SUMMARY
Crossing over between homologous chromosomes during meiosis repairs programmed DNA double-strand breaks, ensures proper segregation at meiosis I [1], shapes the genomic distribution of nucleotide variability in populations, and enhances the efficacy of natural selection among genetically linked sites [2]. Between closely related Drosophila species, large differences exist in the rate and chromosomal distribution of crossing over. Little, however, is known about the molecular genetic changes or population genetic forces that mediate evolved differences in recombination between species [3, 4]. Here we show that a meiosis gene with a history of rapid evolution acts as a trans-acting modifier of species differences in crossing over. In transgenic flies, the dicistronic gene, mei-217/mei-218, recapitulates a large part of the species differences in the rate and chromosomal distribution of crossing over. These phenotypic differences appear to result from changes in protein sequence not gene expression. Our population genetics analyses show that the protein-coding sequence of mei-218, but not mei-217, has a history of recurrent positive natural selection. By modulating the intensity of centromeric and telomeric suppression of crossing over, evolution at mei-217/-218 has incidentally shaped gross differences in the chromosomal distribution of nucleotide variability between species. We speculate that recurrent bouts of adaptive evolution at mei-217/-218 might reflect a history of coevolution with selfish genetic elements.
RESULTS AND DISCUSSION
Despite its functional and evolutionary benefits [1, 5], crossing over entails risks. First, selfish repetitive DNA sequences (e.g., transposons) distributed throughout the genome present the risk of non-homologous ectopic exchange [6, 7] which can give rise to deleterious de novo duplications and deletions in ≥2% of meioses in D. melanogaster [8]. Second, crossovers in centromere- and telomere-proximal regions can increase the risk of improper chromosomal segregation, resulting in breakage and non-disjunction [9]. The optimal rate and distribution of crossing over may therefore evolve to balance the benefits of recombination against the costs of ectopic exchange and missegregation.
Between Drosophila melanogaster and its closely related species, D. mauritiana, appreciable differences in the rate and chromosomal distribution of crossing over have evolved despite comparable genome sizes and karyotypes [10]. In D. mauritiana, the total genetic map lengths of the three major chromosomes, X, 2, and 3, are 1.7-, 1.5-, and 2.1-fold longer, respectively, than those in D. melanogaster [10]. Some of these differences in genetic map length are attributable to differences in the chromosomal distribution of recombination: crossing over is suppressed at considerable distances from telomere- and especially centromere-proximal regions in D. melanogaster [11], whereas the range of these effects is narrower in D. mauritiana [10]. How and why genetic maps evolve is almost entirely unknown [3].
We sought to determine the genetic basis and evolutionary causes of these species differences in crossover rate and distribution. To identify candidate genes, we surveyed the molecular evolution of genes previously identified in classical screens for mutations that disrupt meiosis in D. melanogaster [11–13]. These mutations disrupt genes that function in synaptonemal complex formation, double-strand break (DSB) formation, DSB repair, establishment of crossover intermediates, and resolution of crossover intermediates [13–15]. We generated sequence alignments for a set of 35 meiosis genes and performed an evolutionary screen for unusually high protein-coding sequence divergence between D. mauritiana and D. melanogaster. Among the 35 genes, mei-218 is an outlier with the highest dN (0.094) and dN/dS (0.632; Table S1), placing it among the most diverged protein-coding sequences in the genome (dN and dN/dS are in the 99.97%- and 97%-percentiles, respectively; Figure S1). Previous analyses have established that the MEI-218 protein has a mini-chromosome maintenance (MCM) domain and interacts with several other meiosis-specific MCM proteins to form a so-called mei-MCM complex [16]. In mei-218 mutant females, synaptonemal complex (SC) formation, DSB formation, and recombination via gene conversion all proceed normally, whereas the rate of crossing over is reduced by ≥90%, the number of spherical recombination nodules is reduced (with those remaining often having abnormal morphology), and the rate of chromosomal nondisjunction is elevated accordingly [12, 14, 15, 17]. During repair of DSBs, mei-218 appears to function after strand invasion but prior to crossover resolution [15, 18]. The MEI-218 protein is thus necessary for the establishment and/or stabilization of heteroduplex crossover intermediates [19]. Its inferred function and rapid sequence evolution together suggest that mei-218 is a reasonable candidate contributor to the evolved species difference in crossing over between D. mauritiana and D. melanogaster.
