Abstract
Objectives
Esophageal microbiota and regulation of adaptive immunity are increasingly being investigated in eosinophilic esophagitis (EoE). Toll-like receptors (TLRs) play a central role in the initiation and maintenance of innate immune activity. Our objective was to characterize the esophageal and duodenal innate immune response in EoE and its modulation by dietary therapy.
Methods
Esophageal and duodenal biopsy samples were collected from 10 adults with untreated EoE, before and after effective treatment with a six-food elimination diet (SFED), and 10 controls with normal esophagus. In all cases, bacterial load (by mRNA expression of 16S), TLRs, mucins, transcription factors, interleukins, components of the NKG2D system, and innate immunity effectors were assessed by qPCR. Protein expression of TLRs were also determined by immunofluorescence.
Results
Bacterial load and TLR1, TLR2, TLR4, and TLR9 were overexpressed on biopsies with active EoE compared with controls. Muc1 and Muc5B genes were downregulated while Muc4 was overexpressed. Upregulation of MyD88 and NFκB was found together with IL-1β, IL-6, IL-8, and IL-10 mediators and PER-1, iNOS, and GRZA effectors. NG-K2D components (KLRK1, IL-15, MICB) were also upregulated. In all cases, changes in active EoE were normalized following SFED and mucosal healing. Duodenal samples also showed increased expressions of TLR-1, TLR-2, and TLR-4, but not 16S or any other mediators nor effectors of inflammation.
Conclusions
Esophageal TLR-dependent signaling pathways in EoE support the potential implication of microbiota and the innate immune system in the pathogenesis of this disease.
Introduction
Eosinophilic esophagitis (EoE) is a chronic, food-triggered, immune-mediated disease of the esophagus, clinically characterized by symptoms referred to esophageal dysfunction, and histologically defined by an eosinophil-rich inflammation of the esophageal mucosa1,2, among other cell types3. The incidence and prevalence of EoE have rapidly increased in children and adults in recent years4, making it today a common cause of chronic dysphagia and food impaction in young patients.
The involvement of an adaptive Th2-type immune response to food antigens in EoE was known from the first descriptions of the disease;5,6 several cytokines, and chemokines derived from T cells present within the inflammatory infiltrate in EoE promote food-specific responses7,8, in which local production of IgE9, but also IgG4 derived from plasma cells located in the esophageal lamina propria of EoE patients10 might play a relevant role. Profibrogenic factors released by inflammatory cells determine fibrous remodeling of the esophageal tissues11,12. Avoiding the consumption of specific food triggers, whenever possible, constitutes a first-line therapy for EoE13,14.
In contrast to the highly specialized adaptive immunity, the innate immune system recognizes and responds to environmental insults and pathogens without the need for an immunoglobulin-driven antigen-specific response. Evidence pointing towards a potential role for the innate immunity in EoE has arisen recently. Esophageal epithelial cells have been revealed as major effectors initiating the inflammatory phenomena in EoE, not just through the release of eotaxin-3 and other chemoattractants for eosinophils15, but also by promoting the recruitment of invariant natural killer T (iNKT) cells toward the esophageal epithelium16, which constitutes a major cytokine source. A specific role for mast cells (MCs) has also been recognized in the pathophysiology and symptoms of EoE which reverse after effective dietary treatment17. Changes in the esophageal microbiome composition in adult and pediatric EoE patients compared to non-EoE controls have also been recently described18,19 while modification of the microbiota caused by antibiotic consumption has been recognized as an early life risk factor for developing EoE20. Together, these evidences give rise to a potential role that the innate immune system in general, and the microbial pattern recognition receptors (PRRs) in particular, might play in EoE pathogenesis.
Among PRRs, Toll-like receptors (TLRs) are type-I transmembrane receptors expressed both on epithelial and lamina propria cells with the capacity to distinguish between pathogen and commensal microbes21. In humans, there are a total of 11 different TLR (named from TLR-1 to TLR-11), each having different specificities which, once stimulated, activate intracellular signal transduction pathways mediated by MAP kinases and NF-κB, ultimately triggering a pro-inflammatory immune response. As a part of the innate immune system22, TLRs activation is responsible, among other functions, for triggering inflammatory responses by acting as a link between innate and adaptive immunity23,24. Indeed, activation and maturation of antigen-presenting cells and regulatory T cells (Tregs) depends on TLR-mediated signaling, highlighting their role on mucosal immune homeostasis.
Numerous studies have evaluated the role of TLRs in inflammatory, autoimmune, and allergic diseases, with the relationship between allergy and TLR activation currently positioned at the frontier of immunology research22,25,26. TLR expression in esophageal epithelial samples, however, has only been demonstrated recently27. Despite this, no study has yet assessed their potential role in EoE. Therefore, in order to get a deeper insight into this mechanism in the context of EoE, here we have characterized the expression of human TLRs, as well as of several immune-mediators and effectors, on esophageal and duodenal samples from healthy controls and patients with EoE, both before and after dietary-induced disease remission.
Methods
Participants and clinical assessment
Adult EoE patients who were naïve to topical or systemic steroids and dietary therapy for EoE were prospectively recruited. Diagnosis for EoE was defined by consensus guidelines28 and consisted in (i) infiltration of esophageal epithelium by 15 or more eosinophil leukocytes per high-powered field (hpf) (ii) absence of eosinophilic infiltration in biopsy specimens from gastric and duodenal mucosa; (iii) lack of histologic response after an 8-week trial of PPI therapy; and (iv) exclusion of drug intake, parasites, esophageal caustications, hematologic neoplasm, or other events in the patient’s medical history as possible causes of esophageal eosinophilia. Esophageal biopsies were obtained from each patient with EoE at baseline and after 6-weeks of an empiric six-food elimination diet (SFED) that induced histologic and clinical remission of EoE. Patients’ support was provided as previously described31. The duration and intensity of dysphagia events, along with the frequency and intensity of heartburn and regurgitation, were assessed structurally, by means of a non-validated score developed for achalasia32 and previously used in adult EoE12,17, at the beginning of the study and after completing the dietary treatment.
Gender-matched control samples were obtained from individuals who consecutively underwent endoscopy under sedation during the study period, because of dyspepsia or a suspected gastroduodenal ulcer. All selected control subjects exhibited a normal endoscopic appearance of the esophagus, in which hiatus hernia, incompetent cardias, and esophageal peptic lesions were excluded, and the analyses of esophageal mucosal biopsies were also reported as normal. Familial and personal background of atopy was identified in all EoE patients and control participants, based on clinical records.
Endoscopic and biopsy-sampling procedure
All endoscopic exams were performed under propofol sedation by a single board-certified gastroenterologist (AJL) with a flexible 9-mm-caliber Pentax EG-2770K gastroscope (Pentax of America, Inc, Montvale, NJ). A minimum of four biopsies were taken from both upper and lower esophageal thirds with the aid of a standard needle biopsy forceps (Endo Jaw FB-220U, Olympus Medical Systems, Tokyo, Japan). As TLR have been described as overexpressed in the duodenal mucosa of several digestive diseases29,30, and even in non-inflamed tissues33, four mucosal biopsies were also taken from the second portion of the duodenum and processed for histopathological analysis. Three additional biopsies from the middle esophageal third and two from the duodenum of each participant were collected during the endoscopic procedure and preserved in an RNA stabilization solution (RNAlater; Ambion, Inc, Austin, Tex) at –80°C until processing for gene expression study.
Histological study
Esophageal samples were fixed in formalin, embedded in paraffin, and routinely processed for hematoxylin and eosin staining. The histological analysis was performed by an experienced pathologist (JMO) blind to the experimental groups. The peak number of eosinophils was counted in the most densely inflamed areas with the aid of Nikon Eclipse 50i (Nikon Corp, Tokyo, Japan) light microscopy in three high-powered field (0.238 mm2). Peak eosinophil count per hpf was calculated in the epithelial strata by averaging the eosinophil counts.
