Abstract
Many of the brain regions, neurotransmitter systems, and behavioral changes that occur after occasional drug use in healthy subjects and after chronic drug abuse in addicted patients are well characterized. An emerging literature suggests that epigenetic processes, those processes that regulate the accessibility of DNA to regulatory proteins within the nucleus, are keys to how addiction develops and how it may be treated. Investigations of the regulation of chromatin, the organizational system of DNA, by histone modification is leading to a new understanding of the cellular and behavioral alterations that occur after drug use. We will describe how, when, and where histone tails are modified and how some of the most recognized histone regulation patterns are involved in the cycle of addiction, including initial and chronic drug intake, withdrawal, abstinence, and relapse. Finally, we consider how an approach that targets histone modifications may promote successful treatment.
1. Introduction
Adapting to a constantly changing environment requires the ability to acquire new behaviors and change old ones in response to environmental contingencies. For this reason, much of our behavior is controlled by its consequences, with response probabilities in given situations changing as a function of the history of reinforcement in those situations. These experiences are learned and remembered through several well-defined neural circuits that are involved in acquisition, consolidation, and expression of memories. In addiction, however, the reinforcing properties of the abused substance can be so powerful that the plasticity mechanisms involved in normal learning and memory are usurped, resulting in repetitive and persistent behaviors that are resistant to change, even in the face of strong negative consequences. One key to understanding addiction is to understand basic mechanisms of learning and memory and how those mechanisms may be altered by abused substances (e.g., Hyman et al., 2006). A great deal is now known about the ways in which memories are formed, from binding neurotransmitters at the receptor level, to the activation of transcriptional machinery needed for the synthesis of new proteins that solidify long-term memories.
A growing literature indicates that long-term memories form at a molecular level as a consequence of changes in gene expression induced by activity-dependent histone modifications (e.g., Levenson et al., 2004; Vecsey et al., 2007). These same mechanisms are involved in acquiring and stabilizing the long-term reinforcing effects of various drugs of abuse. In this chapter, we examine the relation between histone modifications and long-term memory in behavioral approaches that model different aspects of addiction. We consider how these modifications may interact with the behavioral experience (acquisition and retrieval of memories for abused substances, as well as extinction of drug-seeking behavior) and review some different theoretical perspectives that have been offered for these effects. We begin this chapter with a brief overview of epigenetics and the general memory processes that may occur in the development and maintenance of addiction. We then describe different histone-mediated mechanisms that may control long-term gene expression and plasticity and review current literature demonstrating how these mechanisms may be involved in different addiction processes. Finally, we end this chapter with a description of currently unresolved issues in the field of histone-mediated epigenetics in addiction and we suggest several future directions that may help to resolve some of these debates.
2. General overview of histone-mediated epigenetics
The term epigenetics refers to the regulation that occurs ‘epi’, or “over” the genomic DNA. Although controversy about the term epigenetics remains (to be discussed later in the chapter), most research supports the idea that chemical modifications applied to the DNA or near DNA (i.e., to histone proteins) help regulate transcription and are modifiable throughout life. Within the nucleus of a cell, DNA is wrapped around multiple nucleosomes. Nucleosomes are composed of and linked together by a collection of histone proteins. Histones are the small and positively charged building blocks that help package and organize DNA into a repeat bead-like structure. To allow for selective and modifiable outcomes through epigenetic processes, each histone is classified into one of two super families (i.e., histone core or histone linker), five families (H2A, H2b, H3, and H4, H1/H5), and multiple subfamilies, each having slightly different functions and cellular distribution patterns (Cheung et al., 2000; Strahl & Allis, 2000). Two copies of each histone core (H2A, H2B, H3, and H4) are bound together by linker histones (H1 and H5) to create one nucleosome (an octomer of core histones) for DNA to be carefully wrapped around, making the basic chromatin structure (i.e., nucleosome + DNA). In this chapter, we will consider how these histones may be modified during different stages of drug taking – acute exposure, chronic drug taking, withdrawal, abstinence, and relapse. Before going into histone alterations in greater detail, we will consider addiction at the level of behavior and how different learning and memory processes contribute to long-term drug intake.
3. Learning processes involved in the development of addiction
One of the reasons that addiction is thought to involve learning and memory circuits is that drug seeking often occurs in the presence of specific cues. These cues can be contextual (e.g., drinking a beer in a favorite bar), social (e.g., using cocaine with a specific group of friends), and temporal (e.g., having a cigarette first thing in the morning), among others. Over time, drug-seeking occurs in different situations, broadening the cues that may be associated with drug intake. After even a relatively few number of experiences, those cues will evoke powerful drug cravings when they are encountered. These cravings create a negative internal state that further motivates drug intake. From a learning and memory perspective, the challenge is understanding how these cues become associated with drugs and what can be done to sever or, at the very least, suppress those powerful associations.
Basic research on learning and memory processes in substance abuse has focused on mechanisms that underlie two very general learning processes: initial acquisition, in which the memory is initially formed and consolidated, and long-term maintenance, in which the memory is retrieved and modulated. Research on epigenetic mechanisms in memory aspects of addiction has largely focused on simple memory processes -- how cue-drug associations are initially encoded, consolidated, and retrieved. This research has revealed the critical importance of histone acetylation and gene expression in mediating several aspects of these memory processes. We begin our review of histone-mediated epigenetics in addiction with a description of some of these memory processes in more detail.
3.1. Initial establishment of drug-associated memories
When patterns of drug use first begin, new associations are encoded between the drug and the user’s environment, consolidated into a memory, and later retrieved when cues associated with drug seeking are encountered. The very first experience with a drug of abuse activates circuits that are involved in reward and in learning and memory. With this first exposure, the initial memory begins to form. This memory likely involves distal contexts and discrete cues associated with some aspect of the drug of abuse. Theoretical approaches to memory have found that upon this initial exposure to the drug, the memory is labile for a period of time before it is stabilized through a time-limited consolidation process (McGaugh, 2000). In some cases, a single experience with a drug of abuse can lead to a lasting memory; in other cases, this memory is established and strengthened more incrementally, with repeated exposures to the drug increasing the strength of the memory and distributing the representation of that memory across different circuits in the brain. As these memories are being established, they can be modified by additional processes that are triggered by drugs of abuse, such as sensitization, tolerance, and withdrawal (Gould & Leach, 2014; Wise et al., 2011). Together, these processes ultimately result in habitual drug seeking that results from an interaction of circuits mediating contextual information (e.g., the hippocampus), response initiation and maintenance (e.g., the striatum), and reward value (e.g., nucleus accumbens and amygdala).
These initial memory processes contribute to the development of the repeated binge/intoxication stage of drug addiction, driven by a collection of brain regions in an excitatory circuit. The first time drugs of abuse are used (e.g., psychostimulants, alcohol, opioids, nicotine, Δ 9 tetrahydro-cannabinol), brain regions in both reward (i.e., ventral tegmental area, striatum, nucleus accumbens core, thalamus) and learning (basolateral and central nucleus of the amygdala, medial prefrontal cortex, hippocampus) centers share excitatory information (reviewed in Koob & Volkow 2010; Marchant et al., 2012). Some of these structures serve a primary role in particular stages of addiction (e.g., binge/intoxication, withdrawal/negative affect, and preoccupation/anticipation/craving; Koob & Volkow, 2010) and memory processes (i.e., encoding, consolidation, and retrieval; e.g., Bernardi et al., 2009; Lalumiere et al., 2012), yet many of these regions can be recruited throughout each process. Importantly, the circuits mediating aspects of reward and aspects of memory overlap, with key processes in the amygdala, ventral tegmental area (VTA), and nucleus accumbens (NAc) controlling consolidation of these drug memories.
Current research on histone-mediated epigenetics in addiction focuses on the role of histone acetylation specifically in memory consolidation. Broadly speaking, this histone modification may create a permissive state in chromatin, promoting access by transcription factors to the genomic DNA, thereby facilitating gene expression. Histone modifications alone do not cause long-term memory – they must be paired with activation of signaling cascades that are triggered by exposure to drugs of abuse. Once formed, these memories can be modulated in several different ways when additional exposure to the drug occurs in specific environments.
3.2. Retrieval of drug-associated memories and extinction of drug-seeking behavior
Once memories between environmental cues and drugs of abuse are established, there are several consequences that occur with subsequent exposure to those cues and drugs. First, these memories may evoke cravings, causing an increase of drug-seeking behavior and drug consumption (e.g., Robinson & Berridge 1993). Repeated cravings and drug administration result in tolerance and withdrawal, two processes that are key to maintaining addiction (Siegel 1983). Second, the act of retrieval will trigger some of the initial processes – encoding and consolidation of the retrieved memory – and will potentially include new contextual components that were absent during initial acquisition. This additional consolidation may simply recapitulate the initial consolidation process (i.e., the idea of reconsolidation; Tronson & Taylor 2013), and, in addition, they almost certainly involve consolidation of new memories specific to this new experience (e.g., Badiani & Robinson 2004). These consolidation and reconsolidation processes are thought to contribute to the long-term maintenance of addiction. Third, if the drug that is expected (based on the retrieval of a previous memory) is not consumed, extinction may begin to develop. If no drug is administered during repeated retrieval episodes, drug-seeking behavior may be extinguished due to the “cue-no drug” association developing alongside the original “cue-drug” association.
The inhibitory learning that occurs during extinction requires similar encoding, consolidation, and retrieval processes as during the excitatory learning associated with initial acquisition. There are important similarities in the systems and molecular steps that are involved in initial memory formation and extinction (e.g., Lattal et al., 2006), but there also are critical differences. For example, initial memory formation and extinction may recruit specific and distinct subregions of the medial prefrontal cortex, amygdala, and nucleus accumbens (e.g., Koob & Volkow 2010; Peters et al., 2009; Stefanik et al., 2013; Tye et al., 2010). It is thought that these excitatory and inhibitory circuits are usurped during the transition from drug use to eventual addiction (e.g., Hyman 2005) and modulate inhibition of drug-seeking behavior during extinction or abstinence. Although extinction treatment (a model of clinical exposure therapy; Nic Dhonnchadha & Kantak, 2011) diminishes drug-seeking behavior, relapse often occurs over time (spontaneous recovery; e.g., Brooks 2000), with the presentation of drug associated cues (reinstatement; e.g., Shaham et al., 1997), or after leaving the extinction context (i.e., renewal; e.g., Crombag & Shaham 2002; Crombag et al., 2008). The mechanisms of extinction and relapse are of particular interest in this chapter as they may apply to the treatment and potential prevention of many disorders where extinction may be impaired, including post-traumatic stress disorder and addiction (see Tipps et al., 2014).
4. Histone-mediated epigenetic mechanisms
We now know a great deal about the learning and memory processes that are involved in the establishment and maintenance of addiction. There are several ways in which histone-mediated changes may impact these learning and memory processes. Much of the work on histone modifications in addiction has focused on histone acetylation, but before reviewing those findings, it is important to consider other ways in which histones can be modulated by different molecular events. The study of histone modifications in addiction is in relative infancy compared to the study of these modifications (and epigenetic events in general) in other biological processes, such as cancer.
While epigenetic modifications to the genome are known to change molecular, cellular, and systemic function, they are being discovered as underlying mechanisms to many complex diseases, including developmental, neurodegenerative, and psychiatric (see Portela & Esteller 2010; Tsankova et al., 2007). Much of what is known about epigenetic mechanisms involved in cell biology comes from studies that have focused on the relation between these mechanisms and the causes of and cures for cancer (Sharma et al., 2009). These basic studies have led to the examination of epigenetic mechanisms in other processes, such as learning, memory, and addiction. The similarities between neural circuits, substrates, and many of the epigenetic factors that create long-term memories and those that cause long-term addiction suggest that common mechanisms are involved (e.g., Malvaez et al., 2009; Robison & Nestler 2011). A major focus of current research is investigating how these mechanisms may contribute to developing persistent cellular and molecular changes that may translate into persistent behavioral changes, including lasting suppression of drug seeking.