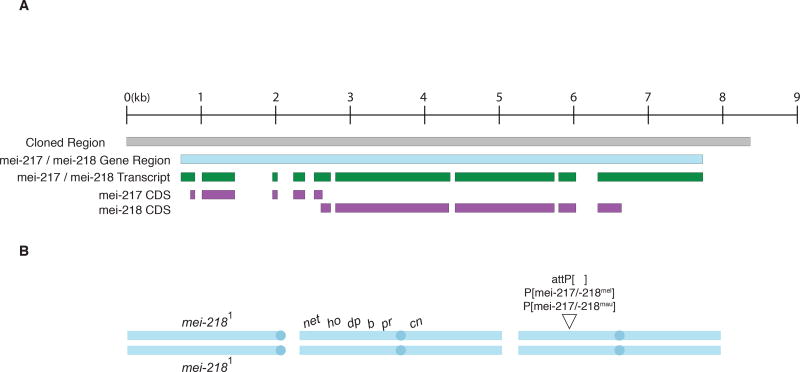
To test the functional consequences of interspecific sequence divergence at mei-218, we assayed the effects of wildtype D. melanogaster and D. mauritiana alleles using a transgenic approach (Figure 1A,B). A dicistronic gene encodes both the MEI-217 and MEI-218 proteins from a single transcriptional unit with open reading frames that overlap by seven codons, in different reading frames, and with separate translation initiation sites (Figure 1A; ref. [20]). We therefore cloned homologous mei-217/mei-218 (hereafter, mei-217/-218) gene regions, including the mei-217 and mei-218 coding sequences and all 5’- and 3’-flanking noncoding sequence, from D. melanogaster and D. mauritiana into separate attB-P[acman] vectors and used site-specific integration to place each transgene construct into a common attP insertion site on chromosome arm 3R (75A10) in D. melanogaster (Figure 1A,B; see STAR Methods for details). We crossed the transgenes into separate but largely identical homozygous mei-2181 loss-of-function mutant genetic backgrounds (Figure S2), yielding two D. melanogaster stocks: mei-2181; P[mei-217/-218mel], which serves as a positive control; and mei-2181; P[mei-217/-218mau] (Figure 1B). We then estimated crossover frequencies among six visible markers that span chromosome arm 2L and the centromere, scoring progeny from replicate mei-2181; net ho dp b pr cn/+ + + + + +; P[mei-217/-218mel]/+ females (n=13 crosses, 2,103 progeny) and, separately, from replicate mei-2181; net ho dp b pr cn / + + + + + +; P[mei-217/-218mau]/+ females (n=13 crosses, 2,369 progeny; Figure 1B; see STAR Methods for details). In wildtype D. mauritiana, the total genetic distance between net and cn is ~1.4-fold longer than in D. melanogaster [10].
Figure 1. The mei-217/mei-218 gene region and genotypes assayed for crossing over.
(A) Within the ~8.5-kb region cloned (gray) the mei-217/-218 gene (light blue) on chromosome X (15E5) gives rise to a single dicistronic transcript (green) that encodes both the MEI217 and MEI218 proteins from different exons and different translation initiation sites (purple; ref. [20]). (B) Three genotypes were used to test if the D. mauritiana and D. melanogaster alleles of mei-217/-218 mediate species differences in rates of crossing over: females with no transgene (a negative control); females with a transgene of a D. melanogaster mei-217/-218 allele (a positive control); and females with a transgene of a D. mauritiana mei-217/-218 allele. The two transgenes were inserted into the same position on chromosome 3L (75A10). The endogenous mei-2181 allele contains a nonsense mutation [14]. Crossover frequencies were scored among six visible markers spanning the left arm of chromosome 2 and the centromere: net (net), decapentaplegic (ho), dumpy (dp), black (b), purple (pr), cinnabar (cn). For additional details on genotype construction, see STAR Methods and Figure S2.
There are three possible outcomes: (i) despite considerable interspecific sequence divergence, the two alleles might be functionally equivalent, such that mei-217/-218mau rescues the mei-218 mutant phenotype and produces D. melanogaster-like rates of crossing over; (ii) the divergent mei-217/-218mau allele might be non-functional in D. melanogaster— a kind of molecular incompatibility between species— and fail to rescue the mei-218 mutant phenotype; or (iii) mei-217/-218mau might rescue the mei-218 mutant phenotype and produce elevated, D. mauritiana-like, rates of crossing over. As expected, crossing over is reduced in mutant mei-218 females (Table 1; refs. [12, 14]), yielding a genetic map length that is reduced by 95% relative to that of the positive control transgene mei-217/-218mel, which fully rescues the mutant mei-218 phenotype (Table 1). This finding confirms that a single copy of the mei-217/-218mel transgene is sufficient to rescue wildtype genetic map distances in otherwise mei-218 mutant females [14]. The mei-217/-218mau transgene also rescues the mei-2181 mutant phenotype but significantly increases the rate of crossing over relative to the positive control. The total genetic map length is increased 1.23-fold in mei-217/-218mau females relative to mei-217/-218mel females (Table 1; 95% confidence intervals = 1.13–1.31; P = 0.0002), accounting for ~43% of the wildtype species difference in the total net to cn map distance.
Table 1.
The mei-217/-218 allele of D. mauritiana alters the rate and patterning crossing over relative to that of D. melanogaster.
| Genetic Interval |
mei-2181 | mei-218mel | mei-218mau |
mel vs. mau fold-change |
P-value [Power] |
|---|---|---|---|---|---|
| net-ho | 0 [0] | 2.01 [1.45] | 3.70 [1.38] | 1.84 | 0.005 [0.72] |
| ho-dp | 0 [0] | 5.28 [1.95] | 6.02 [1.94] | 1.14 | 0.349 [0.15] |
| dp-b | 0.77 [1.07] | 22.20 [4.13] | 24.27 [5.04] | 1.09 | 0.250 [0.20] |
| b-pr | 0.41 [0.57] | 10.65 [2.62] | 14.53 [2.93] | 1.36 | 0.001 [0.83] |
| pr-cn | 0.84 [1.15] | 4.13 [1.32] | 5.49 [1.94] | 1.33 | 0.034 [0.41] |
| Total Map Length | 2.02 [1.72] | 44.27 [6.00] | 54.01 [5.38] | 1.23 | 0.0002 [0.90] |
| Total Progeny | 330 | 2103 | 2369 | -- | |
| E0 | 0.958 | 0.205 | 0.130 | 0.64 | |
| E1 | 0.042 | 0.685 | 0.686 | 1.00 | |
| E2 | 0 | 0.107 | 0.160 | 1.51 | |
| E3 | 0 | 0.004 | 0.024 | 6.21 |
For each genotype, the means and standard deviations (in brackets) of crossover frequencies for five genetic intervals measured in mei-2181 mutant and two transgenic genotypes (see STAR Methods and Figure S2). P-values are derived from unpaired t-tests, and we estimated the power (in brackets) associated with each test. E0, E1, E2 and E3 are the estimated frequencies of tetrads with zero, one, two, and three inferred crossovers, respectively (see STAR Methods for details).
