Abstract
The small subunit processome is the first precursor of the small eukaryotic ribosomal subunit. During its assembly in the nucleolus, many ribosome biogenesis factors, an RNA chaperone, and ribosomal proteins associate with the nascent pre-rRNA. Biochemical studies have elucidated the rRNA-subdomain dependent recruitment of these factors during SSU processome assembly and have been complemented by structural studies of the assembled particle. Ribosome biogenesis factors encapsulate and guide subdomains of pre-ribosomal RNA in distinct compartments. This prevents uncoordinated maturation and enables processing of regions not accessible in the mature subunit. By sequentially reducing conformational freedom, flexible proteins facilitate the incorporation of dynamic subcomplexes into a globular particle. Large rearrangements within the SSU processome are required for compaction into the mature small ribosomal subunit.
Graphical abstract
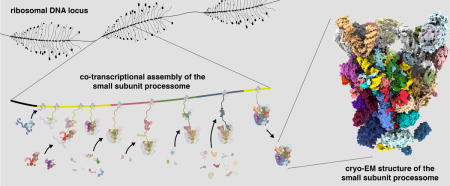
Introduction
The synthesis and maturation of eukaryotic ribosomal subunits is an intricate, multidimensional and energy-consuming process involving a large set of small nucleolar RNAs (snoRNAs), more than 200 conserved assembly factors and all three RNA polymerases [1]. RNA polymerase II generates mRNAs of ribosomal proteins and ribosome assembly factors whereas RNA polymerases I and III transcribe ribosomal RNA (rRNA) precursors from 100-200 rDNA loci. In yeast, RNA polymerase I synthesizes an initial 35S pre-rRNA transcript, which contains rRNA of the small subunit (SSU; 18S RNA) as well as the large subunit (LSU; 5.8S and 25S RNA) separated by several external and internal transcribed spacers (5′ ETS, ITS1, ITS2, 3′ ETS). Cleavage events within ITS1 (at sites A2 or A3) separate the precursors of the small and large ribosomal subunit. While co-transcriptional processing of the 35S pre-rRNA is associated with RNA cleavage at site A2, post-transcriptional processing occurs via RNA cleavage at site A3 [2]. In parallel, RNA polymerase III transcribes the 5S rRNA that later becomes part of the LSU.
Early visualizations of actively transcribed chromatin on Miller spreads by electron microscopy (EM) revealed the presence of large terminal knobs that are associated with nascent pre-ribosomal RNA [3]. The molecular identity of some of these particles was uncovered approximately three decades later, when a large U3-snoRNA-containing complex, the small subunit (SSU) processome, was identified [4]. A particle containing the entire 35S pre-rRNA was termed the 90S pre-ribosome [5].
A 23S pre-rRNA species (containing the 5′ ETS, 18S rRNA and ITS1 cut at site A3) has been associated with the SSU processome [6]. This intermediate was also found to accumulate as a result of mutations and depletions of small subunit ribosomal proteins [7], ribosome assembly factors [8] or components of the exosome [9]. More recently, the accumulation of the 23S pre-rRNA species was also observed in response to cellular stress, nutrient starvation or as a result of mTOR inhibition [10-12].
Here we discuss how advances in our biochemical understanding of many SSU processome factors combined with recent cryo-EM reconstructions have facilitated a mechanistic understanding of the assembly and structure of this pre-ribosomal particle.
Early events in small subunit assembly
Current assembly models of the SSU processome are based on the biochemical identification, purification and characterization of multi-protein complexes [13-16], protein depletion analyses [17,18] and studies in which aptamer-tagged pre-rRNA mimics were purified to analyze associated proteins [19,20].
SSU processome assembly proceeds in a chronological and co-transcriptional manner (Figure 1). The 5′ external transcribed spacer (5′ ETS), which precedes the 18S rRNA, enables the recruitment of several protein factors and the U3 snoRNP and leads to the formation of an early assembly intermediate termed the 5′ ETS particle [19].
Figure 1.
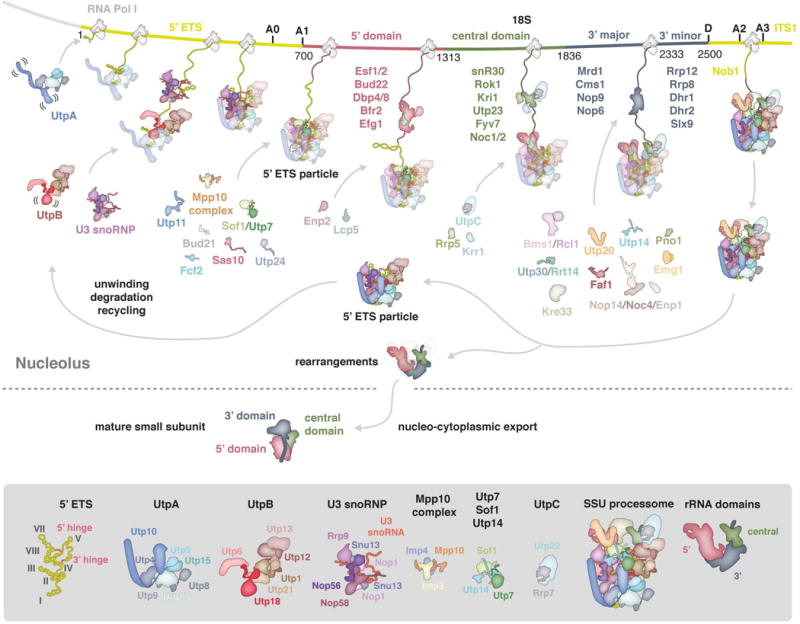
Co-transcriptional assembly of the small subunit processome. Schematic representation of early events of small subunit assembly in the nucleolus and subsequent maturation resulting in the mature cytoplasmic SSU. A section of the rDNA locus is shown with 5′ ETS and ITS1 colored in yellow and 18S colored in red (5′ domain), green (central domain) and slate (3′ domain). Factors visualized in the assembled SSU processome [30] are shown as schematic outline while transient components are listed and colored according to the rRNA domain they are associated with [19,20]. The SSU processome and its major components are shown in detail below.
