Abstract
Class I knox genes play an important role in shoot meristem function and are thus involved in the ordered development of stems, leaves, and reproductive organs. To elucidate the mechanism underlying the expression pattern of these homeobox genes, we studied a spontaneous tomato (Lycopersicon esculentum) mutant that phenotypically resembles, though is more extreme than, transgenic plants misexpressing class I knox genes. This mutant was found to carry a recessive allele, denoted clausa:shootyleaf (clau:shl)—a newly identified allele of clausa. Mutant plants exhibited abnormal leaf and flower morphology, epiphyllus inflorescences, fusion of organs, calyx asymmetry, and navel-like fruits. Analysis by scanning electron microscopy revealed that such fruits carried ectopic ovules, various vegetative primordia, as well as “forests” of stalked glandular trichomes. In situ RNA hybridization showed a peculiar expression pattern of the class I knox gene LeT6/TKn2; expression was restricted to the vascular system and palisade layer of mature leaves and to the inner part of ovules integuments. We conclude that CLAUSA regulates various aspects of tomato plant development, at least partly, by rendering the LeT6/TKn2 gene silent in specific tissues during development. Considering the expression pattern of LeT6/TKn2 in the clausa mutant, we suggest that the control over a given homeobox gene is maintained by several different regulatory mechanisms, in a cell type-dependent manner.
Many plants tend to have indeterminate growth and are capable of producing new organs and tissues throughout their life. This capability is largely retained by the activity of the two apical meristems: the shoot apical meristem, which continuously generates cells for the growth of the shoot system (leaf and bud primordia), and the root apical meristem, which generates cells for the development of the root system (Steeves and Sussex, 1989). New leaves and buds are initiated on the flanks of the apical meristem in a species-specific succession that gives the plant its particular phyllotactic arrangement and general architecture.
Homeobox-containing genes are involved in pattern formation in multicellular organisms and share a conserved sequence that encodes a DNA-binding homeodomain (Gehring, 1987; Hayashi and Scott, 1990). These homeodomain proteins function as transcription factors, thus controlling gene expression. Various plant homeobox genes were isolated from a variety of plant species and, based on their sequence homology, were subdivided into different families, each consisting of several members (for review, see Chan et al., 1998). The first identified plant homeobox gene, KNOTTED1 (KN1; Vollbrecht et al., 1991), isolated from maize, provided evidence that plant homeobox genes, similar to those of animals, play an important role in regulating developmental processes. On the basis of sequence homology and expression pattern, Kn1-like homeobox (knox) genes were grouped into two classes, I and II (Kerstetter et al., 1994). Whereas class II knox genes are differentially expressed in all plant organs (Serikawa et al., 1997), class I genes are mainly expressed in vegetative and inflorescence meristems and are involved in shoot meristem function and in leaf and flower morphology (Hake et al., 1995; Long et al., 1996; McSteen and Hake, 1998; Frugis et al., 1999). Overexpression of the maize KN1 gene in tobacco and of the class I knox gene KNAT1 in Arabidopsis led to changes in leaf morphology and formation of ectopic meristems (Hake et al., 1995, and refs. therein). In tomato (Lycopersicon esculentum), misexpression of class I knox genes had a profound effect on leaf morphology, giving rise to excessive proliferation of leaflets and abnormal development of reproductive organs (Hareven et al., 1996; Janssen et al., 1998a). Hence the genetic control over homeobox genes is of prime importance for plant development.
In Drosophila melanogaster, the expression pattern of developmental genes such as homeobox genes is maintained in an elaborated manner involving the antagonistic action of the Polycomb (PcG) and the trithorax (trxG) groups of genes. Whereas PcG proteins are necessary for stable repression of homeotic genes, trxG proteins are required for the maintenance of their active state (for review, see Gould, 1997; Schumacher and Magnuson, 1997; Cavalli and Paro, 1998; Jenuwein et al., 1998). The PcG and trxG gene families contain the SET domain, an evolutionarily conserved motif originally identified in three chromosomal proteins [Su(var)3-9, enhancer-of-zeste, and trithorax] that modulate gene expression, at least partly, by affecting chromatin structure (Cavalli and Paro, 1998; Jenuwein et al., 1998). Several PcG genes that control the development of vegetative and reproductive organs in Arabidopsis were recently identified (Goodrich et al., 1997; Grossniklaus et al., 1998; Kiyosue et al., 1999; Luo et al., 1999; Ohad et al., 1999). The recessive curly leaf-2 (clf-2) mutation pleiotropically affects leaf and flower morphology as well as flowering time. The CLF gene encodes a PcG protein that negatively regulates the expression of the floral homeotic gene AGAMOUS (AG) in leaves (Goodrich et al., 1997). Several recessive mutations in maize that alter leaf morphology are involved in the regulation of knox gene expression, e.g. leafbladeless1 (lbl1), narrow sheath (ns), and rough sheath2 (rs2) (Scanlon et al., 1996; Timmermans et al., 1998; Schneeberger et al., 1998). The ROUGH SHEAT2 (RS2) gene was isolated by DNA tagging as well as by phenotypic similarities to Antirrhinum majus plants that are mutated in the PHANTASTICA (PHAN) gene. Similar to the PHAN gene, RS2 was found to encode a Myb protein that represses the expression of homeobox genes such as ROUGH SHEAT1 (RS1) and KN1 (Waiters et al., 1998; Timmermans et al., 1999; Tsiantis et al., 1999).
We studied a recessive tomato mutant, clausa:shootyleaf (clau:shl), that partly phenocopies transgenic plants overexpressing class I knox genes. We hypothesized that such a mutant is defective in its ability to properly control the expression pattern of homeotic genes. The clau:shl mutation affects the development of vegetative and reproductive organs, giving rise to altered leaf and carpel morphology, ectopic meristems, and fusion of organs. Misexpression of the class I knox gene LeT6/TKn2 was observed in distinct regions of leaves and carpels. The significance of the CLAUSA gene to plant growth and development is discussed.