Notably, the mei-217/-218mau-mediated increase in genetic map length is not uniform across genetic marker intervals. Those intervals with significantly increased crossover rates occur in telomere- and centromere-proximal regions (1.84-fold for net-ho and 1.36-fold for b-pr) or span the centromere (1.33-fold for pr-cn; Table 1). No difference is expected in crossover rates in the medial regions of 2L (ref. [10]) and, while the two medial intervals scored have higher rates of crossing over in mei-217/-218mau than mei-217/-218mel females, neither differs significantly (P≥0.2501, Table 1). (We note, however, that our statistical power is relatively weak for these two non-significant intervals, ≤0.20; Table 1). We next tested if species differences in mei-217/-218 gene expression might mediate these differences in crossing over using quantitative reverse transcription PCR (qRT-PCR). Assaying expression in ovaries from 3–5 day-old females, we find no difference in gene expression between wildtype D. melanogaster and D. mauritiana females or mei-217/-218mel and mei-217/-218mau transgenic females (Figure S3). These findings suggest that the observed differences in the rate and distribution of crossing over are attributable to evolution of the mei-217/-218 protein-coding sequence, not to its gene expression level.
The number of crossovers formed among homologous chromosomes of a tetrad is highly regulated [13, 21]. Crossover assurance mechanisms promote the formation of one crossover per tetrad, and crossover interference mechanisms inhibit the formation of multiple crossovers in close proximity on a chromosome arm [22]. Consistent with regulation, we find that the distributions of the number of crossovers per tetrad are under-dispersed relative to Poisson expectations for both mei-217/-218mel and mei-217/-218mau transgenes (χ2-test, df=5, P≤e−200, Table 1). The number of crossovers per tetrad also differs between mei-217/-218mel and mei-217/-218mau females (χ2-test, df=3, P≤e−80; Table 1). An average of 1.08 crossovers per tetrad occurs in mei-217/-218mau females versus 0.91 in mei-217/-218mel females. We tested if the mei-217/-218mau-mediated increase in the average number of crossovers per tetrad is achieved by decreasing the incidence of tetrads with no crossovers (E0), increasing the incidence of those with single crossovers (E1) or multiple crossovers (E≥2), or a combination [23]. We find that the incidence of E1 tetrads is the same for mei-217/-218mau and mei-217/-218mel females (Table 1). However, the incidence of E0 tetrads in mei-217/-218mau females is only 0.64-fold that in mei-218mel females, whereas the incidences of E2 and E3 tetrads are 1.5- and 6.2-fold higher, respectively (Table 1). The resulting increase in the occurrence of multiple crossovers accounts for ~59% of the observed increase in genetic map length. Estimating crossover interference for the two largest adjacent intervals (dp-b-pr) shows that interference is ~36% weaker for mei-217/-218mau than for mei-217/-218mel females (0.508 versus 0.793; Mann-Whitney P=0.0085). These results show that the mei-217/-218mau transgene simultaneously strengthens crossover assurance and weakens crossover interference.
As our transgenic flies are genetically identical (or nearly so), the observed differences in crossover rate and distribution are not readily attributable to differences in genetic background or to any aspect of meiosis not affected by mei-217/-218. How mei-217/-218 regulates the number and distribution of crossovers is not known [14–16]. One possibility is that, just as the canonical MCM complex functions as a holoenzyme to facilitate DNA synthesis into replication forks [24], the mei-MCM complex might facilitate DNA synthesis into the forks of heteroduplex DNA structures as required for the formation and stabilization of crossover intermediates. If heteroduplex structures are stabilized more effectively in mei-217/-218mau females, then more heteroduplexes might achieve second-end capture and be resolved as crossover events versus dissolve and result in non-crossover gene conversion events. Given the shared genetic backgrounds of our transgenic flies, we infer that mei-217/-218mau increases the probability that a DSB will be repaired as a crossover (versus a non-crossover gene conversion) than mei-217/-218mel. As a result, crossover assurance is strengthened (fewer E0 tetrads) whereas the intensities of crossover interference and centromere (telomere) suppression are diminished (see above; ref. [22])
Why mei-218 has evolved so rapidly between these closely related species is unclear. Rapid sequence evolution can result from relaxed functional constraints or from divergent positive natural selection. To investigate the population genetic forces responsible for the rapid evolution of mei-218, we studied nucleotide polymorphism and divergence in resequence data obtained from 20 D. melanogaster samples from Rwanda and 8 D. mauritiana samples from Mauritius. There is no evidence for recent hard selective sweeps in the mei-217/-218 gene regions, as levels of polymorphism and the site frequency spectra are typical for these species (Table 2). However, two analyses provide evidence for a history of recurrent positive natural selection. First, using lineage-specific McDonald-Kreitman tests [25], we find that D. melanogaster mei-218, but not mei-217, has an excess of nonsynonymous substitutions (Table 2). Second, to localize the signals of positive selection, we implemented gammaMap [26], a powerful phylogenetics-population genetics method that combines information from lineage-specific substitutions and the site frequency spectrum from each species to infer the posterior probability of positive selection at individual codons. The gammaMap results show that the probability of positive selection is >0.5 for 99 and 130 codons of mei-218 in D. melanogaster and D. mauritiana lineages, respectively (Figure 2A; Table 2). These signals of positive selection are restricted to mei-218 almost exclusively, as only one codon in mei-217 shows evidence of positive selection (Figure 2A; Table 2). Within mei-218, positively selected codons are concentrated in regions encoding the N-terminal basic region and the middle acidic region but appear absent from the C-terminal MCM-domain region (Figure 2A). The small MCM-domain itself has no detected positively selected substitutions in either lineage.
Table 2.
