ABSTRACT
Nitrogenase catalyzes the reduction of dinitrogen (N2) using low-potential electrons from ferredoxin (Fd) or flavodoxin (Fld) through an ATP-dependent process. Since its emergence in an anaerobic chemoautotroph, this oxygen (O2)-sensitive enzyme complex has evolved to operate in a variety of genomic and metabolic backgrounds, including those of aerobes, anaerobes, chemotrophs, and phototrophs. However, whether pathways of electron delivery to nitrogenase are influenced by these different metabolic backgrounds is not well understood. Here, we report the distribution of homologs of Fds, Flds, and Fd-/Fld-reducing enzymes in 359 genomes of putative N2 fixers (diazotrophs). Six distinct lineages of nitrogenase were identified, and their distributions largely corresponded to differences in the host cells' ability to integrate O2 or light into energy metabolism. The predicted pathways of electron transfer to nitrogenase in aerobes, facultative anaerobes, and phototrophs varied from those in anaerobes at the levels of Fds/Flds used to reduce nitrogenase, the enzymes that generate reduced Fds/Flds, and the putative substrates of these enzymes. Proteins that putatively reduce Fd with hydrogen or pyruvate were enriched in anaerobes, while those that reduce Fd with NADH/NADPH were enriched in aerobes, facultative anaerobes, and anoxygenic phototrophs. The energy metabolism of aerobic, facultatively anaerobic, and anoxygenic phototrophic diazotrophs often yields reduced NADH/NADPH that is not sufficiently reduced to drive N2 reduction. At least two mechanisms have been acquired by these taxa to overcome this limitation and to generate electrons with potentials capable of reducing Fd. These include the bifurcation of electrons or the coupling of Fd reduction to reverse ion translocation.
IMPORTANCE Nitrogen fixation supplies fixed nitrogen to cells from a variety of genomic and metabolic backgrounds, including those of aerobes, facultative anaerobes, chemotrophs, and phototrophs. Here, using informatics approaches applied to genomic data, we show that pathways of electron transfer to nitrogenase in metabolically diverse diazotrophic taxa have diversified primarily in response to host cells' acquired ability to integrate O2 or light into their energy metabolism. The acquisition of two key enzyme complexes enabled aerobic and facultatively anaerobic phototrophic taxa to generate electrons of sufficiently low potential to reduce nitrogenase: the bifurcation of electrons via the Fix complex or the coupling of Fd reduction to reverse ion translocation via the Rhodobacter nitrogen fixation (Rnf) complex.
KEYWORDS: fix, nitrogenase, Rnf, ferredoxin, flavodoxin, hydrogen, nitrogen fixation, oxygen, photosynthesis, pyruvate, bifurcation
INTRODUCTION
Nitrogenase is an oxygen-sensitive enzyme that catalyzes the reduction of dinitrogen (N2) to ammonia (NH3), accounting for nearly two-thirds of fixed nitrogen (N) on earth today (1, 2) and thereby modulating the global fixed N supply (3, 4). Nitrogenase is comprised of two components, a homodimeric iron protein (H subunit) and a heterotetrameric dinitrogenase reductase protein complex (D and K subunits) (5, 6). Homologs of nitrogenase are widely distributed among Bacteria but have only been identified in one group of taxa within the archaeal Euryarchaeota phylum, methanogens. Nitrogenase homologs have not been identified among eukarya (1, 7–12). Three types of nitrogenase have been described that are evolutionarily and structurally related but differ in the metallic composition of their active site: molybdenum (Mo)-containing nitrogenases (Nif), vanadium (V)-containing nitrogenases (Vnf), and iron (Fe)-containing nitrogenases (Anf) (13–16).
Phylogenetic evidence indicates that Nif emerged prior to Anf and Vnf and that the earliest-evolving Nif lineages are from anaerobic, hyperthermophilic methanogens, implicating a chemotrophic origin for N2 fixation in an anoxic environment (7, 9, 13, 16, 17). N2-fixing microorganisms (i.e., diazotrophs) have since diversified to function in a wide variety of genomic and metabolic backgrounds, including those of aerobes, facultative anaerobes, and phototrophs. Given the O2 sensitivity of Nif, aerobic, facultatively anaerobic, and oxygenic phototrophic diazotrophs have had to evolve or acquire one of several mechanisms to mitigate the toxicity of O2 to this enzyme during N2 fixation (17). These mechanisms include fixing N2 only during dark hours when oxygenic photosynthesis ceases and when heterotrophic respiration keeps O2 tensions low (18), using specialized cells called heterocysts in filamentous Cyanobacteria to spatially localize nitrogenase in anaerobic cellular compartments (19), maintaining an anoxic environment in the cytoplasm through increased O2-dependent respiratory activity (20), or fixing N2 only during anaerobic growth. A recent study showed that the diversification of Nif in aerobes and facultative anaerobes was also accompanied by the recruitment and loss of genes that are involved in regulating and protecting Nif against oxidative stress (17).
The primary electron donors to Nif are reduced ferredoxin (Fd) (21–24) and flavodoxin (Fld) (25–27). The transfer of eight electrons from Fd or Fld to NifH and ultimately to NifDK is an ATP-dependent process, requiring at a minimum 16 mol ATP per mol N2 reduced (28–30). Less is known of the stoichiometry of ATP hydrolysis per mol N2 reduced by Anf or Vnf (31–34). The reduction of Fd or Fld in diazotrophs that occupy anoxic environments (e.g., clostridia and methanogens) occurs via the activity of pyruvate-flavodoxin oxidoreductase (PFOR) (35–38), a select subset (groups 3c, 3d, 4d, and 4e) of [NiFe]-hydrogenases, or [FeFe]-hydrogenases (39–45). However, the reduction of Fd (standard electrode potential [Eo′] ∼ −420 mV) in aerobic and some anoxygenic phototrophic diazotrophs that inhabit less-reducing environments is more of a challenge. Rather than producing reduced Fd during their primary energy metabolism, aerobic bacteria and some anoxygenic phototrophs, in particular facultatively anaerobic purple sulfur and nonsulfur bacteria, generate reduced NADH or NADPH (Eo′ = −320 mV) (46, 47), which are not of low enough potential to drive N2 reduction (48–50). Anaerobic purple sulfur and facultatively anaerobic nonsulfur anoxygenic phototrophic bacteria utilize photosystems to drive cyclic electron transfer. The reduction of NAD+/NADP+ is typically accomplished with electrons supplied by the oxidation of an inorganic substrate (e.g., sulfide, thiosulfate, or hydrogen [H2]) or by the oxidation of organic compounds and energy from reverse electron transport if the inorganic electron donor is not of low-enough potential to reduce NAD+/NADP+ (51, 52). It has been proposed that these taxa acquired the Fix complex (encoded by fixABCX) and/or the Rhodobacter nitrogen fixation [Rnf, encoded by rnfABCDEG(H)] complex in order to generate reduced Fd from NADH/NADPH (46). Fix catalyzes the oxidation of two NADH to generate a reduced quinone, which fuels the respiratory chain, and a reduced Fd (27, 53, 54), whereas Rnf catalyzes the NADH-dependent reduction of Fd by coupling it to the depletion of the electrochemical gradient (55–58).
Oxygenic phototrophic Cyanobacteria utilize photosystem I to energize electrons to potentials negative enough to drive the reduction of Fd (51, 52). However, this Fd is not available to Nif, since it must be temporally or spatially separated from oxygenic photosynthesis due to inhibition of Nif by O2. Rather than coding for [FeFe]-hydrogenase, [NiFe]-hydrogenase, Fix, or Rnf and using these enzymes to reduce Fd, Cyanobacteria encode ferredoxin-NADP+ oxidoreductase (FNR), which can function in reverse to reduce Fd or Fld with NADPH generated by carbohydrate oxidation in heterocysts or when O2 tensions are low (35, 59–62). Some Cyanobacteria also encode PFOR, which might be expected to contribute to Fd reduction in these cells, and this might be used by Nif. Like Cyanobacteria anaerobic anoxygenic green sulfur bacteria use a type I photosystem that is distantly related to photosystem I to generate reduced Fd as a component of photosystem-driven cyclic electron transfer (51, 52); however, unlike Cyanobacteria it is possible that this serves as a reductant for N2 fixation (63). Green sulfur bacteria also encode an FNR, which is phylogenetically and structurally unrelated to conventional FNR found in Cyanobacteria and this can also be used to drive reduction of Fd (64, 65). Thus, at least seven enzyme complexes have evolved to provide reduced Fd for N2 fixation: PFOR, [NiFe]-hydrogenase, [FeFe]-hydrogenase, Rnf, Fix, and both forms of FNR (35); however, their distribution in the genomes of diazotrophs is not known.
Different Fds and Flds are also likely to be involved in the delivery of electrons to nitrogenase in aerobes, anaerobes, and phototrophs, and these Fds and Flds may vary alongside the primary enzymes that are involved in reducing these electron carriers. Fds are sensitive to O2 due to the lability of their iron sulfur (FeS) clusters (66–69). In contrast, Flds contain flavin mononucleotide as the prosthetic group involved in electron transfers instead of FeS clusters and, hence, are thought to be less sensitive to O2 (67, 70, 71). Previous bioinformatics analyses have shown that NifF (17), an Fld that can donate electrons to Nif (24, 72, 73), was recruited to nif operons during the diversification of Nif from anaerobic to aerobic taxa (17), which may point to the use of Flds as an adaptive strategy to fix N2 in oxic environments. Moreover, under the iron-deficient conditions that characterize most circumneutral oxic environments, diazotrophs tend to synthesize Flds preferentially as primary electron donors to Nif (22–24, 26, 74). Studies have also shown that electron delivery by an Fd or Fld can be complemented by other Fds or Flds that are encoded in the genomes of diazotrophs (24, 35, 74–76). These observations suggest that pathways that mediate electron flow to nitrogenase are flexible and vary according to the genomic and metabolic background of taxa.
To better define the systems of electron transfer to nitrogenase in diverse microbes, we compiled all Fd and Fld homologs in Nif-encoding genomes and classified them using homology-based methods. In addition, we compiled homologs of all enzymes that have been shown to reduce Fd or Fld for use in the reduction of N2 by nitrogenase. These include PFOR, [NiFe]-hydrogenase, [FeFe]-hydrogenase, Fix, Rnf, and both forms of FNR. Since Nif is encoded in all genomes that encode Anf and Vnf (1), we also examined the systems of electron transfer to these alternative nitrogenase isoforms. Organisms encoding Nif, Anf, and Vnf were classified phylogenetically and physiologically as aerobes, anaerobes, or facultative anaerobes and as chemotrophs, anoxygenic phototrophs, or oxygenic phototrophs based on published data. Statistical analyses were then applied to this curated data set to identify patterns of cooccurrence between the distribution of nitrogenase lineages/isoforms, enzymes that putatively reduce Fd/Fld, and Fds/Flds. The results are discussed in the context of the metabolism of the cells, specifically the influence of O2 and light on putative pathways of electron delivery to nitrogenase.
RESULTS AND DISCUSSION
Taxonomic distribution and phylogeny of Nif/Anf/Vnf HDK homologs.
Nitrogenases were identified in the genomes of diverse Bacteria and Archaea that included obligate aerobes, facultative anaerobes, obligate anaerobes, phototrophs and chemotrophs, and autotrophs and heterotrophs. The identification of Nif in organisms with diverse metabolisms is consistent with a complex evolutionary history that has been described previously for N2 fixation (see Table S1A in the supplemental material) (1, 7, 8, 13, 16). Of the total 4,588 publicly available genomes in our database, 359 genomes (7.8% of the total) encoded the minimal set of proteins for nitrogen fixation (i.e., homologs of NifHDKENB) (7). Forty-six of the 359 taxa with genomes that encode nitrogenase complements have been experimentally shown to grow with atmospheric N2 as their sole N source (Table S1B). Of these 359 nitrogenase-encoding genomes, 48 belonged to obligately chemotrophic and anaerobic Archaea, all of which were affiliated with methanogens within the phylum Euryarchaeota. The remaining 311 diazotrophic genomes were identified in the bacterial domain, with the majority identified as members of the Proteobacteria (n = 191) and Firmicutes (n = 68) (Table S1A). Of the 311 diazotrophic bacterial genomes, 31% were from aerobes, 40% were from facultative anaerobes, and 28% were from obligate anaerobes. Furthermore, 79% of the diazotrophic bacterial genomes were from chemotrophs and 21% were from phototrophs (both oxygenic and anoxygenic).