RESULTS
Genetic Analysis of the clausa:shootyleaf (clau:shl) Mutant
A spontaneous tomato mutant in which shoot-like structures emerge from the rachis, hence denoted shootyleaf (shl), was found to be phenotypically similar to clau mutants of tomato. Crosses were carried out between the clau:shl mutant and tomato (cv M82; referred to as wild type) to define the Mendelian character of CLAU. All F1 progeny showed wild-type phenotype, whereas the F2 population segregated at a ratio of nearly 1:3 (mutant phenotype was evident in 208 of 865 F2 plants) indicating that tomato plants homozygous for the recessive mutation at the CLAU locus have a mutant phenotype. A test for allelism confirmed that shl is allelic to clau:ff and clau:vc (D. Zamir, personal communication) and is therefore referred to as clau:shl. The CLAU gene is located on the short arm of chromosome 4 (Khush and Rick, 1967).
Morphological Analysis
Cultivated tomato plants carry compound unipinnate leaves that exhibit a basipetal order of leaflet initiation and maturation (Chandra-Sekhar and Sawhney, 1990). Upon maturation each leaf carries major and minor leaflets (Dengler, 1984), most of which are lobed to various degree, which exhibit plagiotropic growth (Fig. 1A). Wild-type flowers have five to six yellow petals and five to six green hairy sepals, both curved backward. Stamens are fused and form a cylindrical cone surrounding the style. The fruit is a fleshy berry consisting of a pericarp, derived from the ovarian walls, which surrounds the placental tissue and the seeds (Hayward, 1938).
Figure 1.
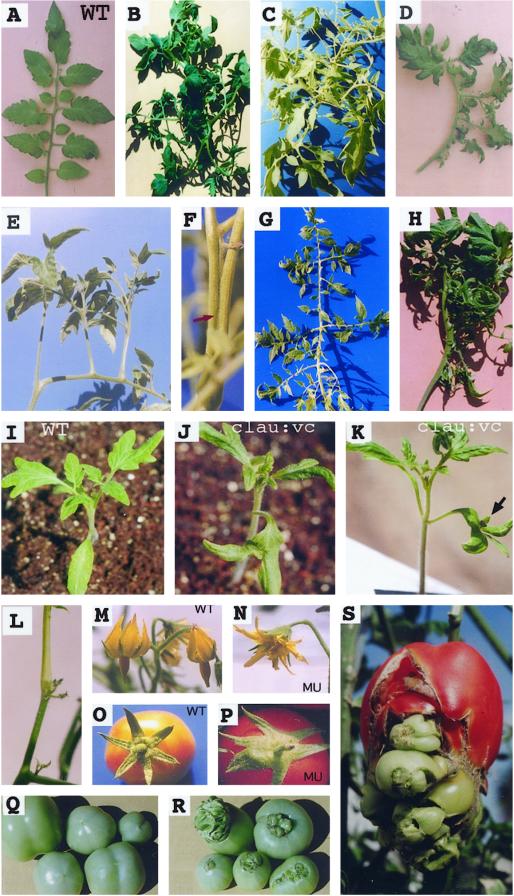
Phenotypic alterations in clau mutants. A, A wild-type (WT) unipinnate leaf with normal plagiotropic growth of leaflets. B through H, Modified leaves common in different clau mutants. Often leaflets exhibit orthotropic growth (such as in C, D, and E) resulting from fusion of petiolules (arrow in F). I, A wild-type seedling with normal cotyledons. J and K, Bifurcated cotyledons in clau:vc. Arrow points to a bud at the notch. L, A suppressed inflorescence typical to clau:ff carrying clusters of undeveloped flowers. M, A wild-type inflorescence carrying flowers with petals and sepals curved backward. N, A typical partly cleistogamous flower of clau mutants (MU) with uncurved petals and sepals. O, A wild-type fruit showing normal, symmetrically arranged sepals. P, A clau mutant fruit exhibiting asymmetrical calyx with partly fused sepals. Q, Wild-type tomato fruits. R and S, Navel-like fruits of clau mutants with fruit-like structures protruding from the stylar end of the fruit.
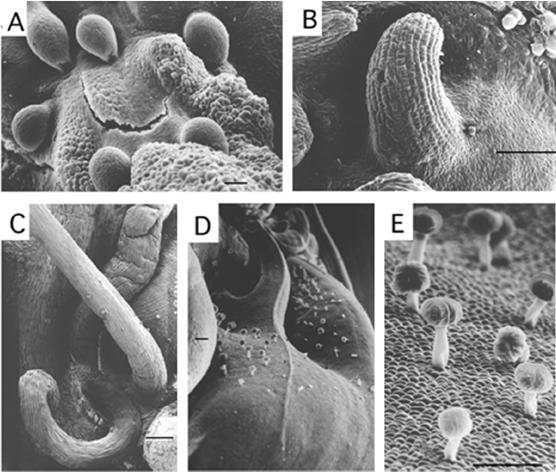
The clau mutant plants exhibit a wide range of phenotypic perturbations including abnormal leaf and flower morphology, epiphyllus inflorescences, fusion of organs, calyx asymmetry, and navel-like fruits. The phenotypes of the different clau mutants are described in Table I and in Figure 1. It is notable that all clau alleles display abnormal, excessively divided leaves, often carrying shoot-like structures on the rachis (Fig. 1, B–H). Such shoot-like organs, which in fact resulted from the fusion of petiolules (Figs. 1F and 4), gave rise to leaflets assuming an orthotropic growth pattern (Fig. 1, C–E) rather than the normal plagiotropic one (Fig. 1A). Seedlings of clau:vc mutant, unlike wild-type seedlings (Fig. 1I), often developed shooty cotyledons, i.e. gave rise to a bud formed at the notch of bifurcated cotyledons (Fig. 1, J and K), which could develop into a mature plant (not shown). The reproductive stage of clau:ff mutant was strongly suppressed; inflorescences were developed late and most of them carried clusters of undeveloped flowers (Fig. 1L). Flowers in mutant plants were partly cleistogamous, i.e. sepals and petals of mutant flowers were not curved backward (Fig. 1N) as did their wild-type counterparts (Fig. 1M). This is reminiscent of flowers found in transgenic tomato plants overexpressing the homeobox LeT6 gene (Janssen et al., 1998a). The fused stamens in clau appeared normal and contained viable pollen (data not shown). Mutant plants invariably had partly fused, asymmetrically arranged sepals (Fig. 1, compare panel O with P). Mutant fruits often displayed a navel-like appearance in which fruit-like structures emerged from the stylar end of the fruit (Fig. 1, compare panel Q with R and S), alluding to indeterminate growth of the flower. In navel fruits the placental tissue occupied most of the volume of the locule and seed production was poor (not shown). Scanning electron microscopy (SEM) analysis at the navel region of mutant fruits showed ectopic development of ovule-like structures (Fig. 2A), various vegetative primordia (Fig. 2, B and C), as well as “forests” of stalked glandular trichomes (Fig. 2, D and E) commonly found on wild-type tomato leaves, stems, and sepals.
