Abstract
Less than 3% of protein-coding genetic variants are predicted to result in loss of protein function through the introduction of a stop codon, frameshift, or the disruption of an essential splice site; however, such predicted loss-of-function (pLOF) variants provide insight into effector transcript and direction of biological effect. In >400,000 UK Biobank participants, we conduct association analyses of 3759 pLOF variants with six metabolic traits, six cardiometabolic diseases, and twelve additional diseases. We identified 18 new low-frequency or rare (allele frequency < 5%) pLOF variant-phenotype associations. pLOF variants in the gene GPR151 protect against obesity and type 2 diabetes, in the gene IL33 against asthma and allergic disease, and in the gene IFIH1 against hypothyroidism. In the gene PDE3B, pLOF variants associate with elevated height, improved body fat distribution and protection from coronary artery disease. Our findings prioritize genes for which pharmacologic mimics of pLOF variants may lower risk for disease.
Examination of predicted loss-of-function (pLOF) genetic variants allows direct identification of genes with therapeutic potential. Here, Emdin et al. perform association analysis for 3759 pLOF variants with 24 traits and highlight protective variants against cardiometabolic and immune phenotypes.
Introduction
A focused investigation of predicted loss-of-function (pLOF) variants provides several advantages when compared with analysis of other types of variants. First, analysis of pLOF variants may allow for the direct identification of a gene rather than a locus containing many candidate genes1. Second, pLOF variants provide directionality of effect, unlike non-coding regulatory variants which may increase or decrease expression of a given gene. Third, identification of pLOF variants which protect against disease may aid with prioritization of therapeutic target genes (e.g., the recent development of inhibitors of PCSK9 or ANGPTL3 which mimic human pLOF mutations protective against cardiovascular disease)2–6.
Here, we analyse pLOF variants in UK Biobank and other datasets to identify genes for which pharmacologic inhibition may protect against disease. We identify associations of pLOF variants with cardiometabolic and immune disease, prioritizing the genes GPR151, IL33, GSDMB, IFIH1, and PDE3B as potential therapeutic targets.
Results
Analysis of loss-of-function variants
In 405,569 individuals in UK Biobank (335,660 individuals of European ancestry and 69,909 individuals of Non-European ancestry, Supplementary Table 1), we analyzed the association of 3759 pLOF variants with six metabolic traits [body mass index (BMI), waist-to-hip ratio adjusted for body mass index (WHRadjBMI), height, systolic blood pressure (SBP), diastolic blood pressure (DBP), forced expiratory volume to forced vital capacity ratio (FEV1/FVC)], six cardiometabolic diseases (coronary artery disease, type 2 diabetes, atrial fibrillation, stroke, heart failure, venous thromboembolism) and twelve diseases with more than 5000 cases (allergic rhinitis, asthma, anxiety, breast cancer, cataract, cholelithiasis, depression, hypothyroidism, gastric reflux, osteoporosis, osteoarthritis, and psoriasis). The Variant Effect Predictor and associated LOFTEE plugin7,8 algorithms were used to annotate variants which were pLOF (1) nonsense mutations that resulted in early termination of a protein; (2) frameshift mutations due to insertions or deletions of DNA; or (3) splice-site mutations which result in an incorrectly spliced protein. For coronary artery disease, we pooled UK Biobank estimates with estimates from the CARDIoGRAM Exome consortium9. For height, we pooled UK Biobank estimates with estimates from the GIANT Exome Consortium10. Variants with a P < 5.5 × 10−7 [0.05/(24 outcomes × 3759 variants) in the combined analysis were considered significant. Quantile–quantile analysis was used to examine for the presence of population stratification. No evidence of inflation was observed (inflation factors <1.1; Supplementary Fig. 1).
We identified 18 new low-frequency or rare (<5%) pLOF variants associated with traits and diseases in UK Biobank (Table 1 and Supplementary Table 2). We also discovered 26 new common frequency (≥5%) pLOF variants associated with UK Biobank phenotypes (Supplementary Table 3). Variants identified within the same locus in prior genome-wide association studies (+500 Kb; 1 Mb total) for the same phenotype are provided (Supplementary Tables 4 and 5).
Table 1.