Eukaryotic ribosome biogenesis is initiated by the seven-subunit UtpA complex (Utp4, Utp5, Utp8, Utp9, Utp10, Utp15 & Utp17). This initially flexible complex was shown to specifically interact with the first half of the 5′ ETS by in vivo pull-down assays as well as UV-crosslinking and analysis of cDNA (CRAC) [16,19,20]. UtpA binding and the presence of additional parts of the 5′ ETS appear to be required for the subsequent recruitment of the six-subunit UtpB complex (Utp1/Pwp2, Utp6, Utp12, Utp13, Utp18 & Utp21) and the U3 snoRNP (U3 snoRNA, Nop1/fibrillarin, Snu13, Nop56 & Nop58) (Figure 1) [14,17]. UtpB chaperones an RNA duplex formed by the 5′ ETS and U3 snoRNA and later parts of the 3′ domain of the 18S rRNA [16].
U3 snoRNA acts as a vital RNA chaperone during the ensuing assembly steps where it base-pairs with two regions of the 5′ ETS and two regions of the 18S rRNA [21-23]. This prevents the pre-mature formation of the central pseudoknot - a tertiary RNA structure that determines the relative positions of rRNA domains in the mature small subunit. U3 snoRNA dictates the topology of the growing particle by first base-pairing with the 5′ ETS through its 3′- and 5′ hinges and then with the 18S rRNA through its Box A (18S nucleotides 9-15) and Box A′ (18S nucleotides 1111-1122). As transcription continues, these base-pairing interactions presumably result in a reduction of conformational freedom of pre-ribosomal RNA, thereby directing RNA folding and the topology of the entire particle.
With the completion of the 5′ ETS, the Mpp10 complex (Imp3, Imp4 & Mpp10), individual assembly factors (Bud21, Fcf2, Utp7, Utp11, Sas10 & Sof1) and later the A1 site nuclease Utp24 [24,25], join UtpA, UtpB and U3 snoRNP to finalize the 5′ ETS particle (Figure 1). With the exception of Utp7, Sof1 and Utp24, these factors lack canonical protein folds and act as multi-functional binding partners, which may, similar to U3 snoRNA, reduce conformational freedom by interacting with multiple other proteins as the assembly of the small subunit processome continues. Conceptually, these peptides and U3 snoRNA could therefore allow an initially flexible array of subdomains to be stabilized in a co-transcriptional manner to result in a rigidified SSU processome.
The 5′ ETS particle is an architectural scaffold onto which individual subdomains of the small ribosomal subunit (5′, central, 3′ major and 3′ minor domains) can be assembled. The purification of affinity-tagged truncated rRNA mimics has shown that each domain is associated with a distinct set of ribosome assembly factors (Figure 1) [19,20]. However, many of these proteins and RNAs such as snR10 and snR30 [26,27], perform transient roles during the assembly of the SSU processome and are not part of the stable particles recently studied by cryo-EM [11,28-31]. Beyond the initial assembly events, flexible regions of Sas10, Utp18 and later Lcp5 contain exosome-interacting motifs, which may facilitate processing, degradation or recycling of SSU processomes [19,32].
Architecture of the small subunit processome
During the last year, three medium resolution cryo-EM reconstructions of the Chaetomium thermophilum and Saccharomyces cerevisiae SSU processomes (7.4 to 4.5 Å) [11,28,29] and very recently near-atomic structures of the C. thermophilum SSU processome [31] and the complete S. cerevisiae SSU processome [30] have been reported. These reconstructions together with protein-protein interaction data [33-35], crystal structures of the yeast small ribosomal subunit [36] and ribosome assembly factors [37-45] have highlighted the structural similarities between particles of these two species. The high degree of similarity in composition, structure and pre-rRNA state (A1 uncut, A0 cut), is further remarkable as these stable intermediates were purified from mid-log [28,31], starved [11,30] or Mtr4-depleted [29] cells.
Here we focus on the most complete near-atomic structure from the model organism S. cerevisiae to describe this particle (Figure 2) [30].
Figure 2.