There are at least five types of chemical modifications (i.e., methylation, phosphorylation, acetylation, poly-ADP-ribosylation, and SUMOylation) that take place on the amino acids of histone tails. These modifications allow access to certain genomic regions to be increased or decreased, which is associated with activation or repression of transcription of specific genes. The enzymes that complete these chemical modifications to DNA, transcription factors, and histones, are typically recruited during development or after some type of stimulation to an organism (e.g., Lv et al., 2013; Vecsey et al., 2007). With activity-dependent depolarization of neurons, activation of inter and intracellular pathways leads to an interaction between these enzymatic coactivators and their substrate (e.g., DNA, transcription factors, or histones), largely regulating gene and protein expression, ultimately altering system function. The steps in this process are also modified by the physiological state of the organism and the type and extent of stimulation applied.
Research pertaining to histone-mediated epigenetic regulation of addiction has focused largely on the induction of factors downstream of histone modifications that are associated with increases or decreases of drug-seeking in rodents. Multiple studies have identified a major role for immediate early genes (IEGs; such as c-fos, c-jun, and fosB), transcription factors and coactivators (such as cAMP response element-binding protein, [CREB] and CREB-binding protein [CBP]), and kinases (such as protein kinase A, C, and Ras) in the plasticity induced during learning or drug use (Darcy et al., 2014; Levenson et al., 2004; Shalin et al., 2006). The involvement of these key regulatory factors and the related gene targets relies on complex chemical modifications made to both DNA and histone proteins. In the last two decades increasing recognition for the necessity of these modifications has led to advancements in the field. While both DNA and histone modifications have been shown to interact and rely on each other (Cedar & Bergman, 2009) each substrate (DNA or histone protein) modification leads to a unique sequence of events.
Multiple types of chemical modifications can be made to histones, each of which is thought to create a novel surface to be recognized by effector proteins and specific downstream events. Although there are many types of modifications made to histones, such as phosphorylation, SUMOylation, ubiquitination, and poly-ADP-ribosylation, the majority of addiction-related research has investigated the effects of altering methylation and acetylation levels on histones 3 and 4 (Strahl & Allis, 2000). Broadly, research suggests that drug use stimulates many chemical modifications that are needed to activate or repress the transcription and translation of DNA into functional proteins.
Some of these marks can even have opposing effects on transcription. For example, methylation and phosphorylation are thought to participate in both closing and opening of chromatin and potentially mediating the repression and activation of transcription (Cheung et al., 2000). Alterations to transcription after histone methylation or phosphorylation is based on many variables (e.g., organism’s developmental stage, the onset and duration of stimulation, the type of tissue, cell, or histone residue that is being targeted for modification; Cheung et al., 2000; Greer & Shi, 2012; Smith & Shilatifard, 2010). In contrast, histone acetylation is primarily associated with activate gene transcription with very few exceptions (e.g., Braunstein et al., 1996). The combination of these marks are in large part thought to be how environmental effects lead to individual variability, and how such great diversity can be created through epigenetic regulation. Yet, it is still unknown whether histone-mediated epigenetics can be used in the clinic to prevent and treat addiction. Here, we will review the research and remaining questions pertaining to histone-mediated epigenetics in addiction.
4.1. Repressive histone modifications
Methylation and phosphorylation
A repressed state is by and large the default structure of chromatin, preventing abnormal changes to DNA’s code (e.g., segregation, recombination, replication, etc.; Grewal & Jia 2007). The most common way that the tails of each histone remain tightly bound within the chromatin structure, repressing future gene transcription, is through methylation. Methylation is the act of adding or subtracting a methyl group to a substrate, or in the case of histone-mediated epigenetics, to an amino acid. Due to the chemical properties of histones and DNA, epigenetic modification by methylation is only possible on lysine and arginine amino acids, and only on amino acids that are bound around or within the tail region of histones 3 and 4. There are three possible degrees of methylation (mono, di, and tri-methylation), providing further regulation and ultimately determining the outcome on transcription (Santos-Rosa et al., 2002). In addition to the state of methylation, the high degree of specificity to function arises from the residue modified and from the type of enzyme that catalyzes lysine and arginine methylation or demethylation (histone methyltransferases; HMT and demethylases; HDMT). The enzyme complexes created largely regulate histone structure and accessibility to promoter or coding regions on individual genes within DNA. For example, repressive type di- or trimethylation often occurs on histone 3 (H3) at the lysine 9 (K9) or 27 (K27) residue, whereas activation is often associated with trimethylation that occurs at H3K4 or H3K36 (Greer & Shi, 2012; Kouzarides, 2007; Lee et al., 2010; Muratani & Tansey, 2003; Zhang et al., 2014). Evidence like this clearly demonstrates how repressive methylation can be used to repress transcription in a complex way. As an example, mono-methylation on histone 4 in the lysine 20 position (i.e., H4K20me1) has been shown to functionally change chromosome condensation and transcription (Wang & Jia, 2009) as well as trimethylation at lysine sites 9 and 27 of histone 3. To be expected, each of these methylation marks similarly binds nucleosomes together into a more stable and condensed structure and primarily represses overall gene transcription, but are associated with different downstream events accomplishing different results through slightly separate avenues. For example, monomethylation of H3K27 and H3K9 may serve to recruit additional silencing machinery and be a substrate for trimethylation which further enhances the repression of noncoding regions of the genome (Mozzetta et al., 2014; Maze et al., 2011).
In addition to the location and type of methylation mark, the timing of histone marks and the host’s current modification state may affect the binding of additional regulatory proteins, such as heterochromatin protein 1 (HP1α). HP1α interacts with the methylated tail of lysine 9 on histone H3 (i.e., H3K9me) and dimerizes with other regulatory proteins. The methylation and additional cross-linking between proteins reinforce the stable and condensed chromatin structure, called heterochromatin. This tightly bound heterochromatin structure increases gene-silencing overall. In addition, HP1α can also be modified, through phosphorylation. Once serine residues on HP1α are phosphorylated the structure will greatly increase its binding efficiency to H3K9me (Alvaro-Bartolome & Garcia-Sevilla, 2013). In this way and many others, methylation and its associated partners (e.g., HP1α) can result in transcriptional inhibition and decreased genome activity. Similar to phosphorylation of HP1α, histones can also be phosphorylated and increase chromatin condensation, potentially decreasing replication and transcription (Castellano-Pozo et al., 2013; Johansen & Johansen, 2006; Wilkins et al., 2014; Zhang et al., 2004). The details related to repressive effects of phosphorylation on histone tails are not yet characterized well in the addiction field.
4.2. Active histone modifications
4.2a. Methylation
In contrast to the common repression of gene transcription by histone methylation, a few studies have demonstrated that methylation can in fact activate gene transcription. Trimethylation at lysine 4 of H3 (Santos-Rosa et al., 2002) and dimethylation at lysine 36 and 79 (Chen et al., 2011; Sims et al., 2008) contribute to transcriptional elongation and euchromatin, the accessible chromatin structure created to enhance gene activation, transcription, and behavioral changes. Modifications like these three activating methylation marks require different catalyzing enzymes (e.g., methyltransferases Set 7/9, Set2, or Dot1, respectively) and binding proteins (Sims et al., 2008). Each enzyme and subsequent mark recruits different downstream effectors and leads to different outcomes on transcription, further demonstrating how histone-mediated regulation is both specific and elaborate. As an example, the methyltransferase and demethylase responsible for di and tri-methylation of H3K4 (i.e., writer MLL1 and eraser kdm5c) are thought to upregulate transcription of the oxytocin receptor and Fos protein in the nucleus accumbens, mediating methamphetamine associated memory development and expression (Aguilar-Valles et al., 2014).
4.2b. Acetylation
Histone acetylation occurs on the nitrogen of lysine amino acids and causes the positive charge of histones to detach from negatively charged DNA in the chromatin structure (Jenuwein & Allis, 2001; Loidl, 1994). Acetylation levels are largely coordinated through histone acetyltransferase (HAT) and deacetylase (HDAC) enzymes. Histone deacetylases (HDACs) are the enzymes that remove acetyl groups from amino acid tails of histones. HDACs can be classified into four classes differing by the types of tissues they reside in, where they are active within the cell, the number and homology of catalytic sites and what substrates and binding partners they interact with to determine functional outcomes (Dokmanovic et al., 2007). For example, Class I HDACs reside within the nucleus of cells, are dispersed ubiquitously throughout the body, have one catalytic site, and have activity on DNA binding transcription factors and nuclear receptors, signaling mediators, and chromatin remodeling substrates. Each class and individual HDAC is thought to serve different functions. By decreasing Class I HDAC function, alterations in cell survival and proliferation occur, whereas knock-out analysis of Class II HDACs may localize effects to specific tissue types (Dokmanovic et al., 2007). Within each class, individual HDACs contribute to a wide range of independent roles, from cardiac function and chondrocyte differentiation to changes in global histone acetylation and gene expression (Dokmanovic et al., 2007). Additionally, the phosphorylation state of HDACs themselves can determine their permissibility to histones as well. For example, HDAC5 is known to be phosphorylated through activity-dependent mechanisms and after cocaine administration and then exported out of the nucleus, decreasing activity at specific histone sites (Dietrich et al., 2012). Correspondingly, the repression of genes, such as NR4A1, a nerve growth factor involved in inflammation and cell survival is increased by dephosphorylation of HDAC7. It is still unclear how HDAC function is selectively recruited and how sequence specificity of histone tails helps determine and coordinate these regulatory factors.
4.2c. Phosphorylation, ubiquitination, SUMOylation and poly-ADP-ribosylation
These post-translational modifications are well understood in other areas of research, but less studied with respect to addiction physiology. Phosphorylation of serine 10 on histone 3 has been shown to induce acetylation at a nearby lysine site (H3K9) and potentially others (e.g., H3K14) and subsequent transcription (Brami-Cherrier et al., 2009; Cheung et al., 2000; Clayton et al., 2000). It is here that histone phosphoacetylation is thought to occur, subserving a “subset of rapid transcriptional responses” for gene induction (Clayton et al., 2000). These are thought to be mediated by mitogen-activated protein (MAP) kinase and often lead to CREB and CBP/p300 activation. Although little research has connected histone phosphorylation or phosphoacetylation to addiction (to be discussed in section 4.2 and 4.3), cancer research recently reported evidence that androgen stimulation can result in histone phosphorylation on histone 3 threonine 11 (H3T11), recruitment of H3K4me3 and H4K16ac, and subsequent activation of androgen receptor target genes and proliferation of prostate cancer (Kim et al., 2014). Similar to in these studies, these phosphorylative steps may be initiating or potentiating other post-translational cascades that are recognized to occur during or after drug use (i.e., acetylation, methylation of histones).
Ubiquitination is a key regulator for several histone modifications. Histone ubiquitination is the addition of a covalently bonded ubiquitin protein to the lysine residue on the N-terminal tails of histones. The ligation of one or more ubiquitin subunits onto lysine residues and the target proteins they reside in will be marked for further protein trafficking or degradation (Weake & Workman, 2008; Welchman et al., 2005). The addition of these bulky ubiquitin moieties to substrates, such as histones, initiates degradation or activity, communication, or location changes. The main catalyst recruited for the ubiquitination system is Rad6, the enzyme dedicated to ubiquitin-like conjugation (e.g., the addition of ubiquitin) of substrates (Muratani & Tansey, 2003). Ubiquitination often works in a phosphorylation-dependent manner to control gene transcription. It may also be a signal for active versus inactive chromatin, and recruit methyl and acetyltransferases to assist in silencing or activating gene transcription at specific loci, respectively. In addition to catalyzing ubiquitination, the Rad6 enzyme likely plays a role in recognizing active versus inactive substrate sites on chromatin (Muratani & Tansey, 2003).