Predicted loss-of-function variants with minor allele frequency <5% which are significantly associated with traits or diseases in UK Biobank
| Outcome | Gene | pLOF variant | Location | EA | RA | AA change | Freq (%) | Beta | SE | P-value | Novel? | MHC locus? |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Asthma | FLG | rs61816761 | 1:152285861 | A | G | p.Arg501Ter | 1.51 | 0.21 | 0.03 | 1.51 × 10−15 | Yes | No |
| Asthma | HLA-DQB1 | rs28688207 | 6:32628660 | C | T | Splice Acceptor c.773−1A > G | 3.14 | −0.17 | 0.02 | 3.11 × 10−15 | Yes | Yes |
| Asthma | IL33 | rs146597587 | 9:6255967 | C | G | Splice Acceptor c.487−1G > C | 0.44 | −0.54 | 0.06 | 7.79 × 10−17 | No15 | No |
| BMI | GPR151 | rs114285050 | 5:145895394 | A | G | p.Arg95Ter | 0.78 | −0.07 | 0.01 | 4.89 × 10−8 | Yes | No |
| BMI | PKHD1L1 | rs533623778 | 8:110523131 | T | C | p.Arg769Ter | 1.0 × 10−4 | 5.30 | 0.99 | 9.45 × 10−8 | Yes | No |
| DBP | ENPEP | rs33966350 | 4:111431444 | A | G | p.Trp413Ter | 1.19 | 0.06 | 0.01 | 8.12 × 10−10 | No39 | No |
| DBP | BTN3A2 | rs58367598 | 6:26370833 | G | T | Splice Donor c.715 + 2T > G | 3.75 | 0.03 | 0.01 | 2.03 × 10−8 | Yes | Yes |
| DBP | TMC3 | rs150843673 | 15:81624929 | T | G | p.Ser1045Ter | 2.14 | 0.05 | 0.01 | 8.16 × 10−9 | Yes | No |
| Height | PDE11A | rs76308115 | 2:178879181 | A | G | p.Arg57Ter | 0.52 | 0.07 | 0.01 | 6.20 × 10−11 | Yes | No |
| Height | CLHC1 | rs114931154 | 2:55407644 | T | A | Splice Donor c.1384 + 2T > A | 1.26 | −0.05 | 0.01 | 1.54 × 10−11 | Yes | No |
| Height | CCDC66 | rs150364083 | 3:56628033 | T | C | p.Arg427Ter | 0.58 | 0.05 | 0.01 | 2.09 × 10−7 | Yes | No |
| Height | DAP | rs201354802 | 5:10761153 | A | C | p.Glu10Ter | 0.24 | 0.13 | 0.02 | 1.68 × 10−8 | Yes | No |
| Height | TRIM40 | rs115651142 | 6:30115320 | T | G | Splice Donor c.602 + 1G > T | 0.63 | −0.08 | 0.01 | 1.16 × 10−9 | Yes | Yes |
| Height | MICA | rs181430930 | 6:31378575 | A | G | Splice Donor c.286 + 1G > A | 0.26 | −0.12 | 0.02 | 7.87 × 10−8 | Yes | Yes |
| Height | PDE3B | rs150090666 | 11:14865399 | T | C | p.Arg783Ter | 0.06 | 0.24 | 0.04 | 9.32 × 10−9 | Yes | No |
| Height | APOLD1 | rs202116412 | 12:12879031 | A | G | Splice Donor c.96 + 1G > A | 0.03 | 0.12 | 0.02 | 3.06 × 10−8 | Yes | No |
| Hypothyroidism | IFIH1 | rs35337543 | 2:163136505 | G | C | Splice Donor c.1641 + 1G > C | 1.45 | −0.27 | 0.04 | 2.95 × 10−9 | Yes | No |
| Psoriasis | ZKSCAN3 | rs73387810 | 6:28318166 | A | G | Splice Donor c.−63 + 1G > A | 0.86 | 0.55 | 0.08 | 4.18 × 10−11 | Yes | Yes |
| Psoriasis | EGFL8 | rs141826798 | 6:32134395 | G | C | p.Arg74Ter | 0.53 | 0.90 | 0.08 | 2.19 × 10−26 | Yes | Yes |
| SBP | ENPEP | rs33966350 | 4:111431444 | A | G | p.Trp413Ter | 1.19 | 0.06 | 0.01 | 3.46 × 10−9 | No39 | No |
| SBP | GEM | rs138582164 | 8:95264265 | A | G | p.Arg199Ter | 0.04 | 0.30 | 0.06 | 1.93 × 10−7 | No40 | No |
| WHRadjBMI | PYGM | rs116987552 | 11:64527223 | A | G | p.Arg50Ter | 0.39 | 0.09 | 0.02 | 1.32 × 10−7 | Yes | No |
Beta in terms of standard deviations and reported for the effect allele
pLOF predicted loss-of-function, EA effect allele, RA reference allele, AA Change amino acid change, Freq(%) Frequency(%); BMI body mass index, DBP diastolic blood pressure, SBP systolic blood pressure, WHRadjBMI waist-to-hip ratio adjusted for body mass index
A locus-wide conditional analysis (±500 kb of the pLOF variant) was performed to determine the extent to which the identified pLOF variant signal was independent of other genetic variation at the locus. Independence of pLOF variants may provide increased confidence for a causal association. Independent variants at the loci of rare and low frequency pLOFs are reported in Supplementary Data 1 and independent variants at the loci of common frequency pLOFs are reported in Supplementary Data 2. Of the 16 low frequency pLOF variants outside of the MHC locus, 14 were identified as independent variants in the locus-wide conditional analysis. These include the IL33 variant rs146597587, the GPR151 variant rs114285050, the IFIH1 variant rs35337543, and the PDE3B variant rs15009066 which are further analyzed in the text of this manuscript. Below, we highlight several of these associations.