Table I.
Phenotypes of tomato clau mutants
| Genotype (background) | clau:shl (unknown) | clau:ff (VFSM) | clau:vc (unknown) |
|---|---|---|---|
| Leaf morphology | Excessively divided; fused petiolules | Excessively divided; fused petiolules | Excessively divided; fused petiolules |
| Flower morphology | Partly cleistogamous | Partly cleistogamous | Partly cleistogamous |
| Sepal arrangement | Asymmetrical; partly fused | Asymmetrical; partly fused | Asymmetrical; partly fused |
| Fruit morphology | Strongly navel | Weakly navel | Moderately navel |
| Reproductive stage | Normal | Strongly suppressed | Normal |
| Cotyledons | Normal | Normal | Often bifurcateda |
A bud located at the notch of bifurcated cotyledons may develop into a mature plant.
Figure 4.
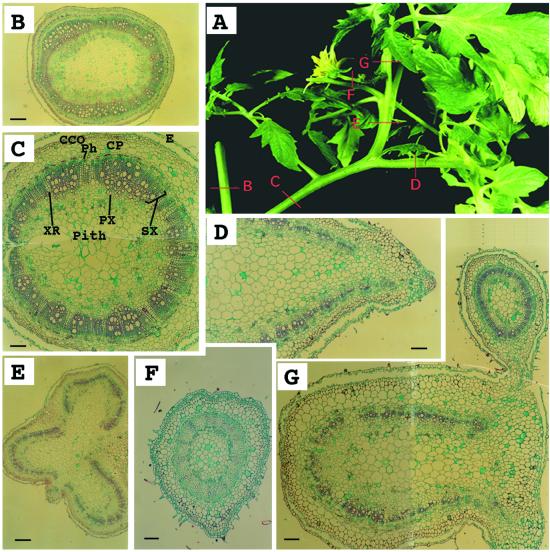
Anatomical analysis of an epiphyllus inflorescence in clau mutant. A, An epiphyllus inflorescence. Red bars B through G correspond to sites of cross sections shown in B through G. B, A cross-section through a stem showing ring-shaped vascular tissues. Bar = 560 μm. C, A cross-section through a petiole-like organ showing ring-shaped vascular tissues characteristic of a stem. Bar = 220 μm. D, A cross-section through a rachis showing U-shaped vascular tissues. Bar = 220 μm. E, A cross-section through a shoot-like structure emerging from a rachis demonstrating fusion of three petiolules, each with U-shaped vascular tissues. Bar = 560 μm. F, A cross-section through a flower-carrying stem (pedicle). Bar = 130 μm. G, A cross-section through the fused structure of a petiolule (U-shaped) and the inflorescence stem (peduncle; ring-shaped). Bar = 220 μm. CCO, Cortex collenchyma; CP, cortex parenchyma; E, epidermis; Ph, phloem; PX, primary xylem; SX, secondary xylem; XR, xylem rays.
Figure 2.
SEM analysis of the navel region of clau fruits showing ectopic meristem activity. A, Ovule-like structures. B, A leaf primordium-like structure. C, Vegetative structures. D and E, Stalked glandular trichomes. Bar = 100 μm.
Anatomical Analysis
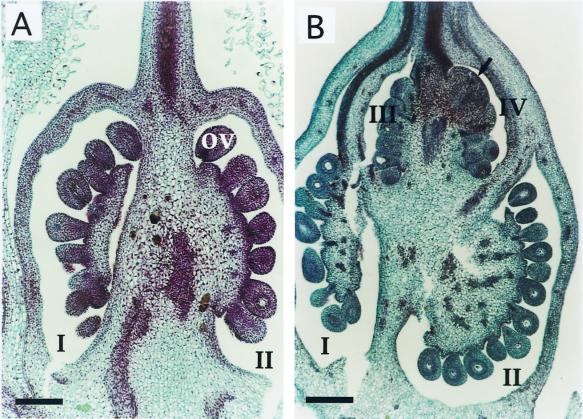
The carpel of cultivated tomato is usually composed of several ovule-containing locules (Hayward, 1938) arranged side by side. A longitudinal section of wild-type carpels revealed one or two locules (depending on the orientation of the section) containing several ovules (Fig. 3A). Mutant carpels containing normal ovules were composed of many locules often arranged in double or triple tiers (Fig. 3B). This abnormal structure may account for the navel-like appearance of fruits. In mutant fruits meristem-like structures are often ectopically initiated in place of ovules (arrow in Fig. 3B).
Figure 3.
