Summary
Biotin is an essential cofactor utilized by all domains of life, but only synthesized by bacteria, fungi and plants, making biotin biosynthesis a target for antimicrobial development. To understand biotin biosynthesis in mycobacteria, we executed a genetic screen in Mycobacterium smegmatis for biotin auxotrophs and identified pyruvate carboxylase (Pyc) as required for biotin biosynthesis. The biotin auxotrophy of the pyc::tn strain is due to failure to transcriptionally induce late stage biotin biosynthetic genes in low biotin conditions. Loss of bioQ, the repressor of biotin biosynthesis, in the pyc::tn strain reverted biotin auxotrophy, as did reconstituting the last step of the pathway through heterologous expression of BioB and provision of its substrate DTB. The role of Pyc in biotin regulation required its catalytic activities and could be supported by M. tuberculosis Pyc. Quantitation of the kinetics of depletion of biotinylated proteins after biotin withdrawal revealed that Pyc is the most rapidly depleted biotinylated protein and metabolomics revealed a broad metabolic shift in wild type cells upon biotin withdrawal which was blunted in cell lacking Pyc. Our data indicate that mycobacterial cells monitor biotin sufficiency through a metabolic signal generated by dysfunction of a biotinylated protein of central metabolism.
Graphical Abstract
Through a combination of genetics, biochemistry, and metabolomics, we identify mycobacterial Pyruvate Carboxylase as necessary to transcriptionally induce biotin biosynthesis through BioQ. Our results suggests that mycobacteria monitor biotin sufficiency through a biotinylated central metabolic enzyme.
INTRODUCTION
Biotin (vitamin B7, or vitamin H) is an essential cofactor used by the carboxylase and decarboxylase family of enzymes. Whereas biotin is vital to and used by members of all domains of life, it is synthesized only by bacteria, plants, and fungi (Lin & Cronan, 2011). For this reason, biotin biosynthesis has been proposed as a promising target for antibiotic development. In the human pathogen M. tuberculosis, depletion of the essential biotin biosynthetic protein BioA leads to bactericidality in vitro and clearance of M. tuberculosis from mice when BioA is depleted either in acute or persistent infection (Woong Park et al., 2011). The antibiotic amiclenomycin and its analogues inhibit BioA and show whole-cell activity against M. tuberculosis (Shi et al., 2011), and a target-based approach identified novel compounds with inhibitory activity against BioA (Park et al., 2015), many of which were also present in the diversity-oriented scaffolds (DOS) collection (Galloway et al., 2010). Further characterization of the biotin biosynthetic pathway may reveal additional antibiotic targets.
The latter stages of biotin biosynthesis are conserved among all organisms proficient for biotin biosynthesis. Pimeloyl-ACP/CoA is converted to biotin via the sequential action of four enzymes: BioF, BioA, BioD, and BioB. Thus far, no alternative pathway has been identified to perform this stage of biotin biosynthesis. In contrast, multiple different mechanisms have been shown to yield pimeloyl-ACP. In E. coli, the methyltransferase BioC allows malonyl-ACP to enter the fatty acid elongation pathway, and after two rounds of elongation, pimeloyl-ACP methyl ester exits the pathway when it is hydrolyzed by the methyl esterase BioH (Lin et al., 2010). B. subtilis has a cytochrome P450 enzyme (BioI) that can convert convert long-chain fatty acids to pimeloyl-ACP, and also possesses a pathway which converts free pimelic acid to pimeloyl-CoA using the enzyme BioW (Cryle, 2010, Lin & Cronan, 2011) (Summarized in Fig. 1). Recent evidence indicates that BioW is the dominant pathway in B. subtilis (Manandhar and Cronan, 2017).
Figure 1. Summary of previously-characterized biotin biosynthetic pathway.
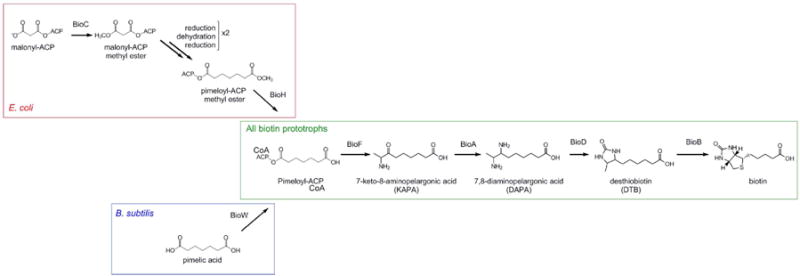
Late (green box) biotin biosynthesis is conserved in all biotin-synthesizing organisms and is catalyzed by the action of four enzymes: BioF, BioA, BioD, and BioB, which convert pimeloyl-ACP/CoA to biotin. Synthesis of the pimeloyl-ACP moiety differs between bacteria. In E. coli (red box), BioC methylates malonyl-ACP to form malonyl-ACP methyl ester, which then enters the fatty-acid elongation pathway. BioH demethylates pimeloyl-ACP methyl ester to form pimeloyl-ACP (Lin et al., 2010). In contrast, B. subtilis (blue box) utilizes BioI to break down long-chain fatty acids into pimeloyl-ACP. A secondary pathway exists in which free pimelic acid is converted to pimeloyl-ACP by BioW (Cryle, 2010).
As biotin biosynthesis is an energetically costly process, many biotin prototrophs have evolved mechanisms to control the transcription of biotin biosynthetic genes such that expression is induced when exogenous biotin is not available. E. coli possesses a bifunctional biotin ligase BirA, which acts as a biotin ligase to covalently attach biotin to biotin-utilizing enzymes, and as a transcriptional regulator of biotin biosynthesis (Cronan, 1988). In biotin-replete conditions, BirA forms a homodimer, which binds the biotin operator and represses the transcription of bioFADBC. In biotin-starved conditions, unbiotinylated biotin binding proteins (AccB) competitively bind to single BirA proteins to form BirA-AccB heterodimers, which do not possess repressor activity (Chakravartty & Cronan, 2012, Solbiati & Cronan, 2010, Weaver et al., 2001). As such, in E. coli, biotin biosynthesis is coupled to the accumulation of an unbiotinylated protein through the bifunctional BirA.
In contrast, Brucella and other Alphaproteobacteria utilize the GntR family protein BioR to repress expression of bioFADBZ and of the biotin transporter bioY during biotin-replete conditions (Feng et al., 2013b, Feng et al., 2013a). The mechanism of this repression is poorly understood, but Brucella (type I, or unifunctional) BirA lacks the DNA-binding domain found in E. coli BirA (Beckett, 2007). Along with many other Actinobacteria, Mycobacteria also possess a type I BirA protein, and the soil bacterium M. smegmatis has recently been discovered to employ the TetR family protein BioQ as a transcriptional regulator of bioFDB expression (Tang et al., 2014). TetR family proteins typically repress transcription in the absence of their ligand, and dissociate from DNA after ligand binding triggers a conformational shift in their helix-turn-helix DNA binding domain (Ramos et al., 2005). As is the case in Alphaproteobacteria, the mechanism of BioQ derepression is poorly characterized, though it has been shown that biotin itself is not the ligand of BioQ (Tang et al., 2014), suggesting that some alternate non-biotin ligand mediates BioQ release from its operator. Although TetR like repressors are abundant in M. tuberculosis, it lacks a predicted ortholog to BioQ and the transcriptional regulation of biotin in M. tuberculosis has not been elucidated.
In this study we undertook a genetic screen in M. smegmatis for biotin auxotrophs. In addition to identifying previously known late stage biotin biosynthetic genes, we identified multiple transposon insertions in pyruvate carboxylase (pyc) as conferring biotin auxotrophy. Analysis of pyc null strains supports a model in which dysfunction of Pyc during biotin depletion, itself a biotin dependent enzyme, generates a metabolic signal that stimulates BioQ derepression.
RESULTS
Disruption of M. smegmatis gene MSmeg_2412 confers biotin auxotrophy
To identify new genes involved in biotin biosynthesis in mycobacteria, we employed a forward genetics approach using the mycobacteriophage φMycoMarT7. We constructed a library of approximately 10,000 transposon insertion mutants in M. smegmatis mc2155 strain, and screened these mutants for inability to grow on medium lacking biotin. This strategy identified insertions in the previously characterized biotin biosynthetic genes bioA, bioB, and bioF as conferring biotin auxotrophy. Three strains, each bearing independent transposon insertions in MSmeg_2412, were unable to grow on medium lacking biotin (Table 1). MSmeg_2412 encodes pyruvate carboxylase, an enzyme of central carbon metabolism not previously implicated in biotin biosynthesis. Pyruvate carboxylase converts pyruvate to oxaloacetate utilizing biotin as an essential cofactor (Jitrapakdee et al., 2008, Yu et al., 2009), and has been shown to have roles in anaplerosis, gluconeogenesis, elongation of fatty acids, and branched-chain amino acid biosynthesis (Peters-Wendisch et al., 1998, Attwood, 1995, Sauer & Eikmanns, 2005, Owen et al., 2002, Mukhopadhyay & Purwantini, 2000).
Table 1.
