ABSTRACT
Deep sequencing and single-chain variable fragment (scFv) yeast display methods are becoming more popular for discovery of therapeutic antibody candidates in mouse B cell repertoires. In this study, we compare a deep sequencing and scFv display method that retains native heavy and light chain pairing with a related method that randomly pairs heavy and light chain. We performed the studies in a humanized mouse, using interleukin 21 receptor (IL-21R) as a test immunogen. We identified 44 high-affinity binder scFv with the native pairing method and 100 high-affinity binder scFv with the random pairing method. 30% of the natively paired scFv binders were also discovered with the randomly paired method, and 13% of the randomly paired binders were also discovered with the natively paired method. Additionally, 33% of the scFv binders discovered only in the randomly paired library were initially present in the natively paired pre-sort library. Thus, a significant proportion of “randomly paired” scFv were actually natively paired. We synthesized and produced 46 of the candidates as full-length antibodies and subjected them to a panel of binding assays to characterize their therapeutic potential. 87% of the antibodies were verified as binding IL-21R by at least one assay. We found that antibodies with native light chains were more likely to bind IL-21R than antibodies with non-native light chains, suggesting a higher false positive rate for antibodies from the randomly paired library. Additionally, the randomly paired method failed to identify nearly half of the true natively paired binders, suggesting a higher false negative rate. We conclude that natively paired libraries have critical advantages in sensitivity and specificity for antibody discovery programs.
KEYWORDS: IL-21R, humanized mouse antibody repertoires, deep sequencing, yeast display
Introduction
Mouse immunization followed by hybridoma screening has long been used successfully for discovery of therapeutic monoclonal antibodies (mAbs) that are approved by the US Food and Drug Administration.1 Using this method, mice are first immunized with an immunogen and adjuvant. Conventionally, wild type mice are used, but the use of mice transgenically engineered to express human immunoglobulin (Ig) V(D)J sequences has recently become more popular.2 After assessing titer and sacrificing the animals, hybridomas are generated by fusing primary B cells with myeloma cells.3 Although hybridoma protocols are cheap, the process of screening polyclonal pools of hybridomas remains expensive and inefficient. To increase hybridoma screening throughput, large-scale antibody discovery groups, including one at Bristol-Myers Squibb (BMS), use costly robotic systems to automate workflows.
“Deep sequencing” genomic technology, which is an alternative for mouse antibody repertoire mining, involves the acquisition of millions of antibody sequences from B-cell RNA.4 However, in any given B cell, pairing between heavy and light chain is unique, and such native pairing is important for antibody function.5 Conventional deep sequencing methods do not link heavy and light chain immunoglobulin sequences at the single cell level. To address this deficiency, we previously developed a novel technology that combines molecular genomics, yeast single-chain variable fragment (scFv) display, fluorescence-activated cell sorting (FACS), and deep sequencing for antibody discovery.6-7 However, no studies have rigorously compared antibody discovery from natively paired with randomly paired antibody libraries.
IL-21R is expressed in many lymphoid cells, including B cells and activated CD4+ T cells, and it is the receptor for IL-21.8 Pathways for proliferation and differentiation are activated upon binding of IL-21 to IL-21R. Therefore, antagonism of IL-21R could modulate inflammatory diseases, such as rheumatoid arthritis, by blocking proliferation and differentiation of B cells and CD4+ T cells.9 We used IL-21R as an antibody target for technical assessment of natively paired versus randomly paired scFv libraries, both generated from the same pool of chimeric humanized mice immunized with recombinant IL-21R. We re-formatted 46 of the IL-21R binding scFv as full-length antibodies, and subjected them to a panel of assays, including kinetic analysis and epitope binning. We found that the natively paired method is more sensitive and specific than the random pairing method, yet using both methods concurrently helps generate additional antibody diversity that might be valuable for downstream development.
Results
Overview of the experimental approach
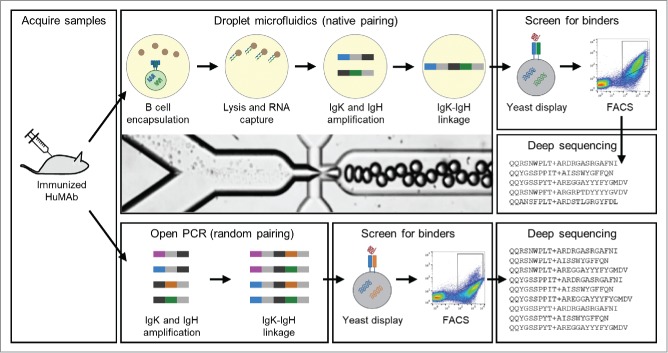
We first immunized six humanized mice with recombinant IL-21R (Fig. 1). Animals with a positive serum titer were sacrificed, and single cell suspensions were generated from the lymph nodes and pooled. We then ran approximately 1.2 million B cells through our emulsion droplet microfluidic platform,6-7 which captures mRNA from single B cells onto oligo-dT beads. The mRNA-bound beads were re-injected into a second emulsion with RT-PCR mix containing primers that generate fusions between heavy and light chain Ig. This process largely preserves cognate pairing between heavy and light chain Ig,6-7 although some mispairing may occur due to factors such as droplets that contain multiple cells. Randomly paired libraries were generated without the second emulsion, but with the same mRNA-bound beads, RT-PCR mix, and primers.
Figure 1.
Overview of the workflow used to generate the scFv libraries from B cells isolated from Medarex humanized mice. B cells were isolated from the lymph nodes of immunized mice then pooled. To make natively paired libraries, B cells were encapsulated into droplets with oligo-dT beads and a lysis solution (top). mRNA-bound beads were purified from the droplets, and then injected into a second emulsion with an OE-RT-PCR amplification mix that generates DNA amplicons that encode scFv with native pairing of heavy and light chain Ig. Libraries of natively paired amplicons were then electroporated into yeast for scFv display. FACS is used to identify high affinity scFv. Finally, deep antibody sequencing was used to identify all clones in the pre- and post-sort scFv libraries. The same process was used for generating randomly paired libraries, except RNA was isolated from a pool of B cells, and emulsions were not used for the OE-RT-PCR amplification step (bottom).
Next, the amplified libraries were expressed as surface scFv in yeast. The yeast display libraries were stained with biotinylated IL-21R antigen, and then subjected to FACS. Binding yeast scFv clones were recovered and subjected to deep antibody sequencing.6-7 Another round of staining, FACS, and deep antibody sequencing was performed to further enrich the libraries and increase confidence in the binders. Antibodies that were low frequency in pre-sort libraries and high frequency in post-sort libraries were then synthesized as full-length mAbs in Chinese hamster ovary (CHO) cells. We then used surface plasmon resonance (SPR) for affinity and epitope binning studies. We also performed FACS binding studies using CHO cells expressing recombinant cell surface IL-21R.
Isolation of anti-IL-21R scFv binders by yeast display
Before FACS sorting for binders, we analyzed the initial diversity of the yeast scFv libraries by obtaining approximately 1.2 million sequence reads for the natively paired library, and 1.3 million reads for the randomly paired library (Supplementary Table S1). We defined antibody “clones” as the consensus of closely related groups of scFv sequences with ≤2 amino acid differences in their complementarity-determining region (CDR)3 sequences. This is an extremely conservative definition of a clone, which corrects for sequencing errors but possibly also masks the long tail of diversity. We estimate that the natively paired scFv library contained 10,200 clones and the randomly paired scFv library contained 25,800 clones. The natively paired scFv library was much more oligoclonal than the randomly paired library (Fig. 2). Of note, the randomly paired library had a very flat distribution (the most abundant clone was only 0.16% of reads), whereas the natively paired library contained several higher abundance clones and a more gradual curve with several higher abundance clones (the most abundant sequence was 1.4% of reads and ∼50 sequences were >0.16% of reads).
Figure 2.
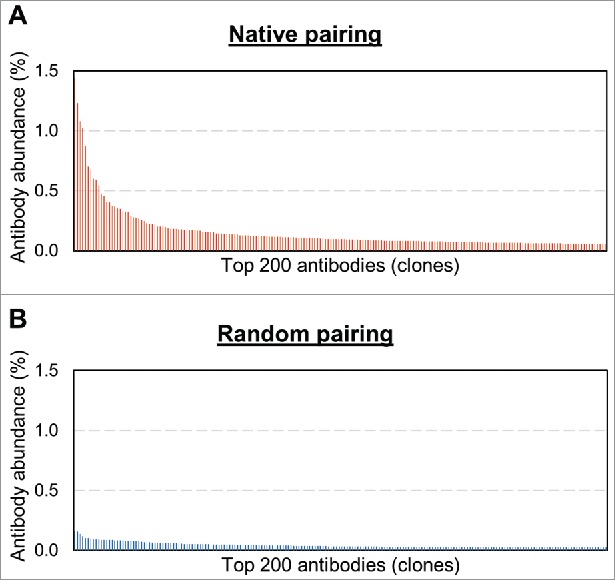
Relative oligoclonality of the two scFv libraries. (A) Natively paired scFv bar plot. (B) Randomly paired scFv bar plot. First, deep sequencing was used to tabulate the frequencies of scFv clones in each repertoire. Frequency is computed as the count of sequence instances of a given scFv clone divided by the total sequence instances in the deep sequencing data set. The 200 most frequent scFv clones from each repertoire were sorted from the most frequent sequence to the least frequent sequence. The frequencies for each scFv repertoire were then plotted. Note that each repertoire comprises a “long tail” of many more than 200 antibody sequences that are not shown.