Anatomy of wild-type and clau carpels. A, A longitudinal section through a wild-type carpel showing two ovule-containing locules arranged side by side (I and II). B, A longitudinal section through a mutant carpel showing four locules arranged in two tiers, i.e. locules III and IV above locules I and II. Arrow in locule IV points to an ectopic meristem. ov, Ovule. Bar = 350 μm.
Some mutant plants produced epiphyllus inflorescences in place of inflorescences (Fig. 4A). The nature of this homeotic phenomenon (Sattler, 1988) was demonstrated by a series of cross-sections. In wild-type tomato the vascular tissues of stems, peduncles (inflorescence stalks), and pedicels are ring-shaped, whereas those of petioles, rachises, and petiolules are U-shaped (data not shown; Howard, 1979). In accordance with this, the epiphyllus inflorescence is carried by a petiole-like organ (Fig. 4A) having features characteristic of a stem, i.e. ring-shaped vascular tissues (Fig. 4, B and C). The single flower is carried by a pedicel (Fig. 4F) attached to a peduncle (Fig. 4G; note the fusion between the peduncle and the petiolule). The pedicel and the peduncle are ectopically expressed on a compound leaf (U-shaped vascular tissues, Fig. 4, A, D, E, and G). The anatomical analysis confirmed that the shoot-like structures emerging from the rachis of mutant leaves resulted from fusion of petiolules (Fig. 4E).
Misexpression of the LeT6/TKn2 Gene in Mature Leaves and in Developing Carpels of clau Mutants
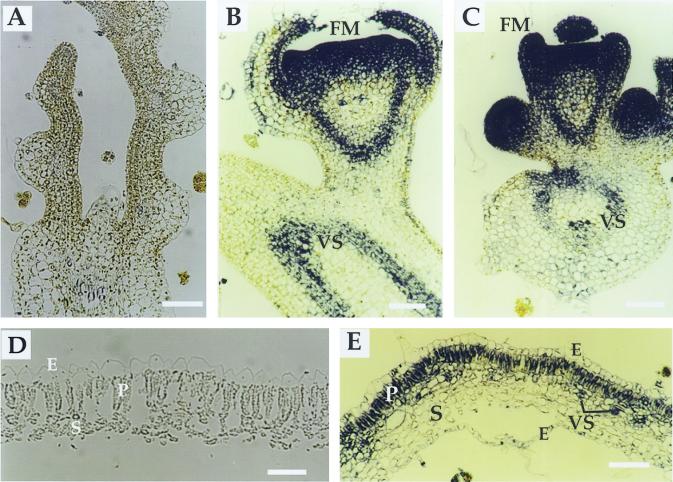
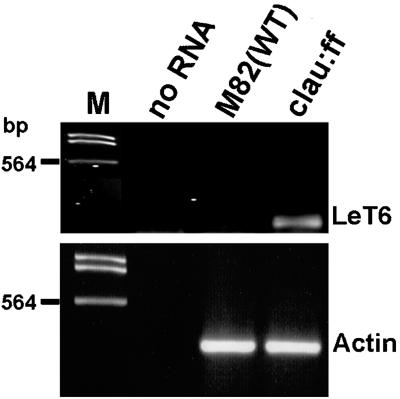
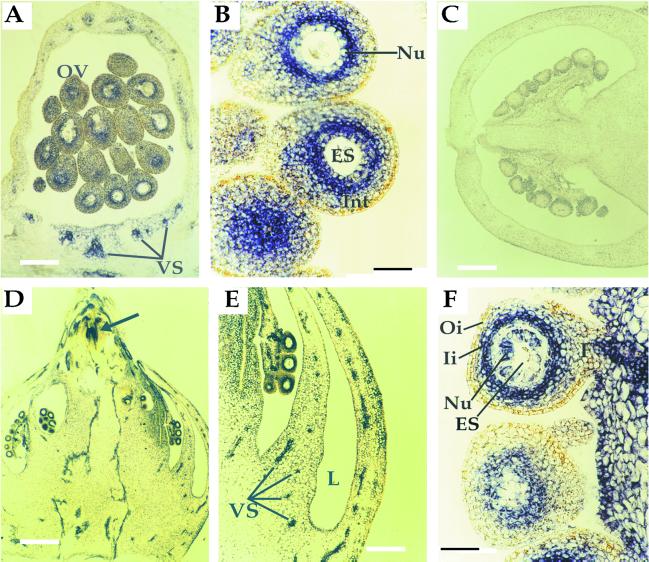
The aforementioned phenotypic alterations suggested that clau mutants are defective, at least in part, in the proper expression of homeobox genes such as LeT6/TKn2. To determine the expression pattern of LeT6 in various tissues, we performed in situ RNA hybridization in shoot apices, mature leaves, and carpels derived from wild-type and clau:shl mutant plants. Consistent with previous reports (Chen et al., 1997; Parnis et al., 1997; Janssen et al., 1998a, 1998b), LeT6 was found to be expressed in wild-type meristems, leaf primordia (not shown), as well as vascular tissues (Fig. 5B), but not in mature wild-type leaflets (Fig. 5D). In mutant plants the expression pattern of LeT6 in vegetative (not shown) and floral meristems (Fig. 5, B and C) was indistinguishable from that of wild type. However, unlike wild-type plants, LeT6 was strongly expressed in mature mutant leaves, specifically in the palisade layer and in the vascular region (Fig. 5E). Reverse transcriptase (RT)-PCR analysis showed that LeT6 is misexpressed in clau:ff, but not in wild-type leaves (Fig. 6). In wild-type carpels at anthesis LeT6 was expressed in vascular tissues and in the inner part of ovule integument, adjacent to the nucellus (Fig. 7, A and B). Consistent with Janssen et al. (1998b), no expression was detected in wild-type carpels post-anthesis (Fig. 7C). In mutant carpels, however, the expression of LeT6 was evident post-anthesis in different cell layers surrounding the embryo sac—stronger in the inner part of the integument and relatively weak in the nucellar layer (Fig. 7, D and F). Mutant plants also exhibited strong expression of LeT6 in ectopic meristem near the stylar end of carpels (see arrow in Fig. 7D).