Biotin auxotrophic transposon mutants.
| Mutant # | MGM # | gene | Gene length (NT) | Nucleotide at Tn insertion | orientation of Tn (relative to coding sequence) |
|---|---|---|---|---|---|
| 10 | 8001 | MSmeg_2412 | 3381 | 2061–2062 | sense |
| 35 | 8002 | bioB | 3186 | 443–444 | sense |
| 43 | 8003 | bioF | 1146 | 132–133 | antisense |
| 67 | 8003 | bioF | 1146 | 132–133 | antisense |
| 86 | 8004 | bioA | 1296 | 1211–1212 | antisense |
| 112 | 8004 | bioA | 1296 | 1211–1212 | antisense |
| 119 | 8005 | MSmeg_2412 | 3381 | 1358–1357 | antisense |
| 130 | 8004 | bioA | 1296 | 1211–1212 | antisense |
| 134 | 8006 | MSmeg_2412 | 3381 | 2470–2471 | antisense |
We proceeded to confirm that disruption of pyruvate carboxylase itself, rather than second-site mutations or polar effects, was responsible for the inability of pyruvate carboxylase insertion mutants to grow on medium lacking biotin. To do so, we complemented these strains with plasmids expressing MSmeg_2412 or expressing the downstream genes MSmeg_2413 and MSmeg_2414. Expression of MSmeg_2412 restored biotin prototrophy to the pyc::tn strain, whereas neither expression of MSmeg_2413 nor MSmeg_2414 had any effect (Fig. 2A).
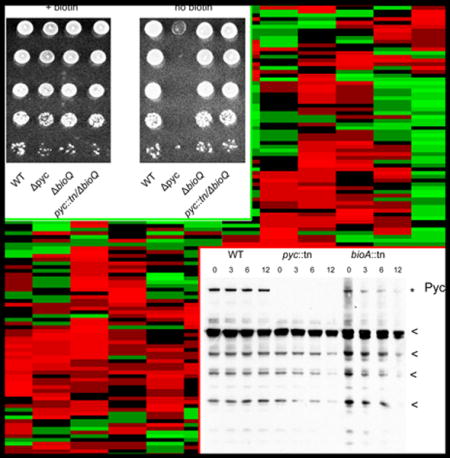
Figure 2. A transposon insertion in MSMEG_2412 (pyruvate carboxylase) impairs growth on medium lacking biotin.
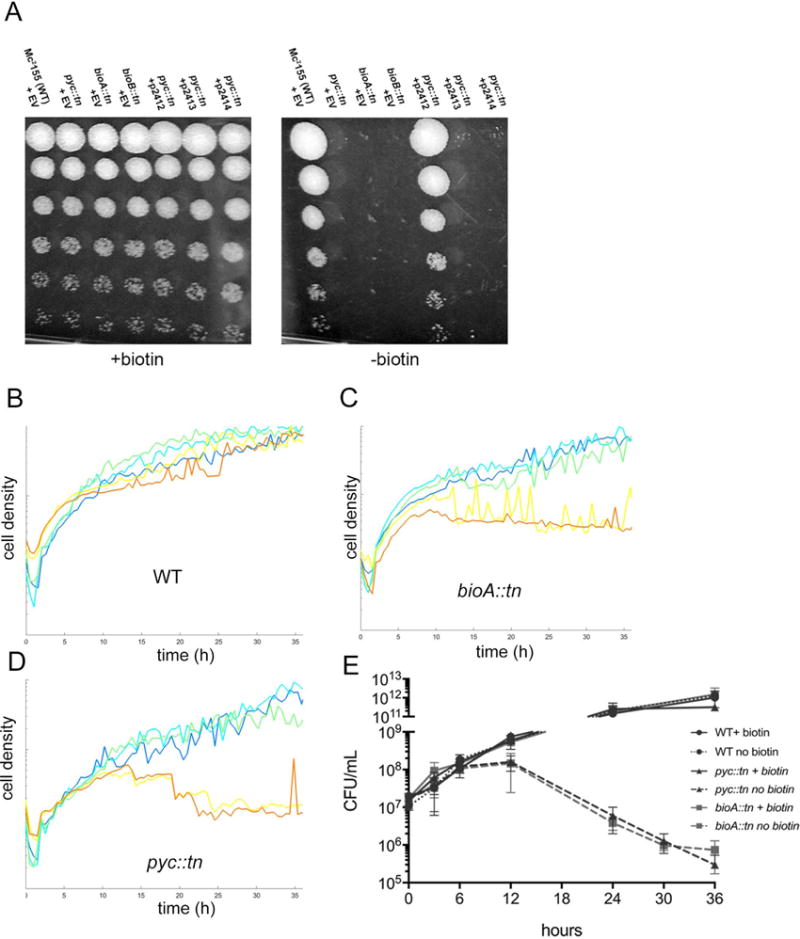
(A) Growth of WT M. smegmatis (MGM8007), pyc::tn (MGM8008) bioA::tn (MGM8009) and bioB::tn (MGM8010), and pyc::tn complemented with MSMEG_2412 (MGM8011), MSMEG_2413 (MGM8012), or MSMEG_2414 (MGM8013) on biotin containing (left) or biotin free (right) media. Rows are ten-fold serial dilutions. EV=empty vector. (B-D) Growth of WT (B), bioA::tn (C), and pyc::tn (D) in biotin-replete medium (500 ng/mL, dark blue), medium with decreasing concentrations of biotin (50 ng/mL: light blue; 5 ng/mL: green; 0.5 ng/mL: yellow), and biotin-deficient medium (orange). Each line is the mean of three biological replicates. (E) Survival during biotin depletion. WT (circles), bioA::tn (squares), and pyc::tn (triangles) were grown in medium containing (solid lines) or lacking biotin (dashed lines), and viable CFU was assessed by culturing serial dilutions on agar media. Error bars are standard deviations of three biological replicates.
To further assess the biotin auxotrophy in the pyc::tn strain, we grew wild type M. smegmatis, pyc::tn, and bioA::tn in liquid medium containing decreasing concentrations of biotin. Wild type M. smegmatis did not show an identifiable defect at any biotin concentration, whereas both pyc::tn and bioA::tn grew similarly well in medium containing 1x (500 ng/mL), 1/10x, or 1/100x biotin, but failed to grow after approximately 12 hours in medium containing either no biotin or 1/1000x biotin (Fig. 2B–D). Quantitation of cell death indicated that both pyc::tn and bioA::tn lose viability with similar kinetics, beginning at 12 hours after biotin withdrawal (Fig. 2E). These data indicate that loss of pyruvate carboxylase confers biotin auxotrophy in mycobacteria with a biotin requirement for viability similar to a known late stage biotin biosynthetic defect.
Pyruvate carboxylase is required for transcriptional derepression of biotin biosynthesis
Given the similar phenotype of the pyc::tn and bioA::tn mutants, we tested whether pyc::tn was defective in the late stages of biotin biosynthesis. Addition of desthiobiotin (DTB), the penultimate biosynthetic intermediate in the biotin biosynthetic pathway (Fig. 1), was unable to restore growth to pyc::tn, indicating a defect in bioB function (Fig. 3A). DTB was unsurprisingly able to restore growth to bioA::tn strain due to intact BioB function (Fig. 3C, right panels). Expression of bioB restored biotin prototrophy to pyc::tn when the growth medium was supplemented with DTB (Fig. 3C, bottom right), but only partially restored growth in the absence of DTB (Fig. 3C, bottom left), indicating that the pool of DTB in the pyc::tn strain is limiting and implying an additional defect in the biotin biosynthetic pathway before the generation of DTB.
Figure 3. pyc::tn is defective in late-stage biotin biosynthesis.
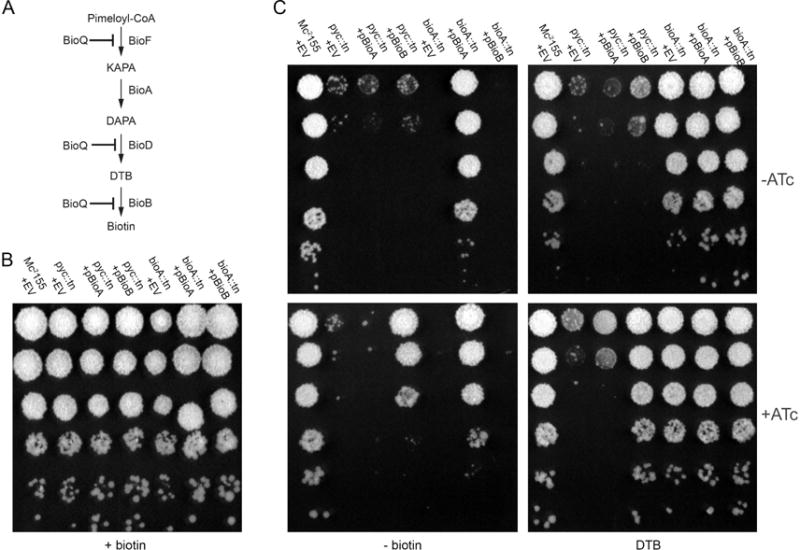
(A) pathway of late-stage biotin biosynthesis. Pimeloyl-CoA is converted to biotin via the action of four biotin biosynthetic enzymes: BioF, BioA, BioD, and BioB. Expression of bioFD and of bioB, but not of bioA, has been shown (Tang et al., 2014) to be repressed by the TetR family repressor BioQ under biotin-replete conditions in M. smegmatis. (B, C) WT (MGM8019), bioA::tn (MGM8021), and pyc::tn (MGM8020) were tested for growth on medium containing (B), or lacking biotin (C, left panels) or containing the biosynthetic intermediate desthiobiotin (DTB) (C; two rightmost panels). bioA::tn and pyc::tn strains expressing bioA (pyc::tn: (MGM8034)); bioA::tn: (MGM8036) or bioB (pyc::tn: (MGM8035); bioA::tn: (MGM8037)) under the control of a heterologous promoter responsive to the inducer anhydrotetracycline (ATc) (C; ATc present in bottom panels) were also tested.