SUMOylation is similar to ubiquitination, as it results in the addition of bulky peptides to substrates such as histones. SUMO refers to a small ubiquitin related modifier, one that can recruit HDACs and HP1 and lead to potent transcriptional repression (Sims et al., 2008). This modification has only been identified on a few histone sites so far, shown to block activating events like acetylation or ubiquitination, but also occurs on transcription factors, indirectly repressing histone acetylation and gene transcription (Sims et al., 2008; Wilkinson & Henley, 2012). In addition, the phosphorylation state of substrates (e.g., various kinases, transcription factors and histones) can inhibit or enhance SUMOylation (Wilkinson & Henley, 2012). While the temporal and spatial details are limited to date, it is clear that complex interactions exist between SUMOylation and other histone marks (e.g., phosphorylation, acetylation, ubuiqitination, etc.) to repress transcription.
Histone poly-ADP-ribosylation is the addition of one or more ADP (adenosine diphosphate) ribose moieties to acceptor sites such as lysine, arginine, glutamic acid, and aspartic acid of histones. Nuclear proteins, such as histones, interact with poly-ADP-ribose polymerase 1 (PARP-1) to get mono or poly-ADP-ribosylated. The activation of PARP-1 can mediate chromatin structure, gene transcription, and environmental stimuli responses of cells, both during development and adulthood (Tulin et al., 2003). Accordingly, it is well established that PARP-1 is required for long-term memory formation by targeting (e.g., p53, fos) and binding to a variety of transcription factors (e.g., NF-kb, AP-2), each involved in plasticity or learning and memory processes (Liu et al., 2012; Salles et al., 2014).
4.3. Combinatorial modifications
As noted briefly above, these modifications may act alone or in concert with other modifications to create a complex code that determines individual gene regulation. For example, H3K9me3 initiates and maintains repression of transcription by antagonizing active modifications (e.g., H3K9ac, H3S10p, H3K4me3; (Chen et al., 2011). In addition, extensive coordination occurs between histones and DNA to modify the epigenome, facilitating, impairing, or neutralizing transcription. By depleting the enzyme that poly-ADP-ribosylates proteins (PARP1), H4K20me3 will decrease, and further deplete ubiquitin ligase UHRF1 and DNMT1 (DNA methyltransferase 1) expression (De Vos et al., 2014). Without the maintenance of this DNA methylation, the nucleosome assumes a euchromatic structure and allows abnormal transcription. Another example of DNA and histone-mediated interactions was demonstrated in fetal rat cortical neurons (Chen et al., 2003). Stimulation and calcium influx of these neurons results in phosphorylation of methyl CpG (cytosine-phosphate-guanine) binding protein 2 (MeCP2), releasing it from methylated CpG sites of DNA and ultimately decreases methylation and increases acetylation at lysine 9. Early results from Jones et al (1998) also demonstrated that MeCP2 binds methylated DNA and recruits HDACs to further stabilize transcriptional repression of chromatin. Therefore, when MeCP2 is bound to DNA, it is responsible for global gene repression, and when released, histone acetyltransferases and demethylases help remove methylation, and acetylate nearby sites (i.e., H3K9) to increase activity at specific gene promoters and increase protein expression (e.g., promoter region of brain-derived neurotrophic factor [BDNF], Chen et al., 2003). While this is just one example of how DNA and histone–mediated epigenetics can regulate chromatin accessibility and relevant learning and memory proteins, many examples of this coordination exist (discussed further in Lv et al., 2013).
5. Histone-mediated actions in drug addiction
With the use of animal models and advancing technology, researchers are trying to uncover the complex environmental and genetic mechanisms that underlie addiction. Although significant differences exist between human and animal conditions of drug-craving, seeking, and taking (Stephens et al., 2013), the research field is focused on delineating each aspect of addiction, in hopes of piecing together a pathway for improved treatment and prevention. Because drugs of abuse often change cellular and systemic activity in animals and humans, simple measures of cellular and locomotor sensitization or tolerance are used to measure changes in physiology (e.g., receptor function) and overall behavior after acute or chronic drug administration. In addition, animal models like conditioned place preference (CPP) and operant self-administration are commonly used to investigate the reinforcing effects of drugs and infer the strength of drug-cue or drug-response associations. Similar to humans, animals will associate the reinforcing effects of drugs with stimuli (i.e., places) and behaviors (i.e., voluntary responses that lead to drug delivery) that are paired with a drug experience. Subsequently, if re-introduced to drug-paired stimuli or the opportunity to obtain drug reinforcement, animals and humans alike will retrieve these memories and demonstrate drug-craving, seeking, or taking behavior.
The various histone-modifications discussed above (i.e., Section 3) contribute to many diseases, including drug addiction, and are initiated and terminated for many reasons (Bohacek & Mansuy, 2013; Lv et al., 2013; Petronis, 2010). Similar to other diseases, histone-mediated changes can influence and be influenced by a patient’s or subject’s genetic background, surrounding environment, and stage of development or disease. Within the drug abuse field, epigenetic researchers have predominantly discovered changes that occur to histone methylation and acetylation status (e.g., Renthal & Nestler 2009).
5.1. Methylation and addiction
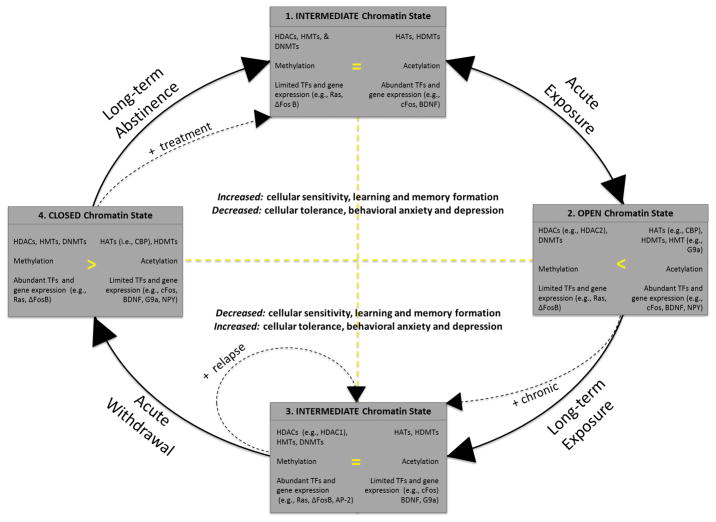
One of the more prominent and recent discoveries on mechanisms underlying addiction described how the epigenetic response to an initial or habitual dose of drug administration is often different (see Figure 1). After just one administration of cocaine (Acute Exposure in Figure 1), the methyltransferase responsible for methylating H3K9 sites, G9a, is increased. Increased methylation at this site results in greater binding of G9a to the immediate early gene fosB, an effect that seems to be counteracted after repeated cocaine use, where G9a levels and fosB binding decrease (Maze et al., 2010). As noted previously, methylation often leads to a heterochromatin, or an inaccessible structure, increasing the likelihood of decreased transcription. While it is known that ΔFosB, a product of the fosB gene, accumulates with repeated cocaine exposure and is associated with increased cocaine reward (Renthal et al., 2008), removing this G9a hindrance at fosB sites, enables ΔFosB to be increased, perpetuating the accumulation of ΔFosB and the addiction cycle (Maze et al., 2010).
Figure 1. Epigenetic changes in the cycle of addiction.
Some of the potential chromatin dynamics are shown for the cycle of addiction, which moves from acute exposure, to chronic drug taking, to withdrawal, to abstinence and recovery, and finally back to acute exposure in cases of relapse, which begins the cycle again. Three potential states of chromatin (i.e., OPEN, CLOSED, or INTERMEDIATE) and their associated nuclear changes are depicted within four small gray boxes (numbered 1–4). These chromatin states create a more accessible (Box 2), inaccessible (Box 3), or intermediate (Box 1 and 3) structure for DNA to be accessed and transcription to take place. The top half of this figure signifies chromatin in a more “adaptive and responsive” state. The bottom half signifies chromatin in a more “inflexible and unresponsive” state (splitting states 2 and 4 into both of these categories evenly). The left half of this figure signifies a more “heterochromatin” state, while the right half signifies a more “euchromatin” state (splitting states 1 and 3 into both of these categories). Dashed lines (e.g. +chronic, +relapse, +treatment) represent the potential for associated changes to be accelerated by rate and/or intensity. Box 1 (INTERMEDIATE chromatin before drug intake) represents a basal chromatin state with normal transcription (determined primarily by genetic and previous environmental interactions). In this state, the chromatin and associated nuclear changes are well balanced and highly regulated. Box 1→Box 2 Transition: With acute exposure to stress or drugs of abuse, brief and reversible changes (see bi-directional arrows) occur to select histone and DNA regions (increased histone acetylatransferases like CREB-binding protein [CBP], acetylation, DNA accessibility, learning and memory related gene transcription). Box 2 (OPEN chromatin) represents the change that occurs with a single or acute insult to the system (e.g., acute stress or drug exposure). Chromatin expands, releasing repressive marks and tipping the balance of epigenetic regulation towards those associated with gene activation. Box 2→Box 3 Transition: With repeated exposure to stress or drugs, a prolonged and less reversible change occurs to select histone and DNA regions. Box 3 (INTERMEDIATE chromatin after drug intake) represents chromatin with dysregulated histone enzymes, marks, transcription factors (TFs) and transcription. In this state, similar process occur as in Box 1, yet the location where histone modifications occur, the type of modification, and the effect that histone modifications have on cellular and behavioral outcomes is altered to positively reinforce this chromatin state. The balance of regulation is shunted away from promoter regions that are associated with learning and adaption (cFos, BDNF) and shifted toward promoter regions that are associated with this altered chromatin state and positively reinforce this altered gene regulation (e.g., Ras and ΔFosB). These changes are thought to induce increased cellular tolerance and maladaptive behavior. Box 3→Box 4 Transition: With acute drug abstinence, brief and reversible changes occur to select histone and DNA regions (increased histone deacetylases, methylation, DNA inaccessibility, decreased gene transcription) in an attempt to rebalance the previous dysregulation. Yet, after chronic or repeated insults to the system recent drug abstinence induces withdrawal associated effects (e.g., anxiety and depression) making the organism increasingly susceptible to relapse rather than recovery and long-term treatment. Box 4 (CLOSED chromatin) represents the change that occurs with acute abstinence (without relapse) and the associated withdrawal from drugs of abuse. Here chromatin begins the process of rebalancing enzyme levels, histone marks, and gene transcription by generally increasing the repression of prior imbalances related to addiction. The previous epigenetic and behavioral changes placed on the system (e.g., positive feedback of ΔFosB and behavioral depression) make this process slow, the system resistant to rebalancing, and deprived of necessary proteins to counteract this state. As chromatin becomes more condensed, regulation is increased (although exceptions to this mechanism exist, such as decreases to repressive methylation with withdrawal, noted in Section 5.3). Transition from Box 4→Box 1: With repeated and long-term abstinence from drug use a prolonged and less reversible change occurs to select chromatin regions, rebalancing the location, type, and effect that histone modifications have on cellular and behavioral outcomes, recovering to a more normal and highly regulated level of transcription.
