Abstract
Post-traumatic stress disorder (PTSD) after exposure to a traumatic event is a highly prevalent psychiatric disorder. Heritability estimates from twin studies as well as from recent molecular data (h2SNP) indicate moderate to high heritability, yet robust genetic variants for PTSD have not yet been identified and the genetic architecture of this polygenic disorder remains largely unknown.
To date, less than ten large-scale genome-wide association studies (GWAS) of PTSD have been published, with findings that highlight the unique challenges for PTSD genomics, including a complex diagnostic entity with contingency of PTSD diagnosis on trauma exposure, and the large genetic diversity of the study populations.
The Psychiatric Genomics Consortium PTSD group (PGC-PTSD) has brought together over 200 scientists with the goal to increase sample size for GWAS and other genomic analyses to sufficient numbers where robust discoveries of molecular signatures can be achieved. The sample currently includes over 32,000 PTSD cases and 100,000 trauma-exposed controls and collection is ongoing. First results found a significant shared genetic risk of PTSD with other psychiatric disorders, and sex-biased heritability estimates with higher heritability in females compared to males.
This review describes the scope and current focus of the PGC-PTSD, and its expansion from the initial GWAS group to nine working groups, including epigenetics, gene expression, imaging, and integrative systems biology. We further briefly outline recent findings and future directions of ‘omics-based’ studies of PTSD, with the ultimate goal of elucidating the molecular architecture of this complex disorder to improve prevention and intervention strategies.
Keywords: Psychiatric genomics consortium (PGC), genetics, epigenetics, gene expression, imaging, systems biology
Genetics of PTSD in the context of other psychiatric disorders
PTSD is a debilitating psychiatric disorder precipitated by traumatic experience, with subsequent pathological re-experiencing, avoidance, negative alterations in cognitions and mood, and hyperarousal symptoms (DSM-5 (1)). While most individuals exposed to trauma are resilient, PTSD prevalence is directly related to the severity and type of trauma, with rape or direct combat conferring very high risk, and lifetime risk in women is twice that in men (2). PTSD prevalence varies by country, but lifetime prevalence in the US is over 7% – making it among the most common psychiatric disorders (3).
Genetic factors influence who develops PTSD; family and twin studies have estimated heritability of PTSD from ~40-70% following trauma (4–7). However, despite over a decade of research on genetic candidate genes, robust genetic variants for PTSD have yet to be identified and the genetic architecture of this polygenic disorder remains largely unknown.
To address this gap in knowledge, the field of psychiatric genomics has moved its focus over the last decade from small studies on specific candidate genes (8) to agnostic, genome-wide association studies (GWA studies) and ultimately to well-powered, large-scale meta-analyses made possible through efforts such as the Psychiatric Genomics Consortium (PGC) (9). Recent success in the identification of robustly associated genetic variants in psychiatric disorders such as schizophrenia (10), bipolar disorder (11) and major depressive disorder (MDD) (12) has confirmed that very large sample sizes are necessary to discover loci with the small genotypic relative risks typically seen in psychiatric disorders. Accordingly, the short-term goal of the PGC is to obtain GWAS data on 100,000 cases for each of its nine disorders (9).
Genome-wide association approaches to PTSD
PGC-PTSD (https://pgc-ptsd.com/) was initiated in 2013 by bringing together four groups with published GWA studies of PTSD (13–16), making it a relative latecomer to the PGC (17). To date, four additional association studies of PTSD with genome-wide data have been published (18–21) and at least one more study in Danish soldiers has been completed (Wang et al., pers. com.). Typically, these studies present a finding that meets criteria for genome-wide significance plus evidence of replication in at least one independent cohort (Table 1). While this has been the gold standard for GWA studies, none of the identified genes in these GWA studies has robustly replicated across multiple studies. With the rapid availability of PGC-PTSD summary data on a large number of studies, best practice guidelines for GWAS replication are currently being discussed (e.g. https://www.cohenveteransbioscience.org/2017/06/29/psychiatric-genomic-consortium-workshop-summary/) and can now include the pre-specified selection of specific replication cohorts matched for example on ancestry, gender, and trauma type.
Table 1.
Genome-wide significant hits and replication in individual PTSD studies
| Study | Hits | Cohort | N PTSD | N controls | Ancestry1 | Sex | Trauma2 | P-value | Comments |
|---|---|---|---|---|---|---|---|---|---|
| Logue et al. (14) | RORA | NCPT | 295 | 196 | EA | pred. M | pred. military | 2.50E-08 | |
| replication replication |
NCPT DNHS |
43 100 |
41 421 |
AA pred. AA |
pred. M F |
pred. military civilian |
0.05 NS |
gene-base test gene-base test |
|
| Guffanti et al. (13) | LINC01090 | DNHS | 94 | 319 | pred. AA | F | civilian | 5.09E-08 | |
| replication | NHSII | 578 | 1963 | EA | F | civilian | 0.07 | marginal | |
| Xie et al. (15) | COBL | GSD | 300 | 1,273 | EA | M+F | civilian | 3.96E-08 | |
| replication | GSD | 207 | 1692 | EA | M+F | civilian | NS | ||
| Xie et al. (15) | TLL1 | GSD | 300 | 1,273 | EA | M+F | civilian | 2.99E-07 | marginal GWAS hit |
| replication | GSD | 207 | 1692 | EA | M+F | civilian | 6.30E-06 | ||
| Solovieff et al. (18) | SLC18A2 | NHSII | 845 | 1,693 | EA | F | civilian | 2.10E-05 | candidate-gene study |
| replication | DNHS | 748 | 748 | pred. AA | M+F | civilian | 0.05 | haplotype | |
| Nievergelt et al. (16) | PRTFDC1 | MRS | 940 | 2,554 | mixed | M | military | 2.04E-09 | |
| replication | NCPT | 313 | 178 | EA | pred. M | military | 0.14 | marginal for SNP in LD | |
| Almli et al. (19) | rs717947 | SBPBC | 63 | 84 | mixed | pred. M | military | 1.28E-08 | |
| replication replication |
GTP GTP |
2006 subjects 862 subjects |
AA AA |
F M |
civilian civilian |
0.005 NS |
PTSD symptoms PTSD symptoms |
||
| Stein et al. (20) | ANKRD55 | NSS | 497 | 815 | AA | pred. M | military | 2.34E-08 | |
| replication | MRS&PPDS&MIREC | 947 | 4969 | AA | pred. M | military | NS | meta-analysis | |
| Stein et al. (20) | ZNF626 | NSS | 2,140 | 2,909 | EA | pred. M | military | 4.59E-08 | |
| replication | PPDS | 672 | 3335 | EA | pred. M | military | NS | ||
Ancestry: EA: European ancestry; AA: African American
Trauma: cohorts have been separated by military and civilian; individual type of trauma is not considered here
The PGC-PTSD has adopted pipelines and protocols established by the PGC (https://data.broadinstitute.org/mpg/ricopili/), which facilitates integration of data across disorders (e.g. (22)). However, PGC-PTSD has to consider some unique challenges not faced by other groups, including the contingency of PTSD diagnosis on trauma exposure, a complex and changing diagnostic entity (23), and very diverse genetic ancestry within and across study cohorts, resulting in considerable heterogeneity (17).