Biotin biosynthetic genes are transcriptionally induced in low biotin conditions such that bioBFD are coordinately regulated. To examine whether transcriptional upregulation of these genes is defective in the pyc::tn strain, we quantitated the mRNAs for bioFADB by RT-qPCR (Fig. 3A). In biotin-replete conditions, WT and pyc::tn M. smegmatis express similar levels of these transcripts. However, after three hours of biotin deprivation in wild type cells, we observed a 16-fold induction of bioB and bioFD (expressed as a single transcript) mRNAs (Fig. 4A). We do not find bioA to be transcriptionally upregulated by low biotin, consistent with previous reports (Tang et al., 2014). In contrast, pyc::tn fails to upregulate bioB or bioFD in low biotin, potentially explaining its defect in growth in this condition (Fig. 4A). Complementation of pyc::tn with a wild type copy of pyc, which complemented biotin auxotrophy (Fig. 2A), restored bioB induction in low biotin and in fact de-repressed bioB even in biotin-replete conditions (Fig. 4A). These data indicate that pyc is required for the transcriptional induction of biotin biosynthetic genes in low biotin conditions.
Figure 4. Pyruvate carboxylase is required for BioQ mediated derepression of bioFDB.
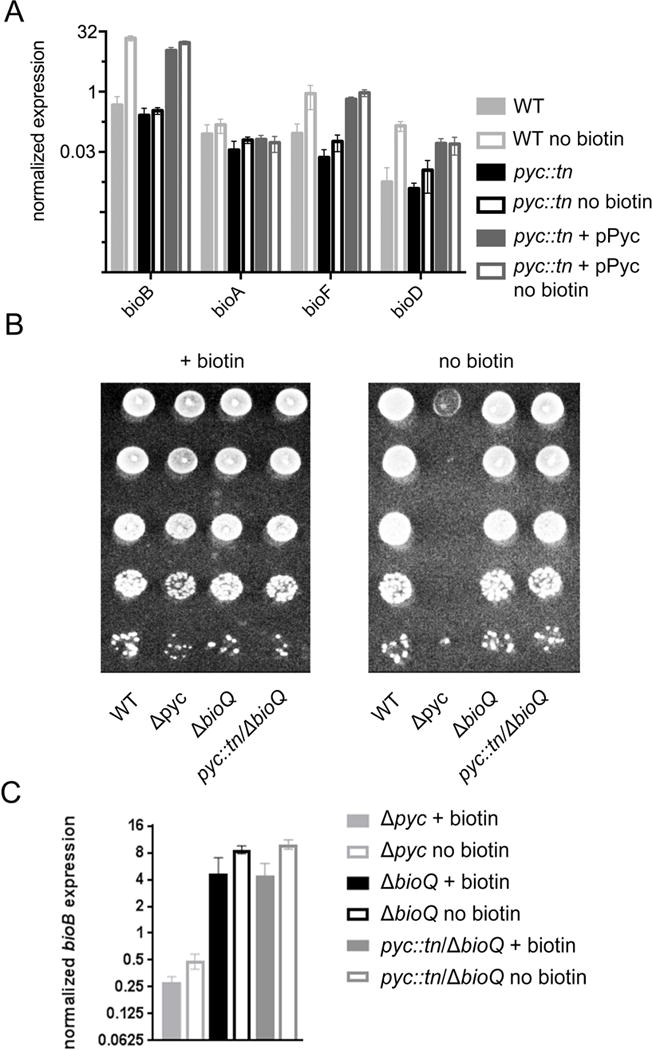
(A) Expression of the biotin biosynthetic genes bioFADB in WT (MGM8007), pyc::tn (MGM 8008) and complemented (MGM8011) strains under biotin-replete and biotin-deficient (-bio) conditions. Strains were grown for three hours with or without biotin, RNA was prepared, and RT-qPCR was performed for the indicated mRNAs. (B) Loss of bioQ suppresses the biotin auxotrophy of pyc::tn. Genetic deletions of pyc (MGM6514) and bioQ (MGM6516 and MGM6520) were introduced via homologous recombination and double counterselection (see strain table), and growth on medium containing or lacking biotin was assessed. (C) Loss of bioQ restores bioB expression to pyc::tn. Error bars are standard deviation of three biological replicates.
Previous work (Tang et al., 2014) has shown that bioB and bioFD—but not bioA—are repressed by the TetR like regulator BioQ, which binds to an operator in the bioB and bioF promoters which overlaps the TSS for these genes (Tang et al., 2014). BioQ dissociates from its operator in low biotin conditions, although the signal for BioQ binding or dissociation is not clear and does not appear to be biotin itself or biotinoyl-AMP (Tang et al., 2014). To test whether the biotin auxotrophy of the pyc::tn strain requires BioQ mediated repression, we introduced null mutations in bioQ in both the WT and pyc::tn backgrounds using homologous recombination and double counterselection (Barkan et al., 2011). We also constructed a null mutation of pyc to confirm that biotin auxotrophy was not specific to the pyc::tn transposon mutant. We tested each of these strains for the ability to grow without biotin and observed that a Δpyc strain was indeed a biotin auxotroph (Fig. 4B) and that deletion of bioQ in the pyc::tn background restored the ability of this strain to grow without biotin (Fig 4B). We also constructed a bioQ null mutant via CRISPR-mediated chromosomal cleavage (see methods) in the pyc::tn background, (Supplementary Fig. S1A). We tested the ability of pyc::tn ΔbioQ and pyc::tn bioQ (frameshift) strains to grow without biotin and found that inactivation of bioQ reverted the biotin auxotrophy of the pyc::tn strain (Fig 4B and data not shown).
RT-qPCR of bioB mRNA showed that loss of BioQ derepressed BioB expression in wild type cells, as reported previously ((Tang et al., 2014), Fig. 4C) and also restored bioB expression in pyc::tn (Fig. 4C, Supplementary Fig. S1B). These results demonstrate that the biotin auxotrophy conferred by inactivation of pyruvate carboxylase is due to inability to derepress biotin biosynthetic genes during biotin deprivation and imply that pyruvate carboxylase is required for generation of the signal that relieves BioQ mediated repression.
Pyruvate carboxylase catalytic activity is required for its regulatory function
M. smegmatis possesses two genes predicted to code for pyruvate carboxylase enzymes: MSmeg_2412, the transposon mutant of which we analyze above, and the additional gene MSmeg_6648. Meanwhile, the predicted proteome of the pathogenic mycobacterium M. tuberculosis contains a single pyruvate carboxylase encoded by the gene Rv2967c, which is most similar in primary amino acid sequence and genomic organization to MSmeg_2412. The two M. smegmatis pyruvate carboxylase proteins share a high degree of similarity (~90%) by amino acid sequence (Supplementary Fig. S2) with conserved amino acid residues found to be necessary for enzymatic activity (biotin carboxylase activity: E288, N290, R292 (Janiyani et al., 2001); carboxytransferase activity: (R532, D533, E566 (Yu et al., 2009, Marchler-Bauer et al., 2017, St Maurice et al., 2007)), or biotin attachment (K1093 (Polyak et al., 2001, Kim et al., 2004)). To determine whether the pyruvate carboxylases MSmeg_6648 or Rv2967c can compensate for loss of pyc (MSmeg_2412), we expressed each of these proteins in pyc::tn. Whereas the M. tuberculosis pyruvate carboxylase encoded by Rv2967c did complement the biotin biosynthetic defect of pyc::tn, MSmeg_6648 failed to complement even when expressed under control of the inducible tetON promoter (Fig. 5A). However, western blotting did not reveal a detectable protein product corresponding to MSmeg_6648 (data not shown). These data demonstrate functional conservation between MSmeg_2412 and M. tuberculosis Rv2967c.
Figure 5. Pyruvate carboxylase catalytic activity is required for biotin prototrophy.