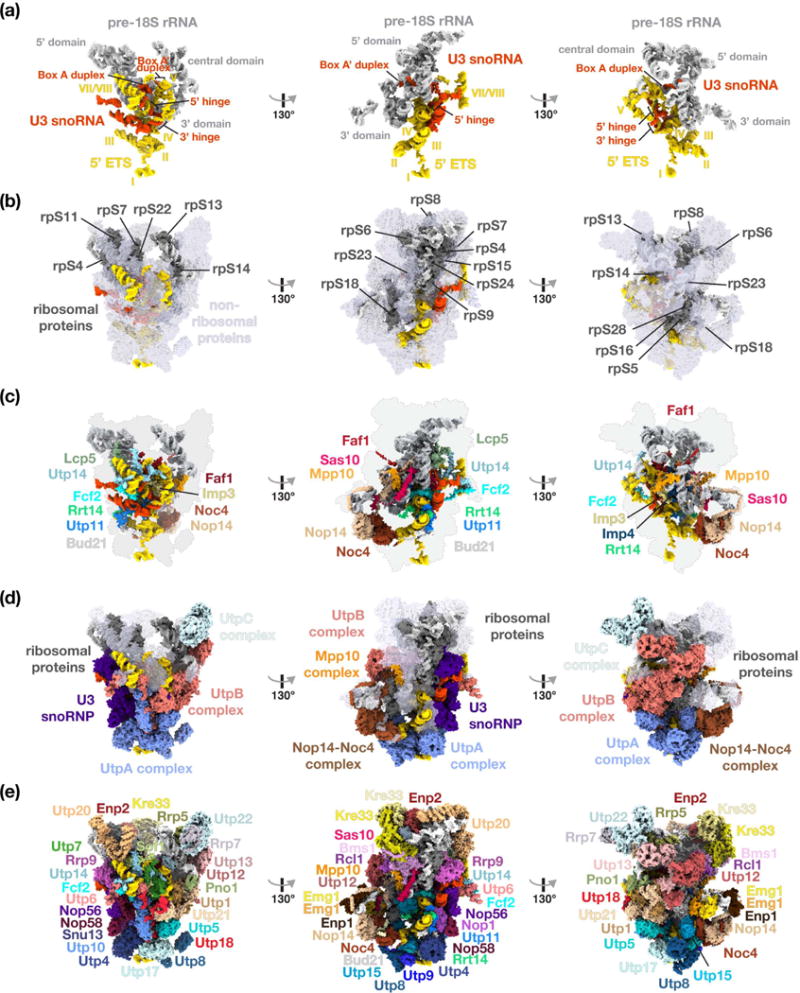
Structural organization of the yeast small subunit processome. (a) RNA molecules of the SSU processome are shown as surfaces with 5′ ETS (yellow), U3 snoRNA (red) and pre-18S (light-grey). Structural elements of RNAs and helices of the 5′ ETS are indicated. (b) Ribosomal proteins are represented in dark-grey, non-ribosomal assembly factors in transparent light-blue, and RNA species as in (a). (c) Surface representation of centrally located ribosome assembly factors. (d) Visualization of the complexes UtpA (blue), UtpB (red), U3 snoRNP (purple), UtpC (light-blue), the Nop14-Noc4 complex (brown) and the Mpp10 complex (orange). (e) Surface representation of all individual components of the small subunit processome.
The core of the SSU processome is composed of the 5′ ETS at the base of the structure, ribosomal RNA on top and U3 snoRNA, which reaches from the outside into the center, where all interactions with the 5′ ETS and 18S rRNA occur (Figure 2a). The tips of the 5′ ETS helices project freely into the solvent. This explains the length variation of this pre-ribosomal spacer which contains under 600 nucleotides in C. thermophilum, 700 nucleotides in yeast and more than 3600 nucleotides in humans. U3 snoRNA not only rigidifies the structure of the 5′ ETS through its 5′ and 3′ hinges, but further outlines the positions of the 5′, central and 3′ major domains by providing spatial constraints of the regions that base-pair with the 5′ end of the 18S rRNA (Box A) and a region between the central and 3′ domains (Box A′).
Fifteen ribosomal proteins are predominantly adopting the same conformations and binding sites as in the mature ribosome (Figure 2b). Only rpS6, rpS18 and rpS23 assume slightly different, yet near-mature conformations [30]. A large shell of more than 51 ribosome assembly factors encapsulates pre-ribosomal RNA and ribosomal proteins. The innermost layer of this shell is formed by extended peptide-like proteins, which weave through the entire particle (Figure 2c). Members of this group include the multimodular proteins Faf1, Lcp5, Mpp10, Sas10, Fcf2, Rrt14, Utp11 and Utp14, which are characterized by their unusual folds and many interaction partners as described later.
