Abstract
Sirtuins are NAD+-dependent protein deacylases capable of cleaving off acetyl as well as other acyl groups from the ɛ-amino group of lysines in histones and other substrate proteins. They have been reported as promising drug targets, and thus modulators of their activity are needed as molecular tools to uncover their biological function and as potential therapeutics. Here, we present new assay formats that complement existing assays for sirtuin biochemistry and cellular target engagement. Firstly, we report the development of a homogeneous fluorescence-based activity assay using unlabelled acylated peptides. Upon deacylation, the free lysine residue reacts with fluorescamine to form a fluorophore. Secondly, using click chemistry with a TAMRA-azide on a propargylated sirtuin inhibitor, we prepared the first fluorescently labelled small-molecule inhibitor of Sirt2. This is used in a binding assay, which is based on fluorescence polarization. We used it successfully to map potential inhibitor-binding sites and also to show cellular Sirt2 engagement. By means of these new assays, we were able to identify and characterize novel Sirt2 inhibitors out of a focused library screen. The binding of the identified Sirt2 inhibitors was rationalized by molecular docking studies. These new chemical tools thus can enhance further sirtuin research.
This article is part of a discussion meeting issue ‘Frontiers in epigenetic chemical biology'.
Keywords: sirtuins, Sirt2, NAD+, assays, epigenetics, deacetylases
1. Introduction
The human genome codes for 18 different lysine deacetylases (KDACs), which have been grouped into four classes, according to their sequence homology to yeast KDACs [1]. These enzymes are capable of removing not only acetyl but also other acyl groups from the ɛ-amino group of acylated lysine residues. Sirtuins, which had been initially described as class III histone deacetylases (HDACs), use NAD+ as a cofactor and constitute the class III KDACs. Seven different human isotypes of sirtuins (Sirt1–7) have been identified. They differ in their catalytic activity as well as subcellular localization [2,3]. In contrast to the Zn2+-dependent KDACs, which are mostly restricted to deacetylation [4], sirtuins can remove, e.g., glutaryl [5], succinyl [6], crotonyl [7], myristoyl [8] and palmitoyl [9] groups. Recently, the catalytic activity of isotypes Sirt6 and Sirt7 was shown to be increased by long-chain fatty acids and nucleic acids, respectively [8,10,11]. Aside from histones, a multitude of non-histone proteins have been discovered as sirtuin substrates in the last few years, e.g. p53 [12], α-tubulin [13], NFκB [14] and BubR1 [15]. By controlling the acylation state of their substrate proteins, sirtuins are involved in the regulation of a variety of cellular processes such as apoptosis [16], transcription [17], ageing [18], metabolic sensing [16] and inflammation [19]. The isotype Sirt2 is predominantly localized in the cytoplasm and was shown to have a significant impact on autophagy [20,21], peripheral myelination [22], cell-cycle regulation [13], and immune and inflammatory response [23–26]. Despite its robust deacetylase activity, a recent study reported an even higher catalytic efficiency (kcat/Km) of Sirt2 for demyristoylation compared with deacetylation [27]. Sirt2 dysregulation has been reported to be associated with the pathogenesis of type II diabetes [28], bacterial infections [24,25], neurodegenerative diseases [29] and cancer [30,31], which highlights Sirt2 as a promising target for pharmaceutical intervention. Yet, for some diseases, e.g. Huntington's disease, there is conflicting evidence whether Sirt2 has to be downregulated, inhibited or upregulated to ameliorate specific disease conditions [32–34]. A number of cell-based and animal studies have tried to answer these questions. However, they have been impeded by the lack of tool compounds that show suitable isotype selectivity as well as pharmacokinetic properties. The urgent need for such compounds, to further interrogate the consequences of Sirt2-dependent deacylation as well as their impact on downstream signalling, resulted in the discovery of several Sirt2-selective small-molecule inhibitors [35–45]. Different assays have been applied for the discovery and the characterization of these inhibitors. However, most of the deacylase activity assays that are used for high-throughput screening for Sirt2 modulators are based on the conversion of fluorophore-labelled model substrates [46–50]. Such fluorophore labels were reported to impair the binding of the acyl-lysine to the active site of sirtuins, leading to an unphysiological binding mode of the model substrate, which may result in misleading assay artefacts. This phenomenon was extensively studied using the example of Sirt1 [51–54], which highlights the importance of activity assays for sirtuins that are based on unlabelled substrates. In contrast to biochemical deacylase activity assays, which serve to characterize the intrinsic activity of potential enzyme modulators, binding assays can provide insights into inhibitor-binding sites and affinities. The sequence of the amino acids that give form to the catalytic core domain of the different sirtuin isotypes is highly conserved. Therefore, all mammalian sirtuins share a catalytic core domain with a high degree of structural similarity. Despite this, some isotype-selective modulators have been discovered [35–45]. One class of these inhibitors is the aminothiazoles (1a/b–3a/b), also referred to as sirtuin rearranging ligands (SirReals, figures 1 and 2) [55–57]. These inhibitors are highly selective Sirt2 inhibitors with IC50 values in the nanomolar range. The SirReals bind to the extended C-site (ECS) as well as to the so-called ‘selectivity pocket', which is present neither in the disengaged apo ‘open conformation' nor in the ‘closed conformation', which is formed upon binding of the substrate. The dimethylmercaptopyrimidine residue triggers the formation of this new binding pocket by rearranging two loops of the hinge region, along with freezing the enzyme in the so-called ‘locked-open conformation' (figure 1b). Recently, Sundriyal et al. [45] discovered Sirt2-selective inhibitors that are structurally different from the SirReals, which gain Sirt2-selectivity by also binding to the ‘selectivity pocket'. Also a myristoylated peptide was shown to bind with the long-chain acyl group in this pocket [58]. These findings highlight the importance of the ‘selectivity pocket’ for the development of novel Sirt2-selective inhibitors. Although having a strong inhibitory effect on deacetylation, the SirReal 2a (figure 2) was shown to not impair the ability of Sirt2 to cleave off longer fatty acids like myristic acid [49]. These studies made use of an assay that uses a fluorescently labelled peptide as deacylation substrate. To validate these findings and to gain further insights into the ‘acyl selectivity profile' of Sirt2 inhibitors, one goal of this study was to provide an activity assay for Sirt2 that uses unlabelled peptide substrates in order to be able to investigate both deacetylation and the removal of longer acyl groups on unlabelled substrates in an assay amenable to high-throughput screening. A second goal was to develop a screening tool that can be used to interrogate binding to the ECS and to the ‘selectivity pocket' of Sirt2, which cannot be studied by existing microplate reader-based methods. Current protocols only allow probe binding to the substrate or cofactor-binding site of Sirt2 using rather time- and work-consuming kinetic competition studies [55].
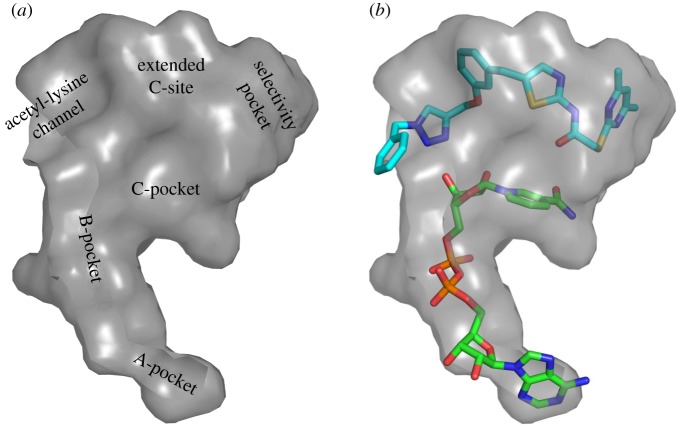
Figure 1.
Surface representation of the active site of Sirt2 in the ‘locked-open conformation'. (a) Labelling of the different binding pockets, (b) orientation of 3a (light blue) and NAD+ (green).
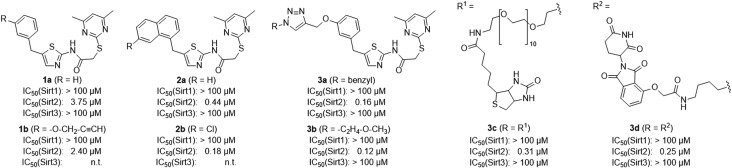
Figure 2.
Chemical structures and inhibition data of selected SirReals (1a/b−3a/b), the SirReal-based affinity probe for Sirt2 (3c) and the Sirt2 degradation probe (3d). n.t., not tested.
2. Assay development
As already mentioned in the Introduction, there is a need for both activity assays that do not rely on fluorescent substrates and binding assays that allow the identification and characterization of new sirtuin inhibitors in a high-throughput manner. In this paper, we first present the development of a homogeneous fluorescence-based fluorescamine assay that uses unlabelled peptide substrates to determine effects on the deacylase activity of sirtuins and then the development of a binding assay based on fluorescence polarization (FP).