Population genetic evidence for positive selection at mei-218, not mei-217
| D. mauritiana | D. melanogaster | |||
|---|---|---|---|---|
|
|
||||
| mei-217 | mei-218 | mei-217 | mei-218 | |
| n | 8 | 20 | ||
| Gene region (kb) | 8.2 | 8.2 | ||
| θw | 0.0064 | 0.0044 | ||
| π | 0.0058 | 0.0059 | ||
| Tajima’s D | −0.530 | −1.044 | ||
| Coding sequence length (bp) | 840 | 3,561 | 840 | 3,561 |
| Nonsynon. polymorphisms | 2 | 30 | 5 | 17 |
| Synonymous polymorphisms | 10 | 15 | 7 | 18 |
| Nonsynon. substitutions | 3 | 67 | 8 | 53 |
| Synonymous substitutions | 7 | 39 | 7 | 22 |
| Lineage-specific MK test, PFET | 0.624 | 0.715 | 0.704 | 0.033 |
| P0.50 (γ > 0) | 0 | 130 | 1 | 99 |
| P0.75 (γ > 0) | 0 | 101 | 0 | 80 |
| P0.95 (γ > 0) | 0 | 43 | 0 | 14 |
Summary statistics for the mei-217/mei-218 gene region and the two coding sequences for D. mauritiana and D. melanogaster samples. The combined gene region was used to obtain summaries of the level of polymorphism (θW and π; [36, 37]) and the site frequency spectra (Tajima’s D; [38]). Lineage-specific McDonald-Kreitman [25] tests were performed with the coding sequences of D. yakuba mei-217 and mei-218 as outgroup sequences to polarize substitutions along the D. melanogaster and D. mauritiana lineages. Consistent with the absence of a gene expression difference (see main text; Figure S3), McDonald-Kreitman tests contrasting polymorphisms and fixed differences from noncoding sequences (5’-UTR, 3’-UTR, and introns) with those at synonymous positions revealed no evidence for recurrent positive selection. Positions with evidence of multiple substitutions were excluded, as the inferred ancestral state is ambiguous by simple parsimony. We used gammaMap to estimate the number, posterior probability, and location of positively selected substitutions in mei-217 and mei-218 (see Figure 2A). For additional details, see STAR Methods, Table S1, and Figure S1.
Figure 2. The distribution of positively selected codons in mei-217 and mei-218 in D. melanogaster and D. mauritiana.
(A) The N-terminal basic and central acidic regions of MEI-218 are encoded by codons 1–500 and 500–800, respectively, and the MCM domain is encoded by codons 1019–1124 (ref. [35]). Nearly all of the positively selected codons fall within the first 800 codons of mei-218, and none occur in the MCM domain. In mei-217, a single codon in D. melanogaster has a 0.52 probability of positive selection, whereas no codons in D. mauritiana mei-217 have a ≥0.50 probability of positive selection. Codon substitutions are indicated as red (nonsynonymous) and blue (synonymous) circles. (B) The standardized density of single nucleotide polymorphisms (SNPs) per site in 50-kb windows across chromosome 2 plotted for D. melanogaster (blue) and D. mauritiana (red) with loess-smoothed curves. For each chromosome arm (2L and 2R) and species, SNP densities were standardized by the respective maximum value. Gray triangles show the positions of the six visible markers used to score crossover frequencies. See STAR methods for more details.
To explain recurrent bouts of adaptive evolution at mei-218, which has accumulated 218 fixed nonsynonymous differences between species, would seem to require a model of adaptation to a moving fitness optimum. One possibility is that adaptive evolution at mei-218 results from selection on a function other than recombination in females. The mei-217/-218 gene is expressed at high levels in testes, although its function in males, which are achiasmate, is unknown (mei-2181 males are fertile [12]). Another possibility is that mei-217/-218-mediated change in recombination rates may have evolved in response to a history of recurrent meiotic drive in the female germline, either increasing or decreasing the rate of crossing over depending on the timing of drive (MI or MII) and the genetic linkage between mei-217/-218 and drive alleles [27]. Finally, mei-217/-218-mediated change in recombination rates could reflect adaptation to species differences in transposon abundance. There are two competing models here. First, as the transposon content of the D. melanogaster genome is several-fold higher than that of D. mauritiana [28], reduced rates of crossing over in D. melanogaster may have evolved to mitigate a higher risk of ectopic exchange between non-homologous transposon insertions [29]. Under this model, the rate and distribution of recombination might evolve frequently to balance the benefits of crossing over versus the risk of ectopic exchange arising from historically fluctuating, species-specific transposon loads [29–31]. Second, and alternatively, once transposon copy numbers reach equilibrium, selection may favor the evolution of increased crossover rates, facilitating the elimination of transposons via ectopic exchange [32].
Whatever the cause(s), mei-217/-218-mediated changes in crossing over have, as an incidental by-product, contributed to species differences in the chromosomal distribution of nucleotide variability. Recurrent positive and negative selection both reduce nucleotide variability at genetically linked sites [33, 34]. These so-called hitchhiking effects are pervasive but ameliorated by recombination, giving rise to genome-wide correlations between nucleotide variability and local recombination rates in many taxa [2]. In D. melanogaster, the domain of crossover suppression extends further from the centromere than in D. mauritiana [10], a difference attributable in part to evolution at mei-217/-218 (Table 1). We find that, consequently, levels of nucleotide variability recover less quickly with physical distance from the centromere in D. melanogaster than in D. mauritiana (Figure 2B; Figure S4). Taken together, our results show that adaptive protein evolution at the mei-217/-218 gene has contributed to change in the recombination landscapes of D. mauritiana and D. melanogaster and incidentally shaped species differences in the chromosomal distribution of nucleotide variability.