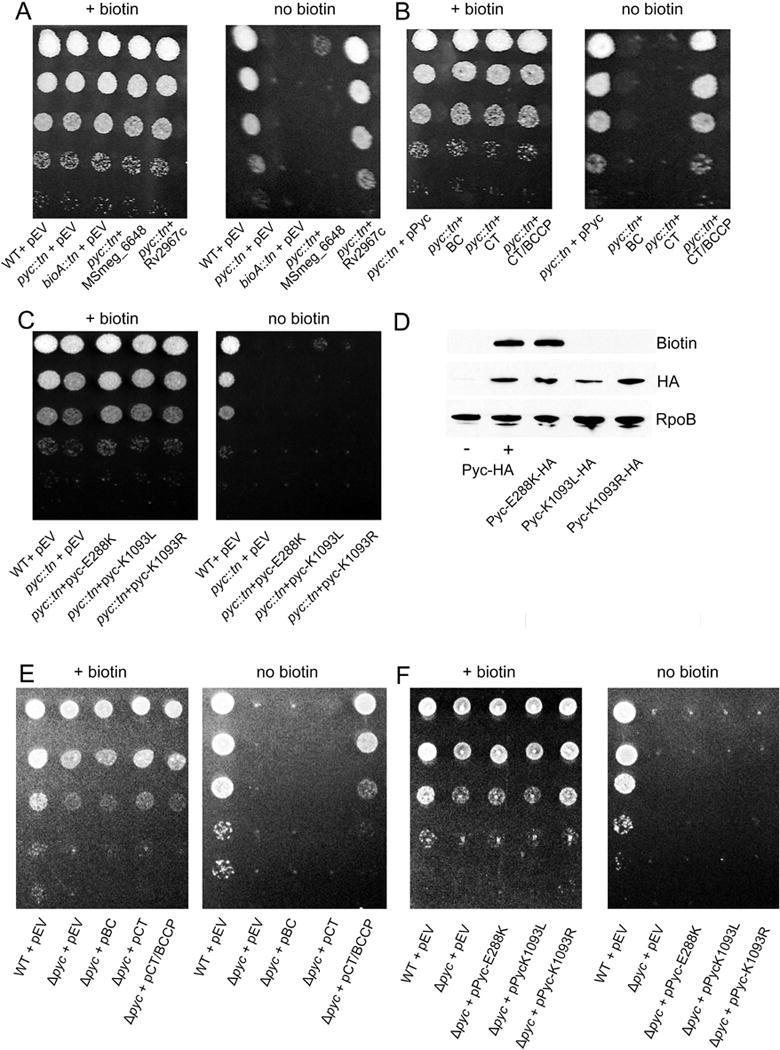
(A) TB Pyruvate carboxylase supports biotin biosynthesis. WT (MGM8019), pyc::tn (MGM8020), bioA::tn (MGM8021), and pyc::tn expressing either the annotated second pyruvate carboxylase in M. smegmatis (MSmeg_6648) (MGM8024) or the M. tuberculosis ortholog Rv2967c (MGM8025) for growth on medium lacking biotin. EV=empty vector (B) Truncation of Pyc. pyc::tn was complemented with plasmids expressing full length pyruvate carboxylase (MGM8023), biotin carboxylase (BC) domain (MGM8026), only the carboxytransferase (CT) domain (MGM8027), or the CT domain and the biotin-binding (BCCP) domains (MGM8028). (C) Pyruvate carboxylase BC active site or biotin attachment sites are required for biotin prototrophy. pyc::tn was complemented with plasmids expressing full length Pyc with a BC catalytic residue mutation (E288K) (MGM8029) or the essential residue for covalent attachment of biotin (K1093L/R) (MGM8030/8031). Each strain was N-terminally HA-tagged and expressed under the control of an ATc-dependent promoter. (D) Each of the Pyc isoforms from (C) as well as native pyruvate carboxylase (MGM8011) were N-terminally HA tagged. Expression of each of the mutants (E288K, K1093L/R) was similar to levels of native pyruvate carboxylase (Pyc-HA+) when induced with ATc. Low levels of pyruvate carboxylase were present in the absence of induction (Pyc-HA-). Detection of protein biotinylation was performed using streptavidin-HRP. RNA polymerase subunit B (RpoB) is shown as a loading control. Native pyruvate carboxylase (Pyc-HA) is the same strain shown in Fig. 2A and Fig. 4A. (E) Δpyc (MGM6521) was complemented with plasmids expressing the BC domain (MGM6522), only the CT domain (MGM6523), or the CT domain and BCCP domains (MGM6524). (F) Pyruvate carboxylase BC active site or biotin attachment sites are required for biotin prototrophy. Δpyc was complemented with plasmids expressing Pyc-E288K (MGM6525) or Pyc-K1093L/R (MGM6526/2527).
To assess whether pyruvate carboxylase activity itself, rather than another property of MSmeg_2412, is required for its biotin regulatory function, we expressed truncations of MSmeg_2412 lacking one or more predicted domains. Pyruvate carboxylase possesses three major structural domains: the N-terminal biotin carboxylase (BC), carboxytransferase (CT), and the C terminal biotin binding (BCCP). The transposon insertions we identified as conferring biotin auxotrophy were located between the BC domain and the CT domain (mutant 119) or in the CT domain (mutants 10 and 134) C-terminal to active site residues (Supplementary Fig. S2, black arrows). The biotin carboxylase domain adds a carboxyl group to biotin in the first step of the pyruvate carboxylase mechanism, and is essential for pyruvate carboxylase activity (Janiyani et al., 2001).
Truncations containing only the MSMEG_2412 biotin carboxylase (BC) or carboxytransferase (CT) domains failed to restore growth to pyc::tn on medium lacking biotin. Surprisingly, a truncation lacking the BC domain but containing both the CT domain and the biotin-binding (BCCP) domain did successfully revert the pyc::tn biotin phenotype (Fig. 5B). However, expression of a BC catalytic mutant (E288K), which we confirmed is both expressed and biotinylated (Fig 5D), failed to restore growth to pyc::tn on medium lacking biotin (Fig. 5C). Expression of each of these constructs in the Δpyc strain, which lacks the entire Pyc protein, revealed identical requirements for complementation (Figure 5E,F).
To further address the question of whether pyruvate carboxylase activity is responsible for the requirement for pyruvate carboxylase function and relief of BioQ repression, we constructed two mutants which lack the MSMEG_2412 biotin attachment site (K1093L (Polyak et al., 2001) and K1093R (Kim et al., 2004)). As expected, expression of either of these mutants failed to restore growth to pyc::tn (Fig. 5C), despite expression of the mutant proteins at wild type levels (Fig. 5D, HA panel). We were unable to detect biotinylation of each of these mutants using streptavidin-HRP, indicating that despite their detectable expression, they each failed to be biotinylated (Fig. 5D, biotin panel). As covalent linkage of biotin to the biotin-attachment lysine is required for pyruvate carboxylase activity (St Maurice et al., 2007), this experiment provides further evidence that pyruvate carboxylase activity of MSMEG_2412, rather than any other function of the protein or its coding sequence, is the key factor which is essential for its regulatory function. As a final attempt to deduce the unique features of the M. smegmatis Pyc enzyme that are required for biotin biosynthesis, we complemented the pyc::tn strain with Pyc from B. subtilis (strain MGM6518). Although we confirmed protein expression and biotinylation of the B. subtilis Pyc in M. smegmatis, this enzyme did not rescue the biotin auxotrophy (data not shown). The failure of a nonbiotinylated Pyc enzyme to induce biotin biosynthesis clearly indicates that it is not simply the accumulation of a biotin free protein that is the signal for BioQ derepression.
Biotinyl-pyruvate carboxylase is depleted more quickly than other biotinylated proteins during biotin deprivation
The data presented above indicates that pyruvate carboxylase activity is required for to induce biotin biosynthetic genes through BioQ. As pyruvate carboxylase is itself a biotinylated protein, we hypothesized that loss of Pyc function, through loss of biotinylation, may be a sensitive indicator of cellular biotin starvation. To determine the effects of biotin deprivation on bacteria which cannot synthesize their own biotin, we used streptavidin-HRP to probe cellular biotinylated proteins in order to assess the kinetics of biotinylated protein depletion in WT, pyc::tn, and bioA::tn M. smegmatis. We identified biotinyl-pyruvate carboxylase (Fig. 6A, top band, absent in pyc::tn, marked with an asterisk) as well as multiple additional biotinylated proteins. The pyc::tn strain lacked any gross defects in biotinylated proteins during biotin-replete (0h) conditions (other than the absence of the ~120 kDa band for pyruvate carboxylase), and showed similar rates of disappearance of biotinylated proteins when compared to the bioA::tn strain (Fig. 6A. Supplementary Fig. S3). A search for homology to the Pyc biotin-binding domain within the M. smegmatis identified the pyruvate carboxylases MSMEG_2412 and MSMEG_6648, as well as the acetyl/propionyl-CoA carboxylases MSMEG_0334, 1807, 5943, and 4716. Each of these acetyl/propionyl-CoA carboxylases are predicted to have a molecular weight between 60 and 75 kDa. The biotinylated proteins we detected with molecular weights of below 60 kDa do not correspond to any protein identifiable via this method, and may either represent degradation products or uncharacterized biotinylated proteins.
Figure 6. Analysis of protein biotinylation during biotin deprivation.