(a). Homogeneous fluorescence-based fluorescamine assay
To determine effects on Sirt2 activity by means of an unlabelled peptide substrate, we developed a homogeneous assay with a fluorescence readout. The spiro-compound fluorescamine acts as a developer to obtain a fluorescence signal. Fluorescamine itself is a non-fluorescent compound, but reacts selectively with primary amines to give a fluorescent product (figure 3) [59]. However, the fluorophore formation is not limited to the primary amines of the deacylated substrate lysines; other primary amines of the enzyme preparation or the buffer are also able to react with fluorescamine. Therefore, an amine-free buffer as well as a highly purified protein preparation are indispensable for a feasible assay set-up. By means of our well-established expression and purification protocol, we are able to obtain Sirt2 and 3 with a purity of over 90%, as analysed by SDS-PAGE. As the developing reaction can hardly be performed under physiological conditions in terms of buffer composition, the fluorophore formation is typically initiated after the enzymatic reaction has been terminated. Thus, fluorescamine-based assays are usually performed as non-continuous endpoint assays. The underlying assay principle has already been reported in a study mainly focused on Zn2+-dependent KDACs, which also included initial data on an application for Sirt1. Long-chain deacylation was not covered [60]. For the set-up of our fluorescamine-based assay, we used a substrate derived from α-tubulin, a physiological sirtuin substrate, either acetylated or myristoylated at the ɛ-nitrogen of its lysine residue. For reasons of comparability with the ZMAL-assay [47], which is a well-established assay for sirtuins and Zn2+-dependent KDACs, we referred during assay development to the conditions (assay volume, NAD+ concentration and incubation temperature) that are applied for the ZMAL-assay. As a major step in assay development, the Km values of the myristoylated peptide for Sirt2 and 3 and of the acetylated peptide for Sirt3 were determined (figure 4a–c). According to a high pressure liquid chromatography (HPLC)-based Sirt2-deacetylation assay which makes use of the same α-tubulin-derived peptides [55], the concentrations of the peptides were fixed at 20 µM for the acetylated and 10 µM for the myristoylated peptide, respectively. The substrate concentration should be in the range of the Km value or slightly below to be sensitive for substrate-competitive inhibitors. Owing to enzyme kinetic measurements, we fixed the concentration of the enzymes at a substrate conversion of up to 30% in 1 h to ensure linear conditions (figure 4d; electronic supplementary material, figure S1A). Fluorescamine requires a non-hydrolytic environment to form a fluorophore with primary amines; otherwise the reagent is deactivated by hydrolysis. Furthermore, the fluorescence intensity of the formed fluorophore is pH-dependent [61]. To screen for conditions that facilitate the greatest possible fluorophore formation rate and fluorescence intensity of the fluorophore in the final developing step after the deacetylation had been stopped, solvent composition and solvent pH were varied. As shown in electronic supplementary material, figure S1C, dimethyl sulfoxide (DMSO), acetone, acetonitrile and dimethyl formamide are suitable solvents for the fluorophore formation. For further studies, we used acetone as the solvent for fluorescamine. A final assay concentration of 33% of acetone or higher is recommended for the final developing step (see electronic supplementary material, figure S1D). To examine the effects of OH− concentration on the total fluorescence signal, we investigated different NaOH concentrations. The highest signal window was obtained by adding 5 µl of 0.1 M NaOH (see electronic supplementary material, figure S1E). For a better physiological imitation, the assay is performed at pH 8.0 and NaOH is added after the incubation is finished. A final fluorescamine assay concentration of 62.5 µM is sufficient to gain maximum fluorescence readout (figure 4e). Variations in DMSO content of the assay buffer used during the enzymatic conversion showed only a minor impact of Sirt2 activity (see electronic supplementary material, figure S1F). However, according to previous studies, a decrease in enzyme activity by increasing DMSO concentrations should be expected [62]. A reason that we only observed a minor impact of DMSO content on Sirt2 activity could be the dependence of the fluorescence intensity of the formed fluorophore on the proportion of DMSO, as shown above. Applying our optimized assay conditions, we were able to determine a Z-factor of 0.65 for the Sirt2-catalysed deacetylation and a Z-factor of 0.67 for the Sirt2-catalysed demyristoylation (figure 4f; electronic supplementary material, figure S1B), which are both considered to be feasible for high-throughput screening [63]. A schematic illustration of our fluorescamine-based sirtuin activity assay is given in figure 3.
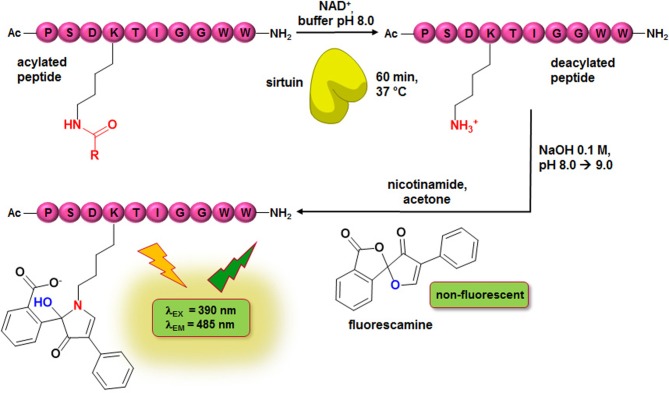
Figure 3.
Scheme of homogeneous fluorescence-based fluorescamine assay (FA-assay). R: CH3, C13H27.
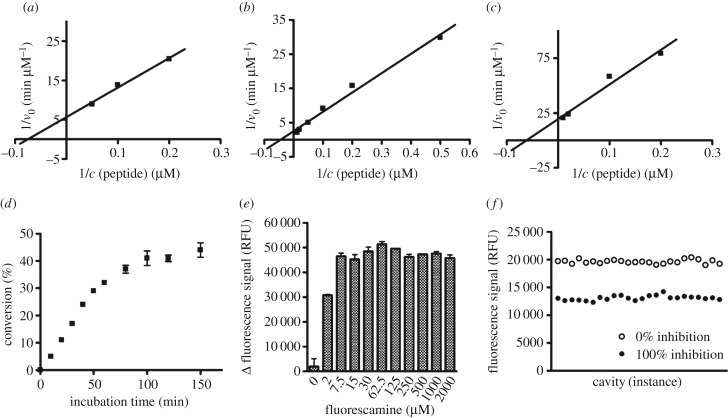
Figure 4.
Set-up of the fluorescamine-based activity assay (FA-assay). (a) Determination of Km value of α-tubulin-myr-peptide on Sirt2 (13.4 µM) by linearization of the Michaelis–Menten plot in a double reciprocal diagram. (b) Determination of Km value of α-tubulin-ac-peptide on Sirt3 (22.2 µM) by linearization of the Michaelis–Menten plot in a double reciprocal diagram. (c) Determination of Km value of α-tubulin-myr-peptide on Sirt3 (16.1 µM) by linearization of the Michaelis–Menten plot in a double reciprocal diagram. (d) Time course of Sirt2-catalysed deacetylation (0–150 min). (e) Effect on signal window of final assay concentration of fluorescamine in a range from 0 to 2000 µM, dissolved in acetone. (f) Determination of Z-factor (0.65) of deacetylation by Sirt2; 24-fold determination of 100% inhibition and 0% inhibition. (Δ fluorescence signal: fluorescence signal of 100% conversion minus 100% inhibition.) v0, velocity; c, concentration; RFU, relative fluorescence units.
(b). Fluorescence polarization assay
In addition to the activity assay described above, we developed a fluorescence polarization (FP)-based assay to complement our existing sirtuin assay platform with a binding assay. The major goals were to create a tool that allows mapping of inhibitors/ligands to different binding pockets of the active site of Sirt2, especially the ‘selectivity pocket', as well as to develop a robust assay that enables screening at high concentrations, which are commonly used in fragment screening. We were able to combine these requirements in one assay by using fluorescence-labelled SirReal in an assay with an FP readout.
(i). Assay principle
If plane polarized light excites a fluorophore, light will be emitted with a certain degree of polarization that is inversely proportional to the rate of molecular rotation. Small fluorescent molecules, e.g. fluorescently labelled probes, largely emit depolarized light owing to the fast reorientation of the fluorophor during the lifetime of its excited state. Fluorophores that are covalently or non-covalently bound to larger structures, e.g. proteins, retain the polarization of the excitation light to a higher degree (see electronic supplementary material, figure S2). FP is determined by detecting the fluorescence intensities of the emitted light from a parallel and from a perpendicular direction in relation to the excitation plane. In a set-up of a fluorescently labelled small-molecule ligand and its targeted protein, the observed FP is a function of the fraction of the ligand that is bound to the protein. Commonly, for competitive ligand screening a reduction of the protein-bound fraction of the fluorescent probe is monitored by a decrease in FP after co-incubation of the potential ligand with a preformed complex of targeted protein and fluorescent probe. However, as the SirReals were shown to have a very slow dissociation rate of the inhibitor–enzyme complex [56], which could be problematic for use of displacement assay protocols, we set up our competition assay to screen for prevented binding of the SirReal-based fluorescent probe when it was added to a preformed complex of Sirt2 and the potential ligand.