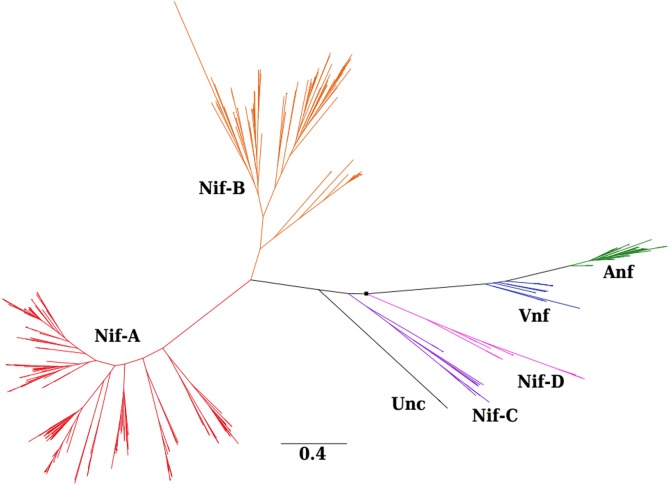
Phylogenetic reconstruction of a concatenation of HDK proteins revealed six distinct lineages of nitrogenases (Fig. 1). These included the four lineages that have been identified in previous studies (1, 8, 13, 14, 16, 77), which include two Nif sublineages (designated Nif-A and Nif-B) and two sublineages designated Anf and Vnf. In addition, previous combined informatics and structural analyses have suggested that two biochemically uncharacterized lineages likely harbor a molybdenum cofactor (9), and organisms that encode them have experimentally been shown to fix N2 (11, 78); hence, we refer to these lineages as Nif-C and Nif-D in this study.
FIG 1.
Phylogenetic reconstruction of a concatenation of H, D, and K subunits of nitrogenase and uncharacterized nitrogenase paralogs (n = 420 concatenated protein sequences). All nodes shown exhibited bootstrap supports of >90% (out of 1,000 bootstrap replicates), except where a black box (>70%) is shown. Nif, molybdenum (Mo) nitrogenase; Anf, iron-only (Fe) nitrogenase; Vnf, vanadium (V) nitrogenase; Unc, uncharacterized nitrogenase-like proteins.
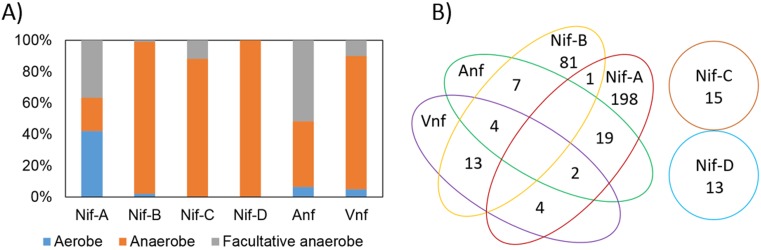
A total of 224 genomes encoded Nif-A, and these were from taxa that are primarily from the Proteobacteria (n = 169), Firmicutes (n = 26), and Cyanobacteria (n = 20) (Table S1A). Thirty of these taxa have been experimentally shown to fix N2 (Table S1B). However, the cyanobacterium Microcoleus chthonoplastes (which is not included in our database since a complete genome is not available) does encode Nif-A and the minimal set of proteins that allow for N2 fixation (i.e., homologs of NifHDKENB) (7). And yet, cultivation studies suggest that this taxon cannot grow with N2 as its sole N source (79), indicating that the presence of a full nif gene complement does not guarantee the ability to fix N2. The majority of the taxa that encode Nif-A are aerobes (42%) or facultative anaerobes (38% of total taxa) (Fig. 2A). In addition, 23% of Nif-A-encoding genomes were from phototrophs and 77% were from chemotrophs (Fig. S1). Among the 23% of Nif-A homologs that are encoded in the genomes of phototrophs, 9% are from Cyanobacteria, 11% are from facultatively anaerobic anoxygenic purple nonsulfur bacteria, and 2% are from anaerobic anoxygenic purple bacteria. However, it is not clear how rigorously O2 usage has been characterized in these anaerobic anoxygenic purple bacteria, which include the following strains: Allochromatium vinosum, Thiocystis violascens, Thioflavicoccus mobilis, and Halorhodospira halophila.
FIG 2.
Homologs of nitrogenase identified in the genomes of putative diazotrophs. (A) Histogram depicting the percentage of diazotrophs within each specified Nif lineage (see Fig. 1) that are aerobic, anaerobic, or facultatively anaerobic, as determined from surveys of the literature for cultivated organisms. (B) Venn diagram representing the numbers of genomes that encode one or more specified lineages of nitrogenase (see Fig. 1). Genomes that encode Nif-C and Nif-D do not encode other isoforms of nitrogenase and are thus depicted as separate.
The majority of the 106 genomes that encode Nif-B were from obligate anaerobes (93% of total taxa) and chemotrophs (87% of the total taxa) and anaerobic anoxygenic green phototrophic bacteria (11% of total). Nif-B-encoding taxa were primarily affiliated with Firmicutes (n = 34), Euryarchaeota (n = 29), and Proteobacteria (n = 22). Of these 106 Nif-B-encoding genomes, 13 were from taxa (primarily from the genus Clostridium) that have been experimentally shown to fix N2 (Table S1B). All of the anaerobic anoxygenic phototrophs were affiliated with the Chlorobi or Chloroflexi (n = 12). Members of the Firmicutes (n = 8) and Euryarchaeota (n = 6) encoded Nif-C (n = 15), while all (n = 13) Nif-D-encoding genomes were methanogens (Euryarchaeota). None of the taxa that encode Nif-C have been shown to fix N2, while three organisms that encode Nif-D (all of which are methanogens) have been experimentally shown to fix N2 (Table S1B). Taxa that encode Nif-C were primarily anaerobes (88% of the total taxa), while taxa that encode Nif-D were all anaerobes. All the Nif-C- and Nif-D-encoding genomes were chemotrophs.
A total of 32 genomes encoded Anf, and these were primarily from the Proteobacteria (n = 16) and Firmicutes (n = 11), while a total of 23 genomes encoded Vnf. Most of the Vnf-encoding genomes were from the Euryarchaeota (n = 11) and Firmicutes (n = 8). A separate lineage comprising HDK proteins from taxa that have not yet been shown to fix N2 were nested among Nif sublineages. These proteins were termed “uncharacterized,” as previously described (13), and were affiliated with members of the Chloroflexi (i.e., Roseiflexus spp.).
Multiple forms of nitrogenase were often detected in the same genome (Fig. 2B). However, the genomes that coded for Nif-C or Nif-D did not code for any other forms of nitrogenases. The genome of the cyanobacterium Pleurocapsa sp. strain PCC 7327 was found to encode both Nif-A and Nif-B. All Anf- and Vnf-encoding genomes also encoded Nif, which is consistent with previous observations (1, 7, 13). Four genomes were identified that coded for Anf, Vnf, and Nif-B, while two genomes encoded Anf, Vnf, and Nif-A. Interestingly, Anf was more commonly detected in Nif-A-encoding genomes (65% of total Anf-encoding genomes), while Vnf was more commonly detected in Nif-B-encoding genomes (77% of total Vnf-encoding genomes). If the root of the nitrogenase phylogeny is within the Nif-D lineage, as has been suggested previously (7, 13, 16), our phylogenetic reconstruction indicates that Anf diverged from a Vnf-like ancestor, both of which emerged from a Nif-C- or Nif-D-like ancestor. Likewise, our phylogenetic reconstruction indicates that Nif-A diversified from a Nif-B-like ancestor.
Taxonomic distribution of ferredoxin and flavodoxin homologs in the genomes of putative diazotrophs. (i) Ferredoxin.
The taxonomic distribution of all 47 Fds identified in the genomes of putative diazotrophs is detailed in Table S2 in the supplemental material. A total of 36 Fds were detected in Nif-A-encoding genomes, of which FdxB and FdxA were the most common; they were present in >40% of those genomes (Fig. S2A). fdxB is encoded near nifQ and nifB in the genome of Azotobacter vinelandii (80, 81) and is expressed at a similar level to these genes under N2-fixing conditions (82). Mutational studies have shown involvement of FdxB in active-site metallocluster biosynthesis in A. vinelandii (83, 84). Furthermore, FdxB was shown to be incapable of serving as an electron donor to nitrogenase in vitro using protein from Rhodobacter capsulatus (83). In contrast, in the cyanobacterium Anabaena sp. strain PCC 7120, FdxB has been shown to complement fdxH mutant strains; FdxH is the preferred donor of electrons to nitrogenase in this taxon (85). These observations suggest that FdxB has multiple roles in diazotrophic cells. The prevalence of FdxA in Nif-A-encoding genomes is consistent with its colocalization near major nif gene clusters, such as in the case of A. vinelandii (24), or within the nif gene cluster itself, such as in Herbaspirillum seropedicae (86). This agrees with findings from multiple studies that have documented the ability of FdxA to donate electrons to nitrogenase (87–90). Several less commonly detected Fds (identified in <40% of the Nif-A-encoding genomes) have also been shown or predicted to be involved in electron transfer to nitrogenase. These include FdxN in Rhodospirillum rubrum and Rhizobium meliloti (91, 92), FdxE in Rhodobacter capsulatus (93), FdxH (85, 94), FdxI in Anabaena sp. PCC 7120 (95, 96), and FdxC in R. capsulatus (Fig. S2A) (97). Together, these results suggest that FdxA and FdxB may have a role in electron delivery to Nif-A-like nitrogenases; however, other Fds could potentially complement the functionality of these Fds.
Two Fds found in Clostridium pasteurianum, CpFd1 (65% of the total Nif-B-encoding genomes) and CpFd4 (48% of the total genomes), correlated with the distribution of Nif-B in diazotrophic genomes (Fig. S2A), suggesting a role for these Fds in electron delivery to Nif-B-like nitrogenases. FdxA (47% of the total Nif-C-encoding genomes) and three Fds found in Caldicellulosiruptor saccharolyticus, CsFd3 (47% of the total genomes), CsFd6 (47% of the total genomes), and CsFd1 (41% of the total genomes), were predominant in Nif-C-encoding genomes, while Nif-D-encoding genomes encoded multiple Fds found in Methanocaldococcus vulcanius (MvFds) (Fig. S2A). The distribution of Anf in genomes was only moderately correlated with the distribution of Fds, with FdxA and FdxB yielding the highest correlations (i.e., >45% of the genomes) (Fig. S2A). Finally, the distribution of Vnf was highly correlated with the distribution of CpFd4 (80% of the total genomes) and, to lesser extents, four Fds found in Methanosarcina barkeri, MbFd1, MbFd2, MbFd3, and MbFd4 (Fig. S2A). While electron transfer to Nif-A-like nitrogenases has been studied extensively, we are unaware of experimental data on the role of Fds in electron transfer to Nif-B, Nif-C, Nif-D, Anf, or Vnf, which precludes a comparison of our informatics-based predictions to experimental data.
The Fds identified in our study varied markedly among genomes coding for the various isoforms of nitrogenase (Fig. S2A). Importantly, genomes encoding Nif-A nitrogenases coded for unique Fds that were either absent or present but rarely detected in the genomes of other diazotrophs (Fig. S2A). For example, of 14 abundant Fds (i.e., present in >40% of the genomes) encoded by Nif-A-encoding genomes, 12 were unique to Nif-A-encoding genomes, while only two were identified in the genomes of other diazotrophs (Fig. S2A). In contrast, considerable overlap was observed in the composition of Fds/Flds encoded in genomes that also encoded Nif-B, Nif-C, or Nif-D.
(ii) Flavodoxin.
We identified a total of five phylogenetically distinct Flds among nitrogenase-encoding genomes. The taxonomic distribution of all five Flds identified is detailed in Table S2 in the supplemental material. Of the five Flds detected in Nif-A-encoding genomes, FldA was the most common and was identified among 26% of the genomes (Fig. S2B). The distribution of CpFld2 and CpFld3 cooccurred with Nif-B in >40% of the genomes, while CpFld3 was detected in >40% of the Nif-C- and Nif-D-encoding genomes (Fig. S2B). Like Nif-A-encoding genomes, FldA was the dominant Fld in Anf-encoding genomes (Fig. S2B). Lastly, CpFld3 and CpFld2 were the dominant Flds in Vnf-encoding genomes (Fig. S2B). There is no experimental evidence that these Flds are involved in electron transfer to nitrogenase, with the exception of NifF (present in a few Nif-A-encoding genomes), which has been shown to transfer electrons to nitrogenase in Klebsiella pneumoniae (98, 99) and A. vinelandii (24, 27).
