Abstract
Endothelial-to-hematopoietic transition (EHT) is an important stage in definitive hematopoietic development. However, the genetic mechanisms underlying human EHT remains poorly characterized. We performed single cell RNA-seq using 55 hemogenic endothelial cells (HE: CD31+CD144+CD41−CD43−CD45−CD73−RUNX1c+), 47 vascular endothelial cells without hematopoietic potential (non-HE: CD31+CD144+CD41−CD43−CD45−CD73−RUNX1c−), and 35 hematopoietic progenitor cells (HP: CD34+CD43+RUNX1c+) derived from human embryonic stem cells (hESCs). HE and HP were enriched in genes implicated in hemogenic endothelial transcriptional networks, such as ERG, GATA2, and FLI. We found transcriptional overlap between individual HE and HP cells; however, these populations were distinct from non-HE. Further analysis revealed novel biomarkers for human HE/HP cells, including TIMP3, ESAM, RHOJ, and DLL4. Collectively, we demonstrate that hESC-derived HE and HP share a common developmental pathway, while non-HE are more heterogeneous and transcriptionally distinct. Our findings provide a novel strategy to test new genetic targets and optimize the production of definitive hematopoietic cells from human pluripotent stem cells.
Keywords: Single cell RNA-seq, Hemogenic endothelium, Human embryonic stem cells, Human hematopoiesis
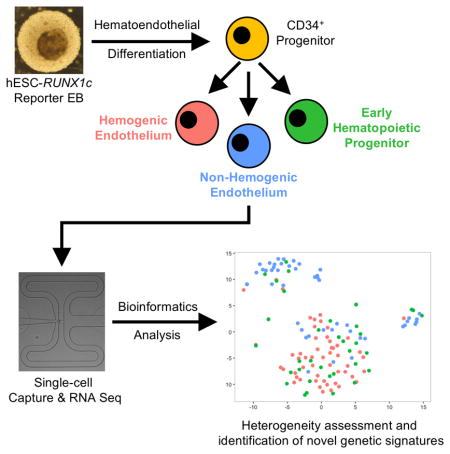
Graphical Abstract
INTRODUCTION
Hemogenic endothelium is a rare and highly specialized subset of vascular endothelial cell that functions as a precursor population to blood development [1–4]. In mammals, hematopoiesis occurs as a series of developmental phases in a defined temporo-spatial manner [5–7]. Hemogenic endothelium is associated with the definitive phase of embryological hematopoiesis, which is characterized by the life-long development of multipotent hematopoietic stem and progenitor cells (HSPCs) [8, 9]. One of the hallmarks of definitive hematopoiesis is the endothelial-to-hematopoietic transition (EHT), a process where hemogenic endothelial cells phenotypically switch from a stationary endothelial cell state to a detached and free-moving HSPC. Although EHT has been visualized both in vitro and in vivo at specific anatomical landmarks via lineage tracing studies [1, 10, 11], the regulation of this fate change at both a cellular and molecular level remains unclear. Confounding this knowledge further is an absence of unique cell surface markers allowing phenotypic identification and isolation of hemogenic endothelium from other developmentally related cell types [12, 13]. Furthermore, specific hemogenic endothelial cell genetic identifiers from humans have yet to be adequately defined.
One candidate identifier of human hemogenic endothelium is RUNX1c, an isoform of the RUNX1 gene. Expression of the +24 intronic enhancer, which drives RUNX1c expression via the P1 promoter, has been shown to be restricted to a subset of endothelial cells where de novo generation of HSPCs occurs in both zebrafish and mouse models [13–17]. RUNX1c expression has also been correlated exclusively to human definitive hematopoietic cells, including CD34+ umbilical cord blood and hematopoietic stem cells [18–24]. Our lab, and others, have previously described human EHT using a human RUNX1c pluripotent stem cell reporter system [22, 23]. As such, RUNX1c can serve as a genetic basis for selecting human hemogenic endothelial cells from other developing endothelial and hematopoietic cell populations.
Human pluripotent stem cells, such as human embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs) serve as a useful platform to understand basic mechanisms underlying human EHT. We, and others, have previously shown differentiation of early hematopoietic progenitor cells from hESC-derived bi-potent endothelial cells capable of developing into cells of the erythroid [24, 25], myeloid [26–29], and lymphoid lineages [30–32]. However, production HSPCs derived from hESCs/hiPSCs that are capable of long-term multi-lineage engraftment has yet to be achieved. One hypothesis is that hESCs are biased toward primitive hematopoietic lineages, and fail to adequately generate hemogenic endothelial cells that produce definitive hematopoietic cells [27, 33–35]. To assess this degree of heterogeneity from an hESC/hiPSCs system, single cell RNA sequencing has emerged as an invaluable tool to discover novel and rare cellular subsets otherwise obscured in bulk RNA-seq experiments [36–39].
In the present study, we utilized hESCs previously engineered to express a RUNX1c-tdTomato reporter construct [22] to investigate the transcriptional signatures of human hemogenic endothelial cells (HE), vascular endothelial cells that lack hematopoietic potential (non-HE), and the earliest definitive hematopoietic progenitor cells (HP). We sequenced and analyzed 137 individual cells and performed comprehensive computational analyses to reveal that endothelial cells derived from hESCs are heterogeneous in nature. Human hemogenic endothelial cells and early hematopoietic progenitors shared similar gene expression signatures, suggesting a common developmental process. Intriguingly, vascular endothelial cells were found to be distinct from HE and HP, with a unique population of cells transformed into a mesenchymal/fibroblast-like cell phenotype. These studies now allow us to define novel genetic biomarkers to distinguish HE from non-HE and serve as a unique strategy to optimize definitive hematopoietic cell differentiation from human pluripotent stem cells.
MATERIALS AND METHODS
Human embryonic stem cell (hESC) culture
hESC-RUNX1c-tdTomato reporter cells were previously engineered and validated in our lab [22]. In brief, a 1 kb fragment of the human RUNX1c P1 promoter and 250 bp conserved intronic region of the +24 enhancer were flanked by tdTomato. Upstream, a constitutively active GFP:zeo fusion protein permitted identification of cells differentiated from hESCs with stable reporter integration. hESC-RUNX1c-tdTomato were TrypLE adapted for single cell culture and maintained on irradiated mouse embryonic fibroblasts (MEF) in ES growth media consisting of Dulbecco’s Modified Eagle’s Media (DMEM)/Ham’s F-12 Media (F12), 15% Knockout Serum Replacement, 1mM L-glutamine, 0.1 mM β-mercaptoethanol, 1% minimum essential medium nonessential amino acids, 1% penicillin-streptomycin (all ThermoFischer Scientific, Carlsbad, CA), and 4 ng/mL basic fibroblast growth factor (R&D Systems, Minneapolis, MN). Cells were cultured in a 37°C air humidified incubator supplemented with 5% CO2 until 70–80% confluency, and passaged using 1x TrypLE Select (ThermoFischer Scientific).
Spin embryoid body (Spin-EB) formation and hemato-endothelial differentiation
Single cell adapted hESCs were harvested with 1x TrypLE Select and aggregated as spin embryoid bodies, as previously described [25, 43]. In brief, hESCs were plated at 3,000 cells/100 μL in a round-bottom 96-well plate using serum-free BPEL media supplemented with 20 ng/mL BMP4, 40 ng/mL SCF, and 20 ng/mL VEGF (all R&D Systems) (Stage I media). Cells were centrifuged for 5 minutes at 1500 rpm to form embryoid bodies (Day 0) and were incubated for 6 additional days to promote mesoderm induction. To differentiate early hemato-endothelial cells, Day 6 spin-EBs were transferred to pre-gelatinized 24-well plates with BPEL media (without polyvinyl alcohol) supplemented with 40 ng/mL SCF, 40 ng/mL VEGF, 30 mg/mL thrombopoietin (R&D Systems), 30 ng/mL IL-3 (PeproTech, Rocky Hill, NJ), and 30 ng/mL IL-6 (PeproTech) (Stage II media). For long-term culture, media was exchanged every 3–4 days with cytokine supplementation. To harvest endothelial cells for analysis, non-adherent (hematopoietic) cell fractions were removed while the remaining adherent fractions were washed and treated with 0.05% trypsin containing 2% chicken serum for 5 minutes. Adherent cells were collected, vortexed, and filtered to generate single cell suspensions suitable for analysis. To harvest hematopoietic cells, only non-adherent cell fractions were harvested and filtered to generate single cell suspensions suitable for analysis.