As described above, the randomly paired library was generated from cell-free mRNA isolated from the same population of cells that was used to generate the natively paired library. Therefore, the diversity of the randomly paired library is the theoretical maximum for the native pairing microfluidics method. To estimate the “yield” of our native pairing microfluidics method, we divided the heavy chain CDR3 diversity observed in the natively paired library (2,800 clones) by the heavy chain CDR3 diversity observed in the randomly paired library (3,700 clones). We conclude that the yield of our native pairing microfluidics method for these samples is at least 76%.
After deep sequencing the initial yeast scFv libraries, we performed two rounds of FACS to identify IL-21R antigen-binding yeast. In a typical FACS dot plot for such experiments, the upper right corner comprises yeast clones that stain for both antigen binding and scFv expression (which contain a C-terminal c-Myc tag). The lower left corner comprises yeast that do not stain for either the antigen or scFv expression. The upper left corner comprises yeast that do not stain for scFv expression, but which stain for antigen binding, presumably representing non-specific binding of the antigen to the yeast cell surface. The lower right corner is made up of yeast that express the scFv but do not bind the antigen. We compute the frequency of antigen-binding yeast by dividing the count of yeast that double stain by the count of yeast that express an scFv (Fig. 3).
Figure 3.
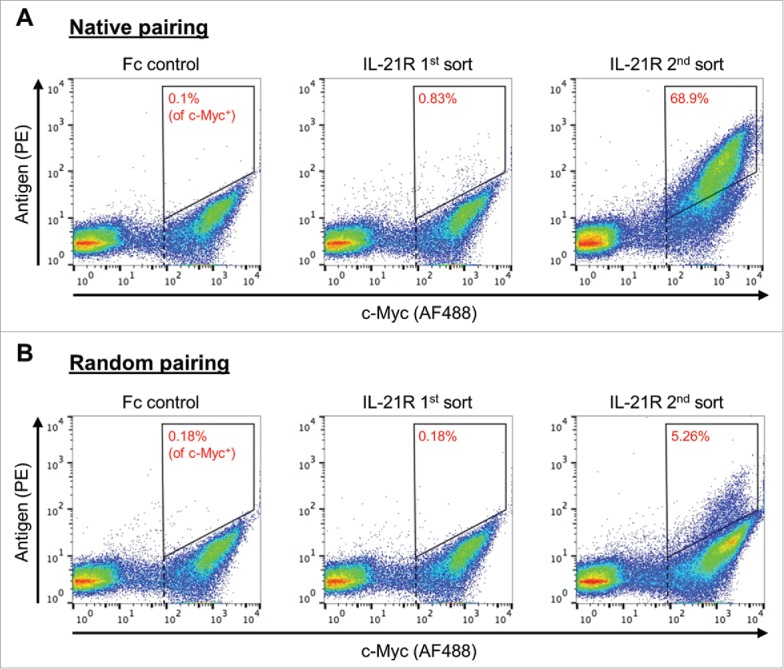
scFv libraries subjected to FACS for IL-21R. Staining for c-Myc (AF488) was used to differentiate yeast cells that express scFv from yeast cells that do not express scFv (x-axis). Staining for biotinylated IL-21R (PE) was used to identify yeast that express scFv that bind to the antigen (y-axis). The Fc negative control was used to set gates that are used to capture yeast clones that express scFv and which also bind antigen (upper right corner of the FACS plot). Gates for yeast selection are indicated by the quadrangle in the upper right corner of each FACS plot. The percentage in each quadrangle (red type) indicates the proportion of yeast that expressed c-Myc and fell within the gate. A vertical dotted line (black) indicates the gate used to determine the number of yeast that express scFv (c-Myc+). (A) 1st and 2nd sort FACS data for the natively paired scFv library. (B) 1st and 2nd sort FACS data for the randomly paired scFv library.
FACS on the natively paired scFv library yielded a higher proportion of antigen-binding yeast clones than FACS on the randomly paired scFv library. In the natively paired library, the proportion of antigen-positive yeast during the first sort was 0.83% (Fig. 3A). In contrast, the proportion of antigen-binding yeast during the first sort for the randomly paired library was no different than the Fc negative control (0.18%; Fig. 3B). During the second FACS, the proportion of antigen-binding yeast clones in the natively paired library was 68.9%, indicating high specificity. The specificity of scFv binder discovery in the randomly paired library was much lower, as indicated by the much lower proportion of binders (5.26%). We hypothesize that sorting the randomly paired library is less specific because random pairing leads to many additional non-binding yeast clones, making antigen-binding yeast rarer and therefore more difficult to detect.
Using the same deep sequencing methods that were used to determine the sequence content of the pre-sort yeast scFv libraries, we assessed the clonal diversity of each population of antigen-binding yeast clones, after the second FACS sort (Supplementary Tables S1-S3). Proportionally, the number of sequences from the pre-sort libraries that also appeared in the post-sort libraries was smaller for the random library than the native library (3.0% versus 5.8%, respectively). The scFv sequences in the post-sort libraries were always oligoclonal, with the top 10 antibodies representing 63.6% of the total sequence count in the natively paired library, and 51.5% of the total sequence count in the randomly paired library.
For expedience, we define a “binder” as any scFv sequence present at 0.1% frequency or greater in the second FACS sort deep sequencing data. By this definition, we identified a total of 44 scFv binders from the natively paired library, versus 100 scFv binders from the randomly paired library (Supplementary Tables S2, S3). Among the binders, the average enrichment after the second FACS was 792-fold in the randomly paired library, and 334-fold in the natively paired library. The largest enrichment was 17,188-fold in the randomly paired library (scFv Random-1), and 1,882-fold in the natively paired library (scFv Native-29 and Native-30). The average frequency of scFv binders in the original randomly paired library was 0.0065%, versus 0.028% for the natively paired library (none of the binders was present at >0.25% frequency in the original library). We conclude that, because binders were rarer in the original randomly paired library, larger enrichments were required to find randomly paired binders versus natively paired binders.
Sequence characteristics of natively paired and randomly paired anti-IL-21R binders
We compared the sequence characteristics of the 44 natively paired and 100 randomly paired scFv binders. A simple overlap analysis revealed that 30% of the natively paired binders were also discovered as binders with the randomly paired method, and 13% of the randomly paired binders were also discovered with the natively paired method as binders. We observed that 29/87 (33%) of the scFv binders discovered only in the randomly paired library were present in the natively paired pre-sort library (Supplementary Table S3). Thus, a significant proportion of “randomly paired” scFv were actually natively paired, but not discovered as binders in the natively paired library.
Alignment with germline V-gene sequences revealed that, generally, the IgKV and IgHV sequences of scFv binders from the natively paired library and the randomly paired library were 98–99% identical to their respective germline V sequences. Across all 144 scFv binders, the most divergent IgKV was 93.3% identical (scFv Random-88) and the most divergent IgHV was 94.5% identical (scFv Random-91). Prior studies of plasma cell repertoires for immunized mice yielded similar rates of divergence.4
Though all multiplex PCR suffers from some non-representative amplification, many investigators have used multiplex PCR to describe V-gene diversity in mouse repertoires.4,6 Our data showed that the diversities of IgKV and IgHV were higher among scFv binders from the randomly paired library (Supplementary Figure S1). One of the most frequent IgKV genes in the scFv binders from the randomly paired library (IGKV1-13*02) was not even present among the scFv binders from the natively paired library. Also of note, the most abundant IgKV gene in both pre-sort libraries was IGKV1D-16*01 for ∼32% of the reads. Only 7% of scFv binders from the natively paired library comprised IGKV1D-16*01, suggesting that this V-gene was not as favorable for binding despite its dominant abundance post-immunization. In contrast, binders from the randomly paired library retained IGKV1D-16*01 at the same pre-sort frequency (36%). In another example, IGKV3-20*01 was present in ∼20% of the pre-sort reads in both libraries, linked to a wide range of heavy chain sequences. IGKV3-20*01 was present in 8% of binders among post-sort randomly paired scFv, whereas the natively paired library increased its share to 27% of binders, suggesting that this V-gene was more favorable for binding when properly paired.
Prior work has shown that certain IgHV and IgKV gene families pair preferentially,5 possibly because certain pairings lead to more stable antibody molecules.10 To investigate this phenomenon, we examined frequencies of IgHV and IgKV pairings at the gene family level (Supplementary Tables S2-S3). Among scFv binders from the natively paired library, 77% were pairings between the IgKV3 and IgHV3 gene families, versus 29% for scFv binders from the randomly paired library. Another common pairing was between IgKV1 and IgHV3, comprising 16% of binders from the natively paired library and 21% of binders from the randomly paired library. The 100 scFv binders from the randomly paired library comprised only nine V-gene family pairings, versus only four V-gene family pairings for the 44 binders from the natively paired library. Taken together, these data suggest that natively paired scFv libraries produce binders with higher V-gene family pairing biases than randomly paired scFv libraries.