The distribution of the five Flds identified in this study varied in diazotrophs with different metabolic backgrounds. Of the five Flds identified among the genomes of diazotrophs, NifF and FldA were only detected in the genomes of aerobes, facultative anaerobes, or phototrophs that encoded Nif-A (Fig. S2B). The sole exception to this observation was in the genome of the anaerobic spirochete Spirochaeta smaragdinae strain DSM 11293, which was found to code for FldA and Nif-B (Table S2). The remaining three Flds were common in the genomes of anaerobic chemotrophs that encoded Nif-B, Nif-C, and Nif-D. This observation is like that made for Fds, where the distribution of Flds in genomes that encode Nif-A were distinct from those that encoded Nif-B, Nif-D, and Nif-D. These collective observations indicate that the Fds/Flds in aerobic/facultatively anaerobic diazotrophs differ from those in anaerobic diazotrophs, leading to the hypothesis that O2 played a role in the diversification of systems for the delivery of electrons to nitrogenase. Moreover, the Fds/Flds are different among phototrophs and chemotrophs, leading to the hypothesis that the integration of light into the energy metabolism of diazotrophs played a key role in the diversification of systems of electron delivery to nitrogenase.
Distribution of ferredoxin- and flavodoxin-reducing protein homologs in the genomes of putative diazotrophs.
The taxonomic distributions of PFOR, [NiFe]-hydrogenase, [FeFe]-hydrogenase, Rnf, Fix, and FNR homologs are detailed in Table S2. Briefly, genomes that encoded Nif-A and Nif-B coded for homologs of six of the seven putative Fd- or Fld-reducing enzymes (Fig. S3); FNR homologs identified in green sulfur bacteria were not identified in Nif-A-encoding genomes. Nif-A-encoding genomes tended to code for Fix, PFOR, and Rnf homologs, while Nif-B-encoding genomes tended to code for [FeFe]-hydrogenase, [NiFe]-hydrogenase (primarily group 4e) (41), and PFOR (Fig. S3). Genomes that coded for Nif-C were found to code for homologs of four of the seven Fd- or Fld-reducing enzymes, with [NiFe]-hydrogenase (primarily group 4e) (41) and PFOR being the most abundant among these. Nif-D-encoding genomes were found to code for only group 4d [NiFe]-hydrogenase (41) and PFOR. Anf- and Vnf-encoding genomes coded for homologs of five of the seven reducing enzymes. Fix, [FeFe]-hydrogenases, and PFOR were enriched in Anf-encoding genomes, while group 4e [NiFe]-hydrogenases (41), [FeFe]-hydrogenases, and PFOR were enriched in Vnf-encoding genomes.
The shift in the distribution of enzyme homologs capable of reducing Fd or Fld in diazotrophic genomes generally corresponded to the diversification of Nif-A from Nif-B/Nif-C/Nif-D (Fig. 1 and 3). Chemotrophic anaerobic diazotrophs that coded for Nif-B/Nif-C/Nif-D also encoded PFOR, [FeFe]-hydrogenase, or [NiFe]-hydrogenase. PFOR couples the oxidation of pyruvate to the reduction of Fd, and its expression is regulated by the availability of N in a variety of diazotrophs (36, 73, 100, 101), whereas specific lineages of hydrogenase (both [FeFe] and [NiFe]) have been shown to couple reversible H2 oxidation to the reduction of Fd (Fig. 3) (41, 102, 103).
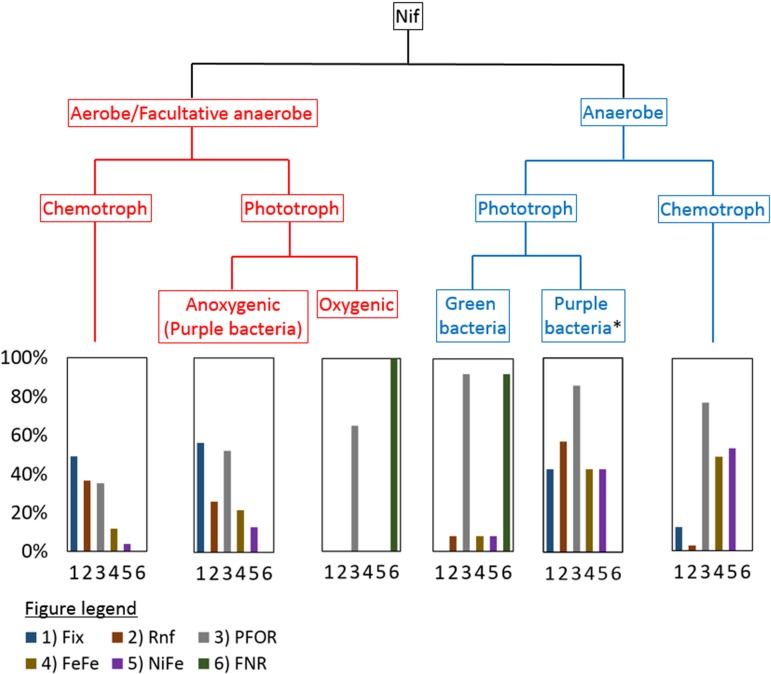
FIG 3.
Flow chart of nitrogenase in different metabolic backgrounds and histograms depicting the percentage of enzyme homologs putatively involved in Fd/Fld reduction identified in the genomes comprising a specified group. Aerobic/facultative anaerobic and anaerobic organisms are further classified as phototrophs or chemotrophs. Phototrophs in aerobes/facultative anaerobes are further classified based on whether they are oxygenic or anoxygenic phototrophs, while anaerobic phototrophs are classified as either purple sulfur/nonsulfur bacteria or green sulfur/nonsulfur bacteria. Importantly, it is not clear from surveys of the literature that anaerobic purple bacterial strains (denoted by an asterisk) were robustly tested for their ability to use O2. FNR, ferredoxin-NADP+ oxidoreductase; PFOR, pyruvate-flavodoxin oxidoreductase; Rnf, Rhodobacter nitrogen fixation protein; FeFe, iron-only hydrogenase; NiFe, nickel-iron hydrogenase; Fix, electron transfer flavoprotein involved in nitrogen fixation.
Like chemotrophic anaerobes, anaerobic anoxygenic phototrophic green (both sulfur and nonsulfur) bacteria and anaerobic anoxygenic purple (both sulfur and nonsulfur) bacteria tended to encode PFOR (92% and 60% of total genomes, respectively). Anaerobic anoxygenic purple bacteria also encoded Fix, Rnf, [NiFe]-hydrogenase, and [FeFe]-hydrogenase, while green sulfur bacteria also encoded a functionally similar but phylogenetically distinct FNR isoform that is found in Cyanobacteria (Fig. 3). Like anaerobic diazotrophs, PFOR was identified in the genomes of aerobic/facultatively anaerobic diazotrophs. However, unlike anaerobic diazotrophs, the genomes of aerobic/facultatively anaerobic diazotrophs also tended to encode Fix, Rnf, and FNR (Fig. 3). Fix and Rnf generate reduced Fd with a more-reduced potential than its substrate NADH via electron bifurcation (27, 56) or by coupling the reaction with reverse ion translocation (either proton or sodium dependent) (55–57), respectively. Fix has been suggested to play a role in supplying reductant to Nif in aerobic diazotrophs (27, 104–106). Consistent with this suggestion, Fix and Rnf are encoded in the genomes of aerobic chemotrophs (50% of these genomes) and anoxygenic phototrophs (68% of these genomes), specifically purple sulfur and nonsulfur bacteria (Fig. 3). Furthermore, homologs of FNR were only identified in the genomes of oxygenic phototrophic and anoxygenic green sulfur phototrophic bacteria, although the FNR homologs identified in green sulfur phototrophic bacteria were phylogenetically distinct from those cyanobacterial FNR. In Cyanobacteria it is thought that FNR catalyzes the reduction of Fd with NADPH generated during carbohydrate oxidation, which is then used to fix N2 (61); it is not yet known if Fd produced by FNR in green sulfur bacteria is involved in N2 fixation. Together, these observations suggest that the enzymes that diazotrophs use to generate reduced Fd or Fld to drive N2 reduction vary in organisms with different metabolisms, in particular those that are able to integrate O2 or light into their energy metabolism compared to those that are not able to take advantage of these metabolic strategies.
Predicted electron transfer pathways to nitrogenase.
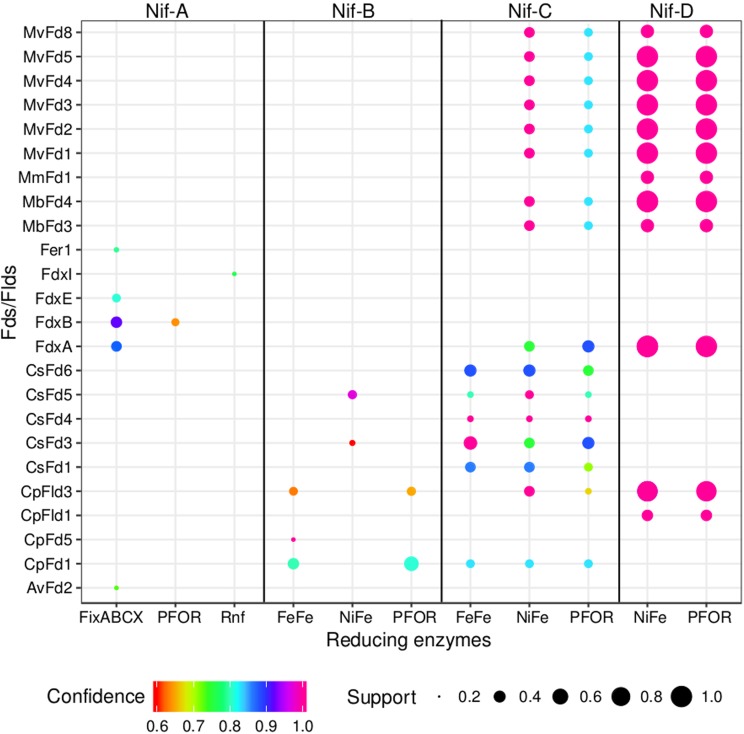
The covariation among the distributions of specified nitrogenase lineage homologs, Fd/Fld homologs, and Fd-/Fld-reducing-enzyme homologs was examined to identify putative systems of electron delivery to Nif in different metabolic backgrounds (Fig. 4). The primary Fd-/Fld-reducing-enzyme homologs in genomes that code for Nif-A are Fix, PFOR, and Rnf; FNR homologs were enriched in the genomes of Nif-A-encoding diazotrophic Cyanobacteria (n = 20). The source of electrons to Nif-A is likely either pyruvate in the case of PFOR (107), NADH generated from central metabolism in the case of Fix and Rnf (55, 56, 58), or NADPH in the case of FNR (60, 108).
FIG 4.
Bubble plot depicting the dominant patterns in the distribution of putative electron carrier protein homologs (Fds/Flds), enzyme homologs that putatively reduce Fd/Fld, and specified Nif lineages, as determined by the Apriori algorithm (124). Each unique pattern is given as a bubble, and the color represents the confidence value, or statistical significance, in the codistribution of specified proteins (only confidence values of ≥0.6 are presented). The size of the bubble represents the support value (≥0.2), or the frequency with which two proteins are identified in the same genome. For simplicity, only the proteins that were present in >20% of the diazotrophic genomes for each specified nitrogenase lineage were considered in this analysis. PFOR, pyruvate-Fld oxidoreductase; Rnf, Rhodobacter nitrogen fixation protein; FeFe, iron-only hydrogenase; NiFe, nickel-iron hydrogenase; FixABCX, electron transfer flavoprotein that is involved in nitrogen fixation. Names of source organisms for Fds and Flds are abbreviated as follows: Mv, Methanocaldococcus vulcanius; Mm, Methanococcus maripaludis; Mb, Methanosarcina barkeri; Cs, Caldicellulosiruptor saccharolyticus; Cp, Clostridium pasteurianum; Av, A. vinelandii. Abbreviations for Fds and Flds are presented in Tables 1 and 2, respectively.
Genomes that code for Nif-B were associated with different Fd/Fld homologs than Nif-A-encoding genomes and were associated with homologs of three different Fd-/Fld-reducing enzymes: PFOR, [FeFe]-hydrogenase, and [NiFe]-hydrogenase (Fig. 4). FNR was detected in all Nif-B-encoding green sulfur bacterial anoxygenic phototrophs (n = 11). To this end, the source of electrons to reduce Nif-B is predicted to be H2 in the case of [FeFe]- and [NiFe]-hydrogenases (109, 110), pyruvate in the case of PFOR, or NADPH in the case of FNR.
Genomes that encoded Nif-C and Nif-D coded for a different suite of Fd/Fld homologs potentially involved in electron delivery to Nif than did Nif-A- and Nif-B-encoding genomes (Fig. 4). Very little variation was observed in the distribution of putative Fd-/Fld-reducing-enzyme homologs and Fd/Fld homologs in Nif-C-encoding genomes compared to their distribution in genomes that encoded Nif-D. However, unlike Nif-C-encoding genomes, Nif-D-encoding genomes, which are derived from methanogens, lack evidence for homologs of [FeFe]-hydrogenase. This is consistent with the absence of genes encoding [FeFe]-hydrogenase in Archaea (102, 103). Thus, like Nif-B, the source of electrons to reduce Nif-C and Nif-D is likely H2 or pyruvate.