Additional studies have demonstrated that chronic exposure to drugs of abuse, such as cocaine and opioids, reduces dimethylation at H3K9 by decreasing G9a and GLP (G9a-Like Protein) enzymes in the nucleus accumbens of mice (Aguilar-Valles et al., 2014; Renthal & Nestler, 2009; Sun et al., 2012). Similar effects occur in the mouse cortex and in cultures of human lymphocytes after repeated nicotine treatment (Chase & Sharma, 2012). In parallel, alterations to these enzyme levels lead to positively correlated responses in drug-seeking and drug sensitizing behavior (as measured by animal models in conditioned place preference and locomotor sensitization paradigms) after acute drug administration. In contrast, G9 levels decrease as drug-taking increases with chronic treatment (i.e., as administered and measured with an animal model of operant drug self-administration, Maze et al., 2010; Sun et al., 2012). This suggests that increased levels of dimethylation at H3K9 impair drug reward initially but are mitigated by repeated drug use.
Trimethylation of H3K9 has also been shown to play an important role in the addiction process. While H3K9me1 and me2 are known to reside in euchromatin, me3 occurs in non-genomic, or heterochromatin regions of the DNA (Greer & Shi, 2012). Repeated cocaine treatment increases the expression of this specific type of methylation as well, resulting in enhanced expression of transposons and typically silenced (heterochromatic) regions of the DNA by decreasing repressive methylation within the nucleus accumbens of mice (Maze et al., 2011). This work and others noted above suggests that repeated treatment with drugs of abuse causes the de-repression of previously silenced DNA regions by inhibiting methylation at key non-genomic and genomic sites. This is likely a key factor contributing to the altered gene expression and impaired physiologic function after long-term use of drugs and with addiction. Importantly, these alterations are thought to release the brakes placed on transcription and lead to a more permissive epigenetic environment (McQuown & Wood, 2011).
Covington et al (2011) further investigated if dimethylation on H3K9 could be related to the depressive-like phenotype expressed after chronic cocaine administration and social defeat stress (a paradigm inducing anxious, stressful and depressive characteristics in animals; Toth & Neumann 2013). As expected, their results suggested that chronic cocaine administration led to increased vulnerability to the detrimental effects of social stressors and that this was due to removal of G9a, GLP, and subsequent demethylation at H3K9me2 and enhanced Ras-CREB signaling. This is likely a key mechanism for chromatin to be opened and the expression of downstream proteins associated with addictive behavior to increase (e.g., Ras G-proteins, ΔFosB transcription factors). In contrast, increased BDNF in rats leads to greater drug reinforcement (Bahi et al., 2008) and is decreased with chronic cocaine use in humans (Corominas-Roso et al., 2013). BDNF’s down regulation in reward-related regions of the brain (e.g., VTA, NAc) after chronic cocaine use, and its replenishment during abstinence (Corominas-Roso et al., 2013) is likely a compensatory mechanisms of cocaine’s effects. Interestingly, if the regulation of this factor contributes to the rewarding properties of cocaine and is increased during abstinence, this may account for slower rates of CPP extinction reported in rats (Bahi et al., 2008) and the positive correlations between BDNF, abstinence, anxiety, and depression during early abstinence (Corominas-Roso et al., 2013). These types of changes in gene regulation are often enabled by the combination of decreased methylation status and subsequent increased histone acetylation (Fuchikami et al., 2010). Coordinated mechanisms between histone methylation and acetylation will likely be targeted for treatments of addiction and other disorders in the future (Kennedy et al., 2013; Sen, 2014).
5.2. Acetylation and addiction
As more diseases are being attributed to novel histone-mediated mechanisms (e.g., palmitoylation, isomerization, deimination; Chavda et al., 2014; Khanal et al., 2013; Kouzarides 2007; Dieker & Muller 2010), alterations involving histone acetylation have been leading the charge in addiction research. Major advances in understand how occasional drugs use can manifest into chronic problems have largely been due to the use of pharmacological agents that target HDACs. Initial investigations with HDAC inhibitors focused on anticancer activity (Wagner et al., 2010; Yoshida et al., 1990; Zhang & Zhong, 2014), but more recent work has examined the potential of HDAC inhibitors in treating a variety of psychiatric disorders (i.e., addiction, PTSD, depression; reviewed in Renthal & Nestler 2009). One of many examples of this is with Trichostatin A (TSA), a general HDAC inhibitor with antitumor activity (Drummond et al., 2005), that has been shown to decrease the motivation for and intake of addictive drugs (Romieu et al., 2008). Such a finding has been observed with other HDAC inhibitors and variations to the time and route of drug administration (Arora et al., 2013; Kim et al., 2014; Raybuck et al., 2013).
In an operant measure of drug-seeking behavior, Romieu et al (2008) demonstrated that rats that voluntarily self-administered high levels of cocaine would decrease responding when administered this nonspecific HDAC inhibitor, TSA. In addition, repeated administration of TSA was shown to stably and selectively decrease cocaine administration and not sucrose intake while reducing cocaine induced locomotor sensitization. This was mediated through decreases in HDAC deacetylation activity in the prefrontal cortex and nucleus accumbens, either with or without cocaine administration. These results and others (Malvaez et al., 2010, 2013) lend to the potential use of HDAC inhibitors in relapsing addicts (Romieu et al., 2008). Although the circuits mediating these effects remain to be fully described, a focus is on the nucleus accumbens, where drugs of abuse increase acetylation, locomotion, reinforcement, and reward (Kumar et al., 2005; Renthal et al., 2009; Schroeder et al., 2008; Sun et al., 2008; Wang et al., 2007).
Work from these groups and others have also demonstrated that chronic drug use (e.g., with cocaine or alcohol abuse) will initiate decreases in HDAC activity in reward and learning-related brain regions (e.g., nucleus accumbens, prefrontal cortex, hippocampus; Romieu et al., 2008; Zou & Crews 2014), creating a more permissive genome for drug regulation to be imparted. Renthal et al. (2007) discovered a potential mediator for the transition from dysfunction or drug abuse to disease and drug addiction with HDAC5. HDAC5 is a class II HDAC that is phosphorylated minutes after activity-dependent stimulation (e.g., initial drug use, stress, etc.) and is exported from the nucleus, allowing gene transcription and behavioral adaption to take place. Without additional insult, HDAC5 will be replaced to the nucleus within 24 hours, capping gene transcription. Chronic stimulation, such as chronic cocaine-administration or chronic social-defeat stress, results in long-term HDAC5 inhibition, increases in cocaine-, or stress-associated gene transcription (i.e., Ras, ΔFosB), and dysregulated sensitivity to subsequent challenges. These data demonstrate the importance of balancing acetylation to maintain flexible behavior.
As occasional drug use turns chronic, many cellular and behavioral changes occur (Long-term Exposure in Figure 1). In light of the role that HDAC5 plays in chronic but not acute drug treatment or stress, the transcription factor ΔFosB is also thought to mark the transition from abuse to addiction (Renthal et al., 2008). With chronic amphetamine treatment, ΔFosb is increased, leading to recruitment of HDAC1 and attenuation of the immediate early gene, c-fos (Renthal et al., 2008). In concert, dimethylation of histone 3 at lysine 9 is increased (recall that this type and location of methylation is repressive in nature). These data further demonstrate that chronic drugs (i.e., amphetamine) not only swap methylation for acetylation at the cfos promoter within the striatum (a region highly important to addictive behavior) but also lead to decreased expression of a methyltransferase (KMT1A) necessary to remedy this imbalance. As ΔFosB increases with repeated drug use, G9a (another methyltransferase) is built to decrease the levels of this addiction marker by binding to the fosB promoter and suppressing its expression (Maze et al., 2010). As one can see though, the coordinated tools put in place to rectify such epigenetic imbalances are in competition with drug effects that are inherently self-perpetuating.
While drug use impairs HDAC activity and positive feedback of ΔFosB (by ΔFosB) is initiated during chronic drug use (reviewed in Maze & Nestler 2011), additional changes compound these imbalances. For example, an acute administration of alcohol or cocaine will briefly increase H4 acetylation and H3 phosphoacetylation (previously associated with active immediate early genes, c-fos and c-jun, Clayton et al., 2000). Kumar et al. (2005) demonstrated mechanistic differences between acute and chronic covalent chemical changes in the brain. They determined that histone acetylation and phosphoacetylation influenced gene regulation in the striatum, an addiction mediating region of the brain, and behavior in mice and rats after acute and chronic cocaine administration (by investigator initiated intraperitoneal injections and subject initiated self-administration infusions). Cocaine induced acetylation (on H4) and phosphoacetylation (on H3) at specific gene promoters, with acute affects at the c-fos and fosB sites on H4 and chronic effects at the fosB, cyclin-dependant kinase 5 (cdk5), and BDNF sites on H3. This creates an overall pattern of hypoacetylation and desensitization on H4 at the c-fos promoter but an exaggerated state of acetylation on H3 at ΔFosB promoters (only partially desensitizing ΔFosB, Kumar et al., 2005; Renthal et al., 2008). Similar patterns of gene regulation have been demonstrated with c-fos and BDNF in the hippocampus (rather than c-fos and fosB in the striatum) after acute and chronic electroconvulsive seizure induction (a procedure shown to decrease long-term plasticity in the hippocampus of rodents and be an effective treatment for depression in humans; Tsankova et al., 2004).
Similar to previous studies mentioned, alcohol can also modify acetylation patterns with temporal and spatial specificity (Sakharkar et al., 2014; Shepard et al., 2008). Acute doses of alcohol can alleviate minor levels of stress or anxiety-like behavior (Starkman et al., 2012), possibly because of alcohol’s ability to decrease HDAC levels in the amygdala (a brain region necessary for affective and emotional associations; Gruber & McDonald, 2012; Pandey et al., 2008; Sakharkar et al., 2012). Pandey et al. (2008) demonstrated that decreases in HDAC activity (i.e., following acute ethanol) correspond to increases in H3 and H4 acetylation and increases in anxiolytic and plasticity-related protein levels (i.e., CBP and neuropeptide Y [NPY]). In contrast, increases in HDAC activity and subsequent decreases, or rebalancing of H3 acetylation, CBP, and NPY likely mediate alcohol withdrawal and the associated anxiety (Pandey et al., 2008). As previous data would predict, inhibition of HDAC activity (by TSA) normalizes the sharp decreases in H3ac, H4ac, NPY expression and the corresponding anxiogenic effects of withdrawal (Pandey et al., 2008).
Although there are nuances, the majority of research thus far supports the idea that drugs of abuse and various stressors generally repress methylation which can be detrimental and lead to addiction and depressive-like behaviors, while acetylation tends to be protective and lead to adaptive behaviors. Interestingly, these two common mechanisms can lead to an increase in transcription, yet the timing and location of these modifications play a critical role in their behavioral outcomes.
5.3. Combinatorial modifications and addiction
While histone modifications are merely a part of the physiological changes that occur with drug addiction, it is becoming accepted that each histone modification plays a role in the development, maintenance or potential treatment of drug addiction. Therefore, understanding the upstream and downstream coordination of each modification is becoming increasingly necessary and elaborate. As one might expect, dopamine receptor agonists elicit rewarding effects, but the combination of dopamine agonists and HDAC inhibitors seem to compound these effects, leading to a synergistic increase in cocaine-induced locomotion or sensitization and CPP (Schroeder et al., 2008). Research by Schroeder et al. (2008) demonstrated that a class I/II HDAC inhibitor (sodium butyrate, NB) and SKF82958 (D1 receptor agonist) increased H3 phosphoacetylation (i.e., phosphorylation at serine 10 and acetylation at lysine 14) in striatal homogenates and increased deacetylation in the substantia nigra/ventral tegmental area at the promoter regions of BDNF and tyrosine hydroxylase (the rate-limiting enzyme for synthesis of catecholamines like dopamine and norepinephrine). In another study, a dopamine D2 receptor antagonist was found to induce similar changes (Li et al., 2004). Importantly, the molecular effects in the Schroeder et al. (2008) study were sensitive to acute versus repeated administration of these drugs and CPP effects only emerged when the drugs were administered directly after cocaine-induced place preference (presumably during consolidation of CPP learning). This highlights two important points, 1) the transient molecular effects of epigenetic modifications that are known to change after repeated drug use and 2) the experience, or activity-dependent nature of histone-modifications.
