusive, with multiple co-existing transcriptional complexes in plants providing various means for fine-tuned control of the flavonoid pigment pathway.
Evidence for multiple TTG2 regulatory models
Indeed, extensive genetic data available from studies in the PA pathway may reveal additional, subtle insights into the WRKY and MBW regulatory mechanisms. It is noteworthy that there seems to be a qualitative difference between the degree of regulation of target genes by TTG2 and TTG1 compared with TT2 and TT8. For example, in ttg2 developing seed, the expression of TT12 is completely repressed. Similarly, the expression of TT12 is undetectable in ttg1 seed coats (Xu et al. 2014, Gonzalez et al. 2016). Although greatly reduced in expression, tt2 and tt8 seed coats still show detectable residual expression of TT12, in contrast to ttg1 and ttg2 (Xu et al. 2014, Gonzalez et al. 2016). As previously mentioned, TTG2 physically interacts with TTG1 but not with MYB or bHLH MBW complex members, including TT2 and TT8 (Pesch et al. 2014, Gonzalez et al. 2016). Additionally, TTG2’s control of TT12 is dependent upon TTG1.
So, what is providing the residual TT12 gene expression in tt2 and tt8 mutant seed coats that is otherwise completely absent in ttg1 and ttg2 seed coats (Xu et al. 2014)? Could it be TTG1 in a complex with TTG2 protein, produced from residual TTG2 gene expression, in tt2 and tt8 mutants? If so, this would suggest that something akin to the WW duo model (Fig. 2B) is operating in tt2 and tt8 mutants to yield detectable TT12 expression. Ectopically expressing TTG2 in ttg1 mutants did not restore TT12 expression at all (Gonzalez et al. 2016), although this result would be predicted given either of the two proposed TTG2 regulatory models. However, it would be interesting to express TTG2 ectopically in tt2 and tt8 mutant seed coats. If TT12 expression is restored in these mutants, this would indicate that TTG2 could function independently of the MYB and bHLH factors, providing evidence that the TTG1–TTG2 model is feasible.
Co-option of Phenylpropanoid-Regulating MBW Complex Members to Regulate Unrelated Pathways
The MBW complex regulates epidermal developmental processes in the Arabidopsis lineage
While the MBW complex has been most extensively studied in the context of the anthocyanin and PA pathways, the complete complex, or complex members, have been co-opted to regulate unrelated pathways in certain plant taxa. As discussed above, in Arabidopsis, in addition to the anthocyanin and PA pathways, the complex is well known to regulate various epidermal cell fate pathways including trichome initiation, root hair/non-root hair cell fate and outer seed coat differentiation. The taxonomic extent of this co-opted regulation extends to at least the closely related Malvaceae family where it regulates the economically important cotton fiber initiation process, another seed coat differentiation process (Wan et al. 2014). In Arabidopsis, this MBW regulation includes both activation of trichome cell fate by the complex and near-neighbor repression of the trichome fate by truncated MYBs descended from full-length activator MYBs (see Feller et al. 2011 for a review).
Co-option of an anthocyanin-regulating MYB to regulate the betalain pigment pathway
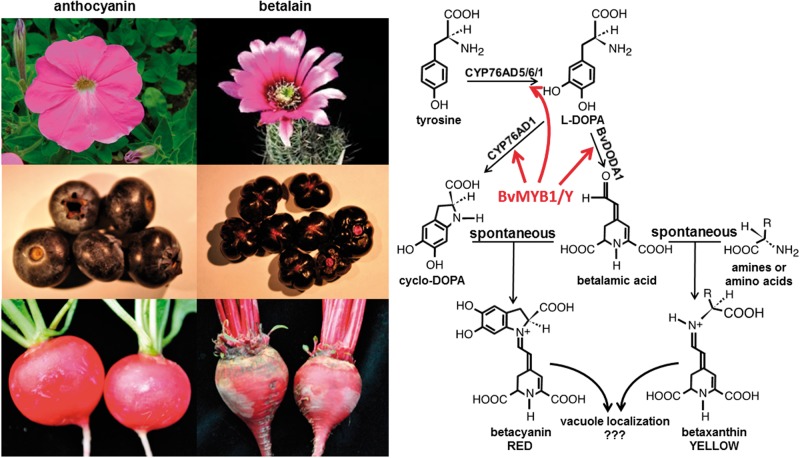
Betalains are red and yellow pigments that are produced in some families of the Caryophyllales order. Where they occur, they are mutually exclusive with the anthocyanins and they appear to provide all of the functions of anthocyanins, pigmenting flowers and fruits and responding to the same environmental stimuli as the anthocyanin pathway (Fig. 3). Betalains and anthocyanins are chemically unrelated, with the betalains being derived from tyrosine while anthocyanins are derived from phenylalanine. Their biological functions as pigments are so similar that betalains were known as ‘nitrogenous anthocyanins’ before the structure was determined by Mabry (Mabry et al. 1962, Mabry 1964). Mabry coined the term betalain naming these pigments after the beet genus, Beta.