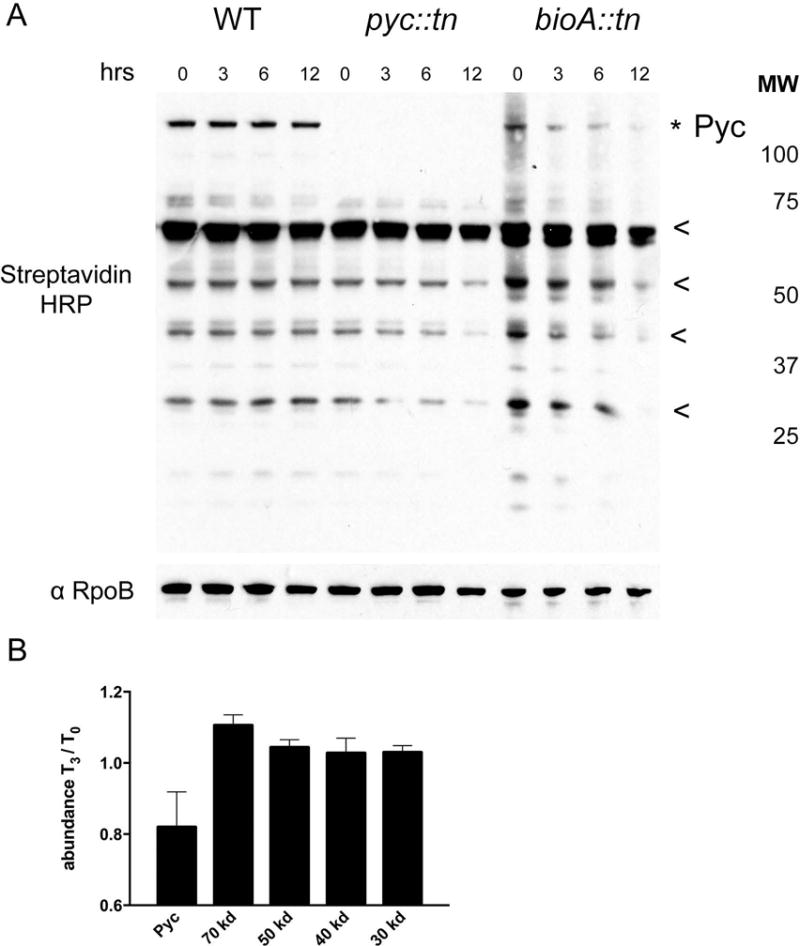
(A) Streptavidin HRP Western blot of total biotinylated protein in WT (MGM8007), pyc::tn (MGM8008), and bioA::tn (MGM8009) during biotin deprivation for 0, 3, 6, and 12 hours. Biotinylated proteins were visualized using streptavidin-HRP with anti-RpoB as a loading control. Pyruvate carboxylase (topmost band, marked with *) was absent in pyc::tn, as expected, but pyc::tn showed no obvious deficiencies in other biotinylated proteins. The biotinylated proteins used for quantitation (see Figure S3) are marked with * or <. (B) Quantitation of the fraction of the indicated proteins remaining at 3 hours of biotin deprivation compared to T0 in a bioA::tn strain. Error bars are standard deviations of three biological replicates.
We repeated this experiment in triplicate to assess the rate at which the biotin auxotroph bioA::tn loses biotinylated proteins (Supplementary Fig. S3), and quantitated the results (Fig. 6B). Biotinyl-pyruvate carboxylase begins to be depleted by three hours of biotin depletion, a time point at which growth of biotin auxotrophs is comparable to WT (Fig. 2C), and other biotinylated proteins have not yet declined in abundance. Depletion of the majority of biotinylated proteins is observed by six hours or more of biotin depletion, a time point that coincides with growth arrest of biotin auxotrophs (Fig. 2E). We conclude that depletion of biotinyl-pyruvate carboxylase is an early marker of biotin deprivation and may generate the signal to upregulate biotin biosynthesis.
Metabolomic analysis identifies candidate inducers of BioQ dependent genes
The requirement for Pyc enzymatic function to support biotin synthesis may suggest that the signal for BioQ depression is a metabolic shift due to Pyc dysfunction. To assess this idea, we pursued a metabolomic approach. We collected metabolites from WT and pyc::tn, either in biotin-replete or biotin depleted conditions for three hours. This short period of biotin withdrawal was chosen to ensure that metabolic changes were indeed a result of biotin deprivation, rather than generalized signals of cell death, which might occur if biotin deprivation had been lengthened until growth arrest occurs. Using this approach, we were able to identify and to quantitate the levels of approximately 150 metabolites with distinct LC-MS peaks and retention times (Supplementary Fig. S4). Statistical analysis identified 16 of these metabolites present at levels that were significantly different between strains and/or conditions (Fig. 7A). Pyruvate, the substrate of pyruvate carboxylase, accumulated in wild type cells with biotin withdrawal, consistent with the kinetics of Pyc protein depletion, but was depleted in all conditions in pyc::tn cells, i.e. both with and without biotin (Fig. 7A). Principal-component analysis (PCA) on the metabolomic data (Fig. 7B) separates the samples primarily by strain on the first principal component, which explains the majority of the variation between samples. The PCA also shows that biotin withdrawal from the wild type strain causes a metabolic shift in both principal components, whereas biotin withdrawal from pyc::tn has a minimal effect (Figure 7B) Additionally, the majority of the differences between metabolites which differed between the strains (Fig. 7A) or metabolites as a whole (Supplementary Fig. S4) are found between strains, rather than between conditions. These metabolomic data indicate that biotin withdrawal from wild type cells causes broad metabolic perturbation, including dysfunction of Pyc. In cells lacking Pyc, these metabolic changes are blunted or absent and the strain, even with biotin, has adapted to the loss of Pyc.
Figure 7. Significantly perturbed metabolites in pyc::tn.
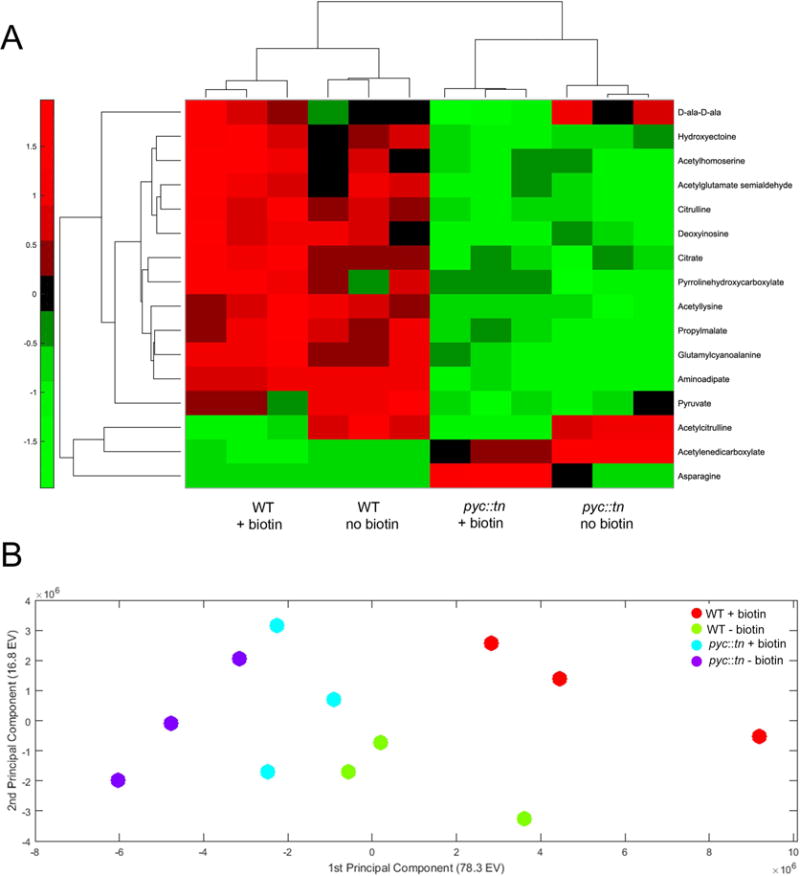
(A) We collected metabolites from three replicates of WT (MGM8007) and pyc::tn (MGM8008) after three hours of growth with or without biotin. Metabolite levels were normalized to total biomass measured in each replicate using a generalized linear regression model. Metabolites which were significantly different between strains or conditions are summarized in the heat map. Higher abundance for a metabolite than expected by the model given the strain’s biomass is indicated in red, and lower abundance in green. (B) Principal-component analysis (PCA) of all metabolites. Each dot is one replicate of the indicated strain/biotin condition.
We performed pathway analysis (Kamburov et al., 2011) and identified alanine/aspartic acid/glutamic acid metabolism, arginine biosynthesis, lysine biosynthesis, the citric acid cycle, and pyruvate metabolism to be overrepresented within our list of significantly perturbed metabolites. However, we were unable to restore growth of pyc::tn on medium lacking biotin by supplementing the media with metabolites found at lower levels in pyc::tn than in WT (data not shown), possibly because these metabolites are not cell-permeable. It is also possible that the factor required to derepress biotin synthesis is a modified species of one of the accumulated metabolites or that our method failed to detect the relevant metabolite.