Immunofluorescence
Formalin-fixed, paraffin-embedded tissues were sectioned at 5 μm. Cuts were deparaffinized and rehydrated following general procedures. Specific antigen retrieval and permeabilization processes were performed depending on the antibody. After treatment with Blocking Solution (Dako Diagnósticos, Barcelona, Spain) for 2 h at room temperature, samples were incubated with the primary antibodies anti-TLR1, TLR2, TLR3, TLR4, TLR6, or TLR9 (Supplementary Table 1) overnight at 4 °C. Incubation with the secondary antibodies Alexa Fluor 594 goat anti-rabbit IgG or Alexa Fluor 488 goat anti-mouse IgG (Life Technologies, Madrid, Spain) was performed for 30 min at room temperature. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). The negative control slides followed the same procedure excluding the addition of the primary antibodies. Fading was controlled using the Prolong anti-fade mounting media (Molecular Probes, Barcelona, Spain). A fluorescence microscope (BX61, Olympus, Barcelona, Spain) was used for visual analysis and images of the epithelium and the lamina propia were taken at high magnification (×400).
Analysis of RNA expression
Total RNA was isolated with the MirVanaTM miRNA Isolation Kit (Ambion), following the manufacturer’s instructions. Gene expression for the different determined genes was evaluated in all samples. Each assay and its assay ID number is available at Applied Biosystems (Madrid, Spain) (Supplementary Table 2). Simultaneous quantitative real-time PCRs (qPCR) were performed with TaqMan Low-Density Arrays (Applied Biosystems) preconfigured in a 384-well format and spotted on a microfluidic card. Each TaqMan Gene Expression Assay consists of a forward and reverse primer at a final concentration of 900 nM and a Taq-Man MGB probe (6-FAM dye-labeled; Applied Biosystems), with a final concentration of 250 nM. The assays are gene specific and have been designed to span an exon–exon junction. Thermal cycling conditions were 2 min at 50 °C, 10 min at 95 °C, followed by 40 cycles of denaturation at 95 °C for 15 s, and annealing and extension at 60 °C for 1 min in an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems). This procedure was replicated twice for each gene and each sample, with water as a negative control.
Relative changes in mRNA expression of human genes were calculated with the cycle threshold (Ct) method35 with the aid of Sequence Detection System 2.1 software (Applied Biosystems). The amount of mRNA for each gene was calculated in each sample using the Ct value. Relative gene expression was calculated as follows: 2ΔΔCt, where ΔΔCt = ΔCttarget gene − ΔCt control genes. The fold change for the treatment was defined as the relative expression compared with the corresponding control and was calculated as follows: 2ΔΔCt, where ΔΔCt = ΔCtpatient − ΔCthealthy, and expressed as arbitrary relative units (rU). Expression levels of all target genes were normalized to 18S, GAPDH, PGK1, GUSB, and b-actin expression.
Bacterial load was determined by using two primers developed against the V4 region of the 16S rRNA, as previously described34. Three replicas were amplified per sample and expression levels were normalized to those of the same eukaryotic genes, thus making them independent of the biopsy size.
In order to identify overlap or cluster formation we performed Principal Component Analysis (PCA) plots and Heatmaps by ClustVis web tool36. ClustVis is written using the Shiny web application framework (R package version 0.10.2.1) for R statistics software, using several R packages internally36,37. Each PCA was calculated using Singular Value Decomposition (SVD) with imputation38. Heatmap is plotted using pheatmap R package (version 0.7.7). The package uses popular clustering distances and methods39 implemented in dist and hclust functions in R. Heatmaps show a data matrix where coloring gives an overview of the numeric differences, and genes and samples are clustered hierarchically.
Statistical analysis
Optimal sample size was calculated based on our previous results17 aimed for a power of 90%. Means and standard deviations were reported for continuous variables and are expressed as mean (standard deviation) throughout the text. Proportions were reported for categorical data. Results are expressed as a median with an interquartile rank (IQR) for scoring clinical symptoms. Comparisons between groups (control subjects and EoE patients) were performed with nonparametric tests: the Mann–Whitney U-test for quantitative variables and the Fisher exact test for nominal variables. For comparison before and after SFED treatment, the nonparametric-paired Wilcoxon signed-rank test was used. To control for multiple testing, post hoc comparisons were performed using Holm-Bonferroni-corrected p values. A nonparametric correlation test (Spearman’s rho) was used for analyzing the association between eosinophils, gene expression, and clinical symptoms. A 0.05 level of significance was used throughout. Statistical analyses were performed with the aid of PASW 18.0 statistical analysis software (SPSS Inc, Chicago, Ill).
Ethics
The study was conducted in accordance with the principles of the Declaration of Helsinki and approved by the institutional review board of La Mancha Centro General Hospital. Informed consent was obtained from all patients prior to all endoscopic exams.
Results
Study population
Of the 14 patients with EoE screened, 10 (8 men and 2 women) achieved histological and clinical remission and were included in this study. Additionally, 10 gender-matched control subjects were also included. The groups had a mean (standard deviation) age of 33.1 (10.1) and 53 (19.9) years, respectively. Individual clinical characteristics of the experimental subjects are given in Table 1 and Supplementary Table 3. Mean duration of symptoms in EoE patients exceeded 4 years (50.8 ± 40.9 months). No difference in clinical manifestations was observed between atopic and non-atopic subjects (Table 2).
Table 1.
Clinical characteristics of EoE patients included in the study
| Patients | Age (years) | Sex | Time of evolution (months) | Symptoms | Endoscopy | Familiar background of atopy | Personal background of atopy | Identified food triggered | |
|---|---|---|---|---|---|---|---|---|---|
| Caliber | Mucosal appearance | ||||||||
| 1 | 25 | M | 12 | FI, Dy | N | LF, Rg | No | No | F&S & Ri |
| 2 | 18 | M | 60 | FI, WL | N | LF, C | Sister: D | AR | Le, Nu & Co |
| 3 | 38 | M | 4 | Dy, AP | R | WP, Rg | No | BA, AR | Mi, Eg, F&S, Le & So |
| 4 | 36 | M | 36 | FI | N | LF, WP, Rg | Brother: FS | BA, AR | Mi, Nu & So |
| 5 | 38 | F | 60 | FI, Dy | N | LF, WP | Sister: AR | BA, AR | Wh, Leg & Nu |
| 6 | 18 | M | 24 | AP, V | N | LF, Rg | No | AR, FS | Mi, Le, Nu & Co |
| 7 | 51 | F | 24 | FI, Dy | N | LF, WP, Rg | No | No | Mi & Egg |
| 8 | 34 | M | 48 | FI, Dy, Ht | R | LF, WP, C, Rg | Father: BA; Brother: AR | No | Mi |
| 9 | 38 | M | 120 | FI, Dy | N | Normal | No | BA, AR, FS | Ri |
| 10 | 35 | M | 120 | Dy, AP | N | Rg, C | Brother: DS | BA, AR, FS | Mi, F&S & Co |
Sex: M male, F female. Symptoms: FI food impaction, Dy dysphagia, AP abdominal pain, V vomiting, Ht heartburn, WL weight loss. Endoscopy: N normal, R reduced, Rg rings, LF longitudinal furrows, C crêpe-paper appearance, WP white plaques. Atopy: BA bronchial asthma, AR allergic rhinitis, FS food sensitivity, D dermatitis, DS drug sensitivity. Food triggers: Mi milk, Ri rice, F&S fish & seafood, Le legumes, Nu nuts, Wh wheat, Co corn, Eg eggs, So soy
Table 2.