Fig. 3.
Betalain pathway with comparision of analogous structures pigmented with anthocyanin or betalain. The six left panels are representative flowers, fruits and roots, with the far left panels pigmented with anthocyanins and the right with betalains. Upper left, Petunia flower; upper right, cactus flower; middle left, blueberries; middle right, malabar spinach berries; lower left, radish roots; lower right, beet roots. The betalain pathway is shown on the right, with the enzymatic steps regulated by BvMYB1 indicated with arrows. The condensation of betalamic acid with cyclo-DOPA to produce red betalains, or amine compounds to produce yellow betalains is spontaneous. Betalains are trafficked to the vacuole by unknown means. Note that betacyanins are usually modified, often by glycosylation on the cyclo-DOPA moiety (not shown).
In 1936, Keller described two loci in beet that affect betalain pigmentation, the R and Yellow (Y) genes (Keller 1936). The R locus has been shown to encode a Cyt P450, CYP76AD1 (Fig. 3; Hatlestad et al. 2012). Mutations at the R locus lead to the loss of red pigmentation and the accumulation of yellow. Keller’s Y locus has been shown to encode a MYB transcriptional regulator, BvMYB1 (Hatlestad et al. 2015). Phylogenetic analysis of BvMYB1 clearly indicates that it is within the anthocyanin MBW complex MYB clade (S6), distinct from the MBW-independent MYB regulators (such as AtMYB11, AtMYB12 and AtMYB111) that regulate early steps of the flavonoid pathway (Liu et al. 2015). However, it has lost the ability to interact with bHLH members of the MBW complex. Furthermore, BvMYB1 cannot up-regulate the anthocyanin pathway in Arabidopsis, and the Arabidopsis PAP MYBs cannot regulate betalains in beet. BvMYB1 has been shown to regulate directly genes in the betalain pigment pathway, CYP76AD1/R and BvDODA1 (Hatlestad et al. 2015; Fig. 3). BvDODA1 encodes the DOPA 4,5-dioxygenase enzyme (DODA) required for all betalain biosynthesis. More recently, CYP76AD5 and CYP76AD6 have been shown to hydroxylate tyrosine to l-DOPA (Fig. 3; Polturak et al. 2015, Sunnadeniya et al. 2016) and that these genes are also up-regulated by BvMYB1, but it is not known if this regulation is direct. It was shown that a regulatory mutation at the Y locus isolated during beet domestication causes up-regulation of BvMYB1 and this mutation is responsible for the intense pigment accumulation in the interior of the beet root. It will be interesting to determine if there are also changes in DNA element motifs bound by the beet MYB. In addition, there are two other closely related beet MYBs that can up-regulate the betalain pathway when overexpressed. Neither of these MYBs is able to interact with bHLH proteins (Lloyd lab, unpublished observations). As discussed elsewhere, the duplicated Arabidopsis PAP MYBs differentially regulate the anthocyanin pathway in varied environmental and developmental contexts (Lea et al. 2007, Gonzalez et al. 2008, Shan et al. 2009, Luo et al. 2012, Shin et al. 2013). It may be that the duplicated beet MYBs have similar differential activities.