To address ancestral diversity, the PGC pipeline was extended to include an ancestry inference module, which allows for stable ancestry determination across studies (https://github.com/nievergeltlab). It was designed to be portable to enable PGC-PTSD studies that cannot share individual-level genotype data (e.g. some US military and non-US cohorts) to generate summary-level results for meta-analysis. The majority of GWA studies to date have been performed in subjects of European (EA) and African American (AA) ancestry groups, while carefully addressing residual population stratification (Figure 1). With the sustained PGC-PTSD efforts to increase sample size, other ancestries such as Latinos and East Asians are reaching considerable size. Trans-ethnic GWAS have been generated using meta-analytical approaches (16, 24), and alternative mega-analysis methods are currently employed to leverage all available samples, irrespective of ancestry (25).
Figure 1. Ancestry composition and main ancestry groups analyzed in the PGC-PTSD.
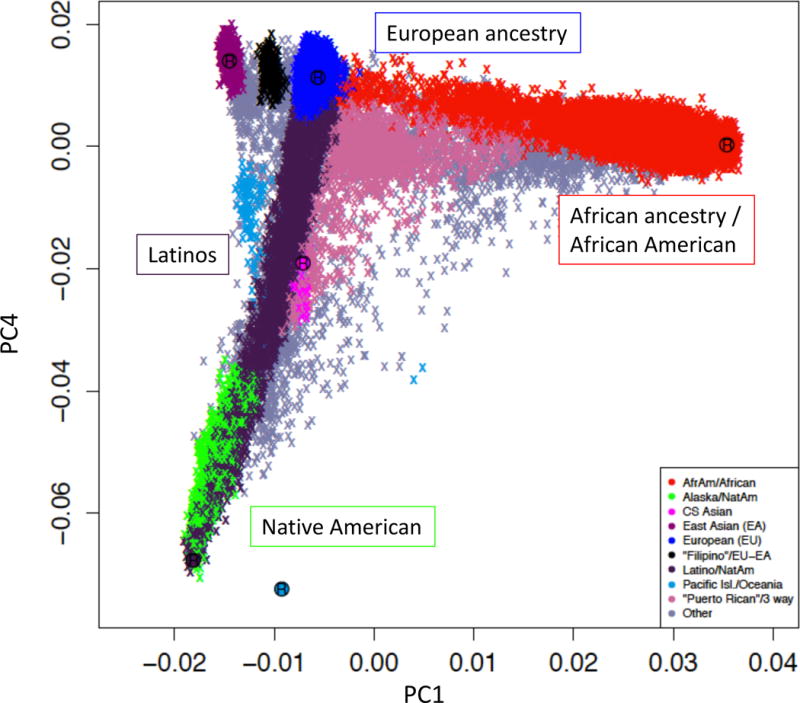
Principal component (PC) plot showing inferred ancestry of > 80,000 subjects from 56 studies. Main population groups analyzed to date include homogeneous subjects of European ancestry (N>40,000), and one-way admixed African Americans (N~20,000) and Latinos, including Native Americans (N>6,500). With expansion of the data collection, additional populations, such as East Asians, will reach optimal sizes for analysis. The plot is based on PC1 and PC4 to highlight the Asian populations.
The first PGC-PTSD publication in 2017 included 11 multi-ethnic studies of 5,000 PTSD cases and 15,000 controls (freeze 1) and is largest published genetic association study of PTSD to date. SNP-based heritability (h2SNP) estimates for PTSD were ~29% in females, but substantially lower in males (24), consistent with lower twin-based heritability estimates in males. In addition, the study found significant shared genetic risk of PTSD with schizophrenia, bipolar disorder and MDD. Investigating the implications of sex-based heritability in PTSD and cross-disorder genetic risks are high priority for future study design.
Expanding the PGC-PTSD scope
Although successful in demonstrating some aspects of the genetic architecture of PTSD, the PGC-PTSD freeze 1 was underpowered to identify genome-wide significant loci (24). Thus, expanding the sample size to sufficient numbers for GWAS remains one of the main goals of the PGC-PTSD. The current freeze 2 includes over 32,000 PTSD cases and 100,000 trauma-exposed controls (Nievergelt et al., manuscript in preparation), approaching the number of cases for which other PGC studies showed first robust discoveries (26).
In addition, future analyses will also explore analytical models that are potentially stronger than conventional case-control analyses, including quantitative symptom scores and clusters as well as trauma exposure, similarly to a recent meta-analysis of GWA studies on anxiety disorder (27). The grouping of sub-clinical PTSD cases with trauma-exposed controls is a potential limitation of the conventional PGC case-control studies.
A promising development in the PGC-PTSD is the expansion from the initial GWAS group to nine integrated working groups (see Figure 2). While some working groups such as the physical health (28), psychophysiology, and imaging groups have extended the phenotype (i.e., PTSD diagnosis) with highly relevant additional phenotypes, other working groups have assembled complimentary ‘omic’ type data such as copy-number variants (CNV), methylation, and gene-expression. A microbiome group has recently been initiated. Finally, the systems biology group is charged with integration of these multiple data types to maximally leverage data resources for discovery. A rational for some of these efforts is discussed below.
Figure 2. Main interactions and dataflow between the nine PGC-PTSD working groups.
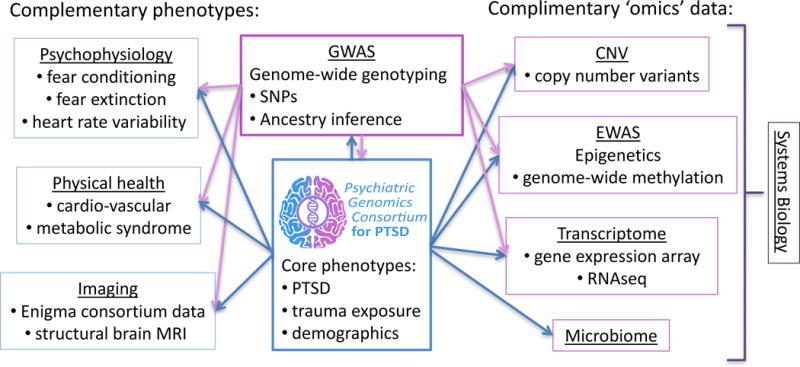
The PGC-PTSD GWAS group has drastically expanded its scope since its initiation in 2013. It currently includes working groups with emphasis on complimentary phenotypes (psychophysiology, physical health, and imaging), working groups contributing complimentary ‘omics’ data (CNV, epigenetics, transcriptome, and microbiome), as well as a systems biology group aiming at integration of the different types of data. Arrows are indicating primary flow of data, but interactions among groups are expanding.