STAR Methods
Contact For Reagent And Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Daven Presgraves (daven.presgraves@rochester.edu).
Experimental Model And Subject Details
We used the following fly strains in this study: the genome reference strains D. mauritiana w (mau12 w) and D. melanogaster (iso-1); a mei-2181 loss-of-function mutation-bearing strain of D. melanogaster (y mei-2181 / FM7c; Spapol/+); and a multiply marked second chromosome strain of D. melanogaster (net dpp[ho] dp b pr cn). The latter two were generously provided by Jeff Sekelsky (University of North Carolina). We refer to the y mei-2181 chromosome as simply mei-2181 below and in Figure S2. All strains and crosses described were set on standard cornmeal-agarose Drosophila medium and kept in an incubator at 24C.
Method Details
Generating transgenic flies
To create transgenic flies we used the ΦC31 integrase-mediated transgenesis system, which allows for site-specific integration [39]. The full-length mei-217/-218 gene, and all of the flanking 5’ and 3’ noncoding regions, was PCR-amplified with Expand Long Range dNTPack PCR System (Sigma-Aldrich Co., Carlsbad, CA) from the genome reference strains, D. mauritiana w (mau12 w) and D. melanogaster (iso-1). Both clones are anchored in the sequences of the neighboring protein codng genes, CG5004 and RpS5a. The PCR products were cloned into a pCR-XL TOPO vector (Invitrogen Inc., Carlsbad, CA). To identify possible PCR-induced mutations, we sequenced clones for each allele and compared them to sequences amplified from genomic DNA. All mutations were corrected using the QuikChange Lightning Multi Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA). Mutation-free clones were confirmed by sequencing. We then cut the mei-217/-218 insert from the pCR-XL TOPO vector with NotI (New England BioLabs, Ipswich, MA) and subcloned into an attB[Pacman]-ApR vector obtained from the Drosophila Genomics Resource Center (Bloomington, IN). The constructs were introduced into D. melanogaster y w; PBac[y+-attP-9A]VK00018 flies, which have an attP transgene landing site at cytological position 75A10 on chromosome 3L, via injections performed by Best Gene Inc. (Chino Hills, CA). The attB-P[w+ mei-218mel]-ApR and attB-P[w+ mei-218mau]-ApR transgenic flies (for simplicity, hereafter referred to as P[mei-217/218mel] and P[mei-217/-218mau], respectively) were then made homozygous and maintained as stocks.
Measuring crossover rates
We estimated crossover rates for a multiply marked second chromosome in three genotypic backgrounds: (1) mei-2181, (2) mei-2181; P[mei-217/-218mel]/+, (3) mei-2181; P[mei-217/-218mau]/+. The crossing scheme used to generate these flies is shown in Figure S2. We first constructed mei-2181/ FM6; net ho dp b pr cn females (Figure S2). Then, in a separate crossing scheme, we introduced the transgene-bearing third chromosomes into a mutant mei-2181 background, constructing mei-2181/Y; P[mei-217/-218mel] and mei-2181/Y; P[mei-217/-218mau] males (Figure S2). Finally, we crossed the mei-2181/ FM6; net ho dp b pr cn females with the mei-2181/Y; P[mei-217/-218mel] or mei-2181/Y; P[mei-217/-218mau] males to generate three female genotypes (Figure S2):
mei-2181; net ho dp b pr cn/ + + + + + +; / PBac[y+-attP-9A]VK00018/+;
mei-2181; net ho dp b pr cn/ + + + + + +; P[mei-217/-218mel]/ +; and
mei-2181; net ho dp b pr cn/ + + + + + +; P[mei-217/-218mau]/ +.
To estimate crossover frequencies, we crossed the three female genotypes above to net ho dp b pr cn males and scored the progeny for all markers. To estimate tetrad frequencies, E0, E1, E2 and E3, we used the algebraic methods of Weinstein [23]. For each cross, we collected ~10 virgin mei-2181; net ho dp b pr cn/ + + + + + +; P[mei-217/-218mau]/+ females, aged them for three to five days, and crossed them to ~10 net ho dp b pr cn males that were aged for at least two days. As a positive control, we followed the same procedure for mei-2181; net ho dp b pr cn/ + + + + + +; P[mei-217/-218mel]/+ females. As a negative control, we followed the same procedure for mei-2181; net ho dp b pr cn/ + + + + + +; PBac[y+-attP-9A]VK00018/+ females lacking the transgene. After five days, parents were dumped and the vials were hydrated with a solution of 0.5% propionic acid.
Expression analysis
We used quantitative reverse transcription PCR (qRT-PCR) to measure mei-217/-218 expression in wildtype D. melanogaster (iso-1) and D. mauritiana (mau12 w) flies as well as transgenic flies bearing either P[mei-217/-218mel] and P[mei-217/-218mau] alleles. Five ovaries from 3-5 day old virgins were dissected into Ringer’s Solution for a total of five biological replicates per genotype. RNA was extracted using the Nucleospin RNA XS kit (Clontech, Mountain View, CA). cDNA was synthesized from the SuperScript III kit (Invitrogen Inc., Carlsbad, CA). All qRT-PCR primers were designed to bind to regions lacking species-specific sequence differences and optimized to 92%–107% efficiency. For all reactions 2µl of cDNA was used in a 20µl qRT-PCR reaction with SYBR-Green I nucleic acid gel stain (Invitrogen Inc., Carlsbad, CA). Two technical replicate qRT-PCR reactions were run for each biological replicate. RpL32 was used as a control gene. All samples were run on a single plate. Ct values were averaged across technical replicate wells for each biological replicate. Normalized Ct values were determined by subtracting mei-218 Ct values from RpL32 values.