(ii). Design and synthesis of the fluorescence-labelled probes
For the design of our fluorescent affinity probes for Sirt2, we referred to a protocol that has recently been successfully applied for the development of a Sirt2-selective pull-down probe (3c) and a proteolysis targeting chimera (PROTAC, 3d), respectively [56,64]. These approaches made use of the highly potent and Sirt2-selective SirReals, which were shown to tolerate the attachment of different functional labels on the benzylmethyl group of SirReal1 (1a) without losing affinity to Sirt2. In order to circumvent potential assay interference by autofluorescent inhibitors, we aimed to develop two probes that are labelled with different fluorescent tags. On the basis of this previous work, we synthesized the envisaged fluorescence probes (4a/b, figure 5a) by conjugation of the propargylated SirReal analogue (1b) with either N-(3-azidopropyl)-3′,6′-bis(dimethylamino)-3-oxo-3H-spiro[isobenzofuran-1,9'-xanthene]-5-carboxamide (5-TAMRA-azide) or N-(3-azidopropyl)-3′,6′-dihydroxy-3-oxo-3H-spiro[isobenzofuran-1,9′-xanthene]-5-carboxamide (5-FAM-azide) via Cu(I)-catalysed Huisgen cycloaddition (for synthesis and characterization data, see electronic supplementary material) [65,66].
Figure 5.
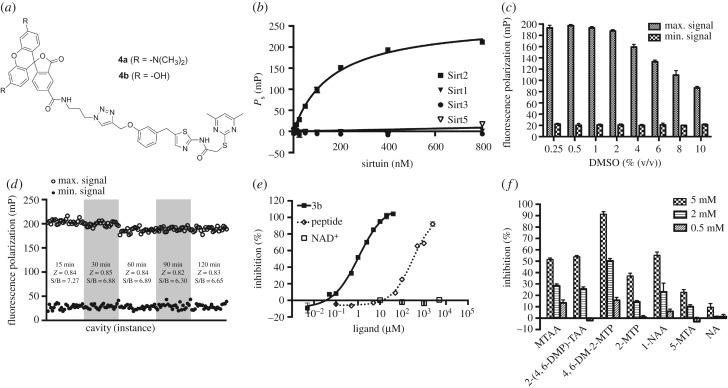
Set-up of the fluorescence polarization-based binding assay (FP-assay) using the TAMRA-labelled SirReal (4a) as a fluorescent affinity probe for Sirt2. (a) Chemical structures of the SirReal-derived fluorescent probes 4a and 4b. (b) KD determination for the interaction between 4a and different sirtuin isotypes. FP due to specific binding (Ps) was plotted against a panel of 10 different sirtuin concentrations (1.56–800 nM) and a one-site binding fit (hyperbola) was applied. Data represent the mean and standard deviation of triplicate measurements. (c) DMSO tolerance of the FP-assay. A panel of DMSO concentrations (0.25–10%) was exposed to the FP-assay mixture and the resulting influence on the maximum- and minimum signal was plotted. Data represent the mean and standard deviation of triplicate measurements. (d) Evaluation of the effect of incubation time on Z-factor. FP signals of 24 microplate wells containing either positive or negative control mixtures were plotted after incubation periods of 15, 30, 60, 90 and 120 min. S/B, signal to baseline ratio. (e) Dose–response inhibition experiments for 3b and the α-tubulin-ac-peptide yielded IC50 values of 0.86 ± 0.09 µM and 344.4 ± 25.9 µM, respectively. NAD+ showed no competition. We used 40 nM 4a for binding to 200 nM Sirt2. Assay mixtures were incubated for 30 min before FP was measurement. Data represent the mean and standard deviation of triplicate measurements. (f) Fragments of SirReal2 prevent 4a from binding to Sirt2. Fragments were tested at concentrations of 5, 2 and 0.5 mM (for conditions, see (e)). MTAA, N-(5-methylthiazol-2-yl)acetamide; 2-(4,6-DMP)-TAA, 2-((4,6-dimethylpyrimidin-2-yl)thio)acetamide; 4,6-DM-2-MTP, 4,6-dimethyl-2-(methylthio)pyrimidine; 2-MTP, 2-(methylthio)pyrimidine; 1-NAA, 1-naphthylacetic acid; 5-MTA, 5-methylthiazol-2-amine; NA, nicotinamide (for structures, see electronic supplementary material, figure S5). (Error bars show standard deviation.) mP, milli-polarization units.
(iii). Assay development and validation
To show the linearity of the fluorescence signals of our fluorescent probes (4a/b), we initially performed a probe titration by serially diluting our probes in assay buffer. The fluorescence intensities were plotted against the respective probe concentration. Linearity of the fluorescence intensity of 4a and 4b was shown in a range from 1.25 to 640 nM (R2 = 0.9993 for 4a, R2 = 0.9989 for 4b, see electronic supplementary material, figures S3A and S4A). Calculation and plotting of FP values against the respective probe concentration indicated a high signal stability at probe concentrations exceeding 20 nM (data not shown). To evaluate the binding affinities of our probes to Sirt2, we titrated serial dilutions of Sirt2 against our probes at a concentration of 40 nM. From the obtained specific binding values (Ps), we determined a KD value of 164 nM for the interaction between 4a and Sirt2. Furthermore, we were able to show that the attachment of a fluorescent label does not alter the isotype selectivity profile of the SirReal-derived ligand (figure 5b). The fluorescein-labelled probe (4b) showed a similar affinity to Sirt2, with a KD value of 147 nM (see electronic supplementary material, figure S4B). Thus, both 4a and 4b are high-affinity fluorescent probes for Sirt2 and suitable for further assay development. Therefore, we set up an assay to screen for potential competitors of our fluorescent probes, which is based on FP. As the probe concentration should not surpass its KD to the targeted protein, we maintained the probe concentration at 40 nM, which is in accordance with the results of our probe titration mentioned above. In consideration of a large assay window, which is ensured by saturated binding of the probe (figure 5b), and a protein consumption that should be as low as possible to make the assay suitable for high-throughput screening, we set the Sirt2 assay concentration to 200 nM. Although DMSO concentrations exceeding 2% were shown to markedly decrease the assay window, sufficient assay windows were provided even at very high concentrations of DMSO (figure 5c; electronic supplementary material, figure S4C). This enables screening of compounds with a low solubility or screening at high compound concentrations. An average Z-factor of 0.84 for the TAMRA-labelled probe (4a) and 0.79 for the FAM-labelled probe (4b) classifies our assays as excellent (Z > 0.5, figure 5d; electronic supplementary material, figure S4D) [63]. Moreover, consistent Z-factors and signal-to-background relations were shown by repeated readouts after 15, 30, 60, 90 and 120 min. For subsequent screening, we chose an incubation time of 30 min prior to the readouts. To examine the effect of potential competitors on the binding of the fluorescent probes to Sirt2, we used both the unlabelled SirReal 3b and the α-tubulin-ac-peptide. The probes were shown to be competitive to the unlabelled SirReal 3b as well as to the peptide substrate. This is consistent with the reported binding mode of 3a (figure 1), which shows that the acyl-lysine channel is efficiently blocked by the triazole linker moiety [56]. However, NAD+, even at high concentrations, did not prevent the probes from binding to Sirt2 (figure 5e; electronic supplementary material, figure S4F), which was expected from our previous structural studies which showed simultaneous binding of SirReals and NAD+ to Sirt2 [55,57]. Thus, our FP-assay based on the fluorescently labelled probes (4a/b) is a very useful tool to distinguish between inhibitors/ligands binding to the substrate and cofactor-binding site of Sirt2. Most importantly, the fluorescently labelled probes (4a/b) enable the mapping of inhibitors/ligands binding to either the ECS or ‘selectivity pocket', which would not necessarily show a competition to the substrate or cofactor in enzyme kinetic analyses. As the TAMRA-labelled probe (4a) provides a larger assay window than the FAM-labelled probe (4b), we focused on the application of 4a for further studies. To validate the suitability of our 4a-based FP-assay for fragment screening, we tested fragments of SirReal2 (2a) to assess their potency to compete with 4a for Sirt2 binding. As expected, fragments of 2a prevented our TAMRA-labelled probe (4a) from binding to Sirt2 (figure 5f). In accordance with the fact that methylation of the pyrimidine moiety in positions 4 and 6 is beneficial for binding to Sirt2 [55], we noted here that the dimethylated fragment (4,6-DM-2-MTP) provokes a significant decrease in 4a binding to Sirt2 as compared with the fragment without methyl groups (2-MTP). No effect was observed for nicotinamide (NA), which was used as a negative control because the SirReals were previously shown not to be bound to the NA-binding site of Sirt2 [55]. Additional information on assay conditions and procedure can be found in the electronic supplementary material.