DISCUSSION
We have uncovered an unanticipated role for pyruvate carboxylase in regulating biotin biosynthesis in mycobacteria. We identified transposon insertions in MSmeg_2412, encoding pyruvate carboxylase, as unable to grow on medium lacking biotin. The role of pyruvate carboxylase in regulating biotin biosynthesis is to mediate transcriptional induction of BioQ repressed biotin biosynthetic genes. In the absence of Pyc, these genes remain repressed, conferring biotin auxotrophy. This function of Pyc in biotin biosynthesis requires the active site residues of the Pyc enzyme as well as enzyme biotinylation, strongly suggesting that the participation of Pyc in the biotin pathway reflects its canonical enzymatic activity in central metabolism. We find that biotinyl-Pyc is the most rapidly depleted biotinylated protein upon biotin withdrawal, supporting the idea that the loss of Pyc activity is an early consequence of biotin starvation. Our primary model to explain these findings is that transient loss of pyruvate carboxylase activity caused by biotin deprivation generates the metabolic signal that derepresses BioQ-regulated genes. This metabolite could be the direct substrate of Pyc, pyruvate, which we find to accumulate with biotin withdrawal in our metabolomic studies. However, it may be unlikely that such a central metabolite, the levels of which could be affected by many factors independent of biotin starvation, would be the signal for BioQ depression. More likely is that the BioQ signal is Pyc dependent, but is a downstream conversion product of a metabolite that accumulates due to Pyc dysfunction, which we did not identify in our metabolomic studies. Further work is therefore required to identify the Pyc dependent BioQ inducer.
At first glance, it may seem paradoxical that transient loss of Pyc activity (through loss of biotinylation during biotin deprivation) induces biotin biosynthesis through BioQ, whereas permanent loss (ie genetic deletion) prevents BioQ derepression. Our data indicate that the Pyc enzyme, itself a biotinylated protein, declines rapidly in abundance during the early hours of biotin withdrawal in a strain that cannot synthesize biotin, before other biotinylated proteins have declined and before cell death occurs. Changes in metabolites caused by the transition of pyruvate carboxylase from holo- to apo- form, and its subsequent loss of catalytic activity, would be predicted to result in acute metabolic shifts that would be different from those experienced by a strain lacking pyruvate carboxylase through genetic deletion. Indeed, our metabolomic analysis strongly supports that 1) biotin withdrawal causes broad metabolic shifts in wild type cells 2) these shifts are much less dramatic or absent in Pyc deficient cells and 3) genetic deletion of pyc causes metabolic perturbation even in the presence of biotin such that this mutant has adapted to the metabolic shifts that accompany acute loss of Pyc function. This effect is obvious when observing the detected levels of pyruvate, the substrate of Pyc. Pyruvate accumulates in wild type cells upon biotin starvation, consistent with Pyc dysfunction. However pyruvate does not accumulate in the Pyc deficient strain in any condition, indicating that the strain has adapted to chronic loss of pyruvate carboxylase.
An alternative explanation for the role of pyruvate carboxylase in regulation of biotin biosynthesis would be that BioQ senses low levels of biotin in a manner similar to E. coli BirA. When biotin levels are low, biotin-binding domains would not be saturated with biotin cofactor, and unbiotinylated BCCP domains could interact directly with BioQ to modulate BioQ’s regulatory function. However, this hypothesis is unlikely to explain the mechanism by which Pyc relives BioQ-mediated repression. Expression of mutant pyruvate carboxylase enzyme lacking the biotin attachment lysine (K1093), and which we have shown is not biotinylated, is unable to restore biotin prototrophy to pyc::tn. As this Pyc enzyme accumulates in its unbiotinylated form, it should directly depress BioQ if the accumulation of biotin free Pyc were the signal, yet it is unable to complement the auxotrophy. Additionally, this mechanism would be atypical of TetR like repressors, which typically bind a small molecule rather than a protein as their ligand, though proteins have been shown to serve as the ligands of TetR like repressors in certain cases (Burkovski, 2007).
A surprising finding of our study is that a Pyc allele lacking the N terminal biotin carboxylase domain (BC) is able to support biotin biosynthesis, yet an amino acid substitution in the BC domain active site (E288K) cannot. Our original suspicion that this effect was due to the N terminal BC fragment that may be expressed in the pyc::tn strain is belied by the ability of PycΔBC to complement the Δpyc strain (in which the entire pyc ORF is deleted). We hypothesize that complementation with the CT+BCCP truncation may occur with the biotin carboxylase domain of another biotin-utilizing enzyme in trans, such as the acetyl/propionyl-CoA carboxylase alpha chain enzymes MSMEG_0334 or MSMEG_1807. Each of these proteins contains a BC and BCCP domain, but lacks a CT domain, which is present in the beta chain present on a separate polypeptide. In the E288K Pyc mutant, the presence of structurally intact but inactive BC domain in the Pyc tetramer may prevent this in trans complementation. Prior work has demonstrated that Pyc truncations retain their enzymatic activity when alone (Sueda et al., 2004) and intermediate transfer between domains has been shown to occur (St Maurice et al., 2007, Islam et al., 2005). However, full explanation of this finding will require further biochemical characterization of the mycobacterial Pyc enzyme.
In summary, we have uncovered a new mechanism of biotin regulation, which relies on the competence of a biotinylated enzyme of central metabolism as a sensor for biotin sufficiency. We anticipate that this mechanism may be applicable to other bacteria which have dissociated biotin ligase function from transcriptional regulation of biotin biosynthesis.
Experimental Procedures
Growth of mycobacteria
M. smegmatis was grown aerobically in liquid culture in 7H9 broth (Difco) supplemented with 0.5% dextrose, 0.5% glycerol, and either 0.05% Tween 80 or 0.02% Tyloxapol (7H9/smeg) and on 7H10 agar plates supplemented with 0.5% dextrose and 0.5% glycerol (7H10/smeg). 7H9 lacking biotin was made according to the listed ingredients: ammonium sulfate 500 mg/L, monopotassium phosphate 1 g/L, disodium phosphate 2.5 g/L, sodium citrate 100 mg/L, magnesium sulfate 50 mg/L, calcium chloride 500 μg/L, zinc sulfate 1 mg/L, copper sulfate 1 mg/L, L-glutamic acid 500 mg/L, ferric ammonium citrate 40 mg/L, pyridoxine hydrochloride 1 mg/L and supplemented with dextrose, glycerol, and Tween 80/Tyloxapol as above. 7H10 lacking biotin was made according to the listed ingredients: ammonium sulfate 500 mg/L, monopotassium phosphate 1.5 g/L, disodium phosphate 1.5 g/L, sodium citrate 400 mg/L, magnesium sulfate 25 mg/L, calcium chloride 500 μg/L, zinc sulfate 1 mg/L, copper sulfate 1 mg/L, L-glutamic acid 500 mg/L, ferric ammonium citrate 40 mg/L, pyridoxine hydrochloride 1 mg/L, malachite green 250 μg/L, agar 15 g/L and supplemented with dextrose and glycerol as above. Antibiotics were added at the following concentrations for mycobacteria: 20 μg/mL kanamycin, 20 μg/mL streptomycin, 50 μg/mL hygromycin. Supplements were added at the following concentrations: 50 ng/mL anhydrotetracycline (ATc), 500 ng/mL desthiobiotin (DTB), 500 ng/mL biotin, 1 μg/mL avidin.
Transposon insertional mutagenesis
A library of transposon insertion mutants in M. smegmatis was generated as described previously (Siegrist & Rubin, 2009). Briefly, a 50 mL culture of mc2155 M. smegmatis was grown aerobically to log phase (OD 0.5-1) in (7H9/smeg). The culture was washed twice in 50 mL of mycobacteriophage buffer (MP Buffer) (50mM Tris [pH 8.0], 150 mM NaCl, 10mM MgCl2, 2 mM CaCl2), and then resuspended in 5mL MP buffer at 37°C. Cells were transduced with φMycoMarT7 at a multiplicity of infection (MOI) of 10 for one hour, and then were transferred to 150 mm diameter 7H10/smeg plates supplemented with kanamycin and incubated for three days at 37°C. A small aliquot was serially diluted 10-fold and cultured on 7H10/smeg plates with kanamycin to determine the overall frequency of tn mutagenesis in the library. 10,000 kanamycin resistant tn mutants were pooled and then frozen at −80°C.
Screening and transposon insertion mapping
Aliquots of the transposon insertional mutant library generated above were diluted to a cell density to yield single colonies on 150 mm 7H10/smeg plates supplemented with kanamycin. Individual colonies were patched onto 7H10/smeg medium lacking biotin but with 1 μg/mL avidin and onto 7H10/smeg medium with biotin. tn mutants which grew on medium containing biotin but not on medium lacking biotin were retested for biotin auxotrophy and confirmed auxotrophs were processed for transposon mapping as described previously (Siegrist & Rubin, 2009). Briefly, biotin auxotrophs were grown to log phase and genomic DNA was prepared. Genomic DNA was digested with SacII restriction endonuclease (NEB) overnight and fragments were self-ligated using T4 DNA ligase (NEB) overnight. The reaction was dialyzed, transformed into competent DH5α λpir E. coli, plated on LB agar supplemented with 40 μg/mL kanamycin, and incubated overnight at 37°C. A single colony was picked and used to inoculate a 5 mL culture of liquid LB broth supplemented with kanamycin. Plasmid DNA was prepared, and the insertion site was sequenced using primers (oNZL003/004) which anneal to the transposon sequence.