Because the set of 144 binders comprised very few V-gene family pairings, we hypothesized that most of the sequence diversity in our data set was derived from somatic hypermutation or affinity maturation in vivo. To investigate clonal lineages among FACS-sorted scFv binders, we clustered natively paired and randomly paired sequences with at most 9 amino acid differences across a concatenation of the IgH and IgK variable components of each scFv binder (Fig. 4). We defined a “major clonal cluster” as a cluster of five or more scFv sequences. We observed five large clonal clusters among the scFv binders. The major clonal clusters were all a mix of both native-paired and randomly paired scFv. Presumably, because the native-paired scFv occurred more frequently in true clonal lineages, the native-paired library yielded fewer unclustered scFv than the randomly paired library (3 vs. 51 scFv, respectively).
Figure 4.
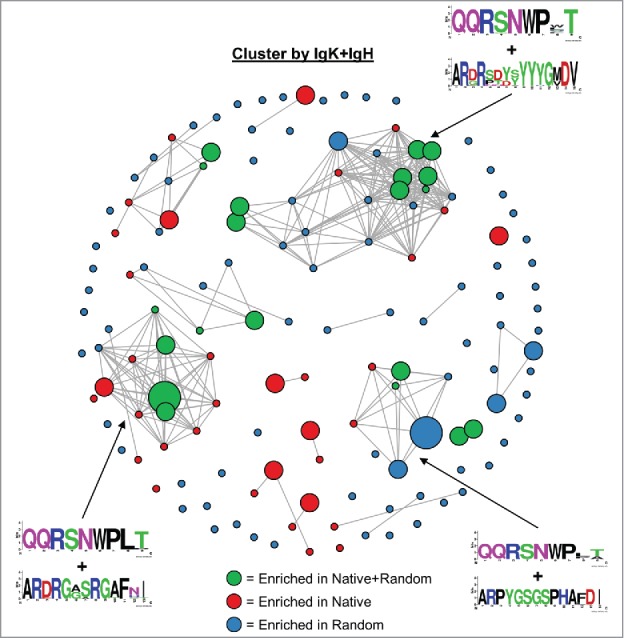
Clonal cluster analysis for FACS-sorted scFv binders. We computed the total number of amino acid differences between each pairwise alignment of FACS-sorted scFv. Edges were drawn only for pairwise alignments with ≤9 amino acid differences. The node for each scFv sequence was sized based on frequency in the FACS-sorted population: small (<1% frequency), medium (1–10% frequency), and large (>10% frequency). scFv isolated from the native+random libraries are indicated with green circles, scFv isolated from the native library only are indicated with red circles, and scFv isolated from the random library only are indicated with blue circles. Web logos of CDR3K + CDR3H sequences of select clusters are shown.
Clustering as a concatenation of IgH and IgK domains masks the sequence diversity contributed by the IgH versus the IgK. To investigate, we clustered IgH (Supplementary Figure S2) and IgK (Supplementary Figure S3) sequences separately, using amino acid difference thresholds of 6 and 3, respectively. Major clonal clusters accounted for a larger percentage of all clones for the IgK clustering analysis than for the IgH and concatenated libraries (84.4% vs. 32.4% and 39.7%, respectively). Also, we only observed four unclustered IgK sequences, all of which were discovered in the randomly paired scFv library. One major clonal cluster in the IgK-only analysis (CDR3K: QQFNSYP.T) was composed exclusively of scFv post second FACS from the randomly paired library (Supplementary Figure S3). A closer examination of this IgK clonal cluster reveals that the 17 IgK sequences pair with 14 distinct IgH sequences. Taken together, these data suggest that, whether randomly or natively paired, the IgK sequences are generally more promiscuous than IgH sequences.
Validation of natively paired and randomly paired anti-IL-21R mAbs
We selected a set of 46 scFv binders to synthesize as full-length antibodies with a human IgHG1 constant domain (Table 1). For both the natively and randomly paired libraries, we selected the 20 most abundant post-sort antibodies, most of the antibodies that had identical CDR3K+CDR3H sequences in both libraries, and several lower abundance antibodies unique to each library. MAbs discovered in both libraries are named using a “Native-#;Random-#” concatenation; for example, the mAb Native-16;Random-19 was synthesized using the identical sequence shared by scFv Native-16 and Random-19. The full-length antibodies were produced in CHO cells. The binding specificity and affinity of each antibody was verified through multiple assays using clarified supernatant.
Table 1.
Characteristics of the 46 full-length anti-IL-21R mAbs. For the Non-native LC (light chain) column, CDR3H that were enriched in the random library but were not found in the native library are indicated by “?” to indicate that it is not known if the enriched pairing is natively paired or not. In the Affinity column, * indicates that the observed data is <50% of the calculated Rmax. The major epitope bins are as defined in Fig. 5.
| Native/Random Enriched | Native Ab # | Random Ab # | CDR3K+CDR3H | Non-native LC | Native Pre-sort (%) | Native Post-sort (%) | Random Pre-sort (%) | Random Post-sort (%) | Affinity to IL-21R (KD) | IL-21R ka (1/Ms) | IL-21R kd (1/s) | Binds surface IL-21R? | Major epitope bin |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Native+Random | Native-1 | Random-6 | QQRSNWPLT+ARDRGASRGAFNI | 0.032 | 17 | 0 | 4.9 | 20nM | 10,350 | 0.000205 | Yes | Bottom | |
| Native+Random | Native-2 | Random-20 | QQRSNWPLT+ARDRGGSRGAFHI | 0.02 | 9.5 | 0 | 0.66 | 49nM | 22,000 | 0.00105 | Yes | Bottom | |
| Native+Random | Native-3 | Random-4 | QQRSNWPLT+ARDRSDYSYYYGMDV | 0.22 | 7.2 | 0.013 | 5.7 | 16nM | 12,500 | 0.0002 | Yes | Top | |
| Native+Random | Native-5 | Random-40 | QQYGSSPYT+AREGGAYYYFYGMDV | 0.058 | 5.6 | 0.0087 | 0.26 | 660nM* | 3,750 | 0.0024 | No | No binning observed | |
| Native+Random | Native-6 | Random-27 | QQRSNWPLT+ARPYGSGSPHAFDI | 0.044 | 5.4 | 0.0035 | 0.39 | 97nM | 24,000 | 0.0023 | Yes | Bottom | |
| Native+Random | Native-7 | Random-10 | QQRSNWPFT+ARGRPTDYYYYGVDV | 0.05 | 3.6 | 0 | 1.5 | 9.25nM | 14,000 | 0.00013 | Yes | Top | |
| Native+Random | Native-9 | Random-3 | QQYGSSPRT+ARHEVWGPFDY | 0.02 | 3.2 | 0.0023 | 6.3 | 170nM | 7,700 | 0.00125 | Yes | Bottom | |
| Native+Random | Native-15 | Random-2 | QQRSNWPWT+ARDRSDYSYYYGMDV | 0.023 | 1.1 | 0.0097 | 8.1 | 11.5nM | 13,000 | 0.000155 | Yes | Top | |
| Native+Random | Native-16 | Random-19 | QQYGSSIFT+AREGGAYYYFYGMDV | 0.048 | 1.1 | 0.0021 | 0.68 | 120nM | 9,000 | 0.0011 | No | Middle | |
| Native+Random | Native-17 | Random-8 | QQYNSYPIT+ASLHY | 0.026 | 0.81 | 0.0014 | 3.1 | 30nM | 4,850 | 0.0001355 | No | Top | |
| Native+Random | Native-23 | Random-5 | QQRSNWPRT+ARDRSDYSYYYGMDV | 0.0052 | 0.24 | 0.0058 | 5.4 | 5.05nM | 13,500 | 0.0000705 | Yes | Top | |
| Native | Native-4 | NA | QQYGSSPPIT+AISSWYGFFQN | 0.031 | 5.8 | 0 | 0.0019 | 22.5nM | 7,350 | 0.00017 | Yes | Bottom | |
| Native | Native-8 | NA | QQANSFPLT+ARDSTLGRGYFDL | 0.016 | 3.2 | 0 | 0.0019 | 9.45nM | 29,500 | 0.000275 | Yes | Bottom | |
| Native | Native-10 | NA | QQYGNSPRDPIT+ARAPTGFFDY | 0.026 | 3.1 | 0 | 0.0037 | 15.5nM | 12,900 | 0.0002 | No | Middle | |
| Native | Native-11 | NA | QQANSFPT+ARIAPRPGWYFDL | 0.044 | 2.7 | 0 | 0.0028 | 6.05nM | 14,000 | 0.0000835 | No | Middle | |
| Native | Native-12 | NA | QQRSNWPLT+ARDRGGSRGAFAI | 0.023 | 1.7 | 0.0021 | 0 | 115nM | 18,000 | 0.00205 | Yes | Bottom | |
| Native | Native-13 | NA | QQRSNWPLT+ARDTLTVFFDY | 0.22 | 1.6 | 0.0021 | 0 | 200nM* | 16,000 | 0.0031 | No | Bottom | |
| Native | Native-14 | NA | QQRSNWPLT+ARDLSHTIRLGNWFDP | 0.045 | 1.4 | 0.00032 | 0.032 | 23nM | 8,700 | 0.0002 | Yes | Bottom | |
| Native | Native-18 | NA | QQRSNWPLT+VRDRSNYYYYHGMDV | 0.077 | 0.75 | 0.0019 | 0 | 35.5nM | 13,500 | 0.00048 | Yes | Top | |