Several large multi-subunit complexes (Figure 2d) as well as individual ribosome assembly factors (Figure 2e) provide the outer shell of the SSU processome. In agreement with previous functional data, UtpA stabilizes the first half of the 5′ ETS and is located at the bottom of the particle. Through multiple interactions with UtpA and the 5′ ETS, UtpB components Utp18 and Utp6 likely make the first contact before the other four subunits of the UtpB complex are fully integrated into the SSU processome [16,30]. Interestingly, while both UtpA and UtpB share a common evolutionary origin as four subunits of each complex form a structurally related tetrameric arrangement with their C-terminal domains [11,29,38], their roles within the SSU processome are different. UtpA serves as a foundation of the particle to initiate its assembly, while UtpB provides a binding platform for the 3′ hinge, stabilizes spatially distant parts of the assembly including RNA elements near the 3′ end of the 18S rRNA. The UtpC complex is positioned at the top of the particle, where it interacts with the tip of helix 44 [29,30,45].
The outermost shell of the SSU processome is formed by many additional ribosome assembly factors (Figure 2e). These include the acetyltransferase/helicase Kre33, which rests on the Bms1-Rcl1 GTPase complex at the top of the structure, and the methyltransferase Emg1, which is positioned on a lateral extension formed by the Nop14/Noc4 complex. Lastly, Utp20, Utp10, Rrp5 and the Nop14/Noc4 complex provide large helical repeat structures to support and bridge distant regions of the particle.
The role of flexible proteins with multiple conserved binding motifs
The SSU processome is a eukaryote-specific particle with an intricate network of elongated peptides containing multiple binding motifs (Figure 3a). Strikingly, these long linkers are used to bridge conserved protein-protein interaction motifs (Figure 3b–d).
Figure 3.
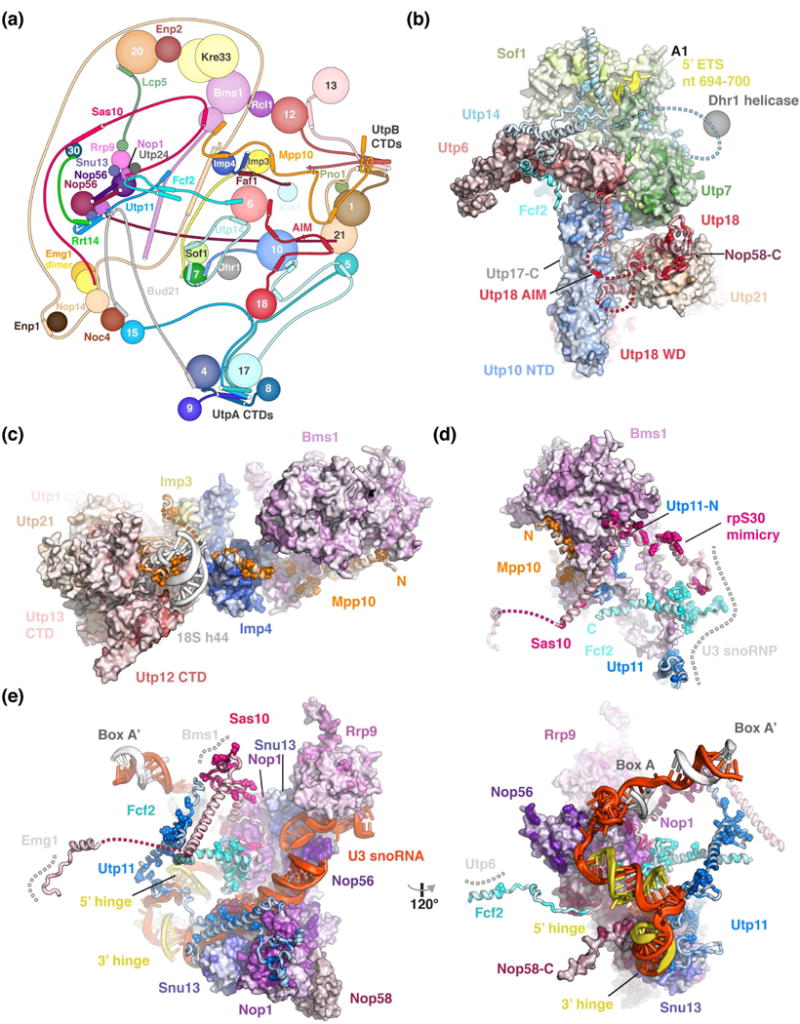
Peptides connect distant sites within the SSU processome via conserved binding motifs. (a) Schematic protein-protein interaction diagram of selected SSU processome components represented as spheres or lines with interacting elements as helices or strands. The Utps (U-three proteins) are labeled with their respective number. (b-e) Detailed views of Utp18 and Utp14 (b), Mpp10 (c), Bms1 (d) and the U3 snoRNP (e) with proteins shown as surface or cartoon, colored according to conservation with residues conserved more than 90 % highlighted as spheres. Direct interaction partners depicted as surface or a grey dashed line. All proteins are colored by conservation from lighter to darker shades. Clustal [49] was used to align manually curated sequences (H. sapiens, S. cerevisiae, G. gallus, D. melanogaster, S. pombe, C. elegans, D. rerio, A. thaliana, A. gambiae, P. troglodytes, R. norvegicus, M. musculus, B. taurus, S. scrofa) and plotted onto the structure using Homolmapper [50].