Mutations leading to the loss of bHLH interaction by the beet myb may have been important for the co-option from anthocyanin regulation, helping to confer betalain specificity. Using site-directed mutagenesis, BvMYB1 was modified so that it interacted with anthocyanin bHLH regulatory proteins. These residue changes all map to a fairly well-characterized region that was previously shown to mediate MYB–bHLH interaction although the analysis expands this region by a single amino acid required for interaction. It was noted that this modified beet MYB is still unable to regulate the anthocyanin pathway when expressed in Arabidopsis. It is likely that the beet MYB no longer binds to the same DNA element, but this has not been verified (Hatlestad et al. 2015).
MBW co-option may partially explain mutual exclusivity of betalains and anthocyanins
Shimada et al. (2005) show that both spinach and pokeweed, two betalain-producing species, encode anthocyanidin synthase (ANS) proteins that are fully functional in producing anthocyanidin, which is required for anthocyanin production. However, they show that the expression of these ANS genes, as well as the requisite dihydroflavonol 4-reductase (DFR) genes, is mostly restricted to the seed, where they may be involved in the production of PA (tannin) pigments. Although it has not been demonstrated, this expression restriction may be the result of co-option of the required anthocyanin MBW MYB to regulate betalain genes and loss of the ability to activate anthocyanin biosynthetic genes in the plant body, flowers or fruit, eliminating the full anthocyanin pathway from these species. It will be interesting to analyze whether there are MBW complex members in betalain-producing species with expression limited to the developing seed coat.
In addition to loss of anthocyanin pathway gene expression, it may be that the latest steps in the anthocyanin pathway, steps such as glycosylation and transport into the vacuole, are simply missing from the betalain species. This is not yet known.
Concluding Remarks
Decades of fruitful research into the regulation of flavonoid plant pigments has taught us a tremendous amount about widely conserved mechanisms of transcriptional control in plants. Much of this work has culminated in the MBW transcriptional complex paradigm regulating not only the flavonoid pigment pathway but also a suite of plant epidermal developmental pathways. Interestingly, recent studies in beets revealed that this regulatory mechanism was partially co-opted to regulate pigment biosynthetic genes in the betalain pathway. In addition, elucidating the TTG2 and PH3 WRKY protein regulatory mechanisms has recently contributed significant, novel modifications to our understanding of the MBW regulatory complex. However, some interesting questions remain regarding the exact molecular make-up, function and conservation of WRKY-containing transcriptional complexes. Do WRKY proteins simply join the MBW complex to modify its transcriptional activity? Alternatively, are there yet to be discovered new complexes centered on TTG2? Besides flower color modification through vacuolar acidification, do WRKY factors such as PH3 in Petunia also regulate the genes encoding transporters of pigment precursors into the tonoplast (similar to TTG2 in the Arabidopsis PA pathway)? Like flavonoid pigments, betalain pigments also accumulate in the vacuole. However, to date, there is no information about how betalains are transported to or enter this organelle. Have betalain-producing species co-opted other aspects of flavonoid pigment regulation such as the WRKY strategy for the regulation of pigment transport or acidification of the vacuole? It seems that ongoing studies of pigment pathways will still teach us much about regulatory mechanisms in plants for years to come.
Funding
This work was supported by a US National Science Foundation grant [IOS-1556348 to A.L.]; Howard Hughes Medical Institute grants [52006985, 52008124]; and the Texas Institute for Discovery in Science (TIDES).
Disclosures
The authors have no conflicts of interest to declare.