Clinical characteristics and gene expression levels of atopic and non-atopic EoE patients
| Atopic vs. non-atopic | p | |
|---|---|---|
| Time of evolution (months) | 60.6 (45.1) vs. 28 (18.3) | 0.250a |
| Symptom score | 9 (5.9) vs. 6 (2) | 0.723a |
| Symptoms | ||
| Dysphagia | 57.1% vs. 100% | 0.475b |
| Food impaction | 57.1% vs. 100% | 0.475b |
| Abdominal pain | 42.9% vs. 0% | 0.475b |
| Heartburn | 0% vs. 33.3% | 0.300b |
| Vomiting | 14.3% vs. 0% | >0.999b |
| Weight loss | 14.3% vs. 0% | >0.999b |
| Endoscopy findings | ||
| Reduced caliber | 14.3% vs. 33.3% | >0.999b |
| Normal mucosa | 14.3% vs. 0% | >0.999b |
| Longitudinal furrows | 57.1% vs. 100% | 0.475b |
| Rings | 71.4% vs. 66.7% | >0.999b |
| Crêpe-paper appearance | 28.6% vs. 33.3% | >0.999b |
| White plaques | 42.9% vs. 66.7% | >0.999b |
| Peak eosinophil count | 55 (30.4) vs. 61 (34.8) | 0.908a |
| TLR1 gene expression | 2.2 (2.3) vs. 1.5 (2.4) | 0.569a |
| TLR2 gene expression | 41.3 (62.9) vs. 44.2 (34.5) | 0.909a |
| TLR3 gene expression | 10.5 (9.5) vs. 3.2 (4.4) | 0.087a |
| TLR4 gene expression | 3.6 (9.3) vs. 3.1 (2.7) | 0.569a |
| TLR6 gene expression | 2.1 (4.6) vs. 2.9 (3.1) | 0.909a |
| TLR9 gene expression | 1.9 (6.7) vs. 3.8 (2.7) | 0.425a |
| 16S gene expression | 0.75 (0.5) vs. 0.54 (0.8) | 0.732a |
| MUC1 gene expression | 1.1 (1.7) vs. 0.3 (0.9) | 0.360a |
| MUC2 gene expression | NA | — |
| MUC4 gene expression | 14.4 (22.1) vs. 20.3 (10.6) | 0.732a |
| MUC5B gene expression | 0.32 (0.51) vs. 0.09 (0.03) | 0.138a |
| MyD88 gene expression | 2 (0.4) vs. 2.3 (0.5) | 0.425a |
| NF-κB gene expression | 2.9 (0.8) vs. 4 (0.8) | 0.305a |
| IL-1α gene expression | 0.13 (0.17) vs. 0.13 (0.06) | 0.909a |
| IL-1β gene expression | 3.3 (5.6) vs. 8.8 (4.3) | 0.125a |
| IL-6 gene expression | 0.21 (0.14) vs. 1.3 (1.2) | 0.138a |
| IL-8 gene expression | 103 .7 (196.6) vs. 179.39 (138.1) | 0.425a |
| IL-10 gene expression | 6.7 (5.7) vs. 29.9 (14.8) | 0.087a |
| TNF-α gene expression | 1.8 (2.2) vs. 1 (2.2) | 0.909a |
| PRF1 gene expression | 10.4 (8.7) vs. 7.9 (5.7) | 0.909a |
| iNOS gene expression | 0.041 (0.03) vs. 0.03 (0.02) | 0.909a |
| GZMA gene expression | 2.3 (1.7) vs. 3 (4.3) | 0.909a |
| GZMB gene expression | 17.3 (18.7) vs. 26.3 (13) | 0.425a |
| IL-15 gene expression | 13.1 (11.8) vs. 15.7 (6.3) | 0.305a |
| MICA gene expression | 1.2 (1.3) vs. 0.8 (1.6) | 0.909a |
| MICB gene expression | 2.8 (2.4) vs. 2.5 (0.6) | 0.305a |
| KLRK1 gene expression | 1.7 (1.2) vs. 4.6 (4.4) | 0.210a |
a Mann–Whitney U-test
b Chi-square test
Intraepithelial eosinophils
In EoE patients, absolute peak intraepithelial eosinophil density was 56.8 (29.9) cells/hpf, which decreased to 3 (4.2) cells/hpf after SFED-based treatment (p < 0.001). No eosinophils were detected in any of the esophageal samples from controls. No differences in eosinophil counts were detected between atopic and non-atopic EoE patients, being 55 (30.4) vs. 61 (34.8) cells/hpf, respectively. No eosinophilic infiltration was found in duodenal samples.
TLR1, TLR2, TLR4, and TLR9 are overexpressed on the inflamed EoE esophagus
Given that it has been recently demonstrated that TLRs are expressed on esophageal epithelial cells40, here we decided to assess their levels on the inflamed mucosa from EoE patients, as well as on the paired non-inflamed mucosa from the same patients after dietary treatment-induced disease remission compared with healthy controls. Our results, showed that mRNA expression of 4 out of the 6 TLR studied was higher in patients with active EoE, compared to healthy controls: TLR1 (2.7-fold increase), TLR2 (3.7-fold increase), TLR4 (4.6-fold increase), and TLR9 (3.4-fold increase) (p < 0.05 for all comparisons). TLR expression in EoE patients returned to normal following dietary therapy-induced remission (Fig. 1a–f) (p < 0.05 regarding baseline conditions), findings confirmed at the protein level by immunofluorescence (Fig. 1g–j). No significant changes were found for TLR3 and TLR6 mRNA or protein expression. No association was observed between age of patients/controls and TLR expression levels (data not shown).
Fig. 1. TLR overexpression in the esophagus of EoE patients.
a–f mRNA expression (in relative units) of TLR1, TLR2, TLR3, TLR4, TLR6, and TLR9 in esophageal biopsies from EoE patients before and after six-food elimination diet (SFED) treatment, and healthy controls. g–j Immunofluorescence expression of TLR1, TLR2, TLR4, and TLR9 was also determined on the same type of samples. Paired t-test compared EoE patients before and after SFED, while EoE patients (both before and after SFED) were compared with the control population by non-paired t-test. Horizontal bars indicate mean values (*p < 0.05; **p < 0.01; ***p < 0.001)
TLR receptors allow the innate immune system to recognize conserved pathogen associated molecular patterns, so we next determined the total mucosa-associated microbiota load in those samples. The average bacterial load detected in esophageal samples of subjects with active EoE was higher (2.85-fold) compared to control non-EoE samples (p < 0.002), thus confirming previous observations on a pediatric cohort18. Microbiota levels were subsequently normalized (1.16-fold increase) following SFED-induced disease remission (p < 0.005) (Fig. 2a), in parallel with the observed TLRs expression.
Fig. 2. Bacterial load and mucin expression in the esophagus of EoE patients.
a Total microbiota load (determined as 16s gene expression) and b–d mRNA expression (in relative units) of Muc1, Muc4, and Muc5B mucins were determined in esophagus biopsies from patients before and after six-food elimination diet (SFED) treatment, and healthy controls. Paired t-test compared EoE patients before and after SFED, while EoE patients (both before and after SFED) were compared with the control population by non-paired t-test. Horizontal bars indicate mean values (*p < 0.05; **p < 0.01; ***p < 0.001)
Given that the microbiota is not usually in direct contact with the epithelium but, instead, embedded on the mucus-layer, we also studied the expression levels of the mucins that have been described to be expressed by the human esophagus41,42. Our results revealed that, while Muc1 and Muc5B were downregulated by 2-fold (p = 0.023) and 21.5-fold decrease (p = 0.003), respectively, Muc4 was expanded on the inflamed mucosa from EoE patients (7.2-fold increase; p = 0.001) with all mucin levels restored to normal following SFED-induced mucosal healing (Fig. 2b–d). As expected, and in agreement with the literature42, Muc2 expression was not found in our samples.