Fluorescent-Activated Cell Sorting
For cell sorting of hESC-derived hemogenic endothelium (HE) and vascular endothelial cells without hematopoietic potential (non-HE), Day 11 hESC-RUNX1c-tdTomato were differentiated and harvested as described above. Cells were stained with anti-human CD41a-APC (BD Biosciences), CD43-APC (BD Biosciences), CD45-APC (BD Biosciences), CD73-APC (BD Biosciences), CD144-PECy7 (eBioscience), and CD31-APC-eFluor 780 (eBioscience) in sterile FACS Buffer for 30 minutes at 4°C. Cells were washed with FACS buffer and dead cells were counterstained with Live/Dead Fixable Aqua (ThermoFisher Scientific) immediately prior to sorting. Live HE (CD31+CD144+CD41a−CD43−CD45−tdTomato+) and non-HE (CD31+CD144+CD41a−CD43−CD45−tdTomato−) populations were sorted using a FACSAria II (BD Biosciences) directly into BPEL media. Early human hematopoietic progenitor cells (HP) were harvested from a parallel differentiation culture at matched time points. Non-adherent hESC-RUNX1c-tdTomato derived cells were harvested as described above and stained with anti-human CD34-PECy7 (BD Biosciences) and CD43-APC (BD Biosciences) for 30 minutes at 4°C. Cells were washed with FACS buffer and dead cells were counterstained with Sytox Blue (ThermoFisher Scientific) immediately prior to sorting. Live HP (CD34+CD43+tdTomato+) were also sorted on a FACSAria II directly into BPEL media. Post-sort flow cytometry sample validation was performed on all samples.
Single cell capture and RNA-Seq
Sorted HE, non-HE, and HP were stained with a Live/Dead viability/cytotoxicity kit (ThermoFisher Scientific) and resuspended at 50 cells/μL. Cells were captured on three, separate medium-sized (10–17 μm cell diameter) chips using the Fluidigm C1 Single Cell Auto Prep System (Fluidigm, San Diego, CA) per the manufacturer protocol. Following capture, cells were visualized using phase-contrast and fluorescent microscopy using a Nikon Inverted Ti-E Deconvolution motorized Microscope (Nikon, Belmont, CA). At this point, only live, single, and GFP+ cells were selected for cDNA library preparation. Additionally, for HE and HP sorted cells, tdTomato expression was also verified; cells absent for tdTomato signal were excluded. Following capture and validation, cDNA was prepared from each cell using the SMARTer Ultra Low RNA kit for the Fluidigm C1 system, according to the manufacturer recommendations (Clontech, Mountain View, CA). cDNA library concentrations were assessed using Quant-iT PicoGreen dsDNA assay Kit (ThermoFischer Scientific). cDNA concentrations that were less than 25 ng/μL were further excluded from subsequent sequencing. mRNA libraries were transcribed using the Illumina Nextera XT preparation kit (Illumina, San Diego, CA) according to the manufacturer protocol and sequenced on an Illumina HiSeq 2500. Sequencing was performed in rapid-mode using 50 bp paired-end reads to a depth of approximately 1.8×106 reads/sample. In parallel from the same differentiation as single cell captures, bulk population RNA controls were also similarly processed and included in subsequent analyses.
Bioinformatic analyses of single cell RNA-Seq data
Illumina reads were processed via a pipeline developed by the University of Minnesota Informatics Institute in collaboration with the Minnesota Supercomputing Institute and the University of Minnesota Genomics Center. Briefly, FastQ files were trimmed using Trimmomatic (Usadel Lab, Max Planck Institute, Germany; parameters used: -phred33 -threads 8, ILLUMINACLIP, LEADING:3 TRAILING:3 SLIDINGWINDOW:4:16 MINLEN:25). After trimming, mapping to the human genome was performed via TopHat (v. 2.0.13, Johns Hopkins University) and Bowtie (v. 2.2.4.0, Johns Hopkins University). Fraction per kilobases per million (FPKM) genes mapped expression was calculated via cuffquant function in Cufflinks (Trapnell Lab, University of Washington). Normalized log-transformed FPKM values were analyzed using the Seurat R toolkit developed by the Satija Lab (https://github.com/satijalab/seurat; New York Genome Center, NYU), which has been extensively published elsewhere [42–45]. Principal component analysis (PCA) and t-distributed stochastic neighbor embedding (t-SNE) analysis were performed in parallel with two gene lists in Seurat; one list of the total genome (“Total”) mapped transcripts (26,257 genes) and another restricted list of blood and endothelial (“Blood & EC”; 2,556 genes) gene subsets. The Blood & EC gene list was exported from a list of genes tagged to “hematopoiesis” and “endothelial” categorizers within Ingenuity Pathway Analysis (IPA) software (Qiagen, Valencia, CA). For t-SNE analyses, only statistically significant principal components (defined as p<0.05, see Supplemental Figure 4) for each gene list were used as function input. Differentially expressed genes between cell populations and clusters were assessed using “ROC” and “t-test” functions in the Seurat package using the default settings. Isoform level quantification of RUNX1c was mapped using Salmon (Patro Lab, Stony Brook University) [46]. FPKM values were averaged between HE and non-HE groups and compared to assess RUNX1c enrichment. Gene ontology enrichment analysis of the total mapped genes between HE and non-HE was performed using IPA.
Additional Materials and Methods
Immunofluorescent imaging, flow cytometry, post-sort HE and non-HE culture conditions, and statistical methods can be found in the Supplemental Methods.
RESULTS
hESC-RUNX1c-tdTomato cells differentiate into early hemato-endothelial cells in defined xenogenic-free culture conditions
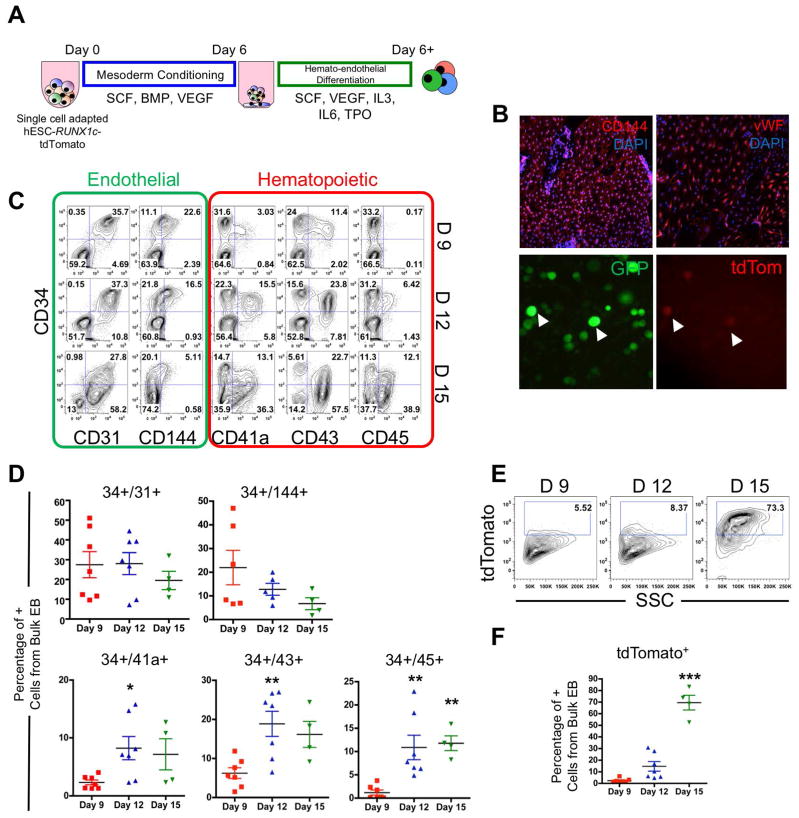
We first assessed the kinetics of hESC-RUNX1c-tdTomato differentiation into early hemato-endothelial cells using defined culture conditions (Figure 1A). Here, we expanded on our previously published data to better identify the emergence of endothelial progenitor cells [22]. As previously demonstrated, hESC-RUNX1c-tdTomato cells differentiated into adherent cells with endothelial characteristics as early as Day 9 of culture. Endothelial cells propagated around the perimeter of each embryoid body, and were positive for two endothelial cell markers: CD144 (VE-Cadherin) and von Willebrand Factor (vWF) (Figure 1B, top panels). At Day 12 of differentiation, rounded, non-adherent hematopoietic progenitor cells could be seen (Figure 1B, bottom panels). These hematopoietic cells all expressed a constitutively active GFP:zeo fusion protein, while a fraction of the them also dually expressed tdTomato, suggesting these cells are of the definitive hematopoietic lineage and developed from EHT.
Figure 1. hESC-RUNX1c-tdTomato cells can model the human endothelial-to-hematopoietic transition (EHT) in vitro.
(A) Schema of hemato-endothelial differentiation from hESC-RUNX1c-tdTomato cells as spin embryoid bodies (spin-EB). (B) Top; Representative immunofluorescent images of hESC-RUNX1c-tdTomato at Day 9 of differentiation for endothelial specific surface antigens CD144 and vWF (red). Original magnification is 40x. Nuclei are counterstained with DAPI (blue). Representative immunofluorescent images of hESC-RUNX1c-tdTomato at Day 12 of differentiation demonstrating development of non-adherent, dual constitutive GFP+ (green) and tdTomato+ (red) reporter hematopoietic cells from spin-EBs (bottom panels). Magnification 200x. (C) Representative flow cytometry plots of endothelial (CD31, CD144) and early hematopoietic surface antigens (CD41a, CD43, CD45) at Day 9 (D9), Day 12 (D12) and Day 15 (D 15) time points over the course of hESC-RUNX1c-tdTomato differentiation. (D) Quantification of flow cytometry as shown in panel C. *p<0.05, **p<0.01 as compared to Day 9 assessed by one-way ANOVA + Tukey-Kramer post-hoc test. Each data point represents a distinct hESC-RUNX1c-tdTomato differentiation and errors bars represent SEM; n=4–7 (E) Representative flow cytometry plots of tdTomato (RUNX1c) expression at Day 9 (D9), Day 12 (D12) and Day 15 (D 15) time points over the course of hESC-RUNX1c-tdTomato differentiation. (F) Quantification of flow cytometry as shown in panel E. ***p<0.001 as compared to Day 9 assessed by one-way ANOVA + Tukey-Kramer post-hoc test, error bars represent SEM; n=4–7 independent, biological replicates.