Utp18 contains an N-terminal extension, which interacts with parts of the UtpB complex (Utp21 & Utp6) as well as the UtpA complex (Utp10), the U3 snoRNP (Nop58) and the exosome-associated helicase Mtr4 via the arch-interacting-motif (AIM) [32] (Figure 3b). These peptides enable an initial flexibility within the yeast UtpB complex [16] followed by a later stable association with the SSU processome [30]. Similar peptide-like interactions are employed by Utp14, which interacts with three globular folds of other SSU processome components (Utp7, Sof1, Utp6) to stabilize nucleotides around the A1 cleavage site [30]. Additionally, Utp14 recruits Dhr1, the helicase required to unwind the U3 snoRNA [46] (Figure 3b).
Mpp10 fulfills many different roles within the SSU processome (Figure 3c). Chemical cross-linking and biochemical studies have shown that its flexible N-terminus interacts with rpS5 and Sas10 [30,47]. In addition, the structured C-terminus of Mpp10 is involved in stabilizing Bms1 (Figure 3d), bridging between Imp3, Imp4 and UtpB and supporting a remodeled pre-rRNA segment near helix 44.
The U3 snoRNP is a central nexus for several peptide-like motifs (Figure 3e) [30]. Here the C-terminal halves of Utp11, Sas10 and Fcf2 bind to conserved surfaces of Nop1 while their N-terminal halves together with an Mpp10 segment are used to stabilize Bms1 upon its incorporation within the SSU processome (Figure 3d). Sas10 also employs molecular mimicry by occupying the same position as rpS30 in the mature small ribosomal subunit (Figure 3d, e).
The observed dynamics of SSU processome assembly together with a complete high-resolution structure of this particle now allow us to propose a three-dimensional assembly model in Saccharomyces cerevisiae (Figure 4a). Conceptually this model takes into account the roles of initially flexible peptide sequences (Utp11, Bud21, Sas10 and Mpp10) and RNA sequences (U3 snoRNA 5′ and 3′ hinges as well as Box A and Box A′), which co-transcriptionally dictate the locations of rRNA and protein folds and reduce conformational freedom to stabilize the maturing SSU processome (Figure 4a). Major enzymatic and structural changes are required to move beyond the SSU processome towards the mature small ribosomal subunit (Figure 4b). Enzymatic steps include the unwinding of the Box A and Box A′ duplexes by RNA helicases such as Dhr1 [46], cleavage at site A1 by the nuclease Utp24 [24,25,48], and the replacement of the GTPase Bms1 by the structurally related factor Tsr1. In addition, structural changes such as rotational and translational movements of the central and 3′ domains with respect to the 5′ domain are required (Figure 4b). The formation of the central pseudoknot and its surrounding elements, the formation of inter-domain base-pairing interactions between the central and 5′ domains and the incorporation of additional ribosomal proteins are further steps requiring control during later stages of the assembly pathway.
Figure 4.
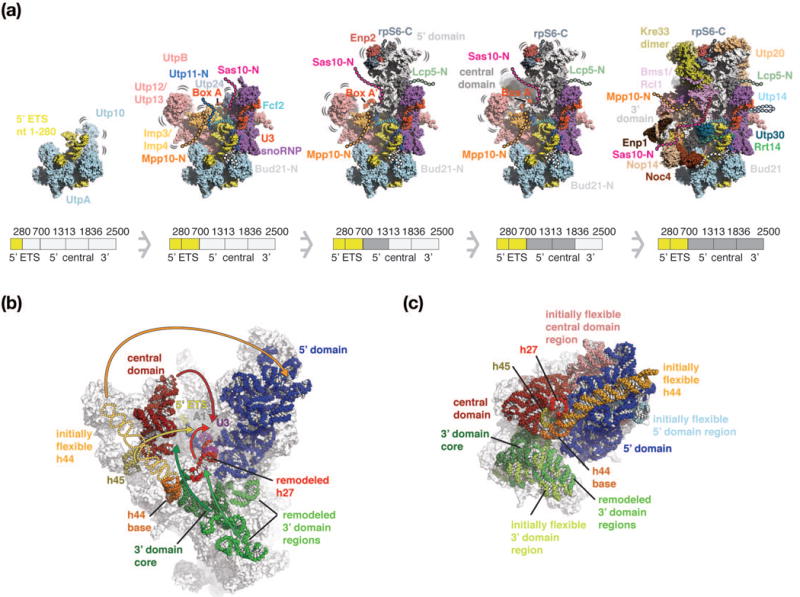
Three-dimensional maturation model of the Saccharomyces cerevisiae small-subunit processome. (a) Co-transcriptional assembly of the SSU processome as a function of transcription of rRNA regions (280 nucleotides of 5′ ETS, 5′ ETS, 5′ domain, central domain and 3′ domain). 5′ ETS (yellow), rRNA domains (white) and complexes such as UtpA (light-blue), UtpB (red), U3 snoRNP (purple, red), the Mpp10 complex (orange) and additional proteins, are shown as surfaces. Bound peptides are shown as cartoon with their initially flexible tails as dashed lines. As maturation progresses, these tails recruit additional factors and become ordered. The pre-rRNA shown in the intermediates is schematically indicated below each particle and colored in darker shades. As this model is based on the mature SSU processome structure, regions that are presumed to be flexible in earlier states are highlighted accordingly. (b, c) Comparative locations of rRNA domains within the SSU processome [PDB 5wlc] (b) and the mature small ribosomal subunit [pdb 4v88] (c). Individual rRNA domains are colored identically with 5′ domain (blue), central domain (red), 3′ domain (green) and shown as spheres superimposed onto transparent outlines of the particles. In the SSU processome, the flexible helix 44 is indicated as schematic outline. Rearrangements of rRNA domains from the SSU processome (b) that are necessary to obtain the positions within the mature small ribosomal subunit (c) are indicated with arrows. The central U3 snoRNA Box A and Box A′ are colored in purple. RNA elements disordered in the SSU processome are indicated in lighter shades in the mature SSU.