The innate immune system is activated in the inflamed mucosa from active EoE patients
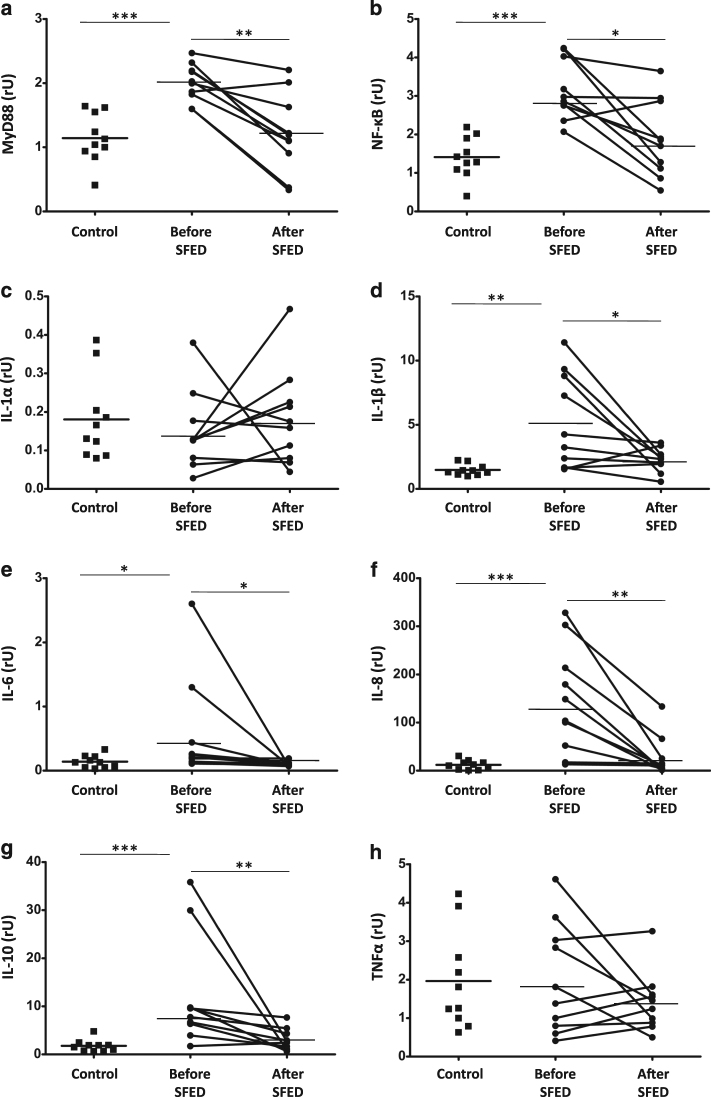
Having identified that the microbiota, TLR receptors, and mucins expression were altered in adult patients with active EoE (Figs. 1 and 2), we next studied whether that could translate to an activated innate immune system in those patients. Every TLR—except TLR3 that was not upregulated in our samples (Fig. 1)—utilizes the adapter protein MyD88 to activate the transcription factor NF-κB43. Therefore, we first assessed the mucosal expression of both transcription factors (Fig. 3a, b), which were upregulated in samples of EoE patients with active disease (1.8- and 2.2-fold increase, respectively; p < 0.001) suggesting that TLR signaling is functional in those patients. In order to further confirm this signaling pathway, we assessed the expression of several NF-κB-induced cytokines. IL-1β (3.5-fold increase; p < 0.01), IL-6 (4-fold increase; p < 0.05), IL-8 (12.2-fold increase; p < 0.001), and IL-10 (6.8-fold increase; p < 0.001) were also upregulated on the inflamed mucosa of EoE patients compared to controls, values that returned to normal following SFED-induced mucosal healing, in parallel to MyD88 and NF-κB (p < 0.001 in both cases). No changes were noted for IL-1α and TNFα, (Fig. 3c–h).
Fig. 3. Innate immune system activation in the esophageal mucosa of EoE patients.
a, b mRNA of transcription factors (MyD88 and NF-κB) and c–h cytokines (IL-1α, IL-1β, IL-6, IL-8, IL-10, and TNFα) expression (in relative units) were determined in esophageal biopsies from patients before and after six-food elimination diet (SFED) treatment, and healthy controls. Paired t-test compared EoE patients before and after SFED, while EoE patients (both before and after SFED) were compared with the control population by non-paired t-test. Horizontal bars indicate mean values (*p < 0.05; **p < 0.01; ***p < 0.001)
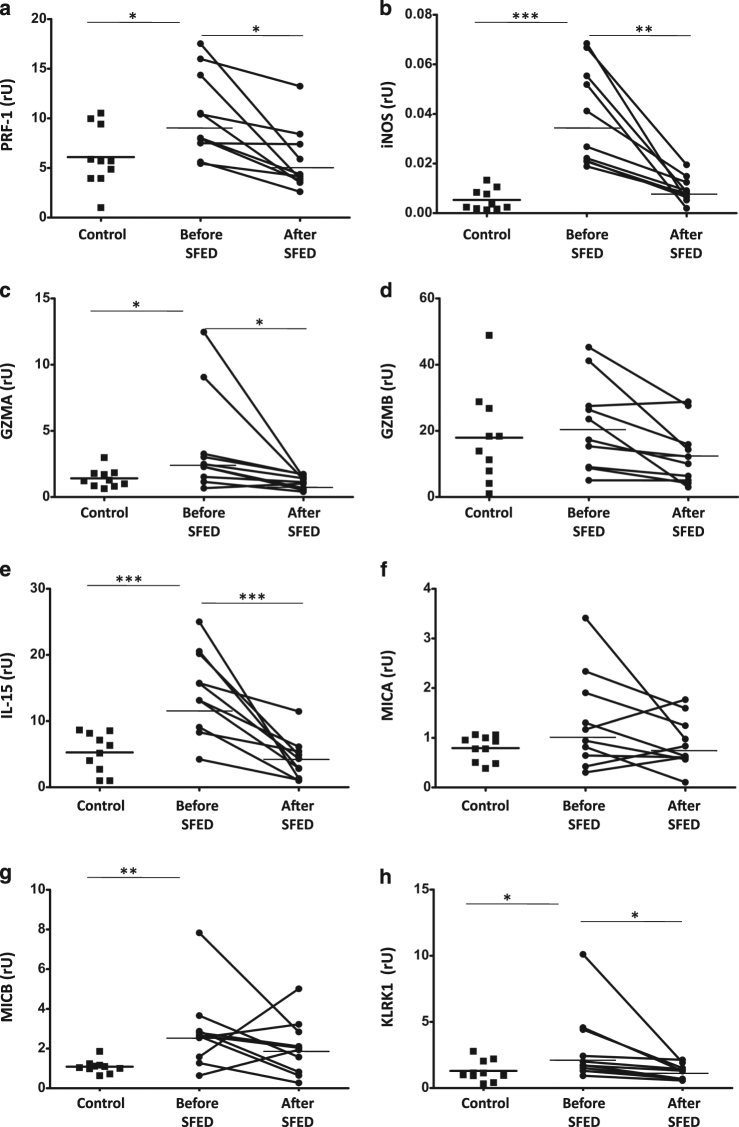
As a consequence of the immune activation displayed by the esophageal mucosa on EoE patients, we further studied whether these changes also correlated with the expression of several innate immune effectors including PRF-1, iNOS, GZMA, and GZMB (Fig. 4a–d) all of them, with the exception of GZMB, were upregulated in the inflamed mucosa from those patients (1.6-, 7.1-, and 2.7-fold increase, respectively; p < 0.05; p < 0.001, and p < 0.05, respectively) compared to controls, with levels being restored to control values following dietary intervention (p < 0.05 compared to baseline).
Fig. 4. Activation of the NKG2D system and expression of effectors of inflammation in the esophageal mucosa of EoE patients.
a–d mRNA of innate immunity effectors (PRF-1, iNOS, GZMA, and GZMB); and e–h the NK-G2D system (IL-15, MIC-A, MIC-B, and KLRK1) expression (in relative units) were determined in esophagus biopsies from patients before and after six-food elimination diet (SFED) treatment, and healthy controls. Paired t-test compared EoE patients before and after SFED, while EoE patients (both before and after SFED) were compared with the control population by non-paired t-test. Horizontal bars indicate mean values (*p < 0.05; **p < 0.01; ***p < 0.001)
Finally, we also assessed expression of the NK-G2D system (IL-15, MICA, MICB, and KLRL1)44, which was also upregulated in the inflamed mucosa from EoE patients (2.8-fold increase for IL-15, 2.6-fold increase for MICB, 2.4-fold increase for KLRK1; p < 0.001, p < 0.01, and p < 0.05, respectively) (Fig. 4e–h). The levels of IL-15 and KLRK1 came back to normal following dietary intervention (p < 0.001 and p < 0.05, respectively). No changes in mRNA expression of MICA were noted. Together, our results confirm that the innate immune system is activated in active EoE patients, hence suggesting that it may participate in its pathogenesis.