GPR151, obesity, and type 2 diabetes
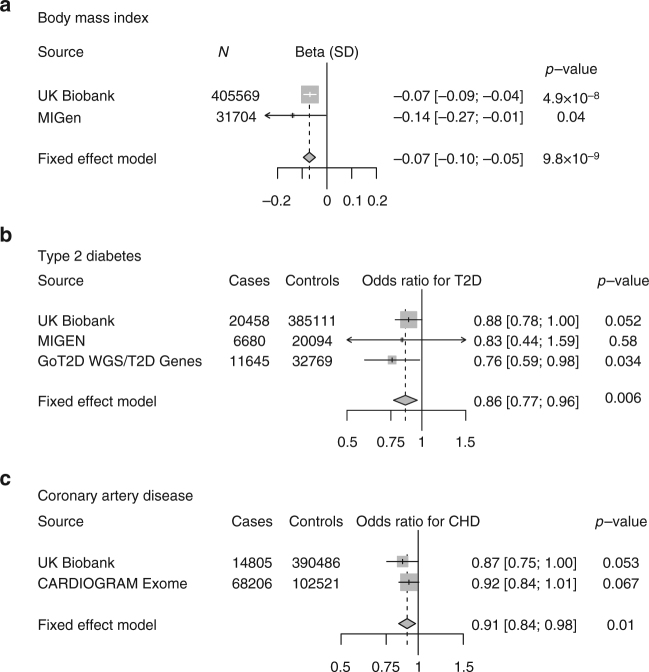
In GPR151 (encoding G-protein coupled receptor 151), the p.Arg95Ter variant (rs114285050, allele frequency 0.8% in European ancestry) was associated with reduced BMI (beta −0.07 standard deviations, −0.36 kg/m2, P = 4.9 × 10−8). We replicated this association in an independent cohort, the Myocardial Infarction Genetics Consortium (MIGen), where p.Arg95Ter carriers had reduced BMI (beta −0.14, P = 0.04; pooled beta −0.07, P = 9.8 × 10−9; Fig. 1). UK Biobank participants who carry one copy of p.Arg95Ter were at 12% lower odds of clinical obesity (BMI > 30 kg/m2). As obesity is a causal risk factor for type 2 diabetes and coronary artery disease, we examined whether p.Arg95Ter may provide protection against both diseases. p.Arg95Ter was associated with 14% lower odds of type 2 diabetes (OR 0.86; P = 0.006) and 9% lower odds of coronary artery disease (OR 0.91; P = 0.01; Fig. 1). Although GPR151 encodes a G-protein coupled receptor of unknown function whose expression is limited to the central nervous system, recent studies tracing the lineage of neurons expressing GRP151 have localized connections to hypothalamic neurons, a region of the brain important in the control of appetite11.
Fig. 1.
Association of a loss-of-function variant (p.Arg95Ter) in GPR151 with a body mass index, b type 2 diabetes, and c coronary artery disease. Estimates were derived in UK Biobank using logistic regression, adjusted for age, sex, ten principal components of ancestry, and array type
IL33, GSDMB, and asthma
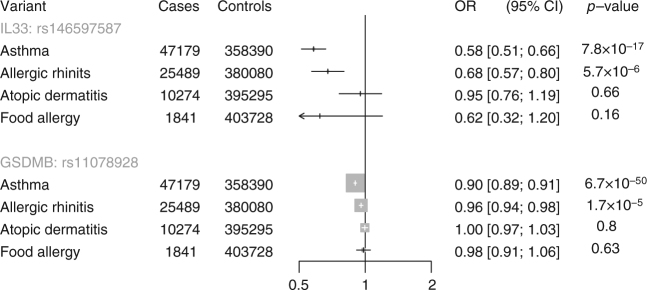
We identified several pLOF variants that associated with lower risk of asthma (Table 1). At GSDMB encoding gasdermin B, splice acceptor variant c.662-2A > G (rs11078928, allele frequency 46% in European ancestry) protected against asthma (OR 0.90 CI 0.89, 0.91, P = 6.7 × 10−50; Supplementary Table 3). This variant is in tight linkage disequilibrium (r2 = 0.99) with a previously identified non-coding variant in the GSDMB locus (rs2305480) associated with lower risk of asthma (P = 9.6 × 10−8)12. GSDMB c.662-2A > G is associated with lower expression of GSDMB transcripts13. Furthermore, overexpression of GSDMB causes airway remodeling and asthma symptoms in a mouse model14, suggesting that loss of GSDMB function may protect against asthma.
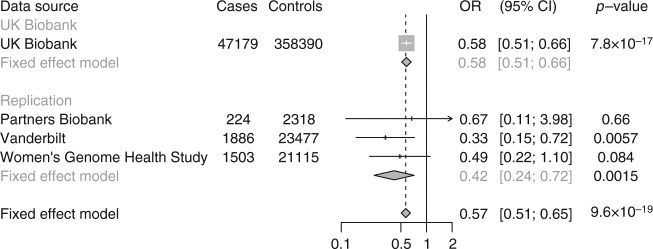
At the IL33 gene, a splice acceptor site variant c.487-1G > C (rs146597587, allele frequency 0.004 in European ancestry) was observed to protect against asthma (OR 0.58 CI 0.51, 0.66, P = 7.8 × 10−17). This variant was recently identified as associated with lower blood eosinophil concentration at genome-wide significance and with lower risk of asthma at more modest levels of significance (P = 1.8 × 10−4)15. To further replicate the association of IL33 c.487-1G > C with asthma, we examined the association of IL33 c.487-1G > C with asthma in individuals from three additional studies (Partners Biobank, the Vanderbilt eMERGE network, and the Women’s Genome Health Study). IL33 c.487-1G > C was associated with a protective effect of asthma in each data set. Overall, IL33 c.487-1G > C was associated with 43% lower odds of asthma (OR 0.57 CI 0.51, 0.65, P = 9.6 × 10−19, Fig. 2), suggesting that IL33 inhibition may be a useful approach for treatment of asthma. Of note, an inhibitor of IL33 is currently under development for treatment of asthma16.
Fig. 2.
Association of IL33 c.487-1G > C with asthma in UK Biobank, Partners Biobank, Vanderbilt eMERGE network and Women’s Genome Health Study. UK Biobank estimates were derived using logistic regression, adjusted for age, sex, ten principal components of ancestry, and array type. Partners Biobank and Vanderbilt estimates were derived using logistic regression, adjusted for age, sex, and principal components of ancestry. Women’s Genome Health Study estimates were derived using logistic regression, adjusted for age and principal components of ancestry
Asthma is often associated with other allergic phenotypes—atopic dermatitis, food allergy, and allergic rhinitis17. We therefore examined whether asthma-associated pLOF variants in GSDMB and IL33 associate with a lower risk of other atopic disorders in UK Biobank. Both pLOF variants also protected against allergic rhinitis (Fig. 3). In contrast, nominal associations with atopic dermatitis and food allergy were not observed, although point estimates for food allergy were similar to asthma.