Quantification And Statistical Analysis
Evolutionary screen of meiosis genes
We performed a screen of 35 meiosis genes to identify those with high protein-coding sequence divergence between D. mauritiana and D. melanogaster (Table S1). Our nucleotide alignments consisted of the FlyBase reference CDS for D. melanogaster and the orthologous sequence from a whole-genome reference assembly of the mau12 w strain of D. mauritiana [40]. From the BAM file of the mau12 w reads mapped to the D. melanogaster genome, we used samtools and bcftools [41] to obtain mau12 w fasta sequence for the gene span coordinates for D. melanogaster in FlyBase. The D. melanogaster and D. mauritiana CDS alignment was compiled by hand in Geneious 6.1.8 (Biomatters, Aukland, New Zealand), and the Jukes-Cantor corrected synonymous and nonsynonymous divergence was obtained in DnaSP v. 5 (ref. [42]) using the method of Nei and Gojobori[43]. In addition, we calculated the “alignment percentage”, as the percent of the alignment length to the full length of the D. melanogaster CDS. Repetitive sequence or genomic regions that are highly divergent between the species may have low read coverage or low mapping quality in the reference-based assembly. These regions would be masked or missing from the D. mauritiana fasta sequence, thereby decreasing the alignment length. Initially, mei-218 had the lowest alignment percentage (77.25%). After performing a local reassembly of the mei-218 region using the D. mauritiana sequence as our reference (see Population genetic analyses section below), the alignment percentage increased to 98.6%.
Population genetic analyses of mei-217/mei-218
We sampled 8 lines of D. mauritiana: 7 isofemale lines collected in 2006 (kindly donated by Maria Ramos-Womack) that were sib-mated for a minimum of 9 generations, and the inbred genome reference strain, mau12 w. Genomic DNA extraction and library preparation were performed as previously described [40, 44]. For each line, we obtained paired-end sequence reads from a single lane on an Illumina Genome Analyzer II. Sequence reads were 75bp long, except for those of mau12 w, which were 86bp long. The number of reads per line ranged from 75.6 million to 89.4 million. We performed an iterative reference assembly using an 8.2 kb region of the X chromosome containing the mei-217/-218 gene region from mau12 w as our reference sequence, obtained by PCR and Sanger sequencing (primers and conditions available by request). For each D. mauritiana line, we mapped the Illumina reads to the reference using the software BWA [41]. After the first round of assembly, we visualized the assembly using the program, Geneious 6.1.8 (Biomatters, Aukland, New Zealand) and identified “low coverage” regions in which the number of reads mapping per base was more than two standard deviation below the mean. We determined that these regions contained indels or copy number variants with respect to the mau12 w reference sequence. With PCR and Sanger sequencing, we obtained sequence contigs across the low coverage regions, which were aligned to the consensus sequence of the first BWA assembly to create a “hybrid consensus” sequence for each line. This “hybrid consensus” sequence was then used as the reference sequence for another round of BWA assembly using the original reads. From the second assembly, we obtained a final consensus sequence for each line using a consensus cut off of 75%.
For D. melanogaster, we obtained fasta sequences for the corresponding region on the X chromosome (bases 17135985-17142990) in 20 Rwandan lines through the Drosophila Population Genetics Project (http://www.dpgp.org/). Although 27 Rwandan genomes are available, we chose the 20 lines with the fewest masked bases in this genomic region. These data were also generated from Illumina reads mapped to the D. melanogaster reference genome using BWA. FastQ sequences were obtained using samtools [41] with a mapping quality cut off of 20; SNPs within 5 bp of indels and heterozygous sites were masked.
Multiple-sequence alignments were obtained separately for the D. mauritiana and D. melanogaster population samples by eye, using blast2seq to help resolve alignment in repetitive regions. In addition, a multiple-species alignment of the CDS regions of mei-218 and mei-217 were obtained for the above samples as well as the FlyBase reference sequence from D. melanogaster and D. yakuba. For mei-218, which is highly divergent between these species, we obtained an amino-acid alignment using the “Geneious aligner” with default settings, whereas the mei-217 CDS was easily aligned by eye. Basic population genetic analyses were performed using DnaSP v5 (ref. [42]).
Implementation of gammaMap
To identify individual codons under selection in D. mauritiana and D. melanogaster, we used the program, gammaMap, which models differential selection among lineages and among sites within a gene. Using the population resequence data from D. melanogaster and D. mauritiana, and setting D. yakuba as an outgroup, the method employs a population genetics-phylogenetics method to estimate selection and population genetic parameters under a model of recurrent selection. We used the method to obtain the probabilities of positive selection for each codon in the sequence, which is estimated using a Bayesian sliding window approach. The posterior probability of model parameters is obtained using Markov Chain Monte Carlo (MCMC) with Metropolis-Hastings parameter updates [26]. We assessed convergence and mixing of the MCMC output using the “coda” package in R [45]. We assessed chain convergence by calculating the potential scale reduction factor (PSRF) using the Gelman and Rubin convergence diagnostic, after excluding the burn-in steps and including log or logit transform [46]. An autocorrelation coefficient was calculated for each parameter using the autocorr.diag function in coda. Priors for model parameters that are shared across genes were based on the posterior probabilities obtained by Wilson et al. [26] for their analysis of 100 X-linked genes in D. melanogaster and D. simulans, with D. yakuba as an outgroup. As D. simulans and D. mauritiana are closely related sister species with similar estimated effective population sizes and identical divergence times from D. melanogaster, we reasoned that using a prior based on a large chromosome-wide dataset, even if based on a different albeit closely related species, was preferable to basing model priors on data from the mei-218 or mei-217 coding sequences alone.