Gene Expression analysis in PTSD
Analysis of gene expression in PTSD presents challenges, as the underlying molecular events causing this disorder likely occur in the central nervous system (CNS), and archival of postmortem brain tissue from individuals affected by PTSD lags behind other CNS disorders such as Parkinson’s and Alzheimer’s Disease. However, considerable data also point to PTSD being characterized by systemic immune and metabolic perturbations caused by stress-responsive changes in the hypothalamic–pituitary–adrenal (HPA) axis (29–31). These systemic changes give rise to differential gene expression signatures in peripheral blood of PTSD cases versus controls that can serve as biomarkers for disease, and can also provide insight into disease-associated systemic effects on immune function and organ pathology. Moreover, the changes occurring in the periphery may be promoting or exacerbating changes in the brain (32). Thus, gene expression analysis of peripheral blood of PTSD cases and controls is being performed by the PGC-PTSD not merely as a matter of convenience, but because it is likely to illuminate critical disease processes and potentially identify individuals most at-risk for PTSD.
A number of gene expression studies in peripheral blood have already been reported. While statistical power of some of these studies has been limited by small sample size (33–38), others have reported gene expression changes that were statistically significant after rigorous correction for multiple testing (39–41). These studies have replicated a few differentially expressed genes: USP48 was identified as differentially expressed in PTSD cases versus controls by two studies (39, 40), and DICER1 was similarly identified by two studies (40, 41). While in general there is little concordance of specific genes between studies, pathway and gene network analyses have consistently and reproducibly identified differential expression of transcripts involved in innate immunity, interferon signaling, and wound healing (42, 43). These studies have provided valuable insights into the pathobiology of PTSD. With a combined sample size of almost 5000 samples, future peripheral blood gene expression studies by the PGC-PTSD should refine and extend these findings. As CNS tissue samples become increasingly available, gene expression data from post-mortem brains can be compared and integrated with findings from peripheral blood.
PTSD Epigenetics
PTSD is unique among psychiatric disorders in that it requires a traumatic environmental event as part of its diagnosis. Among the different epigenetic modifications, DNA methylation has received the most attention by researchers studying psychosocial stress, childhood trauma, and PTSD due to its relative stability and its ability to be assessed with microarrays that facilitate replication within and between studies (40, 44–50). Immune dysregulation figured prominently among the biologic pathways associated with PTSD and are replicable between studies (44, 46). A recent study examined DNA methylation along with microRNA (miRNA), another epigenetic modification, in a small group of PTSD cases and controls. The authors noted reductions in miRNA levels in PTSD cases and proposed that epigenetic changes may contribute to systemic inflammation in PTSD (51). These studies echo those of other psychiatric disorders that emphasize the cross-talk between the peripheral immune system and the brain (32, 52).
Other studies suggest more widespread mechanisms for epigenetic dysregulation in PTSD. For example, Maddox and colleagues reported DNA methylation differences in HDAC4, a histone deacetylase, in the blood of women with PTSD and went on to show that variation in genetic and epigenetic predictors of HDAC4 expression associated with fear-potentiated startle response and functional connectivity differences in the amygdala (53). Similarly, lower expression of DICER1, which is required for processing mature miRNAs, is associated with PTSD cases with comorbid depression and increased amygdala activation in response to fearful stimuli (41, 54), a neural phenotype strongly associated with risk for PTSD even prior to trauma exposure (reviewed in (55)).
In addition to these genome-scale approaches, epigenetic summary measures may be particularly informative. The most widely used measure is the ‘epigenetic clock’ (56, 57) for assessing age acceleration, which is associated with psychosocial stress and higher mortality risk (57, 58). The phenomenon describes methylation-based prediction of age that exceeds chronological age. Of note, a relatively high proportion – almost 25% of epigenetic clock-related CpG sites are located in glucocorticoid response elements (GREs) – a genomic region in which methylation levels vary in relation trauma exposure (59) and dexamethasone suppression (57). These environmentally sensitive genomic sites have been explicitly linked to traumatic stress, neural integrity and mortality (60), discussed in more detail below, further demonstrating the utility of using epigenetic summary measures as an index of the biologic impact of lived experience, including trauma exposure.
There are numerous challenges in conducting epigenetic studies of PTSD. Similar to gene expression discussed above, DNA methylation patterns vary by tissue type, with the majority of studies to date conducted in blood, whereas the most relevant tissue is brain. As new postmortem samples and single-cell technologies become available, there will be substantial advancement in identification of genes whose regulation is altered in those with PTSD. This may further support the identification of peripheral biomarkers or may restrict the scope of peripheral tissues. A second challenge is limited platforms, and thus limited coverage, for DNA methylation or other arrays that are widely used for population-scale studies. Epigenome-wide investigations require cost-effective and highly reproducible methods to achieve the sample sizes required to detect associations that withstand multiple testing correction. The PGC-PTSD EWAS group (Figure 2) has the goal to assemble such data to perform meta-analyses across cohorts with a common multi-site analysis pipeline (61). In some ways, these platform limitations increase opportunities for replication, but also complicate linking genome-wide discoveries with those based on sequencing or targeted assays.
PTSD Imaging Genetics
Elevated risk of psychopathology may be more powerfully investigated with intermediate phenotypes (or endophenotypes) than clinical diagnoses. Brain measures from MRI may have a simpler underlying genetic architecture involving fewer individual genes or pathways than the polygenicity driving overall risk for psychopathology (62), and offer a more precise and reproducible phenotype than clinical diagnostic scales (63). A GWAS of continuous brain measures may be statistically more powerful and more efficient than binary traits (diagnosis), which may disguise complexities such as co-morbidity and syndromal heterogeneity (64). Furthermore, brain phenotypes may provide common pathways for the combined effects of environmental and genetic risk factors that may underlie multiple diagnoses (62). However, there are two important caveats (1) the effect sizes for gene effects on neuroimaging phenotypes are unlikely to be greater than for behavioral traits or psychiatric disorders (65), and (2) genetics of brain phenotypes may reveal common mechanistic pathways for a number of psychiatric disorders resulting in a loss of specificity when moving from psychiatric disorder to brain phenotype – different disorders may possess nearly identical brain abnormalities (66). This loss of specificity may prove advantageous for drug development, by facilitating the design of a target-specific intervention that is effective for multiple neuropsychiatric disorders.