Fig. 3.
Association of predicted loss-of-function variant in GSDMB and IL33 with allergic disease in UK Biobank. Estimates were derived in UK Biobank using logistic regression, adjusted for age, sex, ten principal components of ancestry, and array type
IFIH1 and autoimmune disorders
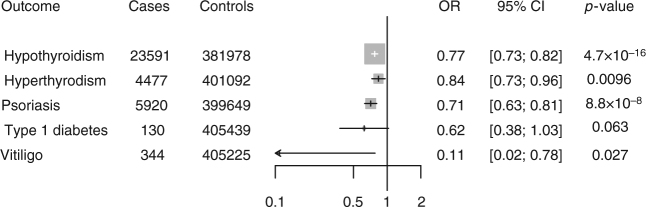
A splice donor variant in IFIH1 (interferon induced with helicase C domain 1), c.1641 + 1G > C (rs35337543, allele frequency 1.5% in European ancestry), is associated with a reduced risk of hypothyroidism in UK Biobank participants (OR 0.77 CI 0.70, 0.85; P = 5 × 10−9; Table 1). A gene-based test combining four additional pLOF variants in IFIH1 (rs35732034, rs201026962, rs35744605, rs148590996) similarly demonstrated protection against hypothyroidism in UK Biobank (OR 0.79 CI 0.72, 0.86; P = 4.4 × 10−8). Carriers of pLOF variants in IFIH1 were also protected against hyperthyroidism (OR 0.84 CI 0.73, 0.96; P = 0.01; Fig. 4).
Fig. 4.
Association of predicted loss-of-function variants in IFIH1 with thyroid disorders, type 1 diabetes, psoriasis, and vitiligo in UK Biobank. Estimates were derived in UK Biobank using logistic regression, adjusted for age, sex, ten principal components of ancestry, and array type
Common variants in the IFIH1 locus have previously been associated with psoriasis18 and vitiligo19, while rare pLOF variants in IFIH1 have been associated with a reduced risk of type 1 diabetes20. We therefore examined whether carriers of pLOF variants in IFIH1 in UK Biobank were protected against these diseases. Carriers of pLOF variants in IFIH1 were protected against type 1 diabetes, psoriasis, and vitiligo in UK Biobank (Fig. 4). These results suggest that IF1H1 pLOF variants alter risk for a range of autoimmune diseases.
In addition, an exploratory analysis demonstrated a nominally lower risk of coronary artery disease among IFIH1 pLOF carriers. Pooling UK Biobank and MIGen, IFIH1 pLOF carriers were protected against coronary artery disease (OR 0.92 CI 0.87, 0.98; P = 0.009). To examine whether this may be a chance finding, we examined whether the common IFIH1 missense variant rs1990760, previously identified as associated with autoimmune disorders, also associated with risk of coronary artery disease. The T allele of rs1990760 (frequency 41%) associated with a reduced risk of hypothyroidism (OR 0.92 CI 0.90, 0.94; P = 9.3 × 10−17) in UK Biobank. Pooling UK Biobank and CARDIOGRAM Exome, the T allele of rs1990760 also associated with a lower risk of coronary artery disease (OR 0.97 CI 0.96, 0.99; P = 2.5 × 10−5), providing complementary evidence that IFIH1 may influence coronary artery disease risk.
PDE3B and body fat distribution
At PDE3B encoding the gene phosphodiesterase 3B, p.Arg783Ter (rs150090666, allele frequency 0.0006 in European ancestry) associated with elevated height (beta 0.24, P = 9.3 × 10−9). Targeted deletion of Pde3b in mice leads to white adipose tissue gaining characteristics of brown adipose tissue21, a reduction in adipocyte size22, smaller fat deposits23 and reduced atherosclerosis24. We therefore studied the association of PDE3B p.Arg783Ter with metabolic phenotypes in UK Biobank and/or MIGen, where 36,581 individuals have been sequenced for the 16 exons of the PDE3B gene. In UK Biobank, which lacks direct measurements of blood lipids, carriers of p.Arg783Ter carriers were at reduced risk of physician-diagnosis of hypercholesterolemia (OR 0.52, P = 0.0002). Pooling UK Biobank and MIGen, pLOF carriers in PDE3B had reduced WHRadjBMI (beta −0.15, P = 0.0005). As genetic predisposition to improved body fat distribution has been associated with a lower risk of coronary artery disease25, we examined whether loss of PDE3B function protects against coronary artery disease. We aggregated rare PDE3B pLOFs in cases and compared this count with that controls. Across 14,805 cases in UK Biobank, the carrier frequency of pLOF in cases was 0.1% and in controls 0.2%. Across 20,186 cases in MIGen, the carrier frequency of pLOF was 0.05% and 0.1% in controls. Collectively, carrier status for PDE3B pLOFs was associated with reduced risk for coronary artery disease (OR 0.65 CI 0.43, 0.97; P = 0.03; Fig. 5).
Fig. 5.
Association of predicted loss-of-function variants in PDE3B with coronary artery disease. Estimates were derived in UK Biobank using logistic regression, adjusted for age, sex, ten principal components of ancestry, and array type. Estimates were derived in MIGEN (Myocardial Infarction Genetics Consortium) using logistic regression, adjusted for sex and five principal components of ancestry
Presence of homozygote individuals for pLOF variants in target genes may provide an in vivo demonstration of safety of pharmacologic inhibition of target genes. We therefore examined whether homozygotes for these pLoF variants were present in UK Biobank and in the gnomAD database26. At least one individual homozygous for a pLOF variant was identified in UK Biobank or the gnomAD database for the genes GPR151, GSDMB, IL33 and IFIH1, and PDE3B (Supplementary Table 6).