(c). Assay validation
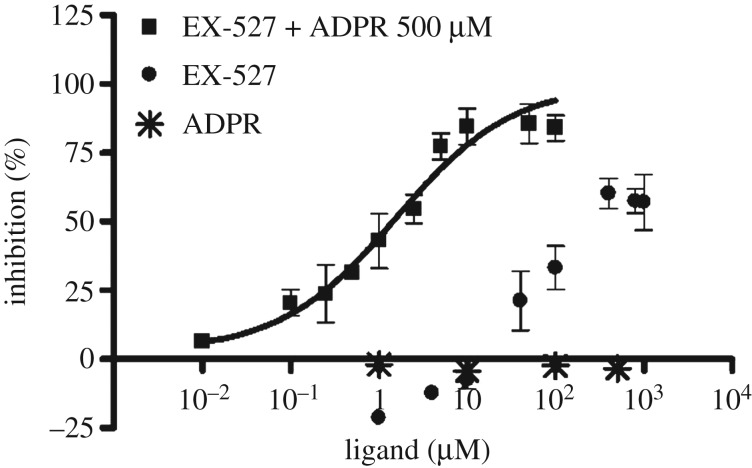
To interrogate the suitability of both above-described assays for orthogonal Sirt2 inhibitor screening, we tested a series of known sirtuin modulators. By comparing the FA-assay results for the inhibition of Sirt2-catalysed deacetylation with the results of the well-established ZMAL-based assay, we aimed to validate the FA-assay for Sirt2 inhibitor screening. Furthermore, we wanted to demonstrate prevention of binding of the rhodamine probe 4a to Sirt2 by pre-incubation with ligands that are known to bind to the ECS or the selectivity pocket (table 1). Overall, the data for the inhibition of Sirt2/3-catalysed deacetylation generated by means of our FA-assay fitted well to the data from the ZMAL-assay. Nicotinamide, the physiological inhibitor of the sirtuin-mediated deacylation [67], gave similar IC50 values for Sirt2- and Sirt3-catalysed deacetylation in both ZMAL- and FA-assay. For nicotinamide, Feldman et al. reported a decrease in Sirt3 inhibition with increasing acyl chain length. For Sirt2, they showed the opposite [58]. Using our FA-assay, we observed a slight decrease in nicotinamide-mediated inhibition by an increase of acyl chain length, which is even less pronounced in the case of Sirt2 compared with Sirt3. As expected, nicotinamide does not prevent 4a from binding to Sirt2 because nicotinamide was shown to bind to the C-pocket [68], which does not interfere with binding to the ECS and the ‘selectivity pocket', respectively [56]. The Sirt2-selective inhibitor AEM2 [43] showed a weaker inhibition of demyristoylation compared with deacetylation. The minor impact of the SirReals on Sirt2-catalysed demyristoylation has already been documented so we speculate the AEM2 might also bind to the selectivity pocket [49]. By means of our FP-assay, we were able to further support this hypothesis. EX-527 is reported as a highly potent Sirt1 inhibitor (IC50 = 0.098 µM) with moderate effects on Sirt2-mediated deacetylation (IC50 = 19.6 µM) [69]. Our Sirt2-deacetylation assays gave similar results for EX-527, whereas the Sirt2-catalysed demyristoylation was shown to be not affected by EX-527. Furthermore, EX-527 is hardly able to prevent the SirReal-derived probe 4a from binding to Sirt2 (figure 6). As described above, the SirReals bind to the ‘selectivity pocket' and ECS of Sirt2 and freeze the protein in the ‘locked-open' conformation. Conversely, EX-527 binds to a conformation of sirtuins, which either requires the cofactor NAD+, its cleavage product 2′-O-acetyl-ADP ribose (2′-OAADPR), or ADPR. EX-527 is not able to bind efficiently in the absence of NAD+/analogues [70]. Consistent with this, EX-527 was shown to potently prevent 4a from binding to Sirt2 (IC50 = 1.63 ± 0.49 µM) in the presence of ADPR (500 µM, figure 6). Thus, our SirReal-based probe (4a) is not only an unprecedented tool to detect binding to the ECS and the ‘selectivity pocket', but also it allows probing of the high flexibility of Sirt2 by exclusively binding to the ‘locked-open' state of Sirt2. The Sirt1-activator resveratrol shows, as expected, no significant effect in all of the assays. SRT1720, a Sirt1-activator and Sirt3-selective inhibitor [71], shows a robust inhibition of Sirt3-catalysed deacetylation and demyristoylation.
Table 1:
Selected sirtuin ligands/inhibitors examined by means of FP-based binding assay using the TAMRA-labelled SirReal (4a) as well as the ZMAL- or FA-based activity assays. n.c. = no competition (inhibition of probe (4a) binding to Sirt2 < 10% at given concentration), n.i. = no inhibition (less than 10% inhibition at 50 µM). Chemical structures of the tested small-molecule ligands/inhibitors are given in figure 2 or electronic supplementary material, figure S6. The biological activity of the inhibitors/ligands used has been validated by cellular investigations in former studies.
| ligands/inhibitors | IC50 (µM) or percentage inhibition |
||||||
|---|---|---|---|---|---|---|---|
| binding assay |
activity assays |
||||||
| Sirt2 |
Sirt2 |
Sirt3 |
|||||
| FP-assay | ZMAL-assay | FA-assay |
ZMAL-assay | FA-assay |
|||
| acetyl | myristoyl | acetyl | myristoyl | ||||
| nicotinamide | n.c. at 400 µM | 49.8 ± 4.6 | 56.8 ± 8.6 | 98.2 ± 28.6 | 67.9 ± 3.3 | 35.1 ± 3.8 | 122.7 ± 44.8 |
| AEM2 | 20.9 ± 0.7 | 2.5 ± 0.2 | 1.1 ± 0.1 | 35.8% at 50 µM | n.i. | n.i. | n.i. |
| SirReal1 (1a) | 2.8 ± 0.2 | 3.7 ± 0.8 | 13.3 ± 2.2 | IC50 > 100 µM | IC50 > 100 µM | IC50 > 100 µM | IC50 > 100 µM |
| SirReal2 (2a) | 0.95 ± 0.07 | 0.38 ± 0.11 | 0.472 ± 0.08 | 17.4% at 50 µM | n.i. | n.i. | 24.8% at 50 µM |
| MZ242 (3b) | 0.86 ± 0.09 | 0.12 ± 0.01 | 0.22 ± 0.10 | 18.1% at 50 µM | n.i. | n.i. | n.i. |
| EX-527 | IC50 > 400 µM | 10.6 ± 1.1 | 10.9 ± 1.6 | n.i. | 19.0% at 50 µM | 20.4% at 50 µM | n.i. |
| SRT1720 | 13.8% at 50 µM | 15.9% at 50 µM | 24.6% at 50 µM | 11.1% at 50 µM | 4.5 ± 0.2 | 1.5 ± 0.1 | 8.1 ± 1.7 |
| resveratrol | n.c. at 100 µM | 11.4% at 50 µM | 29.7% at 50 µM | 14.9% at 50 µM | 10.1% at 50 µM | 23.8% at 50 µM | 26.0% at 50 µM |
Figure 6.
Dose–response inhibition experiment examined by means of FP-based binding assay for EX-527 in combination with ADPR (500 µM) yielded an IC50-value of 1.63 ± 0.49 µM. ADPR showed no competition up to 500 µM. EX-527 without ADPR showed a maximum competition of 60% at 400 µM or higher.