An international collaboration of investigators (17) within the PGC and Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA) plans to investigate the genetic effects of complex brain traits (66). The first major analysis of the PGC-ENIGMA PTSD Working Group with 1,868 samples has demonstrated that PTSD is associated with smaller hippocampus and amygdala volume (67). Exposure to childhood trauma was negatively associated with hippocampal and amygdala volume when adjusting for age, sex, and intracranial volume (67). Both structures have ample a priori evidence implicating their role in PTSD starting with the report of reduced hippocampal volume in a small PTSD sample over 20 years ago (68). However, we confirmed this finding across a large number of demographically and clinically heterogeneous cohorts analyzed with a standardized segmentation technique, and a harmonized analysis protocol across all sites. Methodological consistency was promoted by using the same statistical models across all samples, making this the largest and most powerful study of subcortical volumes in PTSD to date. The analysis of 12 hippocampal subfield volumes, DTI measures of 26 white matter tracts, the cortical thickness of 78 regions-of-interest, and whole-brain vertex-based analyses are currently underway using ENIGMA pipelines, which can be downloaded at https://pgc-ptsd.com/methods-tools/imaging-pipeline/.
Forthcoming analyses gene-by-environment (GxE) GWAS of relevant structural brain phenotypes with childhood trauma as major risk factor are planned with the long-term goal of identifying genetic modulators of brain structure that help early prediction and treatment for a range of psychiatric disorders, followed by deep sequencing in a subset of samples to identify potential causal variants within the coding and/or regulatory regions of implicated risk loci. Over 40 participating sites have coalesced around the common goal to form ENIGMA-PGC-PTSD, which has already received 4,000 samples among which 3,000 samples have been aggregated and analyzed. Nevertheless, the analyses are expected to be woefully underpowered with the large number of phenotypes available in neuroimaging data. Two approaches address the shortcomings of previous neuroimaging-genetics studies that have been plagued by small sample size due to the large expense of MRI acquisition, and the use of candidate genes that have been criticized for being susceptible to population stratification and fueling information bottlenecks. First, no candidate gene analyses are planned. Second, replication samples of neuroimaging data such as the UK Biobank and the Million Veteran Program (MVP) will be leveraged. Third, we will focus on polygenic risk score (PRS) calculation and PRS x E interaction analyses. Discovery samples for calculating PRS include (1) the GWAS of the PTSD diagnosis from the PGC from 80,000 samples, (2) the GWAS of subcortical volumetry performed from 31,000 normative neuroimaging and genomic samples that provided several SNP associations (69), and (3) the GWAS of cortical thickness and surface area in which preliminary results from 30,000 samples has generated 120 SNPs that show robust genome-wide significant associations after correction for 78 cortical structures. The testing of PRS that are calculated in discovery samples, which have yielded robust genome wide associations, in our sample avoids the criticisms previously leveled against neuroimaging studies (70).
Addressing the inaccessibility of brain tissue
To date, EWAS studies of PTSD have been limited to accessible peripheral tissues, specifically whole blood (40, 44, 46). While potentially informative as biomarkers of the disorder, the extent to which PTSD-associated DNA methylation patterns in blood or other peripheral tissues reflect patterns that may exist in the brain remains unknown. As the target organ of most interest to the disorder, the brain remains a challenge to access in living individuals, and brain-based epigenetic predictors of PTSD have yet to be identified.
Despite these challenges, recent work has attempted to bridge the link between brain and periphery by examining peripherally derived epigenetic biomarkers of neuroimaging-based phenotypes and endophenotypes of PTSD. Much of this work to date has adopted a candidate gene approach (e.g. (71, 72)). A notable exception is a study conducted by Wolf and colleagues (60) which tested the hypothesis that PTSD is associated with accelerated cellular age and degraded neural integrity, as well as reduced performance on executive function tasks, in a sample of U.S. veterans. Lifetime PTSD severity was found to be positively associated with DNA methylation-derived age, and these age estimates were negatively associated with neural integrity in the genu of the corpus callosum and working memory performance. Of note, the mean age in this sample was ~32 years, suggesting that the neurobiological effects of traumatic stress may impair neuropsychological functioning of veteran populations. This genome-scale study represents a genomic approach to PTSD with high public health impact, as it holds the potential to identify individuals who may be most in need of intervention by leveraging peripherally-derived, polygenic epigenetic measurements that are predictive of neural integrity and memory performance. Cohorts within the PGC-PTSD that include both neuroimaging and EWAS data are optimally positioned to combine efforts and pursue similar studies in the future, augmented by the meta-analytic framework currently being employed by both the EWAS and the neuroimaging working groups within the PGC-PTSD.
Data integration and systems biology approaches
Based on current findings, the underlying etiology of PTSD is likely the result of a complex interplay between various molecular systems (73), and to delineate this, a holistic (systems biology) approach which integrates different ‘omics layers’ amongst PTSD cases and trauma-exposed controls may be the next logical step. One of the strengths of the PGC-PTSD is that multiple forms of genomic data have been generated for many datasets, which will allow cross-platform analyses to be performed (Figure 2). Several methods have been proposed for meta-analysis across platforms (74–77), as well as for imputing data to improve the ability to combine datasets (78–82). Such joint analyses of multiple –omics datasets have been previously termed “genomic convergence” and have great potential to inform the genetic architecture of PTSD (83).
Through the co-ordination of the various PGC-PTSD working groups (Figure 2), there is potential to identify DNA methylation patterns that may be giving rise to altered gene expression, and sufficient power to conduct eQTL and meQTL analyses, which assesses whether particular genetic variants alter the levels of expression and methylation of specific genes, respectively. For example, previous studies for PTSD have shown that the risk variant, rs363276 located within an intronic region of SLC18A2, is an eQTL, significantly associated with decreased expression of the genes SLC18A2 and PDZD8 in the dorsolateral prefrontal cortex (DLPFC) of post-mortem human brains (84). Another PTSD risk variant rs717947, located at chromosome 4p15, was shown to be a meQTL (85). These types of analyses provide clues on the etiology of PTSD; however, identifying definitive causal risk factors requires alternate methods, such as Mendelian Randomization (MR), which quantifies causality between a risk factor and a disease outcome by utilizing SNP data as an instrumental variable (86). This technique has been applied to PTSD whereby a causal relationship was identified between plasma dopamine beta-hydroxylase (DBH), an enzyme which catalyzes the synthesis of norepinephrine, on symptoms of re-experiencing (87). Another study investigated the relationship between BMI adjusted weight circumference (WCadj) and PTSD in women and found that an increase in WCadj results in a relative decrease in the risk of developing PTSD (88).