Discussion
In this study, we identified pLOF variants that protect against obesity (GPR151), asthma (GSDMB, IL33), autoimmune disorders (IFIH1), and coronary artery disease (PDE3B), prioritizing genes and pathways for which pharmacologic attempts to mimic these protective mutations might ameliorate disease.
Identification of protective loss-of-function variants has led to the development of therapeutics. Most notably, the discovery of missense and loss-of-function variants in PCSK9 that lower LDL cholesterol and protect against coronary artery disease suggested that inhibition of PCSK9 may be a useful therapeutic strategy for prevention and treatment of cardiovascular disease3,27. These genetic studies were validated by a large-scale randomized trial demonstrating that a monoclonal antibody directed against PCSK9 reduced the risk of recurrent cardiovascular events4. More recently, the discovery of loss-of-function variants in ANGPTL3 that lower blood triglyceride levels and protect against coronary artery disease has suggested that ANGPTL3 inhibition may reduce blood triglyceride levels and risk of coronary artery disease28,29. ANGTPL3 inhibitors are in clinical development and have now been demonstrated to reduce blood triglyceride levels5,6.
Our findings identify putative therapeutic targets that, similar to PCSK9 and ANGPTL3, may be useful for prevention of disease. For obesity and type 2 diabetes, these results hightlight GPR151 as a potential therapeutic target. GPR151 encodes a largely uncharacterized G protein-coupled receptor. The mechanism by which it influences risk of obesity and type 2 diabetes is unclear. However, it is highly expressed in the hypothalamus, a region of the brain known to be involved in appetite regulation11. Indeed, genetic variation in MC4R, which encodes the melanocortin 4 receptor, strongly influences obesity risk at a population level30. Similar to GPR151, MC4Ris highly expressed in the hypothalamus and is involved in appetite regulation31.
PDE3B inhibition may be a useful therapeutic strategy to improve body fat distribution and reduce risk of coronary artery disease. Unlike GPR151, for which no pharmacologic inhibitor is currently in clinical use, an inhibitor of PDE3B is available. Cilostazol is a non-selective pharmacologic inhibitor of both phosphodiesterase 3B and the related isoform phosphodiesterase 3A32. In a small randomized trial including 211 participant, cilostazol significantly reduced restenosis after percutaneous coronary balloon angioplasty33. The association of PDE3B pLOFs with improved body fat distribution, reduced risk of hypercholesterolemia and reduced risk of coronary artery disease suggests that selective inhibition of PDE3B may be useful for multiple features of metabolic syndrome.
We identified pLOF variants in IL33 (encoding interleukin 33) and GSDMB (encoding gasdermin B) as associated with a lower risk of asthma. The IL33 variant rs146597587 was recently found to be associated with lower blood eosinophil concentration at genome-wide significance and with lower risk of asthma at more modest levels of significance (P = 1.8 × 10−4)15. Consistent with these findings, induction of antibodies against IL33 by vaccination induces protection against airway inflammation in a mouse model of asthma34. An inhibitor of IL33 is currently under development for treatment of asthma16. In contrast to IL33, no inhibitor for GSDMB is in clinical development. These findings suggest that IL33 and GSDMB inhibition may both be useful therapeutic strategies for treatment of asthma and allergic disease.
Although our restriction of the present analysis to pLOF variants increases the likelihood of identifying causal variants substantially, it remains possible that a highly correlated nearby variant could be driving the association in some cases. Future functional studies may permit additional validation of causal variants.
In summary, we associated pLOF variants with a range of biomarker and disease phenotypes in a large, national biobank and identified several new genes in which pLOF variants protect against disease, prioritizing these genes for therapeutic targeting. More generally, large-scale analysis of pLOF variants is emerging as a useful tool for therapeutic target identification and validation.
Methods
Gene and variant annotation
Variants in hg19 coordinates were annotated with information from Ensembl release 82 using Variant Effect Predictor (VEP)7. Only pLOFs, defined as premature stop (nonsense), canonical splice-sites (splice-donor or splice-acceptor) or insertion/deletion variants that shifted frame (frameshift) were annotated as predicted loss-of-function (pLOF), using the “--pick-allele” annotation. PLOFs as defined by VEP were then merged with publicly available data from the Exome Aggregation Consortium (ExAC), Version 0.3.1, to confirm consistency in variant annotation26.
We identified 3759 pLOF variants in UK Biobank with an info score greater than 0.3 (Supplementary Table 1). We used a Bonferroni corrected P value of 5.5 × 10−7 to denote significance [0.05/(3759 variants × 24 outcomes) = 5.5 × 10−7] in our primary pLOF analysis.
Study design
We analyzed the association of pLOF variants with 24 phenotypes: cardiovascular, metabolic and pulmonary phenotypes: six metabolic traits (body mass index, waist-to-hip ratio, height, systolic blood pressure, diastolic blood pressure, and forced expiratory volume to forced vital capacity ratio), six cardiometabolic diseases (coronary artery disease, type 2 diabetes, atrial fibrillation, stroke, heart failure, and venous thromboembolism) and 12 diseases with more than 5000 cases (allergic rhinitis, asthma, anxiety, breast cancer, cataract, cholethiasis, depression, hypothyroidism, gastric reflux, osteoporosis, osteoarthritis, and psoriasis; Supplementary Table 4). All six metabolic traits were inverse normalized prior to analysis, with adjustment for age and sex. Forced expiratory volume to forced vital capacity ratio was additionally adjusted for height. To adjust for the presence of antihypertensive medication, we added 15 mm Hg to systolic blood pressure and 10 mm Hg to diastolic blood pressure of individuals on antihypertensive medication at baseline, as in the International Consortium for Blood Pressure genome-wide association study35. Definitions for disease outcomes in UK Biobank are provided (Supplementary Table 7).