Glossary
Abreviations
- AN
Anthocyanin
- ANS
anthocyanidin synthase
- B
Booster
- BAN
Banyuls
- bHLH
basic helix–loop–helix protein
- BiFC
bimolecular fluorescence complementation
- C1
Colorless1
- CYP
Cyt P450
- DFR
dihydroflavonol 4-reductase
- DODA
DOPA 4,5-dioxygenase
- EGL3
ENHANCER of GLABRA3
- GL
GLABRA
- MATE
multidrug and toxin extrusion
- MBW
MYB–bHLH–WD repeat protein
- MYB
myeloblastosis protein
- PAC1
Pale aleurone color1
- PA
proanthocyanidin
- PAP
production of anthocyanin pigment
- PH
increased pH of petunia petal homogenates
- Pl
Purple Plant
- R
in maize or beet = Red
- SPL9
Squamous promoter binding-protein-like 9
- TCP
protein family named after teosinte branched1, cycloidea, proliferating cell nuclear antigen factor proteins
- TT
transparent testa
- TTG
transparent testa glabra
- WD
protein domain containing a tryptophan–aspartate amino acid motif
- WDR
WD repeat
- WIP
class of zinc finger domain containing a tryptophan–isoleucine–proline amino acid motif
- WRKY
protein domain containing a tryptophan–arginine-lysine–tyrosine amino acid motif
- Y
Yellow
- ZIM
ZO-1 interaction motif
References
- Appelhagen I., Lu G.H., Huep G., Schmelzer E., Weisshaar B., Sagasser M. (2011) TRANSPARENT TESTA1 interacts with R2R3-MYB factors and affects early and late steps of flavonoid biosynthesis in the endothelium of Arabidopsis thaliana seeds. Plant J. 67: 406–419. [DOI] [PubMed] [Google Scholar]
- Appelhagen I., Nordholt N., Seidel T., Spelt K., Koes R., Quattrochio F.. et al. (2014) Update on transparent testa mutants from Arabidopsis thaliana: characterisation of new alleles from an isogenic collection. Planta 240: 955–970. [DOI] [PubMed] [Google Scholar]
- Appelhagen I., Thiedig K., Nordholt N., Schmidt N., Huep G., Sagasser M.. et al. (2015) TRANSPARENT TESTA 13 is a tonoplast P3A-ATPase required for vacuolar deposition of proanthocyanidins in Arabidopsis thaliana seeds. Plant J. 82: 840–849. [DOI] [PubMed] [Google Scholar]
- Baudry A., Heim M.A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39: 366–380. [DOI] [PubMed] [Google Scholar]
- Baxter I.R., Young J.C., Armstrong G., Foster N., Bogenschutz N., Cordova T.. et al. (2005) A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102: 2649–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt C., Lee M.M., Gonzalez A., Zhang F., Lloyd A., Schiefelbein J. (2003) The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130: 6431–6439. [DOI] [PubMed] [Google Scholar]
- Bernhardt C., Zhao M., Gonzalez A., Lloyd A., Schiefelbein J. (2005) The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132: 291–298. [DOI] [PubMed] [Google Scholar]
- Borevitz J.O., Xia Y., Blount J., Dixon R.A., Lamb C. (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Can’ani A., Spitzer-Rimon B., Ravid J., Farhi M., Masci T., Aravena-Calvo J.. et al. (2015) Two showy traits, scent emission and pigmentation, are finely coregulated by the MYB transcription factor PH4 in petunia flowers. New Phytol. 208: 708–714. [DOI] [PubMed] [Google Scholar]
- Carey C.C., Strahle J.T., Selinger D.A., Chandler V. (2004) Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotype relative to the functionally similar Transparent Testa Glabra1 gene in Arabidopsis thaliana. Plant Cell 16: 450–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler V.L., Radicella J.P., Robbins T.P., Chen J., Turks D. (1989) Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R genomic sequences. Plant Cell 1: 1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone K.C., Burr F.A., Benjamin B. (1986) Molecular analysis of the maize anthocyanin regulatory locus C1. Proc. Natl. Acad. Sci. USA 83: 9631–9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone K.C., Cocciolone S.M., Burr F.A., Burr B. (1993) Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell 5: 1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies K.M., Albert N.W., Schwinn K.E. (2012) From landing lights to mimicry: the molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 39: 619–638. [DOI] [PubMed] [Google Scholar]
- de Vetten N., Quattrocchio F., Mol J., Koes R. (1997) The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 11: 1422–1434. [DOI] [PubMed] [Google Scholar]
- de Vlaming P., Schram A.W., Wiering H. (1983) Genes affecting flower colour and pH of flower limb homogenates in Petunia hybrida. Theor. Appl. Genet. 66: 271–278. [DOI] [PubMed] [Google Scholar]