| Native | Native-19 | NA | QQRSNWPLT+ARDRGGSRGAFGI | 0 | 0.34 | 0 | 0 | 32nM | 16,500 | 0.00053 | Yes | Bottom | |
| Native | Native-20 | NA | QQYGSSPP+AREGGDILTGYYSNYYYYGMDV | 0.017 | 0.31 | 0 | 0 | Does not bind | — | — | No | — | |
| Native | Native-22 | NA | QQRSNWPLT+ARDRNDYYYYYGMDV | 0.0074 | 0.26 | 0 | 0.012 | 15.15nM | 8,850 | 0.0001335 | Yes | Top | |
| Native | Native-28 | NA | QQRSNWPLT+ARDRGGSRGAFNI | 0.011 | 0.17 | 0.0013 | 0.033 | 200nM | 11,000 | 0.00225 | Yes | Bottom | |
| Native | Native-34 | NA | QQYNSYPLT+AALDY | 0.026 | 0.14 | 0 | 0.014 | 9.2nM | 10,450 | 0.000098 | No | Top | |
| Native | Native-35 | NA | QQRSNWPLT+ARERSDYSYYYGMDV | 0.0015 | 0.13 | 0 | 0.08 | 3.1nM | 10,000 | 0.000032 | Yes | No binning observed | |
| Native | Native-37 | NA | QQYGSSPIT+ARFSNWGFDY | 8.50E-05 | 0.12 | 0.0014 | 0 | 23nM | 10,000 | 0.00022 | No | Middle | |
| Random | NA | Random-1 | QQRSNWPPT+ARPYGSGSPHAFDI | 0.0011 | 0.018 | 0.00064 | 11 | 56nM | 20,000 | 0.00115 | Yes | Bottom | |
| Random | NA | Random-7 | QQYNSYPIT+ATMGT | ? | 0 | 0.0021 | 0 | 3.9 | 4.15nM | 24,500 | 0.0001015 | Yes | Top |
| Random | NA | Random-9 | QQRSNWPPA+ARPYGSGSPHAFDI | Yes | 0 | 0.0021 | 0 | 1.6 | 103.5nM | 29,000 | 0.0031 | Yes | Bottom |
| Random | NA | Random-11 | QQRSNWPWT+ARDRNDYYYYYGMDV | Yes | 0 | 0 | 0 | 1.3 | 26.5nM | 9,200 | 0.000245 | Yes | Top |
| Random | NA | Random-12 | QQYNSYPLT+ATMGT | ? | 0 | 0 | 0.0021 | 1.2 | 6.8nM | 25,500 | 0.00017 | Yes | Top |
| Random | NA | Random-13 | QQFNSYPQT+ARDGGSGAFDI | Yes | 0 | 0 | 0.0021 | 0.95 | Does not bind | — | — | No | — |
| Random | NA | Random-14 | QQRSNWPLT+ARGRPTDYYYYGVDV | 0.00068 | 0.048 | 0.0017 | 0.92 | 17.5nM | 15,000 | 0.00026 | Yes | Top | |
| Random | NA | Random-15 | QQRSNWPT+ARDLSHTIRLGNWFDP | Yes | 0 | 0 | 0 | 0.86 | 1.9nM | 5,650 | 0.00001 | Yes | Bottom |
| Random | NA | Random-16 | QQYGSSPIT+VRFSNWGFDY | 0.012 | 0 | 0 | 0.77 | 36nM | 7,500 | 0.000275 | No | Middle | |
| Random | NA | Random-17 | QQRSNWPLT+ARPYGSGSPHALDI | Yes | 0 | 0.026 | 0 | 0.72 | 137nM | 15,000 | 0.002 | Yes | Bottom |
| Random | NA | Random-18 | QQRSNWPWT+ARDRSNYYYYYGMDV | Yes | 0 | 0 | 0.001 | 0.71 | 25.5nM | 9,200 | 0.00025 | Yes | Top |
| Random | NA | Random-21 | QQRSNWPYT+ARGRPTDYYYYGVDV | Yes | 0 | 0 | 7.90E-05 | 0.6 | 19.5nM | 17,000 | 0.000335 | Yes | Top |
| Random | NA | Random-23 | QQYNSYPLT+VNMDV | ? | 0 | 0 | 0.0014 | 0.54 | 55nM | 14,500 | 0.00078 | Yes | Top |
| Random | NA | Random-30 | QQYGSSLT+AREGGDILTGYYSNYYYYGMDV | Yes | 0 | 0 | 0 | 0.33 | Does not bind | — | — | No | — |
| Random | NA | Random-32 | QQYGSSRT+ARHEVWGPFDY | Yes | 0 | 0 | 0 | 0.32 | 98.5nM | 33,500 | 0.0035 | Yes | Bottom |
| Random | NA | Random-35 | QQYNSYPLT+ASLHY | 0.0027 | 0 | 0.0052 | 0.29 | 67.5nM | 8,500 | 0.000575 | No | Top | |
| Random | NA | Random-48 | QQYGSSPLT+ARDLSHTIRLGNWFDP | 0.00042 | 0 | 0.0012 | 0.21 | Does not bind | — | — | No | — | |
| Random | NA | Random-68 | QRTYNAPLT+ASLTGDYYYGMDV | Yes | 0 | 0 | 0 | 0.15 | Does not bind | — | — | No | — |
| Random | NA | Random-96 | QQYNSYPLT+ARVGLTGDYYYGIDV | Yes | 0 | 0 | 0 | 0.1 | Does not bind | — | — | No | — |
| Random | NA | Random-97 | QQRSNWPWT+ARDRSDYAYYYGMDV | Yes | 0 | 0.013 | 0 | 0.1 | 10.8nM | 10,800 | 0.00011 | Yes | Top |
Next, we analyzed the binding kinetics of the mAbs on an array SPR instrument. We tethered the mAbs to the chip surface via the Fc region. IL-21R protein was injected at 5 concentrations, ranging from 2 nM to 500 nM, and binding was globally fit to estimate the ka (association rate constant, or “on-rate”), kd (dissociation rate constant, or “off-rate”), and KD (affinity). Of the 46 clones, 40 showed clear binding to the soluble IL-21R and were suitable for kinetic evaluation (Table 1; Supplementary Figure S4). We observed substantial kinetic diversity, with affinities ranging from 1.9 nM to >500 nM. All 11 mAbs that were enriched from both libraries bound to soluble IL-21R. The percentage of antibodies that bound from the natively paired library (93%) was higher than from the randomly paired library (75%), although the difference was not statistically significant (Chi-squared test, α = 0.05). The average KD was also not significantly different between the native+random enriched, native only enriched, and random only enriched mAbs (108 nM vs. 50.7 nM vs. 44.4 nM, respectively; t-test, α = 0.05). However, if the binders with non-native light chains are separated from those with native light chains (Table 1), the percentage of binders with native light chains (94%) is significantly higher than binders with non-native light chains (67%; Chi-squared test, α = 0.05).
Using the affinity data, we investigated whether the efficiency of recovery of natively paired mAbs that bind soluble antigen is different between the randomly and natively paired libraries. First, we examined the scFv second FACS data to categorize each of the full-length antibodies as true positive (TP), false positive (FP), or false negative (FN) for both the native and randomly paired libraries (Supplementary Table S4). In the context of the randomly paired library, we counted non-natively paired antibodies as neither TP nor FP because they could not have been discovered in the natively paired library. We then computed False Discovery Rate (FDR = FP/(TP+FP)) and False Negative Rate (FNR = FN/(TP+FN)). FNR was significantly higher in the randomly paired library than the natively paired library (44% versus 22%, respectively; binomial proportion test, α = 0.05). FDR was not significantly higher in the randomly paired library than the natively paired library (5% versus 4%, respectively; binomial proportion test, α = 0.05). We conclude that the randomly paired method fails to detect nearly half of true natively paired binders, resulting in a higher rate of false negatives.
Antibodies that bind soluble antigen may not bind endogenously expressed antigen on the cell membrane. To investigate, we used FACS to test whether each mAb binds to surface-expressed IL-21R. Recombinant IL-21R was stably expressed on the surface of CHO cells. These CHO cells were then incubated with each individual mAb supernatant and then co-stained with fluorescently labeled anti-human IgG antibody. CHO cells stably expressing programmed cell death 1 (PD-1)6 were used as a negative control. We then performed FACS to measure the ability of the antibody to bind the IL-21R-expressing cells versus the PD-1 negative control (Table 1; Supplementary Figure S5). Of 46 mAbs, 30 (65%) specifically bound the IL-21R-expressing cells. The fraction of positive cell surface binders was not significantly different between native+random enriched, native only enriched, and random only enriched mAbs (73% vs. 60% vs. 65%, respectively; Chi-squared test, α = 0.05).