Research by Qiang et al. (2011) demonstrates how methylation (i.e., repressive di- and tri-states) and acetylation coordinate to compound the effects of histone modifications. Neuron cultures of mice had large increases in activating acetylation at H3K9 that coincided with large decreases in repressive methylation during a time of withdrawal from ethanol. Surprisingly, global and local downregulation of histone demethylases (i.e., G9a) at the NMDA receptor gene (NR2B) rather than changes in global or local HATs or HDACs seemed to underlie this effect, underscoring the idea that these modifications work in concert to regulate or dysregulate the system at specific chromatin sites. Correspondingly, Sheng et al. (2011) demonstrated that intracranial administration of an NMDA antagonist (MK-801) into the nucleus accumbens (but not the medial prefrontal cortex) decreased H3 phosphoacetylation and acquisition of heroin place preference, while heroin CPP dose dependently increased H3 phosphoacetylation in the nucleus accumbens (but not the medial prefrontal cortex). TSA (HDAC inhibitor) infusions prior to each CPP acquisition session facilitated heroin’s effects, further increasing H3 phoshoacetylation in the nucleus accumbens and CPP. Research like this is clarifying ideas that certain brain and chromatin regions, and types of histone modifications are targeted based on drug type and administration patterns.
As we better understand how cocaine and alcohol affect methylation and acetylation, appreciation grows for modifications involved with other drugs of abuse. For example, hints are emerging that poly-ADP-ribosylation and ubiquitination may also contribute to the cellular changes that occur with drug abuse. Cigarette smoke extract leads to an overall increase in H4 acetylation, due to ubiquitination and subsequent degradation of HDAC2 (Adenuga et al., 2009). In addition, the dopamine transporter (DAT), implicated in cognition, affect, behavioral reinforcement, and motor control, can be tagged by ubiquitination (Schmitt & Reith, 2010) and lead to altered endocytosis and degradation levels. Although it is known that drugs of abuse alter the function of DATs, the full mechanism and histone modifications likely behind these alterations are clearly still being discovered. In addition, it is established that the primary polymerase instigating poly-ADP-ribosylation, PARP, is a coactivator of AP-2-mediated transcriptional activation (Kannan et al., 1999), but no direct link has been formed between histone ribosylation and transcriptional changes by drugs of abuse thus far. However, the overexpression of AP-2 recruits the coactivator PARP and recent evident attests to the idea that acute and repeated morphine exposure locally increases AP-2 in neurons of mice hippocampi (i.e., specifically at the post-synaptic density). This is thought to contribute to the dysregulation of hippocampal plasticity, by specifically targeting glutamatergic synapses (those with AMPA and NMDA receptors). Although many factors contribute to the synaptic changes noted in this and other studies, PARP and increased histone poly-ADP-ribosylation may be another mechanism of drug-induced changes usurping learning and memory functions. While many of the links between ubiquitination, ribosylation, histones, and addiction have yet to be uncovered, these marks (like others) presents plausible mechanisms by which drugs of abuse, epigenetic processes, and long-term changes take place.
5.4. Summary of histone-mediated epigenetic regulation in addiction
Fundamental theories about addiction evolved from the epigenetic research summarized here and elsewhere (Biliński et al., 2012; Malvaez et al., 2009; Robison & Nestler, 2011; Starkman et al., 2012). This system of regulation enables an infinite level of control and flexibility on gene regulation. The interaction between specific sequences of DNA, specific histone proteins, and other regulatory factors induced by the environment, lead to highly controlled genomic outcomes, including increased or decreased transcription of selected genes. While initial drug use is now thought to prompt the brief opening and closing of chromatin and adaptive behavior, repeated drug use seems to decrease the system’s plasticity as cellular tolerance takes over. This broadens the time and resources needed for each epigenetic modification and narrows the adaptiveness of the system. Without treatment, epigenetic modifications will become perpetually dysregulated and behavioral vulnerability will linger.
6. Debates and considerations
Because histone-mediated epigenetics in addiction is such a new field, there are many unresolved issues that are currently being debated in the literature. Some of these are general debates that are common in all fields in which epigenetic mechanisms are studied. Other debates are more specific to the addiction and learning and memory fields, where very different theoretical approaches have been offered for defining epigenetic mechanisms, the nature of cellular memory and how it relates to psychological memory, and the nature of psychological processes in general, which are open to many different interpretations. Our goal in this section is to present a broad overview of some of these debates. We cannot provide resolution for most of these issues, but we can survey the literature and provide suggestions about the direction of research in these areas.
6.1. Are histone marks causal or correlational to downstream processes and do histone modifications change the structure and function of the genome?
Histone modifications are a contributing factor in a larger dynamic process that regulates accessibility of DNA and transcription (Henikoff & Shilatifard, 2011). By changing the charge between nucleosomes and nearby DNA, histone modifications help maintain an unraveled or raveled chromatin structure and docking area for regulatory modules. This account of histone marks highlights their many correlated and few causative roles in transcriptional regulation (Zhang et al., 2014). As partial justification for a correlative role, histone modifying enzymes often have effects on other substrates besides histones, leading to multiple effects on the genome, rather than direct effects on transcription only. In addition, it seems that histone modifications play a larger role in stabilizing nucleosome occupancy and position rather than recruiting regulatory factors to DNA. It is unlikely that modifications, like methylation or acetylation, recruit, organize, and direct downstream binding and function of other molecules, since they have minimal binding affinity to most binding modules or regulatory factors.
On the other hand, modifications likely help maintain a particular chromatin state and stability, or instability by altering nucleosome dynamics. This presumably helps to ensure high or low accessibility of DNA by other factors to influence transcription. Nucleosomes are thought to impede the binding of elements to DNA. Therefore, by maintaining the occupancy and position of nucleosomes, histone modifications contribute to changes in chromatin landscape, being one of many events that alter transcription regulation. This is in contrast to the idea, or overgeneralization perhaps, that histone modifications themselves increase and decrease DNA accessibility and are directly activating and repressing transcription (reviewed in Smith & Shilatifard, 2010). This logic corresponds to the idea that histone modifications are not just creating a simple change of charge between DNA and histones to maintain the overall structure of nucleosomes, but they actually create binding sites for regulatory elements necessary for transcription, with specific location and marks combining to recruit specific regulatory proteins and outcomes (Kouzarides, 2007; Liu et al., 2012; Nakamura et al., 2007).
6.2. What is the relation between memory at the cellular, organismal, and transgenerational levels?
One of the reasons that epigenetic approaches to addiction are potentially so promising is that successful treatment of addiction requires long-term changes in behavior. Epigenetic changes are associated with long-term changes at the cellular and molecular levels, such as cell fate and cellular memory. But what is the relation between cellular memory and long-term memory that is distributed among different neurobiological circuits? What is the relation between memory at the neurobiological level and memory at the transgenerational level? These are key questions that are not yet resolved.
At the level of the cell, histone-mediated epigenetics may create tags that allow the transcriptional machinery to operate on certain DNA sequences. This creates a specific cellular memory that may result in increased rates and levels of transcription and translation the next time the circuit is activated. This cellular memory need not correspond directly to memory on the level of the whole organism because psychological memories are widely distributed and are controlled by many molecular processes. But the cellular memory that is created by histone marks in various neurobiological circuits triggers the downstream events that are needed to solidify and maintain these organismal level memory circuits.
Can a histone-mediated epigenetic memory be transferred across generations? This is a critical question that requires an agreed upon definition of epigenetics. This is often disputed, largely on the basis of whether post-translational changes can be lasting and inherited from cell to daughter cell or parent to offspring (Brumfiel, 2008). While the term genetics implies the necessity of inheritance, less traditional views are incorporating the idea that changes to the transcription and translation of the genetic code may be inherited along with the DNA code itself. Although some studies have demonstrated that histone-mediated alterations in parents can lead to effects in subsequent offspring during development (Hammoud et al., 2009; Heard & Martienssen, 2014) few addiction-related studies have tested if offspring can inherit increased risk or protection from parental modifications due to drug use (Bohacek & Mansuy 2013).
In addition, the definitions of trans- and intergenerational inheritance have been broad. As defined by Heard & Martienssen (2014), intergenerational effects are those inherited in utero through parental effects or stimulus exposure. This is in contrast to epigenetic effects that are inherited generations later, without exposure to the stimuli that epigenetically altered gene expression or function initially, generations earlier. Few studies to date have demonstrated that drug use alters the information inherited by offspring and subsequent brain function (e.g., Vassoler et al., 2011, 2013) and that this may be mediated through histone modifications (Vassoler et al., 2013). In this last study, a cocaine-resistant phenotype (i.e., slower rate of acquisition and decreased motivation to administer high doses of cocaine as measured by a progressive ratio schedule of reinforcement) was inherited by male, but not female offspring of parents with extensive and voluntary use of cocaine. The authors showed that this effect was reliant on H3-mediated increases in acetylation levels on the BDNF promoter, subsequent increases in BDNF mRNA in the medial prefrontal cortex, and BDNF protein expression. A review by the same group emphasized the likelihood of epigenetic inheritance involved in many diseases, but reiterated that further research on the inheritance of post-translational modifications and drug abuse was needed (Vassoler & Sadri-Vakili, 2014).
Evidence for human trans- and intergeneration epigenetic inheritance is still lagging and a key challenge is to demonstrate that experience-dependent changes, such as those outlined in this chapter, can be passed through multiple generations. One study by Norrholm et al. (2013) demonstrated that PTSD patients with an alleged PTSD risk genotype (i.e., Met/Met single nucleotide polymorphism) had greater fear to an experimental safety signal and were unable to extinguish this fear after training compared to PTSD patients without the risk genotype. This Met/Met versus Val/Met or Val/Val single nucleotide polymorphism at the catechol-o-methyltransferase [COMT] gene likely contributes to increased forebrain dopamine levels (Bilder et al., 2004; Matsumoto et al., 2003) in carriers. Accordingly, the Met/Met genotype was associated with greater DNA methylation at CpG sites that were also associated with patients experiencing enhanced fear to safety signals. Another study in rodents demonstrated that fear conditioning in an F0 generation mouse can lead to rapid and specific fear learning in F1 and F2 generation mice (Dias & Ressler, 2014). Studies like these (e.g., Vassoler et al., 2013; Norrholm et al., 2013) will contribute to understanding how epigenetic and DNA based modifications can be inherited and influence health and behavior in subsequent generations.
6.3. What theoretical processes are altered by drugs that target histone modifications during treatment of addiction?
Much of the work reviewed here on histone-mediated epigenetics and addiction points to lasting effects on the levels of the cell and behavior. The most widely demonstrated effects on addiction-related memory processes come from studies showing that administration of HDAC inhibitors can promote memory and synaptic plasticity. When we think of treatments for addiction, however, we are often interested in ways in which memories associated with drugs can be weakened by some pharmacological treatment.
As noted in Section 3.2, when memories are retrieved, there are multiple theoretical processes that may be triggered. The memory itself may become labile, necessitating a post-retrieval reconsolidation process that centers on the original drug-associated memory. However, if the expected drug is not administered, the behavior may begin to show extinction – the absence of expected drug leads to a weakening of behavior through the development of an inhibitory memory. A consistent finding from the literature on histone-mediated epigenetics is that HDAC inhibitors paired with drug-related memory retrieval will decrease drug-seeking behavior and weaken subsequent relapse. The issue at a theoretical level is, why does this occur? The answer to this question has focused on both extinction and reconsolidation, with the distinction between them often boiling down to assumptions that are made about these theoretical processes (Lattal & Wood, 2013).