Figure 5.
In situ localization of LeT6 RNA in leaves of wild-type and clau mutant plants. A, A negative control depicting a longitudinal section of wild-type shoot apical meristem probed with LeT6 sense RNA. Bar = 250 μm. Longitudinal sections of wild-type (B) and clau (C) floral meristems probed with LeT6 antisense RNA. Bar = 60 μm. D, A cross-section of a mature wild-type leaflet showing no expression of LeT6. Bar = 250 μm. E, A cross-section of a mature clau leaflet showing LeT6 RNA restricted to the palisade layer and vascular regions. Bar = 350 μm. E, Epidermis; FM, floral meristem; P, palisade cells; S, spongy layer; VS, vascular system.
Figure 6.
RT-PCR analysis of LeT6 expression in mature leaves of clau:ff and wild-type plants. Poly(A)+ RNA was used as a template. Actin was used as a reference RNA. Note that 60% of LeT6 PCR reaction was loaded on the gel (1.5% [w/v] agarose gel) compared with 20% of the actin reaction. M indicates DNA size marker.
Figure 7.
In situ localization of LeT6 RNA in wild-type (A–C) and clau mutant (D–F) carpels. A, A longitudinal section of a wild-type carpel at anthesis showing expression of LeT6 in vascular tissues and in a distinct region of the ovule integument. Bar = 250 μm. B, A higher magnification of wild-type ovules at anthesis showing the confinement of LeT6 RNA to the inner part of the integument. Bar = 50 μm. C, A longitudinal section of a wild-type carpel post-anthesis. Bar = 400 μm. D, A longitudinal section of a clau mutant carpel post-anthesis showing LeT6 RNA in ovules and vascular tissues. Arrow indicates an ectopic meristem near the stylar end. Note the typical multiloculed ovary arranged in tiers. Bar = 400 μm. E, A higher magnification of the mutant ovary wall showing expression in vascular tissues. Bar = 250 μm. F, A higher magnification of mutant ovules post-anthesis showing LeT6 RNA in the nucellar layer and the inner integument. Bar = 50 μm. ES, Embryo sac; F, funiculus; Ii, inner integument; Int, integument; L, locule; Nu, nucellus; Oi, outer integument; OV, ovule; VS, vascular system.
DISCUSSION
To study the regulation of homeobox gene expression in plants we analyzed a recessive mutant of tomato, clau:shl, that phenocopies several features of transgenic plants overexpressing class I knox genes (for review, see Hake et al., 1995; Tamaoki et al., 1997; Janssen et al., 1998a; McSteen and Hake, 1998, and refs. therein). Similar to such transgenic plants, clau mutants exhibited abnormal leaf morphology, epiphyllus inflorescences, and ectopic meristems. Mutant plants, however, also displayed fusion of organs, particularly of petiolules, calyx asymmetry, altered carpel morphology, and navel-like fruits carrying fruit-like appendages. Such fruits can be interpreted as representing indeterminate growth of the flower, which normally terminates by the production of the ovary in the innermost whorl. The abnormal development of vegetative and reproductive organs is accompanied by misexpression of the class I knox gene LeT6/TKn2, but the wide range of phenotypic changes point to other developmental genes being affected by the clausa mutation.
Several genes were reported to negatively regulate homeotic gene expression. The Arabidopsis CLF gene is required to repress the floral homeotic gene AG (Goodrich et al., 1997). In maize, the RS2 encodes a Myb protein that represses the expression of the class I knox genes ROUGH SHEATH1 (RS1) and KN1 during leaf development (Schneeberger et al., 1998; Timmermans et al., 1999; Tsiantis et al., 1999). Because CLAUSA is located on chromosome 4 (Khush and Rick, 1967) and LeT6 on chromosome 2 (Janssen et al., 1998b), the deregulated expression of LeT6 in clau mutants cannot be attributed to a mutation within its transcription regulatory regions. We assume that the wide range of homeotic phenomena displayed by the recessive clau mutant reflect loss-of-function of a factor, such as PcG or Myb genes, which negatively regulates the expression of the homeobox gene LeT6/TKn2 and probably other homeotic/developmental genes in tomato.
The differential expression pattern of the LeT6 gene in wild-type and mutant plants provides insight into the mechanism regulating the expression pattern of homeotic genes during plant development. In mature wild-type leaves, the LeT6 gene is kept silent. In contrast, in mature leaves of clau, LeT6 is expressed in a tissue-specific manner, e.g. in the palisade layer and in the vascular system. These findings suggest that stable repression of LeT6 in various tissues of mature leaves is maintained by different regulatory mechanisms. We propose that the CLAUSA gene determines the pattern of LeT6 gene expression by rendering it silent in the palisade layer and the vascular tissues; in other tissues, e.g. the spongy layer, stable repression of LeT6 is CLAUSA-independent.
Consistent with a previous report (Janssen et al., 1998b), the expression pattern of LeT6 in wild-type carpels at anthesis is confined to the vascular system and to distinct regions of the ovule; expression of LeT6 in carpels is dramatically reduced post-anthesis (Fig. 7C). Janssen et al. (1998b) suggested that the expression of LeT6 is localized to the nucellus, but careful analysis (Fig. 7B) indicates that the expression of this gene is confined to the inner part of the integument. This interpretation is supported by a morphological study (Cooper, 1931) showing that the ovule of tomato develops a one cell-layer nucellus surrounded by a single, massive integument. Our results suggest that this single integument can be biochemically dissected into two parts: an inner integument that expresses LeT6, and the outer integument that does not. In carpels of clau mutants the expression of LeT6 is evident post-anthesis not only in the inner integument, but also in the nucellar layer. Hence the expression of LeT6 in ovules is spatially and temporally regulated by the CLAUSA gene product: at anthesis, CLAUSA is not active in repressing the expression of the LeT6 gene in the inner integument, but becomes active post-anthesis in the inner integument and in the nucellus.