Drug development programs often require antibodies that bind to a diversity of epitopes to increase the probability of finding candidates with specific pharmacological properties. To investigate the epitope diversity of our set of mAbs, we performed high-throughput epitope binning using array SPR in a modified classical sandwich protocol. Antibodies were covalently linked to the chip (as “ligands”), and then antigen and competitive antibodies (as “analytes”) were individually bound to the ligands. Whereas all mAbs were tested both as analyte and ligand, not all ligands remained active following repeated regeneration cycles. This reduced the useful data to an 18 ligand × 46 analyte competitive binding matrix. We observed six non-identical epitope bins that group into three distinct communities (Fig. 5; Supplementary Figure S6; Table 1). The two major communities, consisting of 17 mAbs (Top) and 16 mAbs (Bottom), were clearly independent of each other. Five other mAbs (Middle) bind an intermediate epitope, and they compete with one another and with a subset of mAbs from both major bins. Antibodies from both the natively paired and randomly paired libraries were represented across the three epitope bins. We also note that all five members of the Middle bin bound to soluble IL-21R but did not bind to the cell surface expressed IL-21R, suggesting that this intermediate epitope is only present in soluble IL-21R but is lost in the natively folded protein expressed on the cell surface.
Figure 5.
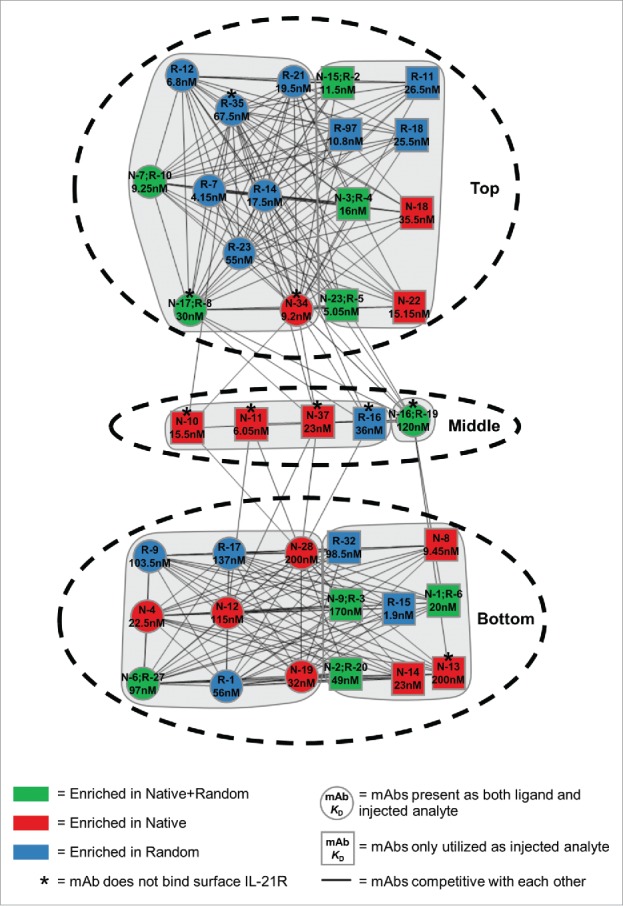
Epitope binning analysis for full-length anti-IL-21R mAbs. MAbs that were competitive with each other are indicated by a connecting line and therefore bind a similar epitope. MAbs with highly similar competition profiles were grouped together within communities (grey regions). The three major, distinct communities were indicated by the black hashed ovals. Circles represent mAbs that regenerated well as immobilized ligands and had both ligand and analyte competition profiles. Squares represent clones where the competition data was only available in the analyte direction. The affinity (KD) of each mAb is indicated inside each shape. The color of the shape indicates whether the mAbs were enriched in both native+random libraries (green), the native only library (red), or the random only library (blue). *, indicates mAbs that did not bind surface expressed IL-21R by FACS.
Comparison of anti-IL-21R mAbs with closely related sequences
The sequences of several mAbs were very similar to each other, and in most cases, they displayed similar binding properties (Table 1). However, there were two examples where small changes led to markedly different functions. First, the mAbs Native-14, Random-15, and Random-48 have very similar heavy chains with only two differences that are both outside of the CDR3H region; the light chains of Native-14 and Random-15 were very similar with only one deletion in CDR3K and one substitution in the J-gene, while Random-48 had an additional nine differences throughout, including four in the CDR3K (Supplementary Figure S7). Random-15 had a 10-fold stronger affinity to IL-21R than Native-14, and Random-48 did not bind IL-21R at all. The heavy chains for Random-15 and Random-48 were identical to each other (and very similar to Native-14), thus the different light chains appeared to be responsible for determining the ability of these pairings to bind IL-21R. Second, the mAbs Native-2;Random-20, Native-12, Native-19, and Native-28 all had identical light chains, and their heavy chains varied by only a few amino acids (Supplementary Figure S8). All four sequences bound to IL-21R, but the strength of binding was variable, ranging from KD of 32 nM to 200 nM. These data suggest that different, yet similar, light chains paired with the same heavy chain can lead to highly divergent binding properties, and minor changes in heavy chain sequences can also alter antibody function.
Discussion
In this study, we discovered anti-IL-21R antibodies of potential therapeutic interest in both natively paired and randomly paired scFv libraries generated from chimeric humanized mice. Screening natively paired libraries identified many of the same antibodies as screening randomly paired libraries. However, the natively paired and randomly paired libraries yielded many distinct antibodies; for example, one major IgK clonal cluster was not present at all among scFv identified in the natively paired library. The randomly paired library yielded more antibody candidates, but a closer look revealed that 33% of the scFv binders discovered only in the randomly paired library were present in the natively paired pre-sort library. Thus, a significant proportion of “randomly paired” scFv were still natively paired, but not discovered in the natively paired library. We found that antibodies with native light chains were more likely to bind IL-21R than antibodies with non-native light chains, suggesting a higher false positive rate for randomly-paired antibodies. Additionally, the randomly paired method failed to identify nearly half of the true binders, suggesting a higher false negative rate. We conclude that natively paired libraries have key advantages in sensitivity and specificity for antibody discovery programs.
We speculate that natively paired antibodies may be more “developable” than randomly paired antibodies. For example, randomly paired antibodies could be inherently less stable than natively paired antibodies because certain IgHV and IgKV pairings result in more stable proteins.5 Future work should therefore compare the stability and pharmacokinetics of randomly paired antibodies versus natively paired antibodies. Also, randomly paired antibodies have not been subjected to in vivo selection against self-reactivity,11-12 so future work should test the self-reactivity of natively paired antibodies versus randomly paired antibodies. We also note that the assays performed in our study measured affinity, rather than avidity, and antibody efficacy and persistence in the lymphoid tissues could be variously enhanced and modulated by avidity to cell surface receptors. Because only natively paired antibodies were subjected to germinal center selection in vivo,13-14 future work should compare the avidity of natively paired antibodies versus randomly paired antibodies.
Previously, targeted biochemical studies of a handful of antibodies have suggested that the association between IgHV and IgKV contribute significantly to antibody structural and functional diversity.15-16 Additionally, studies of IgHV-IgKV pairing in tens to hundreds of peripheral B cells5,17 suggested a strong bias toward a limited number of IgHV-IgKV pairings. However, such pairing biases could be driven by initial association during B cell development, selection of biochemically stable pairings, de-selection of self-reactive antibodies, or exposure to antigen. Other studies of IgHV-IgKV pairing in naïve versus antigen-experienced B cell repertoires18-19 suggested that pairing is initially random, and then becomes biased after antigen exposure. Given that we specifically selected antigen-exposed IgHV-IgKV pairs, our data lend additional evidence to the idea that antigen exposure biases repertoires toward a limited set of pairings. We also found significant light chain promiscuity, even among natively paired antibodies, which is consistent with prior evidence of light chain promiscuity in both naïve and antigen-experienced B cells.19-20
We are not aware of any prior studies of chimeric mouse repertoires using deep sequencing. Generally, the Medarex mice that we used produced a robust diversity of fully human anti-IL-21R antibody sequences. However, the mice yielded a low diversity of light chain V-genes; for example, >50% of scFv binders in the natively paired library were IgKV3-11*01. Additionally, the V sequences of scFv binders from both libraries were 98–99% identical to germline V sequences, suggesting little if any affinity maturation in vivo. Our analysis of highly similar mAb sequences demonstrated that a few amino acid substitutions can have major impacts on function (Supplementary Figure S7-S8). Thus, immunization programs that increase sequence diversity are also likely to increase functional diversity. We are currently exploring alternative methods for mouse immunization to increase affinity maturation in vivo, including longer immunization protocols21 and fusion of antigens to T cell epitope-containing sequences.22 The methods used in this study likely result in antigen-binding antibodies mainly from plasmablasts, whereas alternative immunization methods might result in formation of memory B cells that produce antigen-binding antibodies. When performing the alternative immunization methods, the resulting memory B cells might yield higher diversity, affinity matured, and higher affinity antibodies. Because the mechanisms influencing antibody diversity in transgenic mice are poorly characterized,23 future work should apply deep sequencing methods to different mouse strains.