Perspectives
During the last five years, our understanding of the small subunit processome has progressed from a list of protein factors to an initial assembly model [19,20] and a near-atomic description of its structure [30,31]. However, the functional integration of this giant particle in the cellular context is still poorly understood. Little is known about the specific signaling that results in either the arrest of ribosome assembly or its resumption. While the SSU processome has been identified as storage particle, the mechanisms leading to its accumulation or subsequent processing remain unclear. Furthermore, structural intermediates that capture earlier conformations or released states of the SSU processome will be required to understand the mechanistic principles of this essential eukaryotic assembly intermediate.
Supplementary Material
Highlights.
A decade after its discovery, the structure of the small subunit processome emerges
More than 50 proteins and a snoRNA remodel pre-ribosomal RNA in the SSU processome
Initially flexible assembly factors guide the formation of a rigid scaffold
rRNA domains are kept separate to facilitate their modification and processing
Rearrangements and compaction of rRNA domains are required to form the mature SSU
Acknowledgments
We are grateful to all members of the Klinge laboratory for helpful discussions and critical reading of this manuscript. J.B. was supported by an EMBO long-term fellowship (ALTF 51-2014) and a Swiss National Science Foundation fellowship (155515). S.K. is supported by the Robertson Foundation, the Irma T. Hirschl Trust, the Alexandrine and Alexander L. Sinsheimer Fund, and an NIH New Innovator Award (1DP2GM123459).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
Nothing declared.
References
- 1.Woolford JL, Baserga SJ. Ribosome Biogenesis in the Yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turowski TW, Tollervey D. Cotranscriptional events in eukaryotic ribosome synthesis. WIREs RNA. 2015;6:129–139. doi: 10.1002/wrna.1263. [DOI] [PubMed] [Google Scholar]
- 3.Miller OL, Beatty BR. Visualization of nucleolar genes. Science. 1969;164:955–957. doi: 10.1126/science.164.3882.955. [DOI] [PubMed] [Google Scholar]
- 4.Dragon F, Gallagher JEG, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schäfer T, Kuster B, Tschochner H, Tollervey D, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein KA, Gallagher JEG, Mitchell BM, Granneman S, Baserga SJ. The small-subunit processome is a ribosome assembly intermediate. Eukaryotic Cell. 2004;3:1619–1626. doi: 10.1128/EC.3.6.1619-1626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira-Cerca S, Pöll G, Kühn H, Neueder A, Jakob S, Tschochner H, Milkereit P. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol Cell. 2007;28:446–457. doi: 10.1016/j.molcel.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 8.Gallagher JEG, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes & Development. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Research. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Talkish J, Biedka S, Jakovljevic J, Zhang J, Tang L, Strahler JR, Andrews PC, Maddock JR, Woolford JL. Disruption of ribosome assembly in yeast blocks cotranscriptional pre-rRNA processing and affects the global hierarchy of ribosome biogenesis. RNA. 2016;22:852–866. doi: 10.1261/rna.055780.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Chaker-Margot M, Barandun J, Hunziker M, Klinge S. Architecture of the yeast small subunit processome. Science. 2017;355:eaal1880. doi: 10.1126/science.aal1880. Along with Refs [28•, 29•] these papers provide the first medium-resolution cryo-EM reconstructions and architectural models of Chaetomium thermophilum and Saccharomyces cerevisiae small subunit processomes. Placement of homology models and crystal structures enables the identification of major folds including the localization of the 3 large complexes UtpA, UtpB and U3 snoRNP that encapsulate the 5′ external transcribed spacer. [DOI] [PubMed] [Google Scholar]
- 12.Kos-Braun IC, Jung I, Koš M. Tor1 and CK2 kinases control a switch between alternative ribosome biogenesis pathways in a growth-dependent manner. PLoS Biol. 2017;15:e2000245. doi: 10.1371/journal.pbio.2000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krogan NJ, Peng W-T, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 14.Dosil M, Bustelo XR. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90 S pre-ribosomal particle. J Biol Chem. 2004;279:37385–37397. doi: 10.1074/jbc.M404909200. [DOI] [PubMed] [Google Scholar]