Considering the expression pattern of LeT6 in leaves and ovules, we propose that the control over a given homeobox gene is maintained by various regulatory mechanisms, in a cell type-dependent manner. Support for this proposition comes from the dominant mutant Curl (Cu) that also affects LeT6/TKn2 gene expression (Parnis et al., 1997). Contrary to clau, in the dominant Cu mutant, TKn2 is expressed in the abaxial spongy mesophyll cells, but not in the palisade tissue, again pointing to the involvement of various mechanisms in the control of LeT6 gene expression in different cell types. The mechanism for the abnormal expression of TKn2 in Cu leaves is yet unknown. Parnis et al. (1997) proposed that overexpression of TKn2 in Cu leaves may result from a mutation within a putative silencer in the long intron 2, or within 0.1 cM (50–100 kb) of the Cu locus. This suggests that the spatiotemporal regulation of knox gene expression requires cis regulatory elements that lie outside the transcribed region of the gene, but within its regulatory region (50–100 kb). Such elements could mediate the repressive action of polycomb proteins on homeotic gene expression, e.g. polycomb response elements (PREs; Brown et al., 1998; Mihaly et al., 1998).
Organ fusion is a conspicuous feature in clau mutants, in particular the fusion of petiolules. Organ fusion often occurs in reproductive organs and is well exemplified by the fusion of carpels in Catharanthus roseus (Walker, 1975). The initially separated carpels become completely fused as they develop side by side at the primordium stage. In C. roseus, carpel fusion involves a diffusible factor/s that promotes redifferentiation of carpel epidermal cells into parenchyma cells, leading to the union of two adjacent carpels (Siegel and Verbeke, 1989). A limited proliferation of epidermal cells may occur in the crinkly4 (cr4) mutant of maize in regions of adherence between leaves (Becraft et al., 1996). Our study shows that the anatomical features of petiolules (U-shaped vascular tissues) are retained in the fused organs (Fig. 4E), suggesting that fusion occurred after organ identity had been determined. That the epidermis identity in the fused region is not retained implies that petiolules union takes place through redifferentiation or dedifferentiation of epidermal cells. This is different from the adherent1 (ad1) mutant of maize in which tissue identity is preserved at the attached region (Sinha and Lynch, 1998). The control over organ fusion in the clau mutant cannot be solely attributed to LeT6, because tomato plants misexpressing this gene do not exhibit fusion of organs (Chen et al., 1997; Janssen et al., 1998a; Parnis et al., 1997). Taken together, the phenotypic alterations exerted by the clau mutation suggest that CLAUSA is involved not only in controlling the expression pattern of LeT6, but also in the control of other developmental genes in tomato plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of clau:shl, a spontaneous tomato (Lycopersicon esculentum) mutant found in a commercial field, clau:ff (TGRC LA0896), clau:vc (TGRC 2–505), and tomato cv M82 were kindly provided by Dr. D. Zamir (The Hebrew University of Jerusalem). Plants were grown during the summer (June through October) in 5-L plastic containers (one plant per pot) in a greenhouse (natural daylight, 28°C–30°C at high humidity) with weekly administration of insecticides.
Histology, in Situ RNA Hybridization, and SEM
Plant tissues for histological examinations were processed essentially as described (Yang et al., 1998). Immediately after harvesting, tissues were fixed (overnight at room temperature) in freshly prepared 4% (w/v) paraformaldehyde and 2% (w/v) glutaraldehyde (Sigma, St. Louis). Samples were dehydrated through ethanol-dilution series, cleared with xylene, and embedded in paraffin. Prepared sections (8-μm thick) were spread on microscope slides, rehydrated, and stained in 1% (w/v) Safranin followed by 0.2% (w/v) Fast-Green. Sections were dehydrated in ethanol series and mounted with Permount (Fisher Scientific, Loughborough, Leicestershire, UK). For in situ RNA hybridization, plant tissues were fixed in freshly prepared 4% (w/v) paraformaldehyde and processed as above. Tissue sections (8-μm thick) were prepared and spread on microscope slides coated with poly-L-Lys (Sigma) and probed with digoxigenin (DIG)-labeled LeT6 sense or antisense RNA, essentially as described (Yang et al., 1998). Slides were examined at 16 to 400 magnification under bright-field using a microscope (Dialux 20, Leitz, Wetzlar, Germany) equipped with a camera (FTN, Nikon, Tokyo). Digital images of photographic prints were generated using a computerized scanner. Composites of individual prints were assembled using Adobe Photoshop (Adobe Systems, Mountain View, CA). For SEM analysis, samples were fixed and dehydrated through ethanol-dilution series, and then dried with CO2 using a Critical Point Dryer (CPD2, Pelco, Redding, CA). Samples were mounted on SEM stubs and coated with gold using a S150 Sputter Coater (Edwards, Crawley, England). Samples were viewed with a JSM 6400 scanning electron microscope (JEOL, Tokyo, Japan) and micrographs were taken with TMAX 120 film (Eastman-Kodak, Rochester, NY) .
RNA Probe Preparation
LeT6 cDNA in pBluescript was kindly provided by N. Sinha (University of California, Davis). The LeT6 plasmid was linearized either with BamHI to generate an antisense RNA or with XhoI to produce a sense RNA probe. Linearized plasmids were subjected to in vitro transcription using the DIG RNA labeling mix (Boehringer Mannheim, Basel) with either T7 or T3 RNA polymerase according to the manufacturer's protocol. Following in vitro transcription, the DNA template was removed by incubation with RQ1 DNase (2 units/μg DNA, Promega, Madison, WI) for 15 min at 37°C and ethanol-precipitated. To allow better penetration into the tissue, DIG-labeled RNAs were hydrolyzed for 20 min at 60°C in hydrolysis carbonate buffer as described (Moench et al., 1985).