We next quantified the development of endothelial and hematopoietic cells. At Day 9 of culture, a majority of the differentiated cells were endothelial cells, defined as CD34+CD31+ (27.5%±6.6) and CD34+CD144+ (22.0%±7.3) with limited numbers of hematopoietic progenitor cells, defined by CD34+CD41a+ (2.3%±0.4), CD34+CD43+ (6.3%±1.4), and CD34+CD45+ (1.2%±0.6) (Figures 1C & 1D). By Day 12, the endothelial cell populations declined, accompanied by a reciprocal increase in hematopoietic progenitor cells. By Day 15, a majority of cells were hematopoietic, with statically significant gains in the percentage of CD34+CD41a+ (7.16%±2.7, p<0.05), CD34+CD43+ (16.17%±3.3, p<0.01), and CD34+CD45+ (11.8%±1.6, p<0.01) phenotypes. To parallel hematopoietic development, tdTomato (RUNX1c) was detected in appreciable quantity at Day 12 (14.7%±4.1), with a majority of the cells later expressing RUNX1c after blood development occurred at Day 15 (69.5%±6.4, p<0.01) (Figures 1E & 1F). As such, hESC-RUNX1c-tdTomato cells serve as an excellent platform to allow us to isolate defined endothelial cell populations that are suitable for single cell genetic analyses.
Combined endothelial cell surface antigen and RUNX1c expression delineate human hemogenic endothelium from vascular endothelium lacking hematopoietic potential
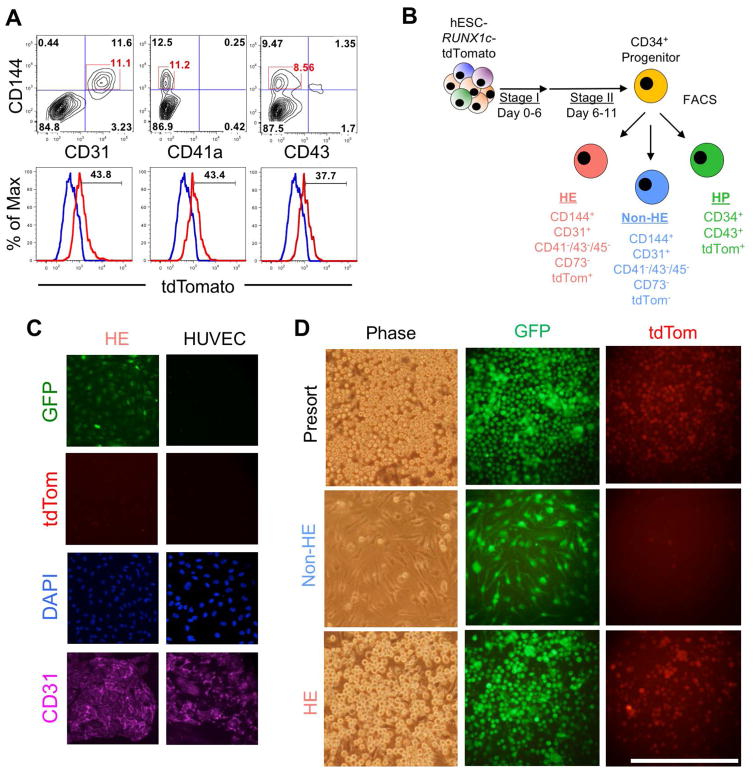
We next assessed for the presence of human hemogenic endothelium from differentiating hESC-RUNX1c-tdTomato cells by using a combination of endothelial cell surface markers and tdTomato+ expression. Because Day 11 was just prior to the onset of detectable RUNX1c hematopoietic cells, we characterized adherent hESC-derived cells at this time point. Here, approximately 10% of the total cells were CD144+CD31+ and negative for CD41a and CD43 expression (Figure 2A, top panels). When sub-gating on these populations, we found approximately 40% of the cells were dually tdTomato+, suggestive of a hemogenic endothelium phenotype (Figure 2A, bottom panels). We next used FACS to sort three populations: 1) putative hemogenic endothelial cells (HE; defined as CD31+CD144+CD41−CD43−CD45−CD73−tdTomato+); 2) vascular endothelial cells lacking hematopoietic potential (non-HE; defined as CD31+CD144+CD41−CD43−CD45−CD73−tdTomato−) and 3) early hematopoietic progenitor cells (HP; defined as CD34+CD43+tdTomato+), and further assessed their phenotypic responses in both endothelial cell and hematopoietic cell culture conditions (Figure 2B and Supplemental Figure 1). Here, we demonstrate HE cells retain endothelial morphology in the absence of pro-hematopoietic growth conditions. hESC-derived HE seeded onto fibronectin coated wells in endothelial growth media were able to generate a confluent, cobblestone monolayer that fully expressed CD31 (PECAM1) at the cellular junctions (Figure 2C). The morphological and phenotypic appearance was similar to that of control human umbilical vein endothelial cells (HUVEC). We next assessed whether HE and/or non-HE would generate tdTomato+ hematopoietic cells in pro-hematopoietic culture conditions. In the span of two days, HE robustly produced non-adherent, tdTomato+ cells similar to pre-sorted cells from the same hESC differentiation (Figure 2D). Additionally, non-HE cells failed to produce tdTomato+ cells, as these cells remained adherent and retained an endothelial-like morphology. Taken together, these results demonstrate that phenotypic HE and non-HE can be sorted and distinguished from hESCs based on this combination of endothelial surface antigen and RUNX1c expression.
Figure 2. Functional human hemogenic endothelium can be phenotypically identified and sorted from hESC-RUNX1c-tdTomato cells.
(A) Representative flow cytometry plots of differentiating hESC-RUNX1c-tdTomato cells at Day 11 expressing typical endothelial surface antigens (CD144+CD31+), but absent for early hematopoietic surface antigens (CD144+CD41a−, CD144+CD43−). Phenotypic endothelial cells were gated (top panel) and assessed for tdTomato expression (bottom panel). A fraction of endothelial cells were tdTomatodim (red histogram) as compared to control hESCs lacking the GFP-RUNX1c-tdTomato reporter (blue histogram), consistent with the identification of hemogenic endothelium. n=2 independent, biological replicates. (B) Schema of FACS sorting hESC-RUNX1c-tdTomato cells into hemogenic endothelium (HE), endothelial cells that lack hematopoietic potential (non-HE), and early hematopoietic progenitor cells (HP). (C) Sorted HE cells seeded on fibronectin-coated wells with EGM-2 media for 5 days and subsequently immunofluorescently imaged for CD31 (left column). HE stained positive for CD31 (purple), GFP (green), and were tdTomatodim (red) while retaining a stereotypical cobblestone morphology of traditional endothelial cells. Untransfected human umbilical vein endothelial cells (HUVECs) are shown as an endogenous control (right column). (D) Presorted hESC-RUNX1c-tdTomato-derived adherent cells, sorted HE, and sorted non-HE were plated in Stage II hemato-endothelial differentiation media for two additional days to promote hematopoiesis. Both Presort and HE populations robustly generated non-adherent GFP+tdTomato+ hematopoietic progenitor cells, while non-HE did not adequately support hematopoietic development. Underlying non-HE cells further retain their endothelial morphology. Bar=100μm.
Single cell RNA-seq reveals similarities in HE and HP gene signatures, but both are transcriptionally distinct from non-HE
Using characterized hESC-derived HE, non-HE, and HP cell populations, we next defined each cell population on a single cell transcriptional level. To do so, we performed single cell RNA-seq to assess individual cells and determine the developmental similarities of each population. We used the Fluidigm C1 Single Cell Autoprep system in conjunction with Illumina Next Generation Sequencing to analyze 55 HE, 47 non-HE, and 35 HP single cells (Supplemental Figure 2A & 2B). To ensure correct identification of single cells, we validated that individual HE and HP cells captured were tdTomato+ within the microfluidic capture chamber, while non-HE cells were tdTomato− (Supplemental Figure 2A). We performed sequencing to a depth of approximately 1.8×106 reads/cell to further verify enrichment of RUNX1c at the isoform level and provide a complete gene expression profile for each individual cell. We first confirmed the representativeness of single cell transcripts to the expression level of the bulk cell populations. As a function of fraction per kilobase per million (FPKM) mapped fragments, we found a strong correlation between gene expression averaged across individual cells as compared to the bulk populations (R2=0.90) (Supplemental Figure 2C). While we did not see statistically significant differences in the expression of all combined transcript variants of the RUNX1 gene across populations, we confirmed HE possessed higher log-transformed FPKM expression of the RUNX1c specific isoform compared to non-HE (9.23±2.61 vs. 1.22±0.67, p<0.01) (Supplemental Figure 2D). We next assessed the ontological pathways significantly enriched between hESC-derived HE and non-HE using Ingenuity Pathway Analysis software. Several of the highest statistically significant ontological pathways were specific to embryological development and hematopoiesis, including “Hematological System Development and Function” (log p<10−15), “Tissue Development” (log p<10−8), “Embryonic Development” (log p<10−7), and “Hematopoiesis” (log p<10−6; Supplemental Figure 2E).