Increased duodenal expression of TLR receptors, but not other immune components, in EoE
The esophagus of EoE patients carries a higher bacterial load, which coupled with altered mucus layers and increased levels of TLR receptors (Figs. 1 and 2) results in an activated innate immune system in those patients (Figs. 3 and 4). Therefore, we next studied whether some of those characteristics could also be displayed in other gastrointestinal tissues where EoE patients do not display inflammation, as is the case with the duodenum.
To our surprise, TLR1 (2.04-fold increase, p = 0.001), TLR2 (1.4-fold increase; p = 0.007), and TLR4 (1.4-fold increase; p = 0.013), but not TLR9, were also upregulated in the non-inflamed duodenum from the same patients, levels that were restored to control values in SFED-induced disease remission (Fig. 5a–f). Nevertheless, total bacteria load (Fig. 5g) as well as mucin levels (Supplementary Figure 1) were normal in the same patients. Indeed, the increased expression of duodenal TLRs does not appear to be functional as it did not result in increased levels of transcription factors triggered by TLRs (Supplementary Figure 2A,B), higher cytokine profile (Supplementary Figure 2C-H), levels of innate immunity effectors (Supplementary Figure 3A-D) or the activation of the NK-G2D system (Supplementary Figure 3E-H) on the duodenum.
Fig. 5. The TLR overexpression in the duodenum from EoE patients it not coupled with increased bacterial load.
a–f mRNA expression (in relative units) of TLRs (TLR1, TLR2, TLR3, TLR4, TLR6, and TLR9); and g microbial 16S were determined in duodenal biopsies from patients before and after six-food elimination diet (SFED) treatment, and healthy controls. Paired t-test compared EoE patients before and after SFED, while EoE patients (both before and after SFED) were compared with the control population by non-paired t-test. Horizontal bars indicate mean values (*p < 0.05; **p < 0.01)
Differential genetic signature in the esophagus and the duodenum
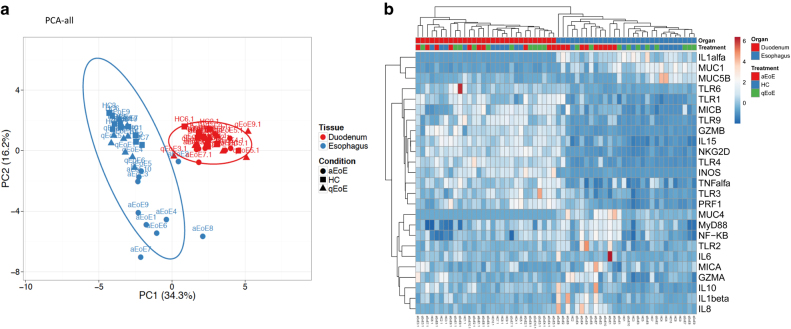
Having detected an altered gene expression profile in samples from patients with active EoE regarding controls with a healthy esophagus, which decreased following SFED-induced remission, we next studied whether that was reflected in a differential gene expression fingerprint for those patients. Given that TLR expression was also higher in the non-inflamed duodenum of EoE patients, we first analyzed all the data revealing that the samples sort together based on the tissue (Fig. 6) irrespective of the source of the patients, as should be expected, given that the esophagus and the duodenum are two different sections of the gastrointestinal tract with different functions and structures.
Fig. 6. The esophagus and the duodenum display a differential gene expression profile.
a Principal component analysis (PCA) and b Heatmaps were determined using all genes detailed in Table 2 from both the esophagus and the duodenum from active (aEoE) and quiescent (qEoE) patients and healthy controls
We also performed a multivariate analysis separating the samples based on the tissue source. Our results revealed how esophageal samples from patients with active EoE display a differential gene expression profile, compared with the EoE samples under remission and from controls with a healthy esophagus, when studied both as a PCA (Supplementary Figure 4A) or as a heatmap analysis (Supplementary Figure 4B), although, the same was not true for the duodenum (Supplementary Figure 5). Therefore, although TLR receptors seem to be constitutively overexpressed throughout the upper gastrointestinal tract during active EoE, their signaling is only functional in the esophagus of these patients, hence keeping the immune response restricted to this segment.
Discussion
This is the first study examining the potential role of TLRs in the pathophysiology of EoE. Our results demonstrate that active EoE is characterized by upregulated esophageal expression of TLRs compared to healthy controls, despite the wide interindividual variability documented in our series of patients. Moreover, transcription factors and subsequent effectors of the TLRs signaling pathway are also upregulated in EoE, and restored to control after effective dietary treatment. In contrast, the duodenal mucosa shows no inflammatory activity despite comparable profile expression of the same TLR genes. This study adds to the cumulative literature investigating the role of TLRs in different gastrointestinal inflammatory conditions, including inflammatory bowel disease29,45,46, celiac disease30,47, food allergy48, and several atopic disorders49,50.
In recent years, multiple studies have investigated the signaling pathways mediated by TLRs in the allergic airway disease51,52, where they regulate immune responses and are connected to the activity of high affinity IgE receptor (FcεRI) expressed on mast cells, acting as a connector between the innate and adaptive immune systems. A predominant role for TLR-2, 4 and 9 has been recognized in bronchial asthma53,54. In contrast, the functioning of TLRs in the gastrointestinal tract has just started to be defined and its role is being increasingly recognized in digestive disorders. However, a map regarding TLR expression by different cell types in different human intestinal segments is still missing, something which is particularly important, as the properties of the immune system change systematically throughout the length of the gastrointestinal tract55. Indeed, focusing on the esophagus, which is particularly exposed to multiple antigens from microbial, alimentary and airborne origin, this organ requires specific mechanisms to protect its mucosa from chronic damage, including an effective peristaltic activity, epithelial tight junctions, and stratified squamous epithelium. Also, the role of esophageal epithelial cells in immune defense and maintenance of tolerance has not yet been fully investigated.
Bronchial asthma and EoE share multiple resemblances, including an altered Th2-type immune response triggered by potentially innocuous antigens, the involvement of eosinophils and mast cells in the pathophysiology17, the transmural inflammation that promotes smooth muscle dysfunction and fibrous remodeling8,56, and clinico-pathological response to topic steroids and avoidance of antigen triggers exposure13,57,58. However, and despite all these similarities, as well as the fact that the prevalence of bronchial asthma among EoE patients is three times higher than in the non-EoE population59, no study has assessed yet the role of TLRs on EoE, as in the case of bronchial asthma52,53. Hence, and given that it has been recently reported that TLR receptors are expressed in the healthy esophagus40,60, we decided to characterize their expression in the context of EoE by describing how TLR-1, TLR-2, TLR-4, and TLR-9 are expanded in the inflamed mucosa from active EoE patients, and its modulation by SFED.