Current data integration efforts will be augmented through the increasing availability of publically available data sets, such as GTex (89) and PsychENCODE (90), which can provide additional annotation for the PTSD-specific findings. For example, recently developed methods such as PrediXcan uses genome-wide variation to predict or “impute” gene expression in test datasets, based on tissue-dependent modeling performed on transcriptome data from reference databases such as GTex. The imputed expression can be tested for association to the phenotype of interest enabling the identification of trait associated loci (91). Additionally, as technologies continue to evolve and become more widely-available for single cell RNA sequencing (92, 93), induced pluripotent stem cell (iPSC)-derived neural progenitor cells (94) and brain organoids (95–97), these technologies can also be integrated into the ongoing PGC-PTSD efforts, particularly with respect to understanding the roles of specific genes and variants in PTSD risk.
Studies of this nature fill a significant gap in the available literature on the complex genetic mechanisms and pathways underlying complex psychiatric disorders, including PTSD. Systems biology approaches will lay the groundwork for future development of more accurate diagnostic methods, improved management and the development of more suitable and individualized treatment strategies for patients.
Conclusion
The promise of finding genetic determinants of psychiatric disorder is identifying etiologic pathways for targeted interventions. However, before this can become a reality, biological validation of genetic findings will be required. Making individual prediction to support the emerging discipline of precision medicine holds the promise of personalized medical decisions driven by an individual’s genetic make-up and environment, other risk factors, and large databases of genotype-phenotype relationships.
Acknowledgments
We thank the research groups contributing to the PGC-PTSD working groups for sharing their data and time to make this work possible, and especially the over 80,000 research participants world-wide who shared personal experiences and biological samples with these research studies.
Financial support was provided by the Stanley Center for Psychiatric Genetics at the Broad Institute, One Mind, Cohen Veterans Bioscience, and the National Institute of Mental Health (NIMH) grants R01-MH111671-01 (RAM, AAK, MAH), and NIMH/U.S. Army Medical Research and Materiel Command support for R01MH106595 (CMN) and R01MH108826 (AKS, MU, CMN). AAK and MAH were supported by the Department of Veterans Affairs’ (VA) Mid-Atlantic Mental Illness Research, Education, and Clinical Center (MIRECC) and the Research & Development and Mental Health Services of the Durham Veterans Affairs Medical Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
The authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.American Psychiatric Association. Trauma- and Stressor-Related Disorders. Diagnostic and statistical manual of mental disorders. (Fifth) 2013 doi: 10.1176/appi.books.9780890425596.991543. [DOI] [Google Scholar]
- 2.Yehuda R, Hoge CW, McFarlane AC, Vermetten E, Lanius RA, Nievergelt CM, et al. Post-traumatic stress disorder. Nat Rev Dis Primers. 2015;1:15057. doi: 10.1038/nrdp.2015.57. [DOI] [PubMed] [Google Scholar]
- 3.Kessler RC, Sonnega A, Bromet E, Hughes M, Nelson CB. Posttraumatic stress disorder in the National Comorbidity Survey. Archives of general psychiatry. 1995;52:1048–1060. doi: 10.1001/archpsyc.1995.03950240066012. [DOI] [PubMed] [Google Scholar]
- 4.True WJ, Rice J, Eisen SA, Heath AC, Goldberg J, Lyons MJ, et al. A twin study of genetic and environmental contributions to liability for posttraumatic stress symptoms. Archives of General Psychiatry. 1993;50:257–264. doi: 10.1001/archpsyc.1993.01820160019002. [DOI] [PubMed] [Google Scholar]
- 5.Stein MB, Jang KJ, Taylor S, Vernon PA, Livesley WJ. Genetic and environmental influences on trauma exposure and posttraumatic stress disorder: A twin study. American Journal of Psychiatry. 2002;159:1675–1681. doi: 10.1176/appi.ajp.159.10.1675. [DOI] [PubMed] [Google Scholar]
- 6.Sartor CE, Grant JD, Lynskey MT, McCutcheon VV, Waldron M, Statham DJ, et al. Common heritable contributions to low-risk trauma, high-risk trauma, posttraumatic stress disorder, and major depression. Archives of general psychiatry. 2012;69:293–299. doi: 10.1001/archgenpsychiatry.2011.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sartor CE, McCutcheon VV, Pommer NE, Nelson EC, Grant JD, Duncan AE, et al. Common genetic and environmental contributions to post-traumatic stress disorder and alcohol dependence in young women. Psychological medicine. 2011;41:1497–1505. doi: 10.1017/S0033291710002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ioannidis JP, Tarone R, McLaughlin JK. The false-positive to false-negative ratio in epidemiologic studies. Epidemiology (Cambridge, Mass) 2011;22:450–456. doi: 10.1097/EDE.0b013e31821b506e. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Borglum AD, Breen G, et al. Psychiatric Genomics: An Update and an Agenda. Am J Psychiatry. 2017 doi: 10.1176/appi.ajp.2017.17030283. appiajp201717030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hyde CL, Nagle MW, Tian C, Chen X, Paciga SA, Wendland JR, et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet. 2016;48:1031–1036. doi: 10.1038/ng.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guffanti G, Galea S, Yan L, Roberts AL, Solovieff N, Aiello AE, et al. Genome-wide association study implicates a novel RNA gene, the lincRNA AC068718.1, as a risk factor for post-traumatic stress disorder in women. Psychoneuroendocrinology. 2013;38:3029–3038. doi: 10.1016/j.psyneuen.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Logue MW, Baldwin C, Guffanti G, Melista E, Wolf EJ, Reardon AF, et al. A genome-wide association study of post-traumatic stress disorder identifies the retinoid-related orphan receptor alpha (RORA) gene as a significant risk locus. Mol Psychiatry. 2013;18:937–942. doi: 10.1038/mp.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie P, Kranzler HR, Yang C, Zhao H, Farrer LA, Gelernter J. Genome-wide association study identifies new susceptibility loci for posttraumatic stress disorder. Biol Psychiatry. 2013;74:656–663. doi: 10.1016/j.biopsych.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nievergelt CM, Maihofer AX, Mustapic M, Yurgil KA, Schork NJ, Miller MW, et al. Genomic predictors of combat stress vulnerability and resilience in U.S. Marines: A genome-wide association study across multiple ancestries implicates PRTFDC1 as a potential PTSD gene. Psychoneuroendocrinology. 