- Debeaujon I., Nesi N., Perez P., Grandjean O., Caboche M., Lepiniec L. (2003) Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 11: 2514–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deboo G.B., Albertsen M.C., Taylor L.P. (1995) Flavonone 3-hydroxylase transcripts and flavonol accumulation are temporally coordinated in maize anthers. Plant J. 7: 703–713. [DOI] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci. 15: 573–581. [DOI] [PubMed] [Google Scholar]
- Feller A., Machemer K., Braun E.L., Grotewold E. (2011) Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66: 94–116. [DOI] [PubMed] [Google Scholar]
- Feyissa D.N., LØvdal T., Olsen K.M., Slimestad R., Lillo C. (2009) The endogenous GL3, but not EGL3, gene is necessary for anthocyanin accumulation as induced by nitrogen depletion in Arabidopsis rosette stage leaves. Planta 230: 747–754. [DOI] [PubMed] [Google Scholar]
- Fong H.K.W., Hurley J.B., Hopkins R.S., Miake-Lye R., Johnson M.S., Doolittle R.F.. et al. (1986) Repetitive segmental structure of the transducin β subunit: homology with the CDC4 gene and identification of related mRNAs. Proc. Natl. Acad. Sci. USA 83: 2162–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.A., Cone K.C., Chandler V.L. (1992) Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 6: 864–875. [DOI] [PubMed] [Google Scholar]
- Goff S.A., Klein T.M., Roth B.A., Fromm M.E., Cone K.C., Radicella J.P.. et al. (1990) Transactivation of anthocyanin biosynthetic genes following transfer of B-regulatory genes into maize tissues. EMBO J. 9: 2517–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A., Brown M., Hatlestad G., Akhavan N., Smith T., Hembd A.. et al. (2016) TTG2 controls the developmental regulation of seed coat tannins in Arabidopsis by regulating vacuolar transport steps in the proanthocyanidin pathway. Dev. Biol. 419: 54–63. [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Mendenhall J., Huo Y., Lloyd A.M. (2009) TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev. Biol. 325: 412–421. [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Zhao M., Leavitt J.M., Lloyd A.M. (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53: 814–827. [DOI] [PubMed] [Google Scholar]
- Goodrich J., Carpenter R., Coen E. (1992) A common gene regulates pigmentation pattern in diverse plant species. Cell 68: 955–964. [DOI] [PubMed] [Google Scholar]
- Gou J.Y., Felippes F.F., Liu C.J., Weigel D., Wang J.W. (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23, 1512–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatlestad G., Sunnadeniya R., Akhavan N., Gonzalez A., Goldman I., McGrath J.. et al. (2012) The beet R locus encodes a new cytochrome P450 required for red betalain production. Nat. Genet. 44: 130–816. [DOI] [PubMed] [Google Scholar]
- Hatlestad G., Akhavan N., Sunnadeniya R., Elam L., Cargile S., Hembd A.. et al. (2015) The beet Y locus encodes an anthocyanin-MYB-like protein that activates the betalain red pigment pathway. Nat. Genet. 47: 92–96. [DOI] [PubMed] [Google Scholar]
- Ishida T., Hattori S., Sano R., Inoue K., Shirano Y., Hayashi H.. et al. (2007) Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 19: 2531–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C.S., Kolevski B., Smyth D.R. (2002) TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14: 1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W. (1936) Inheritance of some major color types in beets. J. Agric. Res. 52: 27–38. [Google Scholar]
- Koornneef M. (1981) The complex syndrome of ttg mutants. Arabidopsis Inf. Serv. 18: 45–51. [Google Scholar]
- Koornneef M. (1990) Mutation affecting the testa color in Arabidopsis. Arabidopsis Inf. Serv. 28: 1–4. [Google Scholar]
- Lea U.S., Slimestad R., Smedvig P., Lillo C. (2007) Nitrogen deficiency enhances expression of specific MYB and bHLH transcription factors and accumulation of end products in the flavonoid pathway. Planta 225: 1245–1253. [DOI] [PubMed] [Google Scholar]
- Lepiniec L., Debeaujon I., Routaboul J., Baudry A., Pourcel L., Nesi N.. et al. (2006) Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57: 405–30. [DOI] [PubMed] [Google Scholar]
- Li S., Zachgo S. (2013) TCP3 interacts with R2R3–MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 76, 901–913. [DOI] [PubMed] [Google Scholar]
- Lipsick J.S. (1996) One billion years of Myb. Oncogene 13: 223–235. [PubMed] [Google Scholar]
- Liu J., Osbourn A., Ma P. (2015) MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8, 689–708. [DOI] [PubMed] [Google Scholar]