3. Screening of focused library (GSK-box)
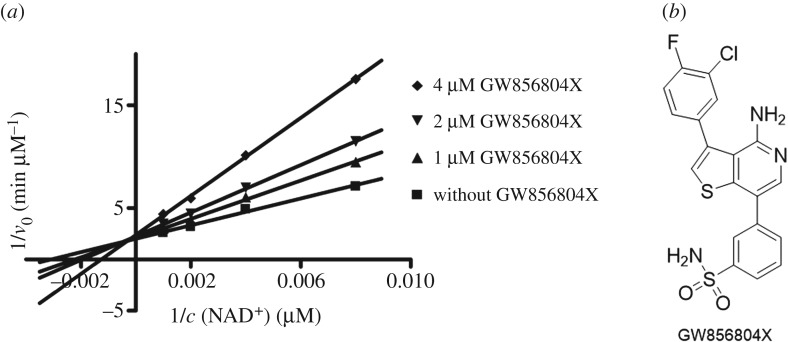
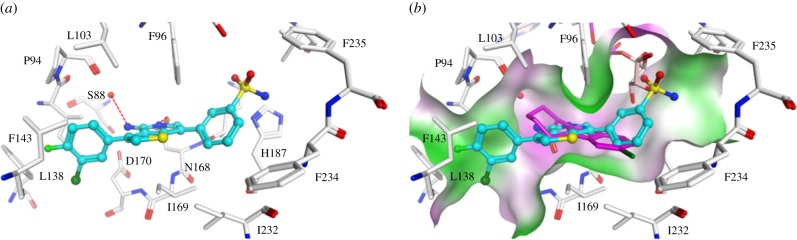
We screened a focused kinase inhibitor library from GlaxoSmithKline (GSK), a published kinase inhibitor set (PKIS) [72], to identify new chemotypes for Sirt2 inhibition. The library contains 367 ATP-competitive protein kinase inhibitors. As previously shown, screening of kinase inhibitor libraries is a quite feasible approach to identify novel sirtuin inhibitors [73,74]. Initially, we screened the library for Sirt2 inhibitors using the ZMAL-assay while the newly established assays described here were still under development. For the most promising hits (inhibition greater than 40% at 50 µM), we determined IC50 values. Furthermore, we tested them on selectivity towards the isotypes Sirt1 and 3. Subsequently, the hits were verified via our FA-assay using the unlabelled substrate and analysed for their behaviour in the FP-assay. As a biophysical method to validate the specificity of the Sirt2–ligand interaction, we applied a thermal shift assay [55] (table 2). Among the identified screening hits, all compounds show a weaker effect on Sirt1 and 3 compared with Sirt2. GW435821X was found to be the least-selective compound in terms of Sirt1–3 inhibition. GW654652X, GW799251X and GW856804X were characterized with IC50 values below 10 µM for the inhibition of Sirt2-catalysed deacetylation in both ZMAL- and FA-assays and were shown to prevent 4a from binding to Sirt2 (GW799251X showed assay interference at concentrations higher than 4 µM). Interestingly, GW799251X has a similar effect on Sirt2-mediated demyristoylation compared with deacetylation and induces a thermal shift of the Sirt2 melting curve of only 0.7°C. In contrast to this, GW654652X and GW856804X show a drastic loss in potency for Sirt2-catalysed demyristoylation and provoke a substantial thermal stabilization (thermal shift greater than 4°C) of Sirt2. Furthermore, these two compounds are Sirt2 selective. Both results imply binding of the latter two to the ‘selectivity pocket'. For further characterization of GW856804X, we performed a competition analysis towards NAD+ (figure 7a). Computational docking studies rationalized the binding of the identified screening hits to Sirt2 (figure 8 or electronic supplementary material, figures S8–S11). The predicted binding mode of GW856804X is consistent with the results of competition analyses that identified this compound as an NAD+-competitive Sirt2 inhibitor. In terms of protein stabilization, Sirt2-deacetylation selectivity and 4a displacement, GW856804X showed a similar behaviour to the SirReals (thermal shift: SirReal1 = approx. 2°C; SirReal2 = approx. 3°C) [55].
Table 2.
Screening results for identified hits from the GSK library, n.i., no inhibition (less than 10% inhibition at 50 µM); n.d., not detectable (assay interference at higher concentrations than the given concentration). Chemical structures are given in figure 7b or electronic supplementary material, figure S7.
| inhibitors | IC50 (µM) or percentual inhibition |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sirt1 |
Sirt2 |
Sirt3 |
|||||||
| ZMAL-assay | ZMAL-assay | FA-assay |
FP-assay | T-shift-assay | ZMAL-assay | FA-assay |
|||
| acetyl | myristoyl | acetyl | myristoyl | ||||||
| GSK237700A | n.i. | 4.3 ± 0.4 µM | 40.9 ± 19.5 µM | 16.2% at 50 µM | 23.3 ± 3.5 | 2.3°C | 35.2% at 50 µM | 39.4% at 50 µM | 43.2% at 50 µM |
| GSK300014A | n.i. | 44.5% at 50 µM IC50-value n.d. |
44.6% at 50 µM IC50-value n.d. |
24.5% at 50 µM | n.d. | 2.7°C | n.i. | n.i. | 42.4% at 50 µM |
| GW435821X | 26.5% at 50 µM | 24.6 ± 2.8 µM | 12.9 ± 2.8 µM | 58.8 ± 6.8 | 16.5 ± 3.5 | 2.8°C | 41.7 ± 2.0 | 15.9 ± 2.0 | 37.6% at 50 µM |
| GW654652X | n.i. | 1.1 ± 0.1 µM | 5.6 ± 0.6 µM | 21.9% at 50 µM | 1.6 ± 0.4 | 4.0°C | 19.6% at 50 µM | 12.8% at 50 µM | 32.4% at 50 µM |
| GW799251X | 15.5% at 50 µM | 1.6 ± 0.3 µM | 8.8 ± 2.2 µM | 10.5 ± 1.1 | 53.2% at 4 µM IC50-value n.d. |
0.7°C | n.i. | 16.7 ± 2.1 | 21.7 ± 4.3 |
| GW856804X | n.i. | 2.2 ± 0.2 µM | 2.8 ± 0.3 µM | 24.9% at 50 µM | 2.7 ± 0.8 | 4.6°C | n.i. | n.i. | 30.2% at 50 µM |
Figure 7.
(a) Competition analysis for the Sirt2 inhibition of GW856804X (0, 1, 2, 4 µM final assay concentration) towards NAD+ (125, 250, 500, 1000 µM final assay concentration). GW856804X is an NAD+-competitive Sirt2 inhibitor. v0, velocity; c, concentration. (b) Chemical structure of GW856804X.
Figure 8.
(a) Interaction of GW856804X (coloured cyan) with Sirt2 (PDB ID 5D7Q). (b) Overlay of the predicted binding mode of GW856804X to Sirt2 with the co-crystallized inhibitor CHIC35 (coloured magenta) and ADPR (coloured salmon). Molecular surface is coloured according to hydrophobicity (magenta = polar region, green = hydrophobic region). Hydrogen bonds are shown as dashed lines. Water molecules are shown as red spheres.
4. Docking
The applied docking protocol used in Glide was first validated and was found to correctly reproduce the location and conformation of the co-crystallized inhibitors in the corresponding X-ray structures (root mean square deviation below 1.2 Å). Subsequently, we docked the inhibitors under study into the NAD+- and substrate-binding pocket of the Sirt2 X-ray structures. A conserved water molecule bridging the interaction between Pro94 and the carbonyl group of SirReal2 as well as a conserved water molecule bridging the interaction between nicotinamide and Asn168 was considered as part of the protein during ligand docking. We docked the inhibitors in all available Sirt2 structures and analysed the Glide docking scores as well as the MM-GB/SA interaction energies (see electronic supplementary material, table S1). Crystal structures of SirReals in complex with Sirt2 showed the binding of the inhibitors to the so-called ‘selectivity pocket' (surrounded by several hydrophobic residues: Phe96, Phe190, Ile169, Leu134, Ile232, Phe119, Leu138, Tyr139, Pro140 and Phe143), which explains the Sirt2 selectivity.
The docking of the GSK compounds showed that they also prefer to bind to this pocket even if they are structurally diverse. The docking of GW654652X (IC50(Sirt2) = 1.1 µM) suggested that the methylindazole overlaps with the dimethylpyrimidine in the so-called ‘selectivity pocket'. The dimethylpyrimidine moiety was found to be crucial for the potency of SirReals (electronic supplementary material, figure S8). The sulfone group of GW654652X accepts a hydrogen bond from Arg97 like the triazole of the triazolo-SirReals, whereas the pyrimidine mimics the aminothiazole system of the SirReals. Also in the case of GW799251X (IC50(Sirt2) = 1.6 µM), a good agreement with the SirReal structure can be recognized: the pyrimidine adopts the same position as the dimethylpyrimidine of SirReal2 and the terminal fluorophenyl mimics the interaction of the naphthyl ring of SirReal2 (electronic supplementary material, figure S9). The aminopyrimidine occupies the nicotinamide subpocket and interacts with Asp170 and a conserved water molecule via hydrogen bonds. Docking of GSK237700A showed that the amide group interacts with the nicotinamide subpocket (Asp170 and conserved water molecule), whereas the chlorophenyl group interacts with the hydrophobic residues of the selectivity pocket like the dimethylpyrimidine of SirReal2 (electronic supplementary material, figure S10). The stilbene derivative GW435821X also mimics the nicotinamide moiety of NAD+ and interacts with Asp170 and the conserved water molecule (electronic supplementary material, figure S11). The dimethylphenol is stabilized by the aromatic residues Tyr139 and Phe190. In the case of the inhibitor GW856804X, the most favourable docking solution was found for the Sirt2 structure in complex with the indole derivative CHIC35 and ADPR (figure 8) [75]. The aminopyridine of GW856804X mimics the interaction of the nicotinamide and amide-part of CHIC35 (H-bonds to Asn168 and the conserved water molecule), whereas the hydrophobic halogenated phenyl ring occupies the selectivity pocket. The sulfonamide group of GW865804X interacts with polar residues in the ribose-binding pocket of Sirt2. This binding mode is consistent with the observed NAD+-competition found in the kinetic experiments.