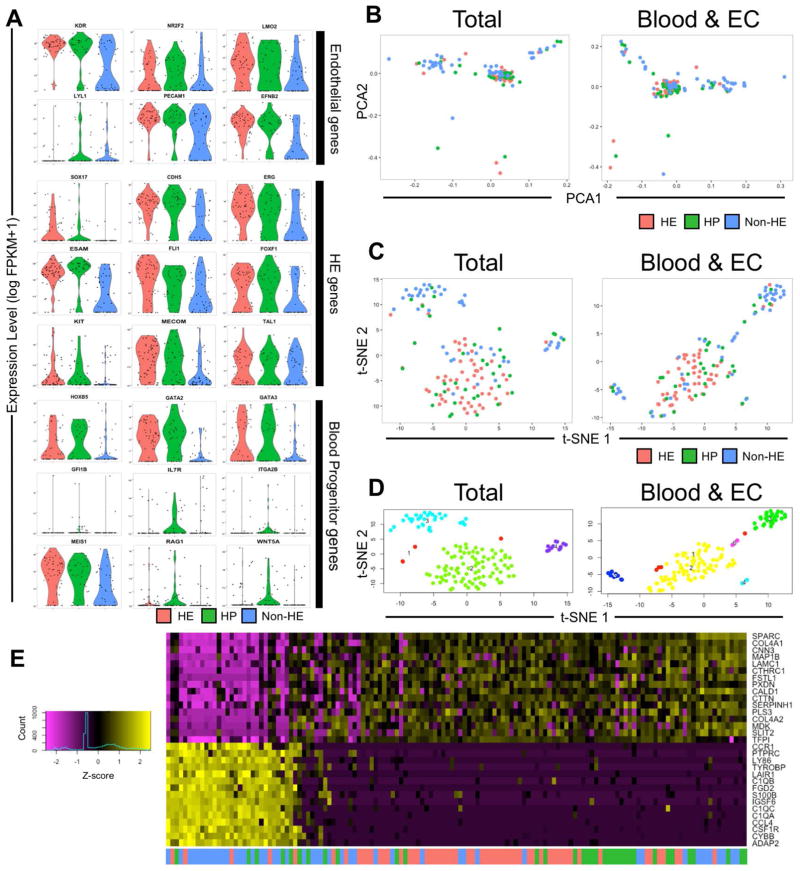
By generating violin plots of log-transformed FPKM values, we next analyzed the expression of key genes known to be associated with vascular endothelial cells, hemogenic endothelium, and hematopoietic progenitor cell phenotypes. These plots provide complementary data regarding total gene expression and the frequency of expression across individual cells in a given population. We found that both HE and HP possessed similar expression and distribution of several endothelial and hemogenic endothelial genes that was higher than those of non-HE. For example, KDR, NR2F2, LMO2, PECAM1, and EFNB2 were significantly increased vascular endothelial cell-related genes in the HE and HP groups as compared to the non-HE group (Figure 3A). SOX17, CDH5, ERG, ESAM, FLI1, FOXF1, KIT, and MECOM were also significantly increased hemogenic endothelium-related genes in HE and HP as compared to non-HE. We also observed HP cells to have overexpression of key hematopoietic genes as compared to HE and non-HE, including IL7R, ITGA2B, RAG1, and WNT5A, while some genes such as HOXB5, GATA2, GATA3, and MEIS1 were also overexpressed in HE. We further confirmed that genes associated with cardiac development and terminal erythroid, myeloid, and lymphoid cell differentiation were not appreciably expressed, while endogenous housekeeper genes, such as GAPDH, ACTB, and GPI possessed uniform expression across all three populations (Supplemental Figure 3). Genes characteristically associated with pluripotency (POU5F1/OCT4, NANOG, DPPA2), primitive streak development (MIXL1, T), lateral plate mesoderm (HAND1, IRX3), ectoderm (PAX6, SOX1, SOX10), and endoderm (FOXA1A, GATA4) did not yield any detectable FPKM expression (data not shown).
Figure 3. Single-Cell RNASeq of hESC-RUNX1c-tdTomato hemato-endothelial cells reveals distinct transcriptional networks between HE and non-HE.
(A) Distribution of the normalized fraction per kilobases per million (FPKM) reads mapped for characteristic endothelial, hemogenic endothelium, and hematopoietic genes from HE, non-HE, and HP cells. Values along the vertical axis represents the normalized log-transformed FPKM expression of each single cell sequenced, while the width of the violin indicates the frequency of cells at a particular FPKM level. (B) Principal component projections of the first and second statistically significant principal components for all 137 cells sequenced. PCA was performed using the entire mapped human genome sequence (Total; left panel) as well as a restricted gene list specific for known endothelial and hematopoietic genes (Blood & EC; right panel). Each data point represents the dimensionally reduced gene expression data for a single cell. (C) t-distributed stochastic neighbor embedding (t-SNE) plots of all 137 cells for Total (left) and Blood & EC (right) gene sets. (D) Statistically distinct clusters that originated from t-SNE dimensionality reduction shown in Panel C were computationally labelled and reclassified as belonging to a transcriptionally distinct population. Cluster 1 (red dots) are cells that failed to orient into one of the defined clusters (Cluster 2: green, Cluster 3: cyan, or Cluster 4: purple for the Total gene set; Cluster 2: yellow, Cluster 3: green, Cluster 4: cyan, or Cluster 5: blue for the Blood & EC gene set). (E) Heat map of Total gene expression defined within projected and statistically significant principal components (see Supplemental Figure 2–5A). Horizontal rows represent z-score expression of normalized log-transformed FPKM values while vertical columns represent each single cell.
We next sought to investigate wide-scale transcriptional profile similarities between HE, non-HE, and HP populations. Using Seurat (see Materials and Methods for details), we performed principal component analysis to reduce the dimensionality of FPKM expression values using: 1) the total mapped gene list (26,257 genes; “Total”) and 2) a hematopoietic and endothelial restricted gene list (2,556 genes; “Blood & EC”). In both analyses, we found at least 3 distinct groups of cells when plotted against the first two most statistically significant principal components (Figure 3B). In either analysis, many of the HE and HP single cells overlapped within a common principal component area, while two clusters primarily composed of non-HE cells were distinctly located in separate areas. We further resolved the composition of these clusters by performing t-distributed stochastic neighbor embedding (t-SNE) analysis using genes assigned only to statistically significant principal components for both the Total (Supplemental Figures 4A & 5A) and Blood & EC (Supplemental Figures 4B & 5B) gene lists. Using this approach, clear resolution of one, centralized cluster of transcriptionally similar HE and HP populations could be ascertained, while a majority of the non-HE cells were segregated into two divergent clusters (Figure 3C). In applying Seurat’s density parameter clustering functions, three main clusters (Clusters 2, 3, and 4, with Cluster 1 representing statistical outlier single cells) were identified using the total gene list (Figure 3D). A similar identification was seen in the Blood & EC gene lists in marking Clusters 2, 3, and 5 (note: Cluster 1 represents statistical outlier single cells and Clusters 4 and 6 are comprised of ≤3 single cells). In analyzing the Total gene list, we observed Cluster 2 was mostly heterogeneous for HE (55.8%) and HP (27.91%), with few non-HE cells (16.28%) (Supplemental Figure 6). Clusters 3 and 4 were mainly composed of non-HE (75.76% and 69.23%, respectively). We further validated separation of non-HE from HE and HP by generating heat maps using unsupervised clustering of genes found within the statistically significant principal components (Figure 3E). Taken together, these data demonstrate that individual HE and HP are transcriptionally related to one another, while non-HE are heterogeneous and transcriptionally distinct from both HE and HP.