- Lloyd A.M., Walbot V., Davis R.W. (1992) Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258: 1773–1775. [DOI] [PubMed] [Google Scholar]
- Ludwig S.R., Habera L.F., Dellaporta S.L., Wessler S.R. (1989) Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc homology region. Proc. Natl. Acad. Sci. USA 86: 7092–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q.J., Mittal A., Jia F., Rock C.D. (2012) An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol. Biol. 80: 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabry T.J., Wyler H., Sassu G., Mercier M., Parikh I., Dreiding A.S. (1962) Die struktur des neobetanidins. Helv. Chim. Acta 45: 640–647. [Google Scholar]
- Mabry T.J. (1964) The betacyanins, a new class of red-violet pigments, and their phylogenetic significance. InTaxonomic Biochemistry and Serology. Edited by Leone C.A. pp. 239–254. Ronald Press, New York. [Google Scholar]
- Marinova K., Pourcel L., Weder B., Schwarz M., Barron D., Routaboul J.M.. et al. (2007). The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+ antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 19: 2023–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Prescott A., Mackay S., Bartlett J., Vrijlandt E. (1991) Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant J. 1: 37–49. [DOI] [PubMed] [Google Scholar]
- Massari M.E., Murre C. (2000). Helix–loop–helix proteins: regulators of transcription in eukaryotic organisms. Mol. Cell. Biol. 20: 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.C., Chezem W.R., Clay N.K. (2016) Ternary WD40 repeat-containing protein complexes: evolution, composition and roles in plant immunity. Front. Plant Sci. 6: 1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y., Saitoh M., Hoshino A., Nitasaka E., Iida S. (2006) Isolation of cDNAs for R2R3–MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese Morning Glory. Plant Cell Physiol. 47: 457–470. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P.S., Baltimore D. (1989) A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56: 777–783. [DOI] [PubMed] [Google Scholar]
- Nesi N., Debeaujon I., Jond C., Pelletier G., Caboche M., Lepiniec L. (2000) The TT8 gene encodes a basic helix–loop–helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12: 1863–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N., Jond C., Debeaujon I., Caboche M., Lepiniec L. (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13: 2099–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer D.G., Herman P.L., Sivakumaran S., Esch J., Marks M.D. (1991) A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67: 483–493. [DOI] [PubMed] [Google Scholar]
- Pang Y., Cheng X., Huhman D.V., Ma J., Peel G.J., Yonekura-Sakakibara K.. et al. (2013) Medicago glucosyltransferase UGT72L1: potential roles in proanthocyanidin biosynthesis. Planta 238: 139–154. [DOI] [PubMed] [Google Scholar]
- Payne C.T., Zhang F., Lloyd A.M. (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares J., Ghosal D., Wienand U., Peterson P.A., Saedler H. (1987) The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcription factors. EMBO J. 6: 3553–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch M., Dartan B., Birkenbihl R., Somssich I.E., Hüslkamp M. (2014) Arabidopsis TTG2 regulates TRY expression through enhancement of activator complex-triggered activation. Plant Cell 26: 4076–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier M.K., Winkel-Shirley B. (1996) Analysis of flavanone 3-hydroxylase in Arabidopsis seedlings. Plant Physiol. 111: 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier M.K., Murrell J.R., Winkel-Shirley B. (1997) Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis. Plant Physiol. 113: 1437–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polturak G., Breitel D., Grossman N., Sarrion-Perdigones A., Weithorn E., Pliner M.. et al. (2015) Elucidation of the first committed step in betalain biosynthesis enables the heterologous engineering of betalain pigments in plants. New Phytol. 210: 269–283. [DOI] [PubMed] [Google Scholar]