Growth on solid medium
Strains were grown in 7H9/smeg medium with appropriate antibiotics to log phase, then washed three times in 7H9/smeg, with avidin substituted for biotin. ODs were normalized to a starting OD of 0.1, and then serial dilutions were spotted onto solid medium (spots 2–5 μL). Agar media cultures were incubated at 37°C for three days.
Growth in liquid medium
Strains were grown in 7H9/smeg medium with appropriate antibiotics to log phase, then washed three times in 7H9/smeg reconstituted using its components, but omitting biotin. Cultures were inoculated to a starting OD of 0.075 at 150 μL per well in a 96 well flat-bottomed clear microplate (Corning) in medium containing 1x (500 ng/mL), 1/10x, 1/100x, 1/1000x, or 0 biotin. Each strain/condition was repeated in triplicate. The microplate was incubated at 37°C for 36 hours in a Tecan M1000 plate reader constantly shaking at a 6 mm amplitude in orbital mode. OD600 readings were taken every 30 minutes for the duration of the experiment.
Western blot analysis
Protein lysates were made from M. smegmatis cultures grown to mid-log OD600, then washed three times in 7H9/smeg broth made without biotin but including avidin, then resuspended to various ODs such that the culture would reach OD 0.6 after biotin deprivation (OD 0.3 for 3h deprivations and 0.15 for 6 and 12 hour deprivations). Cultures were pelleted by centrifugation at 3000g for ten minutes, then supernatant was aspirated and the pellets were frozen overnight at −80°C. Pellets were resuspended in lysis buffer (50 mM Tris-HCl [pH 8.0], 50 mM NaCl, 2% SDS, 2 mM EDTA, 1 mM DTT, 1 mM PMSF) and lysed by bead beating with 0.1 mm zirconia beads in a Biospec Mini-Beadbeater-16 3 times for 45s each. Samples were centrifuged at 3000g for 10 minutes and soluble protein concentration in the supernatant was quantified by absorbance at 280 nm in a NanoDrop 1000 (Thermo). Samples were added 1:1 to 2x SDS sample buffer (20% glycerol, 125 mM Tris [pH 6.8], 4% SDS, 0.2% bromphenol blue, 100 mM DTT), boiled for 10 minutes, then cooled on ice before loading equal quantities of protein into a 4–12% NuPage Bis-Tris protein gradient gel (Thermo Fisher). Gels were run using MOPS SDS running buffer (Thermo Fisher) for 80 minutes at 150 V before being transferred to a Protran nitrocellulose membrane (Whatman) using a Trans-Blot semi-dry apparatus (Bio-Rad). Biotinylated protein was detected using streptavidin-HRP.
Metabolomics
Metabolites were detected using LC-MS on samples prepared from M. smegmatis cultures grown on nitrocellulose filters as performed previously (Puckett et al., 2017, Eoh & Rhee, 2013, de Carvalho et al., 2010). Strains were grown to mid-log OD in 7H9/smeg and then inoculated onto 0.22 μM nitrocellulose filters under vacuum filtration. Experiments were done in triplicate. Filters were placed on top of 7H10/smeg agar plates and allowed to grow at 37°C for two days, followed by transfer to 7H10/smeg agar plates made with biotin or without biotin and with avidin for three hours. Filters were transferred to methanol/acetonitrile/water (2:2:1) precooled on dry ice to metabolically quench the bacteria and bacteria were scraped off of filters. Cells were lysed by bead beating with zirconia beads, centrifuged, and supernatants were filtered across 0.22 μM filters. LC-MS was performed with an Agilent Accurate Mass 6220 TOF coupled with an Agilent 1200 Liquid Chromatography system using a Cogent Diamond Hydride Type C as described previously (Eoh & Rhee, 2013). Metabolites were identified based on unique accurate mass-retention time identifiers for masses exhibiting the expected distribution of accompanying isotopomers.
Metabolites were normalized to overall biomass using a generalized linear regression model which takes into account that most metabolites remain unchanged (Boyle et al., under review). Statistical analysis was performed using ANOVA for fixed effects on a linear mixed-effects model. The threshold of significance was adjusted for multiple hypotheses testing using the Bonferroni correction.
Reverse transcription-quantitative PCR
Relative mRNA levels were identified using reverse transcriptase-quantitative PCR (RT-qPCR). M. smegmatis cultures were grown to mid log OD and either not treated (+ biotin) or washed three times in 7H9/smeg lacking biotin and containing avidin before being resuspended in medium lacking biotin. All cultures were inoculated in triplicate to an OD of 0.3 and incubated at 37°C for three hours before being centrifuged at 3000g. The supernatant was aspirated and cells were frozen overnight at −80°C. Cells were lysed and RNA prepared using a GeneJET RNA Purification kit (Thermo) following the bacteria total RNA purification protocol with alterations. Before lysis, cells were resuspended in 200 μL Tris-EDTA (TE) buffer supplemented with 1 mg/mL lysozyme. Cells were disrupted by beat beating with zirconia beads, and protein was digested with proteinase K for 5 minutes at room temperature. Lysis buffer was added and the mixture was vortexed, then ethanol was added and the mixture was centrifuged at 3000g. The supernatant was added to a GeneJET RNA purification column and the protocol followed as written. cDNA was synthesized using a Thermo Maxima First Strand cDNA synthesis kit with dsDNAse, using 500 ng of RNA per sample. qPCR was performed using a Thermo Scientific DyNAmo Flash Probe qPCR kit and TaqMan reagents on an Applied Biosystems 7500 quantitative PCR machine. Single amplifications were confirmed by the presence of a melting curve with a single peak, and the absence of contaminating genomic DNA was confirmed by the absence of amplification in control cDNA synthesis reactions performed without RT. The cycle threshold (CT) value observed for each sample was normalized to the endogenous control value observed for the housekeeping gene sigA using the formula ΔCT = CT; gene - CT; sigA. Normalized expression levels for each gene and condition were calculated using the formula .
Construction of mutants using CRISPR
Human codon-optimized Cas9 from pX330 (Cong et al., 2013) was cloned using a “dual control” system (Kim et al., 2013) under a tight anhydrotetracycline-dependent promoter with a mutated ribosome-binding site (SD5) (Woong Park et al., 2011) to produce an inducible plasmid (pAJF658) and integrated into the M. smegmatis genome at the L5 phage attB site. crRNA containing 20 nucleotides of homology to the desired cut site was fused to tracrRNA present on an episomal plasmid (pAJF619) to form a single guide RNA (sgRNA). The resultant vector was transformed into strains already containing pAJF658. Parent strains containing both pAJF658 and pAJF619 were grown to mid-log OD, induced with ATc for three hours or not induced for controls, and was cultured on 7H10/smeg plates containing and lacking ATc. Individual surviving colonies were picked for inductions which produced approximately 10-fold or more killing and colony PCR was performed to amplify the genomic DNA region surrounding the sgRNA targeted site. PCR products were treated with ExoSAP-IT (Thermo) and sequenced.
Construction of mutants using homologous recombination and double counterselection
Mutants were constructed using the method described in (Barkan et al., 2011). Briefly, plasmids were constructed containing 500 bp flanking regions 5′ and 3′ of the gene to be deleted. Plasmids also contained a hygromycin resistance cassette, as well as galK and sacB genes to be used for double counterselection. The plasmids lacked a mycobacterial origin of replication to prevent replication of the plasmid without integration via homologous recombination. Plasmids were transformed into parent strains and intermediates were selected by resistance to hygromycin. Integrated plasmids were counterselected using 0.2% 2-deoxy-galactose (2-DOG) and 5% sucrose. Recombinations which produced genetic deletions were confirmed via southern blot.
Supplementary Material
Acknowledgments
This work was supported by NIH grant P30 CA008748 and 1R01AI080628. The authors thank Dirk Schnappinger for helpful suggestions during the course of this work and sharing of reagents.
Footnotes
The authors declare that no conflict of interest exists in relation to this work.
Author Contributions
The conception or design of the study (MSG, KR, MN, NL)
The acquisition, analysis, or interpretation of the data (NL, AF, MN, KEB, JX, KR, MSG)
Writing of the manuscript (NL, MSG).