Anf- and Vnf-encoding diazotrophic genomes also encode Nif, which in the case of Anf was typically Nif-A and for Vnf was typically Nif-B. As such, patterns in the distribution of Fds/Flds and Fd/Fld-reducing enzymes in Anf- and Vnf-encoding genomes are like Nif-A and Nif-B, respectively. The organisms that encode Anf are likely to rely on three putative Fd-/Fld-reducing enzymes: [FeFe]-hydrogenase, Fix, or PFOR (Fig. S4). As in Nif-A-encoding organisms, the source of electrons to Anf is predicted to be from pyruvate or from central metabolism in the form of NADH. The organisms that encode Vnf are likely to rely on three putative Fd-/Fld-reducing enzymes: PFOR, [FeFe]-hydrogenase, and [NiFe]-hydrogenase (Fig. S4). As such, the source of electrons used to reduce Vnf is likely from H2 or pyruvate.
Intriguingly, the genomes of several putative diazotrophs (n = 25) lacked a homolog of PFOR, [NiFe]-hydrogenase, [FeFe]-hydrogenase, Rnf, Fix, or either isoform of FNR (Table S2). The inability to detect homologs of these seven enzymes was not due to these genomes being incomplete, since all of them were closed. Of the 25 genomes that lacked a homolog of these enzymes, 14 encoded Nif-A-like nitrogenases, while 11 encoded Nif-B-like nitrogenases. All 14 genomes that encoded Nif-A-like nitrogenases were classified as either aerobic or facultatively anaerobic, while all 11 genomes that encoded Nif-B-like nitrogenases were classified as strictly anaerobic. These data suggest the presence of at least one additional enzymatic mechanism for generating Fds or Flds that can be used to reduce N2.
Possible drivers in the diversification of pathways for electron transfer to nitrogenase.
Previous studies have shown that Nif diversified in large part in response to O2, in particular the integration of O2 into cellular metabolism (17, 111). By extension, this indicates that O2 would have been at least temporarily available in the local habitat of the ancestors of aerobic/facultatively anaerobic diazotrophs, which would increase the oxidation state of their local habitat. Previous studies have noted a strong correlation between the average oxidation state of carbon in archaeal and bacterial proteomes (inferred from metagenomic data) and the oxidation state of the local environment (112). Likewise, extracellular proteins from yeast have been shown to have a higher average oxidation state of carbon than cytoplasmic proteins, an observation that was attributed to a higher oxidation state on the exterior of the cell than in the interior (113). As has been suggested previously, a shift in the oxidation state of a system to a more-oxidizing potential favors the formation of products (e.g., proteins) that themselves are more oxidized (113). Since the oxidation state of the cytoplasm of a cell should be related to that of the external environment of a cell, and cells that are under selection should be under selection to minimize the cost of protein synthesis to the extent that those proteins remain functional, we hypothesized that the integration of O2 into the metabolism of diazotrophs would be accompanied by an overall shift in the oxidation state of the proteome of those cells that may in turn influence the functionality or stability of Fd/Flds, the enzymes that function to reduce these cofactors, or the availability of substrates for these enzymes. Observations supporting this hypothesis would serve as evidence that O2 had a fundamental role in the evolution of diazotrophs, beyond what has been recognized previously, which includes physiological mechanisms of spatial/temporal decoupling of nitrogenase with O2 metabolism (18, 19), increased consumption of O2 through respiratory activity (20), and recruitment and loss of genes involved in regulating nitrogenase expression and activity (17).
To test this hypothesis, we calculated the average oxidation state of carbon in inferred proteomes for the taxa that comprised organisms encoding each of the four Nif sublineages (Fig. 5). Welch's t test showed that the average oxidation state of carbon in inferred proteomes was significantly (P < 2.2 × 10−16) more positive (−0.024 ± 0.018) for Nif-A-encoding genomes than for Nif-B-encoding genomes (−0.043 ± 0.019), the most closely related lineage (Fig. 1). Likewise, the average oxidation state of carbon in inferred proteomes was significantly (P = 1.9 × 10−8 and 1.8 × 10−8, respectively) more positive for Nif-A-encoding genomes than for genomes encoding Nif-C (−0.078 ± 0.033) and Nif-D (−0.058 ± 0.016). These observations are consistent with the observation that most of the Nif-A-encoding genomes were from aerobic or facultatively anaerobic taxa, whereas the majority of Nif-B-, Nif-C-, and Nif-D-encoding genomes were from strict anaerobes and, to a lesser extent, facultative anaerobes (Fig. 2A). These observations may be due to Nif-A-encoding diazotrophs inhabiting, on average, a more oxidized environment than Nif-B-, Nif-C-, and Nif-D-encoding diazotrophs. If true, this finding would suggest that Nif-A-encoding cells also may harbor a more oxidized cytoplasm than strictly anaerobic diazotrophs. A caveat to this analysis is that it assumes that all proteins in a diazotrophic cell are expressed at the same level, which is unlikely to be the case. Further experiments are needed to examine whether the average oxidation state of a cellular proteome is sensitive to growth conditions (e.g., aerobic versus anaerobic) and whether such a response reflects acclimatization to minimize energetic costs associated with protein biosynthesis.
FIG 5.
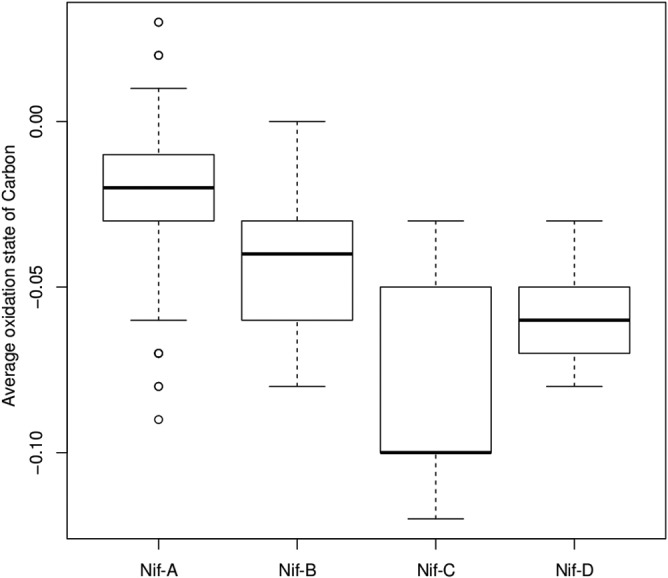
Box plot representing the average oxidation state of carbon in proteomes (inferred from genomes) for the taxa comprising organisms encoding the specified nitrogenase isoform. Here, the box represents the interquartile range and the whiskers show the full range of the data. Outlier values are represented as circles, and the line within the box represents the median value.
We hypothesized that differences in the oxidation state of the external environment of cells and their cytoplasm would affect the functionality of proteins involved in electron transfer to nitrogenase and might account for the differences in the Fds/Flds used by aerobic/facultative anaerobic and strictly anaerobic diazotrophs. To test this hypothesis, we reconstructed the evolutionary history of several Fds/Flds that appear to be commonly used as electron donors to the various lineages of Nif. The Fds FdxA and FdxB, which are commonly identified in Nif-A-encoding genomes, branch among (are nested within) a variety of Fd lineages commonly identified in anaerobic diazotrophs (i.e., Nif-B-, Nif-C-, and Nif-D-encoding taxa). These include MvFd4, MvFd5, CsFd6, MvFd2, and CpFd4 (Fig. S5). Parsimony would suggest that FdxA and FdxB are recently evolved and that these Fds evolved from Fds that likely functioned in an anaerobe. Similarly, NifF and FldA were nested among Flds commonly identified in anaerobic diazotroph genomes, including CpFld1, CpFld3, and CpFld2 (Fig. S6). Among Flds, NifF and FldA formed a monophyletic lineage and were sister to CpFld2, suggesting that they diverged from an ancestor of these proteins and that this ancestral protein most likely functioned in an anaerobe.
A previous study attempted to identify structural features associated with Fds from Anabaena variabilis that lend stability in oxic versus anoxic environments (114). The authors showed that slight changes in residues near the active-site FeS cluster protected it from reactive oxygen species. This indicates that the Fds that are active in aerobes inhabiting oxic environments likely have different amino acid compositions than Fds found in anaerobes, a finding that is supported by our informatics and phylogenetics analyses. As such, the results presented here provide a suite of Fds/Flds that appear to have evolved to function optimally in aerobic versus anaerobic diazotrophs and, thus, provide a template for downstream studies aimed at further elucidating the structural features that enable Fd/Fld function in more-oxidized environments.
The more-positive oxidation state of carbon in the inferred proteomes of Nif-A-encoding diazotrophs, which are largely from aerobic or facultatively anaerobic taxa, may indicate that the cytoplasm or local environment of those cells is more oxidizing. In general, H2 (which itself has a low oxidation-reduction potential) would be expected to be at low abundance in an oxidized environment (112, 113, 115, 116). It follows that the diversification of Nif into more-oxidizing environments may have been accompanied by a decrease in available H2, which in turn may have represented a selective pressure to evolve mechanisms to generate reduced Fd/Fld other than those that are dependent on H2. In response, Nif-A-encoding diazotrophs would have continued to use PFOR, if available, to meet these demands. In other aerobic or facultatively anaerobic and anoxygenic phototrophic Nif-A-encoding diazotrophs, the primary electron carrier involved in central metabolism likely switched to NADH (51, 52). Under such conditions, diazotrophic cells would have been under selective pressure to evolve mechanisms to drive the formation of reduced Fd from NADH, such as Fix and Rnf.
Concluding remarks.
Diazotrophs have evolved elegant mechanisms to overcome O2 toxicity to nitrogenase during the transition from anaerobic to aerobic metabolism, including spatial and temporal decoupling of N2 fixation activity from O2 respiration (18, 19), increased O2 respiration to maintain an anoxic cytoplasm (20), and recruitment and loss of genes involved in regulating nitrogenase expression and activity (17). Our data show that the average oxidation state of carbon in the inferred proteomes of diazotrophs also likely increased during the transition from anaerobic to aerobic metabolism, which would have driven large-scale changes in the composition of proteins and enzymes that drive the energy metabolism of diazotrophic cells, as well as the availability of substrates for those enzymes. Indeed, our analysis of the inferred systems of electron delivery to Nif reveals that substantial changes took place during the diversification of diazotrophs. These changes include those at the level of the primary electron donors that provide reductant to Nif, with early-evolving chemotrophic anaerobic diazotrophs likely supporting N2 reduction with electrons derived from oxidation of H2 or pyruvate. Later-evolving aerobic/facultatively anaerobic diazotrophs likely support this activity with electrons primarily in the form of NADH/NADPH derived from central metabolism or cyclic electron pathways in anoxygenic phototrophs. Similarly, a shift in the enzymes used to generate reduced Fd was observed, with chemotrophic anaerobic taxa inferred to be primarily dependent on [NiFe]-hydrogenase and PFOR and aerobic, facultative anaerobic or anoxygenic phototrophic taxa largely dependent on Fix and, to a lesser extent, PFOR and Rnf. Finally, a nearly complete turnover in the putative Fds/Flds in aerobic/facultatively anaerobic versus anaerobic diazotrophic taxa was observed. Collectively, these data are consistent with previous reports suggesting that O2 had a profound influence on the evolution of nitrogenase (7, 13, 16, 17, 111) and further suggest that these changes may have been a global adaptive response to an increased oxidation state of the local environment.
The ability of cells to harness light energy likely also impacted systems of electron delivery to Nif. Due to the O2 sensitivity of Nif, oxygenic phototrophs likely utilize FNR to generate reduced Fd from NADPH produced during carbohydrate fermentation at night or in specialized cells. The photosystems of anoxygenic green sulfur bacteria that also encode FNR energize electrons and shuttle them through a cyclic electron transport chain that involves Fd. It is possible that this Fd can be used to reduce Nif directly. Alternatively, these cells may reduce Fd with PFOR, which utilizes pyruvate produced from the oxidation of glycogen. In contrast, anoxygenic phototrophic bacteria, in particular purple sulfur and nonsulfur bacteria, generate reduced quinones during light-driven electron transport, with those electrons supplied by the oxidation of exogenous inorganic or organic electron donors. The potential of these electrons can be further reduced through reverse electron transfer such that it is low enough to reduce NADH. These phototrophs often then utilize Fix or Rnf to drive the reduction of Fd with NADH.
The collective insights described herein suggest that the genomic and metabolic backgrounds of diazotrophs are associated with wholesale changes in the source of electron donors, enzymes involved in coupled oxidation of electron donors to the reduction of Fds/Flds, and the Fds/Flds used to reduce Nif. These changes were associated with differences in the ability to integrate O2 and light into the energy metabolism of the cells. While the informatics-based data that are presented here are predictions of potential systems of electron delivery to Nif, in the case of Nif-A-encoding taxa, they are largely consistent with available biochemical data. However, the validity of the predictions made for pathways of electron delivery to Nif-B-, Nif-C-, and Nif-D-encoding taxa is unknown, since biochemical data have yet to be compiled for model taxa encoding homologs of these enzymes. These include diazotrophs that represent the most ancestral forms of Nif in both chemotrophic and phototrophic genomic and metabolic backgrounds.