On one level, behavioral evidence in favor of an impaired reconsolidation account often comes in the form of persistently weakened behavior. Because extinction is often transient, with the response showing spontaneous recovery with time, renewal with context changes, or reestablishment with exposure to drug, any persistent effect is interpreted as an effect on reconsolidation, rather than extinction. Work with HDAC inhibitors, however, challenges this behavioral definition. Because HDAC inhibitors have been shown to promote memory in a variety of behavioral approaches, it is not difficult to imagine how long-term suppression of drug-seeking behavior could result from an enhanced extinction process. Increased histone acetylation and the permissive state of chromatin that it creates should strengthen the consequence of extinction, thereby transforming a potentially weak and transient behavioral experience into one that is long-lasting. Thus, a long-lasting effect on extinction is entirely consistent with an enhanced extinction effect.
On another level, one might examine the molecular processes that are thought to control reconsolidation and extinction. The thought here is that because extinction enhancements involve new memory formation and reconsolidation impairments involve memory elimination, molecular events associated with memory storage and erasure should be uniquely associated with extinction and reconsolidation, respectively. This is a reasonable way to deal with this issue, but the challenge is that we as a field do not really know which molecular processes are specifically associated with memory storage or erasure. Identical molecular evidence has been offered for enhanced extinction and impaired reconsolidation (see Stafford & Lattal 2011) suggesting that the field does not yet have a handle on how these processes underlie memory.
Perhaps the most promising level of analysis is to examine the circuits that are activated by histone-mediated enhancements in extinction. The line of thinking with this approach is that if circuits that regulate extinction are hyperactivated by a drug such as an HDAC inhibitor, then this could be taken as evidence that extinction has been enhanced. Indeed, research from our laboratory has used this approach to identify how extinction circuits are regulated by HDAC inhibitors (Stafford et al., 2012). Again, the challenge here is that the circuits that mediate initial consolidation, reconsolidation, and extinction are highly overlapping and interactive. As more is understood about these circuits (e.g., specific amygdala projections that mediate extinction; neural substrates recruited by different contextual and developmental influences) this approach will become even more powerful in distinguishing between enhanced extinction and impaired reconsolidation processes.
Beyond extinction and reconsolidation, future work on histone-mediated changes in addiction-related behaviors will need to identify other psychological processes that are altered by these mechanisms. For example, behavioral research has identified several critical variables in the establishment and maintenance of drug-seeking behavior. One critical variable is the prediction error between the abused substance and the cues in the environment, with unexpected outcomes (or absences of outcomes in the case of extinction) having a large impact on the effects of the drug itself (Siegel, 1983) and the strength of the cue-drug association (Schultz, 2007). A second variable is the context in which drug seeking occurs – even after long periods of abstinence, exposure to a drug-associated context is enough to trigger relapse of drug seeking. Although there is a great deal known about how these and other behavioral variables influence drug-seeking behavior, very little is known about how epigenetic events contribute to prediction error and contextual modulation of drug-seeking behavior.
7. Considerations for histone-mediated treatment of addiction
Although researchers are starting to understand how histone modifications lead to downstream changes in transcription, there are considerable gaps that still need to be understood before complete treatment can be accomplished. The exact mechanism of modification recruitment, coordination, and differences between diseases is largely unknown. These gaps in knowledge limit the potential for therapeutics to target aberrant modifications with spatial and temporal specificity. For example, most HDAC and HDMT inhibitors affect multiple isoforms and other non-histone proteins with similar activity. This would limit the control that clinicians have over side effects.
At a psychological level, a great deal of attention has been directed toward the idea that histone deacetylase inhibitors may work in a variety of disorders by improving cognitive function. These drugs hold tremendous promise in treatment of any disorder that involves some type of impaired cognitive function. Research from the rodent laboratory has demonstrated that these drugs are often ineffective on their own – they need to be paired with a behavioral experience, such as extinction. When combined with behavioral interventions, these drugs have great potential to promote treatment outcomes (Davis et al., 2006; Kiefer & Dinter, 2013). However, this promise comes with potential peril, as a cognitive enhancing drug could strengthen the impact of an episode of relapse by promoting the transcriptional events associated with the relapse episode. There is therefore a need to administer these drugs under close clinical supervision, during which the clinician has some control over the experiences that the patient has while on the medication.
These basic mechanistic and psychological questions will need to be addressed in future work. What is clear from the literature reviewed in this chapter is that histone modifications play important roles in the development, maintenance, and treatment for addiction. There are many questions that are open and many debates that are unresolved, but there is little doubt that a thorough understanding of the mechanisms of histone modification, transcription, and translation will lead to important scientific discoveries with a high likelihood for clinical translation.
Acknowledgments
Preparation of this chapter was supported by National Institutes of Health grant R01DA025922 (K.M.L.) and T32DA007262 (L.N.H.) and Department of Defense grant W81XWH-12-2-0048 (K.M.L.).
References
- Adenuga D, Yao H, March TH, Seagrave J, Rahman I. Histone deacetylase 2 is phosphorylated, ubiquitinated, and degraded by cigarette smoke. American Journal of Respiratory Cell and Molecular Biology. 2009;40:464–473. doi: 10.1165/rcmb.2008-0255OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Valles A, Vaissière T, Griggs EM, Mikaelsson MA, Takács IF, Young EJ, … Miller CA. Methamphetamine-associated memory is regulated by a writer and an eraser of permissive histone methylation. Biological Psychiatry. 2014;76(1):57–65. doi: 10.1016/j.biopsych.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvaro-Bartolome M, Garcia-Sevilla JA. Dysregulation of cannabinoid CB1 receptor and associated signaling networks in brains of cocaine addicts and cocaine-treated rodents. Neuroscience. 2013;247:294–308. doi: 10.1016/j.neuroscience.2013.05.035. [DOI] [PubMed] [Google Scholar]
- Arora DS, Nimitvilai S, Teppen TL, McElvain Ma, Sakharkar AJ, You C, … Brodie MS. Hyposensitivity to gamma-aminobutyric acid in the ventral tegmental area during alcohol withdrawal: reversal by histone deacetylase inhibitors. Neuropsychopharmacology: Official Publication of the American College of Neuropsychopharmacology. 2013;38(9):1674–84. doi: 10.1038/npp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badiani A, Robinson TE. Drug-induced neurobehavioral plasticity: the role of environmental context. Behavioural Pharmacology. 2004;15:327–339. doi: 10.1097/00008877-200409000-00004. [DOI] [PubMed] [Google Scholar]
- Bahi A, Boyer F, Dreyer JL. Role of accumbens BDNF and TrkB in cocaine-induced psychomotor sensitization, conditioned-place preference, and reinstatement in rats. Psychopharmacology. 2008;199(2):169–182. doi: 10.1007/s00213-008-1164-1. [DOI] [PubMed] [Google Scholar]
- Bernardi RE, Ryabinin AE, Berger SP, Lattal KM. Post-retrieval disruption of a cocaine conditioned place preference by systemic and intrabasolateral amygdala beta2- and alpha1-adrenergic antagonists. Learning & Memory (Cold Spring Harbor, NY) 2009;16:777–789. doi: 10.1101/lm.1648509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilder RM, Volavka J, Lachman HM, Grace AA. The catechol-O-methyltransferase polymorphism: relations to the tonic-phasic dopamine hypothesis and neuropsychiatric phenotypes. Neuropsychopharmacology: Official Publication of the American College of Neuropsychopharmacology. 2004;29(11):1943–61. doi: 10.1038/sj.npp.1300542. [DOI] [PubMed] [Google Scholar]
- Biliński P, Wojtyła A, Kapka-Skrzypczak L, Chwedorowicz R, Cyranka M, Studziński T. Epigenetic regulation in drug addiction. Annals of Agricultural and Environmental Medicine: AAEM. 2012;19(3):491–6. [PubMed] [Google Scholar]
- Bohacek J, Mansuy IM. Epigenetic inheritance of disease and disease risk. Neuropsychopharmacology: Official Publication of the American College of Neuropsychopharmacology. 2013;38(1):220–36. doi: 10.1038/npp.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brami-Cherrier K, Roze E, Girault Ja, Betuing S, Caboche J. Role of the ERK/MSK1 signaling pathway in chromatin remodelling and brain responses to drugs of abuse. J Neurochem. 2009;108(6):1323–35. doi: 10.1111/j.1471-4159.2009.05879.x. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Sobel RE, Allis CD, Turner BM, Broach JR. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Molecular and Cellular Biology. 1996;16(8):4349–56. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DC. Recent and remote extinction cues reduce spontaneous recovery. The Quarterly Journal of Experimental Psychology. B, Comparative and Physiological Psychology. 2000;53:25–58. doi: 10.1080/027249900392986. [DOI] [PubMed] [Google Scholar]
- Brumfiel G. Disputed definitions. Nature. 2008 Oct;455:1023–1028. doi: 10.1038/4551023a. [DOI] [PubMed] [Google Scholar]
- Castellano-Pozo M, Santos-Pereira JM, Rondón AG, Barroso S, Andújar E, Pérez-Alegre M, … Aguilera A. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Molecular Cell. 2013;52(4):583–90. doi: 10.1016/j.molcel.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature Reviews. Genetics. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Chase KA, Sharma RP. Nicotine induces chromatin remodelling through decreases in the methyltransferases GLP, G9a, Setdb1 and levels of H3K9me2. The International Journal of Neuropsychopharmacology. 2012;2:1–10. doi: 10.1017/S1461145712001101. [DOI] [PubMed] [Google Scholar]
- Chavda B, Arnott JA, Planey SL. Targeting protein palmitoylation: selective inhibitors and implications in disease. Expert Opinion on Drug Discovery. 2014:1–15. doi: 10.1517/17460441.2014.933802. [DOI] [PubMed] [Google Scholar]
- Chen F, Kan H, Castranova V. Handbook of Epigenetics. Handbook of Epigenetics. 1. Elsevier; 2011. pp. 149–157. [DOI] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, … Greenberg ME. Science (New York, N.Y.) Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. 2003;302(5646):885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103(2):263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- Clayton aL, Rose S, Barratt MJ, Mahadevan LC. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. The EMBO Journal. 2000;19(14):3714–3726. doi: 10.1093/emboj/19.14.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corominas-Roso M, Roncero C, Eiroa-Orosa FJ, Gonzalvo B, Grau-Lopez L, Ribases M, … Casas M. Brain-derived neurotrophic factor serum levels in cocaine-dependent patients during early abstinence. European Neuropsychopharmacology. 2013;23(9):1078–1084. doi: 10.1016/j.euroneuro.2012.08.016. [DOI] [PubMed] [Google Scholar]
- Covington HE, Maze I, Sun H, Bomze HM, DeMaio KD, Wu EY, … Nestler EJ. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron. 2011;71(4):656–670. doi: 10.1016/j.neuron.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombag HS, Bossert JM, Koya E, Shaham Y. Review. Context-induced relapse to drug seeking: a review. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 2008;363:3233–3243. doi: 10.1098/rstb.2008.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombag HS, Shaham Y. Renewal of drug seeking by contextual cues after prolonged extinction in rats. Behavioral Neuroscience. 2002;116:169–173. doi: 10.1037/0735-7044.116.1.169. [DOI] [PubMed] [Google Scholar]
- Darcy MJ, Trouche S, Jin SX, Feig LA. Ras-GRF2 mediates long-term potentiation, survival, and response to an enriched environment of newborn neurons in the hippocampus. Hippocampus. 2014;13:1–13. doi: 10.1002/hipo.22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M, Myers KM, Chhatwal J, Ressler KJ. Pharmacological treatments that facilitate extinction of fear: relevance to psychotherapy. NeuroRx: The Journal of the American Society for Experimental NeuroTherapeutics. 2006;3(1):82–96. doi: 10.1016/j.nurx.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M, El Ramy R, Quénet D, Wolf P, Spada F, Magroun N, … Dantzer F. Poly(ADP-ribose) polymerase 1 (PARP1) associates with E3 ubiquitine-protein ligase UHRF1 and modulates UHRF1 biological functions. The Journal of Biological Chemistry. 2014;1(23):16223–16238. doi: 10.1074/jbc.M113.527424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias BG, Ressler KJ. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nature neuroscience. 2014;17(1):89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieker J, Muller S. Epigenetic histone code and autoimmunity. Clinical Reviews in Allergy & Immunology. 2010;39(1):78–84. doi: 10.1007/s12016-009-8173-7. [DOI] [PubMed] [Google Scholar]
- Dietrich J-B, Takemori H, Grosch-Dirrig S, Bertorello A, Zwiller J. Cocaine induces the expression of MEF2C transcription factor in rat striatum through activation of SIK1 and phosphorylation of the histone deacetylase HDAC5. Synapse (New York, NY) 2012;66(1):61–70. doi: 10.1002/syn.20988. [DOI] [PubMed] [Google Scholar]
- Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Molecular Cancer Research: MCR. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- Drummond DC, Noble CO, Kirpotin DB, Guo Z, Scott GK, Benz CC. Clinical development of histone deacetylase inhibitors as anticancer agents. Annual Review of Pharmacology and Toxicology. 2005;45:495–528. doi: 10.1146/annurev.pharmtox.45.120403.095825. [DOI] [PubMed] [Google Scholar]
- Fuchikami M, Yamamoto S, Morinobu S, Takei S, Yamawaki S. Epigenetic regulation of BDNF gene in response to stress. Psychiatry Investig. 2010;7(4):251–256. doi: 10.4306/pi.2010.7.4.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould TJ, Leach PT. Cellular, molecular, and genetic substrates underlying the impact of nicotine on learning. Neurobiology of Learning and Memory. 2014;107:108–32. doi: 10.1016/j.nlm.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nature Reviews Genetics. 2012 doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SIS, Jia S. Heterochromatin revisited. Nature Reviews. Genetics. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Gruber AJ, McDonald RJ. Context, emotion, and the strategic pursuit of goals: interactions among multiple brain systems controlling motivated behavior. Frontiers in Behavioral Neuroscience. 2012;6:50. doi: 10.3389/fnbeh.2012.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Martienssen RA. Transgenerational epigenetic inheritance: Myths and mechanisms. Cell. 2014 doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends in Genetics: TIG. 2011;27(10):389–96. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Hyman SE. Addiction: a disease of learning and memory. The American Journal of Psychiatry. 2005;162:1414–1422. doi: 10.1176/appi.ajp.162.8.1414. [DOI] [PubMed] [Google Scholar]
- Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annual Review of Neuroscience. 2006;29:565–598. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science (New York, NY) 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Johansen KM, Johansen J. Regulation of chromatin structure by histone H3S10 phosphorylation. Chromosome Research: An International Journal on the Molecular, Supramolecular and Evolutionary Aspects of Chromosome Biology. 2006;14(4):393–404. doi: 10.1007/s10577-006-1063-4. [DOI] [PubMed] [Google Scholar]
- Kannan P, Yu Y, Wankhade S, Tainsky MA. PolyADP-ribose polymerase is a coactivator for AP-2-mediated transcriptional activation. Nucleic Acids Research. 1999;27(3):866–874. doi: 10.1093/nar/27.3.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy PJ, Feng J, Robison aJ, Maze I, Badimon A, Mouzon E, … Nestler EJ. Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation. Nature Neuroscience. 2013;16(4):434–40. doi: 10.1038/nn.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanal P, Kim G, Lim SC, Yun HJ, Lee KY, Choi HK, Choi HS. Prolyl isomerase Pin1 negatively regulates the stability of SUV39H1 to promote tumorigenesis in breast cancer. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 2013;27(11):4606–18. doi: 10.1096/fj.13-236851. [DOI] [PubMed] [Google Scholar]
- Kiefer F, Dinter C. New approaches to addiction treatment based on learning and memory. Current Topics in Behavioral Neurosciences. 2013;13:671–84. doi: 10.1007/7854_2011_147. [DOI] [PubMed] [Google Scholar]
- Kim JY, Banerjee T, Vinckevicius A, Luo Q, Parker JB, Baker MR, … Chakravarti D. A Role for WDR5 in Integrating Threonine 11 Phosphorylation to Lysine 4 Methylation on Histone H3 during Androgen Signaling and in Prostate Cancer. Molecular Cell. 2014;54:1–13. doi: 10.1016/j.molcel.2014.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology: Official Publication of the American College of Neuropsychopharmacology. 2010;35:217–238. doi: 10.1038/npp.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin Modifications and Their Function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DEH, Truong HT, … Nestler EJ. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48(2):303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Lalumiere RT, Smith KC, Kalivas PW. Neural circuit competition in cocaine-seeking: Roles of the infralimbic cortex and nucleus accumbens shell. European Journal of Neuroscience. 2012;35:614–622. doi: 10.1111/j.1460-9568.2012.07991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattal KM, Radulovic J, Lukowiak K. Extinction: [corrected] does it or doesn’t it? The requirement of altered gene activity and new protein synthesis. Biological Psychiatry. 2006;60:344–351. doi: 10.1016/j.biopsych.2006.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattal KM, Wood MA. Epigenetics and persistent memory: implications for reconsolidation and silent extinction beyond the zero. Nature Neuroscience. 2013;16:124–9. doi: 10.1038/nn.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Smith E, Shilatifard A. The Language of Histone Crosstalk. Cell. 2010;142(5):682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. The Journal of Biological Chemistry. 2004;279(39):40545–59. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Li J, Guo Y, Schroeder FA, Youngs RM, Schmidt TW, Ferris C, … Akbarian S. Dopamine D2-like antagonists induce chromatin remodeling in striatal neurons through cyclic AMP-protein kinase A and NMDA receptor signaling. Journal of Neurochemistry. 2004;90(5):1117–31. doi: 10.1111/j.1471-4159.2004.02569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Lei JX, Luo C, Lan X, Chi L, Deng P, … Liu QY. Increased EID1 nuclear translocation impairs synaptic plasticity and memory function associated with pathogenesis of Alzheimer’s disease. Neurobiol Dis. 2012;45(3):902–912. doi: 10.1016/j.nbd.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl P. Histone acetylation: Facts and questions. Chromosoma. 1994 doi: 10.1007/s004120050053. [DOI] [PubMed] [Google Scholar]
- Lv J, Xin Y, Zhou W, Qiu Z. The epigenetic switches for neural development and psychiatric disorders. Journal of Genetics and Genomics. 2013;40(7):339–346. doi: 10.1016/j.jgg.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Malvaez M, Barrett RM, Wood MA, Sanchis-Segura C. Epigenetic mechanisms underlying extinction of memory and drug-seeking behavior. Mammalian Genome. 2009 doi: 10.1007/s00335-009-9224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvaez M, McQuown SC, Rogge Ga, Astarabadi M, Jacques V, Carreiro S, … Wood MA. HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2647–52. doi: 10.1073/pnas.1213364110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvaez M, Sanchis-Segura C, Vo D, Lattal KM, Wood MA. Modulation of Chromatin Modification Facilitates Extinction of Cocaine-Induced Conditioned Place Preference. Biological Psychiatry. 2010;67:36–43. doi: 10.1016/j.biopsych.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant NJ, Millan EZ, McNally GP. The hypothalamus and the neurobiology of drug seeking. Cellular and Molecular Life Sciences: CMLS. 2012;69(4):581–97. doi: 10.1007/s00018-011-0817-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Weickert CS, Akil M, Lipska BK, Hyde TM, Herman MM, … Weinberger DR. Catechol O-methyltransferase mRNA expression in human and rat brain: evidence for a role in cortical neuronal function. Neuroscience. 2003;116(1):127–137. doi: 10.1016/S0306-4522(02)00556-0. [DOI] [PubMed] [Google Scholar]
- Maze I, Covington HE, Dietz DM, LaPlant Q, Renthal W, Russo SJ, … Nestler EJ. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science (New York, NY) 2010;327(5962):213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Feng J, Wilkinson MB, Sun H, Shen L, Nestler EJ. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(7):3035–3040. doi: 10.1073/pnas.1015483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Nestler EJ. The epigenetic landscape of addiction. Annals of the New York Academy of Sciences. 2011;1216:99–113. doi: 10.1111/j.1749-6632.2010.05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D, Zhu J, Darnell S, Sangrey G, Bhide P, Sadri-Vakili G. In utero cocaine exposure alters behavior, neuronal migration, and BDNF expression in a trans-generational epigenetic study. Washington, DC: Society for Neuroscience; 2011. [Google Scholar]
- McGaugh JL. Memory--a century of consolidation. Science (New York, NY) 2000;287:248–251. doi: 10.1126/science.287.5451.248. [DOI] [PubMed] [Google Scholar]
- McQuown SC, Wood MA. HDAC3 and the molecular brake pad hypothesis. Neurobiology of Learning and Memory. 2011;96:27–34. doi: 10.1016/j.nlm.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozzetta C, Pontis J, Fritsch L, Robin P, Portoso M, Proux C, … Ait-Si-Ali S. The Histone H3 Lysine 9 Methyltransferases G9a and GLP Regulate Polycomb Repressive Complex 2-Mediated Gene Silencing. Molecular Cell. 2014;53(2):277–289. doi: 10.1016/j.molcel.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nature Reviews. Molecular Cell Biology. 2003;4(3):192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Umehara T, Nakano K, Jang MK, Shirouzu M, Morita S, … Yokoyama S. Crystal structure of the human BRD2 bromodomain: insights into dimerization and recognition of acetylated histone H4. The Journal of Biological Chemistry. 2007;282:4193–4201. doi: 10.1074/jbc.M605971200. [DOI] [PubMed] [Google Scholar]
- Nic Dhonnchadha BÁ, Kantak KM. Cognitive enhancers for facilitating drug cue extinction: insights from animal models. Pharmacology, Biochemistry, and Behavior. 2011;99(2):229–44. doi: 10.1016/j.pbb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrholm SD, Jovanovic T, Smith AK, Binder E, Klengel T, Conneely K, … Ressler KJ. Differential Genetic and Epigenetic Regulation of catechol-O-methyltransferase is Associated with Impaired Fear Inhibition in Posttraumatic Stress Disorder. Frontiers in Behavioral Neuroscience. 2013 Apr;7:30. doi: 10.3389/fnbeh.2013.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Ugale R, Zhang H, Tang L, Prakash A. Brain chromatin remodeling: a novel mechanism of alcoholism. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2008;28(14):3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J, Kalivas PW, Quirk GJ. Extinction circuits for fear and addiction overlap in prefrontal cortex. Learning & Memory (Cold Spring Harbor, NY) 2009;16:279–288. doi: 10.1101/lm.1041309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010 Jun;465:721–727. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- Portela A, Esteller M. Epigenetic modifications and human disease. Nature Biotechnology. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- Qiang M, Denny A, Lieu M, Carreon S, Li J. Histone H3K9 modifications are a local chromatin event involved in ethanol-induced neuroadaptation of the NR2B gene. Epigenetics: Official Journal of the DNA Methylation Society. 2011;6(9):1095–104. doi: 10.4161/epi.6.9.16924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybuck JD, McCleery EJ, Cunningham CL, Wood MA, Lattal KM. The histone deacetylase inhibitor sodium butyrate modulates acquisition and extinction of cocaine-induced conditioned place preference. Pharmacology, Biochemistry, and Behavior. 2013;106:109–16. doi: 10.1016/j.pbb.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Carle TL, Maze I, Covington HE, Truong HT, Alibhai I, … Nestler EJ. Delta FosB mediates epigenetic desensitization of the c-fos gene after chronic amphetamine exposure. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2008;28(29):7344–7349. doi: 10.1523/JNEUROSCI.1043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE, Maze I, … Nestler EJ. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62(3):335–48. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE, Xiao G, Kumar A, … Nestler EJ. Histone Deacetylase 5 Epigenetically Controls Behavioral Adaptations to Chronic Emotional Stimuli. Neuron. 2007;56(3):517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Renthal W, Nestler EJ. Histone acetylation in drug addiction. Seminars in Cell & Developmental Biology. 2009;20(4):387–94. doi: 10.1016/j.semcdb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson TE, Berridge KC. The neural basis of drug craving: An incentive-sensitization theory of addiction. Brain Research Reviews. 1993 doi: 10.1016/0165-0173(93)90013-P. [DOI] [PubMed] [Google Scholar]
- Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nature Reviews Neuroscience. 2011 doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romieu P, Host L, Gobaille S, Sandner G, Aunis D, Zwiller J. Histone deacetylase inhibitors decrease cocaine but not sucrose self-administration in rats. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2008;28(38):9342–9348. doi: 10.1523/JNEUROSCI.0379-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar AJ, Tang L, Zhang H, Chen Y, Grayson DR, Pandey SC. Effects of acute ethanol exposure on anxiety measures and epigenetic modi fi ers in the extended amygdala of adolescent rats. International Journal of Neuropsychopharmacology. 2014 doi: 10.1017/S1461145714001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar AJ, Zhang H, Tang L, Shi G, Pandey SC. Histone deacetylases (HDAC)-induced histone modifications in the amygdala: a role in rapid tolerance to the anxiolytic effects of ethanol. Alcoholism, Clinical and Experimental Research. 2012;36(1):61–71. doi: 10.1111/j.1530-0277.2011.01581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles A, Romano A, Freudenthal R. Synaptic NF-kappa B pathway in neuronal plasticity and memory. Journal of Physiology, Paris. 2014;50 doi: 10.1016/j.jphysparis.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NCT, … Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419(6905):407–411. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- Schmitt KC, Reith MEA. Regulation of the dopamine transporter: Aspects relevant to psychostimulant drugs of abuse. Annals of the New York Academy of Sciences. 2010;1187:316–340. doi: 10.1111/j.1749-6632.2009.05148.x. [DOI] [PubMed] [Google Scholar]
- Schroeder Fa, Penta KL, Matevossian A, Jones SR, Konradi C, Tapper AR, Akbarian S. Drug-induced activation of dopamine D(1) receptor signaling and inhibition of class I/II histone deacetylase induce chromatin remodeling in reward circuitry and modulate cocaine-related behaviors. Neuropsychopharmacology: Official Publication of the American College of Neuropsychopharmacology. 2008;33:2981–2992. doi: 10.1038/npp.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz W. Behavioral dopamine signals. Trends in Neurosciences. 2007 doi: 10.1016/j.tins.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Sen N. Epigenetic Regulation of Memory by Acetylation and Methylation of Chromatin: Implications in Neurological Disorders, Aging, and Addiction. Neuromolecular Medicine. 2014 doi: 10.1007/s12017-014-8306-x. [DOI] [PubMed] [Google Scholar]
- Shaham Y, Adamson LK, Grocki S, Corrigall WA. Reinstatement and spontaneous recovery of nicotine seeking in rats. Psychopharmacology. 1997;130:396–403. doi: 10.1007/s002130050256. [DOI] [PubMed] [Google Scholar]
- Shalin SC, Hernandez CM, Dougherty MK, Morrison DK, Sweatt JD. Kinase suppressor of Ras1 compartmentalizes hippocampal signal transduction and subserves synaptic plasticity and memory formation. Neuron. 2006;50(5):765–79. doi: 10.1016/j.neuron.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2009 doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J, Lv ZG, Wang L, Zhou Y, Hui B. Histone H3 phosphoacetylation is critical for heroin-induced place preference. Neuroreport. 2011;22(12):575–580. doi: 10.1097/WNR.0b013e328348e6aa. [DOI] [PubMed] [Google Scholar]
- Shepard BD, Joseph RA, Kannarkat GT, Rutledge TM, Tuma DJ, Tuma PL. Alcohol-induced alterations in hepatic microtubule dynamics can be explained by impaired histone deacetylase 6 function. Hepatology (Baltimore, Md ) 2008;48(5):1671–9. doi: 10.1002/hep.22481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel S. Classical Conditioning, Drug Tolerance, and Drug Dependence. In: Reginald WS, Smart G, Glaser Frederick B, Israel Yedy, Kalant Harold, Popham Robert E, editors. Research Advances in Alcohol and Drug Problems. 7. Boston, MA: Springer US; 1983. pp. 207–246. [DOI] [Google Scholar]
- Sims JK, Magazinnik T, Houston SI, Wu S, Rice JC. In: Epigenetics. Tost J, editor. Norkolk: Horizon Scientific Press; 2008. p. 407. [Google Scholar]
- Smith E, Shilatifard A. The Chromatin Signaling Pathway: Diverse Mechanisms of Recruitment of Histone-Modifying Enzymes and Varied Biological Outcomes. Molecular Cell. 2010 doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford JM, Lattal KM. Is an epigenetic switch the key to persistent extinction? Neurobiology of Learning and Memory. 2011;96:35–40. doi: 10.1016/j.nlm.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford JM, Raybuck JD, Ryabinin AE, Lattal KM. Increasing histone acetylation in the hippocampus-infralimbic network enhances fear extinction. Biological Psychiatry. 2012;72:25–33. doi: 10.1016/j.biopsych.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkman B, Sakharkar AJ, Ph D, Pandey S. Epigenetics — Beyond the genome in Alcoholism. 2012. pp. 293–305. [PMC free article] [PubMed] [Google Scholar]
- Stefanik MT, Kupchik YM, Brown RM, Kalivas PW. Optogenetic evidence that pallidal projections, not nigral projections, from the nucleus accumbens core are necessary for reinstating cocaine seeking. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2013;33(34):13654–62. doi: 10.1523/JNEUROSCI.1570-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens DN, Crombag HS, Duka T. The challenge of studying parallel behaviors in humans and animal models. Current Topics in Behavioral Neurosciences. 2013;13:611–45. doi: 10.1007/7854_2011_133. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000 Jan;403:41– 45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sun H, Maze I, Dietz DM, Scobie KN, Kennedy PJ, Damez-Werno D, … Nestler EJ. Morphine Epigenomically Regulates Behavior through Alterations in Histone H3 Lysine 9 Dimethylation in the Nucleus Accumbens. The Journal of Neuroscience. 2012;32(48):17454–17464. doi: 10.1523/JNEUROSCI.1357-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Wang L, Jiang B, Hui B, Lv Z, Ma L. The effects of sodium butyrate, an inhibitor of histone deacetylase, on the cocaine- and sucrose-maintained self-administration in rats. Neuroscience Letters. 2008;441(1):72–6. doi: 10.1016/j.neulet.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Tipps ME, Raybuck JD, Lattal KM. Substance abuse, memory, and post-traumatic stress disorder. Neurobiology of Learning and Memory. 2014;112C:87–100. doi: 10.1016/j.nlm.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth I, Neumann ID. Animal models of social avoidance and social fear. Cell and Tissue Research. 2013;354(1):107–18. doi: 10.1007/s00441-013-1636-4. [DOI] [PubMed] [Google Scholar]
- Tronson NC, Taylor JR. Addiction: A drug-induced disorder of memory reconsolidation. Current Opinion in Neurobiology. 2013 doi: 10.1016/j.conb.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova NM, Kumar a, Nestler EJ. Histone modifications at gene promoter regions in rat hippocampus after acute and chronic electroconvulsive seizures. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2004;24(24):5603–5610. doi: 10.1523/JNEUROSCI.0589-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Neuroscience. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Tulin A, Chinenov Y, Spradling A. Current Topics in Developmental Biology. Regulation of Chromatin Structure and Gene Activity by Poly(ADP-Ribose) Polymerases. 2003;56:55–83. doi: 10.1016/s0070-2153(03)01007-x. [DOI] [PubMed] [Google Scholar]
- Tye KM, Cone JJ, Schairer WW, Janak PH. Amygdala neural encoding of the absence of reward during extinction. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2010;30:116–125. doi: 10.1523/JNEUROSCI.4240-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassoler FM, Johnson-collins NL, Carini LM, Byrnes EM. Next generation effects of female adolescent morphine exposure: sex-specific alterations in response to acute morphine emerge before puberty. 2011:173–181. doi: 10.1097/FBP.0000000000000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassoler FM, White SL, Schmidt HD, Sadri-Vakili G, Pierce RC. Epigenetic inheritance of a cocaine-resistance phenotype. Nature Neuroscience. 2013;16:42–7. doi: 10.1038/nn.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, … Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JM, Hackanson B, L??bbert M, Jung M. Histone deacetylase (HDAC) inhibitors in recent clinical trials for cancer therapy. Clinical Epigenetics. 2010;1:117–136. doi: 10.1007/s13148-010-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jia S. Degrees make all the difference: the multifunctionality of histone H4 lysine 20 methylation. Epigenetics: Official Journal of the DNA Methylation Society. 2009;4(5):273–276. doi: 10.4161/epi.4.5.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Krishnan HR, Ghezzi A, Yin JCP, Atkinson NS. Drug-induced epigenetic changes produce drug tolerance. PLoS Biology. 2007;5:2342–2353. doi: 10.1371/journal.pbio.0050265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone Ubiquitination: Triggering Gene Activity. Molecular Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nature Reviews. Molecular Cell Biology. 2005 Aug;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- Wilkins BJ, Rall NA, Ostwal Y, Kruitwagen T, Hiragami-Hamada K, Winkler M, … Neumann H. A cascade of histone modifications induces chromatin condensation in mitosis. Science (New York, NY) 2014;343(6166):77–80. doi: 10.1126/science.1244508. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochemical Journal. 2012;428(2):133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise LE, Varvel SA, Selley DE, Wiebelhaus JM, Long KA, Middleton LS, … Lichtman AH. delta(9)-Tetrahydrocannabinol-dependent mice undergoing withdrawal display impaired spatial memory. Psychopharmacology. 2011;217(4):485–94. doi: 10.1007/s00213-011-2305-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Hoshikawa Y, Koseki K, Mori K, Beppu T. Structural specificity for biological activity of trichostatin A, a specific inhibitor of mammalian cell cycle with potent differentiation-inducing activity in Friend leukemia cells. The Journal of Antibiotics. 1990;43(9):1101–1106. doi: 10.7164/antibiotics.43.1101. [DOI] [PubMed] [Google Scholar]
- Zhang H, Gao L, Anandhakumar J, Gross DS. Uncoupling transcription from covalent histone modification. PLoS Genetics. 2014;10:e1004202. doi: 10.1371/journal.pgen.1004202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhong Q. Histone deacetylase inhibitors and cell death. Cell and Molecular Life Sciences. 2014 doi: 10.1007/s00018-014-1656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Griffin K, Mondal N, Parvin JD. Phosphorylation of histone H2A inhibits transcription on chromatin templates. The Journal of Biological Chemistry. 2004;279(21):21866–21872. doi: 10.1074/jbc.M400099200. [DOI] [PubMed] [Google Scholar]
- Zou JY, Crews FT. Release of Neuronal HMGB1 by Ethanol through Decreased HDAC Activity Activates Brain Neuroimmune Signaling. PLoS ONE. 2014;9(2):e87915. doi: 10.1371/journal.pone.0087915. [DOI] [PMC free article] [PubMed] [Google Scholar]