As previously reported, deep sequencing and random Ig pairing has been successfully used to identify high affinity mAbs, with FDRs of 0–40%.4,24-28 Generally, these groups first used bioinformatics to identify the most common antibodies in the animals, and then paired frequent heavy chains with frequent light chains. Because these methods only consider high frequency antibodies, it seems likely that conventional hybridoma methods would yield similar candidates. Tellingly, one study compared randomly paired phage display with algorithmic selection in deep sequencing data, and found that both methods identified antigen-specific binders with low FDR, but phage display yielded a different set of antibodies.24 The authors observed that most of the clones identified by phage display were present at very low frequencies in the deep sequencing data. Therefore, prior literature is consistent with our finding that antibody repertoire frequency is not necessarily correlated with binding affinity, so ignoring lower frequency antibodies surely results in much higher FNRs.
Of the scFv that we converted to full-length mAbs, 13% were not functional in any binding assay. Presumably, scFv do not always reflect the function of full-length mAbs, which sometimes results in false positive scFv. We note that 83% of the false positives were randomly paired, suggesting that natively paired scFv are more likely to become developable mAbs. In the future, we may explore alternative methods, such as antigen-binding fragment (Fab) display, which reflect the function of the full-length mAb more accurately. False positives may also have resulted from an overly inclusive FACS gating strategy. Such issues could have resulted in collection of non-specific binders or non-binders, even after a second round of FACS. Finally, future work that integrates FACS data with computational sequence analysis may decrease the rate of false positives. Prior computational work has focused on choosing the most frequent binders,4,24-25,27 or identifying antibodies shared between replicate mice.28 Other approaches for future study include computational structural analysis of putative binders and computational methods for detection of affinity maturation.
In this study, we identified >100 high-confidence scFv binders. Our yield was comparable to prior work using similar technology in immunized wild type mice and primary human samples.6-7 We acknowledge that development of any of the mAbs identified in this study would require a substantial number of non-clinical studies before clinical studies could be considered. The entire scFv binder discovery process, non-inclusive of mouse immunizations, required only three weeks of effort and a single technician. The randomly paired library required substantially more effort than the natively paired library because additional rounds of FACS were required to isolate binders. We used robotics to automate portions of the process of scFv re-engineering and mAb expression to further minimize technician hours. Though natively paired libraries clearly have critical advantages, a good approach might be to use both natively paired and randomly paired protocols concurrently, in combination with other discovery methods, to yield the richest possible intellectual property portfolio and the safest and most efficacious candidates.
Materials and methods
Mouse immunization and sample preparation
All mouse work was overseen by a licensed veterinarian. Six Medarex (BMS) Human Ig transgenic mice were immunized with recombinant IL-21R-ECD-TVMV-His protein (amino acids 1–232 expressed in 293–6E cells and purified using Ni column followed by 26/60 Superdex 200 column), using Ribi (Sigma) as an adjuvant. The immunogen (7.5 μg) was injected into each footpad every 7 days for 33 days. Titer was assessed by ELISA on a 1:2 dilution series of each animal's serum, starting at a 1:50 dilution. A final tail boost of 10 μg without adjuvant was given to each animal before harvest. Lymph nodes (popliteal, inguinal, axillary, and mesenteric) were surgically removed after sacrifice. Single cell suspensions for each animal were made by manual disruption followed by passage through a 40 μM filter, and then combined into a single pool of cells.
The EasySep™ Mouse Pan-B Cell Isolation (Stemcell Technologies) negative selection kit was used to isolate B cells from the single cell suspension. The lymph node B cell populations were quantified by counting on a C-Chip hemocytometer (Incyto) and assessed for viability using Trypan blue. The cells were then diluted to 6,000 cells/μL in phosphate-buffered saline with 12% OptiPrep™ Density Gradient Medium (Sigma). This cell mixture was used for microfluidic encapsulation as described in the next section.
Generating paired heavy and light chain libraries
Generation of libraries was performed in three steps: 1) poly(A)+ mRNA capture, 2) multiplexed overlap extension reverse transcriptase polymerase chain reaction (OE-RT-PCR), and 3) nested PCR to remove artifacts and add adapters for deep sequencing or yeast display libraries. These steps were identical for the natively paired and randomly paired libraries, except that OE-RT-PCR was performed in emulsion microdroplets for the natively paired library, whereas for the randomly paired library OE-RT-PCR was not performed in emulsion microdroplets.
Briefly, we encapsulated 1.2 million B cells into fluorocarbon oil (Dolomite) emulsion microdroplets with lysis buffer (20 mM Tris pH 7.5, 0.5 M NaCl, 1 mM EDTA, 0.5% Tween-20, and 20 mM DTT) and oligo-dT beads (New England BioLabs), using a custom co-flow emulsion droplet microfluidic chip, as described previously.6-7 The beads were extracted from the droplets using Pico-Break (Dolomite).
Next, for natively paired libraries, we performed multiplex OE-RT-PCR in emulsions, using purified RNA-bound beads as template. Glass Telos microfluidic chips (Dolomite) were used to encapsulate the beads into mineral oil emulsion microdroplets. For randomly paired libraries, we performed multiplex OE-RT-PCR in standard PCR tubes without emulsions, using an aliquot of the same RNA-bound beads as template. OE-RT-PCR molecular biology was identical for natively paired and randomly paired library generation, and has been described previously.6-7
Nested PCR was identical for both natively paired and randomly paired libraries. First, the OE-RT-PCR product was gel purified. Then, PCR was performed to add adapters for Illumina sequencing or yeast display; for sequencing, a randomer of seven nucleotides was added to increase base calling accuracy in subsequent next generation sequencing steps. Nested PCR is performed with 2 × NEBNext High-Fidelity amplification mix (New England BioLabs) with either Illumina adapter containing primers or primers for cloning into the yeast expression vector.
Yeast library screening
Human IgG1-Fc (Thermo Fisher Scientific) and human IL-21R-ECD-TVMV-His (described above) proteins were biotinylated using the EZ-Link Micro Sulfo-NHS-LC-Biotinylation kit (Thermo Fisher Scientific). The biotinylation reagent was resuspended to 9 mM and added to the protein at a 50-fold molar excess. The reaction was incubated on ice for 2 hours, then the biotinylation reagent was removed using Zeba desalting columns (Thermo Fisher Scientific). The final protein concentration was calculated with a Bradford assay.
Saccharomyces cerevisiae cells (ATCC) were electroporated (Bio-Rad Gene Pulser II; 0.54 kV, 25 uF, resistance set to infinity) with gel-purified nested PCR product and linearized pYD vector6-7 for homologous recombination in vivo. Transformed cells were expanded and induced with galactose to generate yeast scFv display libraries.
Expanded scFv libraries were then stained with anti-c-Myc (Thermo Fisher Scientific A21281) and an AF488-conjugated secondary antibody (Thermo Fisher Scientific A11039). To select scFv-expressing cells that bind to IL-21R, biotinylated antigens were added to the yeast culture (70 nM final concentration) and then stained with PE-streptavidin (Thermo Fisher Scientific). Approximately two million cells were then flow sorted on a BD Influx (Stanford Shared FACS Facility) for double-positive cells (AF488+/PE+), in parallel with positive and negative gating controls. The first round FACS clones were recovered, expanded, and then subjected to a second round of FACS with the same antigen at 70 nM final concentration.
Sequence analysis
Libraries were sequenced on a MiSeq (Illumina) using a 500 cycle MiSeq Reagent Kit v2, as described previously.6-7 We performed sequencing in two separate runs. In the first run, we directly sequenced the scFv libraries to obtain a forward read of 340 cycles for the light chain V-gene and CDR3, and a reverse read of 162 cycles that covers the heavy chain CDR3 and part of the heavy chain V-gene. In the second run, we first used the scFv library as a template for PCR to separately amplify heavy and light chain V-genes. We then obtained a forward read of 340 cycles and a reverse read of 162 cycles for the heavy and light chain Ig separately, providing full-length V(D)J sequences with some overlapping sequence.
Error correction, reading frame identification, and FR/CDR junction calls were performed as described previously.6-7 By default, reads with E>1 (E is the expected number of errors) were discarded, leaving reads for which the most probable number of base call errors is zero. As an additional quality filter, singleton nucleotide reads were discarded because sequences found two or more times have a high probability of being correct.29 V and J gene families were identified through nucleotide sequence alignment with the IMGT database.30
We defined “clones” conservatively (Supplementary Table S1). First, we concatenated the CDR3K and CDR3H amino acid sequences from each scFv sequence into a single contiguous amino acid sequence. Next, we used USEARCH31 to compute the total number of amino acid differences in all pairwise alignments between each concatenated sequence in each data set. Groups of sequences with ≤2 amino acid differences in the concatenated CDR3s were counted as a single clone. Finally, we used the majority amino acid identity at each residue position to generate the consensus amino acid sequence of the clone from sequences of the members of the group.