TLR-1 responds to triacyl lipopeptides and TLR-2 to lipotecoic acid and peptidoglycan, both being components of the bacterial wall61. Both are involved in reducing the activation of FcεRI54,62, which results in a reduced IgE-mediated mast cell degranulation. TLR-4, on the contrary, is stimulated upon exposure to lipopolysaccharide present in Gram-negative bacteria. Some allergens (such as the major house dust mite allergen or Derp2) show a structural homology with MD2 protein, which is a co-mediator of TLR-4 activation54,63, and could activate TLR-4-mediated response by a molecular mimicry mechanism. In contrast to TLR-1 and 2, stimulation of TLR-4 increases the activation of FcεRI and promotes Th2-type cytokines involved in eosinophilic responses, as documented in respiratory tract allergy64. In resting conditions, TLR-4 expression is reduced in bronchial mucosal referred to TLR-2, with the TLR-2/4 ratio determining the final sense of the FcεRI activation65,66. In the particular case of our EoE samples, expression of TLR-2 was 10-fold higher that TLR-1, in agreement with the lack of a significant role for IgE in EoE patients. Indeed, IgE plays a limited role in the pathophysiology of EoE, and although it binds to mast cells in the epithelium of atopic patients with this disease67, it does not constitute its main route of activation17. Hence, EoE patients do not develop rapid inflammatory responses following exposure to triggering foods68, and treatment with anti-IgE monoclonal antibodies has been shown to be completely ineffective improving esophageal symptoms and inflammation in patients with EoE10. Finally, TLR-9 is an intracellular receptor activated by bacterial CpG-DNA binding, promoting Th1-type immune responses with increased production of IFNα-b. The stimulation of FcεRI by allergens suppresses the activation of TLR-9, with the consequent reduction of Th1 responses and the promotion of Th2 ones leading to the appearance of allergic reactions.
The increased expression of TLR in active EoE was accompanied by higher bacterial load detected in the same samples and by a downregulation in Muc1 and Muc5B genes, probably determined by epithelial cell damage and dysfunction, impaired mucosal integrity, and increased permeability69 with exposition to bacterial components, and enhanced activation of the mucosal innate immunity mediated by TLRs, which is restored after avoiding exposure to food antigens. Despite constituting a plausible explanation for our results, some other findings point towards the hypothesis that TLRs may play a primary role in EoE. To begin with, an increased expression of Muc4 gene was found, potentially to compensate for the decrease in Muc1 and Muc5B, which suggests that the mucous integrity in active EoE is preserved enough to limit a direct contact of the esophageal microbiota with the mucosal surface. In addition, signaling pathways specific for TLR activation (IL-8, MyD88), together with increased production of several effectors of direct cytotoxicity (PRF-1, GZMA, iNOS) make it hard to consider TLR activation as an epiphenomenon. Notably, increased TLR expression was also found in the duodenum from the same patients despite having a non-inflamed mucosa (as confirmed both during endoscopy and histological assessment) while displaying no changes in bacterial load or upregulation of mediators of inflammation. All together, our results suggest that TLRs are primarily involved in EoE pathogenesis. It can be speculated therefore that an overexpression of TLRs in non-inflamed segments of the gastrointestinal tract of EoE patients could parallel the increase of proinflammatory cytokines also in non-inflamed tissue from patients with IBD33. The question remains, however, why if TLRs are also overexpressed on the non-inflamed mucosa from these patients, disease is nevertheless restricted to the esophagus. One possibility is that increased mucosa-associated microbiota load (or its metabolic activity) in the esophagus (but not in the duodenum) may be the trigger for inflammation (either as a direct effect or by mimicking dietary components), an issue which we are currently studying. Indeed, and given the study approach we used (qPCR), cell types responsible for mucosal TLRs expression were not defined; ongoing work is trying to address this point, by defining the exact immune or epithelial cells that overexpress TLRs, in agreement with recent observations27,40. Last, but not least, current work is also characterizing the mucosa-associated microbiota from those patients, with the overall aim of unraveling the specific microbiota contribution to EoE pathogenesis.
We are aware, however, of the limitations from our study, the main one being the limited sample size (only 10 subjects per group) This was as a consequence of the difficulty in recruiting patients who are naïve to therapy and who also responded to a SFED. Despite the significant differences in gene expression levels between EoE and control samples, a wide variability in expression levels from patient to patient was documented, which prohibits a simplified interpretation of the data. The small number of patients included in this study therefore prevented deeper analysis of the source of such variability. We are also aware that our control group included gender but not aged-matched healthy individuals. This is due to the fact that, according to current guidelines for managing dyspepsia, endoscopic exams can be avoided in young patients who do not present alarming symptoms70. Nevertheless, we feel that these limitations are lessened by the fact that we have only included patients with EoE at the moment of diagnosis, hence, eliminating the effect of previous exposure to topical steroids or any other anti-inflammatory drugs. As such, baseline eosinophil densities and gene expression levels are a true reflection of the pathophysiological changes associated with EoE.
In summary, we here provide evidence, for the first time to our knowledge, that TLR-dependent signaling pathways are activated in the esophageal mucosa of adult patients with EoE, strongly suggesting a role in the pathophysiology of the disease. The exact mechanisms however that mediate the complex interactions between esophageal microbiota, the innate immune system and food-specific inflammatory responses in the pathophysiology of EoE warrants further research.
Study Highlights
What is current knowledge
EoE is a predominantly Th2-type inflammatory esophageal condition, in which preliminary evidence points to a potential role for innate immunity.
The role of TLRs has been evaluated in several inflammatory, autoimmune, and allergic diseases. However, a possible involvement of TLR-mediated signaling in the pathophysiology of EoE has not yet been documented.
What is new here
Active EoE is characterized by an upregulated expression of TLRs in the esophageal mucosa compared to healthy controls, which returns to normal after dietary therapy-induced remission.
Activation of TLRs in the esophageal mucosa of patients with EoE supports a relevant role for microbiota in the pathophysiology of the disease.
TLR-mediated signaling pathways are functional in the esophageal mucosa of patients with active EoE, promoting an activation of the innate immune system that is restricted to the esophagus and contributes to cell damage.
Electronic supplementary material
Competing interests
Guarantor of the article: AJ Lucendo
Specific author contributions: Á.A.: Study conception and design, collection, analysis, and interpretation of data, drafting and revision of the manuscript and approval of the final version of the manuscript. M.V.: Study conception and design, immunofluorescence analysis of esophageal samples, interpretation of data, drafting of the manuscript, and approval of the final version of the manuscript. D.B.: Study conception and design, analysis and interpretation of data, drafting and revision of the manuscript, and approval of the final version of the manuscript. J.M.O.: Histological analysis of esophageal samples, interpretation of data, and approval of the final version of the manuscript. M.F.: Immunofluorescence analysis of esophageal samples, interpretation of data, and approval of the final version of the manuscript. A.M.-A.: Analysis and interpretation of data, and approval of the final version of the manuscript. P.M.-F.: Study conception and design, collection, analysis, and interpretation of data, and approval of the final version of the manuscript. A.M.G.-C.: Immunofluorescence analysis of esophageal samples, interpretation of data, and approval of the final version of the manuscript. T.M.-H.: Histological processing of esophageal samples, interpretation of data, and approval of the final version of the manuscript. L.A.-G.: Interpretation of data and approval of the final version of the manuscript. A.J.L.: Study conception and design, patient diagnosis (performance of endoscopic examinations and esophageal biopsies) and follow-up, interpretation of data, drafting of the manuscript, and approval of the final version of the manuscript.
Financial support: This work was supported by grants from The Castilla-La Mancha Health Research Foundation (Fundación para la Investigación Sanitaria de Castilla-La Mancha or FISCAM) grant PI-2010/038 (A.J.L.); Vall d’Hebron Institut de Recerca, Programa de becas predoctorales “Amics de Vall d’Hebron”: PRED-VHIR-2014-018 (M.F.), and Fondo Europeo de Desarrollo Regional (FEDER), Fondo de Investigación en Salud and Centro de Investigación Biomédica en Red de Enfermedades Hepáticas y Digestivas-CIBEREHD, Instituto de Salud Carlos III, Subdirección General de Investigación Sanitaria, Ministerio de Economía y Competitividad: CPII16/00031 and PI16/00583 (M.V.) and CIBEREHD EDH16PI02 (M.V. and D.B.).
Potential competing interests: None to report.