References
- Attwood PV. The structure and the mechanism of action of pyruvate carboxylase. Int J Biochem Cell Biol. 1995;27:231–249. doi: 10.1016/1357-2725(94)00087-r. [DOI] [PubMed] [Google Scholar]
- Barkan D, Stallings CL, Glickman MS. An improved counterselectable marker system for mycobacterial recombination using galK and 2-deoxy-galactose. Gene. 2011;470:31–36. doi: 10.1016/j.gene.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett D. Biotin sensing: universal influence of biotin status on transcription. Annu Rev Genet. 2007;41:443–464. doi: 10.1146/annurev.genet.41.042007.170450. [DOI] [PubMed] [Google Scholar]
- Boyle KE, Monaco HT, Deforet M, Yan J, Wang W, Rhee KY, Xavier JB. Metabolism and the evolution of social behavior. Submitted. doi: 10.1093/molbev/msx174. (under review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkovski A. Nitrogen control in Corynebacterium glutamicum: proteins, mechanisms, signals. J Microbiol Biotechnol. 2007;17:187–194. [PubMed] [Google Scholar]
- Chakravartty V, Cronan JE. Altered regulation of Escherichia coli biotin biosynthesis in BirA superrepressor mutant strains. J Bacteriol. 2012;194:1113–1126. doi: 10.1128/JB.06549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE., Jr Expression of the biotin biosynthetic operon of Escherichia coli is regulated by the rate of protein biotination. J Biol Chem. 1988;263:10332–10336. [PubMed] [Google Scholar]
- Cryle MJ. Selectivity in a barren landscape: the P450(BioI)-ACP complex. Biochem Soc Trans. 2010;38:934–939. doi: 10.1042/BST0380934. [DOI] [PubMed] [Google Scholar]
- de Carvalho LP, Fischer SM, Marrero J, Nathan C, Ehrt S, Rhee KY. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem Biol. 2010;17:1122–1131. doi: 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Eoh H, Rhee KY. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2013;110:6554–6559. doi: 10.1073/pnas.1219375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Xu J, Zhang H, Chen Z, Srinivas S. Brucella BioR regulator defines a complex regulatory mechanism for bacterial biotin metabolism. J Bacteriol. 2013a;195:3451–3467. doi: 10.1128/JB.00378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Zhang H, Cronan JE. Profligate biotin synthesis in alpha-proteobacteria - a developing or degenerating regulatory system? Mol Microbiol. 2013b;88:77–92. doi: 10.1111/mmi.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway WR, Isidro-Llobet A, Spring DR. Diversity-oriented synthesis as a tool for the discovery of novel biologically active small molecules. Nat Commun. 2010;1:80. doi: 10.1038/ncomms1081. [DOI] [PubMed] [Google Scholar]
- Islam MN, Sueda S, Kondo H. Construction of new forms of pyruvate carboxylase to assess the allosteric regulation by acetyl-CoA. Protein Eng Des Sel. 2005;18:71–78. doi: 10.1093/protein/gzi011. [DOI] [PubMed] [Google Scholar]
- Janiyani K, Bordelon T, Waldrop GL, Cronan JE., Jr Function of Escherichia coli biotin carboxylase requires catalytic activity of both subunits of the homodimer. J Biol Chem. 2001;276:29864–29870. doi: 10.1074/jbc.M104102200. [DOI] [PubMed] [Google Scholar]
- Jitrapakdee S, St Maurice M, Rayment I, Cleland WW, Wallace JC, Attwood PV. Structure, mechanism and regulation of pyruvate carboxylase. Biochem J. 2008;413:369–387. doi: 10.1042/BJ20080709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamburov A, Cavill R, Ebbels TM, Herwig R, Keun HC. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics. 2011;27:2917–2918. doi: 10.1093/bioinformatics/btr499. [DOI] [PubMed] [Google Scholar]
- Kim HS, Hoja U, Stolz J, Sauer G, Schweizer E. Identification of the tRNA-binding protein Arc1p as a novel target of in vivo biotinylation in Saccharomyces cerevisiae. J Biol Chem. 2004;279:42445–42452. doi: 10.1074/jbc.M407137200. [DOI] [PubMed] [Google Scholar]
- Kim JH, O’Brien KM, Sharma R, Boshoff HI, Rehren G, Chakraborty S, Wallach JB, Monteleone M, Wilson DJ, Aldrich CC, Barry CE, 3rd, Rhee KY, Ehrt S, Schnappinger D. A genetic strategy to identify targets for the development of drugs that prevent bacterial persistence. Proc Natl Acad Sci U S A. 2013;110:19095–19100. doi: 10.1073/pnas.1315860110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Cronan JE. Closing in on complete pathways of biotin biosynthesis. Mol Biosyst. 2011;7:1811–1821. doi: 10.1039/c1mb05022b. [DOI] [PubMed] [Google Scholar]
- Lin S, Hanson RE, Cronan JE. Biotin synthesis begins by hijacking the fatty acid synthetic pathway. Nat Chem Biol. 2010;6:682–688. doi: 10.1038/nchembio.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manandhar M, Cronan JE. Pimelic acid, the first precursor of the Bacillus subtilis biotin synthesis pathway, exists as the free acid and is assembled by fatty acid synthesis. Mol Microbiol. 2017;104:595–607. doi: 10.1111/mmi.13648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 2013;41:W597–600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay B, Purwantini E. Pyruvate carboxylase from Mycobacterium smegmatis: stabilization, rapid purification, molecular and biochemical characterization and regulation of the cellular level. Biochim Biophys Acta. 2000;1475:191–206. doi: 10.1016/s0304-4165(00)00064-7. [DOI] [PubMed] [Google Scholar]
- Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- Park SW, Casalena DE, Wilson DJ, Dai R, Nag PP, Liu F, Boyce JP, Bittker JA, Schreiber SL, Finzel BC, Schnappinger D, Aldrich CC. Target-based identification of whole-cell active inhibitors of biotin biosynthesis in Mycobacterium tuberculosis. Chem Biol. 2015;22:76–86. doi: 10.1016/j.chembiol.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters-Wendisch PG, Kreutzer C, Kalinowski J, Patek M, Sahm H, Eikmanns BJ. Pyruvate carboxylase from Corynebacterium glutamicum: characterization, expression and inactivation of the pyc gene. Microbiology. 1998;144(Pt 4):915–927. doi: 10.1099/00221287-144-4-915. [DOI] [PubMed] [Google Scholar]
- Polyak SW, Chapman-Smith A, Mulhern TD, Cronan JE, Jr, Wallace JC. Mutational analysis of protein substrate presentation in the post-translational attachment of biotin to biotin domains. J Biol Chem. 2001;276:3037–3045. doi: 10.1074/jbc.M003968200. [DOI] [PubMed] [Google Scholar]
- Puckett S, Trujillo C, Wang Z, Eoh H, Ioerger TR, Krieger I, Sacchettini J, Schnappinger D, Rhee KY, Ehrt S. Glyoxylate detoxification is an essential function of malate synthase required for carbon assimilation in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2017;114:E2225–E2232. doi: 10.1073/pnas.1617655114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U, Eikmanns BJ. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol Rev. 2005;29:765–794. doi: 10.1016/j.femsre.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Shi C, Geders TW, Park SW, Wilson DJ, Boshoff HI, Abayomi O, Barry CE, 3rd, Schnappinger D, Finzel BC, Aldrich CC. Mechanism-based inactivation by aromatization of the transaminase BioA involved in biotin biosynthesis in Mycobaterium tuberculosis. J Am Chem Soc. 2011;133:18194–18201. doi: 10.1021/ja204036t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist MS, Rubin EJ. Phage transposon mutagenesis. Methods in molecular biology. 2009;465:311–323. doi: 10.1007/978-1-59745-207-6_21. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solbiati J, Cronan JE. The switch regulating transcription of the Escherichia coli biotin operon does not require extensive protein-protein interactions. Chem Biol. 2010;17:11–17. doi: 10.1016/j.chembiol.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Maurice M, Reinhardt L, Surinya KH, Attwood PV, Wallace JC, Cleland WW, Rayment I. Domain architecture of pyruvate carboxylase, a biotin-dependent multifunctional enzyme. Science. 2007;317:1076–1079. doi: 10.1126/science.1144504. [DOI] [PubMed] [Google Scholar]
- Sueda S, Islam MN, Kondo H. Protein engineering of pyruvate carboxylase: investigation on the function of acetyl-CoA and the quaternary structure. Eur J Biochem. 2004;271:1391–1400. doi: 10.1111/j.1432-1033.2004.04051.x. [DOI] [PubMed] [Google Scholar]
- Tang Q, Li X, Zou T, Zhang H, Wang Y, Gao R, Li Z, He J, Feng Y. Mycobacterium smegmatis BioQ defines a new regulatory network for biotin metabolism. Mol Microbiol. 2014 doi: 10.1111/mmi.12817. [DOI] [PubMed] [Google Scholar]
- Weaver LH, Kwon K, Beckett D, Matthews BW. Competing protein:protein interactions are proposed to control the biological switch of the E coli biotin repressor. Protein Sci. 2001;10:2618–2622. doi: 10.1110/ps.32701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woong Park S, Klotzsche M, Wilson DJ, Boshoff HI, Eoh H, Manjunatha U, Blumenthal A, Rhee K, Barry CE, 3rd, Aldrich CC, Ehrt S, Schnappinger D. Evaluating the sensitivity of Mycobacterium tuberculosis to biotin deprivation using regulated gene expression. PLoS Pathog. 2011;7:e1002264. doi: 10.1371/journal.ppat.1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu LP, Xiang S, Lasso G, Gil D, Valle M, Tong L. A symmetrical tetramer for S. aureus pyruvate carboxylase in complex with coenzyme A. Structure. 2009;17:823–832. doi: 10.1016/j.str.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