MATERIALS AND METHODS
Identification and compilation of nitrogenase homologs.
Genomes that encode the alpha (D) subunit of nitrogenase (i.e., NifD, AnfD and VnfD) were compiled as previously described (17, 117). Briefly, NifD homologs that were extracted using BLASTp were first aligned along with paralogs using Clustal Omega (118). The Nif-/Anf-/VnfD paralogs identified included ChlN from Chlamydomonas reinhardtii (ACJ50143) and Synechocystis sp. strain PCC 6803 (WP_010874227), BchN from Acidiphilium rubrum (BAA76536) and Chloroflexus aurantiacus (WP_012258416), and NflD from Methanocaldococcus jannaschii (WP_010870941) and Methanosarcina mazei (AAM30211). The aligned sequences were subjected to maximum-likelihood phylogenetic reconstruction with RAxML (version 7.3.0) (119), specifying the LG substitution matrix, the PROTGAMMA option, and 1,000 bootstrap iterations. Only Anf-/Vnf-/NifD protein homologs that clustered with previously characterized Nif/-Anf-/VnfD lineages (1, 13, 16) were extracted for downstream analysis. Furthermore, only genomes that encode the minimum set of proteins hypothesized to be required for N2 fixation via Nif (i.e., nifHDKENB) or Anf/Vnf (i.e., anfHDK/vnfHDK) were retained, as previously described (7). In addition, genomes that encode homologs of just nifHDKEB were also retained, given that physiological studies suggest that organisms with this minimum gene complement can assimilate N2 (11, 78). A total of 359 genomes from putative diazotrophs were compiled. To retrieve corresponding homologs of the iron protein (i.e., NifH, AnfH, and VnfH) and the beta subunit (i.e., NifK, AnfK, and VnfK), we performed a BLASTp search with NifH (ACO76403) and NifK (ACO76405) from Azotobacter vinelandii as queries, specifying cutoffs of 30% percent amino acid sequence identities and 60% sequence coverage. Only subunits that colocalized (e.g., nifHDK in an apparent operon) were retained. Each of the three nitrogenase subunits (i.e., H, D, and K subunits of Anf, Vnf, and Nif) was aligned individually with Clustal Omega (118), using the default settings, and the resultant alignment blocks were concatenated. The concatenated HDK sequences were subjected to phylogenetic reconstruction as described above.
Identification of homologs of Fd, Fld, and putative Fd- or Fld-reducing enzymes in the genomes of diazotrophs.
To identify Fds and Flds in the genomes of putative diazotrophs, we first identified all the Fds and Flds in representative organisms for each Nif sublineage. These included Azotobacter vinelandii, Klebsiella pneumoniae, Rhodopseudomonas palustris, and Rhodobacter capsulatus for Nif-A, Clostridium pasteurianum and Methanosarcina barkeri for Nif-B, Caldicellulosiruptor saccharolyticus and Methanocaldococcus vulcanius for Nif-C, and Methanococcus maripaludis for Nif-D. All protein sequences that were annotated as Fds or Flds in the genomes of the above-named taxa were compiled. These protein sequences were subjected to Conserved Domain Database (120) BLAST to identify the type of iron-sulfur clusters present in the Fd and to confirm that the extracted Fld contained a flavin binding motif(s). All the compiled and curated Fds and Flds identified in these representative genomes were separately aligned using Clustal Omega (118). The aligned Fd or Fld sequences were subjected to maximum-likelihood phylogenetic reconstruction with RaxML (version 7.3.0) (119), specifying the LG substitution matrix and the PROTGAMMA option to empirically cluster the sequences into unique groups. In total, 48 Fds (Table 1) and five unique Flds (Table 2) were identified among diazotrophic genomes. The amino acid sequences of all the Fds and Flds were then used as bait sequences to identify homologs in the genomes of putative diazotrophs using BLASTp.
TABLE 1.
Ferredoxin homologs identified in the genomes of putative diazotrophsa
| Annotation | Protein accession no. | Type of cluster | Reference(s) or source |
|---|---|---|---|
| asl2914 | WP_013190310 | 2[4Fe-4S] | 126 |
| FdxH | WP_013190616 | [2Fe-2S] | 127 |
| PetF | WP_013189829 | [2Fe-2S] | 128 |
| FdxA | ACO80005 | [3Fe-4S] [4Fe-4S] | 129 |
| FdxI | ACO76607 | [4Fe-4S] | 130 |
| FdxN | ACO81189 | 2[4Fe-4S] | 82, 131 |
| VnfF | ACO76526 | [4Fe-4S] | 132 |
| XylT | ACO77112 | CXXXXCXXXXCXXC | 130 |
| AvFd1 | AGK16019 | [2Fe-2S] | This study |
| AvFd2 | AGK16622 | [2Fe-2S] | This study |
| AvFd3 | AGK13276 | 2[4Fe-4S] | This study |
| CsFd1 | ABP66178 | 2[4Fe-4S] | This study |
| CsFd2 | ABP66040 | 2[4Fe-4S] | This study |
| CsFd3 | ABP66582 | 2[4Fe-4S] | This study |
| CsFd4 | ABP67884 | 2[4Fe-4S] | This study |
| CsFd5 | ABP67132 | 2[4Fe-4S] | This study |
| CsFd6 | ABP67141 | 2[4Fe-4S] | This study |
| CpFd1 | AJA47502 | [2Fe-2S] | This study |
| CpFd2 | AJA47129 | 2[4Fe-4S] | This study |
| CpFd3 | AJA49513 | 2[4Fe-4S] | This study |
| CpFd4 | AJA46278 | [4Fe-4S] | This study |
| CpFd5 | AJA49585 | 2[4Fe-4S] | This study |
| KpFd1 | BAS34286 | [2Fe-2S] | This study |
| KpFd2 | BAS37351 | [2Fe-2S] | This study |
| KpFd3 | BAS33241 | [2Fe-2S] | This study |
| KpFd4 | BAS35880 | CXXCXXCC | This study |
| MvFd1 | ACX73019 | 2[4Fe-4S] | This study |
| MvFd2 | ACX72493 | 2[4Fe-4S] | This study |
| MvFd3 | ACX73480 | 2[4Fe-4S] | This study |
| MvFd4 | ACX72328 | 2[4Fe-4S] | This study |
| MvFd5 | ACX73314 | 2[4Fe-4S] | This study |
| MvFd6 | ACX73502 | 2[4Fe-4S] | This study |
| MvFd7 | ACX72560 | 2[4Fe-4S] | This study |
| MvFd8 | ACX72349 | 2[4Fe-4S] | This study |
| MmFd1 | CAF29654 | 2[4Fe-4S] | This study |
| MbFd1 | AKB59153 | 2[4Fe-4S] | This study |
| MbFd2 | AKB57181 | 2[4Fe-4S] | This study |
| MbFd3 | AKB57361 | 2[4Fe-4S] | This study |
| MbFd4 | AKB58903 | 2[4Fe-4S] | This study |
| MbFd5 | AKB59009 | 2[4Fe-4S] | This study |
| FdxC | ADE87009 | [2Fe-2S] | 97, 133 |
| FdxD | ADE84338 | [2Fe-2S] | 80 |
| FdxE | ADE86134 | [2Fe-2S] | 93 |
| FdxB | ABJ08445 | 2[4Fe-4S] | 81 |
| Fer1 | ADU46354 | [4Fe-4S] | 134 |
| FerN | ACF03599 | [4Fe-4S] | 134 |
| RpFd1 | ABJ04561 | 2[4Fe-4S] | This study |
The protein annotation, accession number for a representative protein homolog, inferred type of [FeS] cluster in a given protein homolog (as predicted via the conserved domain database [120]), and a literature reference, if available, are provided for each Fd identified. Av, A. vinelandii; Cs, Caldicellulosiruptor saccharolyticus; Cp, Clostridium pasteurianum; Kp, Klebsiella pneumoniae; Mv, Methanocaldococcus vulcanius; Mm, Methanococcus maripaludis; Mb, Methanosarcina barkeri; Rp, Rhodopseudomonas palustris.
TABLE 2.
Flavodoxin homologs identified in the genomes of putative diazotrophs
| Annotation | Accession no. | Reference or source |
|---|---|---|
| NifF | ACO76434 | 36 |
| CpFld1 | AJA46461 | This study |
| CpFld2 | AJA47463 | This study |
| CpFld3 | AJA47660 | This study |
| FldA | WP_011157672 | 93 |
The protein annotation, accession number for a representative protein homolog, and a literature reference, if available, is provided for each Fld identified. Cp, Clostridium pasteurianum.
Putative enzymes that have been implicated in providing reductant to nitrogenase in the form of reduced Fd and/or Fld include the following: (i) pyruvate-flavodoxin oxidoreductase (PFOR) (35–38), (ii) ferredoxin-NADP+ oxidoreductase (FNR) (35, 62), (iii) Rhodobacter nitrogen fixation protein (Rnf) (81), and (iv) electron transfer flavoprotein (Fix) (27, 53, 54). Homologs of these enzymes were compiled from genomes of putative diazotrophs using BLASTp. The amino acid sequences of PFOR from Klebsiella pneumoniae (WP_064371580) and Rhodopseudomonas palustris (CAE30161), FNR from Anabaena azollae (WP_013193003) and Chlorobium tepidum strain TLS (AAM72739), Rnf from Azotobacter vinelandii (ACO81179 to ACO81185), and Fix from Rhodopseudomonas palustris (WP_011665892 to WP_011665895) were used as query sequences. In addition, [FeFe]-hydrogenase has been suggested to provide reductant for N2 fixation in Clostridium pasteurianum (39). Thus, the catalytic subunit of [FeFe] hydrogenase (HydA) from Chlamydomonas reinhardtii (AAL23572) was used as the query in a BLASTp search. Finally, group 3c, 3d, 4d, and 4e [NiFe]-hydrogenases catalyze the reversible reduction of Fd with H2 (41, 42, 102). A group 3c representative ([NiFe]-hydrogenase from Methanothermobacter marburgensis [WP_013296467]) and a group 4 representative ([NiFe]-hydrogenase from Pyrococcus abyssi [WP_010867842.1]) were used as BLASTp queries to identify all group 3 and 4 [NiFe]-hydrogenase large-subunit sequences. Compiled sequences were aligned as described above and manually curated to include only those enzymes that had proximal and distal Cys-Cys pairs, the ligands for the [NiFe] active site (121, 122). Extracted sequences homologous to [NiFe] were then subjected to phylogenetic reconstruction, as described above, and assigned to their respective groups based on phylogenetic coherence, as we have described previously (121, 122). Only [NiFe]-hydrogenase homologs belonging to groups 3c, 3d, 4d, and 4e that have been proposed to be capable of reducing Fd were considered in this study (40–44).
Identifying cooccurrence patterns in the distribution of Fds/Flds and putative Fd/Fld-reducing enzymes.
A binary table was created based on the presence or absence of identified Fds, Flds, and Fd- or Fld-reducing enzymes in diazotrophic genomes. To identify the frequent patterns in this data set, the Apriori algorithm was used as implemented with the arules package in R (123, 124). For simplicity, only the Fds, Flds, and Fd- or Fld-reducing enzymes present in >20% of the genomes were considered for this analysis. We applied the Apriori algorithm to our binary database while specifying the following parameters: a confidence threshold of 0.60 and a support threshold of 0.2. The confidence threshold is the measure of the statistical significance of an identified pattern within a data set, while the support threshold is indicative of the relative abundance of a given pattern in a data set. The Apriori algorithm works in two phases, with the first phase consisting of a scan of the entire database to objectively identify and extract frequently observed proteins. In the second phase, the algorithm uses those frequently occurring proteins to identify the dominant patterns in the distribution of proteins or combinations of those proteins. In doing so, the algorithm identifies correlations (within the user-defined confidence and support thresholds) between and among proteins in the database.
Oxidation state of carbon.
The oxidation state of carbon for each amino acid in each protein sequence encoded in 359 putative diazotrophic genomes was calculated following the algorithm mentioned in reference 112 using a custom python script (version 2.7.12). The calculated oxidation number for each amino acid was then normalized to the total number of amino acids encoded in a genome to determine the average oxidation state of the proteome. This number was determined for each genome that encoded a specific Nif isoform. These numbers were summed and then divided by the total number of genomes to arrive at the average oxidation state of carbon in a proteome inferred for a genome that encodes a given nitrogenase lineage. Welch's t test was conducted to determine the statistical significance of differences between the average oxidation states of carbon in proteomes from different lineages using the stats package in R (125). Using R, box plots were produced to show the distribution of the oxidation state of carbon in proteomes inferred for genomes that encoded specific isoforms of nitrogenases (125).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported as part of the Biological Electron Transfer and Catalysis Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, and Basic Energy Sciences under award number DE-SC0012518.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00757-17.