2015;51:459–471. doi: 10.1016/j.psyneuen.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 17.Logue MW, Amstadter AB, Baker DG, Duncan L, Koenen KC, Liberzon I, et al. The Psychiatric Genomics Consortium Posttraumatic Stress Disorder Workgroup: Posttraumatic stress disorder enters the age of large-scale genomic collaboration. Neuropsychopharmacology. 2015 doi: 10.1038/npp.2015.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solovieff N, Roberts AL, Ratanatharathorn A, Haloosim M, De Vivo I, King AP, et al. Genetic Association Analysis of 300 Genes Identifies a Risk Haplotype in SLC18A2 for Post-traumatic Stress Disorder in Two Independent Samples. Neuropsychopharmacology. 2014 doi: 10.1038/npp.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almli LM, Srivastava A, Fani N, Kerley K, Mercer KB, Feng H, et al. Follow-up and Extension of a Prior Genome-wide Association Study of Posttraumatic Stress Disorder: Gene x Environment Associations and Structural Magnetic Resonance Imaging in a Highly Traumatized African-American Civilian Population. Biol Psychiatry. 2014 doi: 10.1016/j.biopsych.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein MB, Chen CY, Ursano RJ, Cai T, Gelernter J, Heeringa SG, et al. Genome-wide Association Studies of Posttraumatic Stress Disorder in 2 Cohorts of US Army Soldiers. JAMA Psychiatry. 2016;73:695–704. doi: 10.1001/jamapsychiatry.2016.0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashley-Koch AE, Garrett ME, Gibson J, Liu Y, Dennis MF, Kimbrel NA, et al. Genome-wide association study of posttraumatic stress disorder in a cohort of Iraq-Afghanistan era veterans. J Affect Disord. 2015;184:225–234. doi: 10.1016/j.jad.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Armour C, Mullerova J, Elhai JD. A systematic literature review of PTSD’s latent structure in the Diagnostic and Statistical Manual of Mental Disorders: DSM-IV to DSM-5. Clin Psychol Rev. 2016;44:60–74. doi: 10.1016/j.cpr.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Duncan LE, Ratanatharathorn A, Aiello AE, Almli LM, Amstadter AB, Ashley-Koch AE, et al. Largest GWAS of PTSD (N=20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol Psychiatry. 2017 doi: 10.1038/mp.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X, Stephens M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat Methods. 2014;11:407–409. doi: 10.1038/nmeth.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gratten J, Wray NR, Keller MC, Visscher PM. Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nat Neurosci. 2014;17:782–790. doi: 10.1038/nn.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otowa T, Hek K, Lee M, Byrne EM, Mirza SS, Nivard MG, et al. Meta-analysis of genome-wide association studies of anxiety disorders. Mol Psychiatry. 2016;21:1391–1399. doi: 10.1038/mp.2015.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sumner JA, Duncan LE, Wolf EJ, Amstadter AB, Baker DG, Beckham JC, et al. Letter to the Editor: Posttraumatic stress disorder has genetic overlap with cardiometabolic traits. Psychological medicine. 2017;47:2036–2039. doi: 10.1017/S0033291717000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michopoulos V, Vester A, Neigh G. Posttraumatic stress disorder: A metabolic disorder in disguise? Experimental neurology. 2016;284:220–229. doi: 10.1016/j.expneurol.2016.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Passos IC, Vasconcelos-Moreno MP, Costa LG, Kunz M, Brietzke E, Quevedo J, et al. Inflammatory markers in post-traumatic stress disorder: a systematic review, meta-analysis, and meta-regression. The lancet Psychiatry. 2015;2:1002–1012. doi: 10.1016/S2215-0366(15)00309-0. [DOI] [PubMed] [Google Scholar]
- 31.Mehta D, Binder EB. Gene x environment vulnerability factors for PTSD: the HPA-axis. Neuropharmacology. 2012;62:654–662. doi: 10.1016/j.neuropharm.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 32.Andrews JA, Neises KD. Cells, biomarkers, and post-traumatic stress disorder: evidence for peripheral involvement in a central disease. Journal of neurochemistry. 2012;120:26–36. doi: 10.1111/j.1471-4159.2011.07545.x. [DOI] [PubMed] [Google Scholar]
- 33.Zieker J, Zieker D, Jatzko A, Dietzsch J, Nieselt K, Schmitt A, et al. Differential gene expression in peripheral blood of patients suffering from post-traumatic stress disorder. Mol Psychiatr. 2007;12:116–116. doi: 10.1038/sj.mp.4001905. [DOI] [PubMed] [Google Scholar]
- 34.Segman R, Shefi N, Goltser-Dubner T, Friedman N, Kaminski N, Shalev A. Peripheral blood mononuclear cell gene expression profiles identify emergent post-traumatic stress disorder among trauma survivors. Mol Psychiatr. 2005;10:500–513. doi: 10.1038/sj.mp.4001636. [DOI] [PubMed] [Google Scholar]
- 35.Sarapas C, Cai G, Bierer LM, Golier JA, Galea S, Ising M, et al. Genetic markers for PTSD risk and resilience among survivors of the World Trade Center attacks. Disease markers. 2011;30:101–110. doi: 10.3233/DMA-2011-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yehuda R, Cai G, Golier JA, Sarapas C, Galea S, Ising M, et al. Gene expression patterns associated with posttraumatic stress disorder following exposure to the World Trade Center attacks. Biological psychiatry. 2009;66:708–711. doi: 10.1016/j.biopsych.2009.02.034. [DOI] [PubMed] [Google Scholar]
- 37.O’Donovan A, Sun B, Cole S, Rempel H, Lenoci M, Pulliam L, et al. Transcriptional control of monocyte gene expression in post-traumatic stress disorder. Disease markers. 2011;30:123–132. doi: 10.3233/DMA-2011-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neylan TC, Sun B, Rempel H, Ross J, Lenoci M, O’Donovan A, et al. Suppressed monocyte gene expression profile in men versus women with PTSD. Brain Behav Immun. 2011;25:524–531. doi: 10.1016/j.bbi.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Logue MW, Smith AK, Baldwin C, Wolf EJ, Guffanti G, Ratanatharathorn A, et al. An analysis of gene expression in PTSD implicates genes involved in the glucocorticoid receptor pathway and neural responses to stress. Psychoneuroendocrinology. 2015;57:1–13. doi: 10.1016/j.psyneuen.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehta D, Klengel T, Conneely KN, Smith AK, Altmann A, Pace TW, et al. Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proceedings of the national academy of sciences. 2013;110:8302–8307. doi: 10.1073/pnas.1217750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wingo AP, Almli LM, Stevens JJ, Klengel T, Uddin M, Li Y, et al. DICER1 and microRNA regulation in post-traumatic stress disorder with comorbid depression. Nature communications. 2015;6 doi: 10.1038/ncomms10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breen MS, Maihofer AX, Glatt SJ, Tylee DS, Chandler SD, Tsuang MT, et al. Gene networks specific for innate immunity define post-traumatic stress disorder. Mol Psychiatry. 