Clustering analysis identifies novel biomarkers of hESC-derived HE and HP with divergent developmental pathways of non-HE
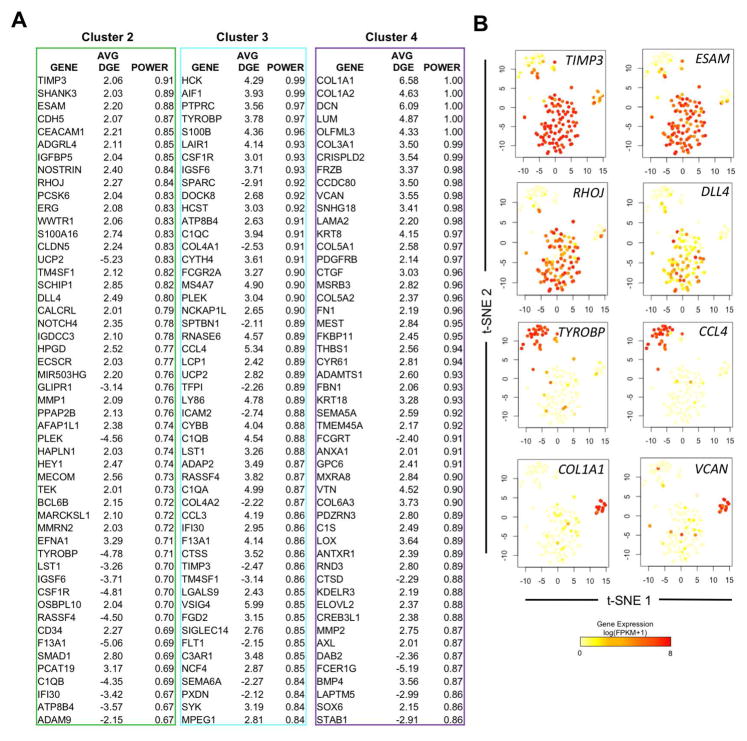
Using individual cells re-classified into transcriptionally distinct groups from the Total gene list, we identified distinguishing biomarkers between HE/HP single cells and the two subsets of non-ECs by performing receiver operating characteristic (ROC) curve analysis in Seurat. ROC analysis is a non-parametric method that is useful in providing the probability (power) that a gene is up- or down-regulated within given group [47]. Cluster 2 yielded 651 statistically significant identifier genes, with the most powerful predictors being previously known hemogenic endothelium related genes such as CDH5 (DGE: 2.07) [48], ERG (DGE: 2.08) [49], CLDN5 (DGE: 2.07) [50], and TEK (DGE: 2.01) [51] (Figure 4A). Cluster 3 and Cluster 4 yielded 916 and 928 statistically significant identifier genes, respectively. Here, we report the top 50 distinguishing genes between groups of cells assigned to Clusters 2, 3, and 4. We next confirmed the specificity of novel biomarkers to each cluster by mapping the gene expression to individual cells on the Total gene list t-SNE plots as shown in Figure 3C. We identified at least four novel gene markers with increased and specific expression to Cluster 2: TIMP3 (DGE: 2.06), ESAM (DGE: 2.20), RHOJ (DGE: 2.27) and DLL4 (DGE: 2.49) (Figure 4B). We also show that TYROBP (DGE: 3.78) and CCL4 (DGE: 5.34) are specific in designating the Cluster 3 subset, which was largely composed of non-HE. Interestingly, Cluster 4 was mostly comprised of genes that encode extracellular matrix protein products. We found significant upregulation of COL1A1 (DGE: 6.58), COL1A2 (DGE: 4.63), DCN (DGE: 6.09), LUM (DGE: 4.87), COL3A1 (DGE: 3.50), VCAN (DGE: 3.55), and several other extracellular matrix components/genes. This would suggest cells of this cluster transformed from an endothelial phenotype into a mesenchymal or fibroblast phenotype. To further validate the uniqueness of each cluster set, we generated a new heat map of gene expression across individual cells using a more stringent approach. We restricted the list of genes to differentially regulated genes with high discriminatory power (Power > 0.80) within each cluster (Supplemental Figure 7). This approach indeed generated clear distinctions in global gene expression that was seen between individual cells assigned to each cluster. Collectively, our approach has established unique genetic biomarkers between hESC-derived HE and non-HE cells, and suggests a subset of non-HE may represent cells transitioning into other mesenchymal lineages.
Figure 4. hESC-RUNX1c-tdTomato-derived non-HE are heterogeneous and can be distinguished from HE and HP using defined gene signatures.
(A) Lists of the top 50 differentially expressed genes that distinguish cells assigned in Cluster 2, 3, and 4 as shown in Figure 3D from the Total gene set. Gene screening was assessed by ROC analysis, with the average differential gene expression (Avg. DGE) and ROC classification power (0=random, 1=perfect correlation) for each gene listed; genes are ranked by their cluster distinguishing potential. (B) Gene expression superimposed onto Total t-SNE plots (Figure 3D) to reflect uniqueness to a cluster subset. Novel identifying biomarkers, such as TIMP3 and ESAM possess high expression within cells of Cluster 2 (mainly HE/HP), while TYROBP and CCL4 are uniquely expressed in Cluster 3 (non-HE), and ECM genes such as COL1A1 and VCAN are expressed in a distinct subset of non-HE cells within Cluster 4.
DISCUSSION
In the present study, we utilized hESCs harboring a RUNX1c-tdTomato reporter to assess the phenotypic and transcriptional profiles of the earliest definitive hemato-endothelial cells at a single cell resolution. In conjunction with endothelial-cell immunophenotyping and RUNX1c expression, we identified hESC-derived HE that not only maintained vascular morphology in endothelial culture conditions, but also differentiated RUNX1c+, non-adherent, hematopoietic progenitors when cultured in the presence of hematopoietic cytokines. Furthermore, through next generation sequencing of individual hemogenic endothelial cells, vascular endothelial cells lacking hematopoietic potential, and the earliest definitive hematopoietic progenitor cells, we determined that hESC-derived HE and HP are transcriptionally similar to each other, but distinct from a heterogeneous population of non-HE. We also identify several novel candidate genes that serve to distinguish HE from non-HE populations.
To date, there have been few reports fully characterizing the complete development of undifferentiated hESCs/hiPSCs through hemogenic endothelium to the generation of HSPCs. Many reports have relied on cell immunophenotyping alone, which has been useful in elucidating key differences between human endothelium and the first formed hematopoietic cells. For example, Nakajima-Takagi et al. [52] separated endothelial cells from the earliest hematopoietic progenitor cells (termed “pre-HPCs”) simply based upon CD34+CD43− and CD34+CD43+CD45−/lo. With this strategy, they discovered SOX17 was overexpressed in human hemogenic endothelium as compared to pre-HPCs and matured hematopoietic cells, and thus a key regulator of hematopoietic development. Indeed, we observed similar overexpression of SOX17 in a majority of cells within our HE population as compared to the HP population. We now provide complementary data that SOX17 is not expressed in endothelial cells lacking hematopoietic potential. Our finding parallels studies in the mouse model, in which knockout of SOX17 abolished the definitive hematopoietic program (loss of T-lymphocyte potential), but did not alter expression levels of EphrinB2 (human homolog: EFNB2) or Coup-TfII (human homolog: NR2F2), genes that are associated with arterial and venous endothelium specification, respectively [53, 54].
Choi et al. [26] and Raffi et al. [10] both used more specific hemato-endothelial phenotypes, relying on CD31+, CD34+, CD117+, CD144+, and CD73− to detect endothelial cells and CD41a+, CD43+, CD235a+ to monitor hematopoietic development. While both studies provide convincing evidence for a precursor endothelial population that directly supports hematopoietic development, it is unclear as to which phase of hematopoiesis (primitive vs. definitive) these earliest cells arise from. Primitive hematopoiesis is thought to primarily originate from hemangioblasts, which are bi-potent cells able to differentiate only into embryonic vasculature cells or transient erythroid/myeloid cells. Hemangioblasts are almost identical in their endothelial surface antigen expression as hemogenic endothelium [55–59]. As such, our described system that is co-dependent on endothelial positive-selection (CD31+CD144+CD73−) and RUNX1c expression provides more precise specification of the definitive hematopoietic program leading to a more resolved understanding of the events preceding HSPC development. Indeed, a related approach has recently been verified in an elegant study by Ditadi, et al [23]. Here, human hESC-derived hemogenic endothelium restricted to an adherent CD34+CD73−CD184−RUNX1c+ population were able to generate RUNX1c+ hematopoietic cells with T-lymphocyte potential. Furthermore, combinations of CD184 (CXCR4), CD73, and RUNX1c could be used as molecular labels in distinguishing the acquisition of either an arterial or venous phenotype from vascular endothelial progenitor cells. Moving forward, it is clear both cell surface and genetic identifiers must be used to adequately define and resolve specialized subsets from a heterogeneous collection of differentiating hemato-endothelial cells.