REFERENCES
- 1.Raymond J, Siefert JL, Staples CR, Blankenship RE. 2004. The natural history of nitrogen fixation. Mol Biol Evol 21:541–554. doi: 10.1093/molbev/msh047. [DOI] [PubMed] [Google Scholar]
- 2.Vitousek PM, Menge DNL, Reed SC, Cleveland CC. 2013. Biological nitrogen fixation: rates, patterns and ecological controls in terrestrial ecosystems. Philos Trans R Soc B Biol Sci 368:20130119. doi: 10.1098/rstb.2013.0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burk D, Lineweaver H, Horner CK. 1934. The specific influence of acidity on the mechanism of nitrogen fixation by Azotobacter. J Bacteriol 27:325–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falkowski PG. 1997. Evolution of the nitrogen cycle and its influence on the biological sequestration of CO2 in the ocean. Nature 387:272–275. doi: 10.1038/387272a0. [DOI] [Google Scholar]
- 5.Bulen W, LeComte J. 1966. The nitrogenase system from Azotobacter: two-enzyme requirement for N2 reduction, ATP-dependent H2 evolution, and ATP hydrolysis. Proc Natl Acad Sci U S A 56:979–986. doi: 10.1073/pnas.56.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters JW, Boyd ES, Hamilton T, Rubio LM (ed). 2011. Biochemistry of Mo-nitrogenase. Caister Academic Press, Poole, United Kingdom. [Google Scholar]
- 7.Boyd ES, Anbar AD, Miller S, Hamilton TL, Lavin M, Peters JW. 2011. A late methanogen origin for molybdenum-dependent nitrogenase. Geobiology 9:221–232. doi: 10.1111/j.1472-4669.2011.00278.x. [DOI] [PubMed] [Google Scholar]
- 8.Dos Santos PC, Fang Z, Mason SW, Setubal JC, Dixon R. 2012. Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics 13:162. doi: 10.1186/1471-2164-13-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGlynn SE, Boyd ES, Peters JW, Orphan VJ. 2013. Classifying the metal dependence of uncharacterized nitrogenases. Front Microbiol 3:419. doi: 10.3389/fmicb.2012.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leigh JA. 2000. Nitrogen fixation in methanogens: the archaeal perspective. Curr Issues Mol Biol 2:125–131. [PubMed] [Google Scholar]
- 11.Mehta MP, Baross JA. 2006. Nitrogen fixation at 92 degrees C by a hydrothermal vent archaeon. Science 314:1783–1786. doi: 10.1126/science.1134772. [DOI] [PubMed] [Google Scholar]
- 12.Nishizawa M, Miyazaki J, Makabe A, Koba K, Takai K. 2014. Physiological and isotopic characteristics of nitrogen fixation by hyperthermophilic methanogens: key insights into nitrogen anabolism of the microbial communities in archean hydrothermal systems. Geochim Cosmochim Acta 138:117–135. doi: 10.1016/j.gca.2014.04.021. [DOI] [Google Scholar]
- 13.Boyd ES, Hamilton TL, Peters JW. 2011. An alternative path for the evolution of biological nitrogen fixation. Front Microbiol 2:205. doi: 10.3389/fmicb.2011.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joerger RD, Bishop PE, Bishop PE. 1988. Bacterial alternative nitrogen fixation systems. Crit Rev Microbiol 16:1–14. doi: 10.3109/10408418809104465. [DOI] [PubMed] [Google Scholar]
- 15.Rubio LM, Ludden PW. 2008. Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu Rev Microbiol 62:93–111. doi: 10.1146/annurev.micro.62.081307.162737. [DOI] [PubMed] [Google Scholar]
- 16.Boyd ES, Peters JW. 2013. New insights into the evolutionary history of biological nitrogen fixation. Front Microbiol 4:201. doi: 10.3389/fmicb.2013.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyd ES, Costas AMG, Hamilton TL, Mus F, Peters JW. 2015. Evolution of molybdenum nitrogenase during the transition from anaerobic to aerobic metabolism. J Bacteriol 197:1690–1699. doi: 10.1128/JB.02611-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stal LJ, Krumbein WE. 1985. Nitrogenase activity in the non-heterocystous cyanobacterium Oscillatoria sp. grown under alternating light-dark cycles. Arch Microbiol 143:67–71. doi: 10.1007/BF00414770. [DOI] [Google Scholar]
- 19.Fay P. 1992. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol Rev 56:340–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poole RK, Hill S. 1997. Respiratory protection of nitrogenase activity in Azotobacter vinelandii: roles of the terminal oxidases. Biosci Rep 17:303–317. doi: 10.1023/A:1027336712748. [DOI] [PubMed] [Google Scholar]
- 21.Egener T, Martin DE, Sarkar A, Reinhold-Hurek B. 2001. Role of a ferredoxin gene cotranscribed with the nifHDK operon in N2 fixation and nitrogenase “switch-off” of Azoarcus sp. strain BH72. J Bacteriol 183:3752–3760. doi: 10.1128/JB.183.12.3752-3760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandmann G, Peleato ML, Fillat MF, Lazaro MC, Gomezmoreno C. 1990. Consequences of the iron-dependent formation of ferredoxin and flavodoxin on photosynthesis and nitrogen fixation on Anabaena strains. Photosynth Res 26:119–125. doi: 10.1007/BF00047083. [DOI] [PubMed] [Google Scholar]
- 23.Yakunin AF, Gennaro G, Hallenbeck PC. 1993. Purification and properties of a nif-specific flavodoxin from the photosynthetic bacterium Rhodobacter capsulatus. J Bacteriol 175:6775–6780. doi: 10.1128/jb.175.21.6775-6780.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin AE, Burgess BK, Iismaa SE, Smartt CT, Jacobson MR, Dean DR. 1989. Construction and characterization of an Azotobacter vinelandii strain with mutations in the genes encoding flavodoxin and ferredoxin I. J Bacteriol 171:3162–3167. doi: 10.1128/jb.171.6.3162-3167.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z-Y, Ledbetter R, Shaw S, Pence N, Tokmina-Lukaszewska M, Eilers B, Guo Q, Pokhrel N, Cash VL, Dean DR. 2016. Evidence that the Pi release event is the rate-limiting step in the nitrogenase catalytic cycle. Biochemistry 55:3625–3635. doi: 10.1021/acs.biochem.6b00421. [DOI] [PubMed] [Google Scholar]
- 26.Gennaro G, Hubner P, Sandmeier U, Yakunin AF, Hallenbeck PC. 1996. Cloning, characterization, and regulation of nifF from Rhodobacter capsulatus. J Bacteriol 178:3949–3952. doi: 10.1128/jb.178.13.3949-3952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ledbetter RN, Garcia Costas AM, Lubner CE, Mulder DW, Tokmina-Lukaszewska M, Artz JH, Patterson A, Magnuson TS, Jay ZJ, Duan HD, Miller J, Plunkett MH, Hoben JP, Barney BM, Carlson RP, Miller AF, Bothner B, King PW, Peters JW, Seefeldt LC. 2017. The electron bifurcating FixABCX protein complex from Azotobacter vinelandii: generation of low-potential reducing equivalents for nitrogenase catalysis. Biochemistry 56:4177–4190. doi: 10.1021/acs.biochem.7b00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simpson FB, Burris RH. 1984. A nitrogen pressure of 50 atmospheres does not prevent evolution of hydrogen by nitrogenase. Science 224:1095–1097. doi: 10.1126/science.6585956. [DOI] [PubMed] [Google Scholar]
- 29.Tso M-YW, Ljones T, Burris R. 1972. Purification of the nitrogenase proteins from Clostridium pasteurianum. Biochim Biophys Acta 267:600–604. doi: 10.1016/0005-2728(72)90193-4. [DOI] [PubMed] [Google Scholar]
- 30.Seefeldt LC, Hoffman BM, Dean DR. 2009. Mechanism of Mo-dependent nitrogenase. Annu Rev Biochem 78:701–722. doi: 10.1146/annurev.biochem.78.070907.103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bergström J, Eady RR, Thorneley RN. 1988. The vanadium- and molybdenum-containing nitrogenases of Azotobacter chroococcum. Comparison of mid-point potentials and kinetics of reduction by sodium dithionite of the iron proteins with bound magnesium adenosine 5′-diphosphate. Biochem J 251:165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dilworth MJ, Eldridge ME, Eady RR. 1992. Correction for creatine interference with the direct indophenol measurement of NH3 in steady-state nitrogenase assays. Anal Biochem 207:6–10. doi: 10.1016/0003-2697(92)90491-O. [DOI] [PubMed] [Google Scholar]
- 33.Dilworth MJ, Eldridge ME, Eady RR. 1993. The molybdenum and vanadium nitrogenases of Azotobacter chroococcum: effect of elevated temperature on N2 reduction. Biochem J 289:395–400. doi: 10.1042/bj2890395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eady RR. 1996. Structure-function relationships of alternative nitrogenases. Chem Rev 96:3013–3030. doi: 10.1021/cr950057h. [DOI] [PubMed] [Google Scholar]
- 35.Saeki K. (ed). 2004. Electron transport to nitrogenase: diverse routes for a common destination. Kluwer, Dordrecht, the Netherlands. [Google Scholar]
- 36.Nieva-Gomez D, Roberts GP, Klevickis S, Brill WJ. 1980. Electron transport to nitrogenase in Klebsiella pneumoniae. Proc Natl Acad Sci U S A 77:2555–2558. doi: 10.1073/pnas.77.5.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wahl RC, Orme-Johnson WH. 1987. Clostridial pyruvate oxidoreductase and the pyruvate-oxidizing enzyme specific to nitrogen fixation in Klebsiella pneumoniae are similar enzymes. J Biol Chem 262:10489–10496. [PubMed] [Google Scholar]
- 38.Yakunin AF, Hallenbeck PC. 1998. Purification and characterization of pyruvate oxidoreductase from the photosynthetic bacterium Rhodobacter capsulatus. Biochim Biophys Acta 1409:39–49. doi: 10.1016/S0005-2728(98)00145-5. [DOI] [PubMed] [Google Scholar]
- 39.Therien J, Artz JH, Poudel S, Hamilton TL, Liu Z, Noone S, Adams M, King PW, Bryant DA, Boyd ES, Peters JW. 2017. The physiological functions and structural determinants of catalytic bias in the [FeFe]-hydrogenases CpI and CpII of Clostridium pasteurianum strain W5. Front Microbiol 8:1305. doi: 10.3389/fmicb.2017.01305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bothe H, Schmitz O, Yates MG, Newton WE. 2010. Nitrogen fixation and hydrogen metabolism in cyanobacteria. Microbiol Mol Biol Rev 74:529–551. doi: 10.1128/MMBR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB, Cook GM, Morales SE. 2016. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J 10:761. doi: 10.1038/ismej.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gutekunst K, Chen X, Schreiber K, Kaspar U, Makam S, Appel J. 2014. The bidirectional NiFe-hydrogenase in Synechocystis sp PCC 6803 is reduced by flavodoxin and ferredoxin and is essential under mixotrophic, nitrate-limiting conditions. J Biol Chem 289:1930–1937. doi: 10.1074/jbc.M113.526376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khanna N, Lindblad P. 2015. Cyanobacterial hydrogenases and hydrogen metabolism revisited: recent progress and future prospects. Int J Mol Sci 16:10537–10561. doi: 10.3390/ijms160510537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitz O, Boison G, Hilscher R, Hundeshagen B, Zimmer W, Lottspeich F, Bothe H. 1995. Molecular biological analysis of a bidirectional hydrogenase from cyanobacteria. Eur J Biochem 233:266–276. doi: 10.1111/j.1432-1033.1995.266_1.x. [DOI] [PubMed] [Google Scholar]
- 45.Rey FE, Oda Y, Harwood CS. 2006. Regulation of uptake hydrogenase and effects of hydrogen utilization on gene expression in Rhodopseudomonas palustris. J Bacteriol 188:6143–6152. doi: 10.1128/JB.00381-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herrmann G, Jayamani E, Mai G, Buckel W. 2008. Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J Bacteriol 190:784–791. doi: 10.1128/JB.01422-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anraku Y. 1988. Bacterial electron-transport chains. Annu Rev Biochem 57:101–132. doi: 10.1146/annurev.bi.57.070188.000533. [DOI] [PubMed] [Google Scholar]