2015;20:1538–1545. doi: 10.1038/mp.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Breen MS, Tylee DS, Maihofer AX, Neylan TC, Mehta D, Binder E, et al. PTSD Blood Transcriptome Mega-Analysis: Shared Inflammatory Pathways Across Biological Sex and Modes of Trauma. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2017 doi: 10.1038/npp.2017.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith AK, Conneely KN, Kilaru V, Mercer KB, Weiss TE, Bradley B, et al. Differential immune system DNA methylation and cytokine regulation in post‐traumatic stress disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2011;156:700–708. doi: 10.1002/ajmg.b.31212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ressler KJ, Mercer KB, Bradley B, Jovanovic T, Mahan A, Kerley K, et al. Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature. 2011;470:492–497. doi: 10.1038/nature09856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de Los Santos R, et al. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proceedings of the National Academy of Sciences. 2010;107:9470–9475. doi: 10.1073/pnas.0910794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mehta D, Bruenig D, Carrillo-Roa T, Lawford B, Harvey W, Morris CP, et al. Genomewide DNA methylation analysis in combat veterans reveals a novel locus for PTSD. Acta psychiatrica Scandinavica. 2017;136:493–505. doi: 10.1111/acps.12778. [DOI] [PubMed] [Google Scholar]
- 48.Hammamieh R, Chakraborty N, Gautam A, Muhie S, Yang R, Donohue D, et al. Whole-genome DNA methylation status associated with clinical PTSD measures of OIF/OEF veterans. Transl Psychiatry. 2017;7:e1169. doi: 10.1038/tp.2017.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuan PF, Waszczuk MA, Kotov R, Marsit CJ, Guffanti G, Gonzalez A, et al. An epigenome-wide DNA methylation study of PTSD and depression in World Trade Center responders. Transl Psychiatry. 2017;7:e1158. doi: 10.1038/tp.2017.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rutten BPF, Vermetten E, Vinkers CH, Ursini G, Daskalakis NP, Pishva E, et al. Longitudinal analyses of the DNA methylome in deployed military servicemen identify susceptibility loci for post-traumatic stress disorder. Mol Psychiatry. 2017 doi: 10.1038/mp.2017.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bam M, Yang X, Zumbrun EE, Zhong Y, Zhou J, Ginsberg JP, et al. Dysregulated immune system networks in war veterans with PTSD is an outcome of altered miRNA expression and DNA methylation. Scientific reports. 2016;6 doi: 10.1038/srep31209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raison CL, Capuron L, Miller AH. Cytokines sing the blues: inflammation and the pathogenesis of depression. Trends in immunology. 2006;27:24–31. doi: 10.1016/j.it.2005.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maddox S, Kilaru V, Shin J, Jovanovic T, Almli L, Dias B, et al. Estrogen-dependent association of HDAC4 with fear in female mice and women with PTSD. Mol Psychiatr. 2017 doi: 10.1038/mp.2016.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wingo AP, Almli LM, Stevens JS, Klengel T, Uddin M, Li Y, et al. Corrigendum: DICER1 and microRNA regulation in post-traumatic stress disorder with comorbid depression. Nature communications. 2016;7:10958. doi: 10.1038/ncomms10958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Admon R, Milad MR, Hendler T. A causal model of post-traumatic stress disorder: disentangling predisposed from acquired neural abnormalities. Trends in cognitive sciences. 2013;17:337–347. doi: 10.1016/j.tics.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 56.Horvath S. DNA methylation age of human tissues and cell types. Genome biology. 2013;14:3156. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zannas AS, Arloth J, Carrillo-Roa T, Iurato S, Röh S, Ressler KJ, et al. Lifetime stress accelerates epigenetic aging in an urban, African American cohort: relevance of glucocorticoid signaling. Genome biology. 2015;16:266. doi: 10.1186/s13059-015-0828-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome biology. 2015;16:25. doi: 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nature neuroscience. 2013;16:33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolf EJ, Logue MW, Hayes JP, Sadeh N, Schichman SA, Stone A, et al. Accelerated DNA methylation age: Associations with PTSD and neural integrity. Psychoneuroendocrinology. 2016;63:155–162. doi: 10.1016/j.psyneuen.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ratanatharathorn A, Boks MP, Maihofer AX, Aiello AE, Amstadter AB, Ashley-Koch AE, et al. Epigenome-wide association of PTSD from heterogeneous cohorts with a common multi-site analysis pipeline. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2017;174:619–630. doi: 10.1002/ajmg.b.32568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geschwind DH, Flint J. Genetics and genomics of psychiatric disease. Science. 2015;349:1489–1494. doi: 10.1126/science.aaa8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Potkin S, Guffanti G, Lakatos A, Turner J, Kruggel F, Fallon J, et al. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer’s disease. 2009 doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. American Journal of Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 65.Flint J, Timpson N, Munafò M. Assessing the utility of intermediate phenotypes for genetic mapping of psychiatric disease. Trends in neurosciences. 2014;37:733–741. doi: 10.1016/j.tins.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson PM, Andreassen OA, Arias-Vasquez A, Bearden CE, Boedhoe PS, Brouwer RM, et al. ENIGMA and the individual: Predicting factors that affect the brain in 35 countries worldwide. Neuroimage. 2015 doi: 10.1016/j.neuroimage.2015.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Logue MW, van Rooij SJH, Dennis EL, Davis SL, Hayes JP, Stevens JS, et al. Smaller Hippocampal Volume in Posttraumatic Stress Disorder: A Multisite ENIGMA-PGC Study: Subcortical Volumetry Results From Posttraumatic Stress Disorder Consortia. Biol Psychiatry. 2017 doi: 10.1016/j.biopsych.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bremner JD, Randall P, Scott TM, Bronen RA, Seibyl JP, Southwick SM, et al. MRI-based measurement of hippocampal volume in patients with combat-related posttraumatic stress disorder. Am J Psychiatry. 1995;152:973–981. doi: 10.1176/ajp.152.7.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivieres S, Jahanshad N, et al. Common genetic variants influence human subcortical brain structures. Nature. 2015;520:224–229. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hayes JP, Logue MW, Sadeh N, Spielberg JM, Verfaellie M, Hayes SM, et al. Mild traumatic brain injury is associated with reduced cortical thickness in those at risk for Alzheimer’s disease. Brain: a journal of neurology. 2017;140:813–825. doi: 10.1093/brain/aww344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vukojevic V, Kolassa IT, Fastenrath M, Gschwind L, Spalek K, Milnik A, et al. Epigenetic modification of the glucocorticoid receptor gene is linked to traumatic memory and post-traumatic stress disorder risk in genocide survivors. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2014;34:10274–10284. doi: 10.1523/JNEUROSCI.1526-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sadeh N, Spielberg JM, Logue MW, Wolf EJ, Smith AK, Lusk J, et al. SKA2 methylation is associated with decreased prefrontal cortical thickness and greater PTSD severity among trauma-exposed veterans. Mol Psychiatry. 2016;21:357–363. doi: 10.1038/mp.2015.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu L, Bhattacharyya A, Kurali E, Anderson A, Menius A, Lee K. Review of Integrative Analysis Challenges in Systems Biology. Statistics in Biopharmaceutical Research. 2011;3:561–568. [Google Scholar]
- 74.Chang LC, Lin HM, Sibille E, Tseng GC. Meta-analysis methods for combining multiple expression profiles: comparisons, statistical characterization and an application guideline. BMC Bioinformatics. 2013;14:368. doi: 10.1186/1471-2105-14-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim J, Patel K, Jung H, Kuo WP, Ohno-Machado L. AnyExpress: integrated toolkit for analysis of cross-platform gene expression data using a fast interval matching algorithm. BMC bioinformatics. 2011;12:75. doi: 10.1186/1471-2105-12-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Campain A, Yang YH. Comparison study of microarray meta-analysis methods. BMC bioinformatics. 2010;11:408. doi: 10.1186/1471-2105-11-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ramasamy A, Mondry A, Holmes CC, Altman DG. Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS medicine. 2008;5:e184. doi: 10.1371/journal.pmed.0050184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chrétien S, Guyeux C, Conesa B, Delage-Mouroux R, Jouvenot M, Huetz P, et al. A Bregman-proximal point algorithm for robust non-negative matrix factorization with possible missing values and outliers-application to gene expression analysis. BMC bioinformatics. 2016;17:284. doi: 10.1186/s12859-016-1120-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang J, Gamazon ER, Pierce BL, Stranger BE, Im HK, Gibbons RD, et al. Imputing gene expression in uncollected tissues within and beyond GTEx. The American Journal of Human Genetics. 2016;98:697–708. doi: 10.1016/j.ajhg.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang Y, Xu Z, Song D. BMC bioinformatics. BioMed Central Ltd; 2016. Missing value imputation for microRNA expression data by using a GO-based similarity measure; p. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H, Zhao C, Shao F, Li GZ, Wang X. A hybrid imputation approach for microarray missing value estimation. BMC Genomics. 2015;16(Suppl 9):S1. doi: 10.1186/1471-2164-16-S9-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liew AW, Law NF, Yan H. Missing value imputation for gene expression data: computational techniques to recover missing data from available information. Brief Bioinform. 2011;12:498–513. doi: 10.1093/bib/bbq080. [DOI] [PubMed] [Google Scholar]
- 83.Noureddine MA, Li YJ, van der Walt JM, Walters R, Jewett RM, Xu H, et al. Genomic convergence to identify candidate genes for Parkinson disease: SAGE analysis of the substantia nigra. Movement disorders. 2005;20:1299–1309. doi: 10.1002/mds.20573. [DOI] [PubMed] [Google Scholar]
- 84.Bharadwaj RA, Jaffe AE, Chen Q, Deep-Soboslay A, Goldman AL, Mighdoll MI, et al. Genetic risk mechanisms of posttraumatic stress disorder in the human brain. Journal of neuroscience research. 2018;96:21–30. doi: 10.1002/jnr.23957. [DOI] [PubMed] [Google Scholar]
- 85.Almli LM, Stevens JS, Smith AK, Kilaru V, Meng Q, Flory J, et al. A genome-wide identified risk variant for PTSD is a methylation quantitative trait locus and confers decreased cortical activation to fearful faces. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2015;168b:327–336. doi: 10.1002/ajmg.b.32315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pasaniuc B, Price AL. Dissecting the genetics of complex traits using summary association statistics. Nat Rev Genet. 2017;18:117–127. doi: 10.1038/nrg.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mustapic M, Maihofer AX, Mahata M, Chen Y, Baker DG, O’Connor DT, et al. The catecholamine biosynthetic enzyme dopamine beta-hydroxylase (DBH): first genome-wide search positions trait-determining variants acting additively in the proximal promoter. Hum Mol Genet. 2014;23:6375–6384. doi: 10.1093/hmg/ddu332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Polimanti R, Amstadter AB, Stein MB, Almli LM, Baker DG, Bierut LJ, et al. A putative causal relationship between genetically determined female body shape and posttraumatic stress disorder. Genome medicine. 2017;9:99. doi: 10.1186/s13073-017-0491-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Consortium G. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, et al. The PsychENCODE project. Nature neuroscience. 2015;18:1707–1712. doi: 10.1038/nn.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cadwell CR, Scala F, Li S, Livrizzi G, Shen S, Sandberg R, et al. Multimodal profiling of single-cell morphology, electrophysiology, and gene expression using Patch-seq. Nat Protoc. 2017;12:2531–2553. doi: 10.1038/nprot.2017.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu YE, Pan L, Zuo Y, Li X, Hong W. Detecting Activated Cell Populations Using Single-Cell RNA-Seq. Neuron. 2017;96:313–329.e316. doi: 10.1016/j.neuron.2017.09.026. [DOI] [PubMed] [Google Scholar]
- 94.Schwartzentruber J, Foskolou S, Kilpinen H, Rodrigues J, Alasoo K, Knights AJ, et al. Molecular and functional variation in iPSC-derived sensory neurons. Nat Genet. 2017 doi: 10.1038/s41588-017-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang P, Mokhtari R, Pedrosa E, Kirschenbaum M, Bayrak C, Zheng D, et al. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells. Molecular autism. 2017;8:11. doi: 10.1186/s13229-017-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Brauninger M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A. 2015;112:15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jo J, Xiao Y, Sun AX, Cukuroglu E, Tran HD, Goke J, et al. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell. 2016;19:248–257. doi: 10.1016/j.stem.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]