Our single cell RNA-Seq experiments revealed several novel candidate markers that were specific to human hemogenic endothelium and definitive hematopoietic progenitor cells. TIMP3, ESAM, RHOJ, and DLL4 were found to possess some of the highest discriminatory powers for distinguishing HE and HP from non-HE (Figure 4B). TIMP3 (tissue inhibitor of metalloproteinase-3), while not previously implicated in developmental hematopoiesis, has previously been shown to play an important role in HSPC proliferation by recruiting quiescent hematopoietic stem cells into the cell cycle [60, 61]. ESAM (endothelial cell selective adhesion molecule) has been identified (albeit in the mouse) as a reliable marker for long-term repopulating hematopoietic stem cells derived from the aorta-gonad-mesonephros region that also possessed lymphoid potential [62, 63]. Moreover, ESAM expression is maintained throughout the transition of hematopoiesis to the bone marrow and its expression is increased during the aging process. Because ESAM was highly expressed in almost every HE and HP single cell we analyzed (Figure 3A), it represents a potential candidate for hemogenic endothelial cell selection and/or overexpression studies to engineer hematopoietic progenitors from hESCs with hemato-lymphoid potential. RHOJ encodes a Rho GTPase that is normally restricted to vascular endothelial cells. RHOJ expression, as well as other key hemogenic endothelial proteins, such as VE-cadherin, endoglin, and vWF, are regulated upstream by ERG, which is further implicated in hESC differentiation into endothelial cells [49, 64–66]. We observed overexpression of both RHOJ and ERG in our HE and HP populations. Interestingly, hyperactivation of RhoJ enhances endothelial cell focal adhesion disassembly and increases the mobility of endothelial cells [67]. Thus, a similar mechanism may be crucial during EHT to convert adherent endothelial cells into non-adherent and free-moving HSPCs. Lastly, EHT and definitive hematopoiesis have been hypothesized to be evolutionary Notch-dependent [23, 68–70]. DLL4 is a downstream mediator of VEGF activity and induces the upregulation of Notch1 and its effector targets. DLL4 is highly enriched in the vascular niche and has recently been shown to support the production of partially engraftable CD34+ cells derived from non-human primate induced pluripotent stem cells [71]. Consequently, enrichment of DLL4 may serve as a useful method for identifying hemogenic endothelial cells derived from hESCs.
We also looked at other key transcriptional regulators of human EHT that have been recently reported from bulk cell sequencing approaches. Dou et al. [72] provided insight that medial HOXA genes are silenced in hESC-derived hemato-endothelial cells, but were critical for fetal-liver HSPC expansion and maintenance. In corresponding fashion, we found no appreciable expression of any HOXA locus genes in our hESC-derived hemato-endothelial cell populations, which is also consistent with our prior analyses [22] (Supplemental Figure 8A). Interestingly, we observed significant increases in HOXB locus gene expression between hESC-derived HE/HP and non-HE populations, specifically in HOXB3, HOXB5, HOXB6, and to an extent, HOXB7 (Supplemental Figure 8B). HOXB5 has been recently described (albeit in mouse) to be expressed during EHT and defines HSCs with long-term engraftment potential [73]. As such, HOXB genes may be of a higher biological importance in conferring hemogenic potential from hESC/hiPSC-derived hemogenic endothelium.
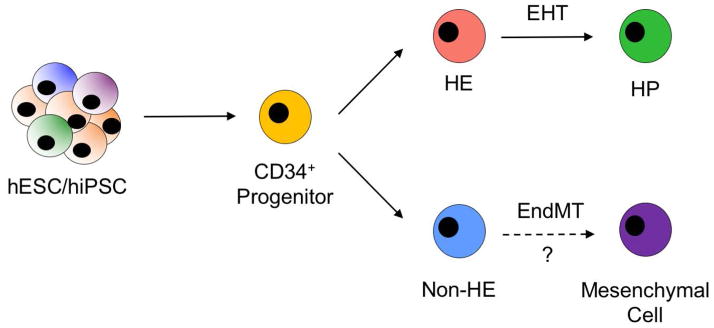
By assessing the transcriptional repertoire of individual cells, we also revealed heterogeneity in non-HE cells. Although all non-HE expressed traditional endothelial cell-specific surface antigens, a small subset of non-HE were heavily enriched with genes encoding extracellular matrix proteins (Cluster 4, Figure 4A). This genetic signature highly suggests transformation from an endothelial lineage to a mesenchymal or stromal cell lineage in a developmental process known as the endothelial-to-mesenchymal transition (EndMT). EndMT primarily is a TGF-β driven process that normally is required for intimal thickening and stabilization of vasculature during embryonic vasculogenesis [74]. Other groups have reported that hESC- and hiPSC-derived endothelial cells are highly susceptible to EndMT, particularly following long-term passaging and cell culture [74–75]. This, in part, may be due to a combination of defined factors within culture media and/or also the cellular origin of hiPSCs used. Our results suggest that a small degree of EndMT occurs even at the earliest development of hemato-endothelial cells from embryoid bodies (Figure 5). Indeed, we observed a small proportion of differentiating cells from spin-EBs exposed to hemato-endothelial growth conditions positively expressed S100A4/FSP1, a fibroblast specific surface antigen that has been previously demonstrated to distinguish EndMT (Supplemental Figure 9) [79, 80]. As such, in future studies it may be imperative to negatively select against these transitioning cells to prevent the out-crowding of true vascular endothelial cells and/or hemogenic endothelial cells.
Figure 5. Working model of human pluripotent stem cell EHT in vitro.
Undifferentiated hESCs/hiPSCs first develop towards mesodermal lineages and generate CD34+ progenitor cells with dual hemato-endothelial potential. CD34+ cells subsequently differentiate into endothelial cells that possess hematopoietic potential (HE) or endothelial cells that lack endothelial potential (non-HE). HE cells can then differentiate further into hematopoietic progenitor cells in a process known as EHT. A subset of non-HE may subsequently transform into other cells of mesenchymal lineages, such as a mesenchymal stromal cell or fibroblast, in a process known as the endothelial-to-mesenchymal transition (EndMT).
Lastly, it is an important distinction that we used a human platform to analyze the transcriptional profiles of single cells as they commit towards hemato-endothelial lineages at the earliest stage of hematopoiesis. Our findings share many commonalities to a related single cell approach investigating mouse hemogenic endothelium [13]. Using a pan-RUNX1 mouse reporter model and single cell qRT-PCR, Swiers et al. demonstrated mouse hemogenic endothelium was also enriched for key endothelial and hematopoietic specific genes simultaneously, similar to what we observed with human HE. Here, they concluded hemogenic endothelium transitions away from an endothelial repertoire and towards a distinct hematopoietic repertoire through analysis of matured hematopoietic cell populations (CD41+ and CD45+). Our findings in which the very first developed human hematopoietic cells (CD34+CD43+RUNX1c+) [81] are transcriptionally similar to hemogenic endothelium complements this working model of EHT. This also lends credence to pursuing future gain and/or loss of function studies using candidate genes described herein to optimize the development of hemogenic endothelium from hESCs. These studies would provide further mechanistic insight into the key molecular drivers that regulate development of multipotent and potentially engraftable human HSPCs.
Supplementary Material
Acknowledgments
Grant Support: National Institutes of Health grants: National Cancer Institute R01CA203348 (D.S.K.), National Institute of Diabetes and Digestive and Kidney Diseases F30DK107071 (M.G.A), National Institute of General Medicine Sciences T32GM113846 (M.G.A and D.S.K.) and T32GM008244 (M.G.A). Other support from the Regenerative Medicine Minnesota program (D.S.K.).
We thank Jerry Daniel and the University of Minnesota Genomic Center for technical assistance with the single cell RNASeq. We thank Gene Yeo (UCSD) for helpful discussion. This work was supported in part by the following National Institutes of Health grants: National Cancer Institute R01CA203348 (D.S.K.), National Institute of Diabetes and Digestive and Kidney Diseases F30DK107071 (M.G.A), National Institute of General Medicine Sciences T32GM113846 (M.G.A and D.S.K.) and T32GM008244 (M.G.A). Other support from the Regenerative Medicine Minnesota program (D.S.K.).
Footnotes
DISCLOSURE OF CONFLICTS OF INTEREST
The authors have nothing to disclose.
Author Contributions:
Mathew G. Angelos: Conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing.