- 48.Watt G, Burns A, Lough S, Tennent D. 1980. Redox and spectroscopic properties of oxidized MoFe protein from Azotobacter vinelandii. Biochemistry 19:4926–4932. doi: 10.1021/bi00562a035. [DOI] [PubMed] [Google Scholar]
- 49.Lanzilotta WN, Ryle MJ, Seefeldt LC. 1995. Nucleotide hydrolysis and protein conformational changes in Azotobacter vinelandii nitrogenase iron protein: defining the function of aspartate 129. Biochemistry 34:10713–10723. doi: 10.1021/bi00034a003. [DOI] [PubMed] [Google Scholar]
- 50.Braaksma A, Haaker H, Grande HJ, Veeger C. 1982. The effect of the redox potential on the activity of the nitrogenase and on the Fe-protein of Azotobacter vinelandi. FEBS J 121:483–491. [DOI] [PubMed] [Google Scholar]
- 51.Dutton PL. 1986. Energy transduction in anoxygenic photosynthesis, p 197–237. In Staehelin A, Arntzen CJ (ed), Photosynthesis III. Photosynthetic membranes and light harvesting systems. Encyclopedia of Plant Physiology (new series), vol 19 Springer, Berlin, Germany. doi: 10.1007/978-3-642-70936-4_5. [DOI] [Google Scholar]
- 52.Jagannathan B, Golbeck J (ed). 2009. Photosynthesis: microbial, p 325–341. In Schaechter M (ed), Encyclopedia of microbiology, 3rd ed Academic press, Cambridge, MA. doi: 10.1016/B978-012373944-5.00352-7. [DOI] [Google Scholar]
- 53.Earl C, Ronson C, Ausubel F. 1987. Genetic and structural analysis of the Rhizobium meliloti fixA, fixB, fixC, and fixX genes. J Bacteriol 169:1127–1136. doi: 10.1128/jb.169.3.1127-1136.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edgren T, Nordlund S. 2004. The fixABCX genes in Rhodospirillum rubrum encode a putative membrane complex participating in electron transfer to nitrogenase. J Bacteriol 186:2052–2060. doi: 10.1128/JB.186.7.2052-2060.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Biegel E, Schmidt S, Gonzalez JM, Muller V. 2011. Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell Mol Life Sci 68:613–634. doi: 10.1007/s00018-010-0555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hess V, Schuchmann K, Muller V. 2013. The ferredoxin: NAD(+) oxidoreductase (Rnf) from the acetogen Acetobacterium woodii requires Na+ and is reversibly coupled to the membrane potential. J Biol Chem 288:31496–31502. doi: 10.1074/jbc.M113.510255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jouanneau Y, Jeong HS, Hugo N, Meyer C, Willison JC. 1998. Overexpression in Escherichia coli of the rnf genes from Rhodobacter capsulatus: characterization of two membrane-bound iron-sulfur proteins. Eur J Biochem 251:54–64. doi: 10.1046/j.1432-1327.1998.2510054.x. [DOI] [PubMed] [Google Scholar]
- 58.Sarkar A, Köhler J, Hurek T, Reinhold-Hurek B. 2012. A novel regulatory role of the Rnf complex of Azoarcus sp. strain BH72. Mol Microbiol 83:408–422. doi: 10.1111/j.1365-2958.2011.07940.x. [DOI] [PubMed] [Google Scholar]
- 59.Schrautemeier B, Böhme H. 1985. A distinct ferredoxin for nitrogen fixation isolated from heterocysts of the cyanobacterium Anabaena variabilis. FEBS Lett 184:304–308. doi: 10.1016/0014-5793(85)80627-X. [DOI] [Google Scholar]
- 60.Apte S, Rowell P, Stewart W. 1978. Electron donation to ferredoxin in heterocysts of the N2-fixing alga Anabaena cylindrica. Proc R Soc B Biol Sci 200:1–25. doi: 10.1098/rspb.1978.0001. [DOI] [Google Scholar]
- 61.Aliverti A, Faber R, Finnerty CM, Ferioli C, Pandini V, Negri A, Karplus PA, Zanetti G. 2001. Biochemical and crystallographic characterization of ferredoxin-NADP(+) reductase from nonphotosynthetic tissues. Biochemistry 40:14501–14508. doi: 10.1021/bi011224c. [DOI] [PubMed] [Google Scholar]
- 62.Isas JM, Yannone SM, Burgess BK. 1995. Azotobacter vinelandii NADPH-rerredoxin reductase cloning, sequencing, and overexpression. J Biol Chem 270:21258–21263. doi: 10.1074/jbc.270.36.21258. [DOI] [PubMed] [Google Scholar]
- 63.Sattley WM, Madigan MT, Swingley WD, Cheung PC, Clocksin KM, Conrad AL, Dejesa LC, Honchak BM, Jung DO, Karbach LE, Kurdoglu A, Lahiri S, Mastrian SD, Page LE, Taylor HL, Wang ZT, Raymond J, Chen M, Blankenship RE, Touchman JW. 2008. The genome of Heliobacterium modesticaldum, a phototrophic representative of the Firmicutes containing the simplest photosynthetic apparatus. J Bacteriol 190:4687–4696. doi: 10.1128/JB.00299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seo D, Sakurai H. 2002. Purification and characterization of ferredoxin-NAD(P)(+) reductase from the green sulfur bacterium Chlorobium tepidum. Biochim Biophys Acta 1597:123–132. doi: 10.1016/S0167-4838(02)00269-8. [DOI] [PubMed] [Google Scholar]
- 65.Muraki N, Seo D, Shiba T, Sakurai T, Kurisu G. 2010. Asymmetric dimeric structure of ferredoxin-NAD(P)+ oxidoreductase from the green sulfur bacterium Chlorobaculum tepidum: implications for binding ferredoxin and NADP+. J Mol Biol 401:403–414. doi: 10.1016/j.jmb.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 66.Petering D, Fee JA, Palmer G. 1971. The oxygen sensitivity of spinach ferredoxin and other iron-sulfur proteins. The formation of protein-bound sulfur-zero. J Biol Chem 246:643–653. [PubMed] [Google Scholar]
- 67.Knight E, Hardy R. 1966. Isolation and characteristics of flavodoxin from nitrogen-fixing Clostridium pasteurianum. J Biol Chem 241:2752–2756. [PubMed] [Google Scholar]
- 68.Lovenberg W, Buchanan BB, Rabinowitz JC. 1963. Studies on the chemical nature of clostridial ferredoxin. J Biol Chem 238:3899–3913. [PubMed] [Google Scholar]
- 69.Imlay JA. 2006. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol 59:1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- 70.Fillat MF, Sandmann G, Gomez-Moreno C. 1988. Flavodoxin from the nitrogen-fixing cyanobacterium Anabaena PCC 7119. Arch Microbiol 150:160–164. doi: 10.1007/BF00425156. [DOI] [Google Scholar]
- 71.Lodeyro AF, Ceccoli RD, Karlusich JJP, Carrillo N. 2012. The importance of flavodoxin for environmental stress tolerance in photosynthetic microorganisms and transgenic plants. Mechanism, evolution and biotechnological potential. FEBS Lett 586:2917–2924. doi: 10.1016/j.febslet.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 72.Bennett LT, Jacobson MR, Dean DR. 1988. Isolation, sequencing, and mutagenesis of the nifF gene encoding flavodoxin from Azotobacter vinelandii. J Biol Chem 263:1364–1369. [PubMed] [Google Scholar]
- 73.Hill S, Kavanagh EP. 1980. Roles of nifF and nifJ gene products in electron transport to nitrogenase in Klebsiella pneumoniae. J Bacteriol 141:470–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shah VK, Stacey G, Brill WJ. 1983. Electron transport to nitrogenase: purification and characterization of pyruvate, flavodoxin oxidoreductase, the nifJ-gene product. J Biol Chem 258:2064–2068. [PubMed] [Google Scholar]
- 75.Yoch DC, Arnon DI. 1972. Two biologically active ferredoxins from the aerobic nitrogen-fixing bacterium, Azotobacter vinelandii. J Biol Chem 247:4514–4520. [PubMed] [Google Scholar]
- 76.Jouanneau Y, Meyer C, Naud I, Klipp W. 1995. Characterization of an fdxN mutant of Rhodobacter capsulatus indicates that ferredoxin-I serves as electron donor to nitrogenase. Biochim Biophys Acta 1232:33–42. doi: 10.1016/0005-2728(95)00106-X. [DOI] [PubMed] [Google Scholar]
- 77.McRose DL, Zhang XN, Kraepiel AML, Morel FMM. 2017. Diversity and activity of alternative nitrogenases in sequenced genomes and coastal environments. Front Microbiol 8:267. doi: 10.3389/fmicb.2017.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dekas AE, Poretsky RS, Orphan VJ. 2009. Deep-sea Archaea fix and share nitrogen in methane-consuming microbial consortia. Science 326:422–426. doi: 10.1126/science.1178223. [DOI] [PubMed] [Google Scholar]
- 79.Bolhuis H, Severin I, Confurius-Guns V, Wollenzien UIA, Stal LJ. 2010. Horizontal transfer of the nitrogen fixation gene cluster in the cyanobacterium Microcoleus chthonoplastes. ISME J 4:121–130. doi: 10.1038/ismej.2009.99. [DOI] [PubMed] [Google Scholar]
- 80.Armengaud J, Meyer C, Jouanneau Y. 1994. Recombinant expression of the fdxD gene of Rhodobacter capsulatus and characterization of its product, a [2Fe-2S] ferredoxin. Biochem J 300:413–418. doi: 10.1042/bj3000413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schmehl M, Jahn A, Vilsendorf AMZ, Hennecke S, Masepohl B, Schuppler M, Marxer M, Oelze J, Klipp W. 1993. Identification of a new class of nitrogen-fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol Gen Genet 241:602–615. doi: 10.1007/BF00279903. [DOI] [PubMed] [Google Scholar]
- 82.Hamilton TL, Ludwig M, Dixon R, Boyd ES, Dos Santos PC, Setubal JC, Bryant DA, Dean DR, Peters JW. 2011. Transcriptional profiling of nitrogen fixation in Azotobacter vinelandii. J Bacteriol 193:4477–4486. doi: 10.1128/JB.05099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jouanneau Y, Meyer C, Gaillard J, Forest E, Gagnon J. 1993. Purification and characterization of a novel dimeric ferredoxin (FdIII) from Rhodobacter capsulatus. J Biol Chem 268:10636–10644. [PubMed] [Google Scholar]
- 84.Moreno-Vivian C, Hennecke S, Pühler A, Klipp W. 1989. Open reading frame 5 (ORF5), encoding a ferredoxin like protein, and nifQ are cotranscribed with nifE, nifN, nifX, and ORF4 in Rhodobacter capsulatus. J Bacteriol 171:2591–2598. doi: 10.1128/jb.171.5.2591-2598.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Masepohl B, Schölisch K, Görlitz K, Kutzki C, Böhme H. 1997. The heterocyst-specific fdxH gene product of the cyanobacterium Anabaena sp. PCC 7120 is important but not essential for nitrogen fixation. Mol Gen Genet 253:770–776. [DOI] [PubMed] [Google Scholar]
- 86.Souza ALF, Invitti AL, Rego FGM, Monteiro RA, Klassen G, Souza EM, Chubatsu LS, Pedrosa FO, Rigo LU. 2010. The involvement of the nif-associated ferredoxin-like genes fdxA and fdxN of Herbaspirillum seropedicae in nitrogen fixation. J Microbiol 48:77–83. doi: 10.1007/s12275-009-0077-y. [DOI] [PubMed] [Google Scholar]
- 87.Hageman RV, Burris RH. 1978. Nitrogenase and nitrogenase reductase associate and dissociate with each catalytic cycle. Proc Natl Acad Sci U S A 75:2699–2702. doi: 10.1073/pnas.75.6.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Malkin R, Aparicio PJ, Arnon DI. 1974. The isolation and characterization of a new iron-sulfur protein from photosynthetic membranes. Proc Natl Acad Sci U S A 71:2362–2366. doi: 10.1073/pnas.71.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hallenbeck PC, Gennaro G. 1998. Stopped-flow kinetic studies of low potential electron carriers of the photosynthetic bacterium, Rhodobacter capsulatus: ferredoxin I and NifF. Biochim Biophys Acta 1365:435–442. doi: 10.1016/S0005-2728(98)00096-6. [DOI] [PubMed] [Google Scholar]
- 90.Hallenbeck PC, Jouanneau Y, Vignais PM. 1982. Purification and molecular properties of a soluble ferredoxin from Rhodopseudomonas capsulata. Biochim Biophys Acta 681:168–176. doi: 10.1016/0005-2728(82)90020-2. [DOI] [Google Scholar]