- 15.Pöll G, Li S, Ohmayer U, Hierlmeier T, Milkereit P, Perez-Fernandez J. In vitro reconstitution of yeast tUTP/UTP A and UTP B subcomplexes provides new insights into their modular architecture. PLoS ONE. 2014;9:e114898. doi: 10.1371/journal.pone.0114898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunziker M, Barandun J, Petfalski E, Tan D, Delan-Forino C, Molloy KR, Kim KH, Dunn-Davies H, Shi Y, Chaker-Margot M, et al. UtpA and UtpB chaperone nascent pre-ribosomal RNA and U3 snoRNA to initiate eukaryotic ribosome assembly. Nat Commun. 2016;7:12090. doi: 10.1038/ncomms12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pérez-Fernández J, Román A, Las Rivas De J, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Molecular and Cellular Biology. 2007;27:5414–5429. doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pérez-Fernández J, Martín-Marcos P, Dosil M. Elucidation of the assembly events required for the recruitment of Utp20, Imp4 and Bms1 onto nascent pre-ribosomes. Nucleic Acids Research. 2011;39:8105–8121. doi: 10.1093/nar/gkr508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Chaker-Margot M, Hunziker M, Barandun J, Dill BD, Klinge S. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat Struct Mol Biol. 2015;22:920–923. doi: 10.1038/nsmb.3111. This paper along with Ref [20••] uses an RNA/protein-tandem affinity purification system to study ribosome assembly as a function of transcription. Many ribosome assembly factors are assigned to particular pre-rRNA domains and the 5′ ETS particle is identified. [DOI] [PubMed] [Google Scholar]
- 20••.Zhang L, Wu C, Cai G, Chen S, Ye K. Stepwise and dynamic assembly of the earliest precursors of small ribosomal subunits in yeast. Genes & Development. 2016;30:718–732. doi: 10.1101/gad.274688.115. See annotation to [• 19] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marmier-Gourrier N, Cléry A, Schlotter F, Senty-Ségault V, Branlant C. A second base pair interaction between U3 small nucleolar RNA and the 5′-ETS region is required for early cleavage of the yeast pre-ribosomal RNA. Nucleic Acids Research. 2011;39:9731–9745. doi: 10.1093/nar/gkr675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kudla G, Granneman S, Hahn D, Beggs JD, Tollervey D. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc Natl Acad Sci USA. 2011;108:10010–10015. doi: 10.1073/pnas.1017386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutca LM, Gallagher JEG, Baserga SJ. The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Research. 2011;39:5164–5180. doi: 10.1093/nar/gkr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomecki R, Labno A, Drazkowska K, Cysewski D, Dziembowski A. hUTP24 is essential for processing of the human ribosomal RNA precursor at site A1, but not at site A0. RNA Biol. 2015;12(9):1010–1029. doi: 10.1080/15476286.2015.1073437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wells GR, Weichmann F, Colvin D, Sloan KE, Kudla G, Tollervey D, Watkins NJ, Schneider C. The PIN domain endonuclease Utp24 cleaves pre-ribosomal RNA at two coupled sites in yeast and humans. Nucleic Acids Research. 2016;44:5399–5409. doi: 10.1093/nar/gkw213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin R, Hackert P, Ruprecht M, Simm S, Brüning L, Mirus O, Sloan KE, Kudla G, Schleiff E, Bohnsack MT. A pre-ribosomal RNA interaction network involving snoRNAs and the Rok1 helicase. RNA. 2014;20:1173–1182. doi: 10.1261/rna.044669.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wells GR, Weichmann F, Sloan KE, Colvin D, Watkins NJ, Schneider C. The ribosome biogenesis factor yUtp23/hUTP23 coordinates key interactions in the yeast and human pre-40S particle and hUTP23 contains an essential PIN domain. Nucleic Acids Research. 2017;45:4796–4809. doi: 10.1093/nar/gkw1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Kornprobst M, Turk M, Kellner N, Cheng J, Flemming D, Koš-Braun I, Koš M, Thoms M, Berninghausen O, Beckmann R, et al. Architecture of the 90S Pre-ribosome: A Structural View on the Birth of the Eukaryotic Ribosome. Cell. 2016;166:380–393. doi: 10.1016/j.cell.2016.06.014. See annotation to Ref [• 11] [DOI] [PubMed] [Google Scholar]
- 29•.Sun Q, Zhu X, Qi J, An W, Lan P, Tan D, Chen R, Wang B, Zheng S, Zhang C, et al. Molecular architecture of the 90S small subunit pre-ribosome. Elife. 2017;6:e22086. doi: 10.7554/eLife.22086. See annotation to Ref [• 11] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Barandun J, Chaker-Margot M, Hunziker M, Molloy KR, Chait BT, Klinge S. The complete structure of the small-subunit processome. Nat Struct Mol Biol. 2017;24:944–953. doi: 10.1038/nsmb.3472. This study and Ref [31••] provide the high-resolution cryo-EM structures of the Saccharomyces cerevisiae and Chaetomium thermophilum small subunit processomes. Near-atomic resolution enables the identification and tracing of many ribosome biogenesis factors including peptide-like motifs, which could not be traced previously. The higher resolution of these reconstructions also reveals how RNA regions close to the central pseudoknot are remodeled in a concerted fashion to prevent pre-mature folding. [DOI] [PubMed] [Google Scholar]