We ran three MCMC chains of 2,000,000 steps, removing 20,000 steps of “burn-in” and recorded parameters every 40 steps. The gammaMap model is parameter rich, with 16 parameters per species shared across genes, and an additional parameter for each codon in each species modeling the selection coefficient. Convergence of the chains was assessed visually by plotting the posterior density of model parameters for each chain and calculating the potential scale reduction factor (PSRF) for each parameter. The 95% CI of the PSRF was less than our cut off of 1.1 for nearly all parameters for mei-218 (3522/3555), (844/848 for mei-217). An autocorrelation coefficient cut off of 0.5 was used and was met by 3,549/3,555 parameters for mei-218 (842/848 for mei-217). Poorly converging or poorly mixing parameters were excluded from any analysis or inference.
SNP density in D. melanogaster and D. mauritiana
To study the distribution of single nucleotide polymorphisms (SNPs) on chromosome 2 we used genome sequence data for 10 lines of D. mauritiana[44] and 10 lines of D. melanogaster sampled from Rwanda as part of the Drosophila Population Genetics Project (http://www.dpgp.org/). The 10 D. melanogaster lines were chosen based on their reported low levels of inferred admixture with cosmopolitan strains [47] : RG22, RG25, RG28, RG3, RG32N, RG36, RG38N, RG5, RG9, RG18N (accession numbers: SRR189383, SRR189385-8, SRR189393, SRR189395, SRR189398, SRR189407, and SRR306619). Sequences were aligned to the D. melanogaster reference genome (r6.13) using BWA, samtools and GATK [48] for variant calls and filtering, and downstream custom perl scripts. For each species (D. melanogaster and D. mauritiana) and chromosome arm (2L and 2R), Figure 2B shows the number of segregating sites per 50-kb window standardized by the maximum value.
Supplementary Material
Highlights.
Two Drosophila species evolved different rates and distributions of crossing over
In transgenic flies, mei-217/mei-218 recapitulates much of the species difference
The mei-217/mei-218 protein-coding sequence evolved by recurrent positive selection
Differences in crossing over alter the genomic patterning of nucleotide variability
Acknowledgments
We thank Shanwu Tang for technical assistance and advice during the early stages of this work; Christina Muirhead for technical assistance with population genomics data; Jeff Sekelsky for kindly sharing fly stocks; Danny Wilson for advice implementing the gammaMap analyses; and Amanda Larracuente and Colin Meiklejohn for comments on earlier drafts of the manuscript. This work was supported by funds to DCP from the Alfred P. Sloan Foundation, the David and Lucile Packard Foundation, the University of Rochester, and the National Institutes of Health (R01 GM111380).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
CLB and DCP designed the study. CLB, MVC generated molecular reagents. CLB performed fly crossing experiments. ELL performed the qPCR experiments. CLB and DCP analyzed the data. SBK and DCP performed the population genetics analyses. CLB and DCP wrote the paper.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- 1.Egel R, Lankenau D-H. Recombination and Meiosis: Crossing-over and Disjunction. Berlin: Springer; 2008. [Google Scholar]
- 2.Cutter AD, Payseur BA. Genomic signatures of selection at linked sites: unifying the disparity among species. Nature Reviews Genetics. 2013;14:262–274. doi: 10.1038/nrg3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dapper AL, Payseur BA. Connecting theory and data to understand recombination rate evolution. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2017;372:20160469. doi: 10.1098/rstb.2016.0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ritz KR, Noor MAF, Singh ND. Variation in recombination rate: adaptive or not? Trends in Genetics. 2017;33:364–374. doi: 10.1016/j.tig.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Fisher RA. The Genetical Theory of Natural Selection. Oxford: Oxford University Press; 1930. [Google Scholar]
- 6.Goldberg ML, Sheen JY, Gehring WJ, Green MM. Unequal crossing-over associated with asymmetrical synapsis between nomadic elements in the Drosophila melanogaster genome. Proceedings of the National Acadamy of Sciences. 1983;80:5017–5021. doi: 10.1073/pnas.80.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrov DA, Fiston-Lavier AS, Lipatov M, Lenkov K, Gonzalez J. Population genomics of transposable elements in Drosophila melanogaster. Molecular Biology and Evolution. 2011;28:1633–1644. doi: 10.1093/molbev/msq337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller DE, Smith CB, Kazemi NY, Cockrell AJ, Arvanitakas AV, Blumenstiel JP, Jaspersen SL, Hawley RS. Whole-Genome Analysis of Individual Meiotic Events in Drosophila melanogaster Reveals That Noncrossover Gene Conversions Are Insensitive to Interference and the Centromere Effect. Genetics. 2016;203:159–171. doi: 10.1534/genetics.115.186486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koehler KE, Boulton CL, Collins HE, French RL, Herman KC, Lacefield SM, Madden LD, Schuetz CD, Hawley RS. Spontaneous X chromosome MI and MII nondisjunction events in Drosophila melanogaster oocytes have different recombinational histories. Nature Genetics. 1996;14:406–414. doi: 10.1038/ng1296-406. [DOI] [PubMed] [Google Scholar]
- 10.True JR, Mercer JM, Laurie CC. Differences in crossover frequency and distribution among three sibling species of Drosophila. Genetics. 1996;142:507–523. doi: 10.1093/genetics/142.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindsley DL, Sandler L. The genetic analysis of meiosis in female Drosophila melanogaster. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 1977;277:295–312. doi: 10.1098/rstb.1977.0019. [DOI] [PubMed] [Google Scholar]
- 12.Baker BS, Carpenter ATC. Genetic analysis of sex chromosomal meiotic mutants in Drosophila melanogaster. Genetics. 1972;71:255–286. doi: 10.1093/genetics/71.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehrotra S, Hawley RS, McKim KS. Synapsis, double-strand breaks, and domains of crossover control in Drosophila females. In: Egel R, Lankenau D, editors. Recombination and meiosis, crossing-over and disjunction. Berlin: Springer-Verlag; 2007. pp. 125–152. [Google Scholar]