- 91.Edgren T, Nordlund S. 2005. Electron transport to nitrogenase in Rhodospirillum rubrum: identification of a new fdxN gene encoding the primary electron donor to nitrogenase. FEMS Microbiol Lett 245:345–351. doi: 10.1016/j.femsle.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 92.Klipp W, Reilander H, Schluter A, Krey R, Puhler A. 1989. The Rhizobium meliloti fdxN gene encoding a ferredoxin-like protein is necessary for nitrogen fixation and is cotranscribed with nifA and nifB. Mol Gen Genet 216:293–302. doi: 10.1007/BF00334368. [DOI] [PubMed] [Google Scholar]
- 93.Armengaud J, Meyer C, Jouanneau Y. 1997. A [2Fe-2S] ferredoxin (FdVI) is essential for growth of the photosynthetic bacterium Rhodobacter capsulatus. J Bacteriol 179:3304–3309. doi: 10.1128/jb.179.10.3304-3309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schrautemeier B, Cassing A, Bohme H. 1994. Characterization of the genome region encoding an FdxH-type ferredoxin and a new 2[4fe-4s] ferredoxin from the nonheterocystous, nitrogen-fixing cyanobacterium Plectonema boryanum PCC-73110. J Bacteriol 176:1037–1046. doi: 10.1128/jb.176.4.1037-1046.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ermakova M, Battchikova N, Richaud P, Leino H, Kosourov S, Isojarvi J, Peltier G, Flores E, Cournac L, Allahverdiyeva Y, Aro EM. 2014. Heterocyst-specific flavodiiron protein Flv3B enables oxic diazotrophic growth of the filamentous cyanobacterium Anabaena sp. PCC 7120. Proc Natl Acad Sci U S A 111:11205–11210. doi: 10.1073/pnas.1407327111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peden EA, Boehm M, Mulder DW, Davis R, Old WM, King PW, Ghirardi ML, Dubini A. 2013. Identification of global ferredoxin interaction networks in Chlamydomonas reinhardtii. J Biol Chem 288:35192–35209. doi: 10.1074/jbc.M113.483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Grabau C, Schatt E, Jouanneau Y, Vignais PM. 1991. A new [2Fe-2S] ferredoxin from Rhodobacter capsulatus. Coexpression with a 2 [4Fe-4S] ferredoxin in Escherichia coli. J Biol Chem 266:3294–3299. [PubMed] [Google Scholar]
- 98.Deistung J, Cannon FC, Cannon MC, Hill S, Thorneley RNF. 1985. Electron transfer to nitrogenase in Klebsiella pneumoniae: nifF gene cloned and the gene product, a flavodoxin, purified. Biochem J 231:743–753. doi: 10.1042/bj2310743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Drummond MH. 1985. The base sequence of the nifF gene of Klebsiella pneumoniae and homology of the predicted amino acid sequence of its protein product to other flavodoxins. Biochem J 232:891–896. doi: 10.1042/bj2320891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Roberts GP, MacNeil T, MacNeil D, Brill WJ. 1978. Regulation and characterization of protein products coded by the nif (nitrogen fixation) genes of Klebsiella pneumoniae. J Bacteriol 136:267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Arnold W, Rump A, Klipp W, Priefer UB, Puhler A. 1988. Nucleotide sequence of a 24,206-base-pair DNA fragment carrying the entire nitrogen fixation gene cluster of Klebsiella pneumoniae. J Mol Biol 203:715–738. doi: 10.1016/0022-2836(88)90205-7. [DOI] [PubMed] [Google Scholar]
- 102.Peters JW, Schut GJ, Boyd ES, Mulder DW, Shepard EM, Broderick JB, King PW, Adams MWW. 2015. [FeFe]- and [NiFe]-hydrogenase diversity, mechanism, and maturation. Biochim Biophys Acta 1853:1350–1369. doi: 10.1016/j.bbamcr.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 103.Poudel S, Tokmina-Lukaszewska M, Colman DR, Refai M, Schut GJ, King PW, Maness PC, Adams MWW, Peters JW, Bothner B, Boyd ES. 2016. Unification of [FeFe]-hydrogenases into three structural and functional groups. Biochim Biophys Acta 1860:1910–1921. doi: 10.1016/j.bbagen.2016.05.034. [DOI] [PubMed] [Google Scholar]
- 104.Kaminski PA, Norel F, Desnoues N, Kush A, Salzano G, Elmerich C. 1988. Characterization of the fixABC region of Azorhizobium caulinodans ors571 and identification of a new nitrogen fixation gene. Mol Gen Genet 214:496–502. doi: 10.1007/BF00330486. [DOI] [PubMed] [Google Scholar]
- 105.Gubler M, Hennecke H. 1986. fixA, B and C genes are essential for symbiotic and free-living, microaerobic nitrogen fixation. FEBS Lett 200:186–192. doi: 10.1016/0014-5793(86)80536-1. [DOI] [Google Scholar]
- 106.Garcia Costas AM, Poudel S, Miller A-F, Schut GJ, Ledbetter RN, Fixen KR, Seefeldt LC, Adams MW, Harwood CS, Boyd ES. 2017. Defining electron bifurcation in the electron transferring flavoprotein family. J Bacteriol 199:e00440-17. doi: 10.1128/JB.00440-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Furdui C, Ragsdale SW. 2000. The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autotrophic growth by the Wood-Ljungdahl pathway. J Biol Chem 275:28494–28499. doi: 10.1074/jbc.M003291200. [DOI] [PubMed] [Google Scholar]
- 108.Neuer G, Bothe H. 1985. Electron donation to nitrogenase in heterocysts of cyanobacteria. Arch Microbiol 143:185–191. doi: 10.1007/BF00411045. [DOI] [Google Scholar]
- 109.Lubitz W, Ogata H, Rudiger O, Reijerse E. 2014. Hydrogenases. Chem Rev 114:4081–4148. doi: 10.1021/cr4005814. [DOI] [PubMed] [Google Scholar]
- 110.Volbeda A, Charon MH, Piras C, Hatchikian EC, Frey M, Fontecilla-Camps JC. 1995. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature 373:580–587. doi: 10.1038/373580a0. [DOI] [PubMed] [Google Scholar]
- 111.Shock EL, Boyd ES. 2015. Principles of geobiochemistry. Elements 11:395–401. doi: 10.2113/gselements.11.6.395. [DOI] [Google Scholar]
- 112.Dick JM, Shock EL. 2011. Calculation of the relative chemical stabilities of proteins as a function of temperature and redox chemistry in a hot spring. PLoS One 6:e22782. doi: 10.1371/journal.pone.0022782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dick JM. 2014. Average oxidation state of carbon in proteins. J R Soc Interface 11:20131095. doi: 10.1098/rsif.2013.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Singh BB, Curdt I, Jakobs C, Schomburg D, Bisen PS, Bohme H. 1999. Identification of amino acids responsible for the oxygen sensitivity of ferredoxins from Anabaena variabilis using site-directed mutagenesis. Biochim Biophys Acta 1412:288–294. doi: 10.1016/S0005-2728(99)00069-9. [DOI] [PubMed] [Google Scholar]
- 115.Jannasch HW, Mottl MJ. 1985. Geomicrobiology of deep-sea hydrothermal vents. Science 229:717–725. doi: 10.1126/science.229.4715.717. [DOI] [PubMed] [Google Scholar]
- 116.Anantharaman K, Breier JA, Sheik CS, Dick GJ. 2013. Evidence for hydrogen oxidation and metabolic plasticity in widespread deep-sea sulfur-oxidizing bacteria. Proc Natl Acad Sci U S A 110:330–335. doi: 10.1073/pnas.1215340110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zheng Y, Harris DF, Yu Z, Fu Y, Poudel S, Ledbetter RN, Fixen KR, Yang Z-Y, Boyd ES, Lidstrom ME, Seefeldt LC, Harwood CS. 2018. A pathway for biological methane production using bacterial iron-only nitrogenase. Nat Microbiol 3:281–186. doi: 10.1038/s41564-017-0091-5. [DOI] [PubMed] [Google Scholar]
- 118.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li WZ, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu SN, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang ZX, Yamashita RA, Zhang DC, Zheng CJ, Bryant SH. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Boyd ES, Schut GJ, Adams MWW, Peters JW. 2014. Hydrogen metabolism and the evolution of biological respiration. Microbe 9:361–367. [Google Scholar]
- 122.Schut GJ, Zadvornyy O, Wu C-H, Peters JW, Boyd ES, Adams MWW. 2016. The role of geochemistry and energetics in the evolution of modern respiratory complexes from a proton-reducing ancestor. Biochim Biophys Acta 1857:958–970. doi: 10.1016/j.bbabio.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 123.Hahsler M, Chelluboina S, Hornik K, Buchta C. 2011. The arules R-package ecosystem: analyzing interesting patterns from large transaction data sets. J Mach Learning Res 12:2021–2025. [Google Scholar]
- 124.Agrawal R, Srikant R. 1994. Fast algorithms for mining association rules, p 487–499. In Bocca JB, Jarke M, Zaniolo C (ed), Proceedings of the 20th International Conference on Very Large Data Bases. Morgan Kaufmann, Burlington, MA. [Google Scholar]
- 125.R Development Core Team. 2010. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 126.Kaneko T, Nakamura Y, Wolk CP, Kuritz T, Sasamoto S, Watanabe A, Iriguchi M, Ishikawa A, Kawashima K, Kimura T, Kishida Y, Kohara M, Matsumoto M, Matsuno A, Muraki A, Nakazaki N, Shimpo S, Sugimoto M, Takazawa M, Yamada M, Yasuda M, Tabata S. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res 8:205–213. doi: 10.1093/dnares/8.5.205. [DOI] [PubMed] [Google Scholar]
- 127.Jacobson BL, Chae YK, Böhme H, Markley JL, Holden HM. 1992. Crystallization and preliminary analysis of oxidized, recombinant, heterocyst [2Fe-2S] ferredoxin from Anabaena 7120. Arch Biochem Biophys 294:279–281. doi: 10.1016/0003-9861(92)90169-W. [DOI] [PubMed] [Google Scholar]
- 128.Rypniewski WR, Breiter DR, Benning MM, Wesenberg G, Oh BH, Markley JL, Rayment I, Holden HM. 1991. Crystallization and structure determination of 2.5 Å resolution of the oxidized [2Fe-2S] ferredoxin isolated from Anabaena 7120. Biochemistry 30:4126–4131. doi: 10.1021/bi00231a003. [DOI] [PubMed] [Google Scholar]
- 129.Jacobson M, Brigle K, Bennett L, Setterquist R, Wilson M, Cash V, Beynon J, Newton W, Dean D. 1989. Physical and genetic map of the major nif gene cluster from Azotobacter vinelandii. J Bacteriol 171:1017–1027. doi: 10.1128/jb.171.2.1017-1027.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Setubal JC, dos Santos P, Goldman BS, Ertesvag H, Espin G, Rubio LM, Valla S, Almeida NF, Balasubramanian D, Cromes L, Curatti L, Du ZJ, Godsy E, Goodner B, Hellner-Burris K, Hernandez JA, Houmiel K, Imperial J, Kennedy C, Larson TJ, Latreille P, Ligon LS, Lu J, Maerk M, Miller NM, Norton S, O'Carroll IP, Paulsen I, Raulfs EC, Roemer R, Rosser J, Segura D, Slater S, Stricklin SL, Studholme DJ, Sun J, Viana CJ, Wallin E, Wang BM, Wheeler C, Zhu HJ, Dean DR, Dixon R, Wood D. 2009. Genome sequence of Azotobacter vinelandii, an obligate aerobe specialized to support diverse anaerobic metabolic processes. J Bacteriol 191:4534–4545. doi: 10.1128/JB.00504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schatt E, Jouanneau Y, Vignais P. 1989. Molecular cloning and sequence analysis of the structural gene of ferredoxin I from the photosynthetic bacterium Rhodobacter capsulatus. J Bacteriol 171:6218–6226. doi: 10.1128/jb.171.11.6218-6226.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Reyntjens B, Jollie DR, Stephens PJ, Gao-Sheridan HS, Burgess BK. 1997. Purification and characterization of a fixABCX-linked 2[4Fe-4S] ferredoxin from Azotobacter vinelandii. J Biol Inorg Chem 2:595–602. doi: 10.1007/s007750050174. [DOI] [Google Scholar]
- 133.Saeki K, Suetsugu Y, Tokuda K-I, Miyatake Y, Young D, Marrs B, Matsubara H. 1991. Genetic analysis of functional differences among distinct ferredoxins in Rhodobacter capsulatus. J Biol Chem 266:12889–12895. [PubMed] [Google Scholar]
- 134.Larimer FW, Chain P, Hauser L, Lamerdin J, Malfatti S, Do L, Land ML, Pelletier DA, Beatty JT, Lang AS, Tabita FR, Gibson JL, Hanson TE, Bobst C, Torres JLTY, Peres C, Harrison FH, Gibson J, Harwood CS. 2004. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat Biotechnol 22:55–61. doi: 10.1038/nbt923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.