RNA Analysis by RT-PCR
Detection of LeT6 by RT-PCR was performed by using the Titan One Tube RT-PCR System (Boehringer Mannheim). Total RNA was isolated from mature leaves using the EZ-RNA kit (Biological Industries, Beit Haemek, Israel). Poly(A)+ RNA was isolated using the PolyATtract mRNA Isolation System according to the manufacturer's protocol (Promega). LeT6 RNA was identified using 1 μg of poly(A)+ RNA as template and the following primers: a sense primer 5′-GGTCAATTGTTGCGTAAGTACAGCGG, and an antisense primer 5′-CCAATCCCGTTGATTCAGCTAGTGC, giving rise to a PCR product of 187 bp. As a reference we analyzed the expression of actin RNA using the following primers: a sense primer 5′-GGTTTTGCTGGGGATGATGC, and an antisense primer 5′-CATGGCTGGACATTGAATGTCTC giving rise to a PCR product of 340 bp. PCR products were run on 1.5% (w/v) agarose gel stained with ethidium bromide.
ACKNOWLEDGMENTS
We thank D. Zamir for providing seeds of clausa:shl and for communicating unpublished data, N. Sinha for providing the LeT6 cDNA clone, and Y. Salts and R. Berg for providing tomato cDNA libraries. We thank E. Galun, R. Fluhr, A. Levy, and G. Galili for their comments on the manuscript, D. Natan for helping with histology, E. Klein for her help with SEM, and A. Mosquna for his help in the greenhouse and the laboratory.
Footnotes
This work was supported by the Ebner Family Foundation for Biomedical Research in Memory of Alfred and Dolfi Ebner and by the Estelle Funk Foundation.
LITERATURE CITED
- Becraft PW, Stinard PS, McCarty DR. CRINKLY4: a TNFR-like receptor kinase involved in maize epidermal differentiation. Science. 1996;273:1406–1409. doi: 10.1126/science.273.5280.1406. [DOI] [PubMed] [Google Scholar]
- Brown JL, Mucci D, Whiteley M, Dirksen M-L, Kassis JA. The Drosophila polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- Cavalli G, Paro R. Chromo-domain proteins: linking chromatin structure to epigenetic regulation. Curr Opin Cell Biol. 1998;10:354–360. doi: 10.1016/s0955-0674(98)80011-2. [DOI] [PubMed] [Google Scholar]
- Chan RL, Gago GM, Palena CM, Gonzalez DH. Homeoboxes in plant development. Biochim Biophys Acta. 1998;1442:1–19. doi: 10.1016/s0167-4781(98)00119-5. [DOI] [PubMed] [Google Scholar]
- Chandra-Sekhar KN, Sawhney VK. Leaf development in the normal and solanifolia mutant of tomato (Lycopersicon esculentum) Am J Bot. 1990;77:46–53. [Google Scholar]
- Chen J-J, Janssen B-J, Williams A, Sinha N. A gene fusion at a homeobox locus: alterations in leaf shape and implications for morphological evolution. Plant Cell. 1997;9:1289–1304. doi: 10.1105/tpc.9.8.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DC. Macrosporogenesis and the development of the macrogametophyte of Lycopersicon esculentum. Am J Bot. 1931;18:739–751. [Google Scholar]
- Dengler NG. Comparison of leaf development in normal (+/+), entire (e/e), and Lanceolate (La/+) plants of tomato, Lycopersicon esculentum “Ailsa Craig.”. Bot Gaz. 1984;145:66–77. [Google Scholar]
- Frugis G, Giannino D, Mele G, Nicolodi C, Innocenti AM, Chiappetta A, Bitonti MB, Dewitte W, Van Onckelen H, Mariotti D. Are homeobox knotted-like genes and cytokinins the leaf architects? Plant Physiol. 1999;119:371–374. doi: 10.1104/pp.119.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ. Homeoboxes in the study of development. Science. 1987;236:1245–1252. doi: 10.1126/science.2884726. [DOI] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G. A polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature. 1997;386:44–51. doi: 10.1038/386044a0. [DOI] [PubMed] [Google Scholar]
- Gould A. Functions of mammalian Polycomb group and trithorax group related genes. Curr Opin Genet Dev. 1997;7:488–494. doi: 10.1016/s0959-437x(97)80075-5. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada J-P, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- Hake S, Char BR, Chuck G, Foster T, Long J, Jackson D. Homeobox genes in the functioning of plant meristems. Phil Trans R Soc Lond B. 1995;350:45–51. doi: 10.1098/rstb.1995.0136. [DOI] [PubMed] [Google Scholar]
- Hareven D, Gutfinger T, Parnis A, Eshed Y, Lifschitz E. The making of a compound leaf: genetic manipulation of leaf architecture in tomato. Cell. 1996;84:735–744. doi: 10.1016/s0092-8674(00)81051-x. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Scott MP. What determines the specificity of action of Drosophila homeodomain proteins? Cell. 1990;63:883–894. doi: 10.1016/0092-8674(90)90492-w. [DOI] [PubMed] [Google Scholar]
- Hayward HE. The Structure of Economic Plants. New York: Macmillan; 1938. Solanaceae: Lycopersicum esculentum. pp. 550–579. [Google Scholar]
- Howard RA. The petiole. In: Metcalfe CR, Chalk L, editors. Anatomy of the Dicotyledons. Oxford: Clarendon Press; 1979. pp. 88–96. [Google Scholar]