- Qi T., Song S., Ren Q., Wu, Huang H., Chen Y.. et al. (2011) The jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F., Wing J.F., Leppen H.T.C., Mol J.N.M., Koes R.E. (1993) Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 5: 1497–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F., Wing J., van der Woude K., Souer E., de Vetten N., Mol J.. et al. (1999) Molecular analysis of the Anthocyanin2 gene of Petunia and its role in the evolution of flower color. Plant Cell 11: 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo I., Fuertes A., Benito M.J., Malpica J.M., Leyva A., Paz-Ares J. (1998) More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 14: 273–284. [DOI] [PubMed] [Google Scholar]
- Schellmann S., Schnittger A., Kirik V., Wada T., Okada K., Beermann A.. et al. (2002) TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21: 5036–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwinn K., Venail J., Shang Y., Mackay S., Alm V., Butelli E.. et al. (2006) A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18: 831–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X., Zhang Y., Peng W., Wang Z., Xie D. (2009) Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J. Exp. Bot. 60: 3849–3860 [DOI] [PubMed] [Google Scholar]
- Shimada S., Otsuki H., Sakuta M. (2006) Transcriptional control of anthocyanin biosynthetic genes in the Caryophyllales. J. Exp. Bot. 58: 957–967. [DOI] [PubMed] [Google Scholar]
- Shin D.H., Choi M., Kim K., Bang G., Cho M., Choi S.. et al. (2013) HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 587: 1543–1547. [DOI] [PubMed] [Google Scholar]
- Smith T.F., Gaitatzes C., Saxena K., Neer E.J. (1999) The WD repeat: a common architecture for diverse function. TIBS 24: 181–185. [DOI] [PubMed] [Google Scholar]
- Spelt C., Quattrocchio F., Mol J.N., Koes R. (2000) Anthocyanin1 of Petunia encodes a basic helix–loop–helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12: 1619–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelt C., Quattrocchio F., Mol J.N., Koes R. (2002) Anthocyanin1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. Plant Cell 14: 2121–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunnadeniya R., Bean A., Brown M., Akhavan N., Hatlestad G., Gonzalez A.. et al. (2016) Tyrosine hydroxylation in betalain pigment biosynthesis is performed by cytochrome P450 enzymes in beets (Beta vulgaris). PLoS One 11: e0149417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L.P., Briggs W.R. (1990) Genetic regulation and photocontrol of anthocyanin accumulation in maize seedlings. Plant Cell 2: 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houwelingen A., Souer E., Spelt K., Kloos D., Mol J., Koes R. (1998) Analysis of flower pigmentation mutants generated by random transposon mutagenesis in Petunia hybrida. Plant J. 13: 39–50. [DOI] [PubMed] [Google Scholar]
- van Nocker S., Ludwig P. (2003) The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics 4: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij W., Spelt C.E., Bliek M., de Vries M., Wit N., Faraco M.. et al. (2016) Functionally similar WRKY proteins regulate vacuolar acidification in Petunia and hair development in Arabidopsis. Plant Cell 28: 786–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A.R., Davison P.A., Bolognesi-Winfield A.C., James C.M., Srinivasan N., Blundell T.L.. et al. (1999) The Transparent Testa Glabra1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q., Zhang H., Ye W., Wu H., Zhang T. (2014) Genome-wide transcriptome profiling revealed cotton fuzz fiber development having a similar molecular model as Arabidopsis trichome. PLoS One 9: e97313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B., Kubasek W.L., Storz G., Bruggemann E., Koornneef M., Ausubel F.M.. et al. (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8: 659–671. [DOI] [PubMed] [Google Scholar]
- Xu W., Dubos C., Lepiniec L. (2015) Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 20: 176–185. [DOI] [PubMed] [Google Scholar]
- Xu W., Grain D., Bobet S., Le Gourrierec J., Thévenin J., Kelemen Z.. et al. (2014) Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB–bHLH–WDR complexes and their targets in Arabidopsis seed. New Phytol. 202: 132–144. [DOI] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C.T., Lloyd A. (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869. [DOI] [PubMed] [Google Scholar]
- Zhao M., Morohashi K., Hatlestad G., Grotewold E., Lloyd A. (2008) The TTG1–bHLH–MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135: 1991–1999. [DOI] [PubMed] [Google Scholar]
- Zhao J., Dixon R.A. (2009) MATE transporters facilitate vacuolar uptake of epicatechin 3′-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 21: 2323–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]