Footnotes
Electronic supplementary material
The online version of this article (10.1038/s41424-018-0017-4) contains supplementary material, which is available to authorized users.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lucendo AJ, et al. Guidelines on eosinophilic esophagitis: evidence-based statements and recommendations for diagnosis and management in children and adults. United Eur. Gastroenterol. J. 2017;5:335–358. doi: 10.1177/2050640616689525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molina-Infante J, et al. Proton pump inhibitor-responsive oesophageal eosinophilia: an entity challenging current diagnostic criteria for eosinophilic oesophagitis. Gut. 2016;65:521–531. doi: 10.1136/gutjnl-2015-310991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucendo AJ, et al. Immunophenotypic characterization and quantification of the epithelial inflammatory infiltrate in eosinophilic esophagitis through stereology: an analysis of the cellular mechanisms of the disease and the immunologic capacity of the esophagus. Am. J. Surg. Pathol. 2007;31:598–606. doi: 10.1097/01.pas.0000213392.49698.8c. [DOI] [PubMed] [Google Scholar]
- 4.Arias A, et al. Systematic review with meta-analysis: The incidence and prevalence of eosinophilic oesophagitis in children and adults in population-based studies. Aliment Pharmacol. Ther. 2016;43:3–15. doi: 10.1111/apt.13441. [DOI] [PubMed] [Google Scholar]
- 5.Straumann A, et al. Idiopathic eosinophilic esophagitis is associated with a T(H)2-type allergic inflammatory response. J. Allergy Clin. Immunol. 2001;108:954–961. doi: 10.1067/mai.2001.119917. [DOI] [PubMed] [Google Scholar]
- 6.Bullock JZ, et al. Interplay of adaptive th2 immunity with eotaxin-3/c-C chemokine receptor 3 in eosinophilic esophagitis. J. Pediatr. Gastroenterol. Nutr. 2007;45:22–31. doi: 10.1097/MPG.0b013e318043c097. [DOI] [PubMed] [Google Scholar]
- 7.Lucendo AJ, Lucendo B. An update on the immunopathogenesis of eosinophilic esophagitis. Expert Rev. Gastroenterol. Hepatol. 2010;4:141–148. doi: 10.1586/egh.10.9. [DOI] [PubMed] [Google Scholar]
- 8.Lucendo AJ. Cellular and molecular immunological mechanisms in eosinophilic esophagitis: an updated overview of their clinical implications. Expert Rev. Gastroenterol. Hepatol. 2014;8:669–685. doi: 10.1586/17474124.2014.909727. [DOI] [PubMed] [Google Scholar]
- 9.Vicario M, et al. Local B cells and IgE production in the oesophageal mucosa in eosinophilic oesophagitis. Gut. 2009;51:12–20. doi: 10.1136/gut.2009.178020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clayton F, et al. Eosinophilic esophagitis in adults is associated with IgG4 and not mediated by IgE. Gastroenterology. 2014;147:602–609. doi: 10.1053/j.gastro.2014.05.036. [DOI] [PubMed] [Google Scholar]
- 11.Aceves SS, et al. Esophageal remodeling in pediatric eosinophilic esophagitis. J. Allergy Clin. Immunol. 2007;119:206–212. doi: 10.1016/j.jaci.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Lucendo AJ, et al. Subepithelial collagen deposition, profibrogenic cytokine gene expression, and changes after prolonged fluticasone propionate treatment in adult eosinophilic esophagitis: a prospective study. J. Allergy Clin. Immunol. 2011;128:1037–1046. doi: 10.1016/j.jaci.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Arias Aacute, et al. Efficacy of dietary interventions for inducing histologic remission in patients with eosinophilic esophagitis: a systematic review and meta-analysis. Gastroenterology. 2014;146:1639–1648. doi: 10.1053/j.gastro.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Molina-Infante, J., et al. Step-up empiric elimination diet for pediatric and adult eosinophilic esophagitis: the 2-4-6 Study. J. Allergy Clin. Immunol.10.1016/j.jaci.2017.08.038 (2017). [DOI] [PubMed]
- 15.Blanchard C, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J. Allergy Clin. Immunol. 2007;120:1292–1300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 16.Lexmond WS, et al. Involvement of the iNKT cell pathway is associated with early-onset eosinophilic esophagitis and response to allergen avoidance therapy. Am. J. Gastroenterol. 2014;109:646–657. doi: 10.1038/ajg.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arias Aacute, et al. Dietary treatment modulates mast cell phenotype, density, and activity in adult eosinophilic oesophagitis. Clin. Exp. Allergy. 2016;46:78–91. doi: 10.1111/cea.12504. [DOI] [PubMed] [Google Scholar]
- 18.Harris JK, et al. Esophageal microbiome in eosinophilic esophagitis. PLoS ONE. 2015;10:e0128346. doi: 10.1371/journal.pone.0128346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benitez AJ, et al. Inflammation-associated microbiota in pediatric eosinophilic esophagitis. Microbiome. 2015;3:23. doi: 10.1186/s40168-015-0085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen ET, et al. Early life exposures as risk factors for pediatric eosinophilic esophagitis. J. Pediatr. Gastroenterol. Nutr. 2013;57:67–71. doi: 10.1097/MPG.0b013e318290d15a. [DOI] [PubMed] [Google Scholar]
- 21.Jiménez-Dalmaroni MJ, Gerswhin ME, Adamopoulos IE. The critical role of toll-like receptors--from microbial recognition to autoimmunity: a comprehensive review. Autoimmun. Rev. 2016;15:1–8. doi: 10.1016/j.autrev.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satoh, T. & Akira, S. Toll-like receptor signaling and its inducible proteins. Microbiol. Spectr. 10.1128/microbiolspec.MCHD-0040-201 (2016). [DOI] [PubMed]
- 23.Kumagai Y, Akira S. Identification and functions of pattern-recognition receptors. J. Allergy Clin. Immunol. 2016;125:985–992. doi: 10.1016/j.jaci.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 24.van Egmond M, Vidarsson G, Bakema JE. Cross-talk between pathogen recognizing Toll-like receptors and immunoglobulin Fc receptors in immunity. Immunol. Rev. 2015;268:311–327. doi: 10.1111/imr.12333. [DOI] [PubMed] [Google Scholar]
- 25.Tunis MC, Marshall JS. Toll-like receptor 2 as a regulator of oral tolerance in the gastrointestinal tract. Mediat. Inflamm. 2014;2014:606383. doi: 10.1155/2014/606383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsybikov NN, et al. Expression of TLR2 and TLR4 on peripheral blood monocytes during exacerbation of atopic dermatitis. Allergy Asthma Proc. 2015;36:140–145. doi: 10.2500/aap.2015.36.3901. [DOI] [PubMed] [Google Scholar]
- 27.Mulder DJ, et al. Expression of Toll-like receptors 2 and 3 on esophageal epithelial cell lines and on eosinophils during esophagitis. Dig. Dis. Sci. 2012;57:630–642. doi: 10.1007/s10620-011-1907-4. [DOI] [PubMed] [Google Scholar]
- 28.Liacouras CA, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J. Allergy Clin. Immunol. 2011;128:3–20. doi: 10.1016/j.jaci.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 29.Hart AL, et al. Characteristics of intestinal dendritic cells in inflammatory bowel diseases. Gastroenterology. 2005;129:50–65. doi: 10.1053/j.gastro.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 30.Szebeni B, et al. Increased mucosal expression of Toll-like receptor (TLR)2 and TLR4 in coeliac disease. J. Pediatr. Gastroenterol. Nutr. 2007;45:187–193. doi: 10.1097/MPG.0b013e318064514a. [DOI] [PubMed] [Google Scholar]
- 31.Lucendo AJ, et al. Empiric 6-food elimination diet induced and maintained prolonged remission in patients with adult eosinophilic esophagitis: a prospective study on the food cause of the disease. J. Allergy Clin. Immunol. 2013;131:797–804. doi: 10.1016/j.jaci.2012.12.664. [DOI] [PubMed] [Google Scholar]
- 32.Zaninotto, G., et al. Treatment of esophageal achalasia with laparoscopic Heller myotomy and Dor partial anterior fundoplication: prospective evaluation of 100 consecutive patients. J. Gastrointest. Surg.4, 282–289 (2000). [DOI] [PubMed]
- 33.León AJ, et al. High levels of proinflammatory cytokines, but not markers of tissue injury, in unaffected intestinal areas from patients with IBD. Mediat. Inflamm. 2009;2009:580450. doi: 10.1155/2009/580450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl Acad. Sci. USA. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Metsalu T, Vilo J. ClustVis. A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ihaka R, Gentleman R. R: a language for data analysis and graphics. J. Comput. Graph Stat. 1996;5:299–314. [Google Scholar]
- 38.Spitzer M, et al. BoxPlotR: a web tool for generation of box plots. Nat. Methods. 2014;11:121–122. doi: 10.1038/nmeth.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rokach, L., Maimon, O. in Data Mining and Knowledge Discovery Handbook (eds Rokach, L. & Maimon, O.) p. 321–352 (Springer, New York, 2005).