In UK Biobank, analysis was performed separately in unrelated individuals of European and Non-European ancestry. Estimates for variants were then pooled using inverse-variance weighted-fixed effects meta-analysis. For coronary artery disease, estimates for variants from UK Biobank were additionally pooled with the effect of variants in the CARDIOGRAM Exome consortium using inverse variance weighted-fixed effects meta-analysis9. For height, estimates of variants in UK Biobank were pooled with the GIANT Height Exome consortium using inverse variance weighted-fixed effects meta-analysis10.
Genotyping and quality control
Phasing and imputation were performed centrally, by UK Biobank, using a reference panel combining UK10k and 1000 Genomes samples. 39,235,157 variants included in the Haplotype Reference Consortium were imputed. As recommended by UK Biobank, we excluded any samples with an imputation quality <0.3 as well as pLOF variants which were not included in the Haplotype Reference Consortium. Mitochondrial genetic data and sex chromosomes were excluded from this analysis. Individual level genetic data was available from individuals in UK Biobank, after excluding one related individual of each related pair of individuals, individuals whose genetic sex did not match self-reported sex and individuals with an excess of missing genotype calls or more heterozygosity than expected.
We analysed 3759 variants identified as pLOF variants in UK Biobank. PLINK 2 software was used to examine the association of these variants with traits and disease in UK Biobank under the assumption of additive effects (https://www.cog-genomics.org/plink/2.0/). Adjustment was performed for age, sex, ten principal components of ancestry, and array type.
Conditional analysis
A locus-wide conditional analysis (±500 kb of the pLOF variant) was performed to determine the extent to which the identified pLOF variant signal was independent of other genetic variation at the locus. We iteratively performed association analyses conditional on the top variants at each locus, until no variants were below the Bonferroni-adjusted threshold for significance (P < 5.5 × 10−7). A statistically significant and independent signal for the pLOF variant provides increased confidence for a causal association.
Analysis of PDE3B association with coronary artery disease
We aimed to analyse the association of pLOF variants in PDE3B with coronary artery disease in a combined analysis of UK Biobank and the Myocardial Infarction Genetics Consortium (MIGen). Replication was performed in MIGen rather than the CARDIOGRAM Exome consortium as rs150090666 was not included in the exome chip analysis of the CARDIOGRAM Exome consortium9. Estimates of the association of rs150090666 with coronary artery disease in UK Biobank were derived as described above, using logistic regression with adjustment for age, sex, ten principal components of ancestry, and a dummy variable for array type. An additional pLOF variant, rs535108921, present in UK Biobank, was also analysed for association with coronary artery disease, as above.
pLOFs variants in PDE3B were identified in the MIGen Consortium using exome sequencing or whole genome sequencing, as previously described36–38. Studies included in the MIGen consortium were: (1) the Italian Atherosclerosis Thrombosis and Vascular Biology (ATVB) study (dbGaP Study Accession phs000814.v1.p1); (2) the Exome Sequencing Project Early-Onset Myocardial Infarction (ESP-EOMI) study(9); (3) a nested case-control cohort from the Jackson Heart Study (JHS); (4) the South German Myocardial Infarction study (dbGaP Study Accession phs000916.v1.p1); (5) the Ottawa Heart Study (OHS) (dbGaP Study Accession phs000806.v1.p1); (6) the Precocious Coronary Artery Disease (PROCARDIS) study (dbGaP Study Accession phs000883.v1.p1); (7) the Pakistan Risk of Myocardial Infarction Study (PROMIS) (dbGaP Study Accession phs000917.v1.p1); (8) the Registre Gironi del COR (Gerona Heart Registry or REGICOR) study (dbGaP Study Accession phs000902.v1.p1); (9) the Leicester Myocardial Infarction study (dbGaP Study Accession phs001000.v1.p1); (10) the BioImage study (dbGaP Study Accession phs001058.v1.p1); (11) the North German Myocardial Infarction study (dbGaP Study Accession phs000990.v1.p1); (12) Multi-Ethnic Study of Atherosclerosis (dbGaP Study Accession: phs000209.v2.p1); (13) Variation In Recovery: Role of Gender on Outcomes of Young AMI cohort; and (14) Taiwan Metabochip Consortium.
The Burrows–Wheeler Aligner algorithm was used to align reads from participants to the reference genome (hg19). The GATK HaploTypeCaller was used to jointly call variants. Metrics including Variant Quality Score Recalibration (VQSR), quality over depth, and strand bias were then used to filter variants. We excluded samples which were related to other samples, which had high ratios of heterozygous to non-reference homozygous genotypes, which had high missing genotypes, which had a discordant genetic gender relative to reports gender, and samples which were discordant relative to genotype data.
After variant calling and quality control, the Variant Effect Predictor7,8 was used to annotate variants which were pLOF: (1) nonsense mutations that resulted in early termination of PDE3B (2) frameshift mutations due to insertions or deletions of DNA; or (3) splice-site mutations which result in an incorrectly spliced protein (Supplementary Table 8). For analysis of rare pLOF variants, we pooled rare pLOF variants in MIGen, testing for the association of a pLOF with coronary artery disease using logistic regression, after adjustment for age, sex, cohort, and five principle components of ancestry. We meta-analysed the association of pLOFs with coronary artery disease in MIGen combined with UK Biobank.