5. SirReal-based fluorescent probe (4a) as an intracellular fluorescent reporter for Sirt2
To examine the suitability of our SirReal-based fluorescent probe (4a) as an intracellular fluorescent reporter for Sirt2, we first aimed to investigate if 4a is able to penetrate the cell membrane to bind to Sirt2, which is mainly localized in the cytoplasm. By means of live cell fluorescence microscopy, we were able to show that the fluorescent probe 4a is taken up by EGFP-Sirt2-transfected HeLa cells (electronic supplementary material, figure S12). It is distributed within the cytoplasm and not transferred into the nucleus. Similar distribution of 4a and EGFP-Sirt2 suggests intracellular co-localization. Furthermore, we proved the intracellular target engagement of 4a by a functional study, which showed that 4a is able to inhibit Sirt2 and thus upregulates the cellular acetylation level of the Sirt2 substrate α-tubulin (electronic supplementary material, figure S13). As a proof of concept study to investigate the suitability of 4a to detect small-molecule binding to Sirt2 in a cellular set-up, we pre-incubated HeLa cells overexpressing EGFP-Sirt2 with our unlabelled SirReal 3a. Pre-incubation with 3a markedly prevented the 4a from binding to its intracellular target, Sirt2 (electronic supplementary material, figure S14). Thus, our SirReal-based fluorescent probe (4a) is the first example of a small-molecule fluorescent reporter for Sirt2 in cells.
6. Conclusion
Owing to some specific characteristics of the sirtuins, e.g. unphysiological binding modes of fluorescently labelled model substrates or rearrangement of the active site upon ligand binding, we identified a need for new assay tools enabling advanced characterization of inhibitors. Our new homogeneous fluorescence-based activity assay uses an unlabelled peptide to circumvent assay artefacts due to impaired binding and conversion of fluorescently labelled model substrates. Such artefacts have been well documented in the literature for the frequently-used aminocoumarin-labelled substrates, especially in the case of sirtuin activators. We were able to show that our new assay protocol can be adapted to long-chain deacylation in order to dissect a potential acyl selectivity of certain inhibitors on an unlabelled substrate. Moreover, we developed a rhodamine-based FP-assay that allows the large structural flexibility of sirtuins, especially Sirt2, to be addressed. This assay can be used to map inhibitor-binding regions that are not easily amenable to competition analysis using peptide substrates or the cosubstrate NAD+. Compared with fluorescence-based assays, protocols that rely on FP show a much higher robustness against fluorescence and quenching interference. This might be interesting either for the characterization of inhibitors that show interference with fluorescence readout or for testing at high concentrations that are required for fragment-based screening. The latter is usually only possible using sophisticated biophysical techniques, which are highly demanding in terms of time and costly equipment. The new activity and affinity assays complement our orthogonal screening platform for sirtuin modulators, which will be useful in further screening approaches. The fluorescent probe also was successfully used to monitor cellular target engagement of a Sir2 inhibitor and represents the first fluorescent small-molecule reporter for Sirt2 in cells to the best of our knowledge. Focused library screening identified new Sirt2 inhibitors. They were verified and mapped by means of this screening platform and can be used as valuable starting points for further inhibitor optimization studies. This is especially true for the potent and Sirt2-selective GW856804X, which can subsequently be modified towards a more potent and selective inhibitor.
The pan-sirtuin inhibitor nicotinamide shows only weak selectivity towards deacetylation and demyristoylation, whereas inhibitors that gain their Sirt2 affinity and selectivity by binding to the ‘selectivity pocket', e.g. the SirReals, have a strong effect on Sirt2-catalysed deacetylation while demyristoylation is nearly unaffected. This was also observed for the Sirt2-selective AEM2 and the newly identified GW654652X and GW856804X. Additionally, these inhibitors showed a competitive binding mode towards our SirReal-based fluorescent probe (4a). Thus, we and others [49] have observed this for different structural classes and we postulate that there is a general inverse correlation of selectivity pocket ligands with regard to the inhibition of the Sirt2-mediated deacetylation versus demyristoylation.
However, the underlying mechanism of this phenomenon is unknown so far. Two different hypotheses can be used to rationalize this phenomenon: (i) The myristoyl residue of the substrate and the small-molecule inhibitor (e.g. SirReal) are able to simultaneously bind to the ‘selectivity pocket’ by inducing an additional change of the conformation of the catalytic domain, so that the catalysis can still proceed. (ii) A considerably stronger binding of the myristoyl substrate to Sirt2's active site than the acetyl substrate owing to additional hydrophobic interactions of the myristoyl moiety with non-polar residues of the ‘selectivity pocket', which would displace a small-molecule inhibitor from the ‘selectivity pocket' and thus reduce its inhibitory effect. We think that the latter hypothesis is a much more reasonable explanation, as simultaneous binding of inhibitor and the myristoyl peptide accompanied by an additional change of the enzyme conformation without a loss of the catalytic reactivity seems rather unlikely. Further investigation of this phenomenon will be part of future studies.
Supplementary Material
Acknowledgements
The PKIS was supplied by GlaxoSmithKline LLC and the Structural Genomics Consortium under an open access Material Transfer and Trust Agreement: http://www.sgc-unc.org. We thank Björn Klaiber for supporting the screening experiments. We thank the COST action CM1406 (Epigenetic Chemical Biology) for support.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
S.Sw. carried out the development and establishment of the fluorescamine-based assay, the inhibitor screening using the fluorescamine assay, participated in data analysis, participated in the design of the study and drafted the manuscript; M.S. carried out the development and establishment of the FP-assay, the inhibitor screening using the FP-assay, and participated in data analysis and in the design of the study and drafted the manuscript; D.M. carried out the IC50-determination using the FP-assay; T.R. supported the development of the fluorescamine-based assay; S.Sz., A.L. and J.O. performed the cellular biology and analysed the cellular data; W.S. rationalized the screening results by molecular docking studies, and participated in the design of the study and drafting of the manuscript; M.J. conceived the study, designed and coordinated the study and participated in drafting the manuscript. All authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
The Jung group thanks the Deutsche Forschungsgemeinschaft (DFG, within GRK1976 and Ju295/14-1) for support. M.S. is supported by the Deutsche Forschungsgemeinschaft (SCHI 1408/1-1). This work was supported by the European Cooperation in Science and Technology COST Action (CM1406 and TD1304) and Hungarian National Scientific Research Fund Grants OTKA (T-101039 and T-112144) to J.O.