To generate clonal cluster plots (Fig. 4, Supplementary Figures S2, S3), we first used USEARCH to compute the total number of amino acid differences between each pairwise alignment of FACS-sorted scFv sequences. Next, we used the igraph R package32 to generate clustering plots for the pairwise alignments. Antibody sequences are represented by “nodes” in the plots. Each node was sized based on the frequency of the antibody clone in the FACS-sorted population: small (<1% frequency), medium (1–10% frequency), and large (>10% frequency). We define “edges” as the links between nodes. The “layout_with_kk” option was used to format the output. Edge lengths are proportional to the number of amino acid differences between linked nodes, i.e., longer edges indicate more amino acid differences.
Monoclonal antibody expression
Monoclonal antibodies were expressed from a variant of the pCDNA5/FRT mammalian expression vector (Thermo Fisher Scientific), as described previously.6-7 Expression constructs were built either using a BioXP™ robotic workstation (SGI DNA) or using gBlocks (IDT) and NEBuilder® HiFi DNA Assembly Master Mix (New England BioLabs). All constructs were synthesized as human IgHG1 isotype. Purified plasmid was used for transient transfection in the ExpiCHO system (Thermo Fisher Scientific). Transfected cells were cultured for 7–9 days in ExpiCHO medium and clarified supernatant was used for SPR and FACS cell surface binding studies. The concentration of antibody in the supernatant was quantified using an IgG ELISA kit (Abcam).
To assess cell surface binding of the anti-IL-21R antibodies, we first generated stable IL-21R-expressing Flp-In CHO cells (Thermo Fisher Scientific). CHO cells stably expressing PD-16 were used as a negative control. One million cells (1:1 mix of IL-21R and PD-1 cells) were stained with 1 μg of each anti-IL-21R antibody in 100 μl MACS Buffer (DPBS with 0.5% bovine serum albumin (BSA) and 2 mM EDTA) for 30 minutes at 4°C. Cells were then co-stained with anti-human IgG Fc-PE (BioLegend M1310G05) and anti-human PD-1-FITC (BioLegend EH12.2H7) antibodies for 30 minutes at 4°C. Cell viability was assessed with DAPI. We then performed FACS analysis on a BD Influx (Stanford Shared FACS Facility). We analyzed the data using FlowJo to determine the intensity of the IL-21R-expressing cells versus the PD-1 negative control (Supplementary Figure S5).
Surface plasmon resonance affinity
We performed affinity studies using array SPR. We amine-coupled a moderate density (∼1,000 Response Units) of an anti-human IgG-Fc reagent (Southern Biotech 2047-01) to a Xantec CMD-50M chip (50nm carboxymethyldextran medium density of functional groups) activated with 133 mM EDC (Sigma) and 33.3 mM S-NHS (ThermoFisher) in 100 mM MES pH 5.5. Then, goat anti-Human IgG Fc (Southern Biotech 2047-01) was coupled for 10 minutes at 25 µg/m L in 10 mM Sodim Acetate pH 4.5 (Carterra Inc.). The surface was then deactivated with 1 M ethanolamine pH 8.5 (Carterra Inc.). Running buffer used for the lawn immobilization was HBS-EP+ (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.05% Tween 20, pH 7.4; Teknova).
The sensor chip was then transferred to a continuous flow microspotter (CFM; Carterra Inc.) for array capturing. The mAb supernatants were diluted 50-fold (3–10 µg/mL final concentration) into HBS-EP+ with 1 mg/mL BSA. The samples were each captured twice with 15-minute and 4-minute capture steps on the first and second prints, respectively, to create multiple densities, using a 65 µL/min flow rate. The running buffer in the CFM was also HBS-EP+.
Next, the sensor chip was loaded onto an SPR reader (MX-96 system; Ibis Technologies) for the kinetic analysis. IL-21R was injected at five increasing concentrations in a four-fold dilution series with concentrations of 1.95, 7.8, 31.25, 125, and 500 nM in running buffer (HBS-EP+ with 1.0 mg/mL BSA). IL-21R injections were 5 minutes with a 15-minute dissociation at 8 µL/second in a non-regenerative kinetic series (Supplementary Figure S4). An injection of the goat anti-Human IgG Fc capture antibody at 75 µg/mL was injected at the end of the series to verify the capture level of each mAb. Binding data was double referenced by subtracting an interspot surface and a blank injection and analyzed for ka (on-rate), kd (off-rate), and KD (affinity) using the Kinetic Interaction Tool software (Carterra Inc.).
Surface plasmon resonance epitope binning
We performed epitope binning using high-throughput Array SPR in a modified classical sandwich approach.33 We functionalized a sensor chip using the Carterra CFM and methods similar to the SPR affinity studies, except a CMD-200M chip type was used (200nm carboxymethyl dextran, Xantec) and mAbs were coupled at 50 µg/mL to create a surface with higher binding capacity (∼3,000 reactive units immobilized). The mAb supernatants were diluted at 1:1 or 1:10 in running buffer, depending on the concentration of the mAb in the supernatant.
The sensor chip was placed in the MX-96 instrument, and the captured mAbs (“ligands”) were crosslinked to the surface using the bivalent amine reactive linker bis(sulfosuccinimidyl)suberate (BS3, ThermoFisher), which was injected for 10 minutes at 0.87 mM in water. Excess activated BS3 was neutralized with 1 M ethanolamine pH 8.5. For each binning cycle, a 7-minute injection of 250 μg/mL human IgG (Jackson ImmunoResearch 009-000-003) was used to block reference surfaces and any remaining capacity of the target spots.
Next, 250 nM IL-21R protein was injected onto the sensor chip, followed by injections of the diluted mAb supernatants (“analytes”) or buffer blanks as negative controls. Thus, the analyte mAb only binds to the antigen if it is not competitive with the ligand mAb. At the end of each cycle, a one minute regeneration injection was performed using a solution of 4 parts Pierce IgG Elution Buffer (ThermoFisher #21004), one part 5 M NaCl (0.83 M final), and 1.25 parts 0.85% H3PO4 (0.17% final). Only 18 of the mAbs remained active as ligands through multiple regenerations, so the binning analysis comprised an 18 × 46 competitive matrix.
We then used a network community plot algorithm in an SPR epitope data analysis software package (Carterra Inc.) to determine epitope bins (Supplementary Figure S6).33-34 Note that the clustering algorithm groups mAbs where only analyte data are available cluster separately from the mAbs where both ligand and analyte data are available. This phenomenon is an artifact of the incomplete competitive matrix. MAbs with both ligand and analyte data have more mAb-mAb measurements, resulting in more mAb-mAb connections, which leads to a closer relationship in the community plot.
Supplementary Material
Funding Statement
This work was partially supported by the National Science Foundation under grant 1230150; National Cancer Institute under grant R44CA187852.
Disclosure statement
ASA, MSA, MAA, RCE, RL, JL, RAM, MJS, and DSJ are employees of GigaGen Inc. and receive both equity shares and salary for their work. DB is an employee of Carterra and receives salary for his work. SRB, HH, PT, and PB are employees of Bristol-Myers Squibb and receive salary for their work.
Acknowledgments
The staff at the Stanford Shared FACS Facility were a valuable resource for flow sorting.