- 40.Lim DM, et al. TLR3-mediated NF-{kappa}B signaling in human esophageal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;297:G1172–G1180. doi: 10.1152/ajpgi.00065.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guillem P, et al. Mucin gene expression and cell differentiation in human normal, premalignant and malignant esophagus. Int J. Cancer. 2000;88:856–861. doi: 10.1002/1097-0215(20001215)88:6<856::AID-IJC3>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 42.Flucke U, et al. Immunoreactivity of cytokeratins (CK7, CK20) and mucin peptide core antigens (MUC1, MUC2, MUC5AC) in adenocarcinomas, normal and metaplastic tissues of the distal oesophagus, oesophago-gastric junction and proximal stomach. Histopathology. 2003;43:127–134. doi: 10.1046/j.1365-2559.2003.01680.x. [DOI] [PubMed] [Google Scholar]
- 43.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 44.Montalban-Arques A, et al. Propionibacterium acnes overabundance and natural killer group 2 member D system activation in corpus-dominant lymphocytic gastritis. J. Pathol. 2016;240:425–436. doi: 10.1002/path.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hausmann M, et al. Toll-like receptors 2 and 4 are up-regulated during intestinal inflammation. Gastroenterology. 2002;122:1987–2000. doi: 10.1053/gast.2002.33662. [DOI] [PubMed] [Google Scholar]
- 46.Cario E. Bacterial interactions with cells of the intestinal mucosa: Toll-like receptors and NOD2. Gut. 2005;54:1182–1193. doi: 10.1136/gut.2004.062794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalliomäki M, et al. Expression of microbiota, Toll-like receptors, and their regulators in the small intestinal mucosa in celiac disease. J. Pediatr. Gastroenterol. Nutr. 2012;54:727–732. doi: 10.1097/MPG.0b013e318241cfa8. [DOI] [PubMed] [Google Scholar]
- 48.Westerholm-Ormio M, et al. Infiltration of Foxp3- and Toll-like receptor-4-positive cells in the intestines of children with food allergy. J. Pediatr. Gastroenterol. Nutr. 2010;50:367–376. doi: 10.1097/MPG.0b013e3181cd2636. [DOI] [PubMed] [Google Scholar]
- 49.Zuo L, et al. Molecular regulation of Toll-like receptors in asthma and COPD. Front. Physiol. 2016;6:312. doi: 10.3389/fphys.2015.00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henmyr V, et al. Characterization of genetic variation in TLR8 in relation to allergic rhinitis. Allergy. 2016;71:333–341. doi: 10.1111/all.12805. [DOI] [PubMed] [Google Scholar]
- 51.Ryu JH, et al. Distinct TLR-mediated pathways regulate house dust mite-induced allergic disease in the upper and lower airways. J. Allergy Clin. Immunol. 2013;131:549–561. doi: 10.1016/j.jaci.2012.07.050. [DOI] [PubMed] [Google Scholar]
- 52.Bezemer GFG, et al. Dual role of Toll-like receptors in asthma and chronic obstructive pulmonary disease. Pharmacol. Rev. 2012;64:337–358. doi: 10.1124/pr.111.004622. [DOI] [PubMed] [Google Scholar]
- 53.Gangloff SC, Guenounou M. Toll-like receptors and immune response in allergic disease. Clin. Rev. Allergy Immunol. 2004;26:115–125. doi: 10.1007/s12016-004-0006-0. [DOI] [PubMed] [Google Scholar]
- 54.Novak N, Bieber T, Peng WM. The immunoglobulin E-Toll-like receptor network. Int Arch. Allergy Immunol. 2010;151:1–7. doi: 10.1159/000232565. [DOI] [PubMed] [Google Scholar]
- 55.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 2014;14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 56.Anto JM, et al. Mechanisms of the development of allergy (MeDALL): introducing novel concepts in allergy phenotypes. J. Allergy Clin. Immunol. 2017;139:388–399. doi: 10.1016/j.jaci.2016.12.940. [DOI] [PubMed] [Google Scholar]
- 57.Rosenberg SR, Kalhan R. Recent advances in the management of chronic obstructive pulmonary disease. F1000Res. 2017;6:863. doi: 10.12688/f1000research.9819.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chuang MYA, et al. Topical steroid therapy for the treatment of eosinophilic esophagitis (EoE): a systematic review and meta-analysis. Clin. Transl. Gastroenterol. 2015;6:e82. doi: 10.1038/ctg.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.González-Cervera J, et al. Association between atopic manifestations and eosinophilic esophagitis: a systematic review and meta-analysis. Ann. Allergy Asthma Immunol. 2017;118:582–590. doi: 10.1016/j.anai.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 60.Uehara A, et al. Various human epithelial cells express functional Toll-like receptors, NOD1 and NOD2 to produce anti-microbial peptides, but not proinflammatory cytokines. Mol. Immunol. 2007;44:3100–3111. doi: 10.1016/j.molimm.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 61.Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem Biophys. Res. Commun. 2009;388:621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- 62.Yoshioka M, et al. Lipoteichoic acid downregulates FcepsilonRI expression on human mast cells through Toll-like receptor 2. J. Allergy Clin. Immunol. 2007;120:452–461. doi: 10.1016/j.jaci.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 63.Trompette A, et al. Allergenicity resulting from functional mimicry of a Toll-like receptor complex protein. Nature. 2009;457:585–588. doi: 10.1038/nature07548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tworek D, et al. Toll-like receptor-induced expression of epithelial cytokine receptors on haemopoietic progenitors is altered in allergic asthma. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2017;47:900–908. doi: 10.1111/cea.12913. [DOI] [PubMed] [Google Scholar]
- 65.Panzer R, et al. TLR2 and TLR4 expression in atopic dermatitis, contact dermatitis and psoriasis. Exp. Dermatol. 2014;23:364–366. doi: 10.1111/exd.12383. [DOI] [PubMed] [Google Scholar]
- 66.Zanin-Zhorov A, Cohen IR. Signaling via TLR2 and TLR4 directly down-regulates T cell effector functions: the regulatory face of danger signals. Front Immunol. 2013;4:211. doi: 10.3389/fimmu.2013.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mulder DJ, et al. Atopic and non-atopic eosinophilic oesophagitis are distinguished by immunoglobulin E-bearing intraepithelial mast cells. Histopathology. 2012;61:810–822. doi: 10.1111/j.1365-2559.2012.4303.x. [DOI] [PubMed] [Google Scholar]
- 68.Simon D, et al. Eosinophilic esophagitis is characterized by a non-IgE-mediated food hypersensitivity. Allergy. 2016;71:611–620. doi: 10.1111/all.12846. [DOI] [PubMed] [Google Scholar]
- 69.van Rhijn BD, et al. Histological response to fluticasone propionate in patients with eosinophilic esophagitis is associated with improved functional esophageal mucosal integrity. Am. J. Gastroenterol. 2015;110:1289–1297. doi: 10.1038/ajg.2015.247. [DOI] [PubMed] [Google Scholar]
- 70.Moayyedi PM, et al. ACG and CAG clinical guideline: management of dyspepsia. Am. J. Gastroenterol. 2017;112:988–1013. doi: 10.1038/ajg.2017.154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.