References
- 1.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. 2003. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370, 737–749. ( 10.1042/bj20021321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schemies J, Uciechowska U, Sippl W, Jung M. 2010. NAD+-dependent histone deacetylases (sirtuins) as novel therapeutic targets. Med. Res. Rev. 30, 861–889. ( 10.1002/med.20178) [DOI] [PubMed] [Google Scholar]
- 3.Schiedel M, Robaa D, Rumpf T, Sippl W, Jung M. 2017. The current state of NAD+-dependent histone deacetylases (sirtuins) as novel therapeutic targets. Med. Res. Rev. 38, 147–200. ( 10.1002/med.21436) [DOI] [PubMed] [Google Scholar]
- 4.Madsen AS, Olsen CA. 2012. Profiling of substrates for zinc-dependent lysine deacylase enzymes: HDAC3 exhibits decrotonylase activity in vitro. Angew. Chem. Int. Edn Engl. 51, 9083–9087. ( 10.1002/anie.201203754) [DOI] [PubMed] [Google Scholar]
- 5.Tan MJ, et al. 2014. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 19, 605–617. ( 10.1016/j.cmet.2014.03.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du J, et al. 2011. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809. ( 10.1126/science.1207861) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao X, et al. 2014. Identification of ‘erasers’ for lysine crotonylated histone marks using a chemical proteomics approach. eLife 3, e02999 ( 10.7554/eLife.02999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldman JL, Baeza J, Denu JM. 2013. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 288, 31 350–31 356. ( 10.1074/jbc.C113.511261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang H, et al. 2013. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496, 110–113. ( 10.1038/nature12038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tong Z, Wang Y, Zhang X, Kim DD, Sadhukhan S, Hao Q, Lin H. 2016. SIRT7 is activated by DNA and deacetylates histone H3 in the chromatin context. ACS Chem. Biol. 11, 742–747. ( 10.1021/acschembio.5b01084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong Z, Wang M, Wang Y, Kim DD, Grenier JK, Cao J, Sadhukhan S, Hao Q, Lin H. 2017. SIRT7 is an RNA-activated protein lysine deacylase. ACS Chem. Biol. 12, 300–310. ( 10.1021/acschembio.6b00954) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. 2001. hSIR2SIRT1 functions as an NAD-dependent p53 deacetylase. Cell 107, 149–159. ( 10.1016/S0092-8674(01)00527-X) [DOI] [PubMed] [Google Scholar]
- 13.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. 2003. The human Sir2 ortholog, SIRT2, is an NAD+ -dependent tubulin deacetylase. Mol. Cell 11, 437–444. ( 10.1016/S1097-2765(03)00038-8) [DOI] [PubMed] [Google Scholar]
- 14.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. 2004. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23, 2369–2380. ( 10.1038/sj.emboj.7600244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.North BJ, et al. 2014. SIRT2 induces the checkpoint kinase BubR1 to increase lifespan. EMBO J. 33, 1438–1453. ( 10.15252/embj.201386907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verdin E, Hirschey MD, Finley LW, Haigis MC. 2010. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem. Sci. 35, 669–675. ( 10.1016/j.tibs.2010.07.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feige JN, Auwerx J. 2008. Transcriptional targets of sirtuins in the coordination of mammalian physiology. Curr. Opin. Cell Biol. 20, 303–309. ( 10.1016/j.ceb.2008.03.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haigis MC, Guarente LP. 2006. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 20, 2913–2921. ( 10.1101/gad.1467506) [DOI] [PubMed] [Google Scholar]
- 19.Liu TF, Vachharajani VT, Yoza BK, McCall CE. 2012. NAD+-dependent sirtuin 1 and 6 proteins coordinate a switch from glucose to fatty acid oxidation during the acute inflammatory response. J. Biol. Chem. 287, 25 758–25 769. ( 10.1074/jbc.M112.362343) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Oliveira RM, Sarkander J, Kazantsev AG, Outeiro TF. 2012. SIRT2 as a therapeutic target for age-related disorders. Front. Pharmacol. 3, 92–100. ( 10.3389/fphar.2012.00082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Oliveira RM, et al. 2017. The mechanism of sirtuin 2-mediated exacerbation of alpha-synuclein toxicity in models of Parkinson disease. PLoS Biol. 15, e2000374 ( 10.1371/journal.pbio.2000374) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beirowski B, et al. 2011. Sir-two-homolog 2 (Sirt2) modulates peripheral myelination through polarity protein Par-3/atypical protein kinase C (aPKC) signaling. Proc. Natl Acad. Sci. USA 108, E952–E961. ( 10.1073/pnas.1104969108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pais TF, Szego EM, Marques O, Miller-Fleming L, Antas P, Guerreiro P, de Oliveira RM, Kasapoglu B, Outeiro TF. 2013. The NAD-dependent deacetylase sirtuin 2 is a suppressor of microglial activation and brain inflammation. EMBO J. 32, 2603–2616. ( 10.1038/emboj.2013.200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao T, Alam HB, Liu B, Bronson RT, Nikolian VC, Wu E, Chong W, Li Y. 2015. Selective inhibition of SIRT2 improves outcomes in a lethal septic model. Curr. Mol. Med. 15, 634–641. ( 10.2174/156652401507150903185852) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eskandarian HA, Impens F, Nahori MA, Soubigou G, Coppee JY, Cossart P, Hamon MA. 2013. A role for SIRT2-dependent histone H3K18 deacetylation in bacterial infection. Science 341, 1238858 ( 10.1126/science.1238858) [DOI] [PubMed] [Google Scholar]
- 26.Buechler N, Wang X, Yoza BK, McCall CE, Vachharajani V. 2017. Sirtuin 2 regulates microvascular inflammation during sepsis. J. Immunol. Res. 2017, 2648946 ( 10.1155/2017/2648946) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng YB, Jing H, Aramsangtienchai P, He B, Khan S, Hu J, Lin H, Hao Q. 2015. Efficient demyristoylase activity of SIRT2 revealed by kinetic and structural studies. Sci. Rep. 5, 8529 ( 10.1038/srep08529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SH, Zhu Y, Ozden O, Kim HS, Jiang H, Deng CX, Gius D, Vassilopoulos A. 2012. SIRT2 is a tumor suppressor that connects aging, acetylome, cell cycle signaling, and carcinogenesis. Transl. Cancer Res. 1, 15–21. ( 10.3978/j.issn.2218-676X.2012.05.01) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donmez G, Outeiro TF. 2013. SIRT1 and SIRT2: emerging targets in neurodegeneration. EMBO Mol. Med. 5, 344–352. ( 10.1002/emmm.201302451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HS, et al. 2011. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 20, 487–499. ( 10.1016/j.ccr.2011.09.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang MH, Laurent G, Bause AS, Spang R, German N, Haigis MC, Haigis KM. 2013. HDAC6 and SIRT2 regulate the acetylation state and oncogenic activity of mutant K-RAS. Mol. Cancer Res. 11, 1072–1077. ( 10.1158/1541-7786.MCR-13-0040-T) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luthi-Carter R, et al. 2010. SIRT2 inhibition achieves neuroprotection by decreasing sterol biosynthesis. Proc. Natl Acad. Sci. USA 107, 7927–7932. ( 10.1073/pnas.1002924107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chopra V, et al. 2012. The sirtuin 2 inhibitor AK-7 is neuroprotective in Huntington's disease mouse models. Cell Rep. 2, 1492–1497. ( 10.1016/j.celrep.2012.11.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bobrowska A, Donmez G, Weiss A, Guarente L, Bates G. 2012. SIRT2 ablation has no effect on tubulin acetylation in brain, cholesterol biosynthesis or the progression of Huntington's disease phenotypes in vivo. PLoS ONE 7, e34805 ( 10.1371/journal.pone.0034805) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui H, Kamal Z, Ai T, Xu Y, More SS, Wilson DJ, Chen L. 2014. Discovery of potent and selective sirtuin 2 (SIRT2) inhibitors using a fragment-based approach. J. Med. Chem. 57, 8340–8357. ( 10.1021/jm500777s) [DOI] [PubMed] [Google Scholar]
- 36.Suzuki T, et al. 2012. Design, synthesis, and biological activity of a novel series of human sirtuin-2-selective Inhibitors. J. Med. Chem. 55, 5760–5773. ( 10.1021/jm3002108) [DOI] [PubMed] [Google Scholar]
- 37.Di Fruscia P, et al. 2015. The discovery of a highly selective 5,6,7,8-tetrahydrobenzo[4,5]thieno-[2,3-d]pyrimidin-4(3H)-one SIRT2 inhibitor that is neuroprotective in an in vitro Parkinson's disease model. ChemMedChem 10, 69–82. ( 10.1002/cmdc.201402431) [DOI] [PubMed] [Google Scholar]
- 38.Outeiro TF, et al. 2007. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science 317, 516–519. ( 10.1126/science.1143780) [DOI] [PubMed] [Google Scholar]