Juan E. Abrahante: Collection and assembly of data
Robert H. Blum: Collection and assembly of data
Dan S. Kaufman: Conception and design, financial support, data analysis and interpretation, manuscript writing, final approval of manuscript
References
- 1.Bertrand JY, Chi NC, Santoso B, et al. Haematopoietic stem cells derive directly from aortic endothelium during development. NATURE. 2010;464(7285):108–11. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boisset J-C, van Cappellen W, Andrieu-Soler C, et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. NATURE. 2010;464(7285):116–20. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- 3.Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. NATURE. 2010;464(7285):112–5. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- 4.Zape JP, Zovein AC. Hemogenic endothelium: origins, regulation, and implications for vascular biology. SEMIN CELL DEV BIOL. 2011;22(9):1036–47. doi: 10.1016/j.semcdb.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Mikkola HKA. The journey of developing hematopoietic stem cells. DEVELOPMENT. 2006;133(19):3733–3744. doi: 10.1242/dev.02568. [DOI] [PubMed] [Google Scholar]
- 6.Palis J, Yoder MC. Yolk-sac hematopoiesis: The first blood cells of mouse and man. EXP HEMATOL. 2001;29(8):927–936. doi: 10.1016/s0301-472x(01)00669-5. [DOI] [PubMed] [Google Scholar]
- 7.Palis J. Primitive and definitive erythropoiesis in mammals. FRONT PHYSIOL. 2014;5(3):1–9. doi: 10.3389/fphys.2014.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jagannathan-Bogdan M, Zon LI. Hematopoiesis. DEVELOPMENT. 2013;140(12):2463–2467. doi: 10.1242/dev.083147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orkin SH, Zon LI. Hematopoiesis: An Evolving Paradigm for Stem Cell Biology. CELL. 2008;132(4):631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rafii S, Kloss CC, Butler JM, et al. Human ESC-derived hemogenic endothelial cells undergo distinct waves of endothelial to hematopoietic transition. BLOOD. 2013;121(5):770–780. doi: 10.1182/blood-2012-07-444208. [DOI] [PubMed] [Google Scholar]
- 11.Eilken HM, Nishikawa S-I, Schroeder T. Continuous single-cell imaging of blood generation from haemogenic endothelium. NATURE. 2009;457(7231):896–900. doi: 10.1038/nature07760. [DOI] [PubMed] [Google Scholar]
- 12.Antas VI, Al-Drees MA, Prudence AJA, et al. Hemogenic endothelium: A vessel for blood production. INT J BIOCHEM CELL BIOL. 2013;45(3):692–695. doi: 10.1016/j.biocel.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 13.Swiers G, Baumann C, O’Rourke J, et al. Early dynamic fate changes in haemogenic endothelium characterized at the single-cell level. NAT COMMUN. 2013;4:2924. doi: 10.1038/ncomms3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng CEL, Yokomizo T, Yamashita N, et al. A Runx1 intronic enhancer marks hemogenic endothelial cells and hematopoietic stem cells. STEM CELLS. 2010;28(10):1869–1881. doi: 10.1002/stem.507. [DOI] [PubMed] [Google Scholar]
- 15.Nottingham WT, Jarratt A, Burgess M, et al. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata / Ets / SCL-regulated enhancer. BLOOD. 2007;110(13):4188–4197. doi: 10.1182/blood-2007-07-100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sroczynska P, Lancrin C, Kouskoff V, et al. The differential activities of Runx1 promoters define milestones during embryonic hematopoiesis. BLOOD. 2009;114(26):5279–5289. doi: 10.1182/blood-2009-05-222307. [DOI] [PubMed] [Google Scholar]
- 17.Bee T, Liddiard K, Swiers G, et al. Alternative Runx1 promoter usage in mouse developmental hematopoiesis. BLOOD CELLS, MOL DIS. 2009;43(1):35–42. doi: 10.1016/j.bcmd.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Zambidis ET, Peault B, Park TS, et al. Hematopoietic differentiation of human embryonic stem cells progresses through sequential hematoendothelial, primitive, and definitive stages resembling human yolk sac development. BLOOD. 2008;106(3):860–870. doi: 10.1182/blood-2004-11-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Challen GA, Goodell MA. Runx1 isoforms show differential expression patterns during hematopoietic development but have similar functional effects in adult hematopoietic stem cells. EXP HEMATOL. 2010;38(5):403–416. doi: 10.1016/j.exphem.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lie-A-Ling M, Marinopoulou E, Li Y, et al. RUNX1 positively regulates a cell adhesion and migration program in murine hemogenic endothelium prior to blood emergence. BLOOD. 2014;124(11):e11–20. doi: 10.1182/blood-2014-04-572958. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka Y, Joshi A, Wilson NK, et al. The transcriptional programme controlled by Runx1 during early embryonic blood development. DEV BIOL. 2012;366(2):404–419. doi: 10.1016/j.ydbio.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen MJ, Yokomizo T, Zeigler B, et al. Runx1 is required for the endothelial to hematopoietic cell transition but not thereafter. NATURE. 2009;457(7231):887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ditadi A, Sturgeon CM, Tober J, et al. Human Definitive Haemogenic Endothelium and Arterial Vascular Endothelium Represent Distinct Lineages. NAT CELL BIOL. 2015;17(5):580–591. doi: 10.1038/ncb3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng ES, Azzola L, Bruveris FF, et al. Differentiation of human embryonic stem cells to HOXA+ hemogenic vasculature that resembles the aorta-gonad-mesonephros. NAT BIOTECHNOL. 2016;34(11):1168–1179. doi: 10.1038/nbt.3702. [DOI] [PubMed] [Google Scholar]
- 25.Ferrell PI, Xi J, Ma C, et al. The RUNX1 +24 enhancer and P1 promoter identify a unique subpopulation of hematopoietic progenitor cells derived from human pluripotent stem cells. STEM CELLS. 2015;33(4):1130–41. doi: 10.1002/stem.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ditadi A, Sturgeon CM, Tober J, et al. Human definitive haemogenic endothelium and arterial vascular endothelium represent distinct lineages. NAT CELL BIOL. 2015;17(5):580–91. doi: 10.1038/ncb3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirose SI, Takayama N, Nakamura S, et al. Immortalization of erythroblasts by c-MYC and BCL-XL enables large-scale erythrocyte production from human pluripotent stem cells. STEM CELL REPORTS. 2013;1(6):499–508. doi: 10.1016/j.stemcr.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobar L, Yates F, Oudrhiri N, et al. Human induced pluripotent stem cells can reach complete terminal maturation: In vivo and in vitro evidence in the erythropoietic differentiation model. HAEMATOLOGICA. 2012;97(12):1795–1803. doi: 10.3324/haematol.2011.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi K-D, Vodyanik Ma, Togarrati PP, et al. Identification of the hemogenic endothelial progenitor and its direct precursor in human pluripotent stem cell differentiation cultures. CELL REP. 2012;2(3):553–67. doi: 10.1016/j.celrep.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vodyanik M, Bork J, Thomson J, et al. Human embryonic stem cell – derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. BLOOD. 2005;105(2):617–626. doi: 10.1182/blood-2004-04-1649. [DOI] [PubMed] [Google Scholar]
- 31.Chadwick K, Wang L, Li L, et al. Cytokines and BMP-4 promote hematopoietic differentiation of human embryonic stem cells. BLOOD. 2003;102(3):906–915. doi: 10.1182/blood-2003-03-0832. [DOI] [PubMed] [Google Scholar]
- 32.Kaufman DS, Hanson ET, Lewis RL, et al. Hematopoietic colony-forming cells derived from human embryonic stem cells. PROC NATL ACAD SCI U S A. 2001;98(19):10716–21. doi: 10.1073/pnas.191362598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knorr DA, Bock A, Brentjens RJ, et al. Engineered human embryonic stem cell-derived lymphocytes to study in vivo trafficking and immunotherapy. STEM CELLS DEV. 2013;22(13):1861–9. doi: 10.1089/scd.2012.0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ni Z, Knorr DA, Kaufman DS. Hematopoietic and Natural Killer Cell Development from Human Pluripotent Stem Cells. EMBRYONIC STEM CELL IMMUNOBIOL METHODS PROTOC METHODS MOL BIOL. 2013;1029:33–41. doi: 10.1007/978-1-62703-478-4_3. [DOI] [PubMed] [Google Scholar]
- 35.Kennedy M, Awong G, Sturgeon CM, et al. T Lymphocyte Potential Marks the Emergence of Definitive Hematopoietic Progenitors in Human Pluripotent Stem Cell Differentiation Cultures. CELL REP. 2012;2(6):1722–1735. doi: 10.1016/j.celrep.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Melichar H, Li O, Ross J, et al. Comparative study of hematopoietic differentiation between human embryonic stem cell lines. PLOS ONE. 2011;6(5) doi: 10.1371/journal.pone.0019854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen B, Mao B, Huang S, et al. Human Embryonic Stem Cell-Derived Primitive and Definitive Hematopoiesis. In: Atwood CS, Vadakkadath Meethal S, editors. Pluripotent Stem Cell Biology - Advanced in Mechanisms, Methods, and Models. InTech; 2014. pp. 87–114. [Google Scholar]
- 38.Kennedy M, D’Souza SL, Lynch-Kattman M, et al. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. BLOOD. 2007;109(7):2679–87. doi: 10.1182/blood-2006-09-047704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shalek AK, Satija R, Adiconis X, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. NATURE. 2013;498(7453):236–40. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan SYS, Krasnow MA. Developmental origin of lung macrophage diversity. DEVELOPMENT. 2016:1318–1327. doi: 10.1242/dev.129122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yvernogeau L, Gautier R, Khoury H, et al. An in vitro model of hemogenic endothelium commitment and hematopoietic production. DEVELOPMENT. 2016:1302–1312. doi: 10.1242/dev.126714. [DOI] [PubMed] [Google Scholar]