Replication of IL33 finding
To replicate the association of rs146597587, a splice site variant in IL33, with asthma, we pooled estimates of the association of rs146597587 with asthma from Partners Biobank, from the Vanderbilt eMERGE network and from the Women’s Genome Health Study. In Partners Biobank, rs146597587 was imputed (info score of 0.77) in 2542 individuals. The association of rs146597587 with asthma (hospitalization for ICD9 code 493) was estimated using logistic regression, adjusted for age, sex, and five principal components of ancestry. In the Vanderbilt eMERGE network, rs146597587 was genotyped in 25,363 individuals using the Illumina Exome BeadChip. The association of rs146597587 with asthma (hospitalization for ICD9 code 493) was estimated using logistic regression, adjusted for age, sex, and principal components of ancestry. In Women’s Genome Health Study, rs14659758 was genotyped in 22,618 individuals using the Illumina Exome. The association of rs14659758 with asthma (hospitalization for ICD9 code 493 or ICD10 code J45) was estimated using logistic regression, adjusted for age and principal components of ancestry.
Data availability
All individual-level data from UK Biobank can be accessed by applying to the UK Biobank Central Access Committee (http://www.ukbiobank.ac.uk/register-apply/).
Electronic supplementary material
Description of Additional Supplementary Files
Acknowledgements
This research has been conducted using the UK Biobank resource, application 7089. The WGHS is supported by the National Heart, Lung, and Blood Institute (HL043851 and HL080467) and the National Cancer Institute (CA047988 and UM1CA182913) with funding for genotyping provided by Amgen. The VIRGO study was supported by grant R01 HL081153-01A1K from the National Heart, Lung, and Blood Institute. The TAICHI study was supported by the National Health Research Institutes, Taiwan (PH-099-PP-03, PH-100-PP-03, PH-101-PP-03), the National Science Council, Taiwan (Grant Nos NSC 101-2314-B-075A-006-MY3, MOST 104-2314-B-075A-006-MY3, MOST 104-2314-B-075A-007, MOST 105-2314-B-075A-003), the Taichung Veterans General Hospital, Taiwan (TCVGH-1020101C, TCVGH-1020102D, TCVGH-1023102B, TCVGH-1023107D, TCVGH-1030101C, TCVGH-1030105D, TCVGH-1033503C, TCVGH-1033102B, TCVGH-1033108D, TCVGH-1040101C, TCVGH-1040102D, TCVGH-1043504C, TCVGH-1043104B), and the National Center for Advancing Translational Sciences, CTSI grant UL1TR001881. The MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts HHSN268201500003I, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420, UL1-TR-001881, and DK063491. Whole genome sequencing of the VIRGO and TAICHI cohorts was funded by grant 5UM1HG008895-02 from the National Human Genome Research Institute’s Center for Common Disease Genomics. Whole genome sequencing of the MESA cohort was funded through the Trans-Omics for Precision Medicine (TOPMed) Program of the National Heart, Lung, and Blood Institute. General study coordination was provided by the TOPMed Data Coordinating Center (3R01HL-12393-02S1). The contributions of the investigators of the NHLBI TOPMed Consortium (https://www.nhlbiwgs.org/topmed-banner-authorship) are gratefully acknowledged. The Atherosclerosis Risk in Communities study is carried out as a collaborative study supported by the National Heart, Lung, and Blood Institute (NHLBI) contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C). The authors thank the staff and participants of the ARIC study for their important contributions. Funding support for “Building on GWAS for NHLBI-diseases: the U.S. CHARGE consortium” was provided by the NIH through the American Recovery and Reinvestment Act of 2009 (ARRA) (5RC2HL102419).
Author contributions
C.A.E., A.V.K., D.K., and S.K. conceived and designed the study. C.A.E., A.V.K., M.C., H.K., D.S., S.G., and S.K. acquired, analysed, and interpreted data. C.A.E., A.V.K., and S.K. drafted the manuscript. All authors revised the manuscript for important intellectual content. S.K. supervised the study.
Competing interests
A.V.K. is supported by a John S. LaDue Memorial Fellowship at Harvard Medical School, and a KL2/Catalyst Medical Research Investigator Training award from Harvard Catalyst funded by the National Institutes of Health (NIH) (TR001100) and has received consulting fees from Merck and Amarin. P.N. reports funding from the John S. LaDue Memorial Fellowship at Harvard Medical School and has received consulting fees from Amarin. S.K. is supported by a research scholar award from Massachusetts General Hospital, the Donovan Family Foundation, and R01 HL127564; he has received a research grant from Bayer Healthcare; and consulting fees from Merck, Novartis, Sanofi, AstraZeneca, Alnylam Pharmaceuticals, Leerink Partners, Noble Insights, MedGenome, Aegerion Pharmaceuticals, Regeneron Pharmaceuticals, Quest Diagnostics, Genomics PLC, and Eli Lilly and Company; and holds equity in San Therapeutics and Catabasis Pharmaceuticals. The remaining authors declare no competing interests.
Footnotes
These authors contributed equally: Connor A. Emdin, Amit V. Khera.
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41467-018-03911-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abifadel M, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 3.Cohen JC, Boerwinkle E, Mosley TH, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 4.Sabatine, M. S. et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N. Engl. J. Med. 376, 1713-1722 (2017). [DOI] [PubMed]
- 5.Dewey FE, et al. Genetic and pharmacologic inactivation of ANGPTL3 and cardiovascular disease. N. Engl. J. Med. 2017;377:211–221. doi: 10.1056/NEJMoa1612790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham MJ, et al. Cardiovascular and metabolic effects of ANGPTL3 antisense oligonucleotides. N. Engl. J. Med. 2017;377:222–232. doi: 10.1056/NEJMoa1701329. [DOI] [PubMed] [Google Scholar]
- 7.McLaren W, et al. Deriving the consequences of genomic variants with the ensemble API and SNP effect predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karczewski, K. J. LOFTEE (Loss-Of-Function Transcript Effect Estimator). https://github.com/konradjk/loftee (2015).