- 39.Lara E, et al. 2009. Salermide, a sirtuin inhibitor with a strong cancer-specific proapoptotic effect. Oncogene 28, 781–791. ( 10.1038/onc.2008.436) [DOI] [PubMed] [Google Scholar]
- 40.Peck B, Chen CY, Ho KK, Di Fruscia P, Myatt SS, Coombes RC, Fuchter MJ, Hsiao CD, Lam EW. 2010. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol. Cancer Ther. 9, 844–855. ( 10.1158/1535-7163.MCT-09-0971) [DOI] [PubMed] [Google Scholar]
- 41.Taylor DM, et al. 2011. A brain-permeable small molecule reduces neuronal cholesterol by inhibiting activity of sirtuin 2 deacetylase. ACS Chem. Biol. 6, 540–546. ( 10.1021/cb100376q) [DOI] [PubMed] [Google Scholar]
- 42.Seifert T, Malo M, Kokkola T, Engen K, Friden-Saxin M, Wallen EA, Lahtela-Kakkonen M, Jarho EM, Luthman K. 2014. Chroman-4-one- and chromone-based sirtuin 2 inhibitors with antiproliferative properties in cancer cells. J. Med. Chem. 57, 9870–9888. ( 10.1021/jm500930h) [DOI] [PubMed] [Google Scholar]
- 43.Hoffmann G, Breitenbucher F, Schuler M, Ehrenhofer-Murray AE. 2014. A novel sirtuin 2 (SIRT2) inhibitor with p53-dependent pro-apoptotic activity in non-small cell lung cancer. J. Biol. Chem. 289, 5208–5216. ( 10.1074/jbc.M113.487736) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang S, et al. 2017. Discovery of new SIRT2 inhibitors by utilizing a consensus docking/scoring strategy and structure–activity relationship analysis. J. Chem. Inf. Model. 57, 669–679. ( 10.1021/acs.jcim.6b00714) [DOI] [PubMed] [Google Scholar]
- 45.Sundriyal S, et al. 2017. Thienopyrimidinone based sirtuin-2 (SIRT2)-selective inhibitors bind in the ligand induced selectivity pocket. J. Med. Chem. 60, 1928–1945. ( 10.1021/acs.jmedchem.6b01690) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roessler C, Tuting C, Meleshin M, Steegborn C, Schutkowski M. 2015. A novel continuous assay for the deacylase sirtuin 5 and other deacetylases. J. Med. Chem. 58, 7217–7223. ( 10.1021/acs.jmedchem.5b00293) [DOI] [PubMed] [Google Scholar]
- 47.Heltweg B, Trapp J, Jung M. 2005. In vitro assays for the determination of histone deacetylase activity. Methods 36, 332–337. ( 10.1016/j.ymeth.2005.03.003) [DOI] [PubMed] [Google Scholar]
- 48.Madsen AS, Olsen CA. 2012. Substrates for efficient fluorometric screening employing the NAD-dependent sirtuin 5 lysine deacylase (KDAC) enzyme. J. Med. Chem. 55, 5582–5590. ( 10.1021/jm300526r) [DOI] [PubMed] [Google Scholar]
- 49.Galleano I, Schiedel M, Jung M, Madsen AS, Olsen CA. 2016. A continuous, fluorogenic sirtuin 2 deacylase assay: substrate screening and inhibitor evaluation. J. Med. Chem. 59, 1021–1031. ( 10.1021/acs.jmedchem.5b01532) [DOI] [PubMed] [Google Scholar]
- 50.Chiang YL, Lin H. 2016. An improved fluorogenic assay for SIRT1, SIRT2, and SIRT3. Org. Biomol. Chem. 14, 2186–2190. ( 10.1039/C5OB02609A) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaeberlein M, et al. 2005. Substrate-specific activation of sirtuins by resveratrol. J. Biol. Chem. 280, 17 038–17 045. ( 10.1074/jbc.M500655200) [DOI] [PubMed] [Google Scholar]
- 52.Pacholec M, et al. 2010. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J. Biol. Chem. 285, 8340–8351. ( 10.1074/jbc.M109.088682) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borra MT, Smith BC, Denu JM. 2005. Mechanism of human SIRT1 activation by resveratrol. J. Biol. Chem. 280, 17 187–17 195. ( 10.1074/jbc.M501250200) [DOI] [PubMed] [Google Scholar]
- 54.Beher D, Wu J, Cumine S, Kim KW, Lu SC, Atangan L, Wang M. 2009. Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem. Biol. Drug Des. 74, 619–624. ( 10.1111/j.1747-0285.2009.00901.x) [DOI] [PubMed] [Google Scholar]
- 55.Rumpf T, et al. 2015. Selective Sirt2 inhibition by ligand-induced rearrangement of the active site. Nat. Commun. 6, 6263 ( 10.1038/ncomms7263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schiedel M, Rumpf T, Karaman B, Lehotzky A, Gerhardt S, Ovadi J, Sippl W, Einsle O, Jung M. 2016. Structure-based development of an affinity probe for sirtuin 2. Angew. Chem. Int. Edn 55, 2252–2256. ( 10.1002/anie.201509843) [DOI] [PubMed] [Google Scholar]
- 57.Schiedel M, et al. 2016. Aminothiazoles as potent and selective Sirt2 inhibitors: a structure–activity relationship study. J. Med. Chem. 59, 1599–1612. ( 10.1021/acs.jmedchem.5b01517) [DOI] [PubMed] [Google Scholar]
- 58.Feldman JL, Dittenhafer-Reed KE, Kudo N, Thelen JN, Ito A, Yoshida M, Denu JM. 2015. Kinetic and structural basis for acyl-group selectivity and NAD+ dependence in sirtuin-catalyzed deacylation. Biochemistry 54, 3037–3050. ( 10.1021/acs.biochem.5b00150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Udenfriend S, Stein S, Böhlen P, Dairman W, Leimgruber W, Weigele M. 1972. Fluorescamine: a reagent for assay of amino acids, peptides, proteins, and primary amines in the picomole range. Science 178, 871–872. ( 10.1126/science.178.4063.871) [DOI] [PubMed] [Google Scholar]
- 60.Toro TB, Watt TJ. 2015. KDAC8 substrate specificity quantified by a biologically relevant, label-free deacetylation assay. Protein Sci. 24, 2020–2032. ( 10.1002/pro.2813) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Bernardo S, Weigele M, Toome V, Manhart K, Leimgruber W, Böhlen P, Stein S, Udenfriend S. 1974. Studies on the reaction of fluorescamine with primary amines. Arch. Biochem. Biophys. 163, 390–399. ( 10.1016/0003-9861(74)90490-1) [DOI] [PubMed] [Google Scholar]
- 62.Schiedel M, et al. 2015. Fluorescence-based screening assays for the NAD+-dependent histone deacetylase smSirt2 from Schistosoma mansoni. J. Biomol. Screen. 20, 112–121. ( 10.1177/1087057114555307) [DOI] [PubMed] [Google Scholar]
- 63.Zhang JH, Chung T, Oldenburg KR. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73. ( 10.1177/108705719900400206) [DOI] [PubMed] [Google Scholar]
- 64.Schiedel M, et al. 2017. Chemically induced degradation of sirtuin 2 (Sirt2) by a proteolysis targeting chimera (PROTAC) based on sirtuin rearranging ligands (SirReals). J. Med. Chem. 61, 482–491. ( 10.1021/acs.jmedchem.6b01872) [DOI] [PubMed] [Google Scholar]
- 65.Huisgen R. 1961. Centenary lecture – 1,3-dipolar cycloadditions. Proc. Chem. Soc. Lond. 1961, 357–396. ( 10.1039/PS9610000357) [DOI] [Google Scholar]
- 66.Chan TR, Hilgraf R, Sharpless KB, Fokin VV. 2004. Polytriazoles as copper(I)-stabilizing ligands in catalysis. Org. Lett. 6, 2853–2855. ( 10.1021/ol0493094) [DOI] [PubMed] [Google Scholar]
- 67.Lombard DB, et al. 2007. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 27, 8807–8814. ( 10.1128/MCB.01636-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guan X, Lin P, Knoll E, Chakrabarti R. 2014. Mechanism of inhibition of the human sirtuin enzyme SIRT3 by nicotinamide: computational and experimental studies. PLoS ONE 9, e107729 ( 10.1371/journal.pone.0107729) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Napper AD, et al. 2005. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J. Med. Chem. 48, 8045–8054. ( 10.1021/jm050522v) [DOI] [PubMed] [Google Scholar]
- 70.Gertz M, Fischer F, Nguyen GTT, Lakshminarasimhan M, Schutkowski M, Weyand M, Steegborn C. 2013. Ex-527 inhibits sirtuins by exploiting their unique NAD+-dependent deacetylation mechanism. Proc. Natl Acad. Sci. USA 110, E2772–E2781. ( 10.1073/pnas.1303628110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin L, et al. 2009. Biochemical characterization, localization, and tissue distribution of the longer form of mouse SIRT3. Protein Sci. 18, 514–525. ( 10.1002/pro.50) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elkins JM, et al. 2016. Comprehensive characterization of the published kinase inhibitor set. Nat. Biotechnol. 34, 95–103. ( 10.1038/nbt.3374) [DOI] [PubMed] [Google Scholar]
- 73.Trapp J, et al. 2006. Adenosine mimetics as inhibitors of NAD+-dependent histone deacetylases, from kinase to sirtuin inhibition. J. Med. Chem. 49, 7307–7316. ( 10.1021/jm060118b) [DOI] [PubMed] [Google Scholar]
- 74.Falenczyk C, Schiedel M, Karaman B, Rumpf T, Kuzmanovic N, Grøtli M, Sippl W, Jung M, König B. 2014. Chromo-pharmacophores: photochromic diarylmaleimide inhibitors for sirtuins. Chem. Sci. 5, 4794–4799. ( 10.1039/C4SC01346H) [DOI] [Google Scholar]
- 75.Rumpf T, Gerhardt S, Einsle O, Jung M. 2015. Seeding for sirtuins: microseed matrix seeding to obtain crystals of human Sirt3 and Sirt2 suitable for soaking. Acta Crystallogr. F Struct. Biol. Commun. 71, 1498–1510. ( 10.1107/S2053230X15019986) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material.