- 14.McKim KS, Dahmus JB, Hawley RS. Cloning of the Drosophila melanogaster meiotic recombination gene mei-218: a genetic and molecular analysis of interval 15E. Genetics. 1996;144:215–228. doi: 10.1093/genetics/144.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhagat R, Manheim EA, Sherizen DE, McKim KS. Studies on crossover-specific mutants and the distribution of crossing over in Drosophila females. Cytogenetics and Genome Research. 2004;107:160–171. doi: 10.1159/000080594. [DOI] [PubMed] [Google Scholar]
- 16.Kohl KP, Jones CD, Sekelsky J. Evolution of an MCM complex in flies that promotes meiotic crossovers by blocking BLM helicase. Science. 2012;338:1363–1365. doi: 10.1126/science.1228190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carpenter AT. Recombination nodules and synaptonemal complex in recombination-defective females of Drosophila melanogaster. Chromosoma. 1979;75:259–292. doi: 10.1007/BF00293472. [DOI] [PubMed] [Google Scholar]
- 18.Sekelsky JJ, McKim KS, Chin GM, Hawley RS. The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics. 1995;141:619–627. doi: 10.1093/genetics/141.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H, Jang JK, Graham J, Nycz K, McKim KS. Two genes required for meiotic recombination in Drosophila are expressed from a dicistronic message. Genetics. 2000;154:1735–1746. doi: 10.1093/genetics/154.4.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawley RS, McKim KS, Arbel T. Meiotic segregation in Drosophila melanogaster females: molecules, mechanisms, and myths. Annual Review of Genetics. 1993;27:281–317. doi: 10.1146/annurev.ge.27.120193.001433. [DOI] [PubMed] [Google Scholar]
- 22.Wang S, Zickler D, Kleckner N, Zhang L. Meiotic crossover patterns: obligatory crossover, interference and homeostasis in a single process. Cell Cycle. 2015;14:305–314. doi: 10.4161/15384101.2014.991185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinstein A. The theory of multiple-strand crossing over. Genetics. 1936;21:155–199. doi: 10.1093/genetics/21.3.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tye BK. MCM proteins in DNA replication. Annual Review of Biochemistry. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- 25.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 26.Wilson DJ, Hernandez RD, Andolfatto P, Przeworski M. A population genetics-phylogenetics approach to inferring natural selection in coding sequences. PLoS Genetics. 2011;7:e1002395. doi: 10.1371/journal.pgen.1002395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brandvain Y, Coop G. Scrambling eggs: meiotic drive and the evolution of female recombination rates. Genetics. 2012;190:709–723. doi: 10.1534/genetics.111.136721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dowsett AP, Young MW. Differering levels of dispersed repetitive DNA among closely related species of Drosophila. Proceedings of the National Acadamy of Sciences. 1982;79:4570–4574. doi: 10.1073/pnas.79.15.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent TV, Uzunovic J, Wright SI. Coevolution between transposable elements and recombination. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2017;372:20160458. doi: 10.1098/rstb.2016.0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montgomery E, Charlesworth B, Langley CH. A Test for the Role of Natural-Selection in the Stabilization of Transposable Element Copy Number in a Population of Drosophila-Melanogaster. Genetical Research. 1987;49:31–41. doi: 10.1017/s0016672300026707. [DOI] [PubMed] [Google Scholar]
- 31.Charlesworth B, Sniegowski P, Stephan W. The Evolutionary Dynamics of Repetitive DNA in Eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 32.Charlesworth B, Barton NH. Recombination load associated with selection for increased recombination. Genetical Research. 1996;67:27–41. doi: 10.1017/s0016672300033450. [DOI] [PubMed] [Google Scholar]
- 33.Maynard Smith J, Haigh J. The hitch-hiking effect of a favourable gene. Genetical Research. 1974;23:23–35. [PubMed] [Google Scholar]
- 34.Charlesworth B, Morgan MT, Charlesworth D. The Effect of Deleterious Mutations on Neutral Molecular Variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manheim EA, Jang JK, Dominic D, McKim KS. Cytoplasmic localization and evolutionary conservation of MEI-218, a protein required for meiotic crossing-over in Drosophila. Molecular Biology of the Cell. 2002;13:84–95. doi: 10.1091/mbc.01-06-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watterson GA. On the number of segregating sites in genetical models without recombination. Theoretical Population Biology. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 37.Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 40.Garrigan D, Kingan SB, Geneva AJ, Andolfatto P, Clark AG, Thornton KR, Presgraves DC. Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Research. 2012;22:1499–1511. doi: 10.1101/gr.130922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 43.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Molecular Biology and Evolution. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 44.Garrigan D, Kingan SB, Geneva AJ, Vedanayagam JP, Presgraves DC. Genome diversity and divergence in Drosophila mauritiana: multiple signatures of faster X evolution. Genome Biology and Evolution. 2014;6:2444–2458. doi: 10.1093/gbe/evu198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plummer M, Best N, Cowles K, Vines K. CODA: Convergence diagnosis and output analysis for MCMC. R News. 2006;6:7–11. [Google Scholar]
- 46.Gelman A, Rubin D. Inference from iterative simulation using multiple sequences. Statistical Science. 1992;7:457–511. [Google Scholar]
- 47.Pool JE, Corbett-Detig RB, Sugino RP, Stevens KA, Cardeno CM, Crepeau MW, Duchen P, Emerson JJ, Saelao P, Begun DJ, et al. Population Genomics of sub-saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genetics. 2012;8:e1003080. doi: 10.1371/journal.pgen.1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.