- 9.Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators. Coding variation in ANGPTL4, LPL, and SVEP1 and the risk of coronary disease. N. Engl. J. Med. 2016;374:1134–1144. doi: 10.1056/NEJMoa1507652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marouli E, et al. Rare and low-frequency coding variants alter human adult height. Nature. 2017;542:186–190. doi: 10.1038/nature21039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broms J, et al. Monosynaptic retrograde tracing of neurons expressing the G-protein coupled receptor Gpr151 in the mouse brain. J. Comp. Neurol. 2017;525:3227–3250. doi: 10.1002/cne.24273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moffatt MF, et al. A large-scale, consortium-based genomewide association study of asthma. N. Engl. J. Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrison FS, et al. The splice site variant rs11078928 may be associated with a genotype-dependent alteration in expression of GSDMB transcripts. BMC Genom. 2013;14:627. doi: 10.1186/1471-2164-14-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das S, et al. GSDMB induces an asthma phenotype characterized by increased airway responsiveness and remodeling without lung inflammation. Proc. Natl Acad. Sci. USA. 2016;113:13132–13137. doi: 10.1073/pnas.1610433113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith D, et al. A rare IL33 loss-of-function mutation reduces blood eosinophil counts and protects from asthma. PLOS Genet. 2017;13:e1006659. doi: 10.1371/journal.pgen.1006659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gangwar RS, Landolina N, Arpinati L, Levi-Schaffer F. Mast cell and eosinophil surface receptors as targets for anti-allergic therapy. Pharmacol. Ther. 2017;170:37–63. doi: 10.1016/j.pharmthera.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 17.Spergel JM. Epidemiology of atopic dermatitis and atopic march in children. Immunol. Allergy Clin. North. Am. 2010;30:269–280. doi: 10.1016/j.iac.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Stuart PE, et al. Genome-wide association analysis of psoriatic arthritis and cutaneous psoriasis reveals differences in their genetic architecture. Am. J. Hum. Genet. 2015;97:816–836. doi: 10.1016/j.ajhg.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin Y, et al. Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants. Nat. Genet. 2016;48:1418–1424. doi: 10.1038/ng.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guirguis E, et al. A role for phosphodiesterase 3B in acquisition of brown fat characteristics by white adipose tissue in male mice. Endocrinology. 2013;154:3152–3167. doi: 10.1210/en.2012-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi YH, et al. Alterations in regulation of energy homeostasis in cyclic nucleotide phosphodiesterase 3B-null mice. J. Clin. Invest. 2006;116:3240–3251. doi: 10.1172/JCI24867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung YW, et al. White to beige conversion in PDE3B KO adipose tissue through activation of AMPK signaling and mitochondrial function. Sci. Rep. 2017;7:40445. doi: 10.1038/srep40445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmad F, et al. Phosphodiesterase 3B (PDE3B) regulates NLRP3 inflammasome in adipose tissue. Sci. Rep. 2016;6:28056. doi: 10.1038/srep28056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emdin C, et al. Genetic association of waist-to-hip ratio with cardiometabolic traits, type 2 diabetes, and coronary heart disease. JAMA: J. Am. Med. Assoc. 2017;317:626–634. doi: 10.1001/jama.2016.21042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen J, et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 28.Musunuru K, et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N. Engl. J. Med. 2010;363:2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stitziel NO, et al. ANGPTL3 deficiency and protection against coronary artery disease. J. Am. Coll. Cardiol. 2017;69:2054–2063. doi: 10.1016/j.jacc.2017.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turcot V, et al. Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat. Genet. 2018;50:26–41. doi: 10.1038/s41588-017-0011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah BP, et al. MC4R-expressing glutamatergic neurons in the paraventricular hypothalamus regulate feeding and are synaptically connected to the parabrachial nucleus. Proc. Natl Acad. Sci. USA. 2014;111:13193–13198. doi: 10.1073/pnas.1407843111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung YW, et al. Targeted disruption of PDE3B, but not PDE3A, protects murine heart from ischemia/reperfusion injury. Proc. Natl Acad. Sci. USA. 2015;112:E2253–E2262. doi: 10.1073/pnas.1416230112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuchikane E, et al. Impact of cilostazol on restenosis after percutaneous coronary balloon angioplasty. Circulation. 1999;100:21–26. doi: 10.1161/01.CIR.100.1.21. [DOI] [PubMed] [Google Scholar]
- 34.Lei Y, et al. Vaccination against IL-33 inhibits airway hyperresponsiveness and inflammation in a house dust mite model of asthma. PLOS One. 2015;10:e0133774. doi: 10.1371/journal.pone.0133774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.International Consortium for Blood Pressure Genome-Wide Association Studies. et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Do R, et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–106. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khera, A. V. et al. Diagnostic yield and clinical utility of sequencing familial hypercholesterolemia genes in patients with severe hypercholesterolemia. J. Am. Coll. Cardiol. 67, 2578-2589 (2016). [DOI] [PMC free article] [PubMed]
- 38.Khera, A. V. et al. Genome-wide polygenic score to identify a monogenic risk-equivalent for coronary disease. Preprint at bioRxiv10.1101/218388 (2017).
- 39.Surendran P, et al. Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat. Genet. 2016;48:1151–1161. doi: 10.1038/ng.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warren HR, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat. Genet. 2017 doi: 10.1038/ng.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All individual-level data from UK Biobank can be accessed by applying to the UK Biobank Central Access Committee (http://www.ukbiobank.ac.uk/register-apply/).