References
- 1.Nelson AL, Dhimolea E, Reichert JM. Development trends for human monoclonal antibody therapeutics. Nat Rev Drug Discov. 2010;9(10):767–74. doi: 10.1038/nrd3229. PMID:20811384. [DOI] [PubMed] [Google Scholar]
- 2.Lonberg N. Fully human antibodies from transgenic mouse and phage display platforms. Curr Opin Immunol. 2008;20(4):450–9. doi: 10.1016/j.coi.2008.06.004. PMID:18606226. [DOI] [PubMed] [Google Scholar]
- 3.Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256(5517):495–7. doi: 10.1038/256495a0. PMID:1172191. [DOI] [PubMed] [Google Scholar]
- 4.Reddy ST, Ge X, Miklos AE, Hughes RA, Kang SH, Hoi KH, Chrysostomou C, Hunicke-Smith SP, Iverson BL, Tucker PW, Ellington AD, Georgiou G. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat Biotechnol. 2010;28(9):965–9. doi: 10.1038/nbt.1673. PMID:20802495. [DOI] [PubMed] [Google Scholar]
- 5.Jayaram N, Bhowmick P, Martin AC. Germline VH/VL pairing in antibodies. Protein Eng Des Sel. 2012;25(10):523–9. doi: 10.1093/protein/gzs043. PMID:22802295. [DOI] [PubMed] [Google Scholar]
- 6.Adler AS, Mizrahi RA, Spindler MJ, Adams MS, Asensio MA, Edgar RC, Leong J, Leong R, Johnson DS. Rare, high-affinity mouse anti-PD-1 antibodies that function in checkpoint blockade, discovered using microfluidics and molecular genomics. MAbs. 2017;9(8):1270–81. Epub 2017/8/28. doi: 10.1080/19420862.2017.1371386. PMID: 28846506; PMCID: PMC5680806.17953510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adler AS, Mizrahi RA, Spindler MJ, Adams MS, Asensio MA, Edgar RC, Leong J, Leong R, Roalfe L, White R, Goldblatt D, Johnson DS. Rare, high-affinity anti-pathogen antibodies from human repertoires, discovered using microfluidics and molecular genomics. MAbs. 2017;9(8):1282–96. Epub 2017/8/28. doi: 10.1080/19420862.2017.1371383. PMID: 28846502; PMCID: PMC5680809.17953510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spolski R, Leonard WJ. Interleukin-21: basic biology and implications for cancer and autoimmunity. Annu Rev Immunol. 2008;26:57–79. doi: 10.1146/annurev.immunol.26.021607.090316. PMID:17953510. [DOI] [PubMed] [Google Scholar]
- 9.Young DA, Hegen M, Ma HL, Whitters MJ, Albert LM, Lowe L, Senices M, Wu PW, Sibley B, Leathurby Y, Brown TP, Nickerson-Nutter C, Keith JC Jr, Collins M. Blockade of the interleukin-21/interleukin-21 receptor pathway ameliorates disease in animal models of rheumatoid arthritis. Arthritis Rheum. 2007;56(4):1152–63. doi: 10.1002/art.22452. PMID:17393408. [DOI] [PubMed] [Google Scholar]
- 10.Narayanan A, Sellers BD, Jacobson MP. Energy-based analysis and prediction of the orientation between light- and heavy-chain antibody variable domains. J Mol Biol. 2009. May 22;388(5):941–53. doi: 10.1016/j.jmb.2009.03.043. PMID:19324053. [DOI] [PubMed] [Google Scholar]
- 11.Goodnow CC, Crosbie J, Jorgensen H, Brink RA, Basten A. Induction of self-tolerance in mature peripheral B lymphocytes. Nature. 1989;342(6248):385–91. doi: 10.1038/342385a0. PMID:2586609. [DOI] [PubMed] [Google Scholar]
- 12.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301(5638):1374–7. doi: 10.1126/science.1086907. PMID:12920303. [DOI] [PubMed] [Google Scholar]
- 13.Chan TD, Brink R. Affinity-based selection and the germinal center response. Immunol Rev. 2012;247(1):11–23. doi: 10.1111/j.1600-065X.2012.01118.x. PMID:22500828. [DOI] [PubMed] [Google Scholar]
- 14.Shlomchik MJ, Weisel F. Germinal center selection and the development of memory B and plasma cells. Immunol Rev. 2012;247(1):52–63. doi: 10.1111/j.1600-065X.2012.01124.x. PMID:22500831. [DOI] [PubMed] [Google Scholar]
- 15.Banfield MJ, King DJ, Mountain A, Brady RL. VL:VH domain rotations in engineered antibodies: crystal structures of the Fab fragments from two murine antitumor antibodies and their engineered human constructs. Proteins. 1997;29(2):161–71. doi: 10.1002/(SICI)1097-0134(199710)29:2%3c161::AID-PROT4%3e3.0.CO;2-G. PMID:9329081. [DOI] [PubMed] [Google Scholar]
- 16.Nakanishi T, Tsumoto K, Yokota A, Kondo H, Kumagai I. Critical contribution of VH-VL interaction to reshaping of an antibody: the case of humanization of anti-lysozyme antibody, HyHEL-10. Protein Sci. 2008;17(2):261–70. doi: 10.1110/ps.073156708. PMID:18227432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Wildt RM, Hoet RM, van Venrooij WJ, Tomlinson IM, Winter G. Analysis of heavy and light chain pairings indicates that receptor editing shapes the human antibody repertoire. J Mol Biol. 1999;285(3):895–901. doi: 10.1006/jmbi.1998.2396. PMID:9887257. [DOI] [PubMed] [Google Scholar]
- 18.Brezinschek HP, Foster SJ, Dörner T, Brezinschek RI, Lipsky PE. Pairing of variable heavy and variable kappa chains in individual naive and memory B cells. J Immunol. 1998;160(10):4762–7. PMID:9590222. [PubMed] [Google Scholar]
- 19.DeKosky BJ, Lungu OI, Park D, Johnson EL, Charab W, Chrysostomou C, Kuroda D, Ellington AD, Ippolito GC, Gray JJ, Georgiou G. Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proc Natl Acad Sci U S A. 2016;113(19):E2636–45. doi: 10.1073/pnas.1525510113. PMID:27114511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, Georgiou G. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat Med. 2015;21:86–91. doi: 10.1038/nm.3743. PMID:25501908. [DOI] [PubMed] [Google Scholar]
- 21.Haessler U, Reddy ST. Using next-generation sequencing for discovery of high-frequency monoclonal antibodies in the variable gene repertoires from immunized mice. Methods Mol Biol. 2014;1131:191–203. doi: 10.1007/978-1-62703-992-5_12. PMID:24515467. [DOI] [PubMed] [Google Scholar]
- 22.Percival-Alwyn JL, England E, Kemp B, Rapley L, Davis NH, McCarthy GR, Majithiya JB, Corkill DJ, Welsted S, Minton K, Cohen ES, Robinson MJ, Dobson C, Wilkinson TC, Vaughan TJ, Groves MA, Tigue NJ. Generation of potent mouse monoclonal antibodies to self-proteins using T-cell epitope “tags”. MAbs. 2015;7(1):129–37. doi: 10.4161/19420862.2014.985489. PMID:25523454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Longo NS, Rogosch T, Zemlin M, Zouali M, Lipsky PE. Mechanisms That Shape Human Antibody Repertoire Development in Mice Transgenic for Human Ig H and L Chain Loci. J Immunol. 2017;198(10):3963–3977. doi: 10.4049/jimmunol.1700133. PMID:28438896. [DOI] [PubMed] [Google Scholar]
- 24.Saggy I, Wine Y, Shefet-Carasso L, Nahary L, Georgiou G, Benhar I. Antibody isolation from immunized animals: comparison of phage display and antibody discovery via V gene repertoire mining. Protein Eng Des Sel. 2012;25(10):539–49. doi: 10.1093/protein/gzs060. PMID:22988130. [DOI] [PubMed] [Google Scholar]
- 25.Wang B, Kluwe CA, Lungu OI, DeKosky BJ, Kerr SA, Johnson EL, Jung J, Rezigh AB, Carroll SM, Reyes AN, Bentz JR, Villanueva I, Altman AL, Davey RA, Ellington AD, Georgiou G. Facile Discovery of a Diverse Panel of Anti-Ebola Virus Antibodies by Immune Repertoire Mining. Sci Rep. 2015;5:13926. doi: 10.1038/srep13926. PMID:26355042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang B, Lee CH, Johnson EL, Kluwe CA, Cunningham JC, Tanno H, Crooks RM, Georgiou G, Ellington AD. Discovery of high affinity anti-ricin antibodies by B cell receptor sequencing and by yeast display of combinatorial VH:VL libraries from immunized animals. MAbs. 2016;8(6):1035–44. doi: 10.1080/19420862.2016.1190059. PMID:27224530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray SA, Moore M, VandenEkart EJ, Roque RP, Bowen RA, Van Hoeven N, Wiley SR, Clegg CH. Selection of therapeutic H5N1 monoclonal antibodies following IgVH repertoire analysis in mice. Antiviral Res. 2016;131:100–8. doi: 10.1016/j.antiviral.2016.04.001. PMID:27109194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kono N, Sun L, Toh H, Shimizu T, Xue H, Numata O, Ato M, Ohnishi K, Itamura S. Deciphering antigen-responding antibody repertoires by using next-generation sequencing and confirming them through antibody-gene synthesis. Biochem Biophys Res Commun. 2017;487(2):300–306. doi: 10.1016/j.bbrc.2017.04.054. PMID:28412367. [DOI] [PubMed] [Google Scholar]
- 29.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–8. doi: 10.1038/nmeth.2604. PMID:23955772. [DOI] [PubMed] [Google Scholar]
- 30.Lefranc MP, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, Regnier L, Ehrenmann F, Lefranc G, Duroux P. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37(Database issue):D1006–12. doi: 10.1093/nar/gkn838. PMID:18978023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–1. doi: 10.1093/bioinformatics/btq461. PMID:20709691. [DOI] [PubMed] [Google Scholar]
- 32.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal Complex Systems. 2006;1695. [Google Scholar]
- 33.Abdiche YN, Miles A, Eckman J, Foletti D, Van Blarcom TJ, Yeung YA, Pons J, Rajpal A. High-throughput epitope binning assays on label-free array-based biosensors can yield exquisite epitope discrimination that facilitates the selection of monoclonal antibodies with functional activity. PLoS One. 2014;9(3):e92451. doi: 10.1371/journal.pone.0092451. PMID:24651868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdiche YN, Yeung AY, Ni I, Stone D, Miles A, Morishige W, Rossi A, Strop P. Antibodies Targeting Closely Adjacent or Minimally Overlapping Epitopes Can Displace One Another. PLoS One. 2017;12(1):e0169535. doi: 10.1371/journal.pone.0169535. PMID:28060885. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.