- 31••.Cheng J, Kellner N, Berninghausen O, Hurt E, Beckmann R. 3.2-Å-resolution structure of the 90S preribosome before A1 pre-rRNA cleavage. Nat Struct Mol Biol. 2017;24:954–964. doi: 10.1038/nsmb.3476. See annotation to Ref [•• 30] [DOI] [PubMed] [Google Scholar]
- 32.Thoms M, Thomson E, Bassler J, Gnädig M, Griesel S, Hurt E. The Exosome Is Recruited to RNA Substrates through Specific Adaptor Proteins. Cell. 2015;162:1029–1038. doi: 10.1016/j.cell.2015.07.060. [DOI] [PubMed] [Google Scholar]
- 33.Lim YH, Charette JM, Baserga SJ. Assembling a protein-protein interaction map of the SSU processome from existing datasets. PLoS ONE. 2011;6:e17701. doi: 10.1371/journal.pone.0017701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bassler J, Ahmed YL, Kallas M, Kornprobst M, Calviño FR, Gnädig M, Thoms M, Stier G, Ismail S, Kharde S, et al. Interaction network of the ribosome assembly machinery from a eukaryotic thermophile. Protein Sci. 2017;26:327–342. doi: 10.1002/pro.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vincent NG, Charette JM, Baserga SJ. The SSU processome interactome in Saccharomyces cerevisiae reveals novel protein subcomplexes. RNA. 2018;24:77–89. doi: 10.1261/rna.062927.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 37.Calviño FR, Kornprobst M, Schermann G, Birkle F, Wild K, Fischer T, Hurt E, Ahmed YL, Sinning I. Structural basis for 5′-ETS recognition by Utp4 at the early stages of ribosome biogenesis. PLoS ONE. 2017;12:e0178752. doi: 10.1371/journal.pone.0178752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang C, Sun Q, Chen R, Chen X, Lin J, Ye K. Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits. Nucleic Acids Research. 2016;44:7475–7486. doi: 10.1093/nar/gkw562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang C, Lin J, Liu W, Chen X, Chen R, Ye K. Structure of Utp21 tandem WD domain provides insight into the organization of the UTPB complex involved in ribosome synthesis. PLoS ONE. 2014;9:e86540. doi: 10.1371/journal.pone.0086540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boissier F, Schmidt CM, Linnemann J, Fribourg S, Perez-Fernandez J. Pwp2 mediates UTP-B assembly via two structurally independent domains. Sci Rep. 2017;7:3169. doi: 10.1038/s41598-017-03034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Lin J, Ye K. Structural and functional analysis of the U3 snoRNA binding protein Rrp9. RNA. 2013;19:701–711. doi: 10.1261/rna.037580.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delprato A, Kadri Al Y, Pérébaskine N, Monfoulet C, Henry Y, Henras AK, Fribourg S. Crucial role of the Rcl1p-Bms1p interaction for yeast pre-ribosomal RNA processing. Nucleic Acids Research. 2014;42:10161–10172. doi: 10.1093/nar/gku682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas SR, Keller CA, Szyk A, Cannon JR, LaRonde-LeBlanc NA. Structural insight into the functional mechanism of Nep1/Emg1 N1-specific pseudouridine methyltransferase in ribosome biogenesis. Nucleic Acids Research. 2011;39:2445–2457. doi: 10.1093/nar/gkq1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng S, Lan P, Liu X, Ye K. Interaction between ribosome assembly factors Krr1 and Faf1 is essential for formation of small ribosomal subunit in yeast. Journal of Biological Chemistry. 2014;289:22692–22703. doi: 10.1074/jbc.M114.584490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin J, Lu J, Feng Y, Sun M, Ye K. An RNA-Binding Complex Involved in Ribosome Biogenesis Contains a Protein with Homology to tRNA CCA-Adding Enzyme. PLoS Biol. 2013;11:e1001669. doi: 10.1371/journal.pbio.1001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sardana R, Liu X, Granneman S, Zhu J, Gill M, Papoulas O, Marcotte EM, Tollervey D, Correll CC, Johnson AW. The DEAH-box helicase Dhr1 dissociates U3 from the pre-rRNA to promote formation of the central pseudoknot. PLoS Biol. 2015;13:e1002083. doi: 10.1371/journal.pbio.1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sá-Moura B, Kornprobst M, Kharde S, Ahmed YL, Stier G, Kunze R, Sinning I, Hurt E. Mpp10 represents a platform for the interaction of multiple factors within the 90S pre-ribosome. PLoS ONE. 2017;12:e0183272. doi: 10.1371/journal.pone.0183272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bleichert F, Granneman S, Osheim YN, Beyer AL, Baserga SJ. The PINc domain protein Utp24, a putative nuclease, is required for the early cleavage steps in 18S rRNA maturation. Proc Natl Acad Sci USA. 2006;103:9464–9469. doi: 10.1073/pnas.0603673103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rockwell NC, Lagarias JC. Flexible mapping of homology onto structure with homolmapper. BMC Bioinformatics. 2007;8:123. doi: 10.1186/1471-2105-8-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