- Janssen B-J, Lund L, Sinha N. Overexpression of a homeobox gene LeT6 reveals indeterminate features in the tomato compound leaf. Plant Physiol. 1998a;117:771–786. doi: 10.1104/pp.117.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen B-J, Williams A, Chen J-J, Mathern J, Hake S, Sinha N. Isolation and characterization of two knotted-like homeobox genes from tomato. Plant Mol Biol. 1998b;36:417–425. doi: 10.1023/a:1005925508579. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Laible G, Dorn R, Reuter G. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter R, Vollbrecht E, Lowe B, Veit B, Yamaguchi J, Hake S. Sequence analysis and expression patterns divide the maize knotted1-like homeobox genes into two classes. Plant Cell. 1994;6:1877–1887. doi: 10.1105/tpc.6.12.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush GS, Rick CM. Studies on the linkage map of chromosome 4 of the tomato and on the transmission of induced deficiencies. Genetica. 1967;38:74–94. [Google Scholar]
- Kiyosue T, Ohad N, Yadegari R, Hannon M, Dinneny J, Wells D, Katz A, Margossian L, Harada JJ, Goldberg RB, Fischer RL. Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:4186–4191. doi: 10.1073/pnas.96.7.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM1 gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM. Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1999;96:296–301. doi: 10.1073/pnas.96.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P, Hake S. Genetic control of plant development. Curr Opin Biotechnol. 1998;9:189–195. [Google Scholar]
- Mihaly J, Mishra RK, Karch F. A conserved sequence motif in polycomb-response elements. Mol Cell. 1998;1:1065–1066. doi: 10.1016/s1097-2765(00)80107-0. [DOI] [PubMed] [Google Scholar]
- Moench TR, Gendelman HE, Clements JE, Narayan O, Griffin DE. Efficiency of in situ hybridization as a function of probe size and fixation technique. J Virol Methods. 1985;11:119–130. doi: 10.1016/0166-0934(85)90035-7. [DOI] [PubMed] [Google Scholar]
- Ohad N, Yadegari R, Margossian L, Hannon M, Michaeli D, Harada JJ, Goldberg RB, Fischer RL. Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell. 1999;11:407–415. doi: 10.1105/tpc.11.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnis A, Cohen O, Gutfinger T, Hareven D, Zamir D, Lifschitz E. The dominant developmental mutants of tomato Mouse-ear and Curl are associated with distinct modes of abnormal transcriptional regulation of a Knotted gene. Plant Cell. 1997;9:2143–2158. doi: 10.1105/tpc.9.12.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler R. Homeosis in plants. Am J Bot. 1988;75:1606–1617. [Google Scholar]
- Scanlon MJ, Schneeberger RG, Freeling M. The maize mutant narrow sheath fails to establish leaf margin identity in a meristematic domain. Development. 1996;122:1683–1691. doi: 10.1242/dev.122.6.1683. [DOI] [PubMed] [Google Scholar]
- Schneeberger R, Tsiantis M, Freeling M, Langdale JA. The rough sheath2 gene negatively regulates homeobox gene expression during maize leaf development. Development. 1998;125:2857–2865. doi: 10.1242/dev.125.15.2857. [DOI] [PubMed] [Google Scholar]
- Schumacher A, Magnuson T. Murine Polycomb- and trithorax-group genes regulate homeotic pathways and beyond. Trends Genetics. 1997;13:167–170. [PubMed] [Google Scholar]
- Serikawa KA, Martinez-Laborda A, Kim H-S, Zambryski PC. Localization of expression of KNAT3, a class 2 knotted1-like gene. Plant J. 1997;11:853–861. doi: 10.1046/j.1365-313x.1997.11040853.x. [DOI] [PubMed] [Google Scholar]
- Siegel BA, Verbeke JA. Diffusible factors essential for epidermal cell redifferentiation in Catharathus roseus. Science. 1989;244:580–582. doi: 10.1126/science.244.4904.580. [DOI] [PubMed] [Google Scholar]
- Sinha N, Lynch M. Fused organs in the adherent1 mutation in maize show altered epidermal walls with no perturbations in tissue identities. Planta. 1998;206:184–195. [Google Scholar]
- Steeves TA, Sussex IM. Patterns in Plant Development. Ed 2. Cambridge, UK: Cambridge University Press; 1989. [Google Scholar]
- Tamaoki M, Kusaba S, Kano-Murakami Y, Matsuoka M. Ectopic expression of a tobacco homeobox gene NTH15 dramatically alters leaf morphology and hormone levels in transgenic tobacco. Plant Cell Physiol. 1997;38:917–927. doi: 10.1093/oxfordjournals.pcp.a029252. [DOI] [PubMed] [Google Scholar]
- Timmermans MCP, Hudson A, Becraft PW, Nelson T. ROUGH SHEATH2: A Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science. 1999;284:151–153. doi: 10.1126/science.284.5411.151. [DOI] [PubMed] [Google Scholar]
- Timmermans MCP, Schultes NP, Jankovsky JP, Nelson T. Leafbladeless1 is required for dorsoventrality of lateral organs in maize. Development. 1998;125:2813–2823. doi: 10.1242/dev.125.15.2813. [DOI] [PubMed] [Google Scholar]
- Tsiantis M, Schneeberger R, Golz JF, Freeling M, Langdale JA. The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science. 1999;284:154–156. doi: 10.1126/science.284.5411.154. [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature. 1991;350:241–243. doi: 10.1038/350241a0. [DOI] [PubMed] [Google Scholar]
- Waites R, Selvadurai HRN, Oliver IR, Hudson A. The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell. 1998;93:779–789. doi: 10.1016/s0092-8674(00)81439-7. [DOI] [PubMed] [Google Scholar]
- Walker DB. Postgenital carpel fusion in Catharanthus roseus: III. Fine structure of the epidermis during and after fusion. Protoplasma. 1975;86:43–63. [Google Scholar]
- Yang T, Lev-Yadun S, Feldman M, Fromm H. Developmentally regulated organ- tissue- and cell-specific expression of calmodulin genes in common wheat. Plant Mol Biol. 1998;37:109–120. doi: 10.1023/a:1005902905512. [DOI] [PubMed] [Google Scholar]