- 42.Björklund ÅK, Forkel M, Picelli S, et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. NAT IMMUNOL. 2016;17(4):451–60. doi: 10.1038/ni.3368. [DOI] [PubMed] [Google Scholar]
- 43.Ng ES, Davis R, Stanley EG, et al. A protocol describing the use of a recombinant protein-based, animal product-free medium (APEL) for human embryonic stem cell differentiation as spin embryoid bodies. NAT PROTOC. 2008;3(5):768–776. doi: 10.1038/nprot.2008.42. [DOI] [PubMed] [Google Scholar]
- 44.Satija R, Farrell JA, Gennert D, et al. Spatial reconstruction of single-cell gene expression data. NAT BIOTECHNOL. 2015;33(5):495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macosko EZ, Basu A, Satija R, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. CELL. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wurtzel O, Cote LE, Poirier A, et al. A Generic and Cell-Type-Specific Wound Response Precedes Regeneration in Planarians. DEV CELL. 2015;35(5):632–645. doi: 10.1016/j.devcel.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Camp JG, Badsha F, Florio M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. PROC NATL ACAD SCI U S A. 2015;112(51):15672–7. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patro R, Duggal G, Salmon Kingsford C. Accurate, Versatile and Ultrafast Quantification from RNA-seq Data using Lightweight-Alignment. BIORXIV. 2015:21592. [Google Scholar]
- 49.Søreide K. Receiver-operating characteristic curve analysis in diagnostic, prognostic and predictive biomarker research. J CLIN PATHOL. 2009;62(1):1–5. doi: 10.1136/jcp.2008.061010. [DOI] [PubMed] [Google Scholar]
- 50.Anderson H, Patch TC, Reddy PNG, et al. Hematopoietic stem cells develop in the absence of endothelial cadherin 5 expression. BLOOD. 2015;126(26):2811–2820. doi: 10.1182/blood-2015-07-659276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nikolova-Krstevski V, Yuan L, Le Bras A, et al. ERG is required for the differentiation of embryonic stem cells along the endothelial lineage. BMC DEV BIOL. 2009;9(1):72. doi: 10.1186/1471-213X-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi KD, Vodyanik MA, Togarrati PP, et al. Identification of the Hemogenic Endothelial Progenitor and Its Direct Precursor in Human Pluripotent Stem Cell Differentiation Cultures. CELL REP. 2012;2(3):553–567. doi: 10.1016/j.celrep.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen MJ, Li Y, De Obaldia ME, et al. Erythroid/myeloid progenitors and hematopoietic stem cells originate from distinct populations of endothelial cells. CELL STEM CELL. 2011;9(6):541–552. doi: 10.1016/j.stem.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakajima-Takagi Y, Osawa M, Oshima M, et al. Role of SOX17 in hematopoietic development from human embryonic stem cells. BLOOD. 2013;121(3):447–58. doi: 10.1182/blood-2012-05-431403. [DOI] [PubMed] [Google Scholar]
- 55.Clarke RL, Yzaguirre AD, Yashiro-Ohtani Y, et al. The expression of Sox17 identifies and regulates haemogenic endothelium. NAT CELL BIOL. 2013;15(5):502–10. doi: 10.1038/ncb2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clarke RL, Robitaille AM, Moon RT, et al. A quantitative proteomic analysis of hemogenic endothelium reveals differential regulation of hematopoiesis by SOX17. STEM CELL REPORTS. 2015;5(2):291–304. doi: 10.1016/j.stemcr.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu S-J, Luo C, Holton K, et al. Robust generation of hemangioblastic progenitors from human embryonic stem cells. REGEN MED. 2008;3(5):693–704. doi: 10.2217/17460751.3.5.693. [DOI] [PubMed] [Google Scholar]
- 58.Lu S, Feng Q, Caballero S, et al. Generation of functional hemangioblasts from human embryonic stem cells. NAT METHODS. 2007;4(6):501–509. doi: 10.1038/nmeth1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kennedy M, D’Souza SL, Lynch-Kattman M, et al. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. BLOOD. 2007;109(7):2679–2687. doi: 10.1182/blood-2006-09-047704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zambidis ET, Sinka L, Tavian M, et al. Emergence of human angiohematopoietic cells in normal development and from cultured embryonic stem cells. ANN N Y ACAD SCI. 2007;1106:223–232. doi: 10.1196/annals.1392.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sturgeon CM, Ditadi A, Awong G, et al. Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. NAT BIOTECHNOL. 2014;32(6):554–61. doi: 10.1038/nbt.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakajima H, Ito M, Smookler DS, et al. TIMP-3 recruits quiescent hematopoietic stem cells into active cell cycle and expands multipotent progenitor pool. BLOOD. 2010;116(22):4474–4482. doi: 10.1182/blood-2010-01-266528. [DOI] [PubMed] [Google Scholar]
- 63.Nakajima H, Shibata F, Fukuchi Y, et al. Immune suppressor factor confers stromal cell line with enhanced supporting activity for hematopoietic stem cells. BIOCHEM BIOPHYS RES COMMUN. 2006;340(1):35–42. doi: 10.1016/j.bbrc.2005.11.146. [DOI] [PubMed] [Google Scholar]
- 64.Yokota T, Oritani K, Butz S, et al. The endothelial antigen ESAM marks primitive hematopoietic progenitors throughout life in mice. BLOOD. 2009;113(13):2914–2923. doi: 10.1182/blood-2008-07-167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ooi aGL, Karsunky H, Majeti R, et al. The adhesion molecule esam1 is a novel hematopoietic stem cell marker. STEM CELLS. 2009;27(3):653–661. doi: 10.1634/stemcells.2008-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yuan L, Sacharidou A, Stratman AN, et al. RhoJ is an endothelial cell-restricted Rho GTPase that mediates vascular morphogenesis and is regulated by the transcription factor ERG. BLOOD. 2011;118(4):1145–1153. doi: 10.1182/blood-2010-10-315275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leszczynska K, Kaur S, Wilson E, et al. The role of RhoJ in endothelial cell biology and angiogenesis. BIOCHEM SOC TRANS. 2011;39(6):1606–1611. doi: 10.1042/BST20110702. [DOI] [PubMed] [Google Scholar]
- 68.McLaughlin F, Ludbrook VJ, Cox J, et al. Combined genomic and antisense analysis reveals that the transcription factor Erg is implicated in endothelial cell differentiation. BLOOD. 2001;98(12):3332–3339. doi: 10.1182/blood.v98.12.3332. [DOI] [PubMed] [Google Scholar]
- 69.Wilson E, Leszczynska K, Poulter NS, et al. RhoJ interacts with the GIT-PIX complex and regulates focal adhesion disassembly. J CELL SCI. 2014;127(Pt 14):3039–51. doi: 10.1242/jcs.140434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim AD, Melick CH, Clements WK, et al. Discrete Notch signaling requirements in the specification of hematopoietic stem cells. EMBO J. 2014;33(20):2363–73. doi: 10.15252/embj.201488784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumano K, Chiba S, Kunisato A, et al. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. IMMUNITY. 2003;18(5):699–711. doi: 10.1016/s1074-7613(03)00117-1. [DOI] [PubMed] [Google Scholar]
- 72.Hadland BK, Huppert SS, Kanungo J, et al. A requirement for Notch1 distinguishes 2 phases of definitive hematopoiesis during development. BLOOD. 2004;104(10):3097–3105. doi: 10.1182/blood-2004-03-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gori JL, Butler JM, Chan YY, et al. Vascular niche promotes hematopoietic multipotent progenitor formation from pluripotent stem cells. J CLIN INVEST. 2015;125(3):1243–1254. doi: 10.1172/JCI79328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dou DR, Calvanese V, Sierra MI, et al. Medial HOXA genes demarcate haematopoietic stem cell fate during human development. NAT CELL BIOL. 2016;18(6) doi: 10.1038/ncb3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen JY, Miyanishi M, Wang SK, et al. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. NATURE. 2016;530(7589):223–227. doi: 10.1038/nature16943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin F, Wang N, Zhang TC. The role of endothelial-mesenchymal transition in development and pathological process. IUBMB LIFE. 2012;64(9):717–723. doi: 10.1002/iub.1059. [DOI] [PubMed] [Google Scholar]
- 77.James D, Nam H, Seandel M, et al. Expansion and maintenance of human embryonic stem cell – derived endothelial cells by TGFb inhibition is Id1 dependent. NAT BIOTECHNOL. 2010;28(2):161–6. doi: 10.1038/nbt.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Z, Hu S, Ghosh Z, et al. Functional characterization and expression profiling of human induced pluripotent stem cell- and embryonic stem cell-derived endothelial cells. STEM CELLS DEV. 2011;20(10):1701–10. doi: 10.1089/scd.2010.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maddaluno L, Rudini N, Cuttano R, et al. EndMT contributes to the onset and progression of cerebral cavernous malformations. NATURE. 2013;498(7455):492–496. doi: 10.1038/nature12207. [DOI] [PubMed] [Google Scholar]
- 80.Evrard SM, Lecce L, Michelis KC, et al. Endothelial to mesenchymal transition is common in atherosclerotic lesions and is associated with plaque instability. NAT COMMUN. 2016;7(11853):1–15. doi: 10.1038/ncomms11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vodyanik MA, Thomson JA, Slukvin II. Leukosialin (CD43) defines hematopoietic progenitors in human embryonic stem cell differentiation cultures. BLOOD. 2006;108(6):2095–105. doi: 10.1182/